Highlights
-
•
Staphylococci are commonly found in the environment of semi-extensive dairy farms and milk is the main source of contamination.
-
•
Manual milking can play a role in the dissemination of Staphylococcus spp. in semi-extensive dairy farms.
-
•
Methicillin-resistant Staphylococcus aureus (MRSA) was detected in milk and milker's hand swabs and might be of public health significance.
Keywords: Antimicrobial resistance, MecA, MRSA, Methicillin-resistant Staphylococcus aureus
Abstract
This study aimed to investigate the occurrence, genotypic relatedness and antimicrobial resistance of Staphylococcus aureus and coagulase-negative Staphylococcus from milk and environmental sources in dairy herds. A total of 110 staphylococci recovered from 147 samples collected at 21 semi-extensive dairy farms in Northeastern Brazil were investigated. Staphylococcus aureus isolates were identified and screened for methicillin resistance by means of a duplex-PCR. The highest frequency of contamination by S. aureus was observed for milk samples (38.1%), while contamination by coagulase-negative staphylococci (CoNS) was most commonly detected in milkers’ hand swabs (52.4%) and environmental samples (29.5%). Two mecA-positive Staphylococcus aureus (2/40; 5%) were detected, while the same gene was found in fourteen (14/70; 20%) CoNS. Clonally related isolates from milk and environmental sources, such as the surface of gates, were detected by PFGE. This study reports the occurrence of MRSA in dairy farms under semi-extensive production practices and reinforces the importance of environment as a source of Staphylococcus contamination in dairy herds.
1. Introduction
Staphylococcus aureus (S. aureus) is a major pathogen causing a variety of diseases in animals, such as mastitis. In humans, S. aureus has been responsible for innumerable cases of food poisoning and invasive infections that are acquired in communities or hospitals. The emergence of antimicrobial resistance in S. aureus poses a risk to public health, especially methicillin-resistant Staphylococcus aureus (MRSA), which are resistant to all β-lactam antimicrobials and many other drugs from different classes. MRSA is recognized worldwide as an important nosocomial pathogen (Kamal et al., 2013, Kwon et al., 2005, Song et al., 2015), but the increase in the number of community-acquired infections (CA-MRSA) is intriguing. Recently, epidemiological studies suggest that CA-MRSA might have originated from livestock. Livestock-associated MRSA strains (LA-MRSA), such as ST398, have been considered a major threat and the epidemiology of those pathogens in animal production systems are still not clarified. Increased resistance to beta-lactams in Staphylococcus isolated from clinical and subclinical mastitis has been reported (Kwon et al., 2005, Li et al., 2015, Vanderhaeghen et al., 2010), which might be linked to the indiscriminate use of antibiotics in animal production.
Despite the extensive knowledge on Staphylococcus as a major cause of mastitis in dairy cattle, little is known about the role of the environment as a source of Staphylococcus contamination in semi-extensive dairy farms. Although the environmental contamination by staphylococci might not play a role in the epidemiology of mastitis, since this pathogen is mainly transmitted from animal-to-animal during milking, information about the epidemiology and antimicrobial resistance of those bacteria in the environment is important for the public health. Therefore, this study aimed to investigate the occurrence, genotypic relatedness and antimicrobial resistance of Staphylococcus aureus and coagulase-negative Staphylococcus isolated from milk and environmental sources in dairy farms under semi-extensive production systems in Northeastern Brazil.
2. Materials and methods
A total of one hundred and forty seven samples including fresh milk, milking bucket, bulk milk tank, rope and gates, as well as from the milkers’ hands were collected from 21 semi-extensive dairy farms. The semi-extensive dairy farming refers to a production system that uses small inputs of labor (normally a family-based sytem), fertilizers, equipments and capital. The farmsa adopt a pasture-based system and use manual milking, which is performed in a barn. Conventional microbial isolation was performed using previously described methods (Oliveira et al., 2016). Typical staphylococcal colonies on Baird-Parker agar were selected, transferred to brain heart infusion broth (BHI, Oxoid, Basingstoke, UK) and incubated at 35–37 °C for 24 h. Confirmation was performed by Gram staining, catalase, oxidase, and coagulase tube test. S. aureus species were confirmed by means of polymerase chain reaction (PCR) targeting nuc gene (270 bp), in a duplex approach targeting the mecA gene (162 bp) to detect MRSA, as previously described by Oliveira and Lencastre (2002); and Oliveira et al. (2016).
