ABSTRACT
The SARS-CoV-2 pandemic has dramatically increased the workload for health systems and a consequent need to optimise resources has arisen, including the selection of patients for swab tests. We retrospectively reviewed consecutive patients presenting to the emergency department with symptoms suggestive of COVID-19 and undergoing swab tests for SARS-CoV-2. Complete blood counts (CBCs) were analysed looking for predictors of test positivity. Eight significant predictors were identified and used to build a ‘complete’ CBC score with a discriminatory power for COVID-19 diagnosis of AUC 92% (p<0.0001). When looking at the weight of individual variables, mean corpuscular volume (MCV), age, platelets and eosinophils (MAPE: MCV ≤90 fL, 65 points; age ≥45 years, 100 points; platelets ≤180×103/μL, 73 points; eosinophils <0.01/μL, 94 points) gave the highest contribution and were used to build a ‘simplified’ MAPE score with a discriminatory power of AUC 88%. By setting the cut-off MAPE score at ≥173 points, sensitivity and specificity for COVID-19 diagnosis were 83% and 82%, respectively, and the actual test positivity rate was 60% as compared to 6% of patients with MAPE score <173 points (odds ratio 23.04, 95% confidence interval [CI] 9.1–58.3, p-value <0.0001). In conclusion, CBC-based scores have potential for optimising the SARS-CoV-2 testing process: if these findings are confirmed in the future, swab tests may be waived for subjects with low score and uncertain symptoms, while they may be considered for asymptomatic or oligosymptomatic patients with high scores.
KEYWORDS: COVID-19, complete blood count, emergency department, swab test
Introduction
The 2019 novel coronavirus (SARS-CoV-2) has been identified as the causative virus of a severe acute respiratory syndrome first detected in Wuhan, China, at the end of 2019, and COVID-19 refers to the disease associated with SARS-CoV-2.1
Since then, SARS-CoV-2 has spread across the globe and the epidemic has reached catastrophic proportions in many countries.2 In March 2020, COVID-19 has been declared a Public Health Emergency of International Concern by the World Health Organization.3
Initial data from Italy suggested a surprisingly high mortality rate (higher than 10%), especially among the elderly.4 The capacity of public health systems has been under exceptional stress5,6 and in particular the workload in emergency departments (EDs) has dramatically increased, with a consequent need to optimise resources, which led to the decision to perform nasopharyngeal/oropharyngeal swab PCR only on symptomatic patients.7–9
The availability of predictors of COVID-19 swab test positivity would be of great use in optimising resource allocation and patient care and in safeguarding healthcare professionals.
The aim of the present analysis was to identify complete blood count (CBC) predictors of test positivity in order to support the management of patients requiring swab tests.
Materials and methods
Consecutive patients presenting to the ED of Tor Vergata University Hospital (Rome, Italy) from 7–19 March 2020 with suspicious symptoms of COVID-19 (fever, dyspnea or cough) and undergoing nasopharyngeal swab tests for SARS-CoV-2 were included in the present retrospective study.
Patients with active hematological disease were excluded. The CBC test was performed in all patients. Nasopharyngeal swab samples were handled as per standard procedure and analysed for SARS-CoV-2 colonisation by using real time PCR for the three genes: gene E, gene N, gene RdRP. As per internal policy, if the first swab test was negative, a second sample, 24 hours apart, was required to confirm the negative result, with the aim of increasing the overall test sensitivity.
12 variables were considered for association with test positivity: gender, age, hemoglobin (g/dL), mean corpuscular volume (MCV, fL), red cell distribution width – coefficient of variation (RDW-CV), and red blood cells, platelets, neutrophils, lymphocytes, monocytes, eosinophils and basophils (all measured as absolute count per μL). Extreme values (inferior and superior fifth percentile) were winsorised to reduce the influence of outliers.
A first round of univariate logistic regression for test positivity was performed for continuous variables with restricted cubic splines with three knots. Variables found not to be linearly associated with test positivity were dichotomised using the Youden Index of receiver operating characteristic (ROC) curve analysis.
A full model including all 12 variables was analysed using the Least Absolute Shrinkage and Selection Operator (LASSO). The Akaike Information Criterion (AIC) was used to eliminate variables with low coefficients (ie coefficient absolute value equal zero) and to identify variables entering the final logistic regression model.
