Abstract
In terms of public health, the 21st century has been characterized by coronavirus pandemics: in 2002-03 the virus SARS-CoV caused SARS; in 2012 MERS-CoV emerged and in 2019 a new human betacoronavirus strain, called SARS-CoV-2, caused the unprecedented COVID-19 outbreak. During the course of the current epidemic, medical challenges to save lives and scientific research aimed to reveal the genetic evolution and the biochemistry of the vital cycle of the new pathogen could lead to new preventive and therapeutic strategies against SARS-CoV-2. Up to now, there is no cure for COVID-19 and waiting for an efficacious vaccine, the development of “savage” protocols, based on “old” anti-inflammatory and anti-viral drugs represents a valid and alternative therapeutic approach. As an alternative or additional therapeutic/preventive option, different in silico and in vitro studies demonstrated that small natural molecules, belonging to polyphenol family, can interfere with various stages of coronavirus entry and replication cycle. Here, we reviewed the capacity of well-known (e.g. quercetin, baicalin, luteolin, hesperetin, gallocatechin gallate, epigallocatechin gallate) and uncommon (e.g. scutellarein, amentoflavone, papyriflavonol A) flavonoids, secondary metabolites widely present in plant tissues with antioxidant and anti-microbial functions, to inhibit key proteins involved in coronavirus infective cycle, such as PLpro, 3CLpro, NTPase/helicase. Due to their pleiotropic activities and lack of systemic toxicity, flavonoids and their derivative may represent target compounds to be tested in future clinical trials to enrich the drug arsenal against coronavirus infections.
Keywords: Coronavirus, Flavonoids, SARS-CoV, MERS-CoV, SARS-CoV-2
Graphical abstract
Abbreviations
- 3CLpro
3C-like-protease
- ACE2
Angiotensin Receptor Enzyme-2
- CC50
50% cytotoxicity concentration
- CLD
C-terminal collectrin
- COVID-19
Coronavirus Disease 2019
- E
Small envelope protein
- EGCG
Epigallocatechin-3-gallate
- GCG
Gallocatechin gallate
- gRNA, sgRNA
Genomic, sub-genomic RNA
- HE
Hemagglutinin esterase
- HR
Heptad regions (HR1 and HR2)
- HSV-1
Herpes simplex virus type 1
- M
Membrane or matrix protein
- MERS-CoV
Middle East Respiratory Syndrome CoronaVirus
- Mpro
Main protease
- N
Nucleocapsid protein
- NSP
Not structural proteins
- ORF
Open reading frame
- PD
N-terminal peptidase domain
- PEDV
Porcine epidemic diarrhea virus
- PLpro
Papain-like cysteine protease
- RBD
Receptor-binding domain structure
- RBM
Receptor binding motif
- RdRp
RNA-dependent-RNA-polymerase
- RSV
Respiratory syncytial virus
- RTC
Replication/transcription complex
- S
Spike glycoprotein
- SAR
Structure–activity relationship
- SARS-CoV
Severe Acute Respiratory Syndrome CoronaVirus
- SARS-CoV-2
Severe Acute Respiratory Syndrome CoronaVirus-2
- SPR
Surface Plasmon resonance
- TCM
Traditional Chinese Medicine
- TfR
Transferrin receptor
- TM
Transmembrane domain
- TMPRSS2
Transmembrane peptidase serine 2
- UTR
Untranslated region
1. Introduction
The 2nd decade of the 21st century began with an unprecedented epidemic in human history: the emergence of a new human betacoronavirus strain, first isolated and sequenced in China in early 2020 [1] and called SARS-CoV-2 (Severe Acute Respiratory Syndrome CoronaVirus-2) as the etiological agent of CoronaVIrus Disease 19 (COVID-19) [2]. Globally, at the time of writing (July 11, 2020), 12,322,395 confirmed cases of COVID-19 have been registered, including 556,335 deaths worldwide except for the Antarctic continent, as reported by WHO (https://covid19.who.int/COVID-19). Although awaited by some microbiologists, this pandemic found the health systems of many Western countries (Italy, Spain, France, UK and USA) largely unprepared. However, at the same time, it mobilized the scientific community with the production of over 6000 peer-reviewed scientific articles on PubMed in the last 5 months. Waiting for a vaccine, there is currently no specific cure against COVID-19. A deeper knowledge of the genetics and biochemistry sustaining SARS-CoV-2 vital cycle and infectivity will certainly lead to the development of new therapeutic protocols.
Recent studies evidenced that SARS-CoV-2 is an efficient killer compared to its close related SARS-CoV, isolated in 2003 since the former acquired an array of “strategic adaptations” [3]. During its evolution in nature, probably decades as meta-genetic analysis are revealing the new strain of coronavirus “jumped” from a bat to an unknown mammalian and then to humans. Here, SARS-CoV-2 can target different tissues at multiple levels, starting from the cells of nose and throat down to the lung, invading, perhaps, through vasal endothelium, kidneys and nervous system where it can cause severe illness and death [4,5].
However, there is still much to learn about SARS-CoV-2 and a deeper knowledge of its biology, comparing metagenomics analysis and biochemical characteristics of previous coronaviruses (SARS-CoV and MERS-CoV) will be crucial to define efficient therapeutic and/or preventive strategies.
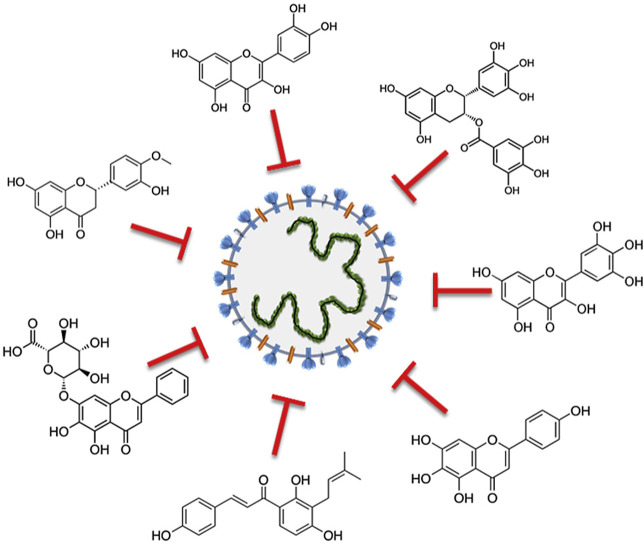
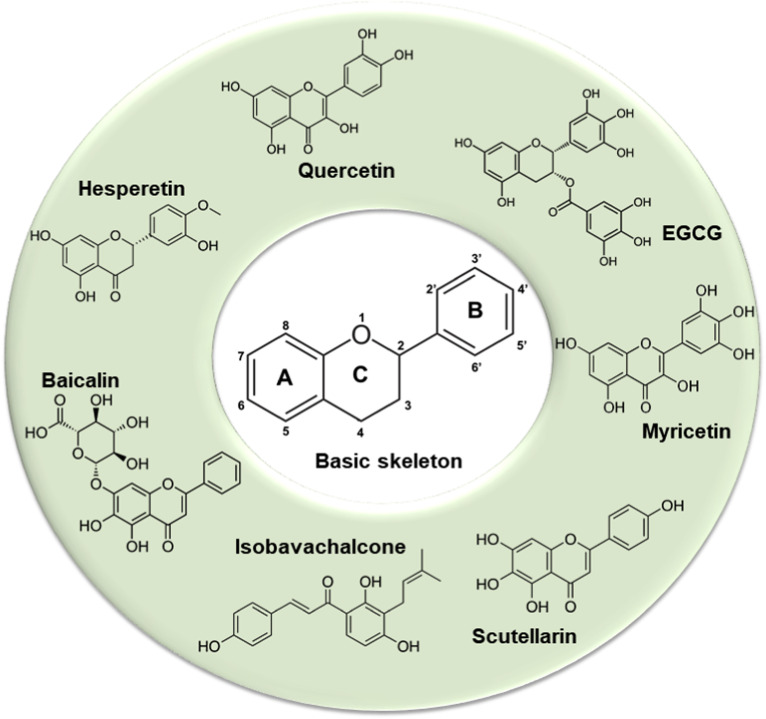
Besides “old” drugs (essentially antivirals, such as favipiravir and ribavirin; anti-HIV protease inhibitors, such as ritonavir and lopinavir, and anti-inflammatory agents, such as tociclizumab or dexamethasone) that clinicians are currently using against the severe cases of the recent COVID-19 outbreak [[6], [7], [8]], natural compounds isolated from the plant kingdom and belonging to the multiple and heterogeneous class of flavonoids (Fig. 1 ) may represent an interesting option. In fact, flavonoids lack systemic toxicity, their ability to synergize with conventional drugs has been largely demonstrated and, finally, they are “pleiotropic” compounds, meaning that their functional groups can interact with different cellular targets and intercept multiple pathways [9,10]. These features make flavonoids potential candidates to interfere with the coronavirus life cycle.
Fig. 1.
Basic skeleton (C6–C3–C6) of flavonoids and representative examples of compounds able to counteract coronavirus infection.
Flavonoids include a large number of secondary metabolites, found in vegetables, seeds, fruit, and beverages, such as red wine and tea [11]. There are more than 6000 structurally identified flavonoid molecules. These compounds are synthesized in plants in response to stressful conditions and play an important role in defending plant cells against pathogens and insects [[12], [13], [14]]. From a chemical point of view, flavonoids are hydroxylated phenolic molecules synthesized by the phenylpropanoid pathway and are distinguished by their structural class, degree of hydroxylation, and polymerization. The hydroxyl functional groups of flavonoids are responsible for their antioxidant activity and are formed by two benzene rings (A-and B-rings), connected via a heterocyclic pyrene ring (C-ring) ( Fig. 1 ). Flavonoids are divided into different classes, such as anthocyanins, chalcones, dihydrochalcones, dihydroflavonols, flavanols, flavanones, flavones, flavonols, isoflavonoids (see Phenol-Explorer database at http://phenol-explorer.eu). The pharmacological properties of flavonoids include antimicrobial, antioxidant, anti-inflammatory, and antiviral functions. Flavonoids have been studied against a wide range of DNA and RNA viruses [15]. For example, apigenin is active against picornavirus (RNA virus), inhibiting protein synthesis by suppressing IRES viral activity [16]. Epigallocatechin-3-gallate (EGCG), active polyphenolic catechin that accounts for approximately 59% of the total catechins from the leaves of the green tea (Camellia sinensis (L.), Kuntze) interferes with the replication cycle of DNA viruses, such as hepatitis B virus, herpes simplex, and adenovirus [17].
To prepare this review article, especially the PubMed database www.ncbi.nlm.nih.gov/pubmed/ (https://pubmed.ncbi.nlm.nih.gov/) was consulted up the end of May 2020, to retrieve articles that included the following combination of terms: “coronavirus” and “flavonoid”. We selected those papers that convincingly focused on the antiviral activity of defined flavonoids against human coronaviruses, excluding some very recent preprint articles on SARS-CoV-2 not certified by peer review that, in our opinion, were of limited quality. We apologize in advance for possible citations omitted due to space limitations.
