Abstract
Objectives
Determine global skin transcriptome patterns of early diffuse systemic sclerosis (SSc) and how they differ from later disease.
Methods
Skin biopsy RNA from 48 patients in the Prospective Registry for Early Systemic Sclerosis (PRESS) cohort (mean disease duration 1.3 years) and 33 matched healthy controls was examined by next-generation RNA sequencing. Data were analysed for cell type-specific signatures and compared with similarly obtained data from 55 previously biopsied patients in Genetics versus Environment in Scleroderma Outcomes Study cohort with longer disease duration (mean 7.4 years) and their matched controls. Correlations with histological features and clinical course were also evaluated.
Results
SSc patients in PRESS had a high prevalence of M2 (96%) and M1 (94%) macrophage and CD8 T cell (65%), CD4 T cell (60%) and B cell (69%) signatures. Immunohistochemical staining of immune cell markers correlated with the gene expression-based immune cell signatures. The prevalence of immune cell signatures in early diffuse SSc patients was higher than in patients with longer disease duration. In the multivariable model, adaptive immune cell signatures were significantly associated with shorter disease duration, while fibroblast and macrophage cell type signatures were associated with higher modified Rodnan Skin Score (mRSS). Immune cell signatures also correlated with skin thickness progression rate prior to biopsy, but did not predict subsequent mRSS progression.
Conclusions
Skin in early diffuse SSc has prominent innate and adaptive immune cell signatures. As a prominently affected end organ, these signatures reflect the preceding rate of disease progression. These findings could have implications in understanding SSc pathogenesis and clinical trial design.
INTRODUCTION
Systemic sclerosis (SSc) is a multi-system autoimmune and fibrotic disease associated with high morbidity and mortality.1,2 Treatment options remain limited, and management is complicated by heterogeneity in clinical course and treatment response.
Whole transcriptome gene expression profiling can yield insights into disease pathogenesis and identify distinct subgroups of patients.3,4 We and others have previously used microarray technology to measure global gene expression in skin biopsies from SSc patients in comparison to healthy controls (HCs),5-12 revealing distinct gene expression patterns in SSc skin. Fibrotic and inflammatory gene expression signatures have been observed in a large percentage of patients, while a subset of patients has ‘normal-like’ gene expression profiles. These studies highlight heterogeneity in SSc skin gene expression. A large-scale study to characterise skin gene expression specifically in early, diffuse SSc in comparison to those with later stage disease has been lacking.
We investigated the transcript expression profiles of skin specimens from a large group of patients with early, diffuse SSc from the Prospective Registry for Early Systemic Sclerosis (PRESS) cohort using next generation RNA sequencing. These data were compared with HC skin and to patients in the Genetics versus Environment in Scleroderma Outcomes Study (GENISOS), in which patients had a longer average disease duration.
METHODS
Patients and control subjects
Patients were recruited from PRESS, an observational cohort of early diffuse SSc patients from 11 US academic medical centres.13 Skin biopsies from 48 patients within 3 years of onset of first non-Raynaud’s symptom were used for RNA sequencing, along with 33 biopsies from HCs matched to patients by age, sex and ethnicity. Ten repeat biopsies from eight SSc patients were also available. Skin biopsy was optional in PRESS, and all available biopsies in the PRESS cohort at the time of study were included. Patients fulfilled the 2013 American College of Rheumatology (ACR)/European League Against Rheumatism (EULAR) classification criteria for SSc and had diffuse skin involvement.14 Modified Rodnan Skin Score (mRSS) and local skin score at the biopsy site were recorded at the time of biopsy. Skin thickness progression rate (STPR) was calculated similarly to what was previously described,15 using the equation mRSS at the time of biopsy/time from first puffy fingers or skin thickening. Participants provided informed and written consent.
Skin biopsy and RNA sequencing
Punch biopsies were obtained from the forearm skin. The methods for RNA sequencing and analysis are described in online supplementary methods. Data from the PRESS cohort were compared with similarly obtained data from the GENISOS cohort that included SSc patients with longer disease duration at the time of biopsy.10 Although microarray technology was used for gene expression profiling in the previously published study, we performed RNA sequencing in these GENISOS samples (n=55) and matched HCs (n=33) for the present study in order to avoid heterogeneity resulting from methodological differences.
Analysis of cell type-specific expression
We performed cell type-specific gene expression analysis using the method we have used previously.10,16 Details are provided in the online supplementary methods.
Assignment of patients to ‘intrinsic subsets’ based on skin gene expression
Fragments per kilobase million (FPKM) values were sent to JMF and MLW who were blinded to all clinical data and assigned each sample to one of four ‘intrinsic subsets’ using previously described methods.17,18
Immunohistochemistry
Immunohistochemical (IHC) analyses of skin biopsies are described in the online supplementary methods.
Statistical analysis
Associations between cell type signatures and clinical or histological features were analysed by Spearman’s rank order correlation. Cell type signature scores were log-transformed and compared between the PRESS and GENISOS cohorts by Student’s t-test. Multivariable regression analyses were performed with pooled data from both cohorts with adjustment for clinical variables noted in the text. mRSS and STPR within the intrinsic subsets were analysed by linear regression analyses, using the normal-like subset as a reference.
RESULTS
Demographics
Demographics and clinical characteristics of participants from the PRESS cohort and matched HCs, along with the GENISOS cohort and their matched HCs, are shown in table 1.
Table 1.
