Abstract
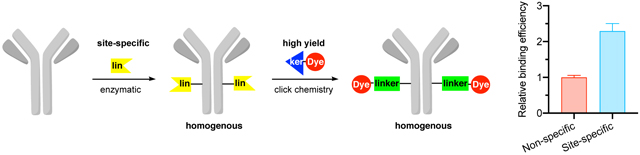
Photosensitizer (PS)–antibody conjugates (photoimmunoconjugates, PICs) enable cancer-cell targeted photodynamic therapy (PDT). Non-specific chemical bioconjugation is widely used to synthesize PICs but gives rise to several shortcomings. The conjugates are heterogenous, and the process is not easily reproducible. Moreover, modifications at or near the binding sites alter both binding affinity and specificity. To overcome these limitations, we introduce convergent assembly of PICs via a chemo-enzymatic site-specific approach. First, an antibody is conjugated to a clickable handle via site-specific modification of glutamine (Gln) residues catalyzed by transglutaminase (TGase, EC 2.3.2.13). Second, the modified antibody intermediate is conjugated to a compatible chromophore via click chemistry. Utilizing cetuximab, we compared this site-specific conjugation protocol to the non-specific chemical acylation of amines using N-hydroxysuccinimide (NHS) chemistry. Both the heavy and light chains were modified via the chemical route, whereas, only a glutamine 295 in the heavy chain was modified via chemo-enzymatic conjugation. Furthermore, a 2.3-fold increase in the number of bound antibodies per cell was observed for the site-specific compared to non-specific method, suggesting that multiple stochastic sites of modification perturbs the antibody-antigen binding. Altogether, site-specific bioconjugation leads to homogenous, reproducible, and well-defined PICs, conferring higher binding efficiency and probability of clinical success.
Keywords: antibody–drug conjugate (ADC), photosensitizer–antibody conjugate (PAC), photoimmunoconjugate (PIC), photodynamic therapy (PDT), photoimmunotherapy (PIT), transglutaminase (TGase), N-hydroxysuccinimide (NHS), click chemistry, convergent synthesis
Graphical Abstract Text:
Photosensitizer-antibody conjugates (PACs or PICs) enable targeted photodynamic therapy. Widely used non-specific chemical bioconjugation methods for PIC synthesis are limited by their heterogeneity, irreproducibility, and crucially, their adverse effect on antibody structure, binding affinity, and specificity. Alternatively, we employ a site-specific, enzymatic conjugation strategy which yields homogenous, reproducible, and well-characterized PICs with a higher binding efficiency compared to their nonspecific counterpart. The process is broadly applicable to a number of antibodies and chromophores (e.g., photosensitizers or fluorophores), thereby enabling rapid generation of diverse libraries of PICs with tailored functionality that retain the native biological activity of the antibody.
INTRODUCTION
Present clinical applications of photodynamic therapy (PDT) to treat cancer typically occur via systemic administration of unmodified photosensitizers (PSs). The efficacy of this treatment strategy is dependent on the selectivity of both the PS pharmacokinetics and the applied light, as only PS molecules activated by light initiate cytotoxic pathways (1, 2). Although effective in many cases, the maximum tolerated dose is limited by off-target toxicity in neighboring organs due to poor selectivity of PSs when applying diffuse light. Targeted approaches using PS–antibody conjugates (PACs) – commonly referred to as photoimmunoconjugates (PICs) – have steadily been developed to improve treatment selectivity (3–15). Activatable PICs—where the PS fluorescence and photocytotoxicity are quenched until target biomarker binding, cellular internalization, and lysosomal proteolysis—enable microscale tumor selectivity (11) and spare local effector immune cells (15).
Like most antibody–drug conjugates (ADCs), PICs target cell surface proteins frequently overexpressed by tumor cells relative to neighboring tissues and show clinical promise (e.g., clinical trial NCT03769506). The primary components in any PIC are a tumor-targeted antibody and a photodynamic chromophore (or chromophores). In most cases, a chemical linker (or linkers) is included to expedite coupling, although direct conjugation of chromophore to antibody is possible. There are a number of clinical antibodies that target a variety of cell-surface antigens for molecular targeted therapy. Biocompatible and clinically approved PSs are also commercially available, and many next-generation PSs are under preclinical and clinical development (14).
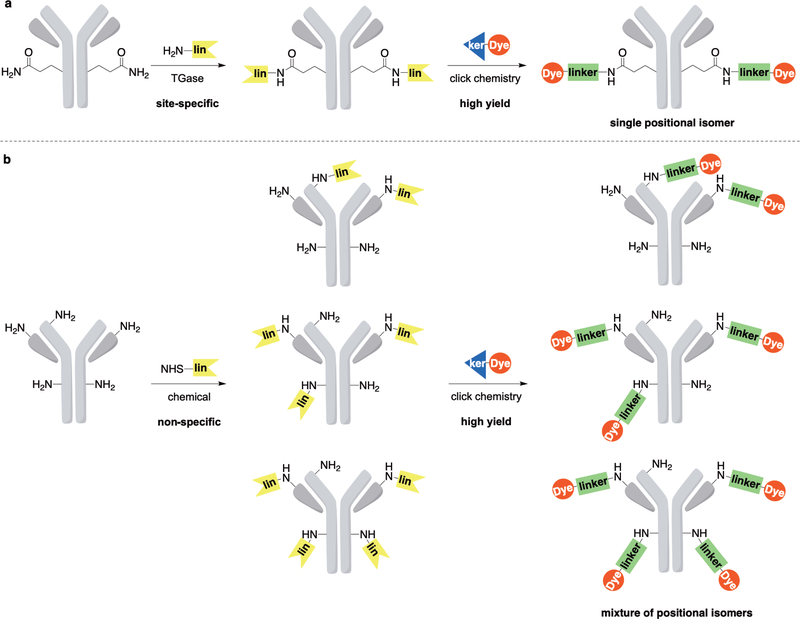
The key challenge to efficient and reliable PIC design and synthesis is the linker chemistry to covalently attach the PS to the antibody in a site-specific and homogeneous manner, without compromising the biological activity and specificity of the antibody. Specifically, a method that avoids modifications on the antigen-binding fragment (Fab) in favor of the crystallizable fragment (Fc) are desirable to limit interference with the Fab–receptor binding kinetics. Here, we introduce a simple and robust approach to facilitate site-specific, convergent (stepwise) PIC synthesis using a single-site modification method integrated with click chemistry (Figure 1a) that is compatible with many antibodies and PSs.
Figure 1.
Convergent assembly of antibody-chromophore conjugates in a stepwise fashion via click chemistry. The linker conjugation reaction may be designed from click chemistries like SPAAC, CuAAC, and tetrazine ligation. (a) Site-specific, transglutaminase (TGase)-catalyzed conjugation to glutamine 295 leads to homogeneous PICs, preserving binding specificity and affinity. (b) Non-specific chemical acylation of amines (i.e., lysine and N-termini) on the antibody yields heterogenous mixtures of PICs, with potential for reduction in binding specificity and affinity.
