As discussed in an earlier section, coronaviruses are a diverse group of viruses capable of causing animal, as well as human disease; from mild illness (low pathogenicity coronaviruses), consistent with what is often referred to as “the common cold1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14,” all the way to severe illness and death (highly pathogenic coronaviruses) from more recently identified SARS, MERS, and now COVID-1915, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34. (Fig. 1).
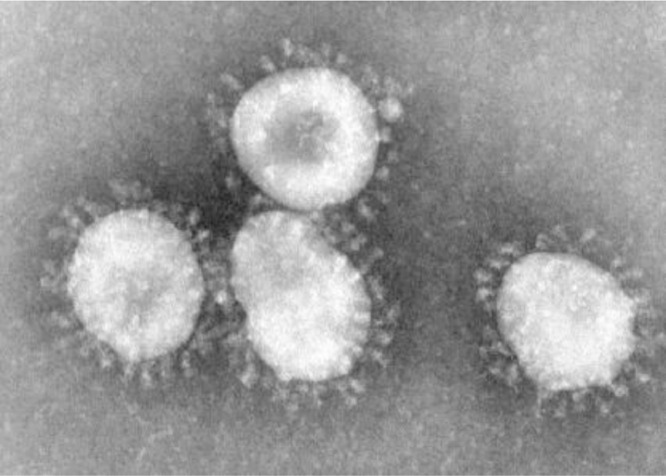
Fig. 1.
Coronavirus. Centers for Disease Control and Prevention (CDC)/Dr. Fred Murphy9.
According to the CDC, the first case of SARS CoV was reported in Asia in early 200335. Further investigation notes November 2002 the first case of an atypical pneumonia emerged in China (Guangdong Province) where ultimately the causative agent was determined to be a newly discovered coronavirus. By 2003 an epidemic of similarly severe, atypical pneumonias, was emerging from Hong Kong, Guangdong, and Toronto, Ontario. The following is the timeline of initial events that transpired referable to SARS CoV. On 11 February 2003 China reported to the World Health Organization (WHO) that 305 cases of atypical pneumonia of unknown etiology had been identified in Guangdong Province since 16 November 2002; five people had died. As of 21 February 2003 a physician from Guangdong Province, who was ill with an atypical pneumonia, travelled to Hong Kong, staying overnight in a hotel. The etiology causing his illness was identified as severe acute respiratory syndrome coronavirus (SARS CoV); it was likely transmitted to at least 10 additional persons. These transmissions/infections subsequently initiated outbreaks in Hong Kong, Singapore, Viet Nam, and Canada20, 21, 22, 23, 24 , 35 , 36
Symptoms characteristic of this aggressive atypical pneumonia included an onset of illness associated with high fever (temperature greater than 100.4°F [>38.0 °C]), headache – often severe, an overall feeling of discomfort, and body aches, again often severe. Some persons may have had respiratory symptoms at the outset. Approximately 10% to 20% percent experienced diarrhea. After 2 to 7 days, SARS patients may develop a dry cough. Most patients develop pneumonia.
Given the prior outbreaks of highly pathogenic avian influenza in that same region of China, it was first considered to be an emerging flu virus. Other pathogens, including members of the Paramyxoviridae family, and human metapneumovirus (hMPV) were considered as causative of this new clinical illness which became known as Severe Acute Respiratory Syndrome or SARS. After international collaboration among multiple research facilities, a previously unknown pathogen was ultimately determined to be causative of SARS - a new coronavirus – SARS CoV20, 21, 22, 23, 24, 25, 26, 27, 28, 29.
Whiles SARS CoV is a significant pathogen capable of causing profound illness, even death, historically coronaviruses were one cause of ‘the common cold.’ Known as endemic human betacoronaviruses HCoV-OC43 and HCoV-HKU1. Coronaviruses affecting humans (HCoVs) historically were associated with mild illness. HCoV-229E and HCoV-OC43 are a widespread cause of mild respiratory illnesses12, although occasionally these CoV cause serious infections of the lower respiratory tract in children and adults, including necrotizing enterocolitis in newborns12, 13, 14, 15, 16. Human coronavirus OC43 (HCoV-OC43) seems to be more predominant than other HCoVs, at least until COVID-19, especially in children and the elderly. An interesting insight into coronavirus persistence, OC43 exhibits high nucleotide substitution rates. It has a capacity for genotype shift based on recombination of HCoV-OC43, which may be an adaptive mechanism allowing to remain a perennial background infection (36b). This level of genetic adaptation potentially poses an additional level of difficulty in vaccine development.
Early research into the SARS Co-V genomic sequence demonstrated that this new CoV does not belong to any of the known groups of coronaviruses, previously described human coronaviruses HCoV-OC43 and HCoV-229E20, 21, 22, 23, 24. In fact it appears SARS CoV is only somewhat related to these HCoV. The SARS-CoV genome appears to be equidistant from those of all known coronaviruses. Moreover, SARS CoV closest relatives appear to be the murine, bovine, porcine, and human coronaviruses in group 2 and avian coronavirus IBV in group 1.
Research on the SARS CoV suggests this new virus represents a fourth group or lineage of coronavirus - Group 423. Genomic sequence analysis seems to support the hypothesis that of SARS-CoV is an animal virus for which the normal host is still unknown and that developed the ability to productively infect humans or has the ability to cross species barriers25. The genome shows that SARS-CoV is neither a mutant of a known coronavirus, nor a recombinant between known coronaviruses. As the virus passes through human beings, SARS-CoV maintains genotype, and is adapted to the human host26. Testing allows genetic analysis to distinguish different strains of SARS-CoV, allowing epidemiological studies28.
Not surprisingly there are also economic, as well as health implications - coronaviruses cause important diseases in domestic animals, as well as in human populations. Toronto during and in the aftermath of their SARS outbreak saw a significant, albeit temporary decline in tourism and business related visits, as well as lost conference and trade show related commerce. Recognizing the importance of animal – human pathogen crossover, opportunities to reduce the spread of contagion, and to identify potential risks is critical to prevent or at least reduce the likelihood of SARS, MERS, and influenza outbreaks such as the avian influenza outbreaks of the 1990′s and early2000’s and the swine flu outbreak in 2009.
SARS Co-V (Fig. 2 )37 can be detected in extracts of lung and kidney tissue by virus isolation or PCR; bronchoalveolar lavage specimens by virus isolation, electron microscopy and PCR; and sputum or upper respiratory tract swab, aspirate, or wash specimens by PCR20 , 21 , 29. SARS-associated coronavirus RNA was detected in nasopharyngeal aspirates by RT-PCR in 32% of persons infected, at initial presentation (mean 3.2 days after onset of illness) and in 68% at day 1430. In stool samples, viral RNA was detected in 97% of patients two weeks after the onset of illness. 42% of urine samples were positive for viral RNA30. Viral RNA was also detected at extremely low concentrations in plasma during the acute phase and in feces during the late convalescent phase, suggesting that the virus may be shed in feces for prolonged periods of time20 , 21.
