Abstract
Background
SARS-CoV2, the agent responsible for the current pandemic, is also causing respiratory distress syndrome (RDS), hyperinflammation and high mortality. It is critical to dissect the pathogenetic mechanisms in order to reach a targeted therapeutic approach.
Methods
In the present investigation, we evaluated the effects of SARS-CoV2 on human bronchial epithelial cells (HBEC). We used RNA-seq datasets available online for identifying SARS-CoV2 potential genes target on human bronchial epithelial cells. RNA expression levels and potential cellular gene pathways have been analyzed. In order to identify possible common strategies among the main pandemic viruses, such as SARS-CoV2, SARS-CoV1, MERS-CoV, and H1N1, we carried out a hypergeometric test of the main genes transcribed in the cells of the respiratory tract exposed to these viruses.
Results
The analysis showed that two mechanisms are highly regulated in HBEC: the innate immunity recruitment and the disassembly of cilia and cytoskeletal structure. The granulocyte colony-stimulating factor (CSF3) and dynein heavy chain 7, axonemal (DNAH7) represented respectively the most upregulated and downregulated genes belonging to the two mechanisms highlighted above. Furthermore, the carcinoembryonic antigen-related cell adhesion molecule 7 (CEACAM7) that codifies for a surface protein is highly specific of SARS-CoV2 and not for SARS-CoV1, MERS-CoV, and H1N1, suggesting a potential role in viral entry. In order to identify potential new drugs, using a machine learning approach, we highlighted Flunisolide, Thalidomide, Lenalidomide, Desoximetasone, xylazine, and salmeterol as potential drugs against SARS-CoV2 infection.
Conclusions
Overall, lung involvement and RDS could be generated by the activation and down regulation of diverse gene pathway involving respiratory cilia and muscle contraction, apoptotic phenomena, matrix destructuration, collagen deposition, neutrophil and macrophages recruitment.
Keywords: Innate immunity, COVID-19, SARS-CoV2, Bioinformatics, CEACAM7, CSF3, DNAH7, c8orf4, Respiratory cilia
Highlights
-
•
SARS-CoV2 causing respiratory distress syndrome, hyperinflammation and high mortality.
-
•
In NHBEC, SARS-CoV2 highly regulated the innate immunity recruitment and the disassembly of cilia and cytoskeletal structure.
-
•
The granulocyte colony-stimulating factor (CSF3) is the most upregulated gene by SARS-CoV2.
-
•
The dynein heavy chain 7, axonemal (DNAH7) represented the most downregulated genes by SARS-CoV2.
-
•
Flunisolide, Thalidomide, Lenalidomide, Desoximetasone, xylazine, and salmeterol as potential drugs against SARS-CoV-2.
1. Introduction
SARS-CoV2, a novel, human-infecting beta-coronavirus, provisionally named 2019 novel coronavirus (2019-nCoV), was identified using of next-generation sequencing. This virus is now pandemic [1]. Most of the infected patients have a high fever and some have dyspnea, with chest radiographs revealing invasive lesions in both lungs, accompanied by hyperinflammation, respiratory distress syndrome and death [2]. SARS-CoV2 infection (COVID-19) is characterized by high mortality especially in older and fragile individuals. This virus induces not only a systemic inflammatory response but also a vigorous lung inflammation that may lead to a difficult-to-treat RDS and death [3]. Human bronchial epithelial cells and pneumocytes express ACE2, the receptor used by the virus to enter the cells and perpetuate its viral life cycle [4]. The infection of such cells ignites molecular mechanisms of defense (cell death and apoptosis) along with the recruitment of the immune system. Cell damage and repair mechanisms contribute to tissue restructuration. Furthermore, other anatomical, physiological and functional barriers (i.e. respiratory cilia) also contribute to viral clearance and tissue regeneration. Knowing the mechanism involved in the pathogenesis of coronavirus induced respiratory and systemic damage is essential to target therapeutic intervention.
In the present study, we investigated the effect of SARS-CoV2 on human bronchial epithelial cells (HBEC). We found that the upregulation and down-regulation of CSF3 and DNAH7, along with other genes overlapping the same paths, are the most modulated genes in NHBE cells infected with COVID-19. We hypothesized that respiratory cilia are impaired by SARS-CoV2 infection of epithelial cells, while apoptosis, matrix destructuration and collagen deposition do occur, potentially leading to respiratory distress syndrome (RDS). These pathogenetic factors could be critical to dissect future clinical and therapeutic interventions.
2. Materials and methods
2.1. Data selection
The aim of our study was to investigate the effect of SARS-CoV2 infection on the bronchial parenchyma. We hypothesized that SARS-CoV2 could modulate bronchial cells of COVID-19 patients at multiple anatomical and physiological levels and regulate the cytoskeletal structures. Furthermore, our hypothesis predicted that these changes were specific to COVID-19 infection and not common to other pandemic virus of airways such as SARS-CoV, MERS-CoV, and H1N1.
In order to test our hypothesis we have collected and analyzed several microarray datasets available on NCBI Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) [[5], [6], [7]]. Mesh terms “coronavirus”, “Human”, and “airway epithelial cells”, were used to identify human potential datasets of interest. Three datasets were selected (GSE147507, GSE47962, GSE81909) (Table 1 ).
Table 1.
Datasets selected.
| Dataset | Virus | Treatment | Exp. | Up | Down | Citation |
|---|---|---|---|---|---|---|
| GSE147507 | SARS-CoV2 | 2 MOI | 24 h | 40 | 56 | [8] |
| GSE47962 | SARS-CoV (icSARS) | 1 MOI | 24 h | 393 | 329 | [9] |
| GSE47962 | H1N1 | 1 MOI | 24 h | 5216 | 7258 | [9] |
| GSE81909 | MERS-CoV (icMERS) | 5 PFU x cells | 24 h | 7039 | 6838 | npaa |
npa: no publication available.
The GSE147507 dataset [8] was composed of Normal Human Bronchial Epithelial cells (NHBE) and transformed lung alveolar (A549) cells treated with MOCK or infected with SARS-CoV2 (USA-WA1/2020) at different MOI (NHBE: 2, A549: 0.2) for 24 h. The authors declared that cDNA libraries were sequenced using an Illumina NextSeq 500 platform (Illumina, CA), following differential expression analysis using DESeq2. As for our investigation, we focused only on the effect of SARS-CoV2 on NHBE cells. The submitter-supplied pre-preprocessed and normalized expression matrix was used for this re-analysis. The GSE147507 RawReadCounts7, subsequently was used for the identification of Differentially Expressed Genes (DEGs).
From GSE47962 we downloaded the data of Human bronchial airway epithelium (HAE) cells, seeded in 6-well plates (1 × 10e6 cells/well) two days prior to infection and then inoculated with wild type infectious clone derived SARS-CoV viruses (icSARS) (MOI = 2) or H1N1 (MOI = 1) [9]. Mock-infected controls were inoculated with culture medium only. For our analysis, we sorted only the data of 24 h treatment.
From GSE81909 we selected the transcriptome of primary human airway epithelial cells infected with a multiplicity of infection of 5 PFU per cell of wild type MERS-coronavirus (MERS-CoV) (icMERS). Furthermore, we sorted only the transcriptome corresponding to 24 h of treatment. Time-matched mocks were collected in parallel with infected samples.
Complete experimental details are available in referenced publications or in the GEODatabase correspondent webpages (Table 1).
2.2. Data processing and experimental design
In order to process and identify Significantly Different Expressed Genes (SDEG) in all selected datasets, we used the MultiExperiment Viewer (MeV) software (The Institute for Genomic Research (TIGR), J. Craig Venter Institute, USA). In cases where multiple genes probes insisted on the same GeneID, we used those with the highest variance. The significance threshold level for all data sets was p < 0.05. Statistically significant genes were selected for further analysis. For GSE47962 and GSE81909 datasets we performed a statistical analysis with GEO2R, applying a Benjamini & Hochberg FDR (False discovery rate) to adjust P values for multiple comparisons [10,11].
For the analysis of GSE147507, we used a conservative approach. We have set a filtering cutoffs of gene-level read counts ≥10, for differential gene expression analysis, in order to reduce artificial variance that results in differential expression or splicing calls, and then standardized the data with z-score transformation [12]. For the identification of the Differentially Expressed Genes (DEGs) in the HNBE cells, the LIMMA (Linear models for microarray data) (MeV) parametric test was used. An adjusted p-value < 0.05 was considered to indicate a statistically significant difference.
2.3. Gene ontology analysis
The genes Ontology analysis was performed using the web utility GeneMANIA (http://genemania.org/, http://genemania.org/) [13] and the GATHER (Gene Annotation Tool to Help Explain Relationships) (http://changlab.uth.tmc.edu/gather/) [14]. The GeneMania was also used to built the weighted gene networks commonly modulated. The database assembles all available interaction data in the dataset by creating large networks, which captures the current knowledge on the functional modularity and interconnectivity of genes in a cell. Gene's annotation was obtained by STRING software (https://string-db.org/). The STRING-combined score was based on data from neighborhood in the genome, gene fusions, co-occurrence across genomes, co-expression, experimental/biochemical data, and association in curated databases [15].
