Abstract
Listeria monocytogenes is an important foodborne pathogen in human and veterinary health, causing significant morbidity and mortality including abortion. It has a particular tropism for the gravid uterus, however, the route of infection in reproductive tissues of ruminants (i.e. placentome), is much less clear. In this study, we aimed to investigate a bovine caruncular epithelial cell (BCEC) line as a model for L. monocytogenes infection of the bovine reproductive tract. The BCEC infection model was used to assess the ability of 14 different L. monocytogenes isolates to infect these cells. Lysozyme sensitivity and bacterial survival in 580 μg lysozyme/ml correlated with attenuated ability to proliferate in BCEC (p = 0.004 and p = 0.02, respectively). Four isolates were significantly attenuated compared to the control strain 10403S. One of these strains (AR008) showed evidence of compromised cell wall leading to increased sensitivity to ß-lactam antibiotics, and another (7644) had compromised cell membrane integrity leading to increased sensitivity to cationic peptides. Whole genome sequencing followed by Multi Locus Sequence Type analysis identified that five invasive isolates had the same sequence type, ST59, despite originating from three different clinical conditions. Virulence gene analysis showed that the attenuated isolate LM4 was lacking two virulence genes (uhpT, virR) known to be involved in intracellular growth and virulence.
In conclusion, the BCEC model was able to differentiate between the infective potential of different isolates. Moreover, resistance to lysozyme correlated with the ability to invade and replicate within BCEC, suggesting co-selection for surviving challenging environments as the abomasum.
Keywords: Bioinformatics, Genetics, Microbiology, Molecular biology, Listeria monocytogenes, Infection model, BCEC, MLST, Sequence typing
Bioinformatics; Genetics; Microbiology; Molecular biology; Listeria monocytogenes; infection model; BCEC; MLST, sequence typing.
1. Introduction
The zoonotic intracellular pathogen Listeria monocytogenes causes a range of clinical presentations including listeriosis, meningitis, septicaemia and abortions, in both cattle and humans. In humans and a number of animals, L. monocytogenes is known to be able to invade the placenta during pregnancy, leading to spontaneous abortion or neonatal infection with high mortality rates [1, 2]. Listeriosis is of major veterinary importance in cattle due to its negative impact on animal health resulting in premature death or reproductive failure responsible for economic losses [3].
The route by which Listeria spp. infect the ruminant placenta is unclear. Most studies have focused on infection of humans and rodents, and distinct species differences in placental structures as well as interhemal barriers mean that making comparisons between ruminants and infection in other species is erroneous [4]. In the placenta, maternal and fetal tissues interact. In hemochorial placentas that are present in humans or guinea pigs, maternal blood comes into direct contact with fetal trophoblast cells. In contrast, in synepitheliochorial placentas found in cattle and sheep, the maternal and fetal blood are separated by several cell/tissue layers which any pathogen must cross to cause fetal infection [5]. In addition, the ruminant placenta is composed of multiple placentomes throughout the uterus, with each placentome formed from fetal cotyledons interdigitating with maternal caruncules. The latter are formed by multiple layers of stromal cells covered in a single layer of carunclar epithelial cells that interact with the fetal trophoblasts [5]. L. monocytogenes have been isolated from infected bovine placentomes post abortion and identified as the causative agent of disease [1]. In addition it has been shown that L. monocytogenes is able to invade and replicate in trophoblastic cells derived from a bovine chorioallantoic membrane explant model [6].
Listeria spp. invasion is primarily mediated by the interaction of the surface Internalin (Inl) proteins A and B with host cell receptors E-cadherin and c-Met tyrosine kinase (c-Met), respectively. For InlA-dependent entry into cells, proline at position 16 of E-cadherin is critical; in rats and mice if this proline is replaced by glutamic acid, then InlA-dependent entry into cells is prevented [7]. Whereas InlB-dependent cell invasion via c-Met does not occur in rabbits and guinea pigs but is functional in both mice and humans [8]. However, InlB also has a secondary role of activating Phosphoinositide 3-kinase (PI3–K), which is required for InlA-dependent invasion in tissues that do not constitutively express this enzyme activity [9]. InlA and InlB may also act in a concerted manner during invasion of the placenta [10]. Given this complex interplay, it is not surprising that the inlA and inlB genes are arranged in an operon and can either be expressed as one bi-cistronic mRNA or independently expressed from promoters [11] and there are multiple promoters which are controlled by both the virulence regulator, PrfA [11], and the stress sigma factor, Sigma B [12]. Generally, inlA mRNA levels are slightly higher than those for inlB, and expression of both genes is higher in the stationary phase of growth or under other environmental stress conditions [13]. However, it is also widely reported that many environmental strains may contain mutations in InlA which result in a less invasive phenotype [14], therefore, sequencing of the gene is required as well as monitoring mRNA levels to fully characterise the virulence potential of strains [15].
Recently, genome wide analysis has led to the identification of InlP as a virulence factor linked to tissue tropism in the gravid uterus [16]. Secreted InlP targets cytosolic afadin and, after host cell invasion mediated by InlA or InlB, facilitates bacterial spread from infected epithelial monolayers into an underlying compartment during placental infection [17].
In this study, we investigated a bovine caruncular epithelial cell (BCEC) line as a model for L. monocytogenes infection of the bovine reproductive tract. BCECs, epithelioid shaped cells, express cytokeratin, zonula occludens-1 and vimentin, while lacking desmin and alpha smooth muscle actin; and form tight junctions with trans epithelial resistance increasing with advancing confluence [18, 19]. To predict the likelihood of interaction of InlA and InlB with bovine E-cadherin and c-Met, respectively, the sequence and mRNA expression of the genes encoding these receptors were analysed. We hypothesized that L. monocytogenes isolates from bovine abortions might more readily infect bovine caruncules and replicate within these cells. Therefore, the ability of a range of L. monocytogenes isolates from different clinical or environmental sources to infect the bovine caruncular epithelial cell lines was investigated. In addition, whole genome sequencing was used to further characterise the genome content of these isolates.
2. Results
2.1. Sequence comparisons of host receptors E-cadherin and c-Met tyrosine kinase receptors in caruncular cells
For this study a bovine caruncular epithelial cell (BCEC) line was chosen as model a for L. monocytogenes infection of the bovine reproductive tract. Since host specificity towards InlA-dependant entry into cells depends on the presence of proline at position 16 of E-cadherin in the first extracellular domain [7], alignment of the E-cad region of a range of species containing residue 16 was performed (Figure 1A). This showed that bovine E-cadherin has proline at position 16 and suggests that bovine and ovine E-cadherin will interact with InlA in a similar way to human and guinea pig E-cadherin and can act as a receptor for L. monocytogenes in ruminant species. Interactions between InlB and c-Met are less well defined; in c-MET the Sema, PSI and Ig1 region have shown to play a role in interaction with InlB [20]. Alignment of amino acids in these c-Met regions derived from bovine, ovine, human, murine, rabbit and guinea pig genome sequences showed that bovine c-Met does not cluster closely to rabbit and guinea pig c-Met (Figure 1C). There were no consistent amino acid substitutions evident in the six amino acids of the Ig1 region that interacts with InlB (Figure 1B), indicating that there is no obvious structural reason why InlB-dependent cell entry would not occur when L. monocytogenes interacts with bovine cells.
Figure 1.
Bovine E-cad and cMet sequence comparisons and expression. (A) Multiple sequence alignment of the Pro16 residue of E-cad correlating with probability of invading the host cell [12], emboldened letters indicate amino acid substitutions previously identified in other studies and red letters indicate amino acid substitution unique to rodents. (B) Multiple sequence protein alignments of the region of c-MET, emboldened letters indicate amino acid substitutions which have previously been identified as a primary interface between InlB and c-MET [19], blue letters indicate relatedness of amino acid substitutions between certain species and red letters indicate amino acid substitutions discovered in this work. (C) Maximum likelihood tree of c-Met. E-cad (D) and c-MET (E) transcript levels in BCEC cells stimulated with 1 μg ml−1 LPS or infected with L. monocytogenes (MOI = 200) for 4, 8 or 24 h.
