Abstract
Human bone marrow-derived mesenchymal stem cells (HBMSCs) have the potential of multidirectional differentiation and self-renewal, which is important for the formation of human bone. It has been reported that long non-coding RNAs (lncRNAs) serve important roles in HBMSC osteogenic differentiation. The current study aimed to investigate the roles of long intergenic non-protein coding RNA 00707 (LINC00707) and microRNA (miR)-103a-3p in the osteogenic differentiation of HBMSCs. Reverse transcription-quantitative PCR (RT-qPCR) was performed to detect the expression levels of LINC00707, miR-103a-3p and osteogenesis-related genes (Alkaline phosphatase, osteocalcin, osteopontin and RUNX family transcription factor 2) in HBMSCs cultured in proliferation medium (PM) and osteogenic medium (OM). Mineralized matrix deposition was measured using Alizarin Red S staining. The protein expression levels of osteogenesis-related genes were detected by western blotting. The relationships between LINC00707, miR-103a-3p and dickkopf WNT signaling pathway inhibitor 1 (DKK1) were predicted using Starbase and TargetScan7.2, and were further assessed with a dual-luciferase reporter assay. After 21 days of cell culture, the results indicated that expression of LINC00707 was downregulated, and those of miR-103a-3p and osteogenesis-related genes were upregulated in OM-cultured HBMSCs. However, there was no significant difference in the aforementioned gene expression levels in PM-cultured HBMSCs. Small interfering (si)LINC00707 increased the deposition of mineralized matrix and promoted the expression levels of osteogenesis-related proteins. Furthermore, miR-103a-3p was predicted to be a target gene of LINC00707, its expression was significantly upregulated by siLINC00707, while overexpression of miR-103a-3p increased the expression levels of osteogenesis-related proteins. DKK1 was also predicted to be a target gene of miR-103a-3p and could inhibit the expression levels of osteogenesis-related proteins, but such effect of DKK1 could be reversed by the miR-103a-3p mimic. In conclusion, the present results suggested that LINC00707 regulated DKK1 expression by targeting miR-103a-3p to regulate osteogenic differentiation.
Keywords: human bone marrow-derived mesenchymal stem cells, microRNA-103a-3p, long intergenic non-protein coding RNA 00707, osteogenic differentiation, dickkopf WNT signaling pathway inhibitor 1
Introduction
Mesenchymal stem cells (MSCs) are pluripotent stem cells derived from the mesoderm, with strong proliferative capacity, multi-directional differentiation potential and low immunogenicity (1). MSCs can differentiate into other cell types, such as osteoblasts, myocytes, hepatocytes and stromal cells, under certain conditions (2,3). In the research field of regenerative medicine, human bone marrow-derived MSCs (HBMSCs) can be easily acquired and cultivated, and are widely used as seed cells for repairing and reconstructing damaged tissues or organs (4). Complex regulatory pathways at both the transcriptional and post-transcriptional levels are involved in the differentiation process (5). However, the exact molecular regulatory mechanism underlying osteogenic differentiation remains unknown. Therefore, the current study aimed to investigate the mechanism of osteogenic differentiation of HBMSCs.
Long-chain non-coding RNAs (lncRNAs) are RNA molecules that are >200 nucleotides in length (6). Moreover, the majority of lncRNAs lack substantial open reading frames for protein translation, and are primarily transcribed by RNA polymerase II (6). Previous studies have reported that lncRNAs serve an important regulatory role in biological processes, such as cell proliferation, metastasis and differentiation (7,8). Long intergenic non-protein coding RNA 00707 (LINC00707) is located on chromosome 10p14 (9,10), and upregulation of LINC00707 is associated with clinical manifestations, such as lymphatic metastasis and tumor size in lung adenocarcinoma (11). In addition, LINC00707 promotes cell proliferation and migration by regulating the target gene cell division cycle 42 (11). A previous study examined the role of LINC00707 in hepatocellular carcinoma, and revealed that it may be involved in the regulation of hepatocellular carcinoma progression by downregulating microRNA (miRNA/miR)-206 to increase cyclin dependent kinase 14 expression (12). In addition, the expression of LINC00707 is upregulated in melanoma, and it could affect the prognosis of patients by regulating inflammation and mitogen-activated protein kinase-related pathways (9). However, the effect of LINC00707 on the osteogenic differentiation of HBMSCs is yet to be fully elucidated.
miRNAs are endogenous non-coding small single-stranded RNAs (13,14), and are involved in the regulation of osteogenic differentiation of HBMSCs (15-17). For instance, miR-33a-5p has been shown to regulate osteogenic differentiation by targeting SATB homeobox 2 expression in HBMSCs (18). Moreover, inhibition of miR-222-3p expression promotes osteoblast differentiation by modulating the Smad5/RUNX family transcription factor 2 (RUNX2) signaling axis (19). It has also been reported that miR-125b regulates osteogenic differentiation of HBMSCs by targeting bone morphogenetic protein receptor type 1B (20). In addition, miR-103a-3p can serve as a reference gene for evaluating osteoarthritis (21).
Dickkopf WNT signaling pathway inhibitor (DKK1) is a potent LDL receptor related protein (LRP)5/6 antagonist that inhibits Wnt signaling pathways (22), and it has been reported that excess glucocorticoids impair osteoblastogenesis by inducing Wnt antagonists, including DKK1, sclerostin and secreted frizzled-related protein 1 (23). Furthermore, silencing DKK1 expression rescues dexamethasone-induced suppression of primary human osteoblast differentiation (24). Li et al (22) also reported that miR-291a-3p promoted BMSCs osteogenic differentiation via targeting DKK1 in dexamethasone-induced osteoporosis.
In the present study, small interfering (si)LINC00707 was constructed in HBMSCs, and the function of LINC00707, miR-103a-3p and target genes were predicted by bioinformatics analysis and further confirmed by performing functional assays. In addition, the effects of these genes on osteogenic differentiation were analyzed.
Materials and methods
Cell culture
HBMSCs (cat. no. PCS-500-012) were purchased from the American Type Culture Collection. The cells were cultured in CTS™ StemPro™ MSC serum-free medium [SFM; proliferation medium (PM); cat. no. A1033201; Thermo Fisher Scientific, Inc.) at 37°C with 5% CO2. After 2 days of culture, the original medium was removed, the cells were washed once or twice with PBS and then 5 ml fresh StemPro™ MSC SFM was added to the cells, which were returned to the CO2 incubator. Cells were subcultured after they reached 60-80% confluence.
HBMSCs were cultured in StemPro™ MSC SFM, passaged to the third generation and osteogenic differentiation was then induced using a StemPro™ Osteogenesis Differentiation kit [osteogenic medium (OM); cat. no. A1007201; Thermo Fisher Scientific, Inc.] after the cells had reached ≥80% confluence. Day 0 of the culture served as a control, and osteoblasts in differentiation groups and RNAs of the undifferentiated cell groups were extracted using TRIzol® reagent (cat. no. 15596026; Thermo Fisher Scientific, Inc.) on day 1, 3, 5, 7, 14 and 21 after the induction.
