Abstract
HIV-1 integrase (IN) catalyzes integration of viral DNA into cell DNA through 30-processing of viral DNA and strand transfer reactions. To learn on binding of IN to DNAs and IN inhibition we applied spectroscopy (circular dichroism, fluorescence) in a simplified model consisting in a peptide analogue (K156) of α4 helix involved in recognition of viral and cell DNA; an oligonucleotide corresponding to the U5' LTR DNA end; and an inhibitor (TB11) of the diketo acid (DKA) family. Results extrapolated to IN show that: the enzyme binds viral DNA with high affinity and specificity, but cell DNA with low affinity and specificity; the affinity of TB11 for IN is high enough to impair the binding of IN to cell DNA, but not to viral DNA. This explains why TB11 is an inhibitor of strand transfer but not of 30-processing. These results can help in the search of new IN inhibitors.
Keywords: HIV-1, Integrase, DNA, Interactions, Inhibitors
Introduction
Integrase (IN) of human immunodeficiency virus type 1 (HIV-1) is an attractive target for anti-AIDS drugs. IN alone is sufficient to catalyze the insertion of virus DNA into host DNA [1, 2]. Insertion involves two steps: the 3'-processing reaction where two nucleotides (GT) are removed from the 30 ends of viral DNA LTRs (long terminal repeats); and the strand transfer reaction, where the newly created 3' ends perform a staggered nucleophilic attack (5 bp) on the target DNA helix (cellular DNA), resulting in an integrated copy of viral DNA [3].
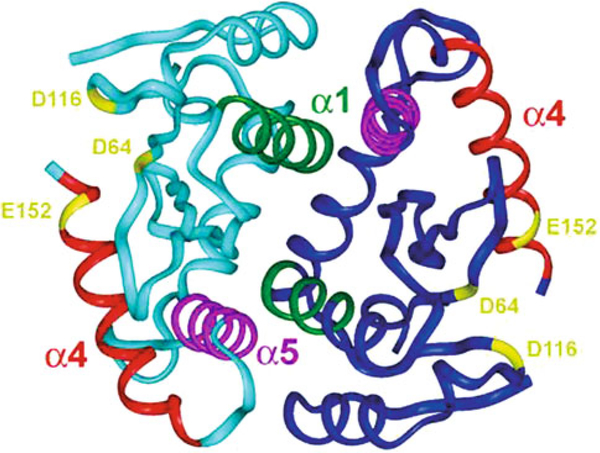
The HIV-1 enzyme contains 288 amino acid residues. It has an N-terminal domain (NTD: 1–50 residues) that includes a conserved HHCC motif that binds zinc and an HTH motif [4]. The C-terminal domain (CTD: 213–288 residues), although less conserved includes an SH3 fold [5]. The central catalytic domain (CCD: 51–212 residues) contains five β strands surrounded by six α helices (including the important amphipathic α4 helix) as well as the highly catalytic D, DX35E motif embedded in a protein RNaseH fold [6–12]. The three domains taken separately form dimers, which are also the case of the two-domain fragments CCD plus CTD [13] and NTD plus CCD [14], but the relative conformation of the entire enzyme and of its active multimeric form is not known. A dimer structure of CCD obtained by crystallography studies is shown in Fig. 1.
Fig. 1.
Dimer structure of IN-CCD obtained by X-ray crystallography [9]. The α4 helix is exposed at the protein surface and interacts with the α5 helix within a HTH motif. The α5 helix of a monomer is further engaged in interactions with the α1' helix of the second monomer
The 30 processing and DNA joining reactions can be achieved in in vitro assays generally in putting together a recombinant IN and a duplex oligonucleotide of 17–21 base-pairs bearing the 30-processing site CA ↓ GT3'. The oligonucleotide reproduces either the U3 or the U5 end of LTRs but behaves in in vitro assays as both DNA substrate (donor) and DNA target (acceptor). Mutations performed within such an oligonucleotide have demonstrated that the six outermost residues are crucial for 3'-processing. This is also true for the four adenine tract (positions 12–15 from the 3' end). However, no experimental structure of a DNA-IN complex is available to date, while there is one published crystal structure of the CCD bound to a lead diketo acid (DKA) inhibitor (5CITEP) [15].
