Abstract
Rectal cancer (RC) is a challenging disease to treat that requires chemotherapy, radiation, and surgery to optimize outcomes for individual patients. No accurate model of RC exists to answer fundamental research questions relevant to patients. We established a biorepository of 65 patient-derived RC organoid cultures (tumoroids) from patients with primary, metastatic, or recurrent disease. RC tumoroids retained molecular features of the tumors from which they were derived, and their ex vivo responses to clinically relevant chemotherapy and radiation treatment correlated with the clinical responses noted in individual patients’ tumors. Upon engraftment into murine rectal mucosa, human RC tumoroids gave rise to invasive RC followed by metastasis to lung and liver. Importantly, engrafted tumors displayed the heterogenous sensitivity to chemotherapy observed clinically. Thus, the biology and drug sensitivity of RC clinical isolates can be efficiently interrogated using an organoid-based, ex vivo platform coupled with in vivo endoluminal propagation in animals.
Introduction
Colon and rectal cancers (RCs) are responsible for > 50,000 deaths per year in the United States1. RC is particularly challenging, as treatment after diagnosis is more complex compared to colon cancer2 due to tumor location in the pelvis and close proximity to critical genitourinary organs. RCs invading the perirectal tissues or lymph nodes are treated with tri-modal therapy, which consists of neoadjuvant chemoradiation (CRT), surgical resection, and 5-fluorouracil (5-FU)-based chemotherapy2. Some RC patients respond completely to CRT alone and can avoid surgery entirely3–6, but others respond poorly and require radical surgery7. Prospective identification of patients who would achieve a complete response7,8 after neoadjuvant therapy alone would enable more tailored individual treatment regimens9,10 and thereby minimize potential harm from overtreatment. The heterogeneity in clinical response and the morbidity associated with radical surgery highlight the need for more sophisticated modeling to predict response to standard therapies.
Few cell lines have been derived from RCs11–15 and whether they were derived from the true anatomic rectum and/or from patients undergoing multimodal therapy is impossible to confirm. Despite the fact that RC is treated differently from colon cancer by using tri-modal therapy in a neoadjuvant context, the preclinical development of treatments for RC has historically relied on colon cancer cell lines16, highlighting the need to develop RC-specific models. Furthermore, efforts to derive organoid “biobanks” have focused primarily on colon cancer specimens17, with a dedicated biorepository of RC tissue or organoids remaining an unmet need in the field.
Given the paucity of xenografts models18 and the complete lack of endoluminal RC models, there is an additional need in RC research for an anatomically accurate in vivo model using patient-derived RC organoids. The rectum has unique venous drainage via the iliac vessels that gives rise predominantly to lung metastases19 (69%), and less frequently liver metastases (20%), which are more commonly seen in colon cancers. Given recent success transplanting mouse colon cancer cells into the colon lumen by our group20 and others21, we set out to derive RC organoids (hereafter “tumoroids”) from resected or biopsied RCs and use them to establish ex vivo and in vivo RC models. We investigate the ability of such tumoroids to model the molecular and histologic features of human RCs, as well as tumor initiation, invasion, and metastasis. We also investigate whether our ex vivo and in vivo platforms could be used to correlate with treatment response in individual patients within a time frame that could potentially inform clinical treatment decisions.
Results
Human RC tumoroid derivation and characterization
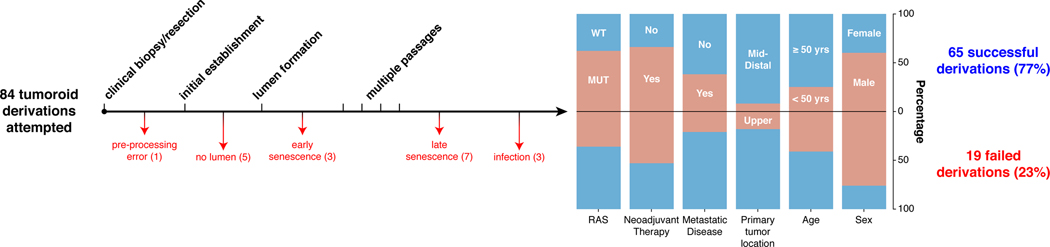
With the goal of generating RC models that would reflect the biology of an individual patient’s tumor, we adapted existing strategies for 3D ex vivo tumor culture22,23 to generate RC tumoroids from pre- and post-treatment patient samples. Basic characteristics of all patients from whom we attempted tumoroid derivation are shown in Extended Data Fig. 1. After 84 tumoroid derivation attempts from 58 individual patients, we established 65 RC tumoroids from 41 patients with an overall success rate of 77% (65/84). In addition to these RC tumoroids, we also generated 51 normal rectal organoids from normal adjacent tissue (see Supplementary Table 1). Of the 19 failed tumoroid attempts, seven were maintained for 6 weeks before senescing. Similar to prior work in colon cancer, RC tumoroids could be cultured in the absence of key growth factors17 (e.g., R-spondin, Wnt-3a, and Noggin), whereas organoids derived from normal rectal mucosa remained growth factor dependent23.
Since endoscopic biopsies are routinely performed in the outpatient setting in the treatment of patients with RC, we asked whether we could derive RC tumoroids from minute amounts of material obtained in the clinic. Of note, 49 of the RC tumoroids were established using tissue obtained with biopsy forceps routinely used in clinical care (Fig. 1a). These data suggest it is possible to use serial biopsies obtained as part of standard care in the pre-treatment or post-therapy settings to generate tumoroid models to assess patient-specific mediators of response and resistance.
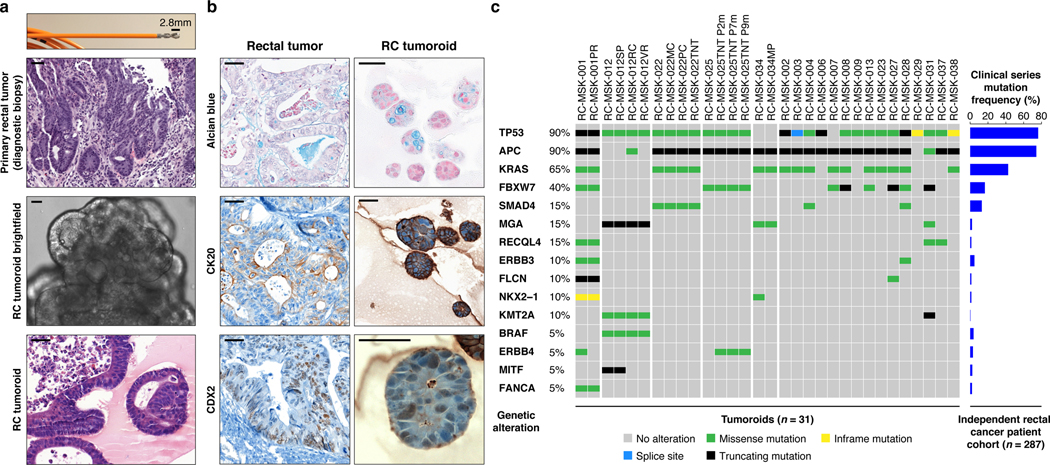
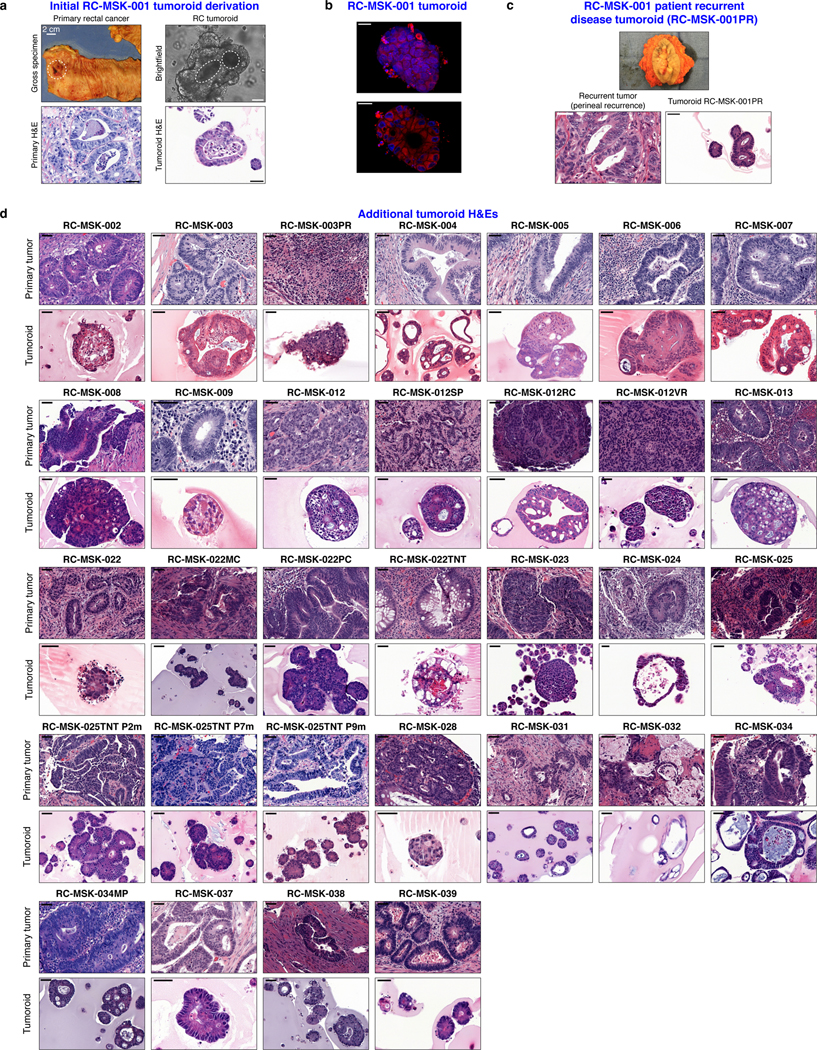
Fig. 1 |. Preservation of rectal cancer histopathology and mutational fingerprint in tumoroids.
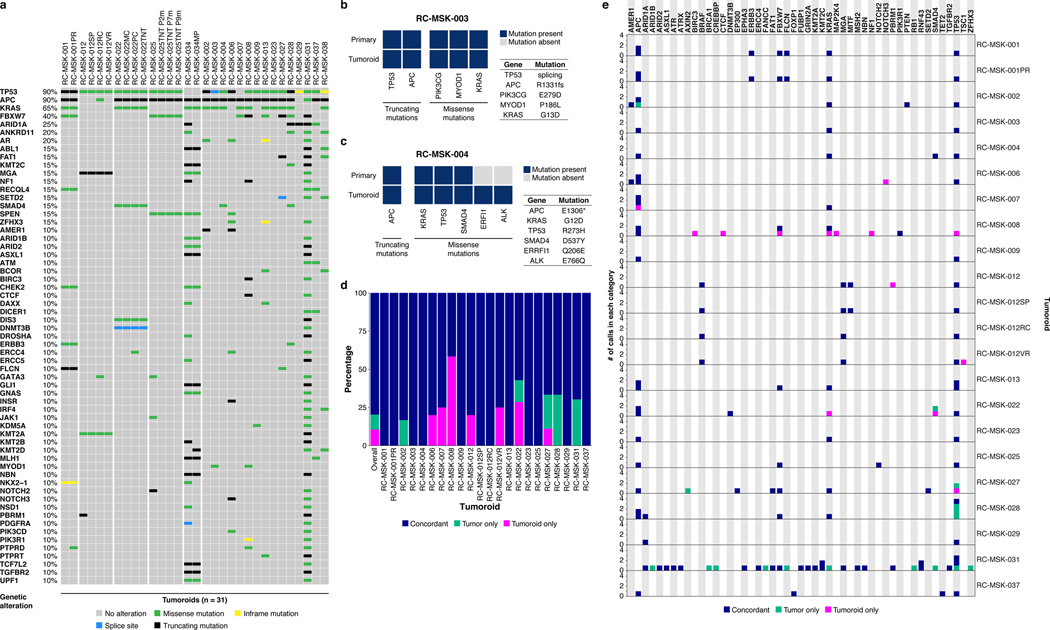
a, Shown at the top is the 2.8 mm cold biopsy forceps (Boston Scientific Corp.™) used for sampling tumor from some of the rectal cancers (RCs) used to derive tumoroids. Also shown is the first primary tumor sampled with this biopsy forceps stained with hematoxylin and eosin (H&E, second panel). The corresponding derived tumoroid, RC-MSK-008, in 3D culture is displayed by brightfield microscopy and H&E (lower 2 panels). Scale bars, 50 μm. b, Histopathologic staining of enterocyte markers (Alcian blue, CK20, and CDX2) of a primary resected rectal tumor (leftmost panels) and the corresponding tumoroid, RC-MSK-001, in 3D culture (right panels). Scale bars, 50 μm. c, The mutation landscape of 31 of the RC tumoroids is displayed (left panel) compared to an independent set of 287 RCs (right panel), both detected by MSK-IMPACT. The frequency of alterations in the RC tumoroids is noted with the type of genetic alteration (noted by color code). Displayed are the top 15 mutated genes observed in the derived tumoroids.
Despite concern that tumor stem cells exposed to prior systemic chemotherapy or radiation would not likely yield viable tumoroids17, we were able to generate RC tumoroids and normal adjacent rectal organoids from patients who had undergone prior chemotherapy and/or radiotherapy, as well as those who had no prior therapeutic intervention before surgical resection. Of the 65 tumoroids, 22 were derived from treatment-naïve patients and 43 were derived from patients undergoing first- or second-line therapy (Supplementary Table 1). Tumoroids were derived from all sites of the rectum (i.e. distal, mid, and upper) and 17 tumoroids were derived from sites of disease recurrence or metastasis. Two of the 41 patients from which tumoroids were derived harbored tumors with mismatch repair (MMR) deficiencies. Tumoroids were also derived from RAS mutant (62%) and wild type tumors and from patients with metastatic disease (38%) and non-metastatic disease (Extended Data Fig. 1). In summary, we sampled a diverse set of RC patients with different stages of disease who were both on or off therapy, had tumors that were both RAS mutant and wild type, and whose clinical variables did not predict success or failure of tumoroid establishment.