Demographics and clinical characteristics of SSc patients from PRESS and GENISOS cohorts and matched healthy controls at the time of skin biopsy
| Characteristic | PRESS SSc (n=48) |
HC matched to PRESS (n=33) |
GENISOS SSc (n=55) |
HC matched to GENISOS (n=33) |
|---|---|---|---|---|
| Age (years) at biopsy, mean (SD) | 48.0 (15.0) | 47.4 (13.4) | 52.8 (12.6) | 46.8 (11.7) |
| Race/ethnicity, n (%) | ||||
| White | 30 (62.5) | 24 (72.7) | 38 (69.1) | 19 (57.6) |
| Black | 5 (10.4) | 4 (12.1) | 8 (14.5) | 8 (24.2) |
| Hispanic | 9 (18.8) | 4 (12.1) | 9 (16.4) | 6 (18.2) |
| Other | 4 (8.3) | 1 (3.0) | 0 | 0 |
| Female, n (%) | 30 (62.5) | 22 (66.7) | 40 (72.7) | 27 (81.1) |
| Disease duration in years, mean (SD) | 1.3 (0.9) | 7.4 (5.2) | ||
| Diffuse skin involvement, n (%) | 48 (100) | 37 (67.3) | ||
| mRSS, mean (SD) | 21.3 (8.7) | 15.3 (10.4) | ||
| Local skin score, mean (SD) | 1.7 (0.8) | 1.1 (0.9) | ||
| FVC % predicted, mean (SD) | 76.0 (19.8) | 77.3 (19.8) | ||
| SSc-associated autoantibody, n (%)* | ||||
| Antitopoisomerase I | 12/41 (29.3) | 15 (27.3) | ||
| Anti-RNA polymerase III | 17/38 (44.7) | 17 (30.9) | ||
| Anticentromere | 1/36 (2.8) | 7 (12.7) | ||
| Mycophenolate, n (%)† | 19 (39.6) | 4 (7.3) | ||
| Methotrexate, n (%)† | 9 (18.8) | 8 (14.5) | ||
| Cyclophosphamide, n (%)† | 1 (2.1) | 1 (1.8) |
indicates positive among those recorded.
Indicates patients taking medication at the time of biopsy.
FVC, forced vital capacity; GENISOS, Genetics versus Environment in Scleroderma Outcomes Study; HC, healthy control; mRSS, modified Rodnan Skin Score;PRESS, Prospective Registry for Early Systemic Sclerosis; SSc, systemic sclerosis.
Transcript expression profile of early diffuse SSc skin
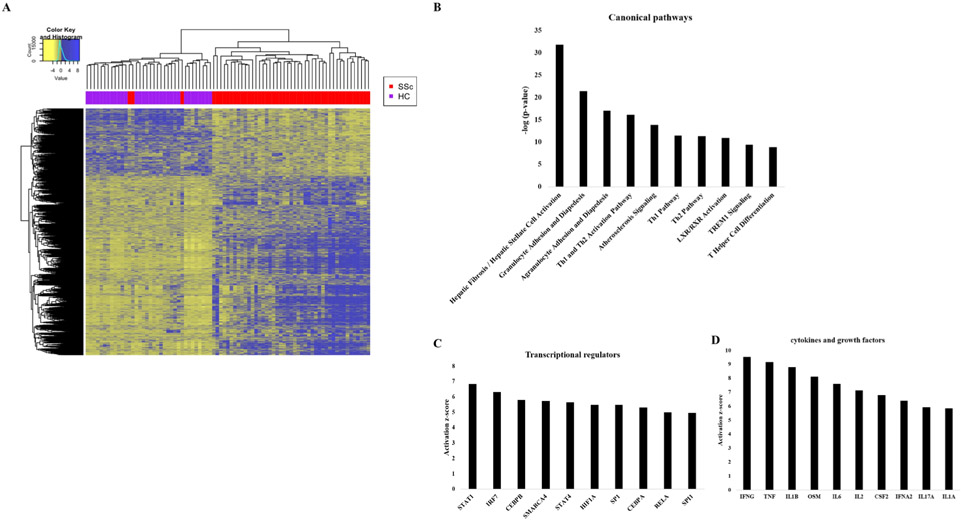
Three thousand eighty seven transcripts were differentially expressed between SSc patients and HCs using a false discovery rate cut-off of 0.05 and fold change cut-off of >1.5 or <0.67, including 927 long non-coding RNAs (lncRNAs). Unsupervised hierarchical clustering revealed nearly complete discrimination between differential transcript expression in HCs and SSc patients, with the exception of three SSc patients whose transcript expression profile largely resembled that of HCs (figure 1A). Lists of differentially expressed transcripts between SSc and HC and associations between transcripts and mRSS or forced vital capacity (FVC) in SSc patients at the time of skin biopsy are included in the supplementary data file on our webpage (https://www.uth.tmc.edu/scleroderma/).The most over-represented pathways in SSc skin based on Ingenuity Pathway Analysis were hepatic fibrosis, granulocyte and agranulocyte adhesion and diapedesis, and Th1 and Th2 activation pathways (figure 1B). Th1 and Th2 activation pathways had not been previously observed in the skin of SSc patients.10 The top activated transcriptional regulators were predicted to be signal transducer and activator of transcription 1, interferon regulatory factor 7, and CCAAT enhancer binding protein beta, while the top activated cytokines/growth factors were interferon gamma, tumour necrosis factor and interleukin 1 beta (IL-1β) (figure 1C, D). Surprisingly, transforming growth factor beta (TGFβ) ranked 15th among upstream cytokines/growth factors (data not shown), in contrast to our prior study of patients with longer disease duration in which it had ranked first.10
Figure 1.