The new PIC synthesis approach introduces a clickable handle (i.e., strained alkyne) to the antibody via site-specific transglutaminase-mediated transamidation of glutamine. The modified antibody intermediate is then linked to the PS with another complementary clickable handle (i.e., azide) via click reaction. This convergent approach allows generation of reproducible conjugates that are easy to characterize and enables generation of libraries of PICs for high-throughput structure—activity relationship studies. In addition, we compared these constructs with the analogous PIC synthesized via non-specific chemical modification of amines followed by click-chemistry conjugation of the chromophore to the stochastically placed handle (Figure 1b). We demonstrate this new approach using anti-epidermal growth factor receptor (EGFR) antibody (cetuximab), and we evaluated the biological activity of these conjugates in three cancer cell lines with varying degrees of EGFR expression. The data indicate that the site-specific technology form well-defined constructs and preserve binding efficiency compared to their non-specific chemical counterpart. Overall, the system is modular, facile to characterize, and enables generation of large and diverse PIC libraries.
Site-specific enzymatic conjugation approaches achieve homogeneous protein modifications (16). Here we utilized microbial transglutaminase (mTGase, E.C. 2.3.2.13) to modify cetuximab at a single site, glutamine (Gln) 295 of the heavy chain (HC) in the Fc region, whereas the light chain (LC) is unmodified (17–19). mTGase catalyzes a transamidation reaction in which an unsubstituted amide in glutaminyl residues is converted into a substituted amide using an amine substrate (20–25). Commercially available and inexpensive, this enzyme has broad specificity towards the amine substrate (26) such as accepting azides (27), strained alkenes and alkynes (28), tetrazines (27), and even bulkier groups such as polyethylene glycol chains (PEG) (29). Therefore, a variety of amine substrates can be attached either in a one-step or stepwise fashion in a reliable and site-specific manner.
The final step in convergent PIC synthesis incorporated click chemistry, which enables biorthogonal, fast, and high yield reactions to prepare conjugate biomolecules (30–32). Remarkably, these features of click chemistry are being tested in humans to synthesize radioimmunoconjugates in vivo—the components are “clicked” together within the body (33). Click chemistry reactions are robust and have been reported to chemically attach various poryphyrin- (34–36) and boron dipyrromethene-class (37) PSs to antibodies. Additionally, diverse chemical moieties of chromophores (e.g., PSs or fluorophores) have been incorporated into antibodies including nanoparticles (38–40), multi-target imaging agents (41), broad wavelength (800 to 1700 nm) dyes (42), and dendrimers or branched polymers to load more PSs (36, 43). Common types of click reactions include copper azide-alkyne cycloadditions (CuAAC), strained promoted azide-alkyne cycloadditions (SPAAC), and tetrazine ligation (i.e., inverse electron-demand Diels–Alder, IEDDA) (44). Because copper may be toxic in vivo and lead to side reactions (e.g., oxidation) (44), SPAAC and IEDDA are preferable alternatives. In order to utilize click reactions, compatible clickable handles need to be conjugated to antibody and as well as the PS. This process has been achieved in proteins using both site specific (i.e., enzymatic) and non-specific (i.e., chemical) methods (45–47). Here, we applied SPAAC click chemistry to assemble the PIC.
In contrast to the methodology presented above, traditional chemical conjugation methods for PIC synthesis involve acylation of amines available on lysine residues and N-termini of protein, or alkylation of thiols on cysteine residues. These strategies have been employed to construct ADCs approved by the FDA (48). Nonetheless, chemically synthesized conjugates are not site-specific, heterogenous, and produce positional isomers in a stochastic distribution (i.e., different sites and ratios) (49). Moreover, some of these chemical reactions also suffer from side-reactions that are often underappreciated (45). The multiple modification sites—particularly at or near the binding sites like the complementarity determining region (CDR) in antibodies—may result in structural changes of the antibody and thus alterations in biological functions, including antigen binding capacity and specificity. For instance, Luo and coworkers have found almost all amines in an IgG (76 total, in both light and heavy chains) were acylated using N-hydroxysuccinimide (NHS) ester chemistry (50). Such reactions yield a mixture of conjugates with a widely distributed antibody loading ratio (ALR) ranging from 0 to 8, and within a given ALR, multiple positional isomers exist. From a practical perspective, since the process is kinetically controlled, considerable optimization is required to ensure reproducibility. Furthermore, new protocols must be re-optimized if either antibody or acylation reagent is changed. The protocols developed here overcome all of the challenges associated with chemical conjugation methods.
MATERIALS AND METHODS
The concentrations of the peptides and proteins were determined using UV absorption at 280 nm and extinction coefficients based on amino acid sequences. All aqueous solutions were prepared using Milli-Q water. All cell culture methods were performed with aseptic technique in a biosafety cabinet.
Deglycosylation of antibody using PNGase F.
The deglycosylation reaction contained 3.5 μM cetuximab (Selleckchem, A2000), 1X GlycoBuffer (New England Biolabs, B0701S), and was initiated with 100 units of PNGase F (New England Biolabs, P0708S) at 37°C and for 9.5 hours. To remove excess unreacted reagents, each reaction mixture was desalted using 50 kDa molecular weight cut-off (MWCO) centrifugal filters (Amicon unit, UFC505024) into 100 mM potassium phosphate (pH 7.6) prior to the transamidation reaction.
Transglutaminase-mediated transamidation reaction.
The transamidation reaction contained 100 mM potassium phosphate (pH 7.6), 5 mM dibenzylcyclooctyne-PEG4-amine (compound 1, Fig. 2; Click Chemistry Tools, A103P), 2 μM deglycosylated cetuximab aforementioned, and was initiated with 6.9 μM microbial transglutaminase (mTGase, Ajinomoto, ACTIVA-TI formulation, Uniprot P81453) and incubated at 37°C for 15.5 hours. To remove excess unreacted reagents, each reaction mixture was desalted using 50 kDa MWCO centrifugal filters into 100 mM potassium phosphate (pH 7.6) prior to the click reaction.
Figure 2.
