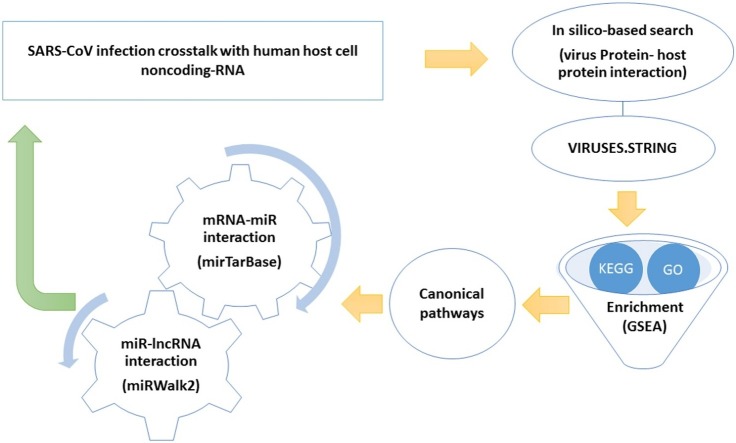
Graphical abstract
Keywords: SARS-CoV; Covid-19; Noncoding RNA, miRNA, lncRNA; Bioinformatics analysis
Highlights
-
•
Protein-protein interaction (PPI) network of the SARS-CoV and Homo sapiens was obtained.
-
•
Based on Bioinformatic analysis, the TGF-beta signaling pathway with the largest numbers of involved genes may play key roles during SARS-CoV infection.
-
•
An integrated network of mRNA/lncRNA/miRNA was created.
-
•
In the network, SMAD2, SMAD3, SMAD4, SMAD7, and TGFBR1 with the highest number of interactions were identified as target hubs.
Abstract
Although 70 % of the genome is transcribed to RNA in humans, only ∼2% of these transcripts are translated into proteins. The rest of the transcripts are defined as noncoding RNAs, including Long noncoding RNAs (LncRNAs) and MicroRNAs (miRNAs) that mostly function post-transcriptionally to regulate the gene expression. The outbreak of a novel coronavirus (SARS-CoV) has caused a major public health concern across the globe. The SARS-CoV is the seventh coronavirus that is known to cause human disease. There are currently no promising antiviral drugs with proven efficacy nor are there vaccines for its prevention. As of August 10, 2020, SARS-CoV has been infected more than 13 million cases in more than 213 countries, with an estimated mortality rate of ∼3 %. Thus, it is of utmost important priority to develop novel therapies for COVID-19. It is not fully investigated whether noncoding RNAs regulate signaling pathways that SARS-CoV involved in. Hence, computational analysis of the noncoding RNA interactions and determining importance of key regulatory noncoding RNAs in antiviral defense mechanisms will likely be helpful in developing new drugs to attack SARS-CoV infection. To elucidate this, we utilized bioinformatic approaches to find the interaction network of SARS-CoV/human proteins, miRNAs, and lncRNAs. We found TGF-beta signaling pathway as one of the potential interactive pathways. Furthermore, potential miRNAs/lncRNAs networks that the virus might engage during infection in human host cells have been shown. Altogether, TGF-beta signaling pathway as well as hub miRNAs, and LncRNAs involve during SARS-CoV pathogenesis can be considered as potential therapeutic targets.
1. Introduction
Viral protein complexity is basically limited by their relatively small genomes; therefore, critical functions are mainly accomplished by cooperation between host and viral protein machinery. This requirement makes the study of host–viral interactions is critical for the understanding of viral pathogenicity [1]. COVID-19 has infected over 20 million people and has killed more than 737,000 people by mid August 2020. This has driven the world into hysteria and panic, causing social and economic disruption. There are currently no promising antiviral drugs or vaccines to combat COVID-19. This is largely due to our limited knowledge about the molecular mechanisms of the virus pathogenesis [2].
