Abstract
Next-generation sequencing (NGS) is increasingly being used in cancer care to identify both somatic tumor driver mutations that can be targeted for therapy, and heritable mutations in the germline associated with increased cancer risk. This report presents a case of a JAK2 V617F mutation falsely identified as a duodenal cancer mutation via NGS. The patient was found to have a history of polycythemia vera, a disorder with a high incidence of JAK2 somatic mutations. Buccal cell DNA showed heterozygosity for the mutation, suggesting that it was potentially germline. However, subsequent resequencing of tumor, adjacent normal tissue, and fingernail DNA confirmed the mutation was somatic, and its presence in tumor and buccal cells resulted from contaminating blood cells. This report highlights important nuances of NGS that can lead to misinterpretation of results with potential clinical implications.
Background
The advent of next-generation sequencing (NGS) has revolutionized the identification of genetic alterations involved with cancer initiation and drug resistance. In oncology, 2 classes of mutation testing are available for patients. Germline testing seeks to identify genetic alterations that predispose to cancer development due to heritable mutations (eg, BRCA1 and BRCA2). In contrast, somatic alterations are genetic changes that occur only in the tumor. For NGS of tumors, the potential advantage of germline DNA sequencing that is performed in parallel with the patient’s tumor is the ability to distinguish somatic mutations from germline variants.1 A major disadvantage is the potential for masking of germline variants of clinical significance (eg, BRCA1 and BRCA2 mutations), because most filtering algorithms simply discard all variants that are present in both germline and tumor DNA without considering whether it may be a germline deleterious mutation. Furthermore, germline testing requires additional sequencing and bioinformatics, which can ultimately increase time and cost. In addition, an underappreciated consideration is the source of germline DNA used for analysis. This report presents an unusual case that highlights potential misinterpretations that can occur when performing NGS on tumor tissues, and the inaccurate information that can result from using inappropriate tissues as the source of germline DNA.
Case Report
A 66-year-old woman experienced 2 weeks of abdominal pain and then noted dark urine, prompting evaluation. Blood work revealed an elevated lipase level, and CT scan results showed an “apple core” lesion in her duodenum. Upper endoscopy demonstrated a stricture at the duodenum, and bloodwork was notable for elevated levels of total bilirubin (4.5 mg/dL), alkaline phosphatase (443 U/L), aspartate transaminase (388 U/L), and alanine transaminase (675 U/L). Her blood counts were within normal limits. A pancreatic CT showed a circumferential mass around the duodenum. She underwent a pancreaticoduodenectomy (Whipple) procedure, with pathology showing a 3.0-cm moderately to poorly differentiated duodenal adenocarcinoma. Of 28 lymph nodes, 6 were positive for disease. The patient initiated adjuvant chemotherapy with folinic acid, 5-fluorouracil, and oxaliplatin (FOLFOX); however, recurrence in her liver was discovered during her adjuvant treatment. Due to side effects, her therapy was changed to capecitabine. Her disease progressed and she was placed on folinic acid, 5-fluorouracil and irinotecan (FOLFIRI). She continued to have progressive disease and was then started on cetuximab; however, her disease worsened. Her primary duodenal cancer was subjected to NGS using a cancer gene panel (FoundationOne; Foundation Medicine, Inc., Cambridge, MA) (see supplemental eAppendix 1, available with this article at JNCCN.org). The Genetic Alterations in Tumors With Actionable Yields (GAITWAY) tumor board at Johns Hopkins received the results and discussed potential mutations that could be targeted for therapy.
