Abstract
Purpose:
The adhesion molecule cluster of differentiation 44v9 interacts with and stabilizes the cystine/glutamate exchanger protein, which functions as a transporter of cystine. Stabilized cystine/glutamate exchanger increases extracellular cystine uptake and enhances glutathione production. Augmented levels of reduced glutathione mitigate reactive oxygen species and protect cancer cells from apoptosis. Salazosulfapyridine blocks cystine/glutamate exchanger activity and mitigates the supply of cystine to increase intracellular ROS production, thereby increasing cell susceptibility to apoptosis. This enhances the effect of anticancer drugs such as cisplatin. Currently, salazosulfapyridine is being developed as a promising anticancer agent. In the present study, we elucidated the molecular mechanism associated with salazosulfapyridine by investigating the salazosulfapyridine-induced changes in glutathione metabolism in cultured gastric cancer cell lines.
Methods:
The effect of salazosulfapyridine treatment on glutathione metabolism was investigated in 4 gastric cancer (AGS, MKN1, MKN45, and MKN74) and 2 colorectal cancer (HCT15 and HCT116) cell lines using metabolomic analyses. In addition, the effect of inhibition of the reduced form of nicotinamide adenine dinucleotide phosphate by 2-deoxyglucose on glutathione metabolism was evaluated.
Results:
Under hypoxia, enhanced glycolysis resulted in lactate accumulation with an associated reduction in nicotinamide adenine dinucleotide phosphate. Salazosulfapyridine treatment decreased the cysteine content and inhibited the formation of glutathione. Combined treatment with salazosulfapyridine and 2-deoxyglucose significantly inhibited cell proliferation. 2-Deoxyglucose, an inhibitor of glycolysis, depleted nicotinamide adenine dinucleotide phosphate required for the formation of glutathione.
Conclusions:
Our results indicate that in cancer cells having a predominant glycolytic pathway, metabolomic analyses under hypoxic conditions enable the profiling of global metabolism. In addition, inhibiting the supply of nicotinamide adenine dinucleotide phosphate by blocking glycolysis is a potential treatment strategy for cancer, in addition to cystine blockade by salazosulfapyridine.
Keywords: salazosulfapyridine (SASP), CD44v9, metabolic profiling, hypoxic condition, glycolysis
Introduction
Surgical resection involving lymph node dissection is currently recognized as a standard treatment procedure for early gastric cancer. However, endoscopic local resection is an alternative treatment option in the absence of lymph node metastasis.1,2 The risk of lymph node metastasis after non-curative endoscopic submucosal dissection is difficult to predict based on clinicopathological factors, necessitating the need for new diagnostic and therapeutic strategies.3-5
Cancer stem cells play an important role in the recurrence, drug resistance, and metastasis of cancer. Cluster of differentiation (CD) 44, an adhesion molecule, is a promising marker for identifying cancer stem cells. CD44v9, an isoform of CD44, is expressed on a wide range of tissues including the lungs, ovaries, and stomach. Overexpression of CD44v9 is observed in primary lesions particularly in advanced gastric cancer with deeper submucosal invasion or lymph node metastasis and in stage III/IV patients.6
In the glutathione metabolic pathway, the cystine/glutamate exchanger (xCT) protein acts a transporter of cystine, the primary substrate required for glutathione synthesis.7,8 CD44v9 interacts with and stabilizes xCT, thereby promoting the extracellular uptake of cystine, resulting in increased glutathione production. Enhanced level of reduced glutathione (GSH) inhibits reactive oxygen species (ROS), preventing cancer cell apoptosis, and suppresses p38 mitogen-activated protein kinase that regulates cell growth and differentiation.9,10 Salazosulfapyridine (SASP) blocks xCT activity and clinical trials on its therapeutic application in treating non-small-cell lung and gastric cancers that are underway.11,12 In theory, SASP-mediated inhibition of cystine uptake increases the intracellular level of ROS and enhances the susceptibility of cells to apoptosis, thereby augmenting the effect of anticancer drugs such as cisplatin. Salazosulfapyridine, first developed in the late 1930s to treat rheumatoid arthritis in multiple joints, is currently used for the treatment of ulcerative colitis.13-15 5-Aminosalicylic acid, the active moiety of SASP, suppresses ROS, inhibits cytokine production, and blocks arachidonic acid signaling.16,17 However, the principal mechanism associated with SASP activity still needs to be elucidated. In patients with glucose-6-phosphate dehydrogenase deficiency, SASP triggers hemolysis and is therefore contraindicated.18 Glucose-6-phosphate dehydrogenase is a glycolytic enzyme that regulates the generation of reduced nicotinamide adenine dinucleotide phosphate (NADPH) required for glutathione metabolism. Deficiency of glucose-6-phosphate dehydrogenase causes depletion of GSH. Consequently, red blood cells fail to eliminate the active oxygen present in the cell membrane, leading to lipid peroxidation-mediated membrane damage and eventual hemolysis.
2-Deoxyglucose (2DG) is a promising agent in many cancers.19-21 It competitively inhibits glucose transport and is phosphorylated by hexokinase to form 2-DG-6-phosphate, which is not metabolized further, thereby reducing NADPH from the pentose–phosphate pathway.22-24
In the present study, we investigated the effect of SASP treatment on glutathione metabolism in cultured gastric cancer cells using metabolomic analyses. In addition, the effect of inhibiting NADPH by 2DG on glutathione metabolism was evaluated.
Materials and Methods
Cell Culture
Four gastric cancer (AGS, MKN1, MKN45, and MKN74) and 2 colorectal cancer (HCT15 and HCT116) cell lines were purchased from the Japanese Collection of Research Bioresources Cell Bank (JCRB) and American Type Culture Collection (ATCC). Specifically, MKN1, MKN45, and MKN74 cells were purchased from JCRB, and AGS, HCT15, and HCT116 cells from ATCC. All cell lines were subcultured in 225 cm2 tissue culture flasks (Corning Glass Works). MKN1, MKN45, MKN74, AGS, and HCT15 cell lines were maintained in Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco/Life Technologies) supplemented with 10% fetal bovine serum (FBS) and 50 mg/mL gentamycin (Gibco/Life Technologies) at 37 °C in water-saturated air with 5% CO2. HCT116 cells were cultured both in Eagle’s Minimal Essential Medium/RPMI1640 (1:1) and in McCoy’s 5a medium (both, Gibco/Life Technologies) supplemented with 10% FBS and 50 mg/mL gentamycin.
Selection of Cell lines by Flow Cytometry
Cell surface expression of CD44v9 was analyzed by flow cytometry. Labeling with anti-human CD44v9 antibody (clone RV3; Cosmo Bio) was performed according to the manufacturer’s instructions, with minor modifications. Briefly, cells were harvested with 0.25% trypsin/EDTA solution and quenched with the medium containing serum. The cells were counted using the Scepter cell counter (Merck), centrifuged, and resuspended at a density of 1 × 105 cells in 0.2% bovine serum albumin (BSA) in phosphate-buffered saline (PBS) in assay tubes. The cells were washed with 0.2% BSA in PBS and incubated with 50 µL of diluted primary antibody (3 μg/mL RV3 or isotype control antibody) for 45 minutes at 4 °C. After incubation, cells were washed 3 times with 1 mL of 0.1% BSA in PBS, centrifuged, and incubated further with 50 µL of phycoerythrin-conjugated secondary antibody solution (anti-rat IgG; Jackson ImmunoResearch) diluted 1:200 in 0.1% BSA in PBS for 30 minutes at 4 °C. Cells were washed 3 times with 0.1% BSA in PBS and probed using a BD Accuri C6 flow cytometer (BD Biosciences). Data were analyzed using the FlowJo software (BD Biosciences).
Preparation of SASP and 2DG Solutions
Salazosulfapyridine was freshly dissolved in 0.1 M NaOH solution and the pH was adjusted to ∼8 with 0.1 M HCl. The solution was then diluted to 30 mM with PBS. The 2DG (Sigma-Aldrich) at a concentration of 600 mM was prepared using sterile water.
Cell Growth Assay
MKN45 cells were resuspended in the respective growth medium at a density of 1.5 × 103 cells/well and incubated overnight to allow cell adhesion at 37 °C in an atmosphere of 5% CO2 or under hypoxic conditions. After incubation, the culture medium was replaced with fresh growth medium containing 0.3 mM SASP, 6 mM 2DG, or 0.3 mM SASP + 6 mM 2DG; these concentrations were optimized in preliminary experiments.25-30 The experiment was based on cell viability data. For 2DG, we also referred to our previous study comprising metabolome experiments.31 After 3 days of culturing, the number of viable cells was determined using a hemocytometer (C-Chip, INCYTO) and a light microscope (CK30, OLYMPUS).