We have previously shown that: (i) the LTR end exhibits a distorted double helix structure, especially at the processing site CA ↓; GT3', that can be selectively recognized by IN [16]; (ii) the α4 helix at the CCD surface is involved in the specific recognition of the 3'-processing site [17]. We show now that the α4 helix recognizes both the viral and cell DNA with, however, completely different affinities, and is a binding site for IN inhibitors of the DKA family.
Experimental Procedure
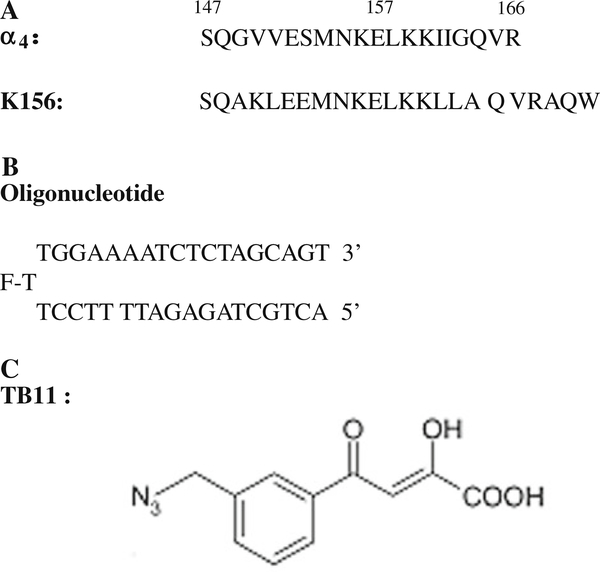
The peptides, oligonucleotides and IN inhibitors used in this work are shown in Fig. 2.
Fig. 2.
The compounds under studies: a Sequences of the α4 peptide and of its structural analog K156; b The oligonucleotide is designed to adopt a hairpin structure with a three thymine (T) loop (the central thymine bears the fluorophore (fluorescein) and a 17-base pair stem). The latter reproduces the end of the U5ĽTR of HIV-1 DNA but plays the role of both virus DNA and cell DNA in in vitro experiments; c The IN inhibitor TB11 includes the critical DKA group and also an aryl ring with an azide substitutent important for biological activity [3, 19]
Peptides
The peptide K156 (Fig. 2a) was synthesized as previously reported [17]. K156 corresponds to a helix stabilized version of the α4 peptide (residues 147 to residues 169 in CCD) [17]. Briefly, several residues have been replaced by more helicogenic ones in parts of the helix deemed not important for DNA recognition. The backbone conformation of the K156 peptide is more native-like compared with the conformation of the α4 helix peptide and is more functional, less aggregation prone and more adapted to the study of specific interactions which are highly conformation dependent.
The Trp (W) aromatic residue was purposely added at the C-terminus in order to enable fluorescence experiments and peptide concentration estimation from absorbance in UV spectra, using molar absorption coefficients of 5,600 M−1 cm−1 at 280 nm for the tryptophane-containing peptide.
Oligonucleotides
The oligonucleotide (LTR34) was purchased from Eurogentec (Belgium) (Fig. 2b). The choice of monomolecular hairpin-forming oligonucleotides rather than bimolecular duplex-forming oligonucleotides was motivated by the need for double helix stability under the low concentrations inherent to the fluorescence and CD experiments (10−9−10−5 M). The central thymine of the three thymine loop bears a fluorescein reporter allowing fluorescence measurements. One assumes that a so located fluorescein does not interfere with the binding of IN to LTR ends.
IN inhibitors
The synthesis of TB11 (C11H9N3O4) (Fig. 1c) has been already reported [18, 19]. TB11 contains the important diketo motif showing affinity for divalent cations (Mg2+, Mn2+…)[20]. The carboxylate group next to the diketo is also involved in the divalent cation chelation [21]. TB11 is an inhibitor of strand transfer, with little effects on 3'-processing.
Fluorescence Measurements
The intrinsic fluorescence quantum yield and fluorescence anisotropy studies were carried out with a Jobin-Yvon Fluoromax II instrument (HORIBA Jobin-Yvon, France) equipped with an Ozone-free 150 W xenon lamp. Samples (800 μl) were placed at 5 C in thermally jacketed 1cm 9 0.5 cm quartz cells. At least ten measurements for each titration point were recorded with an integration time of 1 s. Fluorophores were either Trp or fluorescein purposely fixed to the peptide or the oligonucleotide, respectively. In some experiments the aromatic ring of TB11 was also used as a fluorophore.
