Abstract
Paracoccidioidomycosis (PCM) is a life-threatening systemic mycosis widely reported in the Gran Chaco ecosystem. The disease is caused by different species from the genus Paracoccidioides, which are all endemic to South and Central America. Here, we sequenced and analyzed 31 isolates of Paracoccidioides across South America, with particular focus on isolates from Argentina and Paraguay. The de novo sequenced isolates were compared with publicly available genomes. Phylogenetics and population genomics revealed that PCM in Argentina and Paraguay is caused by three distinct Paracoccidioides genotypes, P. brasiliensis (S1a and S1b) and P. restrepiensis (PS3). P. brasiliensis S1a isolates from Argentina are frequently associated with chronic forms of the disease. Our results suggest the existence of extensive molecular polymorphism among Paracoccidioides species, and provide a framework to begin to dissect the connection between genotypic differences in the pathogen and the clinical outcomes of the disease.
Keywords: Paracoccidioides, Gran Chaco, Phylogenomics, Population genetics
1. Introduction
Paracoccidioidomycosis (PCM) is a systemic mycosis endemic to Latin America that occurs in locations from Mideastern Mexico to Argentina and Uruguay. The disease is caused by fungi in the genus Paracoccidioides (Onygenales, Ascomycota), which are thermally dimorphic fungi that transition between mycelium and yeast (Martinez et al., 2017; Restrepo et al., 2001). The saprophytic stage grows as multifilament mycelia that produce airborne infectious conidia, which may be inhaled by susceptible hosts. Upon inhalation, aerosolized propagules transform to a multi-budding yeast phase that can colonize alveolar macrophages in a mammal host (Martinez et al., 2017; Restrepo et al., 2001). Clinical forms of PCM vary from a mild pneumonia to a chronic and debilitating disease. The acute form is less common, has been associated with the depression of cellular immunity, and can eventually lead to death. Severe dissemination may occur in patients due to comorbidities like HIV/AIDS, hematological neoplasia, and pregnancy (Martinez et al., 2017; Shikanai-Yasuda et al., 2017). PCM is at least 10 times more common in adult males, and causes an average of 150 deaths per year in Brazil with new annual cases estimated to be over 3,000 (Martinez et al., 2017; Prado et al., 2009). Other countries have PCM patients, but morbidity has not been systematically measured outside of Brazil.
Paracoccidioides is composed of at least five species, which differ both genotypically and phenotypically (Matute et al., 2006; Munoz et al., 2016; Teixeira Mde et al., 2014; Teixeira et al., 2009; Teixeira et al., 2014; Turissini et al., 2017): P. lutzii, P. brasiliensis sensu stricto, P. americana, P. restrepiensis, and P. venezuelensis. Divergence times between species pairs range between 0.03 and 33.00 million years. This range of divergence times makes Paracoccidioides an intriguing model for assessing how genome differentiation accrues as speciation proceeds in fungal pathogens. P. brasiliensis sensu stricto also shows strong population structure, which has led to the proposal of two strongly isolated demes (S1a and S1b) (Munoz et al., 2016). Notably, the species of Paracoccidioides show geographic range overlap. Both Brazil and Venezuela are thought to harbor more than one species of Paracoccidioides, opening the possibility of gene exchange between these species. P. americana and P. venezuelensis coexist in Venezuela (Teixeira et al., 2014); and P. americana, P. brasiliensis sensu stricto, and P. lutzii co-occur in Brazil (Munoz et al., 2016; Teixeira et al., 2014). In some cases, isolates from two different species have been collected from the same mammal carrier (Arantes et al., 2016; Hrycyk et al., 2018).
Given the relatively limited number of samples from most of the South and Central American countries, the precise range of Paracoccidioides species remains relatively unexplored. Brazil and Colombia have led the collection efforts for Paracoccidioides, and most of the known isolates come from these two countries (Matute et al., 2006; Teixeira et al., 2014; Turissini et al., 2017). However, Paracoccidioides is also common in Argentina. Paracoccidioidin skin test surveys performed in Argentina demonstrated infection rates ranging from 1.6% to 21.3% of the total reactive population depending on the region (Mangiaterra et al., 1996; van Gelderen de Komaid et al., 1999). Thus, the prevalence of the disease is comparable with Brazil (Rodrigues and de Resende, 1996), and higher than in Colombia (Restrepo et al., 1968). In Argentina, Paracoccidioides has been isolated from soil (Martinez et al., 2017), and PCM is distributed in two well-defined endemic areas (Giusiano et al., 2018b). The first is located in northwestern Argentina (NWA), including the predominantly warm and subtropical Yunga ecoregion of the Salta, Jujuy, and Tucumán provinces with a predominance of acute forms (up to 40%) of the disease (Davel and Canteros, 2007; Giusiano et al., 2018a). The second and more extensive area is located in northeastern Argentina (NEA), including portions of seven different state provinces with a high predominance of chronic forms of the disease (Tracogna et al., 2018). This second endemic area extends into Paraguay (Araujo et al., 2016; Rolón, 2004). The early 2010s saw an increase in the acute and chronic forms of PCM in Argentina (Giusiano et al., 2018a; Giusiano et al., 2018b) and Paraguay. Chaco province records the highest incidence of cases in Argentina, a four-fold increase of PCM rates compared to previous years was reported during 2013 and 2014 (Tracogna et al., 2018).
