Abstract
Metritis is a frequently occurring diseases in postpartum cows and is one of the important reasons for the infertility of dairy cows, accounting for 20–30% of dairy cow diseases and has serious implications for the dairy industry. It has been reported in the literature that the bacterial balance of genital tracts is directly related to the maintenance of physiological function and the development of various diseases of the reproductive system. By analyzing the changes in abundance and diversity of bacteria in the cow uterus from 1 to 35 days postpartum, the objective was to reveal the mechanism of metritis in cows and provide the basis for diagnosis, treatment and prevention of metritis in postpartum dairy cows. Uterine contents were taken from six cows (three healthy and three with metritis) on 1, 7, 14, 21 and 35 days after parturition. DNA genomes extracted from the samples were primed with 515F5′-GTGCCAGCMGCCGCGG-3′ and 907R5′-CCGTCAATTCMTTRAGTTT-3′ for PCR amplification of the V4+V5 regions of the 16S rDNA genes and construction of a gene library. The sequence of the bacterial structure of the cow uterine contents was analyzed using 16S rDNA high-throughput sequencing technology.
A total of 30 samples were tested by PCR, and 29 samples qualified. The results of cluster analysis showed that except for one sample, the number of OTUs in the healthy cows was above 200, while in the cows with metritis, except for three samples, OTUs were below 200. The Chao1 and Shannon indices showed that the abundance of bacteria in the cow uterus was lower than that of healthy cows.
Analysis of the relative abundance of bacteria in the cow uterus showed that there were six phyla present, including Bacteroidetes, Firmicutes, Fusobacteria, Proteobacteria, Actinobacteria and Tenericutes. There were 10 dominant genera in healthy cows, including Bacteroides, Clostridium sensu stricto 1, Escherichia-Shigella, Fusobacterium, Halomonas, Helcococcus, Porphyromonas, Prevotella 6, Rikenellaceae RC9 gut group and Streptococcus. There were nine dominant genera in cows with metritis, including Bacteroides, Caviibacter, Clostridium sensu stricto 1, Falsiporphyromonas, Fusobacterium, Halomonas, Helcococcus, Porphyromonas and Prevotella 7.
Phylogenetic tree analysis showed that uterine contents from 29 samples could be separated into two clusters. Eleven samples from the cows with metritis were clustered with one sample from the healthy group, and 13 samples from the healthy cows were clustered together with four samples from the metritis group. Principal co-ordinate analysis showed that the points representing healthy cows and those representing the metritis group were concentrated in two distinct regions, which shows that there were significant differences in the structure evolution between healthy cows and cows with metritis.
The above results indicate that bacterial diversity declines with time postpartum in healthy cows and is lower in cows with metritis, with characteristic changes in the relative abundances, including increases in Bacteroidetes and Fusobacteria, decreases in Firmicutes and Proteobacteria, increases in Porphyromonas, Bacteroides and Fusobacterium, and a decrease in Clostridium sensu stricto 1.
Keywords: 16S rDNA sequencing, Metritis, Dairy cows, Uterine bacterial community
1. Introduction
Metritis is one of the most common reproductive disorders in dairy cows and has brought huge economic losses to the dairy industry. The incidence of metritis in dairy cows is 20–40% in China, and the economic loss caused by metritis in the USA is 650 million dollars per year, while the European Union it is 1.411 billion euros per year (Machado et al., 2012; Sheldon, Price, Cronin, Gilbert & Gadsby, 2009; Wang, Ametaj, Ambrose & Ganzle, 2013).
Through a large number of studies, scholars have found that reproductive tract microecology plays a crucial role in the occurrence and development of metritis in cows (Turnbaugh et al., 2009). It has been shown that many animal species live in close association with commensal and symbiotic microbiomes. Disease-related alterations in the composition of the bacterial communities in various organisms have been described both by suppression of existing populations and by colonization of new bacterial populations (Fredricks, Fiedler & Marrazzo, 2005; Manichanh et al., 2006; Ott et al., 2004). Turnbaugh researched individual intestinal microbial diversity using 454 high-throughput sequencing technology; the results showed that changes in obesity and intestinal microbial populations were closely related (Turnbaugh et al., 2009). In 2011, Cornell University for the first time compared the bacterial diversity in the uterus of healthy cows and cows with metritis using 16S rDNA high-throughput sequencing technology combined with PCR-denaturing gradient gel electrophoresis (DGGE) technology, and their studies showed that uterine bacterial community composition in healthy and diseased cows was significantly different, and the bacterial diversity in the postpartum cow uterus plays a key role in the occurrence of metritis (Santos, Gilbert & Bicalho, 2011).
Previous studies have shown that comparing the bacterial community composition profiles gathered from the uterine mucosa of cows with genital disease or reproductive disorders with those of control animals showed significant differences. It is not only helpful for us to reveal the role of microecology in the reproductive tract in the occurrence of metritis, but also to provide a reference for analyzing the pathogenesis of metritis in dairy cows to set a theoretical foundation for the diagnosis and prevention of the disease.
