Coronaviruses (CoV) are a diverse group of viruses capable of infecting humans, and a wide range of animals. CoV affect multiple systems, and can cause respiratory, gastrointestinal, hepatic, and neurological illnesses, ranging from mild sickness to death. CoV are classified into multiple genera, including Alpha, Beta, Gamma and Delta coronaviruses.1, 2, 3, 4, 5, 6, 7, 8, 9, 10 Of note, CoV seem to be able to adapt to new hosts and changing environments; this may be related to CoV ability to mutate and recombine,1 , 3 , 5 , 7 perhaps contributing to novel viruses with varying human pathogenicity.
Coronaviruses (order Nidovirales, family Coronaviridae, genus Coronavirus) are large, enveloped, single stranded, positive-sense RNA viruses, capable of infecting a variety of animals, including bats, mice, birds, dogs, pigs, cattle, and humans.
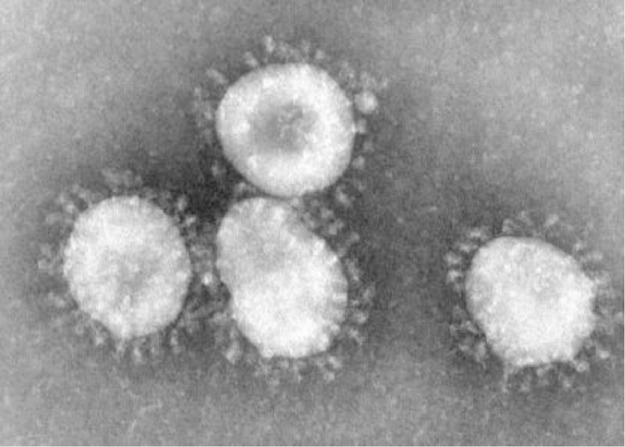
Identified many decades ago, Coronavirus (Fig. 1 )9 – from the Latin corona (translation “crown” or “halo”) represents the appearance of CoV virions as they are viewed through an electron microscope.2 , 7, 8, 9, 10 The virus appearance is created by viral spikes (S), peplomers that populate the surface and determine host tropism (Fig. 1).8, 9, 10
Fig. 1.
Coronavirus. Centers for Disease Control and Prevention (CDC)/Dr. Fred Murphy.9
Typically CoV are considered to be highly species-specific. In immunocompetent hosts, infection elicits the immune response of neutralizing antibodies and cell-mediated immune responses that attempt to kill infected cells.1 , 10 , 11
Coronaviruses, members of the Coronaviridae family were identified and grouped based upon their serological cross-reactivity, and genomic sequence homology. Host ranges are diverse, and can include canines, felines, swine, mice, camels, bats, birds, and humans.1 , 2 , 7 , 10, 11, 12, 13, 14
Across the four genera of coronaviruses are Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus2 , 5 , 10 , 11 there is a high frequency of recombination and rate of mutation which are believed to allow CoVs to adapt to new hosts and environments.3 , 11, 12, 13, 14, 15, 16 SARS Cov2 , 5 , 6 , 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34 is a good illustration of this; studies revealed it originated from animals – bats as the natural reservoir,5, 6, 7, 8 , 10 , 11 , 25 , 26 , 27 and palm civet as the intermediate host.11, 12, 13 This underscores the infection risk in human animal interactions – occupational, or adventure or travel, as well as environmental incursion and changing habitats expected with climate change, which can pose significant risks to human, as well as animal health.
