ABSTRACT
SARS-CoV-2 serological tests are a subject of intense interest and have the potential to significantly enhance the diagnostic capability of healthcare services in the current pandemic. However, as with all novel assays, significant validation is required to understand the clinical relevance of results. We present the first study to assess clinician interpretation of SARS-CoV-2 serology scenarios. We identify common key assumptions regarding patient infectivity and protection that are not currently supported by the SARS-CoV-2 evidence base. In this rapidly developing field, we therefore strongly recommend serological assay results are accompanied by clear interpretive support from laboratory and infectious diseases specialists.
KEYWORDS: COVID-19, SARS-CoV-2, serology, interpretation of laboratory results
Serological testing in SARS-CoV-2
The novel coronavirus SARS-CoV-2, manifesting clinically as the disease named COVID-19, has caused a global pandemic. As of 4 May 2020 there have been 3,442,234 confirmed cases and 239,740 fatalities reported across 215 countries.1
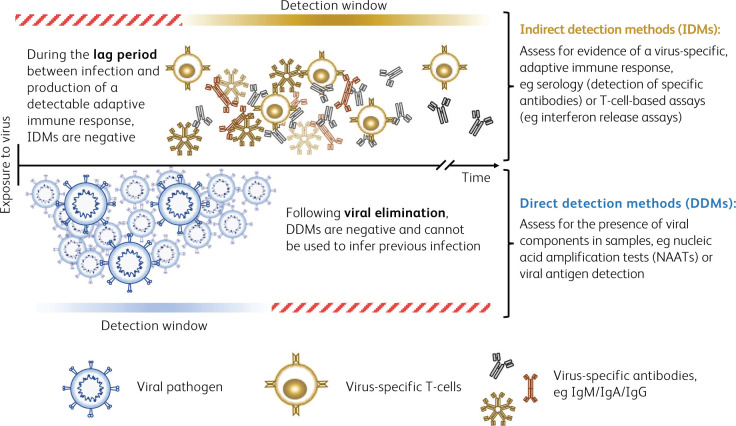
Diagnostic assays for SARS-CoV-2 have been in rapid development, and large studies examining sensitivity and specificity for all platforms are understandably lacking at this early stage. Currently, nucleic acid amplification tests (NAATs) provide a direct method to detect the presence of the SARS-CoV-2 RNA genome. These tests are in widespread diagnostic use to identify active infection. However, in isolation, these assays are not comprehensive. There is an urgent need to expand diagnostic capability to include indirect detection methods, which may be applicable both during active infection and for the identification of previously infected individuals who were not tested at the time of their acute illness. This has led to intense interest in the potential of serological assays (Fig 1).2–4
Fig 1.
Summary of direct and indirect detection methods.
Serological testing identifies host humoral immune responses to an infection. In principle, this has the potential for broad clinical applications, including studying the immune response, epidemiological applications (such as establishing rates of infection and fatality, identifying asymptomatic cases, and carrying out contact tracing, transmission pattern analysis and patient contact studies), and identifying those in the population who may be immune.5 However, this relies on an understanding of the basic immunobiology of an infection, coupled with robust assay validation. It is perhaps unsurprising that even well-established assays, for example those for acute Epstein–Barr virus, hepatitis and cytomegalovirus infections, can present challenges in the interpretation of results for clinical application. The ability to correctly identify all cases of infection (sensitivity) and to discriminate between cross-reactive viruses and other antigens (specificity) vary widely between tests, meaning that it is not possible to make global assumptions regarding the interpretation of vaccine serology.6,7 Long-recognised conditions further benefit from established diagnostic algorithms, such as those detailed in the Public Health England Standards for Microbiology Investigations (UK SMI). However, such interpretive support does not yet exist for SARS-CoV-2.8
Early studies of SARS-CoV-2 immunobiology have identified the emergence of specific immunoglobulin M (IgM)/IgA and IgG SARS-CoV-2 antibodies at approximately day 5 and day 14 of infection respectively.9 However, recent data have challenged the assumed principles of sequential virus-specific antibody seroconversion from an early IgM response followed by a later emergence of IgG. Long et al instead describe three distinct patient groups:10
synchronous seroconversion of IgG and IgM
IgM seroconversion earlier than that of IgG
IgM seroconversion later than that of IgG.
