Abstract
Background
The function of LINC00501, a long-non-coding RNA (lncRNA), is unclear at present. According to the Cancer Genome Atlas (TCGA), LINC00501 is highly expressed in lung cancer (LC), but whether it can be adopted as a potential therapy target for LC still needs further research.
Methods
The expression of LINC00501 in LC was analyzed based on the TCGA, and a real-time fluorescent quantitative PCR assay was carried out to quantify LINC00501 in non-small-cell lung cancer (NSCLC). Additionally, bioinformatics analysis, luciferase reporter gene technique, and RNA immunoprecipitation (RIP) were employed to analyze the direct interaction between LINC00501 and miR-129-5p, and CCK-8 and Transwell assays and flow cytometry were employed to analyze the effects of LINC00501 on cell proliferation, invasion, and apoptosis. Furthermore, a Western blot assay was carried out to determine the protein level of HMGB1.
Results
LINC00501 was highly expressed in LC according to the database, and it was found that LINC00501 was upregulated in NSCLC specimens and cells, and the up-regulation indicated an unfavorable prognosis. Besides, knockdown of LINC00501 hindered the proliferation and invasion of NSCLC cells and intensified their apoptosis, and LINC00501 could be adopted as competitive endogenous RNA to regulate HMGB1 and tumorigenesis through miR-129-5p.
Conclusion
LINC00501 is overexpressed in LC and the overexpression indicates poor prognosis of patients. In addition, LINC00501 can inhibit the invasion and migration of LC by mediating miR-129-5p/HMGB1.
Keywords: LINC00501, HMGB1, miR-129-5p, lung cancer, invasion, migration
Introduction
LC can be classified into small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC), of which NSCLC accounts for more than 85% of LC.1–3 NSCLC is relatively insidious in onset, with inconspicuous clinical manifestations at the initial stage, and there is a lack of good diagnostic indicators for it in clinical practice, so patients are mostly already at the middle or late stage at diagnosis and have missed the optimal operation timing. Moreover, the treatment time of the middle and advanced LC is long, and the prognosis is poor, which increases the pressure of patients and economic burden on the family.4,5 Therefore, it is imperative to study the relevant mechanisms of NSCLC to provide potential diagnostic indicators for clinical practice.
With the deepening of research in recent years, people’s cognition of non-coding RNAs (ncRNAs) has been consistently improved.6 Common ncRNAs include short-chain ncRNAs (microRNA), long-chain ncRNAs (lncRNA), and circular RNAs (circRNAs),7–9 among which research on lncRNA in cancer has made a considerable progress.LncRNA is an ncRNA with a length of more than 200nt. It was previously considered as a metabolic product during transcription due to its inability to directly code proteins. However, it has been reported that lncRNA participates in the proliferation, apoptosis, and migration of various cells,10–12 and it is strongly linked to the occurrence of tumors.13 LINC00501 is a newly discovered lncRNA. One previous study has revealed that LINC00501 expression is high in LC expression profile,14 and one other study has found that LINC00501 was highly expressed in lung adenocarcinoma.15 In this study, we found through the Cancer Genome Atlas (TCGA) that LINC00501 expression was high in LC, indicating that it has a certain potential value in LC, but whether it can be a potential therapeutic target for LC is still under investigation.
Therefore, this study investigated the relevant mechanism of LINC00501 in LC, with the goal of providing potential targets for clinical treatment of LC.
Methods and Materials
Data
Data About Patients
A total of 50 NSCLC patients who underwent surgical treatment in Xuanwu Hospital Capital Medical University between April 2013 and April 2014 were enrolled as a patient group, and tumor tissues were extracted from them during operation, and then transported in liquid nitrogen, and saved at −80°C. The inclusion criteria of patients: Patients diagnosed with NSCLC according to pathological examination and imaging, patients who signed informed consent forms after understanding this study or whose family members signed them, patients with complete pathological data, and those willing to receive follow-up.The exclusion criteria of them: Patients with other comorbid malignant tumors, or liver or kidney function diseases, patients with infection before admission, patients with previous treatment history (surgery, chemotherapy, radiotherapy or antibiotic treatment) before this study, and those with expected survival time < 3 months.This study was conducted with permission from the Medical Ethics Committee of Xuanwu Hospital Capital Medical University, and it was in conformity with the Helsinki Declaration.
Acquisition of GEO and TCGA Data
Data about the expression of factors in lung adenocarcinoma and squamous cell carcinoma were obtained for analysis by logging in TCGA, selecting Access TCGA Data and accessing the database. The data were collected from the core sample database of TCGA, and sequenced and analyzed based on a standardized processing scheme (http://can-cergenome.nih.gov/cancergenomics/tissuesamples). The data involved in 1051 patients in total, including 478 squamous cell carcinoma specimens and 45 paired specimens, and 483 lung adenocarcinoma and 45 paired specimens.
Cell Culturing
Lung cancer cell (LCC) lines H1299, H460, SPC-A1, A549, and HEK293T (CRL-5803, HTB-177, CRL-5803, CCL-185, and CRL-11268, ATCC, United States) and normal lung cells 16HBE (PCS-300-010, ATCC, United States) were purchased from ATCC. All the cells were cultured in the dulbecco’s modified eagle medium (DMEM,10566024, Gibco, United States) containing penicillin-streptomycin double antibody and 10% fetal bovine serum (FBS; 10437028, Gibco, United States) in a constant temperature 5%CO2incubatorat 37°C.
Cell Transfection
The specific short hairpin RNA (shRNA) directed against human lncRNA LINC00501 was cloned into pENTR TM/U6 plasmid (GenePharma, Shanghai, China) and called sh-LINC00501, and non-targeted shRNA (sh-NC, GenePharma) was adopted as negative control.MiR-129-5p mimics (miR-129-5p-mimics), inhibitors (miR-129-5p-inhibit), and their respective controls (miR-NC) were all processed by RiboBio (Guangzhou, China). For overexpression of HMGB1, the full-length HMGB1 sequence was transfected into pcDNA vector (ThermoFisher, China), called pcDNA-LINC00501, and pcDNA-3.1 was adopted as blank control. Transfection operations were all carried out with Lipofectamine 3000 reagent (Life Technologies Corp., Carlsbad, United State) under the manufacturer’s instruction. Stable transfected H1299 and A549 cells were collected, and cultured in medium containing 0.5 mg/mL G418 (Sigma-Aldrich, st Louis, Missouri, Untied States), and stable transfected cells were adopted for later analyses.
