Abstract
The need for detailed structural characterization of glycerophospholipids (GPLs) for many types of biologically motivated applications has led to the development of novel mass spectrometry-based methodologies that utilize alternative ion activation methods. Ultraviolet photodissociation (UVPD) has shown great utility for localizing sites of unsaturation within acyl chains and to date has predominantly been used for positive mode analysis of GPLs. In the present work, UVPD is used to localize sites of unsaturation in GPL anions. Similar to UVPD mass spectra of GPL cations, UVPD of deprotonated or formate-adducted GPLs yields diagnostic fragment ions spaced 24 Da apart. This method was integrated into a liquid chromatography workflow and used to evaluate profiles of sites of unsaturation of lipids in Escherichia coli (E. coli) and Acinetobacter baumannii (A. baumannii). When assigning sites of unsaturation, E. coli was found to contain all unsaturation elements at the same position relative to the terminal methyl carbon of the acyl chain; the first carbon participating in a site of unsaturation was consistently seven carbons along the acyl chain when counting carbons from the terminal methyl carbon. GPLs from A. baumannii exhibited more variability in locations of unsaturation. For GPLs containing sites of unsaturation in both acyl chains, an MS3 method was devised to assign sites to specific acyl chains.
Graphical Abstract
Lipids are a highly diverse class of molecules that participate in a variety of cellular functions including from serving as structural components and signaling precursors and molecules of cells, to functioning as cofactors for protein function.1–4 Lipidomics, the large scale study of lipids, aims to identify and quantify the lipid composition within a biological system with the ultimate goal of correlating differences in lipid profiles and abundances with variations in metabolic function.4–6 While the structural diversity of lipids has presented challenges to broad-scale lipidomics analysis, advances in mass spectrometry technology and innovative methods have resulted in improvements in identification and quantitation of lipids in both targeted and global studies.7–13 There exist two main ex vivo mass-spectrometry-based lipidomic strategies: (1) methods involving liquid chromatography that enable separation of lipids and deeper analysis14–16 and (2) shotgun methods entailing direct infusion of lipid extracts without any online chromatographic separation.6,17–25 Characterization of glycerophospholipids (GPL), the main component of cell membranes, is particularly taxing owing to the combinations of substituents that modify the glycerol backbone. With either of the two lipidomic strategies, acquisition of MS1 spectra, even with high resolution and high mass accuracy, provides only the sum composition of putative GPLs based on mass values that fit chemical formulas.26 Therefore, tandem mass spectrometry (MS/MS) is needed to confirm specific structures of lipids.27
Low-energy collision-based activation methods, by far the most popular and widely available, provide information about the acyl chain compositions and head groups in negative ion mode and positive ion mode, respectively.27–30 However, these MS/MS methods typically fail to reveal the full array of subtle structural details that have significant biological implications, including acyl chain stereochemistry, double bond position, and double bond stereochemistry.31–36 In particular, double bond positions have been shown to impact lipid–protein binding and influence membrane fluidity, thickness, and hydration.3,34,35 A number of mass-spectrometry-based methods that use derivatization or metal complexation have been developed to enable double bond localization in conjunction with low-energy collision-induced dissociation (CID).37–44 These include derivatization with N-(4-aminomethylphenyl) pyridinium,37 reactions with acetonitrile-derived ions via atmospheric pressure covalent adduct chemical ionization,38 epoxidation reactions,39,40 and Paternò–Büchi reactions.41–44 In each case, collisional activation of the resulting modified lipids produced diagnostic fragment ions, thus allowing determination of double bond positions. Multistage collisional activation of lithium-adducted lipids generates fragment ions resulting from vinylic and allylic cleavage that are also diagnostic for double bond positions.45 Additionally, some methods have relied on intentional oxidation of unsaturated lipids to determine double bond positions.46–48 For example, Harris et al. recently reacted phospholipids with ozone online after chromatographic separation using a novel flow-cell device.48 This oxidation strategy allows spectra to be acquired in a data-independent manner and therefore only requires collection of MS1 spectra; however, the breadth of information is more limited than that afforded by other MS/MS methods.
Innovative ion activation techniques that exploit gas-phase ion–neutral, ion–ion, ion–electron, or ion–photon chemistry have also been developed that allow localization of double bonds.49–63 Radical-directed dissociation (RDD), which entails collisional activation of odd-electron fragment ions derived from irradiation of precursor ions containing a photocleavable adduct or tag, results in a diagnostic series of product ions originating from cleavage of carbon–carbon bonds all along the acyl chains.49–51 The spectra generated are clean and highly informative, although the need for derivatization or adduction and the use of multiple stages of analysis add an element of complexity to the method. A number of other alternative ion activation methods including high-energy CID,52 ozone-induced dissociation (OzID),53–57 electron-induced dissociation (EID/EIEIO),58–61 and meta-stable atom-activated dissociation (He-MAD),62,63 have been shown to produce informative spectra for underivatized lipids.
Ultraviolet photodissociation (UVPD) at 193 nm has also shown utility for the structural characterization of lipids.64–71 Recently, UVPD has been used to determine double bond positions within phosphatidylcholines (PC) and sphingolipids.68,69 Both studies were conducted in positive mode owing to the fixed positive charge on the head groups of PC and sphingomyelin. Consequently, diagnostic fatty acid product ions were not observed, and acyl chain compositions were determined either by tracking low-abundance neutral losses or from parallel acquisition of negative mode HCD spectra of anion-adducted PCs.68,69 Without derivatization of GPLs, positive mode methods generally have limited utility for a number of GPL subclasses that preferentially ionize in negative mode, such as phosphatidylglycerols (PG) and phosphatidylinositols (PI).27,72 Williams et al. showed that sodium adduction using infusion of a sodium salt via a tee (i.e., a postcolumn LC setup) allowed determination of acyl chain and double bond positions for a variety of GPLs via UVPD–MS; however, introduction of a nonvolatile salt is not practical for all workflows.70 To address these limitations, a 193 nm UVPD method for the analysis of GPLs in the negative mode is reported in the present study. This method enables determination of the headgroup, acyl chain composition, and double bond positions in a single MS/MS event. Additionally, this method has been integrated with chromatographic separations to reduce complexity in spectra obtained from a shotgun approach resulting from the occurrence of isomeric lipids or other isobaric species. The developed LC–UVPD method was applied to analysis of bacterial GPLs. Interestingly, this method permitted detection of differences in double bond positions between different species of bacteria. This method is therefore another tool that can be used to investigative the mechanisms by which different species of bacteria incorporate sites of unsaturation in acyl chains.
EXPERIMENTAL SECTION
Materials
All GPL standards (synthetic) and the E. coli total lipid extract were purchased from Avanti Polar Lipids (Alabaster, AL) and were used without further purification. Water (H2O) and acetonitrile (ACN) were purchased from EMD Millipore (Billerica, MA). Isopropyl alcohol (IPA), ammonium formate, and formic acid were purchased from Fisher Scientific (Fairlawn, NJ). Chloroform (CHCl3) was purchased from Sigma-Aldrich (St. Louis, MO). GPLs were extracted from A. baumannii following the method of Bligh and Dyer73 as previously described.74 Prior to extraction, A. baumannii was grown to an OD600 of ∼1.0 at 37 °C, harvested at 10 000g for 10 min, and washed with phosphate buffered saline.