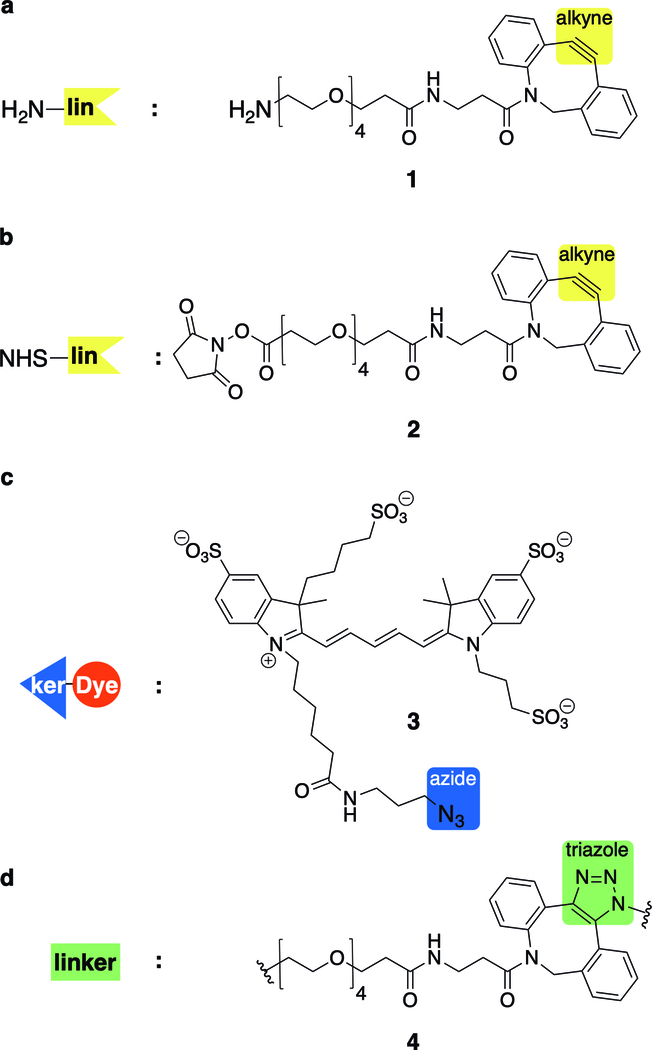
Structures of clickable handles for convergent assembly of PICs via SPAAC click chemistry. The antibody is conjugated through (a) site-specific TGase mediated glutamine transamidation using amine or (b) stochastic amine acylation using NHS ester, each tethered with a strained alkyne for subsequent SPAAC click reaction. (c) Each modified antibody (intermediate) is conjugated to AF647 tethered with a complementary azide clickable handle. (d) The cyclooctyne and azide click together to form a triazole in the final linker structure.
Amine reactive acylation reaction.
The reaction contained 100 mM potassium phosphate (pH 7.5), 155 μM dibenzylcyclooctyne-PEG4-N-hydroxysuccinimide (compound 2, Fig. 2, Broad Pharm, BP-22288), 2.7 μM cetuximab (without deglycosylation), and incubated at 22°C for 3.5 hours. The reaction was then quenched by 2 mM Tris (pH 8). To remove excess unreacted reagents, each reaction mixture was desalted using 30 kDa MWCO centrifugal filters (Amicon unit, UFC503096) into 100 mM potassium phosphate (pH 7.6) prior to the cycloaddition reaction.
Strain promoted cycloaddition click reaction (SPAAC).
The reaction contained 100 mM potassium phosphate (pH 7.5), 121 μM Alexa Fluor® 647 azide (AF647, compound 3, Fig. 2, Click Chemistry Tools, 1299), 1.5 μM cetuximab either modified by mTGase or acylated as aforementioned, and incubated at 37°C for 3.5 hours. To remove excess unreacted reagents, each reaction mixture was desalted using 30 kDa MWCO centrifugal filters into 100 mM potassium phosphate (pH 7.6) prior to analysis.
Determination of chromophore-antibody loading ratio.
The loading ratio was calculated based on the ratio of the absorption of the chromophore and antibody in similarity to prior literature (7, 11). Briefly, the absorbance spectra of PIC solutions were collected from 220 to 750 nm (NanoDrop spectrophotometer, ND-1000). The concentration of the chromophore AF647 was calculated using the absorbance of the conjugate at 650 nm and extinction coefficient of 270,000 M−1cm−1 provided by the vendor. The protein concentration was estimated by subtracting the chromophore AF647 contribution at 280 nm, and the calculated extinction coefficient based on its amino acid sequence (217,440 M−1cm−1). For the non-specific chemical acylation method, the chromophore and antibody concentration were 20.7 ± 0.3 μM and 4.90 ± 0.03 μM, respectively (a ratio of 4.22 ± 0.07). For the site-specific transglutaminase-mediated method, the chromophore and antibody concentration were 2.20 ± 0.10 μM and 2.5 ± 0.00 μM, respectively (a ratio of 0.88 ± 0.04). A summary of each construct used in this study is provided in Table 1.
Table 1.
Antibody chromophore conjugate characteristics. ALR: Antibody Loading Ratio.
| Site-specific | Non-specific | |
|---|---|---|
| Target (antibody) | EGFR (cetuximab) | |
| Chromophore (Ex/Em (nm)) | Alexa Fluor® 647 (648/671) | |
| Conjugation Method | Glutamine | Amine |
| Step 1 | Transglutaminase | NHS ester |
| Number of modification sites | 2 | 76 |
| Step 2 | Click Chemistry | |
| ALR (Chromophore:Antibody) | 0.88 ± 0.04 | 4.22 ± 0.07 |
Characterization of the conjugates by SDS-PAGE.
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was performed using a Bio-rad Mini-PROTEAN 3 system. First, the reaction mixture was incubated with SDS-Sample Buffer at 80°C for 10 minutes. For reducing and non-reducing gels, 4× reducing SDS sample buffer (Boston Bioproducts, BP-110R) and 2× nonreducing SDS sample buffer (Bio-rad, 1610737) were used, respectively. Second, the samples were loaded into 12% tris-tricine precast protein gels (Biorad, 4561044). Precision Plus Protein™ Dual Xtra Prestained Protein Standards (Bio-rad, 1610377) were used for mass calibration. Electrophoresis was then performed at 200 V for 20 min. The gel was stained by Coomassie R250 and then destained using 10% acetic acid and 40% methanol. The gels were imaged using an iBright FL1000 Imaging system (Thermo Fisher Scientific).
Characterization of conjugates by isoelectric focusing (IEF).
Isoelectric focusing was performed using a Bio-rad Criterion system. First, the reaction mixture was mixed in 1:1 dilution with IEF sample buffer (Bio-rad, 1610763). Second, the samples were loaded into pH 3–10 Criterion IEF precast gel and placed into the cell. Then, 1× Anode buffer (Biorad, 1610761) and 1× Cathode buffer (Biorad, 1610762) were placed in the upper and lower chamber of the criterion cell, respectively. Third, IEF was run initially at 100 V for 1 hour to initiate desalting of the sample, followed by higher voltage at 250 V for 1 hour to mobilize the antibody, and lastly 500V for 30 minutes to complete electrofocusing. The gel was stained by Coomassie R250 and Crocein Scarlet, and then destained using 10% acetic acid and 40% methanol. The gels were imaged using an iBright FL1000 Imaging system.
Cancer cell models.
