Abstract
Cell division cycle 5 (Cdc5) is a highly conserved nucleic acid binding protein among eukaryotes and plays critical roles in development. Cdc5 can simultaneously bind to DNA and RNA by its N-terminal DNA-binding domain (DBD), but molecular mechanisms describing its nucleic acid recognition and the regulation of development through its nucleic acid binding remain unclear. Herein, we present a crystal structure of the N-terminal DBD of MoCdc5 (MoCdc5-DBD) from the rice blast fungus Magnaporthe oryzae. Residue K100 of MoCdc5 is on the periphery of a positively charged groove that is formed by K42, K45, R47, and N92 and is evolutionally conserved. Mutation of K100 significantly reduces the affinity of MoCdc5-DBD to a Cdc5-binding element but not to a conventional myeloblastosis (Myb) domain-binding element, suggesting that K100 is a key residue of the high binding affinity to Cdc5-binding element. Another conserved residue (R31) is located close to the U6 RNA in the structure of the spliceosome, and its mutation dramatically reduces the binding capacity of MoCdc5-DBD for U6 RNA. Importantly, mutations in these key residues, including R31, K42, and K100 in AtCDC5, an Arabidopsis thaliana ortholog of MoCdc5, greatly impair the functions of AtCDC5, resulting in pleiotropic development defects and reduced levels of primary microRNA transcripts. Taken together, our findings suggest that Cdc5-DBD binds nucleic acids with two distinct binding surfaces, one for DNA and another for RNA, which together contribute to establishing the regulation mechanism of Cdc5 on development through nucleic acid binding.
Introduction
Cell division cycle 5 (Cdc5) is a highly functionally and structurally conserved eukaryotic protein and participates in diverse molecular processes, such as cell cycle, DNA repair, RNA transcription, am intron splicing [1-4]. During the cell cycle and DNA repair, Cdc5 physically interacts with the cell cycle checkpoint kinase ataxia-telangiectasia and rad3-related (ATR) and directs ATR to rapidly anchor to DNA damage sites [5,6]. Cdc5 also participates in intron splicing as a core component of the Prp19 complex [7,8]. In Arabidopsis thaliana, AtCdc5 regulates the G2/M phase transition in the cell cycle, as well as being involved in the shoot apical meristem development [9]. Loss-of-function mutation of AtCdc5 also reduces immunity to bacterial infections and leads to diverse developmental defects, including reduced fertility and delayed plant growth [2,10]. Furthermore, AtCdc5 positively regulates the transcription of miRNAs by interacting with the DNA-dependent RNA polymerase II complex (Pol II) [2]. In plants, most genes encoding miRNAs (MIRs) are transcribed by Pol II to generate primary transcripts of miRNAs (pri-miRNAs) [11,12]. During the transcription of miRNAs Cdc5 interacts with both the promoters of MIRs and Pol II to promote the occupancy of Pol II on the promoters of MIRs [2,13]. pri-miRNAs are then processed to mature miRNAs by an RNase III domain-containing protein named DICER-LIKE1 (DCL1) [14,15]. But how Cdc5 binds MIR promoters and regulates MIR transcription remains unclear.
Cdc5 contains a myeloblastosis (Myb)-like domain with two DNA-binding domains (DBD) in its N-terminus, suggesting that it has potential roles in DNA-binding [16,17]. Myb domains usually consist of one to four repeats of ~52 amino acids, and each repeat forms three α-helices. The second and third helices of each repeat form a helix–turn–helix (HTH) structure with three regularly spaced tryptophan or hydrophobic residues, resulting in a hydrophobic core in the structure of the Myb domain [18,19]. In Myb–DNA complex structures (PDB:3OSF and 3ZQC), the α3 and α6 helices of Myb directly make contact with DNA and intercalate into its major groove [20-22]. During DNA binding, Myb repeats are closely packed into the major groove, so that the two recognition helices bind cooperatively to a specific AAC(G/T)G DNA motif termed the Myb domain-binding element (MSE) [23,24]. Several positively charged residues on the two helixes, including R84, R87, R89, K138, and T143 in TvMyb2, and K128, E132, R133, N179, and T188 in c-Myb, play important roles in DNA binding [23,24] (Supplementary Figure S1A). However, Myb domain proteins have been extensively studied in A. thaliana, and the DBD of AtCdc5 (AtCdc5-DBD) specifically interacts with a CTCAGCG motif but not an AAC(G/T)G motif [16] (Supplementary Figure S1B), suggesting that Cdc5 may not be a canonical Myb domain protein.
Cdc5 is also a key member of the Prp19 complex in the spliceosome and directly interacts with U6 RNA [25]. The structures of different complexes of the spliceosome from human and yeast have provided clues with regard to the roles of Cdc5 in RNA binding [4,8,26-30]. Moreover, two NMR structures (PDB code: 2DIM and 2DIN) of partial regions of human HsCdc5-DBD have been solved and published online. However, they only contain a single HTH motif from the DBD and thus cannot provide a complete DNA-binding mechanism. Therefore, a detailed crystal structure of Cdc5-DBD should shed light on the differences between Cdc5-DBD and a typical Myb domain in terms of DNA or RNA binding.
In this study, we find that a recombinant DBD of MoCdc5 (MoCdc5-DBD) from the rice blast fungus Magnaporthe oryzae has a higher binding capacity for the CTCAGCG motif, which exists in MIR promoters, than for the MSE motif (AAC(G/T)G), demonstrating that Cdc5-DBD and typical Myb proteins may have different DNA sequence preferences. Furthermore, we present a crystal structure of MoCdc5-DBD at 2.4 Å resolution, which consists of six helices forming two repeats of the HTH motif. Compared with a typical Myb domain, MoCdc5-DBD had a positively charged groove with five amino acid residues, K42, K45, R47, N92, and K100. Mutation analyses of these residues and two Cdc5-specific basic residues, R31 and R38 located around the positively charged groove, show that K42, K45, and N92 were essential for the DNA-binding capacity of Cdc5, while the K100 residue was responsible for the high binding affinity to the Cdc5-binding element. Moreover, an in vitro RNA-binding assay and structural analysis of the cryogenic electron microscopy (Cryo-EM) structure of the spliceosome show that R31 is a key residue at the interface for RNA binding in MoCdc5-DBD. Interestingly, mutations of the counterparts of K42 and R31 in AtCdc5-DBD from A. thaliana disrupt the function of AtCdc5, causing developmental defects and significantly reducing the transcript levels of pri-miRNAs. Taken together, we suggest that Cdc5-DBD regulates development through two distinct nucleic acid binding surfaces, one for DNA and one for RNA.
Materials and methods
Plasmid construction
The nucleotide sequence encoding MoCdc5-DBD (KLU93089, 1–105 aa) was amplified from cDNA templates of the M. oryzae strain P131 using a sticky-end cloning approach with four primers [31]. The resulting PCR product was cloned into the pETM13 vector with a C-terminal 6×His tag (kindly provided by Dr Arie Geerlof, EMBO) using NcoI and XhoI restriction sites. The mutated alleles of MoCdc5-DBD were generated using a one-step single-site plasmid mutagenesis protocol [32]. AtCDC5 cDNA was amplified by RT-PCR and cloned into the pEarlyGate203 vector. Mutations were generated using the QuikChange II Site-Directed Mutagenesis Kit (Agilent). For generating a mosaic allele of MoCdc5-DBD-AtCdc5, the cDNA from MoCDC5-DBD (1–105 aa of MoCDC5; kindly provided by Dr Richard Wilson, University of Nebraska-Lincoln) was amplified by RT-PCR and cloned into the pENTR/D-TOPO vector. The resulting plasmid was linearized with StuI and ligated with a truncated AtCDC5 cDNA encoding 107–844 aa of AtCDC5 with the In-Fusion HD Cloning Kit. All of the PCR primers used in this study were listed in Supplementary Table S3.
