Abstract
Tissue-resident macrophages (TRMs) maintain tissue homeostasis, but they can also provide a replicative niche for intracellular pathogens such as Leishmania. How dermal TRMs proliferate and maintain their M2 properties even in the strong TH1 environment of the L. major infected dermis is not clear. Here, we show that in infected mice lacking IL-4/IL-13 from eosinophils, dermal TRMs shifted to a pro-inflammatory state, their numbers declined, and disease was attenuated. Intravital microscopy revealed a rapid infiltration of eosinophils followed by their tight interaction with dermal TRMs. IL-4-stimulated dermal TRMs, in concert with IL-10, produced a large amount of CCL24, which functioned to amplify eosinophil influx and their interaction with dermal TRMs. An intraperitoneal helminth infection model also demonstrated a requirement for eosinophil-derived IL-4 to maintain tissue macrophages through a CCL24-mediated amplification loop. CCL24 secretion was confined to resident macrophages in other tissues, implicating eosinophil-TRM cooperative interactions in diverse inflammatory settings.
One Sentence Summary
IL-4/CCL24 mediated interaction with eosinophils maintains dermis-resident macrophages as replicative niches for Leishmania major.
Introduction
Most tissues are populated by tissue-resident macrophages (TRM) that perform anatomical niche-specific roles to help maintain tissue homeostasis, integrity and function(1, 2). It is now understood that all TRMs at birth are embryonically seeded, but are gradually replaced by hematopoietic stem cell-derived progenitors with age to a greater (intestine and MHCIIhigh dermal macrophages), lesser (heart and pancreas), or negligible (brain, epidermis, lung, and liver) extent(3). Their ontogeny as well as tissue specific cues contribute to the heterogeneity of TRMs(4, 5). Despite their functional diversity, TRMs in certain settings have been classified as alternatively activated (M2-like) cells with fundamental roles in the resolution of inflammation and tissue repair (1, 6). Embryonic-derived, cardiac TRMs, for example, drive cardiomyocyte proliferation and angiogenesis following sterile heart injury, while monocyte-derived cells exhibit pro-inflammatory activities(7, 8). In a helminth-bacterial co-infection model, the functional plasticity of the nematode expanded, peritoneal TRMs were limited when compared to monocytes(9). In the lung, embryonic-derived alveolar macrophages were shown to have M2-like characteristics that better supported the growth of Mycobacterium tuberculosis compared to monocyte-derived interstitial macrophages(10). Similarly, we have reported that M2-like, embryonic-derived MHCIIlow dermal TRMs better support the growth of L. major compared to monocyte-derived cells and can be preferentially infected by certain strains of L. major to produce non-healing lesions in conventionally resistant C57BL/6 mice(11).
Whereas most innate cells accumulate in the blood during inflammation, embryonic-derived TRMs can increase their population density beyond homeostatic levels through a distinct mechanism that involves IL-4- or IL-13-mediated self-proliferation, first described in the context of helminth driven, TH2 pathology(12, 13). A more recent study showed that IL-4/13 together with recognition of apoptotic cells by TRMs was required to induce their local proliferation in the lung after helminth infection or in the gut after induction of colitis(14). We also demonstrated a role for IL-4 in the local proliferation of M2-like dermal macrophages, exceptional in this case because it functioned in the context of the strong TH1 immune environment characteristically seen in L. major infected C57BL/6 mice(11). The type 2 cytokines required for the maintenance of TRMs has been investigated in adipose tissue, with IL-4 and eosinophils shown to sustain alternatively activated macrophages, although whether or not eosinophils provide an essential source of IL-4 was not addressed (15, 16). Our understanding of the maintenance of TRMs under pathological conditions, such as infection, is still limited, and has yet to be investigated in the skin.
Eosinophils are terminally differentiated, granule-containing effector cells mainly involved in helminth infection and allergy. These cells are now considered as tissue-resident leukocytes having broad tissue distribution and homeostatic functions, such as tissue development/regeneration and metabolic/immune homeostasis(17). IL-5 and eotaxins have been recognized as important regulators of eosinophil differentiation and tissue accumulation(18). In particular, IL-5 produced by ILC2 and eotaxin-1 are critical for steady-state homing of eosinophils into the small intestine(19, 20). The signals that regulate eosinophils homing to the steady state dermis are not known. Interestingly, in vitro-generated macrophages from monocytes and bone marrow were shown to be potent producers of eotaxin-2 following IL-4 and IL-10 stimulation, and exogenous IL-4 activated macrophages promoted CCR3-dependent eosinophilia in filarial helminth infected mice (21), suggesting that tissue myeloid cells could be the source of eotaxins regulating the homing and functions of tissue-dwelling eosinophils(22, 23).
In this study, we document a cooperative interaction between eosinophils and TRMs in the L. major infected dermis. During infection, eosinophils are the major source of IL-4 required to mediate the local proliferation of dermal TRMs and maintain their M2-like phenotype. The IL-4-stimulated dermal TRMs, in concert with IL-10, produce high amounts of CCL24 which amplifies the recruitment of eosinophils and their interaction with TRMs. Intravital imaging confirmed their tight cellular association which was dampened by CCL24 neutralization. Notably, the production of CCL24 was confined to TRMs in other tissues, suggesting that eosinophil-TRM cooperative interactions can be generalized to other tissues and inflammatory settings.
Results
IL-4 competency in innate cells is required for maintaining dermal TRMs during L. major infection
We previously reported that IL-4 maintains dermal TRMs as replicative niches for L. major by inducing their local proliferation during TH1 immunity(11). To identify the nature (innate versus adaptive) of the cellular source of IL-4, we generated mixed bone marrow chimeras with selective IL-4 competency within the hematopoietic compartment (Fig. 1A). Reconstitution of lethally irradiated il4−/− mice with a 4:1 mixture of CD45.1+ rag1−/− and CD45.2+ il4−/− bone marrow cells (BMCs) yielded mice with IL-4 competency solely in innate cells (rag1−/− + il4−/− → il4−/− chimeras; hereafter referred to as “innate” IL-4+ chimeras). Conversely, reconstitution of il4−/− mice with a 4:1 mixture of CD45.2+ rag1−/−il4−/− and WT BMCs produced chimeras with IL-4 competent T and B lymphocytes but a predominantly IL-4 incompetent innate immune compartment (rag1−/−il4−/− + WT → il4−/− chimeras; “adaptive” IL-4+ chimeras). Control chimeras with IL-4 competency in both innate and adaptive compartments (WT → il4−/− chimeras; “both” IL-4+ innate and adaptive chimeras) or neither compartment (il4−/− → il4−/− chimeras; “neither” IL-4+ innate or adaptive chimeras) were also generated. The expected reconstitutions of mixed chimeras generated using this approach were confirmed by CD45.1 and 2 congenic markers staining of ear isolates (Fig. 1B). As we previously reported, the majority of dermal macrophages were radioresistant, not replaced by BMCs, and remained of host origin.
Fig. 1.
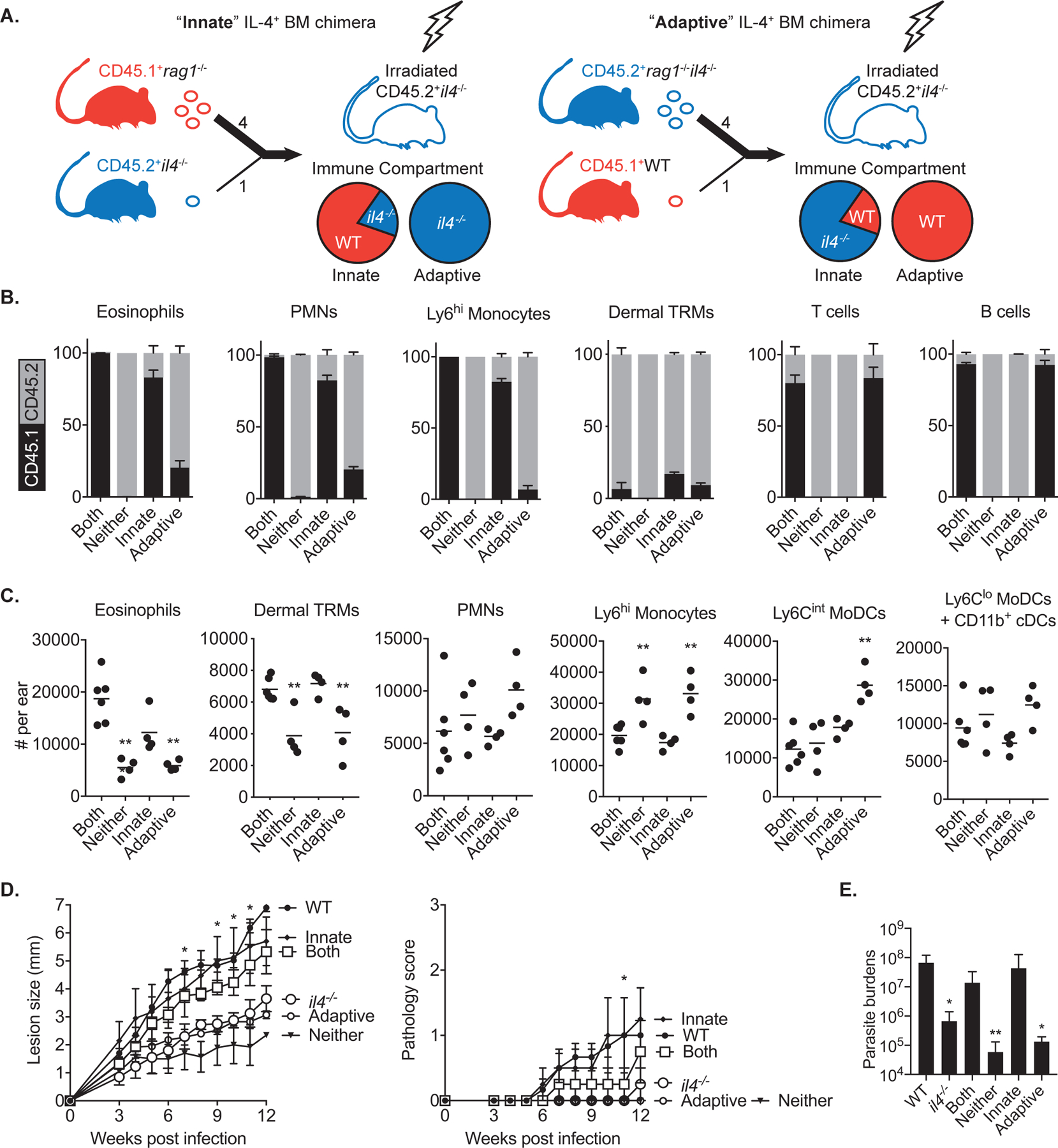
Innate cells are the source of IL-4 to maintain dermal TRMs during L. major infection. (A) Transplantation strategy used to generate mixed bone marrow chimeras with selective IL-4 competency within innate or adaptive compartments. BM cells mixed in a ratio of 4:1 were transferred to irradiated recipients. (B) Mixed bone marrow chimeras (n = 4–6) with selective IL-4 expression in innate or adaptive immune cells were generated by reconstitution of irradiated CD45.2+ il4−/− mice with various mixtures of BM cells from CD45.1+ WT, CD45.2+ il4−/−, CD45.1+ rag1−/−, and CD45.2+ rag1−/−il4−/− mice, as described in Results. (C) The absolute numbers of myeloid subsets recovered from ears of mixed BM chimeras at day 12 p.i. with 2 × 105 LmSd. The gating strategy is depicted in Fig. S1. (D) Lesion development and pathology scores over the course of infection with 103 LmSd metacyclic promastigotes in the ear dermis of WT, il4−/−, and mixed BM chimeras (n = 4–6). Parasite burdens were quantified at 12 weeks p.i. (E). Values represent mean +/− standard deviation. *P < 0.05 and **P < 0.01 by one-way ANOVA with Dunn’s post-test compared to ‘both’ chimeras (C, D, E). Data are representative of two independent experiments (B, C, D, E).
