Abstract
Nogo-B receptor (NgBR) is a specific receptor of Nogo-B that regulates vascular remodeling and angiogenesis. Previously, we found that NgBR promotes the membrane translocation and activation of Ras in breast cancer cells and enhances the chemoresistance of hepatocellular carcinoma cells to 5-fluorouracil. However, the role of NgBR in lung cancer has not yet been elucidated. In the present study, we found that NgBR knockdown inhibited epithelial—mesenchymal transition (EMT) in non-small cell lung cancer (NSCLC) cells in vitro and metastasis of NSCLC cells in vivo. In contrast, NgBR overexpression promoted EMT in and lung metastasis of NSCLC cells. At the molecular level, NgBR modulated the expression of EMT-related proteins and enhanced the protein expression of Snaill, a crucial transcription factor that represses epithelial cell protein marker E-cadherin. Moreover, we found that NgBR overexpression promoted the membrane localization of Ras and activation of downstream MEK/ERK signaling pathway and that NgBR knockdown by using a specific shRNA inversely affected the expression of EMT-related proteins in NSCLC cells. Thus, our results provide novel insights on the regulatory role of NgBR in the metastasis of NSCLC that should be investigated further for developing a therapeutic strategy for treating patients with NSCLC.
Keywords: Nogo-B receptor, Non-small cell lung cancer, Epithelial–mesenchymal transition, Metastasis, Snail1
1. Introduction
Lung cancer is one of the most critical health problems worldwide, with high incidence and poor survival rates [1,2]. Histologically, lung cancer is classified into small cell lung cancer and non-small cell lung cancer (NSCLC), which accounts for 85% of all lung cancer cases [3,4]. The overall 5-year survival rate of patients with NSCLC is low (approximately 18%) [1,2] because majority of patients with lung cancer (57%) are diagnosed at advanced stages of the disease [3], Clinically, early NSCLC is usually asymptomatic, and only 16% patients with NSCLC are diagnosed at an early stage [3]. Therefore, a better understanding of molecular mechanisms underlying NSCLC development and progression could provide helpful insights for NSCLC prevention, early detection, and effective treatment.
During lung carcinogenesis, normal lung epithelial cells undergo malignant transformation through an undefined etiology and mechanism; moreover, NSCLC progression may involve tumor epithelial–mesenchymal transition (EMT) to promote tumor cell invasion and metastasis [4,5]. During metastasis, tumor cells gain access to and spread through the blood and lymphatic systems to metastasize to distant sites [4,5].
EMT is a relatively complex process during which epithelial cells gradually lose their polarity and are transformed into mesenchymal-like cells with enhanced mobility and invasion capacity [6]. EMT is usually associated with the downregulation of epithelial phenotypic protein (E-cadherin) expression and upregulation of mesenchymal phenotypic protein (N-cadherin, vimentin, Snail1, and Twistl) expression [4,5]. Expression of EMT-related proteins is tightly regulated by multiple gene pathways [4,7.8].
Nogo-B receptor (NgBR) is a specific receptor of Nogo-B that regulates vascular remodeling and angiogenesis [9], We previously showed that NgBR is an important gene in cerebral vasculature development and is essential for angiogenesis, which is mediated by the vascular endothelial growth factor/Akt pathway, in zebrafish [10,11]. NgBR expression is upregulated in breast cancer and enhances tumor progression by coordinating with estrogen receptor alpha and survivin to promote the transition of tumor cells to mesenchymal stem cells [12]. NgBR is also overexpressed in hepatocellular carcinoma tissues, which decreases the survival and chemoresistance of patients with hepatocellular carcinoma to 5fluorouracil because of the ubiquitination of p53 [13], At the molecular level, NgBR functions as a docking site for prenylated Ras isoforms (H-Ras and K-Ras) and promotes their localization to the plasma membrane and subsequent activation [14]. However, the role of NgBR and NgBR-mediated signaling pathway in NSCLC is still unknown. Therefore, in this study, we assessed NgBR expression in NSCLC cells and tissues and investigated the effects of NgBR overexpression or knockdown in NSCLC cells in vitro and in a mouse metastasis model.
2. Materials and methods
2.1. Cell lines and culture
Human NSCLC A549, H1299, H460, H520, SPC-A-1, and Anip973 cell lines and normal lung cell CCL-153 cell line were obtained from the American Type Culture Collection (Manassas, VA, USA), where the cell lines were authenticated by STR profiling before distribution. The cell lines were maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum (Gibco, Shanghai, China) in a humidified incubator with 5% CO2 at 37 °C. The cells with or without gene transfection were treated with or without 10 μM MEK inhibitor, U0126, which was purchased from Selleck Chemicals (Houston, TX, USA).
2.2. Knockdown and overexpression of NgBR in NSCLC cells
NSCLC cells were transiently transfected with All-Star non-silencing siRNA (NS, 5'-UUCUCCGAACGUGUCACGUTT-3' and 5'ACGUGACACGUUCGGAGAATT-3') or three different NgBR siRNA oligonucleotides with 3'dTdT overhangs (S1, 5'-GGAAAUACAUAGACCUACA-3' and 5'-UGUAGGUCUAUGUAUUUC-3’; S2, 5'GUAUGGAAAUAAACU UAUA-3' and 5'-UAUAAGUUUAUUUCCAUAC3’; S3, 5'-GCUGAUUCUUAGAUAGAAA-3' and 5'-UUUCUAUCUAAGAAUCAGC-3') as described previously [13]. These oligonucleotides were synthesized by GenePharma (Shanghai, China).
Furthermore, NSCLC cells were grown and transiently transfected with pIRES-NgBR plasmid as described previously [9,11 ]. NgBR expression was stably knocked down in H1299 and A549 cell lines using NgBR shRNA (the target sequences were 5'-CGGTCAATAAGTTGTAATCTTG-3') with puromycin selection.
2.3. Western blot analysis
Cells were lysed in the radioimmunoprecipitation assay buffer containing protease inhibitors and the concentration of protein samples was measured using the BCA Protein Assay Kit (Beyotime, Shanghai, China). An equal amount of protein samples was subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto nitrocellulose membranes (Millipore, Billerica, MA, USA). The membranes were blocked with 5% non-fat milk and then incubated at 4°C overnight with primary antibodies including rabbit polyclonal antiNgBR and anti-MEK1/2 (Abeam, Cambridge, USA), rabbit polyclonal anti-phospho-Akt (Ser473), total Akt, phospho-p44/42 MAPK (ERK1/2; Thr202/Tyr204), p44/42 MAPK (ERK1/2), p-MEKl/ 2(Ser217/221), Na and K-ATPase antibodies (Cell Signaling Technology, Danvers, MA, USA), and monoclonal antibodies anti-Ecadherin, N-cadherin, Vimentin, Twistl, Snaill, β-actin, K-Ras, H-Ras, and EGFR antibodies (ProteinTech, Wuhan, China). Next, the membranes were incubated with the anti-mouse or anti-rabbit IgG-conjugated with horseradish peroxidase and the protein bands were visualized using Western bright ECL kit (Advansta, Menlo Park, CA, USA).
2.4. Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
Total cellular RNA was isolated from cells using Trizol reagent (TaKaRa Bio, Dalian, China) and was then reversely transcribed into cDNA using PrimeScript RT Reagent Kit (TaKaRa) according to the manufacturer’s instructions. qPCR was then performed using QuantiTect SYBR Green PCR kit (TaKaRa) in Stratagene MX3000P (Agilent Technologies, Santa Clara, CA, USA). The primers used were NgBR (5'-TGCCAGTTAGTAGCCCAGAAGCAA-3' and 5'-TGATGTGCCAGGGAAGAAAGCCTA-3'), Snaill (5'-CGAGTGGTTCTTCTGCGCTA3' and 5'-CTGCTGGAAGGTAAACTCTGGA-3'), Twistl (5'TTCTGCCTCTTTCGAGCACC-3' and 5'-TACAACGACCCAGTCTGACG3'), and β-actin (5'-TTCTACAATGAGCTGCGTGTGGCT-3' and 5'TAGCACAGCCTGGATAGCAACGTA-3'). β-actin was used as a normalized control.
2.5. Cell viability CCK-8 assay
Stably transfected NSCLC cells were seeded into 96-well plates at a density of 5000 cells/well and grown for up to 72 h. The medium was removed from each well and replaced with a mixture of 10 μl Cell Counting Kit-8 (CCK-8; Dojindo, Beijing, China) and 100 μl RPMI-1640 and then incubated in 37 °C for 2 h. After that, the optical density of each well was measured using an EnSpire™ 2300 Multilabel Reader (Perkin Elmer, Waltham, MA, USA) at 450 nm. Each experiment was in quintuple repeats.
