Summary
Epstein-Barr virus (EBV) is a cancer-associated pathogen for which there is no vaccine. Successful anti-viral vaccines elicit antibodies that neutralize infectivity; however, it is unknown whether neutralizing antibodies prevent EBV acquisition. Here we assessed whether passively delivered AMMO1, a monoclonal antibody that neutralizes EBV in a cell-type-independent manner, could protect against experimental EBV challenge in two animal infection models. When present prior to a high-dose intravenous EBV challenge, AMMO1 prevented viremia and reduced viral loads to nearly undetectable levels in humanized mice. AMMO1 conferred sterilizing immunity to three of four macaques challenged orally with rhesus lymphocryptovirus, the EBV ortholog that infects rhesus macaques. The infected macaque had lower plasma neutralizing activity than the protected animals. These results indicate that a vaccine capable of eliciting adequate titers of neutralizing antibodies targeting the AMMO1 epitope may protect against EBV acquisition and are therefore highly relevant to the design of an effective EBV vaccine.
Graphical Abstract
Highlights
An anti-EBV mAb, AMMO1, limits viral replication following challenge in humanized mice
AMMO1 cross-reacts with and neutralizes rhesus lymphocryptovirus
Adequate levels of AMMO1 prevent oral acquisition of rhLCV in macaques
Protection afforded by neutralizing antibody provides proof of concept for EBV vaccines
Epstein-Barr virus is a cancer-associated lymphocryptovirus for which there is no vaccine. Singh et al. demonstrate that passive delivery of a potent neutralizing antibody protects against lymphocryptovirus challenge in two animal models. These results indicate that neutralizing antibodies may be an important component of an effective EBV vaccine.
Introduction
Epstein-Barr virus (EBV), a member of the Lymphocryptovirus genus, is associated with 200,000 new cases of cancer and 140,000 deaths annually.1 EBV is also a causative agent of infectious mononucleosis (IM) and can lead to lymphoproliferative disease in immunocompromised individuals, such as those undergoing organ transplant or persons living with AIDS.2 In addition to its contribution to oncogenesis, EBV infection is also associated with multiple sclerosis3,4 and rheumatoid arthritis.5 Thus, a safe and effective vaccine that protects against EBV infection and/or pathogenesis would be of clinical benefit, particularly to those in resource-poor settings where the EBV-associated cancer burden is high.6, 7, 8
Most effective vaccines elicit antibodies that neutralize infection.9 However, it is not clear whether pre-existing neutralizing antibodies can protect against EBV infection in vivo. Infants generally remain EBV negative for 6–8 months until the presence of maternal antibody wanes, suggesting that neutralizing antibodies are protective.10, 11, 12, 13, 14 However, several studies have demonstrated the presence of multiple viral variants in infected individuals,15, 16, 17, 18, 19, 20 which could indicate that an antibody response elicited by primary infection is not sufficient to protect against superinfection with additional EBV strains.
Primary infection occurs in the nasopharynx, where B cells and epithelial cells are susceptible to infection. Host cell entry is a complex process mediated by several viral glycoproteins that define tropism and mediate membrane fusion.21 Sera from infected individuals can neutralize EBV infection of B cells and epithelial cells in vitro.22, 23, 24, 25 Multiple viral antigens have been shown to be targeted by serum neutralizing antibodies,25, 26, 27 but it is not clear which are ideal candidates for an EBV vaccine.
Passive transfer of monoclonal antibodies (mAbs) followed by controlled challenge in susceptible animal models has demonstrated the protective efficacy of neutralizing antibodies and defined relevant protective epitopes against several other viral pathogens.28, 29, 30, 31, 32, 33, 34, 35, 36, 37 Conducting similar proof-of-concept studies with EBV antibodies has been hampered by the near-obligate tropism of the virus for humans and a dearth of mAbs with well-defined neutralizing potentials.
Human B cells in immunocompromised mice engrafted with human lymphocytes can be infected by EBV and provide a small animal model to study EBV infection in vivo.38, 39, 40, 41, 42, 43 A high dose of EBV delivered intravenously to humanized mice leads to development of splenomegaly, splenic tumors, and expansion of activated CD8 T cells.39,44
The viral glycoprotein gp350 facilitates EBV attachment to complement receptors 1 and 2 (CR1 and CR2, respectively).45,46 The anti-gp350 72A1 mAb neutralizes EBV infection of B cells in vitro by disrupting the gp350-CR interaction45, 46, 47 but is ineffective at blocking infection of CR− epithelial cells.48 Repeated transfer of 72A1 into severe combined immunodeficiency (SCID) mice engrafted with peripheral blood mononuclear cells (PBMCs) from EBV-seronegative donors prevented tumor formation following intravenous (i.v.) challenge with EBV, implying that pre-existing neutralizing antibodies may protect against EBV infection in vivo.49
Rhesus lymphocryptovirus (rhLCV) is an ortholog of EBV that naturally infects rhesus macaques.50, 51, 52 RhLCV infection of macaques is considered the best animal model for EBV infection because of the high level of homology between EBV and rhLCV, oral transmission of the virus, and the similar courses of infection in their respective hosts.52 Therefore, rhLCV infection of rhesus macaques is a highly relevant animal model to evaluate the protective efficacy of neutralizing antibodies in a controlled setting. However, no neutralizing mAbs specific for rhLCV have been isolated. Two mAbs, CL40 and E1D1, which bind the EBV gH/gL glycoprotein complex, an essential component of the viral fusion machinery, also bind to rhLCV gH/gL, although it is not known whether they neutralize rhLCV.53 The 72A1 mAb, which showed protection in mice, does not cross-react with the rhesus gp350 protein.54 A chimeric rhLCV virus expressing gp350 from EBV is infectious in macaques and susceptible to neutralization by 72A1,54 but passive transfer studies of neutralizing mAbs against rhLCV have not been reported to date.
We previously isolated an anti-gH/gL mAb, AMMO1, that neutralizes EBV infection of B cells and epithelial cells.55 Here we show that AMMO1 cross-reacts with and neutralizes rhLCV. We took advantage of the cross-reactivity of AMMO1 between EBV and rhLCV to evaluate its protective efficacy against Lymphocryptovirus challenge in complementary animal models, humanized mice, and rhesus macaques.
Results
AMMO1 Confers Protection against High-Dose EBV Challenge in Humanized Mice
72A1 and AMMO1 neutralize EBV infection of B cells with comparable potency.55 Because 72A1 has been shown previously to protect against EBV-driven tumor formation in humanized mice,49 we sought to address whether AMMO1 could also prevent lymphoproliferation in a similar humanized mouse model. To this end, non-obese diabetic (NOD)-scid Il2rgnull (NSG) mice were engrafted with healthy human donor-derived mobilized peripheral blood hematopoietic stem cells (PBSCs), referred to here as humanized mice. 12 weeks post-transplant, ~10%–15% of peripheral blood mononuclear cells were of human origin (Figures S1A and S1B). Among these, ~80% were hCD19+ B cells and very few hCD4+ or hCD8+ T cells (Figures S1A and S1B).
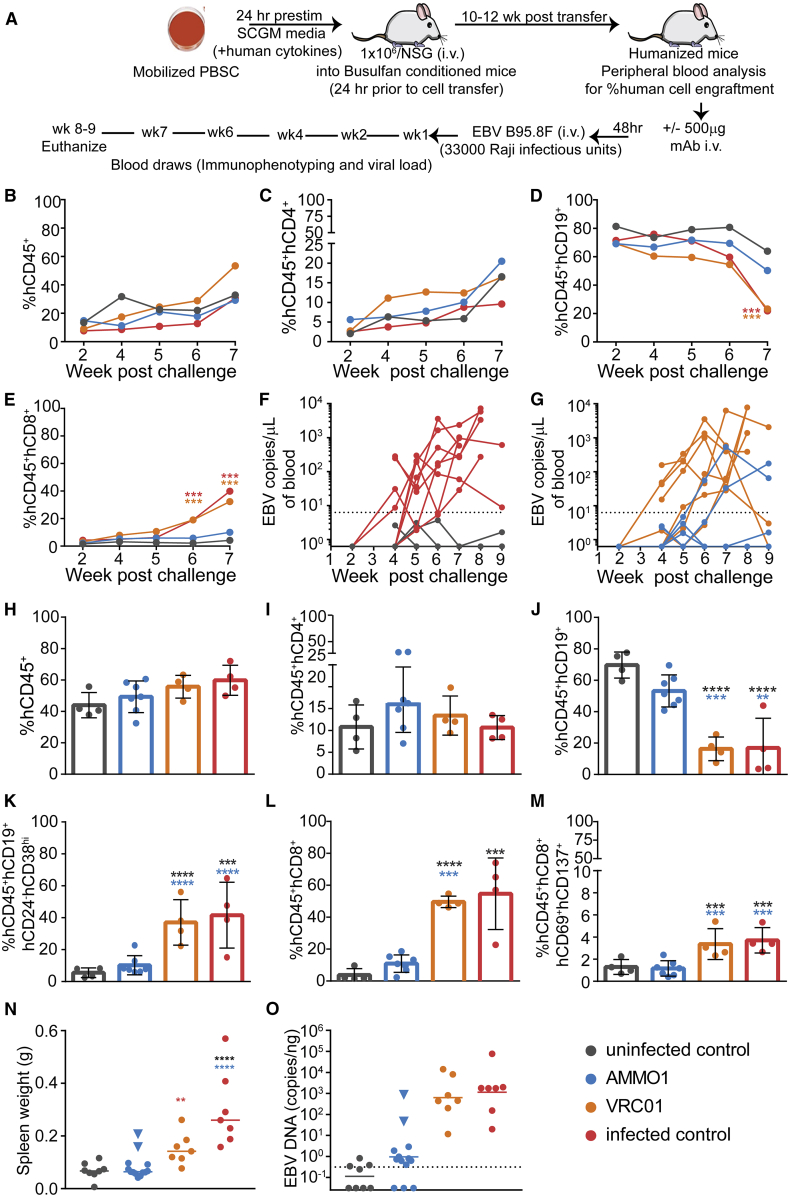
As shown in Figure 1A, humanized mice received an i.v. injection containing 500 μg of purified recombinant AMMO1 or, as a control, the anti-HIV-1 mAb VRC01,56 followed at 48 h by an i.v. injection of EBV B95.8/F22 equivalent to ~33,000 Raji infectious units. Infected control mice received the same dose of virus without antibody pre-treatment, and uninfected control mice did not receive antibody or viral challenge. Mice were bled weekly and euthanized 8–9 weeks following challenge. 6–7 weeks following challenge, although all mice retained equivalent proportions of total human cells and hCD4+ cells (Figures 1B and 1C), mice from the infected control and VRC01 groups showed a marked decrease in the frequency of peripheral hCD19+ B cells (Figure 1D), concurrent with an increase in hCD8+ T cells (Figure 1E), compared with uninfected control mice and mice that received AMMO1. Viremia was detectable in the peripheral blood of the infected control and VRC01 groups and in 2 of 13 mice that received AMMO1 4–6 weeks post-challenge (Figures 1F and 1G). The remaining animals from the AMMO1 group and the uninfected controls remained aviremic (Figures 1F and 1G).
Figure 1.
AMMO1 Inhibits EBV Infection in Humanized Mice
(A) Experimental timeline. Humanized mice received a dose of 0.5 mg of AMMO1 or VRC01 via intravenous (i.v.) injection 48 h prior to i.v. challenge of EBV B95.8/F equivalent to 33,000 Raji infectious units. Infected control mice received the virus but no antibody, and uninfected control mice received neither.
(B–E) The frequency of (B) hCD45+, (C) hCD45+hCD4+, (D) hCD45+hCD19+, and (E) hCD45+hCD8+ cells in peripheral blood at the indicated time points post-challenge. Data points represent the mean of 7 AMMO1, 4 VRC01, 4 infected control mice, and 4 uninfected control mice. A representative flow plot illustrating the gating strategy is shown in Figure S1.
(F and G) Viral DNA was quantitated in the peripheral blood of infected control mice (n = 8), uninfected control mice (n = 8), and (G) mice that received AMMO1 (n = 13) or VRC01 (n = 7) prior to challenge. Each dot represents an individual mouse, and the dashed line indicates the limit of detection.