However, alterations in uterine bacterial composition at different postpartum time points remain poorly documented, as well as in cows with metritis. The uterine bacterial community can be studied by culture-based techniques or by some advanced molecular techniques (DGGE and ribosomal RNA clone libraries). However, traditional culture methods are able to study only 0.1–15% of naturally occurring microbes, as only a small proportion of uterine bacteria can be cultured (Lamont et al., 2011). In addition, given the low sequencing depth of previous approaches, bacterial community analyses reported previously represent a mere snapshot of the diversity within the community (Bibby, Viau & Peccia, 2010; Fouts et al., 2012). With the advent of next-generation sequencing technologies, conducting in-depth sequencing on samples from specific environments, including the complex uterine bacterial community is feasible and allows the description of unculturable bacteria (Bergmann et al., 2010; O et al., 2011; Singh et al., 2009). Both group-specific 16S rDNA PCR-DGGE and clone library sequencing of broad-range 16S rDNA PCR revealed that the diversity of the uterine bacterial community is more complicated than previously discovered using traditional culture methods (Santos, Gilbert & Bicalho, 2011), as cultured uterine bacteria made up only a small proportion of the whole. Further study of the uterine bacterial community by 16S rDNA sequencing avoids the disadvantages of traditional culture methods and makes it possible to identify other bacterial species, thereby improving the completeness of bacterial community studies (Hamady, Walker, Harris, Gold & Knight, 2008).
The aim of our present study was to characterize the uterine bacterial community in dairy cows within their postpartum periods, as well as in cows with metritis, using Illumina amplicon sequencing of 16S rDNA. We determined the relationships between these conditions and compared them with microbiota in normal cows, so allowing for the development of optimal prevention and intervention strategies.
2. Materials and methods
2.1. Experimental animals
The study was conducted on Yisheng Dairy Farm in Yantai (China) between October 2016 and August 2017. The cows were fed Total Mixed Rations (TMR). Concentrates were fed automatically according to individual milk yield. Cows were bred by artificial insemination and calved throughout the year in a straw-bedded group maternity pen. Among the 629 dairy cows examined and diagnosed clinically, cows that had a history of disease or had received antibiotic treatment were excluded; a total of 60 cows were selected and uterine contents were collected at 1, 7, 14, 21 and 35 d postpartum.
2.2. Sample collection
The vulva of the cows were cleaned with bromogeramine alcohol. Using a cow uterine cleaner connected to a syringe 3 mL of uterine contents were extracted. When on 21 d and 35 d postpartum samples of uterine contents were taken, the uterus was infused with 250–500 mL normal saline, the hands placed in the rectum and fully pressed on the uterus, extracting 3 mL uterine irrigating fluid. The samples were placed in a nitrogen canister for transportation back to the laboratory (lag time of approximately 4 h) and stored at −80 °C until used.
Cows with metritis were identified according to clinical signs, including: fever >39.5 °C within 21 days after parturition; postpartum lochia discharge and the first estrus delay; the volume of the uterine cavity abnormally enlarged, presence of liquid, a fetid, watery red-brown uterine discharge; abnormal neutrophils and other signs of toxemia in uterine inflammation in dairy cows (Richard, Hopper & Diplomate, 2014). After that, a total of six cows were selected and divided evenly into healthy group (H) and metritis group (M). A total of 30 samples of uterine content from the six cows were used for the subsequent experiments.
2.3. Total bacterial genomic DNA extraction
Total genomic DNA was extracted from the uterine contents using TIANamp Bacteria DNA Kits (Tiangen Technology. Co. Ltd. China) according to the manufacturer's instructions. DNA concentration and purity were monitored on 1% agarose gels and normalized to 1 ng/μL using sterile water.
2.4. PCR amplification of 16S rDNA
The V4+V5 regions of 16S rDNA genes from bacteria were amplified using the barcoded primer set 515F 5′-GTGCCAGCMGCCGCGG-3′ and 907R 5′-CCGTCAATTCMTTRAGTTT-3′ primers. PCR was by Trans Gen AP221-02:Trans Start Fastpfu DNA Polymerase, 20 μL reaction system as follows: an initial denaturation step at 95 °C for 5 min; 27 cycles of 95 °C for 30 s, 55 °C for 30 s, 72 °C for 45 s; and a final extension step at 72 °C for 10 min, 10 °C until halted by the user. Each 20 μL reaction consisted of 4 μL 5 × FastPfu Buffer, 2 μL, 0.8 μL 2.5 mM dNTPs Forward Primer (5 μM), 0.8 μL Reverse Primer (5 μM), 0.4 μL FastPfu Polymerase, 10 ng Template DNA and 2 μL of ddH2O. The reaction system used ABI Gene Amp®9700 PCR instrumentation. Three repeats for each sample.
From the same sample, PCR products were mixed in equal density ratios. The same volume of 1 × loading buffer, which contained SYBR Green, was mixed with PCR products and subjected to gel electrophoresis on 1.0% agarose gel before visualization under UV light to detect PCR products. Additionally, optical density was used to evaluate DNA concentration and purity using a Nano Drop ND-1000 spectrophotometer (Nano Drop Technologies) at wavelengths of 230, 260 and 280 nm. Samples with a bright band between 400 and 450 bp were chosen for further experiments. Then, mixed PCR products were purified with Axy Prep DNA Extraction Kit (AXYGEN, US). The barcoded 16S rDNA V4+V5 PCR products were then pooled with other samples and sequenced using Illumina HiSeq PE250, 250 bp from both ends of the Biozeron (Shanghai, China).
2.5. Data analysis
Data were prepared and tables and figures produced using Microsoft Excel and the R software environment (Version 2.15.3). All analyses from clustering to alpha and beta diversity were performed with QIIME (Version7.1) (Caporaso et al., 2010). Statistical analysis of the relative abundance of genera, as well as the diversity indices and estimators, was performed using GraphPad Prism version 5.01. For all statistical analyses, sequences sharing more than 97% sequence identity were considered a single phylotype. UCLUST was used to cluster the sequences using default parameters, with the identity parameter set to 97%. The RDP classifier was used to classify these sequences into specific taxa using the default data base (Wang, Garrity, Tiedje & Cole, 2007). The Shannon index and Chao1 index were applied to evaluate the alpha-diversity, and UniFrac distance was used to analyze the β-diversity (multiple alignments were performed by using PyNAST, Green genes core set was used as the template, and two single-ended sequences of each gapped sequence were aligned separately before the alignments were merged) (Lozupone et al., 2011). Principal co-ordinates analysis (PCoA) was implemented using QIIME based on UniFrac distance and included the following processes: representative sequences of each OTU were aligned using PyNAST with the Green genes core set as the template; and phylogenetic tree relating the OTUs was generated using FastTree, which was used to calculate UniFrac distance (Hamady, Lozupone & Knight, 2010).