MicroRNAs (miRNAs) are short non-coding RNAs that inhibit the expression of their target genes by directly binding to target mRNAs [3]. Long noncoding RNAs (lncRNAs) are a category of cellular RNAs that are longer than 200 nucleotides in length. Accumulating evidence suggests that miRNAs and lncRNAs play key roles in different disease pathogenesis [4,5]. More specifically, miRNA have common binding sites for both mRNAs and lncRNAs; thus, lncRNAs can act as competitive endogenous RNA (ceRNA) to suppress the miRNA function or act as precursors and encode miRNAs [4]. These miRNA regulatory functions have been estimated to regulate around 60 % of mammalian genes [6]. LncRNAs/ miRNAs play an important regulatory role in the battle between virus and host, especially in regulating the transcription of virus and host genes, stability and translation of mRNAs, and host antiviral response [6]. Developing functional computational models and networks to predict potential SARS-CoV -miRNA/lncRNA association may benefit not only the understanding of COVID-19 mechanism at the noncoding RNA level, but also the detection of disease biomarkers for disease diagnosis, treatment, prognosis, and prevention. Surprisingly, known experimentally confirmed disease-lncRNA associations are still very limited. Since multiple ways of interaction between miRNAs, lncRNAs, and mRNA have been reported to play key roles in determining the cellular functions during viral infection, it is essential to discover these interactions in an integrated fashion to comprehensively decipher the networks and key regulatory noncoding-RNA hubs underpinning the pathology of SARS-CoV.
Although we learned a great deal of coronavirus infection, the effective approaches for treatment are still scarce. Unsurprisingly, there is little knowledge of the detailed molecular interactions for the SARS-CoV infection crosstalk with noncoding RNAs network in the cell. Several signaling pathways regulate miRNAs and lncRNAs that affect the viral function and the infection progress, including replication, translation, and even modulating the host expression. Since it is also important to consider the virus-host protein-protein interaction in order to understand its pathogenesis during infection [7,8]; we used Viruses- STRING, a protein–protein interaction database specifically designed to analyze the virus-host interactions [9]. Subsequently, the Gene Set Enrichment Analysis (GSEA) was used to assess and interpret the coordinate pathways that might be potentially involved in these interactions [10]. Furthermore, we used the miRWalk2.0 database to generate a network for a comprehensive overview of miRNA-lncRNA interactions using experimentally verified data [11]. Our findings revealed a network of miRNAs/lncRNAs which regulate these pathways. Based on this, we designed a network-based model to propose the most important regulatory factors and hubs with a functional role in regulating the viral infection process using a noncoding RNA perspective.
2. Methods
2.1. Bioinformatic analyses
To generate an interactome network of Human SARS coronavirus (SARS-CoV) with human proteins, we used the STRING database http://viruses.string-db.org/ (version 10.5) [9]. Next, the Gene Set Enrichment Analysis (GSEA) (https://www.gsea-msigdb.org/) was performed for the Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotations [10,12]. Then, the most important canonical pathways with the highest number of genes were selected. In order to disclose the most possible miRNAs regulating the selected mRNAs, the MiRTarBase (http://miRTarBase.cuhk.edu.cn/), one of the most important experimentally validated miRNA-target interactions (MTIs) databases, was employed. For the sake of accuracy, only functional MTIs were chosen for further studies [12]. Further, miRNA-lncRNA interactions were determined using the miRWalk2.0 database (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/), a database on predicted and published noncoding RNAs; and seed length (SL) ≥ 13 were selected as the most significant interactions [11]. Finally, the lncRNA /miRNA/mRNA network was virtualized by Cytoscape 3.8.0. Hub targets were analyzed by cytoHubba package, a Cytoscape plug-in for finding hub objects in a network, and 5 top hub nodes with higher degrees were recognized.