QRT-PCR Detection
Total RNA was obtained from collected cells and tissues with the TRIzol Kit, and its concentration, integrity, and purity were determined using an ultraviolet spectrophotometer and agarose gel electrophoresis.MiR-129-5p was determined as follows: Reverse transcription was performed to the total RNA through the TransScript® miRNA RT Enzyme Mix and 2×TS miRNA Reaction Mix under the original kit instructions (AQ202-01 and AQ301-01, TransGen Biotech, Beijing, China).PCR amplification was conducted under the PCR system (7500, ABI Company, United States) consisted of 20 μL total volume containing 1μL cDNA, 0.4μL upstream and downstream primers, 10μL 2×TransTaq® Tip Green qPCR SuperMix, 0.4μL Passive Reference Dye (50X) and ddH2O added to adjust the volume and the PCR conditions indicated in the kit instructions. Three duplicate wells were provided for each sample, and the assay was repeated three times. The data were analyzed using2-ΔΔct with U6 as an internal reference. Furthermore, LINC00501 and HMGB1 were determined as follows: Reverse transcription was performed using the 5X TransScript® II All-in-One SpuperMix for qPCR and gDNA Remover Kit on the basis of original kit guidelines, and then PCR amplification was carried out through a PCR system consisted of 20 μL total volume containing 1 μL cDNA, 0.4 μL upstream and downstream primers, 10 μL 2X TransScript® Tip Green qPCR SuperMix, 0.4 μL Passive Reference Dye (50X) and nuclease-free water added to adjust the volume under conditions indicated in the kit instructions. Three duplicate wells were provided for each sample, and the assay was repeated three times. In the assay, the data were analyzed using 2-ΔΔct, with GAPDH as an internal reference.16
Western Blot (WB) Assay
Cultured cells of each group were collected, and their total protein was obtained via the RIPA lysis method with RIPA kit (89900, Thermo Scientific™). The protein concentration was determined using the BCA kit (35055, Thermo Scientific™), and adjusted to 4μg/μL. The total protein was isolated using SDS-PAGE, and then transferred to polyvinylidene fluoride (PVDF) membrane, dyed with Ponceau’s stain liquid, soaked in phosphate buffer saline with Tween (PBST) for 5 min for washing, blocked in 5% skim milk for 2 hour, and then added with HMGB1 and primary antibody (1: 1000), Caspase3and primary antibody (1: 1000), Bcl-2and primary antibody (1: 1000), Bax and primary antibody (1: 1000), β-catenin and primary antibody (1: 1000), c-myc and primary antibody (1: 1000), cyclin D1and primary antibody (1: 1000), GAPDH (ab79823, ab13847, ab182858, ab32503, ab32572, ab190026, ab134175, Abcam Company, United States), and sealed at 4°C overnight. The membrane was washed to remove the primary antibody, and then put with horseradish peroxidase (HRP)-labeled goat anti-rabbit immunoglobulin E (IgE) secondary antibody (ab6721, Abcam Company, United States) (1: 5000), cultured at 37°C for 1 h, washed with phosphate buffer saline (PBS;14190250, Gibco, United States) 3 times, 5 min/time, developed in a darkroom, absorbed with filter paper to remove excess liquid on the membrane, and made to be luminescent with electrochemiluminescence (ECL) Kit (35055, Thermo Scientific™), and developed. The protein band was scanned, and the gray value was studied in Quantity One software. The relative expression of protein was recorded as the gray value of target protein band/the gray value of β-Act in protein band.
CCK-8 Assay
Cells transfected for 24 hours were harvested and transferred to a 96-well plate at 4×106/well, and each well was put with 10 μL CCK-8 (C0037, Beyotime Biotechnology, Shanghai, China) solution and 90 μL basic DMEM at 24 h, 48 h, 72h, and 96 h after culturing, respectively. After addition of the solution each time, the plate was incubated at 37°C for 2 hours. The optical density (OD) of each well at 450nm wavelength was detected using the microplate reader (PerkinElmer, BioTek Company, United States).
Transwell Assay
In this study, Transwell kit (A1142802, Gibco, United States) was adopted for detection. Cells transfected for 24h were harvested, transferred to a 6-well plate at 5×104 cells/well, and washed with PBS twice. The upper compartment was put with 200μL DMEM, and the lower compartment was put with 500mL DMEM with 20% FBS The plate was incubated at 37°C for 48h, and the substrate and cells not penetrating the membrane in the upper compartment were wiped off. Afterwards, the plate was cleaned three times, immobilized with paraformaldehyde for 10 min, and cleaned with double distilled water three times, and dyed with 0.5% crystal violet after being dry. Finally, the cell invasion in the plate was analyzed using a microscope.
Flow Cytometry
The transfected cells were trypsinized with 0.25% trypsin (90058, Thermo Scientific™). After digestion, the cells were washed with PBS twice, and then added with 100 μL binding buffer to prepare 1×106 cells/mL suspension. It was added with AnnexinV-FITC (40302ES20, Yeasen Biotechnology Co., Ltd., Shanghai, China) and PI in order, cultured at room temperature in the dark for 5min, and finally detected via the FC500MCL flow cytometer (FACS Canto II, BD Company, United States) system. The experiment was repeated three times, and the obtained data were averaged.
DLR Assay
DNA oligonucleotide and pMiR-reporter vector were adopted to construct report vectors, and LINC00501 wild type/mutant type (LINC00501-WT/MUT) and HMGB1 wild type/mutant type (HMGB1-WT/MUT) were constructed. Then, the vectors were co-transfected with miR-129-5p-mimics and negative control (miR-NC) into HEK293 cells. Cells were collected after incubation for 24 hours and the luciferase activity was determined using the Dual Luciferase Reporter (DLR) Gene Assay Kit (D0010, Solarbio Company, Beijing, China).
PIR Assay
The RIP assay was carried out through the EZ-Magna RIP Kit (Millipore, United States) strictly in accordance with the kit instructions as follows: RCC cells were collected, and lyzed using RIPA. Subsequently, the whole cell protein extract was cultured with RIP washing buffering containing magnetic beads bound to anti-Ago2 antibody (Millipool) or mouse IgG controls. The protein sample was digested using protease K to extract immunoprecipitated RNA. Finally, the purified RNA was analyzed through a qRT-PCR assay to verify the existence of binding targets.