When infected with non-healing L. major Seidman strain (LmSd), “neither” IL-4+ mice showed significantly decreased number of dermal TRMs compared to “both” IL-4+ chimeras, confirming the role of IL-4 for their expansion during infection (Fig. 1C). The number of infiltrating eosinophils was also significantly reduced in “neither” IL-4+ chimeras. We found a similar decrease of both dermal TRMs and eosinophils in mixed BM chimeras with IL-4 competency largely restricted to the adaptive immune compartment. By contrast, the “innate” IL-4+ chimeras maintained their numbers of dermal TRMs and eosinophils during infection. The “adaptive” as well as “neither” IL-4+ chimeras showed increased infiltration of inflammatory Ly6Chi monocytes and/or moDCs compared to “both” or “innate” IL-4+ chimeras (Fig. 1C and S1). When challenged with a low dose of LmSd, “both” and “innate” IL-4+ chimeric mice developed non-healing, ulcerative lesions similar C57BL/6 WT mice, whereas lesion progression, pathology and parasite burdens were substantially ameliorated in “neither” and “adaptive” IL-4+ chimeric mice, as well as in il4−/− mice (Fig. 1D and 1E).
Eosinophils are a critical source of IL-4 to maintain dermal TRMs during L. major infection
Because neither intracellular cytokine staining for IL-4 nor cytokine measurements by ELISA has allowed us to detect IL-4 production by ear lesional cells from LmSd-infected mice(11), we used IL-4 dual-reporter mice (4get x KN2) which permit the simultaneous analysis of cells that are eGFP+ for IL-4 transcription and positive for recent IL-4 protein secretion by staining for human CD2(24). As expected, the frequency of IL-4 secreting cells among the live cells recovered from the ear was extremely low, 4.64% hCD2+ of 4.38% GFP+ cells, or 0.2% in total (Fig. 2A). Of these, roughly 20% were identified as eosinophils and 30% as CD4+ T cells. CD90.2+CD2+CD3− cells, most likely ILC2s, and CD11b+FcεRIα+ basophils were also hCD2 and GFP-positive (Fig. S2). Since the results of the bone marrow chimeras implicated innate cells as a critical source of IL-4 to maintain dermal TRMs, we looked specifically at the number of dermal TRMs in anti-SiglecF antibody treated, eosinophil-depleted animals during the course of infection (Fig. 2B). The number of dermal TRMs was reduced by more than half in the eosinophil-depleted mice, while more inflammatory moDCs and CD4+ T cells were recovered from the lesion at 12 days post-infection. The reduction of dermal TRMs in eosinophil-depleted animals was comparable to that observed in il4−/− animals, and eosinophil depletion in il4−/− mice had no further effect on reducing the number of these cells (Fig. 2C). Similar results were observed in mice lacking IL-5, which is a key mediator of eosinophil maturation and release in the bone marrow (Fig. 2D). Similar to IL-4 deficient mice, il5−/− mice showed smaller lesions and reduced pathology compared to WT mice (Fig. 2E). By intracellular cytokine staining, we identified type 2 innate lymphoid cells (ILC2) as the main source of IL-5 and IL-13, while αβ T cells were the main producers of IL-10 and IL-13 (Fig. 2F).
Fig. 2.
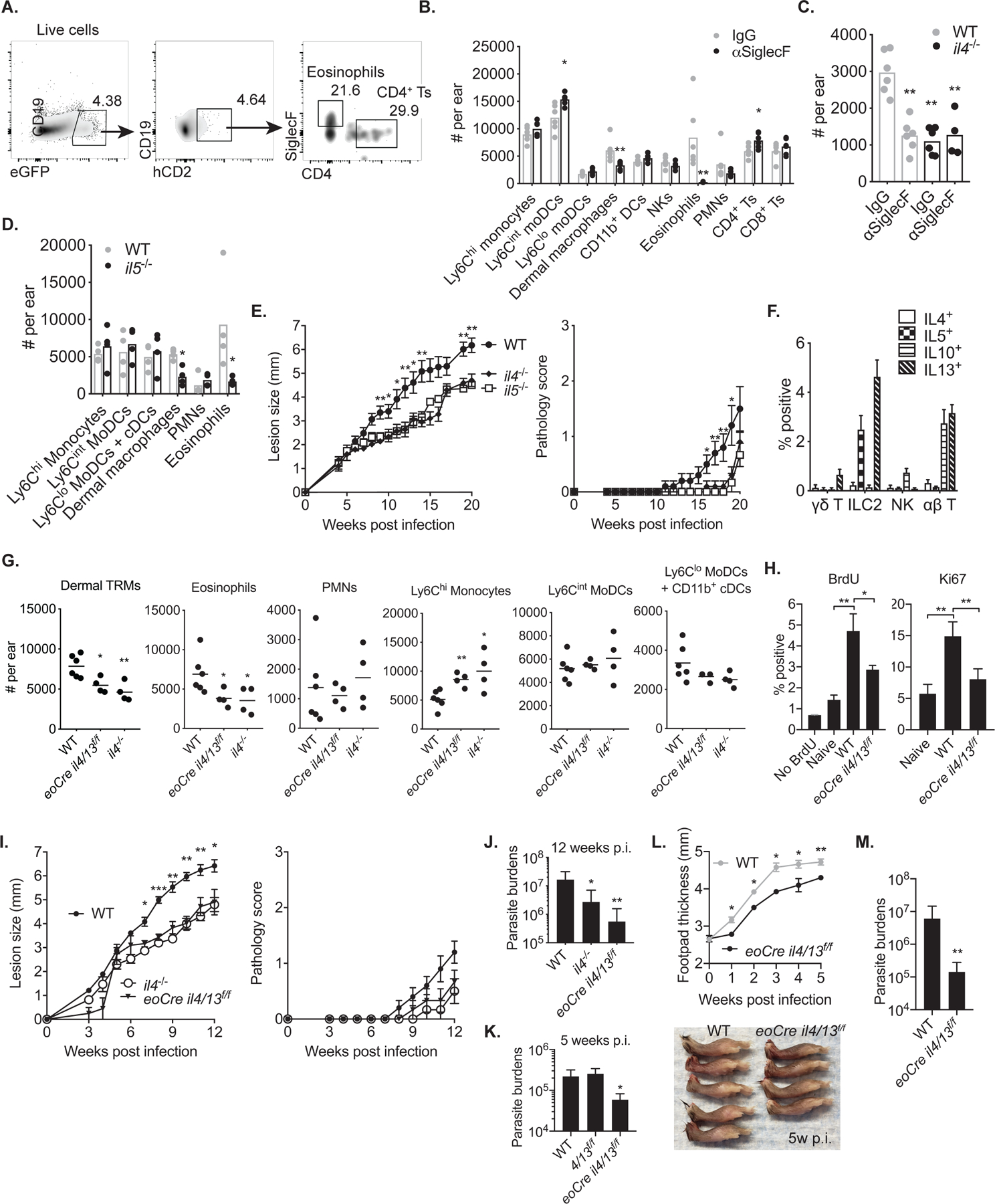
IL-4 production by eosinophils is necessary to maintain dermal TRMs during L. major infection. (A) Representative flow cytometry plots showing eGFP+ and hCD2+ cells from IL-4 dual-reporter mice (4get/KN2) at day 12 p.i. with 2 × 105 LmSd. (B, C) The absolute numbers of lymphoid and myeloid subsets (B) or dermal macrophages (C) recovered from eosinophils-depleted animals using anti-SiglecF antibodies at day 12 p.i. with 2 × 105 LmSd. (D) The absolute numbers of myeloid subsets recovered from il5−/− mice at day 12 p.i. with 2 × 105 LmSd. (E) Lesion development and pathology scores over the course of infection with 103 LmSd metacyclic promastigotes in the ear dermis of C57BL/6, il4−/−, and Il5−/− mice (n = 6–10). (F) Intracellular staining of IL-4, IL-5, IL-10, and IL-13 from indicated populations of ear dermal cells recovered from mice (n = 6) infected with 2 × 105 LmSd for 12 days. (G) The absolute numbers of myeloid subsets recovered from WT, eoCre il4/13f/f, and il4−/− mice at day 12 p.i. with 2 × 105 LmSd. (H) Ki67 expression or BrdU incorporation by dermal TRMs from naive or infected WT and eoCre il4/13f/f mice at 12 d p.i. with 2 × 105 LmSd (n = 4–6). (I) Lesion development and pathology scores over the course of infection with 103 LmSd metacyclic promastigotes in the ear dermis of WT, eoCre il4/13f/f, and il4−/− mice (n = 6–10). Parasite burdens were measured at week 12 p.i. (J). (K) Parasite burdens in WT, il4/13f/f, and il4−/− mice were measured at week 5 p.i. with 103 LmSd metacyclic promastigotes (n=6). (L) Measurement of footpad thickness over the course of infection with 106 LmSd metacyclic promastigotes in the footpads of WT (n=5) and eoCre il4/13f/f (n=4). For reference, the side view of footpads at 5 weeks p.i. is shown in lower panel. (M) Parasite burdens were measured at week 5 p.i. Values represent mean +/− standard deviation. *P < 0.05, **P < 0.01, and ***P < 0.001 by non-parametric Mann-Whitney test (B, D, H, L, M) and by one way ANOVA with Dunn’s post-test compared to IgG control (C) or WT infected with LmSd (E, G, I, J, K). Data are representative of two independent experiments (A, B, C, D, E, F, G, I, J).
To confirm eosinophils as a critical source of IL-4, mice expressing Cre protein under the control of the endogenous eosinophil peroxidase promoter (eoCre) were crossed to il4/13f/f mice to obtain a selective deficiency of IL-4 (and IL-13) in eosinophils(25, 26). The resulting eoCre il4/13f/f mice displayed a reduction in the number of lesional eosinophils and dermal TRMs similar to il4−/− animals, while they showed increased infiltration of Ly6Chi inflammatory monocytes (Fig. 2G). At day 12 0. p.i., dermal TRMs exhibited a 2–3 fold increase in the frequencies of Ki67+ and BrdU+ cells compared to naïve animals. These increased frequencies of dividing cells during infection were abrogated in the eoCre il4/13f/f mice, demonstrating that the expansion of dermal TRMs depends on IL-4 from eosinophils (Fig. 2H). Furthermore, the eoCre il4/13f/f mice ameliorated ear lesion progression and pathology identical to il4−/− mice (Fig. 2I), and controlled parasite burdens at least as well, with a 30-fold reduction compared to WT mice at 12 weeks post-infection (Fig. 2J). In an independent experiment in which the mice were sacrificed at 5 weeks p.i., the eoCre il4/13f/f mice showed 4-fold lower parasite burdens compared to WT and il4/13f/f mice (Fig. 2K). The eoCre il4/13f/f mice also showed less footpad swelling and 40-fold lower parasite burdens compared to WT mice at 5 weeks after subcutaneous challenge in the footpad (Fig. 2L and M).
IL-4/10 stimulated dermal TRMs secrete CCL24 to recruit eosinophils
Stained tissue sections from a lesion revealed significant infiltration of eosinophils around dermal macrophages harboring parasites at day12 p.i. (Fig. 3A; inset 1 and 3, Mov. S1). Eosinophils also infiltrated into a proximal region across the auricular cartilage, opposite to the side of parasite inoculation, and they were tightly associated with dermal TRMs even in the absence of parasites (Fig. 3A, inset 1 and 4; Mov. S2). Quantitatively, more than 80% of eosinophils interacted with dermal TRMs in the proximal area of the injection site compared to roughly 55% in either naïve ear or non-proximal region of the infected ear (Fig. 3B).