2.6. Colony formation assay
Stably transfected NSCLC cells were seeded into 96-well plates at a density of 1000cells/well and grown for at 37 °C for 2 weeks and the medium was replaced with a fresh one every three days. The medium was then removed, and cells were washed twice with PBS, fixed with 4% paraformaldehyde, and stained with Crystal Violet (Sigma-Aldrich, St. Louis, MO, USA). Cell colonies with 50 cells or more were counted under an inverted light microscope (Olympus, Japan). The data were expressed as mean ± SD of quintuple repeats.
2.7. Tumor cell migration and invasion assays
In the wound-healing assay, transfected cells were seeded into the six-well plates and grown to reach 90%–100% confluency. Multiple wounds were created by using a 200-μl pipette tip. The cells were washed with PBS and then cultured for 24 h in a medium containing 1% FBS. Images were recorded at 0,12, and 24 h and the wound healing was measured from these images. The experiments were performed in triplicates and the results were expressed as mean ± SD.
In Transwell assay, cells were seeded in 24-well-Transwell inserts (Costar/Sigma, St Louis, MO, USA) at a density of 50,000 cells and the bottom of the inserts was filled with 500 μl medium containing 10% FBS. These 24-well plates were cultured for 24 h. For Transwell invasion assay, the filter in the insert was precoated with Matrigel (BD Biosciences, San Jose, CA, USA). Cells remained on the top surface of the filter were removed using a cotton swab, while cells migrated or invaded into the bottom surface were washed with PBS, fixed with 4% paraformaldehyde for 15 min and stained with Crystal Violet solution for 10 min. Cells were then counted under an inverted light microscope (Olympus).
2.8. Immunofluorescence staining
Transfected NSCLC cells were seeded on to cover slips and grown to reach 50%–70% confluency. Cells were then washed with PBS, fixed with 4% paraformaldehyde, permeabilized with or without 0.3% Triton-X, and blocked with 5% normal goat serum at 37 °C for 30 min. After that, the cover slips were incubated with a rabbit polyclonal NgBR antibody that was generated in our lab [12] overnight at 4 °C. On the next day, cells were washed with PBST and incubated with a fluorescence secondary antibody at 37 °C for 1 h. After washing with PBS, the cells were stained with 4',6-diamidino2-phenylindole (DAPI; Sigma) for 5 min and reviewed and scored under a light microscope (Nikon, Japan).
2.9. Active Ras pull-down assay
The active Ras pull-down assay was performed using the assay kit from Thermo Fisher Scientific (Waltham, MA, USA). Specifically, cells were grown to 80–90% confluency and added with the lysis/ binding/wash buffer containing protease inhibitors to extract total cellular protein. After quantitation, the samples were mixed with GST-Raf1-RBD from the kit and then with Glutathione Resin and agarose beads and incubated at 4°C for 1 h and the mixture was centrifuged at 15,000 rpm for 15 min and the samples were eluted with dithiothreitol solution, and the resulted pull-down samples were subjected to Western blotting analysis of active Ras.
2.10. Tissue microarray and immunohistochemistry
NSCLC tissue microarrays (TMA), including 52 NSCLC and their corresponding adjacent lung tissues as well as positive lymph nodes were purchased from Shanghai Outdo Biotech (Shanghai, China). TMA and mouse tissue sections were deparaffinized in xylene and rehydrated in series of ethanol solutions and then boiled in a citrate buffer (pH6.0) in a microwave oven for antigen retrieval and incubated in 3% hydrogen peroxide (H2O2) to deactivate potential endogenous peroxide activity. The sections were subsequently incubated with 5% normal goat serum and then with a rabbit polyclonal NgBR antibody [12] overnight at 4 °C in a wet box. On the next day, the sections were washed with PBS thrice and then incubated with pv-9000 kit (ZSGB-Bio, Beijing, China) and were subjected to color development using 3,3-diaminobenzidine (ZSGB-Bio, Beijing, China) and counterstaining with hematoxylin. The proportion of immunostained tumor or epithelial cells and the staining intensity were assessed, i.e„ % of staining was set to be 0 (<5%), 1 (5–25%), 2 (>25–50%), 3 (>50–75%), or 4 (>75%), while the staining intensity was 0, negative; 1, weak; 2, moderate; or 3, strong according to a previous study [13], These two scores were then multiplied to obtain a staining index. A low NgBR expression was defined as a staining index of <6, whereas a high NgBR was ≥6 as described in our previous study [12].
2.11. In vivo lung metastasis assay
The protocol was approved by the Animal Care and Ethics Committee of Dalian Medical University. Male BALB/c nude mice (four weeks old) were purchased from Charles River Laboratories (Beijing, China) and maintained in a specific-pathogen-free environment in an animal facility of Dalian Medical University. The mice were divided into five groups (n = 5) and 100 μl A549 cells with stable NgBR overexpression or NC and H1299 cells with stable NgBR knockdown or NC (1 × 106 cells per mouse) were injected into the mouse tail vein. The mice were then monitored for 60 days and sacrificed and the lung tissues were resected. The morphology of the lung was inspected and processed for sectioning and staining with hematoxylin and eosin (HE).
2.12. Kaplan-Meier survival analysis
NgBR (NUS1) mRNA expression values were retrieved from lung cancer profiling dataset deposited in a Kaplan-Meier Plotter (probe ID:215207_x_at NUS1, Version 2015, n = 2437), the complete analysis tool can be accessed online at: www.kmplot.com/lung [15]. The cumulative survival probability was evaluated by using the Kaplan-Meier method and split patients by median expression. And the online setting for the analysis was followed the previous report [16].
2.13. Statistical analysis
All experiments were repeated for at least three times and data were summarized as mean ± SD. A two-tailed paired t-test was performed for two preselected groups and the comparison among Patients’ clinicopathological characteristics was analyzed using the χ2-test. A p < .05 was defined as statistically significant. All statistical analyses were performed by using SPSS 23.0 (SPSS, Chicago, IL, USA) or GraphPad Prism 7.0 software (GraphPad Software, La Jolla, CA, USA). The survival curve was generated using the Kaplan-Meier method. The median survival comparison between groups was calculated using the log-rank test. P < .05 was considered to indicate a statistically significant difference.
3. Results
3.1. NgBR expression is associated with NSCLC development and metastasis
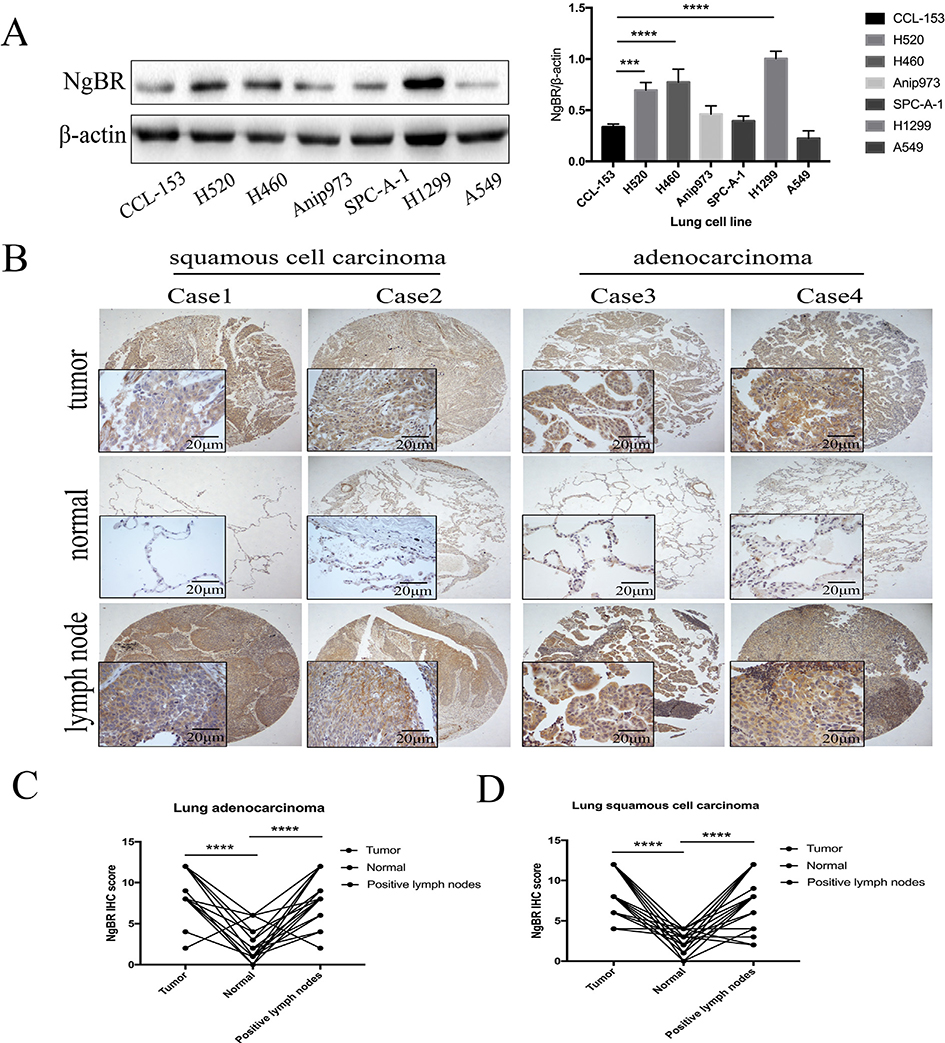
To explore the role of NgBR in NSCLC, we first assessed NgBR expression in normal lung CCL-153 cells and six NSCLC cell lines. Three out of the six NSCLC cell lines, namely, lung squamous cell carcinoma cell line H520, large cell lung cancer cell line H460, and lung adenocarcinoma cell line H1299, showed significantly higher NgBR expression than the CCL-153 cell line (Fig. 1A). Results of immunohistochemistry analysis showed that NgBR was present in both the cell membrane and cytoplasm of NSCLC cells. In adjacent lung tissues, almost no NgBR expression was observed in alveolar epithelial cells but evident NgBR expression was detected in bronchial epithelial cells and stromal cells. In the lymph nodes, no NgBR expression was detected in lymphocytes but high NgBR expression was detected in metastatic tumor cells (Figs. 1B, C, and D and S1A). Immunohistochemistry score indicated that NgBR was highly expressed in NSCLC tissues and in their corresponding tumor-positive lymph nodes compared with that in adjacent lung tissues (Fig. 1B, C, and D). Furthermore, high NgBR expression in NSCLC tissues was associated with tumor lymph node metastasis (p = .024; Table 1). However, no association was observed between NgBR expression and age and gender of patients and pathological grade of NSCLC (Table S1). Next, we obtained NgBR (NUS1) mRNA expression values from a lung cancer profiling dataset deposited in Kaplan–Meier Plotter (probe ID: 215207_x_at NUS1) [15]. Overall survival analysis indicated that patients with high NgBR expression showed significantly lower survival rates than patients with low NgBR expression (p = 9.5e-10; Fig. S1B). These results indicate that NgBR protein expression is upregulated in NSCLC tissues and cell lines and suggest that NgBR upregulation promotes lung tumorigenesis and metastasis.