LASSO-selected variables were used for the final multivariate logistic regression model and a nomogram and a score chart were created providing a probability of test positivity for each patient (‘complete’ CBC score). A ROC curve analysis was then performed to find the optimal CBC score/risk cutoff to divide patients with high versus low probability of COVID-19 posititvity, and sensitivity and specificity were calculated. Odds ratios (ORs) for test positivity and internal calibration of the model were also estimated.
In a second step, the formulation of a ‘simplified’ score was attempted by selecting the variables with the highest weight in the ‘complete’ CBC multivariate logistic regression model (highest contribution to the Wald test chi-square) and by dichotomising all continuous variables of interest. The process followed for the formulation of the ‘simplified’ score was the same as that of the ‘complete’ score. Bootstrap resampling with 5,000 iterations was performed to correct for model optimism. Variable interactions were not investigated as the main emphasis was placed on the independent predictive value of variables found significant at the multivariate analysis.
Because of the urgent need to report data on the COVID-19 outbreak and the retrospective nature of the study, Institutional Review Board approval and informed consent were waived. Only anonymised data were analysed. All analyses were performed by using R software version 3.6.3. All tests were considered statistically significant for two-tail p values <0.05.
Results
171 patients were included: COVID-19 test positivity rate was 24.6% (42 patients) (supplementary material S1). Univariate logistic regression analysis with restricted cubic splines revealed that the following variables were not linearly associated with the test result: age, red blood cells, MCV, RDW-CV, platelets and monocytes (supplementary material S2). Therefore, they were respectively dichotomised as follows: <45 years vs ≥45 years, ≤4.5×106/μL vs >4.5×106/μL, <90 fL vs ≥90 fL, <14.5 vs ≥14.5, <180×103/μL vs ≥180×103/μL and <0.7×103/μL vs ≥0.7×103/μL. Hemoglobin, neutrophils, lymphocytes, eosinophils and basophils were kept as continuous variables (supplementary material S3).
The 11 above-mentioned variables plus gender were analysed using the LASSO technique (supplementary material S4, S5). The selection process identified eight significant predictors (age, hemoglobin, MCV, RDW-CV, platelets, monocytes, eosinophils and basophils) that were used to build the final multivariate logistic regression predictive model (supplementary material S6).
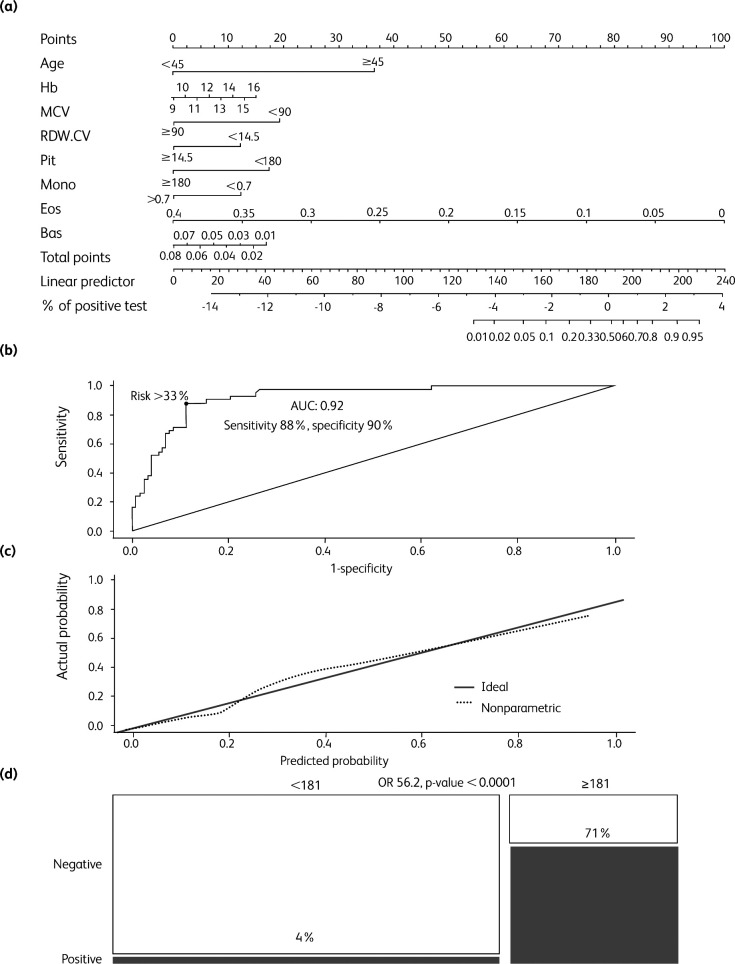
The model had a highly significant predictive value: C-statistics 0.92, p value <0.0001. The model was translated into a predictive score and e nomogram (‘complete’ CBC score, ranging from 0 to ≥ 200) for SARS-CoV-2 test positivity, with a predicted probability of testing positive ≥70% for a score ≥200 (Table 1, Fig 1a). The discriminatory power of the score was AUC 92% according to a ROC curve analysis (Fig 1b), and internal calibration of the model was also good (Fig 1c).