3. Results
3.1. DNA qualification control of DNA extraction and PCR amplification
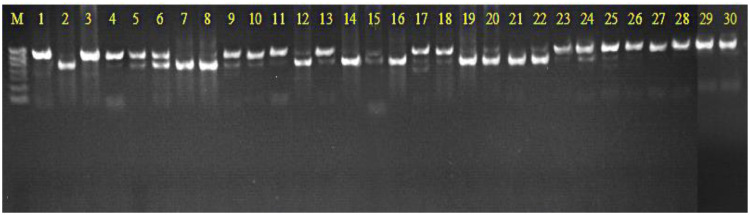
Bacterial DNA was extracted from samples using aTIANamp Bacteria DNA Kit and its concentration and quality measured using a NanoDrop2000. A260/A280 ratios were between 1.8 and 2.0, which indicated that pure DNA was obtained. The template was the extraction of the total bacterial genome DNA and amplification of 16S rDNA V4+V5 region sequence with universal primer 515F and 907R. Extracted PCR amplification products from 2 μL were detected by 2% agarose gel electrophoresis (Fig. 1). The length of the amplified product was about 400 bp, and the brightness was clearly visible. This met the requirements of the following experiment, except for sample No.15. Three PCR products with a clear band and non-specific amplification were selected from each group and used for further sequencing.
Fig. 1.
Quality inspection of cow uterine bacteria 16S rDNA V4+V5 gene fragments of PCR amplification, M: 750 bp maker, 100 bp DNA ladder, Lanes 1–15: H group, Lanes 16–30: M group.
3.2. Data statistics and optimization
A total of 29 individuals were recruited in the present study including healthy (H) group (n = 14) and metritis (M) group (n = 15). With Illumina PE250 sequencing, we determined a total of 706,906,500 bases. After excluding overlaps and removing low-quality reads and chimeras, 471,979,485 clean 16S rDNA V4+V5 bases and 1310,662 clean 16S rDNA V4+V5 sequences were obtained, with an average length of 360.11 bp per sequence. The coverage of all samples was higher than 99%, indicating that our rarified sequencing depth was sufficient to evaluate the diversity in the uterine samples obtained for this study (Table 1).
Table 1.
Sequencing for the 29 prepared samples.
| Sample | Effective Tags | Bases (bp) | Average Length (bp) | Coverage (%) |
|---|---|---|---|---|
| 1H1 | 41,379 | 11,108,937 | 268.47 | 99.45% |
| 1H2 | 40,708 | 15,209,536 | 373.63 | 99.76% |
| 1H3 | 34,449 | 12,909,261 | 374.74 | 99.27% |
| 1H4 | 56,586 | 21,192,183 | 374.51 | 99.96% |
| 1H5 | 55,281 | 20,647,281 | 373.5 | 99.98% |
| 2H1 | 36,482 | 10,130,643 | 277.69 | 99.59% |
| 2H2 | 57,338 | 20,170,148 | 351.78 | 99.86% |
| 2H3 | 39,421 | 14,693,274 | 372.73 | 99.71% |
| 2H4 | 40,734 | 14,677,006 | 360.31 | 99.94% |
| 2H5 | 54,996 | 20,330,630 | 369.67 | 99.97% |
| 3H1 | 31,695 | 9351,464 | 295.05 | 99.98% |
| 3H2 | 55,860 | 20,684,376 | 370.29 | 99.94% |
| 3H3 | 55,337 | 20,543,026 | 371.23 | 99.97% |
| 3H4 | 42,630 | 15,859,692 | 372.03 | 99.84% |
| 3H5 | ## | ## | ## | ## |
| 2M1 | 48,965 | 17,927,620 | 366.13 | 99.96% |
| 2M2 | 37,423 | 13,956,505 | 372.94 | 99.89% |
| 2M3 | 36,548 | 13,643,193 | 373.3 | 99.77% |
| 2M4 | 54,086 | 20,180,034 | 373.11 | 99.86% |
| 2M5 | 46,985 | 17,108,304 | 364.12 | 99.92% |
| 1M1 | 50,923 | 18,998,004 | 373.07 | 99.40% |
| 1M2 | 47,738 | 17,848,379 | 373.88 | 99.62% |
| 1M3 | 59,110 | 22,039,807 | 372.86 | 99.88% |
| 1M4 | 53,445 | 19,896,499 | 372.28 | 99.95% |
| 1M5 | 37,766 | 10,222,701 | 270.69 | 99.90% |
| 3M1 | 32,347 | 12,133,219 | 375.1 | 99.82% |
| 3M2 | 37,460 | 13,964,473 | 372.78 | 99.83% |
| 3M3 | 39,949 | 14,902,049 | 373.03 | 99.96% |
| 3M4 | 30,026 | 11,175,834 | 372.21 | 99.82% |
| 3M5 | 54,995 | 20,475,407 | 372.31 | 99.80% |
Sequencing indices were shown from the cows in the H group and M group. The coverage of all samples was used to determine the effectiveness of rarified sequencing depth; “##” represents missing data.
3.3. OTU-based cluster
Using Usearch (version 7.1) software, the optimized sequence was clustered with 97% similarity according to the similarity of OTU, and a total of 11,487 OTU were obtained. Then the Chao1 indices and Shannon indices of the samples were calculated (Table 2).