Liquid Chromatography and Mass Spectrometry
All experiments were performed using a Thermo Fisher Orbitrap Fusion Lumos mass spectrometer (San Jose, CA) modified with a Coherent Excistar XS excimer laser (Santa Clara, CA) to enable 193 nm UVPD–MS, as previously described.67 GPL standards (10 μM in 50:50 CHCl3/MeOH) were directly infused using a static nanoelectrospray ionization (nanoESI) source (Thermo Fisher) with an applied voltage of 0.8–1 kV. All direct infusion spectra were collected at a resolving power of 120 000 at m/z 200 and were based on an average of 100 scans. For MS/MS experiments, higher-energy collisional dissociation (HCD) was performed with a normalized collision energy (NCE) of 25, and UVPD was performed using 15 laser pulses and 6 mJ per pulse unless indicated otherwise. For MS3 experiments, collision-induced dissociation (CID) was performed at an NCE that provided optimal ion current for neutral ketene losses. Ketene loss fragment ions were isolated in the ion trap with an isolation width of 1 m/z prior to irradiation with 10 laser pulses at 2.5 mJ per pulse. MS3 spectra are an average of 200–300 transients. All UVPD experiments were performed in the high-pressure trap of the dual linear ion trap.
For online separations, the chromatographic method is a variation of the method developed by Damen et al.75 Separations were performed on a Waters CSH C18 column (2.1 × 100 mm, 1.7 μm particle size) (Milford, MA) using a Dionex Ultimate 3000 microflow liquid chromatography system (Sunnyvale, CA) coupled to an Orbitrap Fusion Lumos via an ESI source. The electrospray voltage was held at a spray voltage of 3.8 kV, with a sheath gas of 5 (arbitrary units), an auxiliary gas of 10 (arbitrary units), and a capillary temperature of 300 °C. The column was heated to 50 °C. Approximately 0.3 μg of each sample was injected per analysis (estimated based on signal abundances relative to those obtained for a known commercial standard E. coli phospholipid extract (Avanti)). Mobile phase A consisted of 40:60 H2O/ACN with 10 mM ammonium formate and 0.1% formic acid, and mobile phase B consisted of 10:90 ACN/IPA with 10 mM ammonium formate and 0.1% formic acid. Phospholipids were separated at a flow rate of 260 μL/min over a 60 min gradient that started 2 min after injection. The gradient was ramped from 10 to 45% B over 4 min, followed by a second step to 60% B over 40 min, before an 8 min wash step at 95% B for 6 min and subsequent re-equilibration at 10% B for 8 min. Data-dependent acquisition was performed in top-speed mode with a cycle time of 6 s with consecutive HCD and UVPD spectra acquired for each selected precursor. MS1 spectra were acquired at a resolving power of 30 000 at m/z 200 with two μscans per scan and an AGC target of 1e6. Chromatographic peak widths of 30 s allowed five to seven MS1 spectra to be collected across the elution of a GPL species. MS2 precursors that satisfied the monoisotopic peak selection (MIPS) filter and 1e6 intensity filter were selected and isolated in the quadrupole using an isolation width of 1 m/z. For HCD, precursors were dissociated using an NCE of 25. For UVPD, precursors were dissociated in the high-pressure trap using 10 laser pulses at 2.5 mJ per pulse. A total of three μscans and seven μscans were acquired per HCD and UVPD spectrum, respectively. All MS2 spectra were acquired at a resolving power of 15 000 at m/z 200. The AGC settings were 2e5 and 1e6 for HCD and UVPD, respectively. Lipid notation is based on the format described by Liebisch et al.26 All spectra were interpreted manually with the aid of ChemDraw (PerkinElmer).
RESULTS AND DISCUSSION
HCD and UVPD were performed on a number of GPL standards in negative ion mode (Figures 1, 2, and S2). The HCD and UVPD mass spectra for deprotonated PG 16:0/18:1(9Z) and the corresponding fragmentation map illustrating the cleavages that result in the dominant product ions are shown in Figure 1. As expected, the HCD spectrum contains prominent palmitate (16 carbon chain; 16:0) and oleate (18 carbon chain with 1 site of unsaturation; 18:1) fatty acid fragment ions of m/z 255.23 and 281.25, respectively. Fragment ions of m/z 491.28 and 465.26, corresponding to neutral losses of 16:0 or 18:1 as fatty acids, respectively, and fragment ions of m/z 509.29 and 483.27, corresponding to neutral losses of 16:0 or 18:1 as ketenes, respectively, confirm the acyl chain composition. Additionally, a headgroup ion of m/z 171.01 confirms this lipid as a PG. The UVPD spectrum contains all of the same fragment ions displayed in the HCD spectrum, in addition to a series of ions arising from cleavages throughout the headgroup. Most importantly, a pair of ions of m/z 633.37 and 609.37 with a mass difference of 24 Da that are diagnostic of the double bond position within the 18:1 acyl chain are produced upon UVPD. Diagnostic ions spaced 24 Da apart have previously been observed in positive mode, thus conveying the versatility of UVPD for localizing sites of unsaturation of lipids in both positive and negative modes.68–71
Figure 1.

(a) HCD (NCE 25) and (b) UVPD (10 pulses, 6 mJ) mass spectra of PG 16:0/18:1 (m/z 747.51, [M – H]−) with corresponding fragment ion map. Diagnostic UVPD ions are labeled with red arrows in the spectrum and red markers in the fragment ion map.
Figure 2.