2. Coronavirus biology
2.1. Morphology and biochemistry
Coronavirus is a family of one strand (+) RNA enveloped virus in the order Nidovirales. They were originally identified in the sixties in the United Kingdom and the United States where scientists isolated two viruses causing common colds in humans [18]. Coronaviruses are spherical or pleomorphic, with a diameter of 80–120 nm. In 1968 electron microscopy images revealed the virus crown-like structures resembling the “solar corona” that give rise to the name of this family derived from Latin word: “coronavirus” [19]. Since then and until last year, two highly pathogenic human strains emerged: SARS-CoV, in 2003 and MERS-CoV (Middle East Respiratory Syndrome coronavirus) in 2012 that caused, according to WHO, severe epidemic outbreaks [20,21]. They are transmitted to humans from market civets and dromedary, respectively and both originated from bats, a natural reserve of hundreds of still unknown coronavirus [22].
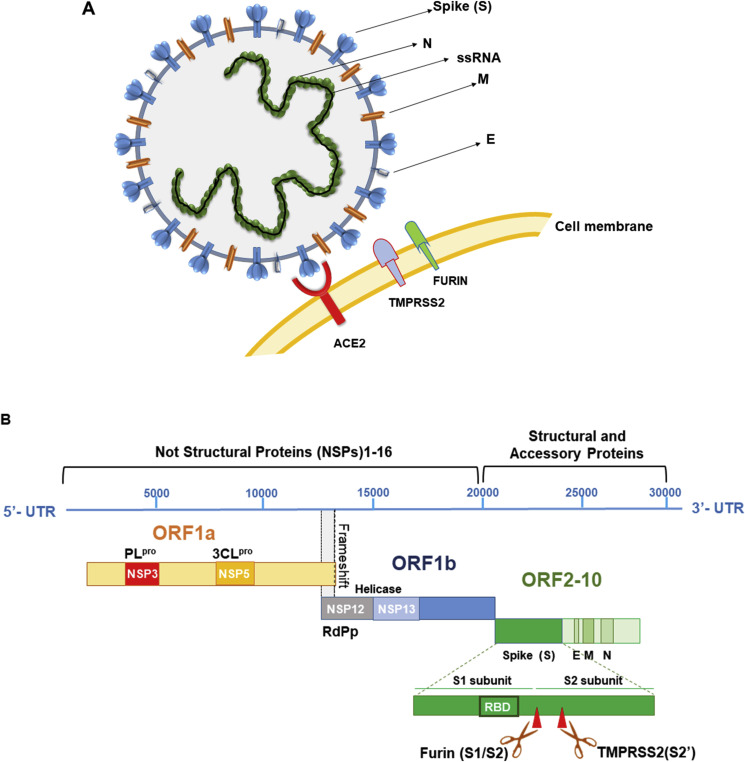
The coronavirus RNA genome is bigger than other RNA viruses with size ranges from 26,000 to 32,000 bases including from 6 to 11 open reading frames (ORF). The first ORF (67% of the genome) encodes not structural proteins (NSP), while the remaining ORFs give rise to accessory and structural proteins [22]. In particular, the first ORF (ORF1a/b) translates two polyproteins: pp1a and pp1ab for the presence of a frameshift between ORF1a and ORF1b. These polyproteins are processed by the main protease (Mpro) also known as 3C-like-protease (3CLpro) and one or two papain–like proteases (PLpro) into 16 NSPs, which produce viral RNA that encodes the four main structural proteins [23] (Fig. 2 ).
Fig. 2.
A. Coronaviruses form enveloped and spherical particles of 100–160 nm in diameter. They contain a positive-sense, single-stranded RNA (ssRNA) genome and nucleocapside proteins (N) that bind to RNA genome forming the nucleocapsid. The trimeric Spike glycoprotein (S) localizes on the surface of virus envelope and is essential for virus entry into the host cells. It recognizes the host receptor protein ACE2 on cell membrane after cleavage and activation by two host serine-proteases: TMPRSS2 and FURIN. Membrane or matrix protein (M) and small envelope protein (E) are both essential for the assembly and release of virions. B. SARS-CoV-2 genome, genes and proteins. There are 10 open reading frames (ORFs). The first ORF (67% of the genome) encodes not structural proteins (NSP), while the remaining ORFs give rise to accessory and structural proteins. ORF1a/b translates two polyproteins: pp1a and pp1b for the presence of a frameshift between ORF1a and ORF1b. These polyproteins are processed by a main protease known as 3C-like-protease (3CLpro) and one or two papain–like proteases (PLpro) into 16 NSPs. NSPs produce replicase complex essential for viral replication: NSP12 encodes RNA dependent RNA Polimerase (RdPd) and NSP13 encodes Helicase. ORFs 2–10 encode viral structural proteins: Spike (S), Envelope (E), Membrane (M), Nucleocapsid (N) and other auxiliary proteins. In particular, Spike protein comprises two regions: S1 with the receptor-binding domain (RBD) essential for the recognition of host receptor and S2, essential for membrane fusion and entry. Between S1 and S2 subunits there is the polybasic sequence recognized by host endo-proteases Furin. The activation site of S protein, is recognized by serine protease TMPRSS2 in region S2′ of S2 domain.
The importance of 3CLpro in the viral cycle and the absence of its human homologue makes this enzyme an attractive target for the development of new drugs directed against coronavirus infection. 3CLpro is a three domains cysteine protease with an active site highly conserved among all coronavirus. Domain I and II are six-stranded antiparallel β barrels very similar to the architecture of chymotrypsin and picornavirus 3C proteinases. The substrate-binding site is located in a cleft between these two domains. A long loop (residues 184 to 199) connects domain II to the C-terminal domain (domain III, residues 200 to 300). This latter domain, a globular cluster of five helices, has been implicated in the proteolytic activity of 3CLpro. Anand et al. [24] and Dai et al. [23] analyzed substrate-binding pocket of SARS-CoV and SARS-CoV-2, respectively to design novel inhibitors for this protease and found that the thiol group of a cysteine residue in the S1′ site is important to anchor molecules by a covalent linkage, obtaining efficacious antiviral activity. The S1′ site represents one of the four sites (S1’, S1, S2 and S4) highly conserved in the catalytic domain of 3CLpro of human coronavirus.
The four major structural proteins of coronavirus are: 1. the trimeric Spike glycoprotein (S) that localizes on the surface of virus envelope and essential for virus entry into the host cells; 2. the membrane or matrix protein (M); 3. the small envelope protein (E), both essential for the assembly and release of virions; 4. the nucleocapsid protein (N), that binds to RNA genome forming the helically symmetric nucleocapsid [19]. In addition, some coronavirus genome encodes for a glycoprotein of approximately 60–65 kDa called hemagglutinin esterase (HE). The role of this protein is still unclear, it possesses an acetyl-esterase enzymatic activity able to disrupt sialic acid receptors on the host cells surface, helping the invasion and attachment of virion [22].
Genetic and comparative analysis of different known coronaviruses represents a powerful strategy to identify potential drug targets against the current outbreak and the 3CLpro protease is a good example. The first three SARS-CoV-2 genomes isolated from bronchoalveolar-lavage fluid and sequenced in Wuhan (China) showed the typical beta coronavirus organization (subgenus sarbecovirus): a 5′ untranslated region (UTR), replicase complex (ORF1a/b), S, E, M and N genes, 3′UTR and several unidentified non-structural ORF [1]. Comparing RNA sequence of 9 Chinese patients affected by COVID-19 with those of other coronaviruses, Lu et al. (2020) performed a phylogenetic analysis to determine the evolutionary history of the virus going back to its likely origin [25]. The authors of this study found that SARS-CoV-2 RNA sequence shared 96% genetic material with a bat virus in a cave of Yunnan (China), but was distant from SARS-CoV (79% identity) and MERS-CoV (50% identity). Homology modelling revealed, however, that the new virus had a similar receptor-binding domain structure (RBD) of spike protein to that of SARS-CoV, despite amino acid variation at few key residues. For this reason, it was hypothesized that SARS-CoV and SARS-CoV-2 shared the same cellular receptor to enter in human cells, the protein Angiotensin Receptor Enzyme-2 (ACE2) widely expressed in lung, heart, kidney, testis and gastrointestinal tract [25].
2.2. Attachment and entry
Several steps are necessary to start and complete the coronavirus infective cycle: 1. recognition and binding to the cellular receptor(s) (one or two); 2. changes in the conformation and proteolysis of S protein; 3. fusion to cellular membrane; 4. entry of the virus into the host cells by endocytosis [26]. In host cells, the virus uses the endogenous cellular machinery to translate its replicase to transcribe and replicate viral RNA. The following steps consist of the translation of the structural proteins by sub-genomic RNAs (sgRNA) generated during genome transcription/replication and, finally, the virion assembly and release [19].
It is well known that the spike glycoprotein S, located on the surface of the viral phospholipidic membrane, is crucial for coronavirus infection and pathogenesis. It includes two functional domains or subunits, S1, the globular head containing the RBD at the N-terminal, and the S2 subunit at the C-terminal responsible for virus-cell membrane fusion, followed by two heptad regions (HR1 and HR2) and the transmembrane domain (TM). The S protein undergoes several post-translational modifications: its ectodomain is heavily glycosylated and, probably, the oligosaccharides could influence priming by host proteases and determine antibody recognition [26]. In particular, membrane fusion depends on S protein cleavage by host cell proteases at S1/S2 and S2’ site responsible for S protein activation (Fig. 2). Hoffman et al. (2020) recently demonstrated that the life cycle of SARS-CoV-2 begins with the RBD of the S protein that makes contact with the ACE2 receptor on the host cells [27,28]. Two host serine proteases participate in this event: the transmembrane TMPRSS2 and the endo-protease Furin. The S1/S2 site in SARS-CoV-2 harbors multiple arginine residues not present on SARS-CoV, but common to other human coronaviruses, like MERS-CoV, that are recognized by Furin protease (Fig. 2). The authors speculate that the presence of this multibasic cleavage between S1/S2 sites may expand SARS-CoV-2 tropism and/or enhance its transmissibility compared to SARS-CoV, due to the ubiquitous presence of Furin-like proteases in human tissues, especially lung [27]. Inhibition of TMPRSS2 and Furin protease activities can be considered an interesting therapeutic option against coronavirus infection, especially COVID-19, allowing the block and/or prevention of SARS-CoV-2 infection, as recently reported [28].