Antimicrobial susceptibility profiles were determined by the disc diffusion method (Kirby-Bauer) according to CLSI (2015) using 12 drugs: ampicillin (Am; 10 µg), erythromycin (Er; 15 µg), streptomycin (St; 30 µg), oxacillin (Ox; 1 µg), tetracycline (Tet; 30 µg), amoxicillin-clavulanic acid (Ax;10 µg), trimethoprim-sulfamethoxazole (Tr; 5 µg), ceftriaxone (Ce; 10 µg), ciprofloxacin (CIP; 5 µg), vancomycin (Vc; 30 µg), penicillin (Pn; 10 µg) and gentamicin (Gm; 10 µg).
3. Results
The highest frequency of contamination by S. aureus was observed for milk samples (38.1%), while CoNS were mainly cultured from the surface of hand swabs (52.4%) and environmental samples. Out of the total 147 collected samples, 110 were positive for staphylococci. The majority of those isolates (70; 47.6%) were coagulase-negative staphylococci (CoNS). A total of 40 (27.2%) was identified as S. aureus, and originated from milk (38.1%), hand swab (14.3%), rope (9.5%), bulk tank milk (4.8%) and gate (4.8%) samples. No S. aureus was cultured from water and milking bucket samples. The frequency of CoNS ranged from 9.5% to 52.4% according to the sample types: 14.3% for milk, 52.4% for hand swabs, 38.1% for milking bucket swabs, 33.3% for rope swabs, 23.8% for water and 9.5% for bulk milk tanks.
Twenty-one different antimicrobial resistance patterns were identified (Table, supplementary material) and PnSt (7; 17.5%) was the most common R-type among S. aureus, while Ox R-type was the most common among CoNS species (8; 11.4%), followed by OxPn (5; 7.14%). Among the 40 S. aureus strains, only two (5%) were confirmed as MRSA and were cultured from a milk sample (farm 59) and from a hand swab (farm 60) (Fig. 1). Not all MRSA isolates were multidrug resistant (MDR).
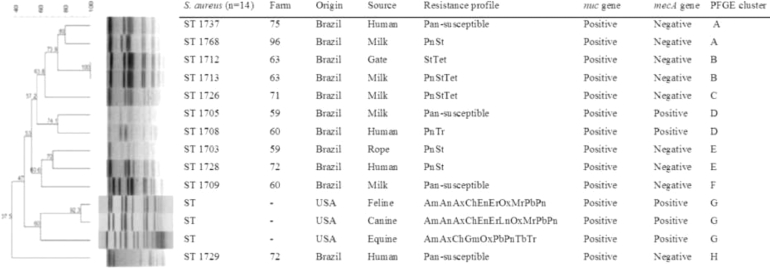
Fig. 1.
Dendrogram showing the genotypic relatedness of 11 S. aureus isolates from north-eastern Brazilian dairy farms by Pulsed-field Gel Electrophoresis (PFGE). As outgroup, 3 non-related S. aureus from feline, canine and equine in USA were included in the analysis.
Ampicillin (Am; 10 µg), Erythromycin (Er; 15 µg), Streptomycin (St; 30 µg), Oxacillin (Ox; 1 µg), Tetracycline (Tet; 30 µg), Amoxicillin-clavulanic acid (Ax;10 µg), Trimethoprim-sulfamethoxazole (Tr; 5 µg), Ceftriaxone (Ce; 10 µg), Ciprofloxacin (CIP; 5 µg), Vancomycin (Vc; 30 µg), Penicillin (Pn; 10 µg), Gentamicin (Gm; 10 µg), Chloramphenicol (Cm; 30 µg), Enrofloxacin (En; 5 µg), Polymixin (Pb; 50 µg/ 300 UI), Amikacin (An; 30 µg) and Ch = cephalothin (30 µg).
Considering CoNS, fourteen strains (14/70; 20%) from various environmental sources (hand, n = 4, milk, n = 3, milking bucket, n = 3, gate, n = 2, rope, n = 1 and water, n = 1 were positive for mecA gene.