Fluorescein-labeled oligonucleotides were diluted to the desired concentration (10 nM) in 800 μl of assay buffer (Na/Na2 phosphate, pH 6, I = 0.1). Peptides were stepwise diluted. The excitation was performed at 488 nm and emission was recorded at 516 nm in the case of LTR34. Fluorescence of K156-W was measured at the 400 nM concentration in 800 ll of reaction buffer. Excitation at 290 nm provided an emission between 300 and 480 nm, using 2 and 5 nm excitation and emission slit widths, respectively. Maximal emission was measured at 355 nm.
CD Spectroscopy
CD spectra were recorded on a CD6 dichrograph (HORIBA Jobin Yvon, France). Measurements were calibrated with (+) − 10-camphorsulfonic acid. Oligonucleotide and peptide concentrations varied from 6 to 12 μM in the phosphate buffer pH 6, I = 0.1. Samples were placed in thermally jacketed cells with a 1 mm path length. To allow the solutions to reach their equilibrium state, these were incubated 10 min at the chosen temperature. Spectra, recorded in 1 nm steps, were averaged over ten scans and corrected for the base line. They were presented as differential molar absorptivity per residue, Δε (M−1 cm−1), as a function of wavelength, between 260 and 185 nm for peptides. The α-helix content of peptides was obtained using the relation: Pα =-[Δε222 × 10] (Pα: percentage of α-helix; Δε222: circular dichroism per residue at 222 nm) [22].
Results
Examination of the CCD surface in a crystal dimer structure shows that the α4 helix exposed at the enzyme surface (Fig. 1) is an amphipathic helix with its hydrophobic face and hydrophilic face oriented inside and outside the protein, respectively. Moreover, the α4 helix is constitutive of a helix turn helix (HTH) motif, where it plays the role of the DNA recognizing helix [23]. The second helix of the HTH, the α5 helix, is further involved in the dimer interface and the stabilization of the α4 helix.
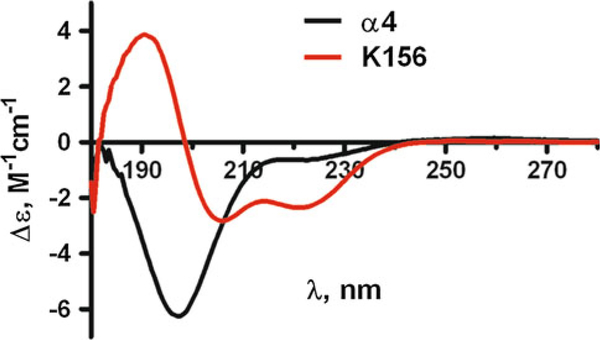
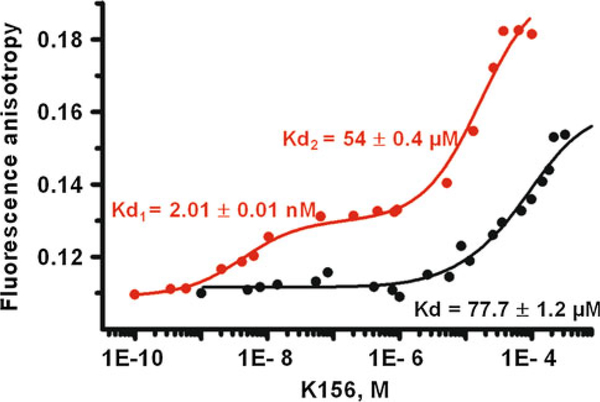
We used a simplified approach based on the use of an analogue (peptide K156) of the α4 helix peptide segment (Fig. 2a) in order to decipher the mechanisms of interaction of IN with viral DNA (Fig. 2b) and to analyze the binding properties of the drug TB11 (Fig. 2c) to the enzyme. In contrast to the α4 peptide that exhibits an unordered structure in solution the structural analogue, that keeps all the residues recognized important for binding to DNA, has an optimized helical content revealed by circular dichroism (Fig. 3). Actually, a high degree of helicity similar to the one found in the protein context is necessary for a minimal loss of free binding energy during the mutual adjustment of molecules and specific complex formation. Fluorescence anisotropy titration experiments indicate that a specific binding (dissociation constant, Kd1 = 2 nM) to viral DNA end occurs only with peptide K156 (Fig. 4). With α4 helix peptide we observe only one binding site characterized by a low affinity (Kd = 77 μM). This site, with Kd in the micromolar range, appears as a second site in K156 (Kd2 = 54 μM) and better corresponds to the binding of IN to cellular DNA, this occurring during the strand transfer reaction.
