Abstract
Multiple myelomas (MM) are the second most common haematological malignancy, for which no curative treatments have been reported to date. MicroRNAs (miRNAs or miRs) have recently been shown to be involved in the proliferation of MM cells. However, the molecular mechanisms through which miRNAs regulate migration and angiogenesis in MM are poorly understood. Accordingly, the present study evaluated the role of miR-144-3p in MM. miR-144-3p exhibited a lower expression in patients with MM and in MM cell lines compared with normal cells (mononuclear cells derived from bone marrow). The transfection of miR-144-3p into MM cells inhibited proliferation, migration and angiogenesis, and induced cell cycle arrest and apoptosis compared to the control cells. Furthermore, miR-144-3p suppressed the transcription and translation of the myocyte enhancer factor 2A (MEF2A) gene and disrupted the expression of vascular endothelial growth factor. The knockdown of MEF2A significantly inhibited the proliferation, migration and angiogenesis of MM cells. However, the overexpression of MEF2A reversed these effects. On the whole, the findings of the present study demonstrate that miR-144-3p exerts antitumour effects by downregulating MEF2A to inhibit the proliferation, migration and angiogenesis of MM cells. This suggests that the miR-144-3p/MEF2A interaction may prove to be a potential therapeutic target for MM.
Keywords: miR-144-3p, myocyte enhancer factor 2A, migration, angiogenesis, multiple myeloma
Introduction
Multiple myelomas (MM) is a type of plasma cell tumour and the second most common haematological malignancy. To date, no curative treatments for MM have been reported (1). MM is characterised by the uncontrolled growth of monoclonal plasma cells in the bone marrow, leading to anaemia, bone lesions, hypercalcaemia, renal failure, and other complications (2). MM cells are located at multiple sites in the bone, and the mechanisms through which MM cells migrate from the primary lesion to distant sites have been studied in detail (3). Angiogenesis is involved in MM progression and is promoted via the production of angiogenic cytokines by plasma cells within the bone marrow microenvironment. Several anti-angiogenic therapeutic strategies, including thalidomide treatment, have been evaluated in patients with myeloma (4). However, the mechanisms of angiogenesis in the pathogenesis of myeloma remain unclear, and effective biomarkers for anti-angiogenic therapy have not been identified.
MicroRNAs (miRNAs or miRs) are a large group of non-coding RNAs. A single miRNA can control multiple genes and molecular pathways. Recently, miRNAs have emerged as instrumental regulators of cellular processes that enable the development and dissemination of MM (5-7). The functions of miRNAs in MM vary. Several miRNAs, including miR-15a and miR-16, are markedly downregulated in MM, suggesting tumour-suppressive roles. By contrast, miR-21 and miR-221 are highly expressed and function as oncogenes (oncomiRs) in MM. In addition, several miRNAs, such as those belonging to the miR-34 family, are transcriptional targets of p53 and mediate its tumour-suppressive functions. miR-34a/b/c, miR-124-1, miR-194-2, miR-192, miR-203, miR-152 and miR-10b-5p are frequently methylated in MM (8).
Recently, the discovery of exosome-mediated transfer of tumour-suppressive miRNAs has demonstrated the need for the deeper understanding of the mechanisms through which tumour cells and the microenvironment communicate. Exosome-associated miR-9 is involved in nasopharyngeal carcinoma tumorigenesis, suggesting potential roles for exosome-based therapies in cancer treatment (9). However, the mechanisms through which miRNAs regulate MM, particularly migration and angiogenesis, remain unclear.
Accordingly, in the present study, the expression of miRNAs was evaluated in patients with MM and the effects of miR-144-3p on the proliferation, migration and angiogenesis of MM cells were examined.
Materials and methods
Patient sample collection
Bone marrow samples were collected from 15 patients with MM and 10 patients with non-haematological diseases at Shengjing Hospital of China Medical University from February, 2015 to November, 2017. The basis clinical information of the study subjects is presented in Table SI. Samples were extracted using CD138 magnetic beads (Miltenyi Biotec GmbH). The purity of the CD138+ plasma cells was at least 90% (data not shown). Mononuclear cells were extracted from bone marrow using Ficoll-Hypaque (lymphocyte separation fluid; Beijing Solarbio Science & Technology Co., Ltd.) density gradient centrifugation. The present study was approved by the Research Ethics Committee of 0Shengjing Hospital of China Medical University (approval no. 2019PS270K) and all patients provided informed consent.
Cell lines and cell culture
The human MM cell lines U266 and RPMI-8226 were purchased from the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences. The cells were stored in RPMI-1640 medium (Cellgro, Mediatech; Corning, Inc.) containing 10% foetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) and cultured at 37°C in a conventional cell culture incubator with a 5% CO2 atmosphere.
Cell transfection
A miR-144-3p mimic, miR negative control (miR-NC), siRNA against myocyte enhancer factor 2A (si-MEF2A) and siRNA negative control (si-NC) were purchased from Hanbio Technology, Ltd. The miR-144-3p mimic and its control sequence were as follows: miR-144-3p mimic sense, 5′-UAC AGU AUA GAU GAU GUA CU-3′ and antisense, 5′-AGU ACA UCA UCU AUA CUG UA-3′; and negative control sense, 5′-UCA CAA CCU CCU AGA AAG AGU AGA -3′ and antisense, 5′-UCU ACU CUU UCU AGG AGG UUG UGA -3′. The siRNA sequences were as follows: si-MEF2A sense, 5′-CCA GAC CCU GAU ACU UCA UdT dT-3′ and antisense, 5′-AUG AAG UAU CAT GGG GCU -3′; and si-NC sense, 5′-UUC UCC GAA CGU GUC ACG UdT d-3′ and antisense, 5′-ACG UGA CAC GUU CGG AGA AdT d-3′. The MEF2A coding sequence was inserted into the pcDNA3.1 vector (Kingsray Biotechnology Co., Ltd.), and an MEF2A overexpression plasmid (pcDNA-MEF2A) was constructed in the cells. Briefly, the transfection mass and concentration of plasmid (Genescript Biotech Corporation) and small molecule RNA (Hanbio Technology, Ltd.) in the 24-well plate/6-well plate were 0.5 µg/5 µg and 100 nM, respectively and were transfected into the cells using Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol, with a final transfection concentration of 100 nM. The transfection efficiency was monitored by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) at 48 h following transfection.