Metabolomic Analyses
MKN45 cells were seeded in 10 cm2 culture dishes and cultured under standard growth conditions to achieve 80% confluence. Each assay was performed with 3 replicates (n = 3) to eliminate the possible effects of time on metabolism. Cells were cultured in culture medium containing 0.3 mM SASP, 6 mM 2DG, or 0.3 mM SASP + 6 mM 2DG or medium without supplement for 24 hours under normoxic and hypoxic conditions. Metabolomic analyses were performed as per the methodology prescribed by Human Metabolome Technologies (HMT). Briefly, cells were washed twice with ice-cold 5% maltose-water solution and the residual solution was removed by aspiration. Cells were then quenched with 1 mL of liquid chromatography (LC)–mass spectrometry (MS)–grade methanol (Sigma-Aldrich), gently detached from the culture dish using a cell scraper (MS-93170; Sumitomo Chemical), and transferred to a 10-mL centrifuge tube for extraction. The cell number was determined for normalization of metabolite concentration. Intracellular metabolites were purified using a 2-phase liquid extraction system. Methanol, chloroform, and water in the ratio 10:10:4 by volume were used for the study. The aqueous phase held water-soluble endogenous low-molecular-weight metabolites and the organic phase held nonpolar metabolites (eg, lipids). The solvent layer between the 2 phases held macromolecules (eg, proteins). For the extraction of endogenous water-soluble low-molecular-weight metabolites, the aqueous phase was collected and centrifugally filtered through a 5-kDa cut-off filter (Millipore) to eliminate macromolecules. Solvent removal was accomplished using a vacuum concentrator (Thermo Electron). The residue was dissolved in 50 µL of Milli-Q water containing internal standards and was immediately subjected to capillary electrophoresis (CE) time-of-flight (TOF) MS analyses on a CE system equipped with 6224 mass spectrometer, 1200 isocratic high-performance LC pump, G1603 CE-MS adapter kit, and G1607 CE-electrospray ionization-MS sprayer kit (all from Agilent Technologies). G2201AA ChemStation and MassHunter software programs (Agilent Technologies) were used for system control/data acquisition in CE and TOFMS, respectively. Cationic and anionic metabolite analyses were performed using the HMT Metabolomics Solution Package as described previously.31,32 Briefly, standard compounds (a kind gift from HMT) were analyzed as references for m/z values, migration times, and quantification. Standard compounds used were glycolic acid, lactic acid, fumaric acid, 2-oxoisovaleric acid, succinic acid, malic acid, phosphoenolpyruvic acid, dihydroxyacetone phosphate, glycerol-3-phosphate, cis-aconitic acid, 3-phosphoglyceric acid, citric acid, isocitric acid, gluconic acid, ribose 5-phosphate, ribulose 5-phosphate, fructose 6-phosphate, glucose 6-phosphate, glucose 1-phosphate, 6-phosphogluconic acid, sedoheptulose 7-phosphate, dTMP, CMP, cAMP, fructose 1,6-diphosphate, cyclic guanosine monophosphate (GMP), adenosine monophosphate (AMP), inosine monophosphate, GMP, phosphoribosyl pyrophosphate, dTDP, CDP, acetyl-coenzyme A, adenosine diphosphate (ADP), guanosine diphosphate (GDP), dCTP, dTTP, CTP, UTP, dATP, ATP, GTP, NAD+, NADP+, Gly, putrescine, β-Ala, Ala, γ-aminobutyric acid, Ser, cytosine, uracil, creatinine, Pro, Val, homoserine, Thr, Cys, hydroxyproline, creatine, Ile, Leu, Asn, ornithine, Asp, adenine, hypoxanthine, anthranilic acid, tyramine, spermidine, Gln, Lys, Glu, Met, guanine, His, Phe, Arg, citrulline, Tyr, DOPA, spermine, Trp, carnosine, cytidine, uridine, adenosine, inosine, guanosine, glutathione divalent, GSH, and S-adenosylmethionine. From the HMT Metabolome database library, reference MS spectra of the following 20 compounds were used for annotation: uric acid, deoxyribose-5′-monophosphate, glucosamine 6-phosphate, N-acetyl d-glucosamine 6-phosphate, deoxycytidine 3-monophosphate, dUMP, UMP, dAMP, UDP, dADP, dGDP, dGTP, UDP-glucose, xanthine, d-glucono-1,5-lactone, d-glucosamine, N-acetyl d-glucosamine, 2′-deoxycytidine, thymidine, and 2′-deoxyadenosine. 2-Deoxy-d-glucose-6-phosphate (Sigma-Aldrich) was used as the standard. Quantification of metabolites was achieved by processing raw data comprising of thousands of peaks using the MasterHands software (Keio University), which has been used in several CE-TOFMS-based profiling studies previously.31,33 The overall data processing flow was as follows: migration time alignment, peak detection, background subtraction, and integration of peak area from a 0.02 m/z-wide slice of the electropherogram. Target metabolites were identified by comparing their m/z values and migration times with those of the standard compounds. A total of 94 compounds were annotated and quantified. For each sample, the measured metabolite concentration was normalized using the cell number to obtain the amount of metabolite (nmol) per 106 cells. Data related to metabolites are shown in Table S1.
Data Visualization
Data were visualized using the open source software Visualization and Analysis of Networks Containing Experimental Data (http://vanted.ipk-gatersleben.de/), which allows the mapping of potential biological pathways.34
Statistical Analysis
Differences among the mean of the groups were evaluated with 2-tailed Student t test. Analyses of metabolomic data were performed using the Welch t test. P < .05 was considered statistically significant and is indicated with a ⋆.
Results
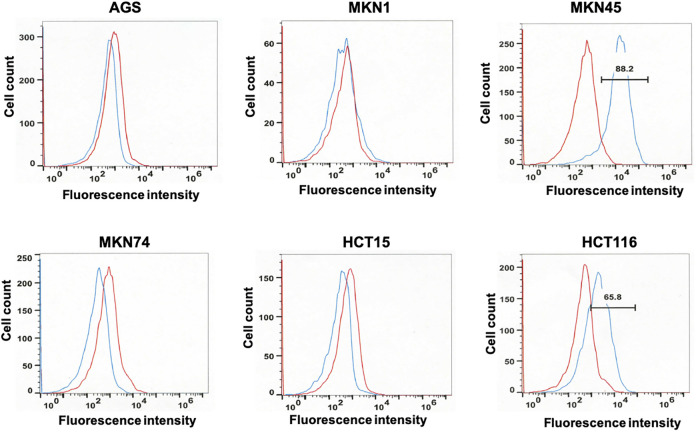
To select an appropriate cell line for SASP treatment, 4 human gastric cancer cell lines (AGS, MKN74, MKN1, and MKN45) and 2 human colorectal cancer cell lines (HCT15 and HCT116) were screened for the expression of CD44v9 by flow cytometry. HCT15 and HCT116, expressing CD44v9 as reported previously,9,35 served as the positive controls. Cultured cells obtained by trypsin treatment and labeled with anti-human CD44v9 antibody were analyzed by flow cytometry. Statistically significant highest expression of CD44v9 was detected in MKN45 cells (Figure 1). Accordingly, MKN45 cells in culture were further subjected to metabolomic analyses.
Figure 1.
Flow cytometry analyses of cell lines labeled with anti-cluster of differentiation (CD)44v9 (blue) or isotype control (red) antibody. Four human gastric cancer cell lines (AGS, MKN1, MKN45, and MKN74) were screened for CD44v9 expression. Two human colorectal cancer cell lines (HCT15 and HCT116) previously reported to express CD44v9 served as positive controls. Harvested cells at a density of 1 × 105 cells were labeled with the primary antibody (3 mg/mL RV3 or isotype control antibody), followed by phycoerythrin-labeled secondary antibody (anti-rat IgG) and analyzed by flow cytometry. The fluorescence intensity of anti-CD44v9 and isotype control antibodies is indicated as blue and red lines, respectively. MKN45 cells showed the highest expression of CD44v9.