The genetic diversity of the Paracoccidioides genus in this region of South America remains to be studied. To date only four strains from Argentina and Paraguay have been genotyped using ten sequenced nuclear loci, and all four isolates belonged to P. brasiliensis sensu stricto (Matute et al., 2006; Turissini et al., 2017). Determining the demographic dynamics of the species might inform if a particular genotype was associated with the increase of PCM cases in the 2010s. We used previously published Paracoccidioides genomes and 31 newly sequenced isolates to resolve the extent of Paracoccidioides’ genetic diversity in the southern portion of South America. Our results suggest that this region harbors multiple Paracoccidioides species, and reveal the importance of systematic sampling in defining the geographic range of a pathogen.
2. Material and methods
2.1. Fungal strains and DNA extraction and sequencing
Thirty-one Paracoccidioides isolates from Argentina, Bolivia, Brazil, Paraguay, Peru and Venezuela were sequenced de novo (Table 1). Cultures of Paracoccidioides were maintained in yeast form in semi-solid Fava-Netto media (1.0% peptone, 0.5% yeast extract, 0.3% proteose peptone, 0.5% beef extract, 0.5% NaCl, 4% glucose, and 1.5% agar, pH 7.2) at 37 °C. DNA extractions were performed (see Supplementary Appendix 1) and the concentration of the resulting DNA measured using a NanoDrop® 1000 (Thermo Fisher Scientific). Approximately 1 μg of purified DNA per sample was used for sequencing library preparation via KAPA library preparation kit for Illumina NGS sequencing (Kapa Biosystems). Each library was indexed with unique 8-bp nucleotide tags and the library concentration was verified using a KAPA library quantification kit (Kapa Biosystems) on a 7900HT Instrument (Life Technologies). All libraries were sequenced to a read length of 100 bp using v3 or v4 chemistries on an Illumina HiSeq 2500AUTHOR instrument (Illumina, San Diego, CA).
Table 1.
Clinical and molecular characteristics of the 31 whole genome-sequenced Paracoccidioides isolates included in this study.
| Strain | Country | Region | Collection Date | Clinical Form | Species (genotype) |
|---|---|---|---|---|---|
| PbA28 | Argentina | NEA | 2014 | Chronic | P. brasiliensis (S1a) |
| Pb33 | Argentina | NEA | 2014 | Chronic | P. brasiliensis (S1a) |
| Pb43 | Argentina | NWA | 2014 | Acute | P. brasiliensis (S1b) |
| PbA48 | Argentina | NEA | 2014 | Chronic | P. brasiliensis (S1a) |
| PbA49 | Argentina | NEA | 2014 | Chronic | P. brasiliensis (S1a) |
| PbA57 | Argentina | NEA | 2014 | Chronic | P. brasiliensis (S1a) |
| Pb65 | Argentina | NEA | 2015 | Chronic | P. brasiliensis (S1a) |
| PbA71 | Argentina | NEA | 2015 | Chronic | P. brasiliensis (S1a) |
| Pb85 | Argentina | NEA | 2015 | Chronic | P. brasiliensis (S1a) |
| PbA93 | Argentina | NEA | 2016 | Unifocal Bone PCM | P. brasiliensis (S1a) |
| Pb100 | Argentina | NEA | Chronic | P. brasiliensis (S1b) | |
| PbA102 | Argentina | NWA | 2004 | Chronic | P. brasiliensis (S1a) |
| Pb103 | Argentina | NEA | 1981 | Acute | P. brasiliensis (S1b) |
| Pb405 | Argentina | NWA | Acute | P. brasiliensis (S1b) | |
| Pb395 | Argentina | 2004 | P. restrepiensis (PS3) | ||
| Pb396 | Argentina | NEA | 2004 | Chronic | P. restrepiensis (PS3) |
| Pb67 | Bolivia | 2015 | Chronic | P. brasiliensis (S1b) | |
| Pb391 | Brazil | Guaranésia | Chronic | P. brasiliensis (S1a) | |
| Pb101 | Paraguay | Chronic | P. brasiliensis (S1b) | ||
| PbP42 | Paraguay | 2014 | Acute | P. brasiliensis (S1b) | |
| Pb59 | Paraguay | 2014 | Chronic | P. brasiliensis (S1b) | |
| Pb98 | Paraguay | 1999 | Chronic | P. brasiliensis (S1b) | |
| Pb111 | Paraguay | PCM | P. brasiliensis (S1b) | ||
| Pb124 | Peru | PCM | P. restrepiensis (PS3) | ||
| Pb304 | Venezuela | Barinas | 1988 | Chronic | P. venezuelensis (PS4) |
| Pb305 | Venezuela | Vargas | 1988 | Chronic | P. venezuelensis (PS4) |
| Pb307 | Venezuela | Vargas | 1991 | Chronic | P. venezuelensis (PS4) |
| Pb309 | Venezuela | Miranda | 1994 | Chronic | P. venezuelensis (PS4) |
| Pb384 | Venezuela | Anzoátegui | 1971 | Chronic | P. venezuelensis (PS4) |
| Pb387 | Venezuela | Miranda | 1983 | Acute | P. venezuelensis (PS4) |
| Pb444 | Venezuela | Miranda | 2004 | Chronic | P. venezuelensis (PS4) |
NEA: northeast of Argentina; NWA: northwest of Argentina; PCM: Paracoccidioidomycosis (without data related to clinical form).