Expression of E-cadherin and c-Met mRNA in BCEC cells was then verified and mRNA for both were detected in these cells in the presence and absence of L. monocytogenes infection (Fig. 1D & E). No difference in E-cadherin and c-Met mRNA expression level was observed when these cells were exposed to four different L. monocytogenes isolates (Fig. 1D & E) indicating that infection with these bacteria did not down-regulate these receptors. Taken together, these results suggest that L. monocytogenes interaction with BCECs was likely to utilise the InlA/B receptor-mediated uptake pathway.
2.2. L. monocytogenes infection of bovine caruncular epithelial cells
The L. monocytogenes strains chosen for this study included the well characterised reference strain 10403S and isolates from different bovine samples (Table 1). These included 6 strains from bovine abortions, 4 strains from bovine eyes, 1 strain from a case of bovine meningitis and 2 strains isolated from bovine milk. Initial experiments carried out to establish an infection method using the BCEC cells used a range of MOIs. This revealed that MOI of at least 200 was required to achieve consistent bacterial recovery from BCEC cells 2 h post infection (data not shown), which is high but not unexpected as placental tissues are not easily or immediately invaded by L. monocytogenes [22]. Thus, all subsequent infections of BCEC cells were carried out using a MOI of 200. After 2 h of infection, very low levels of intracellular bacteria (mean 0.78–1.5 log10 CFU per 2 × 105 BCEC cells per well) were recovered and high levels of variability were observed between replicate infections. Although, the level detected was close to the detection limit (0.7 log10 CFU per 2 × 105 BCEC cells per well), all isolates tested were able to invade BCEC 2h post infections to a similar extent (Fig S1 A). A preliminary time course of 4–24 h incubation post-infection showed that 24h yielded the most consistent and reproducible levels of bacterial recovery (Fig S1 B), therefore this was used for further experiments.
Table 1.
Listeria monocytogenes isolates used in this study.
| Strain number | Sourcea | PCR Serotypec | PCR Lineaged | ST/CCe | Generation time [min]f | Source/Reference | |
|---|---|---|---|---|---|---|---|
| 10403S | Skin Lesion | 1/2a | II | 85 | CC7 | 50 ± 7.5 | (Bishop and Hinrichs, 1987) [55] |
| AR008 | Healthy eye | 1/2a, 3ab | IIb | 12 | CC7 | 83.33 ± 13.89g | (Warren et al., 2015) [21] |
| C00938 | Kerato-conjunctivitis | 1/2a, 3ab | IIb | 20 | CC20 | 43.48 ± 0.76 | APHA |
| R06262 | Kerato-conjunctivitis | 1/2b, 3bb | Ib | 59 | CC59 | 50 ± 10 | APHA |
| C02118 | Kerato-conjunctivitis | 4bb | Ib | 6 | CC6 | 45.45 ± 2.07 | APHA |
| LM7644 | Abortion | 1/2a, 3ab | IIb | 122 | CC9 | 62.5 ± 11.72h | APHA |
| C08389 | Abortion | 1/2a, 3ab | IIb | 7 | CC7 | 58.82 ± 17.3 | APHA |
| C08078 | Abortion | 1/2b, 3b | I | 59 | CC59 | 52.63 ± 5.54 | APHA |
| C07872 | Abortion | 1/2b, 3b | I | 59 | CC59 | 55.56 ± 6.17 | APHA |
| C04949 | Abortion | 1/2b, 3b | I | 59 | CC59 | 38.46 ± 8.88 | APHA |
| C07754 | Abortion | 1/2a, 3a | II | 91 | CC14 | 38.46 ± 2.96 | APHA |
| G03652 | Meningitis | 1/2b, 3bb | Ib | 59 | CC59 | 52.63 ± 5.54 | APHA |
| LM4 | Milk | 1/2b, 3bb | Ib | 1009 | CC5 | 66.67 ± 4.44i | (Lawrence et al., 1995) [56] |
| LM6 | Milk | 4bb | Ib | 1 | CC1 | 52.63 ± 5.54 | (Lawrence et al., 1995) [56] |
APHA: Animal and Plant Health Agency.
All isolates are from bovine sources except for the human isolate 10403S.
Warren et al., 2015 [21].
Serotypes were determined using the PCR-based method of Doumith et al (2004). This method in conjunction with the lineage typing cannot distinguish between serotypes 1/2a and 3a or 1/2b and 3b. However, serotypes 3a and 3b are not commonly isolated.
Lineages were determined using the PCR-based method of Ward et al. (2004).
Institute Pasteur Listeria MLST data base.
Growth rates in broth culture presented as mean +/- SD (n = 3).
Growth rate reduced compared to C04949 (p = 0.001).
Growth rate reduced compared to C04949 (p = 0.024).
Growth rate reduced compared to 10403S (p = 0.039).
Using this infection model, the ability of the different L. monocytogenes isolates to invade BCEC cells was investigated. Of the 14 isolates tested, four were significantly attenuated compared to the control strain (10403S). These were an isolate from a healthy bovine eye (AR008, P < 0.001), an isolate from a milk processing plant (LM4 P < 0.01) and two abortion isolates (7644 P < 0.05, C07754 P < 0.001) (Figure 2) and the percentage of intracellular bacteria recovered compared to the control were 1.9 %, 8 %, 0.5% and 33 %, respectively (Figure 2). No induction of extensive cell death was observed microscopically that would account for these low invasion rates.
Figure 2.
Infection of BCEC cells. BCEC cells were infected with an MOI = 200 with L. monocytogenes isolates for 24 h at 37 °C; For each isolate 5 independent experiments were performed, as represented by each symbol. Dark blue indicates isolates from cases of bovine abortion, pale blue indicates isolates from bovine keratoconjunctivitis, orange indicates isolates from an environmental source, green indicates isolates from a case of meningitis, pink indicates an isolate from a healthy eye and red indicates the control strain 10403S originally a human isolate from a skin lesion. All data points and mean are shown. Statistical significance is shown compared to 10403S: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, (Kruskal-Wallis test followed by Dunn's multiple comparisons test).
Differences in the ability of Listeria strains to internalise may be due to the variation in inlA or inlB expression levels therefore inlA and inlB mRNA levels were measured in the cultures used to inoculate the BCEC cells. All isolates expressed inlA and inlB mRNA and this was not found to vary between strains (Figure 3). As expected, inlA and inlB mRNA levels were positively correlated (r = 0.58, p = 0.03) and as previously reported the levels of inlB mRNA were consistently lower than those of inlA [23]. Thus, there was no evidence that differences in InlA or InlB levels in these strains would account for the differences in levels of intracellular bacteria recovered, although differences in gene sequences could not be ruled out at this stage.
Figure 3.
InlA and InlB expression.inlA and inlB transcript levels in L. monocytogenes isolates grown to late log phase in HI medium. For each isolate 3 independent experiments were performed, as represented by each symbol. Dark blue indicates isolates from cases of bovine abortion, pale blue indicates isolates from bovine keratoconjunctivitis, orange indicates isolates from an environmental source, green indicates isolates from a case of meningitis, pink indicates an isolate from a healthy eye and red indicates the control strain 10403S originally a human isolate from a skin lesion (Kruskal-Wallis test followed by Dunn's multiple comparisons test).
2.3. Sensitivity to lysozyme correlates with attenuated ability to proliferate in BCECs
We have previously reported the lysozyme sensitivity of these strains [21]. This was found to be an important factor in determining the ability of most L. monocytogenes strains to infect a bovine conjunctiva explant model [21]. Interestingly, the number of recovered bacteria from BCEC cells after 24 h of infection again correlated strongly with levels of lysozyme resistance (MIC [r = 0.82, p = 0.004]; bacterial survival in 580 μg lysozyme/ml [r = 0.72, p = 0.02]). Growth rate of the strains was determined in laboratory culture to see if this was a factor that might affect the number of intracellular bacteria recovered. However, there was no correlation with growth rate of these strains (r = 0.49, p > 0.05) or with the ability of the these strains to infect conjunctiva explants (r = 0.55, p > 0.05) (Table 2; for MIC and survival in lysozyme see [21]).