Histopathologic and molecular characteristics are preserved in rectal cancer tumoroids
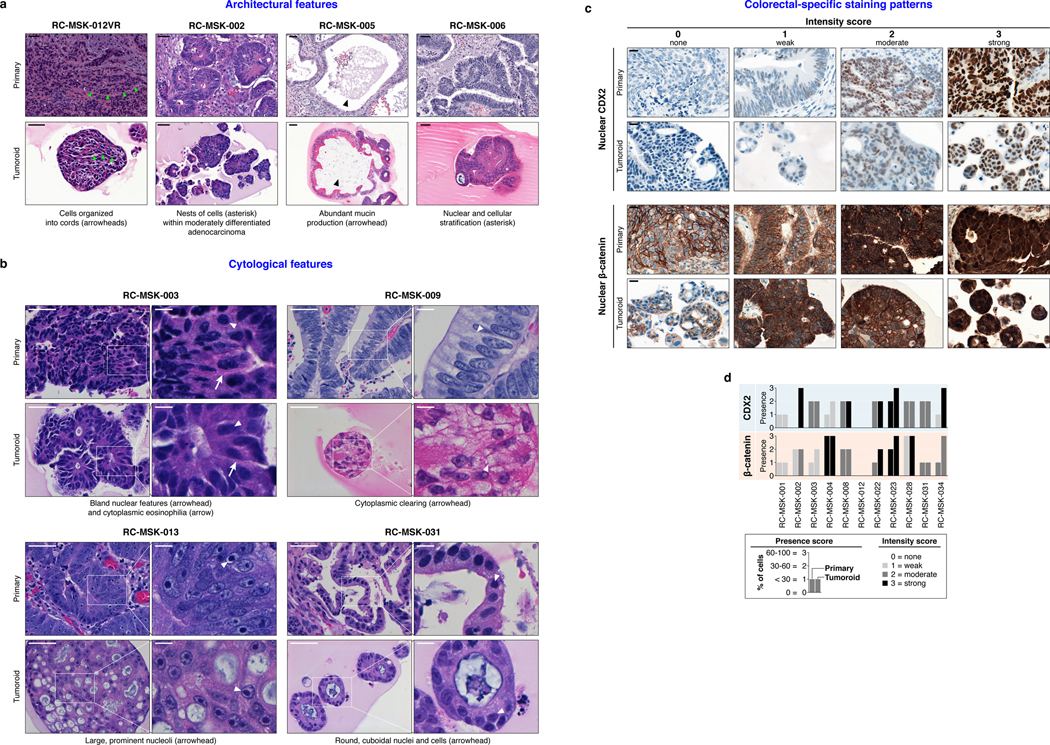
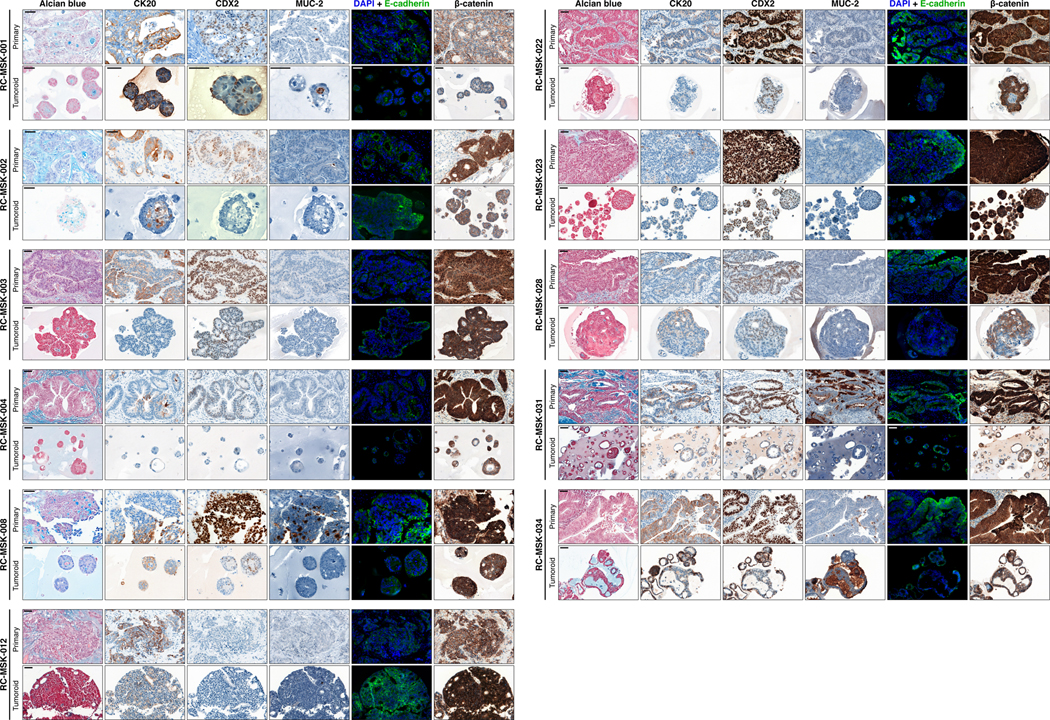
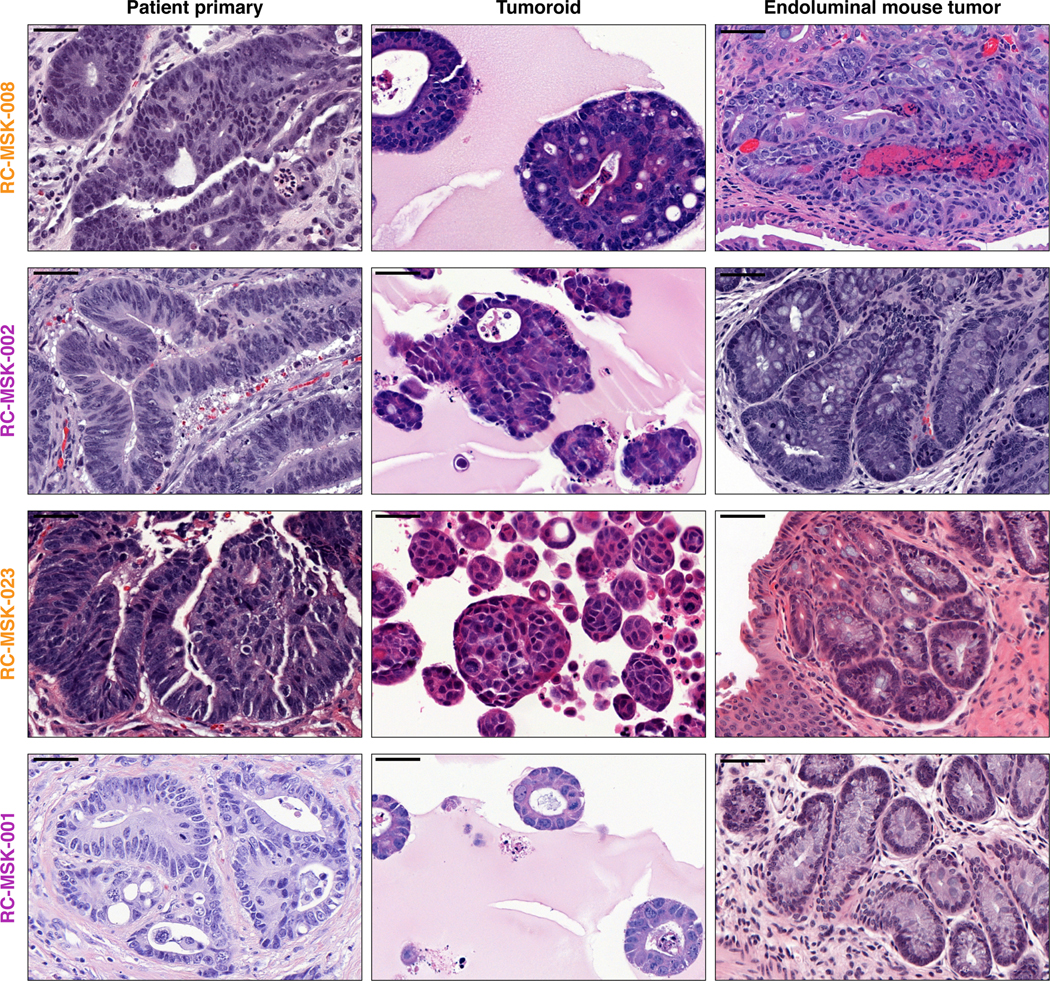
As with cancer organoids derived from other tissues, RC tumoroids retained histopathologic features of the primary tumors from which they were derived (Fig. 1a, Extended Data Fig. 2, Supplementary Table 1). Regarding architecture, tumoroids retain specific glandular features as noted in the corresponding patient tumors, including cord- and nest-like growth patterns, pooled mucin production, nuclear stratification, and similar degrees of differentiation as their corresponding primary tumors, as reviewed by a gastrointestinal pathologist (Extended Data Fig. 3a and b). Tumoroids also preserve subtle cytologic features, including bland nuclear detail, cytoplasmic clearing and cytoplasmic eosinophilia, prominent nucleoli, and cuboidal nuclear morphology (Extended Data Fig. 3 a and b). When colorectal-specific nuclear marker CDX2, as well as nuclear β-catenin were quantified and compared between matched tumoroids and patient tumors, tumoroids retained similar presence and intensity of these markers, thus reflecting the tumors from which they were derived (Extended Data Fig. 3c and d). Further, RC tumoroids retained Alcian blue-positive and MUC-2-positive goblet cells, CK20 and CDX2-positive enterocytes, robust expression of E-cadherin (epithelial marker), and cytoplasmic/nuclear patterns of β-catenin staining, which mirror the tumors from which they were derived (Fig. 1b, Extended Data Fig. 4). Specific pathologist notes regarding β-catenin staining patterns are presented in Supplementary Table 1. In addition, tumoroids derived from mismatch repair deficient patients retain the specific defects observed in the primary tumor (Extended Data Fig. 5). These data indicate that features found in the primary tumor are retained in the derived tumoroid and reflect individual patient tumors.
In addition, the RC tumoroid biorepository reflects the histopathologic heterogeneity observed clinically. Of the 41 patients successfully sampled, 75% have moderately differentiated tumors and tumoroids, while 25% have well or poorly differentiated tumors and tumoroids. This is similar to the prevalence observed in all colorectal cancers: 70% moderately differentiated and 10–20% well or poorly differentiated24. As noted, two tumoroids have also been derived from patients with MMR deficiencies, representing 5% of the patients sampled, which is similar to the 5–6% of MMR-deficient patients observed clinically25.
To determine whether the RC tumoroids also reflect the mutational fingerprint in the corresponding primary tumors—as well as the larger landscape of molecular alterations in RC—we performed massively parallel sequencing using the custom, FDA-cleared tumor profiling panel MSK Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT)26. We sequenced genomic DNA isolated from 31 RC tumoroid lines (Fig. 1c left panel and Extended Data Fig. 6a). Two representative comparisons between examples of tumoroids and the matching primary tumor is shown in Extended Data Fig. 6b and c. Among tumor mutations likely to be oncogenic, 92% (range 0.66–1.00; Extended Data Fig. 6d) were present in their corresponding tumoroids, which is comparable to an 88% concordance (range 0.62–1.00) previously reported for colon organoids17 and organoids from colon metastases27 (no rectal cancers). The degree of conservation of mutations is shown for all available tumoroid-patient comparisons (n = 22) in Extended Data Fig. 6e. Of the clonal oncogenic mutations present in the matched tumor samples, 77% were also present as clonal in tumoroid samples. We then compared these tumoroids to a clinical cohort of 287 rectal cancers resected at Memorial Hospital that had been sequenced by MSK-IMPACT. The top mutations in the 287 RC patients were also noted in the RC tumoroids, with the most common alterations in the genes APC, TP53, KRAS, and FBXW7 (Fig. 1c right panel).
RC tumoroids reveal a diversity of responses to chemotherapy that correlate with clinical response
We next examined the RC tumoroids as a platform for preclinical studies in the context of the RC tri-modal treatment approach. 5-FU is the backbone of RC chemotherapy as part of systemic therapy or in combination with radiation28,29. 21 different RC tumoroids were separately treated with 5-FU and FOLFOX (chemotherapy regimen of 5-FU, leucovorin, and oxaliplatin), the standard of care chemotherapy regimens used to treat RC in the frontline30, yielding a spectrum of dose-responses, displayed as both dose-response curves and area under the curve (AUC) calculations (Fig. 2a). Thus, the RC tumoroids are heterogeneous in their response to physiologic 5-FU and FOLFOX doses.
