Abstract
SARS-CoV-2 is the causative agent of the 2019–2020 pandemic. The SARS-CoV-2 genome is replicated and transcribed by the RNA-dependent RNA polymerase holoenzyme (subunits nsp7/nsp82/nsp12) along with a cast of accessory factors. One of these factors is the nsp13 helicase. Both the holo-RdRp and nsp13 are essential for viral replication and are targets for treating the disease COVID-19. Here we present cryoelectron microscopic structures of the SARS-CoV-2 holo-RdRp with an RNA template product in complex with two molecules of the nsp13 helicase. The Nidovirales order-specific N-terminal domains of each nsp13 interact with the N-terminal extension of each copy of nsp8. One nsp13 also contacts the nsp12 thumb. The structure places the nucleic acid-binding ATPase domains of the helicase directly in front of the replicating-transcribing holo-RdRp, constraining models for nsp13 function. We also observe ADP-Mg2+ bound in the nsp12 N-terminal nidovirus RdRp-associated nucleotidyltransferase domain, detailing a new pocket for anti-viral therapy development.
Graphical Abstract
Chen et al. present cryo-EM structures of the SARS-CoV-2 RNA-dependent RNA polymerase (RdRp) holoenzyme (nsp7/nsp8/nsp12) containing an RNA template product in complex with the viral helicase (nsp13). The work provides insight into assembly and function of the multi-subunit protein machine and how it might be targeted therapeutically to treat COVID-19.
Introduction
Coronaviruses (CoVs) are positive-strand RNA (+RNA) viruses belonging to the order Nidovirales (de Groot et al., 2012). These viruses are responsible for several zoonotic infections (Snijder et al., 2016). Deadly occurrences include the 2003 severe acute respiratory syndrome (SARS) pandemic caused by SARS-CoV-1 and Middle Eastern respiratory syndrome (MERS) outbreaks caused by MERS-CoV (Hilgenfeld and Peiris, 2013). SARS-CoV-2, a β-CoV, has been identified as the cause of the current globally devastating CoV disease 2019 (COVID-19) pandemic (Wu et al., 2020; Zhou et al., 2020).
The Nidovirales order of viruses contains the largest known RNA genomes (Nga et al., 2011; Saberi et al., 2018). The nidovirus RNA-dependent RNA polymerase (RdRp; encoded by non-structural protein 12 [nsp12]) functions in a holo-RdRp (comprising nsp7/nsp82/nsp12) to synthesize all viral RNA molecules (Kirchdoerfer and Ward, 2019; Subissi et al., 2014). The RdRp is the target for antiviral agents such as remdesivir (Agostini et al., 2018; Yin et al., 2020). In addition to its central role in replication and transcription of the viral genome, the RdRp contains an N-terminal nidovirus RdRp-associated nucleotidyltransferase (NiRAN) domain with unknown function but with enzymatic activity that is also essential for viral propagation (Lehmann et al., 2015a).
The holo-RdRp is thought to coordinate with a number of accessory factors during the viral life cycle (Snijder et al., 2016; Sola et al., 2015). One of these accessory factors is nsp13, a superfamily 1B (SF1B) helicase that can unwind DNA or RNA in an NTP-dependent manner with a 5′ > 3′ polarity (Ivanov and Ziebuhr, 2004; Lee et al., 2010; Seybert et al., 2000a, 2000b; Tanner et al., 2003). The helicase is essential for replication in the nidovirus equine arteritis virus (EAV) (van Dinten et al., 2000; Seybert et al., 2000a, 2005) and the β-CoV murine hepatitis virus (Zhang et al., 2015) and is presumed to be essential in all nidoviruses (Lehmann et al., 2015b). The helicase is thought to play crucial roles in many aspects of the viral life cycle, some of which can be uncoupled by point mutations (van Dinten et al., 1996; van Marle et al., 1999). Besides helicase activities, nsp13 harbors RNA 5′-triphosphatase activity, which may play a role in mRNA capping (Ivanov and Ziebuhr, 2004; Ivanov et al., 2004).
In addition to the two canonical RecA ATPase domains of SF1 helicases (Saikrishnan et al., 2009; Singleton et al., 2007), nsp13 contains three domains unique to nidovirus helicases: an N-terminal zinc-binding domain (ZBD), a stalk, and a 1B domain (Hao et al., 2017; Jia et al., 2019; Lehmann et al., 2015b). The role of these domains in nsp13 function in vivo is unclear, but substitutions in these domains or the interfaces between them have disruptive effects on in vitro helicase activity as well as on viral propagation (van Dinten et al., 1996, 2000; Hao et al., 2017; van Marle et al., 1999; Seybert et al., 2005).
Biochemical and biophysical studies suggest that nsp13 and nsp12 interact (Adedeji et al., 2012; Jia et al., 2019), but a stable complex has not been biochemically isolated or structurally characterized, and the mechanistic role of the helicase in SARS-CoV-2 replication-transcription is unknown. Here we generate a stable nsp13:holo-RdRp:RNA complex and determine its structure by cryoelectron microscopy (cryo-EM) to 3.5-Å nominal resolution. The architecture of the complex provides constraints on models for nsp13 function and suggests a possible role of nsp13 in generating backtracked replication-transcription complexes for proofreading, template switching during sub-genomic RNA transcription, or both. Our structure also resolves ADP-Mg2+ in the active site of the nidovirus-specific NiRAN domain, defining a new pocket for anti-viral therapy development.
Results
The nsp13 Helicase Forms a Stable Complex with the Holo-RdRp Elongation Complex
To investigate the potential nsp13:holo-RdRp complex and its role in replication-transcription, we prepared active recombinant SARS-CoV-2 holo-RdRp and nsp13 (Figures S1 and S2 A). Our measured kcat (∼15 s−1) for SARS-CoV-2 nsp13 (Figure S2) is similar to reported kcat values for SARS-CoV hexahistidine (His6)-nsp13 (19 s−1; Tanner et al., 2003) or SARS-CoV MBP-nsp13 (2.3 s−1; Ivanov et al., 2004) (there is one conservative amino acid substitution between SARS-CoV and SARS-CoV-2 nsp13 proteins, SARS-CoV-2 nsp13-Val570 is Ile in SARS-CoV nsp13) but not to kcat values for SARS-CoV His6-nsp13 and glutathione S-transferase (GST)-nsp13 (0.2 s−1 and 104 s−1, respectively; Adedeji et al., 2012). Differences in expression and purification protocols as well as different ATPase assays used could explain the variability.
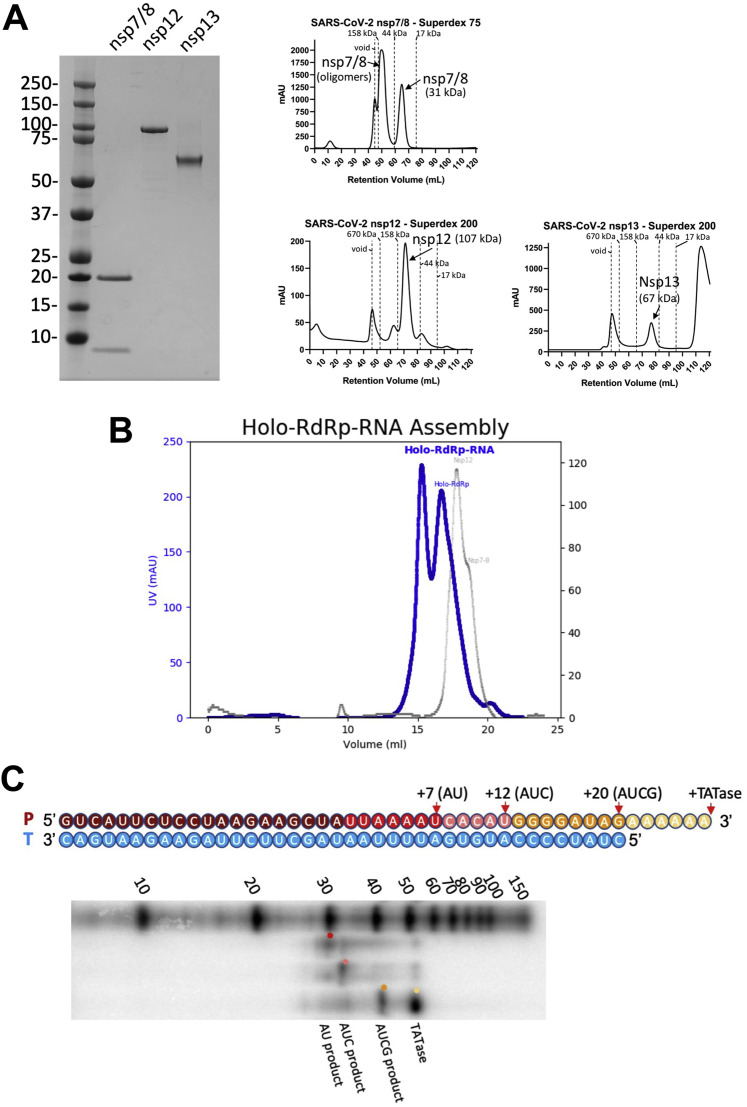
Figure S1.
Purification and Assembly of nsp13 and the RTC, Related to Figure 1
A. (left) SDS-PAGE of purified SARS-CoV-2 nsp7/8, nsp12, and nsp13.
(right) Size exclusion chromatography profiles for the purified nsp7/8, nsp12, and nsp13. Nsp7/8 are known to form high molecular weight complexes (Xiao et al., 2012; Zhai et al., 2005), which we observed in equilibrium with the 31 kDa nsp7/nsp8 heterodimer. For assembly of the holo-RdRp, we isolated the peak corresponding to the 31 kDa heterodimer.
B. Purification of RTC by size exclusion chromatography.
(left) Chromatogram of RTC (blue trace) and individual components (gray trace) labeled.
C. Holo-RdRp elongates the primer-strand of the RNA scaffold shown in the presence of NTPs. Original primer (20-mer, dark red), 27-mer AU elongated product (red), 32-mer AUC product (pink), 40-mer AUCG product (orange) and the product of nsp8-mediated TATAse activity (yellow; Tvarogová et al., 2019). Products, taken at a 30-minute time point, are shown for a representative gel (n = 2) and are visualized alongside Decade RNA ladder (Invitrogen).
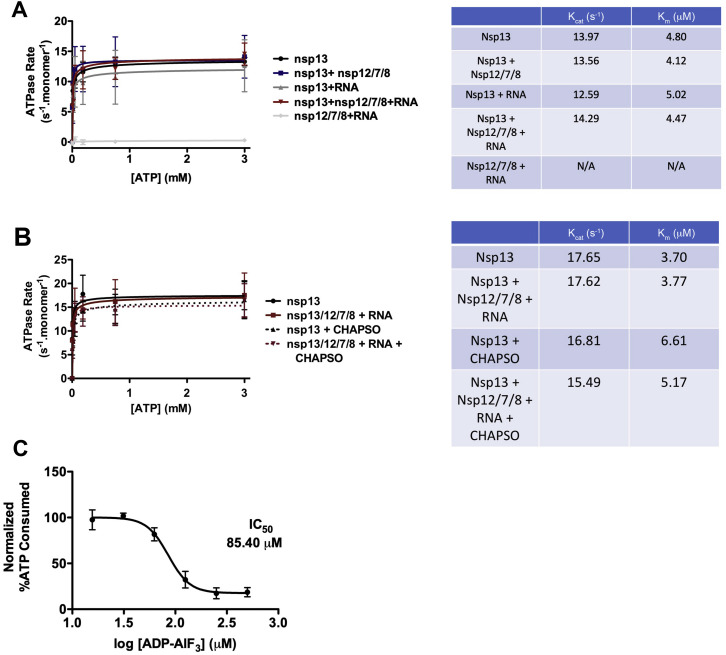
Figure S2.
Nsp13 Activities, Related to Figure 1
A. (left) ATPase assay comparing nsp13 alone, nsp13 + holo-RdRp (nsp12/7/8), nsp13 + RNA scaffold alone, nsp13-RTC (nsp13/12/7/8 + RNA), and the RTC alone (nsp12/7/8 + RNA). Error bars indicate the range for two independent measurements.
(right) Calculated Kcat and Km values for the ATPase assay.
B. (left) ATPase assay comparing nsp13 alone and nsp13-RTC (nsp13/12/7/8 + RNA) ± 8 mM CHAPSO. Error bars indicate the range for two independent measurements.
(right) Calculated Kcat and Km values for the ATPase assay.
C. Inhibitory effect of ADP-AlF3 on nsp13 ATPase activity (N = 6). Error bars denote standard deviation.
We used native electrophoretic mobility shift assays to test binding of holo-RdRp and nsp13 to an RNA scaffold (Figures 1A and 1B). Nsp12 required nsp7:nsp8 to bind the RNA (Figure 1B, compare lanes 2 and 3), and addition of nsp13 to the holo-RdRp:RNA complex caused a super-shift, indicating stable complex formation (Figure 1B, lane 6). Addition of ADP-AlF3 (Chabre, 1990), which inhibits nsp13 ATPase activity (Figure S2C), sharpened the super-shifted band (Figure 1C, lane 7), suggesting formation of a more stable, homogeneous complex. A stable complex was also detected using native mass spectrometry (nMS), revealing a stoichiometry of nsp7:nsp82:nsp12:nsp13 + RNA scaffold (Figure 1C). The ATPase activity of nsp13 was maintained within the complex (Figures S2A and S2B).
Figure 1.
SARS-CoV-2 nsp13 Helicase Forms a Stable Complex with the Holo-RdRp and an RNA Scaffold
(A) The RNA scaffold used for biochemistry, native mass spectrometry (nMS), and cryo-EM.
(B) A native gel electrophoretic mobility shift assay reveals that nsp13 forms a stable complex with holo-RdRp:RNA. The 4.5% polyacrylamide gel was visualized with Gel Red to stain the RNA.
(C) nMS spectra and the corresponding deconvolved spectra for the holo-RdRp containing the RNA scaffold (A) with and without nsp13. The measured mass for the holo-RdRp:RNA complex corroborates the established stoichiometry of 1:2:1:1 for nsp7:nsp8:nsp12:RNA (Hillen et al., 2020; Kirchdoerfer and Ward, 2019; Wang et al., 2020; Yin et al., 2020), respectively (bottom). Addition of the 67.5-kDa nsp13 helicase to the RNA-bound holo-RdRp sample forms a transcription complex/helicase assembly with 1:1 stoichiometry (top).
See also Figures S1 and S2.
Structure of the nsp13:holo-RdRp:RNA Complex
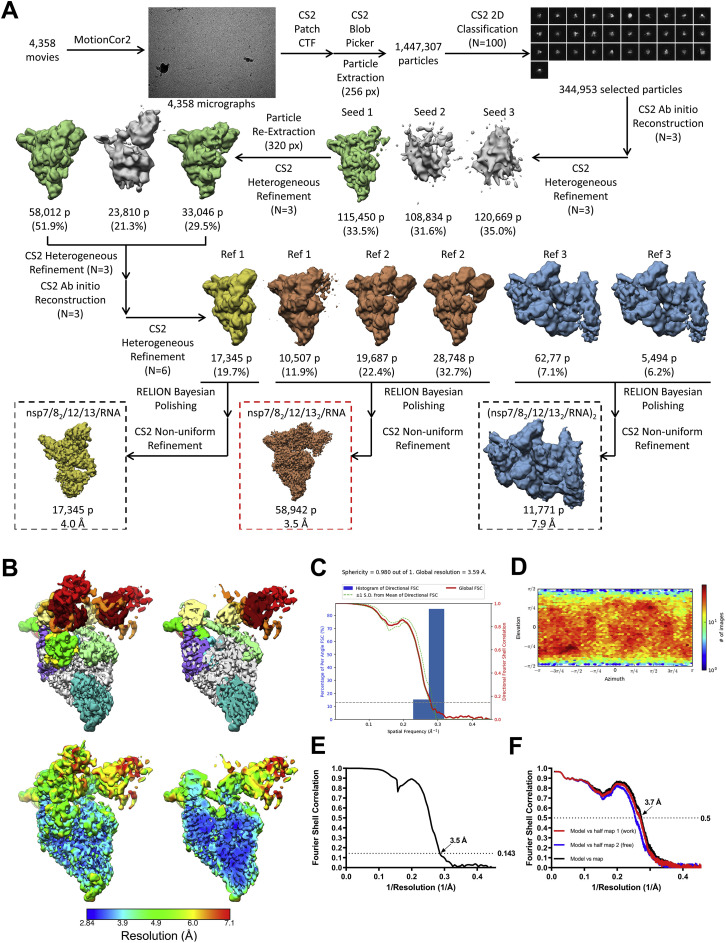
To determine the structural organization of the nsp13(ADP-AlF3):holo-RdRp:RNA complex (hereafter called the nsp13-replication-transcription complex [nsp13-RTC]), we analyzed the samples by single-particle cryo-EM. An initial dataset collected in the absence of detergent (Figure S3 A) yielded poor maps because of severe particle orientation bias (Figures S3B–S3D). The particle orientation bias was eliminated by adding the detergent 3-([3-cholamidopropyl]dimethylammonio)-2-hydroxy-1-propanesulfonate (CHAPSO), shown previously to reduce preferred particle orientation by limiting particle adsorption to the air/water interface (Chen et al., 2019), yielding isotropic maps (Figures S4 and S5 ).