Fig. 3.
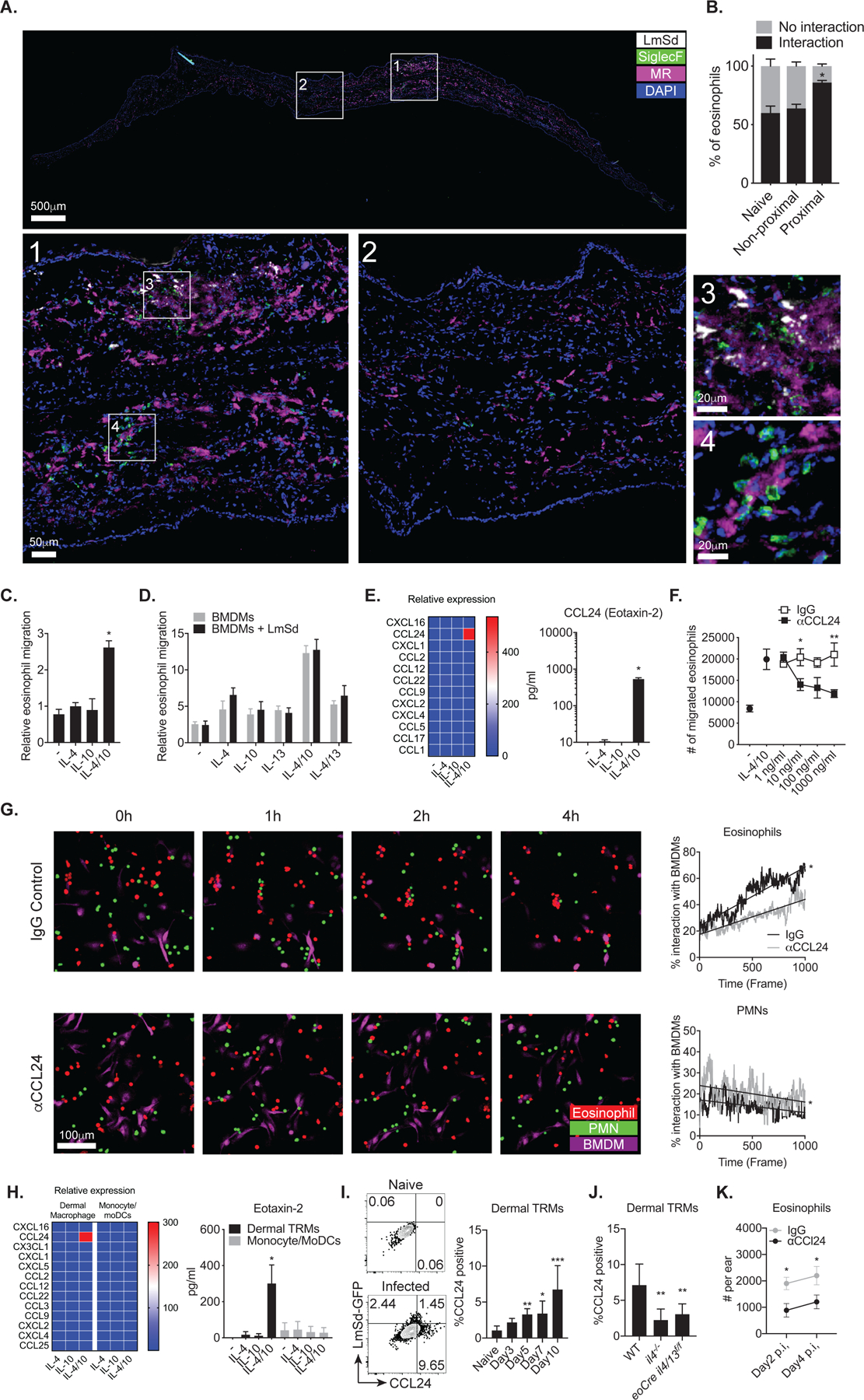
CCL24 produced by dermal macrophages mediates in vitro interaction with eosinophils. (A) Immunofluorescence staining and confocal microscopy on vertical sections of an infected ear showing LmSd-RFP (white), SiglecF (green), MR (magenta) and DAPI (blue). Images in 1–4 show boxed areas at higher magnification. (B) Percentages of eosinophils interacting with dermal macrophages in naïve, non-inflamed, and inflamed regions (n = 6) of ear at 8 days p.i. with 2 × 105 RFP+ LmSd. (C) Trans-well migration of eosinophils from upper chamber to lower chamber containing BMDM treated or non-treated for 72 hours with either IL-4, IL-10, or IL-4/10, compared to cytokine-only controls (n = 6). (D) Trans-well migration of eosinophils from upper chamber to lower chamber containing BMDM treated or non-treated with either IL-4, IL-10, IL-13, IL-4/10, or IL-4/13, compared to cytokine-only controls, and infected or uninfected with LmSd (n = 4). (E) Chemokine array from supernatants of BMDM treated or non-treated for 72 hours with either IL-4, IL-10, or IL-4/10 (n = 6). (F) The number of migrating eosinophils from upper chamber to lower chamber containing IL-4/10 treated BMDMs in the presence of increasing concentrations of anti-CCL24 neutralizing antibodies (n = 6). (G) Representative in vitro live imaging and quantification of interactions between co-cultured eosinophils (red), neutrophils (green), and IL-4/10 stimulated BMDMs (purple) treated with anti-CCL24 antibody or control IgG. (H) Chemokine array from supernatants of dermal macrophages and monocytes/moDCs sorted from naïve wild type mice and mice (n = 4) treated for 72 hr with either IL-4, IL-10, or IL-4/10. (I, J) Representative dot plots and graphs showing the percent of dermal TRMs with intracellular staining of CCL24 in mice (n = 4–6) infected with 2 × 105 LmSd-GFP. (K) The number of eosinophils at days 2 and 4 p.i. with 2 × 105 LmSd in wild type mice (n = 4) treated with anti-CCL24 antibody or control IgG. Values represent mean +/− standard deviation, *P < 0.05, **P < 0.01, and ***P < 0.001 by non-parametric Mann-Whitney test (F, G, K) and by one-way ANOVA with Dunn’s post-test compared to non-treated (C, E, H), naïve (I), or WT (J). Data are representative of over five (A, B), two (C, D, E, F, G, H, I, J, K) independent experiments.
To investigate whether macrophages directly attract eosinophils upon stimulation, we measured eosinophils transmigration into chambers containing BMDMs pre-treated with type 2 cytokines, including IL-4, IL-10, or IL-4/10, which we previously reported as necessary cytokines to maintain dermal TRMs during L. major infection(11). When incubated with IL-4/10, BMDMs induced about 2.6-fold increase in the transmigration of eosinophils compared to the respective cytokines without BMDM (Fig. 3C). IL-13 had no effect, and LmSd infection of the BMDMs did not alter the eosinophil transmigration compared to treatment of BMDMs with cytokines alone (Fig. 3D). Strikingly, a chemokine array using supernatants from the treated BMDMs, demonstrated that IL-4/10 stimulation significantly upregulated the production of a single chemokine, CCL24, also known as Eotaxin-2, up to 540 pg/ml (Fig. 3E). Neutralizing antibodies against CCL24 decreased eosinophil transmigration into wells containing IL-4/10 stimulated BMDMs in a dose dependent manner (Fig. 3F). Live imaging of cultures containing eosinophils, neutrophils, and IL-4/10-stimulated BMDMs revealed the selective association of dermal TRMs and eosinophils (Fig. 3G, Mov. S3 and S4). During a 4-hour incubation, eosinophils showed increasing degrees of interactions with BMDMs in contrast to neutrophils. CCL24 neutralization significantly diminished these interactions, while the neutrophil interactions with BMDMs were slightly increased.
To address whether dermal TRMs secrete CCL24 upon cytokine stimulation, we sorted and stimulated dermal TRMs, Ly6Chi monocytes, and Ly6Cint/lo moDCs ex vivo (Fig. 3H). Dermal TRMs produced a large amount of CCL24 exclusively in response to IL-4/10. No upregulation of the other chemokines was detected, and we were unable to detect CCL24 secretion from monocytes or monocyte-derived cells. These data are consistent with transcriptional profiles of dermal myeloid cells deposited in the Gene Expression (GEO) database (accession number GSE49358) which showed that MHCII+/− dermal TRMs express significantly higher levels of Ccl24 in comparison to the other myeloid cells (Fig. S3). We also detected increased in vivo production of CCL24 in dermal TRMs beginning at 5 days after infection (Fig. 3I), that was significantly reduced in il4−/− and eoCre il4/13f/f mice (Fig. 3J). There was significantly decreased eosinophil recruitment after in vivo CCL24 neutralization, confirming its essential role in the intralesional accumulation of these cells (Fig. 3K).
Eosinophils co-localize with dermal TRMs in the steady state
To visualize the association of dermal TRMs and eosinophils in vivo during steady state, we performed intravital imaging (IVM) on the upper dermis using Manocept labeling, previously shown to selectively bind the mannose receptor on the surface of dermal TRMs(11). Non-migratory, perivascular dermal TRMs were readily visible within one hour after Manocept injection (Fig. 4A; Mov. S5), and showed active, continuous morphological changes involving rapid protrusion of pseudopods (Fig. 4B; Mov. S6 and S7).
Fig. 4.