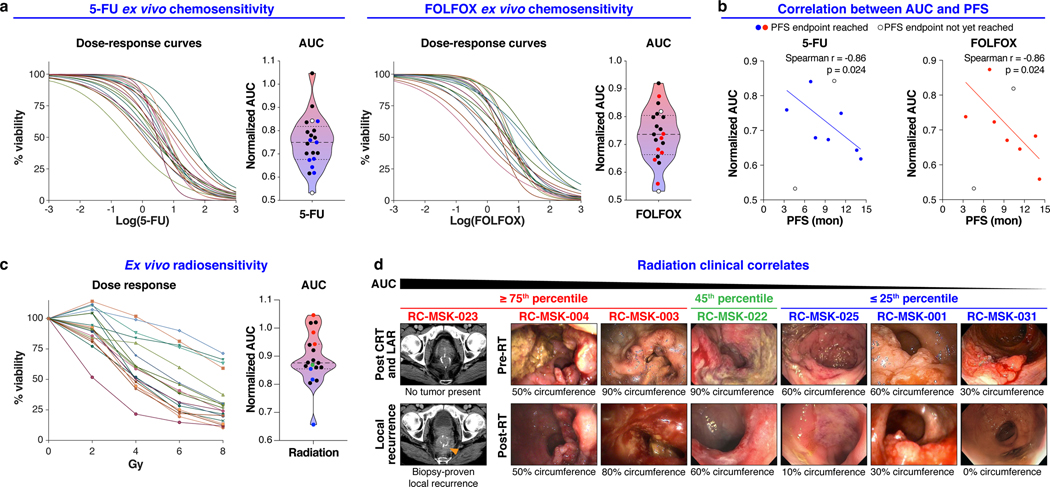
Fig. 2 |. Clinically relevant responses to chemotherapy and radiation in rectal cancer tumoroids ex vivo.
a, Ex vivo chemosensitivity of 21 RC tumoroids to 5-FU and FOLFOX in the form of dose response curves are displayed for each tumoroid (n=2 or 3 independent experiments for each). Area under the curve (AUC) was calculated from the raw dose response data and is displayed as a violin plot; dashed line and dotted lines represent mean and upper/lower quartiles, respectively. Colored data points indicate those tumoroids referenced in b. b, Correlation between AUC and progression free survival (PFS) for the seven patients (n=7) who have a PFS endpoint are displayed (Two-tailed Spearman correlation: Spearman r = 0.86, p = 0.024 for both treatment conditions). Data for both 5-FU and FOLFOX is shown. The linear regression line is plotted. c, Ex vivo radiosensitivity of 19 RC tumoroids (n=2 or 3 independent experiments for each tumoroid) is shown with corresponding AUC calculated and displayed as in a. Colored data points indicate those tumoroids referenced in d. d, Endoscopic clinical responses to radiation are displayed for each patient and ranked by AUC, descending left-to-right as indicated. The percent of the bowel circumference involved by tumor pre- and post-radiation (pre- and post-RT) is displayed as assessed endoscopically by a colorectal surgeon. The RC-MSK-023 tumor is shown radiographically in the left-most panel as a local recurrence following chemoradiation (CRT) and low anterior resection (LAR). The tumoroids are categorized from left to right as ≥ 75th percentile (red), 45th percentile (green), and ≤ 25th percentile (blue) with corresponding color-coded data points indicated in c.
We then asked whether differential ex vivo 5-FU or FOLFOX sensitivity correlates with clinical outcome, specifically progression-free survival (PFS) as measured from the 5-FU-based chemotherapy start date until first evidence of disease progression by radiographic study or endoscopy. Seven patients had sufficient clinical follow-up to yield a PFS interval for correlative analyses. The complete clinical course for each of these patients is displayed in a swimmer’s plot in Extended Data Fig. 7. AUC for both 5-FU and FOLFOX ex vivo treatments correlated with PFS of the corresponding patient (Fig. 2b, Spearman r = 0.86, p = 0.024 for both treatment conditions). Although larger numbers are needed to validate our findings, these data, reflecting mature PFS, suggest that chemosensitivity determined ex vivo could have utility as a predictive tool in identifying RC patients at risk of disease progression.
Ex vivo radiation response correlates with clinical response
19 tumoroids were radiated ex vivo and also yielded a heterogenous response to radiation (Fig. 2c). We next examined the relationship between ex vivo and clinical radiation sensitivity. In order to isolate the clinical effects of radiation and not the summative effects of chemotherapy and radiation, we elected to compare endoscopic tumor assessment immediately before and after radiation to isolate the radiotherapy response (Fig. 2d). Tumoroids that displayed resistance ex vivo (≥ 75th percentile) were derived from patients who either recurred following radiation and surgery (RC-MSK-023) or who had no/minimal response to radiation clinically (RC-MSK-004 and RC-MSK-003). More sensitive tumoroids (≤ 25th percentile) were derived from patients who had at least a 50% reduction in tumor circumference endoscopically (RC-MSK-001) or a near complete or a clinical complete response31 (RC-MSK-025 and RC-MSK-031). The RC-MSK-022 tumoroid (45th percentile) had a partial clinical response. Taken together, these data demonstrate that RC tumoroids display varying sensitivity to ionizing radiation, which corresponds to clinical radiotherapy responses.
Proof-of-principle targeted therapy studies in RC tumoroids
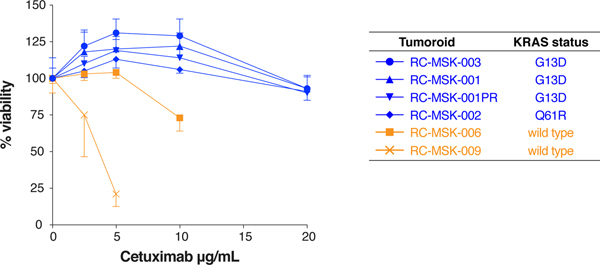
In colorectal cancers, the presence of KRAS mutation predicts resistance to EGFR-targeted therapy; therefore, we asked whether RC tumoroids would recapitulate KRAS mutation-mediated resistance to the anti-EGFR monoclonal antibody cetuximab32. Consistent with these clinical trial data, KRAS mutant tumoroids were resistant to cetuximab, whereas KRAS wild type tumoroids were sensitive (Extended Data Fig. 8). Even though cetuximab is not routinely used clinically in RC, these data provide preliminary evidence that a cell-autonomous response in RC tumoroids reflects a biologically relevant response to a targeted therapy.
Establishing an endoluminal model using human rectal cancers
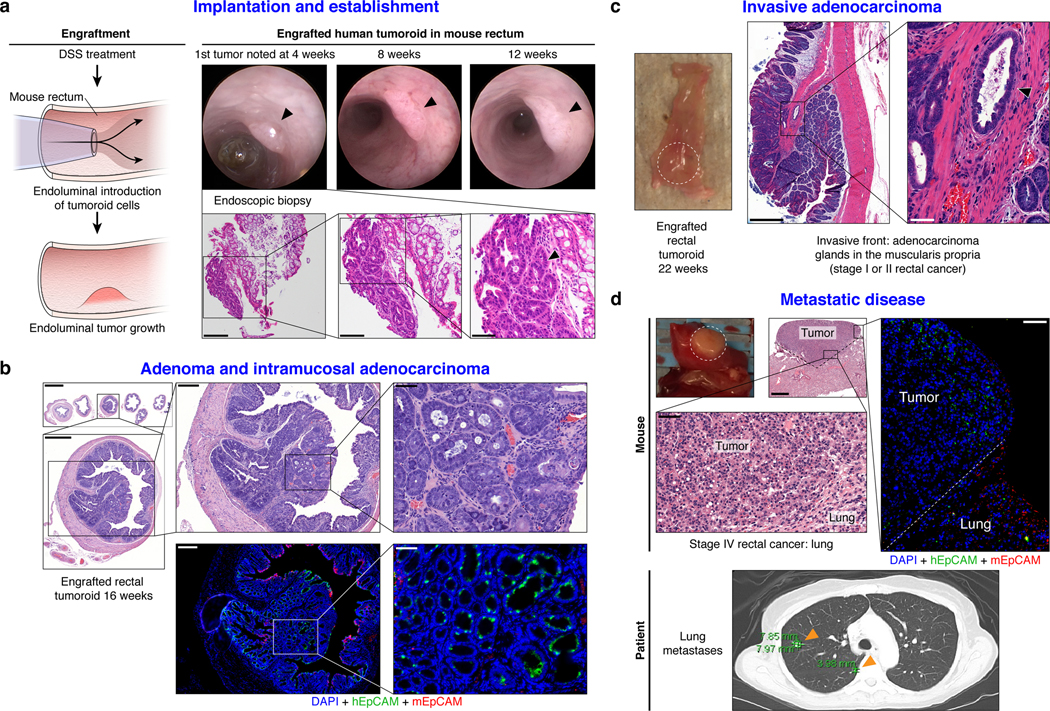
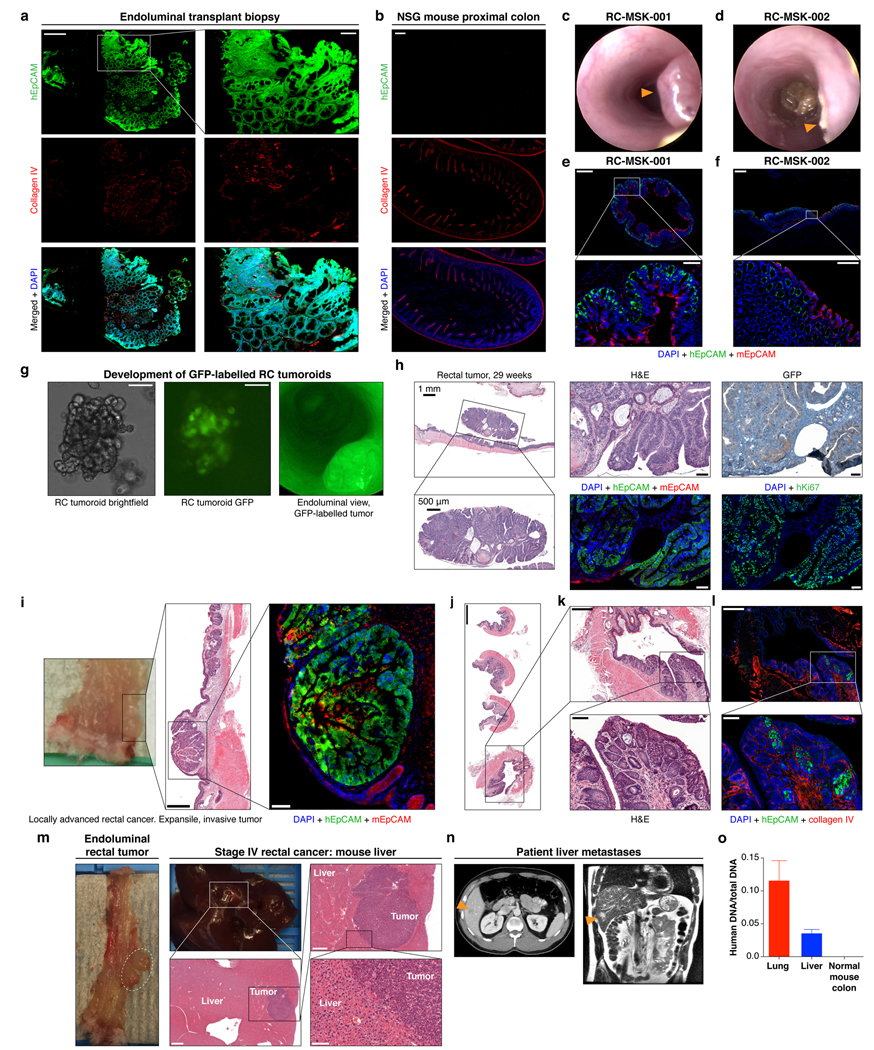
We next sought to establish an orthotopic, patient-derived xenograft model of RC that could reflect the initiation, invasion, and metastatic potential of tumors that develop in the unique anatomy of the rectum. Fig. 3a shows the methodology20 and documents engrafted human rectal cancer in a NOD scid gamma (NSG) mouse rectum after injecting 200,000 cells from tumoroids (Supplementary Video 1). Successful transplantation of human tumoroids was confirmed by human EpCAM (hEpCAM) staining of an endoscopic biopsy specimen taken from the live mouse compared with negative staining in the NSG proximal mouse colon as a control (Extended Data Fig. 9a and b). Fig. 3a also highlights serial assessment of engrafted rectal tumors at 8 and 12 weeks by endoscopy. We noted progression to intramucosal adenocarcinoma at 16 weeks post-transplantation (Fig. 3b top panel). On histopathological review of the transplanted murine rectums, an intact mesorectum was observed that is similar to resected human rectal specimens (Fig. 3b top panel). The endoluminal RC model was reproducible in both male and female mice using cells from eight different RC tumoroid lines. Gross tumor formation ranged from 20%−100% per experiment (Extended Data Fig. 9c and d). Even though we did not observe gross tumors in all mice, 100% of the mice transplanted showed engraftment of cells positive for hEpCAM (Extended Data Fig. 9e and f). We also demonstrated that tumoroids can be genetically modified prior to transplantation, which permits tracing or mechanistic studies, as demonstrated by GFP-labeling of tumoroids in vitro prior to endoluminal transplantation (Extended Data Fig. 9g and h).
Fig. 3 |. Establishment of an endoluminal rectal cancer assay in mice.
a, Cartoon of the implantation process and an endoscopic view of the first rectal tumoroid (RC-MSK-001) implanted in a NSG mouse; mouse at 4, 8, and 12 weeks (top panels). H&E of the engrafted tumoroid biopsied at 4 weeks via endoscopic channel (bottom panels) demonstrates high nuclear/cytoplasmic ratio, poor differentiation, and neoplastic glands (arrowhead). Adjacent normal mouse colonic epithelium and stroma is seen. Scale bars from low to high magnification are as follows: 200 μm, 100 μm, 50 μm. b, H&E, axial view of the rectal tumor within the NSG mouse rectum with adjacent serial sections (upper panels). Inset demonstrates intramucosal adenocarcinoma with atypical neoplastic cells, poorly differentiated tumor cells, and high nuclear to cytoplasmic ratio. IF (lower panels): DAPI (blue), hEpCAM (green), mEpCAM (red). Scale bars from low to high magnification: 2,000 μm, 500 μm, 200 μm, 50 μm. c, NSG mouse rectum (gross rectum with tumor, white dashed circle) and H&E shows evidence of a moderately differentiated tumor engrafted from RC-MSK-001 tumoroids with invasion into the muscularis (inset, arrowhead). Scale bars: 500 μm; inset, 50 μm. d, Gross lung metastasis (white dashed circle) from the same mouse presented in c engrafted endoluminally with RC-MSK-001 tumoroids. Corresponding H&Es demonstrate poorly differentiated architecture of the metastasis. IF serial section demonstrates engraftment of the human tumoroids: DAPI (blue), hEpCAM (green), mEpCAM (red). Lower panel shows axial thoracic CT imaging of the patient from which the tumoroid was derived, demonstrating lung metastases (orange arrows indicate two of multiple lung metastases). Scale bars: 500 μm; H&E and IF insets, 50 μm. Images are representative of 5 tumoroid engraftments into mouse rectum.