Fig. 1. NgBR expression in NSCLC tissues and cell lines.
A, expression of NgBR protein was assessed in normal lung cells and NSCLC cell lines using Western blot. Level of β-actin was used as a loading control (left panel). The intensity of protein levels was quantified using the Image Lab 5.2.1 software and normalized to β-actin (right panel). Error bar, SD of three independent experiments. ***p < .001 and ****p < .0001 vs. normal cells. B, representative immunostained tissue microarray shows high NgBR expression in NSCLC tissues and the corresponding tumor cell-positive lymph nodes vs. the adjacent lung tissues. Scale bar, 20 or 200 μm. C and D, Immunostaining score of NgBR expression in 52 cases of NSCLC tissues, their corresponding adjacent lung tissues, and tumor cell-positive lymph nodes. ****p < .0001 vs. normal tissues.
Table 1.
NgBR expression in tumor tissues and tumor-positive lymph nodes (χ2 test).
| Positive lymph nodes |
n | P | |||
|---|---|---|---|---|---|
| Low | High | ||||
| Tumor tissues | Low | 3 | 3 | 6 | 0.024* |
| High | 6 | 40 | 46 | ||
| n | 9 | 43 | 52 | ||
3.2. NgBR is necessary for EMT in NSCLC cells
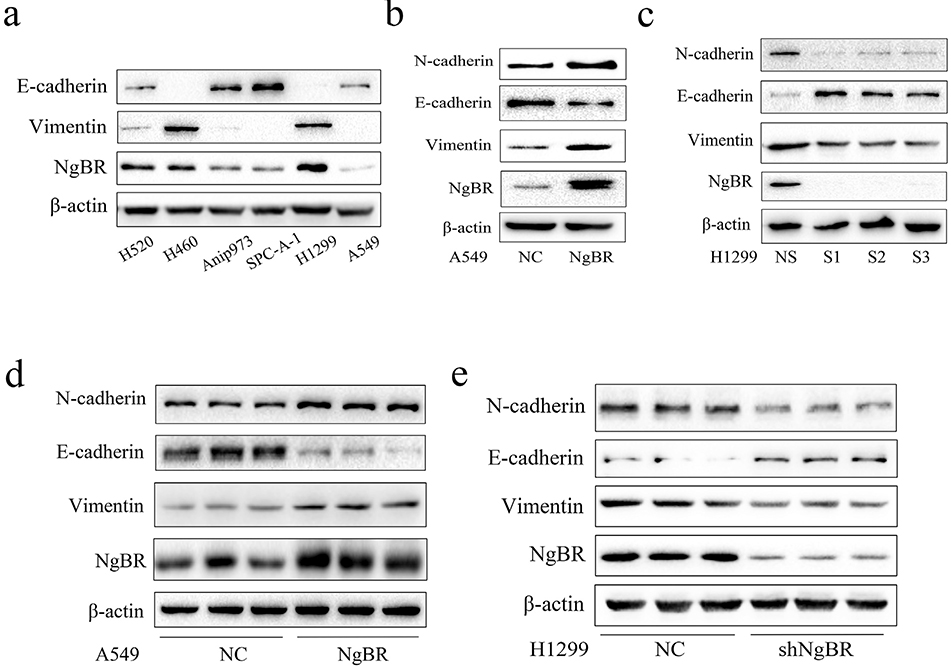
EMT is a driver of tumor metastasis and is involved in the invasion and migration of tumor cells [4,6]. Therefore, we evaluated the expression of EMT-related proteins in parental NSCLC cell lines. We found that parental H520, H460, and H1299 cells, which showed high NgBR expression, showed relatively high expression of vimentin, a mesenchymal phenotypic biomarker (Fig. 2A). In contrast, Anip973, SPC-A-1, and A549 cells, which showed low NgBR expression, showed relatively low expression of vimentin and relatively high expression of E-cadherin, an epithelial phenotypic biomarker (Fig. 2A). Furthermore, NgBR overexpression in A549 cells significantly increased N-cadherin and vimentin expression but decreased E-cadherin expression (Fig. 2B). In contrast, NgBR knockdown in H1299 cells decreased N-cadherin and vimentin expression but increased E-cadherin expression (Fig. 2C). Similar results were obtained using stable NSCLC cell lines compared with those obtained using transiently transfected NSCLC cell lines (Figs. 2D and E and S2C and D). Because A549 and H1299 cells are lung adenocarcinoma cells, we determined the role of NgBR in EMT regulation in other types of NSCLC cells such as lung squamous cell carcinoma H520 cells and large cell lung cancer H460 cells by transiently knocking down NgBR expression. Results of NgBR knockdown in H520 and H460 cells were consistent with those obtained using H1299 cells (Fig. S2A and B). Thus, these results indicate that elevated NgBR expression increases N-cadherin and vimentin expression and decreases E-cadherin expression in NSCLC cells. Moreover, these findings suggest that NgBR is essential for EMT in NSCLC cells.
Fig. 2. NgBR regulates EMT in NSCLC cell lines.
A, expression of NgBR, E-cadherin, and Vimentin in NSCLC cells using Western blot analysis. β-actin was used as a loading control. B, A549 cells were transfected with plRES-NgBR (NgBR) or plRES-NC (NC) for 72 h and subjected to Western blot analysis of different proteins. C, H1299 cells were transfected with NgBR siRNAs (SI, S2, and S3) or All-Star non-silencing siRNA (NS) for 72 h and then subjected to whole-cell lysate extraction and Western blot analysis. D and E, A549 cells were stably transfected with plRES-NC (NC) or plRES-NgBR (NgBR), while HI299 cells were stably transfected with NgBR shRNA (shNgBR) or nonspecific control (NC) and then subjected to Western blot analysis.