The genes overlapping was represented with Venn diagram using a public online tool (http://bioinformatics.psb.ugent.be/webtools/Venn/) and the images readapted for our data with CorelDraw.
The Gene set enrichment analysis (GSE) and gene ontology (GO), were expressed in weighted percentage and graphically rendered in a circular diagram format using freely available CIRCOS software (http://circos.ca/) [7,16,17]. Ribbon size encodes gene number and FDR associated row/column segments with high significance. CIRCOS can be applied to the exploration of data sets involving complex relationships between large numbers of factors. The FDR has been transformed for graphic representation purposes into 2^ (-log10FDR).
2.4. Drugs analysis prediction (DAP)
In order to identify potential novel pharmacological strategies for the treatment of SARS-CoV2 infection, we used The L1000fwd, Large Scale Visualization of Drug Induced Transcriptomic Signatures web-based utility [18]. L1000fwd calculates the similarity between an input gene expression signature and the LINCS-L1000 data, in order to rank drugs potentially able to reverse the transcriptional signature [18]. The L1000 transcriptomic database belonging to the Library of Integrated Network-based Cellular Signatures (LINCS) project, a NIH Common Fund program, that extended the Connectivity Map project and includes the large transcriptional profiles of approximately 50 human cell lines upon exposure to about 20,000 compounds, over a range of concentrations and time [18]. An adjusted p-value (q-value) of 0.05 has been considered as threshold for statistical significance. Combined score is calculated by multiplying the logarithm of the p-value from the Fisher exact test and the Z-score as a composite index:
2.5. Statistical analysis
For statistical analysis, Prism 8.0.2 software (GraphPad Software, USA) was used. Based on Shapiro-Wilk test, almost all data were normal, so parametric tests were used. Significant differences between groups were assessed using the Ordinary one-way ANOVA test, and Tukey's multiple comparisons test was performed to compare data between all groups. Correlations were determined using Pearson correlation. All tests were two-sided and significance was determined at P < 0.05. All MD selected were transformed for the analysis in Z-score intensity signal. Z score is constructed by taking the ratio of weighted mean difference and combined standard deviation according to Box and Tiao (1992) [19]. The application of a classical method of data normalization, z-score transformation, provides a way of standardizing data across a wide range of experiments and allows the comparison of microarray data independent of the original hybridization intensities. The z-score it is considered a reliable procedure for this type of analysis and can be considered a state-of-the-art methods, as demonstrated by the numerous bibliography [[20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31]].
Differential expression analysis was performed using the MeV 4.9 TM4 software, which used R v.2.11.1 and LIMMA v3.4.5.
Principal Component Analysis (PCA) was performed to evaluate the segregation of the genes according to the cells treatment. The PCA was performed with PAST-4 a free software for scientific data analysis (https://folk.uio.no/ohammer/past/), with functions for data manipulation, plotting, univariate and multivariate statistics, ecological analysis, time series and spatial analysis, morphometric and stratigraphy. The efficiency of each biomarker was assessed by the receiver operating characteristic (ROC) curve analyses. The area under the ROC curve (AUC) and its 95% confidence interval (95% CI) indicate diagnostic efficiency. The accuracy of the test with the percent error is reported [32].
3. Results
3.1. Extracellular matrix, collagen metabolism, actin cytoskeleton and muscle contraction are highly modulated by SARS-CoV2 infection
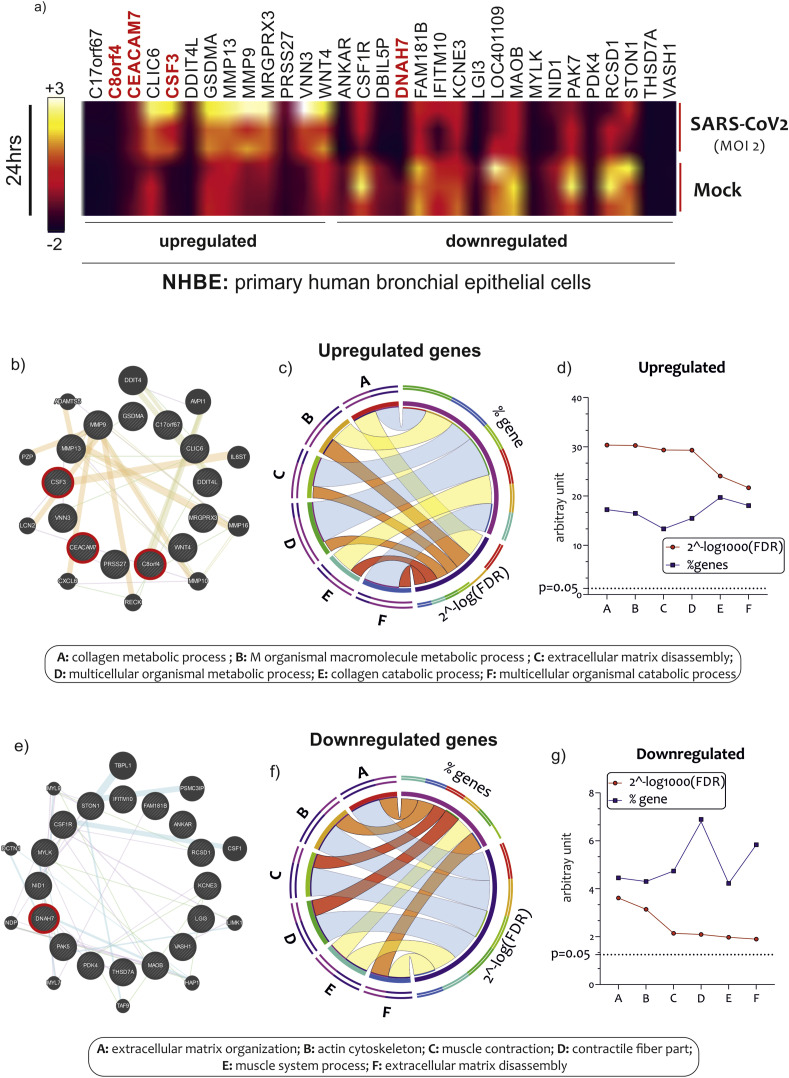
In order to identify a specific gene signature characterizing bronchial epithelial cells (NHBE) infected with SARS-CoV2 for 24 h, we first interrogated the GSE147507 dataset. We identified 105 DEGs in NHBE cells infected with SARS-CoV2 as compared to MOCK controls (40 upregulated and 56 downregulated genes) (Table S1). By carrying out a restrictive analysis (gene-level read counts ≥10, pvalue<0.01), we highlighted 13 upregulated and 18 downregulated genes (Fig. 1 a) (Table S1). Among these genes, we have excluded for future analysis the c17orf67, ANKAR, LOC401109, and DBIL5P genes, currently without a characterized function.
Fig. 1.
GSEA of NHBE infected by SARS-CoV2. Heatmap of most upregulated and downregulated genes in NHBE infected with SARS-CoV2 MOI 2, for 24 h. Highlighted in red bold the genes c8orf4, CEACAM7, CSF3, and DNAH7, highly and significantly modulated by the invention (a). GO analysis of 13 and 17 genes statistically significantly upregulated (b) and downregulated (c) in NHBE cells infected with SARS-CoV2. Among these genes, we have excluded for future analysis the c17orf67, ANKAR, LOC401109, and DBIL5P genes because they do not yet have characterized functions. Data are expressed as a string network (GeneMania), Circos graph and Line chart.
GO analysis revealed a partial overlapping of enriched biological processes among the upregulated DEGs in NHBE cells infected with SARS-CoV2, that included, among the first six significant one, the “collagen metabolic process”, “multicellular organismal macromolecule metabolic process”, “extracellular matrix disassembly”, “multicellular organismal metabolic process”, “collagen catabolic process”, and “multicellular organismal catabolic process” (Fig. 1b/c/d) (Table S2). As regard the downregulated DEGs in NHBE cells infected with SARS-CoV2, we highlighted several biological overlapped processes, among which the first six significant were “extracellular matrix organization”, “actin cytoskeleton”, “muscle contraction”, “contractile fiber part”, “muscle system process”, and “extracellular matrix disassembly” (Fig. 1e/f/g) (Table S2).
3.2. CSF3 upregulation and DNAH7 down-regulation may induce granulocytes, the monocytes-macrophages differentiation, and the reduction of function of respiratory cilia, respectively
The MeV Performed SDEGs analysis showed 12 genes high modulated in NHBE cells infected with SARS-CoV2 (RNA count >10 reads, p < 0.01). Among these genes, granulocyte colony-stimulating factor (CSF3) was the top up modulated. Its expression is linked to the production, differentiation, and function of two related white cell populations of the blood, the granulocytes, and the monocytes-macrophages. As regards the 17 most downregulated genes, dynein heavy chain 7, axonemal (DNAH7) was the top down-modulated. This gene produces force towards the minus ends of microtubules, and consequently generate the force of respiratory cilia.