Expression and purification of mutant and wild-type MoCdc5-DBD
Recombinant plasmids were introduced into Escherichia coli BL21 (DE3) for protein expression. The harvested cells were disrupted by sonication in 20 mM 2-(N-Morpholino) ethane sulfonic acid (MES) (pH 6.0) buffer, 500 mM NaCl, and 1 mM Dithiothreitol, and cell debris was removed by centrifugation. The supernatant was then applied to a Ni-Sepharose FF column (GE Healthcare). Target proteins were eluted with MES buffer containing 400 mM imidazole and kept on ice. Eluted proteins were then further purified by passage through a Superdex 75 10/30 column (GE) with 20 mM MES (pH 6.0) buffer, 150 mM NaCl, using a flow rate of 0.75 ml/min. The proteins were concentrated to 8 mg/ml. After a flash freeze in liquid nitrogen, they were stored at −80°C prior to crystallization and other experiments.
Crystallization, data collection, and structure determination
Random crystallization screening was performed using the sitting-drop vapor diffusion method at 16°C. Crystallization screening kits were purchased from Hampton Research. The sample and well solutions were mixed at a 1:1 volume ratio using an Oryx4 crystallization robot (Douglas Instrument Ltd.). The crystallization condition contained 20% (w/v) MPD equilibrated at 293 K. The crystals were then immersed in a reservoir solution containing 20% (v/v) glycerol as a cryo-protectant, before being flash-cooled by transfer into liquid nitrogen and stored for X-ray diffraction experiments. Diffraction datasets were collected at the Shanghai Synchrotron Radiation Facility on beamline BL-17U and processed with HKL-2000 software [33]. The crystal structure of MoCdc5-DBD was solved by molecular replacement using Phaser with 3JB9 as the initial model [34]. The model was subsequently improved in Coot and further refined using PHENIX with TLS (Translation/Libration/Screw) restraints [35]. The electrostatic potential field was evaluated using the DelPhi Web Server [36].
Circular dichroism
Circular dichroism (CD) analysis was performed on a Chirascan-plus spectropolarimeter (Applied Photophysics, Leatherhead, U.K.) to estimate the protein structure at room temperature. Proteins were prepared at a final concentration of 0.1–0.2 mg/ml in a 1 mm path length quartz cell with 20 mM MES (pH 6.0) and 150 mM NaCl. Spectra were recorded from 200 to 260 nm at a scan speed of 60 nm/min for the corresponding buffer for each sample.
RNA sample preparation for BLI
S. cerevisiae U6 RNA was transcribed in vitro using the T7 RiboMAX™ Express Large-Scale RNA Production System (Promega, Madison, WI, USA) according to the manufacturer’s instructions. A DNA template (Accession: 3JB9_N) was prepared through the ligation of short complementary, overlapping oligonucleotides harboring a T7 promoter. The DNA was then treated with Proteinase K (100 μg/ml) and sodium dodecyl sulfate (0.5%) in 50 mM Tris–HCl (pH 7.5) and 5 mM CaCl2 for 30 min at 37°C. The DNA was further purified by extraction with TE-saturated (pH 8.0) phenol:chloroform:isoamyl alcohol (25:24:1) and ethanol precipitated. In vitro reaction components contained 10 μl RiboMAX™ Express T7 2× Buffer, 8 μl DNA template, and 2 μl Enzyme Mix. The mixture was incubated at 37°C for 30 min. The DNA template was then removed by digestion with RNase-free DNase as per the manufacturer’s recommendations. The U6 RNA was then purified by phenol (pH 4–5):chloroform:isoamyl alcohol (25:24:1) extraction and ethanol precipitation. RNA samples were then re-suspended in nuclease-free water and concentrations were estimated using UV absorption.
Biolayer interferometry
Biolayer interferometry (BLI) experiments were performed on an Octet RED 96 system (FortéBio) at 25°C. Data were processed and analyzed using Data Analysis Software 8.5 (FortéBio, Menlo Park, CA, U.S.A.), with curve fitting using a 1:1 binding model [37]. For protein-DNA BLI assays, streptavidin sensors were pre-wet for 10 min before being loaded with 5′-biotinylated DNA oligonucleotides (MIR167a, MSE) for 300 s. Analyte samples were then diluted in MES buffer at various concentrations, including 4, 2, 1, 0.5, and 0.25 μM. The association phase (for kon) and dissociation phase (for koff) were both recorded for 60 s. For protein-RNA BLI assays, nitrilotriacetic acid sensors were pre-wet for 10 min before being loaded with His-fusion Proteins for 180 s. U6 RNA samples were diluted in PBS buffer at various concentrations, including 1, 0.5, 0.25, 0.125, and 0.0625 μM. The association phase was then recorded for 120 s, and the dissociation phase was recorded for 180 s.
Plant materials and growth conditions
A cdc5-2 (CS426613) mutant was obtained from the Arabidopsis Biological Resources Center (ABRC). This mutant was in the Columbia-0 (Col-0) genetic background. All plants were grown at 22°C with a 16-h light and 8-h dark cycle.
Motif analysis of the promoters of genes encoding miRNAs (MiR)
DNA sequences 1000 bp upstream of the transcription start site from 133 MIRs were extracted and submitted to PlantPan database (http://plantpan2.itps.ncku.edu.tw/) to search for potential transcription factor-binding sites. Thirty seed sequences containing putative Cdc5-binding motifs were selected for co-occurrence analysis with a confidence interval of over 95% [38].
RNA analysis
RNA was extracted using Trizol Reagent (Roche, Beverly, MA, USA). Quantitative RT-PCR (qRT-PCR) analysis of the transcription levels of AtCdc5-targeted primary miRNA transcripts (pri-miRNAs) and mature miRNAs was performed as described previously [39].
Protein immunoblotting
Harvested plant samples were ground into powder in liquid nitrogen and then buffered with protein lysing buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, and 1% Triton X-100) and agitated three times before being centrifugation at 12 000 rpm for 20 min at 4°C. The supernatant was retained and used for standard western blot assays with an anti-Myc antibody and an anti-Hsc70 antibody (Sigma, St. Louis, MO, USA) as previously described [39].
