Abstract
Objectives
A wide range of duration of viral RNA shedding in patients infected with Severe Acute Respiratory Syndrome Coronavirus 2 (SARS‐CoV‐2) has been observed. We aimed to investigate factors associated with prolonged and intermittent viral RNA shedding in a retrospective cohort of symptomatic COVID‐19 patients.
Methods
Demographic, clinical and laboratory data from hospitalised COVID‐19 patients from a single centre with two consecutive negative respiratory reverse transcription‐polymerase chain reaction (RT‐PCR) results were extracted from electronic medical records. Kaplan–Meier survival curve analysis was used to assess the effect of clinical characteristics on the duration and pattern of shedding. Plasma levels of immune mediators were measured using Luminex multiplex microbead‐based immunoassay.
Results
There were 201 symptomatic patients included. Median age was 49 years (interquartile range 16–61), and 52.2% were male. Median RNA shedding was 14 days (IQR 9–18). Intermittent shedding was observed in 77 (38.3%). We did not identify any factor associated with prolonged or intermittent viral RNA shedding. Duration of shedding was inversely correlated with plasma levels of T‐cell cytokines IL‐1β and IL‐17A at the initial phase of infection, and patients had lower levels of pro‐inflammatory cytokines during intermittent shedding.
Conclusions
Less active T‐cell responses at the initial phase of infection were associated with prolonged viral RNA shedding, suggesting that early immune responses are beneficial to control viral load and prevent viral RNA shedding. Intermittent shedding is common and may explain re‐detection of viral RNA in recovered patients.
Keywords: COVID‐19, cytokines, immune responses, SARS‐CoV‐2, viral RNA shedding
We studied 201 patients with PCR‐confirmed COVID‐19 infection. We found median RNA shedding was 14 days and intermittent RNA shedding was observed in 38.3%. The only associated clinical factor with prolonged RNA shedding was invasive mechanical ventilation. Importantly, we observed in a subset of patients with cytokine analysis, that prolonged RNA shedding was associated with EGF, FGF‐2, GRO‐α and RANTES at the initial phase of infection. Intermittent RNA shedding was associated with lower levels of pro‐inflammatory cytokines.
Introduction
The global tally for coronavirus disease 2019 (COVID‐19) had crossed the 4.5‐millon mark and accounted for more than 300 000 deaths by 17 May 2000. 1 The impact of the pandemic has placed an extra burden on healthcare systems, causing capacity limitations. 2 In Singapore, all individuals with confirmed SARS‐CoV‐2 infection were initially admitted into airborne infection isolation rooms and attending staff wore personal protective equipment in accordance with the United States Centers for Disease Control and Prevention guidelines. 3 More than 25 000 COVID‐19 cases have been reported in Singapore as of 16 May 2020. 4 Of these, approximately 1095 (4.3%) were still hospitalised while 17 881 (71%) who were clinically well but still tested positive for severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), the virus causing COVID‐19, had been discharged to community isolation facilities.
A large proportion of patients with COVID‐19 experience only mild disease and do not require inpatient hospital care. 5 However, several countries such as Singapore have a policy to hospitalise or isolate all patients with confirmed infection to mitigate the risk of secondary community transmission. However, the infectious period of COVID‐19 infection is at present still unclear, and the recommended duration of isolation required has varied across regulatory authorities. 6 , 7 , 8 In a cohort of 191 patients in China, the median duration of viral ribonucleic acid (RNA) shedding by RT‐PCR was 20 days. 9 Therefore, the length of stay in healthcare or isolation facilities can be much longer than medically required.
In a study of 113 patients in China, risk factors associated with prolonged SARS‐CoV‐2 RNA shedding included male gender, delayed admission to hospital after illness onset and requirement for invasive mechanical ventilation. 10 At our centre, the National Centre for Infectious Diseases (NCID), we observed patients with prolonged and intermittent viral RNA shedding of SARS‐CoV‐2 RNA by RT‐PCR which precluded discharge from our healthcare or isolation facilities. In this report, we describe the prevalence of prolonged and intermittent viral RNA shedding and investigated associated factors as well as correlation with host immune responses.
Results
Baseline characteristics and clinical outcomes
We identified 205 patients who were hospitalised with at least one positive RT‐PCR for SAR‐CoV2‐2 and discharge followed by two consecutive negative results. As four patients were asymptomatic, they were excluded from analysis. Of the remaining 201 patients, 105 (52.2%) were male. The median age was 49 years (Interquartile range [IQR] 16–61 years). The common comorbidities were hypertension (23.4%), hyperlipidaemia (23.4%) and diabetes mellitus (13.9%). Median duration from illness onset to hospital admission was 5 days (IQR, 3–8 days). One hundred and six patients (52.7%) had pneumonia on chest radiograph, 43 (21.4%) required supplemental oxygen for hypoxia (oxygen saturation <94%), and 10 (5.0%) needed invasive mechanical ventilation. Death occurred in two patients. The patient characteristics are summarised in Table 1.
Table 1.