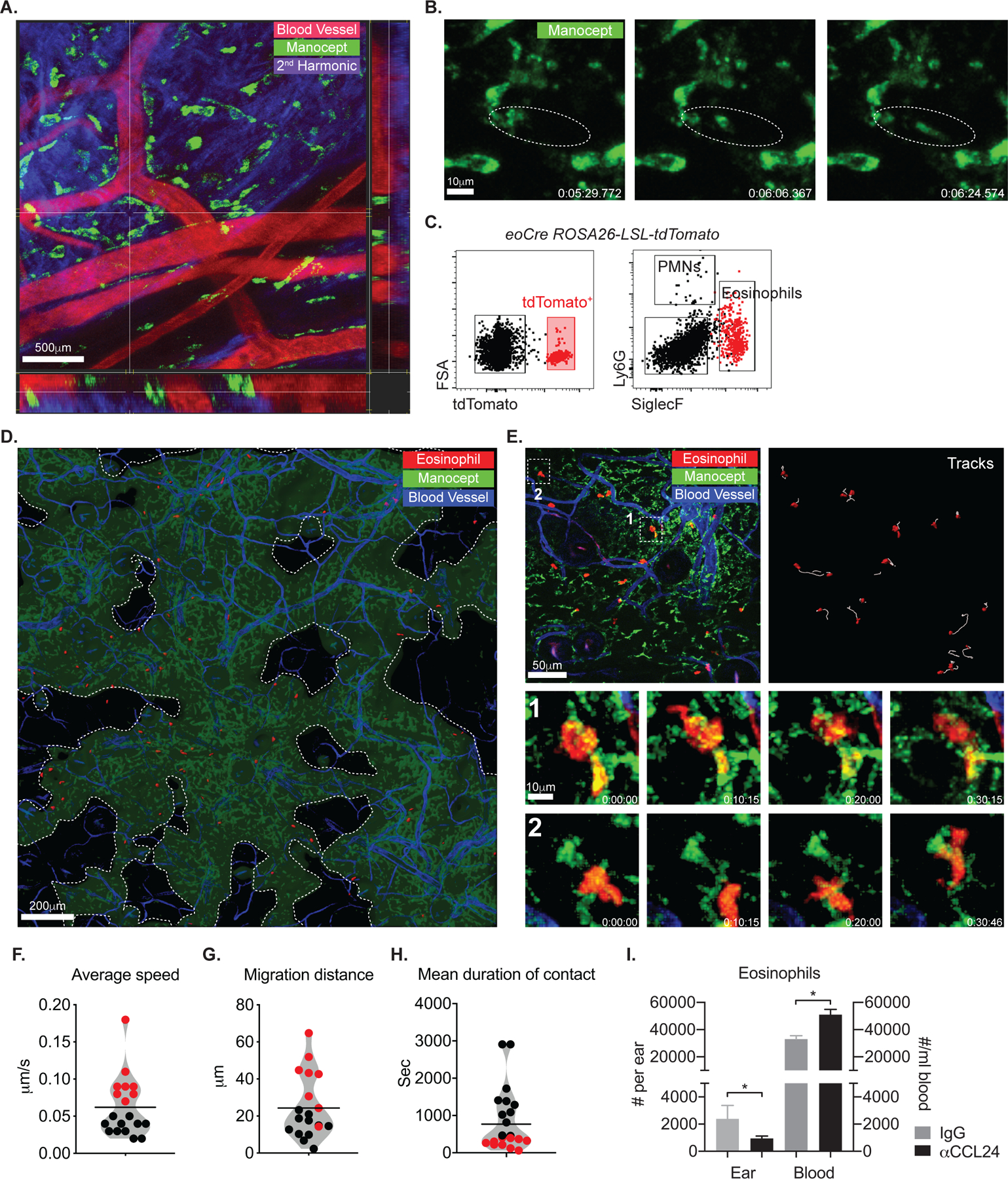
Eosinophils show stable interaction with dermal TRMs in steady state. (A, B) IVM time-lapse image from the ear of a C57BL/6 mouse injected with manocept-Alexa488 to label MRhi dermal TRMs and Evans blue for blood vessels. Data are representative of over ten independent mouse experiments. (C) Representative flow cytometry plots showing tdTomato+ eosinophils from eoCre ROSA-LSL-tdTomato reporter mouse. (D) Image obtained from a IVM of the ear of naïve eoCre ROSA-LSL-tdTomato reporter mouse. Areas devoid of TRMs are demarcated by dotted lines. (E) IVM images from the ears of a naïve eoCre ROSA-LSL-tdTomato mouse. Panel labeled “Tracks” show the paths followed by tdTomato+ eosinophils over 50 min. Bottom panels labeled “1” and “2” are time-lapse images from boxed region in upper left panel. (F, G, and H) violin plots showing the average speed, migration distance, and mean duration of contact (with dermal TRMs) of eosinophils. Stratification of the cells according to their bimodal distribution of average speed (colored red or black), was applied to the other behavior parameters. Data are representative of over four independent mouse experiments (D, E, F, G, H), (I) The number of eosinophils in ear and peripheral blood in mice (n = 4) following administration of either isotype control or anti-CCL24 Abs for 3 consecutive days. Values represent mean +/− S.D., *P < 0.05 by non-parametric Mann-Whitney test (I). Data are representative of two independent experiments (I).
For visualizing eosinophils, we crossed eoCre mice with ROSA26-LSL-tdTomato reporter mice which express robust tdTomato fluorescence following Cre-mediated recombination. tdTomato+ cells recovered from the skin of naïve animals were virtually all CD11b+SiglecF+Ly6G− eosinophils (Fig. 4C). Combining intravital microscopy with eoCre reporter mice revealed low numbers of eosinophils patrolling normal skin (Fig. 4D and E), as previously described(27). Low magnification, IVM imaging of Manocept labeled tissue indicated that the dermal TRMs were not evenly distributed throughout the ear dermis, and that the tissue-resident eosinophils were largely confined to areas populated by dermal TRMs (Fig. 3A and 4D). More than 50% of eosinophils in the steady state continuously interacted with or were in close proximity to dermal TRMs (Fig. 4E, inset 1 and 2; Mov. S8). This was reflected in the bimodal distribution of various parameters, including the instantaneous velocity of crawling eosinophils (0.02 to 18 μm/s, mean 0.059 μm/s) (Fig. 4F), the migration distance (mean 24.65 μm during one-hour imaging) (Fig. 4G), and the duration of contact (mean 778 seconds) (Fig. 4H). The speed and migration distance of the eosinophils were inversely correlated with their contact duration with dermal TRMs.
To test the possibility that CCL24 is produced to control eosinophil migration into the steady state dermis, we neutralized CCL24 and monitored the change in eosinophil numbers. CCL24 neutralization significantly reduced the number of dermal eosinophils while resulting in blood eosinophilia (Fig 4I).
CCL24 regulates eosinophil migration, morphology and interaction with dermal TRMs during L. major infection
As we previously reported(11), a progressive infiltration of eosinophils into the inoculation site was observed during the first 6 days of infection (Fig. S4). Since the number of eosinophils at day 6 post infection exceeded the capability of the imaging software to track individual eosinophils, we focused on earlier events to visualize the behavior of these cells. As early as hour after challenge with 104 LmSd, eosinophils began to localize around the injection site (Fig. 5A; Mov. S9 and S10). The average speed and migration distance of eosinophils at 1h p.i. were significantly increased compared to the steady state, 0.102 μm/s and 96.8 μm vs. 0.059 μm/s and 24.6 μm, respectively (Fig. 4F, 4G, 5C, and 5D). Compared to steady state, the eosinophils at 1h p.i. were also in shorter duration contact with dermal TRMs, 115 seconds vs. 778 seconds (Fig 4H and 5F), with highly fluctuating areas of colocalization between the cells (Fig. 5G, left panel). CCL24 neutralization failed to change any early behavior of the eosinophils, suggesting that other factors related to the tissue damage and mechanical stress associated to the needle injection might control the immediate behavior of eosinophils at the site.
Fig. 5.
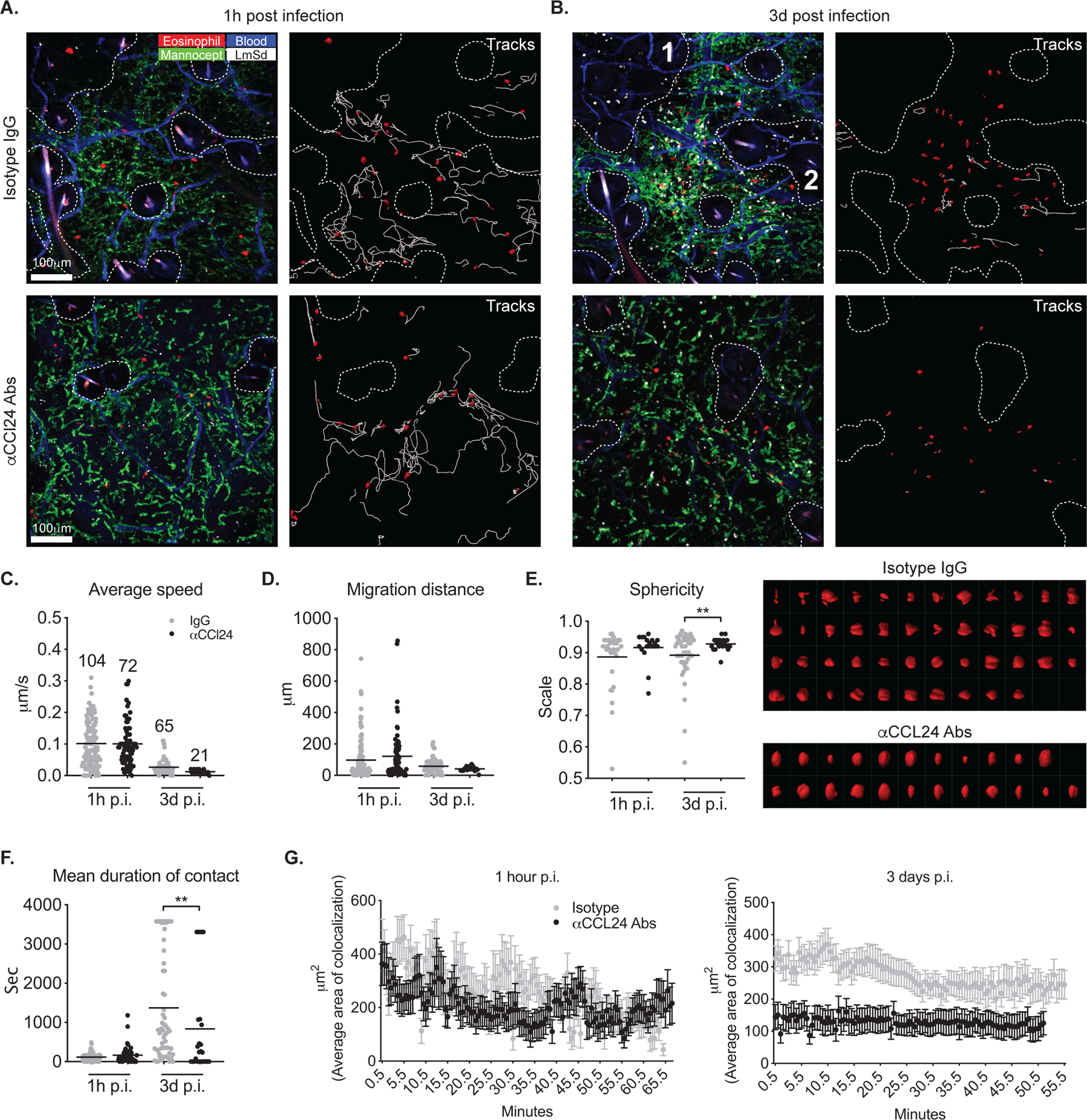
CCL24 promotes eosinophil migration, morphological changes, and interaction with dermal TRMs during infection. (A, B) IVM images from the ears of eoCre ROSA-LSL-tdTomato mice infected intradermally with 2 × 105 LmSd and treated with CCL24 neutralizing antibodies or control IgG. Panels labeled “Tracks” show the paths followed by tdTomato+ eosinophils over 50 min. The areas devoid of dermal TRMs are demarcated by dotted lines. (C–G) Eosinophil average speed of movement, migration distance, sphericity, mean duration of contact with dermal TRMs, and average of colocalization with dermal TRMs were analyzed from IVM images represented in panels (A) and (B). The numbers in panel (C) indicate the total number of trackable eosinophils in one field of view. Surface rendered eosinophils at day 3 post infection were plotted in right panel of (E) in order of low to high sphericity. Values represent mean +/− standard deviation. *P < 0.05 and **P < 0.01 by non-parametric Mann-Whitney test (E, F) to compare two samples. Data are representative of three independent experiments (A–G).
After 3 days, eosinophils in both groups became sessile, shown as significantly decreased average speed (Fig. 5B and C, Mov. S11 and S12). Several lines of evidence suggested that CCL24 produced by the dermal TRMs was involved in the migration and phenotypic changes of eosinophils at this time. First, the number of tracked eosinophils in the field of view was three-fold fewer in CCL24-neutralized animals compared to IgG controls (21 vs. 65 tracked eosinophils; Fig. 5C). This was reflected in the reduced total number of ear dermis eosinophils in the CCL24 neutralized mice quantified by flow analysis at days 2 and 4 p.i. (Fig. 3K). Note that while the number of eosinophils tracked by the imaging software appeared greater at 1 hour compared to 3 days, this reflects an overestimate at the early time point due to cells migrating in and out of the imaging plane and being counted multiple times. Second, while dermal eosinophils in the control mice displayed an activated morphology with irregular shapes, multiple pseudopods and spreading, eosinophils in CCL24 neutralized mice assumed a more uniformly smaller size and more rounded shape, which was quantified by measuring how much the shape approximates a sphere (Fig. 5E). Third, the average contact duration and area between eosinophils and dermal TRMs in a CCL24 neutralized animal was also significantly decreased (Fig. 5F and G). Importantly, eosinophils failed to infiltrate into the areas where dermal TRMs were absent, even where many viable parasites were observed (Fig. 5B; area 1 and 2 demarcated by dotted line).