Results
MoCdc5-DBD has a stronger binding capacity for the CTCAGCG motif than the Myb site-binding element
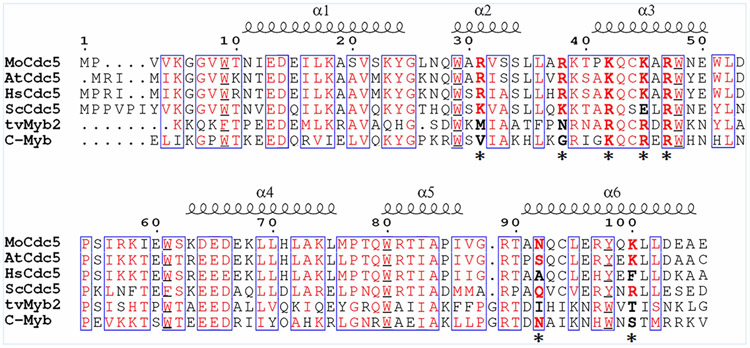
To investigate the differences in DNA binding between Cdc5-DBD and typical Myb domains, we firstly compared their amino acid sequences by selecting Cdc5 proteins from M. oryzae, A. thaliana, Homo sapiens, Saccharomyces cerevisiae, and Myb2 proteins from Trichomonas vaginalis, Mus musculus. The overall identity and similarity were high, with several amino acids being conserved in these species (Figure 1). Secondary structure prediction showed that all of these proteins contained six α-helixes, which form two HTH motifs. Each HTH motif was characterized by conserved aromatic group amino acids, such as Trp and Tyr, separated by ~19 residues. These analyses suggested that the Cdc5-DBD was highly similar to the typical Myb domain. However, the sixth conserved aromatic group amino acid in Cdc5-DBD proteins was Y at aa position 98, which was usually W in typical Myb domains. Moreover, two arginine residues, that is, R31 and R38, were well conserved among Cdc5-DBD proteins but not in typical Myb domains (Figure 1).
Figure 1. Sequence alignment of DNA-binding domains of Cdc5 and Myb.
Cdc5-DBD from Magnaporthe oryzae (MoCdc5), Arabidopsis thaliana (AtCdc5), Homo sapiens (HsCdc5), Saccharomyces cerevisiae (ScCdc5), and Myb-DBD from Trichomonas vaginalis (TvMyb2) and Mus musculus (c-Myb), respectively. Conserved aromatic residues are boxed and marked in red. Sequence alignment was performed with ClustalW. The six α-helixes (α1–α6) are indicated above the sequence, and the seven amino acid residues responsible for DNA binding are indicated with asterisks.
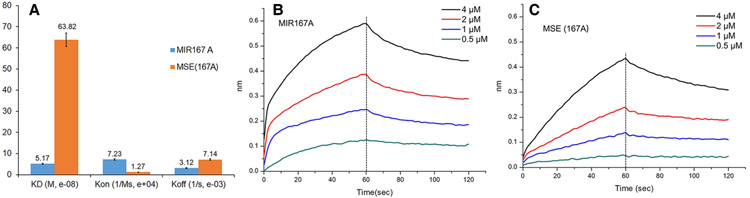
The Myb domain was previously shown to interact specifically with the Myb site-binding element (MSE), AAC (G/T)G, whereas AtCdc5 was shown to bind to CTCAGCG, a sequence often found in the promoters of miRNAs [2,24]. To validate this difference, we expressed and purified a recombinant M. oryzae MoCdc5-DBD from E. coli, and compared its binding capacity using a biolayer interferometry (BLI) approach for a 16-bp DNA fragment containing a CTCAGCG motif to the promoter sequence of MIR167a (designed as MIR167a as following) and to a 16-bp DNA fragment harboring an MSE. The dissociation constant (Kd) of MIR167a to MoCdc5-DBD was 5.17 × 10−8 M, which was 12-fold lower than the 63.82 × 10−8 M of an MSE to MoCdc5-DBD (Figure 2). Meanwhile, the on-rate (kon) and off-rate (koff) constants of MIR167a to MoCdc5-DBD (7.23 × 104 M−1s−1 for kon, 3.12 × 10−3 s−1 for koff) were higher and lower than those of MSE to MoCdc5-DBD (1.27 × 104 M−1s−1 for kon, 7.14×10−3 s−1 for koff), respectively (Figure 2). These results indicated that MoCdc5-DBD had stronger binding affinities to MIR167a than an MSE did. These findings also suggested that MoCdc5-DBD possessed different DNA-binding capacity as compared with the Myb domain. Moreover, we generated three types of mutant alleles of CTCAGCG in MIR167a and found that the interaction capacity between MoCdc5-DBD and mutated alleles of MIR167a was greatly attenuated (Supplementary Table S1). Similar results were obtained by analyzing mutated alleles of MIR167a and AtCdc5-DBD or TvMyb-DBD (Supplementary Table S1). These results further confirmed that MoCdc5-DBD had a strong binding affinity for MIR167a.
Figure 2. Binding capacity between MoCdc5-DBD and MIR167A is stronger than that between MoCdc5-DBD and an Myb site-binding element (MSE).
(A) Representative binding parameters of MoCdc5-DBD to biotin-labeled MIR167A or MSE examined by BLI assays. MoCdc5 bound to MIR167A with higher affinity than MSE. MIR167a had better binding affinities (higher kon, lower koff, and lower Kd) to MoCdc5-DBD than that to MSE(167A). (B,C) Biotin-labeled DNA was coupled to streptavidin-coated biosensors and monitored for binding to purified MoCdc5-DBD (B) and MSE(167A) (C) at a concentration of 0.5, 1, 2, and 4 μM. The data were analyzed based on a 1:1 binding model using BLI Pro software. The BLI signals for association at titrated concentrations and dissociation are shown.
Crystal structure of MoCdc5-DBD
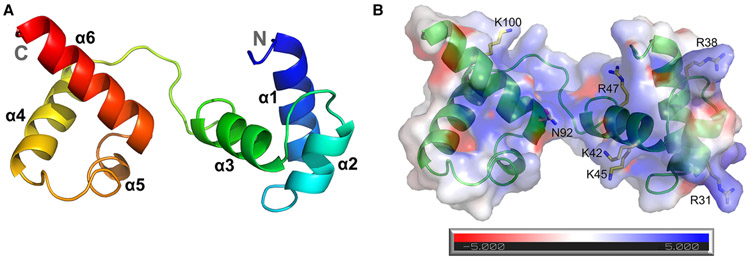
To compare the molecular basis of the differences in DNA binding between MoCdc5-DBD and typical Myb domains, we solved the MoCdc5-DBD crystal structure. This crystal structure was determined by molecular replacement and refined at a resolution of 2.4 Å and belongs to the space group C2 2 21 (Table 1). The MoCdc5-DBD crystal consists of six helices that form two repeats of the HTH motif (Figure 3). The first repeat contained H1 (Asn11 to Tyr24), H2 (Trp29 to Leu35), and H3 (Pro41 to Glu49), while the second repeat contained H4 (Lys63 to Leu75), H5 (Ala84 to Ile86), and H6 (Ala91 to Ala105). The two triple-helix repeats were connected by a 12 residues linker (Trp51 to Ser62). Structural similarity searches in the PDB with free MoCdc5-DBD using the secondary structure matching server identified the strongest matches with the Myb protein from T. vaginalis (TvMyb2-DNA complex; 3OSF, Q score = 0.87) and M. musculus (c-Myb-DNA complex; 1MSE, Q score = 0.51). The root-mean-square deviation and sequence identity compared with TvMyb2 and c-Myb were 0.81 Å, 33.7% and 2.69 Å, 34.4%, respectively [21,24,40].
Table 1.