Fig. 2.
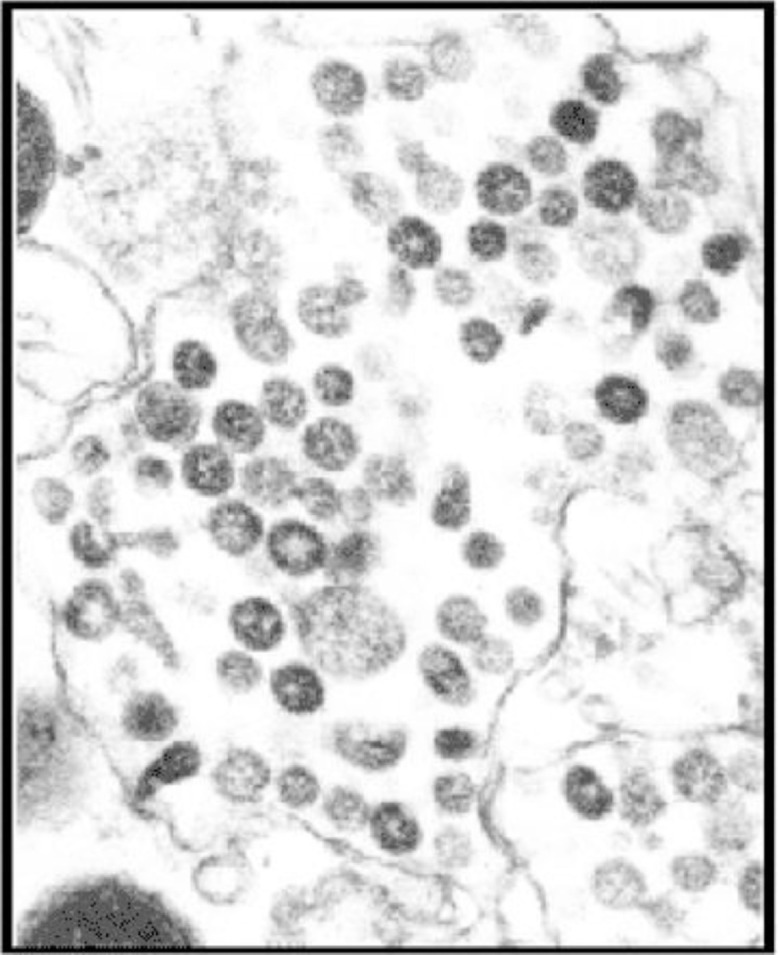
SARS CoV – CDC National Center for Immunization and Respiratory Disease. Division of Viral Diseases37.
The timelines of events as noted by CDC concluded towards the end of 2003 with removal of travel warnings to China and Ontario. By the end of 2003, according to the CDC report of WHO data, reports of SARS infections from 29 countries and regions revealed 8096 persons with probable SARS resulting in 774 deaths – with an estimated case fatality rate just below 10% (higher in elderly, infirm patients). In the United States, eight SARS infections were documented by laboratory testing and an additional 19 probable SARS infections were reported. By 2004 the CDC issued a “Notice of Embargo of Civets” as a SARS-like virus had been isolated from civets (captured in areas of China where the SARS outbreak originated). CDC also banned the importation of civets. The civet is a mammal with a catlike body, long legs, a long tail, and a masked face resembling a raccoon or weasel. SARS CoV was detected in animal handlers of civets. The ban on civets is currently still in effect. By 2012 The National Select Agent Registry Program declared SARS-coronavirus a select agent. A select agent is a bacterium, virus or toxin that has the potential to pose a severe threat to public health and safety32 , 35.
Radiographic features SARS
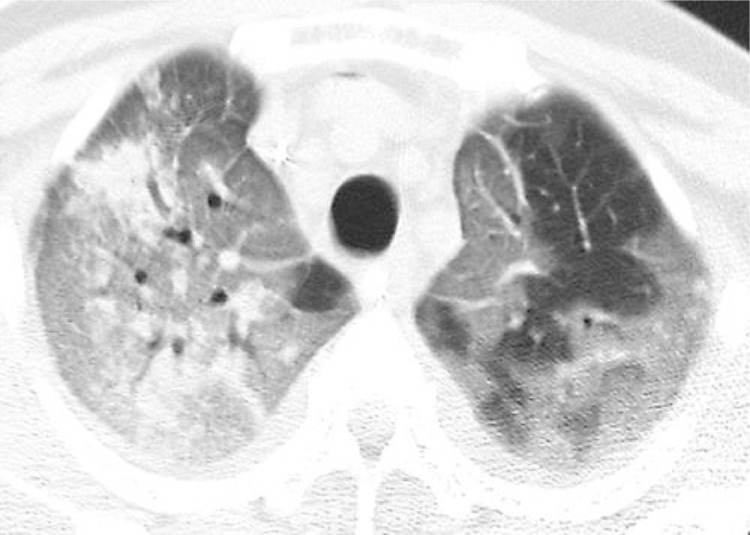
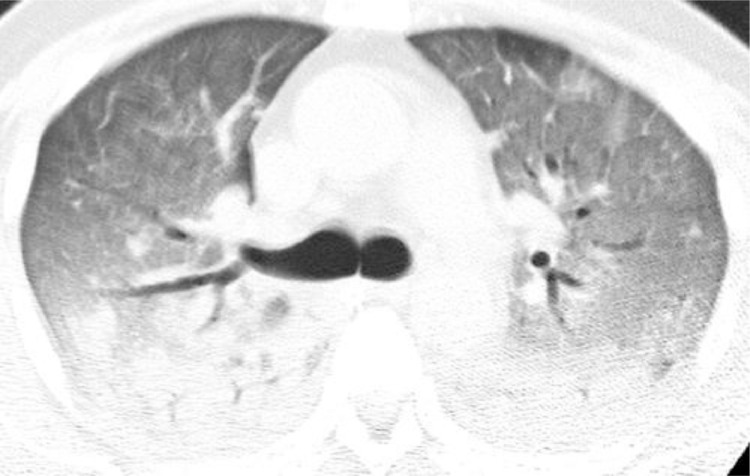
SARS infection can result in rapidly progressive respiratory illness as Fig. 3, Fig. 4, Fig. 5, Fig. 6 reveal38, 39, 40, 41, 42. Initial radiographic studies on chest XRays (CXR) (Figs. 3 and 4) often show small effusions, unilateral or bilateral patchy or confluent areas of consolidation, or ground glass opacities. Similar to other etiologies of ARDS, SARS can cause rapid progression of findings with both CXR and Chest CT (Figs. 5– 6) are common, and reflective of deteriorating lung function38 , 39.
Fig. 3.