Fig. 3.
Circular dichroism spectra of the α4 and K156 peptides in buffer (see "Experimental Procedure") at 10 C in the far UV region (180–250 nm). The α4 peptide spectrum with its deep negative band at ≈195 nm is characteristic of random coil structures. The K156 spectrum is mostly of helix type with its two negative bands at ≈222 nm and ≈208 nm and its positive band at ≈190 nm
Fig. 4.
Fluorescence anisotropy titration of the oligonucleotide labeled with fluorescein by the α4 peptide (thick solid line) and the K156 peptide (thin solid line). Kds were obtained through curve treatment
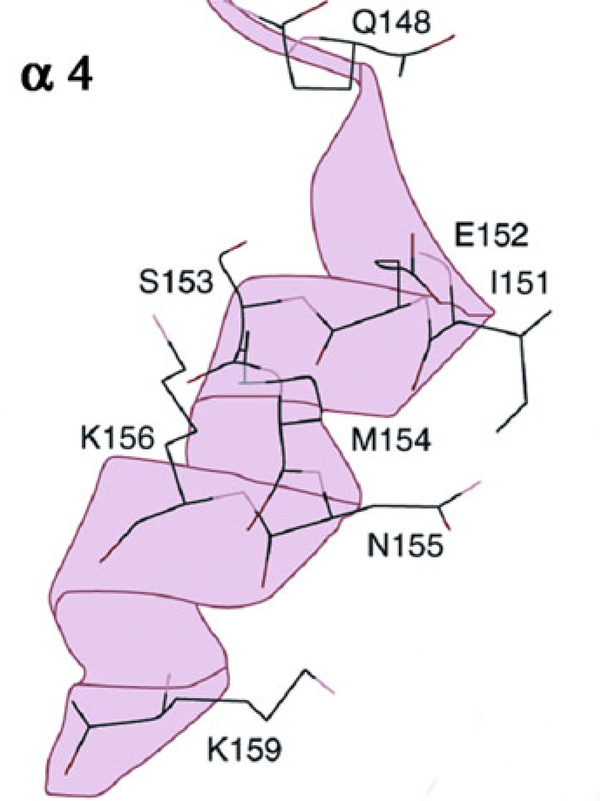
Drugs as the DKAs or related to the DKAs have emerged as specifics inhibitors of IN [3, 15, 24]. One of these, raltegravir, has been licensed for the AIDS treatment. Mutations associated with resistance to raltegravir and other inhibitors have been analyzed through in vivo and in vitro studies [25–27]. Most of these mutations have been identified on the a4 helix (Fig. 5) showing that the α4 helix is an important drug target, which is in agreement with the results provided by the co-crystal structure of 5CITEP–CCD of [15], where the inhibitor is bound to five aminoacid side chains of the α4 helix.
Fig. 5.