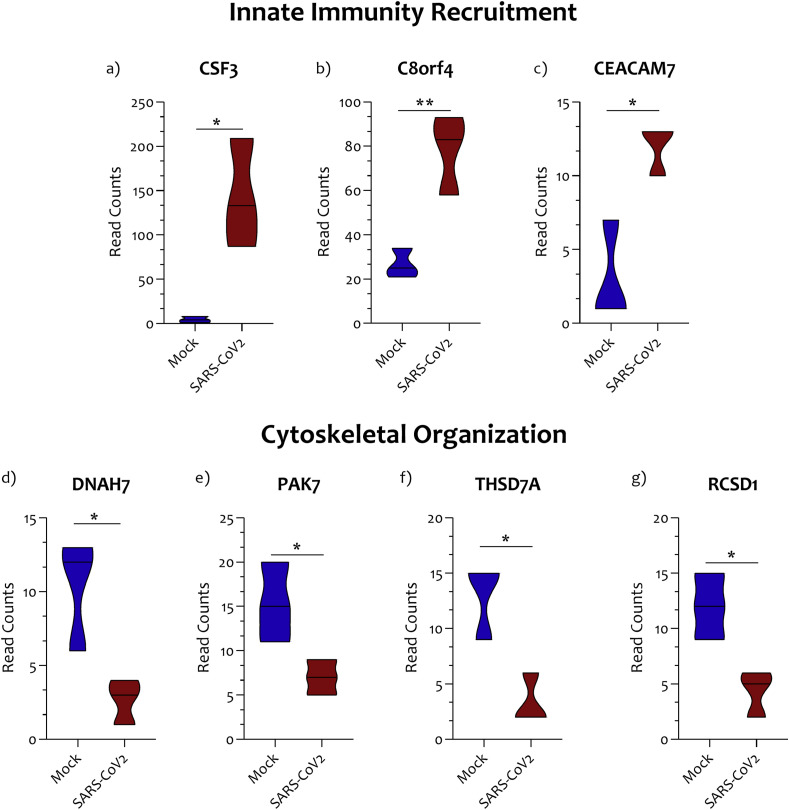
The 12 genes significantly upregulated in NHBE infected cells, has been segregated according to the gene functional characteristics, returned by STRING and GeneMania (Table S2). The groups obtained were heterogeneous. Several mechanisms appear to be related to the infection of SARS-CoV2 virus on NHBE cells. In particular, we have targeted our investigation on the cluster that included CSF3. This gene cluster has been identified as “innate immunity recruitment” group. Three genes out of 12 belong to this group, and were: the CSF3 (Fig. 2 a), the transcriptional and immune response regulator (c8orf4) (Fig. 2b), and the carcinoembryonic antigen-related cell adhesion molecule 7 (CEACAM7) (Fig. 2c).
Fig. 2.
Innate immunity recruitment and Cytoskeleton-related genes modulated in NHBE cells infected with SARS-CoV2. Three genes out of 12 belong to the “innate immunity recruitment” group, namely the CSF3(a), the transcriptional and immune response regulator (c8orf4) (b), and the carcinoembryonic antigen-related cell adhesion molecule 7 (c). Four gene out of 15 belong to the “mechanisms of cytoskeletal organization”, and were Dynein heavy chain 7 (DNAH7) (d), the P21 (RAC1) activated kinase 5 (PAK7) (e), the thrombospondin type-1 domain-containing protein 7A (THSD7A) (f), and the RCSD domain containing 1 (RCSD1) genes (g). Data are expressed as RNA Read count and presented as violin plots. P values < 0.05 were considered to be statistically significant (*p < 0.05).
When we segregated the 15-downregulated genes highlighted during NHBE cell infection, mechanisms were consistent with the GO analysis previously carried out. We selected a genes cluster composed by the Dynein heavy chain 7 (DNAH7) (Fig. 3 a), the P21 (RAC1) activated kinase 5 (PAK7) (Fig. 3b), the thrombospondin type-1 domain-containing protein 7A (THSD7A) (Fig. 3c), and the RCSD domain containing 1 (RCSD1) genes (Fig. 3d), belonging to the “mechanisms of cytoskeletal organization” (Table S2).
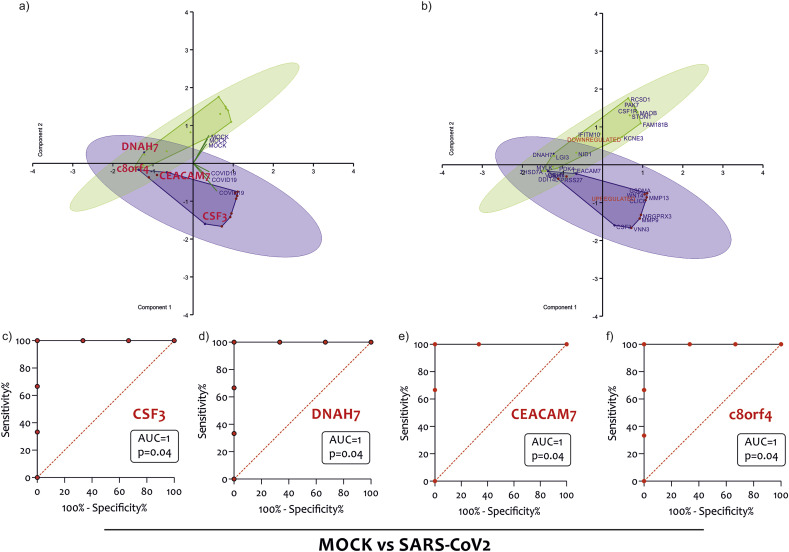
Fig. 3.
Gene segregation according to the NHBE cells treatment. Scatterplot of Principal Component Analysis (PCA) using the DEGs highlighted in NHBE treated with COVID-19 from the GSE147507 dataset with (a) and without (b) genes depicted. ROC curve of CSF3 (c), DNAH7 (d), CEACAM7 (e), and c8orf4 (f).
The 27 SDEG identified were used to perform a PCA analysis on samples of NHBE cell, MOCK treated or infected with SARS-CoV2, obtained from the GSE147507 dataset. As shown in Fig. 3a and b, strong separation of samples from MOCK and infected SARS-CoV2 was observed. We highlighted in red CEACM7, CSF3, DNAH7 and c8orf4, which were strongly modulated in our analysis. In order to evaluate the potential diagnostic ability of these genes to discriminate against the SARS-CoV2 virus infection, we performed a Receiver operating characteristic (ROC) analysis. We confirmed the diagnostic ability of CSF3 (p = 0.04) (Fig. 3c), DNAH7 (p = 0.04) (Fig. 3d), CEACAM7 (p = 0.04) (Fig. 3e), and c8orf4 (p = 0.04) (Fig. 3f) to discriminate the NHBE cells infected by SARS-CoV2 from MOCK treated.
3.3. Gene signature similarity between SARS-CoV2, SARS-CoV, MERS-CoV and H1N1
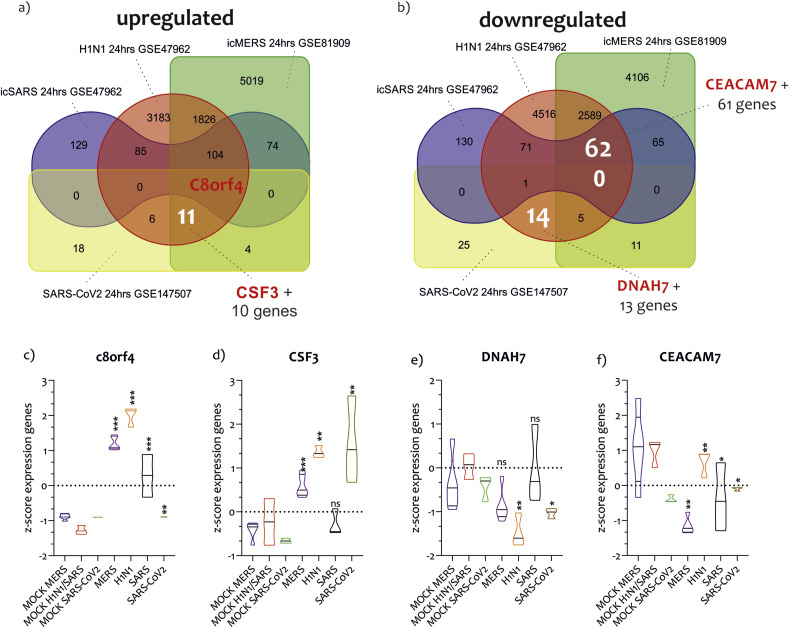
In order to identify possible common strategies among the main pandemic viruses, such as SARS-CoV2, SARS-CoV1, MERS-CoV, and H1N1, we carried out a Venn analysis of the main genes transcribed in the cells of the respiratory tract exposed to these viruses. We have downloaded two further datasets from GEO, GSE47962 composed of the transcriptome of Human bronchial airway epithelium cells (HAE) exposed to SARS-CoV (MOI 1 for 24 h) or H1N1 (MOI 1 for 24 h), and the GSE81909 composed of HAE infected cells with the MERS-CoV virus (5 PFU x cells for 24 h) (Table 1). The statistical analysis with GEO2R of GSE47962 highlighted 393 upregulated genes and 329 downregulated genes in HAE cells exposed to the SARS-CoV virus, and 5216 upregulated genes and 7258 downregulated genes in HAE cells infected with H1N1 (Table 1) (Table S3) (Fig. 4 a and b).