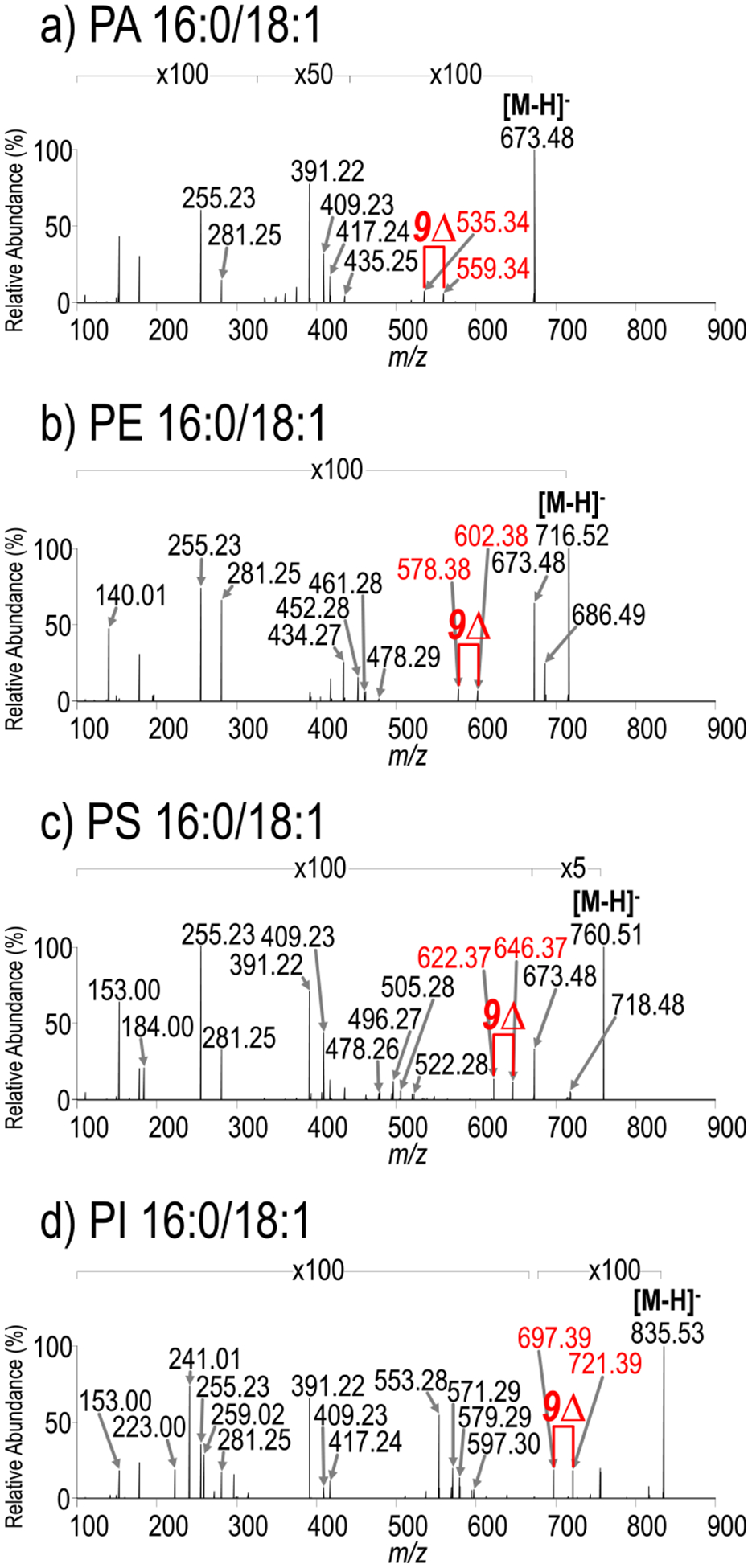
UVPD (15 pulses, 6 mJ) mass spectra of (a) PA 16:0/18:1 (m/z 673.48, [M – H]−), (b) PE 16:0/18:1 (m/z 716.52, [M – H]−), (c) PS 16:0/18:1 (m/z 760.51, [M – H]−), and (d) PI 16:0/18:1 (m/z 835.53, [M – H]−). HCD mass spectra and corresponding fragment ion maps are shown in Figures S1 and S2, respectively. Diagnostic UVPD ions are labeled with red arrows in the mass spectra.
Fragmentation is one of a number of possible outcomes upon ion activation. To gain insight into the extent of other processes occurring upon photoactivation, including electron photodetachment, the total ion current and precursor abundance were monitored for consecutive periods with and without laser irradiation during direct infusion of PG 16:0/18:1(9Z) (m/z 747.51, [M – H]−). Figure S1a shows the total ion chromatogram for alternating sets of 25 scans (laser on/laser off), and Figure S1b shows averaged precursor spectra for each set of 25 scans. Both the total ion current and the precursor abundance drop by approximately 40% upon photoactivation using 10 pulses (nominally 6 mJ per pulse), which suggests that a large portion of the precursor is depleted during photoirradiation. In addition to the formation of meaningful low-abundance fragment ions described in this study, the significant depletion of the precursor suggests that there are other nondetectable processes, such as the formation of small fragment ions that fall below the detectable m/z range (<m/z 100) or electron photodetachment, a process resulting in nondetectable neutral species owing to the fact that the precursor is a singly charged ion.
To assess the broader applicability of UVPD to other subclasses of GPLs, including phosphatidic acid (PA), phosphatidylethanolamine (PE), phosphatidylserine (PS), and phosphatidylinositol (PI), UVPD mass spectra were collected for deprotonated PA 16:0/18:1(9Z), PE 16:0/18:1(9Z), PS 16:0/18:1(9Z), and PI 16:0/18:1(9Z) (Figure 2). The corresponding HCD mass spectra and fragment ion maps are shown in Figures S2 and S3, respectively. Similar to the features displayed in the UVPD mass spectrum of PG 16:0/18:1(9Z), the UVPD mass spectra obtained for the other GPLs exhibit ions that identify the acyl chain compositions, the head groups, and the double bond positions within the acyl chains. The aforementioned GPLs represent subclasses of lipids that readily produce molecular ions in negative mode via deprotonation. However, PCs, owing to the fixed positive charge on their headgroup, typically only produce ions in the negative mode via anion adduction. The HCD and UVPD mass spectra for PC 16:0/18:1(9Z) as a formate adduct are shown in Figure S4. The HCD mass spectrum displays a characteristic loss of a methyl group plus the formate adduct in addition to product ions that identify the acyl chain composition. The UVPD mass spectrum exhibits all of the same ions seen in the HCD mass spectrum in addition to two pairs of diagnostic ions spaced 24 Da apart that both retain and lose the formate adduct and are diagnostic for the double bond position. Acyl chain neutral loss ions that retain the formate adduct and forgo loss of a methyl group are also observed. These mass spectra suggest that the negative mode could be used for acquisition of MS/MS spectra for a wide range of GPL subclasses without the use of a nonvolatile salt. The prevalence of diagnostic pairs of ions with a mass difference of 24 Da mass in both negative mode and positive mode further suggests this fragmentation pathway is independent of charge site or charge type. Ryan et al. have proposed a cis-1,2-elimination as the mechanism by which this diagnostic pair of ions is generated.69 Tables S1–S12 contain lists of all identified fragment ions with calculated ppm error.