Table 2.
Statistical table of species diversity index for the 29 prepared samples.
| Healthy Group | OTU | Chao1 | Shannon | Metritis Group | OTU | Chao1 | Shannon |
|---|---|---|---|---|---|---|---|
| 1H1 | 530 | 787 | 1.03 | 1M1 | 24 | 102 | 0.28 |
| 1H2 | 932 | 994 | 4.79 | 1M2 | 121 | 155 | 2.68 |
| 1H3 | 874 | 1091 | 3.06 | 1M3 | 150 | 246 | 2.47 |
| 1H4 | 433 | 456 | 3.58 | 1M4 | 230 | 460 | 1.65 |
| 1H5 | 236 | 241 | 2.54 | 1M5 | 156 | 199 | 0.98 |
| 2H1 | 799 | 989 | 1.56 | 2M1 | 290 | 310 | 1.54 |
| 2H2 | 1046 | 1229 | 4.42 | 2M2 | 74 | 78 | 2.44 |
| 2H3 | 95 | 210 | 1.43 | 2M3 | 87 | 123 | 2.65 |
| 2H4 | 309 | 326 | 2.96 | 2M4 | 45 | 62 | 1.38 |
| 2H5 | 668 | 690 | 3.06 | 2M5 | 828 | 873 | 4.21 |
| 3H1 | 540 | 620 | 2.28 | 3M1 | 102 | 202 | 0.71 |
| 3H2 | 656 | 696 | 3.85 | 3M2 | 167 | 248 | 2.29 |
| 3H3 | 932 | 1062 | 5.05 | 3M3 | 120 | 132 | 2.9 |
| 3H4 | 318 | 332 | 2.75 | 3M4 | 171 | 216 | 2.11 |
| 3H5 | ## | ## | ## | 3M5 | 554 | 612 | 2.53 |
Diversity indices were shown from the cows in the H group and M group. The Chao1 and Shannon indices of all samples was used to determine the diversity of uterine bacteria, “##” represented missing data.
3.4. Species accumulation curves
Concomitant with an increase in sample size, species accumulation curves gradually slowed after a certain range, indicating that our rarified sequencing sample size was sufficient to evaluate the diversity of uterine bacterial communities in cows (Fig. 2).
Fig. 2.
Species accumulation curves of 29 cow uterine content samples. The number of samples is indicated on the horizontal axis, and OTUs are indicated on the vertical axis. The curved slope represents previously undetected OTUs (new species) as sample size increases.
3.5. Rarefaction curve
Concomitant with an increase in sequencing depth, species reasonableness curves gradually slowed after a certain range, indicating that our rarified sequencing depth was sufficient to evaluate the diversity of uterine bacterial communities in cows (Fig. 3).
Fig. 3.
The reasonableness test of sequencing depth of 29 cow uterine content samples. Number of reads sampled on the horizontal axis and OTU number on the vertical; 1–3: H group, 4–6: M group. The curved slope represents previously undetected OTUs (new species) as sample size increases.
3.6. Distribution of bacterial OTUs in the uterus of dairy cows
The statistical results of the common and unique OTU numbers of each sample were used to make up the overlapping graph shown in Fig. 4. The results showed that there were 1496, 1486 and 1364 OTUs in the H group and 372, 909 and 653 OTUs in the M group, indicating that the bacterial diversity in the H group was higher than that in the M group.
Fig. 4.
OTU distribution of uterine microflora in postpartum dairy cows. Each ellipse represents a cow, 1–3 represents H group, 4–6 represents M group, and overlapping and non-overlapping portions represent mutual and specific OTUs, respectively. One OTU represents one bacterial species. The numbers in the overlapped parts represent the total number of OTUs shared between groups, and the number that does not overlap part of the number represented the unique OTU number of the cow.
3.7. Bacterial diversity in the uterus of dairy cows
According to the results of OTU clustering, the Chao1 and Shannon indices of 29 samples were calculated and the Chao1 and Shannon indices analyzed statistically (Fig. 5). The Chao1 and Shannon indices showed that the bacterial diversity in the uterus of cows with metritis was significantly lower than that in healthy cows at 1–21 d postpartum; however, the difference was disappearing at 35 d postpartum. The above results showed that the bacterial diversity in the uterus of cow with metritis was reduced.
Fig. 5.
Changes in diversity index of uterine bacteria from H group and M group with time. The diversity of bacterial 16S rDNA was estimated through Chao1 index (A) and Shannon index (B).
3.8. Comparison of dominant uterine bacterial composition
Taxonomy's RDP classifier Bayes algorithm was used to annotate OTUs. We further compared the detailed community structure between the selected groups.
At the level of phylum (Fig. 6), there were six dominant bacteria in the uteruses of both healthy cows and cows with metritis; these included Bacteroidetes, Firmicutes, Fusobacteria, Proteobacteria, Actinobacteria and Tenericutes.
Fig. 6.
Uterine microbial community structures of H group (A) and M group (B). The relative abundance of bacterial 16S rDNA was estimated through classification at the phylum level. The color-coded bar plot displays the relative uterine bacterial composition in the H and M groups. The community composition of the two groups was diverse and complex. Color codes were only given for phyla that made up greater than 1% of the relative abundance.
At the level of genus (Fig. 7), there were 10 dominant bacteria in the uterus of healthy cows, including Bacteroides, Clostridium sensu stricto 1, Escherichia-Shigella, Fusobacterium, Halomonas, Helcococcus, Porphyromonas, Prevotella 6, Rikenellaceae RC9 gut group and Streptococcus. There were nine dominant bacteria in the uterus of cows with metritis, including Bacteroides, Caviibacter, Clostridium sensu stricto 1, Falsiporphyromonas, Fusobacterium, Halomonas, Helcococcus, Porphyromonas and Prevotella
Fig. 7.