The patient’s NGS results contained CCND3, CDK4, ERBB3, VEGFA, GLI1, and MDM2 amplifications and SMAD4 E538* and JAK2 V617F mutations. OF these alterations, the JAK2 mutation was noted to be potentially actionable given that this mutation is found in the vast majority of patients with polycythemia vera (PV) and is predictive of response to the targeted kinase inhibitor ruxolitinib.2 After con erring with the re erring physician, it was discovered that the patient had a history of JAK2-positive PV treated for many years exclusively with phlebotomy. At this point, we reasoned that the JAK2 mutation was likely blood contamination of the duodenal tissue. However, there was still the remote possibility that this was instead a heritable, germline mutation, which cannot be differentiated from somatic mutations when only tumor tissue is sequenced. Although the JAK2 V617F mutation has never been reported in the germline of patients with sporadic or hereditary PV, there have been reports of other JAK2 mutations, including V617I, in the germline of patients with the related myeloproliferative disorder essential thrombocytosis.3–5 There ore, we wanted to definitively rule this out, because it would have implications for treatment and genetic counseling. Because blood could not be used in this instance as a source of germline DNA, we performed Sanger sequencing using DNA extracted from a buccal swab. Surprisingly, this analysis showed an approximately 50% allelic fraction, suggesting the mutation was potentially germline (Figure 1). Although saliva and buccal swab samples can contain contaminating hematopoietic cells,6 we had reasoned that DNA from blood cells would contribute only a minor allelic fraction that would be below the limit of detection on standard Sanger sequencing.7 To address this further, we obtained the primary tumor and adjacent normal tissues and repeated NGS testing. The JAK2 V617F mutation was present in both tumor and normal samples, but at low allelic frequencies of 13% and 4%, respectively (Figure 2A). This strongly suggested that the mutation arose from the blood cells and was not germline, because germline mutations would be expected to have an allelic frequency close to 50%.
