Abstract
A total of 32 Pasteurella multocida isolates were obtained from 60 cases of swine pneumonic lungs collected in “Castilla y León” (northwestern Spain) between November 2017 and April 2018. Capsular type A isolates were isolated from 96.9% cases and capsular type D from the remaining 3.1%. All isolates were characterized for their susceptibilities to eight antimicrobial agents and the presence of eight resistance genes. The frequency of susceptibility was lower than 60% in four of the drugs, 84.4% of the isolates showed resistance to at least two compounds, and 46.9% to a combination of three drugs. The resistance patterns suggested that enrofloxacin, chloramphenicol, tetracycline and cefotaxime were the compounds most likely active to P. multocida. The usage of PCR revealed that ermC, blaROB1, tetB and msrE genes occurred in more than 37.0% isolates, that suggested its putative accountability in the resistance of the strains harbor them. However, most were detected in susceptible strains and only a genetic explanation for the resistance could be linked to erythromycin. Therefore, the resistances to clyndamicin, cotrimoxazol, β-lactams and tetracyclin observed by phenotypic testing remains genetically unexplained and further investigations are required.
Keywords: Pasteurella multocida, Swine pneumonia, Antimicrobial agents, Disk diffusion testing, Resistance genes
1. Introduction
Pasteurella multocida is a small nonmotile gram-negative coccobacillus located in the upper portion of the porcine respiratory tract which, although is part of the commensal organisms, also can induce pneumonia in growing and finishing pigs. In this respect, P. multocida is regarded as a secondary opportunistic agent of the porcine respiratory disease complex (PRDC), which is one of the most common diseases of porcine intensive production (Blackall et al., 2000). Besides, it is one of the organisms most commonly isolated from pneumonic lesions in pigs as primary agent of haemorrhagic septicaemia (De Oliveira Filho et al., 2015) caused by P. multocida B:2 (Kachooel, Ranjbar, & Kachooel, 2017) or E:5 (de Alwis, 1992). In addition, P. multocida may induce atrophic rhinitis along with Bordetella bronchiseptica (Gois, Barnes, & Ross, 1983). It has been separated into four subspecies on the basis of fermentation of sorbitol and dulcitol (Mutters, Ihm, & Pohl, 1985), being P. multocida subsp. multocida the only species of interest in diseases of swine.
The usage of antimicrobial agents, such as β-lactams, sulphonamide + trimethoprim (cotrimoxazol), florfenicol, macrolides and tetracyclines remains to be the best treatment to control PRDC (Dayao, Gibson, Blackall and Turni, 2014, Karriker, Coetzee, Friendship and Prescott, 2013). This application has the capability to select for these compounds and gives rise to the development of resistances (Barton, Pratt, & Hart, 2013). So, resistance to antimicrobial agents used to inhibit the growth of P. multocida has been determined previously in disorders caused by this pathogen isolated from different countries (Gutiérrez Martín and Rodríguez Ferri, 1992, San Millán et al., 2009, Tang et al., 2009, Vera Lizarazo, Rodríguez Ferri, Martín de la Fuente and Gutiérrez Martín, 2006). In this respect, tetB gene has been previously detected respectively in 25% and 20% isolates (Dayao, Gibson, Blackall and Turni, 2016, Kehrenber, Salmon and Watts, 2001) and the presence of blaROB-1 and blaTEM-1 genes has been also reported (Chander, Oliveira and Goyal, 2011, Dayao, Gibson, Blackall and Turni, 2016). However, ermA, ermC, msrE or mphE genes were not found among 20 Australian isolates (Dayao et al., 2016).
The objective of this study were to determine the in vitro antimicrobial susceptibility of clinical isolates of P. multocida recovered in Spain from November 2017 to April 2018. from slaughterhouses and to characterize the potential presence of resistant genes to antimicrobials on the part of the isolated strains and their putative association to resistance.
2. Materials and methods
2.1. Bacterial isolates
The P. multocida isolates tested were obtained from 60 lungs with pneumonic lesions coming from pigs from “Castilla y León” which were slaughtered between November 2017 and April 2018 at the abattoir of the “Embutidos Rodríguez” corporation, placed in “Soto de la Vega” (León, northwestern Spain). P. multocida isolates were recovered on Columbia agar with 5% sheep blood (Oxoid). The strains were identified according to standard phenotypic procedures -gram stain, catalase, cytochrome oxydase and API 20-NE strips- (Larivière, Leblanc, Mittal, & Martineau, 1992) and they were then genetically confirmed by a specific species PCR assay (Townsend, Frost, Lee, Papadimitriou, & Dawkins, 1998), which also allowed the determination of the capsular type A or D (Townsend, Boyce, Chung, Frost, & Adler, 2001).
