Abstract
Objective
Although severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA was detected in faeces of patients with COVID-19, the activity and infectivity of the virus in the GI tract during disease course is largely unknown. We investigated temporal transcriptional activity of SARS-CoV-2 and its association with longitudinal faecal microbiome alterations in patients with COVID-19.
Design
We performed RNA shotgun metagenomics sequencing on serial faecal viral extractions from 15 hospitalised patients with COVID-19. Sequencing coverage of the SARS-CoV-2 genome was quantified. We assessed faecal microbiome composition and microbiome functionality in association with signatures of faecal SARS-CoV-2 infectivity.
Results
Seven (46.7%) of 15 patients with COVID-19 had stool positivity for SARS-CoV-2 by viral RNA metagenomic sequencing. Even in the absence of GI manifestations, all seven patients showed strikingly higher coverage (p=0.0261) and density (p=0.0094) of the 3’ vs 5’ end of SARS-CoV-2 genome in their faecal viral metagenome profile. Faecal viral metagenome of three patients continued to display active viral infection signature (higher 3’ vs 5’ end coverage) up to 6 days after clearance of SARS-CoV-2 from respiratory samples. Faecal samples with signature of high SARS-CoV-2 infectivity had higher abundances of bacterial species Collinsella aerofaciens, Collinsella tanakaei, Streptococcus infantis, Morganella morganii, and higher functional capacity for nucleotide de novo biosynthesis, amino acid biosynthesis and glycolysis, whereas faecal samples with signature of low-to-none SARS-CoV-2 infectivity had higher abundances of short-chain fatty acid producing bacteria, Parabacteroides merdae, Bacteroides stercoris, Alistipes onderdonkii and Lachnospiraceae bacterium 1_1_57FAA.
Conclusion
This pilot study provides evidence for active and prolonged ‘quiescent’ GI infection even in the absence of GI manifestations and after recovery from respiratory infection of SARS-CoV-2. Gut microbiota of patients with active SARS-CoV-2 GI infection was characterised by enrichment of opportunistic pathogens, loss of salutary bacteria and increased functional capacity for nucleotide and amino acid biosynthesis and carbohydrate metabolism.
Keywords: gut inflammation, infectious disease, diagnostic virology
Significance of this study.
What is already known on this subject?
GI symptoms are present in a substantial proportion of patients with COVID-19.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA has been detected and remained positive in the faecal samples of some patients with COVID-19 even after respiratory specimens were negative for viral RNA.
In vitro transcriptional analysis on SARS-CoV-2-infected cell model showed that the 3’ end of SARS-CoV-2 genome was substantially highly covered than the 5’ end indicating a signature of active viral replication and infection.
What are the new findings?
We found for the first time a signature of active gut viral infection in a subset (47%) of patients with COVID-19 even in the absence of GI symptoms, suggesting ‘quiescent’ GI infection of SARS-CoV-2.
The transcriptional activity of viral infection and replication persisted in the gut even after respiratory clearance of SARS-CoV-2.
Faecal samples with a signature of high SARS-CoV-2 infectivity harboured a higher abundance of opportunistic pathogens, Collinsella aerofaciens, Collinsella tanakaei, Streptococcus infantis, Morganella morganii and an enhanced capacity for biosynthesis of nucleotide and amino acid and carbohydrate metabolism (glycolysis), whereas faecal samples with a signature of low-to-none SARS-CoV-2 infectivity had a higher abundance of short-chain fatty acid producing bacteria, Parabacteroides merdae, Bacteroides stercoris, Alistipes onderdonkii and Lachnospiraceae bacterium 1_1_57FAA.
Significance of this study.
How might it impact on clinical practice in the foreseeable future?
Active and prolonged SARS-CoV-2 activity in the gut of patients with COVID-19, even in the absence of GI manifestations and after recovery highlights the importance of long-term coronavirus and health surveillance and the threat of potential faecal-oral viral transmission.
Therapeutic approaches including nullifying gut SARS-CoV-2 activity and modulating gut microbiome composition and functionality should be explored.
Introduction
COVID-19, an acute respiratory illness caused by novel coronavirus (severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)), is characterised by active virus replication in the upper respiratory tract.1 Early reports from Wuhan showed that 2%–10% of patients with COVID-19 had GI symptoms including diarrhoea, but a recent meta-analysis reported that up to 20% had GI symptoms.2–5 Moreover, faecal calprotectin, an indicator of inflammatory responses in the gut, was found to be elevated in patients with COVID-19 with diarrhoea.6 These evidence suggest that the digestive tract might be an extrapulmonary site for SARS-CoV-2 infection in patients with COVID-19 with GI manifestations. However, viral activity and infectivity of SARS-CoV-2 in the GI tract of patients with COVID-19 during disease course and after disease resolution is largely unknown.
SARS-CoV-2 RNA has been detected in anal swabs and stool samples based on RT-PCR in a substantial proportion of patients with COVID-19.1 7 In addition, viral particles of SARS-CoV-2 were observed from faeces of patients with COVID-19 through electron microscopy.8 The presence of SARS-CoV-2 RNA in faeces can last longer after respiratory specimens became viral negative in a subset of patients with COVID-197 9. However, it is unknown about the activity of SARS-CoV-2 virus in the gut of those patients with COVID-19 with yet faecal positivity for SARS-CoV-2 after respiratory clearance of SARS-CoV-2. Currently, the viral activity of SARS-CoV-2 in the gut was mostly extrapolated from indirect observations of findings based on intestinal organoid and mammalian cell models: (1) SARS-CoV-2 receptor, ACE2, is highly expressed in intestinal enterocytes and colonocytes10 11 and (2) SARS-CoV-2 infects enterocyte lineage cells in human and bat intestinal organoids.12 13 To date, there is a lack of data of replication-competent and infection-competent SARS-CoV-2 virus in the human gut. Deeper understanding of the life cycle and pathogenicity of SARS-CoV-2 in the human gut is an urgent unmet need.
In this pilot observational study, we postulate that SARS-CoV-2 is active in the gut of patients with COVID-19, and therefore depicted the temporal transcriptional activity and infectivity of SARS-CoV-2 in the gut of hospitalised patients with COVID-19 both during disease course and after disease clearance. We prospectively included 15 hospitalised patients with COVID-19 admitted between 16 February 2020 and 2 March 2020 in Hong Kong, China, followed from hospital admission until discharge. Faecal viral RNA was extracted, followed by shotgun metagenomics sequencing and profiling to investigate SARS-CoV-2 transcriptional activity.