3.3. NgBR promotes the migration and invasion of NSCLC cells in vitro
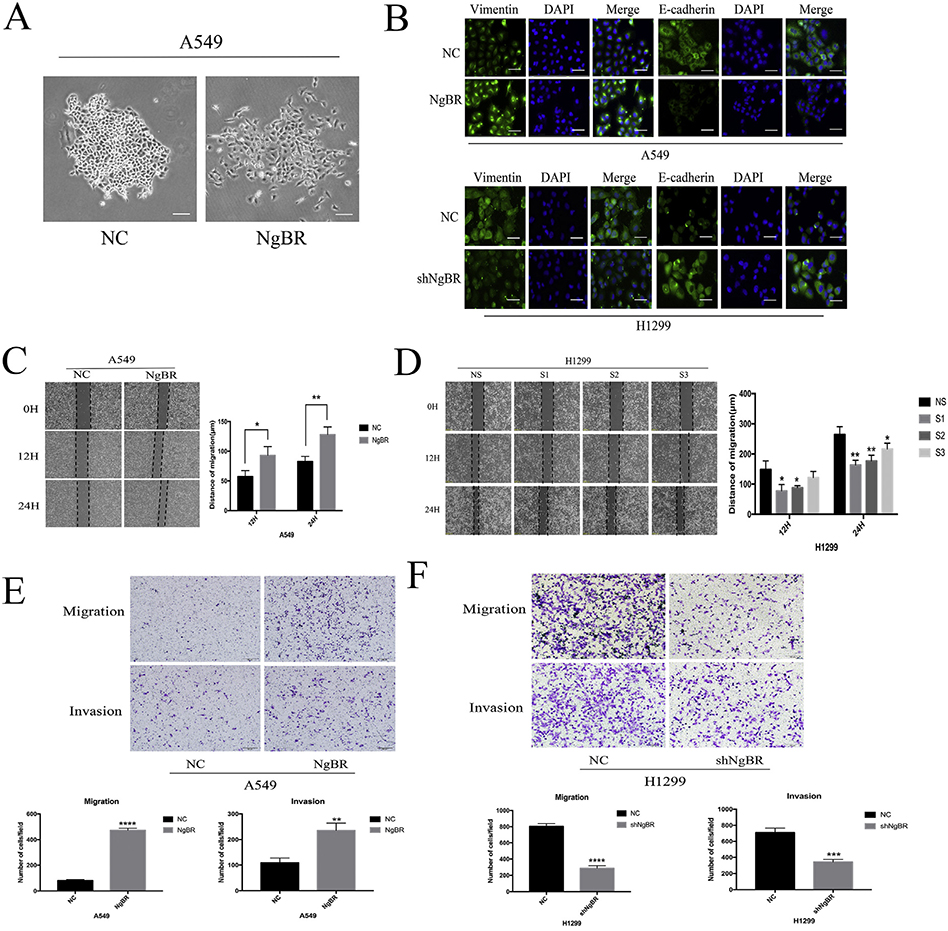
To determine the role of NgBR in regulating the invasion and migration of NSCLC cells, we examined the effect of NgBR overexpression on NSCLC cells. We found that stable NgBR-overexpressing A549 cells showed dispersed morphology without cell-to-cell adhesion, lost their polarity, and showed significantly weak ability to form clusters compared with negative control (NC) cells (Fig. 3A). Results of immunofluorescence staining indicated that NgBR overexpression decreased E-cadherin expression but increased vimentin expression in A549 cells (Fig. 3B), whereas NgBR knockdown exerted opposite effects in H1299 cells (Fig. 3B). These results indicate that NgBR is necessary for maintaining the mesenchymal phenotype of NSCLC cells. Moreover, results of wound healing and Transwell assays showed that NgBR overexpression significantly enhanced the migration and invasion of both A549 (Fig. 3C and E) and H1299 cells (Fig. S3A and D). Moreover, NgBR knockdown suppressed NgBR-induced migration and invasion of H1299 cells (Figs. 3D and F and S3B and C), thus confirming the above findings. However, NgBR overexpression or knockdown did not affect the proliferation and colony formation capacities of A549 and H1299 cells, respectively (Fig. S4A, B, and C). Thus, these results indicate that NgBR increases the invasion and migration capacities of NSCLC cells by promoting EMT.
Fig. 3. NgBR expression regulates NSCLC cells migration and invasion capacity in vitro.
A, cell morphology in stable NgBR-overexpressed A549 cells. Scale bar, 200 μm. B, A549 and H1299 cells were stably transfected with NgBR cDNA or shRNA and then subjected to immunostaining of Vimentin and E-cadherin antibodies, while cell nuclei were visualized with DIPA staining. Scale bar, 37 μm. C, A549 cells were transfected with pIRES-NgBR (NgBR) or pIRES-NC (NC) for 72 h and then subjected to the wound-healing assay. Images were taken at 0 h, 12 h, and 24 h. Scale bar, 200 μm. D, H1299 cells were transfected with NgBR siRNAs (S1, S2 and S3) or All-Star non-silencing siRNA (NS) for 72 h and then subjected to the wound-healing assay. Images were taken at 0 h, 12 h, and 24 h. Scale bar, 200 pm. Error bar, SD of three independent experiments. *p < .05 and **p < .01. E and F, transwell assays assessed tumor cell migration and invasion capacity in stable NgBR overexpressed or knocked down A549 and HI299 cell lines. Images were taken at 24 h in A549 cells and at 8 h (migration), 12 h (invasion) in H1299 cells, respectively. Scale bar, 100 μm. Error bar, SD of three independent experiments. **p < .01, ***p < .001, and ****p < .0001.
3.4. NgBR enhances EMT in NSCLC cells by upregulating Snail1 expression
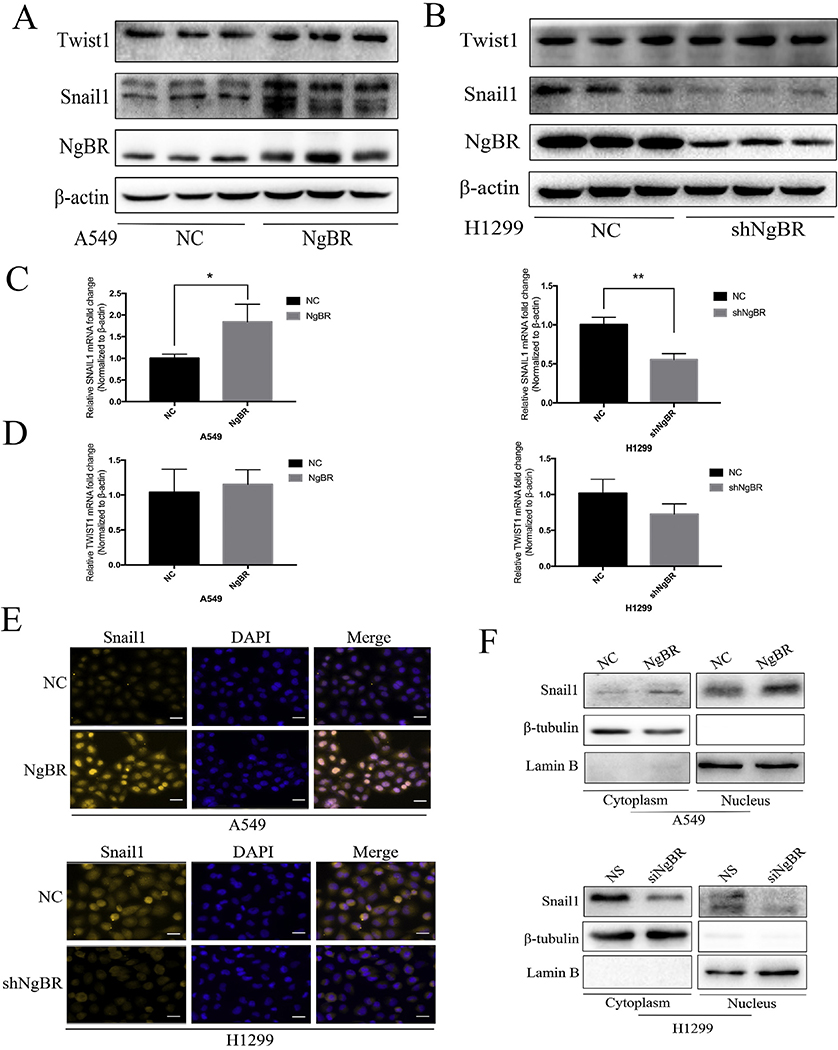
In tumor cells, EMT is modulated at different levels, including epigenetic modification, transcriptional regulation, alternative gene splicing, and/or protein stability alteration and subcellular localization [6]. In the present study, we first explored the role of NgBR in regulating gene expression at the transcriptional level and analyzed the expression of transcription factors Twistl and Snaill, which are evolutionarily conserved repressors of E-cadherin and play an important role in EMT regulation [17,18]. We found that NgBR overexpression significantly increased snaill protein level but did not increase Twist1 protein level in A549 and H1299 cells (Figs. 4A and S5B and C). Stable NgBR knockdown in H1299 cells by using an shRNA (Fig. 4B) and transient NgBR knockdown in A549 and H1299 cells (Fig. S5A) decreased Snaill expression level but did not affect Twistl expression level. Results of qRT-PCR showed that NgBR overexpression increased Snaill mRNA level in both A549 and H1299 cells (Figs. 4C and S5D), whereas NgBR knockdown decreased the Snaill mRNA level but did not significantly change the Twistl mRNA level in H1299 cells (Figs. 4C and D and S5D). Results of immunofluorescence staining and western blotting analysis with subcellular fractionation showed that Snaill expression increased in both the nucleus and cytoplasm of NgBR-overexpressing A549 cells (Fig. 4E and F, upper panel) but decreased in the nucleus and cytoplasm of NgBR-knockout H1299 cells (Fig. 4E and F, lower panel). This finding suggests that NgBR enhances EMT by inducing Snaill expression but not Twistl expression.