MRSA and MRCoNS were genotyped by pulsed field gel electrophoresis (PFGE) according to Ribot et al. 2006. Fig. 1 shows that seven major genotypic clusters (A-G) were identified. A high genetic similarity (greater than 70%) was observed among isolates from different sources in a given farm (Cluster B). MRSA isolates were highly similar, even though they were isolated from different sources (hand swabs and milk) and from different farms (Cluster D). A similar finding was also observed in cluster A for mecA negative Staphylococcus aureus. We observed two clonally related isolates (Cluster B) from different sources (milk and gate) in a same farm. Two isolates sharing the same PFGE pattern (D) harboured the mecA gene and originated from hand swabs and milk. Clusters A (n = 1) and E (n = 2) were comprised mainly by isolates showing the same R type (PnSt).
4. Discussion
The results show that staphylococci are commonly found in the environment of semi-intensive dairy herds. The high recovery rate of S. aureus from milkers hand samples (14.3%) suggest the importance of manual milking in the spread of Staphylococcus in dairy herds. Furthermore, the detection of genotypically-related S. aureus (Fig. 1, clusters A and D) in hand swabs and milk illustrates the potential role of manual milking in the transfer of S. aureus between humans and animals. This reinforces previous findings showing that dairy cows are potential sources of MRSA to humans (Oliveira et al., 2016). Therefore, adequate cleaning of hands before and after milking practices must be considered a key practice to promote health in dairy farms, especially in developing regions where manual milking is a common practice.
We observed different levels of contamination by S. aureus in environmental samples, such as 4.8% for bulk milk tank swabs and gate swabs, and 9.5% for rope swabs. Importantly, those isolates were genotypically related to Staphylococcus aureus from milk (Fig. 1, clusters B and E). These results suggest the presence of clonally-related staphylococci over the environment of extensive dairy farms. The rope is used to restrain the cows for milking and the presence of genotypic related Staphylococcus aureus in rope swabs and hand swabs (cluster E). Again, these findings suggest that milking utensils commonly used for milking might be associated with the spread of staphylococci within a farm. The rope is sometimes used to hobble the cows during milking, and is often in contact with the feces and soil, and the animal's skin.
Under routine daily practices on dairy farms, barn gates are very important means to manage animals. Gates are frequently in contact to animal surfaces and human hands and cleaning of those type of devices are virtually never done. Our results showed an undistinguishable genotypic pattern (Fig. 1, cluster B) between staphylococci from gate and milk in a given farm, reinforcing that Staphylococcus aureus from milk could be spread from the environment.
Water was not contaminated by Staphylococcus aureus, although the water quality is generally poor in the investigated region. Therefore, the results suggest that water does not play a role as a source of Staphylococcus aureus in non-intensive dairy herds, even though it is well known that poor water quality used to clean milking utensils can affect the overall microbial quality of milk. It is noteworthy to mention that the present study was focused on staphylococcal contamination only and water quality is a key factor in the epidemiology of many other diseases. Therefore, in general, water quality must not be neglected to achieve desirable quality and safety in milk production systems. A total of 23.8% of the water samples were contaminated by CoNS, which was expected since those are ubiquitous bacteria. We also reported the occurrence of CoNS in the environmental samples at higher frequencies than S. aureus, such as hand swabs (52.4%), bulk bucket (9.5%), gate (33.3%), and collecting bucket and rope (38.1%). Only milk has a higher contamination by S. aureus compared to CoNS. This could be related to the fact that S. aureus is an important intramammary infectious agent in dairy cows and normally found in milk. Similar results were reported by Beuron et al. (2014). However, the importance of CoNS as mastitis-causing agent has increased worldwide over the last years (Hosseinzadeh & Saei, 2014), which suggests that the high environmental contamination by CoNS must not be neglected in production systems under manual milking.
Regarding the antimicrobial resistance findings, similar studies also reported high resistance rates of Staphylococcus against penicillin (Kwon et al., 2005, Li et al., 2015). The occurrence of MRSA in dairy herds is not common but the frequency of MRSA in milk samples in the present study is similar to those reported elsewhere (Ciftci, Findik, Onuk, & Savasan, 2009; Haran et al., 2011; Kamal et al., 2013, Kumar et al., 2011). More recently, Li et al. (2015) reported one MRSA out of 120 S. aureus from bovine milk in China and Oliveira et al. (2016) detected the presence of mecA gene in twenty-one S. aureus isolates from milk and in one isolate from a hand swab in semi-extensive dairy cows in Brazil. Although resistance to methicillin has been traditionally linked to the expression of the mecA gene, novel resistance mechanism conferring multiresistance against methicillin and other beta-lactams have been identified, such as the homologue mecC which is also located on the staphylococcal cassette chromosome mec (SCCmec) (Li et al., 2015, Paterson et al., 2014, Petersen et al., 2013). Therefore, this should be considered in further studies to investigate the epidemiology of MRSA in animal production systems. (Table 1)
Table 1.