Data collection and refinement statistics for the MoCdc5-DBD crystal
| Data collection | Cdc5-DBD |
|---|---|
| Wavelength (Å) | 0.979 |
| Resolution range (Å) | 38.14–2.402 (2.488–2.402)a |
| Space group | C 2 2 21 |
| Unit cell | |
| a, b, c (Å) | 72.78, 76.277, 54.409 |
| α, β, γ (°) | 90, 90, 90 |
| Total reflections | 45 266 (4565) |
| Unique reflections | 6154 (593) |
| Multiplicity | 7.4 (7.6) |
| Completeness (%) | 99.29 (97.83) |
| Mean I/σ(I) | 22.10 (2.61) |
| Wilson B-factor | 59.37 |
| 0.05466 (0.8814) | |
| Rmeas | 0.0589 (0.9457) |
| CC1/2 | 1 (0.923) |
| 0.2471 (0.3884) | |
| Rfree | 0.2904 (0.4512) |
| Number of non-hydrogen atoms | 840 |
| Macromolecules | 836 |
| Solvent | 4 |
| Protein residues | 102 |
| RMS (bonds) | 0.006 |
| RMS (angles) | 1.01 |
| Ramachandran favored (%)d | 93.00 |
| Ramachandran allowed (%) | 7.00 |
| Ramachandran outliers (%) | 0.00 |
| Rotamer outliers (%) | 0.00 |
| Clashscore | 8.91 |
| Average B-factor | 80.21 |
| Macromolecules | 80.37 |
| Solvent | 47.81 |
| Number of TLS groups | 6 |
Numbers in parenthesis are for the highest resolution data shell.
Rmerge = ∑hkl∑i(∣Ii(hkl) − 〈I(hkl)〉∣)/∑hkl∑iIi(hkl).
Rwork = ∑hkl(∥Fobs∣−∣Fcalc∥)/∑Ihkl∣Fobs∣.
As evaluated by MolProbity.
Figure 3. Structure of MoCdc5-DBD.
(A) Ribbon diagram of the crystal structure of MoCdc5-DBD. Each helix is represented by a different colored ribbon. The N- and C-terminals of the domain are labeled. (B) Model of electrostatic surfaces of MoCdc5-DBD calculated from −5 kT/e to +5 kT/e using DelPhi. Its seven amino acid residues, R31, R38, K42, K45, R47, N92, and K100, are predicted to be involved in nucleic acid binding.
The CD spectrum of MoCdc5-DBD showed standard characteristics of α-helices, such as minimal peaks at 208 and 222 nm (Supplementary Figure S2) [41], which was consistent with the observed α-helix structure in the MoCdc5-DBD crystal. As predicted, six conserved aromatic amino acids, including Trp9, Trp29, Trp48, Trp61, Trp80, and Tyr98 (Supplementary Figure S3A), were arranged regularly and formed hydrophobic cores that were responsible for the stability of the two HTH repeats. When any of these aromatic amino acids were mutated, the CD spectra of the mutated MoCdc5-DBD proteins were visibly changed (Supplementary Figure S3B), indicating that the α-helix structure in MoCdc5-DBD was unstabilized. Our results were consistent with previous findings that the mutation of any one tryptophan residue in c-Myb can destroy the hydrophobic core and results in a loss of binding activity [42,43].
Key residues in MoCdc5-DBD involved in DNA binding
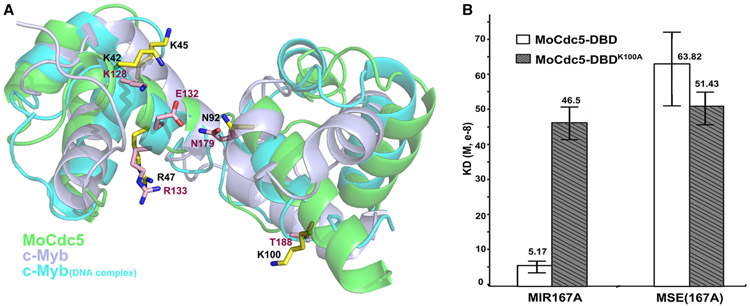
In a comparison of the typical Myb domain of c-Myb (PDB: 1GV2) and the DNA-complexed c-Myb (PDB: 1MSE) [24], structure superposition of MoCdc5-DBD demonstrated the high similarity between these two proteins and indicated that MoCdc5-DBD was more alike to the DNA-complexed c-Myb (Figure 4A). In c-Myb/DNA complex structures, Myb proteins interact with DNA by positioning the two long helixes, α3 and α6, into the major groove using the HTH, and the two long helixes were lined with several positively charged residues. Five key amino acids, K128, E132, R133, N179, and T188 had previously been determined to play important roles in DNA binding [24]. By homologous structural simulation, five conserved residues, K42, K45, R47, N92, and K100, in MoCdc5-DBD were predicted to form a positively charged groove for DNA binding (Figure 4A). Moreover, two basic amino acid residues, R31 and R38, were well conserved among Cdc5-DBD proteins but not in typical Myb domains (Figure 1) and were located around the positively charged groove (Figure 3B).
Figure 4. MoCdc5-DBD displays a similar DNA-binding surface as Myb domain.
(A) Residues involved in intermolecular interactions are represented as sticks on a cartoon model of superimposed structures of MoCdc5-DBD with an Myb domain of c-Myb (PDB: 1GV2) and DNA-complexed c-Myb (PDB: 1MSE). The five amino acid residues, K42, K45, R47, N92, and K100, in MoCdc5-DBD form a DNA-binding surface. MoCdc5-DBD, Myb domains of c-Myb (PDB: 1GV2), and DNA-complexed c-Myb (PDB: 1MSE) are shown in green, light silver, and blue, respectively. (B) The K100 in MoCdc5-DBD is a key residue for high binding affinity. The BLI results with the calculated Kd on the binding of MoCdc5-DBD or its R31M mutant allele with MIR167a (CTCAGCG) or MSE(167A) (AAC(G/T)G), respectively. Error bars indicate SD from three replicates.
To validate the roles of these seven amino acid residues responsible for DNA binding, we independently generated point mutations, K42A, K45A, R47A, N92A, K100A, R31M, and R38N, in MoCdc5-DBD. Analysis of these point mutants by CD showed that all seven point mutations exhibited a nearly superimposable pattern from 195 to 260 nm, suggesting that these mutations did not alter the fold structure of MoCdc5-DBD (Supplementary Figure S2). We then measured their DNA-binding affinities by BLI. Compared with the wild-type MoCdc5-DBD, these seven mutant alleles had reduced binding affinities to MIR167a to varying degrees (Table 2). The R47A, R31M, and R38N alleles only exhibited a slightly reduced capacity for DNA binding to the MIR167a (Table 2).
Table 2.
Binding parameters for the interaction between MoCdc5-DBD or its mutated alleles and the MIR167a or Myb site-binding element (MSE) using BLI
|
MIR167a |
MSE(167A) |
|||||
|---|---|---|---|---|---|---|
| kon (1/Ms, 104) | koff (1/s, 10−3) | kon (1/Ms, 104) | koff (1/s, 10−3) | |||
| MoCdc5-DBD | 5.17 ± 2.13 | 7.23 ± 1.96 | 3.12 ± 0.75 | 63.82 ± 17.39 | 1.27 ± 0.67 | 7.14 ± 3.38 |
| MoCdc5-DBDK42A | NDb | NDb | NDb | NDb | NDb | NDb |
| MoCdc5-DBDK45A | 111.83 ± 67.23 | 2.43 ± 1.75 | 13.34 ± 5.2 | 299.53 ± 65.57 | 0.85 ± 0.26 | 23.34 ± 2.85 |
| MoCdc5-DBDN92A | 154.8 ± 14.13 | 1.17 ± 0.07 | 18.03 ± 0.64 | 231.87 ± 22.15 | 1.07 ± 0.18 | 24.48 ± 2.51 |
| MoCdc5-DBDK100A | 46.5 ± 5.26 | 4.02 ± 0.3 | 18.49 ± 0.94 | 51.43 ± 4.64 | 3.36 ± 0.23 | 17.22 ± 1.47 |
| MoCdc5-DBDR47A | 18.32 ± 3.16 | 7.47 ± 1.05 | 13.42 ± 2.31 | 35.1 ± 4.24 | 4.19 ± 0.16 | 14.63 ± 1.56 |
| MoCdc5-DBDR31M | 19.43 ± 0.64 | 18.75 ± 2.31 | 36.56 ± 5.5 | 46.17 ± 5.49 | 11.41 ± 0.59 | 52.34 ± 3.78 |
| MoCdc5-DBDR38N | 17.04 ± 3.65 | 18.25 ± 1.05 | 30.73 ± 5.26 | 32.53 ± 3.16 | 8.36 ± 0.71 | 27.52 ± 5.11 |
| AtCdc5-DBD | 8.98 ± 0.9 | 25.93 ± 0.82 | 23.27 ± 2.08 | 27.73 ± 0.24 | 26.28 ± 0.32 | 72.88 ± 1.5 |
| tvMyb-DBD | 66.76 ± 3.44 | 1.60 ± 0.04 | 10.74 ± 0.41 | 70.81 ± 3.32 | 1.62 ± 0.042 | 11.51 ± 0.45 |
Kd is the equilibrium dissociation constant. The experiments were repeated three times.