Total RNA extraction RT-qPCR
Total RNA was extracted from each sample using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. The quantity and quality of the extracted RNA were measured with a NanoDrop spectrophotometer (NanoVne, GE Healthcare). The reverse transcription of miRNA was performed using a tailing reverse kit (Sangon Biotech), and mRNA was reversed transcribed into first-strand cDNA using a PrimeScriptTM RT kit (Takara Bio, Inc.). The conditions of reverse transcription were as follows: 42°C for 2 min, 37°C for 15 min and 85°C for 5 sec. The expression of MEF2A and miR-144-3p was detected with SYBR Premix Ex Taq™ (Takara Bio, Inc.) using the Bio-Rad CFX96 Real-Time PCR System (Bio-Rad Laboratories, Inc.). The denaturing, annealing and extension conditions of each PCR cycle were 40 cycles of 95°C for 5 sec and 60°C for 34 sec, respectively. All primers were designed and synthesised by Sangon Biotech. The primer sequences were as follows: miR-144-3p forward, 5′-GCG CGC GTA CAG TAT AGA TGA-3′ and reverse, 5′-AGT GCA GGG TCC GAG GTA TT-3′; U6 forward, 5′-GCT TCG GCA GCA CAT ATA CTA AAA T-3′ and reverse, 5′-CGC TTC ACG AAT TTG CGT GTC AT-3′; MEF2A forward, 5′-ACG TCC AGT GTG GCA TGG AG-3′ and reverse, 5′-AGG CTG GTT TCC ACC CAG AG-3′; and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) forward, 5′-CCA CCC ATG GCA AAT TCC ATG GCA -3′ and reverse, 5′-TCT AGA CGG CAG GTC AGG TCC ACC -3′. The expression of the target genes was calculated using the 2−ΔΔcq method (10).
Cell proliferation assay
A Cell Titer-Glo chemiluminescence cell viability assay (Promega Corp.) was used to detect cell proliferation (11). The transfected MM cells were seeded into 96-well plates (10,000 cells/well). Following culture at 37°C in a 5% CO2 incubator for various periods of time (12, 24, 36, 48 and 60 h), luminescence was added to each well and the substrate was detected at 25 µl and incubated at 37°C for 1 h. The absorbance (OD) value was measured with a Multifunctional enzyme labelling instrument (Synergy2, BioTek Instruments, Inc.) at a wavelength of 560 nm.
Apoptosis and cell cycle analyses
At 48 h following transfection, at least 105 cells were collected from each group and washed twice with PBS. According to the instructions provided with the apoptotic kit (BD Biosciences), dyes were added to the cells and protected from light for 10 min, after which flow cytometry (ACEA Biosciences) was performed to analyse the cell cycle distribution with nova express software (ACEA Biosciences). Cells from the different groups were collected, with at least 105 cells for each group. The cells were then washed twice with PBS and fixed with 75% ethanol overnight, according to the instructions provided with the cell cycle kit (KeyGEN Biotech). Dyes were added sequentially, the samples were protected from light for 30 min, and flow cytometric analysis was then performed.
Transwell assay
Cell suspensions were prepared using serum-free medium and diluted to 105 cells/ml. Subsequently, 200 µl of the cell suspension was added to the supporting chambers of 24-well plates (Corning, Inc.). The medium in the lower well was Dulbecco's modified Eagle's medium/F12 (HyClone; GE Healthcare Life Sciences) containing 10% foetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.). Following incubation for 24 h, the cells were fixed with paraformaldehyde (Bioshap), subjected to crystal violet staining (Beyotime Institute of Biotechnology) at room temperature for 30 min and imaged using an inverted microscope (Eclipse Ci; Nikon Corp.). ImageJ software (version 1.6; National Institutes of Health) was used to analyse the number of cells in each image and perform statistical analysis.
Dual-luciferase reporter assay
miR-144-3p target gene prediction was performed using the online bioinformatics databases, databases, StarBase (http://starbase.sysu.edu.cn/), miRDB (http://mirdb.org/miRDB/), TargetScan (http://www.targetscan.org/), PicTar (https://pictar.mdc-berlin.de/) and miRanda (http://www.microrna.org/microrna/home.do) (12-14). The results of the above 5 databases were integrated, the potential mRNA targets of differentially expressed miR-144-3p were identified, the possible binding sites of MEF2A and miR-144-3p were predicted, and the double luciferase reporter gene was analysed. The 3′ untranslated region (UTR) of MEF2A containing the miR-144-3p target sequence was inserted into the pmirGLO vector (Promega Corp.) to obtain the luciferase reporter plasmid wild-type MEF2A (MEF2A-WT) and antisense mutation of the predicted binding site to obtain the mutant MEF2A-MUT vector. 293T cells (FH0244; Shanghai Fuheng Biotechnology Co., Ltd.) were co-transfected with wild-type or mutant 3′UTR containing and miR-144-3p mimics or NC control. At 36 h following transfection, the luciferase activity was detected, and the results were analysed according to the manufacturer's protocol using the Dual-Luciferase ®Reporter Assay system (E1901; Promega Corp.) experimental procedures.
