Abstract
Mfd couples transcription to nucleotide excision repair, and acts on RNA polymerases when elongation is impeded. Depending on impediment severity, this action results in either transcription termination or elongation rescue, which rely on ATP-dependent Mfd translocation on DNA. Due to its role in antibiotic resistance, Mfd is also emerging as a prime target for developing anti-evolution drugs. Here we report the structure of DNA-bound Mfd, which reveals large DNA-induced structural changes that are linked to the active site via ATPase motif VI. These changes relieve autoinhibitory contacts between the N- and C-termini and unmask UvrA recognition determinants. We also demonstrate that translocation relies on a threonine in motif Ic, widely conserved in translocases, and a family-specific histidine near motif IVa, reminiscent of the “arginine clamp” of RNA helicases. Thus, Mfd employs a mode of DNA recognition that at its core is common to ss/ds translocases that act on DNA or RNA.
Subject terms: DNA repair enzymes, Cryoelectron microscopy
Transcription-repair coupling factors (TRCFs) are large ATPases that mediate the preferential repair of the transcribed DNA strand. Here the authors reveal the cryo-EM structure of DNA-bound Mfd, the bacterial TRCF, and provide molecular insights into its mode of action.
Introduction
Given that DNA serves as a track for multiple essential molecular machines, including those carrying out replication, transcription, and repair, the movement of nucleic-acid translocases along single-stranded (ss) or double-stranded (ds) DNA is highly regulated spatiotemporally1. This control has a central role in development, neurodegeneration, aging, and the diseased state2, and despite advances, still remains insufficiently understood. Translocation on dsDNA has been challenging to study structurally because many DNA motors have little sequence specificity, are difficult to trap within a crystal, and translocation on dsDNA, as opposed to on ssDNA, leads to no detectable product. These challenges are compounded by the finding that translocases often function while coupled to macromolecular machines. They also display varied processivity and coupling to ATP hydrolysis, and while featuring conserved sequence motifs, have mechanisms of action that are modulated by accessory domains in family-specific ways, evading facile generalizations.
Transcription-repair coupling factors (TRCFs) are large superfamily 2 ATPases that mediate the preferential repair of the transcribed DNA strand (aka transcription-coupled DNA repair, TCR) in organisms ranging from bacteria to humans3–5. These factors provide a complex but poorly understood example of regulated translocation on dsDNA. At its core, TCR appears universally conserved, and relies on (1) the ability of transcription-repair coupling ATPases to release damage-stalled RNA polymerases (RNAPs) off the nucleic-acid template owing to ATP-driven translocation on dsDNA upstream of the transcription bubble, and (2) on their ability to recruit nucleotide excision repair (NER) machinery6. In bacteria, a single protein, Mfd (aka TRCF) is necessary and sufficient for the coupling process7,8. Although additional pathways have been implicated in TCR9, Mfd remains the only bacterial factor for which both RNAP release and repair enzyme recruitment functions have been demonstrated3,8,10–14. Critically, Mfd associates with RNAP in cells even in the absence of exogenous DNA damage15, it decreases class II transcriptional pausing16, promotes strand-specific repair “at a distance” downstream of a transcriptional pause site17, and dissociates transcription elongation complexes (TECs) stalled not only by DNA damage, but also by protein roadblocks18, including replication forks colliding head-on with the transcription machinery19. Thus, given these different contexts in which it acts on TECs (Fig. 1), Mfd is more appropriately viewed as a general transcription factor. Paradoxically, under certain conditions, Mfd acts as an evolvability factor, promoting hypermutation, the accelerated evolution of lagging-strand genes20,21 and the rapid development of resistance to multiple, unrelated classes of antibiotics22–25. This makes Mfd an attractive target for the development of a broad-spectrum anti-evolution drug, which could be administered in combination with well-characterized antibiotics to curtail the worldwide crisis of antimicrobial resistance.
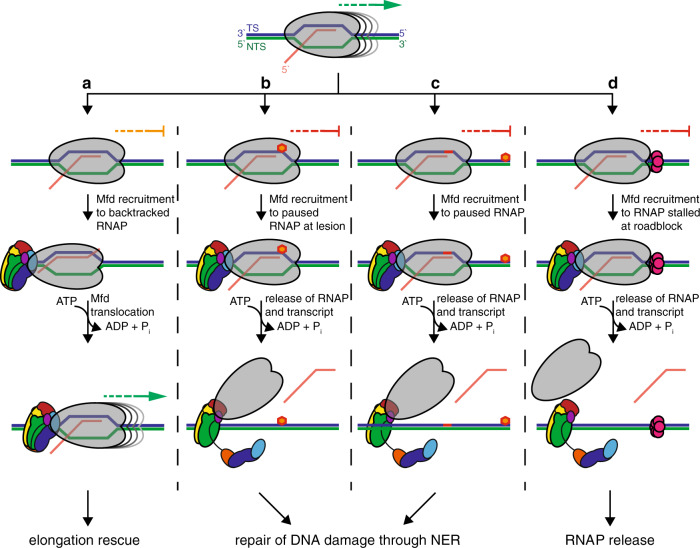
Fig. 1. All Mfd functions are dependent on translocation on DNA or action upstream of transcription elongation complexes.
Shown at the top is a schematic of an elongating RNAP (gray) with labeled nucleic-acid moieties (RNA, red; NTS, non-template strand, green; TS, template strand, blue) that can either become stalled or temporarily paused as indicated in a–d. a In Mfd-dependent rescue of class II transcriptional pausing, Mfd binds to backtracked RNAPs and promotes their forward translocation thereby rescuing transcript elongation16,26. b In canonical TCR, RNAP becomes stalled at a lesion (orange hexagon) on the TS and recruits Mfd (colored by domain, with D1a and D1b in blue, D2 in cyan, D3 in orange, D4 (RID) in magenta, D5 in yellow, D6 in green and D7 in red), either through 3D-diffusion or 1D-diffusion, enabled by a catch-up-then release mechanism dependent on ATP hydrolysis16, disruption of the D2–D7 clamp, RNA release, and UvrAB recruitment29,31. c In repair “at a distance”, Mfd is recruited to RNAP paused at class II pause signals (red line), which is first released off the nucleic-acid chains and acts as a processivity factor for Mfd to translocate toward downstream lesions in the TS, and initiate strand-specific repair17. d When RNAPs collides with protein roadblocks head-on, including replisomes, Mfd will release RNAP off the nucleic-acid chains, thereby freeing the DNA for other DNA-based processes18,19.
Mfd translocation on dsDNA is central to all Mfd functions. This is an ATP-dependent13,26–29 and regulatable process16, which it shares with chromatin remodelers belonging to the same superfamily 2 (SF2) of ATPases. Like chromatin remodelers, Mfd binds and remodels a large macromolecular assembly—the TEC, composed of core RNAP, DNA scaffold, and nascent RNA. TECs activate the Mfd translocase to a level at which processive translocation can readily be detected using classic biochemistry26,30. The structural changes underlying this activation are likely complex and may involve multiple steps27 and interlocked structural elements29,31. Mfd is composed of an ATP-dependent motor core composed of domains D5 and D6 as well as six ancillary domains, including D2 and D7 that pack against each other as an inhibitory “clamp” to restrain the ATPase (Fig. 2a–c) and mask binding determinants for recruitment of UvrA, a component of early NER29,31. Conformational changes in full-length Escherichia coli Mfd occurring during its functional cycle have remained speculative since only a single structure—that of nucleotide-free E. coli Mfd31—has been reported in peer-reviewed literature. Clamp opening was originally proposed to be prerequisite for translocation and dependent on the interaction with RNAP30 (Fig. 1). Recent and more-sensitive single-molecule studies have demonstrated that in fact Mfd can also translocate on naked DNA, albeit with limited processivity16, suggesting that Mfd exists in a dynamic conformational equilibrium that can be shifted by TEC binding. However, provided that ATP is supplied, Mfd can make excursions to the translocation-competent form even in the absence of RNAP16, suggesting that Mfd locates its targets not only by 3D diffusion, but also a more efficient 1-D search along dsDNA, possibly colliding with stalled/paused TECs ahead of it16. This scenario can lead to two outcomes: RNAP rescue by forward translocation (Fig. 1a) or dissociation (Fig. 1b–d), occurring when RNAP encounters severe hindrances to forward movement16. It is important to note that Mfd processivity on naked DNA is substantially lower than that of Mfd bound to a RNAP that has been released off the DNA chains, but remains associated with Mfd via interactions with domain D4, and possibly other unknown binding sites16.