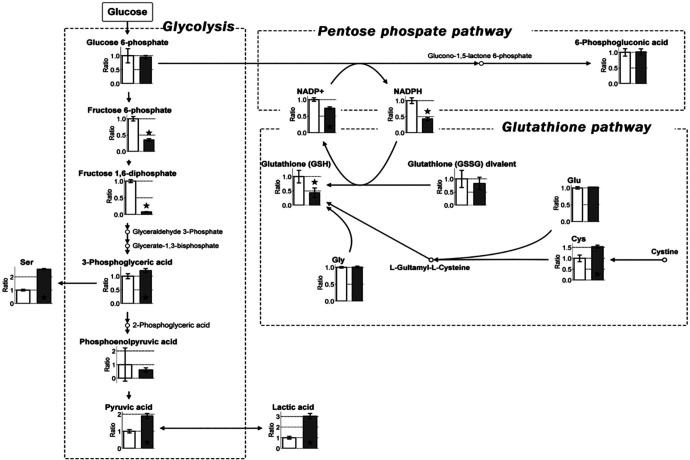
To investigate the effect of hypoxia on metabolism, MKN45 cells were cultured for 24 hours in 20% or 1% O2 and the intracellular metabolites were isolated for further analyses by CE-TOFMS. Metabolomic analyses were carried out for 102 different types of metabolites and the identified peaks were semi-quantified using the reference to standard compounds for 94 of the 102 metabolites (Table 1). Figure 2 summarizes the metabolites associated with glycolysis, the pentose phosphate pathway (PPP), and the glutathione pathway. Under hypoxic conditions, the production of lactic acid was enhanced, serving as an indicator of augmented glycolysis. Accordingly, NADPH, which is essential for reduction in glutathione, was significantly decreased together with diminished levels of GSH. Thus, diverse metabolites were detected in hypoxic cultures compared with those in normoxic cultures. These results emphasize the importance of maintaining hypoxic conditions while investigating the role of cancer cell metabolites in vivo. Therefore, in the present study, all subsequent experiments involving drug treatment were performed using hypoxic cultures.
Table 1.
Relative Ratios of Metabolite Concentrations Under Normoxia and Hypoxia (1% O2 and 5% CO2).
| Metabolite | Normoxia | Hypoxia | Metabolite | Normoxia | Hypoxia | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Average | SD | CV | Average | SD | CV | Average | SD | CV | Average | SD | CV | ||
| Glycolic acid | 1.00 | 0.09 | 0.09 | 1.74 | 0.55 | 0.32 | Gly | 1.00 | 0.02 | 0.02 | 1.01 | 0.03 | 0.03 |
| Pyruvic acid | 1.00 | 0.10 | 0.10 | 1.89 | 0.15 | 0.08 | Putrescine | 1.00 | 0.16 | 0.16 | 0.33 | 0.03 | 0.09 |
| Lactic acid | 1.00 | 0.12 | 0.12 | 3.02 | 0.26 | 0.09 | β-Ala | 1.00 | 0.05 | 0.05 | 0.68 | 0.03 | 0.05 |
| Fumaric acid | 1.00 | 0.07 | 0.07 | 0.75 | 0.02 | 0.02 | Ala | 1.00 | 0.04 | 0.04 | 0.89 | 0.03 | 0.03 |
| 2-Oxoisovaleric acid | 1.00 | 0.13 | 0.13 | 4.39 | 0.36 | 0.08 | γ-Aminobutyric acid | 1.00 | 0.05 | 0.05 | 0.33 | 0.01 | 0.03 |
| Succinic acid | 1.00 | 0.16 | 0.16 | 1.54 | 0.12 | 0.08 | Ser | 1.00 | 0.06 | 0.06 | 2.58 | 0.05 | 0.02 |
| Malic acid | 1.00 | 0.07 | 0.07 | 1.03 | 0.05 | 0.04 | Cytosine | 1.00 | 0.12 | 0.12 | 0.75 | 0.03 | 0.03 |
| 2-Oxoglutaric acid | 1.00 | 0.11 | 0.11 | 1.36 | 0.15 | 0.11 | Uracil | 1.00 | 0.16 | 0.16 | 0.68 | 0.02 | 0.04 |
| Phosphoenolpyruvic acid | 1.00 | 1.23 | 1.23 | 0.61 | 0.15 | 0.24 | Creatinine | 1.00 | 0.00 | 0.00 | 0.88 | 0.02 | 0.02 |
| Dihydroxyacetone phosphate | 1.00 | 0.05 | 0.05 | 0.05 | 0.01 | 0.21 | Pro | 1.00 | 0.03 | 0.03 | 0.80 | 0.01 | 0.02 |
| Glycerol-3-phosphate | 1.00 | 0.07 | 0.07 | 2.04 | 0.28 | 0.14 | Val | 1.00 | 0.06 | 0.06 | 1.86 | 0.07 | 0.04 |
| cis-Aconitic acid | 1.00 | 0.05 | 0.05 | 0.31 | 0.02 | 0.06 | Homoserine | 1.00 | 0.18 | 0.18 | 1.20 | 0.20 | 0.17 |
| 3-Phosphoglyceric acid | 1.00 | 0.09 | 0.09 | 1.21 | 0.08 | 0.06 | Thr | 1.00 | 0.03 | 0.03 | 1.34 | 0.07 | 0.05 |
| Citric acid | 1.00 | 0.09 | 0.09 | 0.33 | 0.07 | 0.22 | Cys | 1.00 | 0.15 | 0.15 | 1.54 | 0.07 | 0.05 |
| Isocitric acid | 1.00 | 0.03 | 0.03 | 0.24 | 0.03 | 0.12 | Hydroxyproline | 1.00 | 0.02 | 0.02 | 0.79 | 0.03 | 0.04 |
| Gluconic acid | 1.00 | 0.07 | 0.07 | 0.81 | 0.07 | 0.09 | Creatine | 1.00 | 0.09 | 0.09 | 0.70 | 0.01 | 0.02 |
| Erythrose 4-phosphate | 1.00 | 0.19 | 0.19 | 0.29 | 0.02 | 0.07 | Ile | 1.00 | 0.04 | 0.04 | 1.21 | 0.02 | 0.02 |
| Ribose 5-phosphate | 1.00 | 0.03 | 0.03 | 0.75 | 0.07 | 0.09 | Leu | 1.00 | 0.02 | 0.02 | 1.33 | 0.07 | 0.05 |
| Ribulose 5-phosphate | 1.00 | 0.11 | 0.11 | 0.11 | 0.04 | 0.35 | Asn | 1.00 | 0.03 | 0.03 | 0.82 | 0.03 | 0.03 |
| Fructose 6-phosphate | 1.00 | 0.07 | 0.07 | 0.35 | 0.04 | 0.11 | Ornithine | 1.00 | 0.05 | 0.05 | 2.99 | 0.33 | 0.11 |
| Glucose 6-phosphate | 1.00 | 0.26 | 0.26 | 0.95 | 0.06 | 0.06 | Asp | 1.00 | 0.12 | 0.12 | 0.74 | 0.05 | 0.07 |
| Glucose 1-phosphate | 1.00 | 0.06 | 0.06 | 0.27 | 0.03 | 0.12 | Adenine | 1.00 | 0.16 | 0.16 | 1.44 | 0.25 | 0.18 |
| 6-Phosphogluconic acid | 1.00 | 0.12 | 0.12 | 1.02 | 0.10 | 0.10 | Anthranilic acid | 1.00 | 0.08 | 0.08 | 0.34 | 0.17 | 0.51 |
| Sedoheptulose 7-phosphate | 1.00 | 0.10 | 0.10 | 0.77 | 0.02 | 0.02 | Tyramine | 1.00 | 0.08 | 0.08 | 1.12 | 0.06 | 0.05 |
| dTMP | 1.00 | 0.26 | 0.26 | 1.15 | 0.34 | 0.30 | Spermidine | 1.00 | 0.23 | 0.23 | 0.81 | 0.40 | 0.49 |
| CMP | 1.00 | 0.16 | 0.16 | 0.29 | 0.02 | 0.08 | Gln | 1.00 | 0.01 | 0.01 | 4.64 | 0.08 | 0.02 |
| Fructose 1,6-diphosphate | 1.00 | 0.04 | 0.04 | 0.08 | 0.01 | 0.15 | Lys | 1.00 | 0.03 | 0.03 | 2.10 | 0.12 | 0.06 |
| AMP | 1.00 | 0.35 | 0.35 | 0.15 | 0.02 | 0.11 | Glu | 1.00 | 0.04 | 0.04 | 1.02 | 0.00 | 0.00 |
| IMP | 1.00 | 0.41 | 0.41 | 0.06 | 0.00 | 0.06 | Met | 1.00 | 0.04 | 0.04 | 1.50 | 0.09 | 0.06 |