2.2. Single nucleotide polymorphism identification
We compared the genomes variation in the newly sequenced Paracoccidioides strains with previously published genomes (Munoz et al., 2016). The Illumina reads were de-multiplexed and single nucleotide polymorphism (SNP) profiling was assessed using the NASP pipeline (Sahl et al., 2016). Poor quality reads and adapters were filtered and removed using Trimmomatic v 0.36 (Bolger et al., 2014). Remaining reads were aligned to P. brasiliensis strain Pb18v2 assembly (ABKI00000000.2) with the Burrows-Wheeler Aligner v0.7.7 (Li and Durbin, 2009; Munoz et al., 2016) – unaligned reads were discarded. The resulting bam files were merged using Samtools 0.1.1 (Li et al., 2009), and the resulting alignment processed with RealignerTarget-Creator and IndelRealigner (GATK v3.3–0; (DePristo et al., 2011; McKenna et al., 2010)) to realign around indels. The GATK UnifiedGenotyper tool was used to call SNPs, and the parameter “het” set to 0.01 to account for a haploid organism. The resulting .vcf files were filtered using the following parameters: QD = 2.0 || FS_filter = 60.0 || MQ_filter = 30.0 || MQ_Rank_Sum_filter = −12.5 || Read_-Pos_Rank_Sum_filter = −8. Finally, all sites with a coverage lower than 10X across all samples, or greater than the 99th quantile of the genomic coverage distribution for the given line, were excluded, as these sites might be duplications or misassembled regions.
2.3. Population structure
Next, we studied whether there was evidence of strong population structure within the Paracoccidioides genus. Initially, we inferred the most likely number of clusters using fastSTRUCTURE v1.0 (Raj et al., 2014). fastSTRUCTURE uses a Bayesian framework to calculate the posterior probabilities of an individual belonging to a given cluster and allows for inference of the most likely number of populations in a sample (K). We used the admixture model and assumed all SNPs are assumed to be unlinked. Likelihood values were calculated for scenarios that ranged from K = 2 to K = 10 populations. Changes in log likelihoods were compared for fastSTRUCTURE runs with sequential values of k using a likelihood ratio test as implemented in the chooseK.py script (Raj et al., 2014). Second, the partition of genetic variation was visualized using Principal Component Analysis (PCA) with the R package adegenet for multivariate analysis of genetic data (Jombart and Ahmed, 2011). Biallelic SNPs were extracted using the function fasta2genlight within adegenet to compute the principal components (PCs) using the function glPca. We only refer to the first two PCs as they explained most of the genetic variance (see results). Finally, we calculated FST as a proxy for genetic differentiation between species using vcftools ((Danecek et al., 2011); See Technical Appendix 1).
2.4. Phylogenetic tree and gene exchange
The resulting .vcf file had 59 fully sequenced P. brasiliensis sensu stricto, P. americana, P. restrepiensis, and P. venezuelensis genomes. Four P. lutzii isolates were included in this dataset as an outgroup. Genealogical relationships among these 63 isolates were inferred using a Maximum Likelihood (ML) approach. To generate the tree, IQ-TREE (Nguyen et al., 2015) using -m MFP option (ModelFinder) to automatically select the best-fitting sequence evolution model (Kalyaanamoorthy et al., 2017) was employed. To calculate individual branch support, we calculated 1,000 ultrafast bootstraps replicates and followed them with a Shimodaira–Hasegawa-like approximate likelihood ratio test (SH-aLRT) (Minh et al., 2013).
Finally, we surveyed for evidence of introgression in the nuclear genome of Paracoccidioides using the model implemented in TreeMix (Pickrell and Pritchard, 2012). TreeMix estimates the most likely evolutionary history of a group of populations by estimating the levels of genetic drift at a set of markers. To select the most likely admixture scenario, five different scenarios (from 1 to 5 migration/admixture events) were inferred, and then selected using sequential Likelihood Ratio Tests (LRT) and Akaike weights (wAIC). See detailed information in the Technical Appendix 1.
3. Results
3.1. Data availability
We sequenced thirty-one new Paracoccidioides genomes from Argentina, Bolivia, Brazil, Paraguay, Peru and Venezuela (Table 1), which increases the total of publicly available Paracoccidioides genomes to sixty-three. The raw Illumina reads for all the sequenced genomes are available at the Sequence Repository Archive under the following: SRASRR9736748-SRR9736778 (Technical Appendix II).