Furthermore, data are not yet available for specific populations who may not mount a specific antibody response, such as those with immunodeficiencies.
The development of serological assays has mainly focused on antibodies directed against the SARS-CoV-2 spike and nucleocapsid proteins. Such antibodies have been shown to neutralise virus in vitro.11,12 However, a significant number of patients experiencing COVID-19 may generate low titres of specific antibodies, presenting a challenge to detection.13 Differing patterns of antibody detection have also been associated with both viral clearance and clinical outcomes.14,15 Overall, at this early stage in the pandemic, the evidence base relating to SARS-CoV-2 remains limited. It is noteworthy that many studies await peer review, with 2,721 preprint articles available on medrxiv and biorxiv websites (www.medrxiv.org, www.biorxiv.org).
Clinician interpretation of IgM and IgG serological results in SARS-CoV-2
With large-scale implementation of novel serology assays likely to be imminent, how the results are used will have implications for both individual patient care and public health measures. To better understand how SARS-CoV-2 IgM and IgG results may be interpreted by clinicians, a survey was designed using the SurveyMonkey web-based platform (SurveyMonkey, San Mateo, USA; www.surveymonkey.com) (supplementary material S1). An online survey link was distributed to clinicians and clinical scientists in the UK via existing professional networks, constituting a ‘snowball’ sampling method. The survey was designed to be appropriate to the responding clinicians by presenting serological results as these may be encountered in routine clinical practise. Due to technological limitations of the survey platform used, this initial survey was closed once a maximum of 100 responses had been received. Grades and specialities of responders are summarised in Table 1. Results were collected between 25 March 2020 and 31 March 2020. During this period, serology testing for SARS-CoV-2 was not generally available in the UK.
Table 1.
Summary of survey responder demographics
| Specialities of clinicians who undertook the survey* | Number of responders |
|---|---|
| Acute medicine | 4 |
| Anaesthetics | 8 |
| Paediatric psychiatry | 1 |
| Clinical immunology | 11 |
| Core medical training | 4 |
| Citical care | 3 |
| GP | 11 |
| Dermatology | 1 |
| Emergency medicine | 1 |
| Endocrinology | 6 |
| ENT | 1 |
| Foundation programme | 4 |
| Gastroenterology | 1 |
| General surgery | 3 |
| Geriatrics | 4 |
| Gynaecology | 1 |
| Haematology | 5 |
| Histopathology | 1 |
| Infectious diseases | 5 |
| International training fellow | 1 |
| Medical microbiology | 1 |
| General internal medicine | 3 |
| Nephrology | 2 |
| Neurosurgery | 2 |
| Specialities of clinicians who undertook the survey* | Number of responders |
| Oncology | 1 |
| Paediatrics | 1 |
| Palliative medicine | 1 |
| Psychiatry | 1 |
| Radiology | 1 |
| Respiratory medicine | 2 |
| Rheumatology | 2 |
| HSST programme | 3 |
| General surgery | 1 |
| Trust grade doctor | 1 |
| Clinical scientist | 2 |
| Reported training grade of clinicians who undertook the survey† | Number of responders |
| Advanced nurse practitioner | 1 |
| Clinical scientist | 6 |
| Core trainee/senior house officer | 21 |
| Consultant | 6 |
| Foundation trainee | 7 |
| GP | 5 |
| Specialist trainee ST3+ (registrar) | 50 |
| Staff grade | 1 |
| Trainee clinical scientist | 3 |
*Clinicians from a total of 35 different specialities provided responses.
†50% of responders were of UK specialist trainee grades ≥ST3.
Responders were asked to interpret four result combinations for SARS-CoV-2-specific IgM and IgG serology, first in isolation and then with the addition of a clinical scenario stating ‘active symptoms consistent with COVID-19’. Responders could select all statements that they felt were appropriate to each scenario. Data were analysed using Graphpad Prism 8 (GraphPad Software, San Diego, California USA, www.graphpad.com) and are summarised in Fig 2. An optional free-text comment box was provided for each scenario and responses recorded (supplementary material S2).
Fig 2.