(H–M) 8–9 weeks after viral challenge, the animals were euthanized, and the frequency of (H) hCD45+, (I) hCD45+hCD4+, (J) hCD45+hCD19+, (K) hCD45+hCD19+hCD24−hCD38hi, (L) hCD45+hCD8+, and (M) hCD45+hCD8+hCD69+hCD137+ cells was measured in splenocytes. Data points represent individual mice, bar graphs indicate the mean, and error bars represent the standard deviation. Representative flow plots illustrating the gating strategy used to analyze splenocytes are shown in Figures S2 and S3.
(N) Spleen weight of study animals at necropsy. Each dot represents an individual mouse, and the bar represents the mean.
(O) qPCR was used to quantitate viral DNA in splenic extracts at necropsy. Each dot represents an individual mouse, the bar represents the mean, and the dashed line indicates the limit of detection.
Data shown in (B)–(E) and (H)–(M) are from one of two independent experiments. Combined data from 2 independent experiments are shown in (F), (G), (N), and (O). Statistical analyses were performed using one-way ANOVA. The color of the asterisks (∗p ≤ 0.032, ∗∗p ≤ 0.002, ∗∗p ≤ 0.0003, ∗∗∗∗p ≤ 0.0002) denotes groups that were significantly different from the control in (B)–(E) or the groups with which there is a significant difference in (H)–(M) using a Sidak multiple comparisons test.
Splenic lymphocytes were analyzed at the study endpoint. Although the proportions of total human cells and hCD4+ T cells were similar in all animals (Figures 1H and 1I), the infected control and VRC01 groups exhibited a decrease in hCD19+ cells (Figure 1J), and this population contained an increased frequency of hCD24−hCD38hi cells (Figure 1K). Concurrent with the decline of hCD19+ cells, there was a significant increase in the frequency of total (Figure 1L) and activated (Figure 1M) hCD8+ T cells in the control-infected and VRC01 groups, consistent with T cell-mediated killing of infected peripheral human B cells.
Spleens from 11 of 13 animals that received AMMO1 were comparable in weight with the uninfected controls, whereas the spleens from the 2 animals that were viremic were enlarged (indicated by blue triangles in Figure 1N), as were the spleens from the infected control and VRC01 groups.
Mice from the infected control and VRC01 groups had 100–1,000-fold higher levels of EBV DNA in the spleen compared with the uninfected controls (Figure 1O). Low amounts of viral DNA were detectable in splenic DNA extracts from 8 of the 11 aviremic mice in the AMMO1 group. Comparable amounts were detected in splenic extracts from half of the uninfected control mice (Figure 1O). In contrast, splenic DNA from the two viremic mice in the AMMO1 group harbored higher levels of EBV DNA (indicated by blue triangles in Figure 1O). Collectively, these data indicate that AMMO1 prevented viremia and splenomegaly in 11 of 13 mice. Because of the trace amount of DNA detected in the spleens of AMMO1-challenged mice and the uninfected controls, we cannot conclude that AMMO1 conferred sterilizing immunity in these animals. However, AMMO1 prevented the splenomegaly, viremia, increase in hCD8+ cells, and decline in hCD19+ cells observed in the positive control and VRC01 groups, which is consistent with high-dose EBV infection in this model.39
Compared with Other Neutralizing mAbs, AMMO1 Provides Superior Protection against EBV Challenge in Humanized Mice
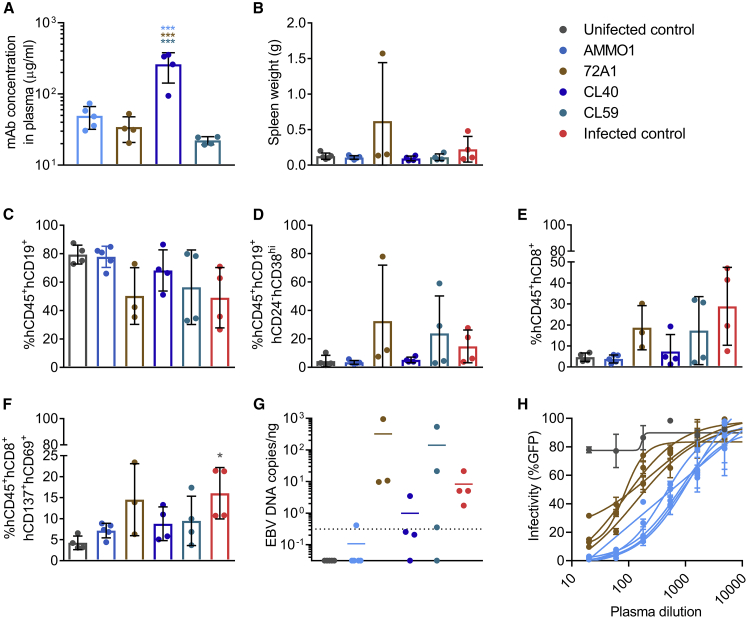
We next evaluated whether passive transfer of the CL40 and CL59 mAbs that target distinct epitopes on gH/gL57,58 could similarly limit or prevent in vivo EBV infection in this humanized mouse model. To bridge our studies with previous work,49 we also included the anti-gp350 72A1 mAb. CL40, CL59, and 72A1 are murine mAbs, whereas AMMO1 is human, so to mitigate potential differences in kinetics and/or effector function, all mAbs were administered as purified recombinant variants with human immunoglobulin G1 (IgG1) constant regions.59 As shown in Figure 1A, mice received 500 μg of each mAb per animal, followed 48 h later by an i.v. injection of ~33,000 Raji infectious units of EBV B95.8/F. 1 week post-viral challenge, the mean concentrations of AMMO1, 72A1, CL40, and CL59 in the plasma were 49 μg/mL, 34 μg/mL, 261 μg/mL, and 22 μg/mL, respectively (Figure 2A).
Figure 2.
AMMO1 Inhibits EBV Infection More Efficiently Than Other Anti-EBV mAbs in Humanized Mice
Humanized mice received an i.v. injection containing 0.5 mg of the mAbs AMMO1 (n = 5), 72A1 (n = 4), CL40 (n = 4), and CL59 (n = 4). Two days later, the mice were challenged with 33,000 Raji infectious units of EBV B95.8/F.
(A) One week following challenge, the concentration of transferred mAbs in plasma was determined by antigen-specific ELISA against gH/gL for AMMO1, CL40 and CL59, and gp350 for 72A1.
(B) Spleen weight of study animals 8 weeks after challenge. Uninfected control mice (n = 4) that did not receive antibody or virus and infected control mice (n = 4) that were challenged with the virus but did not receive antibody pre-treatment are included.
(C–F) At necropsy, the frequency of (C) hCD45+hCD19+, (D) hCD45+hCD19+hCD24−hCD38hi, (E) hCD45+hCD8+, and (F) hCD8+hCD69+hCD137+ cells were measured in splenocytes.
(G) qPCR was used to quantitate viral DNA in splenic extracts at necropsy. Each data point represents an individual mouse, the bar indicates the mean, and the dashed line indicates the limit of detection.
(H) The EBV-neutralizing titers in plasma from AMMO1- or 72A1-infused mice were measured 1 week following challenge.
Bar graphs indicate the mean, error bars represent the standard deviation, and each data point represents an individual mouse in (A)–(F). One of the animals in the 72A1 group died prior to necropsy; therefore, only 3 mice are shown in (B)–(G). Statistical analyses were performed using one-way ANOVA. The color of the asterisks (*p ≤ 0.0332, ***p ≤ 0.0002) denotes the group with which there is a significant difference, determined by a Sidak multiple comparisons test.
4 weeks post-viral challenge, one of the animals in the 72A1 group died. 9 weeks post-viral challenge, the study endpoint, mice from the infected control group had larger spleens than mice in the uninfected control, AMMO1, CL40, and CL59 groups (Figure 2B). One of the remaining animals from the 72A1 group displayed extreme splenomegaly (Figure 2B) and a large splenic tumor (data not shown). Relative to the uninfected controls, the frequency of hCD19+ cells was lower (Figure 2C), whereas the frequencies of hCD19+hCD24−hCD38hi B cells (Figure 2D) as well as total and activated hCD8+ T cells (Figures 2E and 2F) were higher in the 72A1, CL59, and infected control groups, a phenotype consistent with infection in prior challenge experiments (Figure 1). In contrast, animals that received AMMO1 or CL40 had frequencies of hCD19+ and hCD8+ cells indistinguishable from uninfected controls (Figures 2C–2F). EBV viral DNA was present in splenic extracts from all mice in the 72A1 and infected control groups, three of four mice in the CL59 group, one mouse from the CL40 group, and one mouse from the AMMO1 group (Figure 2G). These data indicate that, among anti-gH/gL mAbs, AMMO1 confers superior protection against i.v. EBV challenge in humanized mice, as predicted by the relative potency of each antibody in a B cell neutralization assay.55 The efficacy of CL40 is likely overestimated in this experiment, considering that it was present at approximately 5× higher levels in the blood than AMMO1 at the time of challenge.
Considering that AMMO1 and 72A1 neutralize B cell infection with comparable potency in vitro,55 the complete lack of protection by 72A1 was unexpected. AMMO1 was present at slightly higher levels than 72A1 in plasma 9 days post-transfer (Figure 2A), and plasma from the AMMO1 group had higher in vitro neutralizing capacity than plasma from the 72A1 group (Figure 2H). Thus, the difference in protection between 72A1 and AMMO1 may reflect differences in the effective in vivo plasma neutralizing titer. However, the previous demonstration that 72A1 prevented EBV-driven tumor formation in vivo utilized repeated mAb dosing,49 suggesting that other factors may explain the differences in in vivo functional activity of 72A1 versus AMMO1. Taken together, our in vivo challenge studies demonstrate that passive transfer of AMMO1 limits EBV to levels detected in control animals in 16 of 18 humanized mice, indicating robust capacity for protection in this setting.
AMMO1 Cross-Reacts with and Neutralizes rhLCV
In addition to B cells, epithelial cells present in the human nasopharynx are susceptible to EBV infection;60 thus, the viral dose and kinetics of infection are likely to differ between i.v. challenge in humanized mice and oral transmission in humans. To address whether AMMO1 can also limit EBV infection in a more physiological challenge setting, we sought to assess whether passively transferred AMMO1 could prevent oral transmission in a non-human primate model. To achieve this goal, we utilized rhLCV in a rhesus macaque model.
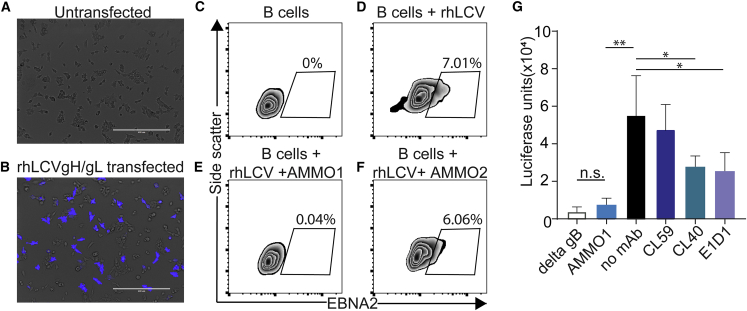
EBV and rhLCV gH and gL share 85.4% and 81.8% amino acid identity, respectively. A population of 293T cells transfected with rhLCVgH/gL stained with fluorescently labeled AMMO1which indicated that the mAb is cross-reactive with the rhesus ortholog (Figures 3A and 3B). To assess whether the neutralizing activity of AMMO1 was conserved, the ability of AMMO1 to neutralize rhLCV infection of rhesus B cells was evaluated using viral expression of EBNA2 in rhesus B cells as a readout. In the absence of antibody or in the presence of the non-neutralizing mAb AMMO2, 6%–7% of rhesus B cells stained positive for EBNA2 following challenge with rhLCV (Figures 3D and 3F). In contrast, EBNA2 expression was absent when the virus was pre-incubated with AMMO1 (Figure 3E).
Figure 3.
AMMO1 Cross-reacts with rhLCV gH/gL
(A and B) AMMO1 conjugated to Dylite-650 was used to stain (A) untransfected 293T cells or (B) 293T cells transfected with plasmids expressing rhLCV gH and rhLCV gL. The scale bar represents 400 μm. The fluorescence channel for Dylite-650 (shown in blue) is overlaid on the transmitted light channel (gray). Images are representative of two independent experiments.
