Abstract
Background
The severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) virus and resulting COVID‐19 pandemic present important diagnostic challenges. Several diagnostic strategies are available to identify current infection, rule out infection, identify people in need of care escalation, or to test for past infection and immune response. Serology tests to detect the presence of antibodies to SARS‐CoV‐2 aim to identify previous SARS‐CoV‐2 infection, and may help to confirm the presence of current infection.
Objectives
To assess the diagnostic accuracy of antibody tests to determine if a person presenting in the community or in primary or secondary care has SARS‐CoV‐2 infection, or has previously had SARS‐CoV‐2 infection, and the accuracy of antibody tests for use in seroprevalence surveys.
Search methods
We undertook electronic searches in the Cochrane COVID‐19 Study Register and the COVID‐19 Living Evidence Database from the University of Bern, which is updated daily with published articles from PubMed and Embase and with preprints from medRxiv and bioRxiv. In addition, we checked repositories of COVID‐19 publications. We did not apply any language restrictions. We conducted searches for this review iteration up to 27 April 2020.
Selection criteria
We included test accuracy studies of any design that evaluated antibody tests (including enzyme‐linked immunosorbent assays, chemiluminescence immunoassays, and lateral flow assays) in people suspected of current or previous SARS‐CoV‐2 infection, or where tests were used to screen for infection. We also included studies of people either known to have, or not to have SARS‐CoV‐2 infection. We included all reference standards to define the presence or absence of SARS‐CoV‐2 (including reverse transcription polymerase chain reaction tests (RT‐PCR) and clinical diagnostic criteria).
Data collection and analysis
We assessed possible bias and applicability of the studies using the QUADAS‐2 tool. We extracted 2x2 contingency table data and present sensitivity and specificity for each antibody (or combination of antibodies) using paired forest plots. We pooled data using random‐effects logistic regression where appropriate, stratifying by time since post‐symptom onset. We tabulated available data by test manufacturer. We have presented uncertainty in estimates of sensitivity and specificity using 95% confidence intervals (CIs).
Main results
We included 57 publications reporting on a total of 54 study cohorts with 15,976 samples, of which 8526 were from cases of SARS‐CoV‐2 infection. Studies were conducted in Asia (n = 38), Europe (n = 15), and the USA and China (n = 1). We identified data from 25 commercial tests and numerous in‐house assays, a small fraction of the 279 antibody assays listed by the Foundation for Innovative Diagnostics. More than half (n = 28) of the studies included were only available as preprints.
We had concerns about risk of bias and applicability. Common issues were use of multi‐group designs (n = 29), inclusion of only COVID‐19 cases (n = 19), lack of blinding of the index test (n = 49) and reference standard (n = 29), differential verification (n = 22), and the lack of clarity about participant numbers, characteristics and study exclusions (n = 47). Most studies (n = 44) only included people hospitalised due to suspected or confirmed COVID‐19 infection. There were no studies exclusively in asymptomatic participants. Two‐thirds of the studies (n = 33) defined COVID‐19 cases based on RT‐PCR results alone, ignoring the potential for false‐negative RT‐PCR results. We observed evidence of selective publication of study findings through omission of the identity of tests (n = 5).
We observed substantial heterogeneity in sensitivities of IgA, IgM and IgG antibodies, or combinations thereof, for results aggregated across different time periods post‐symptom onset (range 0% to 100% for all target antibodies). We thus based the main results of the review on the 38 studies that stratified results by time since symptom onset. The numbers of individuals contributing data within each study each week are small and are usually not based on tracking the same groups of patients over time.
Pooled results for IgG, IgM, IgA, total antibodies and IgG/IgM all showed low sensitivity during the first week since onset of symptoms (all less than 30.1%), rising in the second week and reaching their highest values in the third week. The combination of IgG/IgM had a sensitivity of 30.1% (95% CI 21.4 to 40.7) for 1 to 7 days, 72.2% (95% CI 63.5 to 79.5) for 8 to 14 days, 91.4% (95% CI 87.0 to 94.4) for 15 to 21 days. Estimates of accuracy beyond three weeks are based on smaller sample sizes and fewer studies. For 21 to 35 days, pooled sensitivities for IgG/IgM were 96.0% (95% CI 90.6 to 98.3). There are insufficient studies to estimate sensitivity of tests beyond 35 days post‐symptom onset. Summary specificities (provided in 35 studies) exceeded 98% for all target antibodies with confidence intervals no more than 2 percentage points wide. False‐positive results were more common where COVID‐19 had been suspected and ruled out, but numbers were small and the difference was within the range expected by chance.
Assuming a prevalence of 50%, a value considered possible in healthcare workers who have suffered respiratory symptoms, we would anticipate that 43 (28 to 65) would be missed and 7 (3 to 14) would be falsely positive in 1000 people undergoing IgG/IgM testing at days 15 to 21 post‐symptom onset. At a prevalence of 20%, a likely value in surveys in high‐risk settings, 17 (11 to 26) would be missed per 1000 people tested and 10 (5 to 22) would be falsely positive. At a lower prevalence of 5%, a likely value in national surveys, 4 (3 to 7) would be missed per 1000 tested, and 12 (6 to 27) would be falsely positive.
Analyses showed small differences in sensitivity between assay type, but methodological concerns and sparse data prevent comparisons between test brands.
Authors' conclusions
The sensitivity of antibody tests is too low in the first week since symptom onset to have a primary role for the diagnosis of COVID‐19, but they may still have a role complementing other testing in individuals presenting later, when RT‐PCR tests are negative, or are not done. Antibody tests are likely to have a useful role for detecting previous SARS‐CoV‐2 infection if used 15 or more days after the onset of symptoms. However, the duration of antibody rises is currently unknown, and we found very little data beyond 35 days post‐symptom onset. We are therefore uncertain about the utility of these tests for seroprevalence surveys for public health management purposes. Concerns about high risk of bias and applicability make it likely that the accuracy of tests when used in clinical care will be lower than reported in the included studies. Sensitivity has mainly been evaluated in hospitalised patients, so it is unclear whether the tests are able to detect lower antibody levels likely seen with milder and asymptomatic COVID‐19 disease.
The design, execution and reporting of studies of the accuracy of COVID‐19 tests requires considerable improvement. Studies must report data on sensitivity disaggregated by time since onset of symptoms. COVID‐19‐positive cases who are RT‐PCR‐negative should be included as well as those confirmed RT‐PCR, in accordance with the World Health Organization (WHO) and China National Health Commission of the People's Republic of China (CDC) case definitions. We were only able to obtain data from a small proportion of available tests, and action is needed to ensure that all results of test evaluations are available in the public domain to prevent selective reporting. This is a fast‐moving field and we plan ongoing updates of this living systematic review.
Plain language summary
What is the diagnostic accuracy of antibody tests for the detection of infection with the COVID‐19 virus?
