Highlights
-
•
This meta-analysis is a comprehensively studies of microRNA in OS.
-
•
MicroRNA clusters helped to distinguish OS and healthy people.
-
•
MicroRNA is a potential noninvasive biomarker in early diagnosis of OS.
Keywords: Osteosarcoma, miRNAs, Blood, Diagnosis, Meta-analysis
Abstract
Osteosarcoma (OS) is one of the most common primary malignant tumors in adolescents. In recent years, multiple studies have reported the value of miRNAs in the diagnosis of OS, but the results were very different from each other. Therefore, we conducted this meta-analysis to determine the accuracy of miRNAs in the diagnosis of OS. The meta-analysis searched for relevant researches including PubMed, EMBASE, Web of Science, Wanfang database and China National Knowledge Infrastructure (CNKI) as of June 1, 2020. We used the quality assessment of Diagnostic Accuracy Study 2 (QUADAS-2) to score the quality of each study. A random effects model was used to pool the sensitivity and specificity. We measured the diagnostic value using positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR) and area under the curve (AUC). Subgroup and meta-regression analysis were used to find potential sources of heterogeneity. The meta-analysis finally included 31 articles about 2634 OS patients and 1715 healthy controls. The pooled estimations showed that the circulating miRNAs has a high accuracy in diagnosing OS, with a sensitivity of 0.79, specificity of 0.89, PLR of 7.3, NLR of 0.23, DOR of 31, and AUC of 0.90. In addition, subgroup and meta-regression analysis showed that miRNA clusters have higher diagnostic accuracy than single miRNA, and miRNAs in plasma were more reliable than those in serum. In conclusion, peripheral blood miRNA is a potential noninvasive biomarker to assist in the early diagnosis of OS, especially young patients with bone pain and/or indeterminate radiology findings.
1. Introduction
Osteosarcoma (OS) is a highly malignant and aggressive primary bone tumor, which is common in adolescents between 10 and 20 years old, and its incidence rate is about 0.04 to 0.05 per million, accounting for about 20% of bone malignant tumor [1], [2]. The most common site is the long bone, usually the distal femur and proximal tibia, followed by the proximal humerus. At present, comprehensive treatment methods such as extensive tumor resection, neo-adjuvant chemotherapy and radiotherapy have increased the 5-year overall survival rate of osteosarcoma patients to 55%-68% [3], [4], However, because of the problems of chemotherapy resistance, recurrence and early lung metastasis, the prognosis of patients is still not optimistic [5], [6]. The pathogenesis of osteosarcoma is very complex, and is related to a variety of genetic and cytogenetic abnormalities, including mutations of oncogenes and tumor suppressor genes, chromosome additions, deletions, ectopias, dysregulation of major signal transduction pathways, abnormal telomerase activity, etc. [7], [8], [9]. At present, the diagnostic procedure for osteosarcoma is based on local symptoms, X-ray examination, and invasive biopsy or open biopsy, etc. At this time, the tumor may have metastasized and spread [10], [11]. If osteosarcoma can achieve early diagnosis and treatment, it will greatly improve the prognosis of patients.
The development of biotherapy technologies such as gene therapy, immunotherapy and molecular targeted therapy has opened up a fresh path for the diagnosis and treatment of osteosarcoma and brought hope to patients [12], [13]. In these respects, microRNA (miRNA) has attracted attention because of its strong specificity, repeatability and accuracy, which have become ideal indicators for evaluating osteosarcoma [14].
miRNAs is a group of non-coding small RNA molecules encoded by the genome, comprising 18 to 22 nucleotides, which is combined with the 3′-untranslated region (UTR) of the corresponding target mRNA in the form of base pairing to regulate the gene expression and influence multiple cell biology processes [15], [16]. Several studies have shown that miRNAs was involved in the development and metastasis of various cancers, and plays an important role in tumorigenesis and homeostasis [17], [18]. In recent years, many literatures pointed out that miRNAs can be used as a biomarker for cancer or tumors in the diagnosis and prognosis of breast cancer [19], ovarian cancer [20], gastric cancer [21], and colorectal cancer [22]. Peripheral blood miRNAs may be derived from the lysis of apoptotic tumor cells, blood cells, macrophages, etc [23]. Studies have shown that, whether as an unprotected ribonucleoprotein complex or as a vesicle in the membrane, the secreted miRNAs was stable in the extracellular environment and can be quantitatively detected in plasma or serum [24]. Quantitative analysis of miRNAs in peripheral blood can be used as a new type of noninvasive examination to diagnose and monitor the changes of patients with osteosarcoma.
In recent years, many scholars have tested the accuracy of peripheral blood miRNAs for the early diagnosis of osteosarcoma. The results are gratifying but inconsistent [25], [26], [27], which may be caused by the small clinical samples, the lack of multi-center data demonstration, and the failure to formulate detection method standards. Therefore, we conducted this meta-analysis to assess the value of peripheral blood miRNAs in the diagnosis and prognosis of osteosarcoma.
2. Materials and methods
2.1. Search strategy and literature selection
This meta-analysis was based on diagnostic meta-analysis guidelines. We systematically searched the literature in PubMed, EMBASE, Web of Science, Wanfang database and China National Knowledge Infrastructure (CNKI) without language restrictions and used the following search terms: “osteosarcoma” or “osteosarcoma tumor” and “microRNA” or “miRNA”. The searches were limited to publications with human subjects, without language restrictions. The last search was conducted on 01/06/2020. The retrieved articles were independently screened and carefully evaluated by two investigators (SSG and GXZ), and the third investigator (WTZ) resolved any disagreements in the search process. We extract the required data from the selected studies and record them in sequence.
2.2. Inclusion and exclusion criteria
Publications that were included in our meta-analysis met the following criteria: (1) all of the studies involved both patients with OS and healthy control groups; (2) the microRNAs for diagnosis of OS patients were detected from serum or plasma; (3) included the use of relevant data, such as specificity, specificity, and AUC values. On the other side, exclusion standards were: (1) literatures were case reports, reviews, letters or comments; (2) duplicated information; (3) the obtained microRNAs were from tumor tissues, cell lines or animal experiments; (4) studies with insufficient data.