Table 2.
Pearson correlations.
| r | 95% conf interval | R2 | p-value | |
|---|---|---|---|---|
| mRNA expression: inlA vs InlB | 0.58 | 0.07 to 0.85 | 0.34 | 0.03 |
| Intracellular bacteria count (log CFU/well with: | ||||
| MIC lysozyme | 0.82 | 0.40 to 0.96 | 0.67 | 0.004 |
| Survival in 580 μg/ml lysozyme (log CFU/ml) | 0.72 | 0.16 to 0.93 | 0.52 | 0.02 |
| Growth rate in HI broth | 0.49 | -0.05 to 0.81 | 0.24 | 0.073 |
| Intracellular bacteria counts in conjunctiva | 0.55 | -0.12 to 0.88 | 0.31 | 0.097 |
p-values <0.05 were deemed significant and highlighted in bold.
To investigate the basis of differences in sensitivity to lysozyme of these strains, isolates were challenged with β-lactam antibiotics (ampicillin, penicillin G and cefuroxime) to test cell wall integrity and also a cationic peptide (mCRAMP) to test membrane integrity. Only one isolate, AR008 (isolated from a healthy bovine eye), showed significant increased sensitivity to ampicillin (p = 0.04), penicillin G (p = 0.004) and cefuroxime (p = 0.0001) compared to the reference strain 10403S, suggesting that a compromised cell wall may contribute to the lysozyme sensitivity of this isolate (Figure 4 A–C). Of the three lysozyme-sensitive isolates, only isolate 7644 showed sensitivity to mCRAMP, indicating that compromised cell membrane integrity may contribute to the lysozyme sensitivity of this isolate (Figure 4 D–F).
Figure 4.
Treatment of L. monocytogenes with cell wall acting antibiotics and CRAMP. To investigate cell wall integrity, overnight cultures were plated on heart infusion agar and disks containing 1U penicillin G (A), 25μg ampicillin (B) or 30μg cefuroxime sodium (C). The plates were incubated overnight at 37 °C, and zones of inhibition were measured. Dark blue indicates isolates from cases of bovine abortion, pale blue indicates isolates from bovine keratoconjunctivitis, orange indicates isolates from an environmental source, green indicates isolates from a case of meningitis, pink indicates an isolate from a healthy eye and red indicates the control strain 10403S originally a human isolate from a skin lesion. For each isolate 5 independent experiments were performed. All data points and mean are shown. Statistical significant increase in susceptibility is shown compared to 10403S using a. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001. (One Way ANOVA followed by Dunnett's multiple comparisons test). To assess cell membrane integrity, duplicate cultures of L. monocytogenes isolates AR008 (D), 7644 (E), C08389 (F) were grown to log phase in HI broth at 37 °C and stimulated with a final concertation of 10 mg ml−1 CRAMP/DMSO (closed circles) or DMSO alone (open circles). The red dashed line indicates time of stimulation. Absorbance at 600 nm was measured in 20 min intervals. Data are representative of at least duplicate experiments.
2.4. Analysis of L. monocytogenes sequence types, core genomes and virulence genes
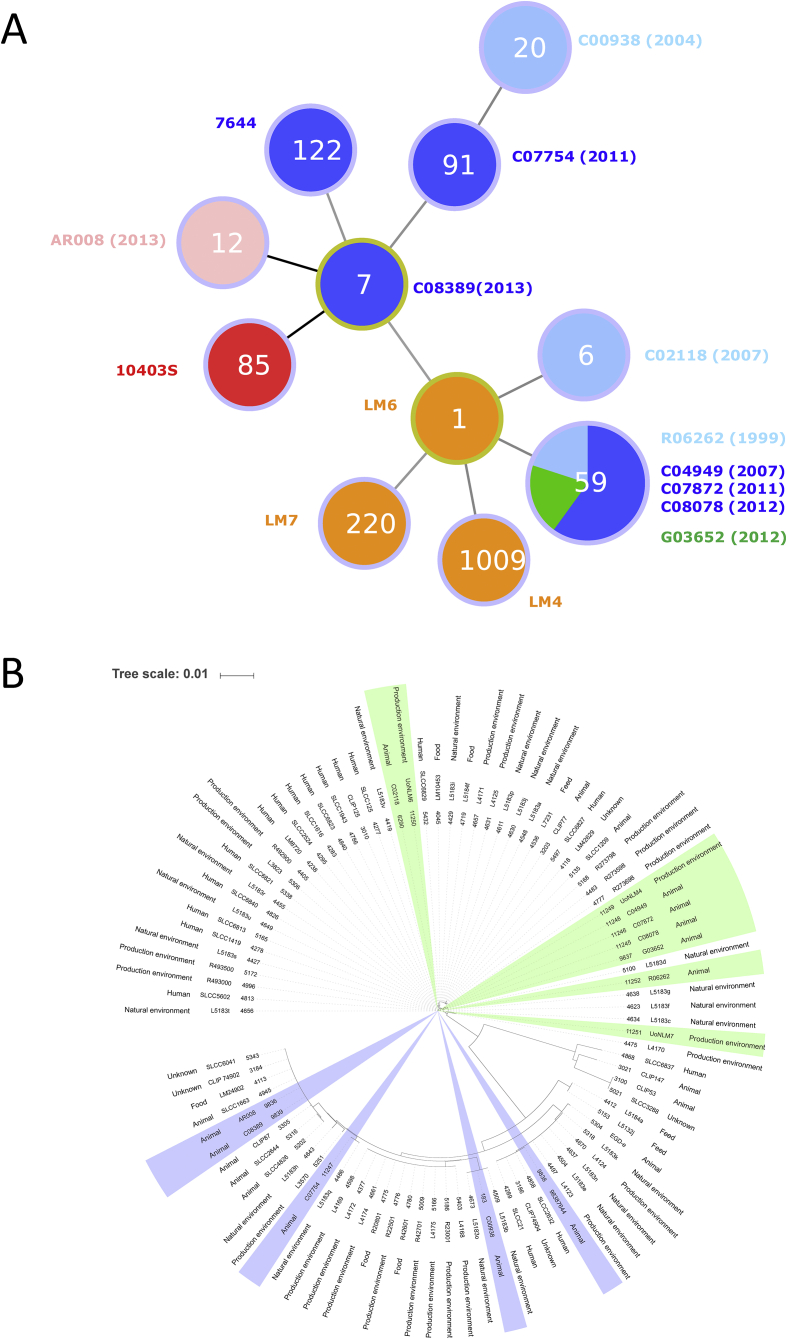
To further characterise these isolates, WGS was carried out on all the uncharacterised isolates and these sequence data were added to the open access Pasteur MLST database to determine sequence types (ST) (Table 1, Figure 5A). From the ten identified, eight were single STs, while five isolates belonged to ST59. Interestingly, while the ST59 isolates were collected over several years (1999–2012) and from three different clinical presentations (keratoconjunctivitis (n = 1), meningitis (n = 1), abortion (n = 3)), they were all able to infect and replicate inside BCEC cells at levels comparable to 10403S (Figures 2, 5).
Figure 5.
Epidemiological analysis of L. monocytogenes isolates based on MLST. (A) goeBURST analysis of the isolates used in this study; dark blue indicates isolates from cases of bovine abortion, pale blue indicates isolates from bovine keratoconjunctivitis, orange indicates isolates from an environmental source, green indicates isolates from a case of meningitis, pink indicates an isolate from a healthy eye and red indicates the control strain 10403S originally a human isolate from a skin lesion. Numbers in the circles denote the sequence type. (B) Maximum likelihood tree of the UK isolates present in the MLST database. Shaded areas correspond to the isolates used in this study, where green identifies linage I and blue identifies linage II.