Background
COVID‐19 is an infectious disease caused by the SARS‐CoV‐2 virus that spreads easily between people in a similar way to the common cold or ‘flu. Most people with COVID‐19 have a mild to moderate respiratory illness, and some may have no symptoms (asymptomatic infection). Others experience severe symptoms and need specialist treatment and intensive care.
The immune system of people who have COVID‐19 responds to infection by developing proteins that can attack the virus (antibodies) in their blood. Tests to detect antibodies in peoples' blood might show whether they currently have COVID‐19 or have had it previously.
Why are accurate tests important?
Accurate testing allows identification of people who might need treatment, or who need to isolate themselves to prevent the spread of infection. Failure to detect people with COVID‐19 when it is present (a false negative result) may delay treatment and risk further spread of infection to others. Incorrect identification of COVID‐19 when it is not present (a false positive result) may lead to unnecessary further testing, treatment, and isolation of the person and close contacts. Correct identification of people who have previously had COVID‐19 is important in measuring disease spread, assessing the success of public health interventions (like isolation), and potentially in identifying individuals with immunity (should antibodies in the future be shown to indicate immunity).
To identify false negative and false positive results, antibody test results are compared in people known to have COVID‐19 and known not to have COVID‐19. Study participants are classified as to whether they are known or not known to have COVID‐19 based on criteria known as the ‘reference standard’. Many studies use samples taken from the nose and throat to identify people with COVID‐19. The samples undergo a test called reverse transcriptase polymerase chain reaction (RT‐PCR). This testing process can sometimes miss infection (false negative result), but additional tests can identify COVID‐19 infection in people with a negative RT‐PCR result. These include measuring clinical symptoms, like coughing or high temperature, or ‘imaging’ tests like chest X‐rays. People known not to have COVID‐19 are sometimes identified from stored blood samples taken before COVID‐19 existed, or from patients with respiratory symptoms found to be caused by other diseases.
What did the review study?
The studies looked at three types of antibody, IgA, IgG and IgM. Most tests measure both IgG and IgM, but some measure a single antibody or combinations of the three antibodies.
Levels of antibodies rise and fall at different times after infection. IgG is the last to rise but lasts longest. Levels of antibodies are usually highest a few weeks after infection.
Some antibody tests need specialist laboratory equipment. Others use disposable devices, similar to pregnancy tests. These tests can be used in laboratories or wherever the patient is (point‐of‐care), in hospital or at home.
We wanted to find out whether antibody tests:
‐ are accurate enough to diagnose infection in people with or without symptoms of COVID‐19, and
‐ can be used to find out if someone has already had COVID‐19.
What did we do?
We looked for studies that measured the accuracy of antibody tests compared with reference standard criteria to detect current or past COVID‐19 infection. Studies could assess any antibody test compared with any reference standard. People could be tested in hospital or the community. Studies could test people known to have – or not to have – or be suspected of having COVID‐19.
Study characteristics
We found 54 relevant studies. Studies took place in Asia (38), Europe (15), and in both USA and China (1).
Forty‐six studies included people who were in hospital with suspected or confirmed COVID‐19 infection only. Twenty‐nine studies compared test results in people with COVID‐19 with test results in healthy people or people with other diseases.
Not all studies provided details about participants’ age and gender. Often, we could not tell whether studies were evaluating current or past infection, as few reported whether participants were recovering. We did not find any studies that tested only asymptomatic people.
Main results
Our findings come mainly from 38 studies that provided results based on the time since people first noticed symptoms.
Antibody tests one week after first symptoms only detected 30% of people who had COVID‐19. Accuracy increased in week 2 with 70% detected, and was highest in week 3 (more than 90% detected). Little evidence was available after week 3. Tests gave false positive results in 2% of those without COVID‐19.
Results from IgG/IgM tests three weeks after symptoms started suggested that if 1000 people had antibody tests, and 50 (5%) of them really had COVID‐19 (as we might expect in a national screening survey):
‐ 58 people would test positive for COVID‐19. Of these, 12 people (21%) would not have COVID‐19 (false positive result).
‐ 942 people would test negative for COVID‐19. Of these, 4 people (0.4%) would actually have COVID‐19 (false negative result).
If we tested 1000 healthcare workers (in a high‐risk setting) who had had symptoms, and 500 (50%) of them really had COVID‐19:
‐ 464 people would test positive for COVID‐19. Of these, 7 people (2%) would not have COVID‐19 (false positive result).
‐ 537 people would test negative for COVID‐19. Of these, 43 (8%) would actually have COVID‐19 (false negative result).
We did not find convincing differences in accuracy for different types of antibody test.
How reliable were the results of the studies of this review?
Our confidence in the evidence is limited for several reasons. In general, studies were small, did not use the most reliable methods and did not report their results fully. Often, they did not include patients with COVID‐19 who may have had a false negative result on PCR, and took their data for people without COVID‐19 from records of tests done before COVID‐19 arose. This may have affected test accuracy, but it is impossible to identify by how much.
Who do the results of this review apply to?
Most participants were in hospital with COVID‐19, so were likely to have more severe disease than people with mild symptoms who were not hospitalised. This means that we don't know how accurate antibody tests are for people with milder disease or no symptoms.
More than half of the studies assessed tests they had developed themselves, most of which are not available to buy. Many studies were published quickly online as ‘preprints’. Preprints do not undergo the normal rigorous checks of published studies, so we are not certain how reliable they are.
As most studies took place in Asia, we don't know whether test results would be similar elsewhere in the world.
What are the implications of this review?
The review shows that antibody tests could have a useful role in detecting if someone has had COVID‐19, but the timing of when the tests are used is important. Antibody tests may help to confirm COVID‐19 infection in people who have had symptoms for more than two weeks and do not have a RT‐PCR test, or have negative RT‐PCR test results. The tests are better at detecting COVID‐19 in people two or more weeks after their symptoms started, but we do not know how well they work more than five weeks after symptoms started. We do not know how well the tests work for people who have milder disease or no symptoms, because the studies in the review were mainly done in people who were in hospital. In time, we will learn whether having previously had COVID‐19 provides individuals with immunity to future infection.
Further research is needed into the use of antibody tests in people recovering from COVID‐19 infection, and in people who have experienced mild symptoms or who never experienced symptoms.
How up‐to‐date is this review?
This review includes evidence published up to 27 April 2020. Because a lot of new research is being published in this field, we will update this review frequently.