We then interrogated our model for the development of invasive cancer and metastatic disease. At 22- and 30-weeks post-transplantation, we noted progression to invasive adenocarcinoma that parallels the features of stage I or II human rectal adenocarcinomas (Fig. 3c and Extended Data Fig. 9i–l). Importantly, we noted evidence of metastases in the lung (Fig. 3d) and liver (Extended Data Fig. 9m) with features of poorly differentiated carcinomas infiltrating normal parenchyma, as confirmed by independent pathological review. Notably, the metastases derived from the endoluminal rectal tumors corresponded to sites of metastases seen in the individual patients from which the tumoroids were derived, namely lung metastases in the case of RC-MSK-001 and liver for RC-MSK-002 (Fig. 3d lower panel, Extended Data Fig. 9n, respectively). The metastases were further confirmed with human-specific qPCR analyses (Extended Data Fig. 9o). These data establish the feasibility of a patient-derived RC endoluminal model and its patterns of clinically relevant metastasis.
The endoluminal model as a platform to investigate chemosensitivity
We next asked if we could mimic chemotherapy response in our in vivo RC model as a parallel, pre-clinical assay to our ex vivo work. To establish safe use of 5-FU in vivo, we first conducted experiments in a subcutaneous injection model and treated the mice systemically with intraperitoneal 5-FU injection. The tumoroids successfully engrafted, the mice tolerated treatment well, and we noted a measurable level of response in two separate tumoroid lines (Supplementary Figure 1).
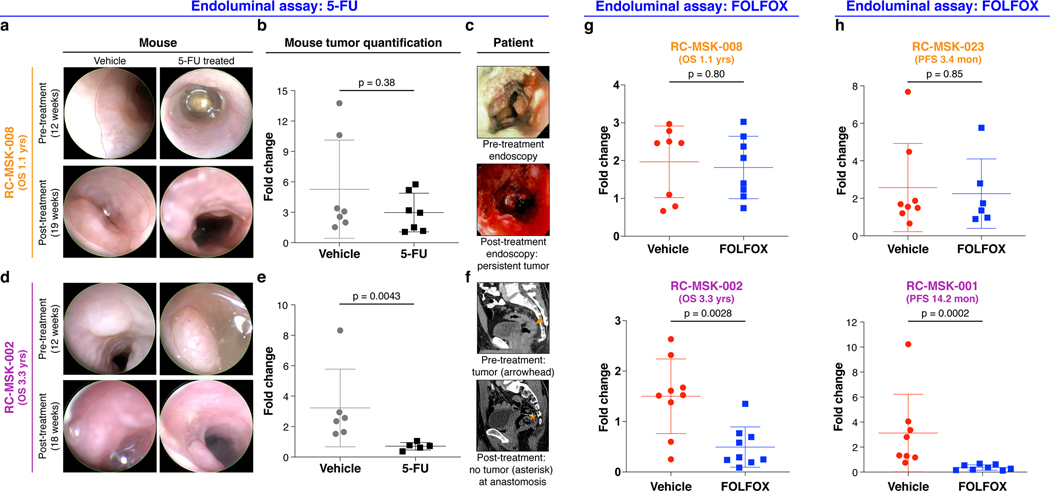
After showing safe use of 5-FU in vivo, we then tested chemosensitivity in our endoluminal RC model. We implanted mice with tumoroids from two lines: RC-MSK-008, a tumoroid derived from a patient with more clinically aggressive disease (overall survival [OS] = 1.1 yrs), and RC-MSK-002, a tumoroid derived from a patient with less clinically aggressive disease (OS = 3.3 yrs). At 12 weeks, 70% of the mice had tumors detectable by endoscopy (Fig. 4a and d, upper panels). All mice were then randomized to 5-FU or vehicle treatment and the tumors were followed endoscopically every 3–4 weeks. Mice engrafted with RC-MSK-008 tumoroids demonstrated a lack of response to treatment with 5-FU (Fig. 4a and b). In contrast, mice engrafted with RC-MSK-002 tumoroids demonstrated significant treatment response to 5-FU (Fig. 4d and e). Of note, these observations were consistent with the individual patient’s clinical course (Fig. 4c and f). The tumoroid-derived endoluminal rectal xenografts were confirmed histologically to represent the glandular morphology of the primary tumors from which they were derived (Extended Data Fig. 10). These experiments were then repeated using FOLFOX chemotherapy. Similarly, the more clinically aggressive RC-MSK-008 tumoroids demonstrated a lack of response to FOLFOX while the less clinically aggressive RC-MSK-002 tumoroids demonstrated response to FOLFOX (Fig. 4g).
Fig. 4 |. The endoluminal rectal cancer model can be used to reflect patient-specific chemoresistance and chemosensitivity.
a, RC-MSK-008 (more clinically aggressive: overall survival [OS]=1.1 yrs) implanted in the mouse rectum is shown endoscopically pre- and post-treatment with vehicle or 5-FU. b, Quantification of RC-MSK-008 tumor area measured endoscopically pre- and post-treatment (n=7 per condition) and presented as fold change in tumor size for each mouse. No difference was observed between vehicle and 5-FU-treated groups (Mann-Whitney: U=17, p=0.38). c, The RC-MSK-008 human rectal tumor viewed endoscopically according to routine clinical care at diagnosis and post 5-FU-based treatment. d, RC-MSK-002 (less clinically aggressive: OS=3.3 yrs) implanted in the mouse rectum is shown endoscopically pre- and post-treatment with vehicle or 5-FU. e, Quantification of RC-MSK-002 tumor area as in b (n=6 for vehicle, n=5 for 5-FU). 5-FU treated tumors had a significantly lower fold change compared to vehicle-treated tumors (Two-tailed Mann-Whitney: U=0, p=0.0043). f, The RC-MSK-002 human rectal tumor viewed by sagittal CT imaging according to routine clinical care pre-treatment (orange arrowhead: tumor) and after both resection and 5-FU based systemic therapy (orange asterisk: anastomosis) with no evidence of recurrence. g, RC-MSK-008 (n=8 mice per condition) and RC-MSK-002 (n=9 mice per condition) tumoroids implanted and measured as in a-f were treated with FOLFOX. Similarly, RC-MSK-008 implanted mice showed no difference between vehicle and FOLFOX-treated groups (Two-tailed Mann-Whitney: U=29, p=0.80), and RC-MSK-002 implanted mice treated with FOLFOX had significantly lower fold change compared to vehicle-treated tumors (Two-tailed Mann-Whitney: U=8, p=0.0028). h, RC-MSK-023 (rapid clinical progression: progression-free survival [PFS] = 3.4 mon) and RC-MSK-001 (slower clinical progression: PFS=14.2 mon) implanted endoluminally into mice as in a-f. RC-MSK-023 (n=8 for vehicle, n=6 for FOLFOX) implanted mice showed no difference was observed between vehicle and FOLFOX-treated groups (Two-tailed Mann-Whitney: U=22, p=0.85). RC-MSK-001 (n=8 per condition) implanted mice with FOLFOX-treated tumors had significantly lower fold change compared to vehicle-treated tumors (Mann-Whitney: U=0, p=0.0002). Error bars in figure: middle line = mean, end lines = s.d.
To further expand upon these observations, separate in vivo experiments were conducted using two different tumoroid lines: RC-MSK-023, a tumoroid derived from a patient with a rapid clinical progression (PFS = 3.4 mon), and RC-MSK-001, a tumoroid derived from a patient with slower clinical progression (PFS = 14.2 mon). Mice endoluminally engrafted with the RC-MSK-023 tumoroid did not respond to FOLFOX while mice engrafted with the RC-MSK-001 tumoroid did respond (Fig. 4h). Again, the glandular morphology was similar between the engrafted tumoroids and primary tumor (Extended Data Fig. 10). Notably, the RC-MSK-023 endoluminal tumor demonstrated moderately differentiated glandular architecture mirroring the primary tumor, even though the RC-MSK-023 tumoroid in 3D culture demonstrated poorly differentiated and scattered glandular histology. These experiments establish the feasibility and reproducibility of an in vivo chemosensitivity assay with the endoluminal model using four independent tumoroid lines.
Discussion
We have established a biorepository of RC models representing the spectrum of genetic diversity and treatment response of RC. This unique resource represents a significant expansion of the reagents for the study of RC and will enable mechanistic and translational studies to be performed on bona fide RCs, rather than surrogate colon cancer cell lines. Importantly, RC tumoroids can be routinely and successfully established from small amounts of starting material using a 2.8 mm biopsy forceps, demonstrating that only a small amount of tissue is required to derive a viable tumoroid line and that samples can be obtained in the outpatient clinical setting. We have demonstrated that RC tumoroids derived from RC patients in the context of multimodal therapy successfully engraft into the rectal mucosa of mice thereby reflecting the anatomical structure of the human rectum (e.g., mesorectum) and that these orthotopic, endoluminal rectal xenograft mouse models can recapitulate patient-specific treatment response in vivo. Notably, we contribute new methodology, a dedicated rectal cancer biorepository and modeling with regard to ex vivo radiation response and patient-specific in vivo chemotherapy response in our endoluminal model. Further, our data suggest that the ex vivo and in vivo models we have derived are a representative analog of individual RC patient response, complementing what has been recently described for other metastatic adenocarcinomas33.
Despite concerns that organoid technology would be unsuccessful with heavily pre-treated tumor specimens34, our findings are especially important as our work shows that this is possible with a success rate of 77%. We have demonstrated that basic histopathology and enterocyte differentiation markers are conserved between RC tumoroids and primary tumors, with specific retention of architectural and cytologic subtleties. The mutational profiles between tumor and derived tumoroid were conserved and we noted similarity between our derived RC tumoroids and an independent MSK-rectal cancer cohort (Fig. 1c).
Recapitulation of patient-specific and clinically relevant responses to chemotherapy, radiation, and targeted therapy may open doors to determine new and more precise treatments for RC patients. Furthermore, the derivation of RC tumoroids and normal rectal organoids from biopsies is critically important, as this means models can be derived at diagnosis, throughout treatment, and at the time of recurrence from minute amounts of biopsy material. Ex vivo and in vivo sensitivity to standard and experimental therapeutics (e.g. trials), as well as radiation, could be fully assayed in 6–12 weeks after establishment (Figs. 2 and 3). If the individual patient fails standard treatment or experiences progressive disease, these assays may provide rationale to attempt new therapies found to be effective both ex vivo and in vivo using the patient’s own tumor tissue as the embedded informant. These data could be gathered within the period it takes a modern-day RC patient to undergo total neoadjuvant therapy (~6 months)35. Larger numbers are needed to fully establish the utility of this approach, including prospective evaluation of the concordance between patient and tumoroid response to standard therapeutics, including both chemotherapy and radiation. Furthermore, if confirmed in larger studies, the ex vivo radiation platform could be used to evaluate patient response before initiating radiation, potentially sparing these resistant patients the toxic side effects of radiation36.
Additional novelty of this work lies in the establishment of the endoluminal RC model that recapitulates clinical response to therapy. This model builds on our recent murine model of transplantation20, but is also the first demonstration that treated and untreated human RC tumors can engraft within the mouse rectum and recapitulate the process of primary disease formation, subsequent invasion, and eventual metastasis. Notably, we observed the development of metastases that may signal tropism of the human RCs from which the tumoroids were derived. While these findings are intriguing, they require prospective validation in a larger cohort. Overall, these data indicate that we have established a working RC platform to better elucidate RC pathogenesis and address questions regarding resistance to chemotherapy and radiation.
In summary, our work addresses an unmet need by establishing a RC-specific ex vivo and in vivo biorepository that markedly increases the ability to study RC. Our model molecularly resembles RC and establishes a relevant framework in which to study the disease. This methodology demonstrates options for eventual drug screening37 in a pre-clinical setting and reflects the clinical outcomes of the patients from which they were derived. Our study demonstrates determination of basic response parameters within weeks of derivation and can serve as a tool for therapeutic response modeling whereby we can study fundamental research questions relevant to individual RC patients.
Methods
Study Design
We developed a 3D organoid system to culture patient-derived RC specimens from treated and untreated patients and dissociated 3D tumoroid and normal adjacent rectal cells (organoids) for long-term use. We developed an in vivo orthotopic, xenograft endoluminal model of RC with the tumoroids and tested initiation, invasion, tumor growth, and chemoresistance in this endorectal endoluminal model.
Statement of compliance with internal review boards
Animal experiments and studies involving human tissues were approved by the following MSKCC Institutional Review Board (IRB) and Institutional Animal Care and Use Committee (IACUC) protocols: #11–083, #16–1071, and 06–07-012. Patients also consented for tissue use and MSK-IMPACT sequencing on the following protocols: #06–107 and #12–245 (see “Life Sciences Reporting Summary” for more information on research participants).