Demographic and clinical characteristics of symptomatic patients with confirmed SARS‐CoV‐2 infection
| N = 201 | Duration of viral RNA shedding from symptom onset | P | ||
|---|---|---|---|---|
| ≤ 14 days (n = 110) | > 14 days (n = 91) | |||
| Median age, years (IQR) | 49 (16–61) | 48 (35–60) | 51 (37–61) | 0.465 |
| Male | 105 (52.2%) | 54 (49.1%) | 51 (56.0%) | 0.395 |
| Median days of hospitalisation (IQR) | 13 (9–17) | 11 (8–13) | 17 (12–20) | <0.001 |
| Co‐morbidities | ||||
| Diabetes mellitus | 28 (13.9%) | 15 (13.6%) | 13 (14.3%) | 1.000 |
| Hypertension | 47 (23.4%) | 26 (23.6%) | 21 (23.1%) | 1.000 |
| Hyperlipidaemia | 47 (23.4%) | 22 (20.0%) | 25 (27.5%) | 0.243 |
| Ischemic heart disease | 10 (5.0%) | 4 (3.6%) | 6 (6.6%) | 0.353 |
| Chronic lung disease | 9 (4.5%) | 3 (2.7%) | 6 (6.6%) | 0.305 |
| Chronic kidney disease | 2 (1.0%) | 2 (1.8%) | 0 (0.0%) | 0.502 |
| Chronic liver disease | 1 (0.5%) | 1 (0.9%) | 0 (0.0%) | 1.000 |
| Cancer | 10 (5.0%) | 7 (6.4%) | 3 (3.3%) | 0.517 |
| Severity | ||||
| Prolonged fevera | 51 (25.4%) | 23 (20.9%) | 28 (30.8%) | 0.143 |
| Pneumonia | 106 (52.7%) | 53 (48.2%) | 53 (58.2%) | 0.160 |
| Required supplemental oxygen | 43 (21.4%) | 23 (20.9%) | 20 (22.0%) | 0.865 |
| Invasive mechanical ventilation | 10 (5.0%) | 2 (1.8%) | 8 (8.8%) | 0.045 |
| Treatment | ||||
| Antiviral agent use | 39 (19.4%) | 17 (15.5%) | 22 (24.2%) | 0.152 |
| Lopinavir/ritonavir | 29 (14.4%) | 10 (9.1%) | 19 (20.9%) | 0.026 |
| Interferon‐beta 1b | 12 (6.0%) | 5 (4.5%) | 7 (7.7%) | 0.384 |
| Remdesivir | 9 (4.5%) | 7 (6.4%) | 2 (2.2%) | 0.188 |
| Hydroxychloroquine | 5 (2.5%) | 2 (1.8%) | 3 (3.3%) | 0.660 |
| Blood results | ||||
| Total white count (min) ×109 L−1 | 4.50 (3.50–5.63) | 4.50 (3.50–5.78) | 4.40 (3.58–5.50) | 0.810 |
| Total white count (max) ×109 L−1 | 6.10 (5.10–7.75) | 6.00 (4.80–7.75) | 6.20 (5.30–7.60) | 0.456 |
| Haemoglobin g dL−1 | 13.40 (12.40–14.6) | 13.50 (12.30–14.50) | 13.40 (12.48–14.70) | 0.976 |
| Platelet (min) ×109 L−1 | 193 (155–250) | 191.00 (154.00–259.50) | 196.00 (157.00–240.00) | 0.996 |
| Lymphocytes (min) ×109 L−1 | 1.10 (0.74–1.57) | 0.96 (0.76–1.53) | 1.10 (0.71–1.38) | 0.922 |
| Lymphocytes (max) ×109 L−1 | 1.57 (1.22–1.91) | 1.56 (1.13–1.86) | 1.57 (1.29–1.94) | 0.477 |
| Neutrophils (min) ×109 L−1 | 2.59 (1.97–3.48) | 2.61 (1.97–3.65) | 2.58 (1.94–3.33) | 0.631 |
| Neutrophils (max) ×109 L−1 | 4.16 (3.22–5.86) | 4.17 (3.16–5.95) | 4.15 (3.34–5.85) | 0.951 |
| Creatinine (max) μmol L−1 | 73.00 (57.75–86.25) | 71.00 (55.00–86.50) | 76.00 (60.00–86.50) | 0.220 |
| ALT (max) U L−1 | 31.00 (19.50–62.50) | 29.50 (20.00–69.50) | 33.00 (19.00–57.00) | 0.962 |
| LDH (max) U L−1 | 507.50 (388.25–681.25) | 510.50 (388.75–659.00) | 494.50 (386.25–686.75) | 0.967 |
| CRP (max) mg L−1 | 17.40 (4.70–74.10) | 16.80 (4.50–77.10) | 19.55 (4.70–74.08) | 0.805 |
Continuous variables reported as median (interquartile range); discrete variables reported as number (percentage). The chi‐square (χ2) test or Fisher's exact test was used for comparison of discrete variables. P < 0.05 was deeemed to be significant.
ALT, Alanine transaminase; CRP, C‐reactive protein; LDH, Lactate dehydrogenase; Max, Maximum; Min, Minimum.≥ 7 days.
Viral RNA shedding patterns and association with clinical characteristics
The median duration of viral RNA shedding in this study was 14 days (IQR 9–18) (Figure 1a). Prolonged viral shedding was thus defined as the duration of SARS‐CoV‐2 RNA shedding being longer than 14 days. The median duration of hospital stay was 13 days (IQR 9–17), and each patient received a median of 7 SARS‐CoV‐2 PCR tests. Duration of viral shedding was not significantly associated with sex, age, presence of comorbidities, baseline investigations, prolonged fever (≥ 7 days), pneumonia, need for supplemental oxygen and use of experimental antiviral agents. Median duration of viral RNA shedding was significantly longer in patients requiring invasive mechanical ventilation (19 days, 95% confidence interval [CI] 17.5–20.2) compared to 14 days (95% CI, 13.1–14.9) in patients not requiring mechanical ventilation, P‐value = 0.01 (Figure 1b). Duration of viral shedding was also significantly associated with lopinavir/ritonavir treatment (median 13 days, 95% CI 12.1–13.9) vs. 16 days (95% CI 14.3–17.8), P‐value = 0.026. However, in the multivariate logistic regression analysis, none of the factors analysed were statistically significant (Table 3).
Figure 1.