Methods
Study subject and design
This prospective study involved 15 patients with COVID-19 hospitalised with laboratory-confirmed SARS-CoV-2 infection (table 1). SARS-CoV-2 infection was confirmed by two consecutive RT-PCR tests targeting different regions of the RdRp gene performed by the local hospitals and Public Health Laboratory Service (Hong Kong, China). All patients with COVID-19 were admitted to the Prince of Wales Hospital or the United Christian Hospital, Hong Kong, between 5 February 2020 and 17 March 2020. They were followed until discharged from hospital or until 4 April 2020. All patients provided informed consent to participate in this study and agreed for publication of the research results. Data including demographic, epidemiological, clinical and laboratory results were extracted from the electronic medical records in Hong Kong Hospital Authority clinical management system. Faecal samples from patients with COVID-19 were collected serially until discharge.
Table 1.
Clinical characteristics of subjects with COVID-19
| Patients with COVID-19 | Disease severity | Age | Sex | Comorbidities | Respiratory symptom | GI symptom | Chest X-ray findings |
| 1 | Critical | 65 | F | Hypertension, chronic hepatitis B carrier | Fever, cough, sputum | Nil | Bilateral lung infiltrates |
| 2 | Moderate | 55 | F | None | Fever, runny nose | Nil | Bilateral lung infiltrates |
| 3 | Critical | 42 | M | None | Fever, cough | Nil | Right lung infiltrates and right lower lobe collapse |
| 4 | Severe | 70 | M | Hyperlipidaemia, duodenal ulcer | Sputum, shortness of breath | Nil | Bilateral lung infiltrates |
| 5 | Moderate | 58 | M | None | Fever, cough | Diarrhoea | Right lung infiltrates |
| 6 | Severe | 71 | M | None | Fever, cough, shortness of breath | Nil | Bilateral lung infiltrates |
| 7 | Moderate | 48 | M | Diabetes, hypertension, hyperlipidaemia | Fever, cough | Nil | Left lung infiltrates |
| 8 | Moderate | 38 | F | None | Fever, cough, sputum, runny nose | Nil | Bilateral lung infiltrates |
| 9 | Mild | 33 | M | None | Fever, cough | Nil | Nil |
| 10 | Moderate | 70 | F | Obesity, hypertension | Cough | Nil | Bilateral lung infiltrates |
| 11 | Severe | 62 | M | Diabetes, hyperlipidaemia, left subclavian artery occlusion | Fever, cough, sputum, shortness of breath | Nil | Bilateral lung infiltrates |
| 12 | Moderate | 71 | F | Hypertension, renal impairment, hyperlipidaemia | Cough | Nil | Bilateral lung infiltrates |
| 13 | Moderate | 47 | F | None | Cough | Nil | Bilateral lung infiltrates |
| 14 | Moderate | 22 | F | None | Fever, runny nose | Nil | Bilateral lung infiltrates |
| 15 | Mild | 46 | F | None | Cough, shortness of breath | Nil | None |
Faecal viral RNA extraction and shotgun metagenomics sequencing
The total viral nucleic acid was extracted from faecal samples, using TaKaRa MiniBEST Viral RNA/DNA Extraction Kit (Takara, Japan) following manufacturer’s instructions. Then extracted total viral nucleic acid was then purified by RNA Clean & Concentrator Kits (Zymo Research, California, USA) to obtain viral RNA. After quality control procedures via Qubit 2.0, agarose gel electrophoresis and Agilent 2100 testing, the qualified RNA was subject to library preparation using KAPA RNA HyperPrep Kit (Illumina, USA). The library preparations were then sequenced on Illumina NextSeq 550 platform (150 bp paired end).
Raw sequence reads were filtered and quality-trimmed using Trimmomatic V.0.3614 as follows: 1) trimming low-quality base (quality score <20); 2) removing reads shorter than 50 bp; 3) tracing and cutting off sequencing adapters. Contaminating human reads were filtering using Kneaddata V.0.5.1 (reference database: GRChg38) with default parameters.
Coverage and density of SARS-CoV-2 genome by metagenomics sequencing
A total of 1666 complete genomes were downloaded from the SAS-CoV-2 genome repository at National Center for Biotechnology Information as of 20 April 2020. Clean faecal viral metagenomic reads were queried against the customised SARS-CoV-2 reference genomes by BBMap V.38.81.15 Mapped reads were manually checked, particularly those mapped to the 3’ end of the SARS-CoV-2 genome, and reads ended with Poly A were discarded.
The 3’ end of the SARS-CoV-2 genome was defined as the genomic region starting from the 25 000 nucleotide base through the full-length end of SARS-CoV-2 genome (~29 9000 nucleotide base), while the remaining genomic region (0–25 000 nucleotide base) was deemed as 5’ end of the SARS-CoV-2 genome. Sequencing coverage was defined as the number of shotgun reads mapped to a given genomic region (per 100 nucleotide base of the SARS-CoV-2 genome). Sequencing density was defined as the frequency of sequenced sites in a given genomic region of SARS-CoV-2 genome (number of metagenomic hits of SARS-CoV-2 per 500 nucleotide base).
Faecal DNA extraction, metagenomics sequencing and bacterial microbiome taxonomic and functional profiling
Approximately 0.1 g faecal sample was prewashed with 1 mL ddH2O and pelleted by centrifugation at 13 000× g for 1 min. The faecal DNA was subsequently extracted from the pellet using Maxwell RSC PureFood GMO and Authentication Kit (Promega, Madison, Wisconsin, USA) following manufacturer’s instructions. Briefly, faecal pellet was added 1 mL of cetyltrimethylammonium bromide (CTAB) buffer and vortexed for 30 s, then heating sample at 95°C for 5 min. After that, the samples were vortexed thoroughly with beads at maximum speed for 15 min. Then 40 µL of proteinase K and 20 µL of RNase A was added into sample and the mixture was incubated at 70°C for 10 min. The supernatant was then obtained by centrifuging at 13 000× g for 5 min and was added in Maxwell RSC machine for DNA extraction. Extracted DNA was subjected to DNA libraries construction, completed through the processes of end repairing, adding A to tails, purification and PCR amplification, using Nextera DNA Flex Library Preparation kit (Illumina). Libraries were subsequently sequenced on our in-house sequencer Illumina NextSeq 550 (150 bp paired-end) at Center for Microbiota Research, The Chinese University of Hong Kong. The ratio of human reads in the faecal metagenome was generated from aligning reads to the reference genome (human release 32, GRCh38.p13, downloaded from Gencode) by Bowtie 2.16
Raw sequence reads were filtered and quality-trimmed using Trimmomatic V.0.3614 as follows: (1) trimming low-quality base (quality score <20); (2) removing reads shorter than 50 bp; 3) removing sequencing adapters. Contaminating human reads were filtering using Kneaddata (reference database: GRCh38.p13) with default parameters. Taxonomic profiling of faecal bacterial communities was performed using MetaPhlAn2 (V 2.9) by mapping reads to clade-specific markers.17 Functional profiling of faecal bacterial communities was performed using HUMAnN2.0.18 Differential bacterial species between faeces with high SARS-CoV-2 infectivity and faeces with low-to-none SARS-CoV-2 infectivity were identified by LefSE across all time-point stools corresponding to respective patients with COVID-19. Only species with linear discriminant analysis(LDA) effect size >2 and false discovery rate (FDR)-corrected p value <0.05 were plotted. High SARS-CoV-2 infectivity was defined as higher 3’ vs 5’ end coverage of SARS-CoV-2 genome in faecal viral RNA metagenome. Low-to-none SARS-CoV-2 infectivity was defined as similar 3’ and 5’ end coverage or no coverage of the SARS-CoV-2 genome in faecal viral RNA metagenome.