(Left) CXR SARS Patient – Consider the extensive bilateral ground-glass opacities and poorly defined nodular pattern. In this case diffuse involvement Rt lung, Lt apical sparing. There is mild air-space consolidation is seen in retro-cardiac region of RLL. Mild cardiomegaly present38,39.
Fig. 4.
(Right) Bedside supine AP CXR – same patient in Fig. 3, radiograph taken 12 hr after initial radiograph – Note progressive disease in SARS patient, consistent with rapidly declining ARDS. Findings: diffuse bilateral air-space consolidation, prominent air bronchograms. Clinical caveat: note the low position of the endotracheal tube (ETT), and gaseous distention of stomach.38,39.
Fig. 5.
CT Scan Transverse unenhanced image obtained at level of apical segments of upper lobes shows extensive bilateral areas of ground-glass attenuation, more severe on right, and focal areas of consolidation in right upper lobe. Note lobular areas of sparing particularly in left upper lobe38,39.
Fig. 6.
CT image obtained at level of right upper lobe bronchus shows diffuse bilateral areas of ground-glass attenuation and dependent areas of consolidation (37b – 37e).
Of note, from the SARS, MERS, and especially early COVID-19 experience, chest CT scanning is an important modality to help characterize the extent of pulmonary disease. Typical findings include ground glass opacities, consolidation; in COVID-19 expect bilateral lung involvement.
The imaging features of SARS and MERS, as well as COVID-19 not surprisingly overlap, but there are some differences (Table 1 )38, 39, 40. According to various reports the initial chest XRays (CXR) will be abnormal in up to 80% patients infected and symptomatic with SARS38, 39, 40, 41, 42. Although it is demonstrated that COVID-19 has bilateral involvement in a significant proportion of patients, with SARS, initial imaging usually reveals abnormalities in one lung, with peripheral distribution and ill-defined areas of airspace opacity in the lower lung zones38, 39, 40, 41, 42. Focal findings on initial studies are found in ~50% patients, with multifocal findings in ~50%. Less than 10% of these studies reveal early diffuse involvement38, 39, 40, 41, 42. Subsequent imaging reveals in the majority of patients progressive multifocal consolidation over a course of 6–12 days, which may at that point involve one or both lungs. It has been noted that in ~25% of patients, visualized opacity will remain focal and unilateral38, 39, 40, 41, 42.
Table 1.
Comparison of clinical and radiologic features of SARS, MERS, and COVID-1940.
| Feature | SARS | MERS | COVID-19 |
|---|---|---|---|
| Clinical sign Or symptom | |||
| Fever or chills | Yes | Yes | Yes |
| Dyspnea | Yes | Yes | Yes |
| Malaise | Yes | Yes | Yes |
| Myalgia | Yes | Yes | Yes |
| Headache | Yes | Yes | Yes |
| Cough | Dry | Dry or productive | Dry |
| Diarrhea | Yes | Yes | Uncommon |
| Nausea or vomiting | Yes | Yes | Uncommon |
| Sore throat | Yes | Uncommon | Uncommon |
| Arthralgia | Yes | Uncommo | |
| Imaging finding | |||
| Acute phase | |||
| Initial imaging | |||
| Normal | 15–20% of patients | 17% of patients | 15–20% of patients |
| Abnormalities | |||
| Common | Peripheral multifocal airspace opacities (GGO, consolidation, or both) on chest radiography and CT | Peripheral multifocal airspace opacities1660, consolidation, or both) on chest radiography and CT | Peripheral multifocal airspace opacities 1660, consolidation, or both) on chest radiography and CT |
| Rare | Pneurmothorex | Pneurmothorex | Pneurmothorex |
| Not seen | Cavitation or lymphadenopathy | Cavitation or lymphadenopathy | Cavitation or lymphadenopathy |
| Appearance | Unilateral, focal (50%)l; multifocal (40%); diffuse (10%) | Bilateral, multifocal basal airspace on chest radiography or CT (80%); isolated unilateral (20%) | Bilateral, multifocal, basal airspace; normal chest radiography findings (15%) |
| Follow-up imaging appearance | Unilateral, focal (25%); progressive (most common, can be unilateral and multifocal or bilateral with multifocal consolidation) | Extension into upper lobes or perihilar areas, pleural effusion (33%) interlobular septet thickening (26%) | Persistent or progressive airspace opacities |
| Indications of poor prognosis | Bilateral (like ARDS), four or more lung zones, progressive involvement after12 d | Greater involvement of the lungs, pleural effusion, pneumothorax | Consolidation (vs GGO) |
| Chronic phase | Unknown, but pleural effusion and interlobar septal thickening have not yet been reported | ||
| Transient reticular opacitiesa | Yes | Yes | |
| Airtrapping | Common I us u a I ly persistent) | ||
| Fibrosis | Rare | One-third of patients | Not yet reported |
Note—SARS = severe acute respiratory syndrome, MERS = Middle East respiratory syndrome, COVID,19 = coronavirus disease 2019. GG0 = ground-glass opacity, ARDS acute respiratory distress syndrome.
Over a period Of weeks or months.
CT studies often reveal patchy areas of ground glass opacity (GGO) and consolidation. Of note, centrilobular nodules and tree-in-bud opacities are not characteristic of the highly pathogenic coronavirus illnesses, possibly suggesting other atypical or opportunistic causes of pneumonia38, 39, 40, 41, 42. Radiologic improvement after recovery is expected in most patients. Poor outcomes are noted in patients with bilateral confluent diffuse airspace opacities, similar to the findings of acute respiratory distress syndrome, involvement of four or more lung zones, bilateral lung involvement, and progressive worsening of airspace consolidation on chest imaging more than 12 days after symptom onset despite treatment38, 39, 40, 41, 42.
With MERS imaging, ~ 83% of patients have abnormal initial chest radiography studies. Multifocal airspace opacities in the lower lung fields are the most commonly reported findings in this patient population38, 39, 40, 41, 42. Abnormalities are noted to extend into other areas, notably the perihilar and upper lobes, associated with disease progression.
CT studies in MERS patients often involve bilateral lungs, where predominantly ground-glass opacities notably in the basilar and peripheral lung regions are seen. Be aware there can be isolated consolidation, interlobular septal thickening, and pleural effusion in MERS as well. Some studies suggest upwards of 20–33% of MERS infected individuals may have these findings. Tree-in-bud opacities and cavitation rarely have been reported. Lymphadenopathy does not seem to be characteristic of MERS infection on radiographic findings. Not surprisingly, pleural effusion, pneumothorax, and greater involvement of the lungs are associated with a poorer prognosis38, 39, 40, 41, 42.
Information gained from SARS and MERS suggests that follow-up imaging should also be obtained in patients recovering from COVID-1940. There remains the potential for chronic involvement of the lungs; examples of which include interlobular thickening, air trapping, or fibrosis38, 40.