2.3. Data collection and study assessment
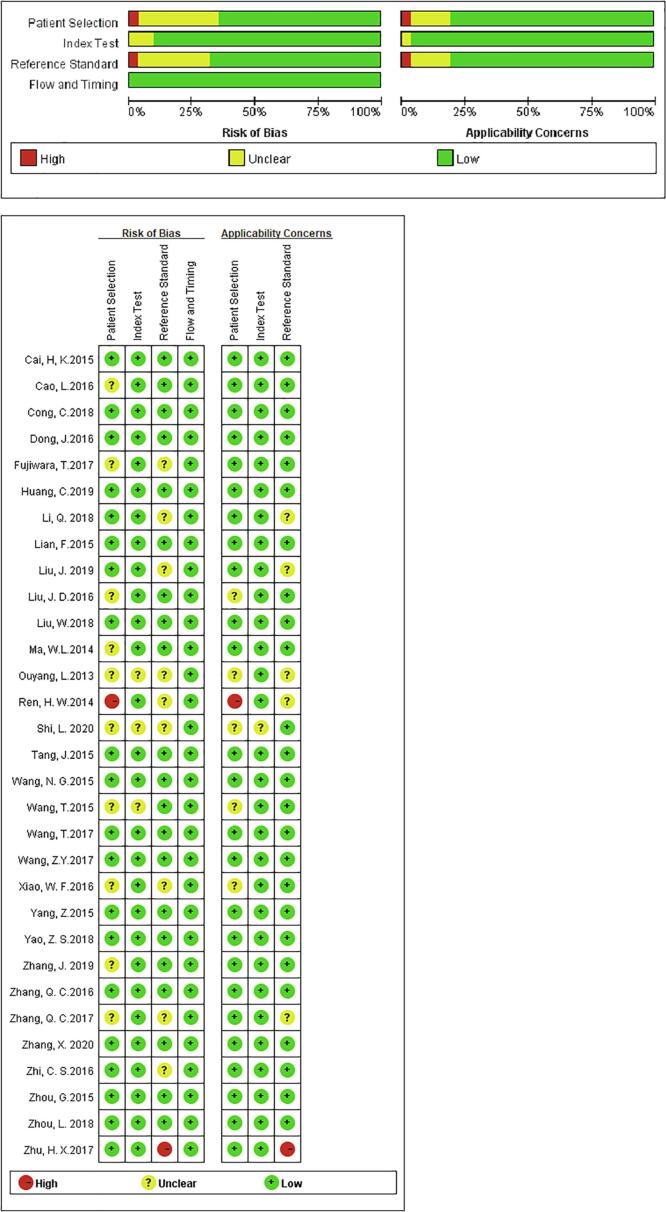
Two investigators selected and screened the relevant studies independently based on the title and abstract, and the full text, which was reviewed for further assessment if the study was collected by either of the investigators. We obtained the following dates from each eligible study: the first author's name, publication year, country, miRNA type, regulation mode (up or downregulated), sample size (number of OS patients/healthy controls), specimen type (serum o plasma), as well as data from two-by-two tables, sensitivity, specificity, AUC, and methodological quality (patient selection, index test, reference standard, flow and timing, patient selection for the applicable concerns, index text for applicable concern, and reference standard for applicable concerns). The quality of included studies were assessed independently by two investigators using diagnostic accuracy studies-2 (QUADAS-2) [28] criteria, and the third investigator resolved any disagreements, and reached a consensus.
2.4. Statistical analysis
The number of true positives, false positives, false negatives, true negatives in patients from each study were extracted. The heterogeneity was evaluated by I2 statistic. The random effects model was conducted if the I2 value was over 50%. The potential sources of heterogeneity were explored through threshold effect analysis, regression analysis, and additional subgroup analysis. Then we calculated sensitivity (SE), specificity (SP), positive likelihood ratios (PLR), negative likelihood ratios (NLR), and diagnostic odds ratio (DOR). In addition, we generated the summary receiver-operating characteristics (SROC) curve and calculated the area under the SROC curve (AUC) for both overall and the subgroup analysis. Assessment criteria for diagnostic efficacy: AUC = 1.00 is perfect, AUC greater than 0.90 is excellent, AUC greater than 0.80 is good, AUC < 0.80 is medium. Finally, a Deeks funnel plot was constructed to detect publication bias, with P < 0.05 indicating publication bias. All this were done by version 13.0 of STATA.
3. Results
3.1. Study selection and literature characteristics
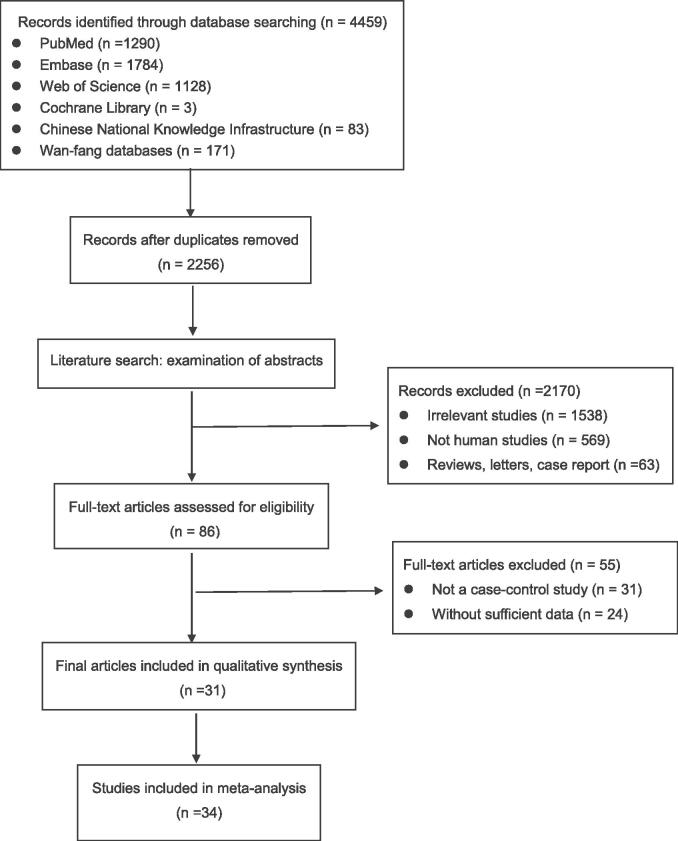
As shown in Fig. 1, the flow chart of the article selection process. According to the literature retrieval strategy, a total of 4459 articles were acquired, of which, 1290 were from PubMed, 1784 were from Embase, 1128 were from Web of Science, 3 were from Cochrane, 83 were from Chinese National Knowledge Infrastructure and 171 were from Wan-fang databases. 2256 articles were left for detection after 2203 duplicates were removed. After that, 1538 irrelevant studies, 569 not human studies, and 63 reviews, letters, case report were removed. Subsequently, the full texts of the remaining 86 articles were read to assess eligibility and 55 articles were excluded. Eventually, 107 studies from 31 articles were included in the current meta-analysis.
Fig. 1.
The flow chart of this meta-analysis to identify inclusion studies.