2.2. Agar disk diffusion method
β-lactams, cephalosporins, fenicols, lincosamides, macrolides, quinolones, tetracyclines and sulfonamides were the groups of antimicrobials agents compared in this study. All the isolates were tested for their antimicrobial susceptibility by the disk diffusion method according to the performance standards M31-A3 of the CLSI (Clinical and Laboratory Standards Institute) (2008). Each strain was tested against eight selected agents: ampicillin (10 μg), cefotaxime (30 μg), chloramphenicol (30 μg), clindamycin (2 μg), erythromycin (15 μg), enrofloxacin (10 μg), tetracycline (30 μg) and sulfamethoxazole + trimethoprim (23.75 μg + 1.25 μg) (cotrimoxazol). Escherichia coli ATCC 25,922 and Staphylococcus aureus ATCC 25,923 were used as quality control (QC) strains. The interpretation of results was based on the breakpoints provided by the CLSI guidelines (CLSI, 2013).
2.3. Detection of antimicrobial resistance genes
The detection of the resistance-associated genes to antimicrobial agents were carried by PCR, as described by Dayao et al. (2016), with all 32 P. multocida recovered. A total of eight antimicrobial resistance genes, belonging to three different families (tetA and tetB, blaROB-1 and blaTEM, and ermA, ermC, msrE and mphE) were tested. The forward and reverse primers used were shown in Table 1.
Table 1.
Primers used in the PCR carried out for the detection of antimicrobial resistance genes in the 32 Pasteurella multocida isolates recovered in Castilla y León, northwestern Spain.
| Resistance gene | Primer | Amplicon size | Annealing temperature |
|---|---|---|---|
| tetA | F: 5′-GTA ATT CTG AGC ACT GTC GC-3′ | 1,057 pb | 62 °C |
| R: 5′-CTG CCT GGA CAA CAT TGT TT-3′ | |||
| tetB | F: 5′CCT TAT CAT GCC AGT CTT GC-3′ | 774 pb | 50 °C |
| R: 5′ ACT GCC GTT TTT TTC GCC-3′ | |||
| blaROB1 | F: 5′ CAT TAA CGG CTT GTT CGC-3′ | 852 pb | 55 °C |
| R: 5′-CTT GCT TTG CTG CAT CTT-3′ | |||
| blaTEM | F: 5′GAG TAT TCA ACA TTT TCG T-3′ | 856 pb | 55 °C |
| R: 5′-ACC AAT GCT TAA TCA GTG A-3′ | |||
| ermA | F: 5′-ACG ATA TTC ACG GTT TAC CCA CTT-A-3′ | 610 pb | 53 °C |
| R: 5-AAC CAG AAA AAC CCT AAA GAC ACG-3′ | |||
| ermC | F: 5′-AAT-CGG CTC AGG AAA AGG-3′ | 562 pb | 55 °C |
| R: 5′-ATC GTC ATT TCC TGC ATG-3′ | |||
| msrE | F: 5′-TAT AGC GAC TTT AGC GCC AA-3′ | 271 pb | 58 °C |
| R: 3′-GCC GTA GAA TAT GAG CTG AT-3′ | |||
| mphE | F: 5’-ATG CCC AGC ATA TAA ATC GC-3’ | 295 pb | 58 °C |
| R:5’-ATA TGG ACA AAG ATAGCC CG-3’ |
F: forward; R: reverse.
3. Results
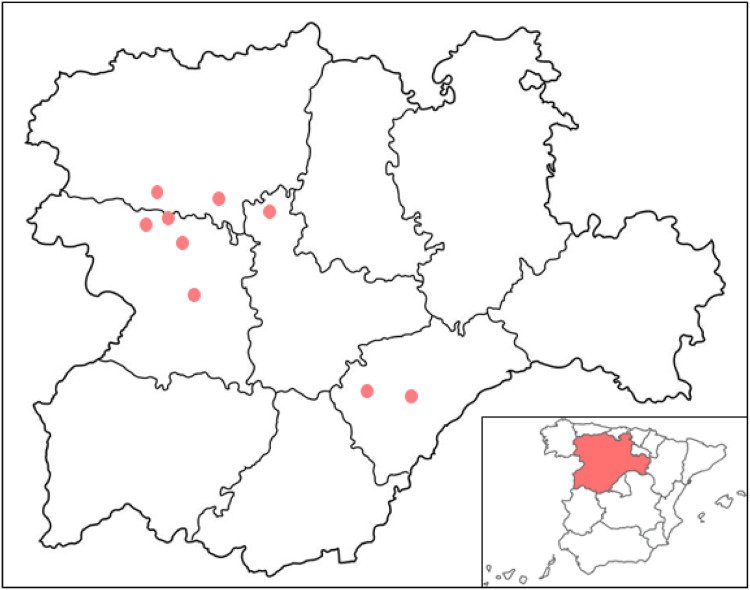
A total of 32 P. multocida clinical isolates were collected, ten (31.2%) of which came from León, eight (25.0%) from Segovia, and seven (21.8%) from each Valladolid and Zamora, all being provinces from “Castilla y León”, Spain (Fig. 1). From these, 31 (96.9%) belonged to capsular type A and only one (3.1%) to capsular type D. The MICs of the E. coli and S. aureus reference strains in each test were within the acceptable QC ranges of antimicrobial disk susceptibility test zone diameters.
