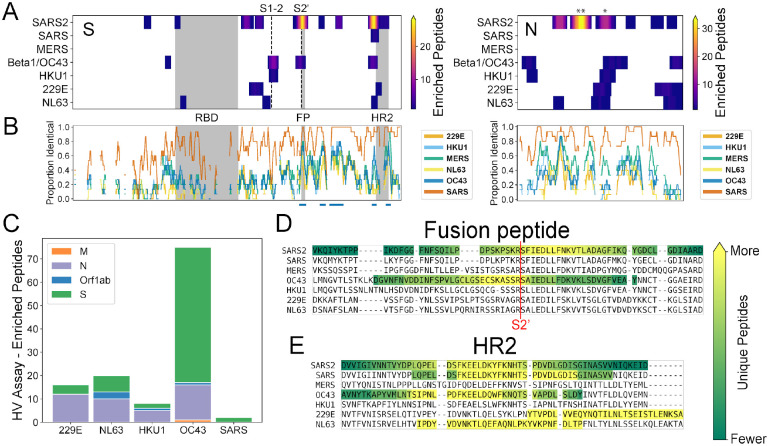
Figure 4. Recurrent SARS-CoV-2 epitopes correspond to conserved regions of Spike S2 that are also targeted in the response to other CoVs.
A) Heat maps illustrating the relative locations of enriched SCV2 (from COVID-19 convalescent samples) and HV (from pre-pandemic controls) library peptides within the S (left) and N (right) proteins and across all human-infecting coronaviruses. Results have been aggregated across all tested samples and the color at each location indicates the number of unique enriched peptides. The vertical dashed lines in the S protein plot represent the S1-S2 and S2’ cleavage sites, respectively. Above the N plot, ‘**’ and ‘*’ indicate the 1st and 2nd most commonly immunogenic regions of this protein in COVID-19 convalescent samples, respectively. B) Comparison of amino acid sequence identity between SARS-CoV-2 and the other six human CoVs across the same S and N alignments used in panel (A). A sliding window of 15 amino acids was used and gaps represent windows with ≥30% indels. Blue bars under the S plot indicate regions ≥15 amino acids long that exhibit ≥70% identity between SARS-CoV-2 and hCoV-OC43 and/or hCoV-HKU1. Grey boxes in panels (A) and (B) indicate selected functional domains: receptor binding domain (RBD), fusion peptide (FP) and heptad repeat 2 (HR2). C) Protein-level distribution of enriched HV library peptides across five HCoVs and 33 pre-pandemic control samples. A single peptide could be counted multiple times if enrichment was independently observed in multiple samples. C-D) Multiple sequence alignments of the immunodominant and most widely-recognized protein regions of SARS-CoV-2, including representative sequences from each of the seven human coronaviruses. Regions containing enriched peptides are highlighted by colored backgrounds, with bright yellow indicating residues contained within the most unique enriched peptides and dark green indicating those contained within the least unique enriched peptides (colorbar scales are distinct among species). SARS-CoV-2 reactivity was determined using the SCV2 peptide library, while reactivity for the other coronaviruses was determined using the HV peptide library.