Results
Patients with COVID-19
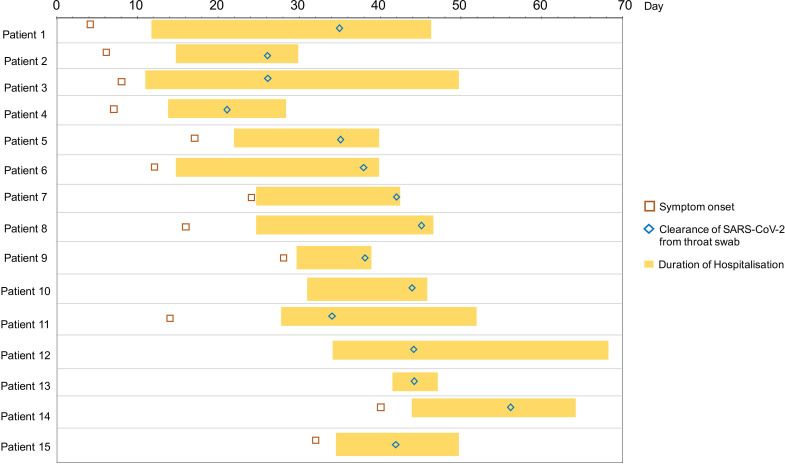
Fifteen hospitalised patients with COVID-19 with laboratory-confirmed SARS-CoV-2 infection were recruited and followed up (figure 1, table 1). Of the 15 patients, 7 were male. The median age of hospitalised patients with COVID-19 was 55 years (IQR: 44–67). Eleven patients had moderate-to-severe COVID-19 and two had critical disease with admission to intensive care unit. All patients presented with respiratory symptoms but only one had concurrent GI symptom of diarrhoea at presentation (patient 5). None of the patients developed GI symptoms during hospitalisation. Serial stool samples were collected from each patient until nasopharyngeal or throat swab tests were negative on two consecutive samples on which patient was discharged. Median duration of hospitalisation was 21±2.4 days (mean±SE).
Figure 1.
Timeline of patient symptom onset, hospitalisation, throat (nasopharyngeal) swab clearance of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and discharge, through the course of disease for 15 patients hospitalised with COVID-19.
Depiction of temporal faecal viral activity of SARS-CoV-2 in patients with COVID-19
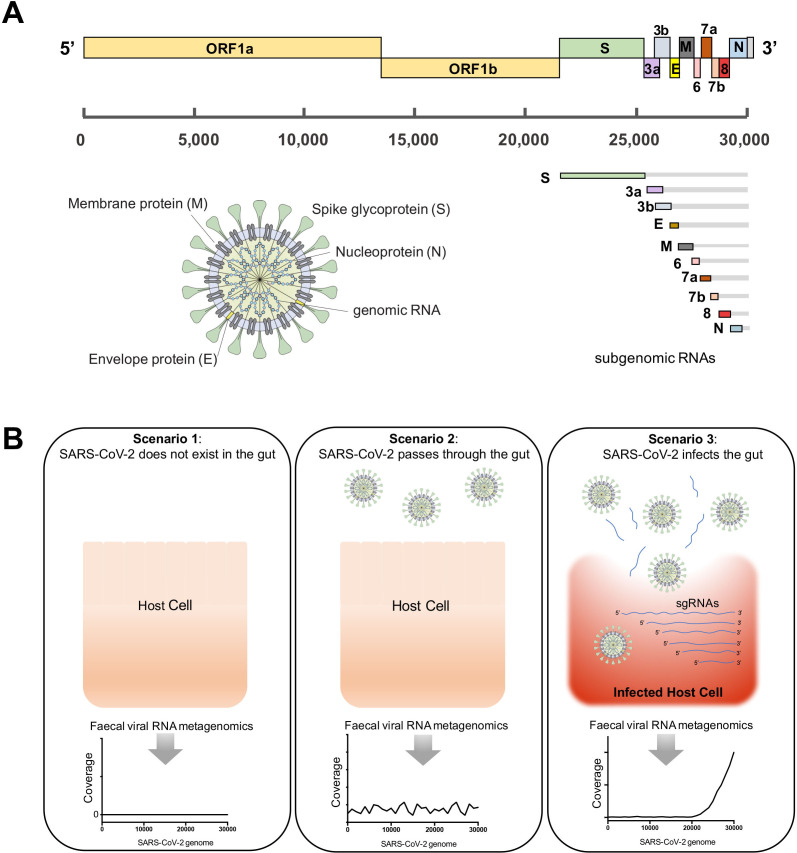
SARS-CoV-2 has a genome size of ~29.9 kb nucleotides (figure 2A).19 20 After entering into the host cell, the full-length genomic RNA that also serves as an mRNA, is translated to ORF1a and ORF1b (5’ end of the SARS-CoV-2 genome). In addition to the full genomic RNA, nine major subgenomic RNAs (sgRNAs), which encode for structural proteins, S, E, M and N, are produced (3’ end of the SARS-CoV-2 genome).20 A recent study demonstrated that active viral replication and transcription of SARS-CoV-2 resulted in substantially higher sequencing coverage of the 3’ end versus the 5’ end of the SARS-CoV-2 genome in an infected cell model (Vero cells) via high-throughput transcriptomic sequencing.20 Such high transcriptional expression of the 3’-end sgRNAs is a signature of active replication and infection of SARS-CoV-2 in host cells.