Differentially expressed transcripts and pathways in Prospective Registry for Early Systemic Sclerosis systemic sclerosis (SSc) patients compared with healthy controls (HCs). (A) Heatmap of differentially expressed transcripts, represented by z-score normalised count values. Unsupervised hierarchical clustering is shown at the top, with HCs represented by purple squares and SSc patients represented by red squares. (B) Top 10 over-represented pathways in SSc compared with HC as determined by Ingenuity Pathway Analysis of differentially expressed transcripts (fold change >1.5 or <0.67 in SSc vs HC, with false discovery rate <0.05). (C) Top 10 predicted upstream transcriptional regulators in SSc compared with HC. (D) Top 10 predicted upstream cytokines/growth factors.
Prominent innate and adaptive immune cell signatures in early diffuse SSc skin
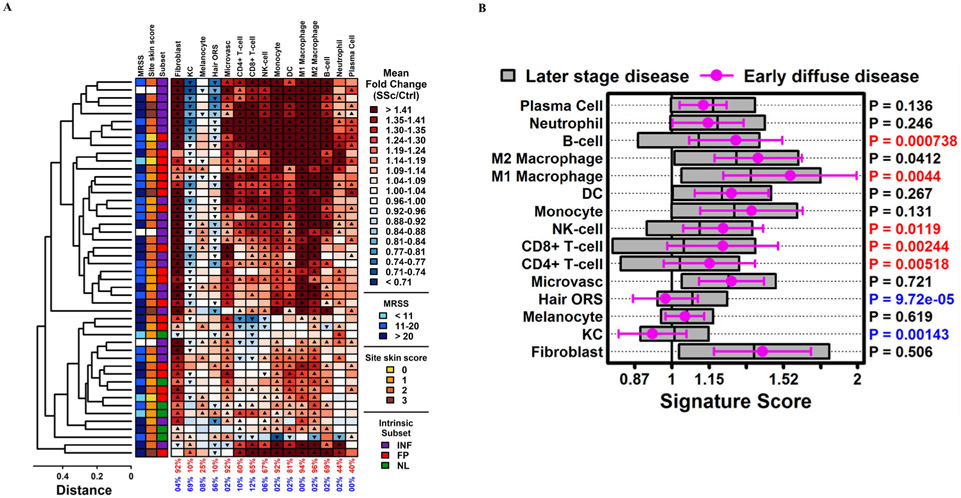
Cell type-specific analysis revealed that most patients had increased innate and adaptive immune cell signatures compared with HCs (figure 2A). The most prevalent signatures upregulated in SSc compared with HC were those of M2 and M1 macrophages (96% and 94% of SSc patients, respectively). A fibroblast signature was present in 92% of patients. Most SSc patients also had CD4 T cell, CD8 T cell and B cell signatures (60%, 65% and 69%, respectively). No significant differences in cell type signatures were observed in male versus female patients or in RNA polymerase III antibody-positive versus topoisomerase I antibody-positive patients (online supplementary tables 1 and 2, respectively).
Figure 2.
Cell type signatures in skin of PRESS SSc patients compared with healthy controls and compared with GENISOS SSc patients. (A) Cell type signature scores for each SSc sample (n=48). Scores represent the average fold-change (SSc/HC) for 125 cell type-specific signature genes (see online supplementary methods). Up-triangles indicate significantly higher scores for signature genes compared with non-signature genes (p<0.05, Wilcoxon rank-sum test). Down-triangles indicate significantly lower scores for signature genes compared with non-signature genes (p<0.05, Wilcoxon rank-sum test). Bottom margin values indicate the percentage of up-triangles (red) and down-triangles (blue), respectively. Patients were clustered based on signature scores (average linkage, Euclidean distance). The coloured boxes to the left of the cell type signature scores indicate the mRSS (left), local skin score at the site of the biopsy (middle) and the intrinsic subset classification, with legends at the right of the figure. White boxes (n=3) indicate no skin scores recorded at the time of the biopsy. (B) Signature scores for PRESS patients (n=48) were compared with those of GENISOS patients (n=55). The mean PRESS score is represented by round symbols with error bars spanning ±1 SD. The mean GENISOS score is represented by the midline for each grey box with boxes spanning ±1 SD. Right margin p values were obtained from a two-sample t-test of PRESS versus GENISOS scores (red: PRESS>GENISOS, FDR<0.05; blue: PRESS<GENISOS, FDR<0.05). Ctrl (HC), healthy control; DC, dendritic cell; GENISOS, Genetics versus Environment in Scleroderma Outcomes Study; hair ORS, hair outer root sheet; KC, keratinocyte; mRSS, modified Rodnan Skin Score; PRESS, Prospective Registry for Early Systemic Sclerosis; SSc, systemic sclerosis. FP, fibroproliferative subset; INF, inflammatory subset; NL, normal-like subset.
We compared the cell type signatures in PRESS patients to those of GENISOS patients for whom we had previously performed skin biopsies and analysed RNA expression by microarray.10 To allow for comparison between the two cohorts, RNA sequencing was performed using the available GENISOS (n=55) and matched HC RNA samples (n=33) from that study. Differences in disease characteristics of the patients whose skin gene expression was analysed in GENISOS and PRESS are shown in table 1. On average, PRESS patients had a shorter disease duration at the time of biopsy than GENISOS patients (1.3 vs 7.4 years, respectively). Compared with GENISOS patients, PRESS patients had higher CD8 T cell, CD4 T cell, B cell and natural killer (NK) cell signatures in addition to M1 and M2 macrophage signatures (figure 2B). Fibroblast signatures were similar between the two cohorts, while hair outer root sheet and keratinocyte signatures were lower in PRESS compared with GENISOS. Restricting the analysis to GENISOS patients with diffuse SSc and >3 years disease duration (n=28), PRESS patients had higher immune cell signatures, although the differences were smaller in this subgroup analysis (online supplementary figure 1). The prevalence of upregulated CD8 T cell, CD4 T cell and B cell signatures was relatively low in GENISOS as a whole (22%, 20% and 22%, respectively), including among the 28 patients with diffuse cutaneous involvement and >3 years disease duration (21%, 18% and 21%, respectively) (online supplementary figure 2).