(C–F) Rhesus PBMC (C) or rhesus PBMC challenged with RhLCV alone (D) or rhLCV pre-incubated with AMMO1 (E) or AMMO2 (F) were cultured in medium. 3 days later, B cells (live, CD19+, CD20+) were monitored for EBNA2 expression by flow cytometry. Values indicate the percentage of EBNA2+ B cells.
(G) CHO-K1 cells were transfected with expression plasmids encoding rhLCV gH, rhLCV gL, rhLCV gB, and luciferase under control of a T7 promoter. 24 h later, the cells were trypsinized and then overlaid on HEK293 cells stably expressing T7 polymerase in wells containing AMMO1, CL59, CL40, E1D1, or no mAb, as indicated. 24 h later, the cells were lysed and assayed for luciferase activity. As a control for non-specific fusion, CHOK1 cells were transfected as above, except the plasmid rhLCV gB was replaced with an empty plasmid (delta gB).
Significant differences (∗p ≤ 0.05 and ∗∗p ≤ 0.01) were determined using an unpaired 2-tailed t test.
AMMO1 disrupts EBV gH/gL- and gB-driven membrane fusion in a virus-free cell fusion assay.55 Here we assessed whether AMMO1 could similarly disrupt rhLCV gH/gL and rhLCV gB-driven fusion using the same assay.61 CHO-K1 cells were transfected with plasmids expressing rhLCV gH, rhLCV gL, and rhLCV gB and with luciferase under control of the T7 promoter. Transfected cells were overlaid on 293 cells stably expressing T7 polymerase. Expression of the entire rhLCV fusion machinery resulted in high levels of luciferase activity that was reduced to background levels in the presence of AMMO1 (Figure 3G). CL40 and E1D1, which also bind rhLCV gH/gL,53 reduced fusion, but to a lesser degree, whereas the CL59 mAb, which does not bind rhLCV gH/gL,53 had no effect. Collectively, these data demonstrate that AMMO1 cross-reacts with rhLCV gH/gL and that its neutralizing activity is conserved against rhLCV.
AMMO1 Confers Sterilizing Immunity against rhLCV Challenge in Macaques
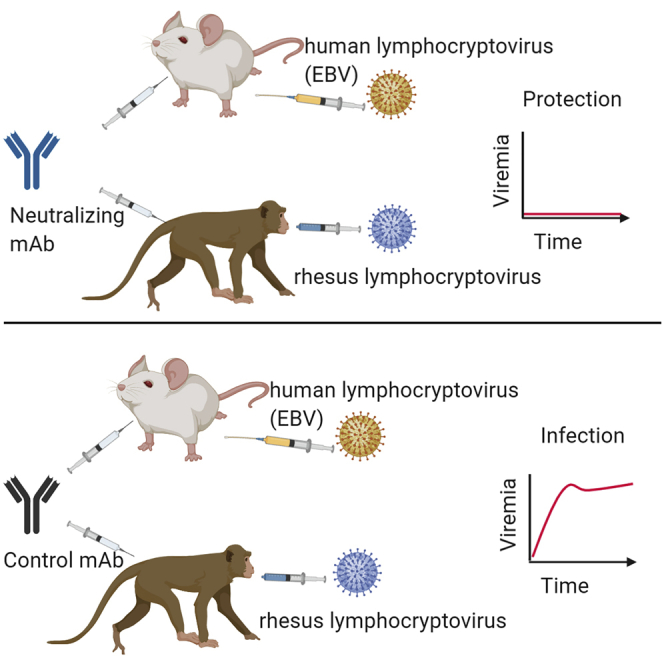
To establish whether AMMO1 could protect against oral rhLCV challenge, 6 infant rhesus macaques were obtained within 48 h of birth and housed separately from other colony animals. Beginning at 2 months of age, the animals were tested for plasma antibodies to the rhLCV viral capsid antigen (VCA)62 and for the presence of rhLCV DNA in PBMCs.63 Maternal antibodies to VCA were detected in 4 of 6 study animals and waned by 6 months of age (Figure S4A). All animals tested negative for rhLCV DNA during this time (Figure S4B). When the animals were ~6 months old, VRC01 (n = 2) or AMMO1 (n = 4) human IgG was administered i.v. at a dose of 20 mg/kg. Three days after antibody transfer, all animals were challenged orally with 1.75 transforming units of rhLCV (Figure S4C). During a 10-week period following challenge, none of the animals seroconverted (Figure S4D) or became viremic (Figure S4E); thus, we performed a dose escalation.
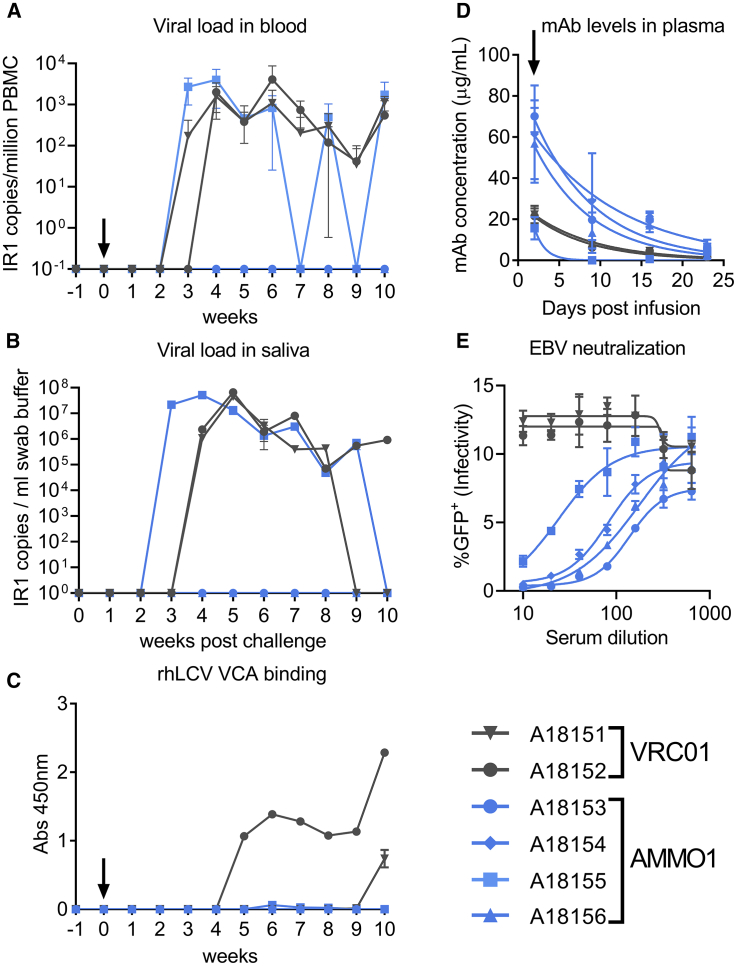
11 weeks after the initial challenge, the study animals were re-infused with VRC01 or AMMO1 and then re-challenged orally 48 h later, with a higher dose of rhLCV (50 transforming units). Blood and oral swabs were collected weekly and assayed for the presence of anti-VCA antibodies in the plasma and for rhLCV DNA in PBMCs and oral swabs. RhLCV DNA was detected in the blood and saliva of animals A18151 and A18152 from the VRC01 group beginning at 2–3 weeks following challenge (Figures 4A and 4B). These animals seroconverted 10 and 5 weeks following the second challenge, respectively (Figure 4C). AMMO1 recipient animal A18155 also tested positive for rhLCV DNA in the blood and saliva (Figures 4A and 4B). In contrast, however, this animal did not seroconvert (Figure 4C). Most strikingly, 3 additional AMMO1 recipient animals (A18153, A18154, and A18156) lacked detectable rhLCV DNA or anti-VCA antibodies at all time points (Figures 4A–4C).
Figure 4.
AMMO1 Prevents rhLCV Infection in Macaques
8-month-old rhLCV-negative rhesus macaques were infused i.v. with 20 mg/kg of VRC01 (gray symbols) or AMMO1 (blue symbols). 2 days later, the macaques were challenged orally with 50 transforming units of rhLCV.
(A and B) Viral DNA was quantitated in the blood (A) and oral swabs (B) using a digital droplet PCR assay with primers and probes specific for the internal repeat (IR1) of rhLCV at the indicated time points. Viral loads from blood were averaged from two aliquots of PBMCs processed independently. Viral loads in swab buffer were determined from 3 independent measurements of the same DNA extract from the swab buffer.
(C) Plasma from the study macaques was diluted 1:10, and reactivity to pooled peptides corresponding to amino acids 117–146 and 147–170 of the rhesus small VCA was measured by ELISA at the indicated time points.
(D) ELISA was used to quantify the levels of AMMO1 and VRC01 in plasma at the indicated time points.
(E) Plasma collected from macaques at the time of challenge was evaluated for its ability of neutralize EBV B95.8F infection of Raji cells.
A black arrow indicates the time of viral challenge in (A), (C), and (D).
We measured plasma levels of passively administered antibodies to determine whether differences in their levels could account for the absence of protection in A18155. AMMO1 was detectable at approximately 60 μg/mL in A18153, A18154, and A18156 at the time of challenge, and the levels waned to nearly undetectable levels by 3 weeks (Figure 4D). In A18155, AMMO1 was present at less than 20 μg/mL at the time of challenge, and it waned to undetectable levels within a week. Plasma from A18153, A18154, and A18156 was able to neutralize EBV infection of Raji cells in vitro with comparable potencies (ID50, ~100), whereas the potency of plasma animal A18155 was ~5-fold lower (ID50, ~20; Figure 4E). In summary, AMMO1 at a plasma levels of ~60 μg/mL provided sterilizing immunity against controlled oral LCV challenge.
Discussion
The correlate of protection for most effective vaccines is elicitation of neutralizing antibodies.9 Although previous subunit vaccines have shown partial efficacy against EBV infection in humans, it remains unclear whether neutralizing antibodies affected the outcome of these trials. Here we evaluated the effect of passively transferred neutralizing antibodies on EBV infection in two animal models of lymphocryptovirus infection: humanized mice and rhesus macaques. Passive delivery of AMMO1 prevented EBV-driven changes in lymphocyte populations and reduced viral loads to nearly undetectable levels in a humanized mouse model following a high-dose i.v. challenge of EBV, whereas other mAbs targeting distinct epitopes on gH/gL and a mAb that binds to gp350 and blocks viral attachment to complement receptors failed to do so. We further demonstrated that AMMO1 conferred sterilizing immunity against controlled oral rhLCV challenge in macaques.
EBV subunit vaccine studies have focused on the gp350 protein and have shown partial efficacy in humans and rhesus macaques. A phase II study of a gp350 subunit vaccine reduced the incidence of IM but failed to protect against acquisition.64 Although the vaccine elicited neutralizing antibodies,65 it is not clear whether these affected the outcome. In another study, a protein subunit vaccine based on the gp350 protein from rhLCV showed partial protection in a non-human primate rhLCV challenge study; however, similarly, the correlate of protection was not identified.63 Here, passive delivery of a recombinant version of the anti-gp350 mAb 72A1 failed to protect against i.v. challenge in humanized mice. This finding contrasts with a previous study demonstrating that repeated delivery of hybridoma-derived 72A1 suppressed tumor formation in a related humanized mouse model of EBV infection.49 Experimental differences, including the human lymphocyte composition arising from engrafted PBMCs versus lymphocytes derived from PBSCs, use of a hybridoma-derived murine mAb versus a recombinant humanized mAb, and the dosing regimen, might account for these distinct observations.