Ovarian cancer cell line NIH:Ovcar-3 (Ovcar3, HTB-161) and breast cancer cell line T-47D (HTB-133) were purchased from the American Type Culture Collection and cultured in RPMI 1640 (Gibco, 11875093) completed with 10% heat-inactivated fetal bovine serum (FBS, Hyclone™ GE Healthcare Life Sciences, SH30071.03HI) and 1% penicillin/streptomycin antibiotic (Pen/Strep, Fisher BioReagents, BP295950). A human primary high-grade serous ovarian cancer line (Powder, Cellaria Biosciences) was cultured in Renaissance Essential Tumor Medium (RETM) and RETM Supplement (Cellaria Biosciences, CM-0001), plus 6.3% FBS and 1% mL Pen/Strep. Cells were grown in a humidified atmosphere at 5% CO2 and 37°C using T75 Flasks (Thermo Scientific, 1256685) according to their recommended culturing protocols. During passaging, cells were lifted with 0.25% trypsin EDTA cocktail (Corning, 25053CI), washed in phosphate buffered saline (PBS, Gibco, 70011069), and resuspended at 10,000 cells/mL. Cells were plated at 3,000 cells per well in a 96-well plate (Perkin Elmer, LLC 6055302) and allowed to grow for 3 days. Three wells were plated for each group.
Antibody binding assessment via flow cytometry.
Cells were plated in triplicate at 3,000 cells per well in a 96-well plate (Perkin Elmer, LLC 6055302) and allowed to grow for 3 days. Cells were lifted with trypsin, washed, and suspended in 50 μL PBS with 0.25 μg of Cet-AF647 per replicate and incubated in non-internalization conditions at 4°C for 30 minutes in the dark. The samples were washed once and resuspended in 300 μL of PBS for analysis via flow cytometry (ThermoFisher, Attune NxT). A 635 nm laser was used to measure the auto-fluorescence of the unstained control group; the PMT voltage in that channel was adjusted so that the mean AF signal was small (~10–50 arbitrary fluorescence units). The rest of the samples were analyzed with identical instrument settings at 500 μL/min flow rate. For each cell line, a gate was placed around the population of cell-sized objects based on forward- vs. side-scatter plot analysis. This population was then displayed as a histogram of fluorescence intensity from the fluorescence measurement channel. The median fluorescence intensity from each replicate was recorded as the measurement of the average brightness of the cells stained with each Cet-AF647 conjugate.
Antibody internalization assessment via confocal microscopy.
Cells were plated at 10,000 cells per well in a 96-well plate and allowed to grow for 24 hours. Wells were stained in-well with 1 μg of Cet-AF647 in 50 μL PBS at 37°C for 1 hour in the dark. After 45 minutes, 50 μL of Hoechst 33342 (Life Technologies, H3570) at 10 μg/mL was added to each well to bring the final staining concentration to 5 μg/mL. The plate was immediately returned to the incubator for the final 15 minutes. The wells were washed twice in PBS before imaging via laser scanning confocal microscope (Zeiss, LSM 800). A 640 nm excitation laser was used to excite AF647, and a 405 nm laser was used for the Hoechst nuclear stain. Images of the AF647 and Hoechst fluorescence were collected using identical laser power and PMT gain using a 10× magnification, 0.3 numerical aperture air objective.
RESULTS AND DIscussion
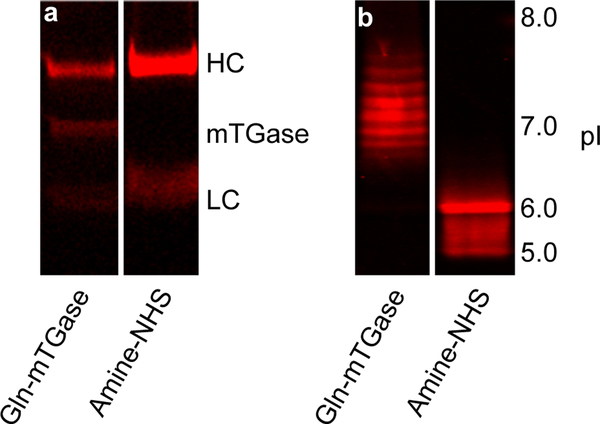
Both site-specific mTGase-mediated and non-specific amine-NHS ester processes successfully conjugated cetuximab with the fluorophore (Fig. 3 and Supporting Information Figures S1–S2). In terms of site-specificity, for the enzymatic method, only the HC was modified, which is consistent with modification at a single site, glutamine 295, as reported by several laboratories (18, 19). Whereas, for the chemical approach, both the HC and LC of the antibody were modified, also consistent with the 76 reactive lysine residues and N-termini amines (50). Interestingly, we observed a faint fluorescent band at 37 kDa indicating mTGase was self-modified (Fig. 3a). Such self-modification has been reported in mammalian and other bacterial TGases (17).
Figure 3.
Analysis of cetuximab–AF647 conjugates synthesized via either transglutaminase-mediated conjugation (Gln-mTGase) or amine-reactive NHS ester (Amine-NHS) chemistries, with (a) reducing SDS-PAGE and (b) IEF. Both fluorescence intensity and migration patterns by IEF indicated the degree of modification. Complete gels with controls are provided in Figures S1–S3. HC, Heavy Chain; LC, Light Chain; pI, isoelectric point.
Under the described reaction conditions, the chromophore–antibody loading ratio for the site-specific enzymatic and non-specific chemical methods were 0.9 and 4.2, respectively, as determined by ultraviolet-visible spectroscopy (Table 1). Further optimization of the enzymatic transamidation reaction is possible by increasing the reaction time, temperature, or enzyme concentration to achieve a maximum ALR of 2 as reported (17–19). The degree of modification is also consistent with the IEF results. Due to the four strong negatively charged sulfonates on AF647, the isoelectric point of the modified antibody via the site-specific method (pI 6.5–7.5) was lower than the unmodified antibody (pI 7.2–8, Fig. S3). Moreover, multiple degrees of antibody modification for the non-specific chemical route (pI 5–6, Fig. 3b) were observed compared to the site-specific approach. It is worth noting that the intrinsic charge heterogeneity of the unmodified antibody contributed to the multiple bands observed for both enzymatic and chemical conjugates. As is well documented in the literature, these antibody charge variants are due to various post-translational modifications, such as deamidation, oxidation, glycosylation, or other reactive metabolites (51–59).