In terms of COVID-19, early evidence suggests that initial chest imaging will show abnormality in at least 85% of patients, with 75% of patients having bilateral lung involvement initially that most often manifests as subpleural and peripheral areas of ground-glass opacity and consolidation. Older age and progressive consolidation might suggest poorer prognosis. Besides the acute phase, CT is recommended for follow-up in individuals who are recovering from COVID-19 to evaluate long-term or permanent lung damage including fibrosis, as is seen with SARS and MERS infection38, 40.
It is worth noting in SARS, even with post infection recovery CT study may still show transient interlobular septal thickening and reticulation for several weeks to months38, 39, 40, 41, 42. Data suggest the reticulation observed usually appears ~ week 2, with peaking expected ~ week 4.38, 39, 40, 41, 42. Of concern, 1/3 of those patients who have persistent respiratory symptoms will have imaging findings of fibrosis, including interlobular and intralobular reticulation, as well as traction bronchiectasis. Although uncommon “honeycombing” has been seen. Air trapping, which is considered the result of damage to the ciliated respiratory epithelium, has been reported as a finding in ~92% of patients who have recovered from pneumonia. It portends chronic pulmonary involvement; these patients are less likely to have complete resolution of their infection38, 39, 40, 41, 42.
Clinical experience with MERS suggests the majority of patients recover fully. However radiographic studies suggest ~33% of MERS survivors will have some evidence of lung fibrosis on follow-up imaging. Demographically, these patients were likely to be older, experienced prolonged ICU care, as well as more significant lung involvement in the acute phase of their illness38, 39, 40.
Discussion
In spite of significant effort to develop countermeasures for coronaviruses, during the SARS and MERS outbreaks there were no specific licensed therapeutics, nor any identified that demonstrated consistent effectiveness against either of these43. The mainstay of medical care was symptomatic and supportive, often involving intensive critical care – providing ventilator, circulatory and other organ system support to preserve renal, hepatic and neurological function, as well as prevention of secondary infection. Newer approaches being trialed against SARS2 COVID-19 will be discussed in that section.
That said, among the available options, including the limited number of medications with potential antiviral capability, various combinations of therapies were trialed43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, but no scaled, controlled approaches were conducted, making recommendations on antivirals employed during the SARS outbreak, with or without interferon combination therapy, of concern, and questionable43, 44, 45, 46, 47, 48, 49, 50, 51, 52.
Immune based therapies were also administered, with equivocal results.
Ribavirin is a potent nucleoside analogue used with varying degrees of success against RNA viruses, but there is the potential for adverse side effects including hemolytic anemia, metabolic derangements. Interferon can also elicit adverse effects, although it has demonstrated value against viral infection43, 44, 45, 46.
One intervention that held promise however during the SARS CoV outbreak was the use of convalescent plasma; hyperimmune globulin was shown to be relatively safe, and possibly effective for reducing mortality50, 51, 52, 53. Convalescent plasma, when administered within 14 days of illness, did decrease mortality in SARS CoV patients, according to one study46. They found this was a time critical issue – administration had to be given within a 2 week period.
Convalescent plasma is showing similar benefit with SARS-2 COVID – 19 patients. The challenges with convalescent plasma are several. First there is the need to identify cases and contacts rapidly, and second, the immediate utilization of such therapies for a chance at optimal effect. Donor supply, technical capacity to obtain and deliver convalescent plasma in regions where SARS, MERS, SARS2-COVID-19 or other emerging threats are likely to occur, safety of end product and other challenges can limit the potential of this therapeutic option.
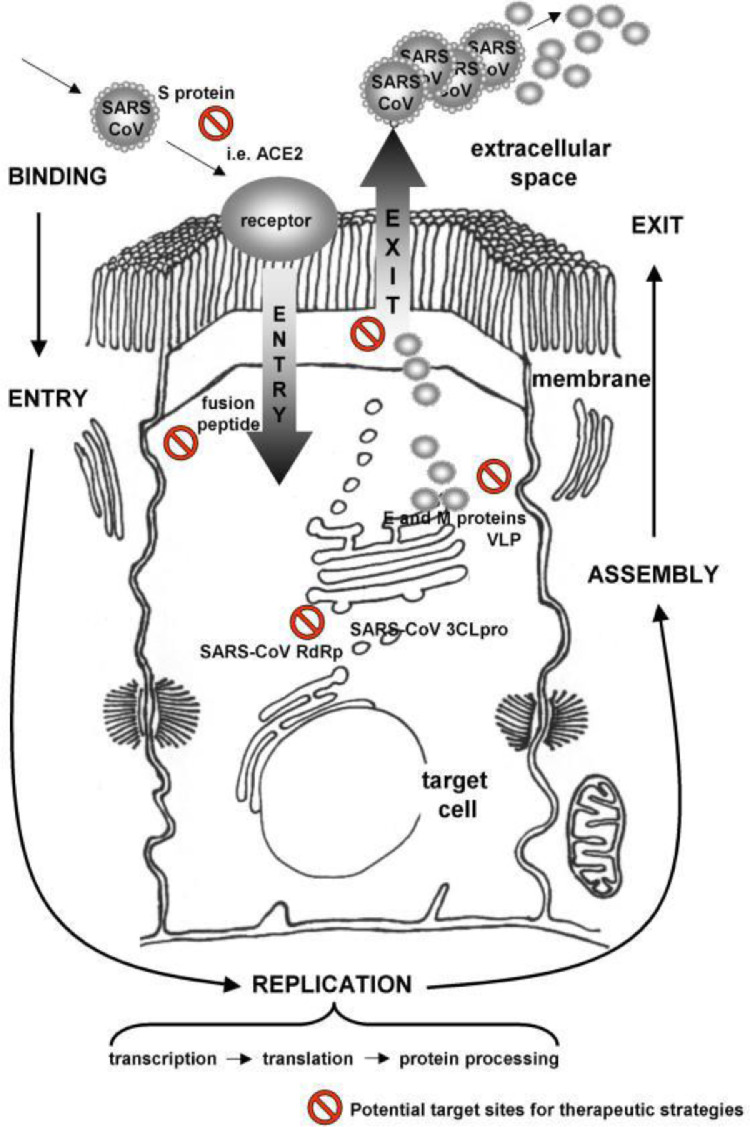
Extensive research is also being conducted towards developing vaccines. Towards that end, not unlike vaccines directed towards other pathogens, research has focused on viral structure56 , 57, and replication mechanisms (Fig. 7 )57/(Table 2 )43.
Fig. 7.