Frequency of resistance patterns in Staphylococcus aureus and coagulase-negative staphylococci (CoNS) from dairy samples collected in north-eastern Brazilian farms.
| Resistance pattern* | S. aureus (n = 40) | % | CoNS (n = 70) | % | Source⁎⁎ | Farm |
|---|---|---|---|---|---|---|
| ErOxPnStTet | 0 | 0.0 | 1 | 1.4 | M | 7 |
| PnTr | 1 | 2.5 | 1 | 1.4 | H; H | 60 |
| CeOxPnSt | 0 | 0.0 | 1 | 1.4 | H | 63 |
| OxPnSt | 0 | 0.0 | 1 | 1.4 | G | 67 |
| ErPnTet | 0 | 0.0 | 1 | 1.4 | R | 71 |
| ErPnTetTr | 0 | 0.0 | 1 | 1.4 | H | 74 |
| ErOxPn | 0 | 0.0 | 1 | 1.4 | R | 96 |
| Er | 0 | 0.0 | 2 | 2.85 | R; M | 94; 13 |
| CeGmOxPnSt | 0 | 0.0 | 2 | 2.85 | BB | 94;94 |
| OxTet | 0 | 0.0 | 2 | 2.85 | CB; W | 59; 91 |
| OxPnTet | 0 | 0.0 | 2 | 2.85 | G; H | 65; 96 |
| OxPn | 0 | 0.0 | 5 | 7.14 | H; G; CB; CB; M | 96; 75; 71; 42 |
| Ox | 2 | 5 | 8 | 11.42 | M; H; R; CB; M; H; G | 71; 74; 73; 79; 88 |
| StTet | 1 | 2.5 | 0 | 0.0 | G | 63 |
| AmErPn | 1 | 2.5 | 0 | 0.0 | R | 72 |
| Tet | 1 | 2.5 | 3 | 4.28 | H; H; G; H | 75; 95; 69; 65 |
| PnTet | 2 | 5 | 0 | 0.0 | M; M | 31; 69 |
| PnStTet | 4 | 10 | 1 | 1.4 | M; M; M; M; H | 8; 33; 63; 71; 67 |
| Pn | 5 | 12.5 | 4 | 5.71 | M | at least five |
| PnSt | 7 | 17.5 | 1 | 1.4 | M; H | at least five |
| Pan-Susceptible | 16 | 40 | 33 | 47.14 | M, HH, BB | at least five |
Resistance pattern: Am = Ampicillin; Er = Erythromycin; St = Streptomycin; Ox = Oxacillin; Tet = Tetracycline. Ax = Amoxicillin-clavulanic acid; Tp = Trimethoprim-sulfamethoxazole; Ce = Ceftriaxone; CIP = Ciprofloxacin. Vc = Vancomycin; Pn = Penicillin and Gm = Gentamicin.
Source: M = milk; H = hand; R = rope; G = gate; BB = bulk bucket; CB = collecting bucket; W = water.
5. Conclusions
The present study indicates that staphylococci are commonly found in the environment of semi-extensive dairy farms in Northeastern Brazil, and the molecular typing findings suggest that most of these may originate from milk. Manual milking could play a role in the spread of staphylococci in the environment. The detection of clonally-related MRSA in milk and on hand swabs reinforces the proposal that manual milking can play a role in the dissemination of MRSA in dairy production systems. As some MRSA infections in humans have been confirmed to originate from animal sources (livestock-associated MRSA), our findings suggest that MRSA could play a role as an occupational agent in semi-extensive dairy herds, mainly in those under manual milking.
Conflict of interest
None of the authors of this paper has a financial or personal relationship with other people or organizations that could inappropriately influence or bias the content of the paper.
Acknowledgments
This study was funded by The National Council for Scientific and Technological Development (CNPq). We thank Tricia H. Gearhart for helping in the sampling collection and also for laboratory technical support.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.vas.2018.07.007.