As an animal pathogen, coronaviruses can lead to highly virulent respiratory, enteric, and neurological diseases, in addition to hepatitis, resulting in epizootics of respiratory diseases and/or gastroenteritis. As a human virus the range of disease is broad, from cold like to severe multisystem involvement (These CoV infections are associated with short incubation periods (2-7 days), such as those found in SARS.2 , 5 , 6 , 17 , 18 , 24 , 25 Several coronaviruses are capable of causing fatal systemic diseases in animals, including feline infectious peritonitis virus (FIPV), swine hemagglutinating encephalomyelitis virus (HEV), some strains of avian infectious bronchitis virus (IBV) and mouse hepatitis virus (MHV). These particular CoV can replicate in liver, lung, kidney, gut, spleen, brain, spinal cord, retina, as well as other tissues.2 , 7 , 13
References
- 1.Siddell S, Wege H, ter Meulen V. The biology of coronaviruses. J Gen Virol. 1983;64(Pt 4):761–776. doi: 10.1099/0022-1317-64-4-761. [DOI] [PubMed] [Google Scholar]
- 2.Perlman S, Netland J. Coronaviruses post SARS: update on replication and pathogenesis. Nature Review Micro. 2009 June:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lau SKP, Lau CCY, Chan KH, Li CPY, et al. Delayed induction of proinflammatory cytokines and suppression of innate antiviral response by the novel Middle Easst respiratory syndrome coronavirus: implications for pathogenesis and treatment. J Gen Virology. 2013;94:2679–2690. doi: 10.1099/vir.0.055533-0. [DOI] [PubMed] [Google Scholar]
- 4.deGroot RJ, Baker SC, Baric R, Enjuanes L, et al. Coronaviridae In Virus Taxonomy; Ninth Report of the International Committee on Taxonomy of Viruses pp. 806-828. Edited byAMQ King, JH Adams, EB Carstens and EJ Lefkowitz. San Diego, Ca. Elsevier Publishing.
- 5.Woo PC, Lau SK, Lam CS, Lau CC, et al. Disocvery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruess as the gene source of alphacoronavirus and betacoronavirus and avian cornnaviruses as the gene source of gammacoronavirus and deltacornavirus. J Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hon CC, Lam TY, Shi ZL, Drummond AJ, et al. Evidence of the recombinant origin of a bat severe acute respiratory syndrome OSARS) like coronavirus and its implications on the direct ancestor of SARS coronavirus. J Virol. 2008;82:1819–1826. doi: 10.1128/JVI.01926-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lau SKP, Lee P, Tsang AKL, Yip CCY, et al. Molecluar epidemiology of human coronavirus OC43 reveals evolution of different genotypes over time and recent emergence of a novle genotype due to natural recombination. J Virol. 2011;85:11325–11337. doi: 10.1128/JVI.05512-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guan Y, Zheng BJ, He Y, Liu XL, et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 9.CDC/National Institutes of Health Public Domain Coronavirus Image. https://www.ncbi.nlm.nih.gov/genomes/SARS/images/sars%20em%20cdc%20rev1.PNGLast accessed 03/06/17.
- 10.Centers for Disease Control and Prevention (CDC) Coronaviruses. https://www.cdc.gov/coronavirus/about/index.htmlLast accessed 03/06/17.
- 11.Lua SK, Woo PC, Li KS, Huang Y, et al. Severe acute respiratory syndrome coronavirus like virus in Chinese horseshoe bats. Proc Natl Acad Sci USA. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Makela MJ, Puhakka T, Ruuskanen O, et al. Viruses and bacteria in the etiology of the common cold. J Clin Microbiol. 1998;36:539–542. doi: 10.1128/jcm.36.2.539-542.1998. http://SARSReference.com/lit.php?id=9466772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McIntosh K. In: Mandell, Douglas, and Bennett's Principles and Practice of Infectious Diseases. 5th ed. Mandell GL, Bennett JE, Dolin R, editors. Churchill Livingstone, Inc.; Philadelphia: 2000. Coronaviruses. [Google Scholar]
- 14.El-Sahly HM, Atmar RL, Glezen WP, Greenberg SB. Spectrum of clinical illness in hospitalized patients with "common cold" virus infections. Clin Infect Dis. 2000;31:96–100. doi: 10.1086/313937. http://SARSReference.com/link.php?id=10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Folz RJ, Elkordy MA. Coronavirus pneumonia following autologous bone marrow transplantation for breast cancer. Chest. 1999;115:901–905. doi: 10.1378/chest.115.3.901. http://www.chestjournal.org/cgi/content/full/115/3/901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sizun J, Yu MW, Talbot PJ. Survival of human coronaviruses 229E and OC43 in suspension after drying on surfaces: a possible source of hospital-acquired infections. J Hosp Infect. 2000;46:55–60. doi: 10.1053/jhin.2000.0795. http://SARSReference.com/lit.php?id=11023724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holmes KV, Enjuanes L. The SARS coronavirus: a postgenomic era. Science. 2003;300:1377–1378. doi: 10.1126/science.1086418. [DOI] [PubMed] [Google Scholar]