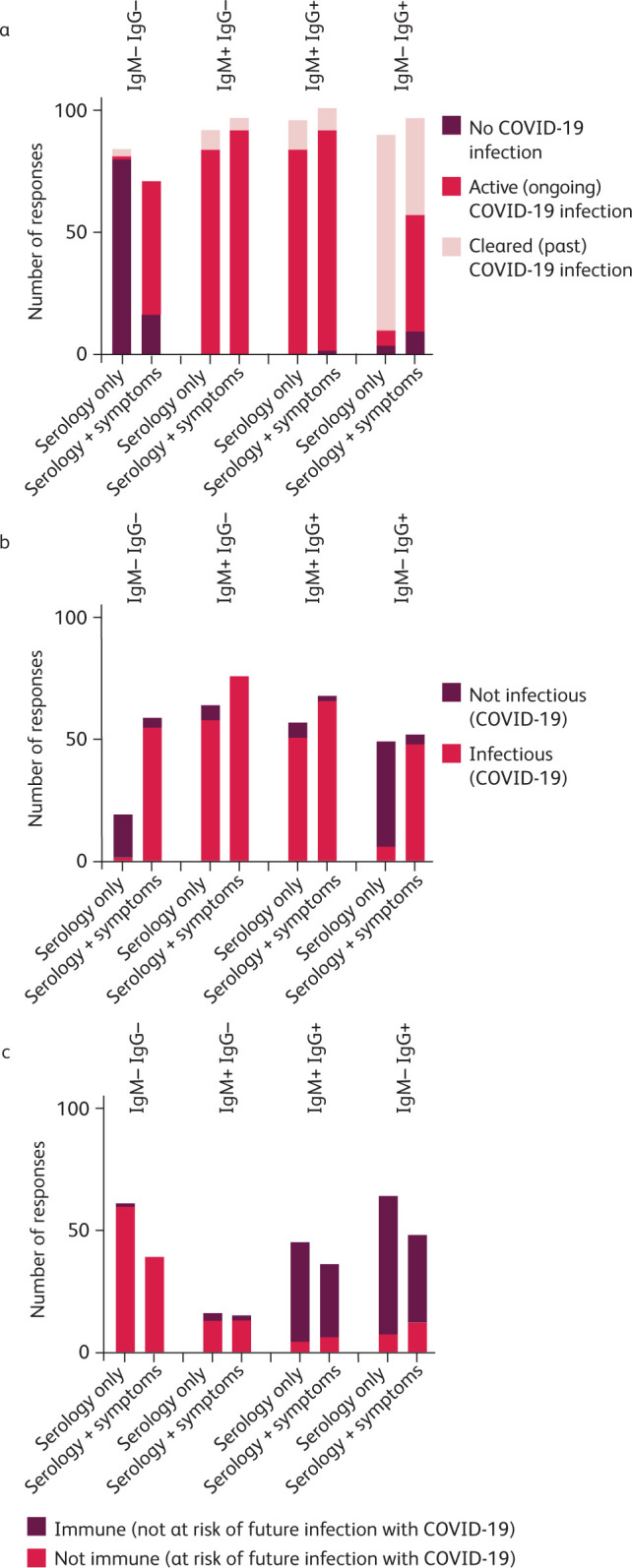
Summary of survey responses. For each scenario, responders were asked to select all statements they felt were appropriate to the serology result with and without associated clinical details of ‘active symptoms consistent with COVID-19’. a) Responses inferring the patient's SARS-CoV-2 infection status. b) Responses inferring the patient's risk of infecting others with SARS-CoV-2. c) Responses inferring the patient's risk of future infection with SARS-CoV-2.
Interpreting serology results alone and in the context of relevant symptoms resulted in notable variation. This was particularly marked for Ig– IgG– and Ig– IgG+ scenarios. 17% of responders classed a patient with negative serology (Ig– IgG–) as having ‘No COVID-19’ despite the presence of active symptoms. Also, 40% considered patients to have ‘cleared COVID-19’ despite active symptoms in the context of serology demonstrating Ig– IgG+.
Links between serology and a patient's risk of infection or their ability to infect others have not been clearly established for SARS-CoV-2. Yet, across all serology and serology plus clinical scenarios, a mean of 57% (SD 17%) of participants selected statements inferring a patient's infectivity status, and 41% (SD 18%) selected statements inferring immunity status. In clinical practice, misplaced confidence in the interpretation of serology could lead to errors of management. 22/91 of the free-text comments queried assay performance, for example wanting to review sensitivity/specificity data.