ND, not determined due to the small binding affinity. Shown values are means ± SD of three experiments.
Notably, no detectable binding signals were measured for the K42A allele, indicating that K42 plays an essential role in MoCdc5-DBD binding to DNA. By sequence alignment as shown in Figure 1, the analogous residue to K42 in MoCdc5-DBD was R84 in TvMyb2 and K128 in c-Myb. Previously, the TvMyb2 R84A mutation was shown to completely abolish TvMyb2 binding to the Ap65-1 promoter [21], and the c-Myb K128A mutant allele losses the capacity to bind cognate DNA [24]. Thus, this result supported these previous findings and predicted a key role for residue K42 in DNA binding in MoCdc5-DBD (Figure 4A).
The K45A and N92A alleles exhibited a 20-fold reduction in binding to MIR167a DNA (Table 2). In previous reports, R87 in TvMyb2, the corresponding residue of K45 in MoCdc5-DBD, interacted with backbone phosphates in DNA and contributed to sequence-specific recognition and binding energy in the TvMyb2-DNA complex [21]. K138 in TvMyb2 and N179 in c-Myb, the counterpart residue to N92 in MoCdc5-DBD, interacted directly with DNA bases by specific hydrogen bonds. Mutation of these residues weakened protein-DNA binding [21,22]. Our results were consistent with previous studies, suggesting that the K45 and N92 in MoCdc5-DBD played vital roles in directly binding to DNA (Figure 4A).
The K100A mutation in MoCdc5-DBD reduced DNA-binding affinity nearly 10-fold (Figure 4B; Table 2). In TvMyb2, the corresponding residue T143 interacted with double phosphate backbones, primarily mediated by two water molecules [21]. Thus, we speculated that MoCdc5-DBD K100 might directly interact with DNA through its long side chain. Interestingly, the K100A mutation resulted in a significantly decreased affinity for binding to MIR167a (CTCAGCG) and no obvious differences in binding to MSE(167A) (AAC(G/T)G) (Figure 4B and Table 2). Therefore, these findings suggest that K100 is a key residue in MoCdc5-DBD for determining its high binding affinity to the CTCAGCG motif.
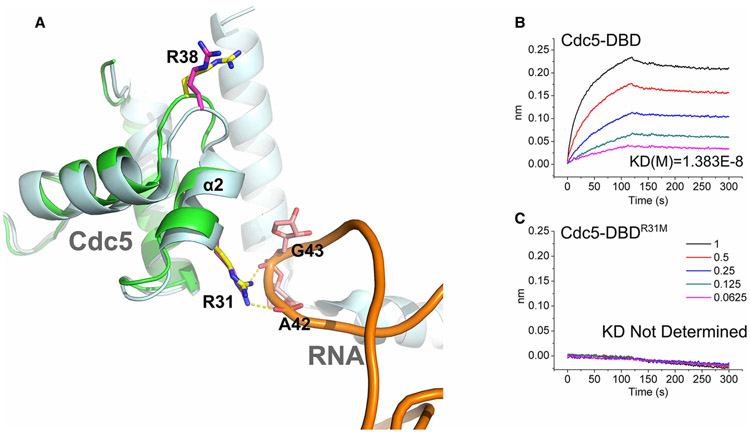
R31 is a key residue for the interaction of MoCdc5-DBD with U6 RNA
Previous findings have reported that the N-terminus of yeast Cdc5 is able to bind U6 RNA [25]. Moreover, R31 in yeast Cdc5 (PDB: 3JB9) was shown to interact with the A42 and G43 bases of U6 RNA [28]. A similar scenario occurred for analogous residues in MoCdc5-DBD (R31) in binding simulation analysis (Figure 5A). To evaluate the role of R31 in RNA binding, the binding capacity between wild-type and mutant MoCdc5-DBD and U6 RNA was measured by BLI. MoCdc5-DBD exhibited a strong binding capacity toward U6 RNA, in which the value of Kd was 1.383 × 10−8 M (Figure 5B and Supplementary Table S2). Intriguingly, the R31M mutation allele completely lost its capacity to bind U6 RNA (Figure 5C and Supplementary Table S2). In contrast, other mutant alleles, including K42A and K100A in the positively charged groove, and R38 M near the R31 residue, only slightly reduced the capacity of MoCdc5-DBD for U6 RNA binding in comparison with wild-type MoCdc5-DBD (Supplementary Table S2). These findings suggest that R31 plays a key role in RNA recognition.
Figure 5. MoCdc5-DBD had a unique nucleic acid binding surface with specific amino acid resides.
(A) Two MoCdc5-specific amino acid residues, R31 and R38, are located on the periphery of the positively charged groove.
The residues are shown in a stick model by structure superposition of MoCdc5-DBD (green) and yeast Cdc5 (PDB code: 3JB9) (light silver). The R31 residue of MoCdc5-DBD interacts with the A42 and G43 of U6 RNA. (B,C) BLI results of MoCdc5-DBD and its R31M mutant allele binding to U6 RNA. The BLI signals for association at titrated concentrations and dissociation and the calculated Kd are shown.
Two distinct nucleic acid binding surfaces are required for Cdc5-DBD function in plant development
The above findings suggested that MoCdc5-DBD could have two distinct nucleic acid binding surfaces. One surface consisted of five amino acid residues that formed a positively charged groove for DNA binding. While K42, K45, and N92 play vital roles in DNA binding, the K100 residue determines the high binding affinity of MoCdc5-DBD to the CTCAGCG motif. The other surface includes the Cdc5-DBD-specific amino acid residue R31, which mainly participates in RNA binding.