Uterine microbial community structures from H group (A) and M group (B). The relative abundance of bacterial 16S rDNA was estimated through classification at the genus level. The color-coded bar plot displays the relative uterine bacterial composition in the H and M groups. The community composition of the two groups was diverse and complex. Color codes were only given for genera that made up greater than 1% of the relative abundance.
The relative abundance of major bacteria at different times postpartum at the level of phylum is shown in Fig. 8. The relative abundance of Bacteroidetes in H group increased from 6.50% to 46.41% from 1 to 14 d postpartum and decreased to 3.05% at 35 d postpartum; in M group it increased from 0.66% to 75.74% from 1 to 7 d postpartum, and at 35 d decreased to 28.44%. The relative abundance of Firmicutes in H group increased from 4.63% to 41.03% from 1 to 21 d postpartum, and at 35 d it decreased to 4.87%; in M group at 1 d postpartum it was 67.64% and between 7 and 35 d postpartum it was about 7.51–19.79%. The relative abundance of Fusobacteria in H group at 7 d was 7.32% and then decreased to less than 1%; in M group, from 1–21 d it increased from 0.07% to 34.27% and at 35 d decreased to 6.71%. Relative abundance of Proteobacteria in H group from 1 to14 d was maintained at about 10%, and from 14–35 d it increased from 10.26% to 84.7%; in M group at 1 d it was 28.29% and at 35 d was 20.50%, less than 1% at other time points. Actinobacteria and Tenericutes membranes were all less than 5% in both H group and M group. Relative abundance of Unclassified in H group at 1 d was 79.1% and at 7 d remained at 3.19%; in M group from 1–21 d it remained below 1% and increased to 32.03% at 35 d The results showed that the relative abundance of Bacteroidetes and Fusobacteria in the uterus of metritis cows increased, while Firmicutes and Proteobacteria decreased.
Fig. 8.
Relative abundance of dominant uterine bacteria from H group (A) and M group (B) at different times postpartum. The relative abundance of bacterial 16S rDNA was estimated by classification at the phylum level. The color-coded bar plot displays the relative uterine bacterial composition in the H and M groups. The community composition of the two groups was diverse and complex.
At the level of the genus (Fig. 9), the relative abundance of Bacteroides in H group at days 1–7 postpartum increased from 0.84% to 10.52% and at 35 d postpartum decreased to 0.63%; in M group, at 1–21 d it increased gradually from 0.19% to 41.18% and decreased to 4.04% at 35 d The relative abundance of Porphyromonas in H group at 1–14 d increased from 0.68% to 23.76% and at 35 d decreased to 0.11%; in M group, at 1–7 d it increased from 0.68% to 29.23%, at 21 d decreased to 9.23%, and at 35 d increased to 20.21%. The relative abundance of Fusobacterium in H group stayed below 7%; in M group, at 1–21 d it increased from 0.07% to 20.81% and at 35 d decreased to 1.65%. The relative abundance of Clostridium sensu stricto 1 in H group at 7 d was 6.71% and less than 1% at other time points; in M group, at 1 d it was as high as 84.18% and less than 1% at other time points. The relative abundance of unclassified genera in H group at 1 d was 79.09% and less than 10% at other time points; in M group, at 1 d it was 3.03%, at 35 d it was 32.03%, and less than 1% at other time points.
Fig. 9.
Relative abundance of dominant uterine bacteria from H group (A) and M group (B). The relative abundance of bacterial 16S rDNA was estimated through classification at the genus level. The color-coded bar plot displays the relative uterine bacterial composition in the H and M groups. The community composition of the two groups is diverse and complex.
The results showed that the relative abundances of Porphyromonas, Fusobacterium and Bacteroides in the uterus of cows with metritis were increased.
3.8.1. Microbial community heatmap analysis
We further compared the detailed community structure among the selected groups. When comparing at either the phylum or genus level, both H and M groups shared the same top 20 phyla and top 30 genera with large differences in overall abundance (Fig. 10). Specifically, at the phylum level, Bacteroidetes, Firmicutes, Proteobacteria, Fusobacteria, Actinobacteria and Tenericutes appeared as the major phyla in both groups. The relative abundances of Gemmatimonadetes, Spirochaetae, Chloroflexi, Planctomycetes, Chlorobi, Acidobacteria, Saccharibacteria, Verrucomicrobia, Euryarchaeota, Deinococcus-Thermus, Lentisphaerae and Nitrospirae in the H group were higher than in the M group. At the genus level, the top 30 bacterial species, including Bacteroides, Porphyromonas, Clostridium sensu stricto 1, Fusobacterium, Halomonas, Coxiella, Falsiporphyromonas, Caviibacter, Achromobacter, Pseudomonas, Prevotella 7, Streptococcus, Escherichia-Shigella, Prevotella 6, Peptostreptococcus, Parvimonas, Peptoniphilus, Anaerosalibacter, Ralstonia, Pelagibacterium, Ruminococcaceae UCG-005, Ruminococcaceae UCG-010 and Staphylococcus, showed little difference in overall abundance. The reason for this was that top 30 genera belong to top 6 phyla. In other words, the composition of uterine microbial communities was more complex in healthy cows than in cows with metritis.
Fig. 10.
Heat map of the relative abundance of dominant uterine bacteria in the community structures of the two groups of cows. The relative abundance of bacterial 16S rDNA was estimated through classification at the phylum (A) and genus (B) level. The color gradation bar plot displays the relative uterine bacterial composition in the H and M groups. The community composition of the two groups was diverse and complex. Color codes were only given for those made up top 20 phyla and top 30 genera in relative abundance.