Fig. 1.
Geographical distribution of the 32 Pasteurella multocida isolates recovered in Castilla y León, northwestern Spain.
Overall, 96.9% isolates (n = 31) were resistant to one or more antimicrobial agents, in such a way that 29.0% (n = 9) showed resistant to only one compound; 42.0% (n = 13), to two drugs and 29.0% (n = 9), to three antimicrobial agents. No strains were resistant to more than three compounds. The capsular type D was resistant to only one antibiotic (clindamycin). A total of 16 antimicrobial resistance patterns could be seen: 84.4% isolates (n = 27) were resistant to at least two antimicrobials, and 46.9% (n = 15) to a combination of three drugs. The 65.6% of strains (n = 21) were resistant simultaneously to the combination of clindamycin and cotrimoxazol, and the 18.7% (6 isolates each) behaved as resistant at the same time to clindamycin and cotrimoxazol along with ampicillin or erythromycin (Table 2).
Table 2.
Patterns of antimicrobial resistance among the 32 Pasteurella multocida isolates recovered in Castilla y León, northwestern Spain.
| No of isolates | No. of antimicrobial agents | Resistance patterns |
|---|---|---|
| 1 | 0 | No antimicrobial resistance |
| 2 | 1 | CLI (S to the remaining antimicrobials) |
| 3 | 1 | CLI (I to ERY) |
| 3 | 1 | CLI (I to AMP + ERY) |
| 1 | 1 | AMP (I to CLI + COT + ERY) |
| 8 | 2 | CLI + COT (I to ERY) |
| 2 | 2 | CLI + COT (I to ERY + AMP) |
| 2 | 2 | CLI + TET (I to COT + ERY) |
| 1 | 2 | CLI + ERY (I to COT) |
| 1 | 2 | CLI + ERY (I to AMP + TET) |
| 1 | 2 | CLI + COT (I to TET + CEF + CLO) |
| 3 | 3 | CLI + COT + AMP (I to ERY) |
| 1 | 3 | CLI + COT + AMP (I to ERY + CEF + CLO) |
| 1 | 3 | CLI + COT + ERY (I to CEF) |
| 1 | 3 | CLI + COT + TET (I to AMP + ERY + CEF) |
| 1 | 3 | CLI + COT + ERY (I to TET) |
CLI, Clindamycin; S, susceptible; I, intermediate behaviour; ERY, erythromycin; AMP, ampicillin; COT, sulphamethoxazole + trimethoprim (cotrimoxazol); TET, tetracycline; CEF, cefotaxime; CLO, chloramphenicol.
All isolates were susceptible to enrofloxacin, followed by chloramphenicol, cefotaxime and tetracycline, to which 93.8%, 87.5% and 81.2% of clinical strains showed susceptibility in vitro, respectively. On the other hand, 96.9% of them were resistant to clindamycin and 68.7% to cotrimoxazol. The most common behaviour of erythromycin to the 32 isolates surveyed was intermediate susceptibility, with inhibition zone diameters comprised between 14 and 21 mm. The rate of resistant or susceptible isolates to this antibiotic was quite similar, with four and three strains, respectively (Table 3).
Table 3.
Antimicrobial susceptibility of 32 Pasteurella multocida isolates recovered in Castilla y León, northwestern Spain, by disk diffusion test.
| Antimicrobial agent | Susceptible (no of isolates) | Intermediate (no of isolates) | Non-susceptible (no of isolates) |
|---|---|---|---|
| CLI | 3.1% (1) | 0 | 96.9% (31) |
| COT | 31.2% (10) | 0 | 68.7% (22) |
| AMP | 59.4% (19) | 0 | 40.6% (13) |
| ERY | 9.4% (3) | 8.1% (25) | 12.5% (4) |
| TET | 81.2% (26) | 0 | 18.8% (6) |
| CEF | 87.5% (28) | 12.5% (4) | 0 |
| CLO | 93.8% (30) | 6.2% (2) | 0 |
| ENR | 100% (32) | 0 | 0 |
CLI, Clindamycin; COT, sulphamethoxazole + trimethoprim (cotrimoxazol); AMP, ampicillin; ERY, erythromycin; TET, tetracycline; CEF, cefotaxime; CLO, chloramphenicol; ENR, enrofloxacin; R, resistant behaviour; I, intermediate behaviour.
Concerning molecular basis of antimicrobial resistance to the 32 P. multocida isolates, the complete patterns are shown in Table 4. No strains were positive for tetracycline resistance tetA gene, being the only one that was not amplified. A percentage of 40.6% of isolates (n = 14) was positive for tetB gene, but none of them were detected in resistant strains. With regard to β-lactam resistance genes, 40.6% of clinical strains (n = 14) were positive to blaROB1 while only 12.5% (n = 4) were to blaTEM. Globally, 53.1% isolates (n = 17) showed some of these two β-lactam genes, and only one showed both of them. From the strains harboring any of the β-lactam resistance genes, only four exhibited resistance to ampicillin.