Figure 2.
Hypothetical scenarios for the fecal viral RNA metagenomic profile of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in association with its infectivity in gut. (A) Schematic presentation of the full-length genome, transcribed subgenomic RNAs (sgRNA) and the virion structure of SARS-CoV-2 virus. (B) Three scenarios hypothesised for the presence and infectivity of SARS-CoV-2 in the gut of patients with COVID-19, and the detection of SARS-CoV-2 virus by faecal viral RNA metegenomics sequencing. If SARS-CoV-2 virus infects the host cells in the gut, its genomic and sgRNAs should be highly expressed and released into the gut lumen on cytolysis, where the 3’ end of SARS-CoV-2 genome should be highly covered by faecal viral RNA metagenomics sequencing.
Based on these transcriptomic patterns of replicative SARS-CoV-2, we hypothesised three potential scenarios of the existence of SARS-CoV-2 virus in the human gut and its detection by faecal viral RNA metagenomics (transcriptomics): (i) scenario 1: if SARS-CoV-2 does not exist in the gut, its RNA genome cannot be detected through metagenomics sequencing; (ii) scenario 2: if free SARS-CoV-2 virions pass through the gut without infecting host cells, the full-length viral RNA genome should be equally covered by metagenomics reads; (iii) scenario 3: if SARS-CoV-2 virus exists in the gut and infects host cells, the 3’ end regions of the viral RNA genome should be more highly covered than the 5’ end genomic regions by metagenomics reads (figure 2B).
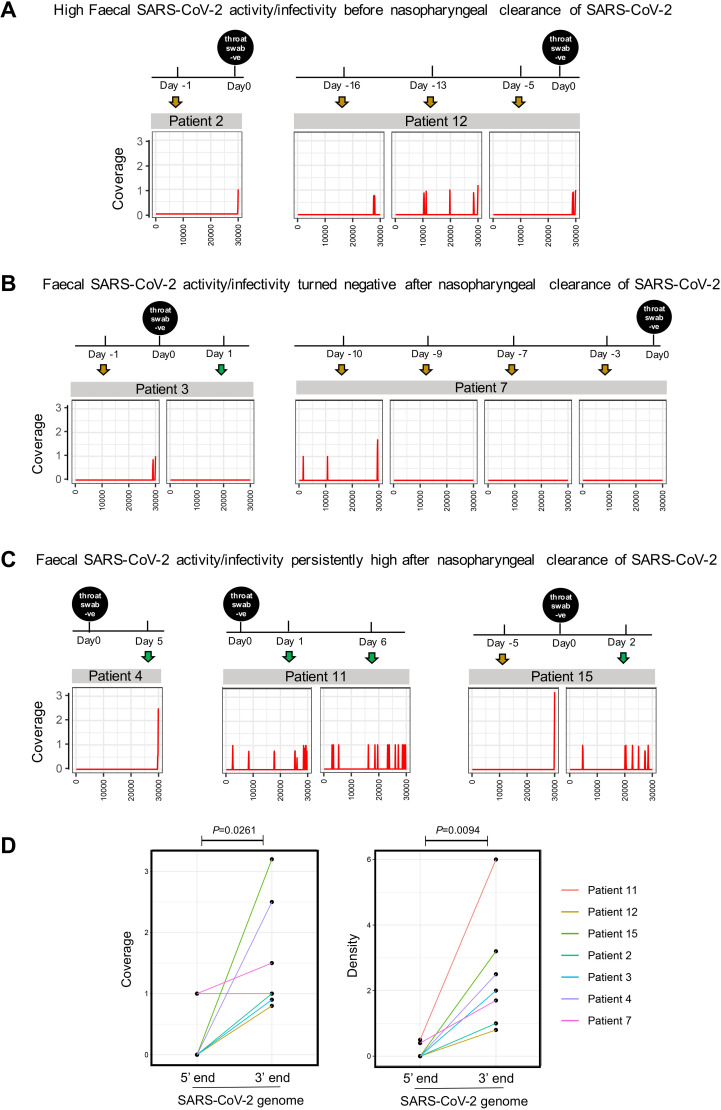
To depict the viral transcriptional activity of SARS-CoV-2 in the gut of patients with COVID-19, we performed faecal viral RNA shotgun metagenomics sequencing on chronological series of faecal samples. Consequently, we obtained 18 773 883±420 617 (mean±SE) reads on average from serial faecal viral RNA preparations of 15 patients with COVID-19. Seven patients were detected positive for SARS-CoV-2 in the faeces at baseline (sampling date of the first stool after hospitalisation), by metagenomics sequencing (figure 3), indicating the presence of SARS-CoV-2 in the gut of a subset of patients with COVID-19. None of these subjects had GI symptoms. Metagenomics sequencing did not detect SARS-CoV-2 in faeces of the other eight patients (online supplementary Figure 1). Although our sequence depth did not allow us to identify the full-length genome of SARS-CoV-2 in faecal samples, interestingly we observed that all seven patients (metagenome positive for SARS-CoV-2) displayed higher coverage and density of the 3’ end region than the 5’ end region of the SARS-CoV-2 genome, at baseline (p=0.0261 and 0.0094, respectively, figure 3D). Patient 12 with moderate COVID-19 and patient 15 with mild COVID-19 (normal chest findings) did not display GI symptoms, yet both showed consistent 3’ end coverage of the SARS-CoV-2 genome in faecal viral metagenome across all time points during the disease course (figure 3A and C). These findings suggest persistent existence of SARS-CoV-2 in the patients’ gut. Given that higher 3’ end versus 5’ end coverage of SARS-CoV-2 genome is a signature of active viral replication and transcription (infection) in host cells,20 our data underscore ongoing active SARS-CoV-2 infection within the gut of patients with COVID-19 despite the absence of GI symptoms and mild COVID-19. In contrast, patients 3 and 7 lost this signature of active viral infection, 1 day after and 10 days before nasopharyngeal clearance of SARS-CoV-2, respectively (figure 3B), suggesting discordance in the timeline of eliminating gut and respiratory SARS-CoV-2 infection among patients with COVID-19 during the disease course.
Figure 3.