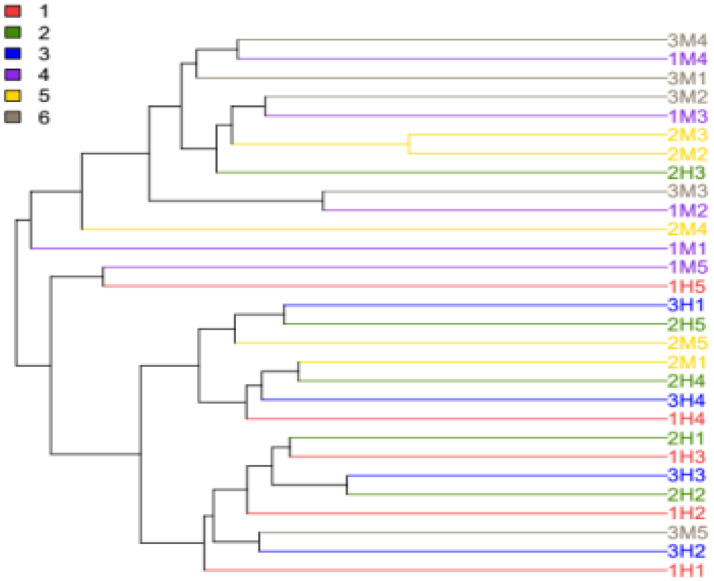
3.9. Phylogenetic tree analysis
The phylogenetic tree built using the UPGMA (unweighted pair group method with arithmetic mean) algorithm is shown in Fig. 11. The samples could be clustered into two clusters: 11 samples from the M group were clustered together with one sample from the H group, and 13 samples from the H group were clustered together with four samples from the M group. The results showed that there were differences in the uterine bacterial structure between the two groups.
Fig. 11.
Phylogenetic tree of uterine bacteria from cows, including healthy cows (1–3) and metritis cows (4–6). The length of the branch represents the distance between the samples.
3.10. PCoA analysis
Using Unifrac PCoA based on the evolutionary distance at the evolutionary level, the potential principal components affecting the community composition of samples were analyzed (Fig. 12).
Fig. 12.
Unifrac PCoA analysis. 1–3: H group; 4–6: M group. The factors explained by horizontal axis (PC1) 45.09%, and factors explained by vertical axis (PC2) 11.32%. Differences between samples are expressed as distance.
On the PC1 axis, the points represented by the postpartum H group were concentrated near the left side of the PC1, while the cows in the M group were concentrated near the right side of the PC1. The results showed that there were significant differences in the structure of uterine flora between healthy cows and those with metritis.
4. Discussion
Modern biology considers the animal body to be a super organism composed of its own eukaryotic cells and microbial communities. The organism's metabolism is regulated both by the host's own genome and the microbial genome, and the common metabolic process between host and microorganism will ultimately regulate the health of the host (O'Hara & Shanahan, 2006). Increasing numbers of in vivo studies have been made on the microbial flora of humans and animals. It has been proved that the imbalance in uterine flora is associated with a variety of diseases, such as vaginitis (Garrett et al., 2007), metritis (Turnbaugh, Backhed, Fulton & Gordon, 2008) and cervical cancer (Vijay-Kumar et al., 2010).
Studies have shown that the diversity of bacterial composition in the uterus of cows may be more complex than previously detected by traditional culture-related methods, and the bacteria that can be cultured in the uterus are only a small part of the flora of the uterus (Santos, Gilbert & Bicalho, 2011). With the development of molecular biology technology, more non-culture and high-throughput methods have been widely applied in the detection of microorganisms in humans and animals. Using the 16S rDNA sequencing technology of the llumina HiSeq PE250 sequencing platform, the diversity, structure and dominant bacteria in the uterus of healthy postpartum cows and of cows with metritis was sequenced. This avoided the disadvantages of traditional bacterial isolation and culture and was more conducive to the identification of species in the low-abundance community, and thus improved the integrity of the microbial community research to explore further the etiology of metritis in dairy cows. The results not only provide a reference for developing prophylactic and therapeutic probiotics in dairy cows, but also lay a foundation for revealing the pathogenesis of the disease.
Generally, more OTUs and a higher diversity index represent more species within a sample. At least 1400 OTUs were observed in the healthy group, indicating a wider range of uterine bacterial species in the healthy postpartum cows compared with 640 OTUs in the metritis group. Chao1 and Shannon indices of the uterus in the metritis group were lower than those in the healthy group. In other words, the bacterial community of the uterus is quite complex in healthy postpartum dairy cows but is comparatively simple in cows afflicted with metritis.
Use PCR-Denaturing Gradient Gel Electrophoresis (DGGE) and DNA pyrosequencing showed that uterine bacteria, regardless of the health of the cows, consist of mainly members of the phyla Bacteroidetes, Fusobacteria, Firmicutes, Proteobacteria and Tenericutes (Santos & Bicalho, 2012). However, our 16S rDNA High-throughput sequencing results showed that the uterine bacterial community in the healthy and metritis postpartum cows was composed of mainly Bacteroidetes, Firmicutes, Fusobacteria, Proteobacteria, Actinobacteria and Tenericutes. In the postpartum, the contents of Bacteroidetes and Fusobacteria were found to be comparatively high, and interestingly in uterine of postpartum health cows, Firmicutes and Proteobacteria showed little presence in cows with metritis. We are using second-generation high-throughput DNA sequencing based on pyrosequencing technology, errors that may be caused by improper manipulation, deletion, rare transcription, and unstable cloned bacteria are eliminated. Differences between this study and previous studies may be due to differences in experimentation technology, sampling position, geographical location or feeding environments.