To characterise clinical correlates of immune cell signatures within both cohorts, we pooled the data and performed multivariable regression analyses where the associations of disease duration, extent of skin involvement (as determined by mRSS), FVC % predicted and immunosuppression (comparing those on no immunosuppression to those on methotrexate, mycophenolate or cyclophosphamide at the time of biopsy) with cell type signatures (dependent variable) were examined. Adaptive immune cell signatures were inversely associated with disease duration after adjustment for mRSS, FVC % predicted and immunosuppression. By contrast, M1 and M2 macrophages and fibroblasts associated with mRSS but did not significantly associate with disease duration after adjustment for other clinical variables (table 2). These associations were similar after additional adjustment for PRESS versus GENISOS cohorts, suggesting that the observations were not driven by batch effects (online supplementary table 3). Of note, the investigated cell type signatures were not associated with immunosuppressive treatment in the univariable analysis (data not shown) or multivariable analysis (table 2).
Table 2.
Multivariable regression analyses of key clinical variables with cell type-specific signatures in pooled PRESS and GENISOS datasets
| Coefficient | 95% CI | P value | |
|---|---|---|---|
| CD8 T cell* | |||
| Disease duration | −0.026 | −0.042 to −0.009 | <0.01 |
| mRSS | 0.006 | −0.002 to 0.014 | 0.12 |
| FVC % pred | −0.001 | −0.005 to 0.002 | 0.54 |
| No immunosuppression | 0.095 | −0.058 to 0.249 | 0.22 |
| CD4 T cell* | |||
| Disease duration | −0.02 | −0.034 to −0.006 | <0.01 |
| mRSS | 0.004 | −0.003 to 0.010 | 0.25 |
| FVC % pred | −0.001 | −0.004 to 0.002 | 0.49 |
| No immunosuppression | 0.06 | −0.069 to 0.190 | 0.36 |
| NK cell* | |||
| Disease duration | −0.019 | −0.031 to −0.007 | <0.01 |
| mRSS | 0.004 | −0.001 to 0.010 | 0.12 |
| FVC % pred | −0.001 | −0.004 to 0.001 | 0.39 |
| No immunosuppression | 0.086 | −0.026 to 0.197 | 0.13 |
| B cell* | |||
| Disease duration | −0.023 | −0.037 to −0.009 | <0.01 |
| mRSS | 0.002 | −0.005 to 0.009 | 0.56 |
| FVC % pred | −0.001 | −0.004 to 0.002 | 0.5 |
| No immunosuppression | −0.014 | −0.146 to 0.119 | 0.84 |
| M1 macrophage* | |||
| Disease duration | −0.013 | −0.030 to 0.004 | 0.13 |
| mRSS | 0.013 | 0.005 to 0.021 | <0.01 |
| FVC % pred | −0.002 | −0.005 to 0.002 | 0.36 |
| No immunosuppression | 0.04 | −0.119 to 0.199 | 0.62 |
| M2 macrophage* | |||
| Disease duration | −0.001 | −0.014 to 0.012 | 0.91 |
| mRSS | 0.014 | 0.007 to 0.020 | <0.01 |
| FVC % pred | −0.001 | −0.003 to 0.002 | 0.61 |
| No immunosuppression | 0.005 | −0.117 to 0.127 | 0.94 |
| Fibroblast* | |||
| Disease duration | 0.001 | −0.015 to 0.016 | 0.93 |
| mRSS | 0.016 | 0.008 to 0.023 | <0.01 |
| FVC % pred | 0.001 | −0.002 to 0.004 | 0.57 |
| No immunosuppression | 0.028 | −0.119 to 0.174 | 0.71 |
Cell type transcript signature used as the dependent variable in the multivariable model. FVC, forced vital capacity; GENISOS, Genetics versus Environment in Scleroderma Outcomes Study; mRSS, modified Rodnan Skin Score; NK, natural killer; PRESS, Prospective Registry for Early Systemic Sclerosis.
Examination of available follow-up samples in the PRESS cohort
The majority of follow-up biopsies showed declines in immune cell signatures compared with their original biopsies (online supplementary figure 3A, B and online supplementary table 4). Fibroblast signatures were more variable at follow-up, with a small decline on average. Keratinocyte signatures were increased in most follow-up biopsies. Most of the patients with follow-up biopsies had a decline in mRSS from baseline to follow-up, and mRSS change correlated with changes in immune cell and fibroblast signatures numerically.
Histological associations with gene expression profiles
Paraffin-embedded skin biopsy samples concurrently collected from a subgroup of PRESS SSc patients were evaluated histologically using standard H&E staining and IHC staining for markers of macrophages (CD68, CD163, AIF1), endothelial cells (CD31) and myofibroblasts (α-smooth muscle actin(SMA)), as well as markers of adaptive immune cells CD3, CD4, CD8, CD20 and CD56 (it should be noted that CD4 is also expressed in monocytes/macrophages, although at a much lower intensity than in CD4 T cells,19 and that CD56 is expressed in a subset but not all NK cells). Demographics for these samples are shown in online supplementary table 5, and representative slides are shown in online supplementary figure 4. As expected, SSc skin had increased collagen thickness, α-SMA expression and macrophage markers compared with HC skin (online supplementary table 6). Markers of adaptive immune cells were also increased in SSc compared with HC skin (online supplementary table 7). Clinical correlates of IHC staining are shown in online supplementary table 8.