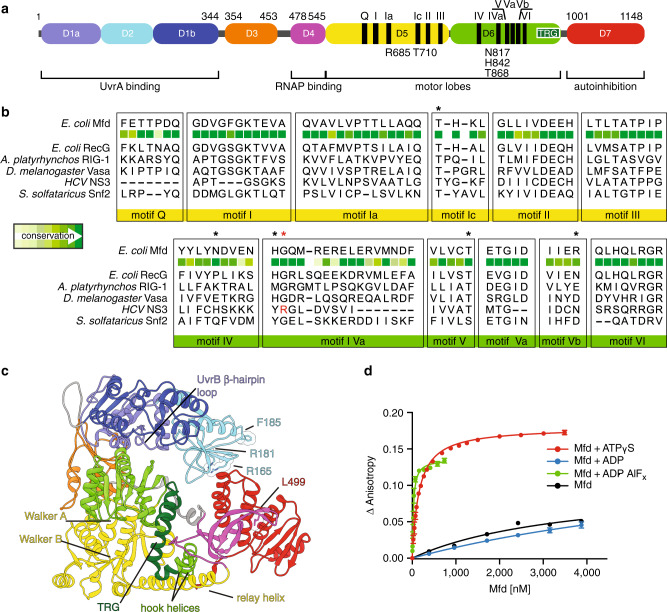
Fig. 2. DNA binding by Mfd becomes tightest in the presence of transition state analog ADP•AlFx.
a Domain organization of E. coli Mfd with annotated conserved sequence motifs and key residues mutated in this study. b Structure-based sequence alignment of the ATPase motifs of Mfd and other dsDNA and ssDNA translocases used for structural modeling purposes. Mutated residues are indicated by asterisks and sequence conservation, based on an alignment of Mfd proteins is color-coded from dark green (strongly conserved) to light green (variable). ATPase domain motifs are shown underneath for Escherichia coli RecG, Sulfolobus solfataricus Swi2/Snf2, Drosophila melanogaster Vasa, HCV NS3, and Anas platyrhynchos RIG-1. The arginine clamp R393 of HCV NS3 is shown in red. c Structural overview of nucleotide-free E. coli Mfd (PDB ID 2EYQ) colored by domain and with annotated ATPase motifs and key structural elements. Shown as CPK models are residues implicated in UvrA and RNAP binding29,31. d Fluorescence anisotropy DNA-binding curves for Mfd in distinct nucleotide states: nucleotide-free (black), with 2 mm ATPγS (red), ADP (blue), or ADP•AlFx (green). Error bars represent S.D.M. (n = 3) and are often smaller than symbols. Curves obtained in the absence/presence of ATPγS and ADP are replotted for convenience from the study by Le et al.16 Source data are provided as a Data Source file.
Despite intense research in the last decade, the mechanochemistry of TRCFs remains poorly defined. How do TRCFs recognize DNA and what are the conformational changes associated with translocation on dsDNA? Here, we shed light on these questions by defining the path of DNA across Mfd using electron cryo-microscopy (cryo-EM), structure-guided mutagenesis, and functional assays. We demonstrate that in the absence of RNAP, independent, single substitutions within the two lobes of the Mfd motor core lead to severe DNA-binding defects, and that translocation relies on the existence of a moderate and a strong binding state that correspond to the ATP- and ADP•AlFx-bound states, respectively. We pinpoint conserved residues within translocase motifs Ic, IV, V, and Vb as contributing to DNA loading, and identify an evolutionary conserved threonine in motif Ic and a family-specific histidine near motif IVa as critical for translocation. We also show that DNA binding to Mfd in the presence of transition state analog ADP•AlFx leads to large-scale rotation of the UvrB homology module, resulting in disruption of the D2–D7 clamp as well as repositioning of domain D3 of hitherto unknown function. This leads to the breaking of contacts of D3 with D5 and D6 and the establishment of new contacts with D7. Our study provides the first glimpse of a substrate-bound bacterial TRCF, and suggests that these ancient ATPases utilize a mode of DNA recognition that at its core, is common to ss/ds nucleic-acid translocases.
Results
Mfd binds dsDNA tightly with transition state analog ADP•AlFx
For processive autonomous movement on dsDNA (i.e., not dependent on tethering to RNAP) to occur, Mfd must cycle through several states, and must possess at least two DNA contact points, utilized differentially during the ATPase cycle. Mfd binding to DNA with non-hydrolysable ground ATP mimics, ATPγS and AMPPNP has been well documented14,32. ATP hydrolysis leads to DNA dissociation, e.g., Mfd translocation towards the end of the fragment3,26,33.
Importantly, although ATP binding is thought to reposition D5/D6 and align the ATPase motifs for catalysis31, there are no reports of a second state tightly associating with DNA. This would be critical because it would provide the necessary powerstroke for unidirectional movement. We have thus asked whether trapping such an alternate state might be possible using ADP and ADP•AlFx. During ATP hydrolysis, the γ-phosphate of ATP passes through a trigonal planar configuration with three equatorial oxygen atoms and two axial ligands, an oxygen from the β-phosphate and the attacking water molecule. To mimic this state, we used Mg2+•ADP•AlFx, the most extensively employed analog of the ATP hydrolysis transition state. This binds as an AlF4− species to most ATPases and adopts an octahedral geometry when complexed to the β-phosphate and the nucleophilic water moiety34. To quantify DNA binding under equilibrium conditions, we used a fluorescence anisotropy assay with a hexachlorofluorescein (HEX)-labeled 40mer dsDNA fragment. In the presence of ATPγS, the equilibrium dissociation constant, Kd, was 151 ± 3 nm (Fig. 2d, Supplementary Table 1), in agreement with previous determinations29. We detected much reduced binding with ADP and enhanced binding with ADP•AlFx (KdADP•AlFx = 24 ± 2 nm). In agreement with several reports in E. coli29, but inconsistent with reports of oligomeric Mycobacterium tuberculosis Mfd35, we observed no oligomerization by gel filtration (Supplementary Fig. 1a), suggesting that the increase in affinity in the presence of the transition state is owing to intramolecular rearrangements rather than oligomerization, as in other SF2 nucleic-acid translocases36.
The primary DNA-binding site in Mfd is its motor core
Both our study and previous work have not detected stable binding of Mfd to dsDNA in the absence of nucleotide14. Modeling of a duplex DNA-Mfd complex based on similarity with other SF2 proteins such as Sulfolobus solfataricus Snf2 is consistent with this finding. In the nucleotide-free form, the motor core of Mfd is more open. dsDNA modeled onto nucleotide-free Mfd contacts a highly conserved and positively charged D5 surface patch centered around invariant K712, but is nevertheless positioned too far away from D6 for direct contacts with this domain (Supplementary Fig. 1b–d), supporting that affinity is dictated by nucleotide status and suggesting that D6 swings down to clamp the DNA (Supplementary Fig. 1b–d). However, the conformation of the ATPase motor module in many translocases/helicases, and by extension, in Mfd, is modulated not only by nucleotide status but also interactions with family-specific accessory domains37, suggesting that changes that are more global rather than restricted to the motor module.
Our observation that ADP•AlFx enhances the affinity for DNA allowed us to reconstitute stable Mfd-DNA complexes, and prepare specimens suitable for single-particle cryo-EM for structure determination. For our DNA substrate, we chose a 21mer blunt dsDNA fragment, consistent with previously determined Mfd footprints of ~26bp16,26,29. This allowed us to restrict binding to one Mfd molecule per DNA fragment. Initial reconstructions obtained from untilted specimens were severely affected by the preferred orientation of the particles at the air-water interface, limiting resolution. We thus acquired images at 30° tilts, which allowed us to obtain a more isotropic map resolved to 5.5 Å (Methods, Supplementary Fig. 2, and Supplementary Table 2). This enabled us to readily dock all the individual domains of Mfd31 into the map and perform real-space refinement to obtain the final model. At this resolution, we do not observe density that can be assigned to the transition state analog moiety. The model (Fig. 3) affords several critical observations.