Tumor Xenotransplantation Models
Ten 5-week-old BALB/c nude mice (20 g) (Vital River Laboratory Animal Technology Co., Ltd., Beijing, China) were assigned to two groups (each n=5).PLC5 cells transfected with sh-LINC00944#2 and sh-NC were injected subcutaneously into the right abdomen of each mouse, respectively (1×106cells for each injection), and on the 7th, 14th, 21st, and 28th day, the tumor volume of each nude mouse was analyzed by the following formula: Volume = (length × width2)/2.All mice were executed under anesthesia and their tumor tissues were collected. The animal experiment was carried out with permission from the Animal Care and Use Committee of Xuanwu Hospital Capital Medical University, and in line with the Laboratory animal—Guideline for ethical review of animal welfare (GB/T 35892–2018).
Statistical Analyses
In the study, the collected data were analyzed statistically, and visualized into required figures using GraphPad 7 (GenePharma, Shanghai, China), and the independent prognostic factors of the patients were analyzed using SPSS20.0. The quantitative data distribution was evaluated via the Kolmogorov–Smirnov (K-S) test. Data in normal distribution are expressed as the mean ± standard deviation (Mean ±SD). Comparison between groups was carried out using the independent-samples T test. Enumeration data are expressed as the percentage (%), and chi-square test was used for them, and expressed by 2. Multi-group comparison was carried out using the one-way ANOVA, and expressed by F. Post hoc pairwise comparison was conducted through the LSD-t test, and expression in multiple time points was compared through the variance of repeated measures, and expressed by F. Post test was carried out using Bonferroni, and Pearson correlation analysis was performed to analyze the correlation of genes. The overall survival of the patients was visualized into Kaplan-Meier (K-M) survival curves and analyzed through the Log rank test, and multivariate Cox regression analysis was carried out for prognosis analysis of patients. P<0.05 implies a significant difference.
Results
LINC00501 Is Highly Expressed in NSCLC Patients and Patients with High LINC00501 Expression Have Unfavorable Prognosis
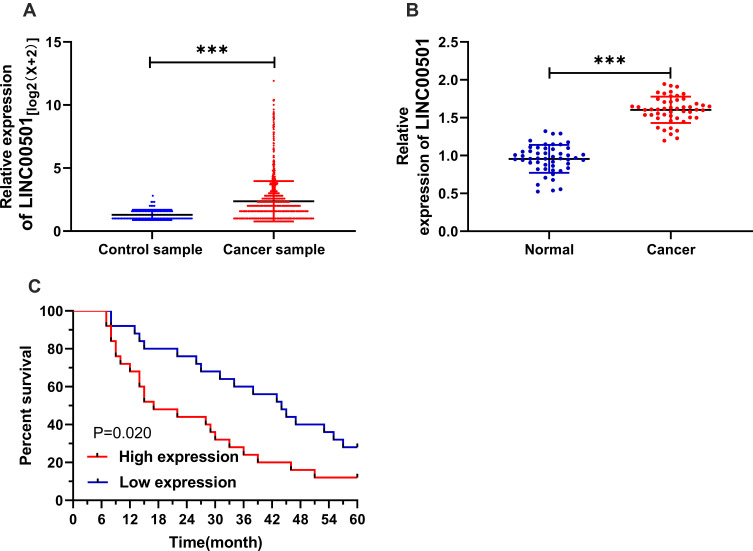
Firstly, we analyzed the expression of LINC00501 based on the TCGA database, and acquired its high expression in LC patients. We also detected the expression of LINC00501 in LC tissues, and also found its high expression in tumor tissues. In order to further verify the value of LINC00501 in LC, we studied the relation between LINC00501 and pathological data of patients, The patients were assigned to high and low expression groups according to the median expression of LINC00501, and it was turned out that patients with high LINC00501 expression showed higher rate of low differentiation, high staging, and distal metastasis. In addition, the survival analysis uncovered a decline in the 5-year survival rate of patients with high LINC00501 expression, and the Cox regression analysis uncovered that differentiation, clinical staging, tumor size, and LINC00501 were independent risk factors for NSCLC patients’ prognosis. Figure 1 and Tables 1 and 2.
Figure 1.
Relationship between LINC00501 expression in NSCLC patients and their survival. (A) Expression of LINC00501 in LC samples based on TCGA. (B) Relative expression of LINC00501 in NSCLC tumor tissue according to the RT-qPCR assay. (C) K-M survival analysis on the 5-year survival of patients with high and low expression of LINC00501. ***indicates P<0.001.
Table 1.
Relation Between LINC00501 and Pathological Data About the Patients
| Factor | Relative Expression of LINC00501 | P-value | ||
|---|---|---|---|---|
| High Expression (n=25) | Low Expression (n=25) | |||
| Sex | ||||
| Male (n=35) | 18 | 17 | 0.758 | |
| Female (n=15) | 7 | 8 | ||
| Age (Y) | ||||
| ≥60 (n=26) | 12 | 14 | 0.571 | |
| <60 (n=24) | 13 | 11 | ||
| Smoking history | ||||
| Yes (n=35) | 18 | 17 | 0.758 | |
| No (n=15) | 7 | 8 | ||
| Differentiation | ||||
| Poor (n=20) | 14 | 6 | 0.021 | |
| Well/moderate (n=30) | 11 | 19 | ||
| Clinical staging | ||||
| I–II stage (n=32) | 12 | 20 | 0.018 | |
| III–IV stage (n=18) | 13 | 5 | ||
| Pathological type | ||||
| Squamous cell carcinoma (n=25) | 12 | 13 | 0.777 | |
| Adenocarcinoma (n=25) | 13 | 12 | ||
| Tumor size (cm) | ||||
| ≥3 (n=24) | 15 | 9 | 0.089 | |
| <3 (n=26) | 10 | 16 | ||
| Distant metastasis | ||||
| Yes (n=23) | 15 | 8 | 0.047 | |
| No (n=27) | 10 | 17 | ||
| Tumor subtypes | ||||
| Adenocarcinoma (n=31) | 14 | 17 | 0.382 | |
| Squamous cell carcinoma (n=19) | 11 | 8 | ||
| EGFR mutation | ||||
| Mutation (n=18) | 10 | 8 | 0.556 | |
| No mutation (n=32) | 15 | 17 | ||
Table 2.