According to reports (Santos & Bicalho, 2012), Firmicutes and Bacteroidetes bacteria are beneficial in the recovery of the endometrium. Our study found that in the metritis group the relative abundance of Firmicutes was decreased, but the relative abundance of Bacteroidetes increased between 7 and 21 d postpartum.
Ron, Tamir & Rahul (2018) Used 16S-rDNA technology to study the microbial communities and inflammatory response in the endometrium differ between normal and metritic dairy cows at 5–10 days post-partum.The results show the most abundant phyla in healthy cows were Proteobacteria (31.8 ± 9.3%), Firmicutes (27.9 ± 8.4%) and Bacteroidetes (19.7 ± 7.2%), while Bacteroidetes (60.3 ± 10.3%), Fusobacteria (13.4 ± 5.9%) and Firmicutes (10.5 ± 3.3%) were most abundant in the endometrial mucosa of metritic cows. And our sampling results on the seventh day also showed that the most abundant phyla in healthy cows is the Bacteroidetes (36.2 ± 5.2%) , Firmicutes (30.8 ± 1.1%) and Proteobacteria (13.9 ± 10.9%) ; the most abundant phyla Bacteroidetes (75.7 ± 2.0%) , Firmicutes (12.3 ± 1.5%) and Fusobacteria (9.4 ± 1.1%) in uterine cows.
Knudsen, Karstrup & Pedersen (2016) sequencing of 16S rRNA V1+V2 gene fragments of bacteria in the uterus of healthy Holstein cows and those infected with metritis found that Porphyromonas, Fusobacterium and Bacteroides were related to the disease. The study by Kolenbrander (2000) indicated that Fusobacterium has a "bridge" effect on the intestinal mucosa and can gather and adhere to bacteria colonized on the intestinal mucosa, and its appearance can promote the growth and colonization of other bacterial groups. In this paper, the relative abundance of Fusobacterium, Porphyromonas and Bacteroides in metritis cows increases from 7 to 21 d postpartum. In summary, significant changes in the uterine bacterial community take place when cows are afflicted with metritis.
Wang, Wang & Li (2017), sequencing the 16S rDNA V4-V6 gene fragments of Holstein cow cervical bacteria, including at the formative, gestational and postpartum stages, and in cows with metritis, found that Firmicutes were the predominant phylum represented. Cervical bacterial diversity decreased in cows with metritis, and the predominant bacterial genera were Porphyromonas and Fusobacterium. The similarity to our study may be due to the similar sampling position of the uterus and the cervix.
This paper reveals a previously unappreciated fraction of the uterine bacterial composition in dairy cows, comparing healthy cows with those with postpartum uterine infection. These results provide a reference to evaluate the uterine microbial community in cows with metritis, thus enabling large-scale cohort studies of the uterine ecosystem. The bacteria observed in this study provide a basis for future detailed in vitro studies to decipher the role of bacteria in uterine health, allowing clarification of the unknown aspects of basic interactions between microbiota and host, and may reveal potential entry points to develop microecological preparations. Our data also showed that the profile of the microbiota was consistent with, but not conclusively demonstrative of, the health status of the cows. Finally, the mechanisms that link microbial composition with these various health statuses remain elusive. The connection between bacterial communities and metritis in dairy cows is likely to be multifactorial, and indeed, our future studies will focus on the biochemical and physiological potential of such microbiota, as well as the effects of the microbiota on host gene expression.
5. Conclusion
Bacterial diversity in the uterus first increased and then decreased in healthy cows; the bacterial diversity in the uterus of the cows with metritis was lower. Characteristic changes in the relative abundance of uterine bacteria in cows with metritis included Bacteroidetes and Fusobacteria increased, decreased Firmicutes and Proteobacteria, increased Porphyromonas, Bacteroides and Fusobacterium, and decreased Clostridium sensu stricto 1.
Declaration of Competing Interest
All authors declare that they have no conflict of interest.
Acknowledgments
This work was supported by a grant from the Shandong Province Natural Science Foundation of China (no. ZR2015CM022), SDAIT-09-14.
Contributor Information
Hao Chen, Email: 794730473@qq.com.
Rongfeng Cao, Email: 794730473@qq.com.