Table 2.
| Mode of action | Drug |
|---|---|
| Virus entry blockers | Anti-S protein monoclonal antibodies Peptides that bind to the heptad repeat on the S (spike) protein Peptides that bind to other regions of S and block oligomerisation, etc. |
| Virus replication blockers | 3C-like protease inhibitors Other viral protease inhibitors, e.g. papain-like cysteine protease nsp1–16 Viral polymerase inhibitors Nelfinavir, lopinavir/ritonavir, ribavirin, RNAi, glycyrrhizin, niclosamide |
| Immune modulators | Type 1 interferons Lopinavir/ritonavir |
Not surprisingly, as with other pathogens, such as Dengue62, 63, 64, 65, 66 where there remain unknowns, such as protective immunity (Table 3)38, 59, 60, 61, cross protection against a variety of strains, and other technical difficulties, there are a variety of challenges to overcome in developing an effective vaccine against SARS or other coronaviruses. For example, in developing a live SARS CoV vaccine, it will be necessary to address the various coronavirus strains to recombine with each other, with the potential of attenuated parts of the genome being replaced with non-attenuated components of the genome, resulting in a pathogenic virus. One approach being considered is the use of reverse genetics; it may eliminate the risk of recombination between coronavirus strains43 , 59, 60, 61 , 63 , 65 , 67
Table 3.
Examples Of Vaccine Strategies For SARS CoV Adapted from Enjuanes et al., Gillim-Ross et al., Lin et al.60 and Martin et al.61
| Vaccine type | Animal studies | Induction of neutralizing antibodies/protection | Human trials |
|---|---|---|---|
| Inactivated virus | Mice | + | + |
| Subunit or expressed protein | Mice | + | − |
| Viral or bacterial expression vectors (S or N protein) | Mice, ferrets, primates | + | − |
| DNA vaccine (S, N, M protein) | Mice, primates | + | + |
| Live attenuated virus | Hamsters | + | − |
The timelines of events as noted by CDC concluded towards the end of 2003 with removal of travel warnings to China and Ontario. By 2004 the CDC issued a “Notice of Embargo of Civets” as a SARS-like virus had been isolated from civets (captured in areas of China where the SARS outbreak originated). CDC also banned the importation of civets. The civet is a mammal with a catlike body, long legs, a long tail, and a masked face resembling a raccoon or weasel. SARS CoV was detected in animal handlers of civets. The ban on civets is currently still in effect. By 2012 The National Select Agent Registry Program declared SARS-coronavirus a select agent. A select agent is a bacterium, virus or toxin that has the potential to pose a severe threat to public health and safety32 , 35.
Not surprisingly there are also economic, as well as health implications - coronaviruses cause important diseases in domestic animals, as well as in human populations. Toronto during and in the aftermath of their SARS outbreak saw a significant, albeit temporary decline in tourism and business related visits, as well as lost conference and trade show related commerce. Recognizing the importance of animal – human pathogen crossover, opportunities to reduce the spread of contagion, and to identify potential risks is critical to prevent or at least reduce the likelihood of SARS, MERS, and influenza outbreaks such as the avian influenza outbreaks of the 1990′s and early2000’s and the swine flu outbreak in 2009.
As suddenly as SARS CoV emerged, it has seemingly gone quiescent. However, there are two new, highly pathogenic, and previously unknown or not described coronaviruses that have emerged.
Treatment
There are as of yet no FDA approved or licensed therapeutics that have shown consistent effectiveness against MERS CoV or SARS CoV38. The emergence of COVID-19 has led to a renewed interest in discovering antiviral treatments – either de novo medications created for this purpose, or repurposing existing medications with potential anti-coronavirus properties are being investigated. These will be discussed in the COVID-19 Therapeutics Section of this article.
To date, whether for SARS, MERS, or COVID-19, Intensive care – providing ventilator, circulatory and other organ system support to preserve renal, hepatic and neurological function, as well as prevention of secondary infection remain the mainstay of care, with early aggressive intervention being critical.
Other interventions, including repurposing currently available therapies were tried. Here are some examples:
During the SARS CoV pandemic of 2003 immune based therapies have been tried, with equivocal results.
Ribavirin and interferon combinations showed some clinical improvements in non human primate studies, but unlike actual clinical experiences with MERS and SARS, where the illness, let alone treatment are rarely initiated rapidly after infection, the trials provided interventions soon after viral challenge38, 39, 40, 41, 42. It has become apparent clinically, especially with certain potential antiviral treatments that this is a time critical step in the life cycle of the virus, as a consideration in medical intervention. This is true for other viruses, as there is often a narrow window of opportunity to interrupt the illness, such in the case with influenza and neuraminidase based therapies such as oseltamivir.
During SARS CoV epidemics, various combinations of therapies were trialed43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, but no scaled, controlled approaches were conducted, making recommendations on current antivirals with or without interferon combination therapy, of concern, and questionable43, 44, 45, 46, 47, 48, 49, 50, 51, 52.
Ribaviran is a potent nucleoside analogue used with varying degrees of success against RNA viruses, but there is the potential for adverse side effects including hemolytic anemia, metabolic derangements. Interferon can also elicit adverse effects, although they have demonstrated value against viral infection43, 44, 45, 46.
Corticosteroids have been tried in SARS CoV infection – they resulted in increased viral load, admissions to intensive care unit, and mortality48 , 49.
During SARS CoV, convalescent plasma, hyperimmune globulin were shown to be relatively safe, and possibly effective for reducing mortality49, 50, 51, 52, 53. Convalescent plasma, when administered within 14 days of illness, did decrease mortality in SARS CoV patients, according to one study46 , 51. They found this was a time critical issue – administration had to be given within a 2 week period. The challenge of course is in identifying cases and contacts rapidly and being able to initiate immediate utilization of such therapies for a chance at optimal effect. Some challenges exist with this approach, depending upon the region. These include donor supply, technical capacity in regions where SARS, MERS or other emerging threats are likely to occur, safety of the end product and other challenges can limit the potential of this therapeutic option. The World Health Organization, and Centers for Disease Control maintain consulting expertise and guidance to assist with these challenges.
Monoclonal antibodies (mAbs) offer promise, and have demonstrated efficacy in the treatment of cancer and autoimmune diseases, as well as respiratory synctial virus (RSV)43 , 52 , 53. Trials are ongoing to determine the use of mAbs for Ebola virus disease, HIV - primary and secondary prevention43. Unfortunately the costs, as well as research and development timelines are longer than for polyclonal antibody preparations. Nevertheless, in spite of rigorous testing, regulatory and cost issues associated with mAbs, their potential as therapies for MERS and other potentially deadly diseases continue to drive research in this area.
Antiviral research into adenine analogues that can disrupt viral RNA replication50 are being developed as well as a nucleoside analogues with the potential to work against filoviruses, coronaviruses, and other RNA viruses56.
Ideally an antiviral that covers a broad range of coronaviruses will be developed based upon the genetic sequence of these viruses and their life cycle. But the development of such an antimicrobial and other interventions still remains in the future.