While new chromatographic methods have been developed that facilitate separation of complex mixtures of GPLs,75,76 differentiation of isomers requires confident characterization by MS/MS. Integrating negative mode UVPD with a chromatographic workflow affords the opportunity to analyze complex mixtures of GPLs with reduced ion suppression effects that can occur during direct infusion shotgun methods and offers the potential to enhance detection of lower-abundance species or species with lower ionization efficiencies. A method employing alternating HCD and UVPD scans was devised to maximize confirmation of lipid identifications. Figure 3a shows a chromatographic separation of an E. coli total lipid extract. Representative HCD and UVPD mass spectra for one selected lipid of m/z 773.53 (retention time of 22.25 min) are shown in Figures S5a and 3d, respectively. A corresponding fragment ion map combining HCD and UVPD fragmentation is shown in Figure S5d. The HCD mass spectrum enables identification of m/z 773.53 as PG 18:1/18:1 via a fatty acid fragment ion at m/z 281.25, acyl chain neutral losses of m/z 509.29 and 478.30, and a headgroup fragment ion at m/z 171.01. The UVPD mass spectrum additionally displays a set of double bond diagnostic ions of m/z 687.41 and 663.42 that allow the double bond in each acyl chain to be localized to the 11-position. This GPL can more specifically be characterized as PG 18:1(11Δ)/18:1(11Δ). Figure 4 shows another set of HCD (Figure 4a) and UVPD (Figure 4b) mass spectra for the lipid of m/z 728.52 also obtained during chromatographic separation of the E. coli lipid extract. The headgroup ion of m/z 140.01 and fatty acid fragment ions of m/z 281.25 and 267.23 in both the HCD and UVPD mass spectra permit characterization of this GPL as PE 17:1_18:1, as seen in the fragment ion map in Figure 4c. The UVPD mass spectrum (Figure 4b) contains a set of ions at m/z 642.40 and 618.40, confirming the presence of a double bond, in addition to a set of ions at m/z 630.41 and 616.39 spaced 14 Da apart. In a recent study by Blevins et al., UVPD was shown to localize cyclopropane rings within acyl chains via diagnostic ions spaced 14 Da apart.77 UVPD of m/z 728.52 provides an example in which both a double bond and a cyclopropane ring are localized within the acyl chains of the same GPL. While UVPD alone enables localization of sites of unsaturation within acyl chains, unambiguous assignment of unsaturation elements to specific acyl chains requires the MSn capabilities of the ion trap, as was recently demonstrated for cardiolipin lipids.78 Figure 4d shows a direct infusion MS3 mass spectrum, whereby the CID neutral loss fragment ion of m/z 464.27 that has lost 18:1 as a ketene is subsequently activated by UVPD. The resulting mass spectrum contains a prominent fragment ion of m/z 267.23, confirming the presence of the 17:1 acyl chain, in addition to a set of ions of m/z 366.16 and 352.15 spaced 14 Da apart, indicating the presence of a cyclopropane ring, as shown in the resulting fragment map in Figure 4e. Therefore, the GPL of m/z 728.52 can be more completely assigned as PE 17:1(c9Δ)_18:1(11Δ) by using the MS3/UVPD approach.
Figure 3.
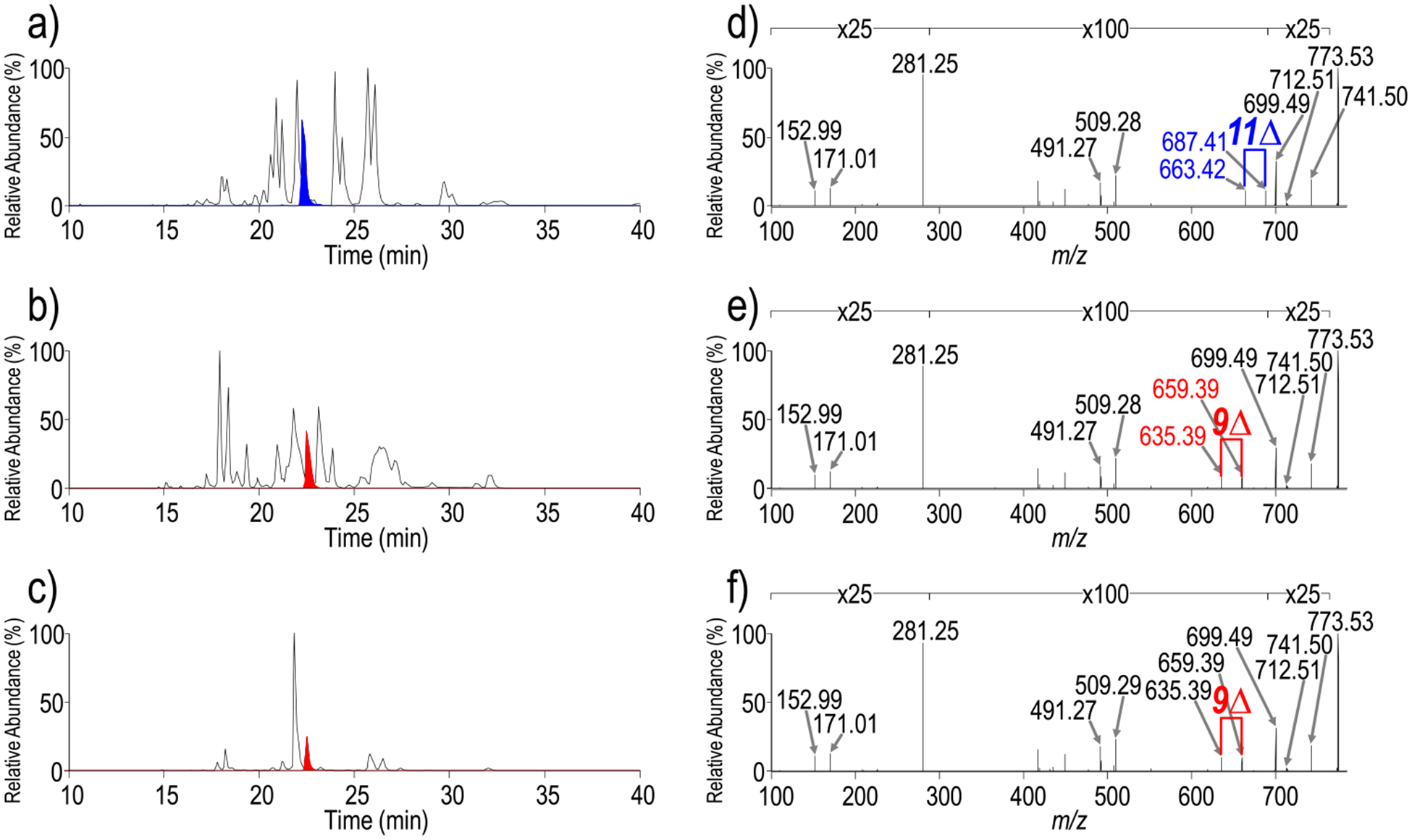
Chromatograms of GPL extracts from (a) E. coli, (b) A. baumannii 17978, and (c) A. baumannii 19606 with extracted ion chromatograms of m/z 773.53 in blue and red. Corresponding UVPD spectra of m/z 773.53 from (d) E. coli, (e) A. baumannii 17978, and (f) A. baumannii 19606. HCD mass spectra and combined HCD and UVPD fragment ion maps are shown in Figure S4. Diagnostic UVPD ions are labeled in blue and red in the mass spectra.
Figure 4.

(a) HCD (NCE 25) and (b) UVPD (10 pulses, 2.5 mJ) spectra of m/z 728.52 identified as PE 17:1(c9Δ)_18:1(11Δ) from the chromatographic separation of an E. coli lipid extract. (c) Combined HCD and UVPD fragment ion map for m/z 728.52. (d) MS3 CID/UVPD spectrum of the m/z 464.27 fragment ion acquired from direct infusion of an E. coli lipid extract. (e) Fragment ion map for CID/UVPD of m/z 464.27. Diagnostic UVPD double bond and cyclopropane ions are labeled with red and green arrows in spectra, respectively, and red and green markers in the fragment ion maps, respectively. MS3 analysis confirms a cyclopropane in 17:1 and therefore a double bond in 18:1.