In contrast to 72A1, passive delivery of AMMO1 provided a clear protective effect in humanized mice. This result indicates that vaccines based on gH/gL may complement or outperform vaccines based on gp350 alone. Indeed, in one study gH/gL-derived vaccines have been shown to elicit higher B cell neutralizing titers than gp350.66 The finding that mAbs targeting other epitopes on gH/gL failed to prevent lymphoproliferation or viremia underscores the importance of the site of vulnerability on gH/gL defined by AMMO1, implying that AMMO1-like antibodies are a desirable vaccine target. In line with this, a gH/gL subunit vaccine presented on self-assembling ferritin nanoparticles elicited antibodies that competed binding of the 769B10 mAb to gH/gL, which binds an epitope similar to AMMO1.26
Although AMMO1 limited i.v. infection in humanized mice, oral transmission is the normal route of lymphocryptovirus infection. The ability of AMMO1 to prevent rhLCV infection in the rhesus macaque model strongly supports the notion that neutralizing antibodies can prevent oral lymphocryptovirus transmission. In addition, our observation that one animal with lower plasma mAb levels became infected suggests that a protective vaccine will likely need to elicit neutralizing antibodies above a threshold titer. Here, a fixed dose of antibody and a dose of virus that was sufficient to infect all control animals was administered. Future work will be required to define the relationship between viral inoculum and the neutralizing antibody titer sufficient for protection.
Collectively, our data demonstrate that AMMO1 is capable of blocking lymphocryptovirus transmission in vivo by limiting infection of human B cells in humanized mice and preventing oral transmission in a non-human primate model. These findings strongly support the concept that gH/gL-based immunogens targeting this epitope may elicit neutralizing antibodies that will be important for an effective EBV vaccine. Moreover, these studies demonstrate that rhLCV infection of rhesus macaques is a suitable model to evaluate the efficacy of EBV vaccines based on the gH/gL glycoprotein.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Recombinant AMMO1 | Snijder et al.55 | N/A |
| Recombinant VRC01 | Dr. John Mascola, NIAID Vaccine Research Center | N/A |
| CL40 with human constant regions | This study | N/A |
| CL59 with human constant regions | This study | N/A |
| 72A1 with human constant regions | This study | N/A |
| hCD45-FITC | ThermoFisher | Cat # 11-9459-41;RRID:AB_1907395 |
| mCD45-APC | ThermoFisher | Cat# 17-0451-82;RRID:AB_469392 |
| hCD33-PE | BD Biosciences | Cat# 555450; RRID:AB_395843 |
| hCD19-PE-Cy7 | ThermoFisher | Cat# MHCD1912;RRID:AB_10373666 |
| hCD4-eFluor450 | ThermoFisher | Cat# 48-0048-42;RRID:AB_2016674 |
| hCD8-PerCP-Cy5.5 | BD Biosciences | Cat# 560662;RRID:AB_1727513 |
| hCD38-PerCP-Cy5.5 | BD Biosciences | Cat# 551400;RRID:AB_394184 |
| hCD24-BV605 | Biolegend | Cat# 311123; RRID:AB_2562287 |
| hCD4-Alexafluor647 | ThermoFisher | Cat# 56-0048-82;RRID:AB_657741 |
| hCD137-BV421 | BD Biosciences | Cat# 564091;RRID:AB_2722503 |
| hCD69-BV605 | Biolegend | Cat# 310937;RRID:AB_2562306 |
| Goat Anti-Human IgG, Monkey ads-HRP | Southern Biotech | Cat# 2049-05; RRID:AB_2795696 |
| hCD19- BV711 | BioLegend | Cat# 302246;RRID:AB_2562065 |
| hCD20-AF700 | BioLegend | Cat# 302322;RRID:AB_493753 |
| EBNA2-FITC | Novus Biologics | Cat# NBP2-50382F |
| Bacterial and Virus Strains | ||
| EBV B95.8/F | Delecluse et al.67 | N/A |
| Rhesus Lymphocryptovirus strain LCL 8664 | From LCL 8664 Cell line, ATCC | CRL-1805 |
| Biological Samples | ||
| human CD34-enriched PBSCs | Fred Hutch Co-operative Center for Excellence in Hematology | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| human recombinant thrombopoietin | PeproTech | Cat# 300-18 |
| stem cell factor | PeproTech | Cat# 300-07 |
| Flt3-Ligand | PeproTech | Cat# 300-19 |
| busulfan | Otsuka America Pharmaceutical | Cat# 516968AA |
| mABSelect SuRe | Millipore Sigma | Cat# GE17-5438-01 |
| Protein A Agarose | GoldBio | Cat# P-400-5 |
| Pierce IgG Elution Buffer | ThermoFisher | Cat# 21009 |
| sheared salmon sperm DNA | ThermoFisher | Cat# AM9680 |
| rhLCV viral capsid antigen amino acids 117-146: AASAPAATP AVSSSISSLRAATSGAAASSA |
Rao et al.62 | Custom synthesis Genscript |
| rhLCV viral capsid antigen amino acids 147-170: AVDTGSGG GAQPQDTSTRGARKKQ |
Rao et al.62 | Custom synthesis Genscript |
| rhLCV viral capsid antigen amino acids 117-146 scramble: ISRP SLSAASSASAAAATGSSVASPTAAAA |
This study | Custom synthesis Genscript |
| rhLCV viral capsid antigen amino acids147-170 scramble: VGGT SAKGQSKRDGARGTQAQPT |
This study | Custom synthesis Genscript |
| HIV-1 Envelope protein 426c.NLGS.TM4.ΔV1-3 | McGuire et al.72 | N/A |
| Dylite-650 NHS Ester | ThermoFisher | Cat# 62265 |
| GeneJuice Transfection Reagent | EMD Millipore | Cat# 70967 |
| 12-O-Tetradecanoylphorbol 13-acetate | Sigma Aldrich | Cat# P8139 |
| sodium butyrate | Sigma Aldrich | Cat# 303410 |
| Cyclosporin A | Fisher Scientific | Cat# AAJ6319103 |
| eBioscience Fixable Viability Dye eFluor 506 | ThermoFisher | Cat# 65-0866-18 |
| Critical Commercial Assays | ||
| DNeasy Blood & Tissue Kit | QIAGEN | Cat# 69504 |
| Allprep DNA/RNA mini kit | QIAGEN | Cat# 80204 |
| QiaAmp DNA blood mini kit | QIAGEN | Cat# 51104 |
| QuantiTect Multiplex PCR Kit | QIAGEN | Cat# 204543 |
| ddPCR Supermix for Probes, no-dUTP | Bio-Rad | Cat# 1863024 |
| AluI restriction-enzyme | New England BioLabs | Cat# R0137S |
| Droplet Generator DG8 Cartridge | Bio-Rad | Cat# 1864008 |
| SureBlue Reserve TMB Microwell Peroxidase substrate | SeraCare | Cat# 5120-0081 |
| Steady-Glo luciferase reagent | Promega | Cat# E2510 |
| Cell Line Nucleofector Kit V | Lonza | Cat# VVCA-1003 |
| eBioscience Foxp3 / Transcription Factor Staining Buffer Set | ThermoFisher | Cat# 00-5523-00 |
| Experimental Models: Cell Lines | ||
| Raji | ATCC | CCL-86 |
| 293E cells | National Research Council, Canada | N/A |
| Freestyle 293-F cells | Thermofisher | Cat# R79007 |
| CHO-K1 | ATCC | CCL-61 |
| 293-T7 | Omerovic et al.75 | N/A |
| LCL 8664 cells | ATCC | CRL-1805 |
| 293T | ATCC | CRL-3216 |
| Experimental Models: Organisms/Strains | ||
| NOD-scid Il2rgnull mice | The Jackson Laboratory | Stock No: 005557 |
| Oligonucleotides | ||
| Forward primer specific for EBV BALF5 gene: CCCTGTTTATCC GATGGAATG |
Kimura et al.71 | N/A |
| Reverse primer specific for EBV BALF5 gene: CGGAAGCCCTC TGGACTTC |
Kimura et al.71 | N/A |
| FAM-labeled probe specific for EBV BALF5 gene: CGCATTTCTC GTGCGTGTACACC |
Kimura et al.71 | N/A |
| TaqMan Copy Number Reference Assay, human, RNase P | Thermofisher | Cat# 4403326 |
| Rhesus LCV IR1 forward primer AAATCTAAACTTTTGAGGCG ATCTG |
Sashihara et al.63 | N/A |
| Rhesus LCV IR1 reverse primer CCAACCATAGACCCGTTTCCT | Sashihara et al.63 | N/A |
| FAM-labeled Rhesus LCV IR1 probe TCTCCGCGTGCGCATAAT GGC |
Sashihara et al.63 | N/A |
| Rhesus RPP30 forward primer GACTTGGACGTGCGAGCG | This study | N/A |
| Rhesus RPP30 reverse primer GCCGCTGTCTCCACAAGT | This study | N/A |
| Hex-labeled Rhesus RPP30 probe TCTGACCTGAAGGCTCTG CGCG |
This study | N/A |
| Recombinant DNA | ||
| Double stranded BALF5 Target DNA for standard: ACCGAGACCCGGCAGGGGGTCCTGCGGTCGAAGGTGCTGG CCTTGAGGGCGCTGAGGACTGCAAACTCCACGTCCAGACCC TGAGGCGCGCTGGCGTAGAAGTAGGCCTGCTGCCCAAACAC GTTCACACACACGCTGGCCCCATCGGCCTTGCGCCGGCCCA GTAGCTTGATGACGATGCCACATGGCACCACATACCCCTGTT TATCCGATGGAATGACGGCGCATTTCTCGTGCGTGTACACCG TCTCGAGTATGTCGTAGACATGGAAGTCCAGAGGGCTTCCGT GGGTGTCTGCCTCCGGCCTTGCCGTGCCCTCTTGGGCACGC TGGCGCCACCACATGCCCTTTCCATCCTCGTCACCCCCCAC CACCGTCAGGGAGTCTTGGTAGAAGCACAGGGGGGGCTGAG GCCCCCGCACATCCACCACCCCTGCGGCGCCTGGTGTCTGG AAACACTTGGGAATGAGACGCAGGTACTCCTTGTCAGGCTTT TTC |
This study | N/A |
| pCAGGS-rhLCVgH | Plate et al.73 | N/A |
| pCAGGS-rhLCVgL | Plate et al.73 | N/A |
| pCAGGS-rhLCVgB | Plate et al.73 | N/A |
| p509 | Delecluse et al.67 | N/A |
| pT7EMCLuc | Okuma et al.74 | N/A |
| Software and Algorithms | ||
| QuantaSoft Analysis Software | Bio-Rad | N/A |
| Prism 7.03 or later software package | Graph Pad Software | N/A |
| Other | ||
| QuantStudio 7 Flex Real Time PCR System | Applied Biosystems | N/A |
| QX200 ddPCR System | Bio-Rad | N/A |
| C1000 Touch Thermal Cycler | Bio-Rad | N/A |
| QX200 Droplet Reader | Bio-Rad | N/A |
| SpectraMax M2 plate reader | Molecular Devices | N/A |
| BD LSRII cytometer | BD Biosciences | N/A |
| Ascent FL fluorimeter | Fluoroskan | N/A |
| EvosFL imaging system | ThermoFisher | N/A |
Resource Availability
Lead Contact
Requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Andrew T. McGuire (amcguire@fredhutch.org).
Materials Availability
Requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact. Expression plasmids derived from pTT3 require an MTA from the National Research Council (Canada).
Data and Code Availability
The published article includes all datasets generated or analyzed during this study
Experimental Model and Subject Details
Cell lines
All cell lines were incubated at 37°C in the presence of 5% CO2 and were not tested for mycoplasma contamination. 293-F and 293E (human female) were maintained in Freestyle 293 media with gentle shaking. CHO-K1 (hamster female) cells were maintained in Ham’s F-12 + 10% FBS, 2 mM L-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin (cF-12). Raji cells (human male) and LCL 8664 (rhesus macaque male) were maintained in RMPI + 10% FBS, 2 mM L-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin (cRPMI). 293T cells (human female) were maintained in DMEM + 10% FBS, 2 mM L-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin (cDMEM). 293-T7 cells (human female) were maintained in cDMEM containing 100μg/ml Zeocin.
Human Subjects
CD34-enriched PBSCs were obtained from granulocyte colony-stimulating factor mobilized healthy donors at the Fred Hutch Co-operative Center for Excellence in Hematology. All healthy donors for mobilized peripheral blood products (PBSCs) provided written informed consent during a face-to-face meeting in compliance with the Declaration of Helsinki. The evaluation for donor eligibility, testing, mobilization, and collection were conducted under an approved protocol overseen by the Fred Hutch Institution Review Board. De-identified CD34 cryopreserved specimens were provided to user after transfer agreement and with the acknowledgment of user approval by the Seattle Children’s Research Institute (SCRI). PBSCs used in this study were obtained from a 35 year old female and a 25 year old female.