An alternative therapeutic/preventive strategy may refer to molecules or antibodies capable of disrupting S protein interaction with ACE2. This interaction, due to natural selection (mutation and probably a recombination event), is 10-20-fold stronger for SARS-CoV-2 respect to SARS-CoV and may explain the higher infectivity of former [28]. ACE2 is a type I transmembrane protein, its physiological role consists in the maturation of the peptide hormone angiotensin, essential for the control of vasoconstriction and blood pressure. From a biochemical point of view, ACE2 is a dipeptidyl carboxypeptidase with the N-terminal peptidase domain (PD) and the C-terminal collectrin (CLD) domain ending with a single transmembrane helix and a ~40-residues intracellular segment [29]. The peptidase activity of ACE2 is not essential for coronavirus infectivity because virus infecting respiratory tissues use this protein essentially as a receptor, being expressed on the cells of the respiratory tract. In the lung, ACE2 is present in alveolar epithelial type II cells and bronchial epithelial and it was first recognized as a receptor for SARS-CoV, MERS-CoV and now for SARS-CoV-2. MERS-CoV also recognizes dipeptidyl peptidase 4 (DPP4) or CD26 as entry receptors [30].
The RBD of S protein has a receptor binding motif (RBM) that makes the primary contact with the carboxyl-peptidase domain of ACE2 receptor. The amino-acidic sequence of RBM is 50% conserved between SARS-CoV and SARS-CoV-2 and structural studies performed by Yan et al. demonstrated that the extracellular PD of ACE2 is recognized by RBD through polar residues [31]. In particular, these researchers found that the most prominent alteration is the substitution of Val404 in the SARS-CoV-RBD with Lys417 in the SARS-CoV-2-RBD. Structural data on co-crystalized proteins demonstrated that the surface of ACE2 contains two “virus-binding hot-spot”, two Lys residues, essential for SARS-CoV binding ACE2 creating positive charges that need to be neutralized by the coronavirus [32]. Two key residues in the RBM of SARS-CoV-2, Gln493 and Leu455, bind to these hotspots leading to considerable stabilization of binding and a higher affinity for ACE2 than SARS-CoV. In addition, SARS-CoV-2 RBD shows a significant higher ACE2 binding affinity due to specific substitutes (residues 482–485: Gly-Val-Glu-Gly) that stabilize the interaction between them. Finally, Phe486, present in the RBM of SARS-CoV-2, is inserted into a hydrophobic pocket of N-terminal α1 helix of ACE2, while a leucine residue present in RBM of SARS-CoV forms probably a weaker contact owing to its smaller chain [32].
2.3. Genome replication/transcription and virion assembly and release
Virus replication takes place at the level of the cytoplasmic membrane and is mediated by a multi-subunit replication/transcription complex (RTC) formed by different viral NSPs. After entry, genomic RNA (gRNA) is translated by host ribosomes in polyprotein pp1a and pp1b, which are auto-cleaved to form NSP. These NSPs induce a rearrangement of cellular membrane to form double-membrane vesicles where the viral replication complexes are anchored [33]. The core component of RTC complex is the catalytic subunit, NSP12 of an RNA-dependent-RNA-polymerase (RdRp). For optimal function, this enzyme requires accessory factors: NSP7 and NSP8 that increase RdRp template binding and processivity. NSP3 and NSP5 encode papain like-protease, PLpro, and 3CLpro, essential, as described above, for the cleavage of polyprotein pp1a and pp1b [19]. Using the gRNA as a template, the coronavirus replicase synthesizes full-length negative sense (−) RNA, which, in turn, serves as a template for the synthesis of new genomic (+) gRNA and a set of different sgRNA, synthetized by discontinuous transcription. These sgRNAs encode viral structural and accessory proteins. Amino acidic residues involved in RNA binding and catalytic active site of RdRp are highly conserved in different RNA virus justifying the use of broad-spectrum antiviral inhibitors, such as Remdesivir. This drug, that very recently showed its efficacy in a placebo-controlled trial in COVID-19 patients [34], is a prodrug of an adenosine analogue that has been proposed as inhibitor of viral RdRp through non-obligate RNA chain termination, a mechanism requiring the conversion of parent compound from mono-phosphate in a tri-phosphate form. In particular, Yin et al. (2020) studied the structure of the nucleotide template in complex with RdRp as a model to understand how these drug categories can inhibit SARS-CoV-2 replicase activity. A new compound, EIDD-2801, showed more potent effects than Remdesivir in blocking virus replication for the capacity to form two extra hydrogen bonds with the key residue Lys545 within the catalytic domain of replicase and N4 hydroxyl group off the cytidine ring, and a guanine base in the template strand [35].
Although genome replication/transcription is mainly mediated by the viral replicase, other host factors have been involved, as an example, coronavirus N protein, known to act as an RNA chaperone to facilitate template switching, and the enzyme glycogen-synthase-kinase-3 (GSK3) [19]. Finally, RNA helicases (NSP13) represent the second most conserved subunit of the RNA synthesis machinery in (+) RNA coronaviruses and are involved in diverse steps of their life cycle. They utilize the energy derived from the hydrolysis of nucleoside triphosphates to unwind double-stranded RNA [33].
The assembly of viral particles takes place in the ER-Golgi intermediate complex under the control of M protein through homotypic interactions. In this phase, M protein acts as a scaffold for virus assembly because the interactions between S and M and M and N proteins allow the recruitment of structural proteins to the assembly site. E protein contributes in this phase interacting with M and inducing membrane curvature [19]. Finally, mature virions are released in smooth-walled vesicles via the secretory pathway and released by exocytosis.
In summary, coronavirus replication takes place in a membrane-protected and nuclease resistant microenvironment that contains (and sequesters) the protein functions required for viral RNA synthesis. This strategy is believed to improve duplication/transcription fidelity and, in parallel, repress host defenses triggered by the presence of double-stranded RNA [33].
3. Flavonoids against coronaviruses
3.1. Early works and effects of flavonoids on veterinarian coronaviruses
The first appearance in PubMed of flavonoids as potential antiviral agents is dated back in 1951 [36] and in 1966 quercetin was indicated among these compounds [37]. In 1977, the viricidal effect of quercetin, together with other flavonoids (apigenin, pelargonidin, procyanidin), was demonstrated on parainfluenza virus Type 3 and herpes simplex virus, but not on poliovirus Type 1 and adenovirus Type 3. The authors concluded that flavonoids may possess a potent viricidal activity, but only a moderate inhibitory effect on virus multiplication [38]. However, later studies indicated that quercetin was both effective in reducing the infectivity and intracellular replication of Herpes Simplex Virus type 1 (HSV–I), polio-virus type 1, Parainfluenza virus type 3 (Pf-3), and Respiratory Syncytial Virus (RSV) [39]. Hesperetin had no effect on infectivity, but reduced intracellular replication, while catechin and naringin had no or limited effects on either virus infectivity or replication [39]. The second half of the eighties saw a proliferation of studies on the antiviral activities of flavonoids, due to the boost of antiviral researches following the HIV emergency. A review article published in 1991 and focused on the capacity of 3-methoxyflavones to inhibit polio- and rhinoviruses infection, concluded that natural products can interfere with several antiviral mechanisms, from “adsorption of the virus to the host cell to release from it” [40].
One of the first paper exploring the antiviral effect of flavonoids on coronaviruses appeared in 1990 [41]. Here, the authors showed that quercetin reduced infectivity of human and bovine coronaviruses, OC43 and NCDCV, respectively, by 50% at a concentration of 60 μg/ml. Other flavonoids, such as kaempferol, were ineffective, although the latter, at 10 μg/ml, reduced virus replication by 65% in NCDCV and 50% in OC43 [41]. Bovine coronavirus, BCV, was also sensitive to a mixture of theaflavins from black tea (theaflavin, theaflavin-3-monogallate, theaflavin-3′-monogallate, and theaflavin-3,3′ digallate) with a mean EC50 of 34.7 μg/ml in infectivity assays on HRT-18 cell line [42]. On a different Coronaviridae of veterinarian interest, the Porcine Epidemic Diarrhea Virus (PEDV), quercetin 7-rhamnoside inhibited PEDV replication in Vero cells with an IC50 of 0.014 μg/ml and a CC50 (cytotoxicity concentration 50%) of 100 μg/ml [43]. Other flavonoids, including quercetin, apigenin, luteolin, catechin also showed significant anti-PEDV activity, but with IC50 values from 10- (apigenin) to 800-fold (catechin) higher than quercetin 7-rhamnoside indicating the importance of the o-dihydroxy functional groups at C-3′ and C-4′ and the rhamnoside at position 7 [43] (Table 1 ).
Table 1.
Studies reporting antiviral activity of natural flavonoids against human and non-human coronaviruses.
| Coronavirus | Compounds | Effects | Methods | Reference |
|---|---|---|---|---|
| Porcine epidemic diarrhea virus (PEDV) | Quercetin 7-rhamnoside | IC50 = 0.014 μg/ml | Vero cells | [43] |
| Bovine coronavirus (BCV) | Theaflavins | EC50 = 34.7 μg/ml | HRT-18 cells | [42] |
| HIV/SARS pseudotyped virus | Quercetin | EC50 = 83.4 μM | Vero E6 cells | [45] |
| HIV/SARS pseudotyped virus | Cinnamomi Cortex extract | IC50 = 37.3 μg/ml | Hep-G2 cells | [46] |
| SARS-CoV | Baicalin | EC50 = 12.5-25 μg/ml | fRhK4 cell line | [44] |
| SARS-CoV | Luteolin | EC50 = 10.6 μM | Vero E6 cells | [45] |
| SARS-CoV | Cinnamomi Cortex extract | IC50 = 7.8 μg/ml | Vero E6 cells | [46] |
| SARS-CoV | Procyanidin A2 Procyanidin B1 Cinnamtannin B1 |
IC50 = 30-40 μM | Vero E6 cells | [46] |
3.2. Flavonoids against SARS-CoV and MERS-CoV
In a formulation of Traditional Chinese Medicine (TMC) for the prevention and treatment of SARS-CoV, one of the components was the flavone glycoside baicalin from Scutellaria baicalensis Georgi. This compound was tested on fRhK4 cell line by a neutralization assay using 10 isolated of SARS-CoV coronavirus from 10 different patients. Baicalin showed an EC50 of 12.5–25 μg/ml at 48 h without significant cytotoxicity. This value was 200-400-fold higher respect to the C max of 60 ng/ml (half-life about 3 h), considering that, in humans, a standard oral dose corresponds to about 1500 mg. This makes hardly practicable baicalin for antiviral prophylaxis or treatment, although after an intravenous administration of 360 mg in humans, the molecule reached a peak serum concentration of 74 μg/ml [44]. Two other flavonoids, luteolin and quercetin, showed the capacity to block the entry of SARS-CoV into host cells [45]. Luteolin inhibited in a dose-dependent manner, SARS-CoV infection of Vero E6 cells with EC50 value of 10.6 μM (CC50 = 155 μM), while quercetin antagonized HIV-luc/SARS pseudotyped virus entry with an EC50 of 83.4 μM. The cytotoxicity of quercetin was very low (CC50 = 3.32 mM) [45] (Table 1).