Summary of findings
Summary of findings 1. What is the diagnostic accuracy of antibody tests, for the diagnosis of current or prior SARS‐CoV‐2 infection?
| Question | What is the diagnostic accuracy of antibody tests, for the diagnosis of current or prior SARS‐CoV‐2 infection? | |||
| Population | Adults or children suspected of
or populations undergoing screening for SARS‐CoV‐2 infection, including
|
|||
| Index test | Any test for detecting antibodies to SARS‐CoV‐2, including:
|
|||
| Target condition | Detection of
|
|||
| Reference standard | RT‐PCR alone, clinical diagnosis of COVID‐19 based on established guidelines or combinations of clinical features and for non‐COVID‐19 cases, the use of pre‐pandemic sources of samples for testing | |||
| Action | The current evidence‐base for antibody tests is inadequate to be clear about their utility (mainly because of small numbers of small studies for each test, few data available outside of acute hospital settings, and many issues in likely bias and applicability of the studies). The sensitivity of antibody tests is too low early in disease for use as a primary test of diagnosis, but they may have value for late diagnosis, for identifying previous infection, and for sero‐prevalence studies. | |||
| Limitations in the evidence | ||||
| Risk of bias |
Participant selection: high risk of bias in 48 studies (89%) Application of index tests: high risk of bias in 14 studies (26%) Reference standard: high risk of bias in 17 studies (31%) Flow and timing: high risk of bias in 29 studies (54%) |
|||
| Concerns about applicability of the evidence |
Participants: high concerns in 44 studies (81%) Index test: high concerns in 17 studies (31%) Reference standard: high concerns in 33 studies (61%) |
|||
| Findings | ||||
| ||||
| Quantity of evidence | Number of studies | Total participants or samples | Total cases | |
| 54 | 15,976 | 8526 | ||
|
Sensitivity (95% CI) Studies (TP/COVID cases) |
Specificity (95%CI) Studies (FP/non‐COVID cases) |
|||
| Days 8‐14 | Days 15‐21 | Days 22‐35 | All time points | |
| IgG | 66.5% (57.9 to 74.2) | 88.2% (83.5 to 91.8) | 80.3% (72.4 to 86.4) | 99.1% (98.3% to 99.6%) |
| 22 (766/1200) | 22 (974/1110) | 12 (417/502) | 44 (159/6136) | |
| IgM | 58.4% (45.5 to 70.3) | 75.4% (64.3 to 83.8) | 68.1% (55.0 to 78.9) | 98.7% (97.4% to 99.3%) |
| 21 (724/1171) | 21 (800/1074) | 11 (378/507) | 41 (183/6103) | |
| IgG/IgM* | 72.2% (63.5 to 79.5) | 91.4% (87.0 to 94.4) | 96.0% (90.6 to 98.3) | 98.7% (97.2% to 99.4%) |
| 9 (441/608) | 9 (636/692) | 5 (146/152) | 23 (78/5761) | |
| Numbers applied to a hypothetical cohort of 1000 patients, using summary data for IgG/IgM at days 15 to 21 as an exemplar (sensitivity 91.4% (87.0 to 94.4) and specificity 98.7% (97.2 to 99.4)) | ||||
| Prevalence of COVID‐19 | TP (95% CI) | FP (95% CI) | FN (95% CI) | TN (95% CI) |
| 2% | 18 (17 to 20) | 13 (6 to 27) | 2 (1 to 3) | 967 (953 to 974) |
| 5% | 46 (44 to 47) | 12 (6 to 27) | 4 (3 to 7) | 938 (923 to 944) |
| 10% | 91 (87 to 94) | 12 (5 to 25) | 9 (6 to 13) | 888 (875 to 895) |
| 20% | 183 (174 to 189) | 10 (5 to 22) | 17 (11 to 26) | 790 (778 to 795) |
| 50% | 457 (435 to 472) | 7 (3 to 14) | 43 (28 to 65) | 494 (486 to 497) |
| CGIA: colloidal gold immunoassays; CI: confidence interval; CLIA: chemiluminescence immunoassays; ELISA: enzyme‐linked immunosorbent assays; FIA: fluorescence‐labelled immunochromatographic assays; FN: false negative; FP: false positive; RT‐PCR: reverse transcription polymerase chain reaction; TN: true negative; TP: true positive; * Positive if either IgG or IgM positive. | ||||
Background
The severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) virus and resulting COVID‐19 pandemic present important diagnostic evaluation challenges. These range from understanding the value of signs and symptoms in predicting possible infection, assessing whether existing biochemical and imaging tests can identify infection and people needing critical care, and evaluating whether new diagnostic tests can allow accurate rapid and point‐of‐care testing, either to identify current infection, rule out infection, identify people in need of care escalation, or to test for past infection and immunity.
We are creating and maintaining a suite of living systematic reviews to cover the roles of tests and characteristics in the diagnosis of COVID‐19. This review summarises evidence of the accuracy of COVID‐19 antibody tests; both laboratory‐based tests and point‐of‐care tests.
Target condition being diagnosed
COVID‐19 is the disease caused by infection with the SARS‐CoV‐2 virus. The key target conditions for this suite of reviews are current SARS‐CoV‐2 infection, current COVID‐19 disease, and past SARS‐CoV‐2 infection.
Antibody tests are being considered and evaluated for both:
identification of past SARS‐CoV‐2 infection, and
current infection.
For current infection the severity of the disease is of importance. SARS‐CoV‐2 infection can be asymptomatic (no symptoms); mild or moderate (symptoms such as fever, cough, aches, lethargy but without difficulty breathing at rest); severe (symptoms with breathlessness and increased respiratory rate indicative of pneumonia); or critical (requiring respiratory support due to severe acute respiratory syndrome (SARS) or acute respiratory distress syndrome (ARDS). People with COVID‐19 pneumonia (severe or critical disease) require different patient management, and it is important to be able to identify them. There is no consideration that antibody tests are able to distinguish severity of disease, thus, in this review, we consider their role for detecting SARS‐CoV‐2 infection of any severity (asymptomatic or symptomatic).
Index test(s)
Antibody tests
This review evaluates serology tests to measure antibodies to the SARS‐CoV‐2 virus. Antibodies are formed by the body's immune system in response to infections, and can be detected in whole blood, plasma or serum. Antibodies are specific to the virus, and therefore can be used to differentiate between different infections. There are three types of antibody created in response to infection: IgA, IgG and IgM; these rise and fall at different times after the onset of infection. IgG is used in most antibody tests as it persists for the longest time and may reflect longer‐term immunity, although it is the last to rise after infection. Many tests assess both IgG and IgM. IgM typically rises quickly with infection and declines soon after an infection is cleared. Alternatively tests may combine IgA with IgG, or measure all antibodies (IgA, IgG and IgM).
Antibody tests are available for laboratory use including enzyme‐linked immunosorbent assay (ELISA) methods, or more advanced chemiluminescence immunoassays (CLIA). There are also laboratory‐independent, point‐of‐care lateral flow assays, which use disposable devices, akin to a pregnancy test, that use a minimal amount of blood on a testing strip. Antibody detection is indicated by visible lines appearing on the test strip, or through fluorescence, which can be detected using a reader device. Many of these tests are known as colloidal gold‐based immunoassays, as they use COVID‐19 antigen conjugated to gold nanoparticles.