Fig. 4.
Gene signature similarity between SARS-CoV2, SARS-CoV, MERS-CoV and H1N1. Overlap of upregulated genes by SARS-CoV2, SARS-CoV, MERS-CoV, and H1N1 in NHBE and HAE. The analysis showed that c8orf4 was the gene commonly regulated in NHBE and HAE under the infection of the four viruses (a and c). Eleven genes, including CSF3, were commonly modulated by SARS-CoV2, MERS-CoV, and H1N1 (a and d). As regards the overlap of downregulated genes, we showed that no genes were modulated by the four viruses. Fourteen genes were shared between SARS-CoV2 and H1N1, including DNAH7(b and e). 62 genes were identified in common between H1N1, MERS-CoV, and SARS-CoV, including CEACAM7(b and f). Data are expressed as RNA Read count and presented as violin plots. P values < 0.05 were considered to be statistically significant (*p < 0.05).
As regards the effect of MERS-CoV on HAE cells transcriptome, we showed 7039 genes upregulated and 6838 genes downregulated (Table 1) (Table S3) (Fig. 4a and b). The Venn diagrams (Fig. 4a/b) showed that the upregulated transcriptome of the cells infected with SARS-CoV2 uniquely overlapped to that of MERS-CoV (10%, 4 genes), H1N1 (15%, 6 genes), and SARS-CoV (2.5%, one gene). Only one gene, c8orf4, was in common between SARS-CoV, H1N1, MERS-CoV, and SARS-CoV2 (Fig. 4a and c). Globally, considering all interactions, SARS-CoV2 had the highest number of genes upregulated in common with H1N1 (45%, 18 genes) (p < 5.18e-0.4, RF = 2.2). As regards with the downregulated transcriptome of NHBE cells under SARS-CoV2 infection, we showed an overlapping to MERS-CoV (19.6%, 11 genes), H1N1 (25%, 14 genes), and SARS-CoV (1.79%, one gene). Also for the downregulated genes, COVID-19 had the highest number of genes in common with H1N1 (35.71%, 20 genes) (p = 0.007, RF = 1.7). Considering both upregulated and downregulated genes, the transcriptome induced by SARS-CoV2 on HNBE cells was overlapped for 39.6% (p = 0.027, RF = 0.8) with that induced by H1N1, for the 33.3% (p = 8.155e-22, RF = 0.4) with that of MERS-CoV, and 2.1% (p = 0.241, RF = 0.5) with that of SARS-CoV (Table 2 ). Furthermore, 11 genes were in common between COVID-19, H1N1, and MERS-CoV, among which we highlighted CSF3 (Fig. 4a and d) (Table S3). DNAH7 together with other 13 genes, was in common between SARS-CoV2, SARS-CoV, and H1N1-induced transcriptomes (Fig. 4a and e). Instead, CEACAM7 was one of the 62 genes in common between SARS-CoV, H1N1, and MERS-CoV-induced transcriptomes (Fig. 4b and f) (Table S3).
Table 2.
The overlapped HEC transcriptomes induced by the infection of SARS-Cov2, MERS-CoV, SARS-CoV, and H1N1.
| Overlapping with SARS-CoV2 | n° Genes | % common Up genes | N° Genes | % common Down genes |
|---|---|---|---|---|
| MERS-CoV | 4 | 10.00 | 11 | 19.64 |
| MERS-CoV/H1N1 | 11 | 27.50 | 5 | 8.93 |
| SARS-CoV/H1N1/MERS-CoV | 1 | 2.50 | 0 | 0.00 |
| H1N1 | 6 | 15.00 | 14 | 25.00 |
| SARS-CoV/H1N1 | 0 | 0.00 | 1 | 1.79 |
3.4. Prediction of Novel drugs for SARS-CoV2 infection
Anti-signature perturbation analysis was performed using the DEGs (GSE147507) identified for the HNBE cells infected with SARS-CoV2 (Fig. 5 a and b) (Table S3).
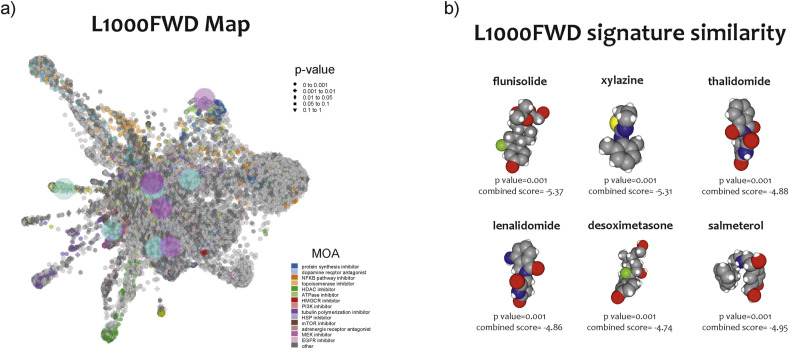
Fig. 5.
Drugs prediction against SARS-CoV2 infection. L1000FDW visualization of drug-induced signature. Input genes are represented by the significantly upregulated and downregulated genes obtained from the analysis of the GSE147507 dataset. Blue and red circles identify drugs with similar and anti-similar signatures. Dots are color-coded based on the Mode of Action (MOA) of the respective drug (a). The drugs with a high significance pvalue (qvalue) and a high combined score (flunisolide, xylazine, thalidomide, lenalidomide, desoximetasone, and salmeterol) were selected (b).
Among the significantly predicted drugs, we only highlighted those that are already in clinical use (Launched) (Table 3 ). We chose to list in Table 3 the potential six anti- SARS-CoV2 drugs identified by the L1000FWD analysis using the HNBE cell model infection. The complete list can be retrieved in Table S3.
Table 3.
Potential anti-AD drugs identified by the L1000FWD analysis.
| n | DRUGS | similarity score | p-value | Z-score | combined score | Target(s) | Indication(s) |
|---|---|---|---|---|---|---|---|
| 1 | flunisolide | −0.102 | 1.33E-03 | 1.87 | −5.37 | cytochrome P450 inhibitor | allergic rhinitis |
| 2 | xylazine | −0.102 | 1.43E-03 | 1.87 | −5.31 | Xylazine HCl is α2 class of adrenergic receptor agonist. | anesthetic |
| 3 | salmeterol | −0.102 | 1.91E-03 | 1.82 | −4.95 | adrenergic receptor agonist | asthma, chronic obstructive pulmonary disease (COPD), bronchospasm |
| 4 | thalidomide | −0.102 | 1.91E-03 | 1.79 | −4.88 | tumor necrosis factor production inhibitor | immunomodulatory agent |
| 5 | lenalidomide | −0.102 | 1.99E-03 | 1.8 | −4.86 | anticancer agent | multiple myeloma, mantle cell lymphoma (MCL), myelodysplastic diseases (MDS) |
| 6 | desoximetasone | −0.102 | 1.93E-03 | 1.74 | −4.74 | glucocorticoid receptor agonist | corticosteroid-responsive dermatoses |
Among them: Flunisolide, a cytochrome P450 inhibitor, used for the allergic rhinitis; Xylazine, a α2 class of adrenergic receptor agonist, used as an anesthetic; Salmeterol, an adrenergic receptor agonist, used for asthma, chronic obstructive pulmonary disease (COPD), bronchospasm; Thalidomide, a tumor necrosis factor production inhibitor, used as an immunomodulatory agent; Lenalidomide, an anticancer agent, used in multiple myeloma, in mantle cell lymphoma (MCL), and in myelodysplastic diseases (MDS); Desoximetasone, a glucocorticoid receptor agonist, used in corticosteroid-responsive dermatoses (Table S3) (Table 3) (Fig. 5b).
4. Discussion
The human respiratory epithelium is the primary target of SARS-CoV2, SARS-CoV, MERS-Cov, and influenza viruses H1N1. The direct and indirect effects of SARS-CoV2 on the respiratory epithelium, and the severity of such process may lead to detrimental anatomical and structural damages, respiratory distress syndrome and death. Viral replication induces a variety of transcriptional events, which pathophysiological consequences need to be dissected.
The integrity of the bronchial epithelium is fundamental for the maintenance of respiratory functions, as well as for limiting the traffic of the immune system locally. Alterations of the bronchial parenchyma cause devastating effects on respiratory functions, [33].