3. Results
3.1. Protein-protein interaction (PPI) network of the SARS-CoV and human host cell
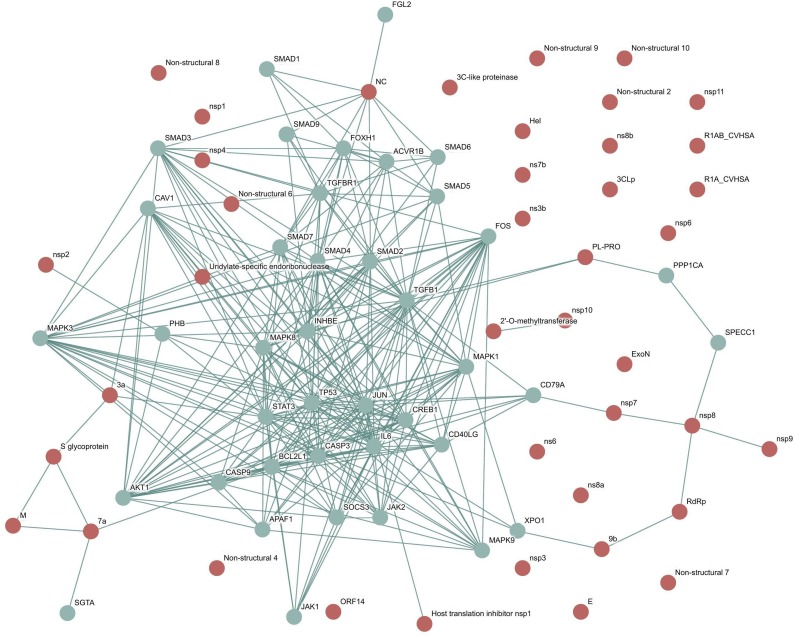
Human SARS coronavirus (SARS-CoV) and the Homo sapiens interaction network was downloaded as a vector image from the viruses STRING dataset. Protein-protein interactions were obtained and shaped a network with median confidence (scores greater than 0.4) between SARS-CoV and human host cells. The obtained protein-protein interactions are summarized in Fig. 1 . As depicted, 293 interactions were obtained and discovered a network, which included 40 proteins from the human host cell and 15 SARS-CoV-interacting proteins.
Fig. 1.
Human SARS coronavirus (SARS-CoV) and Homo sapiens interaction network obtained from the viruses. STRING database. The human host cell (connector proteins in light green color) and SARS-CoV-interacting proteins (red proteins).
3.2. Gene Ontology (GO) enrichment and canonical pathway analysis
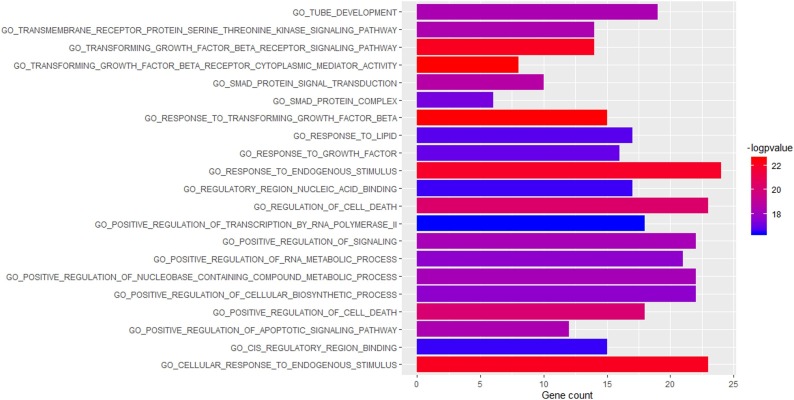
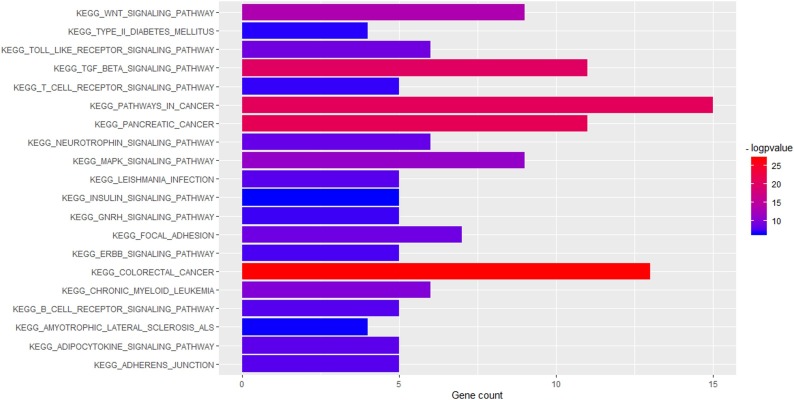
As many as 100 terms were examined in the canonical pathway analysis. It was found that many of these pathways are central pathways, meaning that most of the genes involved in this network are associated with canonical pathways. 20 top canonical pathway terms such as TGF-beta signaling pathway (P value = 6.99E-21; gene count = 11), Wnt signaling pathway (P value = 4.09E-14; gene count = 9), MAPK signaling pathway (P value = 7.51E-12; gene count = 9) are shown in Fig. 2 and the genes involved in these pathways are shown in Table 1 . Among these 20 pathways, the TGF-beta signaling pathway with the highest number of genes was selected for further study. In addition, based on gene ontology, 100 terms were selected such as those associated with response to transforming growth factor-beta (P value = 3.22E-23; gene count = 15), transforming growth factor-beta receptor cytoplasmic mediator activity (P value = 2.95E-23; gene count = 8), cellular response to endogenous stimulus (P value = 8.03E-23; gene count = 23), transforming growth factor-beta receptor signaling pathway (P value = 7.79E-23; gene count = 14). The 20 top enriched GO functions are shown in Fig. 3 .