Mice
NOD-scid Il2rgnull (NSG) mice were housed in a specific pathogen-free facility at SCRI. The facility is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care. Mice were handled in accordance with the NIH Guide for the Care and Use of Laboratory Animals, and experiments were approved by the SCRI Institutional Animal Care and Use Committee and Institutional Review Board. All mice used in this study were female and were 8 weeks old when experiments were initiated.
Rhesus Macaques
All rhesus macaque work was approved by the University of Washington and Fred Hutchinson Cancer Research Center IACUCs. 4 male and 2 female infant rhesus macaques were obtained from the Oregon National Primate Research Center within 24-48 hours of birth and hand-reared in a nursery. 2-3 weeks later the animals were transported to the Washington National Primate Research Center and housed in pairs until they were 6 months of age. From 2 months of age to 6 months of age the animals we bled monthly and tested for serum responses to viral capsid antigen and assayed for viral DNA by qPCR. Prior to the start of the challenge study the animals were housed individually and remained so for the duration of the study.
Method Details
EBV infection in humanized mice
Frozen human CD34-enriched PBSCs were thawed then cultured at a cell density of 1 × 106 cells/mL in a humidified 37°C incubator with 5% CO2. Culture media was SCGM (CellGenix) with 100ng/mL each of human recombinant Thrombopoietin, stem cell factor, and Flt3-ligand. After 24 hours in culture, cells were washed in normal saline and 1 × 106 CD34+ cells delivered per busulfan-conditioned NSG mouse by retro-orbital injection. Busulfan conditioning was performed by intraperitoneal (i.p.) injection of 35 mg/kg clinical grade busulfan into 8 week-old NSG mice 24 hours prior to human PBSC transfer. 10-12 weeks post-cell transfer, successful human cell engraftment was confirmed by the presence of human CD45+ cells in peripheral blood using flow cytometry.12-13 weeks post-human HSPC transfer, 500 μg of experimental or control antibodies were injected per humanized NSG mouse intravenously (i.v.). 48 hours later the mice received a dose of EBV B95.8/F67 equivalent to 33,000 infectious units as determined by infection of Raji cells,68 via retro-orbital injection. Each group of mice receiving the same mAb and/or EBV were housed separately from unchallenged mice to avoid contamination. Beginning at one week post-infection, peripheral blood samples were collected to determine levels of transferred mAbs in plasma, the presence of EBV DNA in whole blood, or were used to assess immunophenotype of circulating lymphocytes using antibodies at a 1:100 dilution unless otherwise noted: hCD45-FITC, mCD45-APC (1:500 dilution), hCD33-PE, hCD19-PE-Cy7, hCD4-eFluor450 and hCD8-PerCP-Cy5.5.
Eight to nine weeks post-challenge, mice were euthanized and single cell suspensions of splenocytes were collected and immunophenotyping of B cells using hCD45-FITC, mCD45-APC, CD19-PE-Cy7, hCD38-PerCP-Cy5.5, and hCD24-BV605, or T cells using hCD45-FITC at a 1:100 dilution, mCD45-APC at a 1:500 dilution, hCD4-Alexafluor647 at a 1:100 dilution, hCD8-PerCP-Cy5.5 at a 1:100 dilution, hCD137-BV421 at a 1:50 dilution and hCD69-BV605 at a 1:50 dilution, was performed by flow cytometry as described previously.69 DNA was extracted from 5 million total splenocytes, utilizing the DNeasy Blood & Tissue Kit (QIAGEN) and according to the manufacturer’s instructions, for subsequent viral load analysis.
Antibody Expression Plasmids
cDNA corresponding to the 72A1 VH (GenBank: KT211017) or VL (GenBank: KT211018)54 were codon optimized and cloned in-frame with the human IgG1 and human lambda constant regions in pTT3-based expression vectors.55 Codon optimized cDNA corresponding to CL40 VH (GenBank: MF104552) and CL59 VH (GenBank: MF104554) was synthesized (Integrated DNA Technologies) and cloned in-frame with the human IgG1 constant region in pTT3-based expression vectors. Codon optimized cDNA corresponding to CL40 VL (GenBank: MF104553) and CL59 VL (GenBank: MF104555) were cloned in-frame with the human kappa constant regions in pTT3-based expression vectors.
Antibody production
The AMMO1 used in passive transfer studies in rhesus macaques was expressed from 293F cells using the Daedalus system70 and purified at the Fred Hutchinson Cancer Research Center Molecular Therapeutics and Design Center. Recombinant AMMO1 (human IgG1) was purified using MabSelect SuRe resin according to the manufacturer’s instructions. AMMO1 IgG1 was dialyzed into PBS, sterile filtered and stored at −80°C until use. AMMO1 was verified to be endotoxin free (< 0.5Eu/mg). GMP grade, recombinant VRC01 IgG was a kind gift from Dr. John Mascola (NIAID, Vaccine Research Center).
pTT3-derived antibody expression plasmids encoding antibody heavy and light chains55 were co-transfected into 293E cells at a density of 106 cells/ml in Freestyle 293 media using the 293Free transfection reagent according to the manufacturer’s instructions. Expression was carried out in Freestyle 293 media for 6 days, after which cells and cellular debris were removed by centrifugation at 4,000 × g followed by filtration through a 0.22 μm filter. Clarified cell supernatant containing recombinant antibodies was passed over Protein A Agarose, followed by extensive washing with PBS, and then eluted with 1 mL of Pierce IgG Elution Buffer, pH 2.0, into 0.1 mL of Tris HCl, pH 8.0. Purified antibodies were then dialyzed overnight into PBS, passed through a 0.2μM filter under sterile conditions and stored at −80°C until use.
Rhesus macaque PBMC isolation
Rhesus macaque blood was drawn by venipuncture and collected in vacutainers containing EDTA. Blood was centrifuged at 1000 × g for 30 min. Plasma was collected and stored at −20°C until use. PBMC were further purified on a Ficoll cushion, subjected to DNA extraction using the Allprep DNA/RNA mini kit (QIAGEN) according to the manufacturer’s instructions, or aliquoted into FBS containing 10% DMSO and stored in liquid nitrogen.
Rhesus macaque oral swabs
A sterile polyester swab was rubbed on the inside of the both cheeks (buccal mucosa), along the upper and lower gum-lines outside of the teeth. Swabs were placed in 1ml of 10mM Tris, 25mM EDTA, 50mM potassium chloride, 1% Igepal CA 630 pH 8.0. Swabs were stored at −20°C until use. After thawing, DNA was extracted from 200μl of buffer was extracted using the QiaAmp blood DNA kit, and eluted in 100 μL of AE buffer (QIAGEN).
Quantitative PCR analysis of human cells in huCD34 engrafted mice
A primer-probe mix specific for the EBV BALF5 gene71 was used to quantify EBV in DNA extracted from blood or spleen in hCD34 engrafted NSG recipient mice at the time points described. Each 25μl qPCR reaction contained 12.5μl of 2 × QuantiTect Probe PCR Master Mix (QIAGEN), 600nM of each primer, 300nM of FAM-labeled probe, 1.25μl of a TaqMan 20 × VIC-labeled RNase-P primer-probe mix. For analysis of splenocytes, reactions contained 200ng DNA extracted from splenocytes as template. To analyze EBV in peripheral blood 70μl of blood collected via cardiac puncture or retro-orbital bleed was mixed with 3μg of sheared salmon sperm DNA using the DNeasy Blood and Tissue Kit (QIAGEN) and eluted in 50μl of Buffer AE (QIAGEN). 10μl of extracted DNA was used as template in qPCR.
Reactions were heated to 95°C for 15 minutes to activate DNA polymerase followed by 50 cycles of 95°C for 15 s 60°C for 60 s, on an Applied Biosystems QuantStudio 7 Flex Real Time PCR System. Synthetic DNA fragments containing the BALF5 target gene as well as flanking genomic regions were synthesized as double stranded DNA gBlocks, and were used to generate a standard curve with known gene copy numbers ranging from 107-100 copies/μl. The copy number in extracted DNA was determined by interpolating from the standard curve. Serial dilutions of reference standard were used to experimentally determine a limit of detection of 6.25 copies, which corresponds to the amount of template that can be detected in > 95% of reactions. For graphical purposes, samples with no amplification or those yielding values below the limit of detection were assigned a value of 0.625 copies.
Digital droplet PCR
Digital droplet PCR (ddPCR) was conducted on DNA extracted from rhesus macaque PBMC or saliva, utilizing Bio-Rad’s QX200 ddPCR system in a 22μl total volume. To quantify rhLCV DNA, ddPCR reactions used primers and a FAM-labeled probe specific for the internal repeat 1 (IR1) region of rhLCV which is present at 5 copies per genome.63 Viral loads in blood were normalized to the number of PBMC in a separate reaction using a primer/probe mix specific for the ribonuclease P/MRP subunit p30 (RPP30) which is present at 2 copies per cell. Each reaction contained 10μl ddPCR Supermix for Probes, no-dUTP, and 5 units of the AluI restriction-enzyme. Reactions which quantified IR1 contained 600nM of primer, 300nM of probe, and 1μg of extracted DNA, while those which quantified RPP30 contained 450nM of primer, 225nM of probe, and 0.05μl DNA extracted from PBMC. Each reaction was adjusted to a final volume of 22μl using dH2O. Each 22μl mixture was loaded into a Droplet Generator DG8 Cartridge (Bio-Rad) and placed in a QX200 Droplet Generator (Bio-Rad). After droplets were made, the droplets were transferred to a 96-well PCR plate, sealed and placed in a C1000 Touch Thermal Cycler (Bio-Rad) and subjected to one cycle of 95°C for 10 minutes for enzyme activation, 40 cycles of 94°C for 30 s for denaturation and 60°C for 1 minute for annealing/extension, one cycle of 98°C for 10 minutes for enzyme deactivation and a 4°C hold. Each step utilized a temperature rate increase or decrease of 2°C per second. After PCR, the droplet plate was placed in a QX200 Droplet Reader (Bio-Rad). The droplets whose fluorescence was measured higher than the threshold (4000 for FAM, 3000 for HEX) were counted as positive droplets. The data were analyzed using QuantaSoft Analysis Software (Bio-Rad) to determine the concentration of the rhLCV in copies per microliter of reaction.
The rhLCV copies per million copies macaque PBMC were determined using the following equation:
rhLCV copies in saliva are expressed as copies/ml of swab buffer.
rhLCV viral capsid antigen ELISA
Peptides corresponding to rhLCV viral capsid antigen (VCA) amino acids 117-146, 147-17062 as well as scramble peptides of each were synthesized by Genscript. 50 ng/well of a 1:1 mixture of to rhLCV VCA 147-170 and rhLCV VCA117-146 or a 1:1 mixture of the respective scramble peptides were adsorbed onto 96 well half-area microplates at 37°C for 1 hour in a solution of 0.1 M NaHCO3 pH 9.4-9.6. Plates were then washed 4 times with ELISA washing buffer (1 × PBS, 0.02% Tween 20) prior to blocking at 37°C for 1 hour with 100 μl per well of PBS containing 3% BSA and 0.02% Tween 20 (blocking buffer). After blocking, plates were washed 4 × with ELISA washing buffer. Serum was diluted 1:10 in 50μl of blocking buffer and added to triplicate wells of pooled rhLCV VCA and scramble peptides followed by a 1 hour incubation at 37°C. Following 4 additional washes with ELISA washing buffer, a 1:4000 dilution of goat anti-human IgG in blocking buffer was added to each well and incubated at 37°C for 1 hour followed by 4 washes with wash buffer. 50 μl/well of SureBlue Reserve TMB Microwell Peroxidase substrate was added. After 3 min, 50 μl/well of 1N sulfuric acid was added and the A450 of each well was read on a Molecular Devices SpectraMax M2 plate reader. Background signal of secondary antibody against peptide coated wells was averaged and subtracted from wells containing rhesus macaque sera. Absorbance values from triplicate wells from each animal’s sera were averaged. The average absorbance of the wells coated with scramble peptides were subtracted from the corresponding pool of rhLCV VCA peptides. Negative values are reported as zero.