Transfection
After culture in OM, cells were incubated in a CO2 incubator for 12-16 h at 37°C to reach 60-70% confluency. Then, the cells were transfected with 20 nM si-negative control (siNC; siN0000001-1-5; 5′-UUC UCC GAA CGU GUC ACG UTT -3′), 20 nM siLINC00707 (siG141020094742-1-5, 5′-GCA GGA ACA UCA CCA UCU UUU -3′), 50 nM miR-103a-3p mimic (M, miR10000101-1-5; 5′-AGC AGC AUU GUA CAG GGC UAUGA-3′), 50 nM mimic NC (MC; miRB160401025525-2-1; 5′-UCA CAA CCU CCU AGA AAG AGU AGA -3′), 50 nM miR-103a-3p inhibitor (I; miR20000101-1-5; 5′-TCA TAG CCC TGT ACA ATG CTG CT-3′) and 50 nM inhibitor NC (IC; miR2N0000001-1-5; 5′-CAG TAC TTT TGT GTA GTA CAA -3′) using Lipofectamine® 3000 (cat. no. L3000015; Invitrogen; Thermo Fisher Scientific, Inc.). si-NC, siLINC00707, M, I, MC and IC were purchased from Guangzhou RiboBio Co., Ltd. (https://www.ribobio.com/). Cells were collected for further experiments 48 h after transfection.
To investigate the effect of overexpression of DKK1, cells were co-transfected with 0.5 µg pcDNA3.1 empty vector (NC; Invitrogen; Thermo Fisher Scientific, Inc.) and M or MC, and co-transfected with 0.5 µg pcDNA3.1-DKK1 vector (DKK1) and M or MC. Cell transfection was performed using Lipofectamine® 3000 transfection reagent Invitrogen kit (cat. no. L3000015; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Cells were prepared at a confluence of 70-90% before transfection, Lipofectamine® 3000 reagent and RNAs (100 ng/well) were diluted in Opti-MEM® medium (cat. no. 31985062; Invitrogen; Thermo Fisher Scientific, Inc.). Then, Lipofectamine® 3000 reagent-mix and the diluted RNAs were added to the diluted Lipofectamine® 3000 reagent. Cells were incubated at room temperature for 10 min. Then, 50 µl RNA-lipid complex was added to cells and incubated for 48 h at 37°C. The transfection efficiency of LINC00707 and miR-103a-3p was analyzed using reverse transcription-quantitative PCR (RT-qPCR).
Alizarin red s staining (ARS)
Cells were fixed with 70% ice ethanol at room temperature for 1 h and washed five times with ddH2O. Cells were then stained with 40 mM ARS (cat. no. ECM815; EMD Millipore) for 10-15 min at room temperature while gently stirring. After staining, cells were washed five times with ddH2O and the orange-red spots were identified as calcified nodules under a light microscope (magnification, ×200).
Bioinformatics prediction
The biological target gene for LINC00707 was predicted using StarBase v2.0 (http://starbase.sysu.edu.cn/index.php). Gene prediction of the miR-103a-3p biological target gene was performed using TargetScan7.2 (http://www.targetscan.org/vert_72/).
Dual-luciferase activity assay
The binding sites of LINC00707-wild-type (WT), LINC00707-mutant (MUT), DKK1-WT and DKK1-MUT DNA were cloned into pmirGLO luciferase vectors (cat. no. E1330; Promega Corporation). To investigate the binding of LINC00707 to miRNA, HBMSCs were transfected with 50 nM I in OM, while the transfection of 50 nM M into HBMSCs was used to determine the binding of miR-103a-3p to the target gene DKK1. The cell transfection was performed with Lipofectamine® 3000 (Thermo Fisher Scientific, Inc.) Cells transfected with IC or MC served as blank control. Luciferase activity was determined using a TransDetect® Double-Luciferase Reporter Assay kit (cat. no. FR201-01; TransGen Biotech Co., Ltd.) according to the manufacturer's instructions. After removing the cell culture medium, cells were carefully rinsed twice in PBS, 100 µl 1X cell lysis buffer was added, cells were fully lysed at room temperature for 10 min and centrifuged (12,000 × g; 4°C; 10 min) to obtain the supernatant. Then, 100 µl Luciferase Reaction reagent (at room temperature) was added, 20 µl cell lysate was carefully pipetted into the tube and mixed gently. The activity of the firefly luciferase reporter gene was measured using a luminescence meter (Molecular Devices, LLC; SpectraMaxL) after 48 h transfection. Then, 100 µl Luciferase Reaction Reagent II (at room temperature) was added to the aforementioned reaction tube, vortexed and the activity of the Renilla luciferase reporter gene was detected using the luminescence meter.
RT-qPCR
The lysis of each group of cells, and the extraction of total RNAs were all conducted at 4°C using TRIzol® (cat. no. 15596-026; Invitrogen; Thermo Fisher Scientific, Inc.). The quality and integrity of RNAs were detected using a NanoDrop One instrument (Thermo Fisher Scientific, Inc.) and 1% modified agarose gel electrophoresis. The reverse transcription reaction of miRNA, lncRNA and mRNA was performed using TaqMan™ MicroRNA RT kit (cat. no. 4366597; Thermo Fisher Scientific, Inc.) and TaqMan™ RT reagents (cat. no. N8080234; Thermo Fisher Scientific, Inc.). The reverse transcription reaction condition were set to 42°C for 30 min, and reverse transcriptase inactivation was conducted at 85°C for 15 min. qPCR was performed using SYBR Premix Ex TaqTM II (Takara Biotechnology Co., Ltd.) in the Opticon RT-PCR detection system (ABI 7500; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The thermocycling conditions were as follows: Initial denaturation at 95°C for 10 min, followed by 45 cycles at 95°C for 30 sec, 60°C for 1 min and 60°C for 1 min, and preserved at 4°C. The relative expression levels were calculated using the 2−ΔΔCq method (25). GAPDH served as an internal control for mRNAs and lncRNAs, and U6 was an internal control for miRNAs.
The sequences of primers were as follows: Alkaline phosphatase (ALP)-forward (F), 5′-ACC ACC ACG AGA GTG AAC CA-3′ and reverse (R), 5′-CGT TGT CTG AGT ACC AGT CCC-3′; osteocalcin (OCN)-F, 5′-GGC GCT ACC TGT ATC AAT GG-3′ and R: 5′-GTG GTC AGC CAA CTC GTC A-3′; osteopontin (OPN)-F, 5′-GGA GTT GAA TGG TGC ATA CAA GG-3′ and R, 5′-CCA CGG CTG TCC CAA TCA G-3′; RUNX2-F, 5′-TGT CAT GGC GGG TAA CGA T-3′ and R, 5′-AAG ACG GTT ATG GTC AAG GTG AA-3′; miR-103a-3p-F, 5′-TGT ACA GGG CTA TGA GTC GT-3′ and R, 5′-TCG TAT CCA GTG CGT G TC G-3′; LINC00707-F, 5′-TGG AAA GTA AGC CTA TTA CAT ATA C-3′ and R, 5′-GGT ATC ACC AAC AAC CCT GA-3′; DKK1-F, 5′-CAG TGC CAC CTT GAA CTC AGT -3′ and R, 5′-CCG CCC TCA TAG AGA ACT CC-3′; GAPDH-F, 5′-GTC AGC CGC ATC TTC TTT TG-3′ and R, 5′-GCG CCC AAT ACG ACC AAA TC-3′; and U6-F, 5′-CTC GCT TCG GCA GCA CA-3′ and R, 5′-AACGCTTCACGAATTTGCGT-3′. Each sample was analyzed in triplicate.