3.2. Population structure
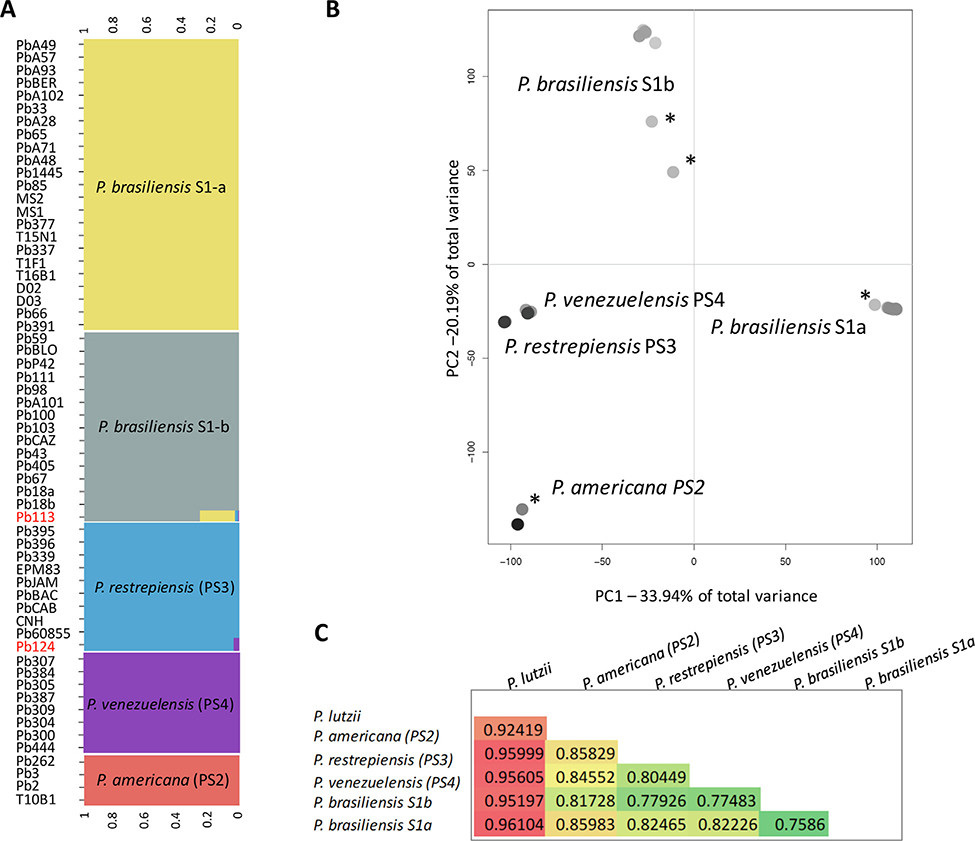
First, we studied how the genetic variation was apportioned in the P. brasiliensis species complex. The approach was twofold, as both fastSTRUCTURE and Principal Coordinate Analysis (PCA) were used in the analysis. First, we ran nine clustering scenarios using polymorphic sites within the P. brasiliensis species complex. Pairs of nested scenarios were compared sequentially using LRT. The results suggest that the most likely scenario involves the existence of five clusters (Fig. 1A). Technical Appendix 1 shows the likelihood values for all scenarios. Three of the five populations are the previously proposed Paracoccidioides species within P. brasiliensis sensu lato: P. americana (PS2), P. restrepiensis (PS3), and P. venezuelensis (PS4) (Fig. 1A). The two additional populations correspond are two strongly structured populations of P. brasiliensis sensu stricto: P. brasiliensis (S1a) and P. brasiliensis (S1b). The vast majority of isolates were classified with a level of confidence over 99% with two exceptions: Pb113 (Brazil) and Pb124 (Peru). The probability of these isolates belonging to P. brasiliensis S1b and P. restrepiensis, respectively, is reasonably high (over 70%). Because fastSTRUCTURE cannot infer the reasons for this uncertainty, we discuss potential causes below.
Fig. 1.
Genome wide genetic variation is portioned across species boundaries in Paracoccidioides. A. Probability of belonging to a cluster when K = 5, the most likely clustering, in Paracoccidioides based on Bayesian algorithm fastSTRUCTURE. Each column represents the genotype of an individual. B. Genetic variation in natural Paracoccidioides populations inferred by Principal Component Analysis (PCA). Only the first two PCs are plotted as they encompass over 50% of the genetic variance. The bar plots of eigenvalues (the inset plot) show the number of retained principal components. PC1 explains 33.94% while PC2 explains 20.19% of the total variance respectively. Individuals marked with asterisks represent potentially admixed strains. C. Triangular matrix showing the mean Fst value in all pairwise comparisons within the P. brasiliensis species complex.
The results from PCA are consistent with the results from fastSTRUCTURE. We focus on the first two PCs as they explain over 50% of the genetic variance. PCA suggests the presence of five distinct lineages among P. brasiliensis sensu lato (Fig. 1B). PC1 explains 33.94% of the total variance and mostly discriminates P. brasiliensis S1a (and to a lesser extent S1b) from the other groups. On the other hand, PC2 explains 20.19% of the total variance and separates P. brasiliensis S1b, P. americana (PS2), P. restrepiensis (PS3), and P. venezuelensis (PS4) (Fig. 1B). Notably, four isolates appear as intermediate in the PCA (marked with asterisks in Fig. 1B): Pb391 (P. brasiliensis S1a), Pb18 and Pb113 (P. brasiliensis S1b), and T10B1 (P. americana) all dispersed along PC2. These results suggest —but do not confirm—that these individuals might represent admixed individuals. As expected, FST values between the most divergent species pairs, P. lutzii, and other Paracoccidioides species, were high (FST = 0.92–0.96) which is in agreement with previous analysis (9). The FST value between the most-recently diverged groups, P. brasiliensis S1a and P. brasiliensis S1b, was the lowest (FST = 0.44–0.79). All pairwise comparisons showed high FST values which were in turn much higher than the any π value observed for any species or group. These results confirm the observation that between species differentiation is much higher than within species diversity (Fig. 1C).