Detection of AMMO1 and VRC01 in plasma
200ng/well of EBV gH/gL55 were adsorbed onto 96 well Immulon 2HB ELISA plates at 37°C for 1 hour in a solution of 0.1 M NaHCO3 pH 9.4-9.6. Plates were then washed 4 times with ELISA washing buffer (1 × PBS, 0.02% Tween 20) prior to blocking at 37°C for 1 hour with 250 μl per well of PBS containing 10% non-fat milk and 0.02% Tween 20 (blocking buffer). After blocking, plates were washed 4 × with ELISA washing buffer. A known concentration of AMMO1 mAb, rhesus macaque serum collected just prior to mAb infusion, or rhesus macaque serum collected 2 days after mAb infusion was diluted 1:100 in blocking buffer and three-fold serial dilutions were performed in duplicate in 100μl volumes followed by a 1 hour incubation at 37°C. Control wells were incubated with blocking buffer not containing mAb or sera. Following 4 additional washes with ELISA washing buffer, a 1:3000 dilution of goat anti-human IgG, cross-adsorbed with rhesus and cynomolgus macaque IgG in blocking buffer was added to each well and incubated at 37°C for 1 hour followed by 4 washes with wash buffer. 100 μl/well of SureBlue Reserve TMB Microwell Peroxidase substrate was added. After 3 min, 100 μl/well of 1N sulfuric acid was added and the A450 of each well was read on a Molecular Devices SpectraMax M2. The background A450 was averaged from control wells and subtracted from all other wells on the plate. To generate a standard curve, the corrected average A450 of duplicate wells were plotted against the known concentration of AMMO1 and the data was fit to a sigmoidal dose-response curve using Prism 7.03 software package (GraphPad Software).
To determine the concentration of AMMO1 in the plasma, the average A450 of wells containing pre-infusion plasma was subtracted from the absorbance of wells containing post-infusion plasma at each dilution. The background-subtracted absorbance values were averaged at each dilution and absorbance readings that fell on the linear (hill slope) part of the AMMO1 standard curve were used to interpolate the concentration of AMMO1 in plasma. The concentration of VRC01 in plasma was determined using the same method, except that the standard curve was generated with VRC01 binding to monomeric 426c.NLGS.TM4.ΔV1-3.72
EBV neutralization assay in B cells
EBV neutralization assays were carried out in Raji cells essentially as described.22 AMMO1 mAb, plasma from humanized mice, or plasma from rhesus macaques, was serially diluted in duplicate wells of 96-well round-bottom plates containing 25μl of cRPMI in duplicate. 12.5μl of B95-8/F virus (diluted to achieve an infection frequency of 1%–10% at the final dilution) was added and incubated at 37°C for 1 hour. 12.5 μl of cRMPI containing 4 × 106 Raji cells/ml was added to each well and incubated for another hour at 37°C. The cells were then pelleted, washed once with cRPMI, and re-suspended in cRMPI. Antibody concentration or serum dilution is reported relative to the final infection volume (50μl). After 3 days at 37°C, cells were fixed in 2% paraformaldehyde. The percentage of GFP+ Raji cells as determined on a BD LSRII cytometer.
To account for any false positive cells due to auto-fluorescence in the GFP channel, the average %GFP+ cells in negative control wells (n = 5-10) was subtracted from each well. The infectivity (%GFP+) for each well was plotted as a function of the log10 of the mAb concentration or serum dilution. The neutralization curve was fit using the log (inhibitor) versus response- variable slope (four parameters) analysis in Prism 7.03 (GraphPad Software).
rhLCV gH/gL cell surface staining
10ml of 293F cells at a density of 1 × 106 cells/ml were transfected with equal amounts of pCAGGS-rhLCVgH, pCAGGS-rhLCVgL,61 or mock transfected using 293 Free according to the manufacturer’s instructions. 1 hour following transfection 500μl of cell culture was added to 500μl of DMEM in a 6-well dish. 24 hours later the cells were fixed in 10% formalin washed 3X with PBS containing 1% BSA and 0.1% Tween-20. The cells were then incubated with 1μg of AMMO1 conjugated to Dylite-650 in 1ml of PBS containing 1% BSA and 0.1% Tween-20 at room temperature for 30min. The cells were then washed 3 times with PBS and visualized on an EvosFL imaging system using a 20 × objective lens and a Cy5 light cube (ThermoFisher).
Virus-free fusion assay
CHO-K1 cells were seeded onto six-well plates at a density of 3 × 105 cells/well. 24 hours later, the cells were transfected with 0.5 μg each of pCAGGS-rhLCVgH, pCAGGS-rhLCVgL, pCAGGS-rhLCVgB73 and 0.8 μg of pT7EMCLuc, which carries a luciferase-containing reporter plasmid under the control of the T7 promoter,74 using GeneJuice Transfection Reagent. As a control for background luciferase activity one well was transfected with the same mix as above, except the pCAGGS-rhLCVgB plasmid was replaced by empty pTT3.
Meanwhile, 293-T7 cells were seeded into a 96 well plate at a density of 1 × 104 cells per well in a volume of 100 μl/well of cF-12 without Zeocin selection.
8 hours later, the transfected CHO cells were trypsinized, washed once with cF-12, and re-suspended at a density of 1 × 105 cells/ml in F-12 media. 100 μl/well of CHO-K1 suspension was added to the plate containing 293-T775 cells. Immediately after the addition of CHO-K1 cells, 2 μg of AMMO1, CL59, E1D1, CL40, or a no antibody control were added to 6 wells in parallel. 24 hours later, the media was aspirated and the cells were lysed in 100 μl of Steady-Glo luciferase reagent. 75 μl of cell lysate was transferred to a white bottom assay plate and luciferase activity was read on a Fluroskan Ascent FL fluorimeter.
rhLCV virus production
Rhesus LCV was isolated from LCL8664 cells using two methods.
TPA/Butyrate
LCL8664 cells were cultured in cRPMI at approximately 0.5-1 × 106 cells/ml. 1 × 109 cells were resuspended at a density of 3 × 106 cells/ml in cRMPI, to which 12-O-Tetradecanoylphorbol 13-acetate was added to a final concentration of 20ng/mL and sodium butyrate was added to a final concentration of 3mM. Cells were then incubated at 37°C overnight. The next day, cells were pelleted by centrifugation at 300 × g for 10 minutes and resuspended at a density of 3 × 106 cells/ml in cRPMI and cultured at 37°C for 6 days and then passed through a 0.45 μM filter. The supernatant was then centrifuged at 10,000 × g for 2 hours at 4°C. The supernatant was aspirated and the pellet was resuspended in ~2ml of RPMI containing 10% FBS. Virus was dispensed into 250μl aliquots and stored at −80°C.
Electroporation
240 million LCL8664 cells were pelleted at 90 X g for 10 minutes. The cell pellet was resuspended in 2.4ml of Nucleofector Solution V containing supplement (Lonza). 0.1ml of cell suspension was added to 24 separate electroporation cuvettes each containing 2.5 μg of p509 an expression plasmid encoding EBV BZLF1. Electroporation was carried out using program X-001 on an Amaxa nucleofector (Lonza). Immediately following electroporation, cells were diluted with 500μl of cRPMI and then transferred to a T225 flask containing 120ml of cRPMI. Cells were cultured at 37°C for 5 days and then passed through a 0.45 μM filter. The supernatant was then centrifuged at 10, 000 X g for 2 hours at 4°C. The supernatant was aspirated and the pellet was resuspended in ~1.5ml of RPMI containing 10% FBS. Virus was dispensed into 250μl aliquots and stored at −80°C.
rhLCV titration
Serial dilutions of rhLCV were added to 96 well plates containing 1 × 105 rhesus PBMC in RPMI containing 10% FBS, 2mM L-glutamine, 100U/ml penicillin, and 100μg/ml streptomycin and 0.5μg cyclosporin A. Each concentration of virus was tested against 20 PBMC-containing wells. Media was replenished weekly with 50-100μl and the cells were cultured for 7-10 weeks. Wells were visually inspected and scored as positive (clumping/growing cells, media turning to yellow) or negative (no cell growth, pink media). Virus concentration was plotted against % transformed wells and fit with a sigmoidal dose-response (variable slope) curve using Prism 7.03 software. 1 Transforming unit is defined as the dilution that yields 62.5% positive wells.76 The transforming units/ml of each stock were determined using PBMC drawn from 2 separate animals and averaged.
rhLCV neutralization assay
2.5 transforming units of RhLCV were incubated with either no antibody or with 5μg of AMMO1 or AMMO2 in a 50μl volume of cRPMI containing 0.5μg cyclosporin A for 30 min at 37°C. Next, 50μl of cRPMI containing 0.5μg cyclosporin A and 1.5 × 105 of fresh rhesus PBMC were added to each well and incubated for an additional hour at 37°C. As a no-infection control 1.5 × 105 fresh rhesus PBMC were diluted into 100μl of cRPMI containing 0.5μg cyclosporin A. The contents of each well were then diluted to 500μl of cRPMI containing 0.5μg cyclosporin A in a 24 well plate. An additional 2μg of AMMO1 or AMMO2 were added to wells containing cells and virus that had been pre-incubated with these Abs. Following a 3 day incubation at 37°C, the PBMC from each well were collected and resuspended in 100ul of PBS containing 3% bovine serum albumin and 1.5μl CD19 and 1μl CD20-AF700 and 0.75μl/sample eBioscience Fixable Viability Dye eFluor 506. The cells were then fixed and permeabilized using the eBioscience Foxp3 / Transcription Factor Staining Buffer Set according to the manufacturer’s instructions and then and stained with a 1:50 dilution of EBNA 2 FITC conjugate derived from the PE2 mAb which reacts with EBNA2 from rhLCV.77 The cells were then washed 2X and then analyzed on a BD-LSII cytometer. rhLCV infected B cells were defined as live, CD19+, CD20+, EBNA2+.
Quantification and Statistical Analysis
Statistical analyses were performed using GraphPad (version 7 and later). Statistical analyses for all humanized mouse studies were conducted using one way ANOVA, differences in the means of each group were determined using the Sidak multiple comparisons test, p values are indicated in the corresponding figure legends. Unless otherwise indicated, analyses were conducted on one representative experimental replicate; ie one group of mice engrafted with the same pool of HPSC on the same day, and also challenged with virus at the same time. No additional methods were used to determine whether the data met assumptions of the statistical approach.
Differences in the cell-free fusion assay were determined using an unpaired 2-tailed t test p values are shown in the corresponding figure legend. Due to the relatively small number of animals, statistical analyses were not performed on rhesus macaque studies.
Acknowledgments
We would like to thank Dr. Leonidas Stamatatos (Fred Hutch) for 426c.NLGSTM4ΔV1-3; Dr. John Mascola (NIH-VRC) for the VRC01 mAb used in the rhesus passive transfer studies; Dr. R. Longnecker for expression plasmids encoding rhLCV gH, rhLCV gL, and rhLCV gB; Meei-Li Huang, Haiying Zhu, and Arun Nalla from the University of Washington Virology Clinical Laboratory for assistance with assay development; Clayton Ferrier, Jesse Day, and Charlotte Hotchkiss from the WANPRC for assistance with the rhesus macaque experiments; and the Fred Hutch CCEH Cell Procurement and Processing Resource for Human CD34-enriched PBSCs. The research reported in this publication was supported by funding from the Office of Research Infrastructure Programs (ORIP) under award P51 OD010425 (to the WANPRC); the National Institute Of Allergy And Infectious Diseases of the National Institutes of Health (NIH) under award R01AI147846 (to A.T.M.); and the National Center for Advancing Translational Sciences (NCATS) under award UL1TR000423. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH, ORIP, or NCATS. Additional support was provided by the Seattle Children’s Research Institute Program for Cell and Gene Therapy, the Children’s Guild Association Endowed Chair in Pediatric Immunology, and the Tom Hansen Investigator in Pediatric Innovation Endowment (to D.J.R.).
Author Contributions
A.T.M., S.S., and D.J.R. conceptualized the study. S.S., L.J.H., N.R.A., S.L., C.M.S., H.M., and Y-H.W. performed experiments and analyzed data. A.T.M., S.S., L.J.H., N.R.A., and D.J.R. wrote the manuscript, and all authors contributed to editing and figure preparation. Funding was secured by A.T.M. and D.J.R.