Table 4.
Resistance genes harboured by the 32 Pasteurella multocida isolated in Castilla y León, northwestern Spain.
| Castilla y León counties and number of isolates (n) | Resistance genes: | |||||||
|---|---|---|---|---|---|---|---|---|
| tetA | tetB | blaROB-1 | blaTEM | ermA | ermC | mphE | msrE | |
| León (10) | 0 | 1 | 7 | 1 | 0 | 3 | 1 | 4 |
| Segovia (8) | 0 | 7 | 0 | 0 | 6 | 6 | 0 | 1 |
| Valladolid (7) | 0 | 2 | 6 | 0 | 0 | 6 | 0 | 7 |
| Zamora (7) | 0 | 4 | 1 | 3 | 2 | 5 | 0 | 0 |
| Total | 0 (0%) | 14 (40.6%) | 14 (40.6%) | 4 (12.5%) | 8 (25.0%) | 20 (62.5%) | 1 (3.1%) | 12 (37.5%) |
A considerably higher number of isolates (84.4%, 27 isolates) showed resistance genes to any of the macrolides. So, although mphE was only amplified in 3.1% of them (1 isolate), 25.0% (n = 8) were positive to ermA, a figure that reached to 37.5% (n = 12) when msrE was tested; finally, 62.5% (20 isolates) were positive to ermC. Only one of the isolates harboring ermA gene was resistant to erythromycin; other harbored msrE gene; a third one harbored ermA and ermC genes, and other one harbored msrE and mphE genes.
The P. multocida capsular type D exhibited only msrE gene. On the other hand, six of the eight strains recovered in Segovia showed the same resistance behaviour, being positive to tetB, ermA and ermC, and negative to the remaining five other genes; the seventh one exhibited only msrE, and the eighth one showed any of the eight resistance genes compared. A rate of 9.4% (3 isolates), two from Valladolid and the other one from Zamora were positive simultaneously to four of the genes tested; 37.5%, (n = 12) to three of them, and 34.4% (n = 11) to two of them both at once. Only 18.7% (6 isolates) showed one of the eight genes tested.
4. Discussion
In examining the results of this study, a number of issues can be taken into account. Firstly, it is necessary to bear in mind that this investigation relies on a collection of 32 P. multocida isolates from northwestern Spain. For this reason, a larger study would be required to gain insight into the national landscape in Spain; however, it must be assessed that this investigation was performed in the same region and slaughterhouses in which the works performed 14 and 20 years ago took place. Secondly, since the samples were obtained at the abattoir, but it is renown enough that Spain is one of the countries in all Europe where the major antibiotic expense takes place (EFSA 2017, EMA 2016), it is highly likely that the pigs (and consequently clinical isolates) used in this study originated from farms exposed to an extensive antimicrobial treatment.
Clindamycin resistance has previously been seen in P. multocida isolates in Spain both 20 and 14 years ago, when 63 and 132 isolates respectively had MIC values higher than the highest concentration tested, thus suggesting a high level of resistance or a completely inactivity to this lincosamide antibiotic (Vera Lizarazo et al., 2006). Similarly, a 100% resistance rate among 29 clinical isolates was reported by other investigators to this drug (Cote, Hare, Higgins, & Jacques, 1991). Other P. multocida has been also found completely resistant to other lincosamide, lincomicycin (Yeh, Lo, Chang, Chou, & Kuo, 2007). Slightly lower resistance rates (80.3%) have been cited for clinical strains recovered in China (Tang et al., 2009). However, some isolates largely susceptible to clindamycin have been detected in recent studies (Niemann et al., 2018).
When comparing the results obtained in the current study for sulfamethoxazole + trimethoprim with those reported previously in Spain, it can be concluded that the resistance percentage is much higher now than 20 years ago. However, it was ten points lower than 14 years ago (Vera Lizarazo et al., 2006), thus indicating that resistance pressure to this combination of two drugs has not been high over the two last decades in Spain. Resistance values for cotrimoxazol reported in earlier studies (Quedi Furian et al., 2016, Tang et al., 2009) were substantially higher (67.5% and 74.2%, respectively) than those observed in the current work. Quite different, hardly or no resistant isolates (2% and 0%, respectively) were found in other studies (Dayao et al., 2014; de Jong et al., 2014).