The characteristics of the 34 articles included are summarized in Table 1, divided into two parts: single miRNA and miRNA clusters, ranging from 2013 to 2020. All together a total of 2634 osteosarcoma patients and 1715 healthy controls were included. In all, 30 miRNAs studies concerned a single miRNA, and 4 studies focused on multiple miRNAs. Apart from that, quantitative real-time reverse transcription-PCR (qRT-PCR) was used to measure the expression of miRNAs from 28 serum specimens and 6 plasmas specimens. Most of the studies come from China, and the dominant ethnicity of the study subjects was Asian. The methodological quality assessments of the included articles according to the Quality Assessment of Diagnostic Accuracy Studies (QUADAS) were shown in a bar graph (Fig. 2).
Table 1.
Characteristics of the included studies.
| Author | Year | Country | microRNAs | Regulation mode | Sample size |
Speci-men | Diagnostic power |
|||
|---|---|---|---|---|---|---|---|---|---|---|
| OS patients | Healthy | Sen (%) | Spe (%) | AUC | ||||||
| Single microRNA | ||||||||||
| Zhu [29] | 2017 | China | miR-17-5p | Upregulated | 62 | 36 | Plasma | 0.500 | 0.972 | 0.793 |
| Fujiwara [30] | 2017 | Japan | miR-17-5p | Upregulated | 14 | 22 | Serum | 0.643 | 0.846 | 0.720 |
| Fujiwara [30] | 2017 | Japan | miR-25-3p | Upregulated | 14 | 22 | Serum | 0.714 | 0.923 | 0.868 |
| Tang [31] | 2015 | China | miR-27a | Upregulated | 166 | 60 | Serum | 0.700 | 0.983 | 0.867 |
| Zhang [32] | 2017 | China | miR-27b | Upregulated | 51 | 51 | Serum | 0.778 | 0.867 | 0.863 |
| Zhi [33] | 2016 | China | miR-34a | Downregulated | 50 | 50 | Serum | 0.867 | 0.686 | 0.830 |
| Wang [34] | 2017 | China | miR-34a | Downregulated | 120 | 87 | Serum | 0.680 | 0.920 | 0.830 |
| Yao [35] | 2018 | China | miR-101 | Downregulated | 152 | 70 | Serum | 0.790 | 0.829 | 0.850 |
| Cong [36] | 2018 | China | miR-124 | Downregulated | 114 | 50 | Serum | 0.798 | 0.860 | 0.846 |
| Xiao [37] | 2016 | China | miR-125b | Downregulated | 20 | 20 | Serum | 0.900 | 0.950 | |
| Zhou [25] | 2018 | China | miR-139-5p | Downregulated | 98 | 50 | Serum | 0.765 | 0.800 | 0.846 |
| Zhang [38] | 2020 | China | miR-144 | Downregulated | 51 | 48 | Serum | 0.738 | 0.882 | 0.852 |
| Ma [39] | 2014 | China | miR-148a | Downregulated | 89 | 89 | Plasma | 0.697 | 0.831 | 0.783 |
| Wang [40] | 2015 | China | miR-152 | Downregulated | 80 | 20 | Serum | 0.925 | 0.962 | 0.956 |
| Wang [41] | 2015 | China | miR-191 | Upregulated | 100 | 20 | Serum | 0.740 | 1 | 0.858 |
| Shi [42] | 2020 | China | miR-194 | Downregulated | 124 | 60 | Serum | 0.842 | 0.791 | 0.855 |
| Cai [43] | 2015 | China | miR-195 | Downregulated | 166 | 60 | Serum | 0.880 | 0.833 | 0.892 |
| Zhou [44] | 2015 | China | miR-199a-5p | Upregulated | 60 | 60 | Serum | 0.883 | 0.767 | 0.861 |
| Yang [45] | 2015 | China | miR-211 | Upregulated | 108 | 50 | Serum | 0.657 | 1 | 0.844 |
| Zhang [46] | 2016 | China | miR-222 | Downregulated | 57 | 57 | Serum | 0.667 | 0.842 | 0.811 |
| Dong [47] | 2016 | China | miR-223 | Downregulated | 122 | 50 | Serum | 0.895 | 0.972 | 0.926 |
| Liu [48] | 2019 | China | miR-223 | Downregulated | 53 | 30 | Serum | 0.736 | 0.600 | 0.747 |
| Zhang [49] | 2019 | China | miR-223 | Downregulated | 38 | 38 | Serum | 0.763 | 0.947 | 0.900 |
| Liu [50] | 2016 | China | miR-300 | Upregulated | 114 | 114 | Serum | 0.842 | 0.886 | 0.885 |
| Cao [51] | 2016 | China | miR-326 | Downregulated | 60 | 20 | Serum | 0.837 | 0.945 | 0.897 |
| Liu [27] | 2018 | China | miR-375 | Downregulated | 95 | 95 | Serum | 0.840 | 0.844 | 0.830 |
| Wang [52] | 2017 | China | miR-491-5p | Downregulated | 72 | 40 | Serum | 0.720 | 0.860 | 0.834 |
| Li [53] | 2018 | China | miR-542-3p | Upregulated | 76 | 76 | Serum | 0.778 | 0.936 | 0.841 |
| Liu [48] | 2019 | China | miR-586 | Upregulated | 53 | 30 | Serum | 0.642 | 0.700 | 0.743 |
| Huang [26] | 2019 | China | miR-663a | Upregulated | 50 | 50 | Plasma | 0.674 | 0.898 | 0.860 |
| microRNA clusters | ||||||||||
| Ouyang [54] | 2013 | China | miRNA clusters (miR-21, miR-199a-3p, miR-143) |
Downregulated | 40 | 40 | Plasma | 0.905 | 0.938 | 0.953 |
| Ren [55] | 2014 | china | miRNA clusters (miR-199b-5p/miR-124) |
Upregulated | 22 | 30 | Plasma | 0.960 | 0.970 | 0.993 |
| Lian [56] | 2015 | China | miRNA clusters (miR-195-5p, miR-199a-3p, miR-320a, miR-374a-5p) |
Upregulated | 90 | 90 | Plasma | 0.911 | 0.944 | 0.961 |
| Liu [48] | 2019 | China | miRNA clusters (miR-586, miR-223) |
Upregulated | 53 | 30 | Serum | 0.887 | 0.634 | 0.813 |
Fig. 2.
Overall methodology quality assessment of included articles using the QUADAS criteria.