Declaration of Interests
A.T.M. holds provisional patents 62/504,447 and 62/560,061 related to this work.
Published: June 23, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.xcrm.2020.100033.
Contributor Information
David J. Rawlings, Email: drawling@uw.edu.
Andrew T. McGuire, Email: amcguire@fredhutch.org.
Supplemental Information
References
- 1.Khan G., Hashim M.J. Global burden of deaths from Epstein-Barr virus attributable malignancies 1990-2010. Infect. Agent. Cancer. 2014;9:38. doi: 10.1186/1750-9378-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taylor G.S., Long H.M., Brooks J.M., Rickinson A.B., Hislop A.D. The immunology of Epstein-Barr virus-induced disease. Annu. Rev. Immunol. 2015;33:787–821. doi: 10.1146/annurev-immunol-032414-112326. [DOI] [PubMed] [Google Scholar]
- 3.Levin L.I., Munger K.L., O’Reilly E.J., Falk K.I., Ascherio A. Primary infection with the Epstein-Barr virus and risk of multiple sclerosis. Ann. Neurol. 2010;67:824–830. doi: 10.1002/ana.21978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Handel A.E., Williamson A.J., Disanto G., Handunnetthi L., Giovannoni G., Ramagopalan S.V. An updated meta-analysis of risk of multiple sclerosis following infectious mononucleosis. PLoS ONE. 2010;5:e12496. doi: 10.1371/journal.pone.0012496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balandraud N., Roudier J. Epstein-Barr virus and rheumatoid arthritis. Joint Bone Spine. 2018;85:165–170. doi: 10.1016/j.jbspin.2017.04.011. [DOI] [PubMed] [Google Scholar]
- 6.Cohen J.I., Mocarski E.S., Raab-Traub N., Corey L., Nabel G.J. The need and challenges for development of an Epstein-Barr virus vaccine. Vaccine. 2013;31(Suppl 2):B194–B196. doi: 10.1016/j.vaccine.2012.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ainsworth C. Building a better lymphoma vaccine. Nature. 2018;563:S52–S54. doi: 10.1038/d41586-018-07366-1. [DOI] [PubMed] [Google Scholar]
- 8.Balfour H.H., Jr., Schmeling D.O., Grimm-Geris J.M. The promise of a prophylactic Epstein-Barr virus vaccine. Pediatr. Res. 2020;87:345–352. doi: 10.1038/s41390-019-0591-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plotkin S.A. Correlates of protection induced by vaccination. Clin. Vaccine Immunol. 2010;17:1055–1065. doi: 10.1128/CVI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biggar R.J., Henle W., Fleisher G., Böcker J., Lennette E.T., Henle G. Primary Epstein-Barr virus infections in African infants. I. Decline of maternal antibodies and time of infection. Int. J. Cancer. 1978;22:239–243. doi: 10.1002/ijc.2910220304. [DOI] [PubMed] [Google Scholar]
- 11.Piriou E., Asito A.S., Sumba P.O., Fiore N., Middeldorp J.M., Moormann A.M., Ploutz-Snyder R., Rochford R. Early age at time of primary Epstein-Barr virus infection results in poorly controlled viral infection in infants from Western Kenya: clues to the etiology of endemic Burkitt lymphoma. J. Infect. Dis. 2012;205:906–913. doi: 10.1093/infdis/jir872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rickinson A.B., Fox C.P. Epstein-Barr virus and infectious mononucleosis: what students can teach us. J. Infect. Dis. 2013;207:6–8. doi: 10.1093/infdis/jis647. [DOI] [PubMed] [Google Scholar]
- 13.Chan K.H., Tam J.S., Peiris J.S., Seto W.H., Ng M.H. Epstein-Barr virus (EBV) infection in infancy. J. Clin. Virol. 2001;21:57–62. doi: 10.1016/s1386-6532(01)00149-4. [DOI] [PubMed] [Google Scholar]
- 14.Slyker J.A., Casper C., Tapia K., Richardson B., Bunts L., Huang M.L., Maleche-Obimbo E., Nduati R., John-Stewart G. Clinical and virologic manifestations of primary Epstein-Barr virus (EBV) infection in Kenyan infants born to HIV-infected women. J. Infect. Dis. 2013;207:1798–1806. doi: 10.1093/infdis/jit093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sitki-Green D., Covington M., Raab-Traub N. Compartmentalization and transmission of multiple epstein-barr virus strains in asymptomatic carriers. J. Virol. 2003;77:1840–1847. doi: 10.1128/JVI.77.3.1840-1847.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brooks J.M., Croom-Carter D.S., Leese A.M., Tierney R.J., Habeshaw G., Rickinson A.B. Cytotoxic T-lymphocyte responses to a polymorphic Epstein-Barr virus epitope identify healthy carriers with coresident viral strains. J. Virol. 2000;74:1801–1809. doi: 10.1128/jvi.74.4.1801-1809.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwok H., Chan K.W., Chan K.H., Chiang A.K. Distribution, persistence and interchange of Epstein-Barr virus strains among PBMC, plasma and saliva of primary infection subjects. PLoS ONE. 2015;10:e0120710. doi: 10.1371/journal.pone.0120710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tierney R.J., Edwards R.H., Sitki-Green D., Croom-Carter D., Roy S., Yao Q.Y., Raab-Traub N., Rickinson A.B. Multiple Epstein-Barr virus strains in patients with infectious mononucleosis: comparison of ex vivo samples with in vitro isolates by use of heteroduplex tracking assays. J. Infect. Dis. 2006;193:287–297. doi: 10.1086/498913. [DOI] [PubMed] [Google Scholar]
- 19.Weiss E.R., Lamers S.L., Henderson J.L., Melnikov A., Somasundaran M., Garber M., Selin L., Nusbaum C., Luzuriaga K. Early Epstein-Barr Virus Genomic Diversity and Convergence toward the B95.8 Genome in Primary Infection. J. Virol. 2018;92 doi: 10.1128/JVI.01466-17. e01466–e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith N.A., Baresel P.C., Jackson C.L., Ogolla S., Toko E.N., Heit S., Piriou E., Sumba O.P., Middeldorp J.M., Colborn K.L., Rochford R. Differences in the Epstein-Barr Virus gp350 IgA Antibody Response Are Associated With Increased Risk for Coinfection With a Second Strain of Epstein-Barr Virus. J. Infect. Dis. 2019;219:955–963. doi: 10.1093/infdis/jiy601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sathiyamoorthy K., Chen J., Longnecker R., Jardetzky T.S. The COMPLEXity in herpesvirus entry. Curr. Opin. Virol. 2017;24:97–104. doi: 10.1016/j.coviro.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sashihara J., Burbelo P.D., Savoldo B., Pierson T.C., Cohen J.I. Human antibody titers to Epstein-Barr Virus (EBV) gp350 correlate with neutralization of infectivity better than antibody titers to EBV gp42 using a rapid flow cytometry-based EBV neutralization assay. Virology. 2009;391:249–256. doi: 10.1016/j.virol.2009.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller G., Niederman J.C., Stitt D.A. Infectious mononucleosis: appearance of neutralizing antibody to Epstein-Barr virus measured by inhibition of formation of lymphoblastoid cell lines. J. Infect. Dis. 1972;125:403–406. doi: 10.1093/infdis/125.4.403. [DOI] [PubMed] [Google Scholar]
- 24.Moss D.J., Pope J.H. Assay of the infectivity of Epstein-Barr virus by transformation of human leucocytes in vitro. J. Gen. Virol. 1972;17:233–236. doi: 10.1099/0022-1317-17-2-233. [DOI] [PubMed] [Google Scholar]
- 25.Tugizov S.M., Berline J.W., Palefsky J.M. Epstein-Barr virus infection of polarized tongue and nasopharyngeal epithelial cells. Nat. Med. 2003;9:307–314. doi: 10.1038/nm830. [DOI] [PubMed] [Google Scholar]
- 26.Bu W., Joyce M.G., Nguyen H., Banh D.V., Aguilar F., Tariq Z., Yap M.L., Tsujimura Y., Gillespie R.A., Tsybovsky Y. Immunization with Components of the Viral Fusion Apparatus Elicits Antibodies That Neutralize Epstein-Barr Virus in B Cells and Epithelial Cells. Immunity. 2019;50:1305–1316.e6. doi: 10.1016/j.immuni.2019.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thorley-Lawson D.A., Poodry C.A. Identification and isolation of the main component (gp350-gp220) of Epstein-Barr virus responsible for generating neutralizing antibodies in vivo. J. Virol. 1982;43:730–736. doi: 10.1128/jvi.43.2.730-736.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mascola J.R., Stiegler G., VanCott T.C., Katinger H., Carpenter C.B., Hanson C.E., Beary H., Hayes D., Frankel S.S., Birx D.L., Lewis M.G. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat. Med. 2000;6:207–210. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 29.Hessell A.J., Hangartner L., Hunter M., Havenith C.E., Beurskens F.J., Bakker J.M., Lanigan C.M., Landucci G., Forthal D.N., Parren P.W. Fc receptor but not complement binding is important in antibody protection against HIV. Nature. 2007;449:101–104. doi: 10.1038/nature06106. [DOI] [PubMed] [Google Scholar]
- 30.Hessell A.J., Poignard P., Hunter M., Hangartner L., Tehrani D.M., Bleeker W.K., Parren P.W., Marx P.A., Burton D.R. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat. Med. 2009;15:951–954. doi: 10.1038/nm.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parren P.W., Marx P.A., Hessell A.J., Luckay A., Harouse J., Cheng-Mayer C., Moore J.P., Burton D.R. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J. Virol. 2001;75:8340–8347. doi: 10.1128/JVI.75.17.8340-8347.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang L., Shi W., Chappell J.D., Joyce M.G., Zhang Y., Kanekiyo M., Becker M.M., van Doremalen N., Fischer R., Wang N. Importance of Neutralizing Monoclonal Antibodies Targeting Multiple Antigenic Sites on the Middle East Respiratory Syndrome Coronavirus Spike Glycoprotein To Avoid Neutralization Escape. J. Virol. 2018;92:e02002–e02017. doi: 10.1128/JVI.02002-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shingai M., Donau O.K., Plishka R.J., Buckler-White A., Mascola J.R., Nabel G.J., Nason M.C., Montefiori D., Moldt B., Poignard P. Passive transfer of modest titers of potent and broadly neutralizing anti-HIV monoclonal antibodies block SHIV infection in macaques. J. Exp. Med. 2014;211:2061–2074. doi: 10.1084/jem.20132494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Magnani D.M., Rogers T.F., Beutler N., Ricciardi M.J., Bailey V.K., Gonzalez-Nieto L., Briney B., Sok D., Le K., Strubel A. Neutralizing human monoclonal antibodies prevent Zika virus infection in macaques. Sci. Transl. Med. 2017;9:eaan8184. doi: 10.1126/scitranslmed.aan8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang K., Tomaras G.D., Jegaskanda S., Moody M.A., Liao H.X., Goodman K.N., Berman P.W., Rerks-Ngarm S., Pitisuttithum P., Nitayapan S. Monoclonal Antibodies, Derived from Humans Vaccinated with the RV144 HIV Vaccine Containing the HVEM Binding Domain of Herpes Simplex Virus (HSV) Glycoprotein D, Neutralize HSV Infection, Mediate Antibody-Dependent Cellular Cytotoxicity, and Protect Mice from Ocular Challenge with HSV-1. J. Virol. 2017;91:e00411–e00417. doi: 10.1128/JVI.00411-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Renegar K.B., Small P.A., Jr. Passive transfer of local immunity to influenza virus infection by IgA antibody. J. Immunol. 1991;146:1972–1978. [PubMed] [Google Scholar]
- 37.Hessell A.J., Rakasz E.G., Poignard P., Hangartner L., Landucci G., Forthal D.N., Koff W.C., Watkins D.I., Burton D.R. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathog. 2009;5:e1000433. doi: 10.1371/journal.ppat.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Traggiai E., Chicha L., Mazzucchelli L., Bronz L., Piffaretti J.C., Lanzavecchia A., Manz M.G. Development of a human adaptive immune system in cord blood cell-transplanted mice. Science. 2004;304:104–107. doi: 10.1126/science.1093933. [DOI] [PubMed] [Google Scholar]