Table 1.
Score chart for the complete cell blood count (CBC) model
| Category | Points | Category | Points |
|---|---|---|---|
| Age | Eosinophils | ||
| <45 | 0 | 0.15 | 63 |
| ≥45 | 36 | 0.2 | 50 |
| Hb | 0.25 | 38 | |
| 9 | 0 | 0.3 | 25 |
| 10 | 2 | 0.35 | 13 |
| 11 | 4 | 0.4 | 0 |
| 12 | 6 | Platelets | |
| 13 | 9 | <180 | 17 |
| 14 | 11 | ≥180 | 0 |
| 15 | 13 | Monocytes | |
| 16 | 15 | <0.7 | 12 |
| MCV | >0.7 | 0 | |
| <90 | 19 | Basophils | |
| ≥90 | 0 | 0.01 | 17 |
| RDW.CV | 0.02 | 14 | |
| <14.5 | 12 | 0.03 | 12 |
| ≥14.5 | 0 | 0.04 | 10 |
| Eosinophils | 0.05 | 7 | |
| 0 | 100 | 0.06 | 5 |
| 0.05 | 88 | 0.07 | 2 |
| 0.1 | 75 | 0.08 | 0 |
| Total points | Predicted probability of positive test | ||
| 133 | 1% | ||
| 153 | 5% | ||
| 162 | 10% | ||
| 172 | 20% | ||
| 179 | 30% | ||
| 181 | 33% | ||
| 184 | 40% | ||
| 189 | 50% | ||
| 194 | 60% | ||
| 200 | 70% | ||
| 207 | 80% | ||
| 217 | 90% | ||
| 226 | 95% | ||
Hb = hemoglobin; MCV=mean corpuscular volume.
Fig 1.
Analysis of the ‘complete’ cell blood count (CBC) score (cutoff score 181, ie cutoff predicted risk 33%). (a) Nomogram for the ‘complete’ CBC score. (b) Receiver operating characteristic (ROC) curve analysis for the complete CBC score predicted probability. Optimal risk cut-off (33%) was calculated according to the Youden index. (c) Calibration plot of the ‘complete’ CBC logistic regression model. (d) Mosaic plot of patients according to test positivity and low/high risk ‘complete’ CBC score (cutoff score 181). AUC = area under the curve; bas = basophils; CBC = complete blood count; eos = eosinophils; Hb = hemoglobin; MCV = mean corpuscular volume; mono = monocytes; plt = platelets; OR = odds ratio; RDW.CV = red cell distribution width – coefficient of variation.
By setting at ≥33% the optimal cut-off risk (score ≥181), model sensitivity and specificity were 88% and 90%, respectively. Patients with a score of ≥181 had an actual frequency of SARS-CoV-2 swab test positivity of 71% (37 out of 52 patients) as compared to 4% of patients with a score of <181 (5 out of 119 patients): OR 56.2, 95% CI 19.1–165.3, p-value <0.0001 (Fig 1d).
By looking at the weight of individual variables, it was found that MCV, age, platelets and eosinophils (MAPE) gave the highest contribution to the model (supplementary material S6). A ‘simplified’ MAPE score was then constructed using these four variables. For further simplicity, eosinophil count was optimally dichotomised by using ROC curve analysis as follows: ≥0.01 vs <0.01.
The MAPE logistic regression model maintained a highly significant discriminatory power with a C-statistics of 0.88, p-value <0.0001) (supplementary material S7).