Proliferation of and tube formation in human umbilical vein endothelial cells (HUVECs)
At 48 h following transfection, the cell supernatants were collected and centrifuged at 500 × g at room temperature for 10 min. The pellets were removed, and supernatants were obtained. HUVECs (obtained from the Cell Resource Center, China; 3142C0001000000138; 2×105 cells/ml) were added to 96-well plates containing mixed medium [tumour cell supernatant (45 µl) + fresh ECM medium (45 µl)]. The cells were cultured in a 5% CO2 incubator at 37°C for 24 h. A Cell Counting kit-8 (CCK-8; Beyotime Institute of Biotechnology) was used to detect HUVEC proliferation. To determine tube formation rates, 96-well plates were precooled at-20°C for 20 min, and the wells were inoculated with 50-70 µl Matrigel [BD Matrigel (Growth factor reduced, cat. no. 356231); BD Biosciences]. The plates were then incubated in a cell incubator for 30 min. HUVECs (2×105 cells/ml; 10 µl/well) were seeded into the wells of the Matrigel-coated plate with 90 µl mixed medium [tumour cell culture supernatant (45 µl) + fresh ECM medium (45 µl); ScienCell]. Tube formation was observed at 1, 4 and 7 h using an inverted microscope (IX71, Nikon Corp.), and the number of tubes was counted in each group.
Western blot analysis
Total protein was extracted from the MM cells using radio-immunoprecipitation assay lysis buffer (Beyotime Institute of Biotechnology) with phenylmethyl-sulphonyl fluoride (Beyotime Institute of Biotechnology). The proteins were isolated by centrifugation at 12,000 × g for 15 min at 4°C. Protein concentrations were quantified using a bicinchoninic acid assay (Beyotime Institute of Biotechnology). All protein samples were boiled with Loading Buffer (Beyotime Institute of Biotechnology) at 100°C for 10 min. Equal amounts of protein (30 µg) in each sample were separated by 10% sodium dodecyl sulphate polyacrylamide gel electrophoresis for 2 h at a constant voltage (110 V) and then transferred onto polyvinylidene fluoride membranes (EMD Millipore). The membranes were blocked for 1 h at room temperature in Tris-buffered saline containing 10% non-fat dried milk and incubated overnight at 4°C with the following primary antibodies: Rabbit polyclonal anti-MEF2A (1:500; cat. no. 9736; Cell Signaling Technology, Inc.), rabbit polyclonal anti-vascular endothelial growth factor (VEGF; 1:500; cat. no. 2463; Cell Signaling Technology, Inc.), and rabbit monoclonal antibody anti-GAPDH (1:1,000; cat. no. ab181602; Abcam). The membranes were washed and then incubated at room temperature for 1 h with horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (IgG, 1:2,000; cat. no. ab205718; Abcam). Chemiluminescent detection was performed using an ECL kit (32132×3; Thermo Fisher Scientific, Waltham, Inc.). Bands were analysed using ImageJ software (version 1.6) to verify the relative expression levels of the target proteins.
Statistical analysis
Data analysis was performed using GraphPad Prism 6.0 software (GraphPad, Inc.). Data are presented as the means ± standard deviation (SD). A Student's t-test was used for comparisons between 2 groups, and a one-way analysis of variance was used for comparisons between multiple groups with Tukey's post hoc test. All experiments were repeated independently at least 3 times. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-144-3p expression is lower in MM than in normal cells
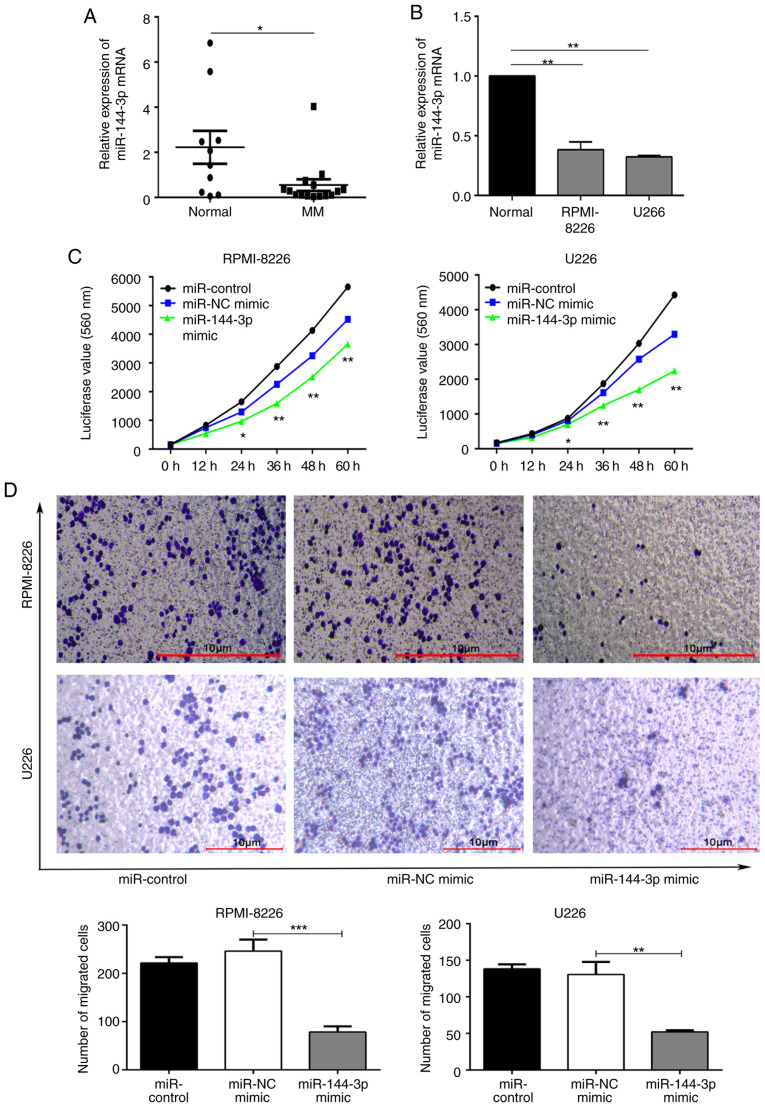
To explore the mechanisms through which miRNAs regulate MM, small RNA expression profiling was first performed in exosomes from bone marrow supernatants from patients with MM and healthy donors. miR-144-3p expression varied between groups, suggesting that miR-144-3p was involved in the pathogenesis of MM (Fig. S1; GEP data were deposited in NCBI SRA; accession ID no. SRP239379). The expression of miR-144-3p in 15 patients with primary MM and 10 normal donors was detected by RT-qPCR. The results revealed that miR-144-3p expression in patients with primary MM was significantly lower than that in normal bone marrow mono-nuclear cells (P<0.05; Fig. 1A). A similar pattern was observed (P<0.01; Fig. 1B) in the MM cell lines, RPMI8226 and U266. These data suggest that miR-144-3p expression is decreased in MM. This finding was consistent with the results of the small RNA sequencing of bone marrow exosomes from patients with MM.