Figure S3.
Cryo-EM Processing Pipeline and Analysis for the nsp13-RTC (No Detergent) Dataset, Related to Figure 2
A. Cryo-EM processing pipeline.
B. Nominal 3.6 Å-resolution cryo-EM reconstruction of nsp132-RTC (no detergent) filtered by local resolution (Cardone et al., 2013) and colored by subunit according to the key on the right.
C. Directional 3D Fourier shell correlation (FSC) for nsp132-RTC (no detergent) calculated by 3DFSC (Tan et al., 2017).
D. Angular distribution plot for reported nsp132-RTC (no detergent) calculated in cryoSPARC. Scale shows the number of particles assigned to a particular angular bin. Blue, a low number of particles; red, a high number of particles.
Figure S4.
Cryo-EM Processing Pipeline and Analysis for the nsp13-RTC (CHAPSO) Dataset, Related to Figure 2
A. Cryo-EM processing pipeline.
B. Nominal 3.5 Å-resolution cryo-EM reconstruction of nsp132-RTC (CHAPSO) filtered by local resolution (Cardone et al., 2013). The view on the right is a cross-section.
(top) Colored by subunit.
(bottom) Color by local resolution (key on the bottom).
C. Directional 3D Fourier shell correlation (FSC) for nsp132-RTC (CHAPSO) calculated by 3DFSC (Tan et al., 2017).
D. Angular distribution plot for reported nsp132-RTC (CHAPSO) calculated in cryoSPARC. Scale shows the number of particles assigned to a particular angular bin. Blue, a low number of particles; red, a high number of particles.
E. Gold-standard FSC plot for nsp132-RTC (CHAPSO), calculated by comparing two independently determined half-maps from cryoSPARC. The dotted line represents the 0.143 FSC cutoff which indicates a nominal resolution of 3.5 Å.
F. FSC calculated between the refined structure and the half map used for refinement (work, red), the other half map (free, blue), and the full map (black).
Figure S5.
Cryo-EM Density Maps, Related to Figure 2
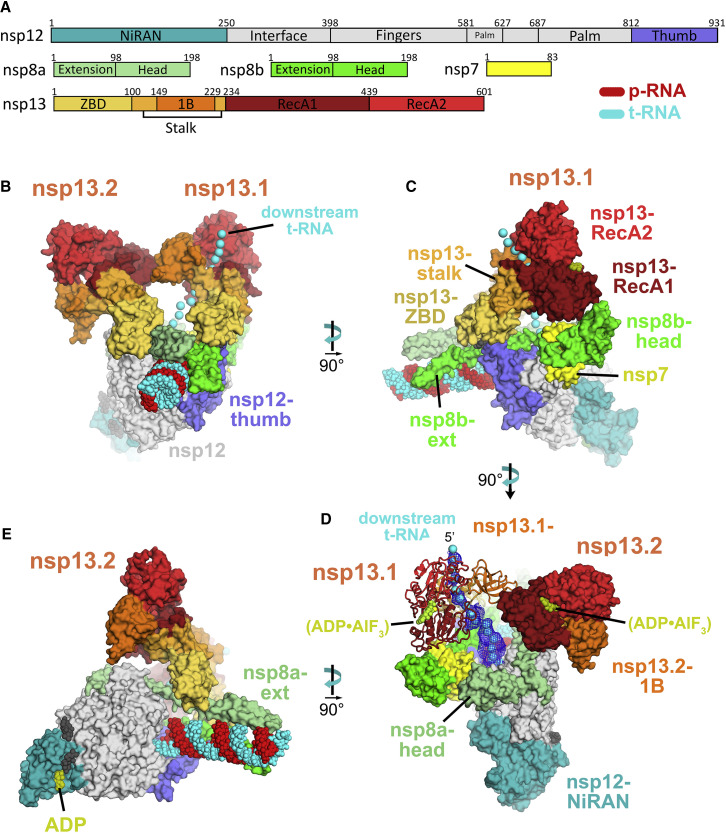
A. Schematic illustrating domain structure of SARS-CoV-2 holo-RdRp (nsp7, nsp8, nsp12) and nsp13. The color-coding corresponds to the figures throughout this manuscript unless otherwise specified.
B-E. Orthogonal views showing the overall architecture of the nsp132-RTC. Shown is the transparent cryo-EM density (local-resolution filtered) with the nsp132-RTC model superimposed. Same views as Figures 2B–2E.
F. View of the nsp12 (RdRp) active site (refined model superimposed onto the cryo-EM density, shown as blue mesh), showing the post-translocated state of the RNA.
G. View of the nsp8b-extension:nsp12-thumb:nsp13-ZBD tripartite interaction (refined model superimposed onto the cryo-EM density, shown as blue mesh). Similar view as Figure 3A.
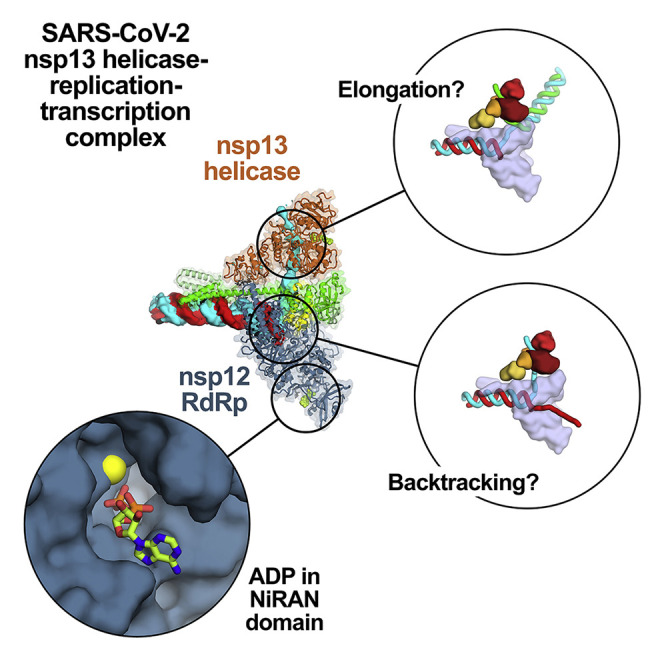
The sample comprised three major classes: 67% of the particles belong to an nsp132-RTC, 20% form nsp131-RTC, and 13% form a dimer of the nsp132-RTC ((nsp132-RTC)2; Figure S4A). The major class (nsp132-RTC) yielded a 3D reconstruction and refined model at a nominal resolution of 3.5 Å (Figure 2 ; Figures S4B–S4F; Table S1). The structure resolves the complete, post-translocated holo-RdRp/RNA complex to roughly between 2.8–3.3 Å resolution (Figure S4B), including the nsp8 N-terminal extensions that interact with the upstream duplex RNA and the complete NiRAN domain (Hillen et al., 2020; Wang et al., 2020).
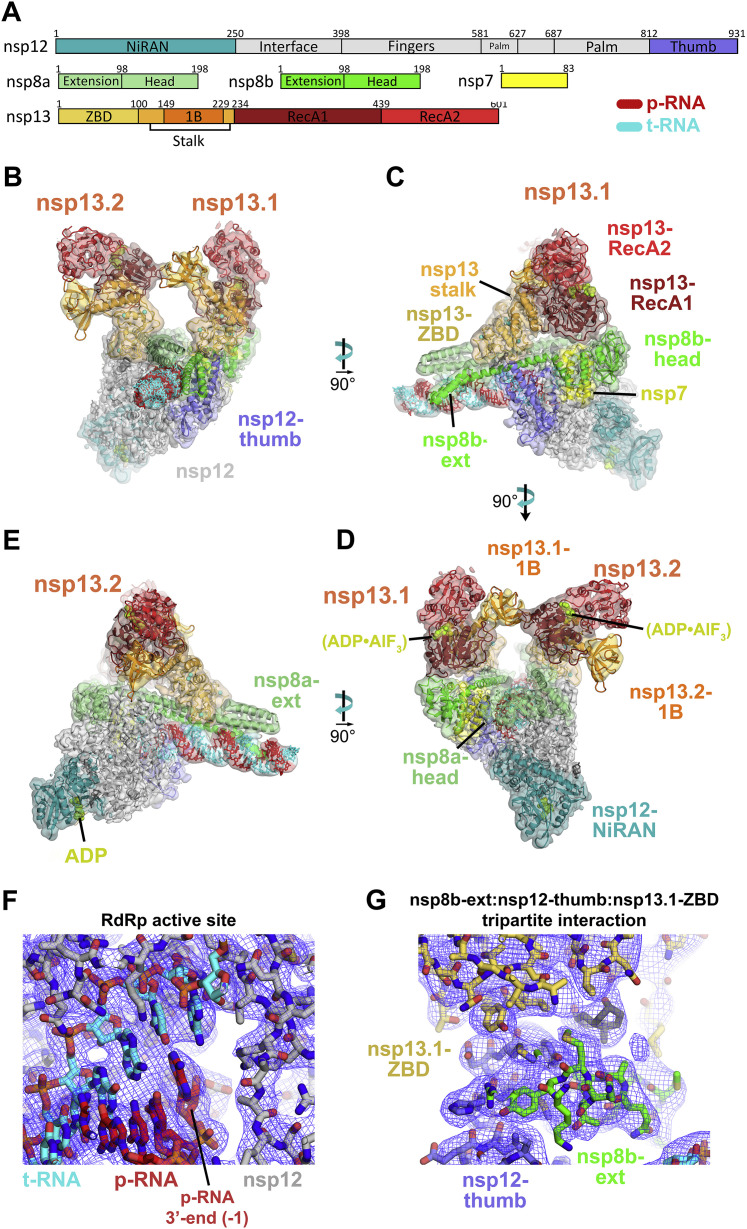
Figure 2.
Overall Structure of the SARS-CoV2 nsp13 Helicase with the Holo-RdRp:RNA Replication-Transcription Complex (RTC)
(A) Schematic illustrating the domain structure of SARS-CoV-2 holo-RdRp (nsp7, nsp8, and nsp12) and nsp13. Structural domains discussed in the text are labeled. The color coding corresponds to the figures throughout this manuscript unless otherwise specified.
(B–E) Orthogonal views showing the overall architecture of the nsp132-RTC. Proteins are shown as molecular surfaces (except nsp13.1 in D) and RNA as atomic spheres. Adventitious CHAPSO detergent molecules are shown as atomic spheres and colored dark gray. The path of downstream tRNA through the nsp13.1 helicase, shown as cyan spheres, is revealed with low-pass-filtered (6 Å) difference density (shown in D).
(B) Two copies of the nsp13 helicase bind to the RTC. Nsp13.1 forms a tripartite interaction with the nsp8b extension and the nsp12 thumb via the nsp13.1-ZBD. The 5′ end of the tRNA extrudes through the nucleic acid binding channel of nsp13.1. The two helicases interact via the nsp13.1-1B domain and the nsp13.2-RecA1 domain.
(C) In addition to the nsp13.1-ZBD:nsp8b extension:nsp12 thumb tripartite interaction, nsp13.1-RecA1 interacts with nsp7 and the nsp8b head.
(D) ADP-AlF3 is modeled in the NTP binding site of each helicase. The low-pass-filtered (6 Å) cryo-EM difference density revealing the path of the downstream t-RNA is shown (dark blue mesh).
(E) The nsp13.2-ZBD interacts with the nsp8a extension. ADP-Mg2+ is bound to the NiRAN domain.
See also Figure S3, Figure S4, Figure S5, Figure S6, Table S1, and Video S1.
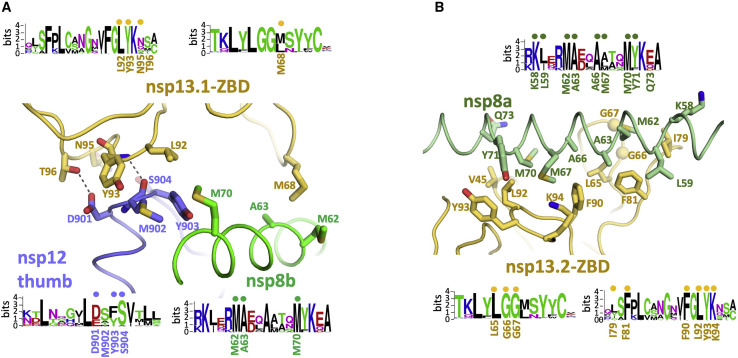
In the nsp132-RTC, the nidovirus-specific nsp13-ZBDs (Figure 2A) interact with the nsp8 N-terminal extensions; one nsp13-ZBD (nsp13.1-ZBD) interacts with nsp8b, whereas the other (nsp13.2-ZBD) interacts with nsp8a (Figure 2). The nsp13.1-ZBD interaction with the holo-RdRp is tripartite; besides nsp8b, the interaction includes the tip of the nsp12 thumb domain (Figure 2C). These interactions involve multiple residues that are universally conserved among α- and β-CoVs (Figure 3 ; Data S1 and S2).
Figure 3.
Interactions of the nsp13-ZBDs with the RTC
(A and B) Views of the nsp13-ZBD:RTC interactions. Proteins are shown as α-carbon backbone worms. Side chains that make protein:protein interactions are shown. Polar interactions are shown as dashed gray lines. Sequence logos (Schneider and Stephens, 1990) from alignments of α- and β-CoV clades for the interacting regions are shown. The residues involved in protein:protein interactions are labeled under the logos. Dots above the logos denote conserved interacting residues.
(A) View of the tripartite nsp13.1-ZBD:nsp8b extension:nsp12 thumb interaction. The adventitiously bound CHAPSO molecule is shown in dark gray.
(B) View of the nsp13.2-ZBD:nsp8a extension interaction.
In addition to the nsp13.1-ZBD:nsp8b:nsp12 thumb tripartite interaction, nsp13.1-RecA1 is braced against nsp7 and the nsp8b head (Figure 2C). Other than the conserved nsp13.2-ZBD:nsp8a interaction (Figure 3B), nsp13.2 only makes additional interactions with nsp13.1 (Figure 2B).