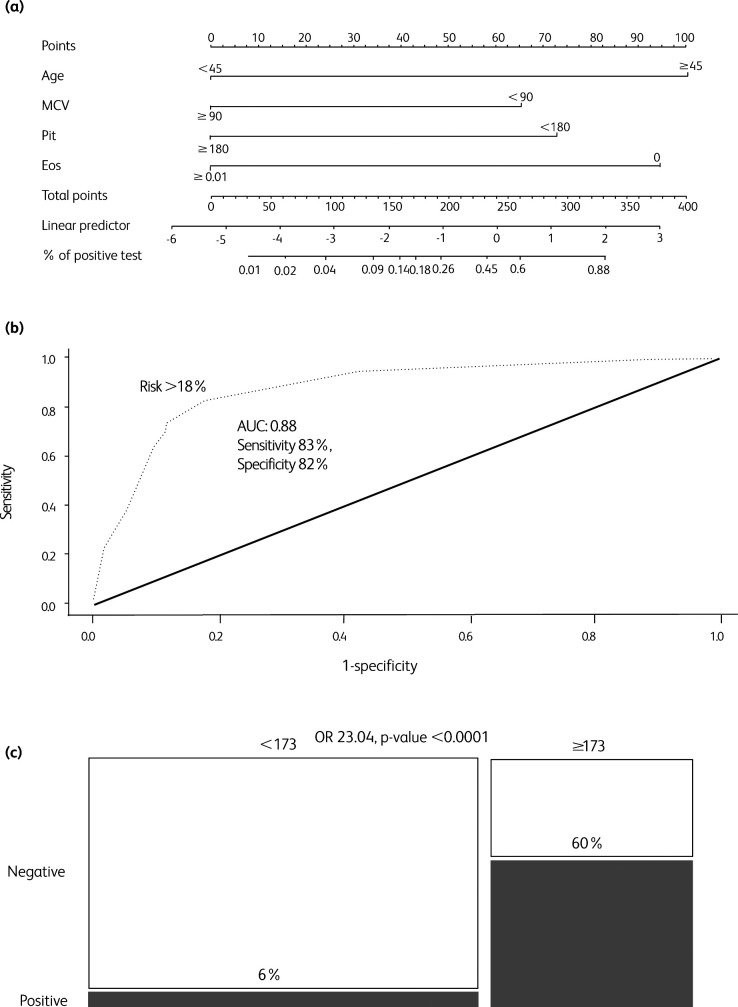
Age ≥45 years, eosinophils <0.01, MCV ≤90 fL and platelets ≤180×103/μL were all independent predictors of test positivity with OR of 9.0, 4.2, 4.9 and 7.9, respectively (p values 0.007, <0.001, 0.007 and 0.001 respectively) (supplementary material S7). The MAPE score ranged from 0 to 332, with test positivity risk ranging from 1% to 88% (Table 2, Fig 2a).
Table 2.
Score chart of the Simplified MAPE (MCV, age, platelets and eosinophils) score
| Age | Points | Platelets | Points |
|---|---|---|---|
| <45 | 0 | ≥180 | 0 |
| ≥45 | 100 | <180 | 73 |
| Eosinophils | Points | MCV | Points |
| ≥0.01 | 0 | ≥90 | 0 |
| 0 | 94 | <90 | 65 |
| Total points | Predicted probability of positive test | ||
| 0 | 1% | ||
| 65–73 | 2% | ||
| 94–100 | 4% | ||
| 138 | 9% | ||
| 159 | 14% | ||
| 165–167 | 16% | ||
| 173 | 18% | ||
| 194 | 26% | ||
| 232 | 45% | ||
| 238 | 48% | ||
| 259 | 60% | ||
| 267 | 64% | ||
| 332 | 88% | ||
Hb = hemoglobin; MCV = mean corpuscular volume.
Fig 2.
Analysis for the simplified MAPE (MCV, age, platelets and eosinophils) score, with optimal risk cut-off (18%, ie score cut-off 173). (a) Nomogram for the simplified MAPE score. (b) Receiver operating characteristic (ROC) curve analysis for the MAPE score predicted probability. Optimal risk cut-off (18%) was calculated according to the Youden index. (c) Mosaic plot for high risk vs low risk MAPE score patients. AUC = area under the curve; eos = eosinophils; MCV = mean corpuscular volume; plt = platelets; OR = odds ratio.