Ampicillin exhibited efficacies over 97.0% two decades ago in Spain (Vera Lizarazo et al., 2006). The fact that in our study the susceptibility decreased to 59.4% clearly states that the wide use of this antibiotic and of β-lactams in general in treatment and prevention of respiratory tract disorders of swine has exerted a high selective resistance pressure on P. multocida isolates. This resistance can be mainly linked with the fact that these isolates would harbour the blaROB1 resistance gene, which is widely carried by different human or animal genera of family Pasteurellaceae, such as P. multocida (Dayao, Gibson, Blackall and Turni, 2016, San Millán et al., 2009), Actinobacillus pleuropneumoniae (Dayao et al., 2016; Matter et al., 2007) or A. porcinotonsillarum (Matter et al., 2007), Haemophilus influenzae (Dayao, Gibson, Blackall and Turni, 2016, Molina et al., 2002) or Haemophilus (Glässerella) parasuis (Dayao, Gibson, Blackall and Turni, 2016, San Millán et al., 2007). As in our study, a low prevalence of strains harboring blaTEM has been previously reported (Dayao et al., 2016).
Most of the isolates recovered during the last century in Spain were susceptible to erythromycin (Vera Lizarazo et al., 2006) and, similarly, only 12.5% of them were resistant to this macrolide in this study. However, the most common pattern in this case was intermediate susceptibility. These data were in accordance with previous findings (Dayao, Gibson, Blackall and Turni, 2014, Gutiérrez Martín and Rodríguez Ferri, 1992, Sellyei et al., 2009, Tang et al., 2009), but other investigators (Salmon et al., 1995) reported limited activity for erythromycin against P. multocida, and even others have found 100% resistance among the isolates recovered from the lungs of diseased pigs from June 2013 to May 2015 (Yeh et al., 2007). In our study, resistance was associated with the presence of ermC, msrE, ermA or mphE genes, while this quality in erythromycin could not be related with any of these genes in Australian isolates (Dayao et al., 2016). According to the data obtained, macrolide resistant genes tested in this work are the only ones that can be linked with the resistance shown for P. multocida isolates, because all harbored some of the four genes compared.
The rate of resistance to tetracyclines has not increased excessively from 1988 to nowadays, because in the former year was only 1.6% and 14.4% in 2004 for oxytetracycline (Vera Lizarazo et al., 2006), while raised to 18.8% 14 years later to tetracycline. Similar results have been obtained in a pan-Europan study (de Jong et al. 2014) and in another one carried out in Poland (Markowska-Daniel, Urbaniak, Stepniewska, & Peisak, 2010), but higher values have been seen in most of the previous reports (Dayao, Gibson, Blackall and Turni, 2014, Quedi Furian et al., 2016, Niemann et al., 2018, Sellyei et al., 2009, Tang et al., 2009, Yeh et al., 2007). The resistance of P. multocida has been related with some genes, such as tetB, tetH and tetM, being tetH gene the most widespread (Hansen, Blanchard and Hirsh, 1996, Kehrenber, Salmon and Watts, 2001, San Millán et al., 2009). Only tetA and tetB genes were tested in our study, but no genes of the former one were found while tetB gene was encountered in 40.6% of isolates. Similar results were reported recently (Dayao et al., 2016), while a considerably lower resistance (25.0%) associated to tetB genes has been already detected (Kehrenber et al., 2001).
A comparable behaviour has been seen in Spain for the last 30 years for cefotaxime, a third-generation cephalosporin (Vera Lizarazo et al., 2006). So, no resistance was observed in any of the isolates recovered in this period, although a total of 12.5% of strains recovered in 2018 were classified as of intermediate susceptibility. Close results were detected among Chinese (Tang et al., 2009), Australian (Dayao et al., 2014), Czechs isolates (Nedbalcova et al., 2014), other ones coming from a pan-European study (de Jong et al., 2014), or among fourth-generation cephalosporins such as cefquinome (Sellyei et al., 2009). However, resistance rates above 22% were detected recently to ceftiofur among Brazilian isolates (Quedi Furian et al., 2016), and to cefazolin among Taiwanese isolates (Yeh et al., 2007). Ceftiofur resistance has been linked to blaTEM-1, in such a way that this gene has been detected both in resistant and susceptible P. multocida isolates (Chander et al., 2011). In this respect, blaTEM gene has been also detected in our study.
Chloramphenicol is known to produce major adverse effects in humans, and its use by mouth or by injection is only recommended when safer antibiotics cannot be used. An excellent effectiveness result was recorded in our study, with no resistant and only two intermediate isolates to it. Similar results were obtained for 36 isolated previously (Sellyei et al., 2009, Tang et al., 2009). Other safer phenicol, florfenicol, that is used exclusively in veterinary patients for the therapy of pneumonia caused by P. multocida (Kehrenberg, Wallmann, & Schwarz, 2008), was equally highly effective against clinical isolates (Dayao et al., 2014; de Jong et al., 2014). Nevertheless, resistance to florfenicol in two German isolates, being associated to plasmid pCCK381 and florR gene, was described ten years ago (Kehrenberg et al., 2008). Besides, resistances beyond 91% were detected for the 62 Taiwanese isolates collected for two years (Yeh et al., 2007).