3.2. Diagnostic accuracy of miRNAs in distinguishing OS from healthy controls
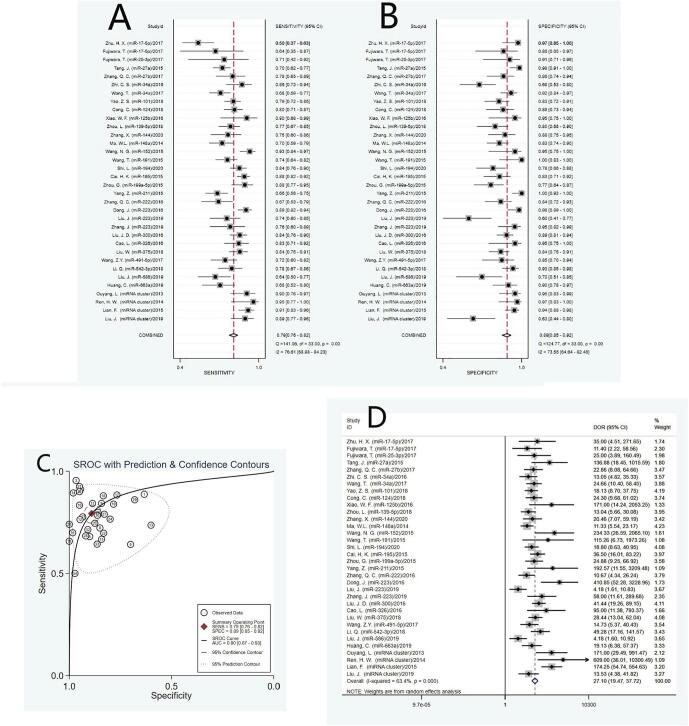
A total of 34 studies involving 4349 participants (2634 OS and 1715 healthy controls) were included in the pooled analysis. The sensitivities and specificities in the peripheral blood circulation of osteosarcoma patients were analyzed by using forest plots. The estimations of sensitivity and specificity (I2 = 76.61% and I2 = 73.55%, respectively, Fig. 3A and B) indicated that there was heterogeneity between the studies’ results. Therefore, the random-effects model was used in our meta-analysis. The pooled results were displayed as follows: sensitivity, 0.79 (95% CI: 0.76–0.82), specificity, 0.89 (95% CI: 0.85–0.92), AUC, 0.90 (95% CI: 0.87–0.93), NLR, 0.23 (95% CI: 0.20–0.27), PLR, 7.3 (95% CI: 5.4–9.7) and DOR, 31 (95% CI: 22–45). (Fig. 3)
Fig. 3.
Forest plots of sensitivity, specificity, area under the curve (AUC) and DOR for diagnosing OS patients from healthy controls among 34 studies. (A) Sensitivity; (B) Specificity; (C) AUC; (D) DOR.
3.3. Diagnostic value of miRNA cluster in OS patients
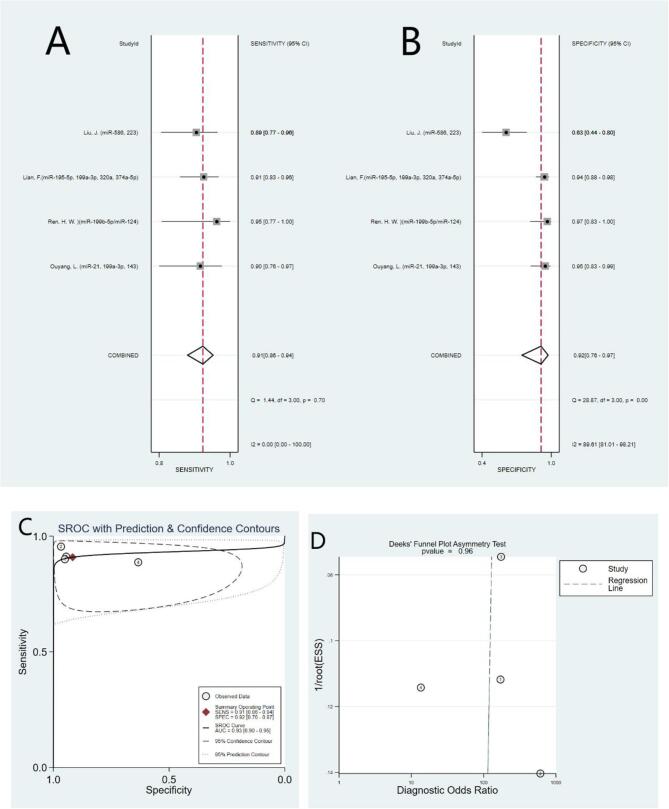
MiRNA cluster were reported in 4 studies of collected studies. The pooled sensitivity was 0.91 (95% CI: 0.86–0.94), specificity was 0.92 (95% CI: 0.76–0.97), AUC was 0.93 (95% CI: 0.90–0.95), the pooled NLR was 0.10 (95% CI: 0.06–0.16), PLR was 10.9 (95% CI: 3.5–34.2) and the pooled DOR was 109(95% CI: 26–465). (Fig. 4)
Fig. 4.
Forest plots of sensitivity, specificity, area under the curve (AUC) and funnel plot for diagnosing OS patients from healthy controls among 34 studies. (A) Sensitivity; (B) Specificity; (C) AUC; (D) Funnel plot.
3.4. Meta-regression analysis
A meta-regression analysis was used to explore possible sources of between-study heterogeneity in sensitivity and specificity. Several confounding covariates were pre-identified, including miRNAs profiling, regulation mode, sample size and specimen types. The meta-regression analysis suggested that regulation mode, sample size and specimen types (P < 0.001) might be potential sources of heterogeneity in sensitivity, and regulation mode, sample size (P < 0.001) might also explain heterogeneity in specificity. The outcomes of the meta-regression are shown in Fig. 5.
Fig. 5.
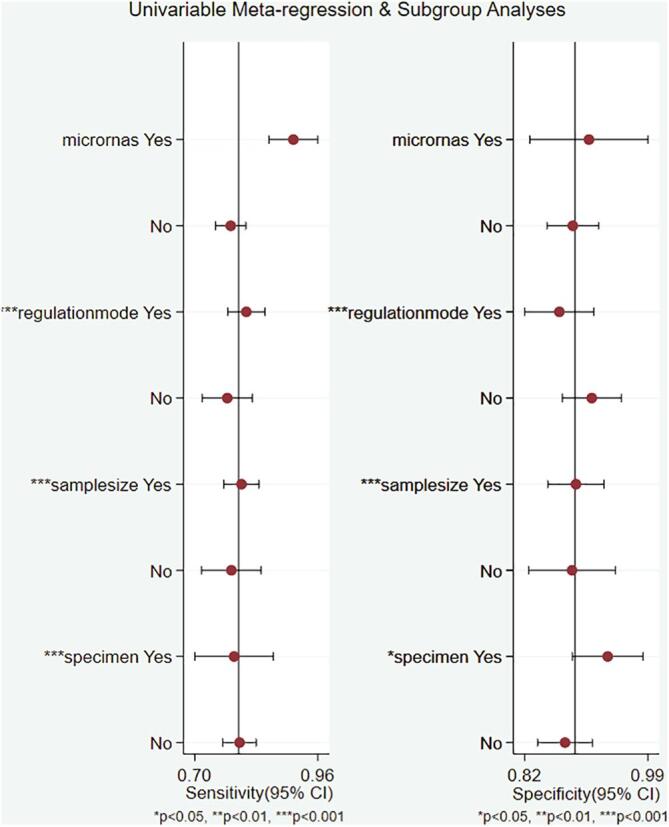
Forest plots of multivariable meta-regression for sensitivity and specificity.