Figure 1.
Expression and biological functions of miR-144-3p in MM. (A) miR-144-3p expression in CD138+ cells isolated and purified from the bone marrow of patients with MM and normal donors, as detected by RT-qPCR. *P<0.05 vs. normal donor. (B) miR-144-3p expression in MM cell lines and normal donor samples, as analysed by RT-qPCR. **P<0.01 vs. normal cells. (C) Proliferation and migration of RPMI-8226 and U266 cells detected by (C) Cell Titer-Glo chemiluminescence assays (CTG) and (D) Transwell assays, respectively, following the transfection of RPMI-8226 and U266 cells with miR-144-3p mimics. *P<0.05, **P<0.01, ***P<0.001 vs. negative control or as indicated. Scale bar, 10 µm. MM, multiple myeloma.
Recovery of miR-144-3p inhibits the proliferation and migration, and induces cell cycle arrest and apoptosis of MM cells
Subsequently, the role of miR-144-3p in MM was examined. miR-144-3p expression in RPMI8226 and U266 cells was induced by transfection with miR-144-3p mimic or miR-NC as a negative control. RT-qPCR was used to detect the expression of miR-144-3p (Fig. S2). Compared with the negative control, the proliferation rate was significantly inhibited by the expression of miR-144-3p in the RPMI-8226 and U266 cells (P<0.01; Fig. 1C). The proportion of cells in the G0/G1 phase was increased, whereas that in the S phase was decreased in the miR-144-3p mimic group. These data demonstrate that this miRNA plays a role in the cell cycle arrest of MM cells (P<0.05; Fig. S3A). Moreover, the results revealed that the recovery of miR-144-3p expression induced the apoptosis (P<0.05; Fig. S3B) and inhibited the migration of MM cells (P<0.001; Fig. 1D). Overall, these data indicate that miR-144-3p affects MM cell progression.
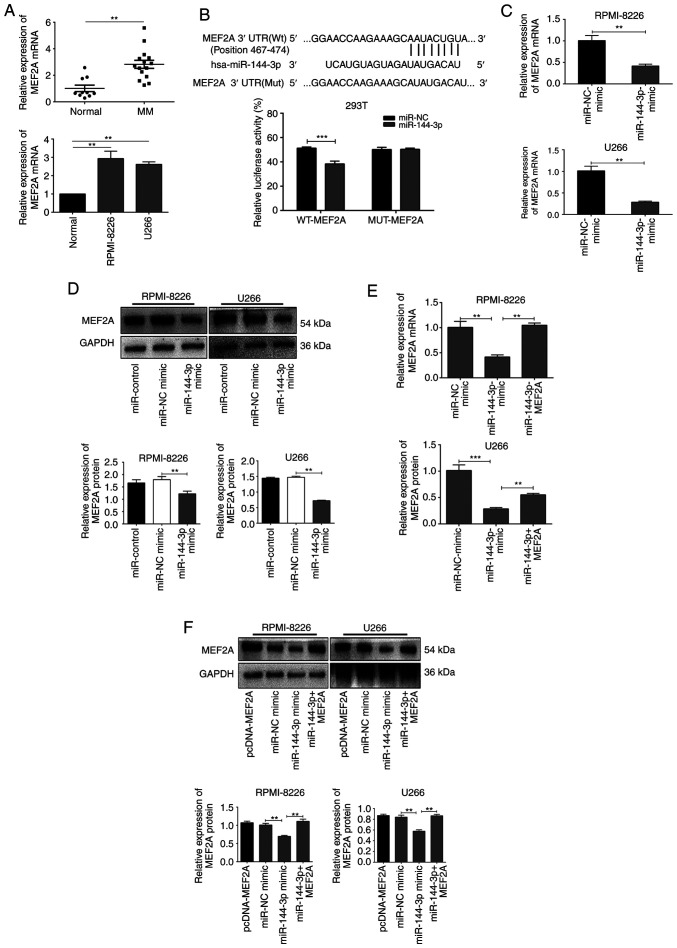
MEF2A is a target ofmiR-144-3p in MM cells
miRNAs specifically bind to the 3′UTRs of mRNAs to promote their degradation or inhibit translation. Bioinformatics databases (StarBase, miRDB, TargetScan, PicTar and miRanda) were used to identify the potential binding sites of miR-144-3p, and MEF2A was found to be a possible target (Fig. 2B). To confirm these findings, the expression of MEF2A was evaluated in patients with primary MM, in MM cell lines, and in healthy donor cells. The results revealed that MEF2A expression was significantly upregulated in MM cells compared to normal cells (P<0.05; Fig. 2A). Moreover, luciferase reporter assays revealed that the luciferase activity of wild-type MEF2A-3′UTR in 293T cells transfected with miR-144-3p was significantly lower than that in cells transfected with miR-NC (P<0.001); importantly, miR-144-3p expression did not affect mutant MEF2A (P>0.05; Fig. 2B). It was also found that the transfection of cells with miR-144-3p mimic decreased the mRNA (P<0.01; Fig. 2C) and protein expression of MEF2A in the RPMI-8226 and U266 cells (P<0.01; Fig. 2D). Finally, MEF2A rescue experiments revealed that the inhibition of MEF2A expression by the miR-144-3p mimic was reversed by transfection with a MEF2A overexpression plasmid (P<0.01; Fig. 2E and F). Collectively, these results indicated that miR-144-3p directly targeted the 3′UTR of MEF2A, resulting in MEF2A inhibition in MM cells.