Conclusions
The rapid development and implementation of a range of diagnostic assays is undoubtedly an essential part of the coordinated response to a new pathogen. However, the limitations of novel assays and of clinicians' understanding of these must be considered.4,5 To our knowledge, this is the first study to investigate clinicians' interpretive response to novel SARS-CoV-2 serology. There are significant limitations to our study design, both in our modest number of survey responses and the necessity for rapid design and implementation due to the evolving nature of the pandemic. As free text comments were optional, analysis of these is also limited. However, we highlight that there is likely to be marked variation in the clinical interpretation of SARS-CoV-2 serology results as they become available. Further research in this area is urgently warranted, as this may have serious implications for ongoing public health efforts to maintain social distancing measures and the isolation of patients affected by COVID-19. Proactive interpretive support, which includes ‘narrative comments’ from laboratory and infectious diseases specialists, is strongly recommended (Box 1).
Box 1.
Examples of interpretative comments that may be useful in reporting SARS-CoV-2 serology
These must be modified to reflect the validation characteristics and specifications of the assay system used.
|
Supplementary material
Additional supplementary material may be found in the online version of this article at www.rcpjournals.org/clinmedicine:
S1 – Survey structure
S2 – Free text comments submitted by survey responders
References
- 1.World Health Organization Coronavirus disease (COVID-19) pandemic. WHO, 2020. Available from www.who.int/emergencies/diseases/novel-coronavirus-2019 [Accessed 4 May 2020].
- 2.World Health Organization Laboratory testing for coronavirus disease 2019 (COVID-19) in suspected human cases. WHO, 2020. www.who.int/publications-detail/laboratory-testing-for-2019-novel-coronavirus-in-suspected-human-cases-20200117 [Accessed 11 April 2020].
- 3.Patel R, Babady E, Theel ES, et al. Report from the American Society for Microbiology COVID-19 International Summit, 23 March 2020: Value of diagnostic testing for SARS-CoV-2/COVID-19. mBio 2020;11:e00722–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Egner W, Beck S, Chopra C, et al. Statement from RCPath's Immunology Specialty Advisory Committee on COVID-19/SARS CoV2 antibody evaluation. Royal College of Pathologists, 2020. www.rcpath.org/uploads/assets/194ed03e-9b0a-4f65-8208563290fb848e/3aeb35a1-97d1-4143-92440dc0aa42c5e8/G211-RCPath-Immunology-SAC-statement-on-COVID-19-SARS-CoV2-antibody-evaluation.pdf [Accessed 11 April 2020]. [Google Scholar]
- 5.Meyer B, Drosten C, Müller MA. Serological assays for emerging coronaviruses: Challenges and pitfalls. Virus Res 2014;194:175–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klutts JS, Ford BA, Perez NR, Gronowski AM. Evidence-based approach for interpretation of Epstein-Barr virus serological patterns. J Clin Microbiol 2009;47:3204–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Landry ML. Immunoglobulin M for acute infection: True or false? Clin Vaccine Immunol 2016;23:540–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Public Health England Standards for microbiology investigations. PHE, 2014. www.gov.uk/government/collections/standards-for-microbiology-investigations-smi#uk-smi-supporting-information [Accessed 4 May 2020].
- 9.Guo L, Ren L, Yang S, et al. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19). Clin Infect Dis 2020;ciaa310 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Long Q-X, Liu B-Z, Deng H-J, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med 2020;26:845–8. [DOI] [PubMed] [Google Scholar]
- 11.Haveri A, Smura T, Kuivanen S, et al. Serological and molecular findings during SARS-CoV-2 infection: the first case study in Finland, January to February 2020. Eur Commun Dis Bull 2020;25:2000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okba Ni MA, Muller MA, Li W, et al. SARS-CoV-2 specific antibody responses in COVID-19 patients. medRxiv 2020;2020.03.18.20038059. [Google Scholar]
- 13.Wu F, Wang A, Liu M, et al. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. medRxiv 2020; 2020.03.30.20047365. [Google Scholar]
- 14.Zhao J, Yuan Q, Wang H, et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. medRxiv 2020;2020.03.02.20030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kai-wang K, Tak O, Tsang Y, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis 2020;20:565–74. [DOI] [PMC free article] [PubMed] [Google Scholar]