The results obtained by us nowadays matched completely with those recorded in both 1987–1988 and 2003–2004 periods for enrofloxacin (Vera Lizarazo et al., 2006). In addition, this quinolone appears to have the highest in vitro activity against the isolates tested in our study, a finding that is in agreement with that previously reported by most authors (de Jong et al., 2014; Dorey, Hobson and Lees, 2017, El Garch et al., 2017, Gutiérrez Martín and Rodríguez Ferri, 1992, Niemann et al., 2018, Sellyei et al., 2009). However, in an investigation by Quedi Furian et al. (2016), 22.5% of strains were found non-susceptible to enrofloxacin, and in other by Yeh et al. (2007) almost 62% of them were non-active to this quinolone.
A total of 16 resistance patterns could be obtained in this study, being CLI + COT the most common of all. A higher spread can be seen in 2018 than in previous investigations conducted in Spain because 56.2% of the profiles were only displayed by one P. multocida isolate while this fact was seen respectively in 42.8% and in 38.5% of the isolates recovered in 1988 and in 2004 (Vera Lizarazo et al., 2006). In a quite similar investigation, only five patterns were detected to 21 P. multocida isolates, but 60% of them were resistant to one combination (Dayao et al., 2014). Five profiles were also encountered in other study, but all were combinations of two antimicrobial agents and each pattern contained more than ten isolates (Glass-Kaastra et al., 2014). Anyway, none of the resistance profiles seen in any of these previous reports overlapped with those 16 patterns showed in the present study. Finally, the fact that 84.4% of isolates has behaved as resistant to at least two of the antimicrobial agents tested evinces the need of restrictive and responsible usage of these compounds in animals, especially in those intended for human consumption, along with susceptibility testing of this pathogen.
5. Conclusion
This study has shown data about the antimicrobial resistance of Spanish P. multocida from swine source and provides a genetic explanation for the resistance to erythromycin. On the other hand, enrofloxacin, chloramphenicol, cefotaxime and tetracycline were highly active in vitro against these isolates and, consequently, they remain useful for treatment of swine pneumonia caused by this pathogen.
Conflict of interest
None of the authors has any financial or personal relationships that could inappropriately influence or bias the content of the paper.
Funding
This study was supported by the project called “Caracterización fenotípica y genética de aislados de Pasteurella multocida en explotaciones porcinas de Castilla y León”, financed by the “Junta de Castilla y León”, Spain.
Contributor Information
M. Petrocchi-Rilo, Email: mpetrr00@estudiantes.unileon.es.
C.B. Gutiérrez-Martín, Email: cbgutm@unileon.es.
J.I. Méndez-Hernández, Email: jmenh@unileon.es.
E.F. Rodríguez-Ferri, Email: ef.rferri@unileon.es.
S. Martínez-Martínez, Email: smarm@unileon.es.
References
- Barton M.D., Pratt R., Hart W.S. Antibiotic resistance in animals. Communicable Diseases Intelligence Quarterly Report. 2013;27(Suppl):S121. doi: 10.33321/cdi.2003.27.34. [DOI] [PubMed] [Google Scholar]
- Blackall P.J., Fegan N., Pahoff J.L., Storie G.J., McIntosh G.B., Cameron R.D. The molecular epidemiology of four outbreaks of porcine pasteurellosis. Veterinary Microbiology. 2000;72:111–120. doi: 10.1016/s0378-1135(99)00192-3. [DOI] [PubMed] [Google Scholar]
- Chander Y., Oliveira S., Goyal S.M. Characterisation of ceftiofur resistance in swine bacterial pathogens. Veterinary Journal. 2011;187:139–141. doi: 10.1016/j.tvjl.2009.10.013. [DOI] [PubMed] [Google Scholar]
- CLSI (Clinical and Laboratory Standards Institute) Clinical and Laboratory Standards Institute; Wayne, PA: 2008. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals – document M31-A3. [Google Scholar]
- CLSI (Clinical and Laboratory Standards Institute) Clinical and Laboratory Standards Institute; Wayne, PA: 2013. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals; second informational supplement – document VET01-S2. [Google Scholar]
- Cote S., Hare J., Higgins R., Jacques M. Resistance to antimicrobial agents and prevalence of R plasmids in Pasteurella multocida from swine. American Journal of Veterinary Research. 1991;52:1653–1657. [PubMed] [Google Scholar]