Following the emergence of COVID‐19 there has been prolific industry activity to develop accurate antibody tests. The Foundation for Innovative Diagnostics (FIND) and Johns Hopkins Centre for Health Security have maintained online lists of these and other molecular‐based tests for COVID‐19. At the time of writing (21 May 2020), FIND listed 279 antibody tests, 196 of which are produced by commercial companies and are commercially available. Reguatory approval in the European Union (EU; CE‐IVD) had been awarded to 185 on the list, whereas in China only seven had been approved, and eight by the FDA (US Food and Drug Administration). For a period of time the FDA allowed commercialisation of antibody tests in the USA without FDA approval, resulting in around 100 tests being placed on the market. Both the content of the list, and these figures will increase over time.
Clinical pathway
Broadly speaking, there are four considered uses of antibody tests.
In diagnosis of acute suspected COVID‐19 in patients who presented with symptoms, particularly where molecular testing had failed to detect the virus.
In assessment of immune response in patients with severe disease.
For individuals to assess whether they have had a SARS‐CoV‐2 infection and have an immune response.
In seroprevalence surveys for public health management purposes.
For 1, the standard approach to diagnosis of COVID‐19 is through a reverse transcription polymerase chain reaction (RT‐PCR) test, which detects the presence of virus in swab samples taken from nose, throat or fluid from the lungs. However, the test is known to give false negative results, and can only detect COVID‐19 in the acute phase of the illness. Both the World Health Organization (WHO) and the China CDC (National Health Commission of the People's Republic of China), have produced case definitions for COVID‐19 that include RT‐PCR‐negative cases that display other convincing clinical evidence (Appendix 1). The most recent case definition from the China CDC includes positive serology tests. Confirming an acute clinical diagnosis using a serology test requires detectable virus‐specific IgM and IgG in serum, or detectable virus‐specific IgG, or a 4‐fold or greater increase in titration to be observed during convalescence compared with the acute phase.
For 2, this is largely a question of monitoring patients, and we will not cover this in this review. Assessment of the accuracy of a test used for assessment of immune response would involve comparison with a reference standard test of antibody response, rather than evidence of infection.
Use 3 involves testing individuals during periods of convalescence (after symptoms have resolved) whereas 4 will involve testing people at a mixture of time points, including long follow‐up. A key difference between 3 and 4 is the likelihood of disease, which is expected to be much higher for 3 than 4.
An extended version of use case scenarios is available in Appendix 2.
Prior test(s)
Prior testing depends on the purpose of the test. For 1 we would anticipate that patients were symptomatic and had most likely undergone RT‐PCR testing and possible computed tomography (CT) imaging. Uses 3 and 4 will most likely include people who have not been tested, and may include people who are asymptomatic as well as symptomatic.
Alternative test(s)
This review is one of six planned reviews that cover the range of tests and characteristics being considered in the management of COVID‐19 (Deeks 2020; McInnes 2020). Full details of the alternative tests and evidence of their accuracy will be summarised in these reviews.
Laboratory‐based molecular tests
Testing for presence of the SARS‐CoV‐2 virus has been undertaken using quantitative RT‐PCR (qRT‐PCR). RT‐PCR tests for SARS‐CoV‐2 identify viral ribonucleic acid (RNA). Reagents for the assay were rapidly produced once the viral RNA sequence was published. Testing is undertaken in central laboratories and can be very labour‐intensive, with several points along the path of performing a single test where errors may occur, although some automation of parts of the process is possible. Although the actual qRT‐PCR test does not take long, the stages of extraction, sample processing and data management mean that test results are typically available in 24 to 48 hours, although faster processes are being implemented. Other nucleic acid amplification methods such as loop‐mediated isothermal amplification (LAMP), or CRISPR‐based nucleic acid detection methods are also being developed, with the potential to reduce the time to produce test results to minutes, but the time for the whole process may still be significant. RT‐PCR tests use upper and lower respiratory samples. Sputum is currently considered better than oropharynx swabs or nasopharynx swabs but is more difficult (and hazardous) to obtain and will only ever be available in a subset of patients.
Laboratory‐independent point‐of‐care and near‐patient molecular and antigen tests
Laboratory‐independent RT‐PCR devices can also be used for identification of infection near patients and even at the bedside. These are small platforms for testing which use matching test cartridges. Several companies have suitable existing technology systems and are producing the required new cartridges for diagnosis of SARS‐CoV‐2 infection. Test results are based on the same samples as those for qRT‐PCR, with results available within minutes or hours. Antigen tests are based on the direct detection of the virus, indicating active infection (i.e. replication of the virus) similar to the detection of RNA. Antigen tests are mainly in the form of lateral flow assays. They will capture the relevant viral antigen using dedicated antibodies, and visualisation is either manual or using a reader device.
Signs and symptoms
Signs and symptoms are used in the initial diagnosis of suspected COVID‐19, and in identifying people with COVID‐19 pneumonia. Key symptoms that have been associated with mild to moderate COVID‐19 include: troublesome dry cough (for example, coughing more than usual over a one‐hour period, or three or more coughing episodes in 24 hours), fever greater than 37.8°C, diarrhoea, headache, breathlessness on light exertion, muscle pain, fatigue, and loss of sense of smell and taste. Red flags indicating possible pneumonia include: breathlessness at rest, increased respiratory rate (above 20 breaths per minute), increased heart rate (above 100 beats per minute), chest tightness, loss of appetite, confusion, pain or pressure in the chest, blue lips or face, and temperature above 38°C. Hypoxia based on measuring pulse oximetry is often used, with various arbitrary thresholds (for example, 93%).
Routinely available biomarkers
Routinely available biomarkers for infection and inflammation may be considered in the investigation of people with possible COVID‐19. For example, many healthcare facilities have access to standard laboratory tests for infection, such as C‐reactive protein (CRP), procalcitonin, measures of anticoagulation, and white blood cell count with different lymphocyte subsets. Evaluation of these commonly available tests, particularly in low‐resource settings, may be helpful for the triage of people with potential COVID‐19.
Imaging tests
Chest X‐ray, ultrasound, and CT are widely used diagnostic imaging tests to identify COVID‐19 pneumonia. Availability and usage varies between settings.
Rationale
It is essential to understand the clinical accuracy of tests and diagnostic features to identify the best way they can be used in different settings to develop effective diagnostic and management pathways. The suite of Cochrane 'living systematic reviews' summarises evidence on the clinical accuracy of different tests and diagnostic features, grouped according to the research questions and settings that we are aware of. Estimates of accuracy from these reviews will help inform diagnosis, screening, isolation, and patient management decisions.
Particularly for antibody tests, new tests are being developed and evidence is emerging at an unprecedented rate during the COVID‐19 pandemic. Tests are being purchased in bulk for seroprevalence studies, and made available for personal purchase online. This review will be updated as often as is feasible to ensure that it provides current evidence about the accuracy of antibody tests.
Objectives
To assess the diagnostic accuracy of antibody tests to determine if a person presenting in the community or in primary or secondary care has SARS‐CoV‐2 infection, or has previously had SARS‐CoV‐2 infection, and the accuracy of antibody tests for use in seroprevalence surveys.