Cox Regression Analysis
| Factor | Univariate Cox | Multivariate Cox | ||||
|---|---|---|---|---|---|---|
| Exp(B) | 95 CI% | Sig. | Exp(B) | 95 CI% | Sig. | |
| Sex (male vs female) | 0.369 | 0.192–0.709 | 0.003 | 0.706 | 0.286–1.741 | 0.449 |
| Age (≥60 years vs.<65 years) | 1.064 | 0.571–1.986 | 0.844 | |||
| Smoking history (yes vs no) | 0.923 | 0.460–1.853 | 0.822 | |||
| Differentiation (low vs moderate and high) | 6.035 | 3.032–12.011 | 0.000 | 3.076 | 1.414–0.294 | 0.005 |
| Clinical staging (I–II vs III–IV) | 0.092 | 0.039–0.218 | 0.000 | 0.270 | 0.089–0.818 | 0.021 |
| Pathological type (squamous cell carcinoma vs adenocarcinoma) | 0.974 | 0.522–1.817 | 0.934 | |||
| Tumor size (≥ 3 vs < 3) | 10.615 | 4.452–25.31 | 0.000 | 0.713 | 0.332–1.528 | 0.014 |
| Distant metastasis (metastasis vs without metastasis) | 3.278 | 1.637–6.565 | 0.001 | 2.035 | 0.874–4.737 | 0.099 |
| LINC00501 (high expression vs low expression) | 0.002 | 0.00–0.028 | 0.000 | 0.014 | 0.001–0.263 | 0.004 |
Down-Regulation of LINC00501 Inhibits the Metastasis and Growth of LCCs
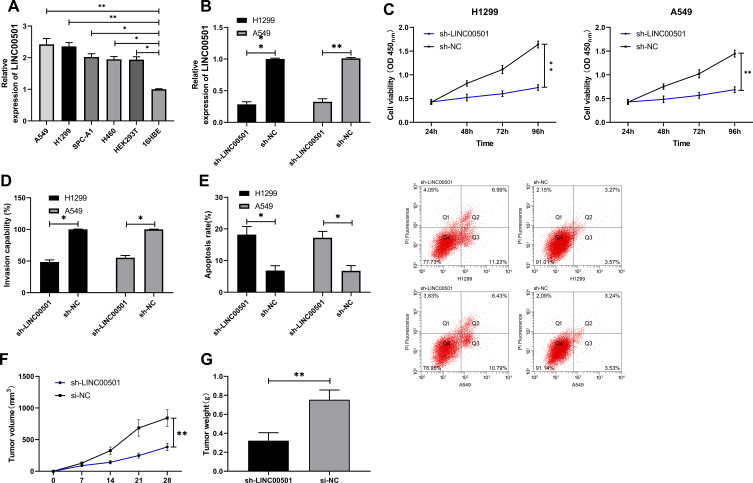
For the purpose of analyzing the effects of LINC00501 on LCCs, we first quantified LINC00501 in LCCs, finding that LINC00501 was also lowly expressed in LCCs. Additionally, we transferred sh-LINC00501 into LCCs, The CCK-8 assay revealed that cells transfected with sh-LINC00501 presented significantly lower proliferation activity than those transfected with sh-NC, and the Transwell assay showed that the cells transfected with sh-LINC00501 also showed significantly lower apoptosis rate than those with sh-NC, and transfection of sh-LINC00501 induced apoptosis of LCCs. Furthermore, the nude mouse tumorigenesis assay revealed that the tumor mass and volume of mice with knock-down of LINC00501 were significantly lower than those of mice in the sh-NC group, and further detection of LINC00501 expression in tumor tissues also revealed that LINC00501 expression was low in them. Through the above research, we can basically confirm that LINC00501 can be adopted as a potential therapy target for LC. Figure 2.
Figure 2.
Growth and metastasis of LCCs after knock-down of LINC00501. (A) Relative expression of LINC00501 in LCCs according to the RT-qPCR assay. (B) Relative expression of LINC00501 in LCCs transfected with sh-LINC00501 according to the RT-qPCR assay. (C) Changes in the proliferation of LCCs after transfection of sh-LINC00501 according to the CCK-8 assay. (D) Transmembrane rate of LCCs transfected with sh-LINC00501 according to the Transwell assay. (E) Apoptosis induction of LCCs after transfection of sh-LINC00501 according to the flow cytometry. (F) Changes in the tumor volume within 28 days after nude mouse modeling. (G) Changes in the tumor mass at 28 days after nude mouse modeling. *indicates P<0.05 and **indicates P<0.01.