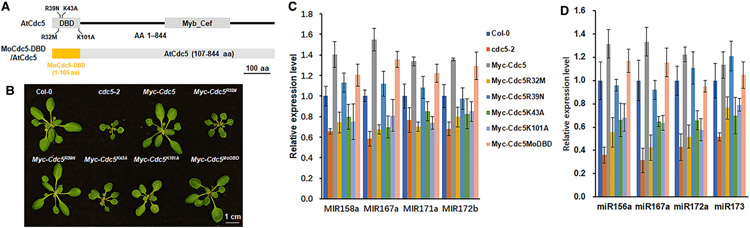
To evaluate the roles of these seven key amino acids of MoCdc5-DBD, we attempted to generate gene deletion mutants of MoCDC5 and then performed complementation assays with point mutant alleles of MoCdc5. However, we failed to obtain the deletion mutants of MoCDC5, which may have been caused by the essential roles of MoCdc5 in the viability of M. oryzae. Recently, a T-DNA knockout mutant (cdc5-2) of AtCdc5 was shown to be viable and exhibited severe defects in plant growth and leaf morphology [2,13]. Thus, we sought to instead use A. thaliana to examine the function of these amino acids. To evaluate if this was feasible, we first evaluated the functional conservation between AtCdc5-DBD and MoCdc5-DBD using a mosaic gene construct, termed MoCdc5-DBD-AtCdc5, in which the DBD (1–106 aa) of AtCdc5 was replaced with its counterpart from MoCdc5-DBD (1–105 aa) (Figure 6A). Constitutive expression of MoCdc5-DBD-AtCdc5 fully complemented the defects in the cdc5-2 line (Figure 6B,C and Supplementary Figure S3), revealing that AtCdc5-DBD and MoCdc5-DBD are functionally interchangeable for development.
Figure 6. The two distinct nucleic acid binding surfaces are both vital to Cdc5-DBD for facilitating full functions in Arabidopsis thaliana.
(A) Schematic structures of site-directed mutagenesis in the AtCdc5-DBD (R32M, R39N, K43A, and K101A), and a mosaic allele of AtCdc5, in which AtCdc5-DBD was replaced with MoCdc5-DBD. Scale bar, 100 aa. (B) Morphological phenotypes of 3-week-old seedlings of the wild-type Col-0, AtCdc5T-DNA knockout mutant cdc5-2, and transgenic cdc5-2 harboring pro35s∷Myc-AtCDC5, pro35s∷Myc-AtCDC5R32M, pro35s∷Myc-AtCDC5R39N, pro35s∷Myc-AtCDC5K43A, pro35s∷Myc-AtCDC5K101A, and pro35s∷Myc-AtCDC5MoDBD. Scale bar, 1 cm. (C,D) Levels of primary miRNA transcripts (pri-miRNAs) (C) and miRNAs (D) in Col-0, cdc5-2, and various transgenic cdc5-2 lines. The pri-miRNA levels in cdc5-2 and various transgenic plants were normalized to those of UBQ5 and compared with that of Col-0 (values were set as 1). The miRNA levels in cdc5-2 and various transgenic plants were normalized to those of U6 RNA and compared with Col-0 (value set as 1). Error bars indicate the SD from three replicates.
Based on the alignment of key amino acids between MoCdc5 and AtCdc5 (Figure 1), we generated four point mutant alleles of AtCdc5, that is, K43A, K101A, R32M, and R39N (Supplementary Figures S1 and S6A), which were counterparts to K42, K100, R31, and R38 of MoCdc5, respectively, and we then examined the effect of these mutations on AtCDC5 function through a complementation assay. These Atcdc5 mutants and a wild-type of AtCDC5 under the control of a constitutive 35S promoter were individually introduced into the cdc5-2 line. In the F2 population, the cdc5-2 plants harboring the transgenes were selected by screening for basta-resistance. Western blot analysis showed that all transgenes displayed similar expression levels in various transgenic cdc5-2 plants (Supplementary Figure S4). We then compared their growth phenotypes with the wild-type Col-0 and cdc5-2 lines. Among transgenic plants, the R39N or AtCdc5 allele fully rescued the developmental defects of the cdc5-2 allele, whereas the K101A allele partially recovered the growth defects of the cdc5-2 allele (Figure 6B). In contrast, the K43A and R32M alleles failed to complement the morphological phenotypes of the cdc5-2 allele (Figure 6B). These findings suggested that K43 and R32, but not R39, were essential for AtCdc5 function, while K101 was important for AtCdc5 activity. We also evaluated the levels of pri-miRNAs and miRNAs in Col-0, cdc5-2, and various transgenic lines with qRT-PCR. Compared with AtCdc5 or R39N, K43A, K101A, and R32M failed to recover the levels of both pri-miRNAs and miRNAs in cdc5-2 (Supplementary Figure S6C,D). These results suggested that both of the two distinct nucleic acid binding surfaces were required for the function of AtCdc5 in controlling plant development and miRNA biogenesis.
Discussion
In this study, we integrated the approaches of protein–DNA interaction analysis, 3-D structural comparison, and functional analysis of key amino acid residues and demonstrated that Cdc5 regulates development via two distinct nucleic acid binding surfaces in its N-terminal DBD. We found that K100 was a key residue for the high binding affinity to the Cdc5-binding element, which was located on the periphery of the positively charged groove and contributed to the higher binding capacity for a Cdc5-binding element (CTCAGCG) than an Myb domain-binding element (AAC(G/T)G). Sequence alignment showed that the 100th amino acids in Cdc5 orthologous proteins were mainly long side-chain amino acids, that is, lysine, arginine, phenylalanine, and tyrosine, which were different from Myb proteins that commonly present amino acids with short side chains, that is, valine, serine, threonine, and alanine (Supplementary Figure S5). T143 in TvMyb2, the counterpart of K100, also interacts with DNA [21]. In our study, mutation of the K100 significantly reduced the binding capacity of MoCdc5-DBD to the CTCAGCG but not to AAC(G/T)G, and lead to loss of selectivity of Cdc5-DBD to CTCAGCG and AAC(G/T)G. Moreover, the mutation line of the K100 counterpart in A. thaliana AtCdc5 exhibited severe defects in plant growth and leaf morphology, as well as having down-regulated transcript levels of targeted miRNA-encoding genes. Thus, these findings suggest that K100 is a key residue for the high binding affinity to a Cdc5-binding element and is important for Cdc5 full functionality. Thus, it plays key roles in the specific recognition of the Cdc5-binding element and differentiating Cdc5-DBD from Myb domains.
We also present here the whole spatial arrangement and high-resolution structure of the HTH tandem repeats of MoCdc5-DBD. Sequence alignment and structure comparison show that Cdc5-DBD shares high similarity to the Myb domain. Both of these contain four basic amino acids, K42, K45, R47, and N92, which are located at the same position in the HTH structure and form a positively charged groove, serving as an interaction interface with DNA [21,24]. The importance of these residues in DNA binding was validated by analysis of point mutants through a BLI assay. Moreover, analyses on the in vivo biological functions and transcript levels of pri-miRNAs further validated the importance of the DNA binding of the counterparts of these four conserved residues in AtCdc5-DBD. Importantly, mutation of K42 in MoCdc5-DBD or its counterpart in AtCdc5-DBD leads to a complete loss of function of Cdc5. In previous reports, the TvMyb2 R84A mutation allele completely abolished protein binding to the Ap65-1 promoter [21], and the c-Myb K128A mutation allele loses the capacity to bind to cognate DNA [24]. Taken together, these findings suggest that Cdc5-DBD is highly similar to the Myb domain at the conserved nucleic acid binding surface used for DNA binding.