Table 1 shows the list of PGs and PEs that were identified in the E. coli lipid extract. Interestingly, all sites of unsaturation in 18:1 acyl chains occur in the 11-position, all sites in the 16:1 acyl chains occur in the 9-position, and the detected cyclopropane ring occurs exclusively at the 9-position. This finding is consistent with reports describing GPL biosynthesis in E. coli whereby the enzyme FabA incorporates a site of unsaturation at the 3-position of a 10-carbon intermediate prior to chain elongation.79 This process leads to acyl chains with a site of unsaturation consistently occurring seven carbons from the terminal methyl carbon.
Table 1.
List of Identified PEs and PGs from an E. coli Lipid Extract
| Precursor ion (m/z) | RT (min) | Lipid identification | Fatty acid fragment ions (m/z) | Diagnostic DB fraqment ions (m/z) | ||
|---|---|---|---|---|---|---|
| 660.4590 | 17.17 | PE 14:1_16:1(9Δ) | 225.1839 | 255.2311 | 550.3466 | 574.3432 |
| 719.4872 | 18.07 | PG 16:0_16:1(9Δ) | 255.2313 | 253.2154 | 609.3839 | 633.3880 |
| 745.5029 | 18.31 | PG 16:1(9Δ)_18:1(11Δ) | 253.2151 | 281.2463 | 635.3823 | 659.3921 |
| 688.4909 | 20.69 | PE 14:0_18:1(11Δ) | 227.1996 | 281.2476 | 578.3745 | 602.3787 |
| 688.4933 | 20.97 | PE 16:0J6:1(9Δ) | 255.2305 | 253.2151 | 578.3766 | 602.3794 |
| 714.5086 | 21.19 | PG 16:0_18:1(11Δ) | 253.2152 | 281.2465 | 604.4000 | 628.4022 |
| 747.5178 | 22.01 | PE 16:1(9Δ)_18:1(11Δ) | 255.2305 | 281.2464 | 637.4058 | 661.3911 |
| 773.5347 | 22.24 | PG 18:1(11Δ)_18:1(11Δ) | 281.2464 | 281.2464 | 663.4206 | 687.4126 |
| 728.5248 | 24.37 | PE 17:1(c9Δ)_18:1(11Δ) | 267.2310 | 281.2463 | 618.4017 | 642.3984 |
| 716.5300 | 25.73 | PE 16:0J8:1(11Δ) | 255.2309 | 281.2459 | 606.4147 | 630.4081 |
| 716.5223 | 26.12 | PE 16:1(9Δ)_18:0 | 253.2154 | 283.2631 | 606.4100 | 630.4100 |
| 742.5406 | 26.08 | PE 18:1(11Δ)J8:1(11Δ) | 281.2461 | 281.2461 | 632.4208 | 656.4300 |
| 775.5500 | 27.27 | PG 18:0_18:1(11Δ) | 281.2460 | 283.2610 | 665.4377 | 689.4350 |
| 744.5530 | 31.76 | PE 18:0_18:1(11Δ) | 281.2464 | 283.2624 | 634.4371 | 658.4405 |
GPL extracts from A. baumannii 19606 and A. baumannii 17978 were also analyzed using the developed alternating HCD and UVPD method. A. baumannii is a nosocomial Gram-negative pathogen that presents a threat to public health owing to multidrug resistance. Therefore, investigations aimed at elucidating A. baumannii lipid biosynthesis pathways provide opportunities for identification of novel drug targets. Figure 3b,c shows chromatographic separations of GPL lipid extracts from A. baumannii 19606 and A. baumannii respectively. Representative HCD mass spectra of the lipid of m/z 773.53 from each extract are shown in Figure S5b,c, respectively. This ion has the same m/z of the ion that was analyzed from the E. coli lipid extract described above (Figure S5a). The HCD mass spectra of m/z 773.53 from E. coli, A. baumannii 19606, and A. baumannii 17978 are nearly and only enable confirmation of this GPL as PG 18:1/18:1 based on headgroup and fatty acid fragment ions. The UVPD mass spectra of the lipids of m/z 773.53 from A. baumannii 19606 and A. baumannii 17978 in Figure 3e,f both have diagnostic sets of ions at m/z 659.39 and 635.39 that localize double bonds to the 9-position. In the A. baumannii samples, UVPD enables characterization of this GPL as PG 18:1(9Δ)/18:1(9Δ), which differs from that of the isomeric GPL from the E. coli sample, in which sites of unsaturation occurred at the 11-position in 18:1 acyl chains. With HCD alone, this difference in double bond position goes undetected.
Tables 2 and 3 show the list of PGs and PEs that identified in the A. baumannii 17978 and A. baumannii 19606 extracts, respectively. In contrast to E. coli, sites of unsaturation are not always located at the same number of carbons from the acyl chain terminal carbon. A. baumannii contains multiple homologues of E. coli FabA as well as multiple homologues of FabB, an enzyme that is required for further elongation of unsaturated fatty acid intermediates (data not shown). This additional enzymatic machinery may contribute to the diversity we observe in double bond location in this organism. While this result is biologically intriguing, spectral interpretation becomes increasingly challenging, as many spectra contain multiple sets of peaks with differences of 24 Da resulting from either chromatographically unresolved isomers or GPLs in which both acyl chains contain different sites of unsaturation relative to the terminal carbon. For spectra with multiple pairs of ions resulting from chromatographically unresolved isomers, improvements in LC separations or implementation of UVPD on an instrument capable of ion mobility separations may facilitate spectral deconvolution. For GPLs with multiple sites of unsaturation, an MS3 strategy, as shown previously, can be used to confidently assign sites to specific acyl chains. Figure S6 shows one example where MS3 from A. baumannii 17978 enables assignment of the lipid of m/z 745.50 as PG 16:1(9Δ)_18:1(9Δ). In this example, both HCD (Figure S6a) and UVPD (Figure S6b) identified the lipid to be PG 16:1_18:1 (fragment map in Figure S6c), based on product ions of m/z 281.25 (18:1 fatty acid), 253.22 (16:1 fatty acid), and 171.01 (PG headgroup). Upon UVPD, however, two pairs of diagnostic products were observed at m/z 607.36/631.36 and 635.39/659.39, precluding localization of the double bond on either of the unsaturated acyl chains. Analogous to the strategy used above to resolve ambiguity in cyclopropyl and double bond acyl chain assignment, CID/UVPD was implemented to promote a ketene neutral loss to yield a monoacylated phospholipid, which upon UVPD, would produce only one set of diagnostic pairs that confidently localize the site of unsaturation. In Figure S6d, PG 16:1_18:1 was deacylated via CID to produce a product of m/z 481.25 composed of only a 16:1 acyl chain. Subsequent UVPD resulted in only one diagnostic pair at m/z 371.14/395.14, allowing the localization of the double bond to the 9-position of 16:1, as shown in the fragment map in Figure S6e, indicating that the pair at m/z 607.36/631.36 was generated by 16:1(9Δ) and the pair at m/z 635.39/659.39 was generated by 18:1(9Δ), confidently localizing the two double bonds. Thus, CID/UVPD provides an avenue to resolve complexities in the MS/MS spectra.