A ribbon view of the α4 helix showing the mutations entailing resistance to DKA drugs (after 26, 27)
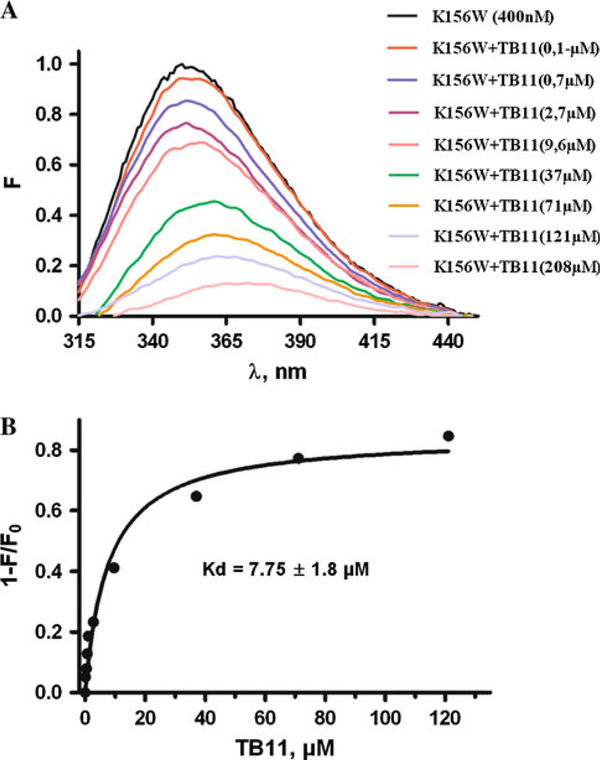
Given the increasing importance of specific IN inhibitors of the DKA family for AIDS therapy we carried out the analysis of TB11 binding to K156. The TB11 drug (Fig. 2C) is mostly an IN-ST inhibitor but is also a weak 30P inhibitor [20]. The fluorescence titration curves reported in Fig. 6 show that TB11 efficiently binds to K156. This binding process is regarded as an equilibrium condition that results from a balance from association and dissociations events. If the binding site of the DKA substrate and of the inhibitor is the same competition can occur. Then, binding affinities of partners can be determinant on the biological response. A quantitative determination provides a ligand-protein dissociation constant (Kdl) of 7.5 μM, that is an intermediate value between the dissociation constant values for specific and non-specific binding of IN to viral DNA and cellular DNA, respectively (Kd1: 2nM < Kdl: 7.5 μM < Kd2: 54 μM or Kd: 77 μM).
Fig. 6.
Quenching fluorescence analysis of the binding of TB11 to K156 using the peptide tryptophane (W) as the fluorophore. a Spectra of unbound K156(W) and bound K156 as a function of added TB11. b Derived binding curve showing calculated Kd
Discussion
Understanding of protein-DNA interaction is crucial for prediction of DNA-binding specificity of IN and design of novel IN inhibitors acting at the DNA-protein interface. Here, thanks to the use of a simple model approach we showed that the amphipathic α4 helix exposed at the CCD surface of IN and binding to a HTH motif is involved in the recognition of both viral DNA and cellular DNA. While the first binding occurs with high affinity (Kd1: 2 nM) and can be qualified of specific, the second occurs with low affinity (Kd2: 54 μM, Kd: 77 μM,) and is non-specific.
The quantitative foundations of these binding processes are mathematical models, so that biological activity can be expressed as the affinity of binding partners for each other and as a thermodynamic equilibrium quantity. How IN inhibitors can interfere with substrate bindings at the protein binding site was still an unanswered question. In fact IN inhibitors are generally divided into: (1) dual inhibitors of 3'-processing and strand transfer (3'P inhibitors); and (2) selective strand transfer inhibitors (IN-ST inhibitors). It is assumed that the first group (3'P inhibitors) docks on IN at the viral DNA-binding site, while the second group occupies the position of cellular DNA [3]. For instance the promising raltegravir drug belongs to the family of IN-ST inhibitors, while 5CITEP, though displaying some of IN-ST inhibitor features, behaves more as a 3'P inhibitor.
Here we propose that the type of inhibition demonstrated 0by DKAs—either 3P'-inhibition or ST-inhibition—is greatly dependent on the binding energy of the inhibitor vs that of the ligand. The affinity of TB11 for the K156 peptide is weaker compared with the specific binding affinity of K156 for viral DNA but significantly stronger than the non-specific binding affinity of K156 for cellular DNA. In fact, these results are in agreement with pharmacological data showing that TB11 is mainly a ST-inhibitor, with only very weak effects on 3'-processing [3]. Finally, our findings will hopefully contribute to the understanding of the mechanism of action of anti-AIDS drugs and the design of more potent members in this family.
Acknowledgments
This work was supported by a grant from SIDACTION (to S.F.) and by the French-Lebanese program CEDRE (05 SF21/L14 to S.F. and R.M.).
Footnotes
Special issue article in honor of Professor Armen Galoyan.