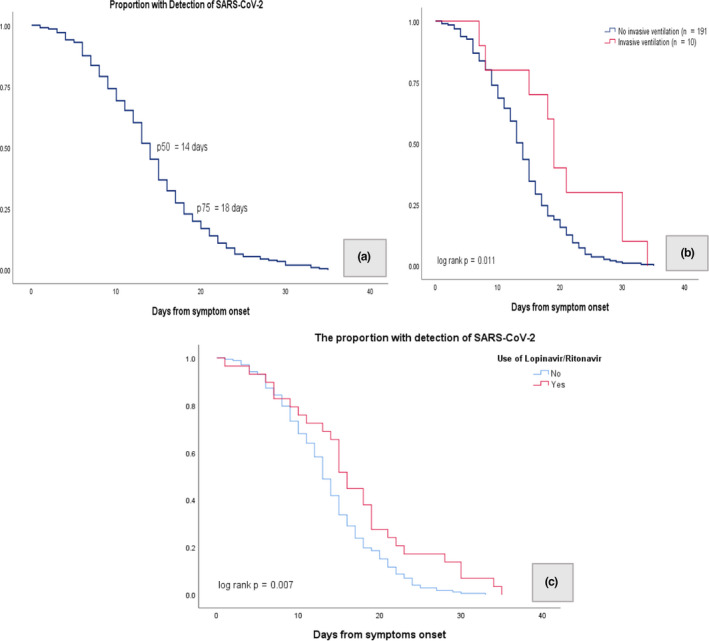
Kaplan–Meier curve. (a) The median duration of viral RNA shedding was 14 days. (b) The proportion of patients with detection of SARS‐CoV‐2 by days from symptom onset between patients who did not have invasive ventilation and patients who had invasive ventilation (log‐rank P‐value = 0.011). (c) The proportion of patients with detection of SARS‐CoV‐2 by days from symptom onset between patients who did not receive lopinavir/ritonavir treatment and patients who received lopinavir/ritonavir treatment (log‐rank P‐value = 0.007).
Table 3.
Multivariate analysis of factors associated with duration of SARS‐CoV‐2 viral RNA detection
| Variable | Multivariable analysis | ||
|---|---|---|---|
| Adjusted odd ratio | 95% CI | P | |
| Age | 1.003 | 0.98–1.02 | 0.770 |
| Gender | 1.201 | 0.67–2.17 | 0.544 |
| Invasive mechanical ventilation | 3.073 | 0.24–17.4 | 0.204 |
| Lopinavir/ritonavir | 1.801 | 0.68–4.77 | 0.237 |
Seventy‐seven patients (38.3%) had intermittent viral RNA shedding, which we defined as alternating between positive and negative PCR tests on serial testing, before cessation of viral shedding with two consecutive negative PCR tests. The median duration from symptom onset to intermittent viral RNA shedding was 13 days (IQR 10–16.5), and the median duration of intermittent viral RNA shedding was 3 days (IQR 2–5) (Figure 2). There was no significant gender difference in duration of viral RNA shedding among the 77 patients. There was also no significant difference in median duration of viral RNA shedding between different age groups, presence of comorbidities, prolonged fever (≥ 7 days), pneumonia, need for supplemental oxygen, use of experimental antiviral agents and invasive ventilation. Comparison of patients with intermittent viral RNA shedding (n = 77) to those without (n = 124) showed that the median duration of persistent viral RNA shedding before a negative RT‐PCR result was longer in patients with intermittent viral RNA shedding than patients without (13 days [95% CI 11.8–14.2] vs. 11 days [95% CI 9.6–12.4], P‐value <0.001). Length of hospital stay was 5 days longer in those with intermittent viral RNA shedding compared with those without (16 days [95% CI 14.6–17.4] vs. 11 days [95% CI 9.6–12.4], P‐value <0.001). There were no other significantly different clinical characteristics (Table 2).
Figure 2.
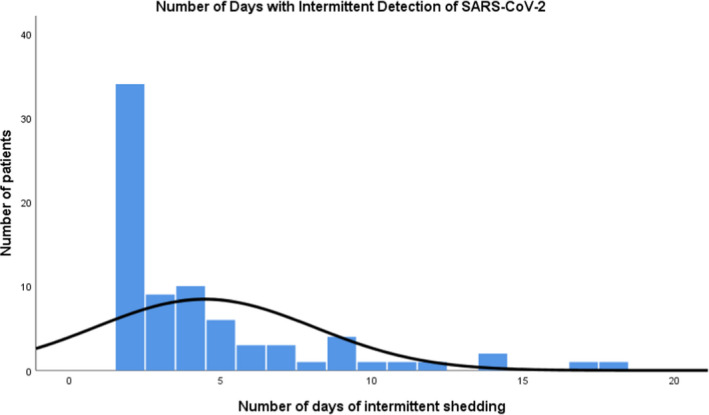
The median duration of intermittent viral RNA shedding was 3 days (n = 77).
Table 2.
Demographic and clinical characteristics of patients with and without intermittent viral RNA shedding
|
Patients with Intermittent Viral RNA Shedding (n = 77) |
Patient without Non‐Intermittent Viral RNA Shedding (n = 124) |
P | |
|---|---|---|---|
| Median days of intermittent viral RNA shedding (IQR) | 3 (2–5) | – | |
| Median days of persistent viral RNA shedding (before negative RT‐PCR result) (IQR) | 13 (10–16.5) | 11 (8–15) | <0.001 |
| Median age, years (IQR) | 48 (33–56) | 51 (36–62) | 0.854 |
| Male | 38 (49.4%) | 67 (54.0%) | 0.563 |
| Median days of hospitalisation (IQR) | 16 (12–19) | 11 (8–15) | <0.001 |
| Co‐morbidities | |||
| Diabetes mellitus | 10 (13.0%) | 18 (14.5%) | 0.836 |
| Hypertension | 16 (20.8%) | 31 (25.0%) | 0.607 |
| Hyperlipidaemia | 16 (20.8%) | 31 (25.0%) | 0.607 |
| Ischemic heart disease | 2 (2.6%) | 8 (6.5%) | 0.323 |
| Chronic lung disease | 5 (6.5%) | 4 (3.2%) | 0.308 |
| Chronic kidney disease | 0 (0.0%) | 2 (1.6%) | 0.525 |
| Chronic liver disease | 0 (0.0%) | 1 (0.8%) | 1.000 |
| Cancer | 1 (1.3%) | 9 (7.3%) | 0.092 |
| Severity | |||
| Prolonged fever a | 18 (23.4%) | 33 (26.6%) | 0.739 |
| Pneumonia | 40 (51.9%) | 66 (53.2%) | 0.885 |
| Required supplemental oxygen | 13 (16.9%) | 30 (24.2%) | 0.288 |
| Invasive mechanical ventilation | 5 (6.5%) | 5 (4.0%) | 0.511 |
| Treatment | |||
| Antiviral agents use | 13 (16.9%) | 26 (21.0%) | 0.583 |
| Lopinavir/ritonavir | 11 (14.3%) | 18 (14.5%) | 1.000 |
| Interferon‐beta | 4 (5.2%) | 8 (6.5%) | 1.000 |
| Remdesivir | 1 (1.3%) | 8 (6.5%) | 0.157 |
| Hydroxychloroquine | 1 (1.3%) | 4 (3.2%) | 0.651 |
| Blood results | |||
| Total white count (min) ×109 L−1 | 4.40 (3.60–5.30) | 4.60 (3.40–5.80) | 0.588 |
| Total white count (max) ×109 L−1 | 6.20 (5.20–7.28) | 6.00 (4.90 – 8.40) | 0.829 |
| Haemoglobin g dL−1 | 13.20 (12.35–14.75) | 13.50 (12.48–14.50) | 0.765 |
| Platelet (min) ×109 L−1 | 185 (155.6–229) | 196.50 (155.25–261.00) | 0.264 |