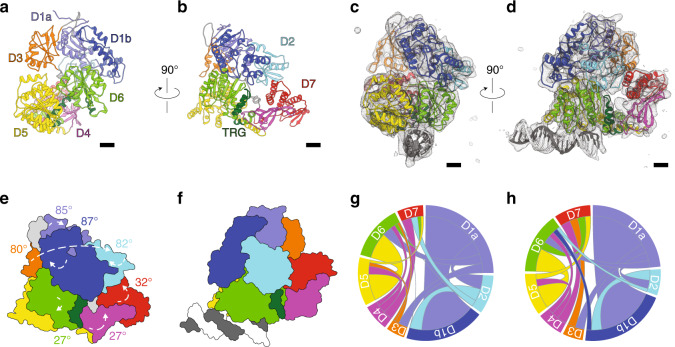
Fig. 3. Cryo-EM reconstruction of dsDNA-bound Mfd reveals a mobile D3 module and the interaction of dsDNA with the motor lobes.
a, b Orthogonal views of nucleotide-free Mfd (PDB ID 2EYQ). Scale bar is equivalent to 10 Å. c, d Orthogonal views of the cryo-EM reconstruction (gray surface) with the fitted Mfd model shown as a cartoon and colored as in a. DNA is shown in gray. e, f Cartoons of Mfd structures in the nucleotide free e and DNA-bound form f after superposition on the Cα trace of D5 and highlighting the relative rotation of Mfd domains in the free and tight DNA-bound state. g, h Chord plots highlighting intramolecular rearrangements in Mfd before g and after h DNA binding. In all panels, Mfd is colored by domain as in Fig. 2a.
First, our reconstruction reveals an unexpected large-scale swiveling motion of the UvrB homology module (D1a-D2-D1b), which results in a ~49 Å displacement and an 82° rotation of D2. Importantly, D1a-D2-D1b appears to move in concert, as a rigid body, which is consistent with previous structures of D1aD2D1b and D1aD2D1bD3 truncations, which assumed the same structure in isolation as in the context of the full-length, nucleotide-free protein (r.m.s.d. of 0.8 Å between PDB ID 2EYQ and PDB ID 2B2N and 0.6 Å between PDB ID 2EYQ and PDB ID 3HJH)32,38. Limited proteolysis of Mfd in the absence/presence of nucleotides and Mg•ADP•AlFx did not reveal differences in protease susceptibility (Supplementary Fig. 3), suggesting that the conformational changes we observe are induced by DNA binding and not by the progression from the ATP ground to the transition state. Recruitment of UvrA might thus preferentially occur in the ADP-Pi state and might promote product release and dissociation of Mfd off the DNA, leaving behind an excinuclease loaded in a strand-specific manner.
Second, particularly striking is the repositioning of domain of unknown function D3, which swings from the ventral side laterally toward D7 upon DNA binding (Fig. 3 and Supplementary Movie 1), establishing new bridging interactions with the UvrB homology module and D7. This represents a large scale ~47 Å displacement of D3 enabled by its flanking flexible linkers connecting D3 to D1b and D4 (Figs. 2a–b, 3). The distance between the termini of the fitted D3 and D4 (36 Å) is fully compatible with the length of this partially disordered D3–D4 linker. Consistent with our data, the sequence corresponding to D3 in the structure of D1aD2D1bD3 was completely disordered32, pointing to it forming a dynamic and separate structural unit. Recent structures of mycobacterial Mfd in the presence of ADP and ADP•Pi also revealed movement of D339, but this is distinct from what we observe. In mycobacterial Mfd, sharing ~33% identity with E. coli Mfd, D3 is repositioned when ADP and Pi are bound such that it contacts D1a and D5 (Fig. 4a–c). These contacts are presumably important for nucleotide binding, as they are accompanied by a flip-flop action of the D3–D4 linker (Fig. 4 and Supplemental Fig. 5)39. Movement of this linker relieves steric occlusion of the conserved phenylalanine of motif Q (F641 mycobacterial Mfd, F599 in Eco Mfd), which becomes free to stack against the adenine base of ADP. In contrast, when Mfd is bound to DNA, D3 makes contacts with D1a and D7 (Fig. 3), swapping position with D2. The UvrA-binding surface on D2, including residues critical for binding such as R165 and R18129 (Fig. 2d–f) becomes fully exposed. Contacts of D2 with conserved residues in D7, D0148, and F1050 are lost.
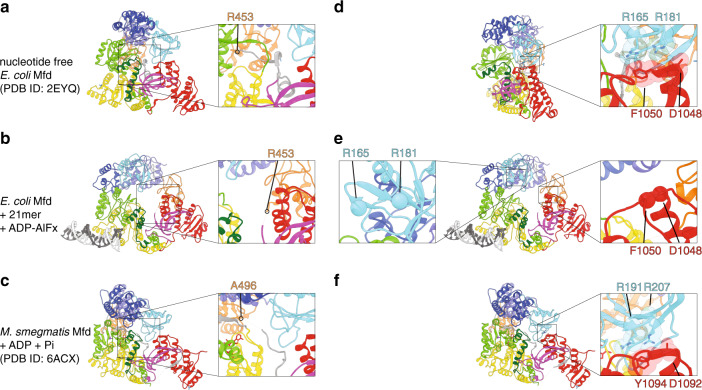
Fig. 4. DNA binding unmasks the UvrA recruitment surface within domain D2 of Mfd.
a–c Top view of nucleotide-free E. coli Mfd a, E. coli Mfd complexed to DNA and ADP-AlFx b and M. smegmatis Mfd bound to ADP and Pi c (PDB ID 6AXC). Insets show the D3–D4 linker, and C-terminal residues in D3 as positional markers. The D3–D4 linker is not visible in our map of DNA-bound Mfd b or in M. smegmatis Mfd c. d–f Overall views of Mfd with insets highlighting the disruption of the D2–D7 interaction upon DNA binding. Amino acids involved in domain–domain interactions (R165, R181, D1048, F1050) are shown as CPK with stick models. Middle inset highlights the unmasking of R165 and R181 (spheres), important for UvrA recruitment. In all panels, Mfd is colored by domain as in Fig. 2a.
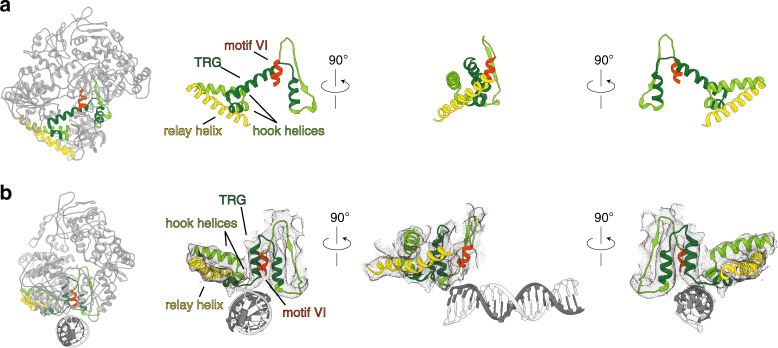
Last, but not least, our reconstruction confirms the long-hypothesized role of the TRG motif in DNA translocation. The TRG (translocation in RecG) motif is common to both the RecG and Mfd subfamilies, and consists of a helical hairpin engaging a helical “hook” C-terminal to it (Fig. 2c). This hook was partially disordered in substrate-bound RecG, but helical in Mfd and wrapped around yet another helix—the relay helix, connecting D4 to the motor core (Fig. 2c). In our reconstruction, the two helices of the TRG snap together and are likely stabilized by ADP•AlFx. binding. In contrast, in nucleotide-free Mfd, the TRG helices are splayed open by motif VI helix ATP sensor31. The closed TRG motif engages part of the hook structure but releases motif VI, which undergoes a 46 Å displacement upon rotation of D6, and indirectly modulates the conformation of the relay helix, which becomes slightly bent and rotates about a hinge located in the loop connecting it to D5 (Fig. 5 and Supplementary Movie 1). Thus, we conclude that domain D3 of hitherto unknown function plays a critical role in unmasking of the UvrA binding site and that the interlocking of motif VI, TRG, hook and relay helices enable the complex and long-range conformational changes associated with DNA binding.
Fig. 5. Structural interlocking of the TRG motif, motif VI, and relay helix.
a View of motif VI (red), TRG motif (dark green), hook helices (green), and relay helix (yellow) in the context of full-length nucleotide-free E. coli Mfd (left) and, for emphasis, as close-ups. b Similar views of DNA-bound E. coli Mfd highlighting conformational changes in this region The TRG helical hairpin closes and a bend is introduced into the relay helix.