The medical herb Cinnamomi Cortex, obtained from the dried bark of Cinnamomum cassia (L.) J. Presl, has been used to prepare several organic fractions enriched in bioactive polyphenols. Among these, the n-butanol fraction was the most active in inhibiting HIV/SARS-CoV pseudovirus infection dose-dependently with an IC50 of 37.3 μg/ml. This result was confirmed also against the wild-type SARS-CoV infection and the measured IC50 for the same fraction was 7.8 μg/ml [46]. The authors attributed the observed antiviral activity to the presence in the extract of procyanidin A2, procyanidin B1 and cinnamtannin B1, which showed an IC50 ranging between 30 and 40 μM in the plaque reduction assay on SARS-CoV. However, none of the procyanidins inhibited the internalization of TfR (transferrin receptor, a marker of clathrin-mediated endocytosis) and they did not affect ACE2 expression which, as reported above, is a SARS-CoV receptor [46] (Table 1).
3.2.1. Flavonoids interaction with SARS-CoV and MERS-CoV proteases
As reported above, SARS-CoV RNA genome encodes for proteinases that are required for replication and transcription. NSP3 and NSP5 are two non-structural regions encoding for PLpro and 3CLpro (also called Mpro, see Section 2), respectively [47]. The former cleaves the NSP1-NSP3 replicase polyproteins [48], while the latter is responsible for the processing of NSP4-NSP16 replicase products into individual polypeptides [24]. Therefore, both the SARS-CoV 3CLpro and SARS-CoV PLpro have been soon considered a potential target for the design and development of antiviral drugs [47]. Using two independent assays measuring SARS-CoV 3CLpro cleavage activity, based on a cell-free and a cell-based method, and aqueous extract from Isatis indigotica Fortune ex Lindl. (synonym of Isatis tinctoria L.) root showed a dose-dependent capacity to inhibit the SARS-CoV 3CLpro proteolytic cleavage activity with an IC50 of 53.8 μg/ml and 191.6 μg/ml on the cell-free and cell-based assay, respectively [49]. In the same experimental models, several herb-derived flavonoids with accredited antiviral effects were tested (hesperetin, quercetin, and naringenin). Among these, only hesperetin dose-dependently inhibited cleavage activity of the 3CLpro in cell-free and cell-based assays (IC50 60 μM and 8.3 μM, respectively) [49]. It is of interest that quercetin did not show any inhibitory effect on SARS-CoV 3CLpro, although it was reported to block the entry of SARS-CoV into host cells [45]. To support the latter conclusion, using a molecular docking approach, it was established that quercetin-3-β-galactoside bound to SARS-CoV 3CLpro with residue Gln189 playing a key role in stabilizing the binding [50]. In an in vitro assay, using His-tag recombinant SARS-CoV 3CLpro, quercetin-3-β-galactoside inhibited the protease activity competitively with IC50 of 42.79 μM. Mutation of Gln189 to alanine did not reduce the enzymatic activity of SARS-CoV 3CLpro, but lowered of about 2-fold the inhibitory potency of quercetin-3-β-galactoside, due to the reduction in binding affinity [50] (see Section 4). Quercetin was also used as a reference compound in a different study where several metabolites isolated from the leaves of Torreya nucifera (L.) Siebold & Zucc were tested for their capacity to inhibit the proteolytic activity of a commercially available form of SARS-CoV 3CLpro using an in vitro assay based on FRET method [51]. The most potent inhibitory effect was attributed to the biflavone amentoflavone with an IC50 of 8.3 μM and a non-competitive inhibition (K i = 13.8 μM). In comparison, quercetin and luteolin showed an IC50 about 3-fold higher, apigenin was the less efficient with an IC50 about 34-fold higher respect to amentoflavone. An in silico docking simulation demonstrated that the biflavone nicely fitted into the SARS-CoV 3CLpro binding pocket [51] (see section 4 below). A different series of flavonoids belonging to the four major groups, e.g. flavonol, flavanonol, isoflavone, and flavan-3-ol were tested in a different work [52]. Here quercetin, gallocatechin gallate (GCG), EGCG resulted the most effective in inhibiting the activity of recombinant SARS-CoV 3CLpro expressed in Pichia pastoris, a methylotrophic yeast, with an IC50 in the range 47–73 μM. Others, such as ampelopsin, puerarin, daidzein showed an IC50 higher than 350 μM. A more detailed characterization of GCG inhibitory capacity indicated a competitive inhibition mechanism (Ki = 25 μM). In addition, a computational docking analysis and hydrophobic and hydrogen bond interactions displayed binding energy of −14 kcal/mol of GCG to the active site of SARS-CoV 3CLpro highlighting the importance of the galloyl moiety at 3-OH position for the inhibitory activity [52] (see section 4). The ethanol extract of Psoralea corylifolia L. (synonym of Cullen corylifolium (L.) Medik) seeds led to the isolation of six flavonoids, namely, bavachinin, neobavaisoflavone, isobavachalcone, 4′-O-methylbavachalcone, psoralidin and corylifol A, all capable to inhibit in vitro SARS-CoV PLpro proteolytic activity using a fluorogenic peptide (Z-Arg-Leu-Arg-Gly-Gly-7-amido-4-methylcoumarin; Z-RLRGG-AMC). All of them inhibited the protease in a dose-dependent manner in the IC50 range of 4.2–38.4 μM with isobavachalcone and psoralidin being the most active [53]. It is of interest that the mode of inhibition is mixed, being the inhibition constants referred to type I, with the inhibitor bound to the free enzyme, or type II, where the enzyme-substrate complex is the target of the inhibitor [53]. Using a similar approach, five new and rare geranylated flavonoids, tomentin A, tomentin B, tomentin C, tomentin D, tomentin E inhibited SARS-CoV PLpro with IC50 raging between 5.0 and 14.4 μM [54]. Among 10 new polyphenols derived from Broussonetia papyrifera (L.) L'Hér. ex Vent and assayed for their capacity to inhibit the proteolytic activity of the two virus proteases 3CLpro and PLpro from both SARS-CoV and MERS-CoV viruses, only the prenylated quercetin derivative, papyriflavonol A, showed a potent, non-competitive inhibitory effect on SARS-CoV PLpro with an IC50 value of 3.7 μM. All the other compounds, presented IC50 values in the tens and/or hundreds micromolar ranges, on all four proteases, although the inhibition was dose-dependent [55]. Surface plasmon resonance (SPR) indicated that the interaction between papyriflavonol A and SARS-CoV PLpro was due to a specific binding event with a K D of 212 μM, stimulating the design of more effective coronavirus inhibitors [55]. A more specific study on the inhibition of MERS-CoV 3CLpro by flavonoids was published later [56]. Here, the authors demonstrated that among 40 flavonoids tested ach at the concentration of 20 μM, four of them, namely herbacetin, isobavachalcone, quercetin 3‐β‐D‐glucoside and helichrysetin were the most effective with and IC50 of 40.59, 35.85, 37.03 and 67.04 μM, respectively. Docking analysis showed the S1 and S2 sites of the protease play a key role in interaction with flavonoids. In fact, the 4‐hydroxyl group of helichrysetin forms a hydrogen bond with the hydroxyl group of Tyr54 of the protease MERS‐CoV 3CLpro; this seems to indicate a better affinity of helichrysetin since Tyr54 is located deep inside of the hydrophobic S2 site [56]. Recently, the same research group tested the 64-flavonoid library in search of potential inhibitors of SARS‐CoV 3CLpro [57]. The screening ended up with the identification of herbacetin, rhoifolin and pectolinarin as the most prominent inhibitors with IC50 values of 33.17, 27.45 and 37.78 μM, respectively. Also in this case, docking studies demonstrated that the better affinity of rhoifolin could be due to the coordinated binding through the polar S1 site, the hydrophobic S2 and the S3′ site with no strong tendency [57] (Table 2 ).
Table 2.
Studies reporting inhibitory activity of natural flavonoids against human coronavirus proteins.
| Viral proteins | Compounds | Effects | Methods | Reference |
|---|---|---|---|---|
| SARS-CoV PLpro | Geranylated flavonoids (tomentin A-E) | IC50 = 5.0–14.4 μM | fluorogenic peptide Z-RLRGG-AMC | [54] |
| SARS-CoV PLpro | Bavachinin | IC50 = 4.2–38.4 μM | fluorogenic peptide Z-RLRGG-AMC | [53] |
| Corylifol A Isobavachalcone | ||||
| 4′-O-methylbavachalcone | ||||
| Neobavaisoflavone | ||||
| Psoralidin | ||||
| SARS-CoV PLpro | Papyriflavonol A | IC50 = 3.7 μM | fluorogenic peptide Z-RLRGG-AMC | [55] |
| SARS-CoV PLpro | Xanthoangelol E | IC50 = 1.2 μM | cell-free method | [58] |
| SARS-CoV 3CLpro | Xanthoangelol E | IC50 = 11.4 μM | cell-free method | [58] |
| IC50 = 7.1 μM | cell-based method | |||
| SARS-CoV 3CLpro | Herbacetin | IC50 = 33.17 | recombinant protein; FRET method | [57] |
| Pectolinarin | IC50 = 27.45 | |||
| Rhoifolin | IC50 = 37.78 μM | |||
| SARS-CoV 3CLpro | Isatis indigotica extract | IC50 = 53.8 μg/ml | cell-free method | [49] |
| Hesperetin | IC50 = 60 μM | |||
| SARS-CoV 3CLpro | Quercetin-3-β-galactoside | IC50 = 42.79 μM | recombinant protein | [50] |
| SARS-CoV 3CLpro | Amentoflavone | IC50 = 8.3 μM | recombinant protein; FRET method | [51] |
| SARS-CoV 3CLpro | Epigallocatechin gallate | IC50 = 47–73 μM. | recombinant protein; FRET method | [52] |
| Gallocatechin gallate | ||||
| Quercetin | ||||
| MERS-CoV 3CLpro | Helichrysetin | IC50 = 40.59 μM | recombinant protein FRET method | [56] |
| Herbacetin | IC50 = 35.85 μM | |||
| Isobavachalcone | IC50 = 37.03 μM | |||
| Quercetin 3‐β‐D‐glucoside | IC50 = 67.04 μM | |||
| SARS-CoV NTPase/helicase | Quercetin | IC50 = 8.1 μM | recombinant protein FRET-based dsDNA unwinding assay | [62] |
| SARS-CoV NTPase/helicase | 7-O-arylmethylquercetin derivatives | IC50 = 2.7–5.2 μM | recombinant protein FRET-based dsDNA unwinding assay | [63] |
| SARS-CoV NTPase/helicase | Myricetin | IC50 = 2.71 μM | recombinant protein FRET-based dsDNA unwinding assay | [64] |
| ATPase activity | Scutellarein | IC50 = 0.86 μM | ATP hydrolysis colorimetric assay | |
| N protein | Catechin gallate | 0.05 μg/ml (40% inhibition) | quantum dots (QDs)-conjugated RNA oligonucleotide on biochip | [60] |
| Gallocatechin gallate |
Chalcones also represented an interesting flavonoid group with significant inhibitory activity against the two main coronavirus proteases. In a study based on nine alkylated chalcones isolated from the Japanese plant Angelica keiskei (Miq.) Koidz, it was demonstrated a significant capacity to inhibit both SARS-CoV 3CLpro and SARS-CoV PLpro protease activity in cell-based and cell-free assays. For the former protease, the cell-free assay indicated a range of IC50 of 11.4–39.4 μM for 7 out of 9 chalcones. The same compounds inhibited SARS-CoV PLpro with an IC50 of 1.2–26.0 μM. The most active resulted xanthoangelol E, containing the perhydroxyl group, that showed an IC50 of 11.4 and 1.2 μM for SARS-CoV 3CLpro and SARS-CoV PLpro, respectively. In the cell-based cleavage, this chalcone resulted to inhibit SARS-CoV 3CLpro with an IC50 of 7.1 μM and a CC50 of 65.6 μM [58] (Table 2).