Importantly, Cdc5-DBD has an extra nucleic acid binding surface, which contains a conserved basic amino acid residue (R31) and is located around the positively charged groove. In fission yeast, the N-terminal region of SpCdc5 contains the R1 and R2 repeats in its DBD, and the D3 repeat is next to the DBD. These three repeats have been recently shown to function as an RNA-binding platform and directly interact with U6 RNA ISL near the catalytic core of the spliceosome [25]. Moreover, a Cryo-EM structure of the spliceosome shows that the counterpart of R31 in yeast Cdc5 (PDB: 3JB9) is close to the U6 RNA [28]. In our study, the MoCdc5-DBD structure also exhibited a similar RNA-binding pattern as yeast Cdc5-DBD does, suggesting that this extra nucleic acid binding surface for RNA binding has been conserved during evolution. Intriguingly, the R31M mutant allele of MoCdc5-DBD lost the capacity to bind U6 RNA. Moreover, the mutation line of the R31 counterpart in AtCdc5 exhibited the same phenotypes as the T-DNA knockout mutant of AtCdc5 in terms of manifesting dwarf plant and small leaves. Taken together, we speculate that this extra nucleic acid binding surface with the key residue R31 collaborates with the D3 domain in Cdc5 to bind RNA and is essential to Cdc5 for fulfilling diverse functions, including plant development.
We also found that the expression of AtCdc5 mutant proteins containing the K43A, K101A, or R32M mutations failed to rescue the pri-miRNA levels in a cdc5 mutant of Arabidopsis. There are at least two explanations for this observation. One is that these Cdc5 mutants may lose their ability to recognize and bind to MIR promoters containing Cdc5-binding motifs, leading to a reduced transcript rate of pri-miRNAs. Supporting this notion, the Cdc5-binding element CTCAGCG exists in promoters of all AtCdc5-targeted MIRs. The other explanation is that these mutant alleles of AtCdc5 may impair the splicing of AtCdc5-targeted pri-miRNAs. We show that these mutations in MoCdc5 abolish its binding to U6 RNA, especially the R31 mutation. In the Prp19 complex, Cdc5 and Cwc2 are the only two components with nucleic acid binding domains and have the potential to interact with snRNAs [44,45]. The N-terminus of SpCdc5 in fission yeast has recently been proposed to play a central role in pre-mRNA processing through its interaction with other components of the Prp19 complex and snRNAs within the catalytic core of the spliceosome [25].
In summary, our findings suggest that the seven amino acid residues, K42, K45, R47, N92, K100, R31, and R38, in Cdc5-DBD form two distinct nucleic acid binding surfaces to participate in nucleic acid binding (Supplementary Figure S6). Similar to Myb domain, the five amino acid residues, K42, K45, R47, N92, and K100 form a positively charged groove and interact with a DNA-binding surface. On the DNA-binding surface, the K42, K45, and N92 residues are essential, and the K100 residue contributes to the high binding specificity for the Cdc5-binding element. On the other RNA-binding surface, R31 is a key residue that is located close to the positively charged groove and binds to U6 RNA. These new findings together with previous findings on Cdc5 contribute to establish a molecular model of its nucleic acid binding and are helpful for explaining how Cdc5 plays diverse important roles during different molecular processes.
Supplementary Material
Acknowledgements
We thank the staff at Shanghai Synchrotron Research Facility (SSRF) beamline BL-17U for help with crystal screening and data collection. We also thank Yuanyuan Chen and Jianhui Li of Institute of Biophysics, Chinese Academy of Sciences for providing the facility support of BLI and CD.
Funding
This work was supported by the National Key Research and Development Plan [2016YFD0300700], the Natural Science Foundation of China [31872916], the Program for Changjiang Scholars and Innovative Research Team in University [IRT1042], the 111 Project [B13006], and the National Institute of Health [GM127414]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- ABRC
Arabidopsis Biological Resources Center
- ATR
ataxia-telangiectasia and rad3-related
- BLI
biolayer interferometry
- Cdc5
cell division cycle 5
- CD
circular dichroism
- Cryo-EM
cryogenic electron microscopy
- DBD
DNA-binding domain
- DCL1
DICER-LIKE 1
- HTH
helix–turn–helix
- MES
2-(N-Morpholino) ethane sulfonic acid
- MIR
miRNA
- MSE
Myb site-binding element
- Myb
myeloblastosis
- Pol II
DNA-dependent RNA polymerase II complex
- pri-miRNAs
primary transcripts of miRNAs
- qRT-PCR
quantitative real-time PCR
Footnotes
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
References
- 1.Zhang N, Kaur R, Akhter S and Legerski RJ (2009) Cdc5l interacts with ATR and is required for the S-phase cell-cycle checkpoint. EMBO Rep. 10, 1029–1035 10.1038/embor.2009.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang S, Xie M, Ren G and Yu B (2013) Cdc5, a DNA binding protein, positively regulates posttranscriptional processing and/or transcription of primary microRNA transcripts. Proc. Natl Acad. Sci. U.S.A 110, 17588–17593 10.1073/pnas.1310644110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lei XH, Shen X, Xu XQ and Bernstein HS (2000) Human Cdc5, a regulator of mitotic entry, can act as a site-specific DNA binding protein. J. Cell Sci 113, 4523–4531 [DOI] [PubMed] [Google Scholar]
- 4.Yan C, Wan R, Bai R, Huang G and Shi Y (2017) Structure of a yeast step II catalytically activated spliceosome. Science 355, 149–155 10.1126/science.aak9979 [DOI] [PubMed] [Google Scholar]
- 5.Maréchal A, Li JM, Ji XY, Wu CS, Yazinski SA, Nguyen HD et al. (2014) Prp19 transforms into a sensor of RPA-ssDNA after DNA damage and drives ATR activation via a ubiquitin-mediated circuitry. Mol. Cell 53, 235–246 10.1016/j.molcel.2013.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wan L and Huang J (2014) The PSO4 complex associates with RPA and modulates the activation of ATR. J. Biol. Chem 289, 6619–6626 10.1074/jbc.M113.543439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burns CG, Ohi R, Krainer AR and Gould KL (1999) Evidence that Myb-related CDC5 proteins are required for pre-mRNA splicing. Proc. Natl Acad. Sci. U.S.A 96, 13789–13794 10.1073/pnas.96.24.13789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan C, Wan R, Bai R, Huang G and Shi Y (2016) Structure of a yeast activated spliceosome at 3.5 Å resolution. Science 353, 904–911 10.1126/science.aag0291 [DOI] [PubMed] [Google Scholar]
- 9.Lin Z, Yin K, Zhu D, Chen Z, Gu H and Qu LJ (2007) Atcdc5 regulates the G2 to M transition of the cell cycle and is critical for the function of Arabidopsis shoot apical meristem. Cell Res. 17, 8–15 [DOI] [PubMed] [Google Scholar]
- 10.Palma K, Zhao Q, Cheng YT, Bi D, Monaghan J, Cheng W et al. (2007) Regulation of plant innate immunity by three proteins in a complex conserved across the plant and animal kingdoms. Genes Dev. 21, 1484–1493 10.1101/gad.1559607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH et al. (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23, 4051–4060 10.1038/sj.emboj.7600385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stegmeier F, Hu G, Rickles RJ, Hannon GJ and Elledge SJ (2005) A lentiviral microRNA-based system for single-copy polymerase II-regulated RNA interference in mammalian cells. Proc. Natl Acad. Sci. U.S.A 102, 13212–13217 10.1073/pnas.0506306102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu Q, Liu Y, Li M and Yu B (2017) Small RNA biogenesis and degradation in plants. Plant Epigenet. 107–127 10.1007/978-3-319-55520-1_6 [DOI] [Google Scholar]