Linchpin residues for DNA loading span both lobes of the motor core
For translocation to occur, Mfd must first load the DNA. Our reconstruction suggests that this involves D5 and D6 exclusively, and no other contacts (Fig. 3a–f), although secondary binding determinants might be unmasked under very specific conditions.
We have thus asked if isolated Mfd domains might contribute to ss or dsDNA binding. We have purified Mfd functional modules as defined by the crystal structure of E. coli Mfd (UvrB homology module MfdD1aD2D1b, domain of unknown function MfdD3, RNAP interaction domain MfdD4, and clamp domain MfdD7 (Fig. 2a and Supplementary Fig. 4) with the exception of MfdD5D6. We then assessed DNA binding with either a 40mer HEX-labeled dsDNA or a shorter Cy3-labeled double- and single-stranded 27mer. As expected, we observed no stable binding to any of the purified modules (Supplementary Fig. 4). However, we cannot rule out lower affinity, secondary binding sites that might require initial contacts with MfdD5D6, a DNA fragment longer than what was used for our EM reconstruction, or a particular DNA structure, akin to an open or partially open bubble. On longer cellular DNA substrates, multiple domains may be required for DNA binding and contacts with individual modules, as defined here, may be too weak to be detected.
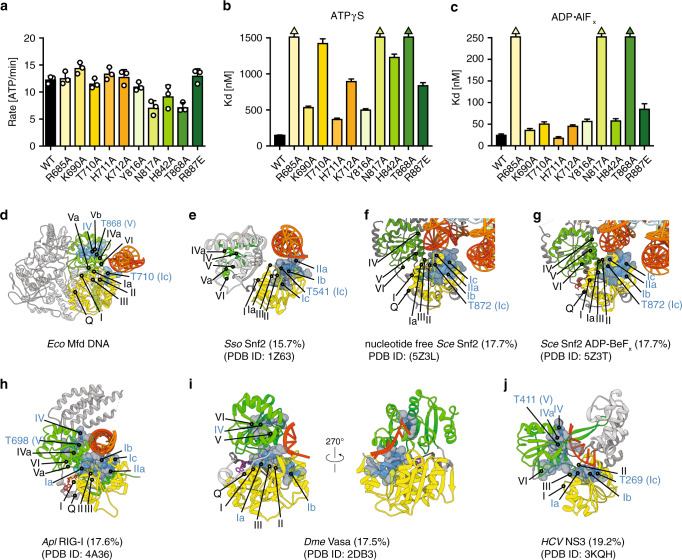
Although the EM reconstruction allowed us to identify residues proximal to DNA, specific contacts with the nucleic acid are not resolved with confidence at this resolution. To identify residues critical for DNA loading in both the ATP- and ADP•AlFx states, we used our EM reconstruction but also took advantage of evolutionary relationships and structural superpositions with well-characterized nucleic acid bound ATPases. Structural alignments with RecG, the helicase most closely related to Mfd, proved unfruitful, as crystallized RecG only contains a short fork DNA that does not reach across the helicase domains40. Instead, we extended our analyses to other ss/ds nucleic-acid translocases of known structure. We focused on the similarity with Snf2, an SF2 chromatin remodeler for which several structural models in different functional states are available. These structures include models of Snf2 bound to dsDNA41, akin to the DNA moiety found in our specimen, but also curved DNA, as found in the structure of Snf2 bound to nucleosomes42,43. We combined the information derived from our EM reconstruction and these superpositions (Supplementary Figure 6), and selected a set of well-conserved residues, highlighted in Fig. 6a and the Supplementary Movie 1, as likely key for DNA recognition. We constructed Mfd variants carrying substitutions in either D5 (MfdR685A, MfdK690A, MfdT710A, MfdH711, MfdK712A) or D6 (MfdY816A, MfdN817A, MfdH842A, MfdT868A, MfdR887E) (Fig. 2a). All were active in ATPase assays at levels comparable to wild type (Fig. 6a), indicating they are competent for nucleotide binding and hydrolysis, and consistent with proper folding, which was assessed using circular dichroism (Supplementary Fig. 7b). We then assessed binding to the same 40mer HEX-labeled dsDNA used above. The variants displayed various degrees of DNA-binding defects in the presence of both ATPγS (Fig. 6b, Supplementary Fig. 7c and Supplementary Table 1) and ADP•AlFx (Fig. 6c, Supplementary Fig. 7d and Supplementary Table 1). Interactions with DNA were more sensitive to salt in the ATPγS than the ADP•AlFx state (Supplementary Fig. 7e, f), pointing to the formation of non-electrostatic contacts as partially accounting for the gain in affinity in the transition state.
Fig. 6. Key roles for R685, N817, and R868 in DNA loading.
a Steady-state ATPase activities of single Mfd variants. ATP hydrolysis rates were measured using an ATP/NADH-coupled ATPase assay. Data shown are the means ± SD (n = 3). Variants are color-coded by domain as in Fig. 2a (D5 variants, yellow hues; D6 variants, green hues). Data for MfdR685A and MfdN817A are replotted for convenience from Le et al.16. b–c Equilibrium dissociation constants for the Mfd-dsDNA interaction derived from fluorescence anisotropy binding curves. D5 and D6 variants are color-coded by domain as in Fig. 2a. Data represent mean values ± SD (n = 3). Error bars are often smaller than symbols. Arrows indicate data sets that could not be reliably fit to a binding model owing to compromised affinity for DNA, and likely represent an overestimation of the true affinity. Binding curves are shown in Supplementary Fig. 7. d–j Structural comparison between double-stranded nucleic-acid translocases and Escherichia coli (Eco) Mfd (d) using superpositions restricted to the Cα trace of D5. Shown are Sulfolobus solfataricus (Sso) Snf2 (r.m.s.d. of 1.1 Å, e), Saccharomyces cerevisiae (Sce) Snf2 in its nucleotide-free (r.m.s.d of 1.4 Å, f) and ADP•BeFx -bound state (r.m.s.d. of 1.4, g), RIG-1 (r.m.s.d. of 1.0 Å, h) and ssRNA translocases, Drosophila melanogaster (Dme) Vasa (r.m.s.d. of 0.9 Å, i) and HCV NS3 (r.m.s.d. of 1.1 Å, j). Bound Mg2+ is shown as a dark green sphere, analog ADP•AlF3− as an aquamarine CPK model, bound nucleotide as magenta sticks, ADP as a red CPK model, BeFx as a dark red CPK model and nucleic-acid strands are colored red and orange. The two lobes of the ATPase core are in yellow (N-terminal lobe) and green (C-terminal lobe), except for DNA-binding residues, which are highlighted as slate CPK models. Other domains are shown as a gray ribbon with annotated ATPase motifs. Those motifs contacting DNA are color-coded in slate font and some are not visible in these views. Indicated in parenthesis is the degree of conservation between the various motor cores and D5 and D6 of E. coli Mfd. Source data are provided as a Source Data file.
MfdR685A, MfdN817A, and MfdT868A were severely impaired with both ATPγS and ADP•AlFx, indicating a critical role in loading DNA. T868 is particularly intriguing, as it is located in conserved helicase motif V (Fig. 2a–b), which in several cases has been shown to contribute to either ATP or oligonucleotide binding44. In the case of Mfd, substitution of T868 affects DNA binding severely, and less so ATP binding or hydrolysis (Fig. 6a–c). This is not without precedent. The structure of duck RIG-I RNA helicase bound to 19mer dsRNA and ADP•AlFx, shows this highly conserved threonine (T698 in duck RIG-I) in direct contact with a phosphate of the RNA backbone (Fig. 6h), making a hydrogen bond with the phosphoryl oxygen atom45. However, mutation of this conserved residue to alanine in human RIG-I eliminates dsRNA binding while retaining ATP hydrolysis activity46, suggesting that this residue confers affinity that is uncoupled from the splitting of the phosphoanhydride bond. In Thermotoga maritima RecG, the mutation of T478 reduces fork reversal (i.e., translocation) without affecting ATP hydrolysis47. A structurally equivalent threonine, T546, also contacts the RNA phosphate backbone in the structure of Vasa RNA helicase bound to ssRNA and AMPPNP (PDB ID 2DB3, Fig. 6i)48. At last, the SF2 helicase domain of virally encoded DExH helicase NS3 also has a similar conserved threonine, and this was seen to contact oligonucleotide in multiple structural studies49,50. As with Mfd, mutation of this threonine (T411 in NS3h, Fig. 6j) does not affect its basal ATPase activity, but abolishes oligonucleotide binding and duplex unwinding activity51.