In our investigation, we focused our attention on two main processes, the immuno-cells recruitment and structure and function perturbation of muscles and bronchial cilia.
We found that SARS-CoV2 induces the modulation of CSF3, DNAH7, CEACAM7 and c8orf4. CSF3 is involved in granulocytes and the monocytes-macrophages differentiation and recruitment to the damaged area, while the other genes contribute to the rearrangement of the extracellular matrix and collagen metabolism. Moreover, actin cytoskeleton and muscle contraction are highly down regulated by SARS-CoV2 infection.
The immunological, structural and physiological perturbations induced by SARS-CoV2 critically contribute to the respiratory distress that characterizes COVID-19. Indeed, tissue destruction, cell recruitment and the inflammatory response, the altered processes of matrix and collagen deposition, and tissue regeneration along with cilia impairment impede the primary function of the respiratory system, namely, to take in oxygen and eliminate carbon dioxide.
We highlighted that three genes, belonging to the processes of immune cells recruitment (CSF3, c8orf4, and CEACAM7), were significantly highly expressed in NHBE cells infected by SARS-CoV2. Furthermore, we showed that four genes that play a fundamental role in the structuring of bronchial cilia, and therefore in the processes of muco-ciliary movement, were significantly downregulated by the effect of viral infection. In addition, these genes were able to discriminate the viral infection compared to controls. The transcriptome overlapping analysis induced on bronchial epithelium cells infected with SARS-CoV2, SARS-CoV, MERS-CoV, and H1N1 has shown that the c8orf4 gene represents a common point of the virus diseases induced in our analysis and that CEACAM7 gene expression is exclusive of SARS-CoV2 infection. Using the new prediction systems L1000FWD has allowed us to highlight that several drugs, already in clinical use (flunisolide, xylazine, salmeterol, thalidomide, lenalidomide, and desoximetasone) for the treatment of other diseases, may have an effect on SARS-CoV2 viral infection.
Paroxysmal immune activation triggered during SARS-CoV2 infection is one of the causes of mortality of this disease. The dysregulation of the immune response allows the development of viral hyper-inflammation. The uncontrolled immuno-activation, in particular the one triggered by innate immunity, mainly represented by the neutrophils and alveolar macrophages, is supported by the genes with chemotactic function. In this light, genes such as CSF3, CEACAM7 and c8orf4 represent a hot spot in this process. In particular, CSF3 is a modulator of neutrophil activation during the antiviral responses to human respiratory syncytial virus (hRSV), a major human pathogen that primarily targets the respiratory epithelium [34]. It is very interesting to note that in a cohort of 452 patients with COVID-19, 286 were diagnosed as severe infections, these patients tended to have lower lymphocytes counts, higher leukocytes count, and neutrophil-lymphocyte-ratio (NLR). This data could indicate that the recruitment of Neutrophils during SARS-CoV2 infection could be due to production of CSF3 from the cells of the bronchial epithelium [35]. In our investigation, it emerged that MERS-CoV and H1N1 could use CSF3 for the recruitment of the immune system. This data is in agreement with the percentages of genomic overlap that we found in our analysis. SARS-CoV did not result in modulation of CSF3. These results surprised us. We would have expected a common line of SARS-CoV2, with MERS-CoV and SARS-CoV and not with H1N1. This high transcriptional overlap with H1N1 (almost 40) and with MERS-CoV (33.3%), represents an important point to explore in the future.
In endothelial cells, the c8orf4 expression (also known as TCIM) gene, it would seem to enhance key inflammatory mediators and inflammatory response through the modulation of NF-kappaB transcription [36]. In addition, the TCIM protein enhances inflammatory parameters such as monocyte-endothelial adhesion and endothelial monolayer permeability, mechanisms present in virus airway infection and most likely in the respiratory tract of COVID-19 patients [37]. These key features in cellular recruitment processes could explain why its expression is the only one in common among the four pandemic viral infections analyzed.
As regards to CEACAM7, this gene belongs to the immunoglobulin superfamily and in epithelial cells plays a role with innate immunity [38]. Noteworthy, the MERS-CoV virus uses the CEACAM5 (belonging to the same family of CEACAM 7) protein to infect the cells of the bronchial epithelium [39]. The entry of MERS-CoV is increased when CEACAM5 is overexpressed in target cells, suggesting that CEACAM5 could facilitate MERS-CoV entry in conjunction with DPP4 despite not being able to support MERS-CoV entry, independently. This result would suggest that CEACAM7 could play the same role in SARS-CoV2 infection. Furthermore, the fact that in the infections caused by SARS-CoV, MERS-CoV, and H1N1 the expression of CEACAM7 was downregulated, would suggest that the action of CEACAM7 is exclusive of the infection caused by SARS-CoV2.
The bronchial epithelium cells mechanisms of cytoskeletal rearrangement could represent a fundamental element in SARS-CoV2 infection. In particular, cytoskeletal changes could significantly undermine the ability of these cells to activate ciliary movement. To confirm this hypothesis, our analysis revealed that the expression levels of the DNAH7, PAK7, THSD7A, and RCSD1 genes were significantly downregulated in the NHBE cells treated with the SARS-CoV2, compared to the MOCK control. These genes could be linked to the “mechanism of respiratory Cilia regulation”. In particular, as stated above the DNAH7 gene plays important role in the movement of bronchial cilia [40], and its reduction is linked to ciliary dysmotility [41]. Interesting to note that in respiratory syncytial virus infection, a major cause of respiratory disease, there is increased ciliary dyskinesia combined with ciliary loss and epithelial damage, likely to result in reduced mucociliary clearance [42]. This condition resembles that caused by SARS-CoV2 [43]. It is interesting to note that DNAH7A expression levels were significantly downregulated in both NHBE cells infected with SARS-COV-2 and in those infected with MERS-CoV and H1N1. Indeed, it has been shown that MERS-CoV, H1N1, but not SARS-CoV, rapidly induces apoptosis of Human Bronchial Epithelial Cells [44,45].
As regards PAK7, it is known that it is involved in a variety of different signaling pathways including cytoskeleton regulation by microtubule protein phosphorylation, cell migration, proliferation or cell survival [46]. The repression of PAK7 expression triggers the apoptotic cascade that leads to apoptosis through the inhibition of BAD phosphorylation [47].
The expression of THSD7A gene, promotes endothelial cell migration and filopodia formation [48]. It is plausible, given its role, that the reduction of THSD7A is associated with a reduction of Bronchial Epithelial Cell Migration Dynamics.
As regards to RCSD1, it may regulate the ability of F-actin-capping protein to remodel actin filament assembly (Fig. 3e) [49]. The importance of actin filaments for the integrity of the epithelial barrier has long been recognized [50].
Our analysis also showed that drugs currently used to treat other diseases could modulate the gene transcription mechanisms activated by the SARS-CoV2 virus on NHBE cells. The finding of novel indications for already approved drugs allows their quick use due to the broad knowledge of toxicity, pharmacokinetic and pharmacodynamics. Recently, the use of computational studies has made it possible to investigate the pharmacological response to a determined disease in a faster way and reducing experimental laboratory costs [24]. Platforms such as L1000FWD are based on the evaluation of the anti-similarity between drugs and disease. Several studies have been carried out based on these procedures [[51], [52], [53], [54]]. Unfortunately, this type of approach has several limitations. The variation of the transcriptome alone cannot accurately predict the effects of the drug on the disease and vice versa. It is also true that the pandemic viral infection we are affected by has prompted us to speed up the investigation time. Even if full of limitations, computational analysis can show new strategies in order to counter the infection. Interestingly, in our study, the thalidomide and lenalidomide (two immunomodulatory drugs) were predicted to be a potential anti- SARS-CoV2 drug, when using in NHBE cells. This is in line with recent evidence [55,56] showing that thalidomide and lenalidomide could reduce the immuno-activation. The flunisolide is a corticosteroid prescribed for the treatment of allergic rhinitis. The flunisolide's main action mechanism is to activate glucocorticoid receptors, and as a consequence, an anti-inflammatory action. Furthermore, the inhibitory effect of inhaled flunisolide on inflammatory functions of alveolar macrophages has already been demonstrated [57]. Interestingly, the HFA-flunisolide effectively suppress eosinophilic inflammation in peripheral and central airways, and these changes are accompanied by improvement in lung function [58].
In addition, our analysis identified salmeterol, xylazine, and desoximetasone, as potential anti- SARS-CoV2 drugs. Salmeterol is used by inhalation to reduce bronchospasm in some pathological conditions such as asthma and other obstructive respiratory diseases [59]. Furthermore, an anti-inflammatory action has been observed at the level of the bronchial epithelium [60]. As regard to desoximetasone, this is a synthetic glucocorticoid receptor agonist with metabolic, anti-inflammatory and immunosuppressive activity. It is used in the treatment of many conditions, including rheumatic problems, severe allergies, asthma, chronic obstructive lung disease, and along with antibiotics in tuberculosis [61]. Among the predictions was xylazine. Most likely, this has been called into question by prediction analysis for its muscle relaxant effect, against bronchospasms caused by respiratory infection. Its choice is extremely controversial, by virtue of the side effects it could cause, including respiratory depression [62].