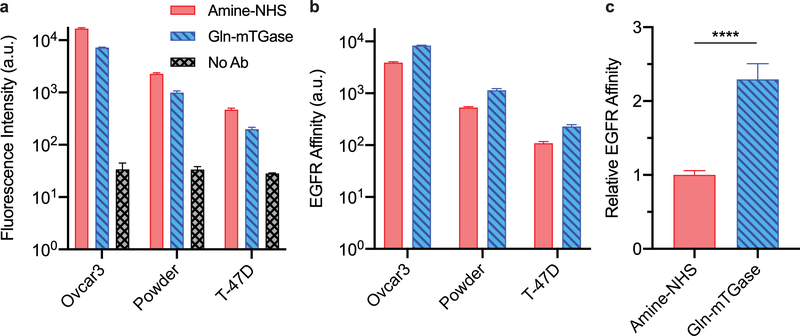
We sought to analyze the binding activity of each construct in three cancer cell lines with varying EGFR expressions: Ovcar3 (high EGFR), Powder (moderate EGFR), and T-47D (low EGFR). Under non-internalization conditions, cells stained with the amine-conjugated constructs were brighter than those stained with glutamine conjugates (Figures 4a and S4). This measurement is the product of both the number of antibodies bound per cell and the number of dyes per antibody. Normalizing by the ALR of each conjugate reveals a relative measurement of each conjugate’s binding capacity to EGFR (Figure 4b), which may then be compared across conjugates and cell lines. Under saturated staining conditions (more antibodies than receptors), this represents a relative measurement of the EGFR expression. The fold difference in relative affinity among cell lines as measured by each conjugate was consistent (p = 0.1116). For example, the 2.15-fold higher binding by the glutamine-conjugated constructs over the lysine-conjugated constructs to Ovcar3 cells was not statistically different from the 2.24-fold increase measured using Powder cells (p = 0.7700). Therefore, we combined the results from all 3 cell lines to assess the overall difference in binding efficiency. We found that the absolute expression of all cell lines measured by the glutamine-conjugated constructs was 2.29 ± 0.21-fold greater (p < 0.0001) than the measurement made by the amine-conjugated constructs (Figure 4c). Clearly, the natural binding properties of EGFR are degraded by non-specific conjugation strategies.
Figure 4.
Binding activity of cetuximab–AF647 conjugates. (a) 3 cell lines with high (Ovcar3), medium (Powder), and low (T-47D) EGFR expression were stained with cetuximab–AF647 conjugates prepared via non-specific chemical (Amine-NHS) or site-specific enzymatic (glutamine-microbial transglutaminase, Gln-mTGase) conjugation. The median fluorescence intensity per cell was recorded by flow cytometric analysis (n = 3 biological replicates, Figure S5). (b) Data from a normalized by ALR (Table 1) reveals the difference in relative EGFR expression of each cell line as measured by each conjugate is the same (p = 0.1116, one-way ANOVA). However, the absolute EFGR expression measured by amine-NHS conjugates is less than that measured by Gln-mTGase conjugates indicating that the site-specific Gln-mTGase chemistry better preserves the binding affinity of the antibody than the non-specific amine-NHS chemistry. (c) The fold-difference in EGFR expression measured by the two conjugates across 3 cell lines was averaged and normalized to the amine-NHS group. The result is a 2.29 ± 0.21-fold higher EGFR affinity (****p < 0.0001, unpaired, two-tailed student’s t-test) of the site-specific conjugates over the amine-NHS group.
The choice of ALR normalization is valid under two important assumptions. First, the surface protein expression should not vary between groups stained with each conjugate. This is achieved simply by culturing and handling cells under identical conditions, i.e., in the same flask or plate. Second, quenching effects arising from antibody conjugation should be negligible. In this case, AF647 conjugation onto the antibody corresponds with increasing fluorescence brightness between ALR ≈ 1–4 (60), so no correction for quenching effects is necessary for a first-order estimate. In general, chromophore quenching as a function of ALR is not predictable and varies by molecule. For example, conjugates of cetuximab and BPD exhibit quenching at ALR ≥ 2 (7). If this is the case, a correction based on the reduced quantum yield of the quenched construct should be included in the analysis scheme.
The biological activity of the antibody was also assessed via confocal fluorescence microscopy. The three aforementioned cell lines were stained under physiological conditions with each antibody conjugate and a nuclear stain (Figure 5). AF647 fluorescence was normalized to the dye-loading ratios of each conjugate. Signal was clearly dependent on the cellular EGFR expression. Fluorescence signal was apparent in the cytoplasm of Ovcar3 cells, indicating cellular uptake of each conjugate. This was also observed for the Powder line, albeit with minimal detectable signal. No signal was observed in the low EGFR-expressing T-47D cell line as expected. Unnormalized images are provided in Figure S5.
Figure 5.
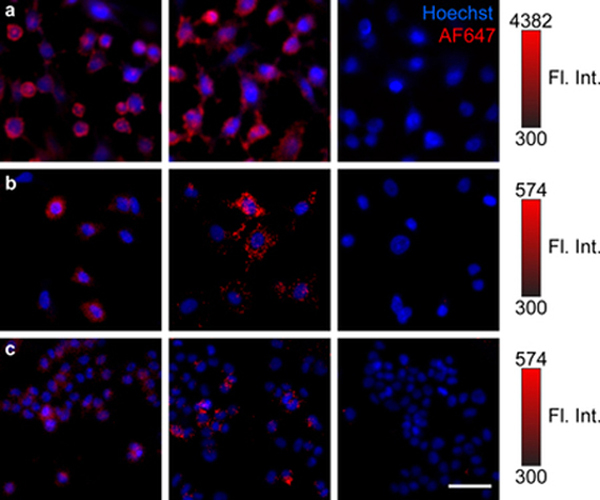
Laser-scanning confocal microscopy of (a) Ovcar3 (EGFR high), (b) Powder (EGFR medium), and (c) T-47D (EGFR (low) cancer cell lines. Cells were incubated with non-specific amine-NHS conjugated (left) or site-specific Glutamine (Gln)-mTGase conjugated (middle) cet–AF647 constructs, or no antibody (right). All cells were stained with Hoechst 33342 nuclear stain. AF647 fluorescence is normalized to the ALR of each conjugate and represents the amount of antibody uptake per cell. AF647 fluorescence intensity for each cell line is indicated via 16-bit color bar. Scale bar, 50 μm.
The data suggest that chemical acylation of amines results in conjugation of multiple copies of the chromophore that may alter the structure of the antibody and thus binding affinity and specificity; moreover, in extreme cases, complete abolishment of the binding activity. Such changes may also lead to nonspecific binding to other cell surface targets and thus off-target toxicity. This further emphasizes the need for site-specific technology to construct PICs.
In summary, there are several attributes for the convergent assembly of PICs in a chemo-enzymatic and site-specific manner that are worth noting. First, the method is widely applicable to other antibodies, particularly IgG1. This synthetic route allows control of the degree of modification and enables reproducible PICs that are easy to characterize. Second, the system is modular. Convergent assembly allows a variety of chromophores to be incorporated including photosensitizers or dyes. Similarly, a variety of IgGs can be evaluated, which enable rapid generation of diverse libraries of PICs for target validation with the ultimate goal of forming clinical candidates. Finally, to increase the payload further, multiple photosensitizers can be appended at a single site; for example using dendrimer (36) or branched polymer (43) technology. The facile, uniform, and site-specific PIC synthesis approach reported here may stimulate new and creative, next-generation multi-PS and bright fluorophore PICs for multi-organelle and hybrid fluorescence-guided surgery–PDT applications.
Supplementary Material
Figure S1. Antibody conjugate characterization: non-reducing SDS-PAGE.
Figure S2.Antibody conjugate characterization: reducing SDS-PAGE.
Figure S3.Antibody conjugate characterization: Isoelectric Focusing.