| Lymphocytes (min) ×109 L−1 | 1.2 (0.76–1.39) | 1.1 (0.70–1.51) | 0.984 |
| Lymphocytes (max) ×109 L−1 | 1.63 (1.30–1.90) | 1.49 (1.11–1.91) | 0.260 |
| Neutrophils (min) ×109 L−1 | 2.58 (2.03–3.31) | 2.61 (1.94–3.65) | 0.809 |
| Neutrophils (max) ×109 L−1 | 4.09 (3.25–5.45) | 4.25 (3.16–6.79) | 0.408 |
| Creatinine (max) μmol L−1 | 67.50 (58.00–84.00) | 75.00 (57.00–88.00) | 0.101 |
| ALT (max) U L−1 | 25.00 (17.25–61.75) | 34.00 (21.50–62.50) | 0.122 |
| LDH (max) U L−1 | 473.00 (391.00–646.00) | 524.00 (388.00–718.00) | 0.257 |
| CRP (max) mg L−1 | 18.80 (5.00–64.80) | 16.80 (4.15–108.35) | 0.795 |
Continuous variables reported as median (interquartile range); discrete variables reported as number (percentage). Chi‐square (χ2) test or Fisher's exact test was used for comparison of discrete variables. P < 0.05was deemed to be significant.
ALT, Alanine transaminase; CRP, C‐reactive protein; LDH, Lactate dehydrogenase; Max, Maximum; Min, Minimum. ≥ 7 days.
Viral RNA shedding and immune responses
Correlations were assessed between levels of immune mediators at median 7 days postillness onset (PIO) (IQR 4–12) and duration of viral RNA shedding in 81 confirmed COVID‐19 patients. The duration of viral RNA shedding was positively correlated with plasma levels of EGF (ρ = 0.29, P‐value = 0.008), FGF‐2 (ρ = 0.27, P‐value = 0.015), GRO‐α (ρ = 0.31, P‐value = 0.005) and RANTES (ρ = 0.30, P‐value = 0.006) and inversely correlated with IL‐1β (ρ = −0.25, P‐value = 0.025) and IL‐17A (ρ = −0.24, P‐value = 0.029). The correlations with epidermal growth factor (EGF), basic fibroblast growth factor (FGF‐2), chemokine (C‐X‐C motif) ligand (CXCL) 1 (GRO‐α) and RANTES(regulated on activation, normal T cell expressed and secreted) were even stronger for patients who required mechanical ventilation (Supplementary table 1). Further stratification of patients based on their duration of viral RNA shedding revealed that patients with prolonged viral RNA shedding (n = 41) had different immune profiles at the acute phase compared with patients with viral RNA shedding ≤ 14 days (n = 40) (Figure 3a) (Supplementary table 2). Systemic levels of cytokines (EGF, FGF‐2, GRO‐α and RANTES) associated with lung inflammation were significantly higher in patients with prolonged viral RNA shedding, and these patients also demonstrated lower levels of T‐cell cytokines (IL‐17A) at median 7 days PIO (IQR 4–12) (Figure 3a).
Figure 3.
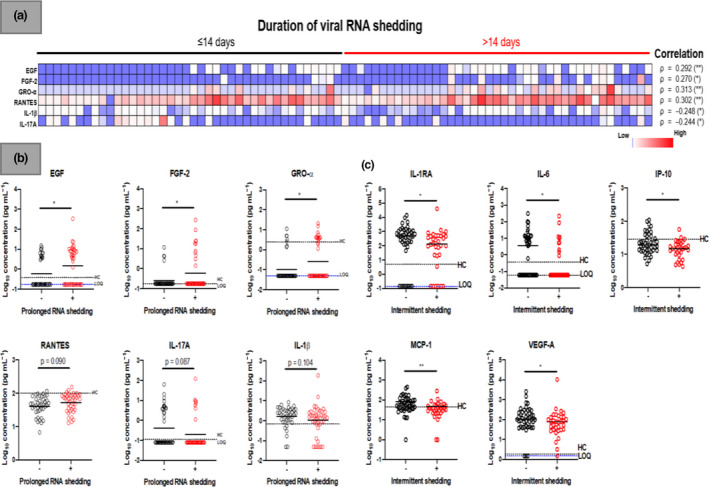
Association of plasma immune mediators levels and viral RNA shedding patterns in COVID‐19 patients. Concentrations of 45 immune mediators were quantified using a 45‐plex microbead‐based immunoassay. (a) Heatmap of significant immune mediators in patients with different duration of viral RNA shedding (≤ 14 days, n = 40; > 14 days, n = 41). Each colour represents the relative concentration of a particular analyte. Blue and red indicate low and high concentrations, respectively. Correlation of immune mediators at the acute phase (median 8 days PIO) with duration of viral RNA shedding in COVID‐19 patients. Spearman rank correlation analysis was conducted on plasma collected at the acute phase (median 8 days PIO) with duration of viral RNA shedding in COVID‐19 patients. Results of the correlation of immune mediators against duration of viral RNA shedding are presented as rho (ρ) and asterisks (**P‐value < 0.01, *P‐value < 0.05). (b) Profiles of immune mediators between patients with or without prolonged viral RNA shedding (> 14 days) and C, patients with or without intermittent viral RNA shedding (without intermittent viral RNA shedding, n = 44; with intermittent viral RNA shedding, n = 33) are illustrated as scatter plots. Statistical analyses were performed with Mann–Whitney U‐test (*P‐value < 0.05). Cytokine level for healthy controls (n = 23) is indicated by the black dotted line. Patient samples that are not detectable are presented as the value of logarithm transformation of limit of quantification (LOQ), indicated by the blue dotted line. EGF, epidermal growth factor; FGF‐2, basic fibroblast growth factor; GRO‐α, chemokine (C‐X‐C motif) ligand (CXCL) 1; IL‐17A, interleukin‐17A, IL‐1RA, interleukin‐1 receptor antagonist; IL‐1β, interleukin‐1 beta; IL‐6, interleukin‐6; IP‐10, interferon gamma‐induced protein 10; MCP‐1, monocyte chemoattractant protein 1; PIO, postillness onset; RANTES, regulated on activation, normal T cell expressed and secreted; VEGF‐A, vascular endothelial growth factors A.