- Dayao D.A.E., Gibson J.S., Blackall P.J., Turni C. Antimicrobial resistance in bacteria associated with porcine respiratory disease in Australia. Veterinary Microbiology. 2014;171:232–235. doi: 10.1016/j.vetmic.2014.03.014. [DOI] [PubMed] [Google Scholar]
- Dayao D.A.E., Gibson J.S., Blackall P.J., Turni C. Antimicrobial resistance genes in Actinobacillus pleuropneumoniae, Haemophilus parasuis and Pasteurella multocida isolated from Australian pigs. Australian Veterinary Journal. 2016;94:227–231. doi: 10.1111/avj.12458. [DOI] [PubMed] [Google Scholar]
- De Alwis M.C. Haemorrhagic septicaemia-a general review. British Veterinary Journal. 1992;148:99–112. doi: 10.1016/0007-1935(92)90101-6. [DOI] [PubMed] [Google Scholar]
- De Oliveira Filho J.X., Morés M.A.Z., Rebelatto R., Agnol A.M.D., Plieski C.L.A., Klein C.S. Pasteurella multocida type A as the primary agent of pneumonia and septicaemia in pigs. Pesquisa Veterinária Brasileira. 2015;35:716–724. [Google Scholar]
- Dorey L., Hobson S., Lees P. Factors influencing the potency of marbofloxacin for pig pneumonia pathogens Actinobacillus pneunomiae and Pasteurella multocida. Research in Veterinary Science. 2017;111:93–98. doi: 10.1016/j.rvsc.2016.11.011. [DOI] [PubMed] [Google Scholar]
- EFSA EMA and EFSA joint scientific opinion on measures to reduce the need to use antimicrobial agents in animal husbandry in the European Union, and the resulting impacts on food safety (RONAFA) EFSA Journal. 2017;15:4666. doi: 10.2903/j.efsa.2017.4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Garch F., Kroemer S., Galland D., Morrissey I., Woehrle F. Survey of susceptibility to marbofloxacin in bacteria isolated from diseased pigs in Europe. Veterinary Record. 2017 doi: 10.1136/vr.103954. [DOI] [PubMed] [Google Scholar]
- EMA . 2016. Sales of veterinary antimicrobial agents in 29 European countries in 2014. Sixth ESVAC report. London. [Google Scholar]
- Glass-Kaastra S.K., Pearl D.L., Smith R.R., McEwen B., Stavic D., Fairles J. Multiple-class antimicrobial resistance surveillance in swine Escherichia coli F4, Pasteurella multocida and Streptococcus suis isolates from Ontario and the impact of the 2004-2006 porcine circovirus type-2 associated disease outbreak. Preventive Veterinary Medicine. 2014;113:159–164. doi: 10.1016/j.prevetmed.2013.11.008. [DOI] [PubMed] [Google Scholar]
- Gois M., Barnes H.J., Ross R.F. Potentiation of turbinate atrophy in pigs by long-term nasal colonization with Pasteurella multocida. American Journal of Veterinary Research. 1983;44:372–378. [PubMed] [Google Scholar]
- Gutiérrez Martín C.B., Rodríguez Ferri E.F. In vitro susceptibility of Pasteurella multocida subspecies multocida strains isolated from swine to 42 antimicrobial agents. Zentralbleat für Bakteriologie. 1992;279:387–393. doi: 10.1016/s0934-8840(11)80371-3. [DOI] [PubMed] [Google Scholar]
- Hansen L.M., Blanchard P.C., Hirsh D.C. Distribution of tetH among Pasteurella isolates from the United States and Canada. Antimicrobrial Agents and Chemotherapy. 1996;40:1558–1560. doi: 10.1128/aac.40.6.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachooel A., Ranjbar M.M., Kachooel S. Evaluation of Pasteurella multocida serotype B:2 resistance to immune serum and complement system. Veterinary Research. 2017;8:179–184. [PMC free article] [PubMed] [Google Scholar]
- Karriker L., Coetzee J., Friendship R., Prescott J. Drug pharmacology, therapy and prophylaxis. In: Zimmerman J., Karriker L., Ramírea A., Schwartz K., Svenson G., editors. Diseases of swine. Wiley Blackwell; West Sussex, USA: 2013. pp. 106–118. [Google Scholar]
- Kehrenber C., Salmon S.A., Watts J.L. Tetracycline resistance genes in isolates of Pasteurella multocida, Mannheimia haemolytica, Mannheimia glucosida and Mannheimia varigena from bovine and swine respiratory disease: Intergeneric spread of the tetH plasmid pMht1. Journal of Antimicrobial Chemotherapy. 2001;48:631–640. doi: 10.1093/jac/48.5.631. [DOI] [PubMed] [Google Scholar]
- Kehrenberg C., Wallmann J., Schwarz S. Molecular analysis of florfenicol-resistant Pasteurella multocida isolates in Germany. Journal of Antimicrobial Chemotherapy. 2008;62:951–955. doi: 10.1093/jac/dkn359. [DOI] [PubMed] [Google Scholar]
- Larivière S., Leblanc L., Mittal K.R., Martineau G.P. Characterization of Pasteurella multocida from nasal cavities of piglets from farms with or without atrophic rhinitis. Journal of Clinical Microbiology. 1992;30:1398–1401. doi: 10.1128/jcm.30.6.1398-1401.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowska-Daniel I., Urbaniak K., Stepniewska K., Peisak Z. Antibiotic susceptibility of bacteria isolated from respiratory tract of pigs in Poland between 2004 and 2008. Poland Journal of Veterinary Science. 2010;13:29–36. [PubMed] [Google Scholar]