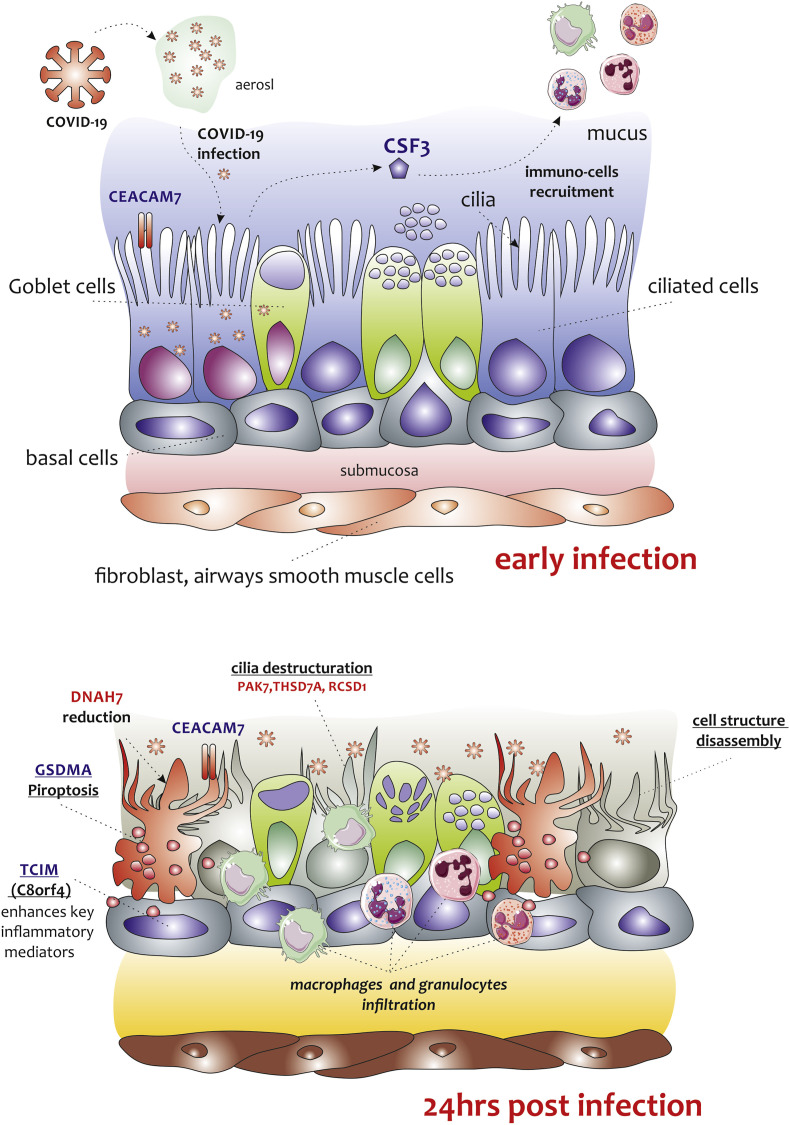
It is conceivable that, in line with the results obtained in our investigation, during the course of SARS-CoV2 infection, the virus descends the respiratory tract and interacts with the cells of the bronchial epithelium through a surface receptor, which could be couple with CEACAM7 as a co-receptor. Once entered, the virus triggers an inflammatory response that passes through several viral containment mechanisms. A series of genes are transcribed that deal with the rearrangement of the extracellular environment, and the recall of innate immunity cells (CSF3 and c8orf4). Alveolar macrophages and neutrophils are recalled locally and activate the paroxysmal response, which results parenchymal destruction and edema. The bronchial epithelium changes its structure. The cytoskeleton of epithelial cells is deconstructed, due to the downregulation of genes such as DNAH7, PAK7, TSHD7A, and RCSD1. The ciliary movement of the bronchial epithelium is lost. Mechanisms of apoptosis are triggered which lead to total ineffectiveness of the respiratory system (Fig. 6 ).
Fig. 6.
Graphical representation of the main process predicted in NHBE cells infected with SARS-CoV2. Panel A and B show our hypothesis of key events in SARS-CoV2 pathogenesis, which is based on extremely limited observations on an in vitro model of NHBE cell infected with SARS-CoV2 at MOI 2 for 24 h. After the inoculation of SARS-CoV2, the NHBE cells were infected. We hypothesize that viral entry could be facilitated by another cell-surface protein, carcinoembryonic antigen-related cell-adhesion molecule 7 (CEACAM7), which is also expressed in gastrointestinal tract. This mechanism could be similar to that used by the MERS-CoV virus. In that case, the virus uses the CEACAM5 protein to infect the cells of the bronchial epithelium. Inflammatory signaling molecules that are released by infected cells (CSF3, c8orf4), recruits.
A weakness of our study is that it is based on an experiment in vitro performed in bronchial cell epithelial obtained from 1 subject (female, 79 years old). The age of this subject is critical because age could dramatically influence response to the virus. It is also true that our study represents a “picture” of a currently evolving event in which all information can make a difference in the idea of a therapeutic strategy. The strength of our manuscript is to be the first to identify the potential genes responsible for muco-ciliary atheration and the potential drugs that can reverse this process.
5. Conclusions
In our analysis, we provided the basis for the identification of potential gene targets for the SARS-CoV2 infection. Both immune recruitment and the destruction of the ciliary parenchyma could represent the two key points of the disease triggered by the SARS-CoV2 infection. Future studies are now warranted in order to further confirm our results in a clinical setting. Furthermore, single and combined administration of potential anti- SARS-CoV2 drugs deserves to be taken into consideration for more in-depth investigations.
Author contributions
Giuseppe Nunnari: Conceptualization, Validation, Writing - original draft, Read and agreed to the published version of the manuscript. Cristina Sanfilippo: Methodology, Validation, Writing - original draft, Read and agreed to the published version of the manuscript. Paola Castrogiovanni: Resources, Read and agreed to the published version of the manuscript. Rosa Imbesi: Resources, Read and agreed to the published version of the manuscript. Giovanni Li Volti: Validation, Writing - review & editing, Read and agreed to the published version of the manuscript. Ignazio Barbagallo: Writing - review & editing, Read and agreed to the published version of the manuscript. Giuseppe Musumeci: Resources, Read and agreed to the published version of the manuscript. Michelino Di Rosa: Conceptualization, Software, Validation, Formal analysis; Investigation, Data curation, Writing - original draft, Visualization, Supervision, Project administration, Writing - review & editing, Read and agreed to the published version of the manuscript.
Ethics approval and consent to participate
Not applicable. This article does not contain any studies with human participants or animals performed by any of the authors.