Depiction of the viral infectivity of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in serial faeces of patients with COVID-19. Infectivity of SARS-CoV-2 virus in gut was investigated by faecal viral RNA metagenomics coverage profile of SARS-CoV-2 genome. (A) The subset of patients who manifested higher 3’ vs 5’ end coverage of the SARS-CoV-2 genome (signature of active SARS-CoV-2 infectivity) before throat swab turned negative for SARS-CoV-2. (B) The subset of patients who manifested higher 3’ vs 5’ end coverage of the SARS-CoV-2 genome (signature of SARS-CoV-2 infectivity) but gradually lost this signature over time of hospitalisation before throat swab turned negative for SARS-CoV-2. (C) The subset of patients who manifested higher 3’ vs 5’ end coverage of the SARS-CoV-2 genome (signature of SARS-CoV-2 infectivity) after throat swab turned negative for SARS-CoV-2. ‘Day 0’ is defined as the date when throat swab turned negative for SARS-CoV-2, as measured by RT-PCR. (D) The coverage and density of the 5’ and 3’ ends of the SARS-CoV-2 genome in COVID-19 faecal viral RNA metagenome. The baseline (the date of first stool collection after hospitalisation) faecal viral RNA metagenomes of the seven patients who were detected faecal positive for SARS-CoV-2 were plotted and subject to comparison. Coverage was defined as the number of shotgun reads mapped to a given genomic region of SARS-CoV-2 genome. Density was defined as the frequency of sequenced sites in a given genomic region of SARS-CoV-2 genome.
gutjnl-2020-322294supp001.pdf (1.6MB, pdf)
Surprisingly, the faecal viral RNA metagenome of patients 4, 11 and 15 continued to display active viral infection signature (higher 3’ vs 5’ end coverage and density of SARS-CoV-2 genome) (figure 3C), at time points ranging from 1 to 6 days after clearance of SARS-CoV-2 from throat swab. In patients 11 and 15, the coverage and density of 5’ end resembled that of the 3’ end in the faecal viral metagenome, after throat swab turned negative (exemplified at days 1 and 6 for patient 11, and day 2 for patient 15, figure 3C). These observations indicate a gradual loss of active viral infection (shifting from scenario 3 to scenario 2) in the gut of these patients after clearance of SARS-CoV-2 from the airway. Our data suggest that SARS-CoV-2 virus is persistently detectable and continued to display features of infectivity in the human gut despite clearance of SARS-CoV-2 from the airway but its transcriptional activity and infectivity may decline gradually afterwards. Such prolonged presence of active SARS-CoV-2 in faeces of patients without enteric involvement as well as recovered patients highlight a potential for faecal-oral transmission.
Faecal microbiome features associated with faecal viral activity of SARS-CoV-2
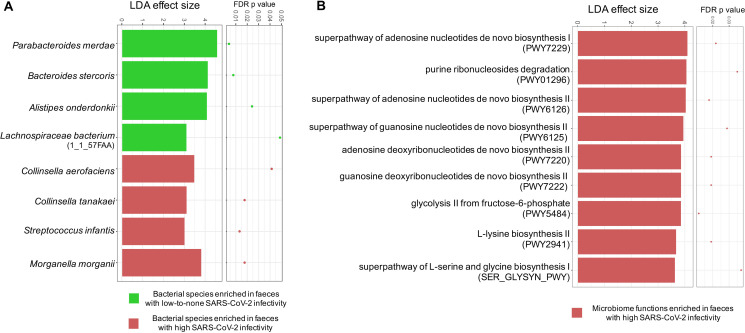
We then investigated the gut microbiome difference between faecal samples with a signature of high SARS-CoV-2 infectivity and those with a signature of low-to-none SARS-CoV-2 infectivity, to gain insights into the interactions between SARS-CoV-2, microbiome and the host. We performed faecal microbiome metagenomic sequencing and compared the bacterial microbiome composition between faeces with high SARS-CoV-2 infectivity and those with low-to-none SARS-CoV-2 infectivity, across all time points in the 15 patients with COVID-19. We found that faecal samples with high SARS-CoV-2 infectivity had a higher abundance of the bacterial species Collinsella aerofaciens, Collinsella tanakaei, Streptococcus infantis, Morganella morganii, compared with samples with low-to-none SARS-CoV-2 infectivity (LefSE analysis, LDA effect size >2, FDR p value <0.05, figure 4A). Among these species, C. aerofaciens and M. morganii have been associated with opportunistic human infections.21 22 S. infantis was an abundant coloniser in the upper respiratory tract and oral cavity.23 Host (human) DNA ratio in the faecal metagenome did not differ between faeces with high SARS-CoV-2 infectivity and those with low-to-none SARS-CoV-2 infectivity (online supplementary figure 2). These tdata together suggest that SARS-CoV-2 actively infected the gut and may pose additional threat to the host.
Figure 4.
Differential bacterial species and functional capacities between faeces with high severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infectivity and faeces with low-to-none SARS-CoV-2 infectivity. Differential bacterial species (A) and functionality (B) were identified via LefSE analysis across all time-point stools of 15 patients with COVID-19. Only species and functional modules with LDA effect size >2 and FDR-corrected p value <0.05 were plotted. High SARS-CoV-2 infectivity was defined as higher 3’ vs 5’ end coverage of SARS-CoV-2 genome in faecal viral RNA metagenome. Low-to-none SARS-CoV-2 infectivity was defined as similar 3’ and 5’ end coverage or no coverage of the SARS-CoV-2 genome in faecal viral RNA metagenome.
gutjnl-2020-322294supp002.pdf (381.5KB, pdf)
In contrast, faecal samples with low-to-none SARS-CoV-2 infectivity had higher abundances of Parabacteroides merdae, Bacteroides stercoris, Alistipes onderdonkii and Lachnspiraceae bacterium 1_1_57FAA, compared with those with high SARS-CoV-2 infectivity (LefSE analysis, LDA effect size >2, FDR p value <0.05, figure 4A). Bacteria members from Parabacteroides, Bacteroides and Lachnospiraceae are known producers of short chain fatty acids (particularly butyrate), which play crucial roles in boosting host immunity.24–27 Alistipes species are indole positive, involved in the serotonin precursor tryptophan metabolism and in maintaining gut immune homeostasis.28 29 In addition, a high abundance of B. stercoris was also marginally associated with low faecal amount of SARS-CoV-2 virus (Spearman’s correlation coefficient rho=−0.26, p=0.06). Our data highlight a potential beneficial role of these intestine-resident salutary bacteria in combating SARS-CoV-2 infection in the gut.