Fig. 2.
Enriched KEGG pathways of the genes involved in SARS-CoV and Homo sapiens interaction network.
Table 1.
The list of the genes involved in canonical pathways.
| Canonical Pathways | Genes |
|---|---|
| KEGG_TGF_BETA_SIGNALING_PATHWAY | SMAD3, SMAD4, SMAD2, MAPK1, TGFB1, TGFBR1, SMAD1, SMAD5, SMAD6, SMAD7, SMAD9 |
| KEGG_WNT_SIGNALING_PATHWAY | SMAD3, SMAD4, SMAD2, MAPK9, MAPK8, MAPK10, JUN, NFATC2, FOSL1 |
| KEGG_MAPK_SIGNALING_PATHWAY | MAPK9, MAPK8, MAPK10, JUN, NFATC2, MAPK1, TGFB1, TGFBR1, FOS |
| KEGG_TOLL_LIKE_RECEPTOR_SIGNALING_PATHWAY | MAPK1, MAPK9, MAPK8, MAPK10, JUN, FOS |
| KEGG_ERBB_SIGNALING_PATHWAY | MAPK1, JUN, MAPK9, MAPK8, MAPK10, FOS, NFATC2, CD79A |
| KEGG_GNRH_SIGNALING_PATHWAY | MAPK1, JUN, MAPK9, MAPK8, MAPK10, FOS, NFATC2, CD79A |
| KEGG_T_CELL_RECEPTOR_SIGNALING_PATHWAY | MAPK1, JUN, MAPK9, FOS, NFATC2, MAPK8, MAPK10, CD79A |
| KEGG_APOPTOSIS | BAX, BCL2L1, CYCS, APAF1 |
| KEGG_CELL_CYCLE | TGFB1, SMAD3, SMAD4, SMAD2 |
| KEGG_RIG_I_LIKE_RECEPTOR_SIGNALING_PATHWAY | MAPK9, MAPK8, MAPK10 |
| KEGG_JAK_STAT_SIGNALING_PATHWAY | BCL2L1, JAK2, STAT3 |
| KEGG_CHEMOKINE_SIGNALING_PATHWAY | JAK2, STAT3, MAPK1 |
| KEGG_P53_SIGNALING_PATHWAY | BAX, CYCS, APAF1 |
Fig. 3.
Enriched GO functions of the genes involved in SARS-CoV and Homo sapiens interaction network.
3.3. Construction of lncRNA-miRNA-mRNA network
In order to establish a lncRNA-miRNA-mRNA network, 11 genes, including SMAD3, SMAD4, SMAD2, MAPK1, TGFB1, TGFBR1, SMAD1, SMAD5, SMAD6, SMAD7, and SMAD9 involved in TGF-beta signaling pathway were selected. MiRNAs and lncRNAs were identified from the MiRTarBase and miRWalk algorithm, respectively. Our data revealed that 709 miRNAs target these mRNAs, among which 145 functional MTIs were found and were subsequently selected as the most important interactions for further examination. In addition, it was predicted that 256,846 specific lncRNAs interact with specific miRNAs and a total of 139 lncRNA-miRNA interactions with SL ≥ 13. To elucidate the functions of the lncRNAs acting as miRNA targets, a network of lncRNAs, miRNAs, and mRNAs was constructed and visualized with Cytoscape. As shown in Fig. 4 , the mRNA/lncRNA/miRNA network was composed of 248 nodes (130 lncRNA nodes, 109 miRNA nodes, 9 mRNA nodes), and 284 edges. The 5 top nodes in the network were recognized by CytoHubba, which include SMAD2, SMAD3, SMAD4, SMAD7, and TGFBR1. All interacting lncRNAs, miRNAs, and mRNAs are summarized in Table 2 .