- 18.Holmes KV. SARS coronavirus: a new challenge for prevention and therapy. J Clin Invest. 2003;111:1605–1609. doi: 10.1172/JCI18819. http://www.jci.org/cgi/content/full/111/11/1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van den Hoogen BG, de Jong JC, Groen J, Kuiken T, de Groot R, Fouchier RA, Osterhaus AD. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med. 2001;7:719–724. doi: 10.1038/89098. http://SARSReference.com/lit.php?id=11385510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drosten C, Gunther S, Preiser W, et al. Identification of a Novel Coronavirus in Patients with Severe Acute Respiratory Syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. http://SARSReference.com/lit.php?id=12690091 Published online Apr 10, 2003. [DOI] [PubMed] [Google Scholar]
- 21.Drosten C, Preiser W, Günther S, Schmitz H, Doerr HW. Severe acute respiratory syndrome: identification of the etiological agent. Trends Mol Med. 2003;9:325–327. doi: 10.1016/S1471-4914(03)00133-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peiris JS, Lai ST, Poon LL, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. http://image.thelancet.com/extras/03art3477web.pdf Published online Apr 8, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marra MA, Jones SJM, Astell CR, et al. The Genome Sequence of the SARS-Associated Coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. http://www.sciencemag.org/cgi/content/abstract/1085953v1 Published online May 1, 2003. [DOI] [PubMed] [Google Scholar]
- 24.Rota PA, Oberste MS, Monroe SS, et al. Characterization of a Novel Coronavirus Associated with Severe Acute Respiratory Syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. http://www.sciencemag.org/cgi/content/abstract/1085952v1 Published online May 1, 2003. [DOI] [PubMed] [Google Scholar]
- 25.Ludwig B, Kraus FB, Allwinn R, Doerr HW, Preiser W. Viral Zoonoses - A Threat under Control? Intervirology. 2003;46:71–78. doi: 10.1159/000069749. http://SARSReference.com/lit.php?id=12684545 [DOI] [PubMed] [Google Scholar]
- 26.Ruan YJ, Wei CL, Ee AL, et al. Comparative full-length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection. Lancet. 2003;361:1779–1785. doi: 10.1016/S0140-6736(03)13414-9. http://image.thelancet.com/extras/03art4454web.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown EG, Tetro JA. Comparative analysis of the SARS coronavirus genome: a good start to a long journey. Lancet. 2003;361:1756–1757. doi: 10.1016/S0140-6736(03)13444-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsui SK, Chim SS, Lo YM, Chinese University of Hong Kong Molecular SARS Research Group Coronavirus genomic-sequence variations and the epidemiology of the severe acute respiratory syndrome. N. Engl. J. Med. 2003;349:187–188. doi: 10.1056/NEJM200307103490216. [DOI] [PubMed] [Google Scholar]
- 29.Ksiazek TG, Erdman D, Goldsmith CS, et al. A Novel Coronavirus Associated with Severe Acute Respiratory Syndrome. New Eng J Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. http://SARSReference.com/lit.php?id=12690092 Published online Apr 10, 2003. [DOI] [PubMed] [Google Scholar]
- 30.Peiris JS, Chu CM, Cheng VC, et al. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. http://image.thelancet.com/extras/03art4432web.pdf Published online May 9, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.http://sarsreference.com/sarsref/virol.htmLast accessed 01/14/17.
- 32.Anderson LJ, Tong S. Update on SARS research and other possibly zoonotic coronaviruses. Int J Antimicrobial Agents. 2010;36S:S21–S25. doi: 10.1016/j.ijantimicag.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peiris JS, Yuen KY, Osterhaus AD, Stohr K. The severe acute respiratory syndrome. N Engl J Med. 2003;349:2431–2441. doi: 10.1056/NEJMra032498. [DOI] [PubMed] [Google Scholar]
- 34.Enjuanes L, Dediego ML, Alvarez E, Deming D, Sheahan T, Baric R. Vaccines to prevent severe acute respiratory syndrome coronavirus-induced disease. Virus Res. 2008;133:45–62. doi: 10.1016/j.virusres.2007.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]