Figure 1.
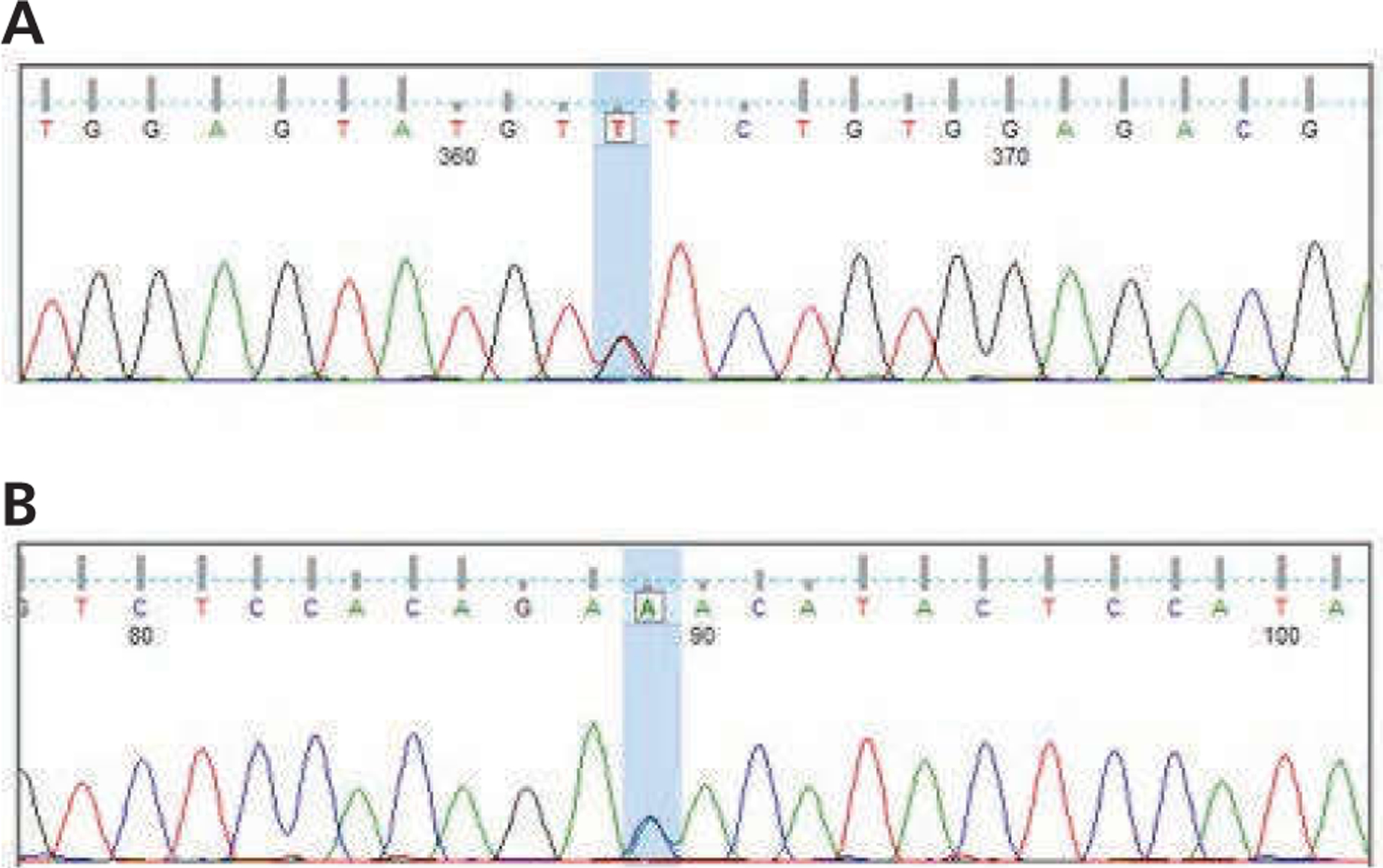
Genomic DNA isolated from buccal swab demonstrates heterozygosity for the JAK2 V617F mutation. DNA was isolated and subjected to polymerase chain reaction amplification and Sanger sequencing. Shown are results from capillary electrophoretic sequencing in (A) forward and (B) reverse directions. The V617 corresponding DNA locus is shown with 2 peaks highlighted in blue, representing the bases G (wild-type) and T (mutant) in the forward direction and C (wild-type) and A (mutant) in the reverse direction.
Figure 2.

False-positive sequencing results due to blood contamination. (A) Schematic showing results from next-generation sequencing (NGS) of (1) the duodenal tumor and (2) normal adjacent tissue, and Sanger sequencing from (3) a buccal swab sample and (4) finger nail clippings. (B) Duodenal adenocarcinoma with blood vessels and hemorrhage (hematoxylin and eosin, original magnification x40). Normal duodenal mucosa (top) with underlying invasive adenocarcinoma (bottom). Note the intraglandular hemorrhage (arrow) and rich vasculature (arrowheads). Neutrophils are noted in the vessels (inset; original magnification x400).
To definitively prove the mutation was not germline, we obtained fingernail clippings as an alternative source of normal tissue. Sanger sequencing analysis demonstrated only wild-type sequence at the JAK2 V617 locus (Figure 3). Unfortunately, the patient’s disease continued to progress, and she ultimately was re erred to hospice care.
Figure 3.
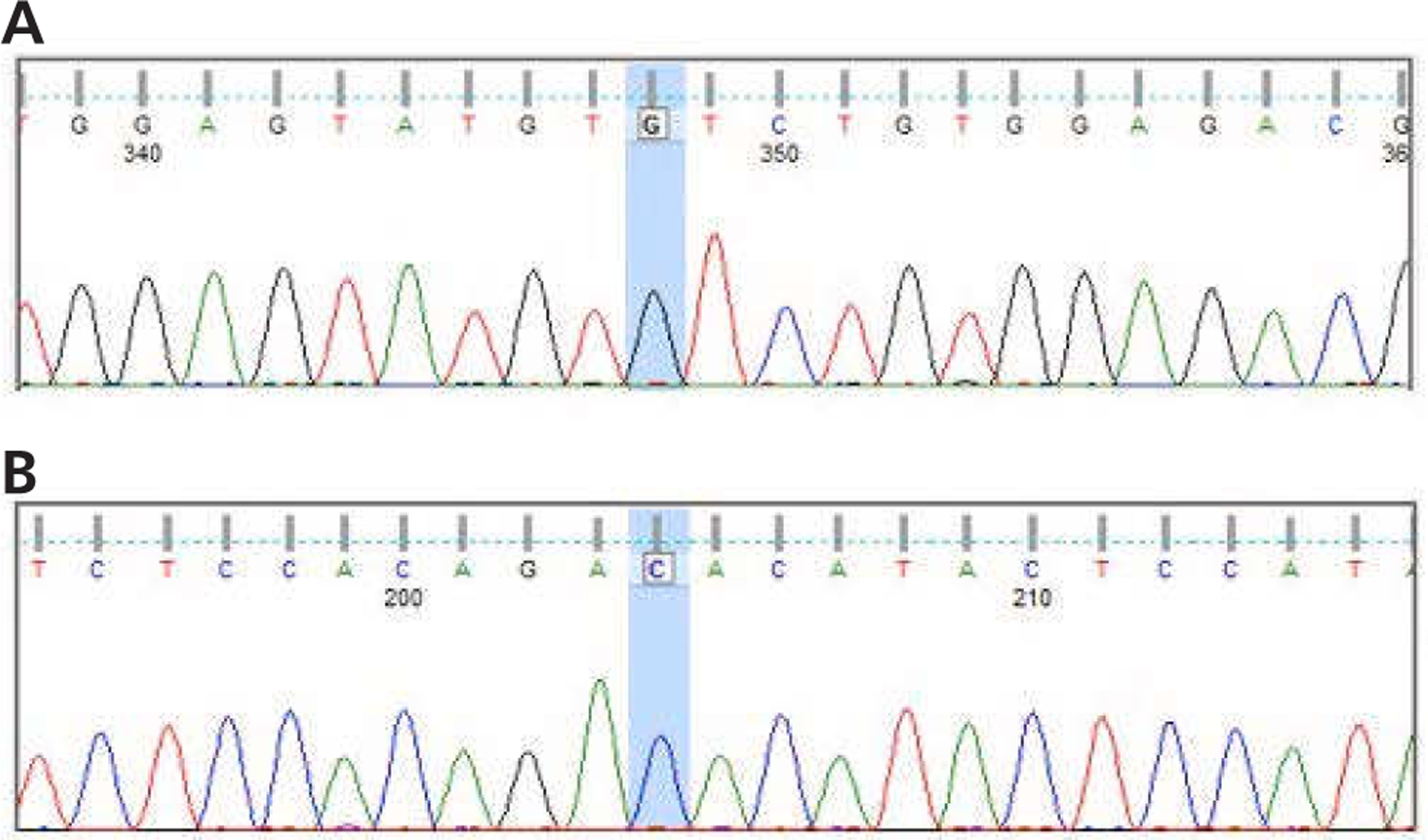
Genomic DNA isolated from fingernails show wild-type sequence for the JAK2 V617 locus. DNA was isolated and subjected to polymerase chain reaction amplification and Sanger sequencing. Shown are results from capillary electrophoretic sequencing in (A) forward and (B) reverse directions. The V617 corresponding DNA locus is highlighted in blue with only wild-type bases present in the forward and reverse direction.
Discussion