Contributor Information
Z. Hobaika, Laboratoire de Biotechnologies et Pharmacologie génétique Appliquée (LBPA), UMR 8113 du CNRS, Ecole Normale Supérieure de Cachan, 61 Avenue du Président Wilson, 94235 Cachan Cedex, France
L. Zargarian, Laboratoire de Biotechnologies et Pharmacologie génétique Appliquée (LBPA), UMR 8113 du CNRS, Ecole Normale Supérieure de Cachan, 61 Avenue du Président Wilson, 94235 Cachan Cedex, France
R. G. Maroun, Département des Sciences de la Vie et de la Terre, Faculté des Sciences, Université Saint-Joseph, CST-Mar Roukos, Beirut, Lebanon
O. Mauffret, Laboratoire de Biotechnologies et Pharmacologie génétique Appliquée (LBPA), UMR 8113 du CNRS, Ecole Normale Supérieure de Cachan, 61 Avenue du Président Wilson, 94235 Cachan Cedex, France
T. R. Burke, Jr., Laboratory of Medicinal Chemistry, Center of Cancer Research, National Cancer Institute, NIH, Frederick, MD 21702, USA
S. Fermandjian, Laboratoire de Biotechnologies et Pharmacologie génétique Appliquée (LBPA), UMR 8113 du CNRS, Ecole Normale Supérieure de Cachan, 61 Avenue du Président Wilson, 94235 Cachan Cedex, France
References
- 1.Craigie R, Mizuuchi K, Bushman FD, Engelman A (1991) A rapid in vitro assay for HIV DNA integration. Nucleic Acids Res 19:2729–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asante-Appiah E, Skalka AM (1997) Molecular mechanisms in retrovirus DNA integration. Antiviral Res 36:139–156 [DOI] [PubMed] [Google Scholar]
- 3.Pommier Y, Johnson AA, Marchand C (2005) Integrase inhibitors to treat HIV/AIDS. Nat Rev Drug Discov 4:236–248 [DOI] [PubMed] [Google Scholar]
- 4.Cai M, Zheng R, Caffrey M, Craigie R, Clore GM, Gronenborn AM (1997) Solution structure of the N-terminal zinc binding domain of HIV-1 integrase. Nat Struct Biol 4:567–577 [DOI] [PubMed] [Google Scholar]
- 5.Musacchio A, Noble M, Pauptit R, Wierenga R, Saraste M (1992) Crystal structure of a Src-homology 3 (SH3) domain. Nature 359:851–855 [DOI] [PubMed] [Google Scholar]
- 6.Engelman A, Craigie R (1992) Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J Virol 66:6361–6369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kulkosky J, Jones KS, Katz RA, Mack JP, Skalka AM (1992) Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol 12:2331–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leavitt AD, Shiue L, Varmus HE (1993) Site-directed mutagenesis of HIV-1 integrase demonstrates differential effects on integrase functions in vitro. J Biol Chem 268:2113–2119 [PubMed] [Google Scholar]
- 9.Dyda F, Hickman AB, Jenkins TM, Engelman A, Craigie R, Davies DR (1994) Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science 266:1981–1986 [DOI] [PubMed] [Google Scholar]
- 10.Bujacz G, Jaskolski M, Alexandratos J, Wlodawer A, Merkel G, Katz RA, Skalka AM (1996) The catalytic domain of avian sarcoma virus integrase: conformation of the active-site residues in the presence of divalent cations. Structure 4:89–96 [DOI] [PubMed] [Google Scholar]
- 11.Davies JF, Hostomska Z, Hostomsky Z, Jordan SR, Matthews DA (1991) Crystal structure of the ribonuclease H domain of HIV-1 reverse transcriptase. Science 252:88–95 [DOI] [PubMed] [Google Scholar]
- 12.Katayanagi K, Okumura M, Morikawa K (1993) Crystal structure of Escherichia coli RNase HI in complex with Mg2+ at 2.8 A resolution: proof for a single Mg(2+)-binding site. Proteins 17:337–346 [DOI] [PubMed] [Google Scholar]
- 13.Chen Z, Yan Y, Munshi S, Li Y, Zugay-Murphy J, Xu B, Witmer M, Felock P, Wolfe A, Sardana V, Emini EA, Hazuda D, Kuo LC (2000) X-ray structure of simian immunodeficiency virus integrase containing the core and C-terminal domain (residues 50–293)—an initial glance of the viral DNA binding platform. J Mol Biol 296:521–533 [DOI] [PubMed] [Google Scholar]