We further explored the difference in faecal microbiome functionality between faeces with a signature of high SARS-CoV-2 infectivity and those with a signature of low-to-none SARS-CoV-2 infectivity. We found that faecal samples with high SARS-CoV-2 infectivity had a higher abundance of functional pathways involved in nucleotide metabolism (superpathway of adenosine nucleotides de novo biosynthesis I, purine ribonucleosides degradation, superpathway of adenosine nucleotides de novo biosynthesis II, superpathway of guanosine nucleotides de novo biosynthesis II, adenosine deoxyribonucleotides de novo biosynthesis II and guanosine deoxyribonucleotides de novo biosynthesis II), carbohydrate metabolism (glycolysis II from fructose-6-phosphate) and amino acid biosynthesis (L-lysine biosynthesis II and superpathway of L-serine and glycine biogenesis), compared with samples with low-to-none SARS-CoV-2 infectivity (LefSE analysis, LDA effect size >2, FDR p value <0.05, figure 4B). The increased nucleotide and amino acid biosynthesis in gut microbiome functionality indicate potential enhanced production of bacterial cellular building blocks for macromolecules, while the increased glycolic activity indicates enhanced energy extraction in bacteria, all essential life activities for gut bacteria under immunopathological conditions. Such alterations in microbiome functionality in association with SARS-CoV-2 infectivity may underlie the pathogenesis and disease course of COVID-19, although the cause versus consequence relationship merits further investigation.
Longitudinal changes of faecal microbiome in association with faecal viral activity of SARS-CoV-2
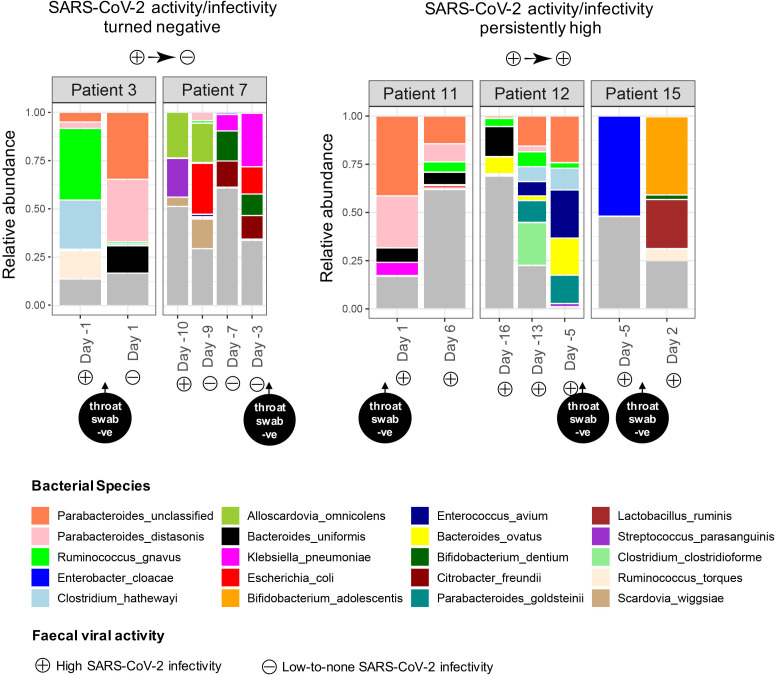
We then investigated longitudinal changes of faecal microbiome in association with clearance of active replicating SARS-CoV-2 virus from the faeces. We followed patients 3 and 7, who had serial stools displaying positive to negative faecal SARS-CoV-2 infectivity, and patients 11, 12 and 15, who had serial stools constantly displaying a signature of high viral infectivity. Overall, all patients showed substantial variations in faecal microbiome composition regardless of presence of faecal viral infectivity (figure 5), indicating an instable gut microbiome during the disease course of COVID-19. After the infectivity signature of SARS-CoV-2 turned negative in faeces, patient 3 had an expansion of the salutary bacteria, Parabacteroides distasonis and Bacteroides uniformis, and a contraction of inflammation-associated bacterium, Ruminococcus gnavus, in the faecal microbiome. However, patient 7 had a transient increase in P. distasonis accompanied by a substantial expansion of the bacterial pathogens, Klebsiella pneumoniae, Citrobacter koseri and Bifidobacterium dentium, following clearance of infective SARS-CoV-2 virus from faeces. Gut inflammation and nosocomial infection-associated bacteria, R. gnavus, Clostridium hathewayi and Enterococcus avium, were prevalent in patients’ faeces who had persistently high SARS-CoV-2 infectivity (figure 5). Our data suggest that although patients with COVID-19 may have amelioration in gut microbiome dysbiosis after faecal clearance of SARS-CoV-2 infectivity, secondary invasion of bacterial pathogens may become paramount during the disease course and clinical surveillance is likely to be warranted.
Figure 5.
Longitudal changes in the faecal microbiome of patients with COVID-19 in association with faecal severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infectivity. Patients 3 and 7 had serial stools displaying positive to negative faecal SARS-CoV-2 infectivity during follow-up, while patients 11, 12 and 15 had serial stools constantly displaying a signature high viral infectivity during follow-up. Only the most abundant 20 species were plotted and shown in relative abundance.
Discussion
We for the first time depicted the SARS-CoV-2 transcriptional activity in gut of patients with COVID-19 over time of hospitalisation. The remarkable active viral infectivity in the gut of patients with COVID-19, characterised by higher 3’ end versus 5’ end coverage of SARS-CoV-2 genome via broad-target shotgun viral metagenomic profiling, indicates active but quiescent GI infection of SARS-CoV-2, considering that most of our patients with COVID-19 did not present with GI symptoms. Such transcriptional feature of active viral infection and replication persisted in the gut even after respiratory clearance of SARS-CoV-2 in a proportion of patients with COVID-19. Our data suggest that there appears to be a lagging window period of up to 1 week, perhaps even longer, between clearance of respiratory infection and GI infection of SARS-CoV-2, in at least a subset of patients with COVID-19. Whether this postrecovery period may be a risk for transmission of the virus is unclear, but the life cycle of SARS-CoV-2 in the gut appears longer than anticipated. Our pilot observational study provides evidence for infective and prolonged SARS-CoV-2 viral activity in the gut of patients with COVID-19 even in the absence of GI manifestations.