This case report highlights several considerations when evaluating tumor sequencing results. First, if germline DNA is being tested for comparative purposes, the source of germline DNA needs to be considered in the context of a patient’s clinical history and disease state. Peripheral blood lymphocytes, buccal swabs, and saliva are o ten used as sources of DNA for germline noncancer controls, but clearly cancer DNA can be overrepresented in these fluid/tissue sources when a patient has a hematologic cancer or myeloproliferative disorder. In this case report, it is likely that there was an extremely high level of blood cells present in the patient’s saliva leading to a 50% allelic fraction.
Second, this case illustrates the need for interpreting NGS results within the context of clinical history, as well as the knowledge of mutations that are strongly associated with a given cancer. Genes frequently mutated in hematologic disorders, including JAK2, IDH1, IDH2, SF3B1, KRAS, NRAS, and others, have also been found to be somatically mutated in a variety of solid tumors.8 Several of these genes are viewed as potential therapeutic targets or predictors of drug response or resistance. In this case, the re erring physician knew of the patient’s PV diagnosis. However, it is possible that for some solid tumors with reported mutations that are typically found in hematologic diseases, the mutation may originate from undiagnosed blood disorders that have contaminated tumor specimens used for NGS, as was the case for our patient (Figure 2B). Oncologists and molecular tumor boards reviewing NGS reports should be vigilant for this possibility.
Third, although tumor cellularity is o ten assessed when evaluating tissue specimens for NGS, there may be utility in reporting hematopoietic cellularity within the tissue specimens used for such analyses, especially when mutations in genes typically associated with hematopoietic disorders are identified. Along these lines, we advocate reporting of mutational allelic frequencies in NGS results, which can aid in interpretation of subclonal populations and contamination by blood cells.