Core genome analysis was carried out for the 128 isolates in the MLST database where a genome sequence was available (accessed 24/7/2017, Table S1). This showed that isolates C00938 (ST20), C07754 (ST91), C02118 (ST6), AR008 (ST12) and LM6 (ST1) all cluster with other isolates from these sequence types (Figure 6). Some clusters contained more than one sequence type, for instance LM7644 (ST122) clustered with sequence types ST9, ST622 and ST441. C08389 (ST7) is part of a cluster that also contains ST58 isolates (including 10403S) and ST98. Isolates LM4 (ST1009) and LM7 (ST220) were the only isolates of that sequence type present in the database but clustered closely with ST5 and ST194, respectively (Figure 6).
Figure 6.
L. monocytogenes core genome comparison. Maximum likelihood tree has been generated using the core genome of isolates in the MLST database. Shaded areas correspond to linages, where green indicates linage I, blue indicates linage II, orange indicates linage III and red indicates linage IV. Darker shading highlights the isolates used in this study.
Further analysis of the WGS data for virulence gene content (presence and absence of genes as well as sequence similarity), showed that they clustered according to their linage and serotype as expected (Figure 7, Table S2). Sequence identity in general was high, between 91.3-100%, except for inlK (90.1–100%) and genes encoding the sRNA family lhrC (90.1–100 %) (Figure 7, Table S2). Five genes were only present in the six isolates of the 1.2a, 3a serotype: inlL (adherence), ami (adherence), vip (invasion) and two genes of the lhrC family (non-coding regulatory sRNA) (Figure 7, Table S2). The absence of those genes did not correlate with attenuation in the context of BCEC infection. In contrast, the attenuated isolate LM4 was the only one in this study that lacked two virulence genes known to be involved in intracellular growth and virulence. These were the uhpT, the sugar phosphate antiporter important for intracellular proliferation [24, 25] and virR, a transcriptional two component response regulator implicated in cell invasion and virulence in vitro and in vivo [26, 27] (Table S2). In addition, LM4 had three other virulence genes (srtB (surface display), sipZ (intracellular survival) and inlC (internalin)) with the lowest reported level of sequence identity to the reference genome (Table S2).
Figure 7.
Virulence gene analysis. Heat map illustrating percentage identity of 87 L monocytogenes virulence genes in comparison to isolate EDG-e determined through virulence finder [64], with crosses denoting the absence of genes. The gene matrix represents from top to bottom, genes involved in teichoic acid biosynthesis (gtcA), located in pathogenicity island LIPI-1 (actA, hly, mpl, plcAB, prfA), genes coding for internalins (inlABCFHLKL) and other genes involved in adherence (ami, dltA, fbpA, lap, lapB), invasion (aut, iap, lpeA, recA, vip), Intracellular survival (clpBCEP, dal, fri, htrA, lplA1, oppA, perR, prsA2, pvcA, relA, sipZ, sod, svpA, tig, uHpt), regulation of transcription and translation (ctsR, fur, gmar, hfg, lhrc, lisKR, mogR, rsbv, sigB, stp, virk, rli55, rli60), surface display (lgt, lsp, sipX, srtAB, secA2), peptidoglycan modification (degU, murA, oatA, pgdA), membrane integrity (ctap, mrpf), motility (flaA, flgCE), anaerobic growth (eut), regulation of metabolism (codY), immunomodulation (chiA, lipA, lnyA, pgl) and bile resistance (bile, bsh).
Analysis of the inlA sequences of these isolates for known changes that would be predicted to reduce levels of InlA (i.e. frameshifts causing premature stop codons or mutations in the promoter region [28]) was also performed. In agreement with the results gained from the inlA mRNA analysis (Figure 3), no differences were identified in the inlA and actA promoter regions that might contribute to the lower levels of cell invasion and intracellular replication recorded. Similarly, InlP had 94% identity at the protein level across all isolates and therefore this did not seem to provide an explanation for the variation seen in the ability of these strains to infect the BCEC cells.
3. Discussion
L. monocytogenes has been isolated from placentomes of infected cattle [1]. The maternal caruncle contains a dense network of blood vessels [29] allowing Listeria access to the maternal side of the placenta through the blood stream. The caruncle is also in close contact with the fetal chorion, meaning infection of the uterus can lead to endotoxaemia, an increased prostaglandin synthesis and subsequent lysis of the corpus luteum, leading to abortion. Alternatively, placentitis itself can disrupt the metabolic exchange of nutrients to the fetus, triggering the abortion [30]. While BCECs have been used for L. monocytogenes infections as a comparison to other tissues previously [31, 32] this study presents BCEC cells as infection model to characterise bovine L. monocytogenes isolates from different clinical presentations and sources. Genome sequencing of the isolates used in this study revealed that within our set of bovine clinical isolates, collected across the UK over several years and from different disease presentations, the MLST sequence type ST59 was over represented, with 5 out of the 10 clinical isolates belonging to that sequence type. Core MLST analysis further confirmed that they are closely related, forming a distinct cluster with other ST59 isolates. This sequence type has been previously reported to be isolated in France predominantly associated with human infections [33]. In this study it was noted that the clones most prevalent in human patients were also those most frequently associated with bovine listeriosis, and the ST59 strains in this study were isolated from cases of Kerato-conjunctivitis, abortion (3 isolates) and meningitis. This suggests that these bovine strains may be transmitted to human through the food chain [33]. Interestingly, one of our cattle isolates has the MLST type ST6 (C02118, keratoconjunctivitis isolate, 2007), which is the sequence type identified in the large outbreaks of human disease in Europe and South Africa during 2017/2018. As sequence type and core genome analyses revealed that isolates from different clinical diseases, as well as from different species (human/cattle) cluster together, this suggests that it is less likely that the ability of L. monocytogenes to infect different host species is due to species-specific virulence factors, but more subtle variation in gene sequence influencing host interactions of this pathogen.
L. monocytogenes infected BCECs at low efficiency and required a high MOI. In other species, such as pregnant guinea pigs, colonisation of the placenta was initially slow with 103-104 fewer bacteria seen in the placenta than in the liver and spleen immediately after intravenous inoculation [22]. This suggests that placental tissues are not easily or immediately invaded by L. monocytogenes. This is consistent with our findings that low numbers of bacteria were recovered from caruncular cells 2 h post infection with a wide range of variation. However, the invasion of a single bacterium into the placenta of guinea pigs can be sufficient to cause an abortion. Once colonised, there is poor bacterial clearance from the placenta and replication allows Listeria spp. to recolonise maternal tissues [22]. This is consistent with our findings that at 24 h post infection, higher numbers of bacteria were recovered from BCECs with less variation between infection experiments. This also suggests that any isolates able to invade BCECs may be able to cause an abortion in utero if they are able to grow within these tissues. Entry points towards systemic infections in cattle may include small breaches of oral mucosa from rough feeding material which may lead to repeat exposure of L. monocytogenes through contaminated silage [34].
The use of the BCEC infection model allowed us to identify strains with different potential to infect this cell type. Surprisingly, given that this isolate originated from a bovine abortion case, isolate 7644 was highly attenuated in the BCEC infection model. However, this strain was found to belong to ST9 and these isolates have been reported to be hypovirulent and are most commonly isolated from food sources rather than clinical infections [15]. Since identification of Listeria from aborted foetuses is problematic, the possibility exists that this isolate was a post-abortive environmental contaminant rather than the causative agent of infection. Alternatively, the animal may have been challenged with a high infectious dose. This assumption was previously proposed for a field isolate from a bovine abortion, which had a truncated PrfA, and was strongly attenuated in infection experiments with a wide range of cell types [32]. However, loss of infectivity may also be due to mutations accumulated during long term culture of the bacteria in a laboratory environment. In our previous study, both LM4 and 7644 were attenuated in a Caco2 infection model, whereas AR008 was able to infect Caco2 cells at similar levels to the control strain 10403S [21]. The fact that AR008 was attenuated in the BCEC model when other strains were able to infect successfully suggests that there are specific factors in these bovine placental cells involved in the interactions with L. monocytogenes.