Western blotting (WB)
An RIPA lysate (cat. no. 89901; Thermo Fisher Scientific, Inc.) containing protease inhibitor and phosphatase inhibitor was used to extract total proteins from HBMSCs, and the protein concentration was detected using a Pierce™ Rapid Gold bicinchoninic acid protein assay kit (cat. no. A53227; Thermo Fisher Scientific, Inc.). A total of 30 µg protein was separated by 10% SDS-PAGE and transferred to PVDF membranes (cat. no. HVLP04700; EMD Millipore), which were rinsed in TBS and blocked with 5% non-fat milk powder solution at 37°C for 1 h. Then, the membranes were incubated with antibodies against ALP (39 kDa; Rabbit; 1:1,000; cat. no. ab83259; Abcam), OCN (12 kDa; Mouse; 1:1,000; cat. no. ab13420; Abcam), OPN (66 kDa; Rabbit; 1:1,000; cat. no. ab8448; Abcam), RUNX2 (57 kDa; Mouse; 1:1,000; cat. no. ab76956; Abcam) or GAPDH (36 kDa; Mouse; 1:1,000; cat. no. ab8245; Abcam) overnight at 4°C. The target bands were then incubated with a horseradish peroxidase-conjugated secondary antibody (1:1,000; cat. nos. ab205718 and ab205719; Abcam) for 2 h at room temperature. SignalFire™ ECL reagent (cat. no. 6883; Cell Signaling Technology, Inc.) was used to detect the signals and the gray values of the bands were analyzed using ImageJ (version 5.0; National Institute of Health).
Statistical analysis
Data are presented as the mean ± SD. Comparisons between the groups were performed using one-way ANOVA, followed by Dunnett test or Tukey's test using GraphPad Prism 7.0 (Graph-Pad Software, Inc.). P<0.05 was considered to indicate a statistically significant difference. All experiments were performed in triplicate.
Results
Expression levels of LINC00707, miR-103a-3p and osteogenesis-related genes in HBMSCs cultured in PM and OM
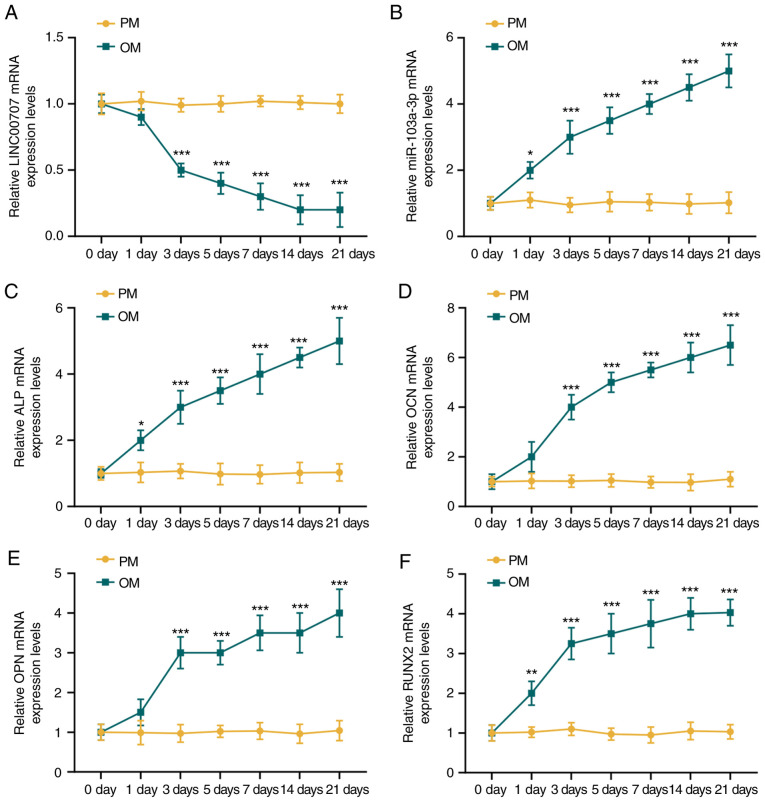
The expression levels of lncRNA LINC00707, miR-103a-3p and osteogenesis-related genes in HBMSCs cultured in PM and OM were detected by RT-qPCR. There was no changes in the expression levels of LINC00707, miR-103a-3p, ALP, OCN, OPN and RUNX2 in HBMSCs cultured in PM over time (P>0.05; Fig. 1). However, the expression of LINC00707 was reduced, while those of miR-103a-3p, ALP, OCN, OPN and RUNX2 were significantly increased in OM-cultured HBMSCs with prolonged culture duration (P<0.001). It was also found that the osteogenesis-related genes were all significantly increased during osteogenic induction from day 3 compared with day 0 (Fig. 1); therefore, in the follow-up experiment, the expression levels of osteogenic genes were detected at 3 days of osteogenic induction.
Figure 1.
Expression levels of LINC00707, miR-103a-3p and osteogenesis-related genes in HBMSCs cultured in PM and OM. (A) LINC00707 and (B) miR-103a-3p expression levels in HBMSCs cultured in PM and OM were measured by RT-qPCR. Expression levels of the osteogenesis-related genes (C) ALP, (D) OCN, (E) OPN and (F) RUNX2 in HBMSCs cultured in PM and OM were measured by RT-qPCR. Each experiment was repeated three times, GAPDH served as an internal control for mRNA and long non-coding RNA, and U6 served as an internal control for miRNAs. *P<0.05, **P<0.01, ***P<0.001 vs. 0 days. HBMSCs, human bone mesenchymal stem cells; PM, proliferation medium; OM, osteogenic medium; RT-qPCR, reverse transcription-quantitative PCR; miR/miRNA, microRNA; LINC00707, Long intergenic non-protein coding RNA 00707; ALP, Alkaline phosphatase; OCN, osteocalcin; OPN, osteo-pontin; RUNX2, RUNX family transcription factor 2.
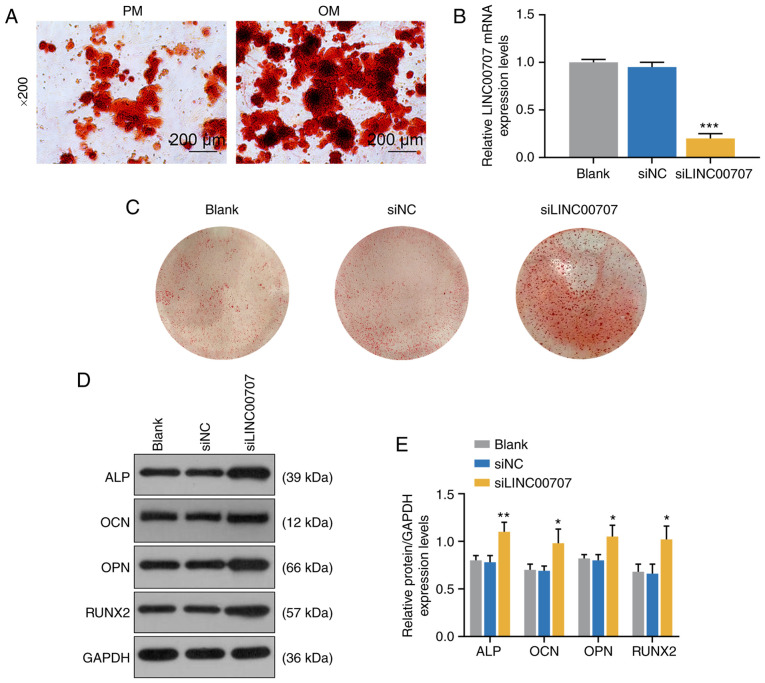
To further investigate the effects of PM and OM on osteogenic differentiation, ARS was performed to detect mineralized matrix deposition in PM and OM, and it was observed that mineralized matrix deposition in OM was notably increased compared with PM (Fig. 2A).
Figure 2.