3.5. Subgroup analysis
Subgroup analyzes were conducted according to miRNAs profiling, regulation mode, sample size and specimen types. The pooled sensitivity, specificity, PLR, NLR, DOR and AUC for each subgroup analysis were listed in Table 2. We found that the assay using multiple miRNAs exhibited a better diagnostic value than single miRNA: sensitivity (0.91 vs. 0.78), specificity (0.92 vs. 0.89), PLR (10.9 vs. 6.8), NLR (0.10 vs. 0.25), DOR (109 vs. 27) and AUC (0.93 vs. 0.89). In addition, plasma types had also a higher diagnostic value than serum types: sensitivity (0.81 vs. 0.79), specificity (0.93 vs. 0.88), PLR (11.0 vs. 6.6), NLR (0.21 vs. 0.23), DOR (53 vs. 28) and AUC (0.95 vs. 0.89).
Table 2.
Summary estimates of diagnostic power and their 95% confidence intervals.
| Subgrupo | Se (95% CI) | Sp (95% CI) | PLR (95% CI) | NLR (95% CI) | DOR (95% CI) | AUC (95% CI) |
|---|---|---|---|---|---|---|
| miRNAs profiling | ||||||
| Single miRNA | 0.78 [0.74–0.81] | 0.89 [0.85–0.92] | 6.8 [5.1–9.1] | 0.25 [0.22–0.29] | 27 [19–38] | 0.89 [0.86–0.92] |
| miRNA clusters | 0.91 [0.86–0.94] | 0.92 [0.76–0.97] | 10.9 [3.5–34.2] | 0.10 [0.06–0.16] | 109 [26–465] | 0.93 [0.90–0.95] |
| Regulation modo | ||||||
| Upregulated | 0.77 [0.71–0.83] | 0.92 [0.86–0.96] | 9.7 [5.6–16.9] | 0.25 [0.19–0.32] | 39 [21–73] | 0.90 [0.88–0.93] |
| Downregulated | 0.81 [0.77–0.84] | 0.87 [0.82–0.90] | 6.1 [4.4–8.5] | 0.22 [0.18–0.27] | 28 [17–45] | 0.90 [0.87–0.92] |
| Sample size | ||||||
| ≥100 | 0.80 [0.76–0.83] | 0.89 [0.85–0.92] | 7.2 [5.3–9.7] | 0.23 [0.19–0.27] | 32 [22–46] | 0.91 [0.88–0.93] |
| <100 | 0.78 [0.70–0.85] | 0.89 [0.81–0.94] | 7.4 [4.0–13.8] | 0.24 [0.17–0.35] | 31 [13–71] | 0.90 [0.87–0.92] |
| Specimen types | ||||||
| Serum | 0.79 [0.76–0.82] | 0.88 [0.84–0.91] | 6.6 [4.8–8.9] | 0.23 [0.20–0.27] | 28 [19–41] | 0.89 [0.86–0.91] |
| Plasma | 0.81 [0.64–0.91] | 0.93 [0.87–0.96] | 11.0 [5.6–21.2] | 0.21 [0.10–0.42] | 53 [16–180] | 0.95 [0.92–0.96] |
Se: sensitivity, Sp specificity, PLR: positive likelihood ratios, NLR: negative likelihood ratios, DOR: diagnostic odds ratio, AUC: area under the curve, CI: confidence interval.
Apart from that, the studies with sample size more than 100 were a little greater than the studies with sample size <100 in the diagnosis of OS patients: sensitivity (0.80 vs. 0.78), specificity (0.89 vs. 0.89), PLR (7.2 vs. 7.4), NLR (0.23 vs. 0.24), DOR (32 vs. 31) and AUC (0.91 vs. 0.90). The regulation mode had no influence on the diagnosis.
3.6. Publication bias
Deeks’ funnel plot test checked the publication bias of the included studies. The pooled Deeks’ test result of all studies was P = 0.85 (Fig. 6), which showed no significant publication bias in this analysis. In additional, the pooled Deeks’ test result of microRNA clusters was P = 0.96 (Fig. 4D), which indicating no publication bias.
Fig. 6.
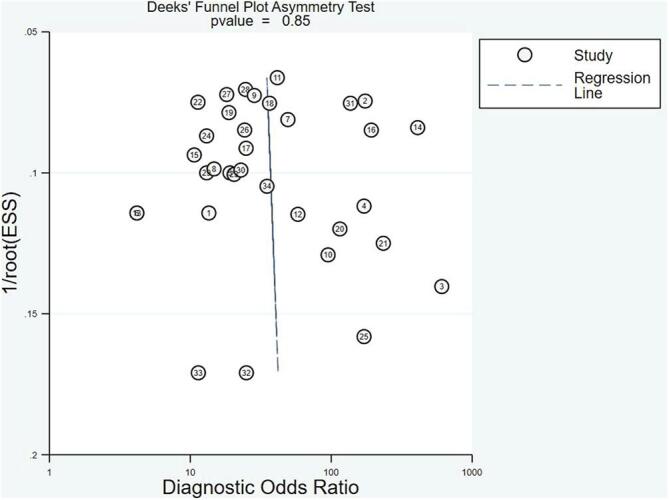
Deeks’ linear regression test of funnel plot asymmetry.
4. Discussion
As the most common primary malignant bone tumor in children and adolescents, osteosarcoma has been the focus of clinicians. Although the treatment of osteosarcoma has made substantial progress in the past few decades. However, for its early diagnosis and treatment of early lung metastasis, we still need a faster and more advanced non-invasive examination method to increase patient survival rate and improve prognosis. The latest research of miRNAs showed that it has strong specificity in the occurrence and metastasis of malignant tumors [57], [58], and has become a new hot spot for many scholars to study early diagnosis and monitor tumors. Several literatures showed that miRNAs in peripheral blood showed a high diagnostic accuracy in distinguishing OS patients from healthy people, but the results were inconsistent. For example, Wang NG et al. [40] reported the sensitivity, and specificity of miR-152 diagnostic accuracy as 92.5, and 96.2%, respectively, while the study of Wang T et al. [41] noted that miR-191 showed 74.0% sensitivity and 100% specificity. Even for the same miRNA, the results may be different because of different research protocols, detection platforms, and sample sizes. Dong J et al. [47] reported that the sensitivity and specificity of the diagnostic accuracy of miRNA-223 were 89.5% and 97.2%, respectively. However, Zhang J [49] et al. pointed out that the sensitivity and specificity exhibited by miR-223 were 76.3% and 94.7%, respectively. Lian F et al. [56] demonstrated that the miRNAs clusters (miR-195-5p, miR-199a-3p, miR-320a, and miR-374a-5p) could provide a high accuracy in separating OS patients from healthy controls with 91.10% sensitivity and 94.40% specificity.