Secondary objectives
Where data are available, we will investigate the accuracy (either by stratified analysis or meta‐regression) according to:
current infection or past infection;
test method and brand;
days since onset of symptoms;
reference standard;
study design;
setting.
Methods
Criteria for considering studies for this review
Types of studies
We applied broad eligibility criteria in order to include all patient groups and all variations of a test (that is, if patient population was unclear, we included the study).
We included studies of all designs that produce estimates of test accuracy or provide data from which estimates can be computed, including the following.
Studies restricted to participants confirmed to have (or to have had) the target condition (to estimate sensitivity) or confirmed not to have (or have had) the target condition (to estimate specificity). These types of studies may be excluded in later review updates.
Single‐group studies, which recruit participants before disease status has been ascertained
Multi‐group studies, where people with and without the target condition are recruited separately (often referred to as two‐gate or diagnostic case‐control studies)
Studies based on either patients or samples
We excluded studies from which we could not extract data to compute either sensitivity or specificity.
We carefully considered the limitations of different study designs in the quality assessment and analyses.
We included studies reported in published articles and as preprints.
Participants
We included studies recruiting people presenting with suspicion of current or prior SARS‐CoV‐2 infection or those recruiting populations where tests were used to screen for disease (for example, contact tracing or community screening).
We also included studies that recruited people either known to have SARS‐CoV‐2 infection or known not to have SARS‐CoV‐2 infection (multi‐group studies).
We excluded small studies with fewer than 10 samples or participants. Although the size threshold of 10 is arbitrary, such small studies are likely to give unreliable estimates of sensitivity or specificity and may be biased.
Index tests
We included studies evaluating any test for detecting antibodies to SARS‐CoV‐2, including laboratory‐based methods and tests designed to be used at point‐of‐care. Test methods include the following.
Laboratory‐based:
enzyme‐linked immunosorbent assays (ELISA)
chemiluminescence immunoassays (CLIA)
other laboratory‐based methods (e.g. indirect immunofluorescence tests (IIFT), luciferase immunoprecipitation system (LIPS)
Rapid diagnostic tests:
lateral flow assays, including both colloidal gold or fluorescence‐labelled immunochromatographic assays (CGIA or FIA).
In this first version of the review we have included both commercially available tests, which have regulatory approval, with in‐house assays and assays in development. Future versions of the review are likely to be restricted to only commercially available assays.
We identified the regulatory status of index tests using two main resources:
WHO: COVID‐19 listing in International Medical Device Regulators Forum (IMDRF) jurisdictions (www.who.int/diagnostics_laboratory/EUL/en/), which includes listings of FDA, Health Canada, Japan, Australia (Therapeutic Goods Administration), Singapore (Health Sciences Authority), Brazil (Agência Nacional de Vigilância Sanitária), South Korea (Ministry of Food and Drug Safety), China (National Medical Products Administration), and Russia (Roszdravnadzor);
FIND: SARS‐COV‐2 Diagnostic Pipeline (www.finddx.org/covid-19/pipeline/), which overlaps with the WHO list, but in addition includes CE‐IVD and IVD India.
In addition, we checked key national websites, including US FDA (www.fda.gov/medical-devices/emergency-situations-medical-devices/emergency-use-authorizations#coronavirus2019) and China FDA (subsites.chinadaily.com.cn/nmpa/2020 03/27/c_465663.htm?bsh_bid=5496527208).
Target conditions
The target conditions were the identification of:
current SARS‐CoV‐2 infection (in symptomatic cases);
past SARS‐CoV‐2 infection (in convalescent (post‐symptomatic) or asymptomatic cases).
Reference standards
We anticipated that studies would use a range of reference standards to define both the presence and absence of SARS‐CoV‐2 infection but were unclear at the start of the review exactly what methods would be encountered. For the QUADAS‐2 (Quality Assessment tool for Diagnostic Accuracy Studies; Whiting 2011), assessment we categorised each method of defining COVID‐19 cases according to the risk of bias (the chances that it would misclassify COVID‐19 participants as non‐COVID‐19) and whether it defined COVID‐19 in an appropriate way that reflected cases encountered in practice. Likewise, we considered the risk of bias in definitions of non‐COVID‐19, and whether the definition reflected those who, in practice, would be tested.
Search methods for identification of studies
Electronic searches
We conducted a single literature search to cover our suite of Cochrane COVID‐19 diagnostic test accuracy (DTA) reviews (Deeks 2020; McInnes 2020).
We conducted electronic searches using two primary sources. Both of these searches aimed to identify all published articles and preprints related to COVID‐19, and were not restricted to those evaluating biomarkers or tests. Thus, there are no test terms, diagnosis terms, or methodological terms in the searches. Searches were limited to 2019 and 2020, and for this version of the review have been conducted to 27 April 2020.
Cochrane COVID‐19 Study Register searches
We used the Cochrane COVID‐19 Study Register (covid-19.cochrane.org/), for searches conducted to 28 March 2020. At that time, the register was populated by searches of PubMed, as well as trials registers at ClinicalTrials.gov and the WHO International Clinical Trials Registry Platform (ICTRP).
Search strategies were designed for maximum sensitivity, to retrieve all human studies on COVID‐19 and with no language limits. See Appendix 3.
COVID‐19 Living Evidence Database from the University of Bern
From 28 March 2020, we used the COVID‐19 Living Evidence database from the Institute of Social and Preventive Medicine (ISPM) at the University of Bern (www.ispm.unibe.ch), as the primary source of records for the Cochrane COVID‐19 DTA reviews. This search includes PubMed, Embase, and preprints indexed in bioRxiv and medRxiv databases. The strategies as described on the ISPM website are described here (ispmbern.github.io/covid-19/). See Appendix 4.
The decision to focus primarily on the 'Bern' feed was due to the exceptionally large numbers of COVID‐19 studies available only as preprints. The Cochrane COVID‐19 Study Register has undergone a number of iterations since the end of March and we anticipate moving back to the Register as the primary source of records for subsequent review updates.
Searching other resources
We identified Embase records obtained through Martha Knuth for the Centers for Disease Control and Prevention (CDC), Stephen B Thacker CDC Library, COVID‐19 Research Articles Downloadable Database (www.cdc.gov/library/researchguides/2019novelcoronavirus/researcharticles.html), and de‐duplicated them against the Cochrane COVID‐19 Study Register up to 1 April 2020. See Appendix 5.
We also checked our search results against two additional repositories of COVID‐19 publications including:
the Evidence for Policy and Practice Information and Co‐ordinating Centre (EPPI‐Centre) 'COVID‐19: Living map of the evidence' (eppi.ioe.ac.uk/COVID19_MAP/covid_map_v4.html);
the Norwegian Institute of Public Health 'NIPH systematic and living map on COVID‐19 evidence' (www.nornesk.no/forskningskart/NIPH_diagnosisMap.html)
Both of these repositories allow their contents to be filtered according to studies potentially relating to diagnosis, and both have agreed to provide us with updates of new diagnosis studies added. For this iteration of the review, we examined all diagnosis studies from either source up to 16 April 2020.