Effects of PM, OM and LINC00707 silencing on osteogenic differentiation of HBMSCs. (A) Alizarin Red Staining of PM and OM were performed on day 21. Scale bar, 200 µm. Magnification, ×200. (B) LINC00707 expression after transfection with blank, siNC or siLINC00707 in HBMSCs cultured in OM was analyzed by reverse transcription-quantitative PCR. Each experiment was repeated three times, GAPDH served as an internal control. (C) Alizarin Red Staining of blank, siNC and siLINC00707 in HBMSCs cultured in OM was performed on day 21. (D) Western blotting results of the (E) protein expression levels of osteogenesis-related genes (ALP, OCN, OPN and RUNX2) in blank, siNC and siLINC00707 group. Each experiment was repeated three times, GAPDH served as an internal control. *P<0.05, **P<0.01, ***P<0.001 vs. siNC. HBMSCs, human bone mesenchymal stem cells; PM, proliferation medium; OM, osteogenic medium; NC, negative control; LINC00707, Long intergenic non-protein coding RNA 00707; ALP, Alkaline phosphatase; OCN, osteocalcin; OPN, osteopontin; RUNX2, RUNX family transcription factor 2; siRNA, small interfering RNA.
Effects of LINC00707 silencing on osteogenic differentiation of HBMSCs
To investigate the effect of LINC00707 on osteogenic differentiation of HBMSCs, blank, siNC and siLINC00707 groups of OM-cultured HBMSCs were established (P<0.001; Fig. 2B), and the mineralization matrix deposition of the three groups were examined by ARS. The results indicated that siLINC00707 markedly increased mineralized matrix deposition (Fig. 2C). In addition, WB was performed to measure the expression levels of osteogenesis-related genes of OM-cultured cells 3 days after the incubation, and it was found that siLINC00707 significantly increased the expression levels of ALP, OCN, OPN and RUNX2 compared with siNC group (P<0.05; Fig. 2D and E).
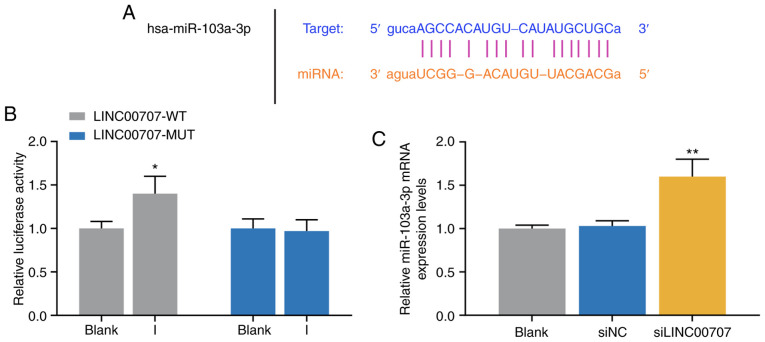
Relationship between LINC00707 and miR-103a-3p in HBMSCs
The mechanism of LINC00707 on osteogenic differentiation of HBMSCs was further investigated. StarBase predicted that miR-103a-3p was the target miRNA of LINC00707 (Fig. 3A), and the prediction was further demonstrated by the dual-luciferase activity assay results (Fig. 3B). The results indicated that cells in the LINC00707-WT and miR-103a-3p I group exhibited significant enhanced luciferase activity, while no significant difference was identified in luciferase activity in cells co-transfected with LINC00707-MUT and miR-103a-3p I compared with the blank group (P<0.05; Fig. 3B). Furthermore, the RT-qPCR assay results suggested that the expression of miR-103a-3p was significantly higher in the siLINC00707 group of HBMSCs compared with the siNC groups, indicating that knocking down LINC00707 upregulated the expression of miR-103a-3p (P<0.01; Fig. 3C).
Figure 3.
Relationship between the expression levels of LINC00707 and miR-103a-3p in HBMSCs. (A) StarBase and (B) dual-luciferase activity were used to identify the LINC00707 binding site in miR-103a-3p. *P<0.05 vs. blank. (C) Expression of miR-103a-3p in blank, siNC and siLINC00707 group was analyzed by reverse transcription-quantitative PCR. Each experiment was repeated three times, and U6 served as an internal control. **P<0.01 vs. siNC. HBMSCs, human bone mesenchymal stem cells; PM, proliferation medium; OM, osteogenic medium; NC, negative control; LINC00707, Long intergenic non-protein coding RNA 00707; siRNA, small interfering RNA; WT, wild-type; MUT, mutant; miR/miRNA, microRNA.
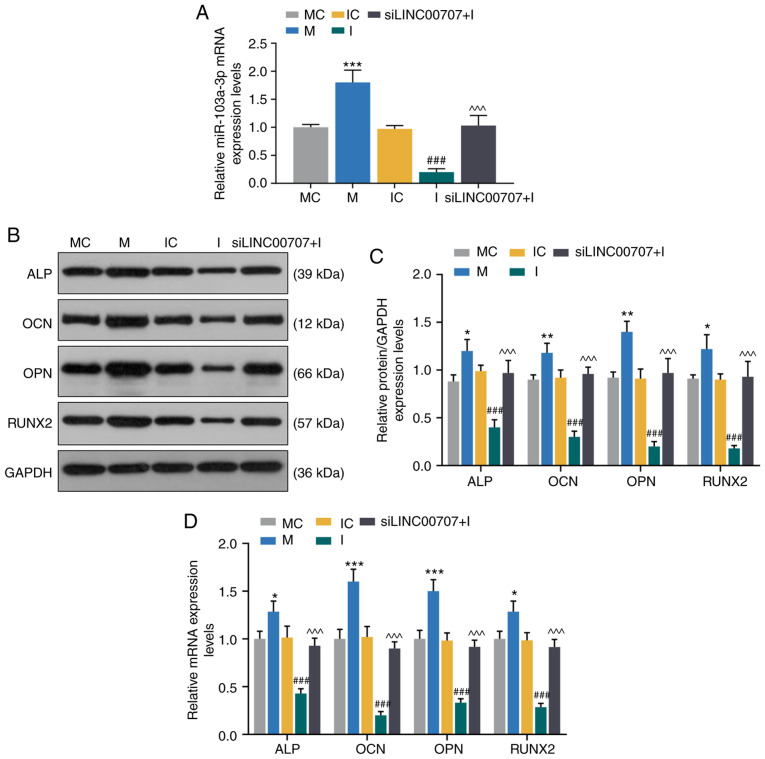
Effects of miR-103a-3p and LINC00707 on expression levels of osteogenesis-related genes of HBMSCs
To further investigate the effect of miR-103a-3p on the expression levels of osteogenesis-related genes, MC, M, IC, I and siLINC00707 + I groups of HBMSCs were established. The RT-qPCR assay results demonstrated that the mimic significantly increased the expression of miR-103a-3p compared with MC group, while the inhibitor downregulated the miR-103a-3p expression compared with IC group, and miR-103a-3p expression was higher in the siLINC00707 + I group compared with the I group (P<0.001; Fig. 4A).
Figure 4.
Effects of miR-103a-3p and LINC00707 on expression levels of osteogenesis-related genes of HBMSCs. (A) Expression of miR-103a-3p after trans-fection with MC, M, IC, I or siLINC00707 + I in HBMSCs was analyzed by RT-qPCR. Each experiment was repeated three times, and U6 served as an internal control. (B) Western blotting results of the (C) protein expression levels of osteogenesis-related genes (ALP, OCN, OPN and RUNX2) in MC, M, IC, I and siLINC00707 + I HBMSCs. (D) mRNA expression levels of osteogenesis-related genes (ALP, OCN, OPN and RUNX2) in MC, M, IC, I and siLINC00707 + I HBMSCs were detected by RT-qPCR. Each experiment was repeated three times, and GAPDH served as an internal control. *P<0.05, **P<0.01, ***P<0.001 vs. MC; ###P<0.001 vs. IC; ^^^P<0.001 vs. I. HBMSCs, human bone mesenchymal stem cells; RT-qPCR, reverse transcription-quantitative PCR; MC, mimic control; IC, inhibitor control; I, inhibitor; M, mimic; LINC00707, Long intergenic non-protein coding RNA 00707; siRNA, small interfering RNA; ALP, Alkaline phosphatase; OCN, osteocalcin; OPN, osteopontin; RUNX2, RUNX family transcription factor 2.