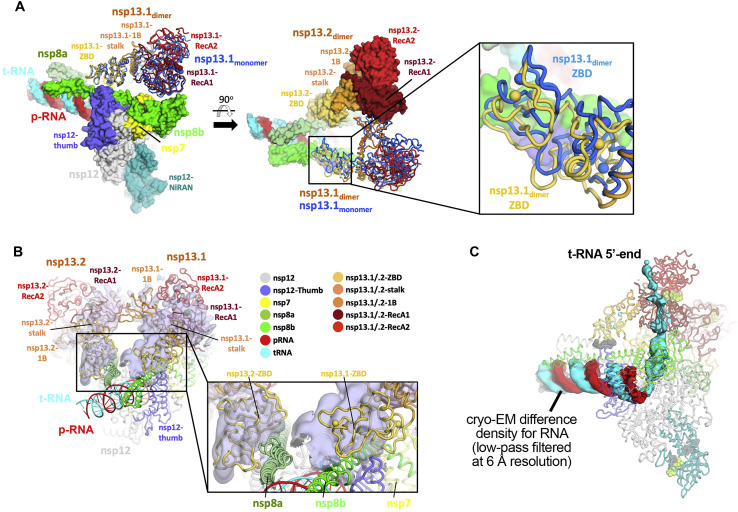
Our cryo-EM analyses show that nsp131-RTC and nsp132-RTC complexes can exist. Nsp12 (the main RTC component) and nsp13 are expressed from the same viral open reading frame (ORF) as a single polyprotein (de Groot et al., 2012) and so are present at equivalent concentrations in vivo (Bouhaddou et al., 2020) (nsp13 was also added at an approximately 1:1 ratio to the RTC to prepare the cryo-EM samples). Because the nsp132-RTC is resolved to much higher resolution (Figure S4), most of our analyses in this manuscript focus on this complex, but both complexes could be physiologically relevant. The disposition of nsp13.1 in nsp131-RTC is nearly identical to its disposition in nsp132-RTC (Figure S6 A), justifying our use of the higher-resolution nsp132-RTC structure as the basis for our analyses. Because nsp13.2 is never present without nsp13.1 (Figures S3A and S4A), nsp13.1 appears to be stably bound, whereas nsp13.2 is dissociable. Only nsp131-RTC was observed in the nMS analysis (Figure 1C), but the nMS solution conditions, electrospray process, and transition to the gas phase may have led to dissociation of nsp13.2, likely explaining the discrepancy with the cryo-EM analyses.
Figure S6.
Comparison of the nsp132-RTC (CHAPSO) Structure with nsp131-RTC (CHAPSO) and nsp132-RTC (No Detergent), Related to Figures 2 and 3
A. Structure of nsp132-RTC (CHAPSO) colored according to key in b and shown as a molecular surface except nsp13.1, which is shown as cartoon tubes. Superimposed on the overall structure is nsp13 (marine) modeled from the nsp131-RTC (CHAPSO). Overall RMSD (calculated using ‘rms_cur’ in PyMOL) between the two nsp13.1 structures is 8.1 Å over 596 Cα atoms. (left) overall structure. (middle) overall structure rotated 90°. (right) zoom-in of boxed region in middle panel, showing region around nsp13.1-ZBD.s RMSD (calculated using ‘rms_cur’ in PyMOL) between the two nsp13.1 ZBDs is 3.6 Å over 100 Cα atoms.
B. Structure of nsp132-RTC (CHAPSO) is shown in cartoon tubes, colored based on key, and superimposed onto the cryo-EM map from the nsp132-RTC (no detergent) dataset (shown as light blue transparent surface). Density map is locally filtered by resolution and difference density for nsp13 is highlighted using ‘isosurf’ command in PyMOL with 10 Å carve buffer. (left) overall structure. (right) zoom-in of boxed region in left panel, showing region around nsp13-ZBDs.
C. Structure of nsp132-RTC (CHAPSO) colored according to key in (b), the view is similar to the view of Figure S6A(left). Protein is shown as pale, transparent backbone worms. The surface shows a cryo-EM different density for the RNA (t-RNA, cyan; p-RNA, red) low-pass filtered to 6 Å resolution. The difference map was generated by calculating a map from the nsp132-RTC coordinates with the RNA removed using the molmap command in Chimera (Pettersen et al., 2004), subtracting this map from the experimental map (Chimera vop command), then low-pass filtering this difference map at 6 Å resolution).
The holo-RdRp:RNA complex and interactions with the nsp13-ZBDs were well resolved in the cryo-EM map (Figures S4B and S5G), but structural heterogeneity in the nsp13-1B and RecA domains gave rise to poorly resolved maps that could not be improved by local refinement procedures. Maps filtered to the local resolution (Cardone et al., 2013) were used to model the overall domain orientations, and difference maps revealed that the added ADP-AlF3 was bound, but the low resolution of the maps prevented detailed modeling.
The nsp13.1-ZBD interface with the holo-RdRp includes an adventitious CHAPSO molecule (Figure 3A). Despite this, the following points speak to the biological relevance of the complex. (1) The ATPase activity of nsp13, a measurable enzymatic property of the complex, was not significantly affected by the presence of CHAPSO (Figure S2B). (2) The nsp13.1:holo-RdRp interface (which includes CHAPSO) involves mutually conserved nsp13.1-ZBD, nsp8b, and nsp12 thumb residues that comprise a subset of the conserved residues participating in the nsp13.2-ZBD:nsp8a interaction that is free from CHAPSO (Figure 3). Thus, the presence of CHAPSO in the nsp13.1:holo-RdRp interface did not generate an entirely new interface. (3) In the absence of CHAPSO, the cryo-EM reconstructions were not suitable for detailed modeling and refinement because of severe particle orientation bias (Figure S3), but domain orientations could be modeled. Comparison with the no-detergent nsp132-RTC map revealed that the presence of CHAPSO slightly altered the disposition of the nsp13.1-ZBD with respect to nsp8b, but the overall architecture of the complex was the same (Figure S6B).
The NiRAN Domain Binds ADP-Mg2+
Nidovirus RdRps contain an N-terminal domain that is lacking in other viral RdRps, the NiRAN domain. The target of NiRAN domain nucleotidylation activity is unknown, but the activity is essential for viral propagation in EAV and SARS-CoV (Lehmann et al., 2015a). In addition to ADP-AlF3 bound in the NTP site of each nsp13 helicase, ADP-Mg2+ was clearly resolved bound to the nsp12-NiRAN domain (Figure 4 ), prompting us to further investigate this domain.
Figure 4.
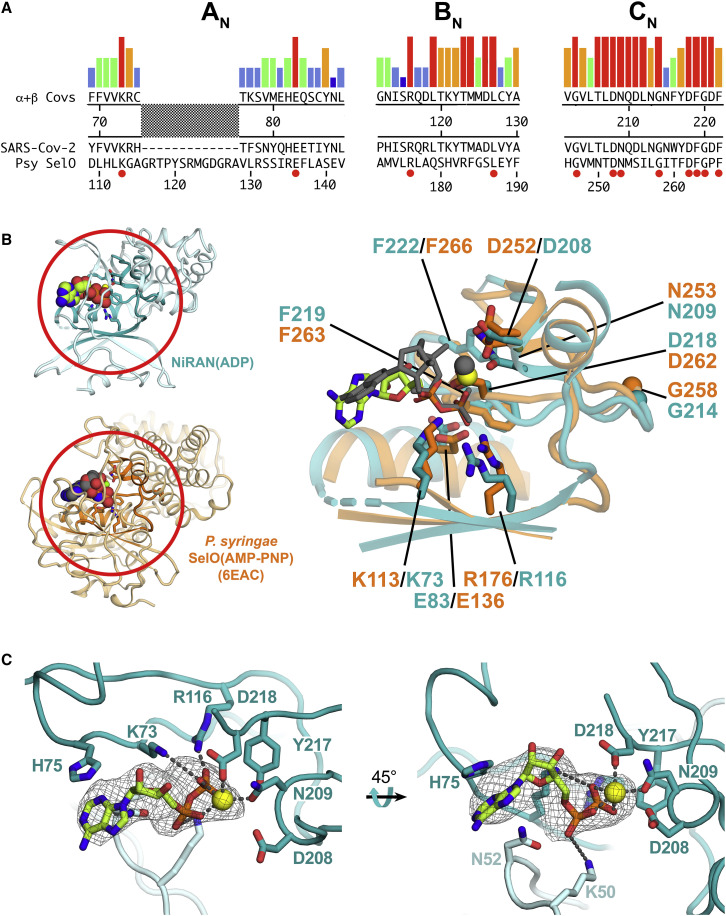
The SARS-CoV-2 nsp12-NiRAN Domain, Pseudokinase SelO, and ADP Binding
(A) The colored histograms denote identity in a sequence alignment of 45 α- and β-CoV nsp12 sequences (red bar, 100% identity; dark blue bar, 20% or less) in the N-terminal signature motifs AN, BN, and CN (Lehmann et al., 2015a) of the NiRAN domain. The consensus sequence is shown below. The SARS-CoV-2 nsp12 and the pseudokinase P. syringae (Psy) SelO (Sreelatha et al., 2018) are aligned below. Residues that are 100% identical in the nsp12 alignment and conserved in SelO are highlighted by a red dot underneath.
(B) Left: structures of the SARS-CoV-2 NiRAN domain (cyan ribbon) with ADP-Mg2+ (spheres) and Psy SelO (orange) with AMP-PNP-Mg2+ (PDB: 6EAC; Sreelatha et al., 2018). The AN, BN, and CN regions are highlighted. Right: structure-based alignment via α-carbons of the AN, BN, and CN regions, with side chains of conserved residues shown. The α- and β-phosphates of the NiRAN domain ADP-Mg2+ (lime carbon atoms and yellow sphere, respectively) superimpose almost exactly with the β- and γ-phosphates of the SelO AMP-PNP-Mg2+ (dark gray), whereas the nucleoside moieties diverge.
(C) Two views of the ADP-Mg2+-bound pocket of the SARS-CoV-2 NiRAN domain. Side chains interacting with the ADP-Mg2+ are shown (polar interactions are denoted by gray dashed lines). D208 likely makes a water-mediated interaction with Mg2+ (Sreelatha et al., 2018). The cryo-EM difference density for ADP-Mg2+ is shown (light gray mesh).
See also Data S3.
Previously, a structure-based search with the partial NiRAN domain resolved in the SARS-CoV holo-RdRp structure revealed significant structural homology to protein kinases (Kirchdoerfer and Ward, 2019). This, combined with the NiRAN domain nucleotidylation activity identified by (Lehmann et al., 2015a), led Kirchdoerfer and Ward (2019) to suggest a functional analogy with the pseudokinase Pseudomonas syringae SelO (D.E. Shaw Research (2020). SelO, with homologs widespread among eukaryotes and also common in prokaryotes (Dudkiewicz et al., 2012), is a pseudokinase containing a kinase fold but lacking key catalytic residues of canonical kinases. Nevertheless, structural studies reveal that AMP-PNP binds in the SelO active site but is flipped relative to the orientation of ATP in canonical kinases. Functional studies show that SelO pseudokinases are, in fact, active enzymes that transfer AMP, rather than a phosphate group, to S, T, and Y residues of protein substrates (AMPylation or adenylylation; Sreelatha et al., 2018). These studies identified key SelO residues involved in catalysis, including K113 and R176 (coordinate ATP γ-phosphate), E136 (stabilizes the position of K113), N253 and D262 (coordinate active-site Mg2+), and D252 (acts as a catalytic base).
A search using our complete NiRAN domain structure (nsp12 residues 1–250) on the DALI webserver (Holm and Laakso, 2016) revealed P. syringae SelO as the most significant hit (PDB: 6EAC; Z score of 10.7; Sreelatha et al., 2018), as suggested by Kirchdoerfer and Ward (2019). Extensive sequence analysis identified three conserved sequence motifs within the nidovirus-specific N-terminal domain: AN, BN, and CN (Lehmann et al., 2015a; Figure 4A). Overall sequence conservation between SelO and the NiRAN domain is essentially non-existent (<10%), but a structure-based alignment reveals that the key SelO catalytic residues are universally conserved within the NiRAN domain sequence motifs (Figure 4A) and conserved in spatial organization within the aligned active-site pocket (Figure 4B; Data S3).
The NiRAN domain in the nsp13-RTC structure is occupied by ADP-Mg2+, which is well resolved (Figure 4C). Remarkably, the α- and β-phosphates of the NiRAN domain ADP coincide almost exactly with the β- and γ-phosphates of AMP-PNP occupying the SelO active site (Figure 4B; root-mean-square deviation [RMSD] = 0.33 Å). All of the conserved active-site residues make interactions with ADP-Mg2+, as expected from the SelO structure, except NiRAN domain K73 and R116 interact with ADP β-phosphate, whereas the corresponding K113 and R176 of SelO interact with the similarly positioned AMP-PNP γ-phosphate.
Roles of SARS-CoV-2 nsp13 in RTC Processivity and Backtracking
The overall architecture of the nsp13-RTC places the nucleic acid binding channel of nsp13.1 (Deng et al., 2014) directly in the path of the downstream tRNA. Indeed, a cryo-EM difference map, low-pass-filtered to 6-Å resolution, reveals the 5′ single-stranded overhang of the tRNA (Figure 1A) engaged between the RecA domains and domain 1B of nsp13.1 before entering the RdRp active site (Figure 2C; Figure S6C).
Nsp13 is an SF1B helicase, which translocate on single-stranded nucleic acid in the 5 > 3′ direction (Saikrishnan et al., 2009). In vitro studies confirm this direction of translocation for nidovirus helicases (Adedeji et al., 2012; Bautista et al., 2002; Ivanov and Ziebuhr, 2004; Seybert et al., 2000a, 2000b; Tanner et al., 2003). Unless the interaction of nsp13 with the holo-RdRp alters the unwinding polarity, which seems unlikely, the structural arrangement observed in the nsp13-RTC (Figure 2D) presents a conundrum. In the observed arrangement, the RdRp would translocate in the 3′ > 5′ direction on the tRNA strand, whereas nsp13 would translocate on the same strand in the opposite direction. Thus, in the proposed scenario, translocation of nsp13 competes with translocation by the RdRp, and if the helicase prevails, then it might be expected to push the RdRp backward on the t-RNA. Reversible backward motion of multi-subunit cellular DNA-dependent RNA polymerases (DdRps), called backtracking, is a well-characterized feature of these enzymes (Komissarova and Kashlev, 1997b, 1997a; Nudler, 2012; Nudler et al., 1997; Wang et al., 2009).
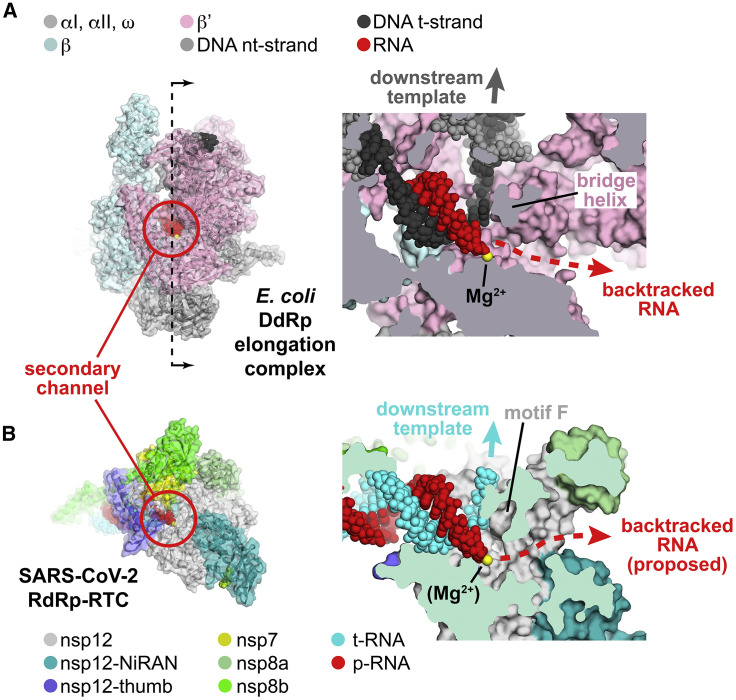
The large active-site cleft of the cellular DdRp is divided into two sections by a conserved structural element called the bridge helix (Figure 5 A provides a cross-section view on the right). The main cleft on top of the bridge helix accommodates the downstream duplex DNA, whereas a pore or channel underneath the bridge helix forms a conduit from solution directly into the active site (Figure 5A, called the secondary channel in bacterial DdRp; Zhang et al., 1999). During normal transcription elongation, the NTP substrates enter the active site through the secondary channel (Westover et al., 2004; Zhang et al., 1999). In backtracking, the DdRp and associated transcription bubble move backward on the DNA, whereas the RNA transcript reverse-threads through the complex to maintain the register of the RNA/DNA hybrid. These movements generate a downstream segment of single-stranded RNA at the 3′ end that is extruded out the secondary channel (Figure 5A; Abdelkareem et al., 2019; Wang et al., 2009).