Figure 2.
MEF2A is a target gene regulated by miR-144-3p. (A) mRNA expression of MEF2A in MM primary cells and cell lines was evaluated by RT-qPCR. **P<0.01 vs. normal donor. (B) Luciferase reporter vectors encoding wild-type (WT) or mutant (MUT) MEF2A-3′UTR were co-transfected into 293T cells with miR-144-3p mimics or NC control to determine the relative luciferase activity. ***P<0.001 vs. negative control. (C and D) RT-qPCR and western blot analysis were used to detect MEF2A mRNA and protein expression in RPMI-8226 and U266 cells transfected with miR-144-3p mimics. **P<0.01 vs. negative control (E and F) pcDNA-MEF2A vector and miR-144-3p mimic were used to co-transfect RPMI-8226 and U266 cells. RT-qPCR and western blot analysis were used to detect the mRNA and protein recovery of MEF2A. **P<0.01, ***P<0.001 vs. miR-144-3p mimic. MM, multiple myeloma.
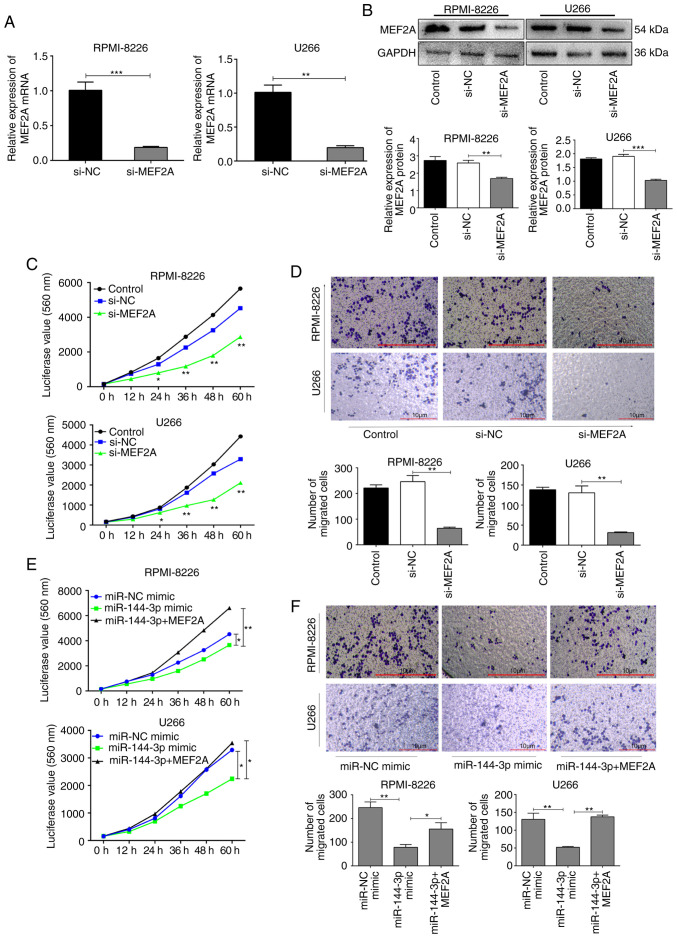
MEF2A regulates proliferation and migration in MM cells
To further explore the biological function of MEF2A, MEF2A was knocked down using short hairpin RNA in RPMI-8226 and U266 cells (Fig. S4). The knockdown efficiency was verified by RT-qPCR (P<0.01; Fig. 3A) and western blot analysis (P<0.001; Fig. 3B). The proliferation and migration of the cells in which MEF2A was knocked down were inhibited compared to the control cells (P<0.01; Fig. 3C and D). By contrast, the overexpression of MEF2A partly reversed the inhibitory effects of miR-144-3p on the proliferation (P<0.01; Fig. 3E) and migration (P<0.05; Fig. 3F) of RPMI-8226 and U266 cells. Moreover, the inhibition of MEF2A also partly induced the cell cycle arrest and apoptosis of MM cells (Fig. S5), whereas the overexpression of MEF2A reversed these effects (Fig. S6).
Figure 3.
Knockdown of MEF2A inhibits the proliferation and migration of MM cells. (A) mRNA and (B) protein expression levels of MEF2A were determined following transfection of RPMI-8226 and U266 cells with siRNA1, which exhibited optimal knockdown effects. **P<0.01, ***P<0.001 vs. negative control. (C) Proliferation of RPMI8226 and U266 cells transfected with si-MEF2A was detected using CTG assays. *P<0.05, **P<0.01 vs. negative control. (D) Transwell assays were used to analyse the migration capacities of RPMI-8226 and U266 cells transfected with different vectors and siRNAs. **P<0.01 vs. negative control. (E and F) RPMI-8226 and U266 cells were co-transfected with miR-144-3p mimic and pcDNA-MEF2A, and cell proliferation and migration were evaluated. *P<0.05, **P<0.01 vs. miR-144-3p mimic. Scale bar, 10 µm. MM, multiple myeloma.