| GMP | 1.00 | 0.17 | 0.17 | 0.18 | 0.02 | 0.11 | Guanine | 1.00 | 0.43 | 0.43 | 0.34 | 0.09 | 0.28 |
| NADPH | 1.00 | 0.09 | 0.09 | 0.43 | 0.05 | 0.12 | His | 1.00 | 0.02 | 0.02 | 1.39 | 0.08 | 0.05 |
| PRPP | 1.00 | 0.21 | 0.21 | 4.66 | 1.09 | 0.23 | Phe | 1.00 | 0.05 | 0.05 | 1.87 | 0.09 | 0.05 |
| FAD | 1.00 | 0.06 | 0.06 | 1.03 | 0.17 | 0.16 | Arg | 1.00 | 0.04 | 0.04 | 1.28 | 0.09 | 0.07 |
| dTDP | 1.00 | 0.12 | 0.12 | 0.25 | 0.17 | 0.66 | Citrulline | 1.00 | 0.05 | 0.05 | 1.25 | 0.08 | 0.06 |
| CDP | 1.00 | 0.16 | 0.16 | 0.44 | 0.01 | 0.03 | Tyr | 1.00 | 0.03 | 0.03 | 1.53 | 0.04 | 0.03 |
| Acetyl-CoA | 1.00 | 0.10 | 0.10 | 6.36 | 0.27 | 0.04 | DOPA | 1.00 | 0.11 | 0.11 | 1.45 | 0.22 | 0.15 |
| Malonyl-CoA | 1.00 | 0.22 | 0.22 | 0.22 | 0.04 | 0.19 | Trp | 1.00 | 0.03 | 0.03 | 1.87 | 0.04 | 0.02 |
| ADP | 1.00 | 0.13 | 0.13 | 0.40 | 0.05 | 0.14 | Carnosine | 1.00 | 0.05 | 0.05 | 0.80 | 0.03 | 0.04 |
| Succinyl-CoA | 1.00 | 0.26 | 0.26 | 0.77 | 0.02 | 0.03 | Cytidine | 1.00 | 0.10 | 0.10 | 0.87 | 0.11 | 0.13 |
| GDP | 1.00 | 0.23 | 0.23 | 0.46 | 0.06 | 0.12 | Adenosine | 1.00 | 0.31 | 0.31 | 0.62 | 0.21 | 0.34 |
| dCTP | 1.00 | 0.07 | 0.07 | 0.88 | 0.11 | 0.12 | Guanosine | 1.00 | 0.09 | 0.09 | 0.03 | 0.01 | 0.39 |
| dTTP | 1.00 | 0.16 | 0.16 | 1.21 | 0.29 | 0.24 | Glutathione (GSSG) divalent | 1.00 | 0.33 | 0.33 | 0.83 | 0.23 | 0.28 |
| CTP | 1.00 | 0.07 | 0.07 | 0.87 | 0.05 | 0.05 | Glutathione (GSH) | 1.00 | 0.22 | 0.22 | 0.43 | 0.17 | 0.39 |
| UTP | 1.00 | 0.09 | 0.09 | 0.54 | 0.04 | 0.08 | S-Adenosylmethionine | 1.00 | 0.02 | 0.02 | 1.74 | 0.18 | 0.10 |
| dATP | 1.00 | 0.13 | 0.13 | 1.38 | 0.16 | 0.11 | |||||||
| ATP | 1.00 | 0.11 | 0.11 | 1.09 | 0.10 | 0.09 | |||||||
| GTP | 1.00 | 0.08 | 0.08 | 1.32 | 0.07 | 0.05 | |||||||
| NAD+ | 1.00 | 0.03 | 0.03 | 1.16 | 0.24 | 0.21 | |||||||
| NADH | 1.00 | 0.03 | 0.03 | 0.33 | 0.04 | 0.11 | |||||||
| NADP+ | 1.00 | 0.05 | 0.05 | 0.74 | 0.04 | 0.06 | |||||||
Abbreviation: CV, coefficient of variation.
Figure 2.
Map depicting the pathway of metabolites in glycolysis, the pentose phosphate pathway (PPP), and glutathione pathway in cultured MKN45 cells under normoxic and hypoxic (1% O2 and 5% CO2) conditions. Cells (at 80% confluence) were incubated for 24 hours under normoxia and hypoxia and subjected to metabolomic analyses. White and dark gray boxes represent the relative ratio of metabolite concentrations under normoxia and hypoxia, respectively. The graph represents enhanced lactic acid, mitigated reduced form of nicotinamide adenine dinucleotide phosphate (NADPH), and reduced glutathione (GSH) under hypoxia. Error bars represent SD (n = 3). P < .05 indicates statistical significance and is represented as ⋆.
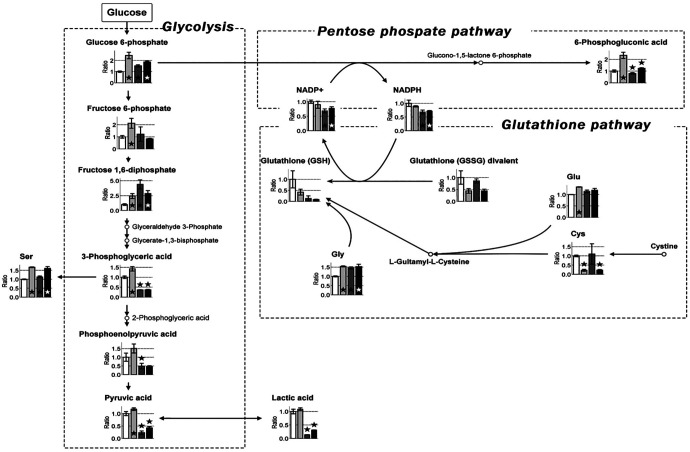
Cultured cells were divided into 4 groups: untreated and treated with 0.3 mM SASP, 6 mM 2DG, or 0.3 mM SASP + 6 mM 2DG. The concentration of SASP and 2DG used in the study was selected based on the results of preliminary experiments and previously published reports.25,30 Accordingly, cells were treated with the respective drugs and cultured for 16 hours. After incubation, the intracellular metabolites were isolated and subjected to CE-TOFMS. The relative ratios of metabolite concentration in SASP- and 2DG-treated cells under hypoxia are summarized in Table 2. Significantly lower levels of Cys (cysteine) and glutathione were detected in the SASP-treated group than in the untreated control cells (Figure 3), suggesting the inhibition of the cystine transporter xCT by SASP. Cystine is transported into cells by xCT and is converted into Cys. Cys serves as the raw material for the synthesis of glutathione, a strong antioxidant. Our results indicate significantly enhanced levels of glycolytic metabolites, suggesting the influence of SASP on glycolysis. Treatment of cells with the glycolysis inhibitor 2DG significantly decreased the levels of 3-phosphoglyceric acid, phosphoenolpyruvic acid, and pyruvic acid, indicating marked suppression of glucose metabolism. In addition, a significant reduction was observed in the level of NADPH, which is produced during the metabolism of glucose to glucono-1-5-lactone 6-phosphate, the branch that leads from glucose 6-phosphate to the PPP, because of the probable effect of 2DG on the PPP. Although 2DG does not directly affect glutathione synthesis, the lack of NADPH required to reduce oxidized glutathione (GSSG) explains the identical level of glutathione observed in 2DG and untreated cells. However, the level of GSH in 2DG-treated cells was significantly lower than that in SASP-treated cells.
Table 2.
Relative Ratios of Metabolite Concentrations in SASP- and 2DG-Treated Cells Under Hypoxia (1% O2 and 5% CO2).