3.3. Phylogenomic analysis
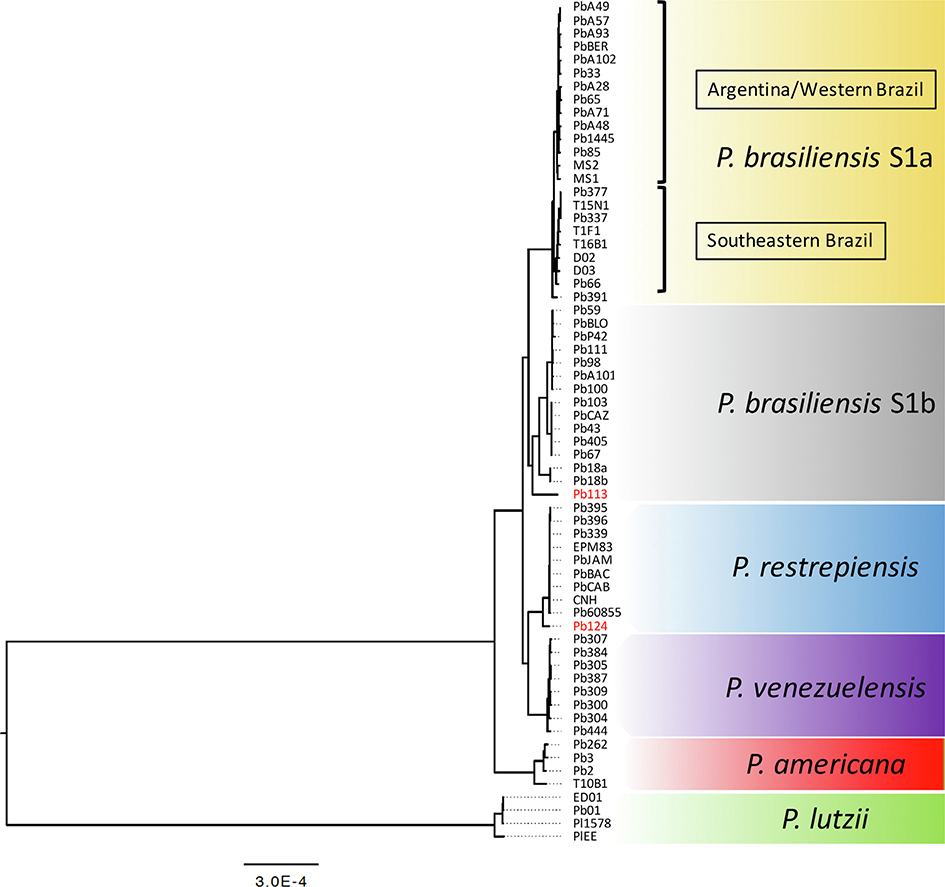
We evaluated the phylogenetic placement of the newly typed strains by adding P. lutzii as outgroup; in total our alignment had 821,941 variable sites. As expected, the longest branch in the tree is the one that separates P. lutzii and all the other species within the P. brasiliensis species complex (Fig. 2). The speciation event that gave rise to P. lutzii and the rest of the Paracoccidioides species is thought to be older than 30 million years ago (Teixeira et al., 2009; Turissini et al., 2017).
Fig. 2.
The genealogical relationships among Paracoccidioides isolates. A. Maximum Likelihood phylogenetic tree generated by whole-genome SNP typing reveals the five genetic clusters: P. brasiliensis (S1a), P. brasiliensis (S1b), P. americana (PS2), P. restrepiensis (PS3), and P. venezuelensis (PS4). A. Branch length is proportional to mutations accumulated for each lineage. P. lutzii was used as the outgroup and the phylogenetic groups are highlighted and supported by both ultrafast bootstraps or Shimodaira–Hasegawa-like approximate likelihood ratio tests (SH-aLRT).
The rest of the genealogy is largely consistent with the clusters inferred by fastSTRUCTURE (Fig. 1A). First, all the Paracoccidioides species appear as reciprocally monophyletic groups (Fig. 2). Second, P. restrepiensis and P. venezuelensis are sister species (consistent with their close arrangement in the PCA plot (Fig. 1B). This dyad is the sister to P. brasiliensis sensu stricto. P. americana is sister to the triad of all the other species in the species complex. These observations match previous species delimitations and reciprocal relationships among Paracoccidioides.
The inferred geographic ranges of P. americana and P. venezuelensis was not expanded with the addition of new samples. However, that was not the case for P. restrepiensis. Two isolates from Argentina (Pb395 and Pb396) and one isolate from Peru (Pb124) are nested within P. restrepiensis suggesting that this species has a broader geographic range than originally thought outside Colombia (Fig. 2).
Additionally, and also consistent with fastSTRUCTURE and PCA, P. brasiliensis sensu stricto is divided into S1b and S1a. The S1a lineage contains Argentinean and Brazilian isolates (N = 23). The tree suggests the existence of two monophyletic clades with P. brasiliensis S1a harboring isolates from Argentina/Western Brazil (S1a-ARG) and Southeastern Brazil (S1a-BR); these clusters were not apparent with fastSTRUCTURE or PCA (Figs. 1 and 2). The FST value between S1a-ARG and S1a-BR is 0.435, which is low relative to other pairwise comparisons (Fig. 1). The S1b clade harbors isolates from São Paulo state (Southeastern Brazil), Argentina, and Paraguay (Fig. 2).
3.4. Paracoccidioides admixture history
Finally, we studied whether there has been genetic exchange between species of Paracoccidioides. The absence, or rarity, of gene exchange is one of the trademarks of speciation. As allele exchange across population boundaries ceases, reproductive isolation builds up more easily (Coyne and Orr, 2004). Studying gene exchange is also important as it addresses the possibility of hybridization leading to interspecific hybrids with new combinations of traits.
Both LRT and wAIC indicate that, of the five tested models, the one with the best fit to the data was m = 2 (LRTm=1vs.m=2 = 11.27, P = 7.89 × 10−4; wAIC = 85.99%; Fig. 3). The two migration edges suggest reciprocal gene exchange between P. brasiliensis sensu stricto and P. americana. This result is puzzling because our expectation was that the majority of gene exchange should occur between populations within P. brasiliensis sensu stricto, but no migration was detected between these populations. Whether or not these potential events of gene exchange constitute true cases of introgression will require studying local ancestry along the genome among multiple species of Paracoccidioides.
Fig. 3.