To assess whether the immune responses correlated with intermittent viral RNA shedding in confirmed COVID‐19 patients, plasma levels of immune mediators were determined from samples collected at the nearest time point to first PCR negative (median 15 days PIO, IQR 10–19) in patients without intermittent viral RNA shedding (n = 44) and compared with samples collected at the nearest time point to first PCR positive (median 16 days PIO, IQR 11–21) during intermittent viral RNA shedding (n = 33). There was no significant difference in the sample collection time points between the two groups (P‐value = 0.47). The levels of pro‐inflammatory cytokines (IL‐6, IP‐10, MCP‐1 and VEGF‐A) were significantly lower during intermittent viral RNA shedding periods compared with those without intermittent viral RNA shedding pattern (Figure 3b) (Supplementary table 2).
Discussion
The duration of viral RNA shedding in COVID‐19 ranged from 1 to 35 days. The factors that correlated significantly with the duration of viral RNA shedding on univariate analysis were the requirement for invasive mechanical ventilation and lopinavir/ritonavir treatment. Age, gender, comorbidities and disease severity were not found to be significantly associated with duration of viral RNA shedding. In the multivariate analysis, none of the factors analysed was statistically significant (Table 3). The duration of viral RNA shedding is similar in SARS and MERS coronavirus infections. In SARS, patients with severe disease had higher viral load and 47% of patients with SARS‐CoV had detectable virus RNA at day 21 of illness. 11 In Middle East respiratory syndrome coronavirus (MERS‐CoV), patients with more severe disease had higher and more prolonged levels of viral RNA tested by RT‐PCR. 12 The median time to negative test was 17 days in MERS. 13 As the period of infectivity of COVID‐19 was unknown, the demonstration of 2 consecutive negative RT‐PCR tests was recommended to de‐isolate patients with COVID‐19, similar to MERS. 14
Xu et al. 10 conducted a retrospective study of 113 patients with COVID‐19 and found that male gender, older age, severe disease, late presentation, use of corticosteroids and invasive mechanical ventilation were associated with prolonged viral RNA shedding. Age, gender distribution, comorbidities and days to hospital presentation were similar to our cohort. However, more patients in this study required invasive mechanical ventilation (15.9% vs 5.0%), which could partially explain the longer median viral RNA shedding duration of 17 days. Additionally, 56.6% patients in the cohort received systemic glucocorticoid therapy compared with 0.5% (1 of 201) in our cohort, which has been associated with longer viral RNA shedding in patients with SARS and MERS. 15 , 16 In another study of 147 patients with COVID‐19, the use of systemic corticosteroids, but not supplemental oxygen requirement, was associated with prolonged viral RNA shedding. 17 In contrast, we found on univariate analysis that patients who required invasive mechanical ventilation and patients who received lopinavir/ritonavir treatment to be significantly associated with longer viral RNA shedding. The association of prolonged viral RNA shedding with invasive mechanical ventilation could be confounded by the testing of endotracheal samples which are known to have higher viral load and sensitivity in patients with COVID‐19 pneumonia. 18 Further, invasive mechanical ventilation was not found to be an independent risk factor of prolonged viral RNA shedding in our multivariable model.
We observed that prolonged viral RNA shedding was associated with elevated levels of cytokines actively involved in pulmonary inflammatory response, 19 , 20 , 21 , 22 particularly in patients who required mechanical ventilation. Interestingly, patients with prolonged viral RNA shedding demonstrated lower levels of IL‐1β and IL‐17A during the acute phase of infection. Both IL‐1β and IL‐17A are pivotal cytokines involved in activation of anti‐viral T‐cell responses. 23 , 24 Exhaustion of T‐cell activation was reported to be associated with increased duration of viral RNA shedding in SARS‐CoV infections. 25 Hence, we postulate that more effective T‐cell activation at the early phase of infection promotes virus clearance and subsequently shortens viral RNA shedding duration in COVID‐19. Further studies are warranted to comprehensively assess the roles of effector T cells in mediating virus clearance during SARS‐CoV‐2 infection.
Recovered COVID‐19 patients who were diagnosed with re‐infection by subsequent positive RT‐PCR have been reported. 26 As a result of RT‐PCR test characteristic, very low levels of RNA at the threshold of detection may cause false‐negative or false‐positive results. 27 This can account for intermittent viral RNA shedding observed in our study. In our cohort, intermittent viral RNA shedding was detected in 77 (38.3%). While most patients had short duration of intermittent viral RNA shedding, 12 (6.0%) had intermittent viral RNA shedding for more than 7 days. In another report, 4 patients who demonstrated 2 negative RT‐PCR tests re‐tested positive again after 5–13 days despite being asymptomatic with no new radiographic changes. 28 While re‐infection is a concern, prolonged intermittent viral RNA shedding may account for re‐detection of viral RNA.