| Metabolite | Hypoxia | SASP | 2DG | SAP+2DG | Metabolite | Hypoxia | SASP | 2DG | SAP+2DG | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| average | SD | CV | Average | SD | CV | average | SD | CV | average | SD | CV | average | SD | CV | average | SD | CV | average | SD | CV | average | SD | CV | ||
| Glycolic acid | 1.00 | 0.32 | 0.32 | 1.04 | 0.19 | 0.19 | 0.78 | 0.17 | 0.21 | 0.67 | 0.07 | 0.11 | Gly | 1.00 | 0.03 | 0.03 | 1.54 | 0.03 | 0.02 | 1.45 | 0.06 | 0.04 | 1.53 | 0.12 | 0.08 |
| Pyruvic acid | 1.00 | 0.08 | 0.08 | 1.18 | 0.05 | 0.05 | 0.23 | 0.07 | 0.31 | 0.42 | 0.05 | 0.12 | Putrescine | 1.00 | 0.09 | 0.09 | 3.18 | 0.13 | 0.04 | 0.37 | 0.01 | 0.02 | 1.01 | 0.14 | 0.14 |
| Lactic acid | 1.00 | 0.09 | 0.09 | 1.08 | 0.05 | 0.05 | 0.14 | 0.01 | 0.04 | 0.30 | 0.01 | 0.04 | β-Ala | 1.00 | 0.05 | 0.05 | 1.40 | 0.04 | 0.03 | 0.72 | 0.05 | 0.07 | 0.95 | 0.10 | 0.10 |
| Fumaric acid | 1.00 | 0.02 | 0.02 | 1.65 | 0.10 | 0.06 | 0.50 | 0.00 | 0.00 | 0.69 | 0.01 | 0.02 | Ala | 1.00 | 0.03 | 0.03 | 1.43 | 0.05 | 0.04 | 0.96 | 0.02 | 0.02 | 1.29 | 0.11 | 0.08 |
| 2-Oxoisovaleric acid | 1.00 | 0.08 | 0.08 | 1.07 | 0.14 | 0.13 | 0.21 | 0.02 | 0.10 | 0.22 | 0.07 | 0.30 | γ-Aminobutyric acid | 1.00 | 0.03 | 0.03 | 2.64 | 0.12 | 0.05 | 0.65 | 0.03 | 0.05 | 1.49 | 0.14 | 0.09 |
| Succinic acid | 1.00 | 0.08 | 0.08 | 1.80 | 0.18 | 0.10 | 0.60 | 0.02 | 0.04 | 0.57 | 0.02 | 0.03 | Ser | 1.00 | 0.02 | 0.02 | 1.68 | 0.02 | 0.01 | 1.13 | 0.05 | 0.05 | 1.61 | 0.10 | 0.06 |
| Malic acid | 1.00 | 0.04 | 0.04 | 1.57 | 0.12 | 0.08 | 0.46 | 0.02 | 0.04 | 0.61 | 0.02 | 0.04 | Cytosine | 1.00 | 0.03 | 0.03 | 1.04 | 0.14 | 0.13 | 0.77 | 0.07 | 0.09 | 1.09 | 0.18 | 0.17 |
| 2-Oxoglutaric acid | 1.00 | 0.11 | 0.11 | 0.97 | 0.09 | 0.09 | 0.16 | 0.03 | 0.20 | 0.32 | 0.03 | 0.09 | Uracil | 1.00 | 0.04 | 0.04 | 1.42 | 0.30 | 0.21 | 0.97 | 0.31 | 0.32 | 1.13 | 0.21 | 0.18 |
| Phosphoenolpyruvic acid | 1.00 | 0.24 | 0.24 | 1.50 | 0.26 | 0.17 | 0.49 | 0.16 | 0.32 | 0.46 | 0.06 | 0.12 | Creatinine | 1.00 | 0.02 | 0.02 | 1.13 | 0.06 | 0.05 | 0.79 | 0.04 | 0.06 | 0.88 | 0.10 | 0.11 |
| Dihydroxyacetone phosphate | 1.00 | 0.21 | 0.21 | 1.87 | 0.22 | 0.12 | 2.39 | 0.21 | 0.09 | 1.72 | 0.22 | 0.13 | Pro | 1.00 | 0.02 | 0.02 | 1.37 | 0.03 | 0.02 | 1.05 | 0.03 | 0.03 | 1.27 | 0.12 | 0.10 |
| Glycerol-3-phosphate | 1.00 | 0.14 | 0.14 | 3.24 | 0.27 | 0.08 | 1.33 | 0.18 | 0.14 | 0.77 | 0.07 | 0.09 | Val | 1.00 | 0.04 | 0.04 | 1.64 | 0.04 | 0.02 | 1.01 | 0.04 | 0.04 | 1.10 | 0.07 | 0.06 |
| cis-Aconitic acid | 1.00 | 0.06 | 0.06 | 1.62 | 0.14 | 0.09 | 0.40 | 0.04 | 0.10 | 0.61 | 0.04 | 0.06 | Homoserine | 1.00 | 0.17 | 0.17 | 1.21 | 0.23 | 0.19 | 0.95 | 0.24 | 0.25 | 1.28 | 0.14 | 0.11 |
| 3-Phosphoglyceric acid | 1.00 | 0.06 | 0.06 | 1.43 | 0.11 | 0.08 | 0.37 | 0.01 | 0.03 | 0.37 | 0.02 | 0.04 | Thr | 1.00 | 0.05 | 0.05 | 1.82 | 0.04 | 0.02 | 1.16 | 0.03 | 0.03 | 1.56 | 0.12 | 0.08 |
| Citric acid | 1.00 | 0.22 | 0.22 | 1.89 | 0.08 | 0.04 | 0.61 | 0.07 | 0.12 | 0.85 | 0.01 | 0.02 | Cys | 1.00 | 0.05 | 0.05 | 0.22 | 0.05 | 0.24 | 1.10 | 0.55 | 0.50 | 0.23 | 0.03 | 0.15 |
| Isocitric acidacid | 1.00 | 0.12 | 0.12 | 1.60 | 0.10 | 0.06 | 0.57 | 0.03 | 0.05 | 0.76 | 0.01 | 0.02 | Hydroxyproline | 1.00 | 0.04 | 0.04 | 1.71 | 0.04 | 0.02 | 1.02 | 0.01 | 0.01 | 1.43 | 0.06 | 0.04 |
| Gluconic acid | 1.00 | 0.09 | 0.09 | 1.08 | 0.17 | 0.16 | 0.33 | 0.01 | 0.03 | 0.30 | 0.01 | 0.02 | Creatine | 1.00 | 0.02 | 0.02 | 0.91 | 0.01 | 0.01 | 1.03 | 0.06 | 0.06 | 0.96 | 0.10 | 0.11 |
| Erythrose 4-phosphate | 1.00 | 0.07 | 0.07 | 1.11 | 0.11 | 0.10 | 7.54 | 0.53 | 0.07 | 7.07 | 0.27 | 0.04 | Ile | 1.00 | 0.02 | 0.02 | 1.39 | 0.04 | 0.03 | 0.99 | 0.04 | 0.04 | 1.10 | 0.08 | 0.07 |
| Ribose 5-phosphate | 1.00 | 0.09 | 0.09 | 1.32 | 0.28 | 0.21 | 2.48 | 0.38 | 0.15 | 1.98 | 0.24 | 0.12 | Leu | 1.00 | 0.05 | 0.05 | 1.32 | 0.04 | 0.03 | 0.98 | 0.06 | 0.06 | 1.13 | 0.08 | 0.07 |
| Ribulose 5-phosphate | 1.00 | 0.35 | 0.35 | 1.95 | 0.27 | 0.14 | 17.62 | 1.47 | 0.08 | 16.36 | 1.06 | 0.06 | Asn | 1.00 | 0.03 | 0.03 | 1.48 | 0.02 | 0.01 | 1.10 | 0.04 | 0.03 | 1.35 | 0.11 | 0.08 |
| Fructose 6-phosphate | 1.00 | 0.11 | 0.11 | 2.12 | 0.41 | 0.19 | 1.22 | 0.60 | 0.49 | 0.84 | 0.04 | 0.05 | Ornithine | 1.00 | 0.11 | 0.11 | 1.26 | 0.11 | 0.09 | 1.03 | 0.13 | 0.12 | 1.04 | 0.06 | 0.06 |
| Glucose 6-phosphate | 1.00 | 0.06 | 0.06 | 2.44 | 0.29 | 0.12 | 1.51 | 0.10 | 0.06 | 1.86 | 0.08 | 0.04 | Asp | 1.00 | 0.07 | 0.07 | 1.71 | 0.09 | 0.05 | 1.46 | 0.13 | 0.09 | 1.57 | 0.12 | 0.08 |
| Glucose 1-phosphate | 1.00 | 0.12 | 0.12 | 1.63 | 0.22 | 0.14 | 0.65 | 0.11 | 0.18 | 0.58 | 0.15 | 0.25 | Adenine | 1.00 | 0.18 | 0.18 | 0.99 | 0.19 | 0.19 | 0.46 | 0.13 | 0.27 | 0.61 | 0.05 | 0.09 |
| 6-Phosphogluconic acid | 1.00 | 0.10 | 0.10 | 2.33 | 0.27 | 0.11 | 0.80 | 0.08 | 0.10 | 1.24 | 0.04 | 0.03 | Anthranilic acid | 1.00 | 0.51 | 0.51 | 2.15 | 0.80 | 0.37 | 1.69 | 0.08 | 0.05 | 1.40 | 0.67 | 0.48 |
| Sedoheptulose 7-phosphate | 1.00 | 0.02 | 0.02 | 1.10 | 0.11 | 0.10 | 0.64 | 0.06 | 0.09 | 0.67 | 0.01 | 0.01 | Tyramine | 1.00 | 0.05 | 0.05 | 1.13 | 0.09 | 0.08 | 1.06 | 0.13 | 0.13 | 1.11 | 0.13 | 0.12 |
| dTMP | 1.00 | 0.30 | 0.30 | 0.90 | 0.35 | 0.39 | 2.66 | 0.21 | 0.08 | 1.88 | 0.07 | 0.03 | Spermidine | 1.00 | 0.49 | 0.49 | 0.85 | 0.23 | 0.27 | 0.85 | 0.40 | 0.46 | 0.91 | 0.09 | 0.10 |