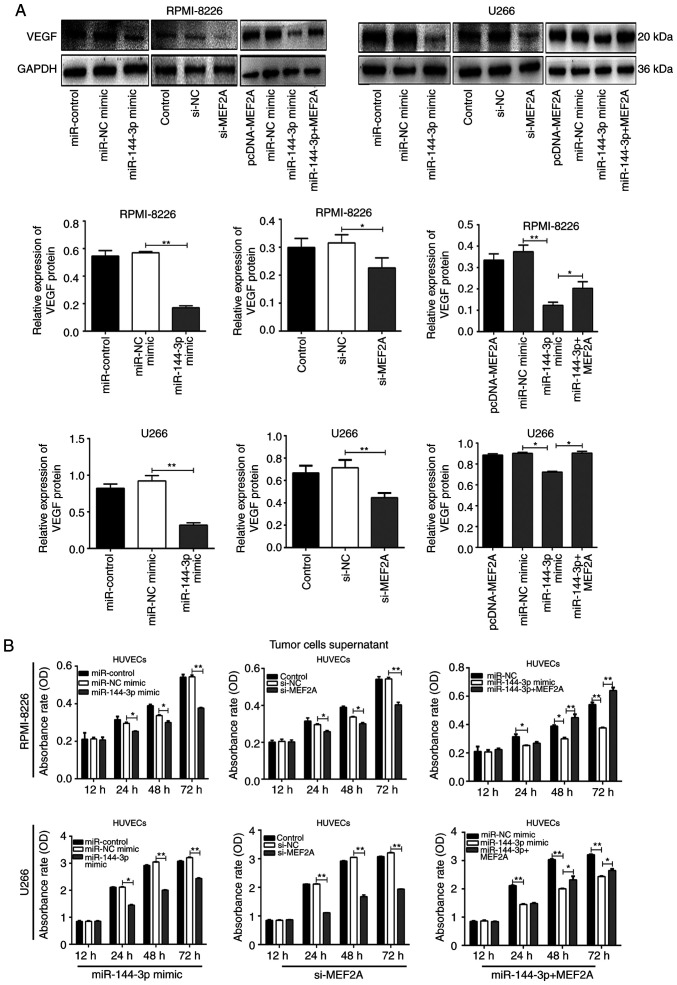
miR-144-3p regulates VEGF expression by targeting MEF2A in MM cells
MEF2A is a DNA-binding protein that regulates transcription. In endothelial cells, MEF2A is involved in sprouting angiogenesis (15). However, the association between MEF2A and VEGF, which regulates angiogenesis, remains unclear. In the present study, it was found that both miR-144-3p overexpression and MEF2A knockdown reduced VEGF expression in MM cells (P<0.01; Fig. 4A). Moreover, co-transfection with pcDNA-MEF2A and miR-144-3p partially restored the expression of VEGF. These data suggest that the reduced expression of miR-144-3p in MM increases VEGF expression by targeting MEF2A.
Figure 4.
miR-144-3p regulates proliferation and angiogenesis by targeting MEF2A in MM. (A) Western blot analysis of VEGF protein expression in RPMI-8226 and U266 cells co-transfected with miR-144-3p mimic or si-MEF2A and pcDNA-MEF2A. *P<0.05, **P<0.01. (B) Proliferation of HUVECs in supernatants from transfected MM cell lines was analysed by CCK-8 assays (CCK-8). *P<0.05, **P<0.01. MM, multiple myeloma.
miR-144-3p inhibits angiogenesis by targeting MEF2A in MM
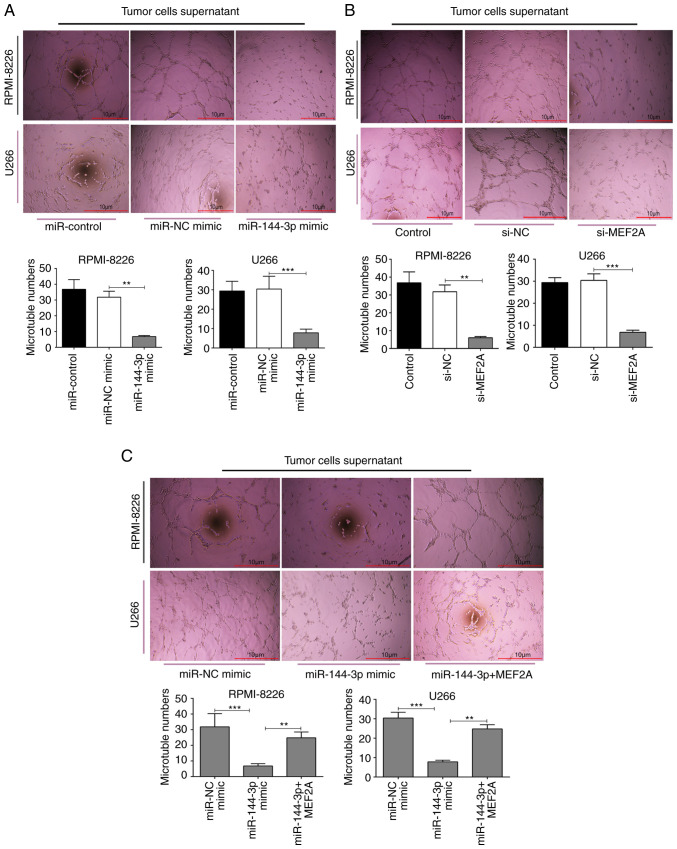
Angiogenesis is a feature of MM and is induced by plasma cells via angiogenic factors released by cells within the tumour microenvironment (4). In the present study, to explore the function of miR-144-3p in angiogenesis, MM cell culture supernatants were prepared and HUVECs were cultured with the different supernatants. The results revealed that the overexpression of miR-144-3p and the knockdown of MEF2A inhibited the proliferation (P<0.01; Fig. 4B) and disrupted the tubular structure of HUVECs (P<0.01; Fig. 5A and B). pcDNA-MEF2A partly restored the inhibitory effects of miR-144-3p on tube formation in HUVECs (P<0.01; Fig. 5C).
Figure 5.