In addition we have used the list of potentially eligible index tests (documented in Criteria for considering studies for this review), to search company and product websites for studies about test accuracy and to contact companies to request further information or studies using their tests. We will include the result of this process in a future iteration of this review.
We have also contacted research groups undertaking test evaluations (for example, UK Public Health England‐funded studies, and FIND studies (www.finddx.org/). We appeal to researchers to supply details of additional published or unpublished studies at the following email address, which we will consider for inclusion in future updates (coviddta@contacts.bham.ac.uk).
We did not apply any language restrictions.
Data collection and analysis
Selection of studies
A team of experienced systematic reviewers from the University of Birmingham screened the titles and abstracts of all records retrieved from the literature searches. Two review authors independently screened studies in Covidence. A third, senior review author resolved any disagreements. We tagged all records selected as potentially eligible according to the Cochrane COVID‐19 DTA review(s) that they might be eligible for and we then exported them to separate Covidence reviews for each review title.
We obtained the full texts for all studies flagged as potentially eligible. Two review authors independently screened the full texts for one of the COVID‐19 molecular or antibody test reviews. We resolved any disagreements on study inclusion through discussion with a third review author.
Data extraction and management
One review author carried out data extraction, which was checked by a second review author. Items that we extracted are listed in Appendix 6. Both review authors independently performed data extraction of 2x2 contingency tables of the number of true positives, false positives, false negatives and true negatives. They resolved disagreements by discussion.
We encourage study authors to contact us regarding missing details on the included studies (coviddta@contacts.bham.ac.uk).
Where possible we extracted 2x2 tables according to time since onset of symptoms. We predefined groups of interest as 1‐7, 8‐14, 15‐21, 22‐35 and over 35 days since onset of symptoms. Where the data presented did not exactly match these categorisations we entered data in the time group that had the greatest overlap with our groupings. Where a study presented data for a group without stating an upper time limit (e.g. more than 21 days) we placed the data in the first category above the stated value (e.g. 22‐35 days).
Where possible, we separately extracted data related to each class of antibody (IgA, IgG and IgM), and combinations of classes (IgA/IgM, IgA/IgG, IgG/IgM, where a positive is defined as either or both classes of antibody being detected). We also extracted data on total antibodies where this was reported.
Assessment of methodological quality
Two review authors independently assessed risk of bias and applicability concerns using the QUADAS‐2 checklist tailored to this review (Appendix 7; Whiting 2011). The two review authors resolved any disagreements by discussion.
Ideally, studies should prospectively recruit a representative sample of participants presenting with signs and symptoms of COVID‐19, either in community or primary care settings or to a hospital setting, and they should clearly record the time of testing after the onset of symptoms. Studies should perform antibody tests in their intended use setting, using appropriate sample types as described in the 'Instructions for use' sheet (e.g. fingerprick blood for tests being evaluated for use as point‐of‐care tests), and tests should be performed by relevant personnel (e.g. healthcare workers), and should be interpreted blinded to the final diagnosis (COVID‐19 or not). Serology samples should be taken at time points that reflect the intended use (either whilst symptomatic for diagnosis of infection, or during a convalescent period (after resolution of symptoms) for diagnosis of previous infection). The reference standard diagnosis should be blinded to the result of the antibody test, and should not incorporate the result of the index test or any other serology test. If the reference standard includes clinical diagnosis of COVID‐19, then established criteria should be used. Studies including samples from participants known not to have COVID‐19 should use pre‐pandemic sources or contemporaneous samples with at least one RT‐PCR‐negative test result. Data should be reported for all study participants, including those where the result of the antibody test was inconclusive, or participants in whom the final diagnosis of COVID‐19 was uncertain. If studies obtained multiple samples for testing over time from the same study participants, then they should disaggregate results by time post‐symptom onset.
Statistical analysis and data synthesis
We grouped data by study and test. Thus studies that evaluated multiple tests in the same participants were included multiple times. We present estimates of sensitivity and specificity for each antibody (or combination of antibodies) using paired forest plots in tables, and also summarise them in tables as appropriate.
For analysis purposes, unlike in most DTA reviews we considered estimates of sensitivity and specificity separately, because many of the included studies presented only estimates of sensitivity. Estimates of specificity were typically exceptionally high, thus the correlation between sensitivity and specificity across studies was unlikely to be high (Macaskill 2010; Takwoingi 2017). We considered the heterogeneity in the study findings through visual inspection of forest plots when deciding to meta‐analyse study estimates, and have not computed summary estimates where they were likely to be regarded as misleading.
Where we pooled results, we fitted random‐effects logistic regression models using the meqrlogit command in Stata v15.1 (Stata). In a small number of instances, the random‐effects logistic regression analyses failed to converge (usually when there were very small numbers of studies), and we have computed estimates and confidence intervals by summing the counts of true positive, false positive, false negative and true negative across 2x2 tables. These analyses are clearly marked in the tables. We present all estimates with 95% confidence intervals.
Investigations of heterogeneity
We investigated sources of heterogeneity in two ways. First, for analysis of sensitivity for time since onset of symptoms, we extracted data by week and extended the random‐effects logistic regression model to include indicator variables for each week. There was a strong relationship between time since onset of symptoms and sensitivity, thus we elected to fit all subsequent models for investigation of heterogeneity in sensitivity stratifying by week. We excluded studies for which stratified data were not available at this stage. For analysis of sensitivity according to the RT‐PCR status of patients (RT‐PCR positive ‘confirmed’ and RT‐PCR negative ‘suspect’), we extracted 2x2 tables stratified by RT‐PCR result (as well as week) and extended the random‐effects logistic regression to include terms for week and RT‐PCR status.
We investigated heterogeneity related to study design, reference standard and test technology by including indicator variables in the random‐effects logistic regression model alongside the variables for week since onset of symptoms. We present estimates from these models by test or reference standard type for the sensitivity of the test in the third week since onset of symptoms (since this is the time point most commonly recommended for post‐infection testing to start to be undertaken).
We did not fit models to compare test brands due to the small number of studies available, but we do report estimates with confidence intervals for each brand.
Sensitivity analyses
We planned to undertake sensitivity analyses by excluding:
unpublished studies;
studies identified only from industry 'Instructions for use' documentation;
studies using sample banks or spiked samples;
studies with inadequate reference standards;
for previous infection, we also planned to assess increasing lengths of time since symptoms cleared.
In this version of the review we did not undertake any of these analyses because the majority of studies were preprints, we did not include any company documents, and no study used spiked samples. We investigated issues with reference standards and time as part of the investigations of heterogeneity.
Assessment of reporting bias
We made no formal assessment of reporting bias. However we were aware of the manner in which results in studies could be suppressed by test developers or manufacturers, and detail where we believe this may have happened.
Summary of findings
We summarised key findings in a 'Summary of findings' table indicating the strength of evidence for each test and findings, and highlighted important gaps in the evidence.