Viral RNA shedding may be intermittent and is affected by the immune status of the patients. Previous studies have reported that virus re‐activation may be provoked by anti‐inflammatory therapies. 29 , 30 We noted that the patients had significantly lower pro‐inflammatory cytokine levels during intermittent viral RNA shedding period, compared with those without intermittent viral RNA shedding after complete virus clearance. A weaker inflammatory response or suppression of inflammatory responses could be triggering virus re‐activation, resulting in intermittent detection in the patients. Whether milder inflammation and virus re‐activation are causally related remains to be explored.
While current de‐isolation strategies generally rely on demonstration of non‐detectable viral RNA, detection by RT‐PCR is only a surrogate marker for infectivity. Positive RT‐PCR test may represent non‐viable viruses or remnant nucleic acid products. A study of 9 patients showed that SARS‐CoV‐2 was readily isolated from respiratory samples within the first week of symptoms, with greater success from sputum than from upper respiratory swabs, and from samples with higher viral loads. Additionally, viral isolation was unsuccessful when patients were beyond day 8 of illness. The authors suggested that patients beyond 10 days of illness with < 100 000 viral RNA copies per mL of sputum could be safely isolated at home. 31 If the suggested de‐isolation strategy can be applied, it can significantly shorten duration of isolation and demand for scarce hospital beds during periods of peak COVID‐19 activity.
There are limitations to our study. As viral culture was not done, we were unable to determine whether the detection of viral RNA by RT‐PCR was related to viable virus or shedding of remnant non‐viable genetic material. This would have public health implications on the period of infectivity and duration of isolation of COVID‐19 cases. The duration of viral RNA shedding relied on negative tests by RT‐PCR which may be influenced by collection technique or the respiratory site from which the sample was obtained. Patients with predominantly lower respiratory infection may have a negative nasopharyngeal test result. The failure to find any clinical correlates of prolonged viral shedding may have been due to inadequate power to detect small differences, including the small number of patients who required invasive mechanical ventilation.
In conclusion, our study observed no independent risk factor associated with prolonged viral RNA shedding. While the duration of viral RNA shedding varied widely, there were no identified demographic or clinical determinants in the duration of viral RNA shedding. COVID‐19 patients with less active T‐cell responses during the initial phase of infection shed viral RNA longer, and these patients also presented lower levels of pro‐inflammatory cytokines during intermittent viral RNA shedding. Intermittent viral RNA shedding is a common phenomenon, and patients with prolonged intermittent viral RNA shedding may explain reports of re‐detection of viral RNA in recovered COVID‐19 patients.
Methods
Patients, clinical data and management
During the study period, all patients with confirmed COVID‐19 infection in Singapore were mandated to be admitted to hospital regardless of illness severity. A total of 201 patients with confirmed COVID‐19 infection admitted in National Centre for Infectious Diseases (NCID) from 22 January 2020 to 5 April 2020 and discharged after obtaining two consecutive negative RT‐PCR results at least 24 h apart were included. The discharge date was censored on 21 April 2020.
Some of the patients received supportive therapy which included supplemental oxygen, empirical antibiotics and/or oral oseltamivir if there was clinical concern of community‐acquired pneumonia (Table 1). Complete blood cell count, renal and liver function, C‐reactive protein (CRP) and lactate dehydrogenase (LDH), and chest radiograph were performed for all patients as standard of care.
Co‐formulated lopinavir–ritonavir, interferon beta‐1b and hydroxychloroquine were prescribed to selected patients at the treating physicians' discretion after shared decision‐making and provision of oral informed consent. Remdesivir was prescribed for patients enrolled in clinical trials in accordance with trial protocols (ClinicalTrials.gov: NCT04280705, NCT04292899, NCT04292730). Corticosteroids were only given for septic shock in the intensive care unit. Demographic, clinical and laboratory data were obtained from the electronic medical records. Data collection was approved under the Infectious Diseases Act with waiver of informed consent. 32
Respiratory samples for SARS‐CoV‐2 testing included nasopharyngeal swab, throat swab, sputum and endotracheal aspirate. The diagnosis of COVID‐19 infection was confirmed through reverse transcription‐polymerase chain reaction (RT‐PCR) testing for SARS‐CoV‐2. Repeat samples were performed on average, every other day after the patients were afebrile and clinically improving. A cycle threshold value (Ct‐value) above 35 indicates low viral load in the sample; this guided the physicians to repeat viral RNA sampling by PCR 24 h later. National Public Health Laboratory developed test targeting the N and ORF1ab genes was used at the start of the outbreak in Singapore in late January 2020. From 6 February 2020, a commercial assay was used. 33 Specific consent for SARS‐CoV‐2 testing was not obtained as all testing was part of routine clinical care. Written informed consent was obtained (approval from the National Healthcare Group Domain Specific Review Board, Study Reference 2012/00917) for measurement of immune mediator serum samples.
Duration of viral RNA shedding was defined as the number of days from symptom onset to the last day of positive RT‐PCR. Duration of persistent viral RNA shedding was defined as the number of days from symptom onset to the first negative RT‐PCR. Duration of intermittent viral RNA shedding, if present, is defined as the number of days from the first negative RT‐PCR test to the last positive RT‐PCR.