Therefore, in this meta-analysis, we collected all the literature on the value of peripheral blood miRNAs in the diagnosis of OS. The pooled sensitivity was 0.79 (95% CI: 0.76–0.82), and the pooled specificity was 0.89 (95% CI: 0.85–0.92). We also drew the SROC curve and obtained the corresponding AUC to assess the overall diagnostic accuracy, with the ideal result of 0.90 in AUC value, meaning that miRNAs reached the high level of evaluation criteria in diagnosis [59]. We also used the PLR, NLR and DOR to further test the discrimination ability of microRNAs, which can provide more meaningful references for clinical usage. In our meta-analysis, the total DOR, the pooled PLR and NLR were 31 (95% CI: 22–45), 7.3 (95% CI: 5.4–9.7) and 0.23 (95% CI: 0.20–0.27), respectively, indicating that the chance of a correct diagnosis of OS individuals was 31 times higher than a false-negative diagnosis of healthy human. However, the PLR was lower than 10 and the NLR was not <0.1, which did not reach the general criterion in ruling in or ruling out decision [60]. All of the results proved that as a diagnostic biomarker, peripheral blood miRNAs obtained relatively high overall accuracy in the diagnosis of OS.
In this study, we conducted a meta-regression to detect the effects of the regulation mode, miRNAs profiling, sample size, specimen types. The result revealed that regulation mode, sample size and specimen types (P < 0.001) might be potential sources of heterogeneity in sensitivity, and regulation mode, sample size (P < 0.001) might also explain heterogeneity in specificity. Furthermore, we measured a subgroup analysis and interestingly, we found that the assay using miRNA cluster exhibited a better diagnostic value than single miRNA: sensitivity (0.91 vs. 0.78), specificity (0.92 vs. 0.89), PLR (10.9 vs. 6.8), NLR (0.10 vs. 0.25), DOR (109 vs. 27) and AUC (0.93 vs. 0.89). This was consistent with Lin Y's research results [61]. The changes in the expression of single miRNA in serum or plasma fluctuate not only in OS, but also in other tumors, infectious diseases, non-specific inflammation, and acute injuries. In other words, a single miRNA lacks specificity in cancer detection. However, for multiple miRNAs with complex molecular mechanisms, such as competing endogenous RNA (ceRNA) networks that intersect during tumorigenesis (such as the occurrence and development of severe tumors), this association may be valuable for early OS detection. Our results also supported the notion that plasma was more accurate than serum: sensitivity (0.81 vs. 0.79), specificity (0.93 vs. 0.88), PLR (11.0 vs. 6.6), NLR (0.21 vs. 0.23), DOR (53 vs. 28) and AUC (0.95 vs. 0.89). Have study found that the concentration of miRNAs was higher in plasma than in serum, so the choice of sample type played an important role in the diagnostic accuracy of peripheral blood miRNAs as a biomarker of disease [62], this may be because plasma retains more protein for co-isolation of miRNAs [63]. Apart from that, the results with sample size more than 100 were a little greater than the studies with sample size <100 in the diagnosis of OS patients: sensitivity (0.80 vs. 0.78), specificity (0.89 vs. 0.89), PLR (7.2 vs. 7.4), NLR (0.23 vs. 0.24), DOR (32 vs. 31) and AUC (0.91 vs. 0.90). This provides support for a larger sample of research in the future.
Throughout the entire article, we could find that our meta-analysis has the following advantages. First, compared with the previous meta-analysis [64], we included the miRNAs detected in the latest study, and our results improved the accuracy and reliability of the assessment of the overall diagnostic value of osteosarcoma. Second, we conducted subgroup and meta-regression analysis to discover potential sources of heterogeneity. Third, sensitivity analysis was also used to confirm the reliability of our results. However, we also recognized that this meta-analysis still has some limitations. First, several valuable studies may be missed in spite of the comprehensive search strategy during our literature search. Secondly, due to limited research data and different standards, we have not extracted cut-off values, different cut-off values may lead to inconsistent conclusions. Third, all the included studies were from Asia, mainly from China, the applicability of peripheral blood miRNAs in the diagnosis of OS is still unknown in other countries and regions. Therefore, a series of large-scale, prospective, multi-center and multi-country clinical trials are needed to provide high-quality evidence. Fourth, peripheral blood miRNAs not only showed high specificity in OS, but also played a key role in the occurrence and development of other malignant tumors. Fifth, in recent years, the profile of microRNA biomarkers has been widely used in different clinical environments due to its stability, such as early detection of disease, prediction of disease, monitoring of disease progression, and response to treatment [65]. However, it seems very unlikely that microRNA would become the mainstay of diagnosis in OS patients, but it could be useful for the preliminary investigation to triage, especially in young patients with bone pain and/or indeterminate radiology findings.
5. Conclusion
In summary, peripheral blood miRNAs could distinguish OS patients from healthy controls with high sensitivity and specificity and may serve as a potential noninvasive biomarker to assist in the early diagnosis, especially in young patients with bone pain and/or indeterminate radiology findings. Selecting a suitable biological specimen (such as plasma) and using miRNA clusters could improve the diagnostic accuracy of OS. Although this seemed to be a boon to OS patients, the actual application to the clinical process requires more data and experiments.