Fig. 4. NgBR expression enhances NSCLC cells EMT progression through upregulating of Snaill expression.
A and B, levels of Snail1 and Twist1 proteins were detected by using Western blotting in stable A549 and H1299 cells. β-actin was used as a loading control. C and D, levels of Snaill and Twist1 mRNA were assessed using qRT-PCR in stable A549 and H1299 cells. (3-actin was used as a normalized control. Error bar, SD of three independent experiments. *p < .05 and **p < .01. E, stable A549 and H1299 cells were immunostained with the Snaill antibody, while cell nuclei were stained with DARI staining. Scale bar, 37 μm. F, A549 cells were transfected with pIRES-NgBR (NgBR) or pIRES-NC (NC) and H1299 cells were transfected with NgBR siRNA (siNgBR) or NS for 72 h and then subjected to extraction of nuclear and cytoplasmic protein and Western blot analysis to assess levels of Snaill subcellular localization. LaminB and β-tubulin were used as loading controls of the nuclear and cytoplasmic proteins, respectively.
3.5. NgBR increases Snail 1 expression in NSCLC cells by activating MEK/ERK pathway
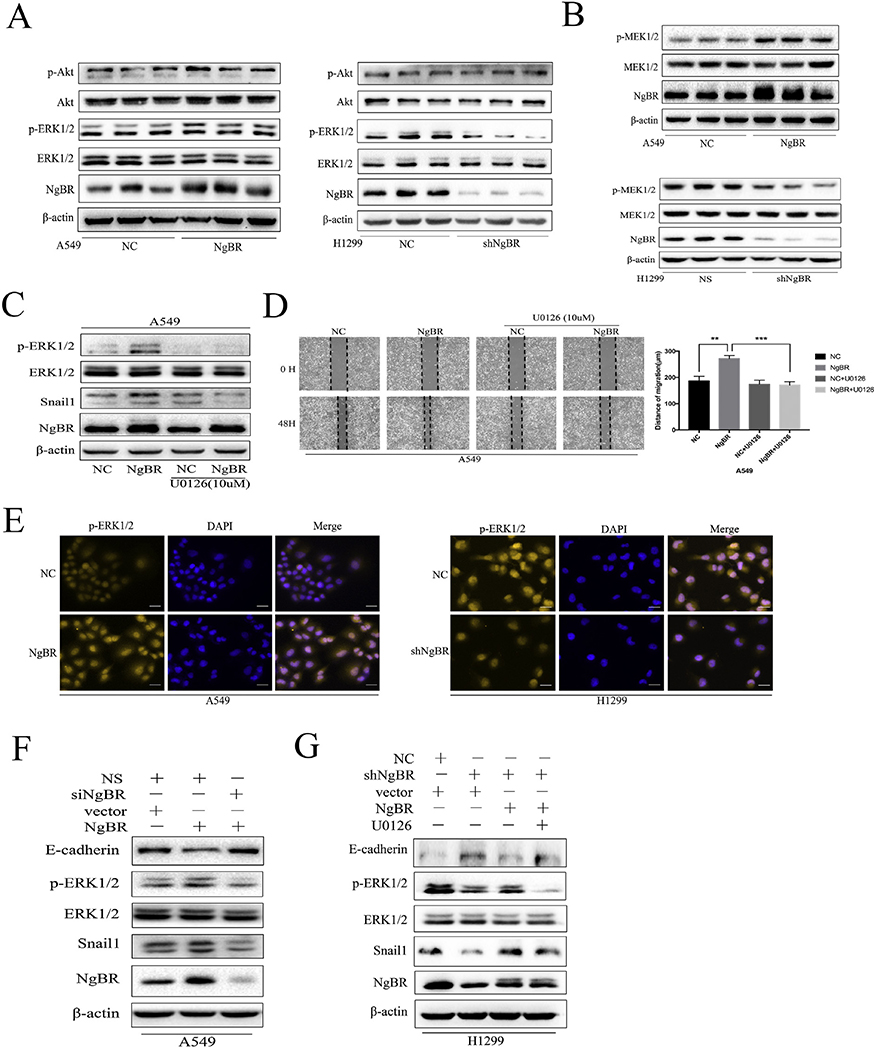
Snail1 expression is driven by the activation of PI3K/Akt and MEK/ERK signaling pathways [19–22]. Therefore, we assessed the activity of these signaling pathways in NgBR-overexpressing A549 and H1299 cells. Interestingly, NgBR overexpression increased the ERK1/2 phosphorylation (p-ERKl/2) level but did not increase Akt phosphorylation in A549 and H1299 cells (Figs. 5A and S6A and C). In contrast, NgBR knockdown decreased p-ERKl/2 level in A549 and H1299 cells (Figs. 5Aand S6B). Similar results were obtained for the MEK1/2 phosphorylation level in NgBR-overexpressing or NgBR-knockout A549 and H1299 cells (Figs. 5B and S6D). These results suggest that NgBR-induced Snaill expression is mediated by the activation of the MEK/ERK signaling pathway. To confirm this, we examined the effect of blocking ERK1/2 activation by using an MEK1/2 inhibitor U0126 on Snaill expression level. Our results showed that treatment with 10 μM U0126 suppressed NgBR overexpression-induced p-ERKl/2 and Snaill levels in A549 (Fig. 5C and H1299 cells (Fig. S6E).
Fig. 5. NgBR expression upregulates Snaill expression through activation of the MEK/ERK/Snaill pathway in NSCLC cells but not the PI3K/Akt pathway.
A, levels of p-ERK1/2 and p-Akt proteins were assessed using Western blotting. Total ERK1/2 and Akt proteins were used as a reference for loading controls in stable A549 and H1299 cells. B, levels of pMEK1/2 protein were assessed using Western blot in stable A549 and HI299 cells. Levels of total MEK1/2 and β-actin proteins were used as loading controls. C, A549 cells were stably transfected with pIRES-NC (NC) and pIRES-NgBR (NgBR) and then treated with or without U0126 (10 μM) for 24 h and then subjected to Western blot analysis of p-ERKl/2, ERK1/2, and Snaill. D, A549 cells were stably transfected with pIRES-NC (NC) and pIRES-NgBR (NgBR) and then incubated with or without U0126 (10 μM) for 24 h and subjected to the wound-healing assay to assess tumor cell migration. Images were taken at 0 h and 48 h. Scale bar, 200 pm (left). Quantitative data of the wound-healing assay (right). Error bar, SD of three independent experiments. **p < .01 and ***P < .001. E, stable A549 and H1299 cells were immunostained with p-ERKl/2 antibody, while cell nuclei were stained with DAPI staining. Scale bar, 37 μm. F and G, stably NgBR or NC transfected A549 cells were transiently transfected with siNgBR or NS for 48 h, while stably shNgBR or NC transfected H1299 cells were transiently transfected with pIRES-NgBR (NgBR) or pIRES-NC (NC) for 48 h. These cells were then subjected to treatment with or without U0126 (10 μM) for additional 24 h and subjected to Western blot analysis of E-cadherin, p-ERKl/2, Snaill protein levels. Total ERK1/2 and β-actin levels were used as loading controls.
Moreover, results of wound healing assay showed that U0126 abrogated NgBR-induced migration of A549 (Fig. 5D) and H1299 cells (Fig. S6F). A study reported that p-ERKl /2 translocates to the nucleus to promote Snaill expression by binding to the minimal promoter of the Snaill gene [22]. Consistently, results of immunofluorescence staining performed in the present study showed subcellular localization of p-ERKl /2, with increased nuclear and cytoplasmic staining of p-ERKl/2 being observed in NgBR-overexpressing A549 cells (Fig. 5E, left panel) and decreased nuclear and cytoplasmic staining of p-ERKl /2 being observed in NgBR-knockout H1299 cells (Fig. 5E, right panel). Results of rescue experiments, which were performed subsequently, confirmed that NgBR promoted snaill expression via the MEK/ERK signaling pathway. NgBR overexpression increased p-ERKl/2 and Snaill levels but decreased E-cadherin level. This effect was reversed in A549 and H1299 cells treated with NgBR siRNA (Figs. 5Fand S6G). However, in H1299 cells, NgBR knockdown inactivated ERK1/2 and downregulated Snaill expression, which upregulated E-cadherin protein expression. Furthermore, NgBR overexpression reversed the effect of NgBR knockdown on ERK1/2, Snail1, and E-cadherin expression. Importantly, these rescue experiments were further inhibited by U0126 treatment in H1299 cells (Fig. 5G). These results suggest that NgBR promotes Snaill expression through the MEK/ ERK signaling pathway in NSCLC cells.