According to a ROC curve analysis, the discriminatory power of MAPE score was AUC 88% (95% CI 0.82–0.94, p<0.0001) and the optimal cutoff risk of ≥18% (score ≥173) was associated with a sensitivity and specificity of 83% and 82%, respectively (Fig 2b).
Patients older than 45 years with either suppressed eosinophils or platelets ≤180×103/μL, or patients at any age with eosinophils <0.01, MCV ≤90 fL and platelets ≤180×103/μL (high risk MAPE score patients, ie score ≥173) had an actual SARS-CoV-2 test positivity rate of 60% (35 out 58 patients) as compared to 6% (7 out of 113) of the remaining patients, OR 23.04, p-value <0.0001 (Fig 2c). Internal validation with 5,000 bootstrap resampling provided minimal need for optimism correction (correction factor 11%, c-statistic lowering from 0.88 to 0.87) (supplementary material S8).
Discussion
The presented ‘complete’ CBC and ‘simplified’ MAPE scores can be of support in the accomplishment of SARS-CoV-2 testing programmes. We acknowledge that these data are preliminary and involve only ED patients. External validation by independent research groups is eagerly awaited, and the ‘COVID-19 specificity’ of the scores also needs to be confirmed. Moreover, the data are based on nasopharyngeal swab PCR positivity and, in order to fully translate them into clinical practice, it would be useful to investigate the scores also in relation to the COVID-19 clinical diagnosis, which includes CT-scan findings.
Data on COVID-19 management in the EDs have so far been scarce.10 In situations where resources are limited and population-wide testing cannot be performed, CBC-based scores may help to pre-evaluate patients referring to ED, or other medical departments, and activate COVID-19-specific pathways and swab testing for patients with high scores independently of symptom severity. In presence of suspected COVID-19 and high scores, logistical management in the ER may also be improved, with prompt activation of isolation procedures even before COVID-19 test results. Moreover, since swab tests have suboptimal sensitivity,11 the scores may be useful to identify ‘false negative’ subjects and decide for second-level diagnostic procedures, thus preventing subclinical disease transmission and unsafe exposure of healthcare professionals.12
In addition to the emergency setting, strong indication for COVID-19 testing may be made for patients with high CBC/MAPE scores in other settings and also for asymptomatic or oligosymptomatic subjects (subclinical COVID-19 transmission), who would not otherwise receive the test.13,14 However, we acknowledge that the scores have exclusively been built using data from symptomatic ED patients and therefore their usefulness in screening asymptomatic or minimally symptomatic individuals outside the ED needs to be confirmed.
Future research should also evaluate whether, within the scope of resource optimisation, swab tests may be waived for uncertain COVID-19 patients with low/zero CBC or MAPE scores, and whether our model may be improved by including non-CBC parameters of routine blood tests (such as liver or renal function tests or inflammatory and coagulation markers).
Variables selected for the MAPE score, in particular older age and low eosinophil count, were reported elsewhere to be associated with COVID-19 diagnosis and prognosis.15,16 Moreover, platelets and eosinophils have been described to be associated with disease severity in previous studies.17,18 Activated eosinophils and platelets have both been found to protect against respiratory virus infection.19,20 It can be hypothesised that they are consumed early during the coronavirus infection and a fall in their circulating pool can be apparent in the CBC.
We could find no clear explanation for the association between microcytosis (MCV <90 fL) and test positivity, though underlying health conditions determining both decrease in MCV and higher risk of developing symptomatic COVID-19 cannot be excluded.
Surprisingly, lymphocytes were not associated with test positivity in our study. Lymphopenia has been found to worsen as the disease progresses (with higher incidence in ICU patients as compared to non-ICU patients).19 It is possible that patients in our cohort were in the early phases of COVID-19 disease trajectory and lymphopenia had not yet fully developed. The pathophysiology behind CBC/MAPE score findings, as well as a possible correlation between score results and disease severity, merit further investigations.
In conclusion, readily available predictive scores derived from CBC, and in particular the simplified and easy-to-use MAPE score, have potential for optimising the SARS-CoV-2 testing process. Within the scope of resource optimisation, swab tests may be waived for subjects with low CBC/MAPE score and uncertain clinical signs, while they may be considered for asymptomatic or oligosymptomatic patients with a high CBC/MAPE score to identify asymptomatic carriers.
Supplementary material
Additional supplementary material may be found in the online version of this article at www.rcpjournals.org/clinmedicine:
S1 – Patient characteristics.