Table 2.
List of Identified PEs and PGs from an A. baumannii 17978 Lipid Extract
| Precursor ion (m/z) | RT (min) | Lipid identification | Fatty acid fragment ions (m/z) | Diagnostic DB fragment ions (m/z) | ||
|---|---|---|---|---|---|---|
| 691.4642 | 14.66 | PG 14:0J6:1(9Δ) | 227.1998 | 253.2150 | 581.3525 | 605.3369 |
| 717.4704 | 15.30 | PG 16:1(9Δ)J6:1(9Δ) | 253.2152 | 253.2152 | 607.3604 | 631.3475 |
| 717.4704 | 15.30 | PG 16:1(7Δ)_16:1(7Δ) | 253.2152 | 253.2152 | 579.3220 | 603.3278 |
| 660.4620 | 16.83 | PE 14:0_16:1(11Δ) | 227.1996 | 253.2152 | 550.3445 | 574.3433 |
| 660.4620 | 16.83 | PE 12:0_18:1(9Δ) | 199.1688 | 281.2477 | 550.3445 | 574.3433 |
| 686.4773 | 17.54 | PE 16:1(9Δ)_16:1(9Δ) | 253.2152 | 253.2152 | 576.3649 | 600.3599 |
| 719.4857 | 17.93 | PG 16:0_1 6:1(9Δ) |
253.2157 | 255.2317 | 609.3869 | 633.3918 |
| 719.4900 | 18.26 | PG 16:0_16:1(7Δ) | 253.2155 | 255.2315 | 581.3253 | 605.3380 |
| 745.5046 | 20.15 | PG 16:1(9Δ)_18:1(9Δ) | 253.2151 | - | 607.3607 | 631.3585 |
| - | 281.2463 | 635.3876 | 659.3928 | |||
| 688.4912 | 21.43 | PE 16:0_16:1(9Δ) | 253.2151 | 255.2307 | 550.3418 | 574.3458 |
| 688.4912 | 21.43 | PE 16:0_16:1(7Δ) | 253.2151 | 255.2307 | 578.3669 | 602.3764 |
| 747.5184 | 21.82 | PG 16:0_18:1(9Δ) | 255.2310 | 281.2470 | 609.3638 | 633.3732 |
| 773.5331 | 22.53 | PG 18:1(9Δ)_18:1(9Δ) | 281.2465 | 281.2465 | 635.3928 | 659.3935 |
| 761.5350 | 24.78 | PG 17:0_18:1(9Δ) | 269.2465 | 281.2463 | 623.3907 | 647.3932 |
| 716.5223 | 25.85 | PE 16:0_18:1(9Δ) | 255.2313 | 281.2462 | 578.3839 | 602.3759 |
| 742.5355 | 26.82 | PE 18:1(9Δ)_18:1(9Δ) | 281.2464 | 281.2464 | 604.4036 | 628.4000 |
| 775.5504 | 27.76 | PG 18:0_18:1(9Δ) | 281.2462 | 283.2618 | 637.4036 | 661.4207 |
| 730.5300 | 28.44 | PE 17:0(9Δ)_18:1(9Δ) | 269.2467 | 281.2464 | 592.3960 | 616.3993 |
| 744.5541 | 31.98 | PE 18:0_18:1(9Δ) | 281.2469 | 283.2627 | 606.4083 | 630.4048 |
Table 3.
List of Identified PEs and PGs from an A. baumannii 19606 Lipid Extract
| Precursor ion (m/z) | RT (min) | Lipid identification | Fatty acid fragment ions (mlz) | Diagnostic DB fragment ions (mlz) | ||
|---|---|---|---|---|---|---|
| 717.4695 | 15.07 | PG 16:1(9Δ)/16:1(9Δ) | 253.2152 | 253.2152 | 607.3607 | 631.3541 |
| 686.4751 | 17.19 | PE 16:1(9Δ)/16:1(9Δ) | 253.2151 | 253.2151 | 576.3596 | 600.3563 |
| 719.4855 | 17.87 | PG 14:0_18:1(9Δ) | 227.2000 | 281.2500 | 581.3370 | 605.3411 |
| 719.4892 | 18.10 | PG 16:0J6:1(9Δ) | 253.2153 | 255.2313 | 609.3834 | 633.3876 |
| 733.5018 | 19.99 | PG 15:0_18:1(10Δ) | 241.2150 | 281.2479 | 609.3696 | 633.3750 |
| 733.5018 | 19.99 | PG 16:0J7:1(9Δ) | 255.2308 | 267.2307 | 609.3710 | 633.3746 |
| 688.4927 | 20.88 | PE 16:0_16:1(9Δ) | 255.2307 | 253.2153 | 578.3740 | 602.3818 |
| 747.5165 | 22.01 | PG 16:0_18:1(9Δ) | 255.2311 | 281.2470 | 609.3639 | 633.3738 |
| 773.5330 | 22.76 | PG 18:1(9Δ)/18:1(9Δ) | 281.2465 | 281.2465 | 635.3925 | 659.3935 |
| 761.5325 | 24.79 | PG 17:0/18:1(9Δ) | 269.2465 | 281.2463 | 623.3923 | 647.3909 |
| 716.5250 | 25.87 | PE 16:0/18:1(9Δ) | 255.2313 | 281.2461 | 578.3824 | 602.3764 |
| 742.5415 | 26.61 | PE 16:0/18:1(9Δ) | 281.2463 | 281.2463 | 604.4012 | 628.3960 |
| 775.5484 | 27.51 | PG 18:0/18:1(9Δ) | 281.2461 | 283.2615 | 637.4040 | 661.4052 |
| 730.5381 | 28.91 | PE 17:0/18:1(9Δ) | 269.2468 | 281.2466 | 592.3951 | 616.4001 |
| 744.5560 | 32.06 | PE 18:0/18:1(9Δ) | 281.2468 | 283.2625 | 606.4075 | 630.4077 |
CONCLUSIONS
UVPD of GPL anions generates double bond diagnostic ions with a mass difference of 24 Da, in addition to the same types of fragment ions generated by collisional activation, confirming the utility of UVPD for GPLs that preferentially ionize in negative mode. The presented work also validates UVPD as a method capable of detecting both double bonds and cyclopropyl modifications within a single workflow. For the first time, untargeted analysis of lipid extracts was performed using this online LC–UVPD–MS method. For GPL structures with sites of unsaturation in each acyl chain, an MS3 infusion-type method enabled localization of sites to specific acyl chains. Application of the LC–UVPD–MS method to bacterial lipid extracts revealed differences in the acyl chain unsaturation profiles between E. coli and A. baumannii. Considering the role unsaturated acyl chains play in the maintenance of membrane structure, enzymes responsible for incorporation of unsaturation elements in bacteria have been considered as antibiotic targets. The devised strategy provides a means to investigate the different mechanisms by which various species of bacteria incorporate unsaturation motifs in acyl chains and has the potential to inform antibiotic development via more detailed lipid structural mapping. While UVPD enables a higher level of structural characterization, it brings attention to the striking complexity of lipid profiles and suggests that additional method development is required to resolve ambiguity in spectra where isomers are not chromatographically resolved. Addition of gas-phase ion-mobility-type separations to the present workflow offers one potential means to further alleviate the occurrence of chimeric spectra.