Tube formation in HUVECs. (A) Evaluation of HUVEC tube formation in supernatants from MM cells transfected with miR-144-3p mimics and (B) with si-MEF2A. **P<0.01, ***P<0.001. (C) Tube formation in HUVECs following treatment with preconditioned medium from miR-144-3p-overex-pressing RPMI-8226 and U266 cells compared with NC. Transfection of MEF2A rescued the angiogenic capabilities of miR-144-3p-overexpressing cells. Representative images of the tubular structure are shown, and quantitative data are expressed as average values of 5 independent measurements from three independent experiments. **P<0.01, ***P<0.001. Scale bar, 10 µm. MM, multiple myeloma.
Discussion
MM is a biologically heterogeneous disease of plasma cells. In recent years, researchers have focused on the roles of non-coding RNAs, such as miRNAs, long non-coding RNAs, siRNAs and piwi-interacting RNAs, in the pathology of MM. Al Masri et al demonstrated that miR-125b, miR-133a, miR-1, miR-124a, miR-15 and miR-16 were downregulated in MM cell lines and samples from patients with MM compared to their normal counterparts (16). In the present study, a lower expression of miR-144-3p was observed in MM compared with normal cells, and the recovery of miR-144-3p expression inhibited the proliferation, and induced the cell cycle arrest and apoptosis of MM cells. Further analysis revealed that miR-144-3p exerted these effects by downregulating MEF2A, suggesting that this miR-144-3p/MEF2A interaction is involved in the mechanism of MM cell proliferation. Zhao et al demonstrated similar results and that miR-144-3p inhibits cell proliferation and induces apoptosis in MM by targeting c-Met (17). Tianhua et al found that the long non-coding RNA Sox2 overlapping transcript (SOX2OT) promoted MM progression via the microRNA-144-3p/c-MET axis (18). However, whether MEF2A interacts with the c-MET pathway to regulate MM proliferation and migration warrants further investigation.
However, the regulation of migration and angiogenesis by miR-144-3p in MM has not yet been investigated to date, at least to the best of our knowledge. Raimondi et al found that miR-199a-5p expression increased the adhesion of MM cells to bone marrow stromal cells under hypoxic conditions (19). These results indicate that miRNA affects the migration of MM. In the present study, it was found that miR-144-3p may directly target the 3′UTR of MEF2A in MM cells to inhibit the metastasis of MM cells. The MEF2 family of transcription factors includes 4 members, i.e., MEF2A, MEF2B, MEF2C and MEF2D, which play key roles in regulating differentiation responses. The roles of MEF2A in promoting muscle differentiation (20) and heart development (21) have been well-established. However, the involvement of MEF2A in cancer has not yet been investigated in detail. In the present study, MEF2A was identified as a target gene regulated by miR-144-3p and it was found that MEF2A expression may promote HUVEC proliferationand induce angiogenesis in MM.
Angiogenesis depends on the balance of positive and negative angiogenic modulators within the vascular microenvironment. MM angiogenesis mainly depends on the release of growth factors, such as VEGF, by neoplastic cells. VEGF is specific for endothelial cells and stimulates the growth of blood vessels (22). Additionally, Roccaro et al (23) demonstrated that miR-15a/-16, which was downregulated in MM plasma cells, exerted anti-angiogenic effects by reducing VEGF secretion from MM cells, thereby suppressing the pro-angiogenic effects of MM plasma cells on endothelial cells by targeting the stromal cell-derived factor 1-α/C-X-C chemokine motif receptor 4 pathway (24). The present study demonstrated a novel possible mechanism through which miR-144-3p targets MEF2A to decrease VEGF expression, thereby blocking angiogenesis in MM.
The BM microenvironment exerts marked effects on MM progression. Gupta et al found that the targeting of stromal versican, which plays a key role in matrix remodelling, malignant transformation and tumour progression, by miR-144/199 may inhibited MM via the downregulation of the FAK/STAT3 signalling pathway (25). This indicates that miR-144 can be used to regulate the BM microenvironment. In the present study, it was found that miR-144-3p expression was lower in exosomes from bone marrow supernatants in patients with MM than in healthy donors. The effects of miR-144-3p on MM cells were then evaluated. However, as miR-144-3p can be transferred through exosomes from MM cells to other cells in the micro-environment, the regulatory effects of miR-144-3p on the BM microenvironment remain unclear. Thus, additional studies and in vivo experiments are warranted to evaluate this mechanism.
In conclusion, the findings of the present study demonstrated that miR-144-3p inhibited the proliferation, migration and angiogenesis of MM cells, and induced cell cycle arrest and apoptosis by targeting MEF2A in MM. Considering the molecular and biological complexity of angiogenesis, these results provide useful insight into the development of novel and effective anti-MM drugs.
Acknowledgments
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (grant no. 81272629), the Natural Science Foundation of Liaoning Province (2019-ZD-0792), the Special Fund for Clinical Medical Research of the Chinese Medical Doctor Association (grant no. 20111210) and the Natural Science Foundation of Liaoning Province (grant no. 20180551257).