Fig. 4.
The lncRNA-miRNA-mRNA network. mRNAs (Green), miRNAs (blue) and lncRNA (orange).
Table 2.
The predicted lncRNAs, miRNAs, and mRNAs associated with SARS-CoV infection.
| mRNA | miRNA | lncRNAs |
|---|---|---|
| SMAD2 | hsa-miR-155-5p | |
| hsa-miR-200a-3p | ||
| hsa-miR-200b-3p | ||
| hsa-miR-484 | RP11-72I8.1 | |
| hsa-miR-148a-3p | AC005154.6 | |
| hsa-let-7g-5p | ||
| hsa-miR-146a-5p | LINC01010 | |
| hsa-miR-18a-5p | ||
| hsa-miR-18b-5p | ||
| hsa-miR-27a-3p | ||
| hsa-miR-27b-3p | ||
| hsa-miR-323a-3p | ||
| hsa-miR-128-3p | ||
| hsa-miR-425-5p | ||
| hsa-miR-486-5p | RAB11B-AS1 | |
| hsa-miR-206 | LINC01531 | |
| CTC-458G6. | ||
| hsa-miR-503-3p | RAP2C-AS1 | |
| hsa-miR-15b-5p | ||
| hsa-miR-205-5p | LINC01451 | |
| hsa-miR-125a-5p | SPACA6P-AS | |
| FLJ42969 | ||
| AC009878.2 | ||
| hsa-miR-145-5p | RP11-240G22.5 | |
| CTB-131B5.2 | ||
| LINC00622 | ||
| RP11-433J8.1 | ||
| RP11-76I14.1 | ||
| RP11-470E16.2 | ||
| RP11-25I9.3 | ||
| MUC19 | ||
| hsa-miR-212-3p | RP11-353N14.2 | |
| hsa-miR-132-3p | RP11-537P22.2 | |
| SMAD3 | hsa-miR-200a-3p | |
| hsa-miR-18a-5p | ||
| hsa-miR-193b-3p | ||
| hsa-miR-145-5p | RP11-240G22.5 | |
| CTB-131B5.2 | ||
| LINC00622 | ||
| RP11-433J8.1 | ||
| RP11-76I14.1 | ||
| RP11-470E16.2 | ||
| RP11-25I9.3 | ||
| MUC19 | ||
| hsa-miR-142-5p | ||
| hsa-miR-491-5p | RP11-317J10.2 | |
| hsa-miR-92b-3p | ||
| hsa-miR-323a-3p | ||
| hsa-miR-708-5p | LINC01193 | |
| RP11-69H14.6 | ||
| RP11-875O11.2 | ||
| hsa-miR-199a-5p | AC144525.1 | |
| RP11-755B10.3 | ||
| LINC00907 | ||
| RP11-338N10.1 | ||
| hsa-miR-145-3p | ||
| hsa-miR-590-5p | SNHG1 | |
| hsa-miR-23a-3p | ||
| hsa-miR-23b-5p | CTD-2560E9.5 | |
| RP11-455I9.1 | ||
| RP11-102K13.5 | ||
| hsa-miR-424-5p | SNHG1 | |
| hsa-miR-29b-1-5p | RP11-114G22.1 | |
| CTC-523E23.1 | ||
| SMAD4 | hsa-miR-26a-5p | RP11-38L15.2 |
| hsa-miR-483-3p | AC002059.10 | |
| RP11-7F17.8 | ||
| RP11-366F6.2 | ||
| hsa-miR-18a-5p | ||
| hsa-miR-17-5p | chr22-38_28785274-29006793.1 | |
| RP11-17M16.2 | ||
| hsa-miR-19a-3p | LINC00261 | |
| RP11-707A18.1 | ||
| hsa-miR-20a-5p | ||
| hsa-miR-203a-3p | ||
| hsa-miR-421 | LINC00862 | |
| hsa-miR-146a-5p | LINC01010 | |
| hsa-miR-224-5p | RP11-318A15.2 | |
| hsa-miR-125a-5p | SPACA6P-AS | |
| FLJ42969 | ||
| AC009878.2 | ||
| hsa-miR-125b-5p | FLJ42969 | |
| AC009878.2 | ||
| RP11-485O10.3 | ||
| RP3-329A5.8 | ||
| SPACA6P-AS | ||
| hsa-miR-199a-5p | AC144525.1 | |
| RP11-755B10.3 | ||
| LINC00907 | ||
| RP11-338N10.1 | ||
| hsa-miR-130a-3p | ||
| hsa-miR-301a-3p | LINC00635 | |
| hsa-miR-454-3p | ||
| hsa-miR-182-5p | ||
| hsa-miR-130b-3p | LINC00608 | |
| hsa-miR-19b-3p | ||
| hsa-miR-204-5p | AC073130.1 | |
| RP11-430L3.1 | ||