Importantly, cell type signature scores for macrophages and adaptive immune cells based on RNA sequencing data correlated with IHC staining for markers of macrophages and adaptive immune cells, respectively (table 3). Histologically, CD68 and CD163 tracked roughly in parallel, consistent with the reported difficulty in discerning M1 from M2 subtypes with these markers in human cells.20 Taken together, the correlations with IHC staining support the validity of the gene expression-based cell type signatures. Moreover, fibroblast gene expression signature scores correlated with α-SMA (Spearman’s rank order correlation coefficient 0.73, p<0.01) and collagen thickness (Spearman’s rank order correlation coefficient 0.76, p<0.01).
Table 3.
Correlation of immune cell gene expression signatures with immunohistochemical staining of immune cell markers
| Cell abundance by IHC staining |
Cell type signature score | Spearman’s r (p value) |
|---|---|---|
| CD68 | M1 macrophage | 0.45 (0.02) |
| CD68 | M2 macrophage | 0.50 (0.01) |
| CD163 | M1 macrophage | 0.47 (0.02) |
| CD163 | M2 macrophage | 0.57 (<0.01) |
| AIF1 | M1 macrophage | 0.66 (<0.01) |
| AIF1 | M2 macrophage | 0.69 (<0.01) |
| CD3 | CD4 T cell | 0.61 (<0.01) |
| CD3 | CD8 T cell | 0.63 (<0.01) |
| CD4 | CD4 T cell | 0.49 (<0.01) |
| CD8 | CD8 T cell | 0.67 (<0.01) |
| CD20 | B cell | 0.54 (<0.01) |
| CD56 | NK cell | 0.24 (0.22) |
IHC, immunohistochemical; NK, natural killer.
Association of cell type signature with disease course
A summary of mRSS, FVC and immunosuppression use 12 months after initial skin biopsy is shown in online supplementary table 9. 78.6% of patients were taking immunosuppressive medication 12 months after initial biopsy, which is expected for a cohort of early diffuse SSc patients. Cell type signatures did not significantly predict change in mRSS 6 or 12 months after biopsy, or change in FVC 12 months after biopsy (online supplementary table 10). Similarly, transcripts recently described as predictive of mRSS progression21 based on samples collected in a phase II study of tocilizumab did not significantly predict postbiopsy mRSS change in this cohort (online supplementary figure 5). Restricting the analysis to those treated with immunosuppressive medications during follow-up also did not show predictive significance for the immune cell signatures (data not shown).
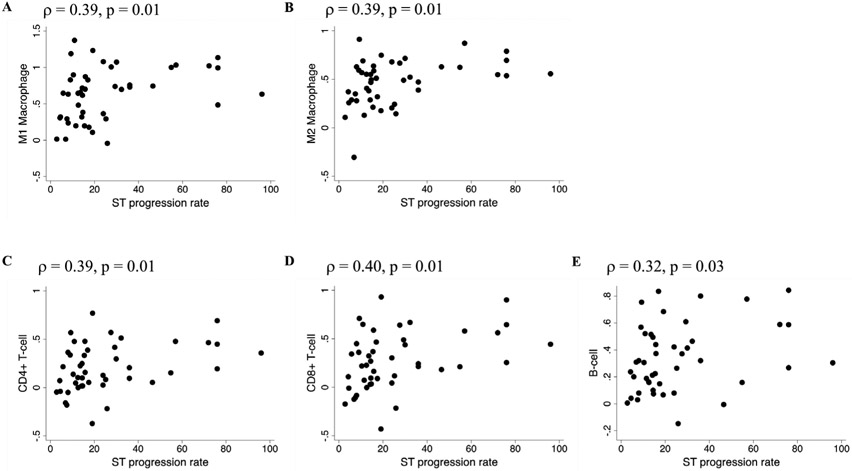
We then looked for associations with the preceding STPR, which was found to be an independent predictor of mortality in patients with early diffuse SSc.15 Significant correlations were seen between immune cell signatures and STPR preceding the biopsy (figure 3 and online supplementary table 10). Thus, immune cell signatures in this cohort were associated with STPR up to the time of biopsy, but did not predict subsequent progression.
Figure 3.
Associations between preceding skin thickness progression rate and skin immune cell type signatures in PRESS SSc patients. Skin thickness progression rate (mRSS at the time of biopsy/years since first skin thickening or puffy fingers) preceding the skin biopsy is plotted on the x-axis. Cell type signature scores for (A) M1 macrophages, (B) M2 macrophages, (C) CD4 cells, (D) CD8 T cells or (E) B cells are plotted on the y-axis. mRSS, modified Rodnan Skin Score; PRESS, Prospective Registry for Early Systemic Sclerosis; SSc, systemic sclerosis; ST, skin thickness.
Comparison to intrinsic subset analysis
The PRESS samples were also assigned to one of four intrinsic subsets (inflammatory, fibroproliferative, limited or normal-like) using previously described methodology by Dr Whitfield’s group.17,18 Thirty-two out of 33 HCs were classified as normal-like, with 1 out of 33 classified as limited (data not shown). Among SSc patients, 23 were classified as inflammatory, 19 as fibroproliferative and 6 as normal-like (figure 2A and online supplementary figure 6). As shown in the figures, there was an over-representation of adaptive immunity cell type signatures in the inflammatory subset of patient samples.
Examination of longitudinal samples revealed that among five samples classified as inflammatory, follow-up biopsies from three of these individuals were classified in non-inflammatory subsets (two fibroproliferative and one normal-like), whereas none of the individuals with biopsies in the fibroproliferative or normal-like subsets on the initial biopsy had a follow-up biopsy in the inflammatory subset ().
Regarding mRSS course, the intrinsic subsets did not significantly predict mRSS change 6 or 12 months postbiopsy in the overall cohort or in the subgroup of patients taking immunosuppressive agents during follow-up (online supplementary table 11). In agreement with the immune cell signature data, STPR preceding the biopsy was significantly higher in the inflammatory subset (online supplementary table 12).