Updating
We are aware that a substantial number of studies have been published since the search date of 27 April 2020 and plan to update this review imminently. We have already completed searches for the update up until 25 May 2020, and report the number of studies that we anticipate will be added to this review in the first update.
Results
Results of the search
We screened 10,965 unique references (published or preprints) for inclusion in the complete suite of reviews to assist in the diagnosis of COVID‐19 (Deeks 2020; McInnes 2020). Of 1430 records selected for further assessment for inclusion in any of the six reviews, we assessed 267 full‐text reports for inclusion in this review. See Figure 1 for the PRISMA flow diagram of search and eligibility results (McInnes 2018; Moher 2009). We included 54 studies from 57 reports in this review, three studies are awaiting assessment including two foreign language papers and one study of neutralising antibodies (Characteristics of studies awaiting classification), 34 are ongoing studies (Characteristics of ongoing studies), and we excluded 172 publications. Exclusions were mainly due to ineligible study designs (n = 84) or index tests (n = 40), or because we could not extract or reconstruct 2x2 data (n = 21). The reasons for exclusion of all 172 publications are provided in Characteristics of excluded studies.
1.

Study flow diagram
The 57 included study reports relate to 54 separate studies, six studies (Gao 2020a; Liu 2020d [A]; Pan 2020a; Okba 2020a; Wang 2020a [A]; Zhao 2020a), having two publications each, and three studies providing data for two separate cohorts of participants (Cassaniti 2020 (A); Cassaniti 2020 (B); Garcia 2020 (A); Garcia 2020 (B); Long 2020 (A); Long 2020 (B)). Of the 57 study reports, 28 studies are available only as preprints and four as preprints with subsequent journal publications. (Please note when naming studies, we use the letters (A), (B), (C) in standard brackets to indicate multiple studies from the same publication, and the letters [A], [B], [C] etc. in square brackets to indicate data on different tests evaluated in the same study).
Description of included studies
The 54 studies include a total of 15,976 samples, with 8526 samples from cases of COVID‐19. Summary study characteristics are presented in Table 2 with further details of study design and index test details in Appendix 8 and Appendix 9. The median sample size across the included studies is 129.5 (interquartile range (IQR) 57 to 347) and median number of COVID‐19 cases included is 62 (IQR 31 to 151). Thirty‐eight studies were conducted in Asia: China (n = 36); Hong Kong (n = 1); or Singapore (n = 1). Fifteen studies were conducted in Europe, and the remaining study included samples from more than one country (Bendavid 2020). Forty‐four studies included only hospital inpatient cases, one included hospital outpatients, two included participants attending emergency departments, two, community screening (including one study of close contacts). Five studies were conducted in mixed or unclear settings.
1. Description of studies.
| Participants |
Studies (percentage) (n=54 studies) |
|
| Sample size | Median (IQR) 129.5 (57 to 347) | Min 10, max 3481 |
| Number of COVID‐19 cases | Median (IQR) 62 (31 to 151) | Min 3, max 555 |
| Setting | Hospital inpatient | 44 (81%) |
| Hospital outpatient | 1 (2%) | |
| Hospital accident and emergency | 2 (4%) | |
| Community | 2 (4%) | |
| Mixed or unclear | 5 (9%) | |
| Patient group | Asymptomatic | 0 (0%) |
| Asymptomatic and acute | 1 (2%) | |
| Acute | 23 (43%) | |
| Acute and convalescent | 22 (41%) | |
| Convalescent | 2 (4%) | |
| Mixed or unclear | 6 (11%) | |
| Study design | ||
| Recruitment structure | Single group, both COVID‐19 and non‐COVID‐19 cases | 6 (11%) |
| Single group, only COVID‐19 cases | 19 (35%) | |
| Two or more groups with COVID‐19 and non‐COVID‐19 cases | 29 (54%) | |
| Reference standard for COVID‐19 cases | All RT‐PCR‐positive | 32 (59%) |
| China CDC criteria including RT‐PCR‐negative patients | 11 (20%) | |
| WHO criteria including RT‐PCR‐negative patients | 1 (2%) | |
| Other criteria including RT‐PCR‐negative patients | 3 (6%) | |
| Other | 2 (4%) | |
| Mixed or unclear | 5 (9%) | |
| Reference standard for non‐COVID‐19 | Pre‐pandemic healthy | 4 (7%) |
| Pre‐pandemic other disease | 3 (6%) | |
| Pre‐pandemic healthy + other disease | 4 (7%) | |
| Current healthy (untested) | 5 (9%) | |
| Current other disease (untested) | 1 (2%) | |
| Current healthy + other disease (untested) | 2 (4%) | |
| Current healthy + other disease (RT‐PCR‐negative) | 2 (4%) | |
| COVID suspects, single RT‐PCR‐negative | 8 (15%) | |
| COVID suspects, two or more RT‐PCR–negative results | 3 (6%) | |
| Mixed/other | 3 (6%) | |
| Tests | ||
| Number of tests per study | 1 | 40 (74%) |
| 2 | 8 (15%) | |
| 3‐5 | 4 (8%) | |
| 6‐10 | 2 (2%) | |
| Test technology (n = 89) | CGIA | 23 (26%) |
| CLIA | 20 (22%) | |
| ELISA | 28 (31%) | |
| FIA | 2 (2%) | |
| IIFT | 1 (1%) | |
| LFA (no details) | 10 (11%) | |
| LIPS | 4 (4%) | |
| S‐flow | 1 (1%) | |
| Test brand (n = 89) | Withheld | 13 (%) |
| Acro Biotech ‐ IgG/IgM | 1 (1%) | |
| Artron Laboratories IgM/IgG | 1 (1%) | |
| Autobio Diagnostics IgM/IgG | 1 (1%) | |
| Beijing Beier Bioengineering CGIA | 1 (1%) | |
| Beijing Beier Bioengineering CLIA | 1 (1%) | |
| Beijing Beier Bioengineering ELISA | 1 (1%) | |
| Beijing Diagreat | 1 (1%) | |
| Beijing Hotgen CGIA | 1 (1%) | |
| Beijing Hotgen ELISA | 2 (3%) | |
| Beijing Wantai CGIA | 1 (1%) | |
| Beijing Wantai ELISA | 3 (3%) | |
| Bioscience Co (Chongqing) | 3 (3%) | |
| CTK Biotech OnSite IgG/IgM | 1 (1%) | |
| Darui Biotech | 1 (1%) | |
| Dynamiker Biotechnology IgG/IgM | 1 (1%) | |
| EUROIMMUN | 3 (3%) | |
| EUROIMMUN Anti‐SARS‐Cov | 1 (1%) | |
| EUROIMMUN Beta | 1 (1%) | |
| Hangzhou Alltest ‐ IgG/IgM | 3 (3%) | |
| Innovita Biological ‐ Ab test (IgM/IgG) | 2 (3%) | |
| Jiangsu Medomics IgG‐IgM | 1 (1%) | |
| Shenzhen YHLO | 7 (8%) | |
| Snibe Diagnostic ‐ MAGLUMI | 2 (3%) | |
| Vivachek ‐ VivaDiag IgM/IgG | 3 (3%) | |
| Xiamen InnodDx Biotech | 1 (1%) | |
| Zhuhai Livzon CGIA | 2 (3%) | |
| Zhuhai Livzon ELISA | 5 (6%) | |
| In‐house, S‐based ELISA | 1 (1%) | |
| In‐house, S‐based LIPS | 1 (1%) | |
| In‐house, rN‐based ELISA | 1 (1%) | |
| In‐house, rS‐based ELISA | 1 (1%) | |
| In‐house CGIA | 2 (2%) | |
| In‐house CLIA | 5 (6%) | |
| In‐house ELISA | 6 (7%) | |
| In‐house FIA | 1 (1%) | |
| In‐house S‐flow | 1 (1%) | |