| hsa-miR-1260b | LINC01359 | |
| RP11-575L7.4 | ||
| AC007952.5 | ||
| LINC00202-1 | ||
| CTD-2515A14.1 | ||
| CTD-2653D5.1 | ||
| RP11-110H1.4 | ||
| hsa-miR-205-5p | LINC01451 | |
| hsa-miR-183-5p | SMC2-AS1 | |
| AC004076.5 | ||
| hsa-miR-34a-5p | RP11-867G23.10 | |
| hsa-miR-27a-3p | ||
| hsa-miR-144-3p | ||
| hsa-miR-574-3p | AC008060.7 | |
| chr22-38_28785274-29006793.1 | ||
| CH17-351M24.1 | ||
| RP4-742C19.13 | ||
| hsa-miR-34a-3p | ||
| hsa-miR-31-5p | RP11-177H2.1 | |
| RP11-863P13.1 | ||
| RP11-426C22.5 | ||
| hsa-miR-144-5p | LINC01555 | |
| ELDR | ||
| SMAD7 | hsa-miR-217 | AC079779.6 |
| hsa-miR-216a-5p | RP11-1080G15.1 | |
| hsa-miR-25-3p | ||
| hsa-miR-21-5p | ||
| hsa-miR-20a-5p | ||
| hsa-miR-106b-5p | EPN2-AS1 | |
| RP11-243E13.2 | ||
| CTC-548K16.2 | ||
| chr22-38_28785274-29006793.1 | ||
| hsa-miR-93-5p | RP11-486M23.3 | |
| hsa-miR-33a-3p | XIST | |
| FAM183CP | ||
| TRPC7-AS1 | ||
| hsa-miR-520g-3p | RP11-315E17.1 | |
| RP11-466A19.4 | ||
| AC144831.1 | ||
| hsa-miR-195-5p | CTBP1-AS2 | |
| SLC7A11-AS1 | ||
| hsa-miR-497-5p | LINC01135 | |
| hsa-miR-424-5p | SNHG1 | |
| hsa-miR-92b-3p | ||
| hsa-miR-185-5p | HMMR-AS1 | |
| RP6-159A1.4 | ||
| KCNQ1OT1 | ||
| MIS18A-AS1 | ||
| hsa-miR-4775 | RP4-671O14.5 | |
| LINC01149 | ||
| LINC01206 | ||
| CTD-2541J13.2 | ||
| hsa-miR-519d-3p | RP11-1099M24.6 | |
| hsa-miR-590-5p | SNHG1 | |
| TGFBR1 | hsa-let-7c-5p | |
| hsa-miR-128-3p | GS1-600G8.5 | |
| hsa-miR-140-5p | ||
| hsa-miR-142-3p | ||
| hsa-miR-181a-5p | ||
| hsa-let-7g-5p | ||
| hsa-let-7b-5p | ||
| hsa-miR-202-5p | CTD-2006H14.2 | |
| RP11-325K19.1 | ||
| hsa-miR-27b-3p | ||
| hsa-miR-376c-3p | BOLA3-AS1 | |
| ERICH3-AS1 | ||
| hsa-miR-135b-5p | RP11-528M18.2 | |
| ZRANB2-AS2 | ||
| CTC-241N9.1 | ||
| FAM25BP | ||
| RP11-96C23.14 | ||
| CTD-2050N2.1 | ||
| CTD-2165H16.4 | ||
| RP4-555D20.2 | ||
| hsa-miR-490-3p | AC009264.1 | |
| RP3-512E2.2 | ||
| hsa-miR-20a-5p | ||
| hsa-miR-181c-5p | RP11-1049A21.2 | |
| hsa-miR-199a-5p | AC144525.1 | |
| RP11-755B10.3 | ||
| LINC00907 | ||
| RP11-338N10.1 |
4. Discussion
Over the last few decades, an overgrowing line of evidence indicated a link of noncoding RNA molecules with the gene regulatory networks including those controlling the disease pathogenesis [6]. Having the tight interaction between viral proteins with their host proteome, it is expected that the viral infection could influence the noncoding RNA network in humans. There is a current urgent need for studying the pathogenesis of human SARS-CoV which has hit the world in a tremendously ascending trend. Thus, a huge number of research groups are trying very hard to find rapid diagnostic methods, studying current