We confirmed the interaction between eosinophils and dermal TRMs using high-resolution confocal microscopy on infected ear sections. While 91% of eosinophils were associated with dermal TRMs in WT mice after infection, this association was significantly decreased to 47% in eoCre il4/13f/f mice (Fig. S5A). As observed by intravital microscopy, the eosinophils infiltrating the dermis in eoCre il4/13f/f mice were smaller in volume and more spherical in shape compared to those in WT mice (Fig. S5B).
Dermal TRMs from infected mice with selective deficiency in eosinophil IL-4/13 exhibit pro-inflammatory transcriptional profiles
We used RNA-seq to identify global changes in the transcriptomes of dermal TRMs comparing cells recovered from WT and eoCre il4/13f/f mice during infection. At day12 p.i., lack of localized production of IL-4/13 by eosinophils induced a marked alteration of dermal TRMs gene expression, with 516 differentially expressed genes (DEGs) significantly up- or downregulated compared to WT mice (Fig. 6A, Table S1). Gene ontology (GO) analysis was used to identify cellular processes that were altered in the dermal TRMs as a consequence of their lack of direct or indirect exposure to IL-4/13 from eosinophils. DEGs more than 1.5 fold, either up- or down-regulated, were used as inputs for the GO analysis, and the enriched categories ranked according to adjusted p-value (cutoff of <0.05) (Table S2). Note that there are only three GO categories enriched among genes that were downregulated in dermal TRMs from infected eoCre il4/13f/f mice, two of which comprise redundant gene sets, and all have weaker p-values than each of the 26 upregulated GO enriched categories. The linkages of the biological GO terms and assigned genes of the three down-regulated and top eight up-regulated categories are depicted as a network (Fig. 6B). Within the upregulated network, the majority of the overrepresented genes are related to pro-inflammatory molecules and signaling pathways, including Tnf, Il6, Il18, IL1b, Il18r1, Nlrp3, Bcl3, Relb, Cd14, Ccl4, and Prdx1. Importantly, the genes within the down-regulated categories included Ccl24 along with genes associated with anti-inflammatory functions, including Ccl17, Ptger4, and Fcer1g. Other genes related to M2 polarization but not assigned to these particular GO categories, including IL13ra, Cd209, Tfam, Tfrc, and Rora, were also significantly down-regulated in the dermal TRMs from the eoCre il4/13f/f mice (Table S1).
Fig. 6.
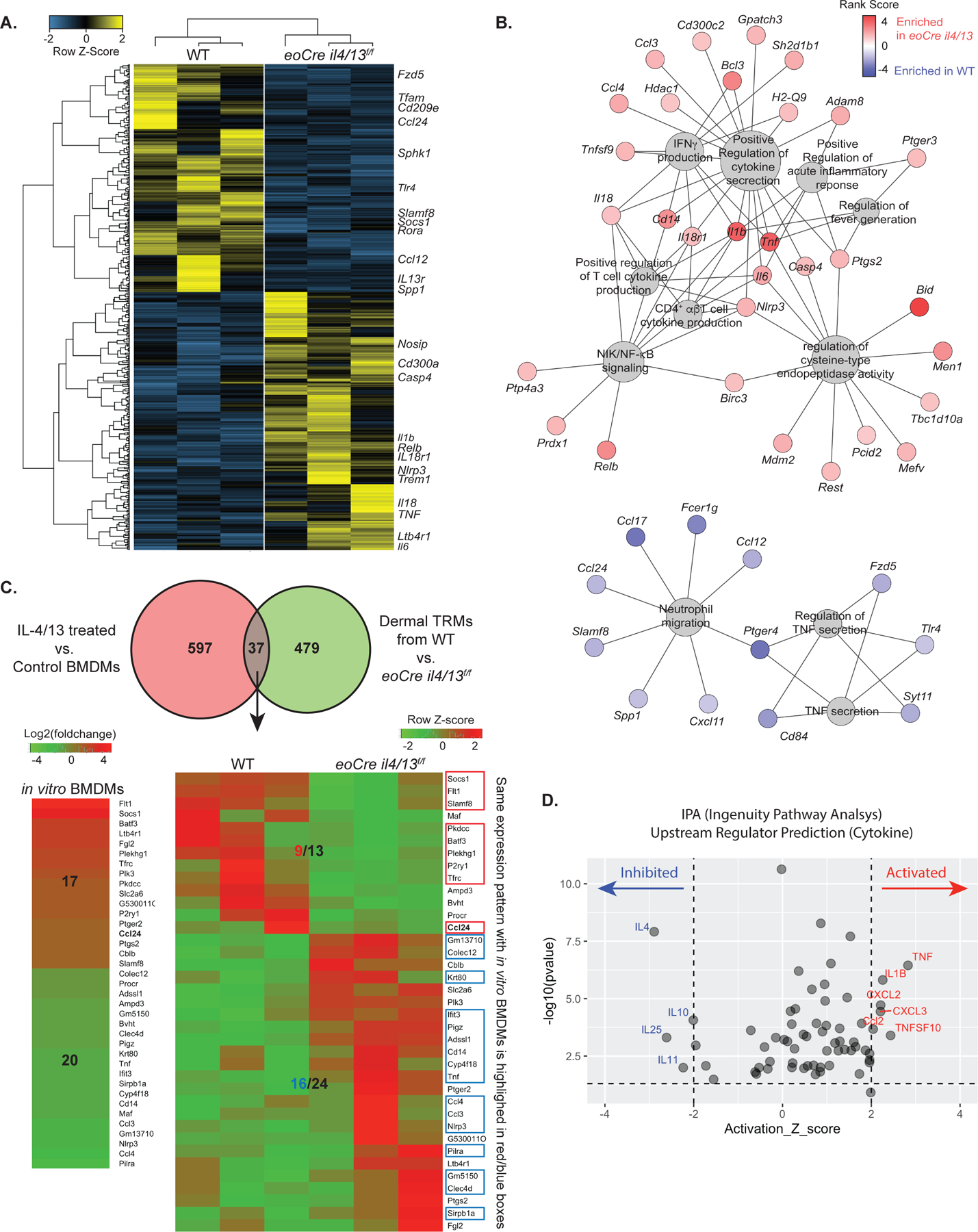
Dermal TRMs from infected eoCre il4/13f/f mice show pro-inflammatory transcriptional profiles compared to dermal TRMs from infected WT mice. (A) Heatmap displaying scaled FPKM expression values of differentially expressed genes (DEGs) of dermal TRMs from L.major infected WT and eoCre il4/13f/f mice. DEGs with absolute fold change > 1.5 and p < 0.05 (total 516 genes) are selected. Genes (row) and samples (column) are clustered using hierarchical clustering based on Pearson Correlation distance. (B) Visualization of results from gene set enrichment analysis using pre-Ranked selected genes (274 upregulated and 242 downregulated DEGs are used). Gene-concept network depicts the linkages of genes and biological GO terms as a network. The size of GO terms (grey) represents the number of genes connected and the color of each genes indicates rank-scores (from −4.79 to 4.79): red, up-regulated and blue, down-regulated in dermal TRMs of eoCre il4/13f/f compared to WT mice. (C) Venn diagram (upper panel) shows 37 overlapping genes between DEGs from IL-4/13 treated vs. non-treated BMDMs and from dermal TRMs of WT vs eoCre il4/13f/f mice. Heatmaps display the differentially expressed values of the overlapping genes from in vitro BMDM on the left (log2(foldchange)) and in vivo dermal TRMs from WT vs eoCre il4/13f/f mice on the right panel (FPKM with Row Z-score). Genes with the same expression pattern between in vitro BMDM and in vivo dermal TRMs are highlighted in red (up) or blue (down) boxes. (D) Plot of upstream regulators predicted using Ingenuity Pathway Analysis (IPA). The 516 DEGs identified between dermal TRMs of infected eoCre il4/13f/f vs WT mice were used as input. Only regulators in the ‘cytokine’ molecular type were selected from the results and used for visualization. Regulators with activation Z score (>2 or <−2, respectively, with 97.72% confidence) and p-value of overlap (<0.05) are considered significantly activated (red) or inhibited (blue) in dermal TRMs from eoCre il4/13f/f mice vs. WT mice.
We also compared DEGs of dermal TRMs recovered from infected WT vs eoCre il4/13f/f mice with previously reported DEGs from IL4/13-treated vs non-treated BMDM in vitro(28). The vast majority of the DEGs were not shared, a finding consistent with the lack of overlap between the transcriptomes of adipose TRMs with those of bone marrow derived, classically activated M1 or M2 macrophages generated in vitro (Fig. 6C). Nonetheless, the differential expression of certain classical M2 markers, such as Chil3 and Retnla, were confined to the IL-4/13 stimulated BMDMs(28). Additionally, we performed ingenuity pathway analysis (IPA) to predict upstream regulators. An overlap p-value was computed based on the significant overlap between the 516 DEGs and known targets of the regulator. The activation z-score was used to make predictions. Critically, IL-4 was identified as the upstream cytokine regulator predicted to be the most inhibited in our dataset (Fig. 6D). In contrast, pro-inflammatory and monocyte chemoattractants such as TNF, IL1β, CXCL2/3, and CCL2 were identified as activated upstream regulators, consistent with the increased infiltration of inflammatory monocytes in the eoCre IL4/13f/f mice (Fig. 2G). Collectively, these data strongly support a role for IL-4/13 from eosinophils not only to induce the local proliferation of dermal TRMs, but to actively maintain their M2-like state during L. major infection.
Eosinophil IL-4 regulates CCL24 expression and proliferation of peritoneal macrophages during experimental Ascaris infection
To evaluate the significance of IL-4-CCL24 mediated interaction between TRMs and eosinophils in other tissue pathologies, we used Ascaris suum, a helminth infection model in the peritoneum which promotes a local eosinophilia(29). Peritoneal fluid from naïve animals already contained a significant amount of CCL24 (300–500 pg/ml), an underestimate considering the fluid was diluted in PBS used to harvest the peritoneal cells (Fig. 7A and C). A single intraperitoneal injection of live Ascaris eggs induced a large amount of CCL24 (1–3 ng/ml) that was significantly reduced in anti-IL-4 treated mice (Fig. 7A) and in eoCre il4/13f/f mice lacking IL-4 production from eosinophils (Fig. 7C). As expected, marked accumulation of peritoneal eosinophils accompanied the increased CCL24 production after Ascaris infection that was ablated by anti-IL-4 treatment (Fig. 7B). The number of peritoneal macrophages was also decreased following IL-4 neutralization, a result consistent with the IL-4 driven, local proliferation of peritoneal macrophages in Brugia malayi, another tissue-invasive nematode(13). Moreover, both CCL24 neutralization as well as selective deficiency of IL-4 from eosinophils strongly reduced peritoneal eosinophil numbers (Fig. 7D). Importantly, both CCL24 neutralized WT and eoCre il4/13f/f mice, CCL24 neutralized or not, had a significantly decreased number of peritoneal TRMs compared to WT mice. While fewer numbers of macrophages were recovered when comparing day 3 Ascaris-infected mice to naïve mice, their adhesion to the parasites and the “macrophage disappearance reaction” that is known to occur after acute inflammation, may have made them more difficult to be recovered in the peritoneal lavage(30, 31). However, increased proliferation of the peritoneal macrophages was confirmed by both Ki67 staining and BrdU incorporation at 72 hours p.i., which was abrogated in the eoCre il4/13f/f mice (Fig. 7E).