LINC00501 Acts as miR-129-5p Sponge to Suppress the Metastasis of LCCs
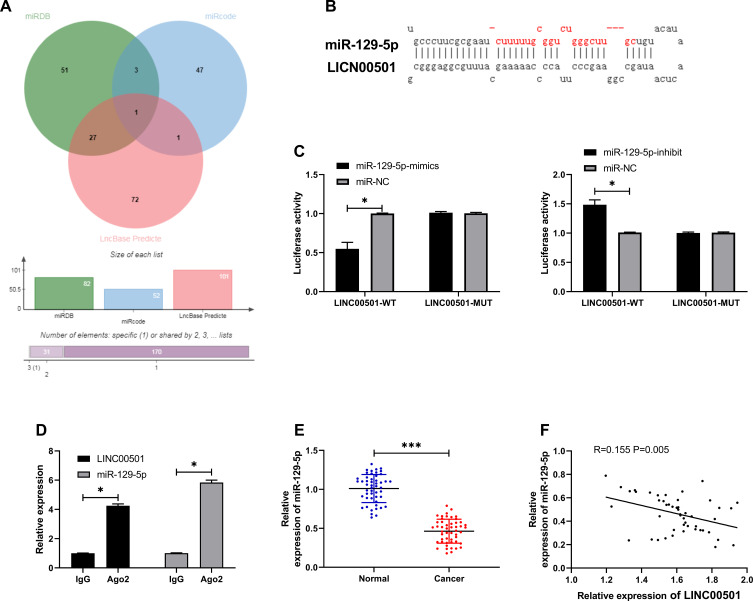
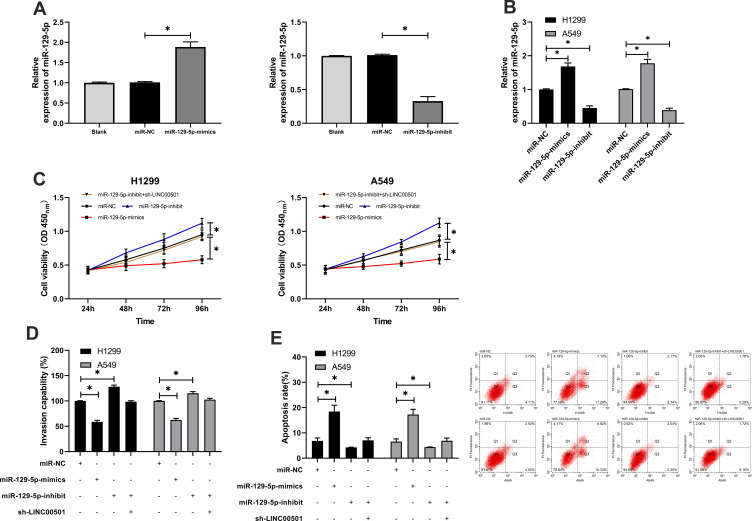
Many studies have confirmed that lncRNA is involved in tumorigenesis by regulating miR. In order to further explore the relevant mechanism of LINC00501, we predicted through miRDB, miRcode, and LncBase Predicted v2 that LINC00501 could bind to miR. It came out that based on the three databases, it was predicted that that there were targeted binding locus between miR-129-5p and LINC00501. With the aim of further determining the relationship between the two, we carried out assays. The DLR assay revealed that miR-129-5p-mimics could inhibit the luciferase activity of LINC00501-WT, while miR-129-5p-inhibit could increase it, and the RIP assay revealed that LINC00501 and miR-129-5p can bind to Ago2 protein, and the expression of LINC00501 and miR-129-5p bound to Ago2 was higher than that of those bound to IgG. In addition, we also detected the miR-129-5p expression in tumor tissues, finding its low expression in them, and correlation analysis revealed that the miR-129-5p expression was negatively related to LINC00501 expression in LC patient tissue. Cell experiments showed that transfection of miR-129-5p-inhibit accelerated cell proliferation, invasion and reduced cell apoptosis, but these biological behaviors were reversed after co-transfection of miR-129-5p-inhibit and sh-LINC00501.It suggests that LINC00501 can regulate miR-129-5p to participate in LC growth. Figures 3 and 4.
Figure 3.
Validation of targeting relationship between LINC00501 and miR-129-5p. (A) Predication of potential miR of LINC00501 through miRDB, miRcode, and LncBase Predicted v2. (B) Targeted binding locus between LINC00501 and miR-129-5p. (C) Binding between LINC00501 and miR-129-5p according to the DLR assay. (D) Binding between LINC00501 and miR-129-5p according to the RIP assay. (E) Relative expression of miR-129-5p in tumor tissues and paracancerous tissues of LC patients according to the RT-qPCR assay. (F) Correlation analysis between LINC00501 and miR-129-5p in tumor tissues of LC patients. *indicates P<0.05 and ***indicates P<0.001.
Figure 4.
Knock-down of LINC00501 can reverse the growth and invasion of LCCs caused by the decrease of miR-129-5p expression. (A) Relative expression of miR-129-5p in constructed vectors according to the RT-qPCR assay. (B) Relative expression of miR-129-5p in transfected LCCs according to the RT-qPCR assay. (C) Changes in the proliferation of LCCs after transfection of miR-129-5p-mimics, miR-129-5p-inhibit, and miR-129-5p-inhibit+sh- LINC00501 according to the CCK-8 assay. (D) Transmembrane rate of LCCs after transfection of miR-129-5p-mimics, miR-129-5p-inhibit, and miR-129-5p-inhibit+sh-LINC00501 according to the Transwell assay. (E) Apoptosis induction of LCCs after transfection of miR-129-5p-mimics, miR-129-5p-inhibit, and miR-129-5p-inhibit+sh-LINC00501 according to the flow cytometry. *indicates P<0.05.
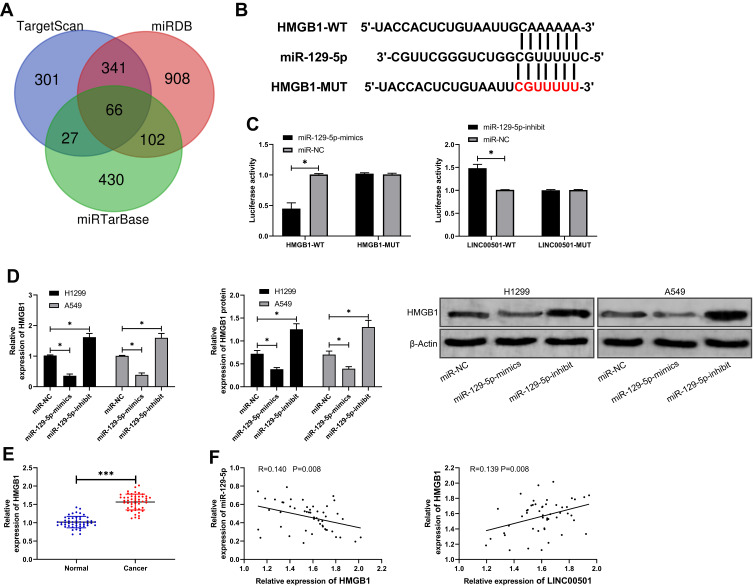
miR-129-5p Suppresses the Metastasis of LC by Targeting HMGB1
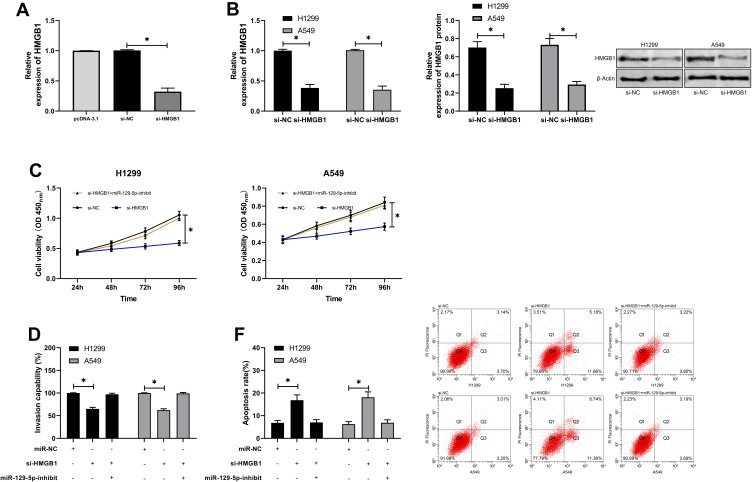
For the purpose of further determining the relevant mechanism of miR-129-5p in LC, we predicted the target genes downstream of miR-129-5p. It was shown that HMGB1 was a potential target of miR-129-5p. In order to verify the relationship between the two, we conducted a DLR assay, finding that miR-129-5p-mimics could inhibit the luciferase activity of HMGB1-WT, while miR-129-5p-inhibit could increase it. Besides, we also quantified HMGB1in tumor tissues of LC patients, finding that its expression increased in the tumor tissues, and correlation analysis revealed that the expression of HMGB1 was negatively linked to that of miR-129-5p in tumor tissues of LC patients, but positively linked to LINC00501 expression. With the aim of further verifying that miR-129-5p can affect HMGB1 to participate in LC, we co-transfected miR-129-5p-inhibit into cells with si-HMGB1. It came out that transfection of si-HMGB1 inhibited cell proliferation and invasion and induced apoptosis, but there were no differences between cells co-transfected with si-HMGB1 and miR-129-5p-inhibit and those transfected with si-NC in cell behavior mentioned above, indicating that the results were reversed, and miR-129-5p targeted HMGB1 to participate in LC. Figures 5 and 6.