The expression levels of osteogenesis-related genes of the OM-cultured cells 3 days after the incubation were detected by WB and RT-qPCR. The results identified that the mimic upregulated the mRNA and protein expression levels of ALP, OCN, OPN and RUNX2 compared with MC group, while the inhibitor downregulated the mRNA and protein expression levels of ALP, OCN, OPN and RUNX2 compared with IC group (P<0.001; Fig. 4B-D). In addition, the mRNA and protein expression levels of ALP, OCN, OPN and RUNX2 were significantly higher in siLINC00707 + I group compared with the I group, indicating that upregulation of miR-103a-3p increased the mRNA and protein expression levels of osteogenesis-related genes (P<0.001; Fig. 4B-D).
miR-103a-3p increases the expression levels of osteogenesis-related genes of HBMSCs by regulating DKK1 expression
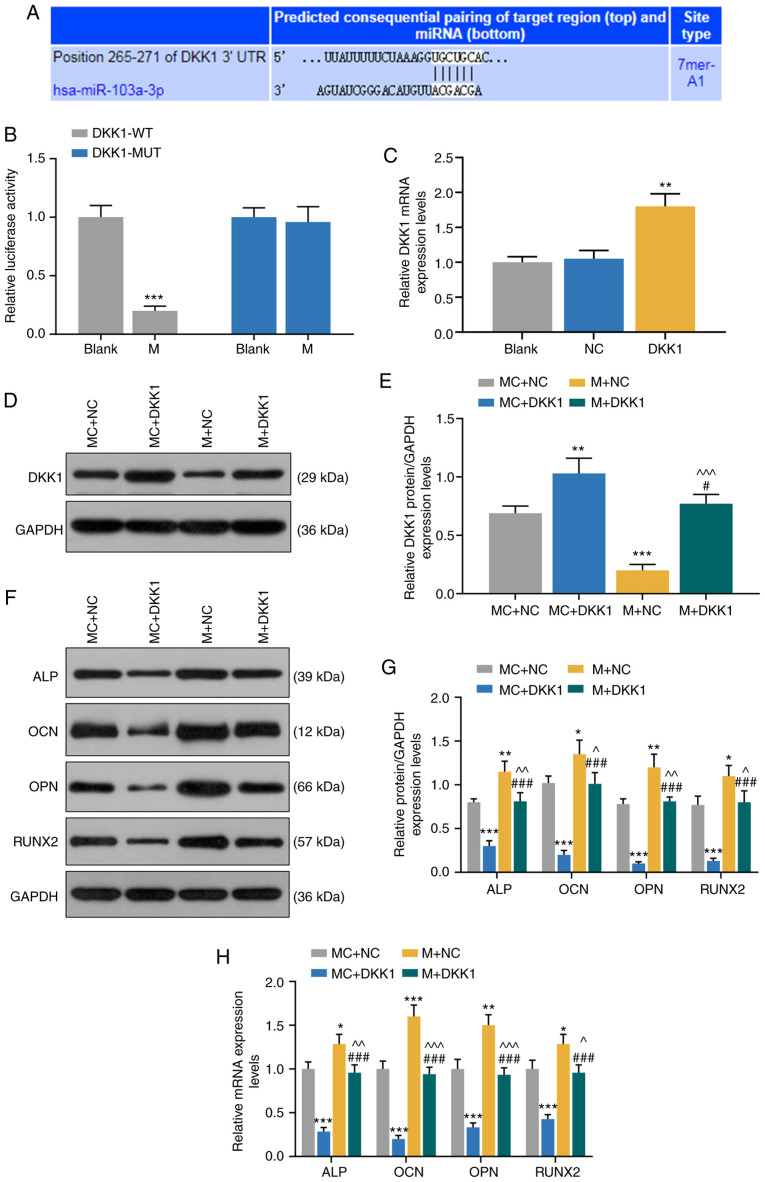
TargetScan7.2 predicted that DKK1 was a target gene for miR-103a-3p in HBMSCs (Fig. 5A), and this was further demonstrated by dual-luciferase activity assay, as it was found that the mimic significantly reduced the luciferase activity of DKK1-WT (P<0.001; Fig. 5B).
Figure 5.
miR-103a-3p promotes expression levels of osteogenesis-related genes in HBMSCs by regulating DKK1. (A) TargetScan7.2 and (B) dual-luciferase activity were performed to identify the miR-103a-3p binding site in DKK1. ***P<0.001 vs. blank. (C) Expression of DKK1 in blank, NC and DKK1 group was detected by RT-qPCR. (D) Western blotting results of (E) DKK1 transfection with MC + NC, MC + DKK1, M + NC or M + DKK1 in HBMSCs. (F) Western blotting results of (G) protein expression levels of osteogenesis-related genes (ALP, OCN, OPN and RUNX2) in MC + NC, MC + DKK1, M + NC and M + DKK1 group. (H) mRNA expression levels of osteogenesis-related genes (ALP, OCN, OPN and RUNX2) in MC, M, IC, I and siLINC00707 + I group were detected by RT-qPCR. Each experiment was repeated three times, and GAPDH served as an internal control. *P<0.05, **P<0.01, ***P<0.001 vs. MC + NC; #P<0.05, ###P<0.001 vs. MC + DKK1; ^P<0.05, ^^P<0.01, ^^^P<0.001 vs. M + NC. HBMSCs, human bone mesenchymal stem cells; RT-qPCR, reverse transcription-quantitative PCR; MC, mimic control; IC, inhibitor control; I, inhibitor; M, mimic; NC, negative control; LINC00707, Long intergenic non-protein coding RNA 00707; siRNA, small interfering RNA; ALP, Alkaline phosphatase; OCN, osteocalcin; OPN, osteopontin; RUNX2, RUNX family transcription factor 2; miR/miRNA, microRNA; WT, wild-type; MUT, mutant; DKK1, dickkopf WNT signaling pathway inhibitor 1.
To further investigate the effects of miR-103a-3p and DKK1 on osteogenic differentiation, MC + NC, MC + DKK1, M + NC and M + DKK1 groups were established, and the transfection efficiency of DKK1 was detected by RT-qPCR. The results identified that DKK1 expression was significantly increased in DKK1 group compared with the NC group (P<0.001; Fig. 5C). Moreover, the expression of DKK1 was higher in MC + DKK1 group but lower in M + NC group, compared with the MC + NC group (P<0.001, Fig. 5D and E). It was also found that DKK1 expression in M + DKK1 group was lower compared with the MC + DKK1 group, but higher compared with the M + NC group (P<0.001; Fig. 5D and E).
The expression levels of osteogenesis-related genes of OM-cultured HBMSCs 3 days after the incubation were detected by WB and RT-qPCR, and it was identified that the mRNA and protein expression levels of ALP, OCN, OPN and RUNX2 were lower in MC + DKK1 group compared with the MC + NC group. Furthermore, the mRNA and protein expression levels of ALP, OCN, OPN and RUNX2 in M + NC group were higher compared with the MC+NC group, while these were reduced in the M + DKK1 group compared with the M + NC group (P<0.001; Fig. 5F-H).