DISCUSSION
Histological and gene expression analyses have demonstrated variable degrees of innate and adaptive immune cells in affected SSc skin.5-11,22-28 In the current study, we measured whole transcriptome expression and cell type signatures in skin specimens in a large cohort of patients specifically with early diffuse SSc and matched HCs. More than half of patients in this cohort had upregulation of CD8 T cell, CD4 T cell and B cell signatures, a higher prevalence than what was observed in patients with longer disease duration from the GENISOS cohort. We also observed a higher prevalence of M1 and M2 macrophage signatures in the skin of early diffuse SSc patients. In patients with longitudinally collected biopsies, immune cell signatures declined on average from initial to follow-up biopsies. These results parallel the clinical observation that early SSc has an edematous, inflammatory phase followed by a more fibrotic phase, and the histological findings in SSc showing an early ‘cellular stage’ characterised by cellular infiltrates in the dermis followed by a later ‘fibrotic stage’ characterised by increased collagen deposition.22,23
Multivariable regression analysis including all samples from the PRESS and GENISOS cohorts showed that adaptive immune cell signatures were significantly associated with shorter disease duration even after adjustment for immunosuppression, severity of skin disease (as assessed by mRSS) and lung disease (as assessed by FVC), whereas macrophage and fibroblast signatures associated predominately with mRSS. These results suggest that the determinants of adaptive versus innate immune cell infiltration in the skin may differ. This can also have implications for target population enrichment strategies in clinical trials, although the observation needs to be confirmed in future studies.
Ingenuity Pathway Analysis suggested that inflammatory cytokines had a more prominent role in driving the dysregulated gene expression in early diffuse SSc compared with later stage disease. Of note, the vast majority of early diffuse SSc patients with a fibroblast signature had a concomitant M1 and/or M2 macrophage signature, and many had concomitant adaptive immune cell signatures, suggesting co-occurrence of dysregulated fibroblast and immune cell function in a majority of early diffuse SSc patients. Our gene expression and IHC data add to the large body of evidence that macrophages are upregulated in SSc.29,30 Macrophages are capable of detecting innate immune stimuli and producing both pro-inflammatory and pro-fibrotic cytokines, including some (eg, IL-6 and TGFβ) that are implicated in SSc pathogenesis. However, the effects of macrophages within the skin and other end organs in SSc require further study.
Taken together, our results indicate that innate and adaptive immune cell activity in the skin is a prominent feature of early diffuse SSc. TGFβ, a key pro-fibrotic cytokine implicated in SSc pathogenesis,31 appears to have a less prominent role in driving the dysregulated gene expression observed during this early, inflammatory phase, in contrast to its prominent role in later-stage disease.
Histological scoring in concurrently collected skin samples supported the gene expression data, demonstrating upregulation of macrophage, adaptive immune cell and fibrotic markers. Immune cell markers correlated with their respective gene expression signatures, and fibrosis markers correlated with fibroblast gene expression signatures. These results support the validity of the gene expression-based cell type signatures.
The RNA processing method used here (ribosomal RNA reduction) enabled the provision of an unbiased comprehensive list of differentially expressed lncRNAs, because this method (unlike poly (A) enrichment) does not remove lncRNAs that do not have a poly(A) tail 32 We have provided a list of differentially expressed lncRNAs expressed in the skin of early diffuse SSc compared with HC, as well as their associations with mRSS. Although our currently available pathway and predicted upstream regulator analytic methods do not include analysis of lncRNAs, the list of disease-relevant lncRNAs represents a resource for follow-up mechanistic studies in this novel area of research.
The carefully collected clinical data in the well-phenotyped PRESS cohort enabled us to examine the correlation of the SSc gene expression profile with the progression rate of skin fibrosis prior to and following skin biopsy. Immune cell signatures were associated with preceding STPR, while they did not have predictive significance for postbiopsy mRSS change. Similarly, transcripts found to be predictive of mRSS progression in previous work21 were not significantly associated with postbiopsy mRSS change in this study. Intrinsic subset classification (normal-like, inflammatory and fibroproliferative)18 did not show predictive significance for mRSS change 6 or 12 months after biopsy. These findings suggest that the use of these previously described gene signatures and subsets for predicting changes in mRSS may not be generalisable to all cohorts. Further research will be needed to determine whether or not a model for prediction of disease progression based on skin gene expression can be universally applied across cohorts, particularly in patients on treatment with commonly used immunosuppressive medications typified in PRESS. The data in this study suggest that skin gene expression signatures in early diffuse SSc are more of a reflection of preceding skin thickness progression than predictors of subsequent progression, supporting the notion that skin is a prominent end organ in SSc rather than an effector organ that drives disease progression.
Our study has several strengths. We examined the transcript expression profile of a relatively large number of skin samples in a well-phenotyped early diffuse SSc cohort using a sensitive, comprehensive RNA sequencing method and compared the results to a later stage SSc transcript expression dataset generated using the same technology. The gene expression-based cell type signatures were validated by IHC staining in concurrently collected samples. There were some limitations to this study that merit discussion. Only a small subgroup of patients (n=8) had follow-up samples available, limiting the ability to analyse changes in gene expression during disease progression. Our future studies will focus on longitudinal collection of early diffuse SSc skin samples. As is common in observational studies and most previous SSc skin gene expression studies, patients enrolled in PRESS were treated according to the standard of care, with the majority being treated with mycophenolate mofetil or methotrexate, which might have affected skin transcript expression.