- 39.Yajima M., Imadome K., Nakagawa A., Watanabe S., Terashima K., Nakamura H., Ito M., Shimizu N., Honda M., Yamamoto N., Fujiwara S. A new humanized mouse model of Epstein-Barr virus infection that reproduces persistent infection, lymphoproliferative disorder, and cell-mediated and humoral immune responses. J. Infect. Dis. 2008;198:673–682. doi: 10.1086/590502. [DOI] [PubMed] [Google Scholar]
- 40.Fujiwara S., Imadome K., Takei M. Modeling EBV infection and pathogenesis in new-generation humanized mice. Exp. Mol. Med. 2015;47:e135. doi: 10.1038/emm.2014.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fujiwara S., Matsuda G., Imadome K. Humanized mouse models of epstein-barr virus infection and associated diseases. Pathogens. 2013;2:153–176. doi: 10.3390/pathogens2010153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Münz C. Humanized mouse models for Epstein Barr virus infection. Curr. Opin. Virol. 2017;25:113–118. doi: 10.1016/j.coviro.2017.07.026. [DOI] [PubMed] [Google Scholar]
- 43.van Zyl D.G., Tsai M.-H., Shumilov A., Schneidt V., Poirey R., Schlehe B., Fluhr H., Mautner J., Delecluse H.-J. Immunogenic particles with a broad antigenic spectrum stimulate cytolytic T cells and offer increased protection against EBV infection ex vivo and in mice. PLoS Pathog. 2018;14:e1007464. doi: 10.1371/journal.ppat.1007464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strowig T., Gurer C., Ploss A., Liu Y.F., Arrey F., Sashihara J., Koo G., Rice C.M., Young J.W., Chadburn A. Priming of protective T cell responses against virus-induced tumors in mice with human immune system components. J. Exp. Med. 2009;206:1423–1434. doi: 10.1084/jem.20081720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ogembo J.G., Kannan L., Ghiran I., Nicholson-Weller A., Finberg R.W., Tsokos G.C., Fingeroth J.D. Human complement receptor type 1/CD35 is an Epstein-Barr Virus receptor. Cell Rep. 2013;3:371–385. doi: 10.1016/j.celrep.2013.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanner J., Weis J., Fearon D., Whang Y., Kieff E. Epstein-Barr virus gp350/220 binding to the B lymphocyte C3d receptor mediates adsorption, capping, and endocytosis. Cell. 1987;50:203–213. doi: 10.1016/0092-8674(87)90216-9. [DOI] [PubMed] [Google Scholar]
- 47.Hoffman G.J., Lazarowitz S.G., Hayward S.D. Monoclonal antibody against a 250,000-dalton glycoprotein of Epstein-Barr virus identifies a membrane antigen and a neutralizing antigen. Proc. Natl. Acad. Sci. USA. 1980;77:2979–2983. doi: 10.1073/pnas.77.5.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Molesworth S.J., Lake C.M., Borza C.M., Turk S.M., Hutt-Fletcher L.M. Epstein-Barr virus gH is essential for penetration of B cells but also plays a role in attachment of virus to epithelial cells. J. Virol. 2000;74:6324–6332. doi: 10.1128/jvi.74.14.6324-6332.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haque T., Johannessen I., Dombagoda D., Sengupta C., Burns D.M., Bird P., Hale G., Mieli-Vergani G., Crawford D.H. A mouse monoclonal antibody against Epstein-Barr virus envelope glycoprotein 350 prevents infection both in vitro and in vivo. J. Infect. Dis. 2006;194:584–587. doi: 10.1086/505912. [DOI] [PubMed] [Google Scholar]
- 50.Rivailler P., Jiang H., Cho Y.G., Quink C., Wang F. Complete nucleotide sequence of the rhesus lymphocryptovirus: genetic validation for an Epstein-Barr virus animal model. J. Virol. 2002;76:421–426. doi: 10.1128/JVI.76.1.421-426.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carville A., Mansfield K.G. Comparative pathobiology of macaque lymphocryptoviruses. Comp. Med. 2008;58:57–67. [PMC free article] [PubMed] [Google Scholar]
- 52.Moghaddam A., Rosenzweig M., Lee-Parritz D., Annis B., Johnson R.P., Wang F. An animal model for acute and persistent Epstein-Barr virus infection. Science. 1997;276:2030–2033. doi: 10.1126/science.276.5321.2030. [DOI] [PubMed] [Google Scholar]
- 53.Wu L., Hutt-Fletcher L.M. Compatibility of the gH homologues of Epstein-Barr virus and related lymphocryptoviruses. J. Gen. Virol. 2007;88:2129–2136. doi: 10.1099/vir.0.82949-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Herrman M., Mühe J., Quink C., Wang F. Epstein-Barr Virus gp350 Can Functionally Replace the Rhesus Lymphocryptovirus Major Membrane Glycoprotein and Does Not Restrict Infection of Rhesus Macaques. J. Virol. 2015;90:1222–1230. doi: 10.1128/JVI.02531-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Snijder J., Ortego M.S., Weidle C., Stuart A.B., Gray M.D., McElrath M.J., Pancera M., Veesler D., McGuire A.T. An Antibody Targeting the Fusion Machinery Neutralizes Dual-Tropic Infection and Defines a Site of Vulnerability on Epstein-Barr Virus. Immunity. 2018;48:799–811.e9. doi: 10.1016/j.immuni.2018.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu X., Yang Z.Y., Li Y., Hogerkorp C.M., Schief W.R., Seaman M.S., Zhou T., Schmidt S.D., Wu L., Xu L. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sathiyamoorthy K., Hu Y.X., Möhl B.S., Chen J., Longnecker R., Jardetzky T.S. Structural basis for Epstein-Barr virus host cell tropism mediated by gp42 and gHgL entry glycoproteins. Nat. Commun. 2016;7:13557. doi: 10.1038/ncomms13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sathiyamoorthy K., Jiang J., Möhl B.S., Chen J., Zhou Z.H., Longnecker R., Jardetzky T.S. Inhibition of EBV-mediated membrane fusion by anti-gHgL antibodies. Proc. Natl. Acad. Sci. USA. 2017;114:E8703–E8710. doi: 10.1073/pnas.1704661114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tiller T., Busse C.E., Wardemann H. Cloning and expression of murine Ig genes from single B cells. J. Immunol. Methods. 2009;350:183–193. doi: 10.1016/j.jim.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 60.Tangye S.G., Palendira U., Edwards E.S. Human immunity against EBV-lessons from the clinic. J. Exp. Med. 2017;214:269–283. doi: 10.1084/jem.20161846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Omerović J., Longnecker R. Functional homology of gHs and gLs from EBV-related gamma-herpesviruses for EBV-induced membrane fusion. Virology. 2007;365:157–165. doi: 10.1016/j.virol.2007.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rao P., Jiang H., Wang F. Cloning of the rhesus lymphocryptovirus viral capsid antigen and Epstein-Barr virus-encoded small RNA homologues and use in diagnosis of acute and persistent infections. J. Clin. Microbiol. 2000;38:3219–3225. doi: 10.1128/jcm.38.9.3219-3225.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sashihara J., Hoshino Y., Bowman J.J., Krogmann T., Burbelo P.D., Coffield V.M., Kamrud K., Cohen J.I. Soluble rhesus lymphocryptovirus gp350 protects against infection and reduces viral loads in animals that become infected with virus after challenge. PLoS Pathog. 2011;7:e1002308. doi: 10.1371/journal.ppat.1002308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sokal E.M., Hoppenbrouwers K., Vandermeulen C., Moutschen M., Léonard P., Moreels A., Haumont M., Bollen A., Smets F., Denis M. Recombinant gp350 vaccine for infectious mononucleosis: a phase 2, randomized, double-blind, placebo-controlled trial to evaluate the safety, immunogenicity, and efficacy of an Epstein-Barr virus vaccine in healthy young adults. J. Infect. Dis. 2007;196:1749–1753. doi: 10.1086/523813. [DOI] [PubMed] [Google Scholar]
- 65.Moutschen M., Léonard P., Sokal E.M., Smets F., Haumont M., Mazzu P., Bollen A., Denamur F., Peeters P., Dubin G., Denis M. Phase I/II studies to evaluate safety and immunogenicity of a recombinant gp350 Epstein-Barr virus vaccine in healthy adults. Vaccine. 2007;25:4697–4705. doi: 10.1016/j.vaccine.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 66.Cui X., Cao Z., Chen Q., Arjunaraja S., Snow A.L., Snapper C.M. Rabbits immunized with Epstein-Barr virus gH/gL or gB recombinant proteins elicit higher serum virus neutralizing activity than gp350. Vaccine. 2016;34:4050–4055. doi: 10.1016/j.vaccine.2016.06.021. [DOI] [PubMed] [Google Scholar]
- 67.Delecluse H.J., Hilsendegen T., Pich D., Zeidler R., Hammerschmidt W. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc. Natl. Acad. Sci. USA. 1998;95:8245–8250. doi: 10.1073/pnas.95.14.8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kalla M., Göbel C., Hammerschmidt W. The lytic phase of epstein-barr virus requires a viral genome with 5-methylcytosine residues in CpG sites. J. Virol. 2012;86:447–458. doi: 10.1128/JVI.06314-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pattabhi S., Lotti S.N., Berger M.P., Singh S., Lux C.T., Jacoby K., Lee C., Negre O., Scharenberg A.M., Rawlings D.J. In Vivo Outcome of Homology-Directed Repair at the HBB Gene in HSC Using Alternative Donor Template Delivery Methods. Mol. Ther. Nucleic Acids. 2019;17:277–288. doi: 10.1016/j.omtn.2019.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bandaranayake A.D., Correnti C., Ryu B.Y., Brault M., Strong R.K., Rawlings D.J. Daedalus: a robust, turnkey platform for rapid production of decigram quantities of active recombinant proteins in human cell lines using novel lentiviral vectors. Nucleic Acids Res. 2011;39:e143. doi: 10.1093/nar/gkr706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kimura H., Morita M., Yabuta Y., Kuzushima K., Kato K., Kojima S., Matsuyama T., Morishima T. Quantitative analysis of Epstein-Barr virus load by using a real-time PCR assay. J. Clin. Microbiol. 1999;37:132–136. doi: 10.1128/jcm.37.1.132-136.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McGuire A.T., Gray M.D., Dosenovic P., Gitlin A.D., Freund N.T., Petersen J., Correnti C., Johnsen W., Kegel R., Stuart A.B. Specifically modified Env immunogens activate B-cell precursors of broadly neutralizing HIV-1 antibodies in transgenic mice. Nat. Commun. 2016;7:10618. doi: 10.1038/ncomms10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Plate A.E., Smajlović J., Jardetzky T.S., Longnecker R. Functional analysis of glycoprotein L (gL) from rhesus lymphocryptovirus in Epstein-Barr virus-mediated cell fusion indicates a direct role of gL in gB-induced membrane fusion. J. Virol. 2009;83:7678–7689. doi: 10.1128/JVI.00457-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Okuma K., Nakamura M., Nakano S., Niho Y., Matsuura Y. Host range of human T-cell leukemia virus type I analyzed by a cell fusion-dependent reporter gene activation assay. Virology. 1999;254:235–244. doi: 10.1006/viro.1998.9530. [DOI] [PubMed] [Google Scholar]
- 75.Omerović J., Lev L., Longnecker R. The amino terminus of Epstein-Barr virus glycoprotein gH is important for fusion with epithelial and B cells. J. Virol. 2005;79:12408–12415. doi: 10.1128/JVI.79.19.12408-12415.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nikitin P.A., Yan C.M., Forte E., Bocedi A., Tourigny J.P., White R.E., Allday M.J., Patel A., Dave S.S., Kim W. An ATM/Chk2-mediated DNA damage-responsive signaling pathway suppresses Epstein-Barr virus transformation of primary human B cells. Cell Host Microbe. 2010;8:510–522. doi: 10.1016/j.chom.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mühe J., Wang F. Host Range Restriction of Epstein-Barr Virus and Related Lymphocryptoviruses. J. Virol. 2015;89:9133–9136. doi: 10.1128/JVI.01235-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all datasets generated or analyzed during this study