Ethics committee
The data comes from literature search and does not require any ethics committee approval and patient informed consent.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.L. Mirabello, R.J. Troisi, S.A. Savage, Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, Epidemiology, and End Results Program. Cancer, 2009. 115(7) 1531–1543.https://doi.org/10.1002/cncr.24121. [DOI] [PMC free article] [PubMed]
- 2.Anderson M.E. Update on survival in osteosarcoma. Orthop. Clin. North Am. 2016;47(1):283–292. doi: 10.1016/j.ocl.2015.08.022. [DOI] [PubMed] [Google Scholar]
- 3.Ottaviani G., Jaffe N. The epidemiology of osteosarcoma. Cancer Treat. Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 4.Reed D.R. Treatment pathway of bone sarcoma in children, adolescents, and young adults. Cancer. 2017;123(12):2206–2218. doi: 10.1002/cncr.30589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y. Progress in the chemotherapeutic treatment of osteosarcoma. Oncol. Lett. 2018;16(5):6228–6237. doi: 10.3892/ol.2018.9434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsuda Y. The outcomes and prognostic factors in patients with osteosarcoma according to age: a Japanese nationwide study with focusing on the age differences. BMC Cancer. 2018;18(1):614. doi: 10.1186/s12885-018-4487-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang N. Osteosarcoma development and stem cell differentiation. Clin. Orthop. Relat. Res. 2008;466(9):2114–2130. doi: 10.1007/s11999-008-0335-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Avril P. Mesenchymal stem cells increase proliferation but do not change quiescent state of osteosarcoma cells: potential implications according to the tumor resection status. J. Bone Oncol. 2016;5(1):5–14. doi: 10.1016/j.jbo.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee M.W. Mesenchymal stem cells in suppression or progression of hematologic malignancy: current status and challenges. Leukemia. 2019;33(3):597–611. doi: 10.1038/s41375-018-0373-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang X. Risk and clinicopathological features of osteosarcoma metastasis to the lung: A population-based study. J. Bone Oncol. 2019;16:100230. doi: 10.1016/j.jbo.2019.100230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durfee R.A., Mohammed M., Luu H.H. Review of osteosarcoma and current management. Rheumatol. Ther. 2016;3:221–243. doi: 10.1007/s40744-016-0046-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.M.M. Rangel-Sosa, E. Aguilar-Córdova, A. Rojas-Martínez, Immunotherapy and gene therapy as novel treatments for cancer. Colombia Medica (Cali, Colombia), 2017. 48(3) 138-147.https://doi.org/10.25100/cm.v48i3.2997. [DOI] [PMC free article] [PubMed]
- 13.Pucci C., Martinelli C., Ciofani G. Innovative approaches for cancer treatment: current perspectives and new challenges. Ecancer Med. Sci. 2019;13 doi: 10.3332/ecancer.2019.961. 961–961. 10.3332/ecancer.2019.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H. Circulating microRNAs as potential cancer biomarkers: the advantage and disadvantage. Clin. Epigenet. 2018;10 doi: 10.1186/s13148-018-0492-1. 59–59. https: //doi.org/10.1186/s13148-018-0492-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwerk J., Savan R. Translating the untranslated region. J. Immunol. 2015;195(7):2963–2971. doi: 10.4049/jimmunol.1500756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palmini G., Marini F., Brandi M.L. What is new in the miRNA world regarding osteosarcoma and chondrosarcoma? Molecules. 2017;22(3) doi: 10.3390/molecules22030417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J. MicroRNAs an active and versatile group in cancers. Int. J. Oral Sci. 2011;3(4):165–175. doi: 10.4248/ijos11063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiao X. microRNA: the impact on cancer stemness and therapeutic resistance. Cells. 2019;9(1):8. doi: 10.3390/cells9010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.G. Bertoli, C. Cava, I. Castiglioni, MicroRNAs: New Biomarkers for Diagnosis, Prognosis, Therapy Prediction and Therapeutic Tools for Breast Cancer, Theranostics, 2015. 5(10) 1122-1143.https://doi.org/10.7150/thno.11543. [DOI] [PMC free article] [PubMed]
- 20.Pal M.K. MicroRNA: a new and promising potential biomarker for diagnosis and prognosis of ovarian cancer. Cancer Biol. Med. 2015;12(4):328–341. doi: 10.7497/j.issn.2095-3941.2015.0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan H.-L., Wang T., Zhang K.-H. MicroRNAs as potential biomarkers for diagnosis, therapy and prognosis of gastric cancer. Oncol. Targets Ther. 2018;11:3891–3900. doi: 10.2147/OTT.S156921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen B. Emerging microRNA biomarkers for colorectal cancer diagnosis and prognosis. Open Biol. 2019;9(1) doi: 10.1098/rsob.180212. 180212–180212. 10.1098/rsob.180212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frank A.-C. Apoptotic tumor cell-derived microRNA-375 uses CD36 to alter the tumor-associated macrophage phenotype. Nat. Commun. 2019;10(1) doi: 10.1038/s41467-019-08989-2. 1135–1135 10.1038/s41467-019-08989-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.M. Cui, et al., Circulating MicroRNAs in Cancer: Potential and Challenge, Front. Genet. 2019. 10 626–626. https://doi.org/10.3389/fgene.2019.00626. [DOI] [PMC free article] [PubMed]
- 25.Zhou L. The diagnostic effect of serum miR-139-5p as an indicator in osteosarcoma. Cancer Biomark. 2018;23(4):561–567. doi: 10.3233/cbm-181744. [DOI] [PubMed] [Google Scholar]
- 26.Huang C. Identification of circulating miR-663a as a potential biomarker for diagnosing osteosarcoma. Pathol. Res. Pract. 2019;215(3):152411. doi: 10.1016/j.prp.2019.04.003. [DOI] [PubMed] [Google Scholar]
- 27.Liu W. MicroRNA-375 as a potential serum biomarker for the diagnosis, prognosis, and chemosensitivity prediction of osteosarcoma. J. Int. Med. Res. 2018;46(3):975–983. doi: 10.1177/0300060517734114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whiting P.F. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 2011;155(8):529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 29.Zhu H.X. Circulating miR-17-5p as a potential biomarker for diagnosis and prognosis in osteosarcoma. Int. J. Clin. Exp. Path. 2017;10(3):3409–3416. [Google Scholar]
- 30.Fujiwara T. Clinical significance of circulating miR-25-3p as a novel diagnostic and prognostic biomarker in osteosarcoma. Oncotarget. 2017;8(20):33375–33392. doi: 10.18632/oncotarget.16498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang J. Diagnostic and prognostic potentials of microRNA-27a in osteosarcoma. Biomed. Pharmacother. 2015;71:222–226. doi: 10.1016/j.biopha.2015.01.025. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Q.C. Expression and clinical significance of circulating microRNA-27b in patients with osteosarcoma. J. Clin. Pathol. Res. 2017;37(12) 2663–2638. Chinese. [Google Scholar]