- 14.Xie Z, Kasschau KD and Carrington JC (2003) Negative feedback regulation of dicer-like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol 13, 784–789 10.1016/S0960-9822(03)00281-1 [DOI] [PubMed] [Google Scholar]
- 15.Zhu H, Zhou Y, Castillo-González C, Lu A, Ge C, Zhao YT et al. (2013) Bidirectional processing of pri-miRNAs with branched terminal loops by Arabidopsis dicer-like1. Nat. Struct. Mol. Biol 20, 1106–1112 10.1038/nsmb.2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirayama T and Shinozaki K (1996) A cdc5+ homolog of a higher plant, Arabidopsis thaliana. Proc. Natl Acad. Sci. U.S.A 93, 13371–13376 10.1073/pnas.93.23.13371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stracke R, Werber M and Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol 4, 447–456 10.1016/S1369-5266(00)00199-0 [DOI] [PubMed] [Google Scholar]
- 18.Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C and Lepiniec L (2010) Myb transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581 10.1016/jrtplants.2010.06.005 [DOI] [PubMed] [Google Scholar]
- 19.Ogata K, Kanei-Ishii C, Sasaki M, Hatanaka H, Nagadoi A, Enari M et al. (1996) The cavity in the hydrophobic core of Myb DNA-binding domain is reserved for DNA recognition and trans-activation. Nat. Struct. Biol 3, 178–182 10.1038/nsb0296-178 [DOI] [PubMed] [Google Scholar]
- 20.Jia L, Clegg MT and Jiang T (2004) Evolutionary dynamics of the DNA-binding domains in putative R2R3-MYB genes identified from rice subspecies indica and japonica genomes. Plant Physiol. 134, 575–585 10.1104/pp.103.027201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang I, Tsai CK, Chen SC, Wang SH, Amiraslanov I, Chang CF et al. (2011) Molecular basis of the recognition of the ap65-1 gene transcription promoter elements by a Myb protein from the protozoan parasite Trichomonas vaginalis. Nucleic Acids Res. 39, 8992–9008 10.1093/nar/gkr558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei SY, Lou YC, Tsai JY, Ho MR, Chou CC, Rajasekaran M et al. (2011) Structure of the Trichomonas vaginalis Myb3 DNA-binding domain bound to a promoter sequence reveals a unique C-terminal β-hairpin conformation. Nucleic Acids Res. 40, 449–460 10.1093/nar/gkr707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanikawa J, Yasukawa T, Enari M, Ogata K, Nishimura Y, Ishii S et al. (1993) Recognition of specific DNA sequences by the c-myb protooncogene product: role of three repeat units in the DNA-binding domain. Proc. Natl Acad. Sci. U.S.A 90, 9320–9324 10.1073/pnas.90.20.9320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogata K, Morikawa S, Nakamura H, Sekikawa A, Inoue T, Kanai H et al. (1994) Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell 79, 639–648 10.1016/0092-8674(94)90549-5 [DOI] [PubMed] [Google Scholar]
- 25.Collier SE, Voehler M, Peng D, Ohi R, Gould KL, Reiter NJ et al. (2014) Structural and functional insights into the N-terminus of Schizosaccharomyces pombe Cdc5. Biochemistry 53, 6439–6451 10.1021/bi5008639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X, Yan C, Zhan X, Li L, Lei J and Shi Y (2018) Structure of the human activated spliceosome in three conformational states. Cell Res. 28, 307–322 10.1038/cr.2018.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wan R, Yan C, Bai R, Huang G and Shi Y (2016) Structure of a yeast catalytic step I spliceosome at 3.4 Å resolution. Science 353, 895–904 10.1126/science.aag2235 [DOI] [PubMed] [Google Scholar]
- 28.Yan C, Hang J, Wan R, Huang M, Wong CC and Shi Y (2015) Structure of a yeast spliceosome at 3.6-angstrom resolution. Science 349, 1182–1191 10.1126/science.aac7629 [DOI] [PubMed] [Google Scholar]
- 29.Hang J, Wan R, Yan C and Shi Y (2015) Structural basis of pre-mRNA splicing. Science 349, 1191–1198 10.1126/science.aac8159 [DOI] [PubMed] [Google Scholar]
- 30.Fica SM, Oubridge C, Wilkinson ME, Newman AJ and Nagai K (2019) A human postcatalytic spliceosome structure reveals essential roles of metazoan factors for exon ligation. Science 363, 710–714 10.1126/science.aaw5569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pham K, LaForge K and Kreek M (1998) Sticky-end PCR: new method for subcloning. Biotechniques 25, 206–208 10.2144/98252bm04 [DOI] [PubMed] [Google Scholar]
- 32.Liu H and Naismith JH (2008) An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 8, 91 10.1186/1472-6750-8-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Otwinowski Z and Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 307–326 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- 34.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC and Read RJ (2007) Phaser crystallographic software. J. Appl. Crystallogr 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N et al. (2010) Phenix: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarkar S, Witham S, Zhang J, Zhenirovskyy M, Rocchia W and Alexov E (2013) Delphi Web server: a comprehensive online suite for electrostatic calculations of biological macromolecules and their complexes. Commun. Comput. Phys 13, 269–284 10.4208/cicp.300611.201011s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sultana A and Lee JE (2015) Measuring protein-protein and protein-nucleic acid interactions by biolayer interferometry. Curr. Protoc. Protein Sci 79, 19.25.1–26 10.1002/0471140864.ps1925s79 [DOI] [PubMed] [Google Scholar]
- 38.Chow CN, Zheng HQ, Wu NY, Chien CH, Huang HD, Lee TY et al. (2015) Plantpan 2.0: an update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Res. 44, 1154–1160 10.1093/nar/gkv1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ren G, Xie M, Dou Y, Zhang S, Zhang C and Yu B (2012) Regulation of miRNA abundance by RNA binding protein tOUGH in Arabidopsis. Proc. Natl Acad. Sci. U.S.A 109, 12817–12821 10.1073/pnas.1204915109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krissinel E and Henrick K (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr 60, 2256–2268 10.1107/S0907444904026460 [DOI] [PubMed] [Google Scholar]
- 41.Sonnichsen F, van Eyk J, Hodges R and Sykes B (1992) Effect of trifluoroethanol on protein secondary structure: an NMR and CD study using a synthetic actin peptide. Biochemistry 31, 8790–8798 10.1021/bi00152a015 [DOI] [PubMed] [Google Scholar]
- 42.Kanei-Ishii C, Sarai A, Sawazaki T, Nakagoshi H, He DN, Ogata K et al. (1990) The tryptophan cluster: a hypothetical structure of the DNA-binding domain of the Myb protooncogene product. J. Biol. Chem 265, 19990–19995 [PubMed] [Google Scholar]
- 43.Saikumar P, Murali R and Reddy EP (1990) Role of tryptophan repeats and flanking amino acids in Myb-DNA interactions. Proc. Natl Acad. Sci. U.S.A. 87, 8452–8456 10.1073/pnas.87.21.8452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohi R, McCollum D, Hirani B, Den Haese G, Zhang X, Burke J et al. (1994) The Schizosaccharomyces pombe cdc5+ gene encodes an essential protein with homology to c-Myb. EMBO J. 13, 471–483 10.1002/j.1460-2075.1994.tb06282.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McGrail JC, Krause A and O’Keefe RT (2009) The RNA binding protein Cwc2 interacts directly with the U6 snRNA to link the nineteen complex to the spliceosome during pre-mRNA splicing. Nucleic Acids Res 37, 4205–4217 10.1093/nar/gkp341 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.