Previously, we have shown that resistance to lysozyme was a positive predictive factor for infection of bovine conjunctiva explant model [21] but genome analysis performed in this study did not reveal any differences between the isolates used in the genes suspected to be involved in lysozyme resistance (pdgA, oatA, degU) [ 21,35, 36]. Interestingly, there was again a strong correlation between L. monocytogenes replication in BCECs 24 h post-infection and their level of lysozyme resistance. In cattle, lysozyme activity in most tissues is relatively low compared to other species, except for the abomasum [37] and tear fluid [38] which both have high levels of lysozyme activity. This may explain the co-selection for high levels of lysozyme resistance found in isolates from conjunctivitis as well as from other sites (reproductive tissues/fetus, brain and milk) that require the bacteria to survive passage through the abomasum. In addition, degradation of L. monocytogenes cell wall by lysozyme leads to the release of peptidoglycan and its breakdown products that are ligands for the pattern recognition receptors of the innate immune system, such as Nod1, Nod2 and Toll-like receptor (TLR) 2 [39, 40, 41, 42]. This is illustrated by L. monocytogenes lacking pgdA, which was not only highly attenuated in its virulence in vivo and in vitro but also elicited a strong TLR2 and Nod1 dependent interferon-ß response [43]. This suggests that lysozyme resistance may contribute to L. monocytogenes virulence in two different manners, by increasing bacterial survival as well as modulating the host response [44].
WGS identified the absence of virulence genes virR and uhpT in LM4 which potentially explains the attenuation of this isolate in the BCEC infection model. VirR is part of a two-component regulator (VirR–VirS) which is required for the virulence of Listeria in vivo [27]. It was also found that virR mutants are affected in their entry into Caco2 cells [26] and we have previously reported that LM4 also has a reduced capacity to invade this cell type [21]. The sugar phosphate antiporter UhpT promotes the uptake of phosphorylated hexoses during cytosolic growth [25] and deletion of this gene also leads to impaired intracellular proliferation in Caco2 cells [24]. Therefore, as LM4 lacks these two genes, it would be predicted that it would be less able to grow in BCEC cells. Interestingly, in our previous study, LM4 was not significantly attenuated in its ability to invade and proliferate in bovine conjunctiva tissues [21] but perhaps in that infection model the high level of resistance to lysozyme may compensate for any reduced intracellular growth. VirRS is also known to control the expression of a set of 17 genes, several of which affect bacterial cell wall and membrane integrity and, virR mutants are reported to be more sensitive to some beta-lactam antibiotics, including penicillin and cefuroxime [45]. However, LM4 did not show increased levels of sensitivity to these two antibiotics, or to challenge with cationic peptides, suggesting in the absence of VirR the genes in this operon are regulated in a different manner.
4. Conclusion
L. monocytogenes is a highly versatile and adaptive bacterium, with the ability to not only infect a wide range of tissues within a host, but also to infect a wide range of physiologically distinct animal hosts. The placentome cell model provides a novel tool to characterise the infection processes carried out by Listeria spp. in a different host, where different host factors may influence the infection process. Correlating WGS analysis with infection models provides more insights into the underlying basis of differences in strain of Listeria.
5. Materials and methods
5.1. Bacterial culture
Listeria monocytogenes strains used in the infection studies are listed in Table 1. Bacteria were cultured overnight (approximately 17h) at 37 °C in 5ml Heart infusion (HI) broth or on HI agar plates (Oxoid, UK). Growth was monitored using optical density (OD600nm) and cultures were diluted depending on the multiplicity of infection (MOI) required for infection experiments. The precise CFU/ml of the inoculum was then determined by serial dilution and plating on HI agar. To determine the growth rates and generation times of the isolates, overnight cultures were diluted in HI broth to OD600 = 0.01. Growth was monitored using optical density (OD600nm) and serial dilution plated on HI agar.
5.2. Multiplex PCR assay for Listeria monocytogenes serotyping
Multiplex PCR was performed in order to separate the four major serovars (1/2a, 1/2b, 1/2c, and 4b) and three main lineages (I, II, III) of L. monocytogenes [46, 47]. To prepare template DNA, three to six colonies resuspended in 1 ml of sterile water were incubated at 90 °C for 10min and then chilled on ice for 10min; 1μl of this was used as template DNA for each PCR reaction.
5.3. Cell culture and infections
Bovine caruncular epithelial cell line BCEC-1 (BCEC), provided by Prof. C. Pfarrer [18], were grown in DMEM/Ham's nutrient mixture F12 1:1 (Sigma-Aldrich, UK) with 10% (v/v) fetal calf serum (Sigma), 2mM L-Glutamine and 100 U/ml penicillin/streptomycin (Gibco) at 37 °C with 5% CO2 [19]. BCECs were seeded into 24-well plates (Thermo Scientific, UK) in 500μl of complete medium and grown to confluence. One hour before infection, the complete medium was replaced with antibiotic-free medium and the plate incubated at 37 °C. Cells were infected with an MOI of 200 (n ≥ 5, for details see result section), and incubated for 1h. Medium was then removed from the wells and replaced with medium containing 100 μg/ml gentamycin (Sigma) to kill extracellular bacteria. After a further 1h incubation, the medium was replaced with medium containing 5 μg/ml gentamycin and incubated for 2–24hr post-infection. To enumerate intracellular bacteria, cells were washed three times with pre-warmed (37 °C) PBS and lysed by addition of 100μl of ice-cold 0.5% (v/v) Triton-X-100 (Fisher Scientific, UK) per well. This was incubated on ice for 20min and the resultant lysate serially diluted in PBS before 10μl samples were plated using the Miles Misra technique onto HI agar and incubated at 37 °C overnight. Then, the CFU/ml of lysates was calculated.
5.4. Antibiotic resistance screening
Samples (100μl) of each of the 14 isolates of L. monocytogenes were spread onto individual HI agar plates. Disks of penicillin G (1U), cefuroxime/sodium (30μg), oxacillin (1μg), ampicillin (25μg) and ciprofloxacin (1μg) (Oxoid Ltd, Basingstoke, UK) were immediately placed on top of the spread culture. The plates were then incubated at 37 °C overnight and the zones of inhibition were measured (mm).
5.5. Antimicrobial peptide challenge assay
The antimicrobial peptide challenge was performed as outlined by Burke et al. (2014) [35] using mouse cathelicidin-related antimicrobial peptide (H-GLLRKGGEKIGEKLKKIGQKIKNFFQKLVPQPEQ-OH; Isca Biochemicals, Exeter, UK) [48] at a final concentration of 10 μg/ml (stock concentration: 1 mg/ml in dimethyl sulfoxide (DMSO)).
5.6. Isolation of RNA, cDNA synthesis and quantitative (q) PCR
Late log phase culture containing approximately 109 CFU/ml was centrifuged at 13000xg for 2 min at room temperature. The pelleted cells were suspended in 1ml RNAlater (Sigma Aldrich) and incubated for 1h at room temperature. The suspension was centrifuged at 13000xg for 5min and the supernatant removed. The pelleted cells were suspended in 375μl of freshly prepared cell wall disruption buffer (30 U/ml mutanolysin, 10 mg/ml lysozyme in 10ml of 10mM Tris, 1mM EDTA buffer, pH 8), incubated at 37 °C for 30 min and then centrifuged at 13000xg for 5 min at room temperature. RNA was extracted using NuceleoSpin®RNA isolation kit (Macherey-Nagel, UK) following manufacturer's instructions.
For BCEC RNA extractions, the supernatant was removed and cells were lysed with 350μl of RNA lysis buffer (Nucleospin®RNA isolation kits, Machery-Nagel, UK) followed by RNA isolation according to manufacturer's instructions. Eluted RNA was quantified using Qubit (Qiagen) and stored at -80 °C. RNA was diluted in water and cDNA was synthesized using MMLV reverse transcriptase (Promega, Madison, USA) according to manufacturer's instructions. The final volume of each reaction was diluted in RNAse/DNAse free water (Fischer Scientific, UK).