| CMP | 1.00 | 0.08 | 0.08 | 1.88 | 0.25 | 0.13 | 1.63 | 0.15 | 0.09 | 1.55 | 0.27 | 0.17 | Gln | 1.00 | 0.02 | 0.02 | 1.51 | 0.02 | 0.01 | 1.05 | 0.06 | 0.06 | 1.29 | 0.06 | 0.04 |
| Fructose 1,6-diphosphate | 1.00 | 0.15 | 0.15 | 2.43 | 0.41 | 0.17 | 4.37 | 0.76 | 0.17 | 2.86 | 0.50 | 0.17 | Lys | 1.00 | 0.06 | 0.06 | 1.51 | 0.07 | 0.05 | 1.09 | 0.11 | 0.10 | 1.12 | 0.05 | 0.04 |
| AMP | 1.00 | 0.11 | 0.11 | 1.65 | 0.08 | 0.05 | 3.12 | 0.26 | 0.08 | 2.19 | 0.38 | 0.17 | Glu | 1.00 | 0.00 | 0.00 | 1.33 | 0.01 | 0.01 | 1.14 | 0.06 | 0.05 | 1.19 | 0.08 | 0.07 |
| IMP | 1.00 | 0.06 | 0.06 | 1.85 | 0.03 | 0.02 | 10.74 | 0.76 | 0.07 | 6.40 | 1.59 | 0.25 | Met | 1.00 | 0.06 | 0.06 | 1.70 | 0.01 | 0.01 | 0.92 | 0.02 | 0.02 | 1.06 | 0.04 | 0.04 |
| GMP | 1.00 | 0.11 | 0.11 | 2.30 | 0.24 | 0.10 | 1.86 | 0.17 | 0.09 | 1.39 | 0.19 | 0.14 | Guanine | 1.00 | 0.28 | 0.28 | 1.42 | 0.28 | 0.20 | 2.89 | 0.75 | 0.26 | 2.50 | 0.16 | 0.06 |
| NADPH | 1.00 | 0.12 | 0.12 | 0.89 | 0.02 | 0.03 | 0.68 | 0.07 | 0.11 | 0.71 | 0.03 | 0.04 | His | 1.00 | 0.05 | 0.05 | 1.61 | 0.04 | 0.02 | 1.04 | 0.07 | 0.06 | 1.18 | 0.10 | 0.08 |
| PRPP | 1.00 | 0.23 | 0.23 | 1.45 | 0.10 | 0.07 | 0.12 | 0.01 | 0.07 | 0.11 | 0.00 | 0.03 | Phe | 1.00 | 0.05 | 0.05 | 1.73 | 0.07 | 0.04 | 0.98 | 0.05 | 0.05 | 1.21 | 0.07 | 0.06 |
| FAD | 1.00 | 0.16 | 0.16 | 0.84 | 0.02 | 0.03 | 0.66 | 0.08 | 0.13 | 0.73 | 0.05 | 0.07 | Arg | 1.00 | 0.07 | 0.07 | 1.33 | 0.05 | 0.04 | 0.94 | 0.10 | 0.11 | 1.07 | 0.05 | 0.05 |
| dTDP | 1.00 | 0.66 | 0.66 | 1.48 | 0.16 | 0.11 | 1.88 | 0.17 | 0.09 | 1.49 | 0.39 | 0.26 | Citrulline | 1.00 | 0.06 | 0.06 | 1.42 | 0.05 | 0.04 | 0.77 | 0.02 | 0.03 | 0.91 | 0.07 | 0.07 |
| CDP | 1.00 | 0.03 | 0.03 | 0.90 | 0.06 | 0.07 | 1.05 | 0.03 | 0.03 | 1.11 | 0.22 | 0.20 | Tyr | 1.00 | 0.03 | 0.03 | 1.63 | 0.06 | 0.04 | 0.93 | 0.03 | 0.04 | 1.11 | 0.08 | 0.07 |
| Acetyl-coA | 1.00 | 0.04 | 0.04 | 1.27 | 0.19 | 0.15 | 0.27 | 0.03 | 0.12 | 0.38 | 0.04 | 0.10 | DOPA | 1.00 | 0.15 | 0.15 | 1.21 | 0.09 | 0.07 | 0.39 | 0.01 | 0.03 | 0.64 | 0.06 | 0.09 |
| Malonyl-CoA | 1.00 | 0.19 | 0.19 | 1.38 | 0.28 | 0.20 | 0.40 | 0.04 | 0.11 | 0.59 | 0.18 | 0.30 | Trp | 1.00 | 0.02 | 0.02 | 1.56 | 0.04 | 0.03 | 0.81 | 0.02 | 0.02 | 1.00 | 0.06 | 0.06 |
| ADP | 1.00 | 0.14 | 0.14 | 0.92 | 0.09 | 0.09 | 1.19 | 0.10 | 0.08 | 1.12 | 0.10 | 0.09 | Carnosine | 1.00 | 0.04 | 0.04 | 1.19 | 0.06 | 0.05 | 0.95 | 0.10 | 0.10 | 1.20 | 0.13 | 0.11 |
| Succinyl-CoA | 1.00 | 0.03 | 0.03 | 0.98 | 0.25 | 0.25 | 0.53 | 0.13 | 0.24 | 0.57 | 0.11 | 0.19 | Cytidine | 1.00 | 0.13 | 0.13 | 0.99 | 0.12 | 0.12 | 0.94 | 0.15 | 0.16 | 0.95 | 0.09 | 0.09 |
| GDP | 1.00 | 0.12 | 0.12 | 1.08 | 0.17 | 0.16 | 1.69 | 0.14 | 0.09 | 1.45 | 0.23 | 0.16 | Adenosine | 1.00 | 0.34 | 0.34 | 0.47 | 0.36 | 0.76 | 0.29 | 0.04 | 0.13 | 0.24 | 0.04 | 0.17 |
| dCTP | 1.00 | 0.12 | 0.12 | 0.75 | 0.02 | 0.03 | 2.22 | 0.34 | 0.15 | 0.43 | 0.10 | 0.22 | Guanosine | 1.00 | 0.39 | 0.39 | 1.30 | 0.23 | 0.18 | 23.76 | 1.37 | 0.06 | 16.08 | 2.51 | 0.16 |
| dTTP | 1.00 | 0.24 | 0.24 | 0.91 | 0.04 | 0.04 | 0.37 | 0.03 | 0.08 | 0.41 | 0.02 | 0.05 | Glutathione (GSSG) divalent | 1.00 | 0.28 | 0.28 | 0.44 | 0.10 | 0.22 | 0.86 | 0.08 | 0.10 | 0.44 | 0.07 | 0.15 |
| CTP | 1.00 | 0.05 | 0.05 | 1.08 | 0.07 | 0.06 | 0.38 | 0.06 | 0.17 | 0.55 | 0.01 | 0.02 | Glutathione (GSH) | 1.00 | 0.39 | 0.39 | 0.42 | 0.13 | 0.32 | 0.13 | 0.12 | 0.86 | 0.09 | 0.02 | 0.19 |
| UTP | 1.00 | 0.08 | 0.08 | 1.20 | 0.08 | 0.07 | 0.60 | 0.06 | 0.09 | 0.77 | 0.03 | 0.04 | S-Adenosylmethionine | 1.00 | 0.10 | 0.10 | 0.89 | 0.06 | 0.07 | 0.41 | 0.05 | 0.12 | 0.46 | 0.03 | 0.07 |
| dATP | 1.00 | 0.11 | 0.11 | 0.64 | 0.02 | 0.03 | 0.22 | 0.01 | 0.03 | 0.26 | 0.01 | 0.03 | |||||||||||||
| ATP | 1.00 | 0.09 | 0.09 | 1.01 | 0.07 | 0.06 | 0.40 | 0.03 | 0.08 | 0.51 | 0.01 | 0.03 | |||||||||||||
| GTP | 1.00 | 0.05 | 0.05 | 0.81 | 0.07 | 0.08 | 0.40 | 0.05 | 0.12 | 0.46 | 0.03 | 0.06 | |||||||||||||
| NAD+ | 1.00 | 0.21 | 0.21 | 0.92 | 0.30 | 0.33 | 0.68 | 0.04 | 0.07 | 0.67 | 0.03 | 0.04 | |||||||||||||
| NADH | 1.00 | 0.11 | 0.11 | 1.66 | 0.21 | 0.12 | 1.10 | 0.06 | 0.05 | 1.25 | 0.13 | 0.11 | |||||||||||||
| NADP+ | 1.00 | 0.06 | 0.06 | 0.90 | 0.12 | 0.14 | 0.68 | 0.07 | 0.10 | 0.77 | 0.06 | 0.08 | |||||||||||||
Abbreviations: 2DG, 2-deoxyglucose; CV, coefficient of variation; SASP, salazosulfapyridine.
Figure 3.
Map depicting the pathway of metabolites in glycolysis, the PPP, and glutathione pathway of SASP- and 2DG-treated cells under hypoxia (1% O2 and 5% CO2). Cells were divided into 4 groups: untreated and treated with 0.3 mM SASP, 6 mM 2DG, or 0.3 mM SASP + 6 mM 2DG. Treated cells were cultured for 16 hours under hypoxia and subjected to metabolomics analyses. White, light gray, dark gray, and black boxes represent relative ratios of untreated and 0.3 mM SASP-, 6 mM 2DG-, and 0.3 mM SASP + 6 mM 2DG-treated cells, respectively, to untreated cells under hypoxia. The graph represents mitigated (Cysteine) Cys in SASP-treated cells. Lower values were detected for lactic acid (glycolysis pathway) and NADPH (PPP pathway) in 2DG-treated cells. Reduced glutathione was decreased in SASP- and SASP + 2DG-treated cells. Error bars represent SD (n = 3). P < .05 indicates statistical significance and is represented as ⋆. 2DG indicates 2-deoxyglucose; NADPH, reduced nicotinamide adenine dinucleotide phosphate; PPP, pentose phosphate pathway; SASP, salazosulfapyridine.