Mfd mechanism of translocation is reminiscent of that used by RNA helicases
In the presence of ADP•AlFx, some of our variants (MfdK690A, MfdT710A, MfdK712A, MfdY816A, MfdH842A) showed only a slight impairment in function, or no DNA-binding defect at all, as was the case for MfdH711A (Fig. 6 and Supplementary Table 1). Most striking are the substitutions of T710A and H842A (Fig. 6a), which lead to severe loss of affinity in the ATPγS state, but had a comparatively minor effect on DNA affinity in the presence of ADP•AlFx. Using optical trapping, we have previously shown that in the context of a nucleotide-starved TEC, Mfd binds to the TEC such that D5 is located in front (adjacent to the TEC), followed by D6, and that upon ATPγS binding D6 moves 12 bp toward a stationary D5, approaching the RNAP footprint16. As ATP hydrolysis is required for processive translocation, this suggested to us that upon hydrolysis and/or product release, D5 may invade the RNAP footprint. We thus reasoned that the residues with nucleotide-dependent roles might be critical for cycling through all the states required for translocation. To probe translocation, we developed a fluorescent triplex displacement assay, based on the ability of dsDNA translocases to displace a fluorescently labeled triplex forming oligonucleotide (TFO) off a DNA duplex. Triplex displacement assays have been well established for constitutively hyperactive Mfd variants, such as MfdD7−AAA, carrying the E1045A, D1048A, and R1049A substitutions in D7 to disrupt the inhibitory clamp interaction (Fig. 7a)52. Given the recently determined short processivity of Mfd of ~200 bp16, we employed a 70 bp substrate, shorter than the Mfd processivity length, in conjunction with the hyperactive MfdD7−AAA variant. We included in our reaction non-labeled ssDNA, for which Mfd has a weaker affinity14, but acts as an efficient trap to inhibit a contaminating endonuclease activity that initially severely reduced the signal corresponding to full-length TFO (not shown). In the presence of MfdD7−AAA, free TFO accumulated over time, and this activity was inhibited by non-hydrolysable analog, AMPPNP and the absence of nucleotide (Fig. 7b–c and Supplementary Fig. 8). Both mutants, MfdD7−AAA;T710A and particularly MfdD7−AAA;H842A displayed defects in translocation and loading, but not ATP hydrolysis (Fig. 7f), and featured circular dichroism spectra similar to MfdD7−AAA (Supplementary Fig. 8b). The loading defects of these complex variants are somewhat different than the defects on the single T710A and H842A variants in the wild-type background (Figs. 6a–c, 7), and suggest that artificial disruption of the clamp interaction partially uncouples DNA binding from nucleotide status, consistent with a now derepressed enzyme30. As such, these variants display severe translocation defects, with MfdH842A and MfdT710 displaying no detectable translocation activity (Fig. 7b, c). At higher protein concentrations used in an attempt to overcome the loss in affinity of the D7-AAA mutants, both in the presence of ATPγS and ADP•AlFx, the apparent translocation activity of both MfdD7−AAA and its two variants decreases (Supplementary Fig. 8), likely owing to multiple loading molecules interfering with translocation, which complicates the analysis. Interestingly, although wild-type Mfd is not stimulated by the addition of herring sperm DNA, MfdD7−AAA and to a lesser extent MfdD7−AAA;T710A, and MfdD7−AAA;H842 show an increase in ATPase activity (Fig. 7f and Supplementary Fig. 8f). Thus, whereas deregulation via clamp disruption hyperactivates Mfd, mutations within motifs Ic and IVa may repress Mfd activity.
Fig. 7. T710 and H842 are critical regulators of translocation by Mfd.
a EM-based model of DNA-bound Mfd with boxed region highlighting the structural environment of T710 and H842 residues. The resolution of our reconstruction precludes precise determination of the conformation of motif IVa loop and of amino-acid rotameric forms. Crucial residues are indicated as Cα spheres, H842 in green, T710A in yellow and E1045, D1048, and R1049, mutated in Mfd D7AAA in red. b–c TFO displacement assays carried out with MfdD7−AAA b and Mfd c variants. Percentage of remaining TFO-Triplex is plotted vs time to show translocation of Mfd variants. Plot shows means ± SD (n = 3), with error bars often smaller than symbols. d–e Electrophoretic separation of TFO displacement assays carried out with MfdD7−AAA. Representative gels for MfdD7−AAA d and Mfd e +/-ATP and +AMPPNP are shown. Example gels for Mfd, MfdD7−AAA;T710A and MfdD7AAA;H842A variants are shown in Supplementary Fig. 8 (n = 3). f Steady-state ATPase activities of Mfd variants normalized to wild-type Mfd activity. Data represent means ± SD (n ≥ 3). **denotes p < 0.01, ****p < 0.0001 and ns, p > 0.05 (unpaired two-tailed t test). The p value for MfdD7AAA;T710 with/without DNA was 0.0012. g, h Equilibrium dissociation constants for the Mfd-dsDNA interaction derived from fluorescence anisotropy binding curves obtained by titrating increasing amounts of protein into a mixture of 10 nm HEX-labeled 40mer DNA and 2.0 mm ATPγS e and ADP•AlFx. Data represent mean values ± SD (n = 3). KD values are tabulated in Supplementary Table 1. Source data are provided as a Source Data file.
H842 maps to a loop N-terminal to motif IVa and is conserved within the Mfd family, but not across translocases (Fig. 2a). However, based on a structural alignment, it is located close to where R393 in HCV NS3 maps (Supplementary Fig. 9). This has been coined the arginine clamp and is key for nucleic-acid binding and translocation53. In agreement with our findings, a set of structures of HCV translocase NS3h bound to DNA alone and in complex with ground-state ATP and transition state mimics revealed contacts of nucleic acid with R393 as well as a series of nucleotide-dependent structural transitions involving the equivalent of T710 (T269 in NS3h, Fig. 6j) as well as NS3h T411, the equivalent of T86850. T269 is in helicase motif Ic (aka TxGx motif), which while not originally identified as a conserved sequence, has been shown to be involved in nucleotide-dependent oligonucleotide binding in a variety of helicases, including NS344,54. The NS3 structure obtained in the absence of ATP shows these threonine residues three nucleotides apart, however they are only two nucleotides apart in the structure obtained with the ATP transition state analog (Supplementary Fig. 9b–d)49,50. T411 maintains contact with the same nucleotide throughout the ATP hydrolysis cycle, whereas T269 releases the DNA in the ATP-bound state and makes a new contact with the DNA one nucleotide away when NS3h is in complex with the ATP transition-state analog. Conservation of these two threonine residues in Mfd, the nucleotide-dependent role of T710 and the confirmation that T868 is critical for DNA binding by Mfd regardless of the status of the phosphoanhydride bond point to common features of translocation in NS3h and Mfd, despite Mfd lacking helicase activity and binding preferentially to DNA rather than RNA.
Discussion
Mfd-dependent processes rely heavily on its translocation on dsDNA. This is important not only for early events required to locate RNAP targets on the chromosome, but also for destabilizing TECs halted by DNA damage, pause signals, or various roadblocks6. Mfd action on transcription complexes effectively builds up torque to reanneal the upstream edge of the transcription bubble, shorten the bubble, and release the RNA transcript26,55. As a first step in shedding light on these events, here we describe the first structure of DNA-bound E. coli Mfd as well as complementary functional analyses. Our data allow for the first model for how nucleotide status and DNA binding control the breaking and formation of interdomain contacts and also reveal the molecular basis for DNA recognition by Mfd proteins.