Fig. 7.
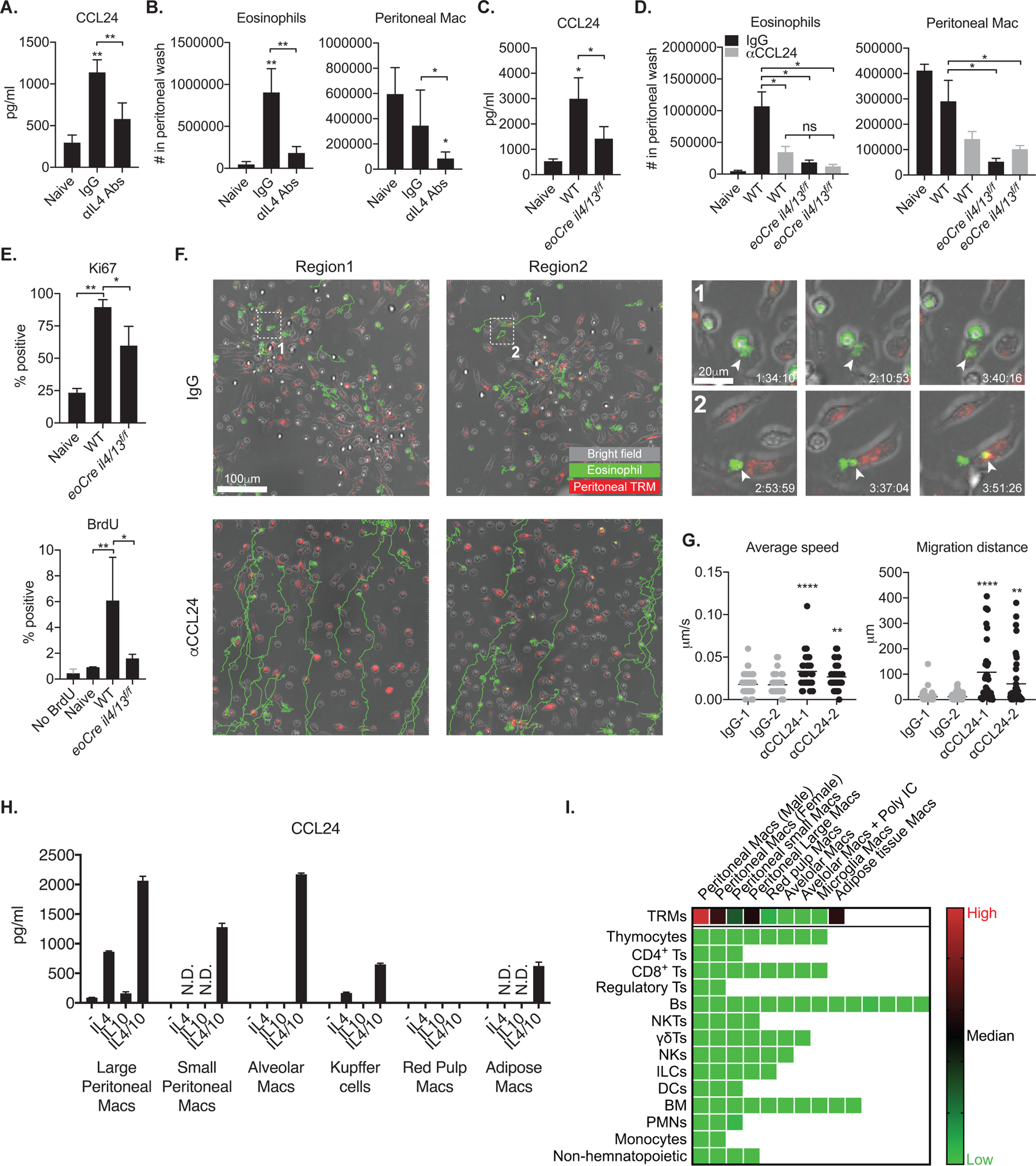
The IL4-CCL24 axis mediates eosinophil influx and interaction with peritoneal macrophages in peritoneal Ascaris infection. (A, C) Quantification of CCL24 from peritoneal fluids harvested from naïve mice (n = 3) or (A) mice (n = 6) infected i.p. with 10,000 Ascaris eggs for 72 hours and treated with IL-4 neutralizing mAbs or control IgG, or (C) wild type and eoCre il4/13f/f infected mice (n = 4). (B, D) The number of eosinophils and peritoneal macrophages in peritoneal fluids harvested from naïve mice (n = 3–4) or (B) Ascaris infected mice (n = 6) treated with IL-4 neutralizing mAbs or control IgG, or (D) wild type and eoCre il4/13f/f infected mice treated with CCL24 neutralizing mAbs or control IgG (n =4). (E) Ki67 expression or BrdU incorporation by dermal TRMs from naive or infected WT and eoCre il4/13f/f mice with 10,000 Ascaris eggs for 72 hours (n = 4). (F) Representative ex vivo live imaging of fluorescence antibody-labeled SiglecF+ eosinophils and F4/80+ peritoneal macrophages from Ascaris-infected peritoneal isolates incubated with anti-CCL24 antibody or control IgG. Images in 1 and 2 show boxed areas at higher magnification. Arrow heads indicate eosinophilic materials transferred to or phagocytosed by F4/80+ peritoneal macrophages. Green-colored tracks show the paths followed by SiglecF+ eosinophils over 4-hour imaging. (G) Eosinophil average speed of movement and migration distance were analyzed from live imaging represented in panel (F). (H) Quantification of CCL24 released from various TRMs sorted from naïve animals (n = 2–3) and cultured for 3 days in vitro with indicated cytokines. (I) Comparison of CCL24 transcription from various immune cells published by Immgen(32). The relative expression of CCL24 gene is shown as a heat map among the indicated populations. Each square represents either a specified subset of TRMs (top row) or populations of the specified cells isolated from different organs. Values represent mean +/− standard deviation. *P < 0.05, **P < 0.01, and ****P < 0.0001 by non-parametric Mann-Whitney test (A, B, C, D, E) to compare two samples and by one-way ANOVA with Dunn’s post-test compared to naïve (A, B, C), and IgG (G). Data are representative of two (A–E, H) or four (F) independent experiments.
To visualize the association of peritoneal TRMs and eosinophils during Ascaris infection, we performed ex vivo live imaging of peritoneal cells harvested from infected mice with or without CCL24 neutralization (Fig. 7F; Mov. S13 and S14). We observed clusters of closely interacting cells containing F4/80+ peritoneal macrophages and eosinophils, which showed minimal motility. Of note, cellular material from eosinophils seemed to be directly transferred to or taken up by peritoneal macrophages during their close interaction (Fig. 7F, inset 1 and 2; Mov. S15 and S16). In contrast, no cluster of cells were formed following CCL24 neutralization, and eosinophils showed increased migration without interaction with peritoneal macrophages (Fig. 7F and G).
To determine whether peritoneal TRMs, as well as TRMs from other tissues, produce CCL24 in response to IL-4/10, we sorted TRMs from various tissues and stimulated them with IL-4 and/or IL-10 (Fig. 7H). Resident macrophages recovered from the peritoneum, lung, liver and adipose tissue all produced a large amount of CCL24 in response to IL-4/10 simulation. IL-4 alone was sufficient to induce CCL24 expression in large peritoneal macrophages and Kupffer cells. In contrast, red pulp macrophages from spleen failed to produce CCL24 under any condition tested.
Finally, we compared CCL24 mRNA expression from gene expression profiles of various hematopoietic populations compiled in the ImmGen RNA-Sequencing database(32). Ccl24 expression is restricted to TRMs, including peritoneal and adipose TRMs (Fig. 7I).
Discussion
In this study, we demonstrate the mechanisms by which the number and activation program of M2-like dermal TRMs are maintained during strong TH1-oriented immunity against cutaneous infection with L. major. Using bone marrow chimeras and conditional knockout mice, innate cells and specifically eosinophils were identified as the main producers of IL-4, required to maintain the population size and M2-like properties of dermal TRMs during infection. In turn, IL-4-stimulated dermal TRMs, in concert with IL-10, released a large amount of CCL24 to sustain eosinophil influx. Intravital imaging of eosinophil behavior in the skin revealed an intimate association between dermal TRMs and infiltrating eosinophils, ensuring highly localized delivery of IL-4 during TH1 inflammation.
While the role of IL-4 produced by innate cells has been mostly studied in the context of TH2 cell differentiation(33, 34), innate sources of IL-4 acting on macrophages have also been described. Basophils were reported to provide IL-4 required for the local proliferation of monocyte-derived macrophages that replaced Kupffer cells lost during bacterial infection(35). In the skin, Ly6C+CCR2+ inflammatory monocytes have been shown to differentiate into M2-like macrophages under the influence of basophil-derived IL-4 after helminth infection or allergen challenge(36, 37). Eosinophils were also found to play an important role in maintaining alternatively activated macrophages in adipose tissue through an IL-4-dependent process(16). The later studies reported very small numbers of adipose- and lung-resident eosinophils producing IL-4. We also found that only a small percentage of eosinophils were identified as active secretors of IL-4. Other cells identified as active IL-4 secretors using the reporter mice were CD90.2+CD11b−CD3−, most likely ILC2s, as well as basophils, and these cells likely contributed to the localized IL-4 production. The role of eosinophils in providing IL-4 to maintain the M2-like, dermal TRMs is supported by the enhanced resistance to infection, accompanied by the reduced numbers of dermal TRMs, in anti-SiglecF treated, IL-5 knockout, and most critically in eoCre il4/13f/f mice lacking IL-4 production from eosinophils. The data support a model whereby the anti-inflammatory programs of dermal TRMs in steady state conditions, linked to their ontogeny and homeostatic cues from tissue microenvironments, are actively maintained by IL-4 producing eosinophils over the course of infection in the TH1 immune environment of the L. major loaded dermis, as evidenced by a shift to a predominant pro-inflammatory transcriptional profile in dermal TRMs from eoCre il4/13f/f mice. By IPA, IL-4 was predicted as the key upstream cytokine regulator whose inhibition/absence could best explain the expression pattern of these cells. Interestingly, an increased infiltration of Ly6Chi inflammatory monocytes was observed in the Leishmania-infected eoCre il4/13f/f mice, consistent with the upregulated expression of CCL3/4 (MIP1α and β) on the dermal TRMs from these mice, known chemoattractants for monocytes expressing CCR1/5(38). The increased number of monocytes and monocyte-derived cells likely contributes to the improved control of infection in the eoCre il4/13f/f mice, as infection of these cells is associated with upregulated iNOS expression and acquired resistance in L. major infected mice(11, 39, 40).
The close physical proximity of eosinophils and dermal TRMs suggests highly localized delivery of IL-4. In live cell-imaging of peritoneal cells recovered from Ascaris-infected animals, we observed that eosinophils directly transferred cellular material to peritoneal TRMs. Considering that eosinophils-derived cytokines are pre-synthesized and can be released by several distinct pathways from their stores within intracellular granules(41), the exact mechanism of IL-4 delivery to TRMs remains to be elucidated. Our studies provide clear evidence that the intimate physical association between eosinophils and TRMs is mediated by CCL24 (Eotaxin-2) produced by dermal TRMs. Over 80% of eosinophils co-localized with dermal TRMs, and both the duration of contact between these cells and their areas of co-localization were significantly reduced following CCL24 neutralization. We show that dermal TRMs were the near exclusive producers of CCL24 among the various innate and adaptive populations in the skin. Even heavily parasitized areas had no eosinophil infiltration unless populated by dermal TRMs. In addition, eosinophils co-localized with dermal TRMs in a naïve animal, with CCL24 neutralization significantly reducing the number of eosinophils in the steady state dermis, consistent with the previously described absence of eosinophils in naïve skin of ccr3−/− mice, lacking the receptor for CCL24(42). Finally, we could observe a similar requirement for eosinophil derived IL-4 for the upregulation of CCL24 secretion which mediates the peritoneal eosinophilia observed in an Ascaris infection model. Ex vivo live imaging of peritoneal cells harvested from infected mice revealed clusters of interacting eosinophils and macrophages that were absent when CCL24 was neutralized. Two recent reports describe macrophages producing CCL24 in other tissue pathologies, including alveolar macrophages in lung melanoma and cardiac macrophages in myocarditis(43, 44).