We next investigated the combined effect of the inhibition of cystine uptake by SASP and the subsequent inhibition of glycolysis and inhibition of NADPH by 2DG in SASP + 2DG-treated cells (Figure 3). Of the 4 experimental groups, the lowest level of GSH was detected in the SASP + 2DG-treated cells compared with that in the untreated SASP, 2DG, and untreated cells. The significant decrease observed in GSSG level was identical in SASP- and SASP + 2DG-treated cells. The level of lactic acid was the second lowest in 2DG-treated cells compared with that in SASP + 2DG-treated cells, indicating that SASP and 2DG treatment exerted their combined effect in an additive manner.
Examination of cell growth in the 4 groups indicated potential suppression of cell proliferation by SASP relative to untreated cells, an effect that was exacerbated by 2DG (P < .0005, SASP vs SASP + 2DG and P < .005, 2DG vs. SASP + 2DG; Figure 4).
Figure 4.
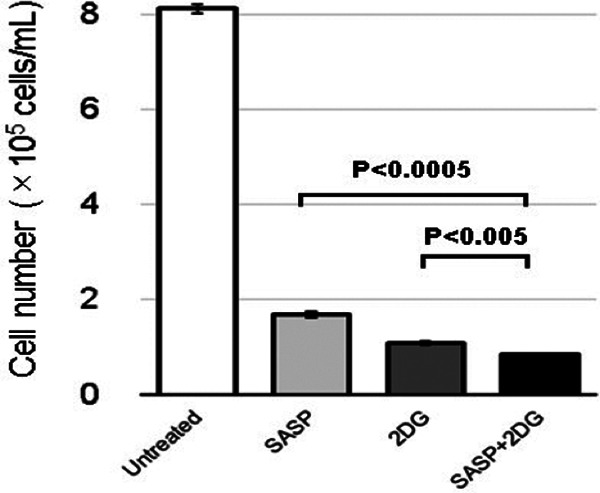
Growth of MKN45 cells treated with SASP, 2DG, or SASP + 2DG. MKN45 cells treated with 0.3 mM SASP, 6 mM 2-DG, or 0.3 mM SASP + 6 mM 2-DG were cultured for 72 hours and the cell number was quantified. Salazosulfapyridine suppressed cell proliferation, which was exacerbated by 2DG. Data are expressed as mean ± SD of at least 3 independent experiments. P < .0005, SASP versus SASP + 2DG and P < .005, 2DG versus SASP + 2DG. 2DG indicates 2-deoxyglucose; SASP, salazosulfapyridine.
Discussion
Rapidly growing cancer cells avoid apoptosis by enhancing the production of GSH, which inhibits the production of ROS. CD44v, a splice variant of CD44 and cancer stem cell biomarker, binds to xCT and increases cystine uptake to maintain its intracellular level, thereby promoting glutathione metabolism.9 In addition, CD44v interacts with and inhibits the activity of pyruvate kinase muscle isozyme M2, promoting glycolysis to sustain the NADPH production pathway, an auxiliary pathway of the PPP.36
Recently, clinical trials were conducted to assess the role of the xCT inhibitor, SASP, in advanced cancer patients. These studies were aimed at evaluating the effect of xCT inhibition in mitigating glutathione and hence, ROS production, to enhance sensitivity to cisplatin and other chemotherapeutic agents. In theory, treatment strategies targeting cancer stem cells ought to prevent recurrence and metastasis and hold the potential to cure cancer.
In the present study, CE-TOFMS-based metabolomic analyses of cultured cancer cells following SASP administration were carried out to assess global changes in the glutathione metabolism pathway. MKN45 cells expressed the highest level of CD44v among the screened cancer cell lines. These cells harbored the wild-type p53, inhibited phosphofructokinase, the rate-limiting enzyme of glycolysis, and were associated with predominant mitochondrial respiration.37 Lactic acid production and glycolysis were significantly enhanced in hypoxic cultures. These results suggest that in p53 wild-type cells, hypoxic conditions may induce a shift toward glycolysis-predominant metabolism by stabilizing hypoxia-inducible factor 1, inducing pyruvate dehydrogenase kinase 1, and inhibiting pyruvate dehydrogenase that forms pyruvic acid from acetyl coenzyme A, thereby blocking the pathway leading to oxidative phosphorylation.38,39
Administration of SASP under hypoxic conditions significantly decreased the levels of Cys and GSH. Although the concentration of metabolites at each step in glycolysis was higher than that of untreated cells, the effect of SASP on glycolysis remains unclear. In addition, significant differences were not detected in the concentration of NADPH produced in the PPP. The glycolysis inhibitor 2DG interrupted the progression of glycolysis to the PPP, leading to the reduced production of NADPH. In addition, glutathione levels were lower in cells treated with SASP and 2DG than in SASP-treated cells. 2-Deoxyglucose is a glucose molecule in which the 2-hydroxyl group is replaced by a hydrogen atom. 2-Deoxyglucose cannot be metabolized by glycolytic enzymes and thus, competitively and non-competitively inhibits phosphoglucose isomerase and hexokinase, respectively.40,41
2-Deoxyglucose- and SASP-mediated inhibition of glutathione suggests increased sensitivity to chemotherapeutic agents. Accordingly, cell growth was significantly inhibited in 2DG- and SASP-treated cells compared with that in SASP treated cells. As evidenced by its use as an optical imaging agent in positron emission tomography, 2DG is selectively taken up by cancer cells where glycolysis predominates. This selectivity could be exploited specifically to target cancer cells when used in combination with SASP.42
In conclusion, the results of our study indicate that in cancer cells where the glycolytic pathway is predominant, metabolomic analyses under hypoxic conditions enable the profiling of global metabolism, including glycolysis. Inhibiting the production of NADPH by blocking glycolysis might be exploited as a new treatment strategy for cancer in addition to cystine blockade by SASP.
Acknowledgment
The authors would like to thank Editage (www.editage.com) for English language editing.
Abbreviations
- ATCC
American Type Culture Collection
- BSA
bovine serum albumin
- CD
cluster of differentiation
- CE
capillary electrophoresis
- xCT
the cystine/glutamate exchanger protein
- Cys
Cysteine
- FBS
fetal bovine serum
- GSH
reduced glutathione
- HMT
Human Metabolome Technologies
- JCRB
Japanese Collection of Research Bioresources Cell Bank
- LC
liquid chromatography
- NADPH
Reduced nicotinamide adenine dinucleotide phosphate
- MS
mass spectrometry
- PBS
phosphate-buffered saline
- ROS
reactive oxygen species
- RPMI
Roswell Park Memorial Institute
- PPP
pentose phosphate pathway
- SASP
salazosulfapyridine
- TOF
time-of-flight
- 2DG
2-deoxyglucose.