S2 – Univariate logistic regression analysis: dichotomised variables.
S3 – Univariate logistic regression analysis: continuous variables.
S4 – Coefficient path of LASSO analysis.
S5 – Akaike's information criterion (AIC) path of LASSO analysis.
S6 – Full multivariate logistic regression analysis (8 predictors) – ‘complete’ CBC score.
S7 – MAPE score simplified multivariate logistic regression model.
S8 – Bootstrap resampling for optimism correction of the ‘simplified’ MAPE score.
Acknowledgements
We thank Dr Roberta Aronica for English language revision.
References
- 1.Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 2020;382:727–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Onder G, Rezza G, Brusaferro S. Case-fatality rate and characteristics of patients dying in relation to COVID-19 in Italy. JAMA 2020, in press ( 10.1001/jama.2020.4683). [DOI] [PubMed] [Google Scholar]
- 3.Sohrabi C, Alsafi Z, O'Neill N, et al. World Health Organization declares global emergency: A review of the 2019 novel coronavirus (COVID-19). Int J Surg 2020;76:71–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang L, He W, Yu X, et al. Coronavirus disease 2019 in elderly patients: characteristics and prognostic factors based on 4-week follow-up. J Infect 2020,80:639–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grasselli G, Pesenti A, Cecconi M. Critical care utilization for the COVID-19 outbreak in Lombardy, Italy: early experience and forecast during an emergency response. JAMA 2020, in press ( 10.1001/jama.2020.4031). [DOI] [PubMed] [Google Scholar]
- 6.Rosenbaum L. Facing Covid-19 in Italy – ethics, logistics, and therapeutics on the epidemic's front line. N Engl J Med 2020;382:1873–5. [DOI] [PubMed] [Google Scholar]
- 7.Adalja AA, Toner E, Inglesby TV. Priorities for the US health community responding to COVID-19. JAMA 2020, in press ( 10.1001/jama.2020.3413). [DOI] [PubMed] [Google Scholar]
- 8.Emanuel EJ, Persad G, Upshur R, et al. Fair allocation of scarce medical resources in the time of Covid-19. N Engl J Med 2020;382:2049–55. [DOI] [PubMed] [Google Scholar]
- 9.Spina S, Marrazzo F, Migliari M, et al. The response of Milan's emergency medical system to the Covid-19 outbreak in Italy. Lancet 2020;395:e49–e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Reilly GM, Mitchell RD, Rajiv P, et al. Epidemiology and clinical features of emergency department patients with suspected COVID-19: Initial results from the COVID-19 Emergency Department Quality Improvement Project (COVED-1). Emerg Med Australas 2020, in press ( 10.1111/1742-6723.13540). [DOI] [PubMed] [Google Scholar]
- 11.Xie X, Zhong Z, Zhao W, et al. Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology 2020, in press ( 10.1148/radiol.2020200343). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song JY, Yun JG, Noh JY, Cheong HJ, Kim WJ. Covid-19 in South Korea – challenges of subclinical manifestations. N Engl J Med 2020;382:1858–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rothe C, Schunk M, Sothmann P, et al. Transmission of 2019-nCoV Infection from an Asymptomatic Contact in Germany. N Engl J Med 2020;382:970–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zou L, Ruan F, Huang M, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med 2020;382:1177–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qin C, Zhou L, Hu Z, et al. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin Infect Dis 2020, in press ( 10.1093/cid/ciaa248). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu F, Xu A, Zhang Y, et al. Patients of COVID-19 may benefit from sustained lopinavir-combined regimen and the increase of eosinophil may predict the outcome of COVID-19 progression. Int J Infect Dis 2020;95:183–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henry BM, de Oliveira MHS, Benoit S, Plebani M, Lippi G. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): a meta-analysis. Clin Chem Lab Med 2020;58:1021–8. [DOI] [PubMed] [Google Scholar]
- 18.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Percopo CM, Dyer KD, Ochkur SI, et al. Activated mouse eosinophils protect against lethal respiratory virus infection. Blood 2014;123:743–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seyoum M, Enawgaw B, Melku M. Human blood platelets and viruses: defense mechanism and role in the removal of viral pathogens. Thromb J 2018;16:16. [DOI] [PMC free article] [PubMed] [Google Scholar]