treatment options, developing novel therapeutic options, and hopefully developing an effective vaccine. However, fine-tuning crosstalk between viral proteins and host noncoding RNA machinery is almost an under appreciated layer in this investigation race. On the other hand, studying the noncoding RNAs in response to SARS-CoV infection using wet-lab strategies or low-scale methods might not fit the current emergency call. Therefore, we aimed to follow a comprehensive bioinformatics approach to dissect the strongest possible changes in the noncoding RNA network in response to the viral infection in humans using the available online databases. Our in-silico analysis has built a network of protein-protein interaction between the Human SARS coronavirus (SARS-CoV) and host proteome (Fig. 1), as well as strong miRNA-mRNA-lncRNA crosstalk (Fig. 4 and Table 2) possibly modulating the human response to the viral infection.
In this study, we have performed multiple layers of bioinformatics analyses in order to obtain a big picture of the most significant events that occurred following the virus-host interaction in terms of changes in cell signaling pathways and gene expression via noncoding RNAs. We applied the viruses. STRING online database as a powerful platform to explore the most significant protein-protein interactions between SARS-CoV viral particles and host proteome. Considering the median confidence scores greater than 0.4, our analysis predicted that 15 viral proteins might be interacting directly with 40 host proteins, as depicted in the network (Fig. 1). In order to gain a functional prediction of the interacting proteins, the list of obtained strong interactions was then reanalyzed using gene ontology and gene set enrichment analysis (GSEA). A more concentrated list of the major pathways involved were provided using these in silico techniques, among which the interacting genes showed a great association with the TGF-β signaling and its receptor cytoplasmic activity. Also, significant GO categories for overlapped genes using the threshold of P < 0.05 and FDR < 0.05 showed meaningful predicted changes in enriched GO functions of the genes involved in interaction network, including those controlling the transforming growth factor-beta, transforming growth factor-beta receptor cytoplasmic mediator activity, and cellular response to endogenous stimulus. Based on our data, it is assumed that SARS-CoV infection in mammals raises a high level of inflammatory and immune responses by direct viral-host protein interactions. There are also studies that have been shown the role of TGF-beta signaling pathway in SARS-CoV infection [[13], [14], [15]].