Multiplex microbead‐based immunoassay
Plasma fractions were extracted from blood samples collected from COVID‐19 patients during hospitalisation. Subsequently, plasma from COVID‐19 patients and healthy controls was treated with Triton™ X‐100 solvent/detergent mix (Thermo Fisher Scientific Waltham, MA USA) for virus inactivation (20 min, RT). 34
Immune mediator levels in inactivated plasma samples were measured with Cytokine/Chemokine/Growth Factor 45‐plex Human ProcartaPlexTM Panel 1 (Thermo Fisher Scientific Waltham, MA USA, #EPX450‐12171‐901), according to the manufacturer's protocol. Kit analyte detection panel included granulocyte–macrophage colony‐stimulating factor (GM‐CSF), epidermal growth factor (EGF), brain‐derived neurotropic factor, beta‐nerve growth factor (bNGF), basic fibroblast growth factor (FGF‐2), hepatocyte growth factor (HGF), monocyte chemoattractant protein (MCP) 1, macrophage inflammatory protein (MIP) 1α, MIP‐1β, RANTES (regulated on activation, normal T cell expressed and secreted), chemokine (C‐X‐C motif) ligand (CXCL) 1 (GRO‐α), stromal cell‐derived factor 1 (SDF‐1α), interferon (IFN) gamma‐induced protein 10 (IP‐10), eotaxin, IFN‐α, IFN‐γ, interleukin (IL) IL‐1α, IL‐1β, IL‐1RA, IL‐2, IL‐4, IL‐5, IL‐6, IL‐7, IL‐8, IL‐9, IL‐10, IL‐12p70, IL‐13, IL‐15, IL‐17A, IL‐18, IL‐21, IL‐22, IL‐23, IL‐27, IL‐31, leukaemia inhibitory factor (LIF), stem cell factor (SCF), tumour necrosis factor (TNF‐α), TNF‐β, vascular endothelial growth factors A and D (VEGF‐A, VEGF‐D), platelet‐derived growth factor (PDGF‐BB) and placental growth factor (PLGF‐1). Samples were obtained from the FLEXMAP® 3D (Luminex) using xPONENT® 4.0 (Luminex) and analysed on Bio‐Plex Manager™ 6.1.1 (Bio‐Rad Hercules, California USA). Standard curves were generated with a 5‐parameter logistic algorithm, reporting values for both mean fluorescent intensity and concentration data. Readout from internal control samples was used to remove any potential plate effects and to obtain a correction factor for normalising assayed plates. The concentrations were logarithmically transformed to ensure normality. Sample with concentration out of measurement range is assigned the value of logarithmic transformation of limit of quantification (LOQ).
Statistical analysis
The Mann–Whitney U‐test was used for comparison of continuous variables, and chi‐square (χ2) or Fisher's exact tests for categorical variables as appropriate. Kaplan–Meier survival curve analysis was used to assess the effect of gender, age, severity of illness and experimental anti‐viral agents on the duration of viral RNA shedding. Significant risk factors identified on univariate analysis were further analysed by adjusted multivariate logistic regression analysis to identify independent risk factors associated with the prolonged duration of viral RNA shedding. Non‐parametric Mann–Whitney U‐tests were conducted on the logarithmically transformed concentration of immune mediators between patients with prolonged or intermittent viral RNA shedding and those without. Correlation analysis was carried out using Spearman's rank correlation. P‐values < 0.05 were considered statistically significant. Data analysis was performed using SPSS, version 26.0 (IBM Corp., Armonk, NY, USA). Plots were generated using GraphPad Prism version 8 (GraphPad Software, San Diego, CA, USA).
Conflict of interest
The authors declare no conflict of interest.
Author contributions
Pei Hua Lee: Investigation; Methodology; Writing‐original draft; Writing‐review & editing. Woo Chiao Tay: Methodology; Project administration; Resources. Stephanie Sutjipto: Investigation; Resources. Siew‐Wai Fong: Formal analysis; Investigation; Methodology; Software; Writing‐original draft; Writing‐review & editing. Sean Wei Xiang Ong: Investigation; Methodology; Writing‐review & editing. Wycliff Enli Wei: Data curation; Resources. Yi‐Hao Chan: Formal analysis; Investigation; Methodology. Li Min Ling: Conceptualization; Resources. Barnaby E Young: Conceptualization; Data curation; Methodology; Resources. Matthias Paul HS Toh: Data curation; Resources; Validation. Laurent Renia: Conceptualization; Resources; Supervision. Lisa FP Ng: Formal analysis; Investigation; Methodology; Supervision; Validation; Writing‐review & editing. Yee‐Sin Leo: Conceptualization; Visualization. David C Lye: Conceptualization; Data curation; Supervision. Tau Hong Lee: Conceptualization; Data curation; Formal analysis; Supervision; Validation; Writing‐original draft; Writing‐review & editing.
Supporting information
Supplementary table 1
Supplementary table 2
Acknowledgments
We thank Professor Olaf Rötzschke, Dr Bernett Lee, Wilson How and Norman Leo Fernandez from the Singapore Immunology Network (SIgN), for their help in running multiplex microbead immunoassay. We are also grateful to Dr Danielle Anderson and her team at Duke‐NUS, for their technical assistance in virus inactivation procedures with Triton™ X‐100. This work was supported by a National Medical Research Council COVID19 Research Fund (Ref: COVID19RF‐001) to Barnaby Young. The multiplex immunoassay studies were supported by the A*STAR COVID‐19 Research funding (H/20/04/g1/006) provided to Singapore Immunology Network (SIgN) and core fund allocated to SIgN by the Biomedical Research Council (BMRC), A*STAR, and by a National Research Foundation grant (#NRF2017_SISFP09) to the SIgN Immunomonitoring platform.