Consent for publication
Not applicable
Availability of data and materials
The datasets analyzed during the current study are available in the GSE147507, GSE47962, GSE47962, and GSE81909 repository, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE17507.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We would like to show our gratitude to the authors of microarray datasets made available online, for consultation and re-analysis. In addition, I would like to express my gratitude to Oliver Di Rosa, an inspiration in my life. Funding This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.yexcr.2020.112204.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Bogoch, Watts A., Thomas-Bachli A., Huber C., Kraemer M.U.G., Khan K. Pneumonia of unknown aetiology in Wuhan, China: potential for international spread via commercial air travel. J. Trav. Med. 2020;27 doi: 10.1093/jtm/taaa008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu T., Pu H. Computed tomography manifestations of 5 cases of the novel coronavirus disease 2019 (COVID-19) pneumonia from patients outside Wuhan. J. Thorac. Imag. 2020;35(3):90–93. doi: 10.1097/RTI.0000000000000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lagunas-Rangel F.A. Neutrophil-to-Lymphocyte ratio and Lymphocyte-to-C-reactive protein ratio in patients with severe coronavirus disease 2019 (COVID-19): a meta-analysis. J. Med. Virol. 2020 doi: 10.1002/jmv.25819. In preparation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.To K.F., Lo A.W. Exploring the pathogenesis of severe acute respiratory syndrome (SARS): the tissue distribution of the coronavirus (SARS-CoV) and its putative receptor, angiotensin-converting enzyme 2 (ACE2) J. Pathol. 2004;203:740–743. doi: 10.1002/path.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clough E., Barrett T. The gene expression Omnibus database. Methods Mol. Biol. 2016;1418:93–110. doi: 10.1007/978-1-4939-3578-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanfilippo C., Castrogiovanni P., Imbesi R., Kazakowa M., Musumeci G., Blennow K., Zetterberg H., Di Rosa M. Sex difference in CHI3L1 expression levels in human brain aging and in Alzheimer's disease. Brain Res. 2019;1720 doi: 10.1016/j.brainres.2019.146305. [DOI] [PubMed] [Google Scholar]
- 7.Sanfilippo C., Castrogiovanni P., Imbesi R., Tibullo D., Li Volti G., Barbagallo I., Vicario N., Musumeci G., Di Rosa M. Middle-aged healthy women and Alzheimer's disease patients present an overlapping of brain cell transcriptional profile. Neuroscience. 2019;406:333–344. doi: 10.1016/j.neuroscience.2019.03.008. [DOI] [PubMed] [Google Scholar]
- 8.Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Moller R., Jordan T.X., Oishi K., Panis M., Sachs D., Wang T.T., Schwartz R.E., Lim J.K., Albrecht R.A., tenOever B.R. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045 e1039. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitchell H.D., Eisfeld A.J., Sims A.C., McDermott J.E., Matzke M.M., Webb-Robertson B.J., Tilton S.C., Tchitchek N., Josset L., Li C., Ellis A.L., Chang J.H., Heegel R.A., Luna M.L., Schepmoes A.A., Shukla A.K., Metz T.O., Neumann G., Benecke A.G., Smith R.D., Baric R.S., Kawaoka Y., Katze M.G., Waters K.M. A network integration approach to predict conserved regulators related to pathogenicity of influenza and SARS-CoV respiratory viruses. PloS One. 2013;8 doi: 10.1371/journal.pone.0069374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao J., Cao H., Chen J. False discovery rate control incorporating phylogenetic tree increases detection power in microbiome-wide multiple testing. Bioinformatics. 2017;33:2873–2881. doi: 10.1093/bioinformatics/btx311. [DOI] [PubMed] [Google Scholar]
- 11.Smyth G.K. vol. 3. 2004. (Linear Models and Empirical Bayes Methods for Assessing Differential Expression in Microarray Experiments, Statistical Applications in Genetics and Molecular Biology). Article3. [DOI] [PubMed] [Google Scholar]
- 12.Lee C., Patil S., Sartor M.A. RNA-Enrich: a cut-off free functional enrichment testing method for RNA-seq with improved detection power. Bioinformatics. 2016;32:1100–1102. doi: 10.1093/bioinformatics/btv694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuberi K., Franz M., Rodriguez H., Montojo J., Lopes C.T., Bader G.D., Morris Q. GeneMANIA prediction server 2013 update. Nucleic Acids Res. 2013;41:W115–W122. doi: 10.1093/nar/gkt533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang J.T., Nevins J.R. GATHER: a systems approach to interpreting genomic signatures. Bioinformatics. 2006;22:2926–2933. doi: 10.1093/bioinformatics/btl483. [DOI] [PubMed] [Google Scholar]
- 15.Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P., Jensen L.J., von Mering C. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castrogiovanni P., Li Volti G., Sanfilippo C., Tibullo D., Galvano F., Vecchio M., Avola R., Barbagallo I., Malaguarnera L., Castorina S., Musumeci G., Imbesi R., Di Rosa M. Fasting and fast food diet play an opposite role in mice brain aging. Mol. Neurobiol. 2018;55:6881–6893. doi: 10.1007/s12035-018-0891-5. [DOI] [PubMed] [Google Scholar]
- 18.Wang Z., Lachmann A., Keenan A.B., Ma'ayan A. L1000FWD: fireworks visualization of drug-induced transcriptomic signatures. Bioinformatics. 2018;34:2150–2152. doi: 10.1093/bioinformatics/bty060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Box G.E.P., Tiao G.C. 6 April 1992. Bayesian Inference in Statistical Analysis. [Google Scholar]
- 20.Cheadle C., Vawter M.P., Freed W.J., Becker K.G. Analysis of microarray data using Z score transformation. J. Mol. Diagn. 2003;5:73–81. doi: 10.1016/S1525-1578(10)60455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scarpino M., Pinzone M.R., Di Rosa M., Madeddu G., Foca E., Martellotta F., Schioppa O., Ceccarelli G., Celesia B.M., d'Ettorre G., Vullo V., Berretta S., Cacopardo B., Nunnari G. Kidney disease in HIV-infected patients. Eur. Rev. Med. Pharmacol. Sci. 2013;17:2660–2667. [PubMed] [Google Scholar]
- 22.Care M.A., Barrans S., Worrillow L., Jack A., Westhead D.R., Tooze R.M. A microarray platform-independent classification tool for cell of origin class allows comparative analysis of gene expression in diffuse large B-cell lymphoma. PloS One. 2013;8 doi: 10.1371/journal.pone.0055895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J., Coombes K.R., Highsmith W.E., Keating M.J., Abruzzo L.V. Differences in gene expression between B-cell chronic lymphocytic leukemia and normal B cells: a meta-analysis of three microarray studies. Bioinformatics. 2004;20:3166–3178. doi: 10.1093/bioinformatics/bth381. [DOI] [PubMed] [Google Scholar]
- 24.Reddy T.B., Riley R., Wymore F., Montgomery P., DeCaprio D., Engels R., Gellesch M., Hubble J., Jen D., Jin H., Koehrsen M., Larson L., Mao M., Nitzberg M., Sisk P., Stolte C., Weiner B., White J., Zachariah Z.K., Sherlock G., Galagan J.E., Ball C.A., Schoolnik G.K. TB database: an integrated platform for tuberculosis research. Nucleic Acids Res. 2009;37:D499–D508. doi: 10.1093/nar/gkn652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le Cao K.A., Rohart F., McHugh L., Korn O., Wells C.A. YuGene: a simple approach to scale gene expression data derived from different platforms for integrated analyses. Genomics. 2014;103:239–251. doi: 10.1016/j.ygeno.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 26.Chen Q.R., Song Y.K., Wei J.S., Bilke S., Asgharzadeh S., Seeger R.C., Khan J. An integrated cross-platform prognosis study on neuroblastoma patients. Genomics. 2008;92:195–203. doi: 10.1016/j.ygeno.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yasrebi H., Sperisen P., Praz V., Bucher P. Can survival prediction be improved by merging gene expression data sets? PloS One. 2009;4 doi: 10.1371/journal.pone.0007431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mehmood R., El-Ashram S., Bie R., Dawood H., Kos A. Clustering by fast search and merge of local density peaks for gene expression microarray data. Sci. Rep. 2017;7:45602. doi: 10.1038/srep45602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheadle C., Cho-Chung Y.S., Becker K.G., Vawter M.P. Application of z-score transformation to Affymetrix data. Appl. Bioinf. 2003;2:209–217. [PubMed] [Google Scholar]
- 30.Feng C., Wu J., Yang F., Qiu M., Hu S., Guo S., Wu J., Ying X., Wang J. Expression of Bcl-2 is a favorable prognostic biomarker in lung squamous cell carcinoma. Oncol. Lett. 2018;15:6925–6930. doi: 10.3892/ol.2018.8198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang C., Huo Y., Xin L., Tian B., Yu B. Feature selection and tumor classification for microarray data using relaxed Lasso and generalized multi-class support vector machine. J. Theor. Biol. 2019;463:77–91. doi: 10.1016/j.jtbi.2018.12.010. [DOI] [PubMed] [Google Scholar]