Figure S4.Antibody conjugate characterization: Flow Cytometry.
Figure S5.Antibody conjugate characterization: Confocal Microscopy.
AcknowledgEments:
This work was supported by the National Institutes of Health award K22CA181611 (to B.Q.S.) and the Richard and Susan Smith Family Foundation Award for Excellence in Biomedical Engineering (to B.Q.S.). The authors thank the Institute for Chemical Imaging of Living Systems at Northeastern University for assistance with confocal microscope image acquisition. A.S. thanks the support of Amgen Wolfgang Goetzinger Scholar Award in Life science Analysis and John Hatsopoulos Scholar Award, Professor Penny Beuning and Jim McIsaac for access to and assistance with the iBright FL1000 Imaging system, Emily Micheloni and the rest of SunnyLanders for helpful discussion.
Footnotes
DISCLOSURES
The authors declare no conflicts of interest.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of the article:
REFERENCES
- 1.Wilson B and Patterson M. (2008) The physics, biophysics and technology of photodynamic therapy. Phys Med Biol 53, R61–109. 10.1088/0031-9155/53/9/r01 . [DOI] [PubMed] [Google Scholar]
- 2.Kessel D. (2015) Apoptosis and associated phenomena as a determinants of the efficacy of photodynamic therapy. Photochem Photobiological Sci Official J European Photochem Assoc European Soc Photobiology 14, 1397–402. 10.1039/c4pp00413b . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mew D, Wat C, Towers G. and Levy J. (1983) Photoimmunotherapy: treatment of animal tumors with tumor-specific monoclonal antibody-hematoporphyrin conjugates. J Immunol Baltim Md 1950 130, 1473–7 [PubMed] [Google Scholar]
- 4.Goff Blake J, ., Bamberg M. and Hasan T (1996) Treatment of ovarian cancer with photodynamic therapy and immunoconjugates in a murine ovarian cancer model. Brit J Cancer 74, 1194–1198. 10.1038/bjc.1996.516 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duska LRR, Hamblin M, Miller JL. and Hasan T. (1999) Combination Photoimmunotherapy and Cisplatin: Effects on Human Ovarian Cancer Ex Vivo. Jnci J National Cancer Inst 91, 1557–1563. 10.1093/jnci/91.18.1557 . [DOI] [PubMed] [Google Scholar]
- 6.Molpus KL, Hamblin MR, Rizvi I. and Hasan T. (2000) Intraperitoneal Photoimmunotherapy of Ovarian Carcinoma Xenografts in Nude Mice Using Charged Photoimmunoconjugates. Gynecol Oncol 76, 397–404. 10.1006/gyno.1999.5705 . [DOI] [PubMed] [Google Scholar]
- 7.Savellano MD and Hasan T. (2003) Targeting Cells That Overexpress the Epidermal Growth Factor Receptor with Polyethylene Glycolated BPD Verteporfin Photosensitizer Immunoconjugates. Photochem Photobiol 77, 431 . [DOI] [PubMed] [Google Scholar]
- 8.Savellano MD and Hasan T. (2005) Photochemical Targeting of Epidermal Growth Factor Receptor: A Mechanistic Study. Clin Cancer Res 11, 1658–1668. 10.1158/1078-0432.ccr-04-1902 . [DOI] [PubMed] [Google Scholar]
- 9.Mitsunaga M, Ogawa M, Kosaka N, Rosenblum LT, Choyke PL and Kobayashi H. (2011) Cancer cell–selective in vivo near infrared photoimmunotherapy targeting specific membrane molecules. Nat Med 17, 1685–1691. 10.1038/nm.2554 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abu-Yousif A, Moor A, Zheng X, Savellano M, Yu W, Selbo P. and Hasan T. (2012) Epidermal growth factor receptor-targeted photosensitizer selectively inhibits EGFR signaling and induces targeted phototoxicity in ovarian cancer cells. Cancer Lett 321, 120–7. 10.1016/j.canlet.2012.01.014 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spring BQ, Abu-Yousif AO, Palanisami A, Rizvi I, Zheng X, Mai Z, ram Anbil, Sears BR, Mensah LB, Goldschmidt R, Erdem SS, Oliva E. and Hasan T. (2014) Selective treatment and monitoring of disseminated cancer micrometastases in vivo using dual-function, activatable immunoconjugates. P Natl Acad Sci Usa 111, E933–42. 10.1073/pnas.1319493111 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe R, Hanaoka H, Sato K, Nagaya T, Harada T, Mitsunaga M, Kim I, Paik CH, Wu AM, Choyke PL and Kobayashi H. (2014) Photoimmunotherapy targeting prostate-specific membrane antigen: are antibody fragments as effective as antibodies? J Nucl Medicine Official Publ Soc Nucl Medicine 56, 140–4. 10.2967/jnumed.114.149526 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Obaid G, Spring BQ, Bano S. and Hasan T. (2017) Activatable clinical fluorophore-quencher antibody pairs as dual molecular probes for the enhanced specificity of image-guided surgery. J Biomed Opt 22, 1 10.1117/1.jbo.22.12.121607 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lan M, Zhao S, Liu W, Lee C, Zhang W. and Wang P. (2019) Photosensitizers for Photodynamic Therapy. Adv Healthc Mater 1900132. 10.1002/adhm.201900132 . [DOI] [PubMed] [Google Scholar]
- 15.Kercher EM, Nath S, Rizvi I. and Spring BQ (2019) Cancer Cell-targeted and Activatable Photoimmunotherapy Spares T Cells in a 3D Co-culture Model. Photochem Photobiol. 10.1111/php.13153 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Park K-Y, Suazo KF and Distefano MD (2018) Recent progress in enzymatic protein labelling techniques and their applications. Chem Soc Rev 47, 9106–9136. 10.1039/c8cs00537k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mindt TL, Jungi V, Wyss S, Friedli A, Pla G, Novak-Hofer I, Grünberg J. and Schibli R. (2008) Modification of Different IgG1 Antibodies via Glutamine and Lysine using Bacterial and Human Tissue Transglutaminase. Bioconjugate Chem 19, 271–278. 10.1021/bc700306n . [DOI] [PubMed] [Google Scholar]
- 18.Jeger S, Zimmermann K, Blanc A, Grünberg J, Honer M, Hunziker P, Struthers H. and Schibli R. (2010) Site-specific and stoichiometric modification of antibodies by bacterial transglutaminase. Angewandte Chemie Int Ed Engl 49, 9995–7. 10.1002/anie.201004243 . [DOI] [PubMed] [Google Scholar]