- 33.Zhi C.S., Wu B. Serum miRNA-34a serves as a diagnostic and prognostic bio-marker in osteosarcoma. Int. J. Clin. Exp. Path. 2016;9(3):3459–3464. [Google Scholar]
- 34.Wang T. Serum miR-34a is a potential diagnostic and prognostic marker for osteosarcoma. Int. J. Clin. Exp. Pathol. 2017;10(9):9683–9689. [PMC free article] [PubMed] [Google Scholar]
- 35.Yao Z.S. Diagnostic and prognostic implications of serum miR-101 in osteosarcoma. Cancer Biomark. 2018;22(1):127–133. doi: 10.3233/cbm-171103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cong C. Identification of serum miR-124 as a biomarker for diagnosis and prognosis in osteosarcoma. Cancer Biomark. 2018;21(2):449–454. doi: 10.3233/cbm-170672. [DOI] [PubMed] [Google Scholar]
- 37.Xiao W.F., Luo W., Xiao K. The expression of microRNA-125b in serum of patients with osteosarcoma and its inhibitory effect on proliferation. Chin. J. Med. Sci. 2016;8(8) 49–52. Chinese. [Google Scholar]
- 38.Zhang X. Influence mechanism of miRNA-144 on proliferation and apoptosis of osteosarcoma cells. Oncol. Lett. 2020;19(2):1530–1536. doi: 10.3892/ol.2019.11197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma W. Circulating miR-148a is a significant diagnostic and prognostic biomarker for patients with osteosarcoma. Tumour Biol. 2014;35(12):12467–12472. doi: 10.1007/s13277-014-2565-x. [DOI] [PubMed] [Google Scholar]
- 40.Wang N.G. Down-regulation of microRNA152 is associated with the diagnosis and prognosis of patients with osteosarcoma. Int. J. Clin. Exp. Pathol. 2015;8(8):9314–9319. [PMC free article] [PubMed] [Google Scholar]
- 41.Wang T. Increased expression of microRNA-191 as a potential serum biomarker for diagnosis and prognosis in human osteosarcoma. Cancer Biomark. 2015;15(5):543–550. doi: 10.3233/cbm-150493. [DOI] [PubMed] [Google Scholar]
- 42.Shi L. Downregulation of serum miR-194 predicts poor prognosis in osteosarcoma patients. Ann. Diagn. Pathol. 2020;46 doi: 10.1016/j.anndiagpath.2020.151488. [DOI] [PubMed] [Google Scholar]
- 43.Cai H. Serum miR-195 is a diagnostic and prognostic marker for osteosarcoma. J. Surg. Res. 2015;194(2):505–510. doi: 10.1016/j.jss.2014.11.025. [DOI] [PubMed] [Google Scholar]
- 44.Zhou G. Identification of miR-199a-5p in serum as noninvasive biomarkers for detecting and monitoring osteosarcoma. Tumour Biol. 2015;36(11):8845–8852. doi: 10.1007/s13277-015-3421-3. [DOI] [PubMed] [Google Scholar]
- 45.Yang Z. Serum microRNA-221 functions as a potential diagnostic and prognostic marker for patients with osteosarcoma. Biomed. Pharmacother. 2015;75:153–158. doi: 10.1016/j.biopha.2015.07.018. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Q.C. Serum level of microRNA-222 acts as a diagnostic and prognostic biomarker for osteosarcoma patients. Int. J. Clin. Exp. Path. 2016;9(4):4843–4848. [Google Scholar]
- 47.Dong J. MiRNA-223 is a potential diagnostic and prognostic marker for osteosarcoma. J. Bone Oncol. 2016;5(2):74–79. doi: 10.1016/j.jbo.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu J., Lv X. Expression and diagnostic value of miR-586 and miR-223 in the peripheral blood of patients with osteosarcoma. Int. J. Clin. Exp. Med. 2019;12(1):836–842. [Google Scholar]
- 49.J. Zhang, Application of combined detection of serum miR-223 and IL-8 in the diagnosis of osteosarcoma, Chin. J. Lab Diagn. 23(4) (2019) 626–628. Chinese. https://doi.org/10.3969/j.issn.1007-4287.2019.04.018.
- 50.Liu J.D. Serum miR-300 as a diagnostic and prognostic biomarker in osteosarcoma. Oncol. Let. 2016;12(5):3912–3918. doi: 10.3892/ol.2016.5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cao L., Wang J., Wang P.Q. MiR-326 is a diagnostic biomarker and regulates cell survival and apoptosis by targeting Bcl-2 in osteosarcoma. Biomed. Pharmacother. 2016;84:828–835. doi: 10.1016/j.biopha.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 52.Wang Z. Low miR-491-5p is an unfavorable prognostic marker for osteosarcoma. 2017;10:3304–3309. [Google Scholar]
- 53.Li Q. Serum miR-542-3p as a prognostic biomarker in osteosarcoma. Cancer Biomark. 2018;21(3) doi: 10.3233/cbm-170255. 521–526. [DOI] [PubMed] [Google Scholar]
- 54.Ouyang L. A three-plasma miRNA signature serves as novel biomarkers for osteosarcoma. Med. Oncol. 2013;30(1):340. doi: 10.1007/s12032-012-0340-7. [DOI] [PubMed] [Google Scholar]
- 55.Ren H.W., Yang C., Su H.W. Predictive effect of microRNA ratio in osteosarcoma. J. Int. Oncol. 2014;41(8):708–711. [Google Scholar]
- 56.Lian F. Identification of a plasma four-microRNA panel as potential noninvasive biomarker for osteosarcoma. PLoS ONE. 2015;10(3) doi: 10.1371/journal.pone.0121499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Condrat C.E. miRNAs as biomarkers in disease: latest findings regarding their role in diagnosis and prognosis. Cells. 2020;9(2):276. doi: 10.3390/cells9020276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y. The roles of circular RNAs in osteosarcoma. Med. Sci. Monit. 2019;25:6378–6382. doi: 10.12659/msm.915559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Swets J.A. Measuring the accuracy of diagnostic systems. Science. 1988;240(4857) doi: 10.1126/science.3287615. 1285–1293. [DOI] [PubMed] [Google Scholar]
- 60.Deeks J.J., Altman D.G. Diagnostic tests 4: likelihood ratios. BMJ. 2004;329(7458) doi: 10.1136/bmj.329.7458.168. 168–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yan L. A comprehensive meta-analysis of MicroRNAs for predicting colorectal cancer. Medicine. 2016;95(9) doi: 10.1097/MD.0000000000002738. pp. e2738–e2738 10.1097/MD.0000000000002738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dufourd T. Plasma or serum? A qualitative study on rodents and humans using high-throughput microRNA sequencing for circulating biomarkers. Biol. Methods Protoc. 2019;4(1) doi: 10.1093/biomethods/bpz006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arroyo J.D. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA. 2011;108(12):5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu H. MicroRNAs as a novel class of diagnostic biomarkers for the detection of osteosarcoma: a meta-analysis. Oncol. Targets Ther. 2017;10:5229–5236. doi: 10.2147/ott.S143974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bottani M., Banfi G., Lombardi G. The clinical potential of circulating miRNAs as biomarkers: present and future applications for diagnosis and prognosis of age-associated bone diseases. Biomolecules. 2020;10(4):589. doi: 10.3390/biom10040589. [DOI] [PMC free article] [PubMed] [Google Scholar]