Of note, extensive research is also being conducted towards developing vaccines. Towards that end, not unlike vaccines directed towards other pathogens, research has focused on viral structure56 , 57, and replication mechanisms (Fig. 7)57/(Table 4 )43.
Table 4.
| Mode of action | Drug |
|---|---|
| Virus entry blockers | Anti-S protein monoclonal antibodies Peptides that bind to the heptad repeat on the S (spike) protein Peptides that bind to other regions of S and block oligomerisation, etc. |
| Virus replication blockers | 3C-like protease inhibitors Other viral protease inhibitors, e.g. papain-like cysteine protease nsp1–16 Viral polymerase inhibitors Nelfinavir, lopinavir/ritonavir, ribavirin, RNAi, glycyrrhizin, niclosamide |
| Immune modulators | Type 1 interferons Lopinavir/ritonavir |
As with other pathogens, such as Dengue57, 58, 59, 60, 61 where there remain unknowns, such as protective immunity, cross protection against a variety of strains, and other technical difficulties, there are a variety of challenges to overcome in developing an effective vaccine against SARS or other coronaviruses. For example, in developing a live SARS CoV vaccine, it will be necessary to address the various coronavirus strains to recombine with each other, with the potential of attenuated parts of the genome being replaced with non-attenuated components of the genome, resulting in a pathogenic virus. One approach being considered is the use of reverse genetics; it may eliminate the risk of recombination between coronavirus strains43 , 59, 60, 61 , 63 , 66 , 67.
Also the question of immune response with SARS, MERS, and COVID-19 remain to be answered, including the sustainability of protection – either through surviving infection, or via vaccination63, 68, 69.
Unlike influenza which is an annual, recurring virus, or the low pathogenic coronaviruses, which are among several pathogens associated with “the common cold” and which seems to linger as a background infectious agent, as suddenly as SARS CoV emerged, it has seemingly gone quiescent. However another significant, and previously unknown or not described coronavirus respiratory disease has emerged.
In the next section another highly pathogenic CoV was discovered that causes human illnes the Middle East Respiratory Syndrome Coronavirus (MERS-CoV, MERS), which is part of the beta group of coronaviruses, and carries a more significant case fatality rate than any coronavirus before it (~30%) . The last sections of this edition will discuss various aspects of the newest highly pathogenic coronavirus – one that shares clinical similarities with the first SARS coronavirus, but seems to cause a more diverse range of sickness, including extrapulmonary disease, and noted as SARS CoV2, or COVID-1968.
References
- 1.Siddell S., Wege H., ter Meulen V. The biology of coronaviruses. J Gen Virol. 1983;64(Pt 4):761–776. doi: 10.1099/0022-1317-64-4-761. [DOI] [PubMed] [Google Scholar]
- 2.Perlman S., Netland J. Coronaviruses post SARS: update on replication and pathogenesis. Nat Rev Micro. 2009 doi: 10.1038/nrmicro2147. June: 439-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lau S.K.P., Lau C.C.Y., Chan K.H., Li C.P.Y., et al. Delayed induction of proinflammatory cytokines and suppression of innate antiviral response by the novel Middle Easst respiratory syndrome coronavirus: implications for pathogenesis and treatment. J Gen Virol. 2013;94:2679–2690. doi: 10.1099/vir.0.055533-0. [DOI] [PubMed] [Google Scholar]
- 4.deGroot R.J., Baker S.C., Baric R., Enjuanes L., et al. Coronaviridae In Virus Taxonomy; Ninth Report of the International Committee On Taxonomy of Viruses pp 806-828. Edited by AMQ King, JH Adams, EB Carstens and EJ Lefkowitz. San Diego, Ca. Elsevier Publishing 2011.
- 5.Woo P.C., Lau S.K., Lam C.S., Lau C.C., et al. Disocvery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruess as the gene source of alphacoronavirus and betacoronavirus and avian cornnaviruses as the gene source of gammacoronavirus and deltacornavirus. J Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hon C.C., Lam T.Y., Shi Z.L., Drummond A.J., et al. Evidence of the recombinant origin of a bat severe acute respiratory synddrome OSARS) like coronavirus and its implications om the direct ancestor of SARS coronavirus. J Virol. 2008;82:1819–1826. doi: 10.1128/JVI.01926-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lau S.K.P., Lee P., Tsang A.K.L., Yip C.C.Y., et al. Molecluar epidemiology of human coronavirus OC43 reveals evolution of different genotypes over time and recent emergence of a novle genotype due to natural recombination. J Virol. 2011;85:11325–11337. doi: 10.1128/JVI.05512-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guan Y., Zheng B.J., He Y., Liu X.L., et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 9.CDC/National institutes of health public domain coronavirus image, 2017, https://www.ncbi.nlm.nih.gov/genomes/SARS/images/sars%20em%20cdc%20rev1.PNG. Last accessed 03/06/17.
- 10.Centers for disease control and prevention (CDC) coronaviruses, 2017, https://www.cdc.gov/coronavirus/about/index.html. Last accessed 03/06/17.