Authors’ Note: Our study did not require an ethical board approval because it did not contain human or animal trials.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Kohei Takizawa, MD  https://orcid.org/0000-0002-7702-0427
https://orcid.org/0000-0002-7702-0427
References
- 1. Gotoda T, Yanagisawa A, Sasako M, et al. Incidence of lymph node metastasis from early gastric cancer: estimation with a large number of cases at two large centers. Gastric Cancer. 2000;3(4):219–225. [DOI] [PubMed] [Google Scholar]
- 2. Japanese Gastric Cancer Association. Japanese gastric cancer treatment guidelines 2010 (ver. 3). Gastric Cancer. 2011;14(2):113–123. [DOI] [PubMed] [Google Scholar]
- 3. Hatta W, Gotoda T, Oyama T, et al. Is radical surgery necessary in all patients who do not meet the curative criteria for endoscopic submucosal dissection in early gastric cancer? A multi-center retrospective study in Japan. J Gastroenterol. 2017;52(2):175–184. [DOI] [PubMed] [Google Scholar]
- 4. Hatta W, Gotoda T, Oyama T, et al. A scoring system to stratify curability after endoscopic submucosal dissection for early gastric cancer: “eCura system”. Am J Gastroenterol. 2017;112(6):874–881. [DOI] [PubMed] [Google Scholar]
- 5. Kawata N, Kakushima N, Takizawa K, et al. Risk factors for lymph node metastasis and long-term outcomes of patients with early gastric cancer after non-curative endoscopic submucosal dissection. Surg Endosc. 2017;31(4):1607–1616. [DOI] [PubMed] [Google Scholar]
- 6. Yasui W, Kudo Y, Naka K, et al. Expression of CD44 containing variant exon 9 (CD44v9) in gastric adenomas and adenocarcinomas: relation to the proliferation and progression. Int J Oncol. 1998;12(6):1253–1258. [DOI] [PubMed] [Google Scholar]
- 7. Bannai S, Kitamura E. Transport interaction of l-cysteine and l-glutamate in human diploid fibroblasts in culture. J Biol Chem. 1980;255(6):2372–2376. [PubMed] [Google Scholar]
- 8. Sato H, Tamba M, Ishii T, Bannai S. Cloning and expression of a plasma membrane cystine/glutamate exchange transporter composed of two distinct proteins. J Biol Chem. 1999; 274(17):11455–11458. [DOI] [PubMed] [Google Scholar]
- 9. Ishimoto T, Nagano O, Yae T, et al. CD44 variant regulates redox status in cancer cells by stabilizing the xCT subunit of system xc(-) and thereby promotes tumor growth. Cancer Cell. 2011;19(3):387–400. [DOI] [PubMed] [Google Scholar]
- 10. Yae T, Tsuchihashi K, Ishimoto T, et al. Alternative splicing of CD44 mRNA by ESRP1 enhances lung colonization of metastatic cancer cell. Nat Commun. 2012;3:883. [DOI] [PubMed] [Google Scholar]
- 11. Otsubo K, Nosaki K, Imamura CK, et al. Phase I study of salazosulfapyridine in combination with cisplatin and pemetrexed for advanced non-small-cell lung cancer. Cancer Sci. 2017;108(9):1843–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shitara K, Doi T, Nagano O, et al. Phase 1 study of sulfasalazine and cisplatin for patients with CD44v-positive gastric cancer refractory to cisplatin (EPOC1407). Gastric Cancer. 2017;20(6):1004–1009. [DOI] [PubMed] [Google Scholar]
- 13. Pullar T, Capell HA. Sulphasalazine: a ‘new’ antirheumatic drug. Br J Rheumatol. 1984;23(1):26–34. [DOI] [PubMed] [Google Scholar]
- 14. Schröder H, Campbell DE. Absorption, metabolism, and excretion of salicylazosulfapyridine in man. Clin Pharmacol Ther. 1972;13(4):539–551. [DOI] [PubMed] [Google Scholar]
- 15. Svartz N. Salazopyrin, a new sulfanilamide preparation. A. Therapeutic results in rheumatic polyarthritis. B. Therapeutic results in ulcerative colitis. C. Toxic manifestations in treatment with sulfanilamide preparations. Acta Med Scand. 1942;110(6):577–598. [Google Scholar]
- 16. Azad Khan AK, Piris J, Truelove SC. An experiment to determine the active therapeutic moiety of sulphasalazine. Lancet. 1977;2(8044):892–895. [DOI] [PubMed] [Google Scholar]
- 17. Hoult JR. Pharmacological and biochemical actions of sulphasalazine. Drugs. 1986;32(suppl 1):18–26. [DOI] [PubMed] [Google Scholar]
- 18. Shahidi NT, Westring DW. Acetylsalicylic acid-induced hemolysis and its mechanism. J Clin Invest. 1970;49(7):1334–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xi H, Kurtoglu M, Liu H, et al. 2-Deoxy-D-glucose activates autophagy via endoplasmic reticulum stress rather than ATP depletion. Cancer Chemother Pharmacol. 2011;67:899–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zagorodna O, Martin SM, Rutkowski DT, Kuwana T, Spitz DR, Knudson CM. 2-deoxyglucose-induced toxicity is regulated by Bcl-2 family members and is enhanced by antagonizing Bcl-2 in lymphoma cell lines. Oncogene. 2012;31:2738–2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ralser M, Wamelink MM, Struys EA, et al. A catabolic block does not sufficiently explain how 2-deoxy-D-glucose inhibits cell growth. Proc Natl Acad Sci USA. 2008;105:17807–17811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Coleman MC, Asbury CR, Daniels D, et al. 2-deoxy-D-glucose causes cytotoxicity, oxidative stress, and radiosensitization in pancreatic cancer. Free Radic Biol Med. 2008;44(3):322–331. [DOI] [PubMed] [Google Scholar]
- 23. Zhang D, Li J, Wang F, Hu J, Wang S, Sun Y. 2-Deoxy-D-glucose targeting of glucose metabolism in cancer cells as a potential therapy. Cancer Lett. 2014;355(2):176–183. [DOI] [PubMed] [Google Scholar]
- 24. Sobhakumari A, Orcutt KP, Love-Homan L, et al. 2-Deoxy-d-glucose suppresses the in vivo antitumor efficacy of erlotinib in head and neck squamous cell carcinoma cells. Oncol Res. 2016;24(1):55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thanee M, Loilome W, Techasen A, et al. CD44 variant-dependent redox status regulation in liver fluke-associated cholangiocarcinoma: a target for cholangiocarcinoma treatment. Cancer Sci. 2016;107(7):991–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sehm T, Fan Z, Ghoochani A, et al. Sulfasalazine impacts on ferroptotic cell death and alleviates the tumor microenvironment and glioma-induced brain edema. Oncotarget. 2016;7(24):36021–36033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Miyoshi S, Tsugawa H, Matsuzaki J, et al. Inhibiting xCT improves 5-fluorouracil resistance of gastric cancer induced by CD44 variant 9 expression. Anticancer Res. 2018;38(11):6163–6170. [DOI] [PubMed] [Google Scholar]
- 28. Kaplan O, Navon G, Lyon RC, et al. Effects of 2-deoxyglucose on drug-sensitive and drug-resistant human breast cancer cells: toxicity and magnetic resonance spectroscopy studies of metabolism. Cancer Res. 1990;50(3):544–551. [PubMed] [Google Scholar]
- 29. Yamaguchi R, Janssen E, Perkins G, et al. Efficient elimination of cancer cells by deoxyglucose-ABT-263/737 combination therapy. PLoS One. 2011;6(9):e24102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lin X, Zhang F, Bradbury CM, et al. 2-Deoxy-D-glucose-induced cytotoxicity and radiosensitization in tumor cells is mediated via disruptions in thiol metabolism. Cancer Res. 2003;63(12):3413–3417. [PubMed] [Google Scholar]
- 31. Urakami K, Zangiacomi V, Yamaguchi K, et al. Impact of 2-deoxy-d-glucose on the target metabolome profile of a human endometrial cancer cell line. Biomed Res. 2013;34(5):221–229. [DOI] [PubMed] [Google Scholar]
- 32. Ohashi Y, Hirayama A, Ishikawa T, et al. Depiction of metabolome changes in histidine-starved Escherichia coli by CE-TOFMS. Mol Biosyst. 2008;4(2):135–147. [DOI] [PubMed] [Google Scholar]
- 33. Todaka A, Umehara R, Sasaki K, et al. Metabolic profiling of gemcitabine- and paclitaxel-treated immortalized human pancreatic cell lines with K-RAS(G12D). Biomed Res. 2017;38(1):29–40. [DOI] [PubMed] [Google Scholar]
- 34. Klukas C, Schreiber F. Integration of -omics data and networks for biomedical research with VANTED. J Integr Bioinform. 2010;7(2):112. [DOI] [PubMed] [Google Scholar]
- 35. Ju SY, Chiou SH, Su Y. Maintenance of the stemness in CD44(+) HCT-15 and HCT-116 human colon cancer cells requires miR-203 suppression. Stem Cell Res. 2014;12(1):86–100. [DOI] [PubMed] [Google Scholar]
- 36. Tamada M, Nagano O, Tateyama S, et al. Modulation of glucose metabolism by CD44 contributes to antioxidant status and drug resistance in cancer cells. Cancer Res. 2012;72(6):1438–1448. [DOI] [PubMed] [Google Scholar]
- 37. Yokozaki H. Molecular characteristics of eight gastric cancer cell lines established in Japan. Pathol Int. 2000;50(10):767–777. [DOI] [PubMed] [Google Scholar]
- 38. Schofield CJ, Ratcliffe PJ. Oxygen sensing by HIF hydroxylases. Nat Rev Mol Cell Biol. 2004;5(5):343–354. [DOI] [PubMed] [Google Scholar]
- 39. Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3(10):721–732. [DOI] [PubMed] [Google Scholar]
- 40. Chen W, Guéron M. The inhibition of bovine heart hexokinase by 2-deoxy-d-glucose-6-phosphate: characterization by 31P NMR and metabolic implications. Biochimie. 1992;74(9-10):867–873. [DOI] [PubMed] [Google Scholar]
- 41. Wick AN, Drury DR, Nakada HI, Wolfe JB. Localization of the primary metabolic block produced by 2-deoxyglucose. J Biol Chem. 1957;224(2):963–969. [PubMed] [Google Scholar]
- 42. Bensinger SJ, Christofk HR. New aspects of the Warburg effect in cancer cell biology. Semin Cell Dev Biol. 2012;23(4):352–361. [DOI] [PubMed] [Google Scholar]