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
AL, FT, HW, HM and YZ conceived and designed the experiments. FT performed the experiments and wrote the manuscript. FT and AL analysed the data and critically revised the manuscript. FT, HM and YZ completed the post-production image processing. AL and HW supervised all research and revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Research Ethics Committee of Shengjing Hospital of China Medical University (approval no. 2019PS270K) and all patients provided informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Liang B, Yin JJ, Zhan XR. MiR-301a promotes cell proliferation by directly targeting TIMP2 in multiple myeloma. Int J Clin Exp Pathol. 2015;8:9168–9174. [PMC free article] [PubMed] [Google Scholar]
- 2.Brigle K, Rogers B. Pathobiology and diagnosis of multiple myeloma. Semin Oncol Nurs. 2017;33:225–236. doi: 10.1016/j.soncn.2017.05.012. [DOI] [PubMed] [Google Scholar]
- 3.Zhao Q, Luo F, Ma J, Yu X. Bone metastasis-related Micrornas: New targets for treatment? Curr Cancer Drug Targets. 2015;15:716–725. doi: 10.2174/1568009615666150629102408. [DOI] [PubMed] [Google Scholar]
- 4.Ribatti D, Vacca A. New insights in anti-angiogenesis in multiple myeloma. Int J Mol Sci. 2018;19:2031. doi: 10.3390/ijms19072031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D'Ario M, Griffiths-Jones S, Kim M. Small RNAs: Big impact on plant development. Trends Plant Sci. 2017;22:1056–1068. doi: 10.1016/j.tplants.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 6.Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biswas S. MicroRNAs as therapeutic agents: The future of the battle against cancer. Curr Top Med Chem. 2018;18:2544–2554. doi: 10.2174/1568026619666181120121830. [DOI] [PubMed] [Google Scholar]
- 8.Misiewicz-Krzeminska I, Krzeminski P, Corchete LA, Quwaider D, Rojas EA, Herrero AB, Gutiérrez NC. Factors regulating microRNA expression and function in multiple myeloma. Noncoding RNA. 2019;5:9. doi: 10.3390/ncrna5010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu J, Liu QH, Wang F, Tan JJ, Deng YQ, Peng XH, Liu X, Zhang B, Xu X, Li XP. Exosomal miR-9 inhibits angiogenesis by targeting MDK and regulating PDK/AKT pathway in nasopharyngeal carcinoma. J Exp Clin Cancer Res. 2018;37:147. doi: 10.1186/s13046-018-0814-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 11.He Y, Li J, Ding N, Wang X, Deng L, Xie Y, Ying Z, Liu W, Ping L, Zhang C, et al. Combination of enzastaurin and ibrutinib synergistically induces anti-tumor effects in diffuse large B cell lymphoma. J Exp Clin Cancer Res. 2019;38:86. doi: 10.1186/s13046-019-1076-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, Piedade ID, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 13.Ritchie W. MicroRNA target prediction. Methods Mol Biol. 2017;1513:193–200. doi: 10.1007/978-1-4939-6539-7_13. [DOI] [PubMed] [Google Scholar]
- 14.van Iterson M, Bervoets S, de Meijer EJ, Buermans HP, Hoen PA, Menezes RX, Boer JM. Integrated analysis of microRNA and mRNA expression: Adding biological significance to microRNA target predictions. Nucleic Acids Res. 2013;41:e146. doi: 10.1093/nar/gkt525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li M, Linseman DA, Allen MP, Meintzer MK, Wang X, Laessig T, Wierman ME, Heidenreich KA. Myocyte enhancer factor 2A and 2D undergo phosphorylation and caspase-mediated degradation during apoptosis of rat cerebellar granule neurons. J Neurosci. 2001;21:6544–6552. doi: 10.1523/JNEUROSCI.21-17-06544.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Al Masri A, Price-Troska T, Chesi M, Chung TH, Kim S, Carpten J, Bergsagel PL, Fonseca R. MicroRNA expression analysis in multiple myeloma. Blood. 2005;106:1554. doi: 10.1182/blood.V106.11.1554.1554. [DOI] [Google Scholar]
- 17.Zhao Y, Xie Z, Lin J, Liu P. MiR-144-3p inhibits cell proliferation and induces apoptosis in multiple myeloma by targeting c-Met. Am J Transl Res. 2017;9:2437–2446. [PMC free article] [PubMed] [Google Scholar]
- 18.Tianhua Y, Dianqiu L, Xuanhe Z, Zhe Z, Dongmei G. Long non-coding RNA Sox2 overlapping transcript (SOX2OT) promotes multiple myeloma progression via microRNA-143-3p/c-MET axis. J Cell Mol Med. 2020;24:5185–5194. doi: 10.1111/jcmm.15171. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Raimondi L, Amodio N, Di Martino MT, Altomare E, Leotta M, Caracciolo D, Gullà A, Neri A, Taverna S, D'Aquila P, et al. Targeting of multiple myeloma-related angiogenesis by miR-199a-5p mimics: In vitro and in vivo anti-tumor activity. Oncotarget. 2014;5:3039–3054. doi: 10.18632/oncotarget.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Estrella NL, Desjardins CA, Nocco SE, Clark AL, Maksimenko Y, Naya FJ. MEF2 transcription factors regulate distinct gene programs in mammalian skeletal muscle differentiation. J Biol Chem. 2015;290:1256–1268. doi: 10.1074/jbc.M114.589838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ewen EP, Snyder CM, Wilson M, Desjardins D, Naya FJ. The Mef2A transcription factor coordinately regulates a costamere gene program in cardiac muscle. J Biol Chem. 2011;286:29644–29653. doi: 10.1074/jbc.M111.268094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ribatti D, Nico B, Crivellato E, Roccaro AM, Vacca A. The history of the angiogenic switch concept. Leukemia. 2007;21:44–52. doi: 10.1038/sj.leu.2404402. [DOI] [PubMed] [Google Scholar]
- 23.Roccaro AM, Sacco A, Thompson B, Leleu X, Azab AK, Azab F, Runnels J, Jia X, Ngo HT, Melhem MR, et al. MicroRNAs 15a and 16 regulate tumor proliferation in multiple myeloma. Blood. 2009;113:6669–6680. doi: 10.1182/blood-2009-01-198408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van de Donk NW, Lokhorst HM, Nijhuis EH, Kamphuis MM, Bloem AC. Geranylgeranylated proteins are involved in the regulation of myeloma cell growth. Clin Cancer Res. 2005;11:429–439. [PubMed] [Google Scholar]
- 25.Gupta N, Kumar R, Seth T, Garg B, Sharma A. Targeting of stromal versican by miR-144/199 inhibits multiple myeloma by downregulating FAK/STAT3 signalling. RNA Biol. 2020;17:98–111. doi: 10.1080/15476286.2019.1669405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.