Phylogenetic network showing the population splits and mixtures within Paracoccidioides deduced by TreeMix analysis. The most likely migration scenario involves two events (m = 2) of gene exchange between P. brasiliensis and P. americana. The length of the black branches is proportional to the genetic drift (branch lengths) of each population and scale bars show ten units of standard error (s.e.). According to the direction of the colored hybridization edges P. venezuelensis, P. restrepiensis, and P. americana have emerged from P. brasiliensis (S1a-ARG, S1a-BR or S1B) populations. The migration edges’ weight is represented by colors and details the proportion of ancestry derived from a given migration edge.
4. Discussion
Frequently, species boundaries in fungi have been defined using a small number of loci. These assessments are limited because gene genealogies and species trees are not equivalent (Matute and Sepulveda, 2019). To date, the most ambitious approach assessed diversity among Paracoccidioides isolates using 15 loci to define species boundaries (Turissini et al., 2017). However, individual gene genealogies reveal the evolutionary history of a gene, but not necessarily that of a species. One way to overcome this issue is to use whole-genome data, which can reveal a clearer perspective of the differentiation among groups. Our results using WGST are consistent with previous approaches and confirm the existence of at least five different species of Paracoccidioides (Munoz et al., 2016; Turissini et al., 2017). Our genome-wide approach also confirms that two P. brasiliensis sensu stricto clusters (S1a and S1b) are reciprocally monophyletic. We argue there is a sore need for assessments of genome concordance, measurements of gene flow, and metadata comparisons before formally describing these populations as isolated species.
One of the features of speciation is the cessation of gene exchange between species through fertile hybrids. Several approaches have proposed the possibility of hybridization between species of Paracoccidioides, mainly based on discordance between gene genealogies (Teixeira et al., 2009; Turissini et al., 2017). Our results suggests that there is a signature of introgression in only one species pair out of ten possible pairs in Paracoccidioides. The results shown here also show that P. restrepiensis coexist with P. brasiliensis sensu stricto in the same localities and thus have the chance to interbreed. Other Paracoccidioides species pairs also show geographic range overlap: P. brasiliensis coexists with P. lutzii and P. americana, while P. venezuelensis coexists with P. americana. This extensive overlap among species suggest that in spite of the opportunities for interbreeding, gene flow between species is not pervasive. Because of the ability to coexist in sympatry, we hypothesize that the Paracoccidioides species must have accrued barriers to gene flow that maintain species boundaries. These results should be considered suggestive, but not conclusive, because Treemix detects deviations from a covariance matrix based on the allele frequencies of each group and does not precisely identify the alleles that have potentially crossed species boundaries.
Hybrid individuals might play an important role on speciation because maladaptive hybridization can complete the speciation process (reviewed in (Brasier, 2000)), and also because hybrids can show fitness advantages over the parentals (hybrid vigor (Lippman and Zamir, 2007)). In the case of fungi, adaptive introgression might have been linked to the transference of alleles involved in antifungal resistance (Maxwell et al., 2018). In some instances, new fungal species can originate through hybridization ((Leducq et al., 2016; Mixao and Gabaldon, 2018) but see (Hibbins and Hahn, 2019)). Systematic analyses that quantify not only whether alleles have been transferred between species of Paracoccidioides but also whether there is variation that segregates before speciation (and thus a source of shared variation across species boundaries) are sorely needed.
Our results have implications beyond the recognition of cryptic species. Our results suggest that the southern part of South America represents a center of diversity for Paracoccidioides in which at least two different species coexist. Investigating the geographic range of each of the species allows us to obtain a genetic portrait of the epidemiology of the different species that cause PCM using whole genome data. Fig. 4 shows the approximate position of each Paracoccidioides strain to each respective country via macro-region (i.e. Argentina) or state (i.e. Brazil) since the precise exposure location is unknown.
Fig. 4.
Geographic distribution of Paracoccidioides species and populations in South America. The map shows the approximate location (green areas of the maps) and the number of clinical and/or environmental isolates sampled in the main endemic areas of the disease in Latin America. The pie chart size is proportional to the number of typed strains. The colors of each pie chart represent the proportion of a given Paracoccidioides genotype to its respective endemic area; color conventions are shown in the inset.
Even though, the assessment of geographic range of the Paracoccidioides species is still in its infancy, several patterns emerge from this biogeographic survey. First, our genome-wide phylogenetic tree shows that Argentina harbors at least two different populations of P. brasiliensis (S1a and S1b) but also P. restrepiensis (isolates Pb 395 and Pb 396; Fig. 1A). The Peruvian strain (Pb124) also clusters with P. restrepiensis. This latter species was initially thought to be endemic to Colombia but our results show the species is more widely distributed than initially thought (Matute et al., 2006; Teixeira et al., 2009). Second, S1a isolates were found in Brazil and Argentina, but not in Paraguay. This biogeographic pattern might be caused by sampling bias, or to particular ecological characteristics of the Paraguayan ecosystems, or both. Our current sampling and patient metadata does not allow us to discern between these options. Third, S1b seems to be more common in Western Brazil and Paraguay compared to Argentina. Only two isolates from NEA (PbA100 and Pb103) clustered within P. brasiliensis S1b. Most of isolates from NEA (13 out of 15–83.3%, Bayesian binomial confidence intervals = 56.4–96.5%) belong to P. brasiliensis S1a, which also occurs in the São Paulo region from Brazil. More work must be done to fully characterize the precise distribution of these species and populations and to determine whether there are differences in the form of PCM they cause.