- 14.Wang JY, Ling H, Yang W, Craigie R (2001) Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein. EMBO J 20:7333–7343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldgur Y, Craigie R, Cohen GH, Fujiwara T, Yoshinaga T, Fujishita T, Sugimoto H, Endo T, Murai H, Davies DR (1999) Structure of the HIV-1 integrase catalytic domain complexed with an inhibitor: a platform for antiviral drug design. Proc Natl Acad Sci U S A 96:13040–13043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Renisio JG, Cosquer S, Cherrak I, El Antri S, Mauffret O, Fermandjian S (2005) Pre-organized structure of viral DNA at the binding-processing site of HIV-1 integrase. Nucleic Acids Res 33:1970–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zargarian L, Benleumi MS, Renisio JG, Merad H, Maroun RG, Wieber F, Mauffret O, Porumb H, Troalen F, Fermandjian S (2003) Strategy to discriminate between high and low affinity bindings of human immunodeficiency virus, type 1 integrase to viral DNA. J Biol Chem 278:19966–19973 [DOI] [PubMed] [Google Scholar]
- 18.Pais GC, Zhang X, Marchand C, Neamati N, Cowansage K, Svarovskaia ES, Pathak VK, Tang Y, Nicklaus M, Pommier Y, Burke TR Jr (2002) Structure activity of 3-aryl-1, 3-diketo-containing compounds as HIV-1 integrase inhibitors. J Med Chem 45:3184–3194 [DOI] [PubMed] [Google Scholar]
- 19.Zhang X, Pais GC, Svarovskaia ES, Marchand C, Johnson AA, Karki RG, Nicklaus MC, Pathak VK, Pommier Y, Burke TR (2003) Azido-containing aryl beta-diketo acid HIV-1 integrase inhibitors. Bioorg Med Chem Lett 13:1215–1219 [DOI] [PubMed] [Google Scholar]
- 20.Marchand C, Johnson AA, Karki RG, Pais GC, Zhang X, Cowansage K, Patel TA, Nicklaus MC, Burke TR Jr, Pommier Y (2003) Metal-dependent inhibition of HIV-1 integrase by beta-diketo acids and resistance of the soluble double-mutant (F185 K/C280S). Mol Pharmacol 64:600–609 [DOI] [PubMed] [Google Scholar]
- 21.Grobler JA, Stillmock K, Hu B, Witmer M, Felock P, Espeseth AS, Wolfe A, Egbertson M, Bourgeois M, Melamed J, Wai JS, Young S, Vacca J, Hazuda DJ (2002) Diketo acid inhibitor mechanism and HIV-1 integrase: implications for metal binding in the active site of phosphotransferase enzymes. Proc Natl Acad Sci U S A 99:6661–6666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhong L, Johnson WC Jr (1992) Environment affects amino acid preference for secondary structure. Proc Natl Acad Sci U S A 89:4462–4465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merad H, Porumb H, Zargarian L, Rene B, Hobaika Z, Maroun RG, Mauffret O, Fermandjian S (2009) An unusual helix turn helix motif in the catalytic core of HIV-1 integrase binds viral DNA and LEDGF. PLoS One 4:e4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hazuda DJ, Felock P, Witmer M, Wolfe A, Stillmock K, Grobler JA, Espeseth A, Gabryelski L, Schleif W, Blau C, Miller MD (2000) Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 287:646–650 [DOI] [PubMed] [Google Scholar]
- 25.Lataillade M, Chiarella J, Kozal MJ (2007) Natural polymorphism of the HIV-1 integrase gene and mutations associated with integrase inhibitor resistance. Antivir Ther 12:563–570 [PubMed] [Google Scholar]
- 26.Rhee SY, Liu TF, Kiuchi M, Zioni R, Gifford RJ, Holmes SP, Shafer RW (2008) Natural variation of HIV-1 group M integrase: implications for a new class of antiretroviral inhibitors. Retrovirology 5:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cotelle P, Queffélec C (2007) Les différents classes des molécules inhibant l'integrase du VIH1. Virologie 11:163–173 [Google Scholar]