References
- 1. Coronavirus disease 2019 Situation Report – 88. World Health Organization; 2020. Available from: https://www.who.int/docs/defaultsource/coronaviruse/situation‐reports/20200417‐sitrep‐88‐covid‐91b6cccd94f8b4f219377bff55719a6ed.pdf?sfvrsn=ebe78315_6 [Google Scholar]
- 2. Zhao J, Yuan Q, Wang H et al Antibody responses to SARS‐CoV‐2 in patients of novel coronavirus disease 2019. SSRN Electron J 2020; 395: 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. United States Centers for Disease Control and Prevention . Using personal protective equipment (PPE) [Internet]. 2020. [cited 18 May 2020]. Available from: https://www.cdc.gov/coronavirus/2019‐ncov/hcp/using‐ppe.html
- 4. Updates on COVID‐19 (Coronavirus Disease 2019) local situation [Internet]. [cited 18 May 2020]. Available from: https://www.moh.gov.sg/covid‐19
- 5. Guan W, Ni Z, Hu Y et al Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 2020; 382: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. WHO . Clinical management of severe acute respiratory infection when novel coronavirus (nCoV) infection is suspected‐ Interim guidance [Internet]. 2020. [cited 5 March 2020]. Available from: https://www.who.int/publications-detail/clinical-management-of-severe-acute-respiratory-infection-when-novel-coronavirus-(ncov)-infection-is-suspected
- 7. European Centre for Disease Prevention and Control ( ECDC ) . Guidance for discharge and ending isolation in the context of widespread community transmission of COVID‐19‐first update Scope of this document. 2020. Available from: https://www.ecdc.europa.eu/sites/default/files/documents/covid‐19‐guidance‐discharge‐and‐ending‐isolation‐first%20update.pdf
- 8. United States Centers for Disease Control and Prevention . Discontinuation of transmission‐based precautions and disposition of patients with COVID‐19 in healthcare settings (interim guidance) [Internet]. [cited 20 April 2020]. Available from: https://www.cdc.gov/coronavirus/2019‐ncov/hcp/disposition‐hospitalized‐patients.html
- 9. Zhou F, Yu T, Du R et al Clinical course and risk factors for mortality of adult inpatients with COVID‐19 in Wuhan, China: a retrospective cohort study. Lancet 2020; 395: 1054–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xu K, Chen Y, Yuan J et al Factors associated with prolonged viral RNA shedding in patients with COVID‐19. Clin Infect Dis 2020; 351: 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Peiris JS, Chu CM, Cheng VC et al Clinical progression and viral load in a community outbreak of coronavirus‐associated SARS pneumonia: a prospective study. Lancet 2003; 361: 1767–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Oh MD, Park WB, Choe PG et al Viral load kinetics of MERS coronavirus infection. N Engl J Med 2016; 375: 1303–1305. [DOI] [PubMed] [Google Scholar]
- 13. Choi WS, Kang C, Kim Y et al Clinical presentation and outcomes of middle east respiratory syndrome in the Republic of Korea. Infect Chemother 2016; 48: 118–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Centers for Disease Control and Prevention . Interim U.S. Guidance for Monitoring and Movement of Persons with Potential Middle East Respiratory Syndrome Coronavirus (MERS‐CoV) Exposure. 2016. Accessed July 13, 2016. [cited September 2012]. Available from: https://www.cdc.gov/coronavirus/mers/hcp/monitoring‐movement‐guidance.pdf
- 15. Lee N, Allen Chan KC, Hui DS et al Effects of early corticosteroid treatment on plasma SARS‐associated Coronavirus RNA concentrations in adult patients. J Clin Virol 2004; 31: 304–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Arabi YM, Mandourah Y, Al‐Hameed F et al Corticosteroid therapy for critically ill patients with middle east respiratory syndrome. Am J Respir Crit Care Med 2018; 197: 757–767. [DOI] [PubMed] [Google Scholar]
- 17. Lin Q, Yong Y, Jiang D et al Factors associated with duration of viral shedding in adults with COVID‐19 outside of Wuhan, China: a retrospective cohort study. Int J Infect Dis 2020; 96: 531–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chang D, Mo G, Yuan X et al Time kinetics of viral clearance and resolution of symptoms in novel coronavirus infection. Am J Respir Crit Care Med 2020; 201: 1150–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Le Cras TD, Acciani TH, Mushaben EM et al Epithelial EGF receptor signaling mediates airway hyperreactivity and remodeling in a mouse model of chronic asthma. Am J Physiol Lung Cell Mol Physiol 2011; 300: 414–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li L, Zhang S, Wei L et al FGF2 and FGFR2 in patients with idiopathic pulmonary fibrosis and lung cancer. Oncol Lett 2018; 16: 2490–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang CH, Liu CY, Wan YL et al Persistence of lung inflammation and lung cytokines with high‐resolution CT abnormalities during recovery from SARS. Respir Res 2005; 6: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Traves SL, Culpitt SV, Russell REK, Barnes PJ, Donnelly LE. Increased levels of the chemokines GROα and MCP‐1 in sputum samples from patients with COPD. Thorax 2002; 57: 590–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nambu A, Nakae S, Iwakura Y. IL‐1β, but not IL‐1α, is required for antigen‐specific T cell activation and the induction of local inflammation in the delayed‐type hypersensitivity responses. Int Immunol 2006; 18: 701–712. [DOI] [PubMed] [Google Scholar]
- 24. Acharya D, Wang P, Paul AM, et al Interleukin‐17A promotes CD8+ T cell cytotoxicity to facilitate west nile virus clearance. J Virol 2017; 91: 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhao J, Zhao J, Legge K, Perlman S. Age‐related increases in PGD 2 expression impair respiratory DC migration, resulting in diminished T cell responses upon respiratory virus infection in mice. J Clin Invest 2011; 121: 4921–4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xiao AT, Tong YX, Zhang S. False‐negative of RT‐PCR and prolonged nucleic acid conversion in COVID‐19: rather than recurrence. J Med Virol 2020. 10.1002/jmv.25855 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mahase E. Covid‐19: WHO and South Korea investigate reconfirmed cases. BMJ 2020; 369: 1498. [DOI] [PubMed] [Google Scholar]
- 28. Lan L, Xu D, Ye G et al Positive RT‐PCR test results in patients recovered from COVID‐19. JAMA 2020; 323: 1502–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Matsumoto T, Marusawa H, Dogaki M, Suginoshita YIT. Adalimumab‐induced lethal hepatitis B virus reactivation in an HBsAg‐negative patient with clinically resolved hepatitis B virus infection. Liver Int 2010; 30: 1241–1242. [DOI] [PubMed] [Google Scholar]
- 30. Liu Y, Li S, Wang Z. The role of cyclooxygenase in multiplication and reactivation of HSV‐1 in vestibular ganglion neurons. Sci World J 2014; 2014: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wölfel R, Corman VM, Guggemos W et al Virological assessment of hospitalized patients with COVID‐2019. Nature 2020; 581: 465–469. [DOI] [PubMed] [Google Scholar]
- 32. Infectious diseases act [Internet]. [cited 5 March 2020]. Available from: https://sso.agc.gov.sg/Act/IDA1976.
- 33. Lee TH, Lin RJ, Lin RTP et al Testing for SARS‐CoV‐2: can we stop at two? Clin Infect Dis 2020. 10.1093/cid/ciaa459 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Darnell MER, Taylor DR. Evaluation of inactivation methods for severe acute respiratory syndrome coronavirus in non‐cellular blood products. Transfusion 2006; 46: 1770–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary table 1
Supplementary table 2