Figure 5.
Verification of targeting relationship between miR-129-3p and HMGB1. (A) Predication of potential mRNA by TargetScan, miRDB, and miRTarBase. (B) Targeted binding locus between miR-129-5p and HMGB1, and mutant locus. (C) Verification of binding between miR-129-5p and HMGB1 by the DLR assay. (D) The mRNA and protein expression of HMGB1 in LCCs after transfection of miR-129-5p-mimics and miR-129-5p-inhibit according to the RT-qPCR and WB assays. (E) Relative expression of HMGB1 in tumor tissues and paracancerous tissues of LC patients according to the RT-qPCR assay. (F) Correlation analysis of HMGB1 with miR-129-5p and LINC00501 in tumor tissues of LC patients. *indicates P<0.05 and ***indicates P<0.001.
Figure 6.
MiR-129-5p inhibits the growth and metastasis of LCCs by targeting HMGB1. (A) Relative expression of si-HMGB1 in constructed vectors according to the RT-qPCR assay. (B) Relative mRNA and protein expression of HMGB1 in transfected LCCs according to the RT-qPCR and WB assays. (C) Changes in the proliferation of LCCs after transfection of si-HMGB1 and si-HMGB1+miR-129-5p-inhibit according to the CCK-8 assay. (D) Transmembrane rate of LCCs transfected with si-HMGB1 and si-HMGB1+miR-129-5p-inhibit according to the CCK-8 assay. (E) Apoptosis induction of LCCs after transfection of si-HMGB1 and si-HMGB1+miR-129-5p-inhibit according to the flow cytometry. *indicates P<0.05.
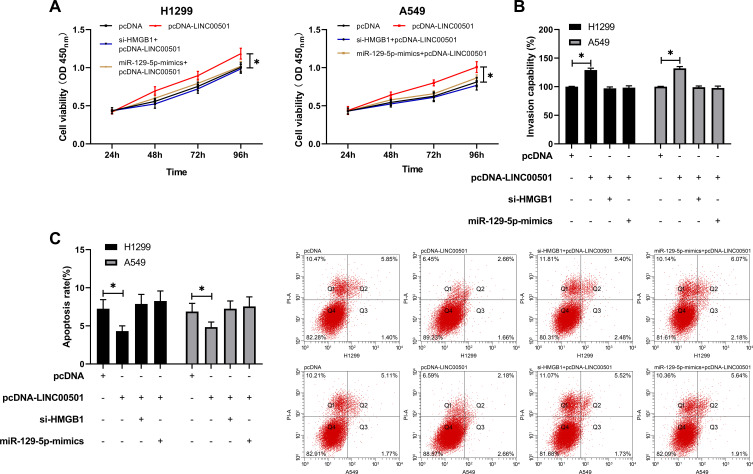
LINC00501 Inhibits the Proliferation of LCCs and Induces Their Apoptosis by Regulating the miR-129-5p/HMGB1 Axis
We speculated that there may be a signal axis relationship among LINC00501/miR-129-5p/HMGB1. In order to further verify our research conclusion, we carried out a rescue experiment. We co-transfected cDNA-LINC00501 in cells with miR-129-5p-mimics and si-HMGB1, respectively, finding that there was no significant difference between cells co-transfected with pcDNA-LINC00501 and those transfected with pcDNA in cell proliferation, invasion, and apoptosis, implying that LINC00501 inhibited the proliferation and invasion of LCCs and induced their apoptosis through regulation on the miR-129-5p/HMGB1 axis. Figure 7.
Figure 7.
LINC00501 inhibits the proliferation and invasion of LCCs and induces their apoptosis by mediating the miR-129-5p/HMGB1 axis. (A) Changes in the proliferation of LCCs after co-transfection according to the CCK8 assay. (B) Changes in the invasion of LCCs after co-transfection according to the Transwell assay. (C) Changes in the apoptosis rate of LCCs after co-transfection according to the flow cytometry assay. *indicates P<0.05.
Discussion
LC is the malignant tumor with the highest incidence worldwide. According to a cancer statistics, the LC-related mortality increased by 200% in 2012 during the past 30 years, ranking first among malignant tumors.17 At present, the main factors affecting the prognosis of LC are as follows. First, early LC is relatively difficult to diagnose, and misdiagnosis or missed diagnosis is prone to result in the missing of optimal surgery timing for patients.18 Second, LC patients have relative resistance against radiotherapy and chemotherapy, and there is no better treatment plan except surgery.19 Therefore, it is required to explore the relevant mechanism of LC to provide more therapeutic targets for clinic practice.