Figure 5.
Correspondence of Structural Determinants for Backtracking between Cellular Multi-subunit DdRp and SARS-CoV-2 RdRp
(A and B) Left: overall views of complexes. Proteins are shown as cartoon ribbons with transparent molecular surfaces. Nucleic acids are shown as atomic spheres. Color coding is shown in the keys. Right: cross-sectional view of the active site region (at the 3′ end of the RNA product).
(A) Left: structure of the E. coli DdRp transcription elongation complex (EC; Kang et al., 2017), viewed down the secondary channel. The DdRp active-site Mg2+-ion is shown as a yellow sphere. The secondary channel is highlighted in the red circle. The thin dashed line illustrated the cut and viewing direction of the cross-section on the right. Right: cross-sectional view showing the RNA/DNA hybrid. The bridge helix (viewed end-on in the cross-section) is denoted. The bridge helix directs the downstream template duplex DNA to the top (dark gray arrow). Under the bridge helix, the secondary channel allows NTP substrates to diffuse into the active site (Westover et al., 2004; Zhang et al., 1999) and accommodates the single-strand RNA transcript 3′ end in backtracked complexes (Abdelkareem et al., 2019; Cheung and Cramer, 2011; Wang et al., 2009).
(B) Left: view down the newly described secondary channel of the SARS-CoV-2 holo-RdRp. The secondary channel is highlighted in the red circle. Right: cross-sectional view showing the t-RNA/p-RNA hybrid. Motif F (viewed end-on in the cross-section) is denoted. Motif F directs the downstream t-RNA to the top (cyan arrow). Under motif F, the secondary channel could accommodate the single-strand p-RNA 3′ end in the event of backtracking.
Although evolutionarily unrelated to the DdRp, the SARS-CoV-2 RdRp active site has a similar general architecture (Figure 5B). In place of the bridge helix, the viral RdRp has conserved motif F (SARS-CoV-2 nsp12 residues 544–555; Bruenn, 2003), which comprises a β-hairpin loop. Motif F directs the tRNA to the top, whereas underneath motif F is a channel that appears able to accommodate single-stranded nucleic acid. The analogous structural arrangement leads us to propose that the SARS-CoV-2 RdRp may backtrack, generating a single-stranded RNA segment at the 3′ end that would extrude out the RdRp secondary channel (Figure 5B).
Discussion
We have established that the critical SARS-CoV-2 replication-transcription components nsp13 and holo-RdRp form a stable complex (Figures 1B and 1C) and determined a nominal 3.5-Å-resolution cryo-EM reconstruction (Figure 2; Figure S4; Table S1; Video S1). In the structure, the primary interaction determinant of the helicase with the RTC occurs between the nsp13-ZBDs and the nsp8 extensions. These structural elements are unique to nidoviruses, and the interaction interfaces are conserved within α- and β-CoV genera (Figure 3), indicating that this interaction is a crucial facet of SARS-CoV-2 replication-transcription. A protein-protein interaction analysis of the SARS-CoV-1 ORFeome (which recapitulates the nsp13-RTC interactions observed in our structure) identified nsp8 as a central hub for viral protein-protein interactions (von Brunn et al., 2007). The structural architecture of nsp8a and nsp8b, with their long N-terminal helical extensions, provide a large binding surface for association of an array of replication-transcription factors (Figures 2C and 2E).
The video highlights features of the nsp132-RTC. The video starts with the experimental nsp132-RTC cryo-EM map [3.5 Å nominal resolution, local-resolution filtered (Cardone et al., 2013)], superimposes the holo-RdRp:RNA structure, nsp13.1, then nsp13.2. Structural features highlighted through the video include: 1)The nsp13.1:holo-RdRp interface, 2) the nsp13.2-nsp8a interface, 3) the ADP-Mg2+ bound in the nsp12-NiRAN domain, and 4) the RdRp secondary channel.
Our structure reveals ADP-Mg2+ occupying the NiRAN domain active site (Figure 4C), presumably because the sample was incubated with ADP-AlF3 prior to grid preparation. The ADP makes no base-specific interactions with the protein; nsp12-NiRAN-H75 forms a cation-π interaction with the adenine base (Figure 4C), but this interaction is not expected to be strongly base specific, and structural modeling does not suggest obvious candidates for base-specific interactions. The position corresponding to H75 in the NiRAN domain An alignment is not conserved (Figure 4A), suggesting that (1) this residue is not a determinant of base specificity for the NiRAN domain active site, (2) the NiRAN domain base specificity varies among different nidoviruses, or (3) NiRAN domains in general do not show base specificity in their activity. The NiRAN domain of the EAV-RdRp appeared to prefer U or G for its activity (Lehmann et al., 2015a). NiRAN domain enzymatic activity is essential for viral propagation, but its target is unknown (Lehmann et al., 2015a). Further experiments will be required to more completely understand NiRAN domain activity, its preferred substrate, and its in vivo targets, and these may vary among different nidoviruses. Our results provide a structural basis for (1) biochemical, biophysical, and genetic experiments to investigate these questions and (2) a platform for anti-viral therapy development.
Our analysis comparing the viral RdRp with cellular DdRps revealed a remarkable structural similarity at the polymerase active sites; immediately downstream of each polymerase active site is a conserved structural element that divides the active site cleft into two compartments, directing the downstream nucleic acid template into one compartment and leaving a relatively open channel (the secondary channel) in the other compartment (Figure 5). In the cellular DdRps, the conserved structural element that divides the active site cleft is the bridge helix (Lane and Darst, 2010), and the secondary channel serves to allow NTP substrates to access the DdRp active site and to also accommodate the single-stranded 3′-RNA fragment generated during backtracking (Figure 5A).
In the viral RdRp, the downstream strand-separating structural element is the motif F β-hairpin loop. As for multi-subunit DdRps, the RdRp secondary channel is perfectly positioned to accommodate backtracked RNA (Figure 5B). Based on this structural analogy, we propose that the viral RdRp may undergo backtracking and that the single-stranded 3′-RNA fragment so generated would extrude out the viral RdRp secondary channel (Figure 5B). Backtracking of Φ6 and poliovirus RdRps has been observed experimentally (Dulin et al., 2015, 2017).
Ignoring sequence variation, the energetics of backtracking by cellular DdRps are close to neutral because the size of the melted transcription bubble and the length of the RNA/DNA hybrid in the active site cleft are maintained (any base pairs disrupted by backtracking are recovered somewhere else). For the SARS-CoV-2 RdRp, the arrangement of single-stranded and duplex nucleic acids during replication-transcription in vivo is not known, but in vitro, the RdRp synthesizes product RNA (p-RNA) from a single-stranded t-RNA, resulting in a persistent upstream p-RNA/t-RNA hybrid. In this case, backtracking is energetically disfavored because it only shortens the product RNA duplex without recovering duplex nucleic acids somewhere else. However, our structural analysis of the nsp13-RTC indicates that nsp13.1 can engage with the downstream single-stranded t-RNA (Figure 2D; Figure S6C). Translocation of the helicase on this RNA strand would proceed in the 5′ > 3′ direction, in opposition to the 3′ > 5′ translocation of the RdRp on the same RNA strand. This aspect of helicase function could provide the NTP-dependent motor activity necessary to backtrack the RdRp. In cellular organisms, DdRp backtracking plays important roles in many processes, including control of pausing during transcription elongation, termination, DNA repair, and fidelity (Nudler, 2012). Two potential roles of backtracking in SARS-CoV-2 replication-transcription include (1) fidelity and (2) template switching during sub-genomic transcription.
Backtracking by cellular DdRps is favored when base-pairing in the RNA/DNA hybrid is weakened by a misincorporated nucleotide in the RNA transcript (Nudler et al., 1997). Bacterial Gre factors or eukaryotic SII (transcription factor IIS, or TFIIS) interact with the backtracked DdRp complex and stimulate endonucleolytic cleavage of the single-stranded 3′-RNA fragment, removing the misincorporated nucleotide and generating a new 3′-OH at the DdRp active site for transcription to resume (Borukhov et al., 1993; Fish and Kane, 2002; Kettenberger et al., 2003; Opalka et al., 2003). In this way, endonucleolytic transcript cleavage factors contribute to transcription fidelity in vivo (Erie et al., 1993; Thomas et al., 1998).
The replication and maintenance of the genomes of the large nidovirus RNA genome (Nga et al., 2011; Saberi et al., 2018) are hallmarks of nidoviruses. These viruses encode a unique set of proofreading activities that are not found in other RNA viruses (Minskaia et al., 2006). This includes the N-terminal exonuclease (ExoN) domain of nsp14, which, together with its co-factor nsp10, forms important RNA proofreading machinery during viral replication (Denison et al., 2011; Smith et al., 2014). The ExoN is conserved in nidoviruses with genome sizes of more than 20 kb (Gorbalenya et al., 2006; Minskaia et al., 2006; Nga et al., 2011; Zirkel et al., 2011), and mutations in the ExoN lead to viruses with a lethal mutator phenotype (Eckerle et al., 2007, 2010; Smith et al., 2013). In vitro, nsp14 3′ > 5′ ExoN activity can efficiently cleave a mismatched 3′ end from an RNA construct thought to mimic an RdRp misincorporation product (Bouvet et al., 2012). The nsp10/nsp14 proofreading machinery is thought to interact with the RTC complex (Subissi et al., 2014), but structural modeling shows that the ExoN active site would not be able to access a mismatched 3′ end in the RdRp active site, which is too narrow to accommodate the nsp10/nsp14-ExoN domain proofreading machinery, a nearly 50-kDa protein. We propose that nsp14 may bind to the holo-RdRp with its ExoN active site at the mouth of the RdRp secondary channel, where it would be encountered by the backtracked mismatched RNA 3′ end. Helicase-powered backtracking could thus play a role in maintaining replication-transcription fidelity by extruding the mismatched RNA 3′ end into the nsp14 ExoN active site for cleavage.
Roadblocks to forward translocation of the cellular DdRps can also induce backtracking (Nudler, 2012). Remdesivir (Rdv) is an adenosine nucleoside triphosphate analog that has been granted emergency use authorization by the US Food and Drug Administration (US Food and Drug Administration, 2020; Warren et al., 2016). Rdv is incorporated into the p-RNA by the RdRp and appears to act by a delayed chain termination mechanism; up to three nucleotides can be added following Rdv incorporation until further forward translocation is stalled (Gordon et al., 2020). Rdv incorporated into the −4 position of the RNA duplex would not be accessible by nsp14 ExoN activity, but CoV mutants lacking nsp14 ExoN activity are more sensitive to Rdv (Agostini et al., 2018), indicating that nsp14 ExoN activity can blunt the effectiveness of the drug (Shannon et al., 2020). The nsp14 ExoN proofreading activity to excise Rdv could be facilitated by backtracking, which would allow nsp14 to degrade the p-RNA from the 3′ end until Rdv is removed.
The efficiency with which the holo-RdRp can negotiate downstream obstacles to elongation is unknown. Our structure suggests that the nsp13 helicase could act in the 5′ > 3′ direction on the tRNA to disrupt stable RNA secondary structures or downstream RNA binding proteins (Figure 6 B), both of which could be significant impediments to RNA elongation (Figure 6B). The helicase may function in this role distributively to avoid interfering with RdRp translocation. Alternatively, in the case of a fully duplex RNA template, the helicase could act processively to unwind the downstream duplex RNA, much like replicative helicases, such as DnaB in Escherichia coli, processively unwind the DNA duplex in front of the replicative DNA polymerase (Kaplan and O’Donnell, 2002).
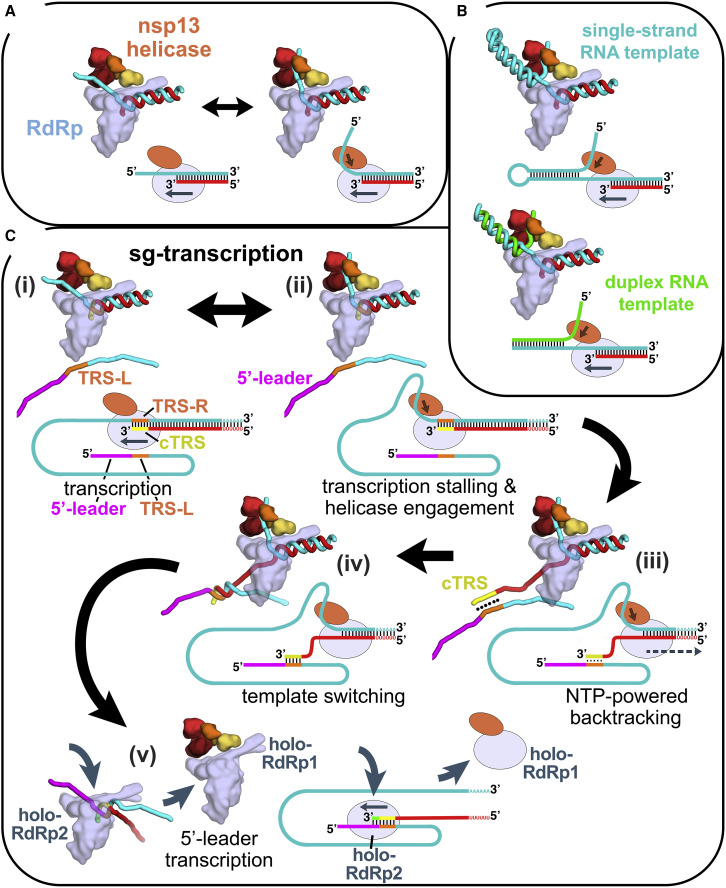
Figure 6.
Structural Basis for Possible nsp13 Helicase Functions during Viral Genome Replication-Transcription
Structural models are shown as cartoons (holo-RdRp, light blue; nsp13.1 helicase, orange shades; RNA strands, colored tubes). The nsp13.2 helicase is not shown for clarity (all models are compatible with the presence of nsp13.2). With each structural diagram is a schematic cartoon illustrating the arrangement of RNA strands. Additional proteins involved in these processes are omitted. The product RNA (p-RNA) being elongated by the RdRp is shown in red.
(A) The SARS-CoV-2 nsp13-RdRp cryo-EM structure likely represents an equilibrium between two states.
(B) During RNA synthesis on a single-stranded RNA template (cyan), nsp13 could function distributively to clear downstream RNA secondary structure (or RNA binding proteins). Similarly, on a duplex RNA template (cyan and green), nsp13 could processively unwind downstream duplex RNA.
(C) Proposed helicase function during template switching associated with sub-genomic transcription (sg-transcription) (Enjuanes et al., 2006; Lehmann et al., 2015b; Pasternak et al., 2001; Snijder et al., 2016; Sola et al., 2015). (i) Negative-strand RNA synthesis proceeds from the genomic 3′ poly(A)-tail until a transcription-regulating sequence (TRS-R, orange) (Alonso et al., 2002) is transcribed (cTRS, yellow). (ii) The TRS causes transcription complex stalling. (iii) Helicase function acting on the + strand RNA (cyan) causes backtracking of the transcription complex, freeing the pRNA 3′ end. (iv) The p-RNA 3′-end cTRS (yellow) hybridizes with the complementary TRS-L (orange) following the genomic 5′ leader sequence (magenta) (Alonso et al., 2002; Pasternak et al., 2001; Zúñiga et al., 2004). (v) Processive helicase function backtracks the RdRp complex and unwinds the p-RNA from the genomic 3′ end. A second RdRp complex (holo-RdRp2) can load into the p-RNA 3′ end and continue transcription using the 5′ leader as a template.