Tissue processing for organoid and tumoroid derivation
For derivation of normal rectal organoids, samples of normal rectal mucosa adjacent to tumor were washed with ice-cold PBS-Abs buffer (phosphate-buffered saline with antibiotic-antimycotic, gentamicin [Gemini Bio Product, West Sacramento, CA, USA] and 5 μg/mL metronidazole [SigmaAldrich, St. Louis, MO, USA]). Samples were then chopped into 1 mm pieces in ice-cold PBS-DTT buffer (PBS with 10 mM DTT [SigmaAldrich]). The tissue fragments were resuspended in PBS-EDTA (PBS with 8 mM EDTA [Invitrogen, Waltham, MA, USA] and 0.5 mM DTT) and were incubated at 4 °C for 1 hr. The suspension was centrifuged at 300×g for 5 min and PBS-EDTA was removed. Tissue fragments were then resuspended vigorously in ice-cold ADF medium (advanced DMEM/F12 plus GlutaMAX, HEPES [Gibco, Waltham, MA, USA] and 5% FBS [SigmaAldrich]). The tissue fragments were allowed to settle under normal gravity for 1 min, and the supernatant was transferred to a new tube for crypt inspection by microscopy. This step was repeated until no crypts were found in supernatants. The supernatants containing crypts were collected, filtered through a 70 μm Cell Strainer, and then centrifuged at 300×g for 5 min. Isolated crypts were embedded in Matrigel (Corning, NY, USA. product# 356231) and culture started.
For derivation of tumoroids, fresh rectal cancer samples were processed as reported38 and as noted here. Surgically resected rectal tissue or biopsy tissue was washed with ice-cold PBS-Abs buffer and then chopped into 1 mm pieces in ice-cold PBS-DTT buffer. The fragments were digested in digestion medium (advanced DMEM/F12 with 2% FBS, Pen/Strep [Gemini Bio Product], 100 U/mL collagenase type XI [SigmaAldrich], and 125 μg/mL dispase type II [Invitrogen]) at 37 °C for 40 min and then further digested for 10 min by adding a half-volume of TrypLE Express (Gibco), and 3 mg of DNase I (SigmaAldrich) per sample. Samples derived from biopsies were then embedded in 800 μL Matrigel and cultured as described below. Samples derived from resected tumors were filtered through a 70 μm Cells Strainer, centrifuged at 300×g for 5 min, and the isolated tumor cells were embedded in 1–2 mL of Matrigel, depending on pellet size.
3D culture conditions for rectal cancer tumoroids
The basal culture medium for healthy tissue-derived organoids and RC tumoroids was modified from the literature38 as follows: advanced DMEM/F12 was supplemented with antibiotic-antimycotic (Gibco), 1×B27, 1×N2 (Gibco), 2 mM GlutaMAX, 10 nM gastrin I, 10 mM HEPES, 1 mM N-acetylcysteine, and 10 mM nicotinamide (SigmaAldrich). The following niche factors were used: 50% Wnt-3A conditioned medium, 20% R-spondin conditioned medium (media collected from HEK293 cell lines expressing recombinant Wnt3a and R-spondin1, kindly provided by Kevin P. O’Rourke and the S. Lowe laboratory)39, 100 ng/mL mouse recombinant noggin (Peprotech, Rocky Hill, NJ, USA), 50 ng/mL human recombinant EGF (Peprotech), 500 nM A83–01 (Tocris, Avonmouth, Bristol, UK), and 10 μM SB 202190 (SigmaAldrich). Upon expansion, RC tumoroids were passaged and then cultured in medium without Wnt-3A, R-spondin, and noggin.
For passage of organoids and tumoroids, the Matrigel surrounding the organoids or tumoroids was depolymerized by cell recovery solution (BD Biosciences, San Jose, CA, USA). The released cell clusters were pelleted. For further dissociation into single cells, the pellets were resuspended in TrypLE Express with pipetting 50 times using a p1000 or p200 pipette, and then incubated at 37 °C for 5 min. Cells were then pipetted several more times to form a homogeneous resuspension. Fivefold volume of culture medium was added to the suspension. Pelleted cells were obtained by centrifuging at 300×g for 5 min and supernatant was discarded. The pellet was resuspended with Matrigel and divided into a 24-well suspension plate (50 μL/well). After the Matrigel balls were polymerized, 500 μL of culture medium was added.
To freeze organoids or tumoroids, Matrigel was dissolved with cell recovery solution and then the cell clusters were pelleted. The pellets were resuspended in ice-cold Recovery Cell Culture Freezing Medium (Gibco,). The pellets also could be dissociated with TrypLE express before Freezing Medium was added. The suspension was frozen down slowly at −80 °C at least overnight followed by storage in liquid nitrogen. The frozen cells were recoverable with standard cell recovery protocol1, could be embedded in Matrigel, and culture started as described above.
Tumoroids were judged to be successful if they met the following criteria: maintenance in culture for > 4 weeks, viability after five serial passages, and ability to resume growth following multiple freeze/thaw cycles.
Endoluminal tumor injections
Adapting the dextran sulfate sodium (DSS) endoluminal protocol40 and taking advantage of the Mouse Hospital in the Precision Modeling Center at Memorial Sloan Kettering Cancer Center (MSK) (https://www.mskcc.org/research-programs/precision-disease-modeling), we injected tumoroids into the rectums of NOD scid gamma (NSG) mice (The Jackson Laboratory, Bar Harbor, ME, USA). As noted, the experimental protocol was approved by MSK’s Institutional Animal Care and Use Committee (IACUC, IRB #06–07-012). The tumoroids were released from Matrigel by using cell recovery solution and then resuspended in ice-cold PBS with 5% Matrigel to a concentration of 4–6×106 viable cells per mL. Mice were pre-treated with 2.5% DSS salt (Affymetrix Inc. Cleveland, OH, USA) in drinking water for 5 days and then were allowed to recover for 2 days. With mice under anesthesia, 50 μL of tumoroid suspension was injected slowly via anus into the mice using a p200 pipette. The anuses were then sealed by 5 μL of Tissue Adhesive (Vetbond) for 4 hrs and then the Vetbond was removed. The progression of endoluminal tumor was checked every 3–4 weeks using small animal endoscopy (Karl Storz Endoscope, El Segundo, CA, USA). Evidence of metastatic disease was assessed by gross examination and microscopic evaluation of resected organs as noted below. The GFP signal was detected by using a 525/45 nm BrightLine® Single-band bandpass filter (IDEX Health & Science, LLC, New York, USA).
Chemotherapy treatment of mice with endoluminally implanted tumors
RC-MSK-008 tumoroids were injected into the rectum (see Fig. 4a) of 15 female (sex matched to tumoroid) mice 6–8 weeks of age. Mice were checked for tumor progression using endoscopy at 3–4-week intervals. Then, the 14 mice that were successfully engrafted as evidenced by tumor formation noted on endoscopy were randomized by the envelope method into two groups for intraperitoneal injection with either 5-FU (SigmaAldrich) (5 mg/mL in PBS, 50 mg/kg/week; n = 7), or PBS (n = 7) as control for the duration of the experiment. After 7 weeks of treatment, the tumors were assessed using endoscopy. All of the mice were sacrificed, and rectum, colon, cecum, liver, lung, and brain were isolated and fixed in formalin overnight. Separately, RC-MSK-002 tumoroids were injected into 15 male (sex matched to tumoroid) mice 6–8 weeks of age (see Fig. 4d). 3 mice did not survive DSS treatment due to severe colitis, and of the remaining 12 mice, 11 successfully engrafted and were treated similarly as with the RC-MSK-008 experiment above with 5-FU (n = 5) and PBS (n = 6).
For the experiments presented in Fig. 4g, RC-MSK-008 tumoroids were injected into the rectum of 20 female (sex matched to tumoroids) mice 6–8 weeks of age. Due to DSS toxicity, four mice were lost. Of the remaining 16 mice, 16 successfully engrafted and were randomized at 10 weeks to FOLFOX (n = 8) and PBS control (n = 8). The treatments were administered intraperitoneally as follows41. FOLFOX treatment was administered in two stages. First, a solution of 6 mg/kg oxaliplatin (Memorial Hospital Pharmacy) in PBS was injected intraperitoneally. 2 hrs later, a solution of 50 mg/kg 5-FU and 90 mg/kg leucovorin (SigmaAldrich) in PBS was injected intraperitoneally. Treatment was repeated in the same fashion weekly. If the mouse lost significant body weight, the oxaliplatin dose was reduced to 4 mg/kg at subsequent treatments, similarly as with patient treatments. After 8 weeks of treatment, the tumors were assessed using endoscopy. All of the mice were sacrificed, and rectum, colon, cecum, liver, lung, and brain were isolated and fixed in formalin overnight. Separately, RC-MSK-002 tumoroids were injected into 20 male (sex matched to tumoroid) mice 6–8 weeks of age. Two mice did not survive DSS treatment due to severe colitis, and of the remaining 18 mice, 18 successfully engrafted and were treated similarly as with the RC-MSK-008 experiment above with 9 mice in the FOLFOX group and 9 mice in the PBS group.
For the experiments presented in Fig. 4h, the RC-MSK-023 and RC-MSK-001 tumoroids were engineered with GFP expression which were utilized for visualization during endoscopy and IHC analyses (Extended Data Fig. 9g and h, further data not shown). RC-MSK-023 tumoroids were injected in the rectum of 18 male mice and RC-MSK-001 tumoroids were injected into the rectum of 18 female mice. Due to DSS toxicity, two mice were lost in the RC-MSK-023 group and one mouse was lost in the RC-MSK-001 group. 16 mice in the RC-MSK-023 group successfully engrafted and were randomized 11 weeks post tumoroid injection to vehicle (n = 8) and FOLFOX (n = 8) groups. 17 mice in the RC-MSK-001 group successfully engrafted and were randomized 15 weeks post tumoroid injection to vehicle (n = 8) and FOLFOX (n = 9) groups. The treatments were administered as above. After 7 weeks of treatment in the RC-MSK-023 group and 8 weeks of treatment in the RC-MSK-001 group, the tumors were measured endoscopically and the mice were sacrificed as above.
To quantify tumor size, two independent observers reviewed all endoscopy videos and took screenshots when the endoscope had the tumor in full view. The field of view and tumor were encircled, and the areas were measured using ImageJ software (NIH). When there were multiple tumors, the sum of the total tumor area was taken. These data were used to calculate the percent of the field of view occupied by the tumor. To calculate fold changes in tumor size, post-treatment percent areas were divided by the pre-treatment percent areas for each mouse. The Mann-Whitney test was used to compare treatment groups.
Ectopic tumor injections
The tumoroids were released from Matrigel by using cell recovery solution and then resuspended in 50% ice-cold PBS and Matrigel (Corning, product #356237) solution to a concentration of 1×106 viable cells per mL. Mice under anesthesia were subcutaneously inoculated with 1×105 viable cells in the both hind flanks (100 μL/site). For the RC-MSK-001 experiment, tumors were grown for 6 weeks and for the RC-MSK-002 experiment, tumors were grown for 7 weeks. After this time, the mice were randomized into two groups and intraperitoneally injected with either 5-FU (50 mg/kg), or PBS as control, twice per week for the duration of the experiment. Tumor sizes were measured weekly using a handheld imaging device (Peira TM900) per the manufacturer’s protocol.
Development of the GFP-labeled tumoroid and endoscopic viewing of GFP
The RC-MSK-001-eGFP/luc tumoroids used for bioluminescent tracking were developed by lentivirally introducing GFP and firefly luciferase. Lentiviral particles were generated by transfecting HEK293T cells (see “Life Sciences Reporting Summary” for cell line source information) with the Ubc-eGFP-Luc, psPAX2, and VSV-G42 constructs. 7.25 × 106 HEK293T cells were seeded into a 10 cm dish. 7.7 μg of the lentiviral construct, 5.8 μg of psPAX2, and 3.9 μg of VSV-G were transfected using Lipofectamine 2000 according to the manufacturer’s protocol (Invitrogen), grown overnight, and then the medium was replaced with standard DMEM supplemented with 10% FBS, GlutaMAX, and PenStrep. Two days after transfection, the virus media was concentrated using PEG-it Virus Precipitation Solution (SBI System Biosciences), and then the lentiviral particles resuspended in 300 μL PBS. For RC-MSK-001 tumoroid infection, tumoroids from three 50μL Matrigel discs per viral construct were dissociated and resuspended in 10 μL of infection medium (tumoroid culture medium plus 8 μg/mL Polybrene [SigmaAldrich] and 10 μM Y27632 [SigmaAldrich]). The tumoroid cluster suspension and viral suspension were combined in a 48-well culture plate, centrifuged at 600×g at room temperature for 60 minutes, and then incubated for 6 hrs at 37 °C and 5% CO2. The contents of each well were homogenized, transferred to a 1.5 mL tube, and centrifuged at 400×g. The supernatant was discarded, the pellet resuspended in 150 μL Matrigel, and the suspension divided into three wells of a 24-well suspension plate. After Matrigel polymerization, 500 μL of culture medium with 10 μM Y27632 was added. Two days after infection, the medium was replaced with tumoroid culture medium. The infected cells were selected for by addition of puromycin (2μg/mL) for 6 days, and then further enriched by fluorescence-activated cell sorting for GFP-positive cells. This protocol was also used to develop eGFP/luc- transfected RC-MSK-002, RC-MSK-004, and RC-MSK-023 (data not shown).
Preparation of tumoroids and tissues for immunohistochemistry and immunofluorescence
After removal of culture medium, the Matrigel disc was washed with PBS, and then fixed with 4% paraformaldehyde for 30 min at room temperature. Then, paraformaldehyde was removed and the fixed sample was stained with eosin and transferred carefully to an embedding cassette. Paraffin embedding and sectioning were processed with standard protocols. Tissues were fixed in formalin and embedded in paraffin. 5 μm sections were used for all analyses.