Discussion
lncRNAs serve important roles in a variety of biological processes, such as embryonic development, angiogenesis and cell differentiation, and participate in a several of cellular regulation activities, such as transcriptional activation, transcriptional interference and intranuclear transport (6,26). For example, Kikuchi et al (27) showed that adipogenesis up-regulated transcript 1 may be involved in the differentiation of MSCs into adipocytes. MSCs are important sources of multifunctional cells for tissue repair, and lncRNAs function in the regulation of HBMSCs (28). Jia et al (29) reported that LINC00707 sponges miR-370-3p to promote osteogenesis of HBMSCs by upregulating Wnt family member 2B. In addition, Cai et al (30) revealed that LINC00707 promoted osteogenic differentiation of HBMSCs via the Wnt/β-catenin pathway activated by the LINC00707/miR-145/LDL receptor related protein 5 axis. The current study investigated LINC00707 and osteogenesis-related genes (31,32) in HBMSCs cultured in PM and OM.
lncRNAs are significantly correlated with MSCs in osteogenic differentiation (33-36). Zhu and Xu (37) reported that Angelman syndrome chromosome region (ANCR) inhibited the expression of RUNX2 by interacting with enhancer of zeste 2 polycomb repressive complex 2 subunit, thus inhibiting the osteogenic differentiation of MSCs. Moreover, Zuo et al (38) studied lncRNA expression profiles in the early osteogenic differentiation of murine MSCs, and found that 116 lncRNAs were differentially expressed in the bone morphoge-netic protein 2-treated group compared with the control group. lncRNA LINC00707 is abnormally expressed in lung adeno-carcinoma and melanoma (9,11), but its effect on osteogenic differentiation of HBMSCs are not fully understood. ARS can be used to measure mineralized matrix deposition (39,40). The current results demonstrated that LINC00707 expression of OM-cultured HBMSCs was significantly reduced, while there was no significant change in LINC00707 expression in PM-cultured HBMSCs, indicating that LINC00707 may be involved in the regulation of osteogenic differentiation. In addition, the mineralized matrix deposition in OM was markedly increased compared with PM. It was also found that knocking down LINC00707 promoted the expression levels of osteogenesis-related genes ALP, OCN, OPN and RUNX2, suggesting that LINC00707 may have similar function to ANCR in inhibiting osteogenic differentiation (37).
Osteoblasts are generally derived from BM-derived stromal stem cells (1). Moreover, significant changes in miRNA profiles during BM-derived morphogenetic protein-2-induced differentiation of BM stromal stem cells into osteoblasts have been observed, and miR-133 and miR-135 are closely related to osteogenic differentiation (41). It has been previously reported that the negative regulation of miR-138 on osteoblasts is caused by mediating the focal adhesion kinase/ERK1/2 signaling pathway (42). Moreover, the miR-196a/homeobox C8 pathway regulates the differentiation and proliferation of adipose stem cells into osteoblasts (43), while miR-103a-3p may play a key role in osteogenic differentiation (21). The role of miR-103a-3p in pancreatic cancer (44), bladder cancer (45) and gastric cancer cells (46) has also been previously revealed, but the specific regulatory mechanism of miR-103a-3p in osteogenic differentiation of HBMSCs remains to be investigated (47). The present results suggested that the expression of miR-103a-3p in OM-cultured HBMSCs was significantly increased, and that miR-103a-3p was a targeted gene for LINC00707, indicating that miR-103a-3p had opposite effects on osteogenic differentiation compared with miR-133 and miR-135 (41). Different miRNAs play diverse roles in osteogenic differentiation (48). Previous studies have shown that miR-210 inhibits the transforming growth factor-β/activin signaling pathway by downregulating the expression of activin A receptor type 1B protein, thus promoting cell osteogenic differentiation (49). In the present study, overexpression of miR-103a-3p promoted the expression levels of osteogenesis-related genes (ALP, OCN, OPN and RUNX2), which could be reversed by inhibition of LINC00707.
DDK1 is an inhibitor of Wnt signaling (50,51). The Wnt pathway induces bone cell differentiation, prevents bone aging and inflammation and is an important pathway in bone metabolism (52). DDK1 binds to low-density lipoprotein receptor-related proteins 5 or 6 and blocks heterodimerization with frizzled, thus affecting osteogenic differentiation (53). Furthermore, Xia et al (54) demonstrated that miR-203 could promote the differentiation of rat MSCs into osteoblast-like cells by regulating the expression of DKK1. In the present study, DDK1 was identified as a target gene of miR-103a-3p, and it inhibited the expression levels of ALP, OCN, OPN and RUNX2, while overexpression of miR-103a-3p reversed the inhibitory effects of DDK1 on the expression levels of osteogenesis-related genes. lncRNAs expressed in cytoplasm can act as a molecular sponge of miRNA and function as competing endogenous RNA (ceRNA) (55). In the current study, LINC00707 acted as a molecular decoy for miR-103a-3p, and LINC00707 also served as a ceRNA to regulate miR-103a-3p and DKK1. However, this study also has some limitations. For example, the effect of LINC00707/miR-103a-3p on the expression levels of genes in the Wnt pathway has yet to be detected, such as JNK and ERK1/2. In addition, LINC00707 knockout mice should be generated to measure the bone mineral density, which will provide evidence for the function of LINC00707 in HBMSCs osteogenic differentiation.
In conclusion, during the osteogenic differentiation of HBMSCs, the expression of LINC00707 was reduced, while that of miR-103a-3p was increased. Moreover, inhibition of LINC00707 promoted osteogenic differentiation gene expression of HBMSCs by targeting miR-103a-3p to regulate the DKK1 signaling pathway. Thus, the current findings provide evidence for further study on the regulatory mechanisms of osteogenic differentiation of HBMSCs.
Acknowledgments
Not applicable.
Funding
This study was supported by the Science and Technology Development Plan Project of Jilin Province (grant no. 20180520128JH).