Finally, CoV transcription includes a discontinuous step during production of sub-genomic RNAs (sg-transcription; Figure 6C) that involves a remarkable template-switching step unique to nidoviruses (Sawicki and Sawicki, 1998). The process produces sgRNAs that are 5′- and 3′ co-terminal with the virus genome. In this process, transcription initiates from the 3′ poly(A) tail of the + strand RNA genome (cyan RNA in Figure 6C(i)). Transcription proceeds until one of several transcription-regulating sequences (TRS-R; orange in Figure 6C) is encountered and transcribed (transcribed cTRS in yellow), resulting in stalling of the RTC (Alonso et al., 2002), which could allow engagement of the helicase with the tRNA (Figure 6C(ii)). The complement to the TRS synthesized at the 3′ end of the p-RNA (cTRS, yellow) is complementary to another TRS near the 5′ leader of the genome (TRS-L, orange; 5′ leader, magenta), and the template switching depends on complete base-pair complementarity between cTRS and TRS-L (Alonso et al., 2002; Pasternak et al., 2001; Zúñiga et al., 2004). Thus, the 6-nt core sequence of cTRS must be available to base-pair with TRS-L (Figure 6D(iv)), which could be made possible by helicase-induced backtracking of the RTC to extrude the cTRL at the p-RNA 3′ end (Figure 6D(iii)). Upon backtracking, the exposed cTRS could form Watson-Crick base pairs with the complementary TRS-L (Figure 6D(iv)). Following hybridization of cTRS with TRS-L, a second holo-RdRp complex (holo-RdRp2) could load onto the free 3′ end of cTRS at the ∼6-bp RNA/RNA hybrid and continue transcription of the 5′ leader (Figure 6D(v)), explaining how template switching may occur. The 6-bp hybrid between TRS-L and cTRS is exactly the length of the t-RNA/p-RNA duplex contacted by the nsp12 subunit of the holo-RdRp (−1 to −7; Figure 1A) and is probably what is required for a minimally stable holo-RdRp:RNA complex. Continued translocation of the helicase on the first RTC might processively backtrack the complex and unwind the p-RNA/t-RNA hybrid left in the wake of the RTC (Figure 6D(v)).
The various possible functional modes of nsp13 (illustrated in Figure 6) would not necessarily operate simultaneously but could be regulated by other components of the RTC (not shown in Figure 6). Thus, the role of nsp13 in facilitating RTC elongation on a single-stranded RNA template (Figure 6A, top), where nsp13 must function distributively, may involve the nsp131-RTC (binding of other RTC components to the nsp8a extension could block binding of nsp13.2, for instance). Nsp13 could function processively to facilitate RTC elongation on a duplex RNA template (Figure 6B, bottom), and this could involve the nsp132-RTC. The backtracking function of nsp13 in the context of sg-transcription (Figure 6C) could be facilitated by pausing, stalling, or inactivation of the RdRp at the TRS, which might be controlled by components of the protein complex presumed to be associated with TRS sequences (Enjuanes et al., 2006; Snijder et al., 2016; Sola et al., 2015).
In addition to nidoviruses, many RNA viruses, including important pathogens such as chikungunya virus, herpesvirus, hepatitis C, Zika virus, and picornaviruses, encode their own helicases (Lehmann et al., 2015b). Although these viral helicases belong to different helicase families and play different and complex roles in viral life cycles, they all have three things in common: (1) the helicases are essential for viral propagation, (2) very little is known about the precise mechanistic roles of the helicases in viral life cycles, and (3) the helicases are thought to function as components of larger macromolecular assemblies. Our structure provides important clues regarding the role of the nsp13-RTC in SARS-CoV-2 replication-transcription (Figure 6) and provides guiding principles to test the roles of the essential helicases in other RNA viruses.
Limitations of Study
The proposal that the holo-RdRp backtracks is based on structural analysis (Figure 5) and needs to be verified experimentally. Following from this, the implications of backtracking by the holo-RdRp in nsp10/nsp14-mediated proofreading and template switching during sg-transcription (Figure 6D(iii–v)) need to be further investigated in vitro (when possible) or in vivo. Finally, the proposed roles of nsp13 in facilitating holo-RdRp elongation through obstacles (Figure 6B) also need to be experimentally verified.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| E. coli BL21(DE3) | NEB | Cat.# C2527I |
| E. coli BL21-CodonPlus | Agilent | Cat.# 230280 |
| E. coli Rosetta (DE3) | Novagen | Cat.# 70954-3 |
| SARS-CoV-2 infected Vero E6 cells | Blanco-Melo et al., 2020 | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 3-([3-Cholamidopropyl]dimethylammonio)-2-hydroxy-1-propanesulfonate | Anatrace | Cat.# C317 |
| Adenosine 5′-diphosphate sodium salt | Sigma-Aldrich | Cat.# A2754-1G |
| Aluminum fluoride | Sigma-Aldrich | Cat.# 449628-10G |
| LDH/PK | Sigma-Aldrich | Cat.# P0294-5ML |
| NADH | Sigma-Aldrich | Cat.# N7410-15VL |
| PEP | Sigma-Aldrich | Cat.# P7127-250MG |
| SARS-CoV-2 nsp12 (biochemistry and cryo-EM) | This paper | N/A |
| SARS-CoV-2 nsp13 (biochemistry and cryo-EM) | This paper | N/A |
| SARS-CoV-2 nsp7/8 (biochemistry and cryo-EM) | This paper | N/A |
| SuperScript III Reverse Transcriptase | Invitrogen | Cat.# 18080051 |
| Critical Commercial Assays | ||
| Kinase-Glo Plus Assay Kit | Promega | Cat.# V3771 |
| Deposited Data | ||
| Coordinates of SARS-CoV-2 nsp132-RTC | This paper | PDB: 6XEZ |
| Cryo-EM map of SARS-CoV-2 (nsp132-RTC)2 | This paper | EMD-22271 |
| Cryo-EM map of SARS-CoV-2 nsp131-RTC | This paper | EMD-22270 |
| Cryo-EM map of SARS-CoV-2 nsp132-RTC | This paper | EMD-22160 |
| Repository of raw data | This paper | https://data.mendeley.com/datasets/njf7w7m5js/draft?a=8a8a4e75-3e95-41f5-9d90-6956c34b5aa3 |
| Oligonucleotides | ||
| The oligonucleotides used in this study are listed in Table S2. | This paper | N/A |
| Recombinant DNA | ||
| pCDFDuet-1 | Novagen | Cat.# 71340-3 |
| pCDFDuet-1-His6-SARS-CoV-2-nsp7_nsp8 | This paper | N/A |
| pET28-His6-SARS-CoV-2-nsp13 | GenScript | N/A |
| pRSFDuet-1 | Novagen | Cat.# 71341-3 |
| pRSFDuet-1-His6-SUMO-SARS-CoV-2-nsp12 | This paper | N/A |
| Software and Algorithms | ||
| blocfilt | Cardone et al., 2013 | https://lsbr.niams.nih.gov/bsoft/programs/blocfilt.html |
| blocres | Cardone et al., 2013 | https://lsbr.niams.nih.gov/bsoft/programs/blocres.html |
| Coot | Emsley and Cowtan, 2004 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot |
| cryoSPARC | Punjani et al., 2017 | https://cryosparc.com/ |
| Dali server | Holm and Laakso, 2016 | http://ekhidna2.biocenter.helsinki.fi/dali/ |
| EMBL-EBI Clustal Omega | Madeira et. Al., 2019 | https://www.ebi.ac.uk/Tools/msa/clustalo/ |
| GraphPad Prism | GraphPad | http://www.graphpad.com/scientific-software/prism/ |
| Leginon | Suloway et al., 2005 | https://emg.nysbc.org/redmine/projects/leginon/wiki/Leginon_Homepage |
| Molprobity | Chen et al., 2010 | http://molprobity.biochem.duke.edu |
| MotionCor2 | Zheng et al., 2017 | https://emcore.ucsf.edu/ucsf-software |
| mtriage | Afonine et al., 2018 | https://www.phenix-online.org/documentation/reference/mtriage.html |
| PHENIX | Adams et al., 2010 | https://www.phenix-online.org/documentation/index.html |
| RELION | Scheres, 2012 | https://www2.mrc-lmb.cam.ac.uk/relion |
| RELION Bayesian Polishing | Zivanov et al., 2018 | https://www2.mrc-lmb.cam.ac.uk/relion |
| SBGrid | Morin et al., 2013 | https://sbgrid.org/ |
| The PyMOL Molecular Graphics System | Schrödinger, LLC | https://pymol.org/2/ |
| UCSF Chimera | Pettersen et al., 2004 | https://www.cgl.ucsf.edu/chimera |
| UniDec | Marty et. al., 2015 | https://github.com/michaelmarty/UniDec/releases |
| WebLogo | Schneider and Stephens, 1990 | https://weblogo.berkeley.edu/ |
| Other | ||
| C-flat CF-1.2/1.3 400 mesh gold grids | Protochips, Inc. | Cat.# CF413-100-Au |
| cOmplete Protease Inhibitor Cocktail | Roche | Cat.# 11697498001 |
| Gibson Assembly Master Mix | NEB | Cat.# E2611S |
| HiLoad 16/600 Superdex 200 pg | GE Healthcare Life Sciences | Cat.# GE28-9893-3 |
| HiLoad 16/600 Superdex 75 pg | GE Healthcare Life Sciences | Cat.# GE28-9893-33 |
| HisTrap HP | GE Healthcare Life Sciences | Cat.# GE17-5248-02 |
| HiTrap Heparin HP | GE Healthcare Life Sciences | Cat.# GE17-0407-03 |
| Isotope [α-32P] ATP | Perkin Elmer | Cat.# BLU003H250UC |
| Superose 6 Increase 10/300 GL column | GE Healthcare Life Sciences | Cat.# GE29-0915-96 |
| Zeba Spin Desalting Columns, 40K MWCO | Thermo Scientific | Cat.# 87765 |
| Zeba Spin Desalting Columns, 7K MWCO | Thermo Scientific | Cat.# 89882 |
Resource Availability
Lead Contact
Further information and requests for resources and reagnets should be directed to and will be fulfilled by the Lead Contact, Seth A. Darst (darst@rockefeller.edu).
Materials Availability
Plasmids generated in this study will be made available on request by the Lead Contact.
Data and Code Availability
The cryo-EM density maps for the nsp131-RTC(CHAPSO), nsp132-RTC(CHAPSO), and (nsp132-RTC)2(CHAPSO) have been deposited in the EMDataBank under accession codes EMD-22270, EMD-22160, and EMD-22271, respectively. Atomic coordinates for nsp132-RTC have been deposited in the Protein Data Bank under accession code 6XEZ. Source data for gel-based images in the paper (Figure 1B; Figure S1C) are available (https://data.mendeley.com/datasets/njf7w7m5js/draft?a=8a8a4e75-3e95-41f5-9d90-6956c34b5aa3)
Experimental Model and Subject Details
Proteins for structural biology and biochemistry
nsp7, nsp8, nsp12, and nsp13 are proteins from the SARS-CoV-2 virus.
Protein expression
For nsp7/nsp8 expression, Eco BL21(DE3) [Eco str. B F– ompT gal dcm lon hsdS B(r B – m B –) λ(DE3 [lacI lacUV5-T7p07 ind1 sam7 nin5]) [malB +]K-12(λS)] were transformed and grown overnight on LB-agar plates containing 50 μg/mL streptomycin (Strep). Single colonies were used to inoculate LB/Strep media and grown overnight at 37°C. Two-liter flasks of LB/Strep were inoculated with 25 mL of overnight culture and ZnCl2 (10 μM final). Cells were grown at 30°C, induced at 0.6 OD600 by the addition of isopropyl β- d-1-thiogalactopyranoside (IPTG, 0.1 mM final), then incubated for 14 hr at 16°C.
For nsp12 expression, Eco BL21-CodonPlus(DE3)-RP [Eco B F– ompT hsdS(r B – m B –) dcm+ Tetr gal λ(DE3) endA Hte [argU proL Camr] (Agilent) were transformed and grown overnight on LB-agar plates containing 50 μg/mL kanamycin (KAN) and 25 μg/mL chloramphenicol (CAM). Single colonies were used to inoculate LB/KAN/CAM media and grown overnight at 37°C. Two-liter flasks of LB/KAN/CAM were inoculated with 20 mL of overnight culture and ZnCl2 (10 μM final). Cells were grown at 37°C, induced at 0.6 OD600 by the addition of IPTG (0.1 mM final), then incubated for 16 hr at 16°C.
For nsp13 expression, Eco Rosetta(DE3) [Eco F- ompT hsdS B(rB - mB -) gal dcm (DE3) pRARE (CamR)] (Novagen) were transformed and grown overnight on LB-agar plates containing 50 μg/mL KAN and 25 μg/mL CAM. Single colonies were used to inoculate LB/KAN/CAM media and grown overnight at 37°C. Two-liter flasks of LB/KAN/CAM were inoculated with 25 mL of overnight culture. Cells were grown at 37°C, induced at 0.7 OD600 by the addition of IPTG (0.2 mM final), then incubated for 17 hr at 16°C.
Method Details
Expression construct cloning
The oligonucleotides used in this study are listed in Table S2. All constructs were verified by sequencing (GeneWiz).
Nsp7/8
The coding sequences of the E. coli codon-optimized SARS-CoV-2 nsp7 and nsp8 genes (gBlocks from Integrated DNA Technologies) were cloned into a pCDFDuet-1 vector (Novagen). Nsp7 bore an N-terminal His6-tag that was cleavable with PreScission protease (GE Healthcare Life Sciences).
Nsp12
SARS-CoV-2 RNA was derived from the supernatant of propagated Vero E6 cells and provided by B.R. tenOever (Blanco-Melo et al., 2020). The sequence encoding nsp12 was reverse transcribed into cDNA using gene-specific primers and SuperScript III Reverse Transcriptase (ThermoFisher). The ORF1a/1b programmed −1 ribosomal frameshift that naturally occurs during ORF1b translation was corrected to express the nsp12 open reading frame without frameshift using the following forward primer 5′-TCAGCTGATGCACAATCGTTTTTAAACCGGGTTTGCGGTGTAAG-3′ (the correction is underlined). The SARs-CoV-2 nsp12 coding sequence was subsequently cloned into a modified pRSFDuet-1 vector (Novagen) bearing an N-terminal His6-SUMO-tag which is cleavable by the ubiquitin-like protease (ULP1).
Nsp13
The OptimumGene method was used to optimize the sequence full-length SARS-CoV-2 nsp13 helicase sequence for insect cell expression. The gene was synthesized and cloned into the pet28(a)+ vector, using NdeI and XhoI restriction sites, by Genescript. We then performed mutagenesis using the Quikchange mutagenesis kit (Agilent) to insert an HRV-3C (PreScission) protease cleavage site and remove the thrombin cleavage site.