We show that in addition to IL-4, IL-10 was required for optimal secretion of CCL24 by dermal TRMs, consistent with the synergistic effects of these cytokines on induction of CCL24 production by BMDM in vitro(23), and the striking loss of dermal TRMs in mice lacking both IL-4 and IL-10(11). We have not addressed the specific cellular source of the IL-10 that induces CCL24 expression in the dermal TRMs. IVM studies using IL-10 reporter mice are planned to help address this question. It should also be noted that as the phenotypes in the eoCre il4/13f/f mice were only partial with respect to infection outcome and loss of dermal TRMs, other sources of IL-4 likely play a role. Nonetheless, as macrophages in the peritoneum, lungs, liver, and adipose tissue all secreted high levels of CCL24 in response to IL-4/10, it would appear that resident macrophages in many different inflammatory settings can be conditioned by a common pathway to promote cooperative interactions with eosinophils required for their maintenance.
Materials and Methods
Study Design
The goal of these studies was to demonstrate how dermal TRMs maintain the population size and M2-like characteristics during classical TH1 immune response against L. major infection. We compared the population size and transcriptional profiles of dermal TRMs between WT and eoCre IL4/13f/f mice. Animals were randomly assigned to different experimental groups. The sample size and reproducibility of experiments are specified in each figure legend. Sample measurements were blinded during experiments or analysis.
Mice
C57BL/6 mice were purchased from Taconic Farms. C57BL/6 CD45.1+ or CD45.2+ rag1−/− mice, C57BL/6 il4−/− mice, and C57BL/6 Lyz2GFP mice were obtained through a supply contract between the National Institute of Allergy and Infectious Diseases (NIAID) and Taconic Farms. ROSA26-LSL-tdTomato mice (also known as Ai14) mice and Actb-DsRed.T3 were purchased from The Jackson Laboratory. Dr. Helene Rosenburg (NIAID) kindly provided C57BL/6 il5−/− mice. eoCre mice, originally developed by Doyle et al.(45), were obtained from Mayo Clinic, Scottsdale, AZ and crossed with ROSA26-LSL-tdTomato mice to generate eosinophil-specific fluorescent reporter animals(27). eoCre il4/13f/f mice with selectively deficiency of IL-4/13 in eosinophils(25, 26), were also obtained from Mayo Clinic, Scottsdale, AZ. All of the mice used in these studies were female, 6–8 weeks old, and were bred and/or maintained in the NIAID animal care facility under specific pathogen-free conditions. They were used under a study protocol approved by the NIAID Animal Care and Use Committee (protocol number LPD 68E). All aspects of the use of animals in this research were monitored for compliance with The Animal Welfare Act, the PHS Policy, the U.S. Government Principles for the Utilization and Care of Vertebrate Animals Used in Testing, Research, and Training, and the NIH Guide for the Care and Use of Laboratory Animals.
In vivo injections
Manocept-Alexa488 or -Cy5 (Navidea Biopharmaceuticals) is a fluorescently labeled derivative of FDA-approved 99MTC-Tilmanocept targeting MR. Naïve mice were injected with 25μg Manocept intravenously with 20μg anti-CD31 Abs (Clone#390, Invitrogen) in total volume not to exceed 100uL per mouse during IVM. For eosinophil depletion, animals were i.p. injected 15μg of SiglecF Abs (Clone# 238047, R&D systems) at day2 p.i. and every two days. In vivo neutralization of CCL24 was carried out by i.p. administration of 100μg of monoclonal CCL24 Abs (Clone# 106521, R&D systems) every day for 3 days beginning one day prior to L. major or Ascaris infection.
Leishmania major and Ascaris suum infection
The L. major Seidman strain (MHOM/SN/74/SD) (LmSd) was maintained as follows: promastigotes were grown at 26°C in medium 199 supplemented with 20% heat-inactivated FCS (Gemini Bio-Products), 100 U/mL penicillin, 100 μg/mL streptomycin, 2 mM L-glutamine, 40 mM Hepes, 0.1 mM adenine (in 50 mM Hepes), 5 mg/mL hemin (in 50% triethanolamine), and 1 mg/mL 6-biotin (M199/S). Parasites expressing a red fluorescent protein (LmSd-RFP) were grown using the identical culture medium supplemented with 50 μg/mL of Geneticin (G418, Gibco, Woodland, CA). Infective-stage, metacyclic promastigotes were isolated from stationary cultures (5–6 days) by density gradient centrifugation, as described previously(46). Mice were then inoculated with metacyclic promastigotes in the ear dermis or footpad by intradermal or subcutaneous injection respectively in a volume of 10μL. Lesion development was monitored weekly by measuring the diameter of the ear nodule with a direct-reading Vernier caliper (Thomas Scientific). Lesion pathology was also evaluated and scored as follows: 0 = no ulceration, 1 = ulcer, 2 = half ear eroded, 3 = ear completely eroded.
For intraperitoneal inoculation of Ascaris suum, live eggs were isolated from adult female uteri through gentle mechanical maceration, purified by straining, and cultured to embryonation in 0.2 M H2SO4 for 100 days, which corresponds to the peak of larvae infectivity as described(47). The fully embryonated eggs (containing the L3 infective stages) were used for experimental infections. Mice were inoculated through the intraperitoneal route with 10,000 fully embryonated eggs to induce a local eosinophilia, as previously demonstrated(29). Three days post-injection, the peritoneal cavity cells and lavage were collected.
Processing of ear tissues and evaluation of L. major parasite burden
Ear tissue was prepared as previously described(48). Briefly, the two sheets of infected ear dermis were separated, deposited in DMEM containing 0.2 mg/mL Liberase TL purified enzyme blend (Roche Diagnostics Corp.), and incubated for 1.5 h at 37°C. Digested tissue was processed in a tissue homogenizer for 3.5 minutes (Medimachine; Becton Dickinson) and filtered through a 70 μm cell strainer (Falcon Products) to obtain single cell suspension. Parasite titrations were performed as previously described(49). Briefly, tissue homogenates were serially diluted in 96-well flat-bottom microtiter plates containing 100 μL M199/S. The number of viable parasites in each ear was determined from the highest dilution at which promastigotes could be grown out after 7–10 days of incubation at 26°C.
Isolation of mouse peritoneal cells
Ascaris-infected mice were i.p. injected with 1ml of PBS. After injection, the peritoneum was gently massged to dislodge any attached cells into the PBS solution. Cell suspension in PBS were collected with 25g needle. After centrifugation, the supernatant was collected for assays, and cells from the first wash were combined with the second lavage with 3ml PBS.
Detection of CCL24 levels by ELISA
CCL24 levels in the peritoneal lavage of Ascaris-inoculated mice were quantified by ELISA kits purchased from R&D Systems, Inc. (Minneapolis, MN, USA). All assays were performed according to the manufacturers’ protocols.
Immunolabeling and flow cytometry analysis
Detailed methods for the surface and intracellular cytokine staining of ear dermal cells for flow cytometry analysis are described in the Supplemental Materials and Methods.
BMDM and BM-derived eosinophil generation, infection, and in vitro assays
Isolated femurs and tibia were flushed with PBS, and for BMDM differentiation precursor cells were cultured in complete RPMI supplemented with 30% L929 cell-conditioned medium. The media was changed every 2 days thereafter. After 7d in culture, mature BMDMs were harvested by washing with cold PBS, incubating on ice for 15 minutes, and pipetting extensively. BMDMs were plated at 5 × 105 /well onto 24 well plates in AIM V serum-free media (Thermofisher Scientific) with 10ng/ml IL4, IL10, or IL13 with or without purified metacyclic promastigotes. After 3d culture, supernatants were harvested and assayed with Quantibody mouse chemokine array Q1 (Raybiotech). For eosinophil differentiation, precursor cells were cultured in IMDM (Iscove’s Dulbecco’s Media) containing Glutamax (Life Technologies), 10% FCS (HyClone), 2mM L-glutamine (Life Technologies), 50μM β-mercaptoethanol (Sigma-Aldrich), Penicillin/Streptomycin (Life Technologies), and 50ng/ml SCF (Peprotech)(50). On day 4, the media was replaced with fresh complete media containing 10ng/ml murine IL-5 (Peprotech). Media with IL-5 was changed every 2 days thereafter until harvesting fully differentiated eosinophils on day 14.
Eosinophil trans-well migration assay
BMDMs were plated 1 × 105/well to lower wells of HTS Transwell-96 well plates (Corning) and stimulated with 10 ng/ml IL-4, IL-10, or IL-13 in the presence of anti-CCL24 Ab or isotype control (R&D systems). After 72 hours, 2 × 105 BM-derived eosinophils were added to the upper wells, and then incubated for 90 min at 37°C. Eosinophil migration was normalized to the respective assays having only IL4, IL10, or IL13 cytokines without BMDMs to exclude chemotactic effect of cytokines.
Confocal microscopy, live cell imaging and intravital microscopy
Detailed methods for immunostaining and confocal microscopy of cryostat sections of infected ears, for live cell imaging of BMDM co-cultured with eosinophils in vitro, for imaging peritoneal cells ex vivo, and for non-invasive intravital imaging of the mouse ear, are described in the Supplemental Materials and Methods.
BM chimeras
Recipient mice were irradiated with a single dose of 1,250 rads and reconstituted with donor bone marrow. The harvested BM cells were injected intravenously at a dose of 2.5 × 106 cells/mouse in 100 µl PBS. Animals were maintained on trimethoprim-sulfamethoxazole (Hi-Tech Pharmacal) antibiotic water for up to 5 weeks after irradiation. Hematopoietic reconstitution of all animals was verified by flow cytometry staining of congenic markers (CD45.1 and 45.2).
RNA sequencing and data analysis
Single cell suspension and antibody staining were performed as described above except for addition of 100 unit/ml SUPERase•InTM RNase Inhibitor (ThermoFisher) to all of the tissue-digestion/staining buffers to inhibit the strong ribonuclease activity in murine skin. One hundred dermal TRMs were sorted directly into lysis buffer included in SMART-Seq V4 Ultra Low Input RNA Kit for Sequencing (Takara), and cDNA was generated according to the commercial protocol. Detailed methods for library preparation, sequencing, and bioinformatic analysis are described in the Supplemental Materials and Methods.
Statistical analyses
The differences in values obtained for two different groups were determined using non-parametric Mann-Whitney test. For comparisons of multiple groups, one-way analysis of variance (ANOVA) followed by Dunn’s post-test was used. Analyses were performed using Prism 8.0 software (GraphPad).
Supplementary Material
Table S1. List of differentially expressed genes (DEGs).
Table S2. List of enriched GO categories.
Movie S1. 3D rotational confocal imaging of inoculation site and the area proximal to the inoculation site.
Movie S2. In vitro live imaging of eosinophils and IL-4/10 stimulated BMDMs with or without CCL24 neutralization.
Supplemental Materials and Methods
Fig. S1. Gating strategy for eosinophils, PMNs, Ly6Chi monocytes, Ly6Cint/Ly6Clo moDCs, and dermal TRMs.