Availability of data and materials
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Authors' contributions
Substantial contributions to conception and design: JL. Data acquisition, data analysis and interpretation: MW, GF, RL, YW and JJ. Drafting the article or critically revising it for important intellectual content: JL. Final approval of the version to be published: All authors. Agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of the work are appropriately investigated and resolved: All authors.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Liu H, Xia X, Li B. Mesenchymal stem cell aging: Mechanisms and influences on skeletal and non-skeletal tissues. Exp Biol Med (Maywood) 2015;240:1099–1106. doi: 10.1177/1535370215591828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ji N, Yu JW, Ni XC, Wu JG, Wang SL, Jiang BJ. Bone marrow-derived mesenchymal stem cells increase drug resistance in CD133-expressing gastric cancer cells by regulating the PI3K/AKT pathway. Tumour Biol. 2016;37:14637–14651. doi: 10.1007/s13277-016-5319-0. [DOI] [PubMed] [Google Scholar]
- 3.Polymeri A, Giannobile WV, Kaigler D. Bone marrow stromal stem cells in tissue engineering and regenerative medicine. Horm Metab Res. 2016;48:700–713. doi: 10.1055/s-0042-118458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joshi R, Goihberg E, Ren W, Pilichowska M, Mathew P. Proteolytic fragments of fibronectin function as matrikines driving the chemotactic affinity of prostate cancer cells to human bone marrow mesenchymal stromal cells via the α5β1 integrin. Cell Adh Migr. 2017;11:305–315. doi: 10.1080/19336918.2016.1212139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takizawa N, Okubo N, Kamo M, Chosa N, Mikami T, Suzuki K, Yokota S, Ibi M, Ohtsuka M, Taira M, et al. Bone marrow-derived mesenchymal stem cells propagate immu-nosuppressive/anti-inflammatory macrophages in cell-to-cell contact-independent and -dependent manners under hypoxic culture. Exp Cell Res. 2017;358:411–420. doi: 10.1016/j.yexcr.2017.07.014. [DOI] [PubMed] [Google Scholar]
- 6.Han P, Chang CP. Long non-coding RNA and chromatin remodeling. RNA Biol. 2015;12:1094–1098. doi: 10.1080/15476286.2015.1063770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jain S, Thakkar N, Chhatai J, Pal Bhadra M, Bhadra U. Long non-coding RNA: Functional agent for disease traits. RNA Biol. 2017;14:522–535. doi: 10.1080/15476286.2016.1172756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kondo Y, Shinjo K, Katsushima K. Long non-coding RNAs as an epigenetic regulator in human cancers. Cancer Sci. 2017;108:1927–1933. doi: 10.1111/cas.13342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang S, Xu J, Zeng X. A six-long non-coding RNA signature predicts prognosis in melanoma patients. Int J Oncol. 2018;52:1178–1188. doi: 10.3892/ijo.2018.4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li T, Li Y, Sun H. MicroRNA-876 is sponged by long noncoding RNA LINC00707 and directly targets metadherin to inhibit breast cancer malignancy. Cancer Manag Res. 2019;11:5255–5269. doi: 10.2147/CMAR.S210845. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Ma T, Ma H, Zou Z, He X, Liu Y, Shuai Y, Xie M, Zhang Z. The long intergenic noncoding RNA 00707 promotes lung adenocarcinoma cell proliferation and migration by regulating Cdc42. Cell Physiol Biochem. 2018;45:1566–1580. doi: 10.1159/000487693. [DOI] [PubMed] [Google Scholar]
- 12.Tu J, Zhao Z, Xu M, Chen M, Weng Q, Wang J, Ji J. LINC00707 contributes to hepatocellular carcinoma progression via sponging miR-206 to increase CDK14. J Cell Physiol. 2019;234:10615–10624. doi: 10.1002/jcp.27737. [DOI] [PubMed] [Google Scholar]
- 13.Tüfekci KU, Oner MG, Meuwissen RL, Genç S. The role of microRNAs in human diseases. Methods Mol Biol. 2014;1107:33–50. doi: 10.1007/978-1-62703-748-8_3. [DOI] [PubMed] [Google Scholar]
- 14.Horsburgh S, Fullard N, Roger M, Degnan A, Todryk S, Przyborski S, O'Reilly S. MicroRNAs in the skin: Role in development, homoeostasis and regeneration. Clin Sci (Lond) 2017;131:1923–1940. doi: 10.1042/CS20170039. [DOI] [PubMed] [Google Scholar]
- 15.Wen D, Peng Y, Liu D, Weizmann Y, Mahato RI. Mesenchymal stem cell and derived exosome as small RNA carrier and Immunomodulator to improve islet transplantation. J Control Release. 2016;238:166–175. doi: 10.1016/j.jconrel.2016.07.044. [DOI] [PubMed] [Google Scholar]
- 16.Ren G, Sun J, Li MM, Zhang YD, Li RH, Li YM. MicroRNA-23a-5p regulates osteogenic differentiation of human bone marrow-derived mesenchymal stem cells by targeting mitogen-activated protein kinase-13. Mol Med Rep. 2018;17:4554–4560. doi: 10.3892/mmr.2018.8452. [DOI] [PubMed] [Google Scholar]
- 17.Jiang C, Gong F. MiR-148a promotes myocardial differentiation of human bone mesenchymal stromal cells via DNA methyltransferase 1 (DNMT1) Cell Biol Int. 2018;42:913–922. doi: 10.1002/cbin.10813. [DOI] [PubMed] [Google Scholar]
- 18.Mi W, Shi Q, Chen X, Wu T, Huang H. miR-33a-5p modulates TNF-α-inhibited osteogenic differentiation by targeting SATB2 expression in hBMSCs. FEBS Lett. 2016;590:396–407. doi: 10.1002/1873-3468.12064. [DOI] [PubMed] [Google Scholar]
- 19.Yan J, Guo D, Yang S, Sun H, Wu B, Zhou D. Inhibition of miR-222-3p activity promoted osteogenic differentiation of hBMSCs by regulating Smad5-RUNX2 signal axis. Biochem Biophys Res Commun. 2016;470:498–503. doi: 10.1016/j.bbrc.2016.01.133. [DOI] [PubMed] [Google Scholar]
- 20.Wang H, Xie Z, Hou T, Li Z, Huang K, Gong J, Zhou W, Tang K, Xu J, Dong S. MiR-125b regulates the osteogenic differentiation of human mesenchymal stem cells by targeting BMPR1b. Cell Physiol Biochem. 2017;41:530–542. doi: 10.1159/000457013. [DOI] [PubMed] [Google Scholar]
- 21.Ragni E, Perucca Orfei C, De Luca P, Colombini A, Viganò M, Lugano G, Bollati V, de Girolamo L. Identification of miRNA reference genes in extracellular vesicles from adipose derived mesenchymal stem cells for studying osteoarthritis. Int J Mol Sci. 2019;20:1108. doi: 10.3390/ijms20051108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li ZH, Hu H, Zhang XY, Liu GD, Ran B, Zhang PG, Liao MM, Wu YC. MiR-291a-3p regulates the BMSCs differentiation via targeting DKK1 in dexamethasone-induced osteoporosis. Kaohsiung J Med Sci. 2020;36:35–42. doi: 10.1002/kjm2.12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komori T. Glucocorticoid signaling and bone biology. Horm Metab Res. 2016;48:755–763. doi: 10.1055/s-0042-110571. [DOI] [PubMed] [Google Scholar]