LncRNA is a long-chain non-coding RNA. LncRNA is reported to be involved in the occurrence of various tumors and can be adopted as an indicator for tumor diagnosis and prognosis of tumors.20 For example, one previous study has found that lncRNA CASC11 can promote the expression of transforming growth factor-β1 to increase cancer stem cells and can be used to predict the postoperative survival of SCLC patients,21 and one other study has revealed that tumor-derived exosome lncRNA GAS5 can be utilized as a biomarker for diagnosis of early NSCLC.22 LINC00501 is a new lncRNA, and there is no previous report on LC and LINC00501.In this study, according to TCGA, we found that LINC00501 was highly expressed in LC, indicating that LINC00501 might be a potential therapy target for LC. With the aim of verifying the expression of LINC00501 in LC, we detected LCCs and tumor tissues of LC patients, finding that LINC00501 expression was significantly high in LC tissues and LCCs. Further analysis revealed that patients with high LINC00501 expression showed higher rates of low differentiation, high staging, and distant metastasis. In addition, it was also acquired that the survival rate of patients with high LINC00501 expression declined and their prognosis was poor. Based on the above research, we can confirm that LINC00501 can be adopted as a potential prognostic indicator of LC, but the role of LINC00501 in LC is still unclear. In order to further explore the role of LINC00501 in LC, we knocked down the expression of LINC00501 in LCCs. It came out that knock-down of LINC00501 inhibited cell proliferation and invasion and promoted cell apoptosis. Additionally, based on in vivo experiments, we have also confirmed that injecting LCCs transfected with sh-LINC00501 subcutaneously into nude mice can effectively inhibit tumor growth in the mice. According to the above results, we can verify that LINC00501 can be a potential therapeutic target for LC, but we are not clear about its downstream mechanism.
The introduction of competing endogenous RNAs accelerates the research on ncRNA.23 Many studies have found that lncRNA can be used as ceRNA of miR to affect miR silencing together with microRNA response elements (MREs).24,25 In order to explore the miR that can bind to LINC00501, we first predicted through three databases that miR-129-5p and LINC00501 had specific binding locus.MiR-129-5p, as an early discovered miR, has been found to have low expression in many tumors.26,27 A recent report has pointed out that miR-129-5p is under-expressed in LC and can suppressLC,28 and our study has also found that miR-129-5p is under-expressed in LC tissues. Moreover, our DLR assay and RIP assay verified specific binding between LINC00501 and miR-129-5p.It implies a regulatory relation between LINC00501 and miR-129-5p. To verify the relation, we carried out cell experiments, which revealed the enhancement of proliferation and invasion of LCCs and decrease in their apoptosis rate caused by transfection of miR-129-5p-inhibit were reversed by co-transfection with sh-LINC00501, verifying that LINC00501 can regulate miR-129-5p to participate in LC development.
At the end of the study, we predicted target genes of miR-129-5p, and found that there were targeted binding locus between HMGB1 and miR-129-5p. HMGB1, as a protein of the superfamily of high mobility group, has encoded nonhistone proteins and nuclear DNA binding proteins and can regulate transcription and participate in DNA tissues.29–31 One recent study has revealed that HMGB1 is highly expressed in LC, and cell invasion can be effectively regulated by suppressing HMGB1.32,33 In this study, the DLR assay revealed that there was a targeting relation between miR-129-5p and HMGB1, and knock-down of HMGB1 strongly inhibited cell invasion, and the inhibition was reversed through co-transfection of miR-129-5p-inhibit, implying that miR-129-5p can inhibit the invasion of LCCs through HMGB1.
In addition, in order to verify the roles of LINC00501 and miR-129-5p/HMGB1 axes in lung cancer, we conducted a rescue experiment, finding that inhibition of miR-129-5p-mimics and si-HMGB1 on the proliferation and invasion of LCCs was reversed by co-transfection with pcDNA-LINC00501, and the co-transfection also inhibited the apoptosis of the cells, implying that LINC00501 participated in the development and progression of lung cancer by mediating miR-129-5p/HMGB1.
Based on the above research, we can preliminarily come to the conclusion that LINC00501 inhibits the growth and metastasis of LCCs via the miR-129-5p/HMGB1 axis, but this study still has certain limitations. Firstly, it is not clear whether LINC00501 can be used as a diagnostic index of LC. Secondly, whether LINC00501 can regulate more miRs to participate in the development of LC still needs further research. Therefore, we hope to increase detection samples, compare diagnostic experiments with conventional tumor diagnostic indexes, and carry out bioinformatics analysis to supplement our research results in the future.
Conclusion
LINC00501 is highly expressed in LC and the high expression indicates poor prognosis of patients. In addition, LINC00501 can inhibit the invasion and migration of LC by mediating miR-129-5p/HMGB1.