- 19.Dennler P, Chiotellis A, Fischer E, Brégeon D, Belmant C, Gauthier L, Lhospice F, Romagne F. and Schibli R. (2014) Transglutaminase-Based Chemo-Enzymatic Conjugation Approach Yields Homogeneous Antibody–Drug Conjugates. Bioconjugate Chem 25, 569–578. 10.1021/bc400574z . [DOI] [PubMed] [Google Scholar]
- 20.Folk J. and Cole P. (1966) Mechanism of action of guinea pig liver transglutaminase. I. Purification and properties of the enzyme: identification of a functional cysteine essential for activity. J Biological Chem 241, 5518–25. [PubMed] [Google Scholar]
- 21.Folk J. (1980) Transglutaminases. Annu Rev Bio chem 49, 517–531. 10.1146/annurev.bi.49.070180.002505 . [DOI] [PubMed] [Google Scholar]
- 22.Ando H, Adachi M, Umeda K, Matsuura A, Nonaka M, Uchio R, Tanaka H. and Motoki M. (1989) Purification and Characteristics of a Novel Transglutaminase Derived from Microorganisms. Agr Biol Chem Tokyo 53, 2613–2617. 10.1080/00021369.1989.10869735 . [DOI] [Google Scholar]
- 23.Strop P. (2014) Versatility of microbial transglutaminase. Bioconjugate Chem 25, 855–62. 10.1021/bc500099v . [DOI] [PubMed] [Google Scholar]
- 24.Stefanetti G, Hu Q-Y, Usera A, Robinson Z, Allan M, Singh A, Imase H, Cobb J, Zhai H, Quinn D, Lei M, Saul A, Adamo R, MacLennan CA and Micoli F. (2015) Sugar-Protein Connectivity Impacts on the Immunogenicity of Site-Selective Salmonella O-Antigen Glycoconjugate Vaccines. Angewandte Chemie Int Ed Engl 54, 13198–203. 10.1002/anie.201506112 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moulton KR, Sadiki A, Koleva BN, Ombelets LJ, Tran TH, Liu S, Wang B, Chen H, Micheloni E, Beuning PJ, O’Doherty GA and Zhou Z. (2019) Site-Specific Reversible Protein and Peptide Modification: Transglutaminase-Catalyzed Glutamine Conjugation and Bioorthogonal Light-Mediated Removal. Bioconjugate Chem 30, 1617–1621. 10.1021/acs.bioconjchem.9b00145 . [DOI] [PubMed] [Google Scholar]
- 26.Gundersen MT, Keillor JW and Pelletier JN (2013) Microbial transglutaminase displays broad acyl-acceptor substrate specificity. Appl Microbiol Biot 98, 219–230. 10.1007/s00253-013-4886-x . [DOI] [PubMed] [Google Scholar]
- 27.Walker JA, Bohn JJ, Ledesma F, Sorkin MR, Kabaria SR, Thornlow DN and Alabi CA (2019) Substrate Design Enables Heterobifunctional, Dual “Click” Antibody Modification via Microbial Transglutaminase. Bioconjugate Chem 30, 2452–2457. 10.1021/acs.bioconjchem.9b00522 [DOI] [PubMed] [Google Scholar]
- 28.Rachel NM, Toulouse JL and Pelletier JN (2017) Transglutaminase-Catalyzed Bioconjugation Using One-Pot Metal-Free Bioorthogonal Chemistry. Bioconjugate Chem 28, 2518–2523. 10.1021/acs.bioconjchem.7b00509 [DOI] [PubMed] [Google Scholar]
- 29.Mero A, Spolaore B, Veronese FM and Fontana A. (2009) Transglutaminase-mediated PEGylation of proteins: direct identification of the sites of protein modification by mass spectrometry using a novel monodisperse PEG. Bioconjugate Chem 20, 384–9. 10.1021/bc800427n . [DOI] [PubMed] [Google Scholar]
- 30.Kolb C, Finn M. and Sharpless BK (2001) Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angewandte Chemie Int Ed 40, 2004–2021. . [DOI] [PubMed] [Google Scholar]
- 31.Rostovtsev VV, Green LG, Fokin VV and Sharpless BK (2002) A Stepwise Huisgen Cycloaddition Process: Copper(I)-Catalyzed Regioselective “Ligation” of Azides and Terminal Alkynes. Angewandte Chemie Int Ed 41, 2596–2599. . [DOI] [PubMed] [Google Scholar]
- 32.Agard NJ, Prescher JA and Bertozzi CR (2004) A Strain-Promoted [3 + 2] Azide−Alkyne Cycloaddition for Covalent Modification of Biomolecules in Living Systems. J Am Chem Soc 126, 15046–15047. 10.1021/ja044996f . [DOI] [PubMed] [Google Scholar]
- 33.Peplow M. (2019) Click chemistry targets antibody-drug conjugates for the clinic. Nat Biotechnol. 10.1038/d41587-019-00017-4 . [DOI] [PubMed] [Google Scholar]
- 34.Bryden F, Maruani A, Savoie H, Chudasama V, Smith ME, Caddick S. and Boyle RW (2014) Regioselective and stoichiometrically controlled conjugation of photodynamic sensitizers to a HER2 targeting antibody fragment. Bioconjugate Chem 25, 611–7. 10.1021/bc5000324 . [DOI] [PubMed] [Google Scholar]
- 35.Maruani A, Savoie H, Bryden F, Caddick S, Boyle R. and Chudasama V. (2015) Site-selective multi-porphyrin attachment enables the formation of a next-generation antibody-based photodynamic therapeutic. Chem Commun 51, 15304–15307. 10.1039/c5cc06985h . [DOI] [PubMed] [Google Scholar]
- 36.Bryden F, Maruani A, Rodrigues JM, Cheng MH, Savoie H, Beeby A, Chudasama V. and Boyle RW (2017) Assembly of High-Potency Photosensitizer-Antibody Conjugates through Application of Dendron Multiplier Technology. Bioconjugate Chem 29, 176–181. 10.1021/acs.bioconjchem.7b00678 . [DOI] [PubMed] [Google Scholar]
- 37.Cheng MH, Maruani A, Savoie H, Chudasama V. and Boyle RW (2018) Synthesis of a novel HER2 targeted aza-BODIPY–antibody conjugate: synthesis, photophysical characterisation and in vitro evaluation. Org Biomol Chem 16, 1144–1149. 10.1039/c7ob02957h . [DOI] [PubMed] [Google Scholar]
- 38.Huang H-C, Pigula M, Fang Y. and Hasan T. (2018) Immobilization of Photo-Immunoconjugates on Nanoparticles Leads to Enhanced Light-Activated Biological Effects. Small 14, 1800236. 10.1002/smll.201800236 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Obaid G, Jin W, Bano S, Kessel D. and Hasan T. (2019) Nanolipid Formulations of Benzoporphyrin Derivative: Exploring the Dependence of Nanoconstruct Photophysics and Photochemistry on Their Therapeutic Index in Ovarian Cancer Cells. Photochem Photobiol 95, 364–377. 10.1111/php.13002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Obaid G, Bano S, valleesha Mallidi, Broekgaarden M, Kuriakose, Silber Z, Bulin A-L., Wang., Mai, Jin., Simeone. and Hasan T. (2019) Impacting Pancreatic Cancer Therapy in Heterotypic in Vitro Organoids and in Vivo Tumors with Specificity-Tuned,NIR-Activable Photoimmunonanoconjugates: Towards Conquering Desmoplasia? Nano Lett 19, 7573–7587. 