Our results come with caveats that should be considered. Clinical isolates might have been acquired in a different region from the place they were diagnosed (Tracogna et al., 2018). This means that the actual environmental origin of the isolates might be obscured by the site of the diagnosis. Because the data are comprised of mostly clinical isolates, the infections might have occurred elsewhere but the diagnoses and isolate recovery occurred in Argentina. For example, the Argentinian isolates of P. restrepiensis could be clinical isolates from patients who came from other countries where P. restrepiensis is known to circulate (e.g., Colombia). This is an unlikely explanation because most PCM patients are rural workers that often reside the same location for decades (Bicalho et al., 2001), but we argue it will be important to consider traveling patterns of patients in isolate metadata. More ecological studies aiming to identify circulating Paracoccidioides genotypes in soil or migrating secondary hosts might help to precisely delimit the geographical range of these species.
Additionally, our sampling may not be completely random and thus might not represent the whole genetic diversity of the species. Most of our Argentinian and Paraguayan isolates came from a case series observed in these two localities in 2010 and 2014, respectively. One possibility is that the isolates are related and that the outbreaks were caused by just a handful of genotypes, as has happened in other mycotic outbreaks (Bryce et al., 1996; Fraser et al., 2005; Teixeira Mde et al., 2015). A proper analysis of cryptic relatedness among isolates is needed to assess whether serial infections of PCM are caused by a few genotypes that are closely related.
4.1. Conclusions and future directions
Molecular epidemiology has substantially evolved in the past decade with the technological advances of next generation DNA sequencing coupled with the ability to precisely identify the infectious disease agents in a given area of exposure. In the case of PCM, epidemiological surveys have revealed the importance of sex and geography, but the genotype of the fungus and of the hosts remain unexplored. For example, different species of Paracoccidioides might show differences in virulence (Macoris et al., 2006; Molinari-Madlum et al., 1999), as occurs in Histoplasma (Sepulveda et al., 2014), Sporothrix (Della Terra et al., 2017) and Cryptococcus (Hagen et al., 2015). If this is the case, then the study of genetic differentiation can also inform the evolution of differences in virulence. Systematic collection of isolates from both clinical and environmental samples and the combination of population genetics with epidemiological studies can answer whether the various species of Paracoccidioides show differences in clinical importance or whether they are similar in the phenomenology of the disease they cause.
Supplementary Material
Acknowledgments
We are thankful to Dr. Anastasia Litvintseva and Centers for Diseases Control and Prevention - Mycotic Disease Branch team for the support with genome sequencing.
Funding
This project was funded by the Conselho Nacional de Ciência e Tecnologia (CNPq), under contract no. 460999/2014-1. B.M.B was supported by NIH/NIAID award R21AI28536. D.R.M and K. I. were supported by NIH/NIGMS award R01GM121750.
Footnotes
Declaration of Competing Interest
The authors declare no conflicts of interest.
Appendix A. Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fgb.2020.103395.
References
- Arantes TD, et al. , 2016. Environmental Mapping of Paracoccidioides spp. in Brazil Reveals New Clues into Genetic Diversity, Biogeography and Wild Host Association. PLoS Negl. Trop. Dis 10, e0004606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo P, et al. , 2016. Paracoccidioidomicosis detectados en el período 2004–2013 en el Laboratorio Central de Salud Pública de Asunción - Paraguay. Revista del Nacional 8, 62–71. [Google Scholar]
- Bicalho RN, et al. , 2001. Oral paracoccidioidomycosis: a retrospective study of 62 Brazilian patients. Oral Dis. 7, 56–60. [PubMed] [Google Scholar]
- Bolger AM, et al. , 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasier C, 2000. The rise of the hybrid fungi. Nature 405, 134–135. [DOI] [PubMed] [Google Scholar]
- Bryce EA, et al. , 1996. An outbreak of cutaneous aspergillosis in a tertiary-care hospital. Infect. Control Hosp. Epidemiol 17, 170–172. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA, 2004. Speciation. Sinauer Associates, Sunderland, Mass. [Google Scholar]
- Danecek P, et al. , 2011. The variant call format and VCFtools. Bioinformatics 27, 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davel G, Canteros CE, 2007. Situación de las micosis en la República Argentina. Rev. Argent. Microbiol 39, 6. [PubMed] [Google Scholar]
- Della Terra PP, et al. , 2017. Exploring virulence and immunogenicity in the emerging pathogen Sporothrix brasiliensis. PLoS Negl. Trop Dis 11, e0005903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, et al. , 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet 43, 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser JA, et al. , 2005. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature 437, 1360–1364. [DOI] [PubMed] [Google Scholar]
- Giusiano G, et al. , 2018a. Emergence of acute/subacute infant-juvenile paracoccidioidomycosis in Northeast Argentina: effect of climatic and anthropogenic changes? Med. Mycol [DOI] [PubMed] [Google Scholar]
- Giusiano G, et al. , 2018b. The Southern Endemic Zone of Paracoccidioidomycosis: epidemiological Approach in Northeast Argentina. Curr. Fungal Infect. Rep 12, 138–143. [Google Scholar]
- Hagen F, et al. , 2015. Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fung. Genet. Biol 78, 16–48. [DOI] [PubMed] [Google Scholar]
- Hibbins MS, Hahn MW, 2019. The timing and direction of introgression under the multispecies network coalescent. Genetics 211, 1059–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrycyk MF et al. , 2018. Ecology of Paracoccidioides brasiliensis, P. lutzii and related species: infection in armadillos, soil occurrence and mycological aspects. Med. Mycol [DOI] [PubMed] [Google Scholar]