We observe that DNA is bound by the two lobes of the motor core, consistent with a previous truncation analysis, which determined that in the presence of ATPγS, the DNA interaction determinants are restricted to D5 and most of D614. In this study, domain boundary definitions were not based on structural information, and notably truncated ATPase motif VI and the TRG motif (Fig. 2a–c), with crucial roles in coupling nucleotide status to translocation33. However, even with our structure-based truncations, we detect no binding of ds/ssDNA to domains flanking the motor core. Previous studies noted a striking similarity between D1aD2D1b and a region in UvrB that contacts DNA during NER, and also locally melts it for damage recognition using a highly conserved beta hairpin at the interface between D1a and D1b38. This beta hairpin extension is absent in Mfd, is replaced by a short loop, which lacks the aromatics involved in nucleotide flipping in UvrB. This loop is located ~50 Å away from the D5/D6 binding site (Fig. 2c), further supporting that the similarity might be strictly architectural. Our reconstruction suggests that on DNA substrates longer than used here, the DNA fragment would likely approach the C-terminal region of the relay helix, where previous studies have identified one substitution (MfdW550A) that enhances DNA binding and motor activity56. This could involve bending of the DNA across the protein surface, and/or modest rearrangements of the hook and TRG motifs, located in close proximity to the DNA.
We thus conceptualize DNA binding by Mfd in terms of two clusters of residues located at the tip of the D5 and D6 pincers that establish contacts with the substrate, primarily the tracking strand, and cycle between a high-affinity state (nucleotide-bound) and a low affinity (nucleotide-free) state. This shares features with findings from the HCV NS3 system, but bears a distinction in that NS3 cycles between a low affinity state when nucleotide-bound to a high-affinity state when nucleotide-free. Cluster I is comprised of R685 and T710, whereas cluster II is comprised of R865, N817, T868, and H842. Within these clusters, we distinguish between residues key for loading (R685, T868) and for translocation (T710 and H842), the latter having markedly nucleotide-dependent roles as also observed in the HCV NS3 system. It will be interesting to determine if Mfd operates via a Brownian ratchet that involves translocation in 1 bp steps (like in NS3), and whether and how the binding of RNAP might rectify this ratchet to achieve high processivity on the order of hundreds of base pairs, detected using single-molecule techniques16,28. We demonstrate that although there is no biochemical evidence for the TRG helices being directly involved in DNA binding, this structural element in direct communication with ATPase motif VI and the hook is mobile and likely used as a pawl during translocation. Here, we present the first structural evidence for the hypothesized interlocking of nucleotide-responsive elements (the TRG, the hook and relay helices, seen in Fig. 2c, 5) that is driven by movement of ATPase motif VI.
Translocases that move along dsDNA often make more extensive interactions with one DNA strand, termed the tracking strand. In the structure of S. solfataricus Snf2, more extensive interactions exist between the tracking strand and motifs I, Ia, Ib, Ic, and IIa41, located in the N-terminal lobe (Fig. 2a, c), whereas recent studies of Snf2 bound to a nucleosome suggest tighter interactions with the motifs in the C-terminal lobe42,43. It is impossible for us to determine at this stage which of the strand represents the “tracking” strand. This remains an important outstanding question since it underlies strand-specific probing of damage, for whose recognition UvrA, UvrB and Mfd are required52.
Our identification of an Mfd family-specific residue important for translocation (H842, located in motif IVa) provides an attractive avenue to pursue in the development of an Mfd inhibitor of high selectivity. Mfd deletion limits the acquisition of antimicrobial resistance in multiple bacteria, including E. coli25, Salmonella tyiphimurium25, Bacillus subtilis25, Helicobacter pylori23, Campylobacter jejuni22, and difficult to treat pathogens such as M. tuberculosis25. Furthermore, in Bacillus cereus and Shigella flexneri, recent experiments have implicated Mfd in resistance to the nitric oxide response generated by the immune response during infection24. Thus, an Mfd inhibitor would provide a novel strategy for antibiotic development. To block function while achieving selectivity, this could target highly variable motif IVa, rather than one of the other, more conserved ATPase motifs. In the NS3 system, the arginine clamp in motif IVa has been shown to be druggable57. We note that at the current resolution of our EM reconstruction, establishing whether H842 directly contacts DNA or may exert its effect indirectly by modulating the conformation of the motif IVa loop remains challenging.
Finally, our work provides the first evidence that domain D3 of unknown function might be key for TCR regulation in E. coli and related species. While D3 is sandwiched between D1a and D5/D6 in nucleotide-free Mfd, it is repositioned in the presence of DNA and transition state analog to make bridging interactions with D7 (Supplementary Movie 1). Motion of D3 is coupled to a peppermill-like motion of the UvrB homology module relative to the C-terminal region of Mfd (D4D5D6D7), which in turn undergoes local rearrangements to close the D5-D7 interdomain cleft, whereas D4–D7 interactions remain largely unperturbed and the UvrA recruitment surface is unmasked. Our work establishes that these conformational changes are coupled to the active site, primarily through motif VI, but also movements of the TRG, hook and relay helices, which remain unresolved at the residue level and await higher resolution structural and mechanistic analyses.
Methods
Cloning, protein expression, and purification
Constructs pMS1-pMS8 were obtained via site-directed mutagenesis of pAD6 (encoding N-terminally His-tagged E. coli Mfd58) using the method of Edelheit et al.59. All constructs were verified by DNA sequencing, and in select cases, purified protein was verified using mass spectrometry. Overexpression and purification was performed according to published protocols by a succession of Ni2+-affinity chromatography, His-tag cleavage with Prescission protease, heparin chromatography and gel filtration59. For constructs pMS2 and pMS5, HiTrap Q-sepharose column (GE Healthcare) was used instead of heparin affinity as these variants failed to bind to heparin affinity matrix. Size-exclusion chromatography was carried out on a Superdex 200 10/30 Increase column (GE Healthcare) using a buffer consisting of 20 mm Tris pH 8.0, 0.10 m NaCl, 5 mm DTT. Mfd truncations were expressed as N-terminal fusions to polyhistidine tags (Supplementary Table 3), which were cleaved using Prescission protease after the initial immobilized metal affinity chromatography step.
Triplex displacement assays
To generate the TFO-Triplex, linear ds 70mer DNA (50 nm) was mixed with 5’ Alexa647-labeled 21mer TFO oligo (25 nm) in an opaque tube in triplex reconstitution buffer (10 mm MES pH 5.5, 12.5 mm MgCl2) and heated to 57 °C for 15 min. The reaction was then placed at 4 °C and allowed to thermally equilibrate overnight. Protein samples were dialyzed against 10 mm Tris-HCl pH 8, 100 mm NaCl, 10 mm MgCl2, 2 mm β-mercaptoethanol. Reactions were assembled by mixing: 10 nm TFO-triplex, 50 nm 44 bp ssDNA trap in reaction buffer (50 mm Tris-HCl pH 8, 10 mm MgCl2, 1 mm DTT) and protein (450 nm for constructs used in Fig. 7b–c; 4.5 μμ for the experiments of Supplementary Fig. 8d). Triplex displacement was initiated by addition of 2 mm ATP. Samples were withdrawn at indicated timepoints, stopped by addition of GSM buffer (15% (w/v) glucose, 3% (w/v) SDS, 250 mm MOPS pH 5.5), and loaded on 3–8% Tris-Acetate gels (Invitrogen, Carlsbad, CA), which were pre-run for 60 min at 75 V using Tris-Acetate Running buffer (40 mm Tris-Acetate pH 5.5, 5 mm sodium acetate, 1 mm MgCl2) at 4 °C. Samples were electrophoresed for 180 min at 75 V and imaged using a ChemiDoc system (Biorad, Hercules, CA). The disappearance of TFO-Triplex over time was quantified using BioRad ImageLab and plotted using the Graphpad Prism software package.
Specimen preparation for cryo-EM
Size-exclusion chromatography (Superdex 200 10/30, GE Healthcare) was performed prior to complex reconstitution in a buffer containing 20 mm Tris-HCl pH 8, 100 mm NaCl, 20 mm MgCl2, 2 mm TCEP. For complex formation, Mfd was incubated with 21mer dsDNA in a 1:1.5 molar excess in the presence of 2 mm ADP, 10 mm NaF, and 1 mm AlCl3. 2.5 µl Mfd-DNA complex at 1.0 mg/mL was applied to UltrAuFoil R1.2/1.3 300 mesh grids (Quantifoil) that were previously plasma-cleaned using a Gatan Solarus (75% argon/25% oxygen atmosphere, 15 W for 7 s), then manually blotted with a Whatman No. 1 filter paper in a cold room with >80% humidity, and plunged into liquid ethane using a manual plunger. The dsDNA used for reconstitution had the following sequence: 5′-ATAGGATACTTACAGCCATCG-3′.