- 32.Zetterberg H., Bozzetta E., Favole A., Corona C., Cavarretta M.C., Ingravalle F., Blennow K., Pocchiari M., Meloni D. Neurofilaments in blood is a new promising preclinical biomarker for the screening of natural scrapie in sheep. PloS One. 2019;14 doi: 10.1371/journal.pone.0226697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yao X.H., Li T.Y., He Z.C., Ping Y.F., Liu H.W., Yu S.C., Mou H.M., Wang L.H., Zhang H.R., Fu W.J., Luo T., Liu F., Chen C., Xiao H.L., Guo H.T., Lin S., Xiang D.F., Shi Y., Li Q.R., Huang X., Cui Y., Li X.Z., Tang W., Pan P.F., Huang X.Q., Ding Y.Q., Bian X.W. [A pathological report of three COVID-19 cases by minimally invasive autopsies] Zhonghua Bing Li Xue Za Zhi. 2020;49:E009. doi: 10.3760/cma.j.cn112151-20200312-00193. [DOI] [PubMed] [Google Scholar]
- 34.Touzelet O., Broadbent L., Armstrong S.D., Aljabr W., Cloutman-Green E., Power U.F., Hiscox J.A. The secretome profiling of a pediatric airway epithelium infected with hRSV identified aberrant apical/basolateral trafficking and novel immune modulating (CXCL6, CXCL16, CSF3) and antiviral (CEACAM1) proteins. Mol. Cell. Proteomics. 2020;19(5):793–807. doi: 10.1074/mcp.RA119.001546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., Xie C., Ma K., Shang K., Wang W., Tian D.S. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin. Infect. Dis. 2020;ciaa248 doi: 10.1093/cid/ciaa248. In preparation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim J., Kim Y., Kim H.T., Kim D.W., Ha Y., Kim J., Kim C.H., Lee I., Song K. TC1(C8orf4) is a novel endothelial inflammatory regulator enhancing NF-kappaB activity. J. Immunol. 2009;183:3996–4002. doi: 10.4049/jimmunol.0900956. [DOI] [PubMed] [Google Scholar]
- 37.Li K., Wohlford-Lenane C.L., Channappanavar R., Park J.E., Earnest J.T., Bair T.B., Bates A.M., Brogden K.A., Flaherty H.A., Gallagher T., Meyerholz D.K., Perlman S., McCray P.B., Jr. Mouse-adapted MERS coronavirus causes lethal lung disease in human DPP4 knockin mice. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E3119–E3128. doi: 10.1073/pnas.1619109114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Czepczynska-Krezel H., Krop-Watorek A. [Human carcinoembryonic antigen family proteins, structure and function] Postepy Hig. Med. Dosw. 2012;66:521–533. doi: 10.5604/17322693.1004113. [DOI] [PubMed] [Google Scholar]
- 39.Chan C.M., Chu H., Wang Y., Wong B.H., Zhao X., Zhou J., Yang D., Leung S.P., Chan J.F., Yeung M.L., Yan J., Lu G., Gao G.F., Yuen K.Y. Carcinoembryonic antigen-related cell adhesion molecule 5 is an important surface attachment factor that facilitates entry of Middle East respiratory syndrome coronavirus. J. Virol. 2016;90:9114–9127. doi: 10.1128/JVI.01133-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones R.T., Abedalthagafi M.S., Brahmandam M., Greenfield E.A., Hoang M.P., Louis D.N., Hornick J.L., Santagata S. Cross-reactivity of the BRAF VE1 antibody with epitopes in axonemal dyneins leads to staining of cilia. Mod. Pathol. 2015;28:596–606. doi: 10.1038/modpathol.2014.150. [DOI] [PubMed] [Google Scholar]
- 41.Horani A., Ferkol T.W., Shoseyov D., Wasserman M.G., Oren Y.S., Kerem B., Amirav I., Cohen-Cymberknoh M., Dutcher S.K., Brody S.L., Elpeleg O., Kerem E. LRRC6 mutation causes primary ciliary dyskinesia with dynein arm defects. PloS One. 2013;8 doi: 10.1371/journal.pone.0059436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith C.M., Kulkarni H., Radhakrishnan P., Rutman A., Bankart M.J., Williams G., Hirst R.A., Easton A.J., Andrew P.W., O'Callaghan C. Ciliary dyskinesia is an early feature of respiratory syncytial virus infection. Eur. Respir. J. 2014;43:485–496. doi: 10.1183/09031936.00205312. [DOI] [PubMed] [Google Scholar]
- 43.Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L., Tai Y., Bai C., Gao T., Song J., Xia P., Dong J., Zhao J., Wang F.S. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tao X., Hill T.E., Morimoto C., Peters C.J., Ksiazek T.G., Tseng C.T. Bilateral entry and release of Middle East respiratory syndrome coronavirus induces profound apoptosis of human bronchial epithelial cells. J. Virol. 2013;87:9953–9958. doi: 10.1128/JVI.01562-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuen K.M., Chan R.W., Mok C.K., Wong A.C., Kang S.S., Nicholls J.M., Chan M.C. Differential onset of apoptosis in avian influenza H5N1 and seasonal H1N1 virus infected human bronchial and alveolar epithelial cells: an in vitro and ex vivo study. Influenza Other Respir. Viruses. 2011;5:437–438. [PMC free article] [PubMed] [Google Scholar]
- 46.Karpov P.A., Nadezhdina E.S., Yemets A.I., Matusov V.G., Nyporko A.Y., Shashina N.Y., Blume Y.B. Bioinformatic search of plant microtubule-and cell cycle related serine-threonine protein kinases. BMC Genom. 2010;11(Suppl 1):S14. doi: 10.1186/1471-2164-11-S1-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Han K., Zhou Y., Gan Z.H., Qi W.X., Zhang J.J., Fen T., Meng W., Jiang L., Shen Z., Min D.L. p21-activated kinase 7 is an oncogene in human osteosarcoma. Cell Biol. Int. 2014;38:1394–1402. doi: 10.1002/cbin.10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuo M.W., Wang C.H., Wu H.C., Chang S.J., Chuang Y.J. Soluble THSD7A is an N-glycoprotein that promotes endothelial cell migration and tube formation in angiogenesis. PloS One. 2011;6 doi: 10.1371/journal.pone.0029000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miyatake K., Kusakabe M., Takahashi C., Nishida E. ERK7 regulates ciliogenesis by phosphorylating the actin regulator CapZIP in cooperation with Dishevelled. Nat. Commun. 2015;6:6666. doi: 10.1038/ncomms7666. [DOI] [PubMed] [Google Scholar]
- 50.Ivanov A.I., Parkos C.A., Nusrat A. Cytoskeletal regulation of epithelial barrier function during inflammation. Am. J. Pathol. 2010;177:512–524. doi: 10.2353/ajpath.2010.100168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu T.P., Hsieh Y.Y., Chou C.J., Yang P.M. Systematic polypharmacology and drug repurposing via an integrated L1000-based Connectivity Map database mining. R. Soc. Open Sci. 2018;5 doi: 10.1098/rsos.181321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Z., Fang H., Reagan K., Xu X., Mendrick D.L., Slikker W., Jr., Tong W. In silico drug repositioning: what we need to know. Drug Discov. Today. 2013;18:110–115. doi: 10.1016/j.drudis.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 53.Hodos R.A., Kidd B.A., Shameer K., Readhead B.P., Dudley J.T. In silico methods for drug repurposing and pharmacology. Wiley Interdiscip. Rev. Syst. Biol. Med. 2016;8:186–210. doi: 10.1002/wsbm.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jin G., Wong S.T. Toward better drug repositioning: prioritizing and integrating existing methods into efficient pipelines. Drug Discov. Today. 2014;19:637–644. doi: 10.1016/j.drudis.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ma J., Xia P., Zhou Y., Liu Z., Zhou X., Wang J., Li T., Yan X., Chen L., Zhang S., Qin Y., Li X. Potential effect of blood purification therapy in reducing cytokine storm as a late complication of severe COVID-19. Clin. Immunol. 2020 doi: 10.1016/j.clim.2020.108408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mihai C., Dobrota R., Schroder M., Garaiman A., Jordan S., Becker M.O., Maurer B., Distler O. COVID-19 in a patient with systemic sclerosis treated with tocilizumab for SSc-ILD. Ann. Rheum. Dis. 2020;79(5):668–669. doi: 10.1136/annrheumdis-2020-217442. [DOI] [PubMed] [Google Scholar]
- 57.Bewig B., Barth J. Inhibitory effects of inhaled flunisolide on inflammatory functions of alveolar macrophages. Eur. J. Clin. Pharmacol. 1993;44:541–544. doi: 10.1007/BF02440855. [DOI] [PubMed] [Google Scholar]
- 58.Hauber H.P., Gotfried M., Newman K., Danda R., Servi R.J., Christodoulopoulos P., Hamid Q. Effect of HFA-flunisolide on peripheral lung inflammation in asthma. J. Allergy Clin. Immunol. 2003;112:58–63. doi: 10.1067/mai.2003.1612. [DOI] [PubMed] [Google Scholar]
- 59.Salpeter S.R., Buckley N.S., Ormiston T.M., Salpeter E.E. Meta-analysis: effect of long-acting beta-agonists on severe asthma exacerbations and asthma-related deaths. Ann. Intern. Med. 2006;144:904–912. doi: 10.7326/0003-4819-144-12-200606200-00126. [DOI] [PubMed] [Google Scholar]
- 60.Siergiejko Z. [The effect of salmeterol on specific and non-specific bronchial response in allergic asthma patients] Pneumonol. Alergol. Pol. 1998;66:440–449. [PubMed] [Google Scholar]
- 61.Rybak M.J., Le J., Lodise T.P., Levine D.P., Bradley J.S., Liu C., Mueller B.A., Pai M.P., Wong-Beringer A., Rotschafer J.C., Rodvold K.A., Maples H.D., Lomaestro B.M. Executive summary: therapeutic monitoring of vancomycin for serious methicillin-resistant Staphylococcus aureus infections: a revised consensus guideline and review of the american society of health-system pharmacists, the infectious diseases society of america, the pediatric infectious diseases society, and the society of infectious diseases pharmacists. Pharmacotherapy. 2020;40(4):363–367. doi: 10.1002/phar.2376. [DOI] [PubMed] [Google Scholar]
- 62.Ruiz-Colon K., Chavez-Arias C., Diaz-Alcala J.E., Martinez M.A. Xylazine intoxication in humans and its importance as an emerging adulterant in abused drugs: a comprehensive review of the literature. Forensic Sci. Int. 2014;240:1–8. doi: 10.1016/j.forsciint.2014.03.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed during the current study are available in the GSE147507, GSE47962, GSE47962, and GSE81909 repository, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE17507.