10.1021/acs.nanolett.9b00859 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karver MR, Weissleder R. and Hilderbrand SA (2011) Bioorthogonal Reaction Pairs Enable Simultaneous, Selective, Multi-Target Imaging. Angewandte Chemie Int Ed 51, 920–922. 10.1002/anie.201104389 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu S, Yang Q, Antaris AL, Yue J, Ma Z, Wang H, Huang W, Wan H, Wang J, Diao S, Zhang B, Li X, Zhong Y, Yu K, Hong G, Luo J, Liang Y. and Dai H. (2017) Molecular imaging of biological systems with a clickable dye in the broad 800- to 1,700-nm near-infrared window. Proc National Acad Sci 114, 962–967. 10.1073/pnas.1617990114 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yurkovetskiy AV, Yin M, Bodyak N, Stevenson CA, Thomas JD, Hammond CE, Qin L, Zhu B, Gumerov DR, Ter-Ovanesyan E, Uttard A. and Lowinger TB (2015) A Polymer-Based Antibody-Vinca Drug Conjugate Platform: Characterization and Preclinical Efficacy. Cancer Res 75, 3365–3372. 10.1158/0008-5472.can-15-0129 . [DOI] [PubMed] [Google Scholar]
- 44.Kim E. and Koo H. (2019) Biomedical applications of copper-free click chemistry: in vitro, in vivo, and ex vivo. Chem Sci 10, 7835–7851. 10.1039/c9sc03368h . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang L, Chumsae C, Kaplan JB, Moulton K, Wang D, Lee DH and Zhou Z. (2017) Detection of Alkynes via Click Chemistry with A Brominated Coumarin Azide by Simultaneous Fluorescence and Isotopic Signatures in Mass Spectrometry. Bioconjugate Chem 28, 2302–2309. 10.1021/acs.bioconjchem.7b00354 . [DOI] [PubMed] [Google Scholar]
- 46.Yamada K. and Ito Y. (2019) Recent Chemical Approaches for Site‐Specific Conjugation of Native Antibodies: Technologies toward Next‐Generation Antibody–Drug Conjugates. Chembiochem 20, 2729–2737. 10.1002/cbic.201900178 . [DOI] [PubMed] [Google Scholar]
- 47.Sandland J. and Boyle RW (2019) Photosensitizer Antibody-Drug Conjugates: Past, Present, and Future. Bioconjugate Chem 30, 975–993. 10.1021/acs.bioconjchem.9b00055 . [DOI] [PubMed] [Google Scholar]
- 48.Carter PJ and Lazar GA (2017) Next generation antibody drugs: pursuit of the ‘high-hanging fruit’. Nat Rev Drug Discov 17, 197–223. 10.1038/nrd.2017.227 . [DOI] [PubMed] [Google Scholar]
- 49.Wang L, Amphlett G, Blättler WA, Lambert JM and Zhang W. (2005) Structural characterization of the maytansinoid-monoclonal antibody immunoconjugate, huN901-DM1, by mass spectrometry. Protein Sci 14, 2436–2446. 10.1110/ps.051478705 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luo Q, Chung H, Borths C, Janson M, Wen J, Joubert MK and Wypych J. (2016) Structural Characterization of a Monoclonal Antibody-Maytansinoid Immunoconjugate. Anal Chem 88, 695–702. 10.1021/acs.analchem.5b03709 . [DOI] [PubMed] [Google Scholar]
- 51.Manning M, Chou DK, Murphy BM, Payne RW and Katayama DS (2010) Stability of protein pharmaceuticals: an update. Pharmaceut Res 27, 544–75. 10.1007/s11095-009-0045-6 . [DOI] [PubMed] [Google Scholar]
- 52.Liu M, Cheetham J, Cauchon N, Ostovic J, Ni W, Ren D. and Zhou Z. (2011) Protein isoaspartate methyltransferase-mediated 18O-labeling of isoaspartic acid for mass spectrometry analysis. Anal Chem 84, 1056–62. 10.1021/ac202652z . [DOI] [PubMed] [Google Scholar]
- 53.Manuilov AV, Radziejewski CH and Lee DH (2011) Comparability analysis of protein therapeutics by bottom-up LC-MS with stable isotope-tagged reference standards. Mabs 3, 387–95. 10.4161/mabs.3.4.16237 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu M, Zhang Z, Zang T, Spahr C, Cheetham J, Ren D. and Zhou Z. (2013) Discovery of undefined protein cross-linking chemistry: a comprehensive methodology utilizing 18O-labeling and mass spectrometry. Anal Chem 85, 5900–8. 10.1021/ac400666p . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chumsae C, Gifford K, Lian W, Liu H, Radziejewski CH and Zhou Z. (2013) Arginine modifications by methylglyoxal: discovery in a recombinant monoclonal antibody and contribution to acidic species. Anal Chem 85, 11401–9. 10.1021/ac402384y . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chumsae C, Zhou L, Shen Y, Wohlgemuth J, Fung E, Burton R, Radziejewski C. and Zhou Z. (2014) Discovery of a chemical modification by citric acid in a recombinant monoclonal antibody. Anal Chem 86, 8932–6. 10.1021/ac502179m . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu M, Zhang Z, Cheetham J, Ren D. and Zhou Z. (2014) Discovery and characterization of a photo-oxidative histidine-histidine cross-link in IgG1 antibody utilizing 18O-labeling and mass spectrometry. Anal Chem 86, 4940–8. 10.1021/ac500334k . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Klaene JJ, Ni W, Alfaro JF and Zhou Z. (2014) Detection and quantitation of succinimide in intact protein via hydrazine trapping and chemical derivatization. J Pharm Sci 103, 3033–42. 10.1002/jps.24074 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chumsae C, Hossler P, Raharimampionona H, Zhou Y, rmott S, Racicot C, Radziejewski C. and Zhou Z. (2015) When Good Intentions Go Awry: Modification of a Recombinant Monoclonal Antibody in Chemically Defined Cell Culture by Xylosone, an Oxidative Product of Ascorbic Acid. Anal Chem 87, 7529–7534. 10.1021/acs.analchem.5b00801 . [DOI] [PubMed] [Google Scholar]
- 60.Johnson I, Spence M. (2010). Molecular Probes Handbook: A Guide to Fluorescent Probes and Labeling Technologies. Eleventh Edition Life Technologies Corporation. ISBN: 9780982927915 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Antibody conjugate characterization: non-reducing SDS-PAGE.
Figure S2.Antibody conjugate characterization: reducing SDS-PAGE.
Figure S3.Antibody conjugate characterization: Isoelectric Focusing.
Figure S4.Antibody conjugate characterization: Flow Cytometry.
Figure S5.Antibody conjugate characterization: Confocal Microscopy.