Supplementary Material
ACKNOWLEDGMENTS
Funding from the NIH (RO1 GM103655 to J.S.B., and AI129940 and AI138576 to M.S.T.) and the Welch Foundation (F-1155) are gratefully acknowledged. Funding from the UT System for support of the UT System Proteomics Core Facility Network is gratefully acknowledged.
Footnotes
The authors declare no competing financial interest.
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.0c00221.
Figure S1. Laser OFF/ON UVPD TIC, spectra, and ion injection times for PG 16:0/18:1(9Z); Figure S2. HCD spectra of PA 16:0/18:1(9Z), PE 16:0/18:1(9Z), PS 16:0/18:1(9Z), and PI 16:0/18:1(9Z); Figure S3. Fragment ion maps; Figure S4. HCD and UVPD spectra for formate-adducted PC 16:0/18:1(9Z); Tables S1– S12. Lists of identified fragment ions with calculated ppm error; Figure S5. HCD spectra of m/z 773.53 and with fragment ion maps from the chromatographic separation of lipid extracts from E. coli, A. baumannii 17989, and A. baumannii 19606; Figure S6. HCD, UVPD, and CID/UVPD spectra of m/z 745.50 from A. baumannii 17989 lipid extract (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.analchem.0c00221
Contributor Information
Martin V. Douglass, Department of Infectious Diseases, The University of Georgia, College of Veterinary Medicine, Athens, Georgia 30602, United States
M. Stephen Trent, Department of Infectious Diseases, The University of Georgia, College of Veterinary Medicine, Athens, Georgia 30602, United States; Department of Microbiology, The University of Georgia, College of Arts and Sciences, Athens, Georgia 30602, United States.
Jennifer S. Brodbelt, Department of Chemistry, The University of Texas at Austin, Austin, Texas 78712, United States.
REFERENCES
- (1).Brügger B Annu. Rev. Biochem 2014, 83, 79–98. [DOI] [PubMed] [Google Scholar]
- (2).Shevchenko A; Simons K Nat. Rev. Mol. Cell Biol 2010, 11, 593–598. [DOI] [PubMed] [Google Scholar]
- (3).Brown SHJ; Mitchell TW; Oakley AJ; Pham HT; Blanksby SJ J. Am. Soc. Mass Spectrom 2012, 23, 1441–1449. [DOI] [PubMed] [Google Scholar]
- (4).Rolim AEH; Henrique-Araújo R; Ferraz EG; de Araújo Alves Dultra FK; Fernandez LG Gene 2015, 554, 131–139. [DOI] [PubMed] [Google Scholar]
- (5).Han X Nat. Rev. Endocrinol 2016, 12, 668–679. [DOI] [PubMed] [Google Scholar]
- (6).Rustam YH; Reid GE Anal. Chem 2018, 90, 374–397. [DOI] [PubMed] [Google Scholar]
- (7).Dennis EA Proc. Natl. Acad. Sci. U. S. A 2009, 106, 2089–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Wenk MR Cell 2010, 143, 888–895. [DOI] [PubMed] [Google Scholar]
- (9).Blanksby SJ; Mitchell TW Annu. Rev. Anal. Chem 2010, 3, 433–465. [DOI] [PubMed] [Google Scholar]
- (10).Earley L; Anderson LC; Bai DL; Mullen C; Syka JEP; English AM; Dunyach J-J; Stafford GC; Shabanowitz J; Hunt DF; Compton PD Anal. Chem 2013, 85, 8385–8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Sandra K; Sandra P Curr. Opin. Chem. Biol 2013, 17, 847–853. [DOI] [PubMed] [Google Scholar]
- (12).Gross RW Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 2017, 1862, 731–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Han X Lipidomics: Comprehensive Mass Spectrometry of Lipids; John Wiley & Sons, Inc, 2016. [Google Scholar]
- (14).Brouwers JF Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 2011, 1811, 763–775. [DOI] [PubMed] [Google Scholar]
- (15).Cajka T; Fiehn O TrAC, Trends Anal. Chem 2014, 61, 192–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Zhao Y-Y; Wu S-P; Liu S; Zhang Y; Lin R-C Chem.-Biol. Interact 2014, 220, 181–192. [DOI] [PubMed] [Google Scholar]
- (17).Han X; Gross RW Mass Spectrom. Rev 2005, 24, 367–412. [DOI] [PubMed] [Google Scholar]
- (18).Han X; Gross RW Expert Rev. Proteomics 2005, 2, 253–264. [DOI] [PubMed] [Google Scholar]
- (19).Ivanova PT; Milne SB; Myers DS; Brown HA Curr. Opin. Chem. Biol 2009, 13, 526–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Han X; Yang K; Gross RW Mass Spectrom. Rev 2012, 31, 134–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Schuhmann K; Herzog R; Schwudke D; Metelmann-Strupat W; Bornstein SR; Shevchenko A Anal. Chem 2011, 83, 5480–5487. [DOI] [PubMed] [Google Scholar]
- (22).Schuhmann K; Almeida R; Baumert M; Herzog R; Bornstein SR; Shevchenko AJ Mass Spectrom 2012, 47, 96–104. [DOI] [PubMed] [Google Scholar]
- (23).Yang K; Han X Trends Biochem. Sci 2016, 41, 954–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Wang M; Wang C; Han RH; Han X Prog. Lipid Res 2016, 61, 83–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Almeida R; Pauling JK; Sokol E; Hannibal-Bach HK; Ejsing CS J. Am. Soc. Mass Spectrom 2015, 26, 133–148. [DOI] [PubMed] [Google Scholar]
- (26).Liebisch G; Vizcaíno JA; Köfeler H; Trötzmüller M; Griffiths WJ; Schmitz G; Spener F; Wakelam MJ O. J. Lipid Res 2013, 54, 1523–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Pulfer M; Murphy RC Mass Spectrom. Rev 2003, 22, 332–364. [DOI] [PubMed] [Google Scholar]
- (28).Hsu F-F; Turk JJ Am. Soc. Mass Spectrom 2000, 11, 892–899. [DOI] [PubMed] [Google Scholar]
- (29).Hsu F.-F. u; Turk JJ Am. Soc. Mass Spectrom 2003, 14, 352–363. [DOI] [PubMed] [Google Scholar]
- (30).Hsu F-F; Turk JJ Chromatogr. B: Anal. Technol. Biomed. Life Sci 2009, 877, 2673–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Zhang Y-M; Rock CO Nat. Rev. Microbiol 2008, 6, 222–233. [DOI] [PubMed] [Google Scholar]
- (32).Hussain NF; Siegel AP; Ge Y; Jordan R; Naumann CA Biophys. J 2013, 104, 2212–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Janmey PA; Kinnunen PK J. Trends Cell Biol 2006, 16, 538–546. [DOI] [PubMed] [Google Scholar]