Fig. S2. Basophils and ILC2s produce IL-4.
Fig. S3. Dermal TRMs are the main producers of Ccl24 among dermal myeloid subsets.
Fig. S4. Eosinophils progressively infiltrate the L. major inoculation site.
Fig. S5. Immunofluorescence staining and confocal microscopy on vertical ear sections of infected eoCre il4/13f/f and WT mice.
Movie S3. Intravital imaging of perivascular dermal TRMs.
Movie S4. Intravital imaging of the active surface movements of dermal TRMs.
Movie S5. Intravital imaging of eosinophils with dermal TRMs in steady state.
Movie S6. Intravital imaging of eosinophils and dermal TRMs at 1h p.i. with or without CCL24 neutralization.
Movie S7. Intravital imaging of eosinophils and dermal TRMs at 3d p.i. with or without CCL24 neutralization.
Movie S8. Ex vivo live imaging of eosinophils and peritoneal TRMs with or without CCL24 neutralization.
Movie S9. Ex vivo live imaging of eosinophils transferring cellular materials to peritoneal TRMs.
Acknowledgements:
We thank Dr. Owen Schwartz (NIAID) and Dr. Sundar Ganesan (NIAID) for help with image acquisition and analysis; Dr. Calvin Eigsti (NIAID) for help with the cell sorting.
Funding: This work was supported in part by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health. F.T.C. core funding is supported by the Swiss National Science Foundation (310030_18475/1).
Footnotes
Competing interests: The authors declare no competing interests.
Data and materials availability: RNA seq data have been deposited in the National Center for Biotechnology Information’s Sequence Read Archive (SRA) and are available under accession number: PRJNA565178. All other data needed to evaluate the conclusions of the paper, including raw data, are present in the paper and the Supplementary Materials. eoCre and eoCre il4/13f/f mice are available upon request but are contingent upon approval of material transfer agreement by the Mayo Clinic, Scottsdale, AZ. Other material used in this study will be provided upon request.
References:
- 1.Davies LC, Jenkins SJ, Allen JE, Taylor PR, Tissue-resident macrophages. Nat Immunol 14, 986–995 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davies LC, Taylor PR, Tissue-resident macrophages: then and now. Immunology 144, 541–548 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ginhoux F, Guilliams M, Tissue-Resident Macrophage Ontogeny and Homeostasis. Immunity 44, 439–449 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Gautier EL et al. , Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol 13, 1118–1128 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lavin Y et al. , Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 159, 1312–1326 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wynn TA, Vannella KM, Macrophages in Tissue Repair, Regeneration, and Fibrosis. Immunity 44, 450–462 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lavine KJ et al. , Distinct macrophage lineages contribute to disparate patterns of cardiac recovery and remodeling in the neonatal and adult heart. Proc Natl Acad Sci U S A 111, 16029–16034 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Epelman S et al. , Embryonic and adult-derived resident cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation. Immunity 40, 91–104 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruckerl D et al. , Macrophage origin limits functional plasticity in helminth-bacterial co-infection. PLoS Pathog 13, e1006233 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang L, Nazarova EV, Tan S, Liu Y, Russell DG, Growth of Mycobacterium tuberculosis in vivo segregates with host macrophage metabolism and ontogeny. J Exp Med 215, 1135–1152 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee SH et al. , Mannose receptor high, M2 dermal macrophages mediate nonhealing Leishmania major infection in a Th1 immune environment. J Exp Med 215, 357–375 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jenkins SJ et al. , IL-4 directly signals tissue-resident macrophages to proliferate beyond homeostatic levels controlled by CSF-1. J Exp Med 210, 2477–2491 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jenkins SJ et al. , Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science 332, 1284–1288 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bosurgi L et al. , Macrophage function in tissue repair and remodeling requires IL-4 or IL-13 with apoptotic cells. Science 356, 1072–1076 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qiu Y et al. , Eosinophils and type 2 cytokine signaling in macrophages orchestrate development of functional beige fat. Cell 157, 1292–1308 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu D et al. , Eosinophils sustain adipose alternatively activated macrophages associated with glucose homeostasis. Science 332, 243–247 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weller PF, Spencer LA, Functions of tissue-resident eosinophils. Nat Rev Immunol 17, 746–760 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JJ et al. , Human versus mouse eosinophils: “that which we call an eosinophil, by any other name would stain as red”. J Allergy Clin Immunol 130, 572–584 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nussbaum JC et al. , Type 2 innate lymphoid cells control eosinophil homeostasis. Nature 502, 245–248 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mishra A, Hogan SP, Lee JJ, Foster PS, Rothenberg ME, Fundamental signals that regulate eosinophil homing to the gastrointestinal tract. J Clin Invest 103, 1719–1727 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turner JD et al. , Interleukin-4 activated macrophages mediate immunity to filarial helminth infection by sustaining CCR3-dependent eosinophilia. PLoS Pathog 14, e1006949 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watanabe K, Jose PJ, Rankin SM, Eotaxin-2 generation is differentially regulated by lipopolysaccharide and IL-4 in monocytes and macrophages. J Immunol 168, 1911–1918 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Makita N, Hizukuri Y, Yamashiro K, Murakawa M, Hayashi Y, IL-10 enhances the phenotype of M2 macrophages induced by IL-4 and confers the ability to increase eosinophil migration. Int Immunol 27, 131–141 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Mohrs K, Wakil AE, Killeen N, Locksley RM, Mohrs M, A two-step process for cytokine production revealed by IL-4 dual-reporter mice. Immunity 23, 419–429 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doyle AD et al. , Eosinophil-derived IL-13 promotes emphysema. Eur Respir J 53, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voehringer D, Wu D, Liang HE, Locksley RM, Efficient generation of long-distance conditional alleles using recombineering and a dual selection strategy in replicate plates. BMC Biotechnol 9, 69 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chojnacki A et al. , Intravital imaging allows real-time characterization of tissue resident eosinophils. Commun Biol 2, 181 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hans CP et al. , Transcriptomics Analysis Reveals New Insights into the Roles of Notch1 Signaling on Macrophage Polarization. Sci Rep 9, 7999 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Archer GT, Coulits N, Jindra J, Robson JE, Eosinophilia, mast cell hyperplasia and antibody production in rats following an intraperitoneal injection of Ascaris cuticle including in-vitro studies of immune eosinophil granule lysis. Pathology 17, 101–107 (1985). [DOI] [PubMed] [Google Scholar]
- 30.Jeska EL, Mouse peritoneal exudate cell reactions to parasitic worms. I. Cell adhesion reactions. Immunology 16, 761–771 (1969). [PMC free article] [PubMed] [Google Scholar]
- 31.Barth MW, Hendrzak JA, Melnicoff MJ, Morahan PS, Review of the macrophage disappearance reaction. J Leukoc Biol 57, 361–367 (1995). [DOI] [PubMed] [Google Scholar]
- 32.Heng TS, Painter MW, C. Immunological Genome Project, The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol 9, 1091–1094 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Spencer LA, Weller PF, Eosinophils and Th2 immunity: contemporary insights. Immunol Cell Biol 88, 250–256 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paul WE, Zhu J, How are T(H)2-type immune responses initiated and amplified? Nat Rev Immunol 10, 225–235 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bleriot C et al. , Liver-resident macrophage necroptosis orchestrates type 1 microbicidal inflammation and type-2-mediated tissue repair during bacterial infection. Immunity 42, 145–158 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Egawa M et al. , Inflammatory monocytes recruited to allergic skin acquire an anti-inflammatory M2 phenotype via basophil-derived interleukin-4. Immunity 38, 570–580 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Obata-Ninomiya K et al. , The skin is an important bulwark of acquired immunity against intestinal helminths. J Exp Med 210, 2583–2595 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolpe SD et al. , Macrophages secrete a novel heparin-binding protein with inflammatory and neutrophil chemokinetic properties. J Exp Med 167, 570–581 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stenger S, Thuring H, Rollinghoff M, Bogdan C, Tissue expression of inducible nitric oxide synthase is closely associated with resistance to Leishmania major. J Exp Med 180, 783–793 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Trez C et al. , iNOS-producing inflammatory dendritic cells constitute the major infected cell type during the chronic Leishmania major infection phase of C57BL/6 resistant mice. PLoS Pathog 5, e1000494 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spencer LA, Bonjour K, Melo RC, Weller PF, Eosinophil secretion of granule-derived cytokines. Front Immunol 5, 496 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma W et al. , CCR3 is essential for skin eosinophilia and airway hyperresponsiveness in a murine model of allergic skin inflammation. J Clin Invest 109, 621–628 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng M et al. , Commensal microbiota maintains alveolar macrophages with a low level of CCL24 production to generate anti-metastatic tumor activity. Sci Rep 7, 7471 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Diny NL et al. , Macrophages and cardiac fibroblasts are the main producers of eotaxins and regulate eosinophil trafficking to the heart. Eur J Immunol 46, 2749–2760 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doyle AD et al. , Homologous recombination into the eosinophil peroxidase locus generates a strain of mice expressing Cre recombinase exclusively in eosinophils. J Leukoc Biol 94, 17–24 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spath GF, Beverley SM, A lipophosphoglycan-independent method for isolation of infective Leishmania metacyclic promastigotes by density gradient centrifugation. Exp Parasitol 99, 97–103 (2001). [DOI] [PubMed] [Google Scholar]
- 47.Gazzinelli-Guimaraes PH et al. , Parasitological and immunological aspects of early Ascaris spp. infection in mice. Int J Parasitol 43, 697–706 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Belkaid Y et al. , A natural model of Leishmania major infection reveals a prolonged “silent” phase of parasite amplification in the skin before the onset of lesion formation and immunity. J Immunol 165, 969–977 (2000). [DOI] [PubMed] [Google Scholar]
- 49.Belkaid Y et al. , Development of a natural model of cutaneous leishmaniasis: powerful effects of vector saliva and saliva preexposure on the long-term outcome of Leishmania major infection in the mouse ear dermis. J Exp Med 188, 1941–1953 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schollaert KL, Stephens MR, Gray JK, Fulkerson PC, Generation of eosinophils from cryopreserved murine bone marrow cells. PLoS One 9, e116141 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of differentially expressed genes (DEGs).
Table S2. List of enriched GO categories.
Movie S1. 3D rotational confocal imaging of inoculation site and the area proximal to the inoculation site.
Movie S2. In vitro live imaging of eosinophils and IL-4/10 stimulated BMDMs with or without CCL24 neutralization.
Supplemental Materials and Methods
Fig. S1. Gating strategy for eosinophils, PMNs, Ly6Chi monocytes, Ly6Cint/Ly6Clo moDCs, and dermal TRMs.
Fig. S2. Basophils and ILC2s produce IL-4.
Fig. S3. Dermal TRMs are the main producers of Ccl24 among dermal myeloid subsets.
Fig. S4. Eosinophils progressively infiltrate the L. major inoculation site.
Fig. S5. Immunofluorescence staining and confocal microscopy on vertical ear sections of infected eoCre il4/13f/f and WT mice.
Movie S3. Intravital imaging of perivascular dermal TRMs.
Movie S4. Intravital imaging of the active surface movements of dermal TRMs.
Movie S5. Intravital imaging of eosinophils with dermal TRMs in steady state.
Movie S6. Intravital imaging of eosinophils and dermal TRMs at 1h p.i. with or without CCL24 neutralization.
Movie S7. Intravital imaging of eosinophils and dermal TRMs at 3d p.i. with or without CCL24 neutralization.
Movie S8. Ex vivo live imaging of eosinophils and peritoneal TRMs with or without CCL24 neutralization.
Movie S9. Ex vivo live imaging of eosinophils transferring cellular materials to peritoneal TRMs.