Cryo-EM data collection
Automated EM image acquisition was performed with Leginon60. Data collection was carried out on a Talos Arctica microscope (FEI) operating at 200 kV and equipped with a K2 Summit direct electron detector (Gatan) at a nominal magnification of ×36,000 and a defocus range from 2.0 μm to 4.0 μm giving a pixel size of 1.15 Å at the specimen level. To ameliorate preferential specimen orientation, images were collected using 30° tilts, as previously described61. Other than setting the nominal tilt angle during data acquisition, standard procedures were employed. Details are in Supplementary Table 2.
Cryo-EM image processing
Movies were aligned and dose-weighted using MotionCor262. CTF estimation and particle picking was performed in Warp63, which resulted in a stack of 69,721 particles that were imported into Relion (version 3.06)64 for 2D classification. One round of 2D classification was performed, and bad particles were excluded. Subsequently, a round of 3D classification, using five classes was run in Relion (Supplementary Fig. 2). The initial model used for 3D classification was generated from a small subset of the data, during data collection using cisTEM65. The best class was selected from 3D classification for moving forward. The selected particle stack from Relion contained 19,083 particles and was then input into cryoSPARC for iterative 2D and 3D classification. This cleaned stack containing 9822 particles was then imported into cisTEM and refined using the Auto-Refine procedure, whereby only frequencies up to 80% of Nyquist were included. This resulted in a final resolution of 5.47 Å, assessed by the conventional Fourier Shell Correlation (FSC) criteria. The refined map was sharpened in cisTEM by flattening the amplitude spectrum between 8 Å and the nominal resolution of 5.5 Å. Directional resolution volumes were generated using the 3D FSC tool61, whereas the local resolution was calculated using sxlocres.py, which is implemented within the Sparx processing package66. Details on model building and refinement are in Supplementary Methods.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
A.M.D. is a recipient of a Medical Research Grant from The Rhode Island Foundation, of RI-INBRE Pilot Project and Career Development Awards under grant P20GM103430 and of R01 grant GM121975 from the National Institute of General Medical Sciences of the National Institutes of Health. Research in the laboratory of D.L. was supported by the Office of the Director grant from the NIH, DP5 OD021396.
Source data
Author contributions
C.B. and M.M.S. performed cloning, protein purification, functional assays, and data analysis. D.D.K. and A.N.S. performed cloning and protein expression and purification. C.Z. carried out cryo-EM work, image analysis, rigid-body docking, and initial refinement. C.B. performed model fitting and real-space refinement. D.L. supervised EM work, and A.M.D. conceptualized and designed the overall study, designed protocols, supervised research, analyzed data, and wrote the manuscript with contributions from M.M.S. and figure contributions from C.B and C.Z. The manuscript was reviewed by all authors.
Data availability
The reconstruction of DNA-bound Mfd was deposited in the EMDB under accession code EMD-22146 and model coordinates into the PDB under PDB ID 6XEO. Other data reported here can be obtained upon request from the authors. Source data are provided with this paper.
Code availability
Computer code used for calculating sampling distributions and the sampling compensation factor value is available upon request from D.L.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Cheng Zhang, Margaret M. Suhanovsky.
Supplementary information
Supplementary information is available for this paper at 10.1038/s41467-020-17457-1.
References
- 1.Lohman TM, Tomko EJ, Wu CG. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat. Rev. Mol. Cell Biol. 2008;9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- 2.Suhasini AN, Brosh RM., Jr. DNA helicases associated with genetic instability, cancer, and aging. Adv. Exp. Med. Biol. 2013;767:123–144. doi: 10.1007/978-1-4614-5037-5_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Selby CP, Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993;260:53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- 4.Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 5.Stantial N, Dumpe J, Pietrosimone K, Baltazar F, Crowley DJ. Transcription-coupled repair of UV damage in the halophilic archaea. DNA Repair (Amst.) 2016;41:63–68. doi: 10.1016/j.dnarep.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 6.Deaconescu AM, Suhanovsky MM. From Mfd to TRCF and back again-A perspective on bacterial transcription-coupled nucleotide excision repair. Photochem. Photobiol. 2017;93:268–279. doi: 10.1111/php.12661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adebali O, Sancar A, Selby CP. Mfd translocase is necessary and sufficient for transcription-coupled repair in Escherichia coli. J. Biol. Chem. 2017;292:18386–18391. doi: 10.1074/jbc.C117.818807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Selby CP, Witkin EM, Sancar A. Escherichia coli mfd mutant deficient in “mutation frequency decline” lacks strand-specific repair: in vitro complementation with purified coupling factor. Proc. Natl Acad. Sci. USA. 1991;88:11574–11578. doi: 10.1073/pnas.88.24.11574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Epshtein V, et al. UvrD facilitates DNA repair by pulling RNA polymerase backwards. Nature. 2014;505:372–377. doi: 10.1038/nature12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Selby CP, Sancar A. Transcription preferentially inhibits nucleotide excision repair of the template DNA strand in vitro. J. Biol. Chem. 1990;265:21330–21336. [PubMed] [Google Scholar]
- 11.Selby CP, Sancar A. Gene- and strand-specific repair in vitro: partial purification of a transcription-repair coupling factor. Proc. Natl. Acad. Sci. USA. 1991;88:8232–8236. doi: 10.1073/pnas.88.18.8232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Selby CP, Sancar A. Mechanisms of transcription-repair coupling and mutation frequency decline. Microbiol. Rev. 1994;58:317–329. doi: 10.1128/mr.58.3.317-329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Selby CP, Sancar A. Structure and function of transcription-repair coupling factor. II. Catalytic properties. J. Biol. Chem. 1995;270:4890–4895. doi: 10.1074/jbc.270.9.4890. [DOI] [PubMed] [Google Scholar]
- 14.Selby CP, Sancar A. Structure and function of transcription-repair coupling factor. I. Structural domains and binding properties. J. Biol. Chem. 1995;270:4882–4889. doi: 10.1074/jbc.270.9.4882. [DOI] [PubMed] [Google Scholar]
- 15.Ho HN, van Oijen AM, Ghodke H. The transcription-repair coupling factor Mfd associates with RNA polymerase in the absence of exogenous damage. Nat. Commun. 2018;9:1570. doi: 10.1038/s41467-018-03790-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le TT, et al. Mfd dynamically regulates transcription via a release and catch-up mechanism. Cell. 2018;172:344–357 e15. doi: 10.1016/j.cell.2017.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haines NM, Kim YI, Smith AJ, Savery NJ. Stalled transcription complexes promote DNA repair at a distance. Proc. Natl. Acad. Sci. USA. 2014;111:4037–4042. doi: 10.1073/pnas.1322350111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Washburn RS, Wang Y, Gottesman ME. Role of E. coli transcription-repair coupling factor Mfd in Nun-mediated transcription termination. J. Mol. Biol. 2003;329:655–662. doi: 10.1016/s0022-2836(03)00465-0. [DOI] [PubMed] [Google Scholar]
- 19.Pomerantz RT, O’Donnell M. Direct restart of a replication fork stalled by a head-on RNA polymerase. Science. 2010;327:590–592. doi: 10.1126/science.1179595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Million-Weaver S, et al. An underlying mechanism for the increased mutagenesis of lagging-strand genes in Bacillus subtilis. Proc. Natl. Acad. Sci. USA. 2015;112:E1096–E1105. doi: 10.1073/pnas.1416651112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paul S, Million-Weaver S, Chattopadhyay S, Sokurenko E, Merrikh H. Accelerated gene evolution through replication-transcription conflicts. Nature. 2013;495:512–515. doi: 10.1038/nature11989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han J, Sahin O, Barton YW, Zhang Q. Key role of Mfd in the development of fluoroquinolone resistance in Campylobacter jejuni. PLoS Pathog. 2008;4:e1000083. doi: 10.1371/journal.ppat.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee GH, et al. The Helicobacter pylori Mfd protein is important for antibiotic resistance and DNA repair. Diagn. Microbiol. Infect. Dis. 2009;65:454–456. doi: 10.1016/j.diagmicrobio.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 24.Guillemet E, et al. The bacterial DNA repair protein Mfd confers resistance to the host nitrogen immune response. Sci. Rep. 2016;6:29349. doi: 10.1038/srep29349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ragheb MN, et al. Inhibiting the evolution of antibiotic resistance. Mol. Cell. 2019;73:157–+. doi: 10.1016/j.molcel.2018.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park J-S, Marr MT, Roberts JW. E. coli transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell. 2002;109:757–767. doi: 10.1016/s0092-8674(02)00769-9. [DOI] [PubMed] [Google Scholar]