- Molina J.M., Córdoba J., Esteban R., Laínez B., Monsoliú A., Gregori V. Study of the β-lactam resistance of Haemophilus influenzae conferred by the bla(ROB-1) gene. Revista Española de Quimioterapia. 2002;15:148–151. [PubMed] [Google Scholar]
- Mutters R., Ihm P., Pohl S. Reclassification of the genus Pasteurella Trevisan 1887 on the basis of deoxyribonucleic acid homology, with proposals for the new species Pasteurella dagmatis, Pasteurella canis, Pasteurella stomatis, Pasteurella anatis and Pasteurella langaa. International Journal of Systematic Bacteriolory. 1985;35:309–322. [Google Scholar]
- Nedbalcova K., Nechvatalova K., Pokludova L., Bures J., Kucerova Z., Koutecka L. Resistance to selected β-lactam antibioticas. Veterinary Microbiology. 2014;171:328–336. doi: 10.1016/j.vetmic.2014.02.004. [DOI] [PubMed] [Google Scholar]
- Niemann L., Müller P., Brauns J., Nathaus R., Schäkel F., Kipschull K. Antimicrobial susceptibility and genetic relatedness of respiratory tract pathogens in weaner pigs over a 12-month-period. Veterinary Microbiology. 2018;219:165–170. doi: 10.1016/j.vetmic.2018.03.030. [DOI] [PubMed] [Google Scholar]
- Quedi Furian T., Apellanis Borges K., Laviniki V., da Silveira Rocha S.L., de Almeida C.N., Pinheiro do Nascimento V. Virulence genes and antimicrobial resistance of Pasteurella multocida isolated from poultry and swine. Brazilian Journal of Microbiology. 2016;47:210–216. doi: 10.1016/j.bjm.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon S.A., Watts J.L., Case C.A., Hoffman L.J., Wegener H.C., Yancey R.J., Jr. Comparison of MICs of ceftiofur and other antimicrobial agents against bacterial pathogens of swine from the United States, Canada, and Denmark. Journal of Clinical Microbiology. 1995;33:2435–2444. doi: 10.1128/jcm.33.9.2435-2444.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Millán A., Escudero J.A., Gutiérrez B., Hidalgo L., García N., Llagostera N. Multiresistance in Pasteurella multocida is mediated by coexistence of small plasmids. Antimicrobioal Agents and Chemotherapy. 2009;53:3399–3404. doi: 10.1128/AAC.01522-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Millán A., Escudero J.A., Catalán A., Nieto S., Farelo F., Gibert M. β-lactam resistance in Haemophilus parasuis is mediated by plasmid pb1000 bearing blaROB-1. Antimicrobial Agents and Chemotherapy. 2007;51:2260–2264. doi: 10.1128/AAC.00242-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellyei B., Varga Z., Szentesi-Samu K., Kaszanyitky E., Magyar T. Antimicrobial susceptibility of Pasteurella multocida isolated from swine and poultry. Acta Veterinaria Hungarica. 2009;57:357–367. doi: 10.1556/AVet.57.2009.3.2. [DOI] [PubMed] [Google Scholar]
- Tang X., Zhao Z., Hu J., Wu B., Cai X., He Q. Isolation, antimicrobial resistance, and virulence genes of Pasteurella multocida strains from swine in China. Journal of Clinical Microbiology. 2009;47:951–958. doi: 10.1128/JCM.02029-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend K.M., Frost A.J., Lee C.W., Papadimitriou J.M., Dawkins H.J. Development of PCR assays for species- and type-specific identification of Pasteurella multocida isolates. Journal of Clinical of Microbiology. 1998;36:1096–1100. doi: 10.1128/jcm.36.4.1096-1100.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend K.M., Boyce J.D., Chung J.Y., Frost A.J., Adler B. Genetic organization of Pasteurella multocida cap Loci and development of a multiplex capsular PCR typing system. Journal of Clinical Microbiology. 2001;39:924–929. doi: 10.1128/JCM.39.3.924-929.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera Lizarazo Y.A., Rodríguez Ferri E.F., Martín de la Fuente A.J., Gutiérrez Martín C.B. Evaluation of changes in antimicrobial susceptibility patterns of Pasteurella multocida subsp. multocida isolates from pigs in Spain in 1987-1988 and 2003-2004. American Journal of Veterinary Research. 2006;67:663–668. doi: 10.2460/ajvr.67.4.663. [DOI] [PubMed] [Google Scholar]
- Yeh J.C., Lo D.Y., Chang S.K., Chou C.C., Kuo H.C. Antimicrobial susceptibility, serotypes and genotypes of Pasteurella multocida isolates associated with swine pneumonia in Taiwan. Veterinary Record. 2007 doi: 10.1136/vr.104023. [DOI] [PubMed] [Google Scholar]