All quantitative real time PCR (qPCR) experiments were designed and performed to comply with the quality controls detailed in the MIQE guidelines (Bustin et al., 2010). Quantitative PCR was performed using a LightCycler® 480 (Roche, Hertfordshire, UK). For primer sequences see Table 3. For bacterial and host gene expression, qPCR was performed in 20μl reactions with 0.25mM of each of the forward and reverse primer, 2X Luminoct SYBR Green qPCR ready mix (Sigma-Aldrich, Dorset, UK), 25ng of cDNA and PCR grade water (Roche, Hertfordshire, UK). An initial denaturation cycle of 95 °C for 10min was used followed by 45 cycles of 10 s at 95 °C, 50 s at 60 °C and 1 min at 72 °C and a final extension of 10 min at 72 °C. Normalized gene expression of each gene was calculated based on the method described by Hughes et al 2007 [49]: Differentiation factor = (overall mean of 40 − CP value for reference gene)/(40 − CP value of reference gene of that sample); Normalised expression = (Mean (40 − CP value for the sample) × target primer slope)/(Differentiation factor for that sample × reference gene primer slope). Fold change = 2(T−C), whereby T = normalised expression level of treated samples; C = s normalised expression level of control samples.
Table 3.
Primers.
| Target | Sequence | Gene reference | Size (bp)/efficiency (%, qPCR) | Reference |
|---|---|---|---|---|
| Bovine primers | ||||
| GAPDH | F = AGTTCAACGGCACAGTCAAG R = AGCAGGGATGATATTCTGGG |
NM_001034034 | 463 bp | This study |
| E-cadherin | F = GGTCAAAGAGCCCTTACTGC R = TGGCTCAAGTCAAAGTCCTG | AY508164.1 | 105 bp | |
| C-met | F = TGAAGGAGGGACAACACTGA R = TAAGGTGCAGCTCTCATTGC | NM_001012999.2 | 112 bp | |
| β actin | F = GAAGGTGACAGCAGTCGGT R = TTTCGCGATATTTGGAATGA | BT030480.1 | 114 bp | |
| Listeria monocytogenes primers (qPCR) | ||||
| 16srRNA | F-CTTCCGCAATGGACGAAAGT R- ACGATCCGAAAACTTCTTCATAC |
95% | Werbrouck et al 2006 [57] | |
| TufA | F- GCTGAAGCTGGCGACAACA R- CTTGACCACGTTGGATATCTTCAC |
102% | ||
| InlA | F- GAACCAGCTAAGCCIGTAAAAG R- CGCCIGTTTGGGCATCA |
95% | ||
| InlB | F- GGAAAAGCAAAAGCAIGATTC R- TCCATCAACATCATAACTTACTGTGTAAA |
92% | ||
5.7. WGS and sequence analysis
DNA was extracted using the Cador Pathogen Minikit (Qiagen) following manufacturer's recommendations. High throughput sequencing was performed at MicrobesNG (Birmingham U.K.). Genomic DNA libraries were prepared using Nextera XT Library Prep Kit (Illumina, San Diego, USA) following the manufacturer's protocol and sequenced using Illumina MiSeq. Raw reads were assembled using the A5-MiSeq pipeline [50] and contigs were uploaded to the Pasteur MLST database were they are publicly available and the MLST sequence type was determined (http://bigsdb.pasteur.fr/listeria/listeria.html).
5.8. Multi sequence alignments
All nucleotide and protein alignments were completed using secondary structure aware high throughput multi-sequence alignment DECIPHER [51] (R script available at https://github.com/ADAC-UoN/DECIPHER-Sequence-Alignment.git). Trees were calculated using maximum likelihood by Fasttree double precision (version 2.1.8) [52] and visualised in iTOL [53].
5.9. Virulence finder
Assembled genome files were uploaded to the virulence finder online Listeria database at the Danish Centre For Genomic Epidemiology (https://cge.cbs.dtu.dk/services/VirulenceFinder/version 1.5) [54] and searches were performed against reference isolate EDG-e, using a minimum of 90% identity along 80% of the coding sequence.
5.10. Statistical analysis
Statistical analysis of data was performed using GraphPad Prism 6.05. To compare the growth rates of L. monocytogenes isolates, a one-way ANOVA was carried out followed by Dunn's multiple comparison test. To compare isolates in an infection context, a Kruskal-Wallis test was carried out, followed by Dunn's multiple comparisons test. Pearson's correlations were performed to compare data sets. L. monocytogenes sequence type distributions were analysed using Fisher's exact test. Significance was reported for p < 0.05.
Declarations
Author contribution statement
Adam M. Blanchard, Sabine Tötemeyer: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Rosemarie Billenness: Performed the experiments; Analyzed and interpreted the data.
Jessica Warren, Amy Glanvill, William Roden, Emma Drinkall, Grazieli Maboni: Performed the experiments.
Robert S Robinson, Catherine E.D. Rees: Conceived and designed the experiments; Wrote the paper.
Christiane Pfarrer: Contributed reagents, materials, analysis tools or data.
Funding statement
This work was supported by the Biotechnology and Biological Sciences Research Council [BB/I024291/1] (BBSRC), to JW (BBSRC Research Experience Placement)and ED (BBSRC Doctoral Training Partnership) and the University of Nottingham; RB was awarded a Microbiology Society Harry Smith Vacation Studentship.
Competing interest statement
The authors declare no conflicts of interest.
Additional information
No additional information is available for this paper.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
Figure S1. Infection of BCEC cells for 2-24h. BCEC cells were infected with an MOI = 200 with L. monocytogenes isolates for 2–24 h at 37 °C. (A) BCEC cells were infected for 2 h with nine different isolates, for each isolate 6–11 independent experiments were performed as represented by each symbol. Dark blue indicates isolates from cases of bovine abortion, pale blue indicates isolates from bovine keratoconjunctivitis, orange indicates an isolate from an environmental source, pink indicates an isolate from a healthy eye and red indicates the control strain 10403S originally a human isolate from a skin lesion. All data points and mean are shown. (B) BCEC cells were infected for 4–24 h with four different isolates, for each isolate 3 independent experiments were performed, and average and standard deviation are shown. Statistical significance is shown: ∗p < 0.05, ∗∗p < 0.01.
Table S1. Multilocus sequence type metadata. Metrics associated with all the isolates held in the MLST database.
Table S2. Determination of virulence associated genes. Whole genome sequences were parsed through virulence finder [64] to identify putative virulence associated genes for the isolates used in this study. Green cells indicate greater than 99% similarity to virulent gene loci, orange cells indicate 95–98% similarity and red indicate between 90-94% similarity.