Immunohistochemistry and immunofluorescence
Hematoxylin and eosin stains were performed on all specimens for initial histopathological evaluation. The immunohistochemistry/immunofluorescence detection for EpCAM, collagen IV, E-cadherin, Muc-2, and Ki67 was performed at Molecular Cytology Core Facility of Memorial Sloan Kettering Cancer Center using a Discovery XT processor (Ventana Medical Systems), as described in Yarilin et al43. Briefly, tissue sections were blocked for 30 minutes, incubated with primary antibody for 4 hrs, and then incubated with biotinylated secondary antibody for 30 minutes. Blocker D and Streptavidin- HRP were used according to the manufacturer instructions (Ventana Medical Systems). Then, for immunohistochemistry, DAB detection kit (Ventana Medical Systems) was used; for immunofluorescence, specimens were incubated with Tyramide-Alexa Fluor 488 (Invitrogen, cat. #T20922) or 568 (Invitrogen, cat. #T20914). Immunohistochemistry for CK20, CDX2, β-catenin, MLH-1, MSH-2, MSH-6, and PMS-2 was performed by the Surgical Pathology Laboratory of Memorial Sloan Kettering Cancer Center using both the Benchmark Ultra (Roche) and Bond III (Leica) automated staining platforms (see “Life Sciences Reporting Summary” for antibody information).
Imaging
Slides were scanned using the Pannoramic Flash slide scanner (3DHISTECH) using a 20x 0.8 NA objective (Carl Zeiss). Images were examined and representative areas exported using CaseViewer 2.2 (3DHISTECH). No gamma changes were made to any immunofluorescence images. All brightfield images are unaltered.
The H&E comparisons reported in Extended Data Fig. 2 and 3 were reviewed by an independent gastrointestinal pathologist who assessed the architecture, cytology, and unique features observed in tumoroids and their corresponding patient tumors. Patient tumors and derived tumoroids stained for β-catenin and CDX2 were reviewed and scored by a pathologist blinded to the clinical data as reported in Extended Data Fig. 3. Scoring was based on the presence of cells with positively-stained nuclei and the intensity of staining. Presence was scored as 0 (none), 1 (<30%), 2 (30–60%), and 3 (>60%); intensity was scored as 0, 1 (weak), 2 (moderate), and 3 (strong). Cohen’s κ44 was used to compare scores between tumor and tumoroid with the κ test statistic and p-values reported in the figure legend. The pathologist’s notes are summarized in Supplementary Table 1.
Alu quantitative PCR
To quantitatively characterize the metastases noted within the mouse lungs and livers as human cells originating from the rectal xenografts, quantitative polymerase chain reaction (q-PCR) was performed with primers specific for human Alu repeats45 and mouse short interspersed nuclear elements (SINE)46. This was done on the lung and liver metastases along with an NSG normal adjacent rectal control and a human colon control. The following primers were used: hAlu-101F: 5’-GGTGAAACCCCGTCTCTACT-3’; hAlu-206R: 5’-GGTTCAAGCGATTCTCCTGC-3’; mSINE-F: 5’-AGATGGCTCAGTGGGTAAAGG-3’; mSINE-R: 5’-GTGGAGGTCAGAGGACAAACTT-3’. The reaction was performed in a final volume of 20 uL using FAST SYBR- Green Master Mix (Applied Biosystems, #4385612) with 0.1 μM of each primer and 20 ng of genomic DNA using an Applied Biosystems 7500 real-time PCR instrument (ABI 7500; Thermo Fisher Scientific). PCR conditions were as follows: 95 °C for 10 min; 45 cycles of 95 °C for 15 s, 56 °C for 30 s, and 72 °C for 30 s. The relative quantity of human Alu sequences was normalized against the relative quantity of mouse SINE sequence for each respective control as ΔCt = CthAlu - CtmSINE and expressed as 2−ΔCt. Human and mouse genomic DNA were used as positive and negative controls, respectively. All Alu Ct values were normalized to human DNA Ct value as 100% (human normal adjacent colon TS62T) (see “Life Sciences Reporting Summary” for software analysis information).
DNA isolation
Tumoroids and normal organoids were released from Matrigel by using cell recovery solution. DNA was extracted using DNeasy Blood and Tissue kit according to the manufacturer’s procedure (Qiagen, Germantown, MD, USA).
Genomic Analysis
DNA derived from the tumor biopsies and the cultured tumoroids was subjected to exon capture sequencing using the MSK Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) platform as previously described47. Matched germline DNA samples, extracted from blood and cultured normal organoids, were also sequenced to identify somatic versus germline nucleotide variants. Somatic variants were called using MuTect (v1.1.4) for single nucleotide variants and Somatic Indel Detector (GATK 2.3–9) for indels. Somatic variants were annotated by Annovar for cDNA and amino acid changes and for their presence in dbSNP (v137), the COSMIC database (v68), and 1,000 Genomes minor allele frequencies. Variants were also annotated for their presence and predicted oncogenic status in OncoKB48 (see “Life Sciences Reporting Summary” for OncoKB software information). Each locus where a variant was called was genotyped in matched timepoint samples from the same patient. If a variant was called in one sample and was present in another sample from the same patient in at least 3 reads and at least 1% of reads, it was marked as ‘detectable’ in the other sample. We considered the most frequently mutated genes in the tumoroids and compared their mutation frequency within the tumoroid cohort with the mutation frequency in a set of 287 prospectively analyzed rectal adenocarcinoma samples from MSK. These data can be viewed on the cBioPortal for Cancer Genomics49,50. In addition, we used the FACETS algorithm to determine allele specific copy number and cancer cell fraction of each mutation in tumoroids and their respective tumors51. Mutations with higher or equal to 85% cancer cell fraction were categorized as clonal. Others were categorized as subclonal.
Drug treatments
Tumoroids were resuspended in Matrigel and embedded in suspension in a 24-well plate (2–5×104 cells/50 μL Matrigel/well) or 48-well plate (2–3 ×103 cells/20 μL Matrigel/well). The cells were allowed to recover for 2 to 3 days. Medium was replaced with fresh culture medium with varying concentrations of the drugs as follows.
For the cetuximab (Memorial Hospital Pharmacy) treatment reported in Extended Data Fig. 8, tumoroids were cultured with cetuximab at the indicated doses in culture media for 72-hours. After 3 days, the drug media was replenished and after 6 days, MTS assay was used to measure cell viability.
For 5-FU treatments, tumoroids were cultured with 5-FU at 50, 10, 5, 1, 0.5, and 0 μM for 6 days, replenishing 5-FU medium on day 3. For FOLFOX treatments, tumoroids were cultured with FOLFOX (The FOLFOX chemotherapy cocktail ratio used was as follows: 5-FU:leucovorin:oxaliplatin, 25:5:1) for 6 days, replenishing FOLFOX media on day 3. The final 5-FU concentrations are 50, 10, 5, 1, 0.5 and 0 μM. At the end of treatment in each experiment, the number of viable cells was determined by CellTiter-Glo assay (Promega, Madison, WI, USA) following the kit protocol. Dose response curves were fit to the data using the two-parameter logistic regression with variable slope and constraints at 100% and 0% viability using GraphPad Prism 8 (La Jolla, CA, USA). Area under the curve (AUC) was calculated using the raw experimental data and normalized uniformly by dividing the AUC by the total maximum area a curve could occupy from 0–100% viability over the range of drug concentrations analyzed (see “Life Sciences Reporting Summary” for additional information).
Extraction of clinical information for correlation analyses to both chemotherapy and radiation
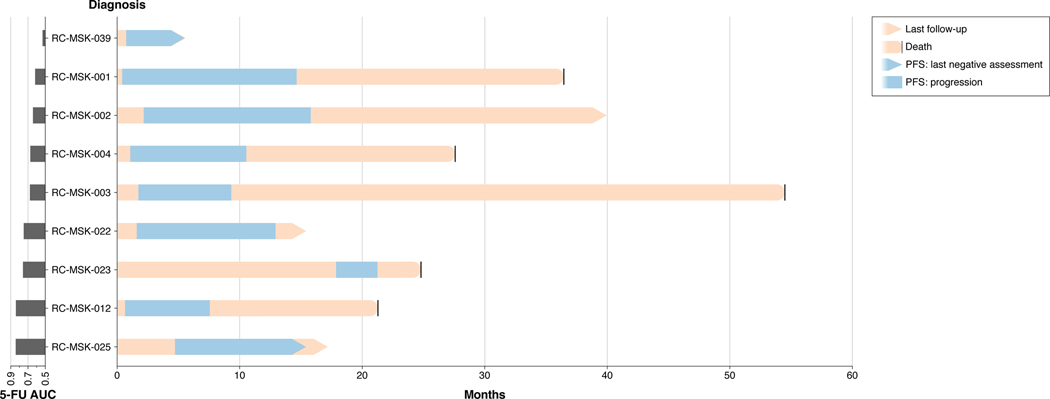
Progression-free survival (PFS) was used to correlate ex vivo chemosensitivity measurements with clinical information as it is used in many oncological drug response studies as a measure of clinical response to therapy. PFS was measured from 5-FU-based chemotherapy start date until the first radiographic or endoscopic evidence of new disease or growth of existing disease. Patients who had not yet progressed were not included in the PFS correlation analysis because an appropriate PFS value could not be assigned. The 21 tumoroids assayed for chemosensitivity were derived from 13 total patients. Nine of these patients received 5-FU-based chemotherapy and of those nine patients, two have not yet progressed, leaving seven patients with complete PFS information available at this time for analysis. Each of these nine patients is displayed in Extended Data Fig. 7. For the two patients who have not yet progressed, RC-MSK-025 is currently 10.4 months post treatment start without evidence of progression and RC-MSK-039 is currently 4.6 months post treatment start without evidence of progression. These two patients are displayed as open data points in Fig. 2b, but were not included in the correlation analysis.
With respect to correlating ex vivo radiation data with clinical information and in order to isolate the clinical effects of radiation and not the summative effects of chemotherapy and radiation, we elected to compare endoscopic tumor assessment immediately before and after radiation to isolate the response specifically to radiotherapy. The percent circumference of the bowel wall involved with tumor is routinely assessed by the clinician throughout the course of clinical care to gauge tumor response to treatment. Percent circumference was reported by the colorectal surgeon performing the endoscopy and documented in the medical record at the time of procedure. This was analyzed for all patients for which pre- and post-radiation endoscopic data was available and whose tumoroids were profiled ex vivo for radiosensitivity.
Radiation experiments
10,000 cells from tumoroids were embedded in 30 μL Matrigel within each well of a 96-well plate and given to our collaborators (P.B.R.) without supplying any information on the individual patient’s clinical course. Radiation was delivered with an X-RAD320 Biological Irradiator (Precision X-Ray, North Branford, CT) at 250 kVp and 12 mA. Tumoroids were allowed to recover for 8 days, with the exception of RC-MSK-031 which recovered for 11 days and RC-MSK-003 for 13 days to allow for appropriate cell numbers to assess cell viability within the range of sensitivity of the cell viability protocol. Cell viability was assessed with the CellTiter-Glo assay following the kit protocol. Data were normalized to the 0 Gy radiation dose wells as 100% viability to calculate cell viability of the remaining radiated wells. AUC was calculated using the raw experimental data and normalized uniformly by dividing the AUC by the total maximum area a curve could occupy from 0–100% viability over the range of radiation doses analyzed.
Statistics
The dose-response curves reported in Fig. 2a were fit using two parameter logistic regression with variable slope and constraints at 100% and 0% viability using GraphPad Prism 8 (La Jolla, CA, USA). Curves with R2 < 0.6 are not reported (1 excluded for 5-FU and 2 excluded for FOLFOX). The correlation analysis reported in Fig. 2b was calculated using Spearman’s correlation with appropriate Spearman r and p-values reported in the figure. Statistical analysis relating to Fig. 4b, e, g, and h were conducted using a two-tailed Mann-Whitney test with appropriate U and p-values reported in the figure legend. Cohen’s κ was used to compare tumor and tumoroid histological scores for similarity. κ statistics and p-values are reported in the Extended Data Fig. 3 legend for each metric. For all statistical testing, α = 0.05. All error bars for graphical analysis are defined in the respective figure legends.
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon reasonable request. The data for the Rectal Cancer (RC) tumoroids on the cBioPortal will be made publicly available on publication.
Code availability
The FACETS algorithm was used to determine allele specific copy number and cancer cell fraction of each mutation in tumoroids and their respective tumors51. Access to this code is available upon reasonable request.
Extended Data
Extended Data Fig. 1 |. Rectal cancer tumoroid derivation and patient characteristics.
The diagram shows the outcome of attempts to derive tumoroids from 84 rectal cancer (RC) tumor samples from 58 individual RC patients. 65 RC tumoroids from 41 patients (77%) were successfully derived. For the 19 failed derivations, the points of failure are shown. Demographics from each group are displayed (RAS status [wild type (WT) or mutant (MUT)], neoadjuvant therapy, metastatic status at derivation, location of the primary tumor [middle/distal or upper rectum], sex, and age). Two patients were mismatch repair deficient (not shown).
Extended Data Fig. 2 |. Preservation of rectal cancer histopathology in tumoroids.
a, Gross resected rectal specimen from which the first RC tumoroid line (RC-MSK-001) was derived and representative brightfield microscopy of the tumoroid in 3D culture two months after processing. Lower panels show hematoxylin and eosin (H&E) staining of the patient tumor (bottom left panel) and the derived RC-MSK-001 tumoroid (bottom right panel) in 3D culture. Scale bars, 50 μm. b, Hoechst and MitoTracker stains of a representative section of the RC-MSK-001 tumoroid demonstrate the luminal and glandular structure. Scale bars, 20 μm. c, Perineal recurrence of the original RC-MSK-001 tumor and the derived tumoroid (RC-MSK-001PR) are shown with H&E staining. Scale bar, 50 μm. d, H&E comparison of 32 additional tumoroid cell lines as noted with the corresponding primary tumor from which they were derived. Scale bars, 50 μm. All representative images are from one patient-specific tumor-to-tumoroid derivation.