3.2.2. Flavonoids interaction with SARS-CoV N protein
As discussed in the previous paragraphs, the genome of SARS-CoV encodes structural proteins, including the N protein, an alkaline protein with a lysine-rich region that suggests a nuclear localization signal. The N factor plays a key role in virion assembly through its interactions with the viral genome and M protein. It also participates in viral RNA synthesis [59]. An interesting approach was established for the screening of potential inhibitors of SARS-CoV N protein using a mimicking on glass chip the direct binding of viral RNA to N protein. The screening showed that among the 23 polyphenolic compounds tested, only (−)-catechin gallate and (−)-GCG were able to displace the binding of N protein to the RNA oligonucleotide. Starting from 0.005 μg/ml, both compounds in a concentration-dependent manner attenuated the binding affinity on the designed biochip and at the concentration of 0.05 μg/ml, they showed up to 40% inhibition activity [60] (Table 2).
3.2.3. Flavonoids interaction with SARS-CoV NTPase/helicase
SARS-CoV NTPase/helicase (also called NSP13) represents an attractive target for anti-SARS therapy since it plays crucial role in the viral life cycle [61]. Quercetin and its derivatives have been proposed as a possible inhibitor of the helicase. Firstly, using the recombinant protein and a FRET-based assay, it was reported that the IC50 of quercetin towards the Duplex DNA-unwinding activity of the SARS-CoV NTPase/helicase was of 8.1 μM, highlighting the importance of the presence of a diketo acid core and a free catechol unit [62]. The same group later published the synthesis of several 7-O-arylmethylquercetin derivatives. Three of them, with 3″-Cl, 3″-CN, and 4″-Cl substituent on arylmethyl group, tested in this study showed inhibitory activity against SARS-CoV NTPase/helicase ranging between 2.7 and 5.2 μM, the same order of magnitude of the parental compound [63]. The NSP13 helicase possesses a dsDNA unwinding activity as well as an ATPase activity allowing the helicase to translocate along with the nucleic acids by hydrolyzing ATP. A screening of about 64 natural compounds, including several flavonoids (strangely quercetin was not included in this study), all tested at the concentration of 10 μM, did not evidence any compound able to significantly inhibit the dsDNA unwinding activity of the NSP13 helicase in a FRET-based assay [64]. However, when the same compounds were tested for their capacity to inhibit the ATPase activity of the helicase, two of them, myricetin and scutellarein, emerged as a potent inhibitor of NSP13 ATPase with IC50 values of 2.71 and 0.86 μM, respectively. It is of interest that these compounds did not inhibit to the same extent the ATP hydrolysis activity of the hepatitis C virus helicase, indicating specificity for the SARS-CoV enzyme [64] (Table 2).
3.3. Flavonoids against SARS-CoV-2
Following the COVID-19 pandemic, the interest of the potential therapeutic use of flavonoids against coronavirus infection focused on SARS-CoV-2 virus. Only few papers have been published up to now, but it is easy to predict that this number will exponentially increase in the next months or weeks. Considering the impact of the COVID-19 outbreak in China, it was expected that TCM could have a primary role in the therapy of the SARS-CoV-2 infection or, at least, in the alleviation of its symptoms. In fact, on February 2020, the rate of TCM treatment for COVID-19 in China was of about 90% with only 5% of patients that manifested worse clinical signs [65]. The formula Qingfei Paidu Decoction (QFPD), consisting of 21 components (herbs and minerals drugs), showed an effectiveness of 92% in patients at all stages including subjects cured and discharged, cases where clinical symptoms were disappeared, remained stable without aggravation or significantly improved [66]. The beneficial effects of QFPD were evident after 6 days of treatment with chest CT results that ameliorated, tracheobronchial shadow was normal, and inflammation was also absorbed accordingly with the theory of TMC that identifies in the lung the primary target of QFPD against COVID-19 [65]. In the attempt to identify the major constituents of QFPD and to investigate its pharmacological mechanism against COVID-19 infection, Yang et al. [66] applied an integrated multidisciplinary approach (in silico technology that included pharmacological network and molecular networking of LC-MS data). They identified 129 compounds clustered in 14 groups, where flavonoids represented 45% of the total. Finally, a recent paper, using a computation approach, demonstrated that narcissoside, an isorhamnetin-3-O-rutinoside flavonoid (glycosyloxyflavone) present in several wild plants, is a potent inhibitor of 6W63, the term that indicates the experimental structure of SARS-CoV-2 3CLpro protease (https://covid-19.uniprot.org/uniprotkb/P0DTD1). The narcissoside showed a higher affinity than the standard inhibitor X77. The two inhibitors shared the same complex, but narcissoside interacted with three amino acids (Leu167, Pro52 and Pro168) different than those interacting with X77 (His172, Leu27 and Thr26). Finally, the better fit of the glycosyloxyflavone into the active site of the 6W63 was also reinforced by thirteen hydrogen bonds compared to the four established by X77 [67].
4. Flavonoids as potential inhibitors of SARS-CoV proteins: In silico studies
From the previous sections, we learned that the bioinformatics approach represents an essential tool to identify antagonist compounds that can specifically target the binding sites of SARS-CoV viral proteins through complex molecular interactions responsible for viral attachment and replication. This goal can be achieved by targeting structurally important binding sites, highly conserved regions in non-structural proteins including NSP12 RdRp, NSP13 helicase, and the proteases 3CLpro and PLpro [68]. 3CLpro is highly conserved in all coronaviruses, it is essential for the polyprotein cleavage and, as reported above, represents an important target for the development of new inhibitory agents [24].
The analysis of SARS-CoV-2 3CLpro crystal structure indicated that the enzyme is a homodimer (chains A and B), composed of 306 amino acids and each monomer consists of three different domains [69,70]. Two catalytic domains, I (residues 8–101) and II (residues 102–184), have an antiparallel β-barrel structure, arranged perpendicular to each other. The active site is located in the cleft between domain I and domain II with a catalytic dyad (His41 and Cys145), connecting to the helical domain III by a long loop region [71]. Domain III (residues 201–303), a globular cluster of five α-helices, is involved in the dimerization of the 3CLpro, essential for the catalytic activity. N-finger of one subunit is involved in the arrangement of the substrate binding-pocket through a salt-bridge interaction between Glu290 and Arg4 of each protomer. It has also been shown that S1, S2, and S4 subsites are involved in substrate recognition. S1 subsite is the polar site of 3CLpro containing a small aliphatic residue (Ser, Gly, Ala) in the P1 position of the proteolytic site. The hydrophobic S2 subsite contains a large hydrophobic residue in P2, whereas the side chain of Val and Ala are at P3 and P4 side, respectively, forming a small hydrophobic pocket [69].
Due to the COVID-19 pandemic, compelling efforts concentrated in identifying and designing new inhibitory compounds targeting the SARS-CoV-2 proteases and, among these, flavonoids attracted scientists’ interest being them promising agents against SARS-CoV infection (see the section above and these reviews [57,72]). Accumulating evidence are going in this direction. The inhibitory activity of flavonoids against SARS-CoV cysteine proteases in the low micromolar range was higher if compared to the effects of peptide-derived inhibitors [55].
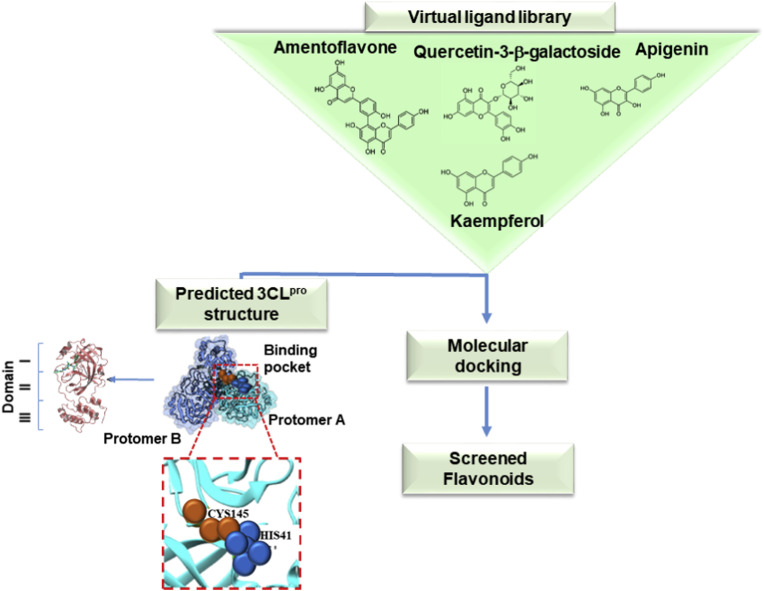
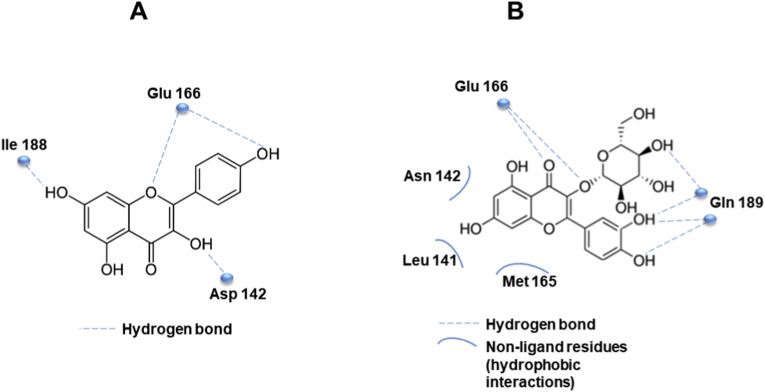
The structural features of flavonoids were responsible for their selectivity, suggesting that the two phenyl groups were associated with inhibitory potency, as resulting by IC50 values of the studies cited above for kaempferol, quercetin, and quercetin-β-galactoside [55]. Docking stimulation screening was performed to predict the binding affinity of flavonoids, showing that apigenin, luteolin, quercetin, daidzein, epigallocatechin, and kaempferol were able to inhibit the proteolytic activity of SARS-CoV 3CLpro [57] (Fig. 3 ). In particular, the molecular dynamics simulations allow to predict that His41, Gly143, and Glu166 formed interactions with common functional groups of flavonoids that showed inhibitory activities [73]. For example, Fig. 4 A shows the predicted interaction between the catalytic site of SARS-CoV 3CLpro and the phenyl group of kaempferol, which locates in the hydrophilic task of SARS-CoV through a hydrogen bond with Glu166. Other hydrogen bonds are formed between the hydroxyl groups and Ile188, Asp142, while the chromen-4-one scaffold was in the hydrophobic S2 site [57]. A similar association between inhibitory effect and molecular interaction established by docking analysis was described for quercetin-3-β-galactoside against SARS-CoV 3CLpro [50]. The analysis of structure–activity relationship highlighted crucial pharmacophore features, showing that the specific interactions between quercetin-3-β-galactoside and the active site of the protease occurred by six hydrogen bonds with residues of the catalytic binding pocket. Indeed, His41, Gly143, Ser144, Cys145, Glu166, and Gln189 residues were located near the pharmacophore spheres. In detail, the inhibitory effect of quercetin-3-β-galactoside was strongly associated to the bind with Gln189 suggesting the formation of four hydrogen bonds with O and N atoms of the Gln189 side chain (Fig. 4B). In addition, the N atom of the main chain of Glu166 was involved in the formation of the other two hydrogen bonds with the flavonoid. Moreover, quercetin-3-β-galactoside formed hydrophobic interactions with residues Leu141, Asn142, Met165, and Glu166 of SARS-CoV 3CLpro (Fig. 4B). According to this structure, it is possible to speculate that the four hydroxyl groups of quercetin are strongly responsible for its inhibitory role and the spatial conformation based on sugar moiety and 7-hydroxy site allowed structural changes through hydrogen bonding interactions [50].