- 24.Butler JS, Queally JM, Devitt BM, Murray DW, Doran PP, O'Byrne JM. Silencing Dkk1 expression rescues dexamethasone-induced suppression of primary human osteoblast differentiation. BMC Musculoskelet Disord. 2010;11:210. doi: 10.1186/1471-2474-11-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Ransohoff JD, Wei Y, Khavari PA. The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol. 2018;19:143–157. doi: 10.1038/nrm.2017.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kikuchi K, Fukuda M, Ito T, Inoue M, Yokoi T, Chiku S, Mitsuyama T, Asai K, Hirose T, Aizawa Y. Transcripts of unknown function in multiple-signaling pathways involved in human stem cell differentiation. Nucleic Acids Res. 2009;37:4987–5000. doi: 10.1093/nar/gkp426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun X, Jia B, Qiu XL, Chu HX, Zhang ZQ, Wang ZP, Zhao JJ. Potential functions of long non-coding RNAs in the osteogenic differentiation of human bone marrow mesenchymal stem cells. Mol Med Rep. 2019;19:103–114. doi: 10.3892/mmr.2018.9674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jia B, Wang Z, Sun X, Chen J, Zhao J, Qiu X. Long noncoding RNA LINC00707 sponges miR-370-3p to promote osteogenesis of human bone marrow-derived mesenchymal stem cells through upregulating WNT2B. Stem Cell Res Ther. 2019;10:67. doi: 10.1186/s13287-019-1161-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cai WL, Zeng W, Liu HH, Zhu BY, Liu JL, Liu Y. LncRNA LINC00707 promotes osteogenic differentiation of hBMSCs through the Wnt/β-catenin pathway activated by LINC00707/miR-145/LRP5 axis. Eur Rev Med Pharmacol Sci. 2020;24:18–28. doi: 10.26355/eurrev_202001_19891. [DOI] [PubMed] [Google Scholar]
- 31.An J, Yang H, Zhang Q, Liu C, Zhao J, Zhang L, Chen B. Natural products for treatment of osteoporosis: The effects and mechanisms on promoting osteoblast-mediated bone formation. Life Sci. 2016;147:46–58. doi: 10.1016/j.lfs.2016.01.024. [DOI] [PubMed] [Google Scholar]
- 32.Wang Q, Li Y, Zhang Y, Ma L, Lin L, Meng J, Jiang L, Wang L, Zhou P, Zhang Y. LncRNA MEG3 inhibited osteogenic differentiation of bone marrow mesenchymal stem cells from postmenopausal osteoporosis by targeting miR-133a-3p. Biomed Pharmacother. 2017;89:1178–1186. doi: 10.1016/j.biopha.2017.02.090. [DOI] [PubMed] [Google Scholar]
- 33.Zhang J, Tao Z, Wang Y. Long non-coding RNA DANCR regulates the proliferation and osteogenic differentiation of human bone-derived marrow mesenchymal stem cells via the p38 MAPK pathway. Int J Mol Med. 2018;41:213–219. doi: 10.3892/ijmm.2017.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cui Y, Lu S, Tan H, Li J, Zhu M, Xu Y. Silencing of long non-coding RNA NONHSAT009968 ameliorates the staphylococcal protein A-inhibited osteogenic differentiation in human bone mesenchymal stem cells. Cell Physiol Biochem. 2016;39:1347–1359. doi: 10.1159/000447839. [DOI] [PubMed] [Google Scholar]
- 35.Gao Y, Xiao F, Wang C, Wang C, Cui P, Zhang X, Chen X. Long noncoding RNA MALAT1 promotes osterix expression to regulate osteogenic differentiation by targeting miRNA-143 in human bone marrow-derived mesenchymal stem cells. J Cell Biochem. 2018;119:6986–6996. doi: 10.1002/jcb.26907. [DOI] [PubMed] [Google Scholar]
- 36.Wang J, Miao J, Meng X, Chen N, Wang Y. Expression of long non-coding RNAs in human bone marrow mesenchymal stem cells co-cultured with human amnion-derived mesenchymal stem cells. Mol Med Rep. 2017;16:6683–6689. doi: 10.3892/mmr.2017.7465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu L, Xu PC. Downregulated LncRNA-ANCR promotes osteoblast differentiation by targeting EZH2 and regulating Runx2 expression. Biochem Biophys Res Commun. 2013;432:612–617. doi: 10.1016/j.bbrc.2013.02.036. [DOI] [PubMed] [Google Scholar]
- 38.Zuo C, Wang Z, Lu H, Dai Z, Liu X, Cui L. Expression profiling of lncRNAs in C3H10T1/2 mesenchymal stem cells undergoing early osteoblast differentiation. Mol Med Rep. 2013;8:463–467. doi: 10.3892/mmr.2013.1540. [DOI] [PubMed] [Google Scholar]
- 39.Puchtler H, Meloan SN, Terry MS. On the history and mechanism of alizarin and alizarin red S stains for calcium. J Histochem Cytochem. 1969;17:110–124. doi: 10.1177/17.2.110. [DOI] [PubMed] [Google Scholar]
- 40.Paul H, Reginato AJ, Schumacher HR. Alizarin red S staining as a screening test to detect calcium compounds in synovial fluid. Arthritis Rheum. 1983;26:191–200. doi: 10.1002/art.1780260211. [DOI] [PubMed] [Google Scholar]
- 41.Li Z, Hassan MQ, Volinia S, van Wijnen AJ, Stein JL, Croce CM, Lian JB, Stein GS. A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc Natl Acad Sci USA. 2008;105:13906–13911. doi: 10.1073/pnas.0804438105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eskildsen T, Taipaleenmäki H, Stenvang J, Abdallah BM, Ditzel N, Nossent AY, Bak M, Kauppinen S, Kassem M. MicroRNA-138 regulates osteogenic differentiation of human stromal (mesenchymal) stem cells in vivo. Proc Natl Acad Sci USA. 2011;108:6139–6144. doi: 10.1073/pnas.1016758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim YJ, Bae SW, Yu SS, Bae YC, Jung JS. miR-196a regulates proliferation and osteogenic differentiation in mesenchymal stem cells derived from human adipose tissue. J Bone Miner Res. 2009;24:816–825. doi: 10.1359/jbmr.081230. [DOI] [PubMed] [Google Scholar]
- 44.Zhou H, Rigoutsos I. MiR-103a-3p targets the 5′ UTR of GPRC5A in pancreatic cells. RNA. 2014;20:1431–1439. doi: 10.1261/rna.045757.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep. 2016;6:30919. doi: 10.1038/srep30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu X, Miao J, Zhang M, Wang X, Wang Z, Han J, Tong D, Huang C. miRNA-103a-3p promotes human gastric cancer cell proliferation by targeting and suppressing ATF7 in vitro. Mol Cells. 2018;41:390–400. doi: 10.14348/molcells.2018.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim DS, Lee SY, Lee JH, Bae YC, Jung JS. MicroRNA-103a-3p controls proliferation and osteogenic differentiation of human adipose tissue-derived stromal cells. Exp Mol Med. 2015;47:e172. doi: 10.1038/emm.2015.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun H, Hu S, Zhang Z, Lun J, Liao W, Zhang Z. Expression of exosomal microRNAs during chondrogenic differentiation of human bone mesenchymal stem cells. J Cell Biochem. 2019;120:171–181. doi: 10.1002/jcb.27289. [DOI] [PubMed] [Google Scholar]
- 49.Mizuno Y, Tokuzawa Y, Ninomiya Y, Yagi K, Yatsuka-Kanesaki Y, Suda T, Fukuda T, Katagiri T, Kondoh Y, Amemiya T, et al. miR-210 promotes osteoblastic differentiation through inhibition of AcvR1b. FEBS Lett. 2009;583:2263–2268. doi: 10.1016/j.febslet.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 50.Schmid A, Sailland J, Novak L, Baumlin N, Fregien N, Salathe M. Modulation of Wnt signaling is essential for the differentiation of ciliated epithelial cells in human airways. FEBS Lett. 2017;591:3493–3506. doi: 10.1002/1873-3468.12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang CY, Chen R, Wang F, Ren C, Zhang P, Li Q, Li HH, Guo KT, Geng DQ, Liu CF. EGb-761 attenuates the anti-proliferative activity of fluoride via DDK1 in PC-12 cells. Neurochem Res. 2017;42:606–614. doi: 10.1007/s11064-016-2115-6. [DOI] [PubMed] [Google Scholar]
- 52.Westendorf JJ, Kahler RA, Schroeder TM. Wnt signaling in osteoblasts and bone diseases. Gene. 2004;341:19–39. doi: 10.1016/j.gene.2004.06.044. [DOI] [PubMed] [Google Scholar]
- 53.Li J, Sarosi I, Cattley RC, Pretorius J, Asuncion F, Grisanti M, Morony S, Adamu S, Geng Z, Qiu W, et al. Dkk1-mediated inhibition of Wnt signaling in bone results in osteopenia. Bone. 2006;39:754–766. doi: 10.1016/j.bone.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 54.Xia ZL, Wang Y, Sun QD, Du XF. MiR-203 is involved in osteoporosis by regulating DKK1 and inhibiting osteogenic differentiation of MSCs. Eur Rev Med Pharmacol Sci. 2018;22:5098–5105. doi: 10.26355/eurrev_201808_15703. [DOI] [PubMed] [Google Scholar]
- 55.Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing MiRNA-LncRNA interactions. Methods Mol Biol. 2016;1402:271–286. doi: 10.1007/978-1-4939-3378-5_21. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.