Extended Data Fig. 3 |. Tumoroids preserve both architecture, cytology, and colorectal-specific staining patterns of the primary tumors from which they were derived.
a, Examples of architecture preservation in tumoroids and primary tumors. Scale bars, 50 μm. b, Examples of cytological preservation in specific tumoroids. Scale bars: low magnification 50 μm, high magnification inset 10 μm. Both architecture and cytology features were identified by an independent gastrointestinal pathologist. c, CDX2 and β-catenin were quantified by both presence and intensity of stain on a 0–3 scale. Presence is defined as the percentage of cells with staining: 0 = 0%, 1 = < 30%, 2 = 30–60%, 3 = >60%. Intensity defines the strength of staining: 0 = none, 1 = weak, 2 = moderate, 3 = strong. Examples for both CDX2 and β-catenin are displayed. Intensity of staining is assessed exclusively within the nuclear compartment. Scale bar, 20 μm. d, The presence and intensity of each tumoroid is shown graphically according to the key. Cohen’s κ was used to assess similarity in score between matched primary and tumoroid samples: β-catenin presence score, κ = 0.51, p = 0.0021; β-catenin intensity score, κ = 0.63, p = 0.00034; CDX2 presence score, κ = 0.45, p = 0.0037; CDX2 intensity score, κ = 0.527, p = 0.00042. All representative images are from one patient-specific tumor-to-tumoroid derivation.
Extended Data Fig. 4 |. Conservation of enterocyte markers.
Eleven tumoroids are compared to their respective primary tumors for Alcian blue, CK20, CDX2, MUC-2, E-cadherin, and β-catenin staining. For immunofluorescent staining: E-cadherin (green), DAPI (blue). See Fig. 1b for another example of RC-MSK-001 Alcian blue, CK20, and CDX2 comparisons. Scale bars, 50 μm. All representative images are from one patient-specific tumor-to-tumoroid derivation.
Extended Data Fig. 5 |. Comparison of nuclear mismatch repair proteins between patient and tumoroid samples.
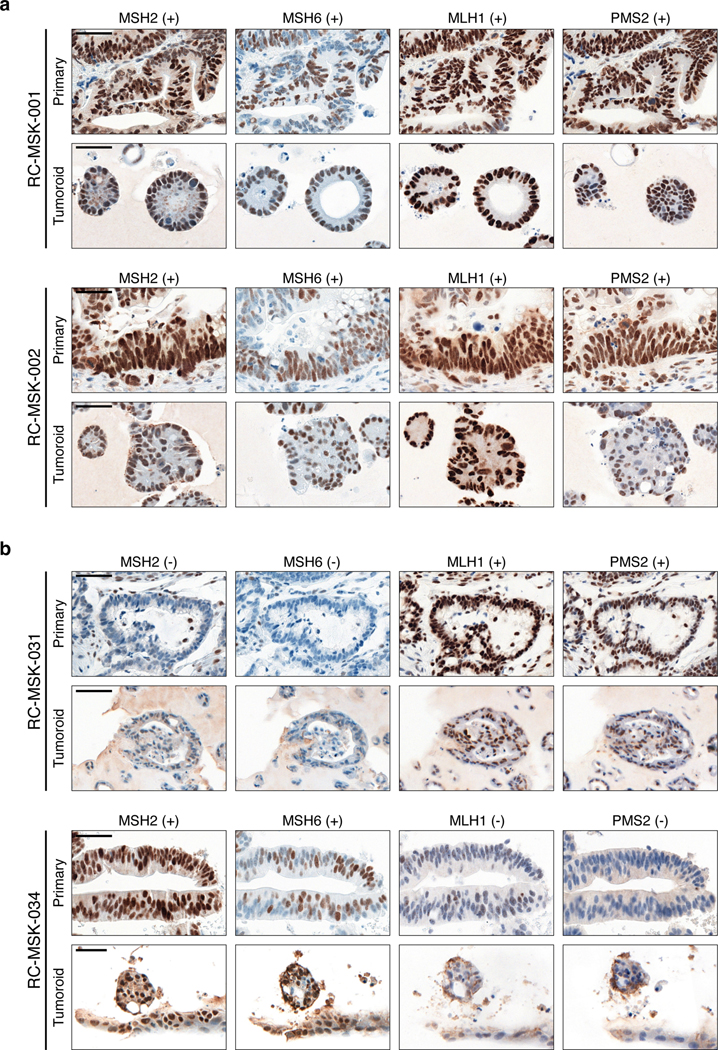
Immunohistochemistry of the nuclear mismatch repair (MMR) proteins MSH2, MSH6, MLH1, and PMS2. The presence of each protein is assessed by nuclear staining verified by pathologic analysis with (+) indicating present and (−) indicating absent staining. a, Displayed are two MMR-proficient tumoroids, RC-MSK-001 and RC-MSK-002. b, Displayed are two MMR-deficient tumors. RC-MSK-031 is deficient in MSH2 and MSH6. RC-MSK-034 is deficient in MLH1 and PMS2. Scale bars, 50 μm. All representative images are from one patient-specific tumor-to-tumoroid derivation.
Extended Data Fig. 6 |. The mutational fingerprint in derived RC tumoroids.
a, The mutational fingerprint of 31 RC tumoroids for the most common alterations as determined by MSK-IMPACT are displayed. The frequency of alteration is noted along with the type of genetic alteration relative to truncating mutation, inframe mutation, missense mutation, or splice site alterations (as noted by the color code). b, Example of a tumoroid (RC-MSK-003) with complete conservation of mutations between the tumoroid and the primary tumor from which it was derived. c, Example of a tumoroid (RC-MSK-004) with conservation of driver mutations and the addition of two secondary mutations noted in the tumoroid in culture only. d, Percentage of concordance between tumoroid and tumor among mutations predicted to be oncogenic overall and by each patient. The mutations represented are those annotated by OncoKB10 as oncogenic or likely oncogenic in each tumoroid and tumor pair. e, All mutations called in the MSK-IMPACT sequencing of tumoroids and primary tumors are shown. The numbers of mutations are displayed with regard to each gene (by column) and each tumoroid and tumor pair (by row). Mutations are colored by concordance status.
Extended Data Fig. 7 |. Swimmer’s plot of each patient treated with 5-FU-based therapy whose tumoroid has been analyzed for chemosensitivity ex vivo.
Data is displayed from top to bottom by descending area under the curve (AUC) calculated from the 5-FU dose-response experiments presented in Fig. 2a. The blue areas denote progression-free survival (PFS) intervals from treatment start date as indicated above. Seven of the nine patients have progressed, with the current status of the two patients who have not progressed indicated (RC-MSK-039 and RC-MSK-025). All patients were treated with FOLFOX, with the exception of RC-MSK-003 (capecitabine [oral 5-FU prodrug] + oxaliplatin + bevacizumab) and RC-MSK-023 (FOLFIRI: 5-FU + leucovorin + irinotecan). Four patients for whom ex vivo chemosensitivity data is presented in Fig. 2a are not shown because they did not receive 5-FU-based therapy.
Extended Data Fig. 8 |. Resistance to a targeted anti-epidermal growth factor receptor therapy, cetuximab, in KRAS mutant compared with KRAS wild type tumoroids.
Resistance to cetuximab is demonstrated in KRAS mutant RC tumoroids (blue) compared with a KRAS wild type tumoroids (orange). Dose range was used as shown and percentage of live cells is displayed for each tumoroid. Results are from two independent experiments done in technical quadruplicate; mean ± s.e.m.
Extended Data Fig. 9 |. Demonstration of endorectally implanted human rectal cancer.
a, The RC-MSK-001 endoluminal mouse biopsy represented in Fig. 3a shows serially sectioned and stained hEpCAM; collagen IV; merged with DAPI. n=5 mice scale bars: 200 μm; inset, 50 μm. b, Colon from an unimplanted NSG mouse as control for a. Scale bars, 200 μm. c-d, 12-week endoscopy of a mouse transplanted with RC-MSK-001 (n=5) or RC-MSK-002 (n=7) tumoroids. e-f, Distinct staining was noted for human and mouse EpCAM for RC-MSK-001 (n=5) and RC-MSK-002 (n=7) engrafted NSG mice. Scale bars: 500 μm; inset, 50 μm. g, RC-MSK-001 tumoroids labeled with GFP and viewed by brightfield (left), intravital GFP imaging (middle; endoscopically (right). Scale bars, 100 μm. h, Invasive rectal tumor after RC-MSK-001 tumoroid implantation (n=7 mice) stained for H&E, GFP (IHC), and IF (hEpCAM, mEpCAM, and hKi67, each merged with DAPI). i, Independent experiment similar to Fig. 3c of one male NSG mouse sacrificed at 22 weeks post-transplantation. Implanted rectal tumor, H&E, and IF (hEpCAM, mEpCAM, DAPI) showing engraftment and invasion of human tumoroids. Scale bars: H&E, 500 μm; IF, 100 μm. j-l, RC-MSK-001 endorectal tumor 16 weeks post-transplantation (n=3 mice). H&E demonstrates invasion at the junction between the columnar and squamous epithelium of anorectal junction. j/k, H&E; l, DAPI + hEpCAM + collagen IV; Scale bars are as follows: j, 1,000 μm; k-l: 400 μm; insets, 100 μm. m, Liver metastasis in an independent experiment (see Fig. 3d) after rectal transplantation in a male NSG mouse sacrificed at 36 weeks. Liver metastasis shows poorly differentiated histology. Scale bars for H&E: 1,000 μm, 500 μm, 100 μm. n, Axial and coronal CT images of liver metastases in the corresponding patient (arrowheads) discussed in m. o, Human-specific Alu qPCR demonstrates that the metastases in Fig. 3d and current panel m arose from implanted human tumoroids (Results based on three independent RNA isolates). Mean ± s.d. Staining: hEpCAM (green), hKi67 (green), mEpCAM (red), Collagen IV (red), and DAPI (blue).
Extended Data Fig. 10 |. Histopathologic conservation of glandular architecture in the endoluminally implanted RC tumoroids.
H&E images are shown for the RC-MSK-008, RC-MSK-002, RC-MSK-023, and RC-MSK-001 tumoroid lines. Left panels display the primary patient tumor from which the tumoroid was derived once per patient. Middle panels display the tumoroids in 3D culture. Right panels display the engrafted tumoroids within the mouse rectum following endoluminal transplantation The number of mice engrafted with indicated tumoroids is 8, 7, 8, and 5 (top to bottom). The H&E photomicrographs demonstrate histopathologic conservation of glandular features as noted in the human adenocarcinomas from which they were derived. Scale bar, 50 μm.
Supplementary Material
Video S1 Endoscopic video of tumor following 200,000 cells from tumoroids injected in NOD scid gamma mice rectum
a, Growth of tumors established from flank injection of RC-MSK-001 tumoroid. The graph displays relative tumor volume over time in the vehicle (n = 4) and 5-FU (n = 6) treated groups. Mean ± s.d. b, RC-MSK-002 tumoroid flank model (similar as in a) for vehicle (n = 2) and 5-FU (n = 3) treated groups. Mean ± s.d. where shown and possible to be calculated.
Acknowledgments
We thank B. Carver for critical review of the manuscript as it developed. We thank A. Jungbluth for initial discussion on tissue fixation and immunostaining of the murine rectal tissues. We thank S. Chandarlapaty for discussions regarding the chemoresistance assays and cetuximab resistance assay. We thank L. Diaz for critical comments as this work developed. We thank N. Kemeny for advice, guidance, and encouragement as this project developed. We thank N. Fan, S. Fujisawa, X. Liu, M. Turkekul, E. Chan, and the Molecular Cytology Core for expert assistance and critical feedback in the design and execution of tissue embedding, immunostaining, and processing of tissues in addition to microscopy assistance. We thank the MSK Molecular Core Cytology Facility for critical technical assistance in performing tissue sections, immunohistochemical stains, scans, and analysis (Institutional Core Grant Number P30CA008748). We thank R. Shah for initial discussions on set-up of the MSK-IMPACT experiments. We thank H. Park and L. Lee for assistance with preparation and processing of tumor and tumoroid specimens for IHC and immunofluorescence. We thank P. Watson of Memorial Sloan Kettering for sharing the Ubc-eGFP-Luc vector. We thank M. Gonzalez and R. Andrade for their work curating patient tissue and slides for analyses.
We also thank members of the Sawyers laboratory for critical review, lively discussion, and helpful comments on this work as it developed. In addition, we thank R. Beauchamp and J. Goldenring for constructive criticism and comments in the development and presentation of this work.
The authors thank J. Novak of Memorial Sloan Kettering for editorial assistance.
Funding Sources:
This work was supported in part by the NIH/NCI Cancer Center Support Grant P30 CA008748. Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Number R25CA020449. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
J.J.S. was supported by NIH/NCI grant 5R01-CA182551–04 and the MSK Colorectal Cancer Research Center. J.J.S. is also supported by the American Society of Colon and Rectal Surgeons Career Development Award, the Joel J. Roslyn Faculty Research Award, the American Society of Colon and Rectal Surgeons Limited Project Grant, the MSK Department of Surgery Junior Faculty Award, and the John Wasserman Colon and Rectal Cancer Fund. J.J.S. is also supported by the Colorectal Cancer Alliance and the Chris4Life Research Award. The work was funded in part by a Stand Up to Cancer (SU2C) Colorectal Cancer Dream Team Translational Research Grant (Grant Number: SU2C: AACR-DR22–17) (L.E.D., K.G.). Stand Up to Cancer is a program of the Entertainment Industry Foundation. Research grants are administered by the American Association of Cancer Research, the scientific partner of SU2C. J.J.S. is partly supported by a loan repayment grant from the NIH via the National Cancer Institute for this work. J.J.S. is also supported in part by funding from the Howard Hughes Medical Institute via C.L.S..