3.6. NgBR facilitates the plasma membrane localization and activation of Ras in NSCLC cells
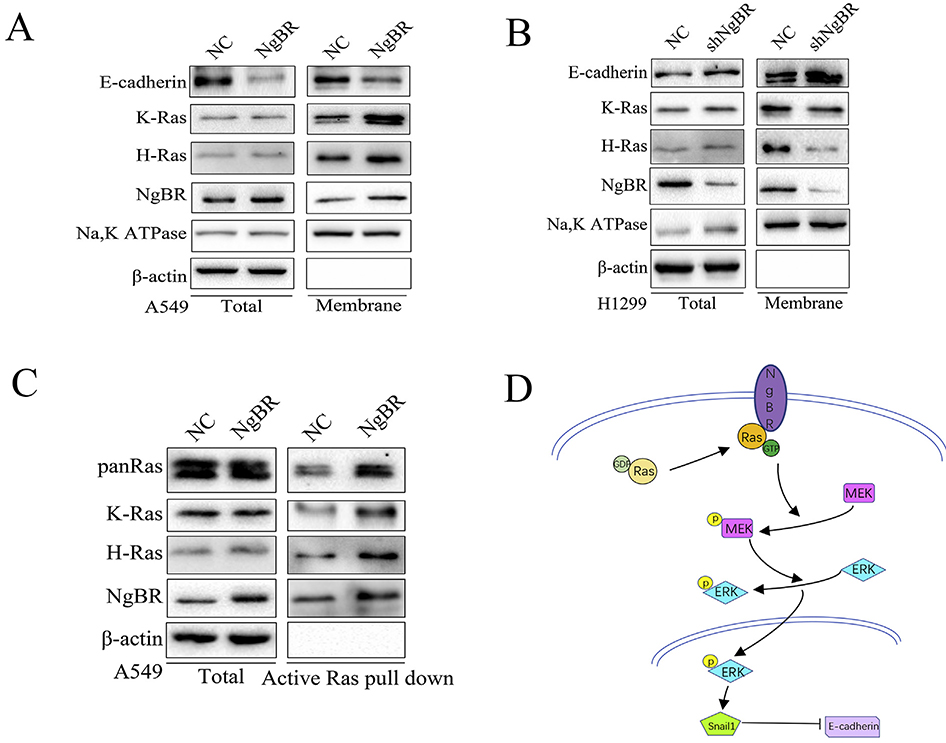
In our previous study [14], we found that NgBR promoted the accumulation of prenylated Ras on the plasma membrane and its subsequent activation, which in turn activated the downstream MEK/ERK signaling pathway [23,24]. In the present study, we found that NgBR overexpression enhanced the subcellular localization of K-Ras and H-Ras to the plasma membrane of A549 and H1299 cells (Figs. 6A and S7), whereas NgBR knockdown reduced the translocation of both K-Ras and H-Ras to the plasma membrane of H1299 cells (Fig. 6B). Membrane localization of Ras was inversely associated with E-cadherin expression in these cells (Fig. 6A and B). Activated Ras exists in a GTP form that binds to the Ras-binding domain of Raf1, leading to its phosphorylation and subsequent activation [23,25,26]. We further assessed NgBR-induced Ras activation by performing an active Ras pulldown assay. As expected, NgBR-overexpressing A549 cells showed increase in the levels of GTP-bound K-Ras, H-Ras, and Pan-Ras but did not show any change in total protein levels of K-Ras, H-Ras, and Pan-Ras (Fig. 6C). Together, these results suggest that NgBR strongly interacts with Ras to promote EMT in NSCLC cells. Moreover, our results indicate that NgBR overexpression induces the plasma membrane localization and subsequent activation of Ras in NSCLC cells, which in turn activates the downstream ERK1/2 pathway to promote Snaill expression and to reduce E-cadherin expression (Fig. 6D).
Fig. 6. NgBR promotes Ras activation and plasma membrane localization in NSCLC cells.
A and B, cell membrane protein was extracted from stable A549 and H1299 cells to evaluate the localization of E-cadherin, K-Ras and H-Ras. Na, K ATPase and β-actin were used as markers for the membrane and cytoplasm, respectively. C, the complex of activated Ras was pulled down by GST-Raf1-RBD beads, and then assessed using Western blotting in A549 cells. Total protein levels were used as the negative control. D, the schematic illustration of NgBR regulation of NSCLC cell EMT.
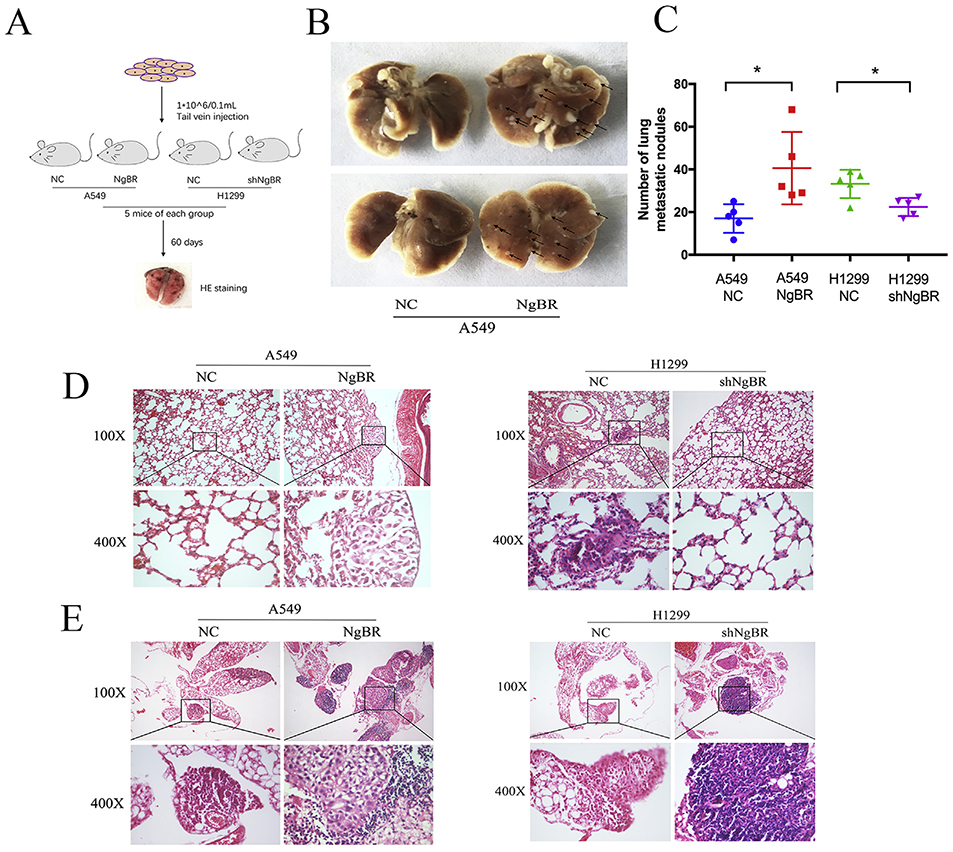
3.7. Increased NgBR expression promotes the invasion and migration of lung tumor cells in vivo
Thus far, we demonstrated the regulatory effect of NgBR on the invasion and metastasis of NSCLC cells in vitro. Next, we determined the effect of NgBR in vivo by using a mouse model of lung metastasis injected with stable NgBR-overexpressing A549 cells (A549NgBR mice), vector-only control cells (A549-NC mice), NgBR-knockout H1299 cells (H1299-shNgBR mice), or nonspecific control cells (H1299-NC mice) (Fig. 7A). After 60 days of tumor cell growth and metastasis, lung tissues were resected from each mouse and were processed for HE staining. Tumor lesions were visible in the lung tissues of A549-NgBR mice but were negligible in the lung tissues of A549-NC mice (Fig. 7B). Microscopically, the number of lung metastatic nodules was higher in A549-NgBR mice than in A549-NC mice. In contrast, H1299-shNgBR mice showed fewer number of lung metastatic nodules than H1299-NC mice (Fig. 7C and D). Furthermore, lung lymph node metastasis was observed in A549-NgBR and H1299-NC mice but not in A549-NC and H1299-shNgBR mice (Fig. 7E). These results indicate that NgBR promotes the lymph node metastasis of NSCLC, which is consistent with our ex vivo results and results obtained by performing NgBR immunostaining of tissue samples (Fig. 7B and C). Collectively, our results suggest that NgBR overexpression promotes the invasion and metastasis of NSCLC cells in vivo.