Protein expression and purification
Nsp12
The pRSFDuet-1 plasmid expressing His6-SUMO-nsp12 was transformed into Eco BL21-CodonPlus cells (Agilent) and grown overnight on LB-agar plates containing 50 μg/mL KAN and 25 μg/mL CAM. Single colonies were used to inoculate LB/KAN/CAM media and grown overnight at 37°C. Two-liter flasks of LB/KAN/CAM were inoculated with 20 mL of overnight culture and ZnCl2 (10 μM final). Cells were grown at 37°C, induced at 0.6 OD600 by the addition of IPTG (0.1 mM final), then incubated for 16 hr at 16°C. Cells were collected by centrifugation, resuspended in 20 mM Tris-HCl, pH 8.0, 0.1 mM ethylenediaminetetraacetic acid (EDTA)-NaOH, pH 8.0, 100 mM NaCl, 5% (v/v) glycerol, 1 mM dithiothreitol (DTT), 10 μM ZnCl2, 1x complete Protease Inhibitor Cocktail (PIC, Roche), 1 mM phenylmethylsulfonyl fluoride (PMSF), and lysed in a continuous-flow French press (Avestin). The lysate was cleared by centrifugation, then loaded onto a HiTrap Heparin HP column (GE Healthcare Life Sciences), and eluted using a gradient from 0.1 M to 1 M NaCl in TGED buffer [20 mM Tris-HCl, pH 8.0, 5% (v/v) glycerol, 0.1 mM EDTA-NaOH, pH 8.0, 1 mM DTT]. The fractions containing nsp12 were pooled and loaded onto a HisTrap HP column (GE Healthcare Life Sciences), washed, and eluted with Nickel elution buffer [20 mM Tris-HCl pH 8.0, 300 mM NaCl, 5% (v/v) glycerol, 250 mM imidazole, 1 mM 2-mercaptoethanol (BME)]. Eluted nsp12 was dialyzed overnight into dialysis buffer [20 mM Tris-HCl pH 8.0, 300 mM NaCl, 5% (v/v) glycerol, 20 mM imidazole, 1 mM BME] in the presence of His6-Ulp1 SUMO protease to cleave the His6-SUMO tag. Cleaved nsp12 was again passed through the HisTrap HP column and the flow-through was collected, concentrated by centrifugal filtration (Amicon), and loaded onto a Superdex 200 Hiload 16/600 (GE Healthcare Life Sciences) that was equilibrated with gel filtration buffer [20 mM Tris-HCl, pH 8.0, 300 mM NaCl, 5 mM MgCl2, 5% (v/v) glycerol, 2 mM DTT]. Purified nsp12 was put into storage buffer [20 mM Tris-HCl, pH 8.0, 300 mM NaCl, 5 mM MgCl2, 20% (v/v) glycerol, 2 mM DTT], aliquoted, flash frozen with liquid N2, and stored at −80°C.
Nsp7/8
The pCDFduet plasmid expressing His6-nsp7/8 was transformed into Eco BL21 (DE3) and grown overnight on LB-agar plates containing 50 μg/mL Strep. Single colonies were used to inoculate LB/Strep media and grown overnight at 37°C. Two-liter flasks of LB/Strep were inoculated with 25 mL of overnight culture and ZnCl2 (10 μM final). Cells were grown at 30°C, induced at 0.6 OD600 by the addition of IPTG (0.1 mM final), then incubated for 14 hr at 16°C. Cells were collected by centrifugation, resuspended in 20 mM Tris-HCl pH 8.0, 0.1 mM EDTA-NaOH, pH 8.0, 300 mM NaCl, 5% (v/v) glycerol, 5 mM imidazole, 1 mM BME, 10 μM ZnCl2, 1x PIC (Roche), 1 mM PMSF, and lysed in a continuous-flow French press (Avestin). The lysate was cleared by centrifugation, then loaded onto a HisTrap HP column (GE Healthcare Life Sciences), washed, and eluted with Nickel elution buffer. Eluted nsp7/8 was dialyzed overnight into dialysis buffer in the presence of His6-Prescission Protease to cleave His6-tag. Cleaved nsp7/8 was passed through the HisTrap HP column and the flow-through was collected, concentrated by centrifugal filtration (Amicon), and loaded onto a Superdex 75 Hiload 16/600 (GE Healthcare Life Sciences) that was equilibrated with gel filtration buffer. Purified nsp7/8 was put into storage buffer, aliquoted, flash frozen with liquid N2, and stored at −80°C.
Nsp13
The pet28 plasmid expressing His6-nsp13 was transformed into Eco Rosetta (DE3) (Novagen) and grown overnight on LB-agar plates containing 50 μg/mL KAN and 25 μg/mL CAM. Single colonies were used to inoculate LB/KAN/CAM media and grown overnight at 37°C. Two-liter flasks of LB/KAN/CAM were inoculated with 25 mL of overnight culture. Cells were grown at 37°C, induced at 0.7 OD600 by the addition of IPTG (0.2 mM final), then incubated for 17 hr at 16°C. Cells were collected by centrifugation, resuspended in 50 mM HEPES-NaOH, pH 7.5, 500 mM NaCl, 4 mM MgCl2, 5% (v/v) glycerol, 20 mM imidazole, 5 mM BME, 1 mM ATP, 1 mM PMSF] and lysed in a continuous-flow French press (Avestin). The lysate was cleared by centrifugation, then loaded onto a HisTrap HP column, washed, and eluted with Nickel elution buffer + 1 mM ATP. Eluted nsp13 was dialyzed overnight into 50 mM HEPES-NaOH pH 7.5, 500 mM NaCl, 4 mM MgCl2, 5% (v/v) glycerol, 20 mM imidazole, 5 mM BME in the presence of His6-Prescission Protease to cleave the His6-tag. Cleaved nsp13 was passed through a HisTrap HP column and the flow-through was collected, concentrated by centrifugal filtration (Amicon), and loaded onto a Superdex 200 Hiload 16/600 (GE Healthcare) that equilibrated with 25 mM HEPES-NaOH pH 7.0, 250 mM KCl, 1 mM MgCl2, 1 mM TCEP. Purified nsp13 was put into 25 mM HEPES-NaOH pH 7.5, 250 mM KCl, 1 mM MgCl2, 20% (v/v) glycerol, 1 mM TCEP, aliquoted, flash frozen with liquid N2, and stored at −80°C.
Native electrophoretic mobility shift assays
Nsp12 (2 μM) or holo-RdRp (nsp12 incubated with three-fold molar excess nsp7/8 and additional three-fold molar excess nsp8) in transcription buffer (120 mM K-acetate, 20 mM HEPES, 10 mM MgCl2, 2 mM DTT) was incubated with 1 μM annealed RNA scaffold (Figure 1A) for 5 minutes at 30°C. Nsp13 (2 μM) was added where indicated. ADP and AlF3 (Sigma-Aldrich) was pre-mixed and added to a final concentration of 2 mM. Reaction mixtures containing nsp13 were incubated an additional 5 minutes at 30°C. Reactions were run on a 4.5% polyacrylamide native gel (37.5:1 acrylamide:bis-acrylamide) in 1x TBE (89 mM Tris, 89 mM boric acid, 1 mM EDTA) at 4°C. The gel was stained with Gel-Red (Biotium).
Native mass spectrometry (nMS) analysis
The reconstituted RNA-bound holo-RdRp (RTC) and the purified nsp13 were buffer exchanged separately into nMS solution (150 mM ammonium acetate, pH 7.5, 0.01% Tween-20) using Zeba microspin desalting columns with a 40-kDa MWCO (Thermo Scientific). The buffer-exchanged samples were mixed yielding a final concentration of 4 μM RTC and 5 μM nsp13, and then incubated for 5 min at RT prior to nMS characterization.
For nMS analysis, 2–3 μL of each sample was loaded into a gold-coated quartz capillary tip that was prepared in-house and then electrosprayed into an Exactive Plus with extended mass range (EMR) instrument (Thermo Fisher Scientific) with a static direct infusion nanospray source (Olinares and Chait, 2019). The MS parameters used include: spray voltage, 1.2 kV; capillary temperature, 125°C; in-source dissociation, 10 V; S-lens RF level, 200; resolving power, 17,500 at m/z of 200; AGC target, 1 × 106; maximum injection time, 200 ms; number of microscans, 5; injection flatapole, 8 V; interflatapole, 4 V; bent flatapole, 4 V; high energy collision dissociation (HCD), 200 V; ultrahigh vacuum pressure, 6 – 7 × 10−10 mbar; total number of scans, at least 100. Mass calibration in positive EMR mode was performed using cesium iodide. For data processing, the acquired MS spectra were visualized using Thermo Xcalibur Qual Browser (v. 4.2.47). MS spectra deconvolution was performed either manually or using the software UniDec v. 4.2.0 (Marty et al., 2015; Reid et al., 2019). The deconvolved spectra were replotted using the m/z software (Proteometrics LLC). Experimental masses were reported as the average mass ± standard deviation (SD) across all the calculated mass values within the observed charge state series. Mass accuracies (indicated in this section in parenthesis after each measured mass) were calculated as the % difference between the measured and expected masses relative to the expected mass. For the reconstituted RTC, we obtained one predominant peak series corresponding to the fully assembled RTC with a mass of 188,984 ± 2 Da (0.008%). When nsp13 was mixed with the RTC, the resulting nMS spectrum showed a predominant peak series corresponding to the supercomplex of nsp13-RTC at 1:1 stoichiometry with a mass of 256,470 ± 10 Da (0.016%). Additional peak series were observed and were assigned to the nsp13-RTC + 490 Da (most likely from one bound Mg-ADP: 454 Da, which was also observed in the nsp13 only sample) with mass of 256,960 ± 10 Da (0.03%), and nsp12 with experimental mass of 106,787 ± 2 Da (0.0017%). The experimental masses of the individual proteins and the RNA scaffold were also determined by native MS and compared to expected masses based on their sequence and known number of zinc ions bound. The following expected masses were used for the component proteins and nucleic acid scaffold—nsp7: 9,739 Da; nsp8 (N-terminal Met lost): 21,881 Da; RNA duplex: 28,681 Da; nsp13 (post-protease cleavage, has three Zn2+ ions coordinated with 3 His and 9 deprotonated Cys residues): 67,463 Da, and nsp12 (has two Zn2+ ions coordinated with 2 His and 6 deprotonated Cys residues): 106,785 Da.
In vitro Transcription Assays
In vitro primer elongation assays were performed using a template-primer RNA scaffold of the following sequence: template (5′- CUAUCCCCAUGUGAUUUUAAUAGCUUCUUAGGAGAAUGAC-3′) and primer (5′- GUCAUUCUCCUAAGAAGCUA-3′) (Figure S1C) (Horizon Discovery Ltd./Dharmacon) annealed in 10 mM Tris-HCl, pH 8.0, 50 mM KCl, 1 mM EDTA. Primer elongation reactions (20 μL) containing 500 nM RNA scaffold, nsp12 (1 μM), nsp7/8 (3 μM) and NTPS (500 μM of UTP, CTP, GTP, 50 μM ATP and 1 μCi α-32P-ATP (Perkin-Elmer) were incubated at 30°C for 30 min prior to quenching with an equal volume of 2x stop solution (Invitrogen-Gel Loading buffer II). Primer elongation products were separated on 15% acrylamide-6M urea denaturing gels and analyzed by phosphorimaging.
Nsp13 Steady-state ATPase Activity Assays
Steady-state ATPase activity of nsp13 was determined using an NADH-coupled assay as described previously (Cupido, et. al. 2019; Pisa, et. al., 2019). Briefly, nsp13 (4.4 nM final) was added in the absence or presence of holo-RdRp (4 nM final), RNA (4.8 nM final), or CHAPSO (8 mM) to wells in a black, clear bottom 384-well microplate (Greiner, 781092), along with NADH (175 μM, Sigma Aldrich), phospho(enol)pyruvic acid monopotassium salt (PEP) (1 mM, Sigma Aldrich) and pyruvate kinase/lactic acid dehydrogenase (PK/LDH) enzymes from rabbit muscle (30 units/ml, Sigma Aldrich) in assay buffer (25 mM HEPES pH 8.0, 150 mM K-acetate, 10 mM MgCl2, 1 mM TCEP, 0.005% Triton X-100, and 0.1 mg/mL BSA). The reaction was initiated by the addition of 6 μL ATP (concentrations of 3 mM to 3 μM, 4-fold serial dilutions), bringing the total reaction volume to 24 μL. The time course of fluorescence decrease, corresponding to the consumption of NADH, was measured using a Synergy NEO Microplate Reader λex = 340 nm, 440 nm emission filter). The rate of fluorescence decrease was calculated after subtraction of background fluorescence from a control reaction containing no ATP and converted to ATPase rate per second per monomer using an ADP calibration curve. These assays were completed with an n = 2 and two biological replicates of nsp13.
To measure inhibition of nsp13 ATPase activity by AlF3, nsp13 ATPase activity was measured using an ATP detection method (Promega Kinase-Glo Plus® kit). The assay was performed in 384-well low-volume white NBS microplates, in 20 μL volumes. Recombinant nsp13 (5 nM) and ATP (50 μM) were added to the assay buffer (20 mM HEPES, pH 7.5, 150 mM KCl, 5 mM MgCl2, 1 mM TCEP, 0.005% Triton X-100, 0.1% BSA and 2% DMSO) and incubated with gentle shaking for 10 min. Following incubation, 10 μL Kinase-Glo Plus® reagent was added to the assay plate and incubated in the dark for 10 min. Relative light unit (RLU) signal was measured on a Synergy Neo2 plate reader (Biotek) in luminescent mode. For inhibition studies, nsp13 was incubated with 0-500 μM ADP:AlF3 (premixed in a 1:2 molar ratio) for 10 min prior to addition of ATP.
Preparation of SARS-CoV-2 nsp13-RTC for Cryo-EM
Purified nsp12 and nsp7/8 were concentrated by centrifugal filtration (Amicon), mixed in a 1:3 molar ratio and dialyzed into 20 mM HEPES-NaOH pH 8.0, 300 mM NaCl, 10 mM MgCl2, 2 mM DTT, for 20 min at 22°C to remove glycerol and to allow for association. Annealed RNA scaffold (Figure 1A) was added to dialyzed nsp7/8/12 mixture and incubated for 15 min at 22°C. Sample was buffer exchanged into S6 buffer [20 mM HEPES-NaOH, pH 8.0, 150 mM K-acetate, 10 mM MgCl2, 2 mM DTT] using Zeba spin desalting columns. After buffer exchange, the sample was further incubated for 20 min at 30°C and then purified over a Superose 6 Increase 10/300 GL column (GE Healthcare Life Sciences) in S6 buffer. The peak corresponding to the RTC was pooled and concentrated by centrifugal filtration (Amicon). Purified nsp13 was concentrated by centrifugal filtration (Amicon) and buffer exchanged into S6 buffer using Zeba spin desalting columns, mixed with ADP and AlF3 (1 mM final), then added to the RTC at a molar ratio of 1:1. Complex was incubated for 5 min at 30°C.
Cryo-EM grid preparation
Prior to grid freezing, 3-([3-cholamidopropyl]dimethylammonio)-2-hydroxy-1-propanesulfonate (CHAPSO, Anatrace) was added to the sample (8 mM final), resulting in a final complex concentration of 8 μM (Chen et al., 2019). The final buffer condition for the cryo-EM sample was 20 mM HEPES-NaOH, pH 8.0, 150 mM K-acetate, 10 mM MgCl2, 2 mM DTT, 1 mM ADP, 1 mM AlF3, 8 mM CHAPSO. C-flat holey carbon grids (CF-1.2/1.3-4Au, Protochips) were glow-discharged for 20 s prior to the application of 3.5 μL of sample. Using a Vitrobot Mark IV (Thermo Fisher Scientific), grids were blotted and plunge-froze into liquid ethane at 90% chamber humidity at 4°C.
Cryo-EM data acquisition and processing
Structural biology software was accessed through the SBGrid consortium (Morin et al., 2013).
Grids were imaged using a 300 kV Titan Krios (Thermo Fisher Scientific) equipped with a GIF BioQuantum and K3 camera (Gatan). Images were recorded with Leginon (Suloway et al., 2005) with a pixel size of 1.1 Å/px (micrograph dimension of 5760 × 4092 px) over a defocus range of −0.8 μm to −2.5 μm and 20 eV slit. Movies were recorded in “counting mode” (native K3 camera binning 2) with ∼30 e-/px/s in dose-fractionation mode with subframes of 50 ms over a 2.5 s exposure (50 frames) to give a total dose of ∼64 e-/Å2. Dose-fractionated movies were gain-normalized, drift-corrected, summed, and dose-weighted using MotionCor2 (Zheng et al., 2017). The contrast transfer function (CTF) was estimated for each summed image using the Patch CTF module in cryoSPARC v2.15.0 (Punjani et al., 2017). Particles were picked and extracted from the dose-weighted images with box size of 256 px using cryoSPARC Blob Picker and Particle Extraction.