P.B.P and M.A. are funded in part by gifts from Corinne Berezuk, Michael Stieber, and Patrick A. Gerschel.
C.L.S. is an investigator of the Howard Hughes Medical Institute. This project was supported by National Institutes of Health grants CA155169, CA193837, CA224079, CA092629, CA160001, CA008748 and Starr Cancer Consortium grant I10–0062.
K.G. was supported by National Institutes of Health grants K08-CA230213 and T32-CA009207, and by an American Cancer Society Postdoctoral Fellowship, AACR Basic Cancer Research Fellowship, Conquer Cancer Foundation of ASCO Young Investigator Award, Shulamit Katzman Endowed Postdoctoral Research Fellowship, and a SU2C Colorectal Cancer Dream Team Translational Research Grant (SU2C: AACR-DR22–17).
J.M. is supported by National Institutes of Health grants CA94060 and CA12924, and by the MSKCC Alan and Sandra Gerry Metastasis and Tumor Ecosystems Center.
S.W.L., L.E.D., and K.P.O. are supported by grants from the NIH (U54 OD020355–01) and by the Starr Cancer Consortium (I8-A8–030). S.W.L. is an investigator of the Howard Hughes Medical Institute.
K.P.O. is supported by an F30 Award from the NIH/NCI (1CA200110–01A1) and by a Medical Scientist Training Program grant from the National Institute of General Medical Sciences of the National Institutes of Health under award number T32GM07739 to the Weill Cornell / Rockefeller / Sloan Kettering Tri-Institutional MD-PhD Program.
L.E.D. is supported by a Stand Up to Cancer Colorectal Cancer Dream Team Translational Research Grant (Grant Number: SU2C: AACR-DR22–17) and was supported by a K22 Career Development Award from the NCI/NIH (CA 181280–01).
P.B.R. is supported by the Memorial Sloan Kettering Cancer Center Imaging and Radiation Sciences Program.
P.B.R. is also supported in part by a K12 Paul Calebresi Career Development Award for Clinical Oncology (K12 CA184746) and an NIH loan repayment program award (LRP).
Footnotes
Competing Interests:
J.J.S. has received travel support from Intuitive Surgical Inc. and has served as a clinical advisor for Guardant Health, Inc.
C.L.S. serves on the Board of Directors of Novartis, is a co-founder of ORIC Pharm, and co-inventor of enzalutamide and apalutamide. He is a science advisor to Agios, Beigene, Blueprint, Column Group, Foghorn, Housey Pharma, Nextech, KSQ, Petra, and PMV. He was a co-founder of Seragon, purchased by Genentech/Roche in 2014.
J.M. is a science advisor and owns company stock in Scholar Rock.
H.C. is an inventor on several patents related to organoid technology.
S.W.L. is a co-founder and scientific advisory board member for ORIC Pharm, Blueprint, and Mirimus. He also serves on the scientific advisory board for Constellation, Petra, and PMV and has recently served as a consultant for Forma, Boehringer Ingelheim, and Aileron.
J.G.-A. has received support from Medtronic (honorarium for consultancy with Medtronic), Johnson & Johnson (honorarium for delivering a talk), and Intuitive Surgical (honorarium for participating in a webinar by Intuitive Surgical).
P.B.R. has received honorarium from Corning to discuss 3D cell culture techniques, has served as a consultant for AstraZeneca, and is a consultant for EMD Serono for work on radiation sensitizers.
R.N.K. is a cofounder of Ceramedix Holding L.L.C. He also has patents unrelated to this work: RK (US7195775B1, US7850984B2, and US10052387B2), RK (US8562993B2, US9592238B2, US20150216971A1, and US20170335014A1), and RK (US20170333413A1 and US20180015183A1).
K.P.O. has received an honorarium from Merck to discuss organoid platforms.
References
- 1.Siegel RL, Miller KD & Jemal A Cancer statistics, 2019. CA. Cancer J. Clin 69, 7–34 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Deborah Freedman-Cass N et al. NCCN Guidelines Version 2.2017 Panel Members Rectal Cancer Continue NCCN Guidelines Panel Disclosures. [Google Scholar]
- 3.Smith JJ et al. Assessment of a Watch-and-Wait Strategy for Rectal Cancer in Patients With a Complete Response After Neoadjuvant Therapy. JAMA Oncol. e185896 (2019). doi: 10.1001/jamaoncol.2018.5896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Valk MJM et al. Long-term outcomes of clinical complete responders after neoadjuvant treatment for rectal cancer in the International Watch & Wait Database (IWWD): an international multicentre registry study. Lancet 391, 2537–2545 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Martens MH et al. Long-term Outcome of an Organ Preservation Program After Neoadjuvant Treatment for Rectal Cancer. J. Natl. Cancer Inst 108, djw171 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Renehan AG et al. Watch-and-wait approach versus surgical resection after chemoradiotherapy for patients with rectal cancer (the OnCoRe project): a propensity-score matched cohort analysis. Lancet. Oncol 17, 174–83 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Park IJ et al. Neoadjuvant treatment response as an early response indicator for patients with rectal cancer. J. Clin. Oncol 30, 1770–6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maas M et al. Long-term outcome in patients with a pathological complete response after chemoradiation for rectal cancer: a pooled analysis of individual patient data. Lancet. Oncol 11, 835–44 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Smith JJ, Chow OS, Eaton A, Widmar M, Nash G, Temple LR, Guillem JG, Weiser MR, Goodman KA, Cercek A, Saltz LB, Gollub MJ, Gonen M, Garcia-Aguilar J, and P. P. Organ preservation in rectal cancer patients with clinical complete response after neoadjuvant therapy. in 2015 Gastrointestinal Cancers Symposium Abstract 509. Presented January 17, 2015 (ed. Armitage James O., MD E-C) February 10, 2015, Volume 6, Issue 2 (The ASCO Post is published by HSP News Service, L.L.C, a subsidiary of Harborside Press, LLC., 2015). [Google Scholar]
- 10.Smith JJ & Garcia-Aguilar J Advances and Challenges in Treatment of Locally Advanced Rectal Cancer. J. Clin. Oncol (2015). doi: 10.1200/JCO.2014.60.1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herold KM & Rothberg PG Evidence for a labile intermediate in the butyrate induced reduction of the level of c-myc RNA in SW837 rectal carcinoma cells. Oncogene 3, 423–8 (1988). [PubMed] [Google Scholar]
- 12.Ye X, Yin H, Lu Y, Zhang H & Wang H Evaluation of Hydrogel Suppositories for Delivery of 5-Aminolevulinic Acid and Hematoporphyrin Monomethyl Ether to Rectal Tumors. Molecules 21, 1347 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emons G et al. Chemoradiotherapy Resistance in Colorectal Cancer Cells is Mediated by Wnt/β-catenin Signaling. Mol. Cancer Res 15, 1481–1490 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kleiman LB, Krebs AM, Kim SY, Hong TS & Haigis KM Comparative Analysis of Radiosensitizers for K-RAS Mutant Rectal Cancers. PLoS One 8, e82982 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujii M et al. A Colorectal Tumor Organoid Library Demonstrates Progressive Loss of Niche Factor Requirements during Tumorigenesis. Cell Stem Cell 18, 827–38 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Hong TS. Midostaurin (PKC412) for Locally Advanced Rectal Cancer - Tabular View - ClinicalTrials.gov. Available at: https://clinicaltrials.gov/ct2/show/record/NCT01282502. (Accessed: 8th May 2018) [Google Scholar]
- 17.van de Wetering, M. et al. Prospective Derivation of a Living Organoid Biobank of Colorectal Cancer Patients. Cell 161, 933–945 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gock M et al. Establishment, functional and genetic characterization of three novel patient-derived rectal cancer cell lines. World J. Gastroenterol 24, 4880–4892 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding P et al. Pulmonary recurrence predominates after combined modality therapy for rectal cancer: an original retrospective study. Ann. Surg 256, 111–6 (2012). [DOI] [PubMed] [Google Scholar]
- 20.O’Rourke KP et al. Transplantation of engineered organoids enables rapid generation of metastatic mouse models of colorectal cancer. Nat. Biotechnol (2017). doi: 10.1038/nbt.3837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roper J et al. In vivo genome editing and organoid transplantation models of colorectal cancer and metastasis. Nat. Biotechnol 35, 569–576 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sato T et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Sato T et al. Long-term Expansion of Epithelial Organoids From Human Colon, Adenoma, Adenocarcinoma, and Barrett’s Epithelium. Gastroenterology 141, 1762–1772 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Fleming M, Ravula S, Tatishchev SF & Wang HL Colorectal carcinoma: Pathologic aspects. Journal of Gastrointestinal Oncology (2012). doi: 10.3978/j.issn.2078-6891.2012.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonneville R et al. Landscape of Microsatellite Instability Across 39 Cancer Types. JCO Precis. Oncol 1–15 (2017). doi: 10.1200/PO.17.00073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zehir A et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med (2017). doi: 10.1038/nm.4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weeber F et al. Preserved genetic diversity in organoids cultured from biopsies of human colorectal cancer metastases. Proc. Natl. Acad. Sci (2015). doi: 10.1073/pnas.1516689112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rödel C et al. Preoperative chemoradiotherapy and postoperative chemotherapy with fluorouracil and oxaliplatin versus fluorouracil alone in locally advanced rectal cancer: initial results of the German CAO/ARO/AIO-04 randomised phase 3 trial. Lancet. Oncol 13, 679–87 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Allegra CJ et al. Neoadjuvant 5-FU or Capecitabine Plus Radiation With or Without Oxaliplatin in Rectal Cancer Patients: A Phase III Randomized Clinical Trial. J. Natl. Cancer Inst 107, djv248 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rödel C et al. Oxaliplatin added to fluorouracil-based preoperative chemoradiotherapy and postoperative chemotherapy of locally advanced rectal cancer (the German CAO/ARO/AIO-04 study): final results of the multicentre, open-label, randomised, phase 3 trial. Lancet Oncol 16, 979–89 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Smith JJ et al. Organ Preservation in Rectal Adenocarcinoma: a phase II randomized controlled trial evaluating 3-year disease-free survival in patients with locally advanced rectal cancer treated with chemoradiation plus induction or consolidation chemotherapy, and total. BMC Cancer 15, 767 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Cutsem E et al. Cetuximab and Chemotherapy as Initial Treatment for Metastatic Colorectal Cancer. N. Engl. J. Med 360, 1408–1417 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Vlachogiannis G et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science (80-. ). 359, 920–926 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van de Wetering M et al. Prospective Derivation of a Living Organoid Biobank of Colorectal Cancer Patients. Cell 161, 933–945 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cercek A et al. Neoadjuvant chemotherapy first, followed by chemoradiation and then surgery, in the management of locally advanced rectal cancer. J. Natl. Compr. Canc. Netw 12, 513–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schrag D et al. Neoadjuvant chemotherapy without routine use of radiation therapy for patients with locally advanced rectal cancer: a pilot trial. J. Clin. Oncol 32, 513–8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roerink SF et al. Intra-tumour diversification in colorectal cancer at the single-cell level. Nature 556, 457–462 (2018). [DOI] [PubMed] [Google Scholar]
Methods-only References:
- 38.Sato T et al. Long-term Expansion of Epithelial Organoids From Human Colon, Adenoma, Adenocarcinoma, and Barrett’s Epithelium. Gastroenterology 141, 1762–1772 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Dow LE et al. Apc Restoration Promotes Cellular Differentiation and Reestablishes Crypt Homeostasis in Colorectal Cancer. Cell (2015). doi: 10.1016/j.cell.2015.05.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Rourke KP et al. Transplantation of engineered organoids enables rapid generation of metastatic mouse models of colorectal cancer. Nat. Biotechnol (2017). doi: 10.1038/nbt.3837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson SM et al. The potential contribution of tumour-related factors to the development of FOLFOX-induced sinusoidal obstruction syndrome. Br. J. Cancer 109, 2396–2403 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanjana NE, Shalem O & Zhang F Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–4 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yarilin D et al. Machine-based method for multiplex in situ molecular characterization of tissues by immunofluorescence detection. Sci. Rep 5, 9534 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Viera AJ & Garrett JM Understanding interobserver agreement: the kappa statistic. Fam. Med 37, 360–3 (2005). [PubMed] [Google Scholar]
- 45.Funakoshi K et al. Highly sensitive and specific Alu-based quantification of human cells among rodent cells. Sci. Rep (2017). doi: 10.1038/s41598-017-13402-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walker JA et al. Quantitative PCR for DNA identification based on genome-specific interspersed repetitive elements. Genomics (2004). doi: 10.1016/j.ygeno.2003.09.003 [DOI] [PubMed] [Google Scholar]
- 47.Zehir A et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med (2017). doi: 10.1038/nm.4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chakravarty D et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis. Oncol (2017). doi: 10.1200/PO.17.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao J et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal (2013). doi: 10.1016/j.expneurol.2012.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cerami E et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data: Figure 1. Cancer Discov. 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen R & Seshan VE FACETS: Allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. (2016). doi: 10.1093/nar/gkw520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1 Endoscopic video of tumor following 200,000 cells from tumoroids injected in NOD scid gamma mice rectum
a, Growth of tumors established from flank injection of RC-MSK-001 tumoroid. The graph displays relative tumor volume over time in the vehicle (n = 4) and 5-FU (n = 6) treated groups. Mean ± s.d. b, RC-MSK-002 tumoroid flank model (similar as in a) for vehicle (n = 2) and 5-FU (n = 3) treated groups. Mean ± s.d. where shown and possible to be calculated.
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon reasonable request. The data for the Rectal Cancer (RC) tumoroids on the cBioPortal will be made publicly available on publication.