Fig. 3.
Graphical representation of computer docking screening indicating the interaction of flavonoids and the binding pocket of SARS-CoV 3CLpro.
Fig. 4.
A. Predicted interaction of kaempferol with the catalytic site of SARS-CoV 3CLpro by hydrogen bond formation. B. Hydrogen bond and hydrophobic interactions between quercetin-3-β-galactoside and the active site of SARS-CoV 3CLpro.
Two other examples can be taken by the works of Nguyen et al. [52] and Ryu et al. [51] commented in Section 3 above. In the former, the GCG interaction with the substrate-binding pocket of 3CLpro involved 7 hydrogen bonds with residues in the catalytic binding pocket and hydrophobic interactions of carbon atoms with His41, Cys145, Met165, Glu166, Asp187, Arg188, and Gln189 of 3CLpro [52]. Interestingly, the methylation of hydroxyl groups at C-7 reduced the inhibitory activity, revealing that the biflavone amentoflavone showed the highest inhibitory activity with an IC50 value of 8.3 μM (Table 2). To support the effective inhibitor role of this molecule, computer-docking analysis indicated the interaction with the S1 site of 3CLpro, forming two hydrogen bonds between the C5 hydroxyl group and the nitrogen atom of the imidazole group of His163 and the -OH of Leu141. An additional hydrogen bond occurred between the hydroxyl group in the B ring with Gln189, involving the S2 site of 3CLpro [51].
The structure-activity relationship analysis of flavones namely apigenin, luteolin and quercetin evidenced that the substitution of C-30 hydroxyl group, as in luteolin, and the hydroxyl group at the C-30 position of quercetin, may play a pivotal role in SARS-CoV 3CLpro inhibition [51]. This observation was supported by a recent study, which explains by molecular docking study that luteolin forms 5 hydrogen bonds with Gln189, Leu4, Asn142, Thr26 and hydrophobic interaction with Met49 and Val3, in agreement with its lower binding energy [68]. Based on these observations, it is not surprising that molecular docking approach, summarized in Fig. 3, supports the role of flavonoids in the inhibition of SARS-CoV 3CLpro by binding His41 and Cys145 of the catalytic site and other active site residues (e.g., Met49, Gly143, His163, His164, Glu166, Pro168, and Gln89), stimulating their validation by in vitro and in vivo studies. A recent paper confirmed that luteolin can also bind and inhibit the SARS-CoV 3CLpro [68].
Another interesting field of investigation is represented by inhibition of RNA viral replication, proposing RdRp as a candidate for targeted drug development. To this purpose, a molecular screening evidenced that theaflavin can interfere with the catalytic pocket of SARS‐CoV‐2 RdRp, showing binding energy of −8.8 kcal/mol. By means of molecular docking, it has been demonstrated that hydrophobic interactions are involved in binding of theaflavin to RdRp. In addition, hydrogen bonds were established between functional moieties of theaflavin and residues Asp452, Arg553 and Arg624 of RdRp [74].
5. Conclusions and future directions
From the data reported in the previous sections, the pleiotropic nature of polyphenols and, among them, the flavonoid class, is emerging as a promising and powerful cornucopia of natural compounds with potential antiviral capacity. We reported and commented on the results of several studies largely based on docking simulations suggesting the possibility that specific flavonoids can interact with and inhibit key factors responsible for the virus life cycle. In different biological fields, the therapeutic applications of flavonoids are usually accompanied by strengthens and weaknesses, generally shared by other bioactive phytochemicals. As an example, being flavonoids present in our diet, it is easy to associate their beneficial effects with the consumption of foods/dietary patterns enriched in this class of compounds. Focusing our analysis on their antiviral properties, recent reviews support this view. In fact, Messina et al. (2020) suggested that dietary intervention may ameliorate COVID-19 outcomes and polyphenols/flavonoids can contribute to these effects reducing inflammation and blocking nuclear NF-κB translocation [75]. Others suggested that “nutraceuticals and functional foods have broad potential for preventing the mechanisms of viral infection and modulating immune responses” [76], and hypothesize a link between the senolytic activity of some flavonoids (e.g. quercetin) and the higher susceptibility of older people to viral infections, including SARS-CoV-2. Similarly, it has been hypothesized that treating patients affected by COVID-19 with senolytics and other anti-aging drugs may represent a prominent approach to prevent viral transmission and quercetin has been included among these potential agents to be tested in clinical trials [77].
We express caution on these interpretations too optimist suggesting that coronavirus viral infections can be ameliorated or prevented by diet. However, although difficult to assess from an experimental and clinical point of view, the concept that strengthening the immune system, reducing inflammation and oxidative stress during pandemic can be achieved by increasing the consumption of food groups and key nutrients remains an attractive hypothesis [78,79]. Obstacles to this assumption are represented by the well-known and largely accepted limits of natural compounds, including flavonoids, in foods and/or in nutraceutical formulations. The extremely low bioavailability, their high bio-transformation due to the intestinal adsorption and complexity of gut microbiota make unlikely that flavonoids and their metabolites can reach blood concentrations in the micromolar range. These limits not only make nutritional supplementation ineffective in humans, but are in clear contradiction with the effective concentrations of flavonoids tested against coronaviruses and viral proteins cited in the previous sections, all in the micromolar range [80] (Table 1, Table 2).
We and others [72] are more prone to believe that the real applicability of flavonoids as antiviral drugs resides in the therapy, more than the prevention. In fact, the large part of the works commented above refers to the direct binding of specific flavonoids to viral targets, although with IC50 of tens of micromolar. This suggests that, based on in vitro assays and docking models, it will be possible to design and chemically synthesize more efficient compounds based on the flavonoid structures, where key active residues are conserved, while others undergo modifications accordingly to the SAR analysis. Of course, both natural and synthetic compounds will need functional validations that cannot be limited to in vitro assays based on recombinant proteins (Table 2). To this aim, the recent COVID-19 pandemic is speeding up enormously the commercialization of new reagents and kits to identify SARS-CoV-2 inhibitors that will probably accelerate the development of innovative methods to assess how coronaviruses infect cells and how to block the infection. The therapeutic application of flavonoids, or their derivatives, as antiviral agents, as pure compounds or in combination with canonical antiviral drugs, presents also the advantage to mitigate the critical issue of their scarce bioavailability. In fact, pharmacological (not nutritional) doses, given for a limited amount of time can be plausibly administered by routes different from oral (e.g., inhalational, intravenous) that preserve the molecules from intestinal metabolism and adsorption, ameliorating their pharmacokinetics and pharmacodynamics parameters. Baicalin, cited above in Section 3, represents an example of a compound that administered intravenously can reach a serum concentration of 74 μg/ml [44].
Of course, clinical studies on infected subjects are the “the great absentee” from the scene, independently if we consider nutritional or pharmacological applications of flavonoids as antiviral agents. To our knowledge, only one trial is present in the database ClinicalTrial.gov [81]. It is in the recruitment status and regards the effect of quercetin on the prophylaxis and treatment of COVID-19. The trial is based on the assumption that quercetin, being a strong scavenger and anti-inflammatory agent, can be effective in COVID-19 cases. The quercetin dosage scheduled is 500–1000 mg for prophylaxis and treatment, respectively [81]. Two other studies based on tannins [82] and an extract obtained from Caesalpinia spinosa (Molina) Kuntze [83] were no yet recruiting.
In conclusion, the interest of scientists for the antiviral capacity of flavonoids against human coronavirus infections can benefit of the enormous amount of resources that governments, health agencies, and private companies are pouring in the field, searching for a cure against SARS-CoV-2. This situation barely resembles what happened in the eighties-nineties following the AIDS pandemic, when the basic knowledge in the immunological mechanisms controlling the response to HIV infection underwent amazing and unpredictable progresses. Waiting for a valuable vaccine against COVID-19, the pharmacological approach remains a priority and flavonoids may contribute to it. In this scenario, the “pleiotropic” properties of flavonoids that we mentioned at the beginning of this review, risks to become the passepartout to counteract coronaviruses since they can be effective on both sides, viral and host cells, to inhibit infection. In fact, recent works hypothesized that flavonoids can inhibit both TMPRSS2 and Furin, which cleave the SARS-CoV-2 Spike protein facilitating SARS-CoV-2 infectivity. Molecular docking-based screening and in vitro assays using recombinant proteins indicated that (−)-epicatechin 3-O-(3′-O-methyl) gallate for TMPRSS2 [84] and baicalein and oroxylin A glycoside for Furin [85] can bind and inhibit their respective proteases blocking virus propagation.
We hope that the hypotheses and discussions presented here can stimulate scientists to design appropriate experiments to prove that naturally occurring flavonoids or their derivatives can ameliorate anti-coronavirus prophylaxis and therapy.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We acknowledge the nurses, the medical doctors and the social and health workers around the world who, with their daily effort, saved lives and gave their lives during the COVID-19 pandemic.