Nsp13-RTC (no detergent)
The entire dataset consisted of 10,423 motion-corrected images with 3,691,022 particles (Figure S3). Particles were sorted using three rounds of cryoSPARC 2D classification (N = 100), resulting in 641,026 curated particles. An initial 3D map was generated using cryoSPARC Ab initio Reconstruction on a subset of the particles (128,437 particles from first 3,501 images). Particles were further curated using the map derived from cryoSPARC Ab initio Reconstruction as a template for cryoSPARC Heterogeneous Refinement (N = 6). Class 1 consisted of 88,918 nsp8/12 particles that were further curated using cryoSPARC Ab initio Reconstruction (N = 2) and refined using cryoSPARC Non-uniform Refinement. The final nsp8/12 map contained 88,918 particles with nominal resolution of 3.7 Å. Class 3 and Class 6 were each classified using Heterogeneous Refinement (N = 3) revealing distinct nsp13-RTCs. Class 3.1 consisted of 29,152 (nsp132-RTC)2 particles (re-extracted with a boxsize of 320 px) that were further curated using cryoSPARC Ab initio Reconstruction (N = 2) and refined using cryoSPARC Non-uniform Refinement. The final (nsp132-RTC)2 map consisted of 17,073 particles with nominal resolution of 4.2 Å. Class 3.3, 6.1, and 6.2 were combined and further curated with two rounds of Heterogeneous Refinement (N = 3) and sorted using cryoSPARC Ab initio Reconstruction (N = 3), revealing two distinct classes: nsp13-RTC and nsp132-RTC. These classes were further curated using cryoSPARC Ab initio Reconstruction (N = 2) and refined using cryoSPARC Non-uniform Refinement. The final nsp131-RTC map was derived from 31,783 particles with nominal resolution of 3.8 Å. The final nsp132-RTC map was derived from 16,521 particles with nominal resolution of 4.3 Å. Local resolution calculations (Figure S3B) were generated using blocres and blocfilt from the Bsoft package (Cardone et al., 2013). The directional FSC (Figure S3C; Tan et al., 2017) and particle orientation distribution map from cryoSPARC (Figure S3D) for the nsp132-RTC particles indicated that the map (and resolution esimations) were corrupted by severe particle orientation bias.
Nsp13-RTC (CHAPSO)
The entire dataset consisted of 4,358 motion-corrected images with 1,447,307 particles (Figure S4A). Particles were sorted using cryoSPARC 2D classification (N = 100), resulting in 344,953 curated particles. Initial models (Seed 1: complex, Seed 2: decoy 1, Seed 3: decoy 2) were generated using cryoSPARC Ab initio Reconstruction on a subset of the particles (10,509 particles from first 903 images). Particles were further curated using Seeds 1-3 as 3D templates for cryoSPARC Heterogeneous Refinement (N = 3), then re-extracted with a boxsize of 320 px, and followed by another round of Heterogeneous Refinement (N = 3) using Seed 1 as a template. The resulting 91,058 curated particles were sorted into three classes using cryoSPARC Heterogeneous Refinement (N = 3). Each class was further sorted using cryoSPARC Ab initio Reconstruction (N = 3) to separate distinct 3D classes. Using these classes as references for Heterogeneous Refinement (N = 6), multi-reference classification was performed on the 91,058 curated particles. Classification revealed three unique classes: (1) nsp13-RTC, (2) nsp132-RTC, (3) (nsp132-RTC)2. Particles within each class were further processed using RELION 3.1-beta Bayesian Polishing (Scheres, 2012; Zivanov et al., 2018). “Polished” particles were refined using cryoSPARC Non-uniform Refinement, resulting in structures with the following particle counts and nominal resolutions: nsp13-RTC (17,345; 4.0 Å), nsp132-RTC (58.942; 3.5 Å), (nsp132-RTC)2 (11,771; 7.9 Å). Regions corresponding to nsp13.1 and nsp13.2 in the nsp132-RTC map were further refined with cryoSPARC Local Refinement. Local resolution calculations (Figure S4B) were generated using blocres and blocfilt from the Bsoft package (Cardone et al., 2013). The directional FSC (Figure S4C; Tan et al., 2017) and particle orientation distribution map from cryoSPARC (Figure S4D) for the nsp132-RTC indicated that the particles did not suffer orientation bias.
Model building and refinement
For initial model of the nsp132-RTC, the initial RTC model was derived from PDB:6YYT (Hillen et al., 2020) and the initial nsp13 model from PDB:6JYT (Jia et al., 2019). The models were manually fit into the cryo-EM density maps using Chimera (Pettersen et al., 2004) and rigid-body and real-space refined using Phenix real-space-refine (Adams et al., 2010). For real-space refinement, rigid body refinement was followed by all-atom and B-factor refinement with Ramachandran and secondary structure restraints. Models were inspected and modified in Coot (Emsley and Cowtan, 2004).
Quantification and Statistical Analysis
The nMS spectra were visualized using Thermo Xcalibur Qual Browser (versions 3.0.63 and 4.2.27), deconvolved using UniDec versions 3.2 and 4.1 (Marty et al., 2015; Reid et al., 2019) and plotted using the m/z software (Proteometrics LLC, New York, NY). Experimental masses (Figure 1C) were reported as the average mass ± standard deviation across all the calculated mass values obtained within the observed charge state distribution.
The local resolution of the cryo-EM maps (Figure S4B) was estimated using blocres (Cardone et al., 2013) with the following parameters: box size 15, sampling 1.1, and cutoff 0.5. Directional 3D FSC (Figures S3C and S4C) were calculated by 3DFSC (Tan et al., 2017). The quantification and statistical analyses for model refinement and validation were generated using MolProbity (Chen et al., 2010) and PHENIX (Adams et al., 2010).
Acknowledgments
We thank A. Aher, J. Berger, R. Landick, and C. Rice for helpful discussions and suggestions; L. Urnavicius (Laboratory of Cell Biology and Laboratory of Chemistry and Cell Biology); M. Ebrahim (The Rockefeller University Evelyn Gruss Lipper Cryo-electron Microscopy Resource Center) and H. Kuang (New York Structural Biology Center [NYSBC]) for help with cryo-EM data collection; T. Tuschl for support; and members of the Chait, Darst/Campbell, and Kapoor laboratories for helpful discussions. We thank B.R. tenOever for SARS-CoV-2 RNA. Some of the work reported here was conducted at the Simons Electron Microscopy Center (SEMC) and the National Resource for Automated Molecular Microscopy (NRAMM) and National Center for CryoEM Access and Training (NCCAT) located at the NYSBC, supported by grants from the NIH National Institute of General Medical Sciences (P41 GM103310), NYSTAR, the Simons Foundation (SF349247), the NIH Common Fund Transformative High Resolution Cryo-Electron Microscopy Program (U24 GM129539), and NY State Assembly Majority. This work was supported by the Pels Family Center for Biochemistry and Structural Biology (The Rockefeller University) and NIH grants P41 GM109824 and P41 GM103314 (to B.T.C.), R35 GM130234 (to T.K.), R35 GM118130 (to S.A.D.), and R01 GM114450 (to E.A.C.).
Author Contributions
Conceptualization, B.M., J.C., T.M.K., S.A.D., and E.A.C.; Cloning, Protein Purification, and Biochemistry, B.M., J.C., E.L., M.G., P.M.M.S., and H.V.; Mass Spectrometry, P.D.B.O.; Cryo-EM Specimen Preparation, B.M., E.L., and J.C.; Cryo-EM Data Collection and Processing, J.C., K.M., and E.T.E.; Model Building and Structural Analysis, S.A.D. and E.A.C.; Funding Acquisition and Supervision, B.T.C., T.M.K., S.A.D., and E.A.C.; Manuscript First Draft, S.A.D. and E.A.C. All authors contributed to finalizing the written manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: July 28, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2020.07.033.
Supplemental Information
References
- Abdelkareem M., Saint-André C., Takacs M., Papai G., Crucifix C., Guo X., Ortiz J., Weixlbaumer A. Structural Basis of Transcription: RNA Polymerase Backtracking and Its Reactivation. Mol. Cell. 2019;75:298–309.e4. doi: 10.1016/j.molcel.2019.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.-W., Kapral G.J., Grosse-Kunstleve R.W. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adedeji A.O., Marchand B., Te Velthuis A.J.W., Snijder E.J., Weiss S., Eoff R.L., Singh K., Sarafianos S.G. Mechanism of nucleic acid unwinding by SARS-CoV helicase. PLoS ONE. 2012;7:e36521. doi: 10.1371/journal.pone.0036521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine P.V., Klaholz B.P., Moriarty N.W., Poon B.K., Sobolev O.V., Terwilliger T.C., Adams P.D., Urzhumtsev A. New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr. D Struct. Biol. 2018;74:814–840. doi: 10.1107/S2059798318009324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostini M.L., Andres E.L., Sims A.C., Graham R.L., Sheahan T.P., Lu X., Smith E.C., Case J.B., Feng J.Y., Jordan R. Coronavirus Susceptibility to the Antiviral Remdesivir (GS-5734) Is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease. MBio. 2018;9 doi: 10.1128/mBio.00221-18. e00221–e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso S., Izeta A., Sola I., Enjuanes L. Transcription regulatory sequences and mRNA expression levels in the coronavirus transmissible gastroenteritis virus. J. Virol. 2002;76:1293–1308. doi: 10.1128/JVI.76.3.1293-1308.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bautista E.M., Faaberg K.S., Mickelson D., McGruder E.D. Functional properties of the predicted helicase of porcine reproductive and respiratory syndrome virus. Virology. 2002;298:258–270. doi: 10.1006/viro.2002.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.-C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borukhov S., Sagitov V., Goldfarb A. Transcript cleavage factors from E. coli. Cell. 1993;72:459–466. doi: 10.1016/0092-8674(93)90121-6. [DOI] [PubMed] [Google Scholar]
- Bouhaddou M., Memon D., Meyer B., White K.M., Rezelj V.V., Marrero M.C., Polacco B.J., Melnyk J.E., Ulferts S., Kaake R.M. The global phosphorylation landscape of SARS-CoV-2 infection. Cell. 2020 doi: 10.1016/j.cell.2020.06.034. Published online June 28, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet M., Imbert I., Subissi L., Gluais L., Canard B., Decroly E. RNA 3′-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc. Natl. Acad. Sci. USA. 2012;109:9372–9377. doi: 10.1073/pnas.1201130109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruenn J.A. A structural and primary sequence comparison of the viral RNA-dependent RNA polymerases. Nucleic Acids Res. 2003;31:1821–1829. doi: 10.1093/nar/gkg277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardone G., Heymann J.B., Steven A.C. One number does not fit all: mapping local variations in resolution in cryo-EM reconstructions. J. Struct. Biol. 2013;184:226–236. doi: 10.1016/j.jsb.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabre M. Aluminofluoride and beryllofluoride complexes: a new phosphate analogs in enzymology. Trends Biochem. Sci. 1990;15:6–10. doi: 10.1016/0968-0004(90)90117-t. [DOI] [PubMed] [Google Scholar]
- Chen V.B., Arendall W.B., 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Noble A.J., Kang J.Y., Darst S.A. Eliminating effects of particle adsorption to the air/water interface in single-particle cryo-electron microscopy: Bacterial RNA polymerase and CHAPSO. J. Struct. Biol. X. 2019;1:100005. doi: 10.1016/j.yjsbx.2019.100005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A.C.M., Cramer P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature. 2011;471:249–253. doi: 10.1038/nature09785. [DOI] [PubMed] [Google Scholar]
- Cupido T., Pisa R., Kelley M.E., Kapoor T.M. Designing a chemical inhibitor for the AAA protein spastin using active site mutations. Nat. Chem. Biol. 2019;15:444–452. doi: 10.1038/s41589-019-0225-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R.J., Baker S.C., Baric R., Enjuanes L., Gorbalenya A.E., Holmes K.V., Perlman S., Poon L., Rottier P.J.M., Talbot P.J. Virus Taxonomy. In: King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J., editors. Family Coronaviridae. Elsevier; 2012. pp. 806–828. [Google Scholar]
- D.E. Shaw Research . 2020. Molecular dynamics simulatiosn related to SARS-CoV-2.https://www.deshawresearch.com/resources_sarscov2.html [Google Scholar]
- Deng Z., Lehmann K.C., Li X., Feng C., Wang G., Zhang Q., Qi X., Yu L., Zhang X., Feng W. Structural basis for the regulatory function of a complex zinc-binding domain in a replicative arterivirus helicase resembling a nonsense-mediated mRNA decay helicase. Nucleic Acids Res. 2014;42:3464–3477. doi: 10.1093/nar/gkt1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison M.R., Graham R.L., Donaldson E.F., Eckerle L.D., Baric R.S. Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 2011;8:270–279. doi: 10.4161/rna.8.2.15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudkiewicz M., Szczepińska T., Grynberg M., Pawłowski K. A novel protein kinase-like domain in a selenoprotein, widespread in the tree of life. PLoS ONE. 2012;7:e32138. doi: 10.1371/journal.pone.0032138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulin D., Vilfan I.D., Berghuis B.A., Poranen M.M., Depken M., Dekker N.H. Backtracking behavior in viral RNA-dependent RNA polymerase provides the basis for a second initiation site. Nucleic Acids Res. 2015;43:10421–10429. doi: 10.1093/nar/gkv1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulin D., Arnold J.J., van Laar T., Oh H.-S., Lee C., Perkins A.L., Harki D.A., Depken M., Cameron C.E., Dekker N.H. Signatures of Nucleotide Analog Incorporation by an RNA-Dependent RNA Polymerase Revealed Using High-Throughput Magnetic Tweezers. Cell Rep. 2017;21:1063–1076. doi: 10.1016/j.celrep.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckerle L.D., Lu X., Sperry S.M., Choi L., Denison M.R. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J. Virol. 2007;81:12135–12144. doi: 10.1128/JVI.01296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckerle L.D., Becker M.M., Halpin R.A., Li K., Venter E., Lu X., Scherbakova S., Graham R.L., Baric R.S., Stockwell T.B. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 2010;6:e1000896. doi: 10.1371/journal.ppat.1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Enjuanes L., Almazán F., Sola I., Zuñiga S. Biochemical aspects of coronavirus replication and virus-host interaction. Annu. Rev. Microbiol. 2006;60:211–230. doi: 10.1146/annurev.micro.60.080805.142157. [DOI] [PubMed] [Google Scholar]
- Erie D.A., Hajiseyedjavadi O., Young M.C., von Hippel P.H. Multiple RNA polymerase conformations and GreA: control of the fidelity of transcription. Science. 1993;262:867–873. doi: 10.1126/science.8235608. [DOI] [PubMed] [Google Scholar]
- Fish R.N., Kane C.M. Promoting elongation with transcript cleavage stimulatory factors. Biochim. Biophys. Acta. 2002;1577:287–307. doi: 10.1016/s0167-4781(02)00459-1. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A.E., Enjuanes L., Ziebuhr J., Snijder E.J. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon C.J., Tchesnokov E.P., Woolner E., Perry J.K., Feng J.Y., Porter D.P., Götte M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020;295:6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao W., Wojdyla J.A., Zhao R., Han R., Das R., Zlatev I., Manoharan M., Wang M., Cui S. Crystal structure of Middle East respiratory syndrome coronavirus helicase. PLoS Pathog. 2017;13:e1006474. doi: 10.1371/journal.ppat.1006474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgenfeld R., Peiris M. From SARS to MERS: 10 years of research on highly pathogenic human coronaviruses. Antiviral Res. 2013;100:286–295. doi: 10.1016/j.antiviral.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillen H.S., Kokic G., Farnung L., Dienemann C., Tegunov D., Cramer P. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020 doi: 10.1038/s41586-020-2368-8. Published online May 21, 2020. [DOI] [PubMed] [Google Scholar]