References
- 1.Johnson C.T., Lupson G.R., Lawrence K.E. The bovine placentome in bacterial and mycotic abortions. Vet. Rec. 1994 Mar 12;134(11):263–266. doi: 10.1136/vr.134.11.263. [DOI] [PubMed] [Google Scholar]
- 2.Yaeger M.J., Holler L.D. Chapter 49 - bacterial causes of bovine infertility and abortion. In: Youngquist R.S., Threlfall W.R., editors. Current Therapy in Large Animal Theriogenology. second ed. W.B. Saunders; Saint Louis: 2007. pp. 389–399.http://www.sciencedirect.com/science/article/pii/B9780721693231500520 [Internet].[cited 2020 Jun 12] Available from: [Google Scholar]
- 3.Cabell E. Bovine abortion: aetiology and investigations. In Pract. 2007 Sep 1;29(8):455–463. [Google Scholar]
- 4.Lecuit M. Listeria monocytogenes, a model in infection biology. Cell Microbiol. 2020 Apr;22(4) doi: 10.1111/cmi.13186. [Internet] [cited 2020 Jun 12] Available from: [DOI] [PubMed] [Google Scholar]
- 5.Robbins J.R., Bakardjiev A.I. Pathogens and the placental fortress. Curr. Opin. Microbiol. 2012 Feb 1;15(1):36–43. doi: 10.1016/j.mib.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rocha C.E., Mol J.P.S., Garcia L.N.N., Costa L.F., Santos R.L., Paixão T.A. Comparative experimental infection of Listeria monocytogenes and Listeria ivanovii in bovine trophoblasts. PloS One. 2017 May 3;12(5) doi: 10.1371/journal.pone.0176911. Lenz LL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lecuit M., Dramsi S., Gottardi C., Fedor-Chaiken M., Gumbiner B., Cossart P. A single amino acid in E-cadherin responsible for host specificity towards the human pathogen Listeria monocytogenes. EMBO J. 1999 Jul 15;18(14):3956–3963. doi: 10.1093/emboj/18.14.3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khelef N., Lecuit M., Bierne H., Cossart P. Species specificity of the Listeria monocytogenes InlB protein. Cell Microbiol. 2006;8(3):457–470. doi: 10.1111/j.1462-5822.2005.00634.x. [DOI] [PubMed] [Google Scholar]
- 9.Gessain G., Tsai Y.-H., Travier L., Bonazzi M., Grayo S., Cossart P. PI3-kinase activation is critical for host barrier permissiveness to Listeria monocytogenes. J. Exp. Med. 2015 Feb 9;212(2):165–183. doi: 10.1084/jem.20141406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Disson O., Grayo S., Huillet E., Nikitas G., Langa-Vives F., Dussurget O. Conjugated action of two species-specific invasion proteins for fetoplacental listeriosis. Nature. 2008 Oct;455(7216):1114–1118. doi: 10.1038/nature07303. [DOI] [PubMed] [Google Scholar]
- 11.Lingnau A., Domann E., Hudel M., Bock M., Nichterlein T., Wehland J. Expression of the Listeria monocytogenes EGD inlA and inlB genes, whose products mediate bacterial entry into tissue culture cell lines, by PrfA-dependent and -independent mechanisms. Infect. Immun. 1995 Oct 1;63(10):3896–3903. doi: 10.1128/iai.63.10.3896-3903.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGann P., Wiedmann M., Boor K.J. The alternative sigma factor σB and the virulence gene regulator PrfA both regulate transcription of Listeria monocytogenes internalins. Appl. Environ. Microbiol. 2007 May 1;73(9):2919–2930. doi: 10.1128/AEM.02664-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim H., Marquis H., Boor K.J. SigmaB contributes to Listeria monocytogenes invasion by controlling expression of inlA and inlB. Microbiology. 2005;151(10):3215–3222. doi: 10.1099/mic.0.28070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Handa-Miya S., Kimura B., Takahashi H., Sato M., Ishikawa T., Igarashi K. Nonsense-mutated inlA and prfA not widely distributed in Listeria monocytogenes isolates from ready-to-eat seafood products in Japan. Int. J. Food Microbiol. 2007 Jul 15;117(3):312–318. doi: 10.1016/j.ijfoodmicro.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 15.Jacquet C., Doumith M., Gordon J.I., Martin P.M.V., Cossart P., Lecuit M. A molecular marker for evaluating the pathogenic potential of foodborne Listeria monocytogenes. J. Infect. Dis. 2004 Jun;189(11):2094–2100. doi: 10.1086/420853. [DOI] [PubMed] [Google Scholar]
- 16.Faralla C., Rizzuto G.A., Lowe D.E., Kim B., Cooke C., Shiow L.R. InlP, a new virulence factor with strong placental tropism. Infect. Immun. 2016 Dec 1;84(12):3584–3596. doi: 10.1128/IAI.00625-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faralla C., Bastounis E.E., Ortega F.E., Light S.H., Rizzuto G., Gao L. Listeria monocytogenes InlP interacts with afadin and facilitates basement membrane crossing. Tsolis RM, editor. PLoS Pathog. 2018 May 30;14(5) doi: 10.1371/journal.ppat.1007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bridger P.S., Haupt S., Klisch K., Leiser R., Tinneberg H.-R., Pfarrer C. Validation of primary epitheloid cell cultures isolated from bovine placental caruncles and cotyledons. Theriogenology. 2007 Sep 1;68(4):592–603. doi: 10.1016/j.theriogenology.2007.05.046. [DOI] [PubMed] [Google Scholar]
- 19.Bridger P.S., Menge C., Leiser R., Tinneberg H.-R., Pfarrer C.D. Bovine caruncular epithelial cell line (BCEC-1) isolated from the placenta forms a functional epithelial barrier in a polarised cell culture model. Placenta. 2007 Nov 1;28(11):1110–1117. doi: 10.1016/j.placenta.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niemann H.H., Jäger V., Butler P.J.G., van den Heuvel J., Schmidt S., Ferraris D. Structure of the human receptor tyrosine kinase met in complex with the Listeria invasion protein InlB. Cell. 2007 Jul;130(2):235–246. doi: 10.1016/j.cell.2007.05.037. [DOI] [PubMed] [Google Scholar]
- 21.Warren J., Owen A.R., Glanvill A., Francis A., Maboni G., Nova R.J. A new bovine conjunctiva model shows that Listeria monocytogenes invasion is associated with lysozyme resistance. Vet. Microbiol. 2015 Aug 31;179(1):76–81. doi: 10.1016/j.vetmic.2015.02.029. [DOI] [PubMed] [Google Scholar]
- 22.Bakardjiev A.I., Theriot J.A., Portnoy D.A. Listeria monocytogenes traffics from maternal organs to the placenta and back. PLoS Pathog. 2006 Jun 30;2(6):e66. doi: 10.1371/journal.ppat.0020066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamburro M., Sammarco M.L., Ammendolia M.G., Fanelli I., Minelli F., Ripabelli G. Evaluation of transcription levels of inlA, inlB, hly, bsh and prfA genes in Listeria monocytogenes strains using quantitative reverse-transcription PCR and ability of invasion into human CaCo-2 cells. FEMS Microbiol. Lett. 2015;362(6) doi: 10.1093/femsle/fnv018. https://academic.oup.com/femsle/article/362/6/fnv018/581427 [Internet] Mar 1 [cited 2020 Jun 12] Available from: [DOI] [PubMed] [Google Scholar]
- 24.Chico-Calero I., Suárez M., González-Zorn B., Scortti M., Slaghuis J., Goebel W. Hpt, a bacterial homolog of the microsomal glucose- 6-phosphate translocase, mediates rapid intracellular proliferation in Listeria. Proc. Natl. Acad. Sci. Unit. States Am. 2002 Jan 8;99(1):431–436. doi: 10.1073/pnas.012363899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chatterjee S.S., Hossain H., Otten S., Kuenne C., Kuchmina K., Machata S. Intracellular gene expression profile of Listeria monocytogenes. Infect. Immun. 2006 Feb 1;74(2):1323–1338. doi: 10.1128/IAI.74.2.1323-1338.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mandin P., Fsihi H., Dussurget O., Vergassola M., Milohanic E., Toledo-Arana A. VirR, a response regulator critical for Listeria monocytogenes virulence. Mol. Microbiol. 2005;57(5):1367–1380. doi: 10.1111/j.1365-2958.2005.04776.x. [DOI] [PubMed] [Google Scholar]
- 27.Camejo A., Buchrieser C., Couvé E., Carvalho F., Reis O., Ferreira P. In vivo transcriptional profiling of Listeria monocytogenes and mutagenesis identify new virulence factors involved in infection. PLoS Pathog. 2009 May 29;5(5) doi: 10.1371/journal.ppat.1000449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moura A., Criscuolo A., Pouseele H., Maury M.M., Leclercq A., Tarr C. Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat Microbiol. 2016 Oct 10;2(2):1–10. doi: 10.1038/nmicrobiol.2016.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Senger P. Pathways to pregnancy and parturition. Psychiatr Rehabil. 2005;35:381. J. [Google Scholar]
- 30.Miller R.B. A summary of some of the pathogenetic mechanisms involved in bovine abortion. Can. Vet. J. 1977 Apr;18(4):87–95. [PMC free article] [PubMed] [Google Scholar]