Finally, we propose that fingernail clippings are a viable source of germline DNA and offer significant advantages over currently used tissue sources, including painless procurement, lack of blood contamination, minimal cost, and ease of obtainment. Although nail clippings are used routinely in forensic medicine, newer extraction techniques have allowed for higher-quality DNA that is now suitable for NGS analysis.9
Conclusions
In the era of precision oncology, there is great enthusiasm for matching targeted therapies to alterations obtained from tumor genomic analysis. This case report highlights the need for caution in evaluating these reports to prevent misdirected therapeutic decisions. Without a priori knowledge of the patient’s history of PV, one could easily envision that therapy with ruxolitinib may have been initiated for this patient, which likely would have had limited therapeutic efficacy. We strongly encourage the use of molecular tumor boards to aid in the interpretation and clinical recommendations derived from NGS and other molecular tests of patients’ cancers.
Supplementary Material
Acknowledgments
The authors would like to thank the entire GAITWAY tumor board for their input and assistance on this case.
Dr. Park is a paid member of the scientific advisory boards of Horizon Discovery, LTD and Loxo Oncology, has ownership interest in Loxo Oncology, and received research support from Genomic Health, Inc. and Foundation Medicine, Inc. Under separate licensing agreements between Horizon Discovery, LTD and The Johns Hopkins University, Dr. Park and Dr. Lauring are entitled to a share of royalties received by the University on sales of products. The terms of this arrangement are being managed by the Johns Hopkins University, in accordance with its conflict of interest policies. None of these conflicts pertains to this manuscript. The remaining authors have disclosed that they have no financial interests, arrangements, affiliations, or commercial interests with the manufacturers of any products discussed in this article or their competitors.
This work was supported in part by The Avon Foundation (B.H.P. and J.L.), NIH CA009071 (K.C. and B.H.P.), and the Sandy Garcia Charitable Foundation (D.C. and B.B.). Support was also received by NIH P30 CA006973, the Commonwealth Foundation, the Santa Fe Foundation, the Breast Cancer Research Foundation (B.H.P.), the ME Foundation, and the Augustine Fellowship (W.B.D.). None of the funding sources influenced the design, interpretation, or submission of this manuscript.
References
- 1.Jones S, Anagnostou V, Lytle K, et al. Personalized genomic analyses for cancer mutation discovery and interpretation. Sci Transl Med 2015;7:283ra53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verstovsek S, Kantarjian H, Mesa RA, et al. Safety and efficacy of INCB018424, a JAK1 and JAK2 inhibitor, in myelofibrosis. N Engl J Med 2010;363:1117–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Etheridge SL, Cosgrove ME, Sangkhae V, et al. A novel activating, germline JAK2 mutation, JAK2R564Q, causes familial essential thrombocytosis. Blood 2014;123:1059–1068. [DOI] [PubMed] [Google Scholar]
- 4.Marty C, Saint-Martin C, Pecquet C, et al. Germ-line JAK2 mutations in the kinase domain are responsible for hereditary thrombocytosis and are resistant to JAK2 and HSP90 inhibitors. Blood 2014;123:1372–1383. [DOI] [PubMed] [Google Scholar]
- 5.Mead AJ, Rugless MJ, Jacobsen SE, Schuh A. Germline JAK2 mutation in a family with hereditary thrombocytosis. N Engl J Med 2012;366:967–969. [DOI] [PubMed] [Google Scholar]
- 6.Thiede C, Prange-Krex G, Freiberg-Richter J, et al. Buccal swabs but not mouthwash samples can be used to obtain pretransplant DNA fingerprints from recipients of allogeneic bone marrow transplants. Bone Marrow Transplant 2000;25:575–577. [DOI] [PubMed] [Google Scholar]
- 7.Tsiatis AC, Norris-Kirby A, Rich RG, et al. Comparison of Sanger sequencing, pyrosequencing, and melting curve analysis for the detection of KRAS mutations: diagnostic and clinical implications. J Mol Diagn 2010;12:425–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raza A, Galili N. The genetic basis of phenotypic heterogeneity in myelodysplastic syndromes. Nat Rev Cancer 2012;12:849–859. [DOI] [PubMed] [Google Scholar]
- 9.Preuner S, Danzer M, Proll J, et al. High-quality DNA from fingernails for genetic analysis. J Mol Diagn 2014;16:459–466. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