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., China Novel Coronavirus I., Research T. A novel coronavirus from patients with pneumonia in China. N. Engl. J. Med. 2019;382(2020):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coronaviridae V. Study Group of the International Committee on Taxonomy of, the species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., Sheng J., Quan L., Xia Z., Tan W., Cheng G., Jiang T. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Puelles V.G., Lutgehetmann M., Lindenmeyer M.T., Sperhake J.P., Wong M.N., Allweiss L., Chilla S., Heinemann A., Wanner N., Liu S., Braun F., Lu S., Pfefferle S., Schroder A.S., Edler C., Gross O., Glatzel M., Wichmann D., Wiech T., Kluge S., Pueschel K., Aepfelbacher M., Huber T.B. Multiorgan and renal tropism of SARS-CoV-2. N. Engl. J. Med. 2020 doi: 10.1056/NEJMc2011400. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Varga Z., Flammer A.J., Steiger P., Haberecker M., Andermatt R., Zinkernagel A.S., Mehra M.R., Schuepbach R.A., Ruschitzka F., Moch H. Endothelial cell infection and endotheliitis in COVID-19. Lancet. 2020;395:1417–1418. doi: 10.1016/S0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guy R.K., DiPaola R.S., Romanelli F., Dutch R.E. Rapid repurposing of drugs for COVID-19. Science. 2020;368:829–830. doi: 10.1126/science.abb9332. [DOI] [PubMed] [Google Scholar]
- 7.Magro G. COVID-19: Review on latest available drugs and therapies against SARS-CoV-2. Coagulation and inflammation cross-talking. Virus Res. 2020;286:198070. doi: 10.1016/j.virusres.2020.198070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson R.M., Vinetz J.M. Dexamethasone in the management of covid -19. BMJ. 2020;370:m2648. doi: 10.1136/bmj.m2648. [DOI] [PubMed] [Google Scholar]
- 9.Russo G.L., Tedesco I., Spagnuolo C., Russo M. Antioxidant polyphenols in cancer treatment: Friend, foe or foil? Semin. Canc. Biol. 2017;46:1–13. doi: 10.1016/j.semcancer.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 10.Spagnuolo C., Moccia S., Russo G.L. Anti-inflammatory effects of flavonoids in neurodegenerative disorders. Eur. J. Med. Chem. 2018;153:105–115. doi: 10.1016/j.ejmech.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Russo M., Spagnuolo C., Tedesco I., Russo G.L. Phytochemicals in cancer prevention and therapy: Truth or dare? Toxins (Basel) 2010;2:517–551. doi: 10.3390/toxins2040517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mierziak J., Kostyn K., Kulma A. Flavonoids as important molecules of plant interactions with the environment. Molecules. 2014;19:16240–16265. doi: 10.3390/molecules191016240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nabavi S.M., Samec D., Tomczyk M., Milella L., Russo D., Habtemariam S., Suntar I., Rastrelli L., Daglia M., Xiao J., Giampieri F., Battino M., Sobarzo-Sanchez E., Nabavi S.F., Yousefi B., Jeandet P., Xu S., Shirooie S. Flavonoid biosynthetic pathways in plants: Versatile targets for metabolic engineering. Biotechnol. Adv. 2020;38:107316. doi: 10.1016/j.biotechadv.2018.11.005. [DOI] [PubMed] [Google Scholar]
- 14.Ferrer J.L., Austin M.B., Stewart C., Jr., Noel J.P. Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiol. Biochem. 2008;46:356–370. doi: 10.1016/j.plaphy.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lalani S., Poh C.L. Flavonoids as antiviral agents for enterovirus A71 (EV-A71) Viruses. 2020:12. doi: 10.3390/v12020184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qian S., Fan W., Qian P., Zhang D., Wei Y., Chen H., Li X. Apigenin restricts FMDV infection and inhibits viral IRES driven translational activity. Viruses. 2015;7:1613–1626. doi: 10.3390/v7041613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steinmann J., Buer J., Pietschmann T., Steinmann E. Anti-infective properties of epigallocatechin-3-gallate (EGCG), a component of green tea. Br. J. Pharmacol. 2013;168:1059–1073. doi: 10.1111/bph.12009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kapikian A.Z., James H.D., Jr., Kelly S.J., Dees J.H., Turner H.C., McIntosh K., Kim H.W., Parrott R.H., Vincent M.M., Chanock R.M. Isolation from man of "avian infectious bronchitis virus-like" viruses (coronaviruses) similar to 229E virus, with some epidemiological observations. J. Infect. Dis. 1969;119:282–290. doi: 10.1093/infdis/119.3.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fung T.S., Liu D.X. Human coronavirus: Host-pathogen interaction. Annu. Rev. Microbiol. 2019;73:529–557. doi: 10.1146/annurev-micro-020518-115759. [DOI] [PubMed] [Google Scholar]
- 20.Zhong N.S., Zheng B.J., Li Y.M., Poon, Xie Z.H., Chan K.H., Li P.H., Tan S.Y., Chang Q., Xie J.P., Liu X.Q., Xu J., Li D.X., Yuen K.Y., Peiris, Guan Y. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. Lancet. 2003;362:1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Groot R.J., Baker S.C., Baric R.S., Brown C.S., Drosten C., Enjuanes L., Fouchier R.A., Galiano M., Gorbalenya A.E., Memish Z.A., Perlman S., Poon L.L., Snijder E.J., Stephens G.M., Woo P.C., Zaki A.M., Zambon M., Ziebuhr J. Middle East respiratory syndrome coronavirus (MERS-CoV): Announcement of the Coronavirus Study Group. J. Virol. 2013;87:7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dai W., Zhang B., Su H., Li J., Zhao Y., Xie X., Jin Z., Liu F., Li C., Li Y., Bai F., Wang H., Cheng X., Cen X., Hu S., Yang X., Wang J., Liu X., Xiao G., Jiang H., Rao Z., Zhang L.K., Xu Y., Yang H., Liu H. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: Basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 25.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pillay T.S. Gene of the month: the 2019-nCoV/SARS-CoV-2 novel coronavirus spike protein. J. Clin. Pathol. 2020;73:366–369. doi: 10.1136/jclinpath-2020-206658. [DOI] [PubMed] [Google Scholar]
- 27.Hoffmann M., Kleine-Weber H., Pohlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78:779–784.e5. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tipnis S.R., Hooper N.M., Hyde R., Karran E., Christie G., Turner A.J. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J. Biol. Chem. 2000;275:33238–33243. doi: 10.1074/jbc.M002615200. [DOI] [PubMed] [Google Scholar]
- 30.Lim Y.X., Ng Y.L., Tam J.P., Liu D.X. vol. 4. Diseases; 2016. (Human Coronaviruses: A Review of Virus-Host Interactions). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ziebuhr J. The coronavirus replicase. Curr. Top. Microbiol. Immunol. 2005;287:57–94. doi: 10.1007/3-540-26765-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lye D.C.B., Hui D.S., Marks K.M., Bruno R., Montejano R., Spinner C.D., Galli M., Ahn M.Y., Nahass R.G., Chen Y.S., SenGupta D., Hyland R.H., Osinusi A.O., Cao H., Blair C., Wei X., Gaggar A., Brainard D.M., Towner W.J., Munoz J., Mullane K.M., Marty F.M., Tashima K.T., Diaz G., Subramanian A., Investigators Remdesivir for 5 or 10 Days in patients with severe covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2015301. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yin W., Mao C., Luan X., Shen D.D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M., Chang S., Xie Y.C., Tian G., Jiang H.W., Tao S.C., Shen J., Jiang Y., Jiang H., Xu Y., Zhang S., Zhang Y., Xu H.E. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–11504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cutting W.C., Dreisbach R.H., Azima M., Neff B.J., Brown B.J., Wray J. Antiviral chemotherapy. V. Further report on flavonoids. Stanford Med. Bull. 1951;9:236–242. [PubMed] [Google Scholar]
- 37.Pusztai R., Beladi I., Bakai M., Mucsi I., Kukan E. Study on the effect of flavonoids and related substances. I. The effect of quercetin on different viruses. Acta Microbiol. Acad. Sci. Hung. 1966;13:113–118. [PubMed] [Google Scholar]
- 38.Beladi I., Pusztai R., Mucsi I., Bakay M., Gabor M. Activity of some flavonoids against viruses. Ann. N. Y. Acad. Sci. 1977;284:358–364. doi: 10.1111/j.1749-6632.1977.tb21971.x. [DOI] [PubMed] [Google Scholar]
- 39.Kaul T.N., Middleton E., Jr., Ogra P.L. Antiviral effect of flavonoids on human viruses. J. Med. Virol. 1985;15:71–79. doi: 10.1002/jmv.1890150110. [DOI] [PubMed] [Google Scholar]
- 40.Vlietinck A.J., Vanden Berghe D.A. Can ethnopharmacology contribute to the development of antiviral drugs? J. Ethnopharmacol. 1991;32:141–153. doi: 10.1016/0378-8741(91)90112-q. [DOI] [PubMed] [Google Scholar]
- 41.Debiaggi M., Tateo F., Pagani L., Luini M., Romero E. Effects of propolis flavonoids on virus infectivity and replication. Microbiol. 1990;13:207–213. [PubMed] [Google Scholar]
- 42.Clark K.J., Grant P.G., Sarr A.B., Belakere J.R., Swaggerty C.L., Phillips T.D., Woode G.N. An in vitro study of theaflavins extracted from black tea to neutralize bovine rotavirus and bovine coronavirus infections. Vet. Microbiol. 1998;63:147–157. doi: 10.1016/S0378-1135(98)00242-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi H.J., Kim J.H., Lee C.H., Ahn Y.J., Song J.H., Baek S.H., Kwon D.H. Antiviral activity of quercetin 7-rhamnoside against porcine epidemic diarrhea virus. Antiviral Res. 2009;81:77–81. doi: 10.1016/j.antiviral.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen F., Chan K.H., Jiang Y., Kao R.Y., Lu H.T., Fan K.W., Cheng V.C., Tsui W.H., Hung I.F., Lee T.S., Guan Y., Peiris J.S., Yuen K.Y. In vitro susceptibility of 10 clinical isolates of SARS coronavirus to selected antiviral compounds. J. Clin. Virol. 2004;31:69–75. doi: 10.1016/j.jcv.2004.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yi L., Li Z., Yuan K., Qu X., Chen J., Wang G., Zhang H., Luo H., Zhu L., Jiang P., Chen L., Shen Y., Luo M., Zuo G., Hu J., Duan D., Nie Y., Shi X., Wang W., Han Y., Li T., Liu Y., Ding M., Deng H., Xu X. Small molecules blocking the entry of severe acute respiratory syndrome coronavirus into host cells. J. Virol. 2004;78:11334–11339. doi: 10.1128/JVI.78.20.11334-11339.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhuang M., Jiang H., Suzuki Y., Li X., Xiao P., Tanaka T., Ling H., Yang B., Saitoh H., Zhang L., Qin C., Sugamura K., Hattori T. Procyanidins and butanol extract of Cinnamomi Cortex inhibit SARS-CoV infection. Antiviral Res. 2009;82:73–81. doi: 10.1016/j.antiviral.