References
- Bergmann I., Mundt K., Sontag M., Baumstark I., Nettmann E., Klocke M. Influence of DNA isolation on Q-PCR-based quantification of methanogenic Archaea in biogas fermenters. Systematic and Applied Microbiology. 2010;33:78. doi: 10.1016/j.syapm.2009.11.004. [DOI] [PubMed] [Google Scholar]
- Bibby K., Viau E., Peccia J. Pyrosequencing of the 16S rRNA gene to reveal bacterial pathogen diversity in biosolids. Water Research. 2010;44:4252. doi: 10.1016/j.watres.2010.05.039. [DOI] [PubMed] [Google Scholar]
- Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouts D.E., Szpakowski S., Purushe J., Torralba M., Waterman R.C., MacNeil M.D. Next generation sequencing to define prokaryotic and fungal diversity in the bovine rumen. PloS one. 2012;7:E48289. doi: 10.1371/journal.pone.0048289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredricks D.N., Fiedler T.L., Marrazzo J.M. Molecular identification of bacteria associated with bacterial vaginosis. New England Journal of Medicine. 2005;353:1899. doi: 10.1056/NEJMoa043802. [DOI] [PubMed] [Google Scholar]
- Garrett W.S., Lord G.M., Punit S., Lugo-Villarino G., Mazmanian S.K., Ito S. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M., Lozupone C., Knight R. Fast Unifrac: Facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and Phylochip data. ISME Journal. 2010;4:17. doi: 10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M., Walker J.J., Harris J.K., Gold N.J., Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5:235. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen L.R., Karstrup C.C., Pedersen H.G. An investigation of the microbiota in uterine flush samples and endometrial biopsies from dairy cows during the first 7 weeks postpartum. Theriogenology. 2016;86(2):642–650. doi: 10.1016/j.theriogenology.2016.02.016. [DOI] [PubMed] [Google Scholar]
- Kolenbrander P.E. Oral microbial communities: Biofilms, interactions, and genetic systems 1. Annual Reviews in Microbiology. 2000;54:413–437. doi: 10.1146/annurev.micro.54.1.413. [DOI] [PubMed] [Google Scholar]
- Lamont R.F., Sobel J.D., Akins R.A., Hassan S.S., Chaiworapongsa T., Kusanovic J.P. The vaginal microbiome: New information about genital tract flora using molecular based techniques. British Journal of Obstetrics and Gynaecology. 2011;118:533. doi: 10.1111/j.1471-0528.2010.02840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C., Lladser M.E., Knights D., Stombaugh J., Knight R. UniFrac: An effective distance metric for microbial community comparison. ISME Journal. 2011;5:169. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado V.S., Oikonomou G., Bicalho M.L., Knauer W.A., Gilbert R., Bicalho R.C. Investigation of postpartum dairy cows' uterine microbial diversity using metagenomic pyrosequencing of the 16S rRNA gene [J] Veterinary Microbiology. 2012;159(3–4):460–469. doi: 10.1016/j.vetmic.2012.04.033. [DOI] [PubMed] [Google Scholar]
- Manichanh C., Rigottier-Gois L., Bonnaud E., Gloux K., Pelletier E., Frangeul L. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut. 2006;55:205. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O C.P., Aguirre D.C.D., Jones M., Klaassens E.S., Worthley D.L., Whitehall V.L. The effects from DNA extraction methods on the evaluation of microbial diversity associated with human colonic tissue. Microbial Ecology. 2011;61:353. doi: 10.1007/s00248-010-9771-x. [DOI] [PubMed] [Google Scholar]
- O'Hara A.M., Shanahan F. The gut flora as a forgotten organ. EMBO Reports. 2006;7(7):688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott S.J., Musfeldt M., Wenderoth D.F., Hampe J., Brant O., Folsch U.R. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53:685. doi: 10.1136/gut.2003.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard M., Hopper D.V.M., Diplomate A.C.T. Bovine reproduction. John Wiley and Sons, Inc; 2014. Postpartum uterine infection[m] pp. 440–448. [Google Scholar]
- Ron Sicsic, Tamir Goshen, Rahul Dutta. Microbial communities and inflammatory response in the endometrium differ between normal and metritic dairy cows at 5-10 days post-partum.[J] Veterinary Research. 2018;49:77. doi: 10.1186/s13567-018-0570-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos T.M., Bicalho R.C. Diversity and succession of bacterial communities in the uterine fluid of postpartum metritic, endometritic and healthy dairy cows. PloS one. 2012;7:E53048. doi: 10.1371/journal.pone.0053048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos T.M., Gilbert R.O., Bicalho R.C. Metagenomic analysis of the uterine bacterial microbiota in healthy and metritic postpartum dairy cows [J] Journal of Dairy Science. 2011;94(1):291–302. doi: 10.3168/jds.2010-3668. [DOI] [PubMed] [Google Scholar]
- Santos T.M., Gilbert R.O., Bicalho R.C. Metagenomic analysis of the uterine bacterial microbiota in healthy and metritic postpartum dairy cows. Journal of Dairy Science. 2011;94:291. doi: 10.3168/jds.2010-3668. [DOI] [PubMed] [Google Scholar]
- Santos T.M., Gilbert R.O., Bicalho R.C. Metagenomic analysis of the uterine bacterial microbiota in healthy and metritic postpartum dairy cows. Journal of Dairy Science. 2011;94(1):291–302. doi: 10.3168/jds.2010-3668. [DOI] [PubMed] [Google Scholar]
- Sheldon I.M., Price S.B., Cronin J., Gilbert R.O., Gadsby J.E. Mechanisms of infertility associated with clinical and subclinical endometritis in high producing dairy cattle [J] Reproduction in Domestic Animals. 2009;44:1–9. doi: 10.1111/j.1439-0531.2009.01465.x. [DOI] [PubMed] [Google Scholar]
- Singh J., Behal A., Singla N., Joshi A., Birbian N., Singh S. Metagenomics: Concept, methodology, ecological inference and recent advances. Biotechnology Journal. 2009;4:480. doi: 10.1002/biot.200800201. [DOI] [PubMed] [Google Scholar]
- Turnbaugh P.J., Backhed F., Fulton L., Gordon J.I. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host & Microbe. 2008;3:213. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P.J., Hamady M., Yatsunenko T., Cantarel B.L., Duncan A., Ley R.E. A core gut microbiome in obese and lean twins [J] Nature. 2009;457(7228):480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay-Kumar M., Aitken J.D., Carvalho F.A., Cullender T.C., Mwangi S., Srinivasan S. Metabolic syndrome and altered gut microbiota in mice lacking toll-like receptor 5. Science (New York, N.Y.) 2010;328:228. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied Environmental Microbiology. 2007;73:5261. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Ametaj B.N., Ambrose D.J., Ganzle M.G. Characterisation of the bacterial microbiota of the vagina of dairy cows and isolation of pediocin-producing Pediococcus acidilactici [J] BMC Microbiology. 2013:13–19. doi: 10.1186/1471-2180-13-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wang J., Li H. Characterization of the cervical bacterial community in dairy cows with metritis and during different physiological phases. [J] Theriogenology. 2017;108:306–313. doi: 10.1016/j.theriogenology.2017.12.028. [DOI] [PubMed] [Google Scholar]