- Holm L., Laakso L.M. Dali server update. Nucleic Acids Res. 2016;44(W1):W351-5. doi: 10.1093/nar/gkw357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov K.A., Ziebuhr J. Human coronavirus 229E nonstructural protein 13: characterization of duplex-unwinding, nucleoside triphosphatase, and RNA 5′-triphosphatase activities. J. Virol. 2004;78:7833–7838. doi: 10.1128/JVI.78.14.7833-7838.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov K.A., Thiel V., Dobbe J.C., van der Meer Y., Snijder E.J., Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 2004;78:5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Z., Yan L., Ren Z., Wu L., Wang J., Guo J., Zheng L., Ming Z., Zhang L., Lou Z., Rao Z. Delicate structural coordination of the Severe Acute Respiratory Syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res. 2019;47:6538–6550. doi: 10.1093/nar/gkz409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J.Y., Olinares P.D.B., Chen J., Campbell E.A., Mustaev A., Chait B.T., Gottesman M.E., Darst S.A. Structural basis of transcription arrest by coliphage HK022 Nun in an Escherichia coli RNA polymerase elongation complex. eLife. 2017;6:e25478. doi: 10.7554/eLife.25478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan D.L., O’Donnell M. DnaB drives DNA branch migration and dislodges proteins while encircling two DNA strands. Mol. Cell. 2002;10:647–657. doi: 10.1016/s1097-2765(02)00642-1. [DOI] [PubMed] [Google Scholar]
- Kettenberger H., Armache K.-J., Cramer P. Architecture of the RNA polymerase II-TFIIS complex and implications for mRNA cleavage. Cell. 2003;114:347–357. doi: 10.1016/s0092-8674(03)00598-1. [DOI] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10:2342–2349. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komissarova N., Kashlev M. Transcriptional arrest: Escherichia coli RNA polymerase translocates backward, leaving the 3′ end of the RNA intact and extruded. Proc. Natl. Acad. Sci. USA. 1997;94:1755–1760. doi: 10.1073/pnas.94.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komissarova N., Kashlev M. RNA polymerase switches between inactivated and activated states By translocating back and forth along the DNA and the RNA. J. Biol. Chem. 1997;272:15329–15338. doi: 10.1074/jbc.272.24.15329. [DOI] [PubMed] [Google Scholar]
- Lane W.J., Darst S.A. Molecular evolution of multisubunit RNA polymerases: structural analysis. J. Mol. Biol. 2010;395:686–704. doi: 10.1016/j.jmb.2009.10.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N.-R., Kwon H.-M., Park K., Oh S., Jeong Y.-J., Kim D.-E. Cooperative translocation enhances the unwinding of duplex DNA by SARS coronavirus helicase nsP13. Nucleic Acids Res. 2010;38:7626–7636. doi: 10.1093/nar/gkq647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann K.C., Gulyaeva A., Zevenhoven-Dobbe J.C., Janssen G.M.C., Ruben M., Overkleeft H.S., van Veelen P.A., Samborskiy D.V., Kravchenko A.A., Leontovich A.M. Discovery of an essential nucleotidylating activity associated with a newly delineated conserved domain in the RNA polymerase-containing protein of all nidoviruses. Nucleic Acids Res. 2015;43:8416–8434. doi: 10.1093/nar/gkv838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann K.C., Snijder E.J., Posthuma C.C., Gorbalenya A.E. What we know but do not understand about nidovirus helicases. Virus Res. 2015;202:12–32. doi: 10.1016/j.virusres.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeira F., Park Y.M., Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A.R.N., Potter S.C., Finn R.D., Lopez R. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47(W1):W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty M.T., Baldwin A.J., Marklund E.G., Hochberg G.K.A., Benesch J.L.P., Robinson C.V. Bayesian deconvolution of mass and ion mobility spectra: from binary interactions to polydisperse ensembles. Anal. Chem. 2015;87:4370–4376. doi: 10.1021/acs.analchem.5b00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minskaia E., Hertzig T., Gorbalenya A.E., Campanacci V., Cambillau C., Canard B., Ziebuhr J. Discovery of an RNA virus 3′->5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. USA. 2006;103:5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin A., Eisenbraun B., Key J., Sanschagrin P.C., Timony M.A., Ottaviano M., Sliz P. Collaboration gets the most out of software. eLife. 2013;2:e01456. doi: 10.7554/eLife.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nga P.T., Parquet M. del C., Lauber C., Parida M., Nabeshima T., Yu F., Thuy N.T., Inoue S., Ito T., Okamoto K. Discovery of the first insect nidovirus, a missing evolutionary link in the emergence of the largest RNA virus genomes. PLoS Pathog. 2011;7:e1002215. doi: 10.1371/journal.ppat.1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudler E. RNA polymerase backtracking in gene regulation and genome instability. Cell. 2012;149:1438–1445. doi: 10.1016/j.cell.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudler E., Mustaev A., Lukhtanov E., Goldfarb A. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- Olinares P.D.B., Chait B.T. Native Mass Spectrometry Analysis of Affinity-Captured Endogenous Yeast RNA Exosome Complexes. Methods Mol. Biol. 2019;2062:357–382. doi: 10.1007/978-1-4939-9822-7_17. [DOI] [PubMed] [Google Scholar]
- Opalka N., Chlenov M., Chacon P., Rice W.J., Wriggers W., Darst S.A. Structure and function of the transcription elongation factor GreB bound to bacterial RNA polymerase. Cell. 2003;114:335–345. doi: 10.1016/s0092-8674(03)00600-7. [DOI] [PubMed] [Google Scholar]
- Pasternak A.O., van den Born E., Spaan W.J.M., Snijder E.J. Sequence requirements for RNA strand transfer during nidovirus discontinuous subgenomic RNA synthesis. EMBO J. 2001;20:7220–7228. doi: 10.1093/emboj/20.24.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pisa R., Cupido T., Steinman J.B., Jones N.H., Kapoor T.M. Analyzing resistance to design selective chemical inhibitors for AAA proteins. Cell Chem. Biol. 2019;26:1263–1273.e5. doi: 10.1016/j.chembiol.2019.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Reid D.J., Diesing J.M., Miller M.A., Perry S.M., Wales J.A., Montfort W.R., Marty M.T. MetaUniDec: High-Throughput Deconvolution of Native Mass Spectra. J. Am. Soc. Mass Spectrom. 2019;30:118–127. doi: 10.1007/s13361-018-1951-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saberi A., Gulyaeva A.A., Brubacher J.L., Newmark P.A., Gorbalenya A.E. A planarian nidovirus expands the limits of RNA genome size. PLoS Pathog. 2018;14:e1007314. doi: 10.1371/journal.ppat.1007314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikrishnan K., Powell B., Cook N.J., Webb M.R., Wigley D.B. Mechanistic basis of 5′-3′ translocation in SF1B helicases. Cell. 2009;137:849–859. doi: 10.1016/j.cell.2009.03.036. [DOI] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L. A new model for coronavirus transcription. Adv. Exp. Med. Biol. 1998;440:215–219. doi: 10.1007/978-1-4615-5331-1_26. [DOI] [PubMed] [Google Scholar]
- Scheres S.H.W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider T.D., Stephens R.M. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seybert A., van Dinten L.C., Snijder E.J., Ziebuhr J. Biochemical characterization of the equine arteritis virus helicase suggests a close functional relationship between arterivirus and coronavirus helicases. J. Virol. 2000;74:9586–9593. doi: 10.1128/jvi.74.20.9586-9593.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seybert A., Hegyi A., Siddell S.G., Ziebuhr J. The human coronavirus 229E superfamily 1 helicase has RNA and DNA duplex-unwinding activities with 5′-to-3′ polarity. RNA. 2000;6:1056–1068. doi: 10.1017/s1355838200000728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seybert A., Posthuma C.C., van Dinten L.C., Snijder E.J., Gorbalenya A.E., Ziebuhr J. A complex zinc finger controls the enzymatic activities of nidovirus helicases. J. Virol. 2005;79:696–704. doi: 10.1128/JVI.79.2.696-704.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon A., Le N.T.T., Selisko B., Eydoux C., Alvarez K., Guillemot J.-C., Decroly E., Peersen O., Ferron F., Canard B. Remdesivir and SARS-CoV-2: Structural requirements at both nsp12 RdRp and nsp14 Exonuclease active-sites. Antiviral Res. 2020;178:104793. doi: 10.1016/j.antiviral.2020.104793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton M.R., Dillingham M.S., Wigley D.B. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- Smith E.C., Blanc H., Surdel M.C., Vignuzzi M., Denison M.R. Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: evidence for proofreading and potential therapeutics. PLoS Pathog. 2013;9:e1003565. doi: 10.1371/journal.ppat.1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.C., Sexton N.R., Denison M.R. Thinking Outside the Triangle: Replication Fidelity of the Largest RNA Viruses. Annu. Rev. Virol. 2014;1:111–132. doi: 10.1146/annurev-virology-031413-085507. [DOI] [PubMed] [Google Scholar]
- Snijder E.J., Decroly E., Ziebuhr J. The Nonstructural Proteins Directing Coronavirus RNA Synthesis and Processing. Adv. Virus Res. 2016;96:59–126. doi: 10.1016/bs.aivir.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola I., Almazán F., Zúñiga S., Enjuanes L. Continuous and Discontinuous RNA Synthesis in Coronaviruses. Annu. Rev. Virol. 2015;2:265–288. doi: 10.1146/annurev-virology-100114-055218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreelatha A., Yee S.S., Lopez V.A., Park B.C., Kinch L.N., Pilch S., Servage K.A., Zhang J., Jiou J., Karasiewicz-Urbańska M. Protein AMPylation by an Evolutionarily Conserved Pseudokinase. Cell. 2018;175:809–821.e19. doi: 10.1016/j.cell.2018.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subissi L., Posthuma C.C., Collet A., Zevenhoven-Dobbe J.C., Gorbalenya A.E., Decroly E., Snijder E.J., Canard B., Imbert I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. USA. 2014;111:E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C., Pulokas J., Fellmann D., Cheng A., Guerra F., Quispe J., Stagg S., Potter C.S., Carragher B. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Tan Y.Z., Baldwin P.R., Davis J.H., Williamson J.R., Potter C.S., Carragher B., Lyumkis D. Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat. Methods. 2017;14:793–796. doi: 10.1038/nmeth.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner J.A., Watt R.M., Chai Y.-B., Lu L.-Y., Lin M.C., Peiris J.S.M., Poon L.L.M., Kung H.-F., Huang J.-D. The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5′ to 3′ viral helicases. J. Biol. Chem. 2003;278:39578–39582. doi: 10.1074/jbc.C300328200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M.J., Platas A.A., Hawley D.K. Transcriptional fidelity and proofreading by RNA polymerase II. Cell. 1998;93:627–637. doi: 10.1016/s0092-8674(00)81191-5. [DOI] [PubMed] [Google Scholar]
- Tvarogová J., Madhugiri R., Bylapudi G., Ferguson L.J., Karl N., Ziebuhr J. Identification and Characterization of a Human Coronavirus 229E Nonstructural Protein 8-Associated RNA 3′-Terminal Adenylyltransferase Activity. J. Virol. 2019;93 doi: 10.1128/JVI.00291-19. e00291-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- US Food and Drug Administration . 2020. Remdesivir EUA Letter of Authorization. Remdesivir-EUA-Letter-Of-Authorization.pdf. [Google Scholar]
- van Dinten L.C., Wassenaar A.L., Gorbalenya A.E., Spaan W.J., Snijder E.J. Processing of the equine arteritis virus replicase ORF1b protein: identification of cleavage products containing the putative viral polymerase and helicase domains. J. Virol. 1996;70:6625–6633. doi: 10.1128/jvi.70.10.6625-6633.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dinten L.C., van Tol H., Gorbalenya A.E., Snijder E.J. The predicted metal-binding region of the arterivirus helicase protein is involved in subgenomic mRNA synthesis, genome replication, and virion biogenesis. J. Virol. 2000;74:5213–5223. doi: 10.1128/jvi.74.11.5213-5223.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Marle G., van Dinten L.C., Spaan W.J.M., Luytjes W., Snijder E.J. Characterization of an equine arteritis virus replicase mutant defective in subgenomic mRNA synthesis. J. Virol. 1999;73:5274–5281. doi: 10.1128/jvi.73.7.5274-5281.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Brunn A., Teepe C., Simpson J.C., Pepperkok R., Friedel C.C., Zimmer R., Roberts R., Baric R., Haas J. Analysis of intraviral protein-protein interactions of the SARS coronavirus ORFeome. PLoS ONE. 2007;2:e459. doi: 10.1371/journal.pone.0000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Bushnell D.A., Huang X., Westover K.D., Levitt M., Kornberg R.D. Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution. Science. 2009;324:1203–1206. doi: 10.1126/science.1168729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Wu J., Wang H., Gao Y., Liu Q., Mu A., Ji W., Yan L., Zhu Y., Zhu C. Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase. Cell. 2020;368:1499–1504. doi: 10.1016/j.cell.2020.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren T.K., Jordan R., Lo M.K., Ray A.S., Mackman R.L., Soloveva V., Siegel D., Perron M., Bannister R., Hui H.C. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westover K.D., Bushnell D.A., Kornberg R.D. Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center. Cell. 2004;119:481–489. doi: 10.1016/j.cell.2004.10.016. [DOI] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Ma Q., Restle T., Shang W., Svergun D.I., Ponnusamy R., Sczakiel G., Hilgenfeld R. Nonstructural proteins 7 and 8 of feline coronavirus form a 2:1 heterotrimer that exhibits primer-independent RNA polymerase activity. J. Virol. 2012;86:4444–4454. doi: 10.1128/JVI.06635-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W., Mao C., Luan X., Shen D.-D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y., Sun F., Li X., Pang H., Xu X., Bartlam M., Rao Z. Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat. Struct. Mol. Biol. 2005;12:980–986. doi: 10.1038/nsmb999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Campbell E.A., Minakhin L., Richter C., Severinov K., Darst S.A. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 A resolution. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- Zhang R., Li Y., Cowley T.J., Steinbrenner A.D., Phillips J.M., Yount B.L., Baric R.S., Weiss S.R. The nsp1, nsp13, and M proteins contribute to the hepatotropism of murine coronavirus JHM.WU. J. Virol. 2015;89:3598–3609. doi: 10.1128/JVI.03535-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S.Q., Palovcak E., Armache J.-P., Verba K.A., Cheng Y., Agard D.A. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirkel F., Kurth A., Quan P.-L., Briese T., Ellerbrok H., Pauli G., Leendertz F.H., Lipkin W.I., Ziebuhr J., Drosten C., Junglen S. An insect nidovirus emerging from a primary tropical rainforest. MBio. 2011;2 doi: 10.1128/mBio.00077-11. e00077–e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J., Nakane T., Forsberg B.O., Kimanius D., Hagen W.J., Lindahl E., Scheres S.H.W. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife. 2018;7:e42166. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zúñiga S., Sola I., Alonso S., Enjuanes L. Sequence motifs involved in the regulation of discontinuous coronavirus subgenomic RNA synthesis. J. Virol. 2004;78:980–994. doi: 10.1128/JVI.78.2.980-994.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The video highlights features of the nsp132-RTC. The video starts with the experimental nsp132-RTC cryo-EM map [3.5 Å nominal resolution, local-resolution filtered (Cardone et al., 2013)], superimposes the holo-RdRp:RNA structure, nsp13.1, then nsp13.2. Structural features highlighted through the video include: 1)The nsp13.1:holo-RdRp interface, 2) the nsp13.2-nsp8a interface, 3) the ADP-Mg2+ bound in the nsp12-NiRAN domain, and 4) the RdRp secondary channel.
Data Availability Statement
The cryo-EM density maps for the nsp131-RTC(CHAPSO), nsp132-RTC(CHAPSO), and (nsp132-RTC)2(CHAPSO) have been deposited in the EMDataBank under accession codes EMD-22270, EMD-22160, and EMD-22271, respectively. Atomic coordinates for nsp132-RTC have been deposited in the Protein Data Bank under accession code 6XEZ. Source data for gel-based images in the paper (Figure 1B; Figure S1C) are available (https://data.mendeley.com/datasets/njf7w7m5js/draft?a=8a8a4e75-3e95-41f5-9d90-6956c34b5aa3)