- (34).Martinez-Seara H; Roóg T; Pasenkiewicz-Gierula M; Vattulainen I; Karttunen M; Reigada R J. Phys. Chem. B 2007, 111, 11162–11168. [DOI] [PubMed] [Google Scholar]
- (35).Martinez-Seara H; Roóg T; Pasenkiewicz-Gierula M; Vattulainen I; Karttunen M; Reigada R Biophys. J 2008, 95, 3295–3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Martinez-Seara H; Roóg T; Karttunen M; Vattulainen I; Reigada R J. Phys. Chem. B 2009, 113, 8347–8356. [DOI] [PubMed] [Google Scholar]
- (37).Yang K; Dilthey BG; Gross RW Anal. Chem 2013, 85, 9742–9750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Xu Y; Brenna JT Anal. Chem 2007, 79, 2525–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Zhao Y; Zhao H; Zhao X; Jia J; Ma Q; Zhang S; Zhang X; Chiba H; Hui S-P; Ma X Anal. Chem 2017, 89, 10270–10278. [DOI] [PubMed] [Google Scholar]
- (40).Feng Y; Chen B; Yu Q; Li L Anal. Chem 2019, 91 (3), 1791–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Ma X; Xia Y Angew. Chem 2014, 126, 2630–2634. [Google Scholar]
- (42).Ma X; Chong L; Tian R; Shi R; Hu TY; Ouyang Z; Xia Y Proc. Natl. Acad. Sci. U. S. A 2016, 113, 2573–2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Stinson CA; Xia Y Analyst 2016, 141, 3696–3704. [DOI] [PubMed] [Google Scholar]
- (44).Wäldchen F; Becher S; Esch P; Kompauer M; Heiles S Analyst 2017, 142, 4744–4755. [DOI] [PubMed] [Google Scholar]
- (45).Hsu F-F; Turk JJ Am. Soc. Mass Spectrom 2008, 19, 1681–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Moe MK; Strøm MB; Jensen E; Claeys M Rapid Commun. Mass Spectrom 2004, 18, 1731–1740. [DOI] [PubMed] [Google Scholar]
- (47).Stinson CA; Zhang W; Xia YJ Am. Soc. Mass Spectrom 2018, 29 (3), 481–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Harris RA; May JC; Stinson CA; Xia Y; McLean JA Anal. Chem 2018, 90, 1915–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Pham HT; Ly T; Trevitt AJ; Mitchell TW; Blanksby SJ Anal. Chem 2012, 84, 7525–7532. [DOI] [PubMed] [Google Scholar]
- (50).Pham HT; Trevitt AJ; Mitchell TW; Blanksby SJ Rapid Commun. Mass Spectrom 2013, 27, 805–815. [DOI] [PubMed] [Google Scholar]
- (51).Pham HT; Julian RR Analyst 2016, 141, 1273–1278. [DOI] [PubMed] [Google Scholar]
- (52).Toyoda M Eur. J. Mass Spectrom 2010, 16, 397–406. [DOI] [PubMed] [Google Scholar]
- (53).Brown SHJ; Mitchell TW; Blanksby SJ Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 2011, 1811, 807–817. [DOI] [PubMed] [Google Scholar]
- (54).Pham HT; Maccarone AT; Thomas MC; Campbell JL; Mitchell TW; Blanksby SJ Analyst 2014, 139, 204–214. [DOI] [PubMed] [Google Scholar]
- (55).Kozlowski R; Mitchell T; Blanksby S Eur. J. Mass Spectrom 2015, 21, 191–200. [DOI] [PubMed] [Google Scholar]
- (56).Poad BLJ; Green MR; Kirk JM; Tomczyk N; Mitchell TW; Blanksby SJ Anal. Chem 2017, 89, 4223–4229. [DOI] [PubMed] [Google Scholar]
- (57).Poad BLJ; Zheng X; Mitchell TW; Smith RD; Baker ES; Blanksby SJ Anal. Chem 2018, 90, 1292–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Campbell JL; Baba T Anal. Chem 2015, 87, 5837–5845. [DOI] [PubMed] [Google Scholar]
- (59).Baba T; Campbell JL; Le Blanc JCY; Baker PR S. J. Lipid Res 2016, 57, 858–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Baba T; Campbell JL; Le Blanc JCY; Baker PR S. Anal. Chem 2017, 89, 7307–7315. [DOI] [PubMed] [Google Scholar]
- (61).Baba T; Campbell JL; Le Blanc JCY; Baker PRS; Ikeda KJ Lipid Res 2018, 59, 910–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Deimler RE; Sander M; Jackson GP Int. J. Mass Spectrom 2015, 390, 178–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Li P; Hoffmann WD; Jackson GP Int. J. Mass Spectrom 2016, 403, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Brodbelt JS Chem. Soc. Rev 2014, 43, 2757–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Madsen JA; Cullen TW; Trent MS; Brodbelt JS Anal. Chem 2011, 83, 5107–5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).O’Brien JP; Brodbelt JS Anal. Chem 2013, 85, 10399–10407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Klein DR; Holden DD; Brodbelt JS Anal. Chem 2016, 88, 1044–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Klein DR; Brodbelt JS Anal. Chem 2017, 89, 1516–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Ryan E; Nguyen CQN; Shiea C; Reid GE J. Am. Soc. Mass Spectrom 2017, 28, 1406–1419. [DOI] [PubMed] [Google Scholar]
- (70).Williams PE; Klein DR; Greer SM; Brodbelt JS J. Am. Chem. Soc 2017, 139, 15681–15690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Klein DR; Feider CL; Garza KY; Lin JQ; Eberlin LS; Brodbelt JS Anal. Chem 2018, 90, 10100–10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Ivanova PT; Milne SB; Byrne MO; Xiang Y; Brown HA Methods Enzymol 2007, 432, 21–57. [DOI] [PubMed] [Google Scholar]
- (73).Bligh EG; Dyer WJ Can. J. Biochem. Physiol 1959, 37, 911–917. [DOI] [PubMed] [Google Scholar]
- (74).Giles DK; Hankins JV; Guan Z; Trent MS Mol. Microbiol 2011, 79, 716–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Damen CWN; Isaac G; Langridge J; Hankemeier T; Vreeken RJ J. Lipid Res 2014, 55, 1772–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Narváez-Rivas M; Zhang QJ Chromatogr. A 2016, 1440, 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Blevins MS; Klein DR; Brodbelt JS Anal. Chem 2019, 91, 6820–6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Macias LA; Feider CL; Eberlin LS; Brodbelt JS Anal. Chem 2019, 91, 12509–12516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Feng Y; Cronan JE J. Biol. Chem 2009, 284, 29526–29535. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