- 11.Lua S.K., Woo P.C., Li K.S., Huang Y. Severe acute respiratory syndrome coronavirus like virus in Chinese horseshoe bats. Proc Natl Acad Sci USA. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Makela M.J., Puhakka T., Ruuskanen O., et al. Viruses and bacteria in the etiology of the common cold. J Clin Microbiol. 1998;36:539–542. doi: 10.1128/jcm.36.2.539-542.1998. http://SARSReference.com/lit.php?id=9466772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coronaviruses McIntosh K. In: 5th ed. Mandell GL, Bennett JE, Dolin R, editors. Churchill Livingstone; Philadelphia: 2000. [Google Scholar]
- 14.El-Sahly H.M., Atmar R.L., Glezen W.P., Greenberg S.B. Spectrum of clinical illness in hospitalized patients with "common cold" virus infections. Clin Infect Dis. 2000;31:96–100. doi: 10.1086/313937. http://SARSReference.com/link.php?id=10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Folz R.J., Elkordy M.A. Coronavirus pneumonia following autologous bone marrow transplantation for breast cancer. Chest. 1999;115:901–905. doi: 10.1378/chest.115.3.901. http://www.chestjournal.org/cgi/conten t/full/115/3/901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sizun J., Yu M.W., Talbot P.J. Survival of human coronaviruses 229E and OC43 in suspension after drying on surfaces: a possible source of hospital-acquired infections. J Hosp Infect. 2000;46:55–60. doi: 10.1053/jhin.2000.0795. http://SARSReference.com/lit.php?id=11023724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holmes K.V., Enjuanes L. The SARS coronavirus: a postgenomic era. Science. 2003;300:1377–1378. doi: 10.1126/science.1086418. [DOI] [PubMed] [Google Scholar]
- 18.Holmes K.V. SARS coronavirus: a new challenge for prevention and therapy. J Clin Invest. 2003;111:1605–1609. doi: 10.1172/JCI18819. http://www.jci.org/cgi/content/full/111/11/1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van den Hoogen B.G., de Jong J.C., Groen J., et al. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med. 2001;7:719–724. doi: 10.1038/89098. http://SARSReference.com/lit.php?id=11385510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drosten C., Gunther S., Preiser W., et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. http://SARSReference.com/lit.php?id=12690091 aPublished online Apr 10, 2003. [DOI] [PubMed] [Google Scholar]
- 21.Drosten C., Preiser W., Günther S., Schmitz H., Doerr H.W. Severe acute respiratory syndrome: identification of the etiological agent. Trends Mol Med. 2003;9:325–327. doi: 10.1016/S1471-4914(03)00133-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peiris J.S., Lai S.T., Poon L.L., et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. http://image.thelancet.com/extras/03art3477web.pdf aPublished online Apr 8, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marra M.A., Jones S.J.M., Astell C.R., et al. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. http://www.sciencemag.org/cgi/content/abstract/1085953v1 Published online May 1, 2003. [DOI] [PubMed] [Google Scholar]
- 24.Rota P.A., Oberste M.S., Monroe S.S., et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. http://www.sciencemag.org/cgi/content/abstract/1085952v1 Published online May 1, 2003. [DOI] [PubMed] [Google Scholar]
- 25.Ludwig B., Kraus F.B., Allwinn R., Doerr H.W., Preiser W. Viral zoonoses - a threat under control? Intervirology. 2003;46:71–78. doi: 10.1159/000069749. http://SARSReference.com/lit.php?id=12684545 [DOI] [PubMed] [Google Scholar]
- 26.Ruan Y.J., Wei C.L., Ee A.L., et al. Comparative full-length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection. Lancet. 2003;361:1779–1785. doi: 10.1016/S0140-6736(03)13414-9. http://image.thelancet.com/extras/03art4454web.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown E.G., Tetro J.A. Comparative analysis of the SARS coronavirus genome: a good start to a long journey. Lancet. 2003;361:1756–1757. doi: 10.1016/S0140-6736(03)13444-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsui S.K., Chim S.S., Lo Y.M. Chinese University of Hong Kong molecular SARS research group. Coronavirus genomic-sequence variations and the epidemiology of the severe acute respiratory syndrome. N.Engl.J.Med. 2003;349:187–188. doi: 10.1056/NEJM200307103490216. [DOI] [PubMed] [Google Scholar]
- 29.Ksiazek T.G., Erdman D., Goldsmith C.S., et al. A novel coronavirus associated with severe acute respiratory syndrome. New Eng J Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. http://SARSReference.com/lit.php?id=12690092 Published online Apr 10, 2003. [DOI] [PubMed] [Google Scholar]
- 30.Peiris J.S., Chu C.M., Cheng V.C., et al. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. http://image.thelancet.com/extras/03art4432web.pdf bPublished online May 9, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.http://sarsreference.com/sarsref/virol.htm, 2003. Last accessed 01/14/17.
- 32.Anderson L.J., Tong S. Update on SARS research and other possibly zoonotic coronaviruses. Int J Antimicrob Agents. 2010;36S:S21–S25. doi: 10.1016/j.ijantimicag.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peiris J.S., Yuen K.Y., Osterhaus A.D., Stohr K. The severe acute respiratory syndrome. N Engl J Med. 2003;349:2431–2441. doi: 10.1056/NEJMra032498. [DOI] [PubMed] [Google Scholar]
- 34.Enjuanes L., Dediego M.L., Alvarez E., Deming D., Sheahan T., Baric R. Vaccines to prevent severe acute respiratory syndrome coronavirus-induced disease. Virus Res. 2008;133:45–62. doi: 10.1016/j.virusres.2007.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Centers for disease control and prevention (CDC) SARS CoV, https://www.cdc.gov/sars/about/index.html. Last accessed 03/12/17.
- 36.World Health Organization (WHO)SARS overview 2003, http://www.who.int/csr/sars/country/table/en/. Last accessed 03/12/17.
- 37.Zhu Y., Li C., Chen L., Xu B., et al. A novel human coxronavirus OC43 genotype detected in mainland. China Emerg Microbes Infect. 2018;7:713. doi: 10.1038/s41426-018-0171-5. www.nature.com/emi Last accessed 06/03/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coronavirus Images from CDC, 2017https://www.cdc.gov/sars/lab/images.htmlLast accessed 03/12/17.
- 39.Nicolaou S., Al-Nakshabandi N.A. Muller NL case report - SARS: imaging of severe acute respiratory syndrome (SARS) Am J Roentgenol. 2003;180(5):1247–1249. doi: 10.2214/ajr.180.5.1801247. May. [DOI] [PubMed] [Google Scholar]
- 40.Hosseiny M., Kooraki S., Gholamrezanezhad A., Reddy S., Myers L., Hosseiny M., et al. Radiology perspective of coronavirus disease 2019 (COVID-19): lessons from severe acute respiratory syndrome and middle east respiratory syndrome. AJR Am J Roentgenol. 2020;214(5):1078–1082. doi: 10.2214/AJR.20.22969. https://www.ajronline.org/doi/full/10.2214/AJR.20.22969 May. Last accessed 06/14/20. [DOI] [PubMed] [Google Scholar]
- 41.Al-Tawfiq J.A., Zumla A., Memish Z.A. Coronaviruses: severe acute respiratory syndrome coronavirus and Middle East respiratory syndrome coronavirus in travelers. Curr Opin Infect Dis. 2014;27:411–417. doi: 10.1097/QCO.0000000000000089. [Crossref] [Medline] [Google Scholar] [DOI] [PubMed] [Google Scholar]
- 42.Das K.M., Lee E.Y., Langer R.D., Larsson S.G. Middle East respiratory syndrome coronavirus: what does a radiologist need to know. AJR. 2016;206:1193–1201. doi: 10.2214/AJR.15.15363. [DOI] [PubMed] [Google Scholar]