- 31.Rupp S., Bärtschi M., Frey J., Oevermann A. Hyperinvasiveness and increased intercellular spread of Listeria monocytogenes sequence type 1 are independent of listeriolysin S, internalin F and internalin J1. J. Med. Microbiol. 2017 Jul;66(7):1053–1062. doi: 10.1099/jmm.0.000529. [DOI] [PubMed] [Google Scholar]
- 32.Rupp S., Aguilar-Bultet L., Jagannathan V., Guldimann C., Drögemüller C., Pfarrer C. A naturally occurring prfA truncation in a Listeria monocytogenes field strain contributes to reduced replication and cell-to-cell spread. Vet. Microbiol. 2015 Aug 31;179(1):91–101. doi: 10.1016/j.vetmic.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 33.Maury M.M., Bracq-Dieye H., Huang L., Vales G., Lavina M., Thouvenot P. Hypervirulent Listeria monocytogenes clones’ adaption to mammalian gut accounts for their association with dairy products. Nat. Commun. 2019 Dec;10(1):2488. doi: 10.1038/s41467-019-10380-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Low J.C., Donachie W. A review of Listeria monocytogenes and listeriosis. Vet. J. 1997 Jan 1;153(1):9–29. doi: 10.1016/s1090-0233(97)80005-6. [DOI] [PubMed] [Google Scholar]
- 35.Burke T.P., Loukitcheva A., Zemansky J., Wheeler R., Boneca I.G., Portnoy D.A. Listeria monocytogenes is resistant to lysozyme through the regulation, not the acquisition, of cell wall-modifying enzymes. J. Bacteriol. 2014 Nov 1;196(21):3756–3767. doi: 10.1128/JB.02053-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aubry C., Corr S.C., Wienerroither S., Goulard C., Jones R., Jamieson A.M. Both TLR2 and TRIF contribute to interferon-β production during Listeria infection. PloS One. 2012 Mar 14;7(3) doi: 10.1371/journal.pone.0033299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prieur D.J. Tissue specific deficiency of lysozyme in ruminants. Comp. Biochem. Physiol. Part B Comp Biochem. 1986 Jan 1;85(2):349–353. doi: 10.1016/0305-0491(86)90011-8. [DOI] [PubMed] [Google Scholar]
- 38.Sotirov L., Semerdjiev V., Maslev T., Draganov B. Breed-related differences in blood lysozyme concentration and complement activity in cows in Bulgaria. Rev. Med. Vet. 2007;6 [Google Scholar]
- 39.Kobayashi K.S., Chamaillard M., Ogura Y., Henegariu O., Inohara N., Nuñez G. Nod2-Dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005 Feb 4;307(5710):731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 40.Kobayashi K., Inohara N., Hernandez L.D., Galán J.E., Núñez G., Janeway C.A. RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature. 2002 Mar;416(6877):194–199. doi: 10.1038/416194a. [DOI] [PubMed] [Google Scholar]
- 41.Opitz B., Püschel A., Beermann W., Hocke A.C., Förster S., Schmeck B. Listeria monocytogenes activated p38 MAPK and induced IL-8 secretion in a nucleotide-binding oligomerization domain 1-dependent manner in endothelial cells. J. Immunol. 2006 Jan 1;176(1):484–490. doi: 10.4049/jimmunol.176.1.484. [DOI] [PubMed] [Google Scholar]
- 42.Torres D., Barrier M., Bihl F., Quesniaux V.J.F., Maillet I., Akira S. Toll-like receptor 2 is required for optimal control of Listeria monocytogenes infection. Infect. Immun. 2004 Apr 1;72(4):2131–2139. doi: 10.1128/IAI.72.4.2131-2139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boneca I.G., Dussurget O., Cabanes D., Nahori M.-A., Sousa S., Lecuit M. A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proc. Natl. Acad. Sci. Unit. States Am. 2007 Jan 16;104(3):997–1002. doi: 10.1073/pnas.0609672104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ragland S.A., Criss A.K. From bacterial killing to immune modulation: recent insights into the functions of lysozyme. PLoS Pathog. 2017 Sep 21;13(9) doi: 10.1371/journal.ppat.1006512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Collins B., Curtis N., Cotter P.D., Hill C., Ross R.P. The ABC transporter AnrAB contributes to the innate resistance of Listeria monocytogenes to nisin, bacitracin, and various β-lactam antibiotics. Antimicrob. Agents Chemother. 2010 Oct 1;54(10):4416–4423. doi: 10.1128/AAC.00503-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doumith M., Buchrieser C., Glaser P., Jacquet C., Martin P. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 2004 Aug;42(8):3819–3822. doi: 10.1128/JCM.42.8.3819-3822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ward T.J., Gorski L., Borucki M.K., Mandrell R.E., Hutchins J., Pupedis K. Intraspecific phylogeny and lineage group identification based on the prfA virulence gene cluster of Listeria monocytogenes†. J. Bacteriol. 2004 Aug 1;186(15):4994–5002. doi: 10.1128/JB.186.15.4994-5002.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fritsche S., Knappe D., Berthold N., Buttlar H von, Hoffmann R., Alber G. Absence of in vitro innate immunomodulation by insect-derived short proline-rich antimicrobial peptides points to direct antibacterial action in vivo. J. Pept. Sci. 2012;18(10):599–608. doi: 10.1002/psc.2440. [DOI] [PubMed] [Google Scholar]
- 49.Hughes S., Poh T.-Y., Bumstead N., Kaiser P. Re-evaluation of the chicken MIP family of chemokines and their receptors suggests that CCL5 is the prototypic MIP family chemokine, and that different species have developed different repertoires of both the CC chemokines and their receptors. Dev. Comp. Immunol. 2007 Jan;31(1):72–86. doi: 10.1016/j.dci.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 50.Coil D., Jospin G., Darling A.E. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 2015 Feb 15;31(4):587–589. doi: 10.1093/bioinformatics/btu661. [DOI] [PubMed] [Google Scholar]
- 51.Wright E.S. DECIPHER: harnessing local sequence context to improve protein multiple sequence alignment. BMC Bioinf. 2015 Oct 6;16(1):322. doi: 10.1186/s12859-015-0749-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Price M.N., Dehal P.S., Arkin A.P. FastTree 2 – approximately maximum-likelihood trees for large alignments. Poon AFY, editor. PloS One. 2010 Mar 10;5(3) doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Letunic I., Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016 Jul 8;44(W1):W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Joensen K.G., Scheutz F., Lund O., Hasman H., Kaas R.S., Nielsen E.M. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 2014 May 1;52(5):1501–1510. doi: 10.1128/JCM.03617-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bishop D.K., Hinrichs D.J. Adoptive transfer of immunity to Listeria monocytogenes. The influence of in vitro stimulation on lymphocyte subset requirements. J. Immunol. 1987;139:2005–2009. [PubMed] [Google Scholar]
- 56.Lawrence L.H., Hill H.A., Linton M. 1995. DNA sequence divergence within the iap gene of L. monocytogenes as a means of characterising strains, XII International Symposium on Problems of Listeriosis, Perth, Australia. [Google Scholar]
- 57.Werbrouck H., Grijspeerdt K., Botteldoorn N. Differential inlA and inlB expression and interaction with human intestinal and liver cells by Listeria monocytogenes strains of different origins. Appl. Environ. Microbiol. 2006;72(6):3862–3871. doi: 10.1128/AEM.02164-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Infection of BCEC cells for 2-24h. BCEC cells were infected with an MOI = 200 with L. monocytogenes isolates for 2–24 h at 37 °C. (A) BCEC cells were infected for 2 h with nine different isolates, for each isolate 6–11 independent experiments were performed as represented by each symbol. Dark blue indicates isolates from cases of bovine abortion, pale blue indicates isolates from bovine keratoconjunctivitis, orange indicates an isolate from an environmental source, pink indicates an isolate from a healthy eye and red indicates the control strain 10403S originally a human isolate from a skin lesion. All data points and mean are shown. (B) BCEC cells were infected for 4–24 h with four different isolates, for each isolate 3 independent experiments were performed, and average and standard deviation are shown. Statistical significance is shown: ∗p < 0.05, ∗∗p < 0.01.
Table S1. Multilocus sequence type metadata. Metrics associated with all the isolates held in the MLST database.
Table S2. Determination of virulence associated genes. Whole genome sequences were parsed through virulence finder [64] to identify putative virulence associated genes for the isolates used in this study. Green cells indicate greater than 99% similarity to virulent gene loci, orange cells indicate 95–98% similarity and red indicate between 90-94% similarity.