In an effort to decipher the probable non-coding RNAs molecules involved, we further analyzed the most significant abovementioned gene sets using the miRTarBase and miRWalk algorithm. This analysis resulted in a network of miRNA-mRNA and miRNA-lncRNA interaction illustrated in Fig. 4. Various mechanisms based on miRNA-mediated regulation of viruses have been reported [16]. This highly coordinated interaction may eventually result in positive outcomes with regard to the host immune response or in favor of viral proliferation [17,18]. This is highly depends on the miRNA target [19]. More precisely, the small noncoding RNA might act directly by guiding an Argonaute protein on the viral mRNA or might target a cellular transcript that is important in the host immune response against viral infection. Consistently, our analysis confirmed that a change in miRNA network targeting the main immune hub signaling pathways responding to SARS-CoV infection potentially act as important immune-modulatory mechanisms in mammals. These findings predict a highly immunogenic viral-host interaction which may be very promising with regard to vaccine development.
Collectively, in this in silico-based study, we reported possible way by which SARS-CoV infection can alter the miRNA network activity, and discussed a scenario in which the host miRNA can function either by modulating the host immune response transcripts or by directly targeting viral transcripts. Thus, it is quite possible that our knowledge of targeting the viral transcript by miRNA machinery can be exploited to develop novel antiviral therapeutic strategies.
Declaration of Competing Interest
The authors declare that there is no conflict of interest.
Acknowledgement
The authors thank LSUHSC School of Medicine and Fred G. Brazda Foudation for financial support.
References
- 1.Gillen J., Nita-Lazar A. Experimental analysis of viral–Host interactions. Front. Physiol. 2019;10 doi: 10.3389/fphys.2019.00425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Omer S.B., Malani P., Del Rio C. The COVID-19 pandemic in the US: a clinical update. JAMA. 2020 doi: 10.1001/jama.2020.5788. [DOI] [PubMed] [Google Scholar]
- 3.Treiber T., Treiber N., Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019;20(1):5–20. doi: 10.1038/s41580-018-0059-1. [DOI] [PubMed] [Google Scholar]
- 4.Yousefi H. Long noncoding RNAs and exosomal lncRNAs: classification, and mechanisms in breast cancer metastasis and drug resistance. Oncogene. 2019:1–22. doi: 10.1038/s41388-019-1040-y. [DOI] [PubMed] [Google Scholar]
- 5.Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011;12(12):861. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 6.Damas N.D., Fossat N. Functional interplay between RNA viruses and non-coding RNA in mammals. Noncoding RNA. 2019;5(1) doi: 10.3390/ncrna5010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gulbahce N. Viral perturbations of host networks reflect disease etiology. PLoS Comput. Biol. 2012;8(6) doi: 10.1371/journal.pcbi.1002531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sadegh S. Exploring the SARS-CoV-2 virus-host-drug interactome for drug repurposing. arXiv preprint arXiv. 2020 doi: 10.1038/s41467-020-17189-2. 2004.12420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cook H.V. Viruses. STRING: A virus-host protein-protein interaction database. Viruses. 2018;10(10):519. doi: 10.3390/v10100519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Subramanian A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dweep H., Gretz N. miRWalk2. 0: a comprehensive atlas of microRNA-target interactions. Nat. Methods. 2015;12(8):697. doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- 12.Mootha V.K. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 13.Li S.-W. SARS coronavirus papain-like protease induces Egr-1-dependent up-regulation of TGF-β1 via ROS/p38 MAPK/STAT3 pathway. Sci. Rep. 2016;6:25754. doi: 10.1038/srep25754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen W. A potential treatment of COVID-19 with TGF-β blockade. Int. J. Biol. Sci. 2020;16(11):1954. doi: 10.7150/ijbs.46891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zuo W., Zhao X., Chen Y.G. SARS coronavirus and lung fibrosis. Mol. Biol. SARS-Coronavirus. 2009;(July (22)):247–258. doi: 10.1007/978-3-642-03683-5_15. [DOI] [Google Scholar]
- 16.Li Q. Cellular microRNA networks regulate host dependency of hepatitis C virus infection. Nat. Commun. 2017;8(1):1789. doi: 10.1038/s41467-017-01954-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jopling C.L. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309(5740):1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 18.Henke J.I. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 2008;27(24):3300–3310. doi: 10.1038/emboj.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Girardi E., López P., Pfeffer S. On the importance of host MicroRNAs during viral infection. Front. Genet. 2018;9:439. doi: 10.3389/fgene.2018.00439. [DOI] [PMC free article] [PubMed] [Google Scholar]