Our prior study showed that 14 out of the 15 patients with COVID-19 (93%) were detected positive for SARS-CoV-2 in faecal samples by RT-PCR.9 In comparison, seven (47%) of them were detected positive for SARS-CoV-2 by faecal viral RNA metagenomics. Only the faeces which had an abundance of >3.2×104 copies per mL inoculum, as measured by RT-PCR, were detected by faecal viral RNA metagenomics. Broad-target sequencing of shotgun metagenomics may yield lower sensitivity than RT-PCR test in detection of targeted virus (SARS-CoV-2), however its upside is broader-spectrum detection of different regions of SARS-CoV-2 genome, which enables us to depict the viral transcriptional activity and infectivity. Although faecal RNA viral metagenomcis may underestimate the proportion of patients with COVID-19 who had presence of SARS-CoV-2 virus in the gut, we found a robust transcriptional signature of SARS-CoV-2 infectivity in faeces of patients with COVID-19 even without GI symptoms, indicating quiescent but active GI infection of SARS-CoV-2. Increasing metagenomics sequencing depth and throughput would identify more patients with active GI infection as well as more viral variants and mutants of SARS-CoV-2. It is possible that the detected viral RNA of SARS-CoV-2 might be residuals from swallowed viruses. However viral RNA was still detected up to 6 days after throat swab had turned negative for SARS-CoV-2 (figure 3C) suggesting that the detection of SARS-CoV-2 in faeces were unlikely to be an artefact. Overall, our data suggest that the human intestinal tract is an extrapulmonary infected site.
Opportunistic bacterial pathogens, Collinsella aerofaciens and Morganella morganii 21 22 were enriched in faecal samples of patients with COVID-19 who had high SARS-CoV-2 infectivity. Streptococcus infantis, an abundant coloniser in the upper respiratory tract and oral cavity,23 was also enriched in these patients’ faecal samples. Its presence suggests the passage or transmission of extraintestinal microbes into the gut in COVID-19 setting. In contrast, short-chain fatty acids and tryptophan producers,24–29 P. merdae, B. stercoris, A. onderdonkii and Lachnospiraceae bacterium 1_1_57FAA, were enriched in the faecal samples with a signature of low-to-none SARS-CoV-2 infectivity. In favour of this finding, our recent study also showed that a high abundance of A. onderdonkii and L. bacterium was associated with a less severe COVID-19.9 B. stercoris is a bacterial species from the phylum Bacteroidetes known to be associated with suppression of colonic expression of ACE2 (a host cell entry point for SARS-CoV-2) in murine model.30 These data highlight a potential beneficial role of salutary bacteria in combating SARS-CoV-2 infection, and a potential detrimental role of opportunistic pathogens in SARS-CoV-2 infection. However, whether or not these species could be harnessed for diagnostic purposes are unclear.
The faeces of high SARS-CoV-2 infectivity had a higher microbiome functional capacity for nucleotide de novo biosynthesis, amino acid biosynthesis and glycolysis. Among them, pathways for synthesising adenosine and guanosine were significantly enriched. Adenosine and guanosine are two important metabolites that are involved in purine nucleotide metabolism.31 Adenosine is physiologically present at low levels, while can rapidly increase in response to pathological conditions, such as hypoxia, ischaemia, inflammation or trauma, which functions as an ‘alarm’ or danger signal.32 Guanosine and its modified derivatives, 8-hydroxydeoxyguanosine (8-OHdG), are endogenous ligands for TLR7.33 Importantly, 8-OHdG is a well-known oxidative DNA damage marker and induced strong cytokine production.33 In addition, the bacterial biosynthesis pathway of L-serine was increased in the faeces of high SARS-CoV-2 infectivity. L-serine could fuel expansion of pathogenic bacteria in the inflamed gut.34 All these increased microbial functionalities in faecal microbiome are essential bioactivities for bacterial survival, growth and metabolism. Such bacterial functional enhancement could be a consequence of SARS-CoV-2 infectivity in the gut, or potentiate the disease course of COVID-19, which remain to be further studied.
Respiratory transmission is still the primary route for SARS-CoV-2 and it is postulated that SARS-CoV-2 may transmit through the faecal-oral route.35 36 Studies have shown that SARS-CoV-2 viruses released into the intestinal lumen were inactivated by colonic fluid and infectious virus was not recovered from the stool specimens of patients with COVID-19.1 37 Although we were not able to titrate or isolate live viruses from the faeces of patients with COVID-19 due to the methodogical limitation that we preemptively added viricidal agent to nullify SARS-CoV-2 virus when handling the faecal samples for safety concerns, the robust high faecal viral transcriptional signature of SARS-CoV-2 indicates its active infectivity in the gut of patients with COVID-19. In accordance, a recent study was able to isolate infectious SARS-CoV-2 virus from the stool specimen of a patient with COVID-19,13 further supporting our findings. Evidence is not yet sufficient to develop practical measures for suppressing viral spreading and infection in the group of patients with negative respiratory tract sample results but positive faecal samples. Further research into revealing the infectivity and pathogenesis of SARS-CoV-2 in the GI tract and measures to combat systemic including GI infection, such as modulation of human gut microbiome, are warranted.
Acknowledgments
The authors would like to thank all healthcare workers working in isolation wards of the Prince of Wales Hospital and the United Christian Hospital, Hong Kong, China. The authors would like to thank Joyce Mak, Apple CM Yeung, Wendy CS Ho, Miu L Chin, Rity Wong and Vickie Li for their contribution in this study. The authors would also like to thank Hui Hoy and Chow Sin Lan Charity Fund Limited, Pine and Crane Company Limited and Mr. Hui Ming for their financial support to this research.
Footnotes
TZ, QL and FZ contributed equally.
Contributors: TZ, QL and FZ performed the experiments, data analyses and drafted the manuscript. YKY, SSB, ZC, FKLC and PC revised the manuscript and provided critical comments. GC-YL and ET recruited patients and clinical data. TZ formulated the concept and hypothesis. SCN designed and supervised the study. TZ, QL and FZ contributed equally to this work.
Funding: This work was supported by The D. H. Chen Foundation and the Health and Medical Research Fund, Hong Kong.
Competing interests: None declared.
Patient consent for publication: Not required.
Ethics approval: This study was approved by the Joint Chinese University of Hong Kong–New Territories East Cluster Clinical Research Ethics Committees (Reference number: 2020.076). This study was conducted in accordance with the Declaration of Helsinki.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: Data are available on reasonable request.