- 27.Fan J, Leroux-Coyau M, Savery NJ, Strick TR. Reconstruction of bacterial transcription-coupled repair at single-molecule resolution. Nature. 2016;536:234–237. doi: 10.1038/nature19080. [DOI] [PubMed] [Google Scholar]
- 28.Graves ET, et al. A dynamic DNA-repair complex observed by correlative single-molecule nanomanipulation and fluorescence. Nat. Struct. Mol. Biol. 2015;22:452–457. doi: 10.1038/nsmb.3019. [DOI] [PubMed] [Google Scholar]
- 29.Deaconescu AM, Sevostyanova A, Artsimovitch I, Grigorieff N. Nucleotide excision repair (NER) machinery recruitment by the transcription-repair coupling factor involves unmasking of a conserved intramolecular interface. Proc. Natl. Acad. Sci. USA. 2012;109:3353–3358. doi: 10.1073/pnas.1115105109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith AJ, Szczelkun MD, Savery NJ. Controlling the motor activity of a transcription-repair coupling factor: autoinhibition and the role of RNA polymerase. Nucleic Acids Res. 2007;35:1802–1811. doi: 10.1093/nar/gkm019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deaconescu AM, et al. Structural basis for bacterial transcription-coupled DNA repair. Cell. 2006;124:507–520. doi: 10.1016/j.cell.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 32.Murphy MN, et al. An N-terminal clamp restrains the motor domains of the bacterial transcription-repair coupling factor Mfd. Nucleic Acids Res. 2009;37:6042–6053. doi: 10.1093/nar/gkp680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chambers AL, Smith AJ, Savery NJ. A DNA translocation motif in the bacterial transcription–repair coupling factor, Mfd. Nucleic Acids Res. 2003;31:6409–6418. doi: 10.1093/nar/gkg868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wittinghofer A. Signaling mechanistics: aluminum fluoride for molecule of the year. Curr. Biol. 1997;7:R682–R685. doi: 10.1016/s0960-9822(06)00355-1. [DOI] [PubMed] [Google Scholar]
- 35.Prabha S, Rao DN, Nagaraja V. Distinct properties of hexameric but functionally conserved Mycobacterium tuberculosis transcription-repair coupling factor. PLoS ONE. 2011;6:e19131. doi: 10.1371/journal.pone.0019131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Racki LR, et al. The chromatin remodeller ACF acts as a dimeric motor to space nucleosomes. Nature. 2009;462:1016–1021. doi: 10.1038/nature08621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 38.Assenmacher N, Wenig K, Lammens A, Hopfner KP. Structural basis for transcription-coupled repair: the N terminus of Mfd resembles UvrB with degenerate ATPase motifs. J. Mol. Biol. 2006;355:675–683. doi: 10.1016/j.jmb.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 39.Sivasankar Putta, S. P., et al. Structural Insights into the molecular mechanisms of the Mycobacterium evolvability factor Mfd. Preprint at https://www.biorxiv.org/content/10.1101/728246v1 (2019).
- 40.Singleton MR, Scaife S, Wigley DB. Structural analysis of DNA replication fork reversal by RecG. Cell. 2001;107:79–89. doi: 10.1016/s0092-8674(01)00501-3. [DOI] [PubMed] [Google Scholar]
- 41.Durr H, Korner C, Muller M, Hickmann V, Hopfner KP. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell. 2005;121:363–373. doi: 10.1016/j.cell.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 42.Liu X, Li M, Xia X, Li X, Chen Z. Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure. Nature. 2017;544:440–445. doi: 10.1038/nature22036. [DOI] [PubMed] [Google Scholar]
- 43.Li M, et al. Mechanism of DNA translocation underlying chromatin remodelling by Snf2. Nature. 2019;567:409–413. doi: 10.1038/s41586-019-1029-2. [DOI] [PubMed] [Google Scholar]
- 44.Caruthers JM, McKay DB. Helicase structure and mechanism. Curr. Opin. Struct. Biol. 2002;12:123–133. doi: 10.1016/s0959-440x(02)00298-1. [DOI] [PubMed] [Google Scholar]
- 45.Kowalinski E, et al. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 46.Bamming D, Horvath CM. Regulation of signal transduction by enzymatically inactive antiviral RNA helicase proteins MDA5, RIG-I, and LGP2. J. Biol. Chem. 2009;284:9700–9712. doi: 10.1074/jbc.M807365200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Warren, G. M., Stein, R. A., McHaourab, H. S. & Eichman, B. F. Movement of the RecG motor domain upon DNA binding is required for efficient fork reversal. Int. J. Mol. Sci.19, 3049 (2018). [DOI] [PMC free article] [PubMed]
- 48.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 49.Gu M, Rice CM. Three conformational snapshots of the hepatitis C virus NS3 helicase reveal a ratchet translocation mechanism. Proc. Natl. Acad. Sci. USA. 2010;107:521–528. doi: 10.1073/pnas.0913380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim JL, et al. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure. 1998;6:89–100. doi: 10.1016/s0969-2126(98)00010-0. [DOI] [PubMed] [Google Scholar]
- 51.Lin C, Kim JL. Structure-based mutagenesis study of hepatitis C virus NS3 helicase. J. Virol. 1999;73:8798–8807. doi: 10.1128/jvi.73.10.8798-8807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Manelyte L, Kim YI, Smith AJ, Smith RM, Savery NJ. Regulation and rate enhancement during transcription-coupled DNA repair. Mol. Cell. 2010;40:714–724. doi: 10.1016/j.molcel.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lam AM, Keeney D, Frick DN. Two novel conserved motifs in the hepatitis C virus NS3 protein critical for helicase action. J. Biol. Chem. 2003;278:44514–44524. doi: 10.1074/jbc.M306444200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr. Opin. Struct. Biol. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park JS, Roberts JW. Role of DNA bubble rewinding in enzymatic transcription termination. Proc. Natl Acad. Sci. USA. 2006;103:4870–4875. doi: 10.1073/pnas.0600145103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith, A. J., Pernstich, C. & Savery, N. J. Multipartite control of the DNA translocase, Mfd. Nucleic Acids Res.40, 10408–10416 (2012). [DOI] [PMC free article] [PubMed]
- 57.Maga G, et al. Specific targeting of hepatitis C virus NS3 RNA helicase. Discovery of the potent and selective competitive nucleotide-mimicking inhibitor QU663. Biochemistry. 2005;44:9637–9644. doi: 10.1021/bi047437u. [DOI] [PubMed] [Google Scholar]
- 58.Deaconescu AM, Darst SA. Crystallization and preliminary structure determination of Escherichia coli Mfd, the transcription-repair coupling factor. Acta Crystallogr Sect. F. Struct. Biol. Cryst. Commun. 2005;61:1062–1064. doi: 10.1107/S1744309105035876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edelheit O, Hanukoglu A, Hanukoglu I. Simple and efficient site-directed mutagenesis using two single-primer reactions in parallel to generate mutants for protein structure-function studies. BMC Biotechnol. 2009;9:61. doi: 10.1186/1472-6750-9-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suloway C, et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 61.Tan YZ, et al. Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat. Methods. 2017;14:793–796. doi: 10.1038/nmeth.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tegunov, D. & Cramer, P. Real-time cryo-electron microscopy data preprocessing with Warp. Nat Methods16, 1146–1152(2019). [DOI] [PMC free article] [PubMed]
- 64.Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife7, e42166 (2018). [DOI] [PMC free article] [PubMed]
- 65.Grant, T., Rohou, A. & Grigorieff, N. cisTEM, user-friendly software for single-particle image processing. Elife7, e35383 (2018). [DOI] [PMC free article] [PubMed]
- 66.Hohn M, et al. SPARX, a new environment for Cryo-EM image processing. J. Struct. Biol. 2007;157:47–55. doi: 10.1016/j.jsb.2006.07.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The reconstruction of DNA-bound Mfd was deposited in the EMDB under accession code EMD-22146 and model coordinates into the PDB under PDB ID 6XEO. Other data reported here can be obtained upon request from the authors. Source data are provided with this paper.
Computer code used for calculating sampling distributions and the sampling compensation factor value is available upon request from D.L.