Core Tip
In this study, based on the data from TCGA and clinical sample detection, we have confirmed that LINC00501 is highly expressed in LC, and the prognosis of patients with high LINC00501 expression is poor. Additionally, we have also confirmed through in vivo and in vitro assays that LINC00501 can suppress the growth and metastasis of lung cancer by mediating miR-129-5p / HMGB1.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 3.Oser MG, Niederst MJ, Sequist LV, Engelman JA. Transformation from non-small-cell lung cancer to small-cell lung cancer: molecular drivers and cells of origin. Lancet Oncol. 2015;16(4):e165–e172. doi: 10.1016/S1470-2045(14)71180-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389(10066):299–311. doi: 10.1016/S0140-6736(16)30958-8 [DOI] [PubMed] [Google Scholar]
- 5.Oberndorfer F, Mullauer L. Molecular pathology of lung cancer: current status and perspectives. Curr Opin Oncol. 2018;30(2):69–76. doi: 10.1097/CCO.0000000000000429 [DOI] [PubMed] [Google Scholar]
- 6.Romano G, Veneziano D, Acunzo M, Croce CM. Small non-coding RNA and cancer. Carcinogenesis. 2017;38(5):485–491. doi: 10.1093/carcin/bgx026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141(4):1202–1207. doi: 10.1016/j.jaci.2017.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jathar S, Kumar V, Srivastava J, Tripathi V. Technological Developments in lncRNA Biology. Adv Exp Med Biol. 2017;1008:283–323. [DOI] [PubMed] [Google Scholar]
- 9.Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25(1):1–7. doi: 10.1007/s12282-017-0793-9 [DOI] [PubMed] [Google Scholar]
- 10.Schmidt K, Joyce CE, Buquicchio F, et al. The lncRNA SLNCR1 mediates melanoma invasion through a conserved SRA1-like region. Cell Rep. 2016;15(9):2025–2037. doi: 10.1016/j.celrep.2016.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H, Huo X, Yang XR, et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol Cancer. 2017;16(1):136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y, Huang W, Sun W, et al. LncRNA MALAT1 promotes cancer metastasis in osteosarcoma via activation of the PI3K-Akt signaling pathway. Cell Physiol Biochem. 2018;51(3):1313–1326. doi: 10.1159/000495550 [DOI] [PubMed] [Google Scholar]
- 13.Renganathan A, Felley-Bosco E. Long noncoding RNAs in cancer and therapeutic potential. Adv Exp Med Biol. 2017;1008:199–222. [DOI] [PubMed] [Google Scholar]
- 14.Lou X, Li J, Yu D, Wei YQ, Feng S, Sun JJ. Comprehensive analysis of five long noncoding RNAs expression as competing endogenous RNAs in regulating hepatoma carcinoma. Cancer Med. 2019;8(12):5735–5749. doi: 10.1002/cam4.2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu B, Chen Y, Yang J. LncRNAs are altered in lung squamous cell carcinoma and lung adenocarcinoma. Oncotarget. 2017;8(15):24275–24291. doi: 10.18632/oncotarget.13651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koul R, Rathod S, Dubey A, Bashir B, Chowdhury A. Comparison of 7th and 8th editions of the UICC/AJCC TNM staging for non-small cell lung cancer in a non-metastatic North American cohort undergoing primary radiation treatment. Lung Cancer. 2018;123:116–120. doi: 10.1016/j.lungcan.2018.06.029 [DOI] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 18.Dela Cruz CS, Tanoue LT, Matthay RA. Lung cancer: epidemiology, etiology, and prevention. Clin Chest Med. 2011;32(4):605–644. doi: 10.1016/j.ccm.2011.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei H, Li G. Serum miR-1826 expression serves as a diagnostic biomarker and inhibits cell proliferation, migration, and invasion of lung cancer. Int J Clin Exp Med. 2019;12(8):10350–10357. [Google Scholar]
- 20.Zhang Z, Zhang K, Yu Q, Shou W. Effects of ginsenoside Rg3 combined with 125I seeds on serum TGF-α, TGF-β1, and VEGF in patients with medium and advanced non-small cell lung cancer. Int J Clin Exp Med. 2019;12(2):1864–1870. [Google Scholar]
- 21.Huang HW, Xie H, Ma X, Zhao F, Gao Y. Upregulation of LncRNA PANDAR predicts poor prognosis and promotes cell proliferation in cervical cancer. Eur Rev Med Pharmacol Sci. 2017;21(20):4529–4535. [PubMed] [Google Scholar]
- 22.Fu Y, Zhang P, Nan H, et al. LncRNA CASC11 promotes TGF-beta1, increases cancer cell stemness and predicts postoperative survival in small cell lung cancer. Gene. 2019;704:91–96. doi: 10.1016/j.gene.2019.04.019 [DOI] [PubMed] [Google Scholar]
- 23.Li C, Lv Y, Shao C, et al. Tumor-derived exosomal lncRNA GAS5 as a biomarker for early-stage non-small-cell lung cancer diagnosis. J Cell Physiol. 2019;234(11):20721–20727. doi: 10.1002/jcp.28678 [DOI] [PubMed] [Google Scholar]
- 24.Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52(10):710–718. doi: 10.1136/jmedgenet-2015-103334 [DOI] [PubMed] [Google Scholar]
- 25.Yu C, Li L, Xie F, et al. LncRNA TUG1 sponges miR-204-5p to promote osteoblast differentiation through upregulating Runx2 in aortic valve calcification. Cardiovasc Res. 2018;114(1):168–179. doi: 10.1093/cvr/cvx180 [DOI] [PubMed] [Google Scholar]
- 26.Lv L, Jia JQ, Chen J. The lncRNA CCAT1 upregulates proliferation and invasion in melanoma cells via suppressing miR-33a. Oncol Res. 2018;26(2):201–208. doi: 10.3727/096504017X14920318811749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao C, Long Z, Zhang X, et al. LncARSR sponges miR-129-5p to promote proliferation and metastasis of bladder cancer cells through increasing SOX4 expression. Int J Biol Sci. 2020;16(1):1–11. doi: 10.7150/ijbs.39461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi Y, Gong W, Lu L, Wang Y, Ren J. Upregulation of miR-129-5p increases the sensitivity to Taxol through inhibiting HMGB1-mediated cell autophagy in breast cancer MCF-7 cells. Braz J Med Biol Res. 2019;52(11):e8657. doi: 10.1590/1414-431x20198657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li G, Xie J, Wang J. Tumor suppressor function of miR-129-5p in lung cancer. Oncol Lett. 2019;17(6):5777–5783. doi: 10.3892/ol.2019.10241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu L, Yang L. The function and mechanism of HMGB1 in lung cancer and its potential therapeutic implications. Oncol Lett. 2018;15(5):6799–6805. doi: 10.3892/ol.2018.8215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang M, Li X, Quan X, Li X, Zhou B. Single nucleotide polymorphisms in HMGB1 correlate with lung cancer risk in the northeast chinese han population. Molecules. 2018;23(4):4. doi: 10.3390/molecules23040832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yusein-Myashkova S, Stoykov I, Gospodinov A, Ugrinova I, Pasheva E. The repair capacity of lung cancer cell lines A549 and H1299 depends on HMGB1 expression level and the p53 status. J Biochem. 2016;160(1):37–47. doi: 10.1093/jb/mvw012 [DOI] [PubMed] [Google Scholar]
- 33.Zheng H, Chen JN, Yu X, et al. HMGB1 enhances drug resistance and promotes in vivo tumor growth of lung cancer cells. DNA Cell Biol. 2016;35(10):622–627. doi: 10.1089/dna.2016.3360 [DOI] [PubMed] [Google Scholar]