- 43.Ketai L., Paul N.S., Wong K.T. Radiology of severe acute respiratory syndrome (SARS): the emerging pathologic-radiologic correlates of an emerging disease. J Thorac Imaging. 2006;21:276–283. doi: 10.1097/01.rti.0000213581.14225.f1. [DOI] [PubMed] [Google Scholar]
- 44.Modjarrad K. Treatment strategies for Middle East respiratory syndrome coronavirus. J Virus Erad. 2016;2:1–4. doi: 10.1016/S2055-6640(20)30696-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Falzarano D., deWit E., Rasmussen A.L., et al. Treatment with interferon-alpha2b and Ribavirin improves outcome in MERS CoV infected rhesus macaques. Nat Med. 2013;19:1313–1317. doi: 10.1038/nm.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shalhoub S., Farahat F., Al – Jiffri A., et al. IFN alpha 2a or IFN beta 1 in combination with Ribavirin to treat Middle East respiratory syndrome coronavirus pneumonia: a retrospective study. J Antimicrob Chemother. 2015;70:2129–2132. doi: 10.1093/jac/dkv085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gross A.E., Bryson M.L. Oral Ribavirin for the treatment of noninfluenza respiratory viral infections: a systematic review. Ann Pharmacother. 2015;49:1125–1135. doi: 10.1177/1060028015597449. [DOI] [PubMed] [Google Scholar]
- 48.Jonasch E., Haluska F.G. Interferon in oncological practice e: review of interferon biology, clinical applicatiosn, and toxicities. Oncologist. 2001;6:34–55. doi: 10.1634/theoncologist.6-1-34. [DOI] [PubMed] [Google Scholar]
- 49.Auyeung T.W., Lee J.S., Lai W.K., et al. The use of corticosteroid aas treatment in SARS was associated with adverse outcomes: a retrospective cohort study. J Infect. 2005;51:98–102. doi: 10.1016/j.jinf.2004.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leff N., Allen Chan K.C., Hui D.S., et al. Effects of early corticosteroid treatment on plasma SARS associated Coronavirus RNS concentrations in adult patients. J Clin Virol. 2004;31:304–309. doi: 10.1016/j.jcv.2004.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arabi Y., Balkhy H., Hajeer A.H., et al. Fesibility, safety, clinical and laboratory effects of convalescent plasma therapy for patients with Middle East respiratory sundrmme coronavirus infection: a study protocol. Springerplus. 2015;4:709. doi: 10.1186/s40064-015-1490-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mair-Jenkins J., Saavedra-Campos M., Baillie J.K., et al. The effectiveness of convalescent plasma and hyperimmune immunoglobulin for the treatment of sever acue trespiratory infectios of viral etiology: a systematic review and exploreatory meta analysis. J Infect Dis. 2015;211:80–90. doi: 10.1093/infdis/jiu396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang L., Want N., Zuo T., et al. Potent neutralization of MERS CoV by human neutralizing monoclonal antibodies to the viral spike glycoprotein. Sci Transl Med. 2014;6 doi: 10.1126/scitranslmed.3008140. 234ra259. [DOI] [PubMed] [Google Scholar]
- 54.Ying T., Du L., Ju T.W., et al. Exceptionally potent neutralization of Middle East respiratory syndrome coronavirus by human monoclonal antibodies. J Virol. 2014;88:7796–7805. doi: 10.1128/JVI.00912-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yount B., Roberts R.S., Lindesmith L., Baric R.S. Rewiring the severe acute respiratory syndrome coronavirus (SARS-CoV) transcription circuit: engineering a recombination-resistant genome. Proc Natl Acad Sci U S A. 2006;103:12546–12551. doi: 10.1073/pnas.0605438103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Warren T., Jordan R., Lo M., et al. San Diego; Ca: 2015. Nucleotide Prodrug GS 5734 is a Broad Spectrum Filovirus Inhibitor that Provides Complete Therapeutic Protection Against the Development of Ebola virus Disease EVD in Infected Non Human Primates ID Week 2015 October 7 - 11. [Google Scholar]
- 57.Bennett R.BioCryst announces Nature publication demonstrating efficacy of BBCX4430 in a non human primate model of filovirus infection2014. Available athttp://globenewsire.com/news-release/2014/03/03/6149090/10070784/4n/biocryst-announces-nature-publication-demonstrating-efficacy–f-bcx4430-in-a-non-human-primate-model-of-filovirus-infection.html.
- 58.Groneberg D.A., Hilgenfeld R., Zabel P Molecular mechanisms of severe acute respiratory syndrome (SARS) Respir Res 2005; 6 (1): 8 Published online 2005 Jan 20. doi: 10.1186/1465-9921-6-8. [DOI] [PMC free article] [PubMed]
- 59.Barnard D.L., Kumaki Y. Recent developments in anti-severe respiratory syndrome coronavirus chemotherapy. Future Virol. 2011 May;6(5):615–631. doi: 10.2217/fvl.11.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gillim-Ross L., Subbarao K. Emerging respiratory viruses: challenges and vaccine strategies. Clin Microbiol Rev. 2006;19:614–636. doi: 10.1128/CMR.00005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin J.T., Zhang J.S., Su N., Xu J.G., Wang N., Chen J.T., et al. Safety and immunogenicity from a phase I trial of inactivated severe acute respiratory syndrome coronavirus vaccine. Antivir Ther. 2007;12:1107–1113. [PubMed] [Google Scholar]
- 62.Martin J.E., Louder M.K., Holman L.A., Gordon I.J., Enama M.E., Larkin B.D., et al. A SARS DNA vaccine induces neutralizing antibody and cellular immune responses in healthy adults in a Phase I clinical trial. Vaccine. 2008;26:6338–6343. doi: 10.1016/j.vaccine.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McFee R.B. Viral hemorrhagic fever viruses in novel viruses, emerging pathogens – the pandemic threat continues. Dis a Month. 2013;59(12):405–448. doi: 10.1016/j.disamonth.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 64.Schmitz J., Roehrig J., Barrett A., Hombach J. Next generation dengue vaccines: a review of candidates in preclinical development. Vaccine. 2011;29(42):7276–7284. doi: 10.1016/j.vaccine.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 65.Hunsperger E.A., Yoksan S., Buchy P., et al. Evaluation of commercially available anti-Dengue virus immunoglobulin M tests. Emerg Infect Dis. 2009;15(30):436–440. doi: 10.3201/eid1503.080923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sabchareon A., Wallace D., Sirivicayakul C., et al. Protective efficacy of the recombinant, live attenuated, CY tetravalent dengue vaccine in Thai schoolchildren: a random controlled phase 2b trial. Lancet. 2012;380(9853):1559–1567. doi: 10.1016/S0140-6736(12)61428-7. [DOI] [PubMed] [Google Scholar]
- 67.Noisakran S., Onlamoon n, Songprakhon P., Hsiao H.M., et al. Cells in dengue virus infection in vivo. Adva Virol. 2010 doi: 10.1155/2010/164878. (ID 164878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van den Worm S.H.E., Eriksson K.K., Zevenhoven J.C., Weber F., et al. Reverse genetics of SARS related coronavirus using vaccinia virus based recombination Published: march 7, 2012. 10.1371/journal.pone.0032857. Last accessed 03/12/17. [DOI] [PMC free article] [PubMed]
- 69.Catanzaro M., Fagiani F., Racchi M., Corsini E., et al. Immune response in COVID-19: addressing a pharmacological challenge by targeting pathways triggered by SARS-CoV-2. Sig Transduct Target Ther. 2020 doi: 10.1038/s41392-020-0191-1. www.nature.com/sigtrans 29 May. Last accessed 06/04/20. [DOI] [PMC free article] [PubMed] [Google Scholar]