References
- 1. Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020;581:465–9. 10.1038/s41586-020-2196-x [DOI] [PubMed] [Google Scholar]
- 2. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet 2020;395:497–506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 2020;395:507–13. 10.1016/S0140-6736(20)30211-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liang W, Feng Z, Rao S, et al. Diarrhoea may be underestimated: a missing link in 2019 novel coronavirus. Gut 2020;69:1141–3. 10.1136/gutjnl-2020-320832 [DOI] [PubMed] [Google Scholar]
- 5. Cheung KS, Hung IFN, Chan PPY, et al. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from a Hong Kong cohort: systematic review and meta-analysis. Gastroenterology 2020. 10.1053/j.gastro.2020.03.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Effenberger M, Grabherr F, Mayr L, et al. Faecal calprotectin indicates intestinal inflammation in COVID-19. Gut 2020;69:1543–4. 10.1136/gutjnl-2020-321388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xu Y, Li X, Zhu B, et al. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat Med 2020;26:502–5. 10.1038/s41591-020-0817-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang W, Xu Y, Gao R, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 2020. 10.1001/jama.2020.3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zuo T, Zhang F, Lui GCY, et al. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology 2020. 10.1053/j.gastro.2020.05.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang J, Zhao S, Liu M, et al. ACE2 expression by colonic epithelial cells is associated with viral infection immunity and energy metabolism. medRxiv 2020. [Google Scholar]
- 11. Xiao F, Tang M, Zheng X, et al. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology 2020;158:1831–3. 10.1053/j.gastro.2020.02.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lamers MM, Beumer J, van der Vaart J, et al. SARS-CoV-2 productively infects human gut enterocytes. Science 2020;369:50–4. 10.1126/science.abc1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou J, Li C, Liu X, et al. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat Med 2020;26:1077–83. 10.1038/s41591-020-0912-6 [DOI] [PubMed] [Google Scholar]
- 14. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114–20. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bushnell B. BBMap: a fast, accurate, splice-aware aligner. Berkeley, CA (United States): Lawrence Berkeley National Lab.(LBNL), 2014. [Google Scholar]
- 16. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9:357–9. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Segata N, Waldron L, Ballarini A, et al. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods 2012;9:811–4. 10.1038/nmeth.2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Franzosa EA, McIver LJ, Rahnavard G, et al. Species-Level functional profiling of metagenomes and metatranscriptomes. Nat Methods 2018;15:962–8. 10.1038/s41592-018-0176-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wu A, Peng Y, Huang B, et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 2020;27:325–8. 10.1016/j.chom.2020.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim D, Lee J-Y, Yang J-S, et al. The architecture of SARS-CoV-2 transcriptome. Cell 2020;181:914–21. 10.1016/j.cell.2020.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shinagawa N, Taniguchi M, Hirata K, et al. [Bacteria isolated from surgical infections and its susceptibilities to antimicrobial agents--special references to bacteria isolated between April 2010 and March 2011]. Jpn J Antibiot 2014;67:293–334. [PubMed] [Google Scholar]
- 22. Liu H, Zhu J, Hu Q, et al. Morganella morganii, a non-negligent opportunistic pathogen. Int J Infect Dis 2016;50:10–17. 10.1016/j.ijid.2016.07.006 [DOI] [PubMed] [Google Scholar]
- 23. Bek-Thomsen M, Tettelin H, Hance I, et al. Population diversity and dynamics of Streptococcus mitis, Streptococcus oralis, and Streptococcus infantis in the upper respiratory tracts of adults, determined by a nonculture strategy. Infect Immun 2008;76:1889–96. 10.1128/IAI.01511-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hwang N, Eom T, Gupta SK, et al. Genes and gut bacteria involved in luminal butyrate reduction caused by diet and loperamide. Genes 2017;8:350. 10.3390/genes8120350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kläring K, Just S, Lagkouvardos I, et al. Murimonas intestini gen. nov., sp. nov., an acetate-producing bacterium of the family Lachnospiraceae isolated from the mouse gut. Int J Syst Evol Microbiol 2015;65:870–8. 10.1099/ijs.0.000030 [DOI] [PubMed] [Google Scholar]
- 26. Remely M, Aumueller E, Merold C, et al. Effects of short chain fatty acid producing bacteria on epigenetic regulation of FFAR3 in type 2 diabetes and obesity. Gene 2014;537:85–92. 10.1016/j.gene.2013.11.081 [DOI] [PubMed] [Google Scholar]
- 27. Ratajczak W, Rył A, Mizerski A, et al. Immunomodulatory potential of gut microbiome-derived short-chain fatty acids (SCFAs). Acta Biochim Pol 2019;66:1–12. 10.18388/abp.2018_2648 [DOI] [PubMed] [Google Scholar]
- 28. Gao J, Xu K, Liu H, et al. Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism. Front Cell Infect Microbiol 2018;8:13. 10.3389/fcimb.2018.00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Verdu EF, Hayes CL, O’Mahony SM. Importance of the microbiota in early life and influence on future health. The Gut-Brain Axis: Elsevier, 2016: 159–84. [Google Scholar]
- 30. Geva-Zatorsky N, Sefik E, Kua L, et al. Mining the human gut microbiota for immunomodulatory organisms. Cell 2017;168:928–43. 10.1016/j.cell.2017.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jimmerson LC, Bushman LR, Ray ML, et al. A LC-MS/MS method for quantifying adenosine, guanosine and inosine nucleotides in human cells. Pharm Res 2017;34:73–83. 10.1007/s11095-016-2040-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Antonioli L, Blandizzi C, Pacher P, et al. Immunity, inflammation and cancer: a leading role for adenosine. Nat Rev Cancer 2013;13:842–57. 10.1038/nrc3613 [DOI] [PubMed] [Google Scholar]
- 33. Shibata T, Ohto U, Nomura S, et al. Guanosine and its modified derivatives are endogenous ligands for TLR7. Int Immunol 2016;28:211–22. 10.1093/intimm/dxv062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kitamoto S, Alteri CJ, Rodrigues M, et al. Dietary L-serine confers a competitive fitness advantage to Enterobacteriaceae in the inflamed gut. Nat Microbiol 2020;5:116–25. 10.1038/s41564-019-0591-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yeo C, Kaushal S, Yeo D. Enteric involvement of coronaviruses: is faecal-oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol Hepatol 2020;5:335–7. 10.1016/S2468-1253(20)30048-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gu J, Han B, Wang J. COVID-19: gastrointestinal manifestations and potential Fecal-Oral transmission. Gastroenterology 2020;158:1518–9. 10.1053/j.gastro.2020.02.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zang R, Gomez Castro MF, McCune BT, et al. Tmprss2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci Immunol 2020;5. 10.1126/sciimmunol.abc3582. [Epub ahead of print: 13 05 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2020-322294supp001.pdf (1.6MB, pdf)
gutjnl-2020-322294supp002.pdf (381.5KB, pdf)