Fig. 7. Increased NgBR expression promotes lung metastasis in vivo.
A, schematic diagram of the lung metastasis model in mice. B, representative images of lung metastatic nodules (the arrows refer to tumor lesions). C, summary of the number of lung metastatic nodules. *p < .05. D and E, HE staining of the lung (D) and lung lymph nodes (E). Scale bar, 20 μm and 100 μm.
4. Discussion
NgBR promotes the plasma membrane translocation and activation of Ras in breast cancer cells and enhances the chemoresistance of human hepatocellular carcinoma cells [12,13,27]. Pula et al. found that significant NgBR mRNA downregulation is associated with large primary tumor size, lymph node involvement, and cancer stage advancement in patients with NSCLC [28],.In the present study, we detected high NgBR protein expression in NSCLC tissues and their corresponding tumor cell-positive lymph nodes. The possible reasons for the contradictory results of NgBR expression patterns in NSCLC tissues are differences in the effect of sex, age, race/ethnicity, socioeconomic status, and geography on the pathogenesis of lung cancer [29].
The present study is the first to highlight the role of NgBR in inducing EMT and metastasis in NSCLC. We found that high NgBR expression was associated with the lymph node metastasis of NSCLC cells. Moreover, lymph node metastasis was observed in A549-NgBR and H1299-NC mice, indicating that NgBR promoted the lymph node metastasis of NSCLC cells, which is the most common form of metastasis in NSCLC or all malignancies of epithelial origin in the clinical setting. A previous study showed that initiation of EMT in tumor cells is the major cause of lung cancer metastasis [18]. Results of the present study also showed that NgBR overexpression remarkably increased the expression of mesenchymal protein markers N-cadherin and vimentin but decreased the expression level of epithelial protein marker E-cadherin, which altered the migration and invasion capacities of NSCLC cells in vitro and lung metastasis of NSCLC cells in the mouse model. However, NgBR knockdown exerted opposite effects on NSCLC cells both in vitro and in vivo, indicating that NgBR is necessary for EMT and for promoting lung cancer metastasis.
EMT is thought to be involved in the pathogenesis of numerous lung diseases, including lung developmental disorders, fibrotic tissue remodeling, and lung cancer [4,6,30]. Loss of E-cadherin expression is considered as a hallmark of EMT in cancer progression. E-Cadherin expression is regulated by different mechanisms such as somatic gene mutation, promoter hypermethylation, transcriptional repression, and histone deacetylation [18,31,32]. Transcriptional repression of E-cadherin expression plays a major role in tumor development.
Moreover, different transcriptional factors such as Snaill and Twistl regulate E-cadherin expression by interacting with specific E-boxes present in the proximal region of the E-cadherin gene promoter [18,31,33]. For example, Snail1, a zinc finger transcription factor, represses E-cadherin expression by binding to the E-cadherin gene promoter, which in turn enhances the migration and invasion of different tumor cells [8,33,34]. Moreover, Twist1, a basic helix-loop-helix transcription factor, plays an important role in breast cancer metastasis by repressing E-cadherin expression [35]. Results of the present study showed that NgBR knockdown upregulated E-cadherin expression by decreasing Snaill expression level but did not significantly alter Twist1 mRNA and protein expression levels. In cells, Snaill is a labile protein with a half-life of approximately 25 min [36]. We examined whether NgBR regulated the ubiquitination and degradation of Snail1 protein by performing a degradation dynamics assay with cycloheximide (CHX), which inhibits protein translation. CHX treatment did not significantly alter Snail1 protein levels in NgBR-overexpressing A549 and H1299 cells (data not shown), indicating that NgBR upregulated Snaill expression at the transcriptional level to suppress E-cadherin expression and to promote EMT in NSCLC cells.
However, Snaill expression is regulated by multiple signaling pathways [8,22,36], including the PI3K/Akt and Ras/MEK/ERK signaling pathways. The Ras family of genes includes N-Ras, H-Ras, and K-Ras genes. Alteration of Ras activity is associated with cancer development and progression [37–39]. For example, mutations in the Ras genes are frequently detected in pancreatic and lung cancers, which result in the constitutive activation of downstream signaling pathways that regulate cell proliferation and apoptosis [40,41]. Ras protein functions as a molecular switch and binds to guanine diphosphate or guanine triphosphate [25,42] to regulated various signaling pathways involved in the proliferation, differentiation, or survival of eukaryotic cells [38,43–46]. Ras signaling pathways also promote cellular metastasis; however, mechanisms underlying these pathways are still not completely understood [46]. We previously showed that NgBRexpression activated Ras by promoting its plasma membrane localization in breast cancer cells [14]. In the present study, we found that this effect of NgBR on Ras localization was not exclusive to breast cancer cells and was also observed in other tumor cells such as NSCLC cells. We found that NgBR knockdown reduced the plasma membrane localization and activation of Ras in NSCLC cells, whereas NgBR overexpression activated the MEK/ERK signaling pathway, which acts downstream of Ras. However, NgBR overexpression or knockdown did not significantly affect the activity of the Akt signaling pathway in NSCLC cells. These results were further confirmed by pharmacologically blocking the MEK/ERK signaling pathway by using the MEK inhibitor U0126 or by performing a rescue experiment with an NgBR cDNA or siRNA.
Thus, the results of the present study indicate the importance of NgBR in NSCLC progression and highlight molecular mechanisms underlying NgBR-induced migration, invasion, and metastasis of NSCLC cells. However, additional clinical investigations should be performed to determine NgBR as a therapeutic target for preventing tumor metastasis in patients with NSCLC.
Supplementary Material
Acknowledgements
This study was supported in part by the National Natural Science Foundation of China (nos. 81470367, 31770893, and 21405153) and Institutions of higher learning of innovation team from Liaoning province (LT2014019). B.Z. would like to thank the “Hundred Talents Program" of CAS for its support. This study was also supported in part by start-up funds from NIH R01HL108938, Institutional Research Grant #86-004-26 from the American Cancer Society, Kathy Duffey Fogarty Award for breast cancer research, and Children’s Hospital of Wisconsin Research Institute Pilot Innovative Research Grant to Q.R.M.
Footnotes
Conflicts of interest
The authors declare that they do not have any conflicts of interest related to this study.
Appendix A. Supplementary data
Supplementary data related to this article can be found at https://doi.Org/10.1016/j.canlet.2018.01.030.