Appendix. Supplementary materials
References
- Beuron D.C., Cortinhas C.S., Botaro B.G., Macedo S.N., Gonçalves J.L., Brito M.A.V.P. Risk factors associated with the antimicrobial resistance of Staphylococcus aureus isolated from bovine mastitis. Brazilian Journal of Veterinary Research. 2014;34:947–952. [Google Scholar]
- Ciftci A., Findik A., Onuk E.E., Savasan S. Detection of methicillin resistance and slime factor production of Staphylococcus aureus in bovine mastitis. Brazilian Journal of Microbiology. 2009;40:254–261. doi: 10.1590/S1517-83822009000200009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI . CLSI document VET01S; Wayne, PA, USA: 2015. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals- Third Edition. [Google Scholar]
- Hosseinzadeh S., Dastmalchi Saei H. Staphylococcal species associated with bovine mastitis in the North West of Iran: Emerging of coagulase-negative staphylococci. International Journal of Veterinary Science and Medicine. 2014;2:27–34. [Google Scholar]
- Kamal R.M., Bayoumi M.A., Abd El Aal S.F.A. MRSA detection in raw milk, some dairy products and hands of dairy workers in Egypt, a mini-survey. Food Control. 2013;33:49–53. [Google Scholar]
- Kumar R., Yadav B.R., Singh R.S. Antibiotic resistance and pathogenicity factors in Staphylococcus aureus isolated from mastitic Sahiwal cattle. Journal of Biosciences. 2011;36:175–188. doi: 10.1007/s12038-011-9004-6. [DOI] [PubMed] [Google Scholar]
- Kwon N.H., Park K.T., Moon J.S., Jung W.K., Kim S.H., Kim J.M. Staphylococcal cassette chromosome mec (SCCmec) characterization and molecular analysis for methicillin-resistant Staphylococcus aureus and novel SCCmec subtype IVg isolated from bovine milk in Korea. Journal of Antimicrobial Chemotherapy. 2005;56:624–632. doi: 10.1093/jac/dki306. [DOI] [PubMed] [Google Scholar]
- Li L., Zhou L., Wang L., Xue H., Zhao X. Characterization of methicillin-resistant and-susceptible staphylococcal isolates from bovine milk in northwestern China. PLoS One. 2015;10 doi: 10.1371/journal.pone.0116699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira C.J.B., Tiao N., de Sousa F.G.C., de Moura J.F.P., Santos Filho L., Gebreyes W.A. Methicillin-resistant Staphylococcus aureus from Brazilian dairy farms and identification of novel sequence types. Zoonoses Public Health. 2016;63:97–105. doi: 10.1111/zph.12209. [DOI] [PubMed] [Google Scholar]
- Oliveira D.C., de Lencastre H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrobial Agents and Chemotherapy. 2002;46:2155–2161. doi: 10.1128/AAC.46.7.2155-2161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson G.K., Morgan F.J., Harrison E.M., Peacock S.J., Parkhill J., Zadoks R.N. Prevalence and properties of mecC methicillin-resistant Staphylococcus aureus (MRSA) in bovine bulk tank milk in Great Britain. Journal of Antimicrobial Chemotherapy. 2014;69:598–602. doi: 10.1093/jac/dkt417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen A., Stegger M., Heltberg O., Christensen J., Zeuthen A., Knudsen L.K. Epidemiology of methicillin-resistant Staphylococcus aureus carrying the novel mecC gene in Denmark corroborates a zoonotic reservoir with transmission to humans. Clinical Microbiology and Infection. 2013;19:E16–E22. doi: 10.1111/1469-0691.12036. [DOI] [PubMed] [Google Scholar]
- Ribot E.M., Fair M.A., Gautom R., Cameron D.N., Hunter S.B., Swaminathan B., Barrett T.J. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7. Salmonella, and Shigella for PulseNet. Foodborne Pathogens and Diseases. 2006;3:59–67. doi: 10.1089/fpd.2006.3.59. [DOI] [PubMed] [Google Scholar]
- Song M., Bai Y., Xu J., Carter M.Q., Shi C., Shi X. Genetic diversity and virulence potential of Staphylococcus aureus isolates from raw and processed food commodities in Shanghai. International Journal of Food Microbiology. 2015;195:1–8. doi: 10.1016/j.ijfoodmicro.2014.11.020. [DOI] [PubMed] [Google Scholar]
- Vanderhaeghen W., Cerpentier T., Adriaensen C., Vicca J., Hermans K., Butaye P. Methicillin-resistant Staphylococcus aureus (MRSA) ST398 associated with clinical and subclinical mastitis in Belgian cows. Veterinary Microbiology. 2010;144:166–171. doi: 10.1016/j.vetmic.2009.12.044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.