- Jombart T, Ahmed I, 2011. adegenet 1.3–1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27, 3070–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, et al. , 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leducq JB, et al. , 2016. Speciation driven by hybridization and chromosomal plasticity in a wild yeast. Nat. Microbiol 1, 15003. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R, 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. , 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman ZB, Zamir D, 2007. Heterosis: revisiting the magic. Trends Genet. 23, 60–66. [DOI] [PubMed] [Google Scholar]
- Macoris S, et al. , 2006. Virulence attenuation and phenotypic variation of Paracoccidioides brasiliensis isolates obtained from armadillos and patients. Memórias do Instituto Oswaldo Cruz. 101, 331–334. [DOI] [PubMed] [Google Scholar]
- Mangiaterra M, et al. , 1996. Histoplasmin and paracoccidioidin skin reactivity in infantile population of northern Argentina (1). Rev Inst Med Trop Sao Paulo. 38, 349–353. [DOI] [PubMed] [Google Scholar]
- Martinez R, 2017. New trends in paracoccidioidomycosis epidemiology. J. Fungi 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matute DR, et al. , 2006. Cryptic speciation and recombination in the fungus Paracoccidioides brasiliensis as revealed by gene genealogies. Mol. Biol. Evol 23, 65–73. [DOI] [PubMed] [Google Scholar]
- Matute DR, Sepulveda VE, 2019. Fungal species boundaries in the genomics era. Fung. Genet Biol 131, 103249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell CS, et al. , 2018. Recent admixture between species of the fungal pathogen Histoplasma. Evol. Lett 2, 210–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, et al. , 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh BQ, et al. , 2013. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol 30, 1188–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mixao V, Gabaldon T, 2018. Hybridization and emergence of virulence in opportunistic human yeast pathogens. Yeast. 35, 5–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari-Madlum EE, et al. , 1999. Virulence of Paracoccidioides brasiliensis isolates can be correlated to groups defined by random amplified polymorphic DNA analysis. Med. Mycol 37, 269–276. [PubMed] [Google Scholar]
- Munoz JF, et al. , 2016. Genome Diversity, Recombination, and Virulence across the Major Lineages of Paracoccidioides. mSphere. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, et al. , 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol 32, 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Pritchard JK, 2012. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8, e1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado M, et al. , 2009. Mortality due to systemic mycoses as a primary cause of death or in association with AIDS in Brazil: a review from 1996 to 2006. Mem. Inst. Oswaldo Cruz 104, 513–521. [DOI] [PubMed] [Google Scholar]
- Raj A, et al. , 2014. fastSTRUCTURE: variational inference of population structure in large SNP data sets. Genetics 197, 573–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo A, et al. , 2001. The habitat of Paracoccidioides brasiliensis: how far from solving the riddle? Med. Mycol 39, 233–241. [DOI] [PubMed] [Google Scholar]
- Restrepo A, et al. , 1968. Distribution of paracoccidioidin sensitivity in Colombia. Am. J. Trop. Med. Hyg 17, 25–37. [PubMed] [Google Scholar]
- Rodrigues MT, de Resende MA, 1996. Epidemiologic skin test survey of sensitivity to paracoccidioidin, histoplasmin and sporotrichin among gold mine workers of Morro Velho Mining, Brazil. Mycopathologia 135, 89–98. [DOI] [PubMed] [Google Scholar]
- Rolón PA, 2004. Paracoccidioidomicosis — Epidemiología en la República del Paraguay. Centro de Sud América. Mycopathologia. 59, 14. [DOI] [PubMed] [Google Scholar]
- Sahl JW, et al. , 2016. NASP: an accurate, rapid method for the identification of SNPs in WGS datasets that supports flexible input and output formats. Microb. Genom 2, e000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulveda VE, et al. , 2014. Comparison of phylogenetically distinct Histoplasma strains reveals evolutionarily divergent virulence strategies. MBio 5, e01376–e1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai-Yasuda MA, et al. , 2017. Brazilian guidelines for the clinical management of paracoccidioidomycosis. Rev. Soc. Bras. Med. Trop [DOI] [PubMed] [Google Scholar]
- Teixeira Mde M, et al. , 2015. Asexual propagation of a virulent clone complex in a human and feline outbreak of sporotrichosis. Eukaryot Cell. 14, 158–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira Mde M, et al. , 2014. Paracoccidioides lutzii sp. nov.: biological and clinical implications. Med. Mycol 52, 19–28. [DOI] [PubMed] [Google Scholar]
- Teixeira MM, et al. , 2009. Phylogenetic analysis reveals a high level of speciation in the Paracoccidioides genus. Mol. Phylogenet. Evol 52, 273–283. [DOI] [PubMed] [Google Scholar]
- Teixeira MM, et al. , 2014. Paracoccidioides species complex: ecology, phylogeny, sexual reproduction, and virulence. PLoS Pathog. 10, e1004397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracogna MF, et al. , 2018. Clinical and epidemiological characteristics of patients with paracoccidioidomycosis diagnosed in a hospital of Resistencia, Chaco. Rev. Argent. Microbiol [DOI] [PubMed] [Google Scholar]
- Turissini DA, et al. , 2017. Species boundaries in the human pathogen Paracoccidioides. Fung. Genet. Biol 106, 9–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gelderen de Komaid A, et al. , 1999. Histoplasmosis and Paracoccidioidomycosis in northwestern Argentina III. Epidemiological survey in Vipos, La Toma, and Choromoro - Trancas, Tucuman, Argentina. Eur J Epidemiol 15, 383–388. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We sequenced thirty-one new Paracoccidioides genomes from Argentina, Bolivia, Brazil, Paraguay, Peru and Venezuela (Table 1), which increases the total of publicly available Paracoccidioides genomes to sixty-three. The raw Illumina reads for all the sequenced genomes are available at the Sequence Repository Archive under the following: SRASRR9736748-SRR9736778 (Technical Appendix II).