In conclusion, this large-scale analysis of whole transcriptome expression in the skin of early diffuse SSc patients revealed a high prevalence of both innate and adaptive immune cell activity. Immune cell signatures were associated with preceding STPR but were not predictive of subsequent mRSS progression. These results shed light on the early pathogenesis of diffuse SSc and could have implications for clinical trials targeting the immune system in SSc patients.
Supplementary Material
Key messages.
What is already known about this subject?
Skin gene expression is altered in patients with systemic sclerosis (SSc) based on data from microarrays, but heterogeneity exists in skin gene expression profiles of SSc patients.
What does this study add?
A large-scale analysis of skin transcript expression specifically in patients with early, diffuse cutaneous SSc and comparison to patients with later disease revealed that innate and adaptive immune cell gene expression is more prominent in early diffuse SSc compared with later disease. After adjustment for key clinical characteristics, adaptive immune cell signatures were associated with shorter disease duration.
Immune cell signatures appeared to reflect preceding skin thickness progression rate but did not predict subsequent modified Rodnan Skin Score progression.
How might this impact on clinical practice or future developments?
The prominence of innate and adaptive immune cell signatures in early diffuse SSc would seem to support the premise of using immune-modulatory therapies in this subgroup of patients.
There appear to be limitations in the use of skin gene expression profiles to predict subsequent disease progression, perhaps related to heterogeneity among SSc patient cohorts.
Acknowledgements
We acknowledge Yoko Takata and Jianjun Shen (MD Anderson Cancer Center, Houston, TX) for performing RNA sequencing. JS is supported by a CPRIT Core Facility Support Award RP170002. We acknowledge Melissa Stephens (UT Health Science Center, Houston) for performing some of the immunohistochemical staining. We thank Sam Theodore (UT Health Science Center, Houston) for assistance with skin biopsies and database management. Part of this work was previously presented orally at the American College of Rheumatology meeting in 2017 (Arthritis Rheumatol. 2017;69 (suppl 10)).
Funding BS is supported by a grant from the Arthritis National Research Foundation. DK is supported by a Scleroderma Foundation SCORE grant and NIH/NIAMS K24 AR 063120. The PRESS Data Coordinating Center at University of Michigan was supported by NIH/NIAMS K24 AR063120 to DK. SA is supported by a Scleroderma Foundation SCORE grant, a DoD grant W81XWH-61-1-0296 and by NIH/NIAMS R01AR073284. MW is supported by a grant from the Scleroderma Foundation. This work was partly supported by the UT Houston Bioinformatics and High Performance Computing Service Center and CPRIT grant RP170668 (WJZ). WJZ is supported by NIH 1UL1 TR003167-01. JMF and MLW are supported by the Scleroderma Research Foundation, Dr Ralph and Marian Falk Medical Research Trust Catalyst and Transformational Award and NIH/NIAMS P50 AR060780-01. JMF is supported by NIH 5T32LM012204-03. EJB is supported by NIH/NIAMS K23 AR075112.
Footnotes
Competing interests DK reports consultancy fees from Acceleron, Actelion, Bayer, Blade Therapeutics, BMS, Galapagos, Genentech/Roche, GSK, Mitsubishi Tanabi, Sanofi-A ventis/Genzyme, and UCB Pharma, and reports grants from Bayer, BMS, and Genentech/Roche outside the submitted work, and reports ownership interest in Eicos Sciences, Inc. WRS reports consulting fees from UT Health Science Center at Houston. JKG reports grants from Corbus Pharmaceuticals, Cumberland Pharmaceuticals and Eicos Pharmaceuticals outside the submitted work. AAS reports support from Sanofi outside the submitted work. MLW reports grants and personal fees from Celdara Medical, LLC and personal fees from Abbvie, Acceleron, BMS, Corbus Pharmaceuticals and Boehringer Ingelheim outside the submitted work. JLB reports grants from Hoffman La Roche and the Scleroderma Foundation outside the submitted work. SB and PB are employees of Boehringer Ingelheim. FVC reports personal fees from Boehringer Ingelheim outside the submitted work. EJB reports a grant from Pfizer and support from Bohringer Ingelheim, Corbus Pharmaceuticals and Eicos outside the submitted work. MDM reports personal fees from Medtelligence, Actelion Pharma, Astellas, Mitsubishi-Tanabe, and Boehringer Ingelheim and grants from Bayer, Reata, Sanofi, Corbus, Eicos and Boehringer Ingelheim outside the submitted work. SA reports personal fees from Boehringer Ingelheim and grants from Boehringer Ingelheim, Bayer and Momenta outside the submitted work.
Patient consent for publication Not required.
Provenance and peer review Not commissioned; externally peer reviewed.
Data availability statement RNA sequencing data are available at NCBi’s Gene Expression Omnibus, accession GSE130955.