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stadler K., Masignani V., Eickmann M., Becker S., Abrignani S., Klenk H.D., Rappuoli R. SARS--beginning to understand a new virus. Nat. Rev. Microbiol. 2003;1:209–218. doi: 10.1038/nrmicro775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harcourt B.H., Jukneliene D., Kanjanahaluethai A., Bechill J., Severson K.M., Smith C.M., Rota P.A., Baker S.C. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 2004;78:13600–13612. doi: 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Soukhova N., Soldin O.P., Soldin S.J. Isotope dilution tandem mass spectrometric method for T4/T3. Clin. Chim. Acta. 2004;343:185–190. doi: 10.1016/j.cccn.2004.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen L., Li J., Luo C., Liu H., Xu W., Chen G., Liew O.W., Zhu W., Puah C.M., Shen X., Jiang H. Binding interaction of quercetin-3-beta-galactoside and its synthetic derivatives with SARS-CoV 3CL(pro): Structure-activity relationship studies reveal salient pharmacophore features. Bioorg. Med. Chem. 2006;14:8295–8306. doi: 10.1016/j.bmc.2006.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ryu Y.B., Jeong H.J., Kim J.H., Kim Y.M., Park J.Y., Kim D., Nguyen T.T., Park S.J., Chang J.S., Park K.H., Rho M.C., Lee W.S. Biflavonoids from Torreya nucifera displaying SARS-CoV 3CL(pro) inhibition. Bioorg. Med. Chem. 2010;18:7940–7947. doi: 10.1016/j.bmc.2010.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nguyen T.T., Woo H.J., Kang H.K., Nguyen V.D., Kim Y.M., Kim D.W., Ahn S.A., Xia Y., Kim D. Flavonoid-mediated inhibition of SARS coronavirus 3C-like protease expressed in Pichia pastoris. Biotechnol. Lett. 2012;34:831–838. doi: 10.1007/s10529-011-0845-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim D.W., Seo K.H., Curtis-Long M.J., Oh K.Y., Oh J.W., Cho J.K., Lee K.H., Park K.H. Phenolic phytochemical displaying SARS-CoV papain-like protease inhibition from the seeds of Psoralea corylifolia. J. Enzyme Inhib. Med. Chem. 2014;29:59–63. doi: 10.3109/14756366.2012.753591. [DOI] [PubMed] [Google Scholar]
- 54.Cho J.K., Curtis-Long M.J., Lee K.H., Kim D.W., Ryu H.W., Yuk H.J., Park K.H. Geranylated flavonoids displaying SARS-CoV papain-like protease inhibition from the fruits of Paulownia tomentosa. Bioorg. Med. Chem. 2013;21:3051–3057. doi: 10.1016/j.bmc.2013.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park J.Y., Yuk H.J., Ryu H.W., Lim S.H., Kim K.S., Park K.H., Ryu Y.B., Lee W.S. Evaluation of polyphenols from Broussonetia papyrifera as coronavirus protease inhibitors. J. Enzym. Inhib. Med. Chem. 2017;32:504–515. doi: 10.1080/14756366.2016.1265519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jo S., Kim H., Kim S., Shin D.H., Kim M.S. Characteristics of flavonoids as potent MERS-CoV 3C-like protease inhibitors. Chem. Biol. Drug Des. 2019;94:2023–2030. doi: 10.1111/cbdd.13604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jo S., Kim S., Shin D.H., Kim M.S. Inhibition of SARS-CoV 3CL protease by flavonoids. J. Enzyme Inhib. Med. Chem. 2020;35:145–151. doi: 10.1080/14756366.2019.1690480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Park J.Y., Ko J.A., Kim D.W., Kim Y.M., Kwon H.J., Jeong H.J., Kim C.Y., Park K.H., Lee W.S., Ryu Y.B. Chalcones isolated from Angelica keiskei inhibit cysteine proteases of SARS-CoV. J. Enzyme Inhib. Med. Chem. 2016;31:23–30. doi: 10.3109/14756366.2014.1003215. [DOI] [PubMed] [Google Scholar]
- 59.Hatakeyama S., Matsuoka Y., Ueshiba H., Komatsu N., Itoh K., Shichijo S., Kanai T., Fukushi M., Ishida I., Kirikae T., Sasazuki T., Miyoshi-Akiyama T. Dissection and identification of regions required to form pseudoparticles by the interaction between the nucleocapsid (N) and membrane (M) proteins of SARS coronavirus. Virology. 2008;380:99–108. doi: 10.1016/j.virol.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roh C. A facile inhibitor screening of SARS coronavirus N protein using nanoparticle-based RNA oligonucleotide. Int. J. Nanomed. 2012;7:2173–2179. doi: 10.2147/IJN.S31379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Keum Y.S., Jeong Y.J. Development of chemical inhibitors of the SARS coronavirus: viral helicase as a potential target. Biochem. Pharmacol. 2012;84:1351–1358. doi: 10.1016/j.bcp.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee C., Lee J.M., Lee N.R., Kim D.E., Jeong Y.J., Chong Y. Investigation of the pharmacophore space of Severe Acute Respiratory Syndrome coronavirus (SARS-CoV) NTPase/helicase by dihydroxychromone derivatives. Bioorg. Med. Chem. Lett. 2009;19:4538–4541. doi: 10.1016/j.bmcl.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park H.R., Yoon H., Kim M.K., Lee S.D., Chong Y. Synthesis and antiviral evaluation of 7-O-arylmethylquercetin derivatives against SARS-associated coronavirus (SCV) and hepatitis C virus (HCV) Arch. Pharm. Res. 2012;35:77–85. doi: 10.1007/s12272-012-0108-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu M.S., Lee J., Lee J.M., Kim Y., Chin Y.W., Jee J.G., Keum Y.S., Jeong Y.J. Identification of myricetin and scutellarein as novel chemical inhibitors of the SARS coronavirus helicase, nsP13. Bioorg. Med. Chem. Lett. 2012;22:4049–4054. doi: 10.1016/j.bmcl.2012.04.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ren J.L., Zhang A.H., Wang X.J. Traditional Chinese medicine for COVID-19 treatment. Pharmacol. Res. 2020;155:104743. doi: 10.1016/j.phrs.2020.104743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang R., Liu H., Bai C., Wang Y., Zhang X., Guo R., Wu S., Wang J., Leung E., Chang H., Li P., Liu T., Wang Y. Chemical composition and pharmacological mechanism of Qingfei Paidu decoction and Ma xing Shi Gan decoction against coronavirus Disease 2019 (COVID-19): in silico and experimental study. Pharmacol. Res. 2020;157:104820. doi: 10.1016/j.phrs.2020.104820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dubey K., Dubey R. Computation screening of narcissoside a glycosyloxyflavone for potential novel coronavirus 2019 (COVID-19) inhibitor. Biomed. J. 2020 doi: 10.1016/j.bj.2020.05.002. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu R., Chen L., Lan R., Shen R., Li P. Computational screening of antagonists against the SARS-CoV-2 (COVID-19) coronavirus by molecular docking. Int. J. Antimicrob. Agents. 2020:106012. doi: 10.1016/j.ijantimicag.2020.106012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 70.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Joshi R.S., Jagdale S.S., Bansode S.B., Shankar S.S., Tellis M.B., Pandya V.K., Chugh A., Giri A.P., Kulkarni M.J. Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. J. Biomol. Struct. Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1760137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mani J.S., Johnson J.B., Steel J.C., Broszczak D.A., Neilsen P.M., Walsh K.B., Naiker M. Natural product-derived phytochemicals as potential agents against coronaviruses: A review. Virus Res. 2020;284:197989. doi: 10.1016/j.virusres.2020.197989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang J. Fast identification of possible drug treatment of coronavirus disease-19 (COVID-19) through computational drug repurposing study. J. Chem. Inf. Model. 2020;73:3277–3286. doi: 10.1021/acs.jcim.0c00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lung J., Lin Y.S., Yang Y.H., Chou Y.L., Shu L.H., Cheng Y.C., Liu H.T., Wu C.Y. The potential chemical structure of anti-SARS-CoV-2 RNA-dependent RNA polymerase. J. Med. Virol. 2020;92:693–697. doi: 10.1002/jmv.25761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Messina G., Polito R., Monda V., Cipolloni L., Di Nunno N., Di Mizio G., Murabito P., Carotenuto M., Messina A., Pisanelli D., Valenzano A., Cibelli G., Scarinci A., Monda M., Sessa F. Functional role of dietary intervention to improve the outcome of COVID-19: A hypothesis of work. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21093104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Haslberger A.G., Jacob U., Hippe B., Karlic H. Mechanisms of selected functional foods against viral infections with a view on COVID-19: Mini review. Funct. Food Health Dis. 2020;5:195–209. [Google Scholar]
- 77.Sargiacomo C., Sotgia F., Lisanti M.P. COVID-19 and chronological aging: senolytics and other anti-aging drugs for the treatment or prevention of corona virus infection? Aging (Albany NY) 2020;12:6511–6517. doi: 10.18632/aging.103001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zabetakis I., Lordan R., Norton C., Tsoupras A. COVID-19: The inflammation link and the role of nutrition in potential mitigation. Nutrients. 2020;12 doi: 10.3390/nu12051466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Iddir M., Brito A., Dingeo G., Fernandez Del Campo S.S., Samouda H., La Frano M.R., Bohn T. Strengthening the immune system and reducing inflammation and oxidative stress through diet and nutrition: Considerations during the COVID-19 crisis. Nutrients. 2020;12 doi: 10.3390/nu12061562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rodriguez-Mateos A., Vauzour D., Krueger C.G., Shanmuganayagam D., Reed J., Calani L., Mena P., Del Rio D., Crozier A. Bioavailability, bioactivity and impact on health of dietary flavonoids and related compounds: An update. Arch. Toxicol. 2014;88:1803–1853. doi: 10.1007/s00204-014-1330-7. [DOI] [PubMed] [Google Scholar]
- 81.Onal H., Semerci S.Y. 2020. Effect of Quercetin on Prophylaxis and Treatment of COVID-19.https://clinicaltrials.gov/ct2/home NCT04377789. [Google Scholar]
- 82.Piskorz M.M. 2020. Tannin Specific Natural Extract for COVID-19 Infection (TaCOVID)https://clinicaltrials.gov/ct2/ NCT04403646. [Google Scholar]
- 83.Manrrique M., Garcia A. 2020. P2Et Extract in the Symptomatic Treatment of Subjects with COVID-19. NCT04410510. [Google Scholar]
- 84.Rahman N., Basharat Z., Yousuf M., Castaldo G., Rastrelli L., Khan H. Virtual screening of natural products against type II transmembrane serine protease (TMPRSS2), the priming agent of coronavirus 2 (SARS-CoV-2) Molecules. 2020:25. doi: 10.3390/molecules25102271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Osadchuk T.V., Shybyryn O.V., Kibirev V.K. Chemical structure and properties of low-molecular furin inhibitors. Ukr. Biochem. J. 2016;88:5–25. doi: 10.15407/ubj88.06.005. [DOI] [PubMed] [Google Scholar]