References
- [1].Siegel RL, Miller KD, Jemal A, Cancer statistics, 2017, CA A Cancer J. Clin. 67 (2017) 7–30. [DOI] [PubMed] [Google Scholar]
- [2].Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J, Cancer statistics in China, 2015, CA A Cancer J. Clin. 66 (2016) 115–132. [DOI] [PubMed] [Google Scholar]
- [3].Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, Stein KD, Alteri R, Jemal A, Cancer treatment and survivorship statistics, 2016, CA Cancer J Clin 66 (2016) 271–289. [DOI] [PubMed] [Google Scholar]
- [4].Bartis D, Mise N, Mahida RY, Eickelberg O, Thickett DR, Epithelial–mesenchymal transition in lung development and disease: does it exist and is it important? Thorax 69 (2014) 760–765. [DOI] [PubMed] [Google Scholar]
- [5].Zeisberg M, Neilson EG, Biomarkers for epithelial-mesenchymal transitions, J. Clin. Invest. 119 (2009) 1429–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Nieto MA, Huang Ruby YJ, Jackson Rebecca A., Thiery Jean P., Emt: 2016, Cell 166 (2016) 21–45. [DOI] [PubMed] [Google Scholar]
- [7].Datto M, Wang XF, Ubiquitin-mediated degradation, Cell 121 (2005) 2–4. [DOI] [PubMed] [Google Scholar]
- [8].Yook JI, Li XY, Ota I, Fearon ER, Weiss SJ, Wnt-dependent regulation of the e-cadherin repressor snail, J. Biol. Chem. 280 (2005) 11740–11748. [DOI] [PubMed] [Google Scholar]
- [9].Miao RQ, Gao Y, Harrison KD, Prendergast J, Acevedo LM, Yu J, Hu F, Strittmatter SM, Sessa WC, Identification of a receptor necessary for Nogo-B stimulated chemotaxis and morphogenesis of endothelial cells, Proc. Natl. Acad. Sci. U.S.A. 103 (2006) 10997–11002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Park EJ, Grabinska KA, Guan Z, Sessa WC, NgBR is essential for endothelial cell glycosylation and vascular development, EMBO Rep. 17 (2016) 167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zhao B, Chun C, Liu Z, Horswill MA, Pramanik K, Wilkinson GA, Ramchandran R, Miao RQ, Nogo-B receptor is essential for angiogenesis in zebrafish via Akt pathway, Blood 116 (2010) 5423–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wang B, Zhao B, North P, Kong A, Huang J, Miao QR, Expression of NgBR is highly associated with estrogen receptor alpha and survivin in breast cancer, PLoS One 8 (2013), e78083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Dong C, Zhao B, Long F, Liu Y, Liu Z, U S, Yang X, Sun D, Wang H, Liu Q, Liang R, Li Y, Gao Z, Shao S, Miao QR, Wang L, Nogo-B receptor promotes the chemoresistance of human hepatocellular carcinoma via the ubiquitination of p53 protein, Oncotarget 7 (2016) 8850–8865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zhao B, Hu W, Kumar S, Gonyo P, Rana U, Liu Z, Wang B, Q Duong W, Yang Z, L Williams C, Miao QR, The Nogo-B receptor promotes Ras plasma membrane localization and activation, Oncogene 36 (2017) 3406–3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Chellappan SP, Gyorffy B, Surowiak P, Budczies J, Lánczky A, Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer, PLoS One 8 (2013), e82241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Shan YS, Hsu HP, Lai MD, Hung YH, Wang CY, Yen MC, Chen YL, Cyclin D1 overexpression correlates with poor tumor differentiation and prognosis in gastric cancer, Oncology letters 14 (2017) 4517–4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Thiery JP, Acloque H, Huang RYJ, Nieto MA, Epithelial-mesenchymal transitions in development and disease, Cell 139 (2009) 871–890. [DOI] [PubMed] [Google Scholar]
- [18].Huber MA, Kraut N, Beug H, Molecular requirements for epithelial-mesenchymal transition during tumor progression, Curr. Opin. Cell Biol. 17 (2005) 548–558. [DOI] [PubMed] [Google Scholar]
- [19].Grille SJ, Bellacosa A, Upson J, Klein-Szanto AJ, van Roy F, Lee-Kwon W, Donowitz M, Tsichlis PN, Larue L, The protein kinase Akt induces epithelial mesenchymal transition and promotes enhanced motility and invasiveness of squamous cell carcinoma lines, Cancer Res 63 (2003) 2172–2178. [PubMed] [Google Scholar]
- [20].Julien S, Puig I, Caretti E, Bonaventure J, Nelles L, van Roy F, Dargemont C, de Herreros AG, Bellacosa A, Larue L, Activation of NF-kappaB by Akt upregulates Snail expression and induces epithelium mesenchyme transition, Oncogene 26 (2007) 7445–7456. [DOI] [PubMed] [Google Scholar]
- [21].Gan Y, Shi C, Inge L, Hibner M, Balducci J, Huang Y, Differential roles of ERK and Akt pathways in regulation of EGFR-mediated signaling and motility in prostate cancer cells, Oncogene 29 (2010) 4947–4958. [DOI] [PubMed] [Google Scholar]
- [22].Barbera MJ, Puig I, Dominguez D, Julien-Grille S, Guaita-Esteruelas S, Peiro S, Baulida J, Franci C, Dedhar S, Larue L, Garcia de Herreros A, Regulation of Snail transcription during epithelial to mesenchymal transition of tumor cells, Oncogene 23 (2004) 7345–7354. [DOI] [PubMed] [Google Scholar]
- [23].L Buday J Downward, Many faces of Ras activation, Biochim. Biophys. Acta 1786 (2008) 178–187. [DOI] [PubMed] [Google Scholar]
- [24].Ahearn IM, K Haigis D Bar-Sagi, M.R. Philips, Regulating the regulator: posttranslational modification of RAS, Nature reviews, MCB (Mol. Cell. Biol.) 13 (2011) 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ehrhardt A, Ehrhardt GR, Guo X, Schrader JW, Ras and relatives–job sharing and networking keep an old family together, Experimental hematology 30 (2002) 1089–1106. [DOI] [PubMed] [Google Scholar]
- [26].Bar-Sagi D, Hall A, Ras and Rho GTPases: a family reunion, Cell 103 (2000) 227–238. [DOI] [PubMed] [Google Scholar]
- [27].Zhao B, Xu B, Hu W, Song C, Wang F, Liu Z, Ye M, Zou H, Miao QR, Comprehensive proteome quantification reveals NgBR as a new regulator for epithelial-mesenchymal transition of breast tumor cells, Journal of proteomics 112 (2015) 38–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Pula B, Olbromski M, Owczarek T, Ambicka A, Witkiewicz W, Ugorski M, Rys J, Zabel M, Dziegiel P, Podhorska-Okolow M, Nogo-B receptor expression correlates negatively with malignancy grade and ki-67 antigen expression in invasive ductal breast carcinoma, Anticancer research 34 (2014) 4819–4828. [PubMed] [Google Scholar]
- [29].Torre LA, Siegel RL, Jemal A, Lung cancer statistics, Adv. Exp. Med. Biol. 893 (2016) 1–19. [DOI] [PubMed] [Google Scholar]
- [30].Larue L, Bellacosa A, Epithelial-mesenchymal transition in development and cancer: role of phosphatidylinositol 3' kinase/AKT pathways, Oncogene 24 (2005) 7443–7454. [DOI] [PubMed] [Google Scholar]
- [31].Peinado H, Portillo F, Cano A, Transcriptional regulation of cadherins during development and carcinogenesis, Int. J. Dev. Biol. 48 (2004) 365–375. [DOI] [PubMed] [Google Scholar]
- [32].Cavallaro U, Christofori G, Cell adhesion and signalling by cadherins and IgCAMs in cancer, Nat. Rev. Cane. 4 (2004) 118–132. [DOI] [PubMed] [Google Scholar]
- [33].Nieto MA, The snail superfamily of zinc-finger transcription factors, Nature reviews, MCB (Mol. Cell. Biol.) 3 (2002) 155–166. [DOI] [PubMed] [Google Scholar]
- [34].Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, Garcia De Herreros A, The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells, Nat. Cell Biol. 2 (2000) 84–89. [DOI] [PubMed] [Google Scholar]
- [35].Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA, Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis, Cell 117 (2004) 927–939. [DOI] [PubMed] [Google Scholar]
- [36].Zhou BP, Deng J, Xia W, Xu J, Li YM, Gunduz M, Hung MC, Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition, Nat. Cell Biol. 6 (2004) 931–940. [DOI] [PubMed] [Google Scholar]
- [37].Wang Y, Kaiser CE, Frett B, Li HY, Targeting mutant KRAS for anticancer therapeutics: a review of novel small molecule modulators, J. Med. Chem. 56 (2013) 5219–5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Spiegel J, Cromm PM, Zimmermann G, Grossmann TN, Waldmann H, Small-molecule modulation of Ras signaling, Nat. Chem. Biol. 10 (2014) 613–622. [DOI] [PubMed] [Google Scholar]
- [39].Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, IC Wong K, Elledge SJ, A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene, Cell 137 (2009) 835–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Meng D, Yuan M, Li X, Chen L, Yang J, Zhao X, Ma W, Xin J, Prognostic value of K-RAS mutations in patients with non-small cell lung cancer: a systematic review with meta-analysis, Lung Canc. 81 (2013) 1–10. [DOI] [PubMed] [Google Scholar]
- [41].Jonckheere N, Vasseur R, Van Seuningen I, The cornerstone K-RAS mutation in pancreatic adenocarcinoma: from cell signaling network, target genes, biological processes to therapeutic targeting, Crit. Rev. Oncol.-Hematol. 111 (2017) 7–19. [DOI] [PubMed] [Google Scholar]
- [42].McCubrey JA, Steelman LS, Chappell WH, L Abrams S, Wong EW, Chang F, Lehmann B, Terrian DM, Milella M, Tafuri A, Stivala F, Libra M, Basecke J, Evangelisti C, Martelli AM, Franklin RA, Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance, Biochim. Biophys. Acta 1773 (2007) 1263–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Nussinov R, Tsai CJ, Chakrabarti M, Jang H, A new view of Ras isoforms in cancers, Cancer Res 76 (2016) 18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Tsai FD, Lopes MS, Zhou M, Court H, Ponce O, Fiordalisi JJ, Gierut JJ, Cox AD, Haigis KM, Philips MR, K-Ras4A splice variant is widely expressed in cancer and uses a hybrid membrane-targeting motif, Proc. Natl. Acad. Sci. U.S.A. 112 (2015) 779–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Omerovic J, Laude AJ, Prior IA, Ras proteins: paradigms for compartmentalised and isoform-specific signalling, Cell. Mol. Life Sci.: CMLS 64 (2007) 2575–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Uekita T, Fujii S, Miyazawa Y, Iwakawa R, Narisawa-Saito M, Nakashima K, Tsuta K, Tsuda H, Kiyono T, Yokota J, Sakai R, Oncogenic Ras/ERK signaling activates CDCP1 to promote tumor invasion and metastasis, Mol. Cane. Res. 12 (2014) 1449–1459. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.