REFERENCES
- 1.Denton CP, Khanna D. Systemic sclerosis. Lancet 2017;390:1685–99. [DOI] [PubMed] [Google Scholar]
- 2.Varga J, Trojanowska M, Kuwana M. Pathogenesis of systemic sclerosis: recent insights of molecular and cellular mechanisms and therapeutic opportunities. J Scleroderma Relat Disord 2017;2:137–52. [Google Scholar]
- 3.Assassi S, Mayes MD. What does global gene expression profiling tell us about the pathogenesis of systemic sclerosis? Curr Opin Rheumatol 2013;25:686–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martyanov V, Whitfield ML. Molecular stratification and precision medicine in systemic sclerosis from genomic and proteomic data. Curr Opin Rheumatol 2016;28:83–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whitfield ML, Finlay DR, Murray Ji, et al. Systemic and cell type-specific gene expression patterns in scleroderma skin. Proc Natl Acad Sci U S A 2003;100:12319–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gardner H, Shearstone JR, Bandaru R, et al. Gene profiling of scleroderma skin reveals robust signatures of disease that are imperfectly reflected in the transcript profiles of explanted fibroblasts. Arthritis Rheum 2006;54:1961–73. [DOI] [PubMed] [Google Scholar]
- 7.Milano A, Pendergrass SA, Sargent JL, et al. Molecular subsets in the gene expression signatures of scleroderma skin. PLoS One 2008;3:e2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pendergrass SA, Lemaire R, Francis iP, et al. intrinsic gene expression subsets of diffuse cutaneous systemic sclerosis are stable in serial skin biopsies. J Invest Dermatol 2012;132:1363–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hinchcliff M, Huang C-C, Wood TA, et al. Molecular signatures in skin associated with clinical improvement during mycophenolate treatment in systemic sclerosis. J Invest Dermatol 2013;133:1979–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Assassi S, Swindell WR, Wu M, et al. Dissecting the heterogeneity of skin gene expression patterns in systemic sclerosis. Arthritis Rheumatol 2015;67:3016–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lofgren S, Hinchcliff M, Carns M, et al. Integrated, multicohort analysis of systemic sclerosis identifies robust transcriptional signature of disease severity. JCI Insight 2016;1:e89073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moon S-J, Bae JM, Park K-S, et al. Compendium of skin molecular signatures identifies key pathological features associated with fibrosis in systemic sclerosis. Ann Rheum Dis 2019;78:817–25. [DOI] [PubMed] [Google Scholar]
- 13.Frech TM, Shanmugam VK, Shah AA, et al. Treatment of early diffuse systemic sclerosis skin disease. Clin Exp Rheumatol 2013;31:166–71. [PMC free article] [PubMed] [Google Scholar]
- 14.van den Hoogen F, Khanna D, Fransen J, et al. 2013 classification criteria for systemic sclerosis: an American College of rheumatology/European League against rheumatism collaborative initiative. Ann Rheum Dis 2013;72:1747–55. [DOI] [PubMed] [Google Scholar]
- 15.Domsic RT, Rodriguez-Reyna T, Lucas M, et al. Skin thickness progression rate: a predictor of mortality and early internal organ involvement in diffuse scleroderma. Ann Rheum Dis 2011;70:104–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swindell WR, Johnston A, Voorhees JJ, et al. Dissecting the psoriasis transcriptome: inflammatory- and cytokine-driven gene expression in lesions from 163 patients. BMC Genomics 2013;14:527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franks JM, Cai G, Whitfield ML. Feature specific quantile normalization enables cross-platform classification of molecular subtypes using gene expression data. Bioinformatics 2018;34:1868–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franks JM, Martyanov V, Cai G, et al. A machine learning classifier for assigning individual patients with systemic sclerosis to intrinsic molecular subsets. Arthritis Rheumatol 2019;71:1701–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wood GS, Warner NL, Warnke RA. Anti-Leu-3/T4 antibodies react with cells of monocyte/macrophage and Langerhans lineage. J Immunol 1983;131:212–6. [PubMed] [Google Scholar]
- 20.Barros MHM, Hauck F, Dreyer JH, et al. Macrophage polarisation: an immunohistochemical approach for identifying M1 and M2 macrophages. PLoS One 2013;8:e80908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stifano G, Sornasse T, Rice LM, et al. Skin gene expression is prognostic for the trajectory of skin disease in patients with diffuse cutaneous systemic sclerosis. Arthritis Rheumatol 2018;70:912–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fleischmajer R, Perlish JS, Reeves JR. Cellular infiltrates in scleroderma skin. Arthritis Rheum 1977;20:975–84. [DOI] [PubMed] [Google Scholar]
- 23.Fleischmajer R, Gay S, Meigel WN, et al. Collagen in the cellular and fibrotic stages of scleroderma. Arthritis Rheum 1978;21:418–28. [DOI] [PubMed] [Google Scholar]
- 24.Roumm AD, Whiteside TL, Medsger TA, et al. Lymphocytes in the skin of patients with progressive systemic sclerosis. quantification, subtyping, and clinical correlations. Arthritis Rheum 1984;27:645–53. [DOI] [PubMed] [Google Scholar]
- 25.Ishikawa O, Ishikawa H. Macrophage infiltration in the skin of patients with systemic sclerosis. J Rheumatol 1992;19:1202–6. [PubMed] [Google Scholar]
- 26.Kraling BM, Maul GG, Jimenez SA. Mononuclear cellular infiltrates in clinically involved skin from patients with systemic sclerosis of recent onset predominantly consist of monocytes/macrophages. Pathobiology 1995;63:48–56. [DOI] [PubMed] [Google Scholar]
- 27.Hinchcliff M, Toledo DM, Taroni JN, et al. Mycophenolate mofetil treatment of systemic sclerosis reduces myeloid cell numbers and attenuates the inflammatory gene signature in skin. J Invest Dermatol 2018;138:1301–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bosello S, Angelucci C, Lama G, et al. Characterization of inflammatory cell infiltrate of scleroderma skin: B cells and skin score progression. Arthritis Res Ther 2018;20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown M, O’Reilly S. The immunopathogenesis of fibrosis in systemic sclerosis. Clin Exp Immunol 2019;195:310–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toledo DM, Pioli PA. Macrophages in systemic sclerosis: novel insights and therapeutic implications. Curr Rheumatol Rep 2019;21:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lafyatis R Transforming growth factor β-at the centre of systemic sclerosis. Nat Rev Rheumatol 2014;10:706–19. [DOI] [PubMed] [Google Scholar]
- 32.Kukurba KR, Montgomery SB. RNA sequencing and analysis. Cold Spring Harb Protoc 2015;2015:951–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.