| In‐house ‐ N‐based ELISA | 1 (1%) | |
| In‐house ‐ N‐based LIPS | 2 (2%) | |
| In‐house ‐ S1‐based LIPS | 1 (1%) | |
| In‐house ‐ tri‐S‐based ELISA | 1 (1%) | |
| In‐house Anti‐SARS‐Cov ELISA | 1 (1%) | |
| Ab: antibody; CDC: Center for Disease Control and Prevention; CGIA: colloidal gold immunoassay; CLIA: chemiluminescence immunoassay; ELISA: enzyme‐linked immunosorbent assay; FIA: fluorescence immunoassay; IQR: interquartile range; IIFT: indirect immunofluorescence assay; LFA: lateral flow assay; LIPS: luciferase immunoprecipitation system; max: maximum; min: minimum; N‐based: nucleocapsid protein; RT‐PCR: reverse transcription polymerase chain reaction; S‐based: spike protein; S‐flow: flow‐cytometry assay; WHO: World Health Organization | ||
Participant characteristics
Twenty‐three studies included cases during the early phase of illness only (< 21 days post‐symptom onset), two only included cases 21 days or more post‐symptom onset, 23 included mixed groups and six did not report days post‐symptom onset. Few studies were clear whether participants were symptomatic or convalescent (i.e. symptoms had resolved) at the time of testing. It is therefore difficult to clearly separate out studies that detected current infection from studies that detected past infection. Thus the two target conditions we defined cannot clearly be distinguished. There were no studies exclusively in asymptomatic participants.
The mean or median age of included COVID‐19 cases ranges from 37 to 76 years (reported in 31 studies), and 26% to 87% of participants were male (reported in 31 studies). Full details are in the Characteristics of included studies table.
Study designs
We identified six studies that recruited suspected COVID‐19 cases before it was ascertained whether the patients did or did not have COVID‐19. These six studies identified people with suspected COVID‐19 based on symptoms or as close contacts of confirmed cases (symptomatic and asymptomatic). Sample sizes of these studies ranged from 50 to 814 with between 3 and 154 COVID‐19 cases. Four of these studies defined the presence or absence of COVID‐19 based on RT‐PCR alone, and two also included clinically confirmed RT‐PCR‐negative cases based on undefined clinical suspicion or CT findings. The absence of SARS‐CoV‐2 infection was confirmed by a single RT‐PCR‐negative result in five of the six and by two or more negative RT‐PCR results in one study.
The other forty‐eight studies retrospectively recruited patients when it was already known whether or not they had COVID‐19.
Twenty‐nine studies used two‐ or multi‐group study designs with separate selection of COVID‐19 cases and healthy participants or non‐COVID‐19 participants with another disease. Sample sizes ranged from 17 to 3481 with between 7 and 276 COVID‐19 cases. Nineteen of these studies defined COVID‐19 cases based on a positive RT‐PCR test, six included clinically defined RT‐PCR‐negative cases in addition to RT‐PCR‐positive cases and the remaining four studies used mixed or unclear criteria to define the presence of COVID‐19. Four of the 29 studies included participants with suspected COVID‐19 but who had subsequently been ruled out on the basis of one (2 studies) or more (2 studies) negative RT‐PCR tests. Ten included contemporaneous non‐COVID‐19 groups, including samples from healthy participants (5 studies), patients with other diseases (one study) or both (4 studies), only two of which used RT‐PCR testing to exclude the presence of SARS‐CoV‐2. Twelve studies included pre‐pandemic non‐COVID 19 groups, using samples from either healthy people (n = 5), participants with other diseases (n = 3), or both (n = 4). The remaining three studies included control samples from mixed sources including pre‐pandemic and contemporaneous samples, with or without RT‐PCR testing.
Nineteen studies included only a single group of only COVID‐19 cases, thus only allowing estimation of sensitivity. They determined COVID‐19 cases based on positive RT‐PCR alone (n = 9), clinically defined criteria including RT‐PCR‐negative cases (n = 8, 7 of which used Chinese government‐issued COVID‐19 guidelines to define cases), one using undefined clinical criteria, and one study that did not report how COVID‐19 cases were defined.
Index tests
Forty‐three studies evaluated only one test, five compared two tests, three compared 3 tests, one 5 tests, one 9 and one 10 tests. In total the 54 studies reported on a total of 89 test evaluations.
There were 52 evaluations of laboratory‐based methods (27 ELISA, 19 CLIA, 6 other methods), including 32 using commercially available laboratory‐based kits produced by 11 different commercial companies (16 ELISAs, 15 CLIAs and 1 IIFT), two where the manufacturer name was withheld, and 20 classified as using in‐house methods (11 ELISA, 4 CLIA and 5 other approaches).
There were 34 evaluations of lateral flow assays, 23 were described as or discovered to be CGIA, two were FIAs and nine were not described. Thirty‐one of the 34 evaluations used commercially available lateral flow assays and three were in‐house (including two CGIA and one FIA). Of the 34 evaluations, only three used whole blood (two using the Vivadiag test), and only two used the assays as point‐of‐care tests rather than in a laboratory setting.
Methodological quality of included studies
We report the overall methodological quality assessed using the QUADAS‐2 tool for all included studies (n = 54) in Figure 2 (Whiting 2011). See Appendix 10 for study‐level ratings by quality.
2.

Risk of bias and applicability concerns graph: review authors' judgements about each domain presented as percentages across included studies
Overall, we judged risk of bias to be high in 48 (89%) studies concerning how participants were selected, 14 (26%) studies related to application of the index test, 17 (31%) through concerns about the reference standard and 29 (54%) for issues related to participant flow and timing. No study had low risk in all domains. We judged that there were high concerns about the applicability of the evidence related to participants in 44 (81%) studies, 17 (31%) related to the index test and 32 (59%) related to the reference standard. Explanations of how we have reached these judgements are given below and in the Characteristics of included studies table.
Participant selection
For participant selection, we judged only one study to be at low risk of bias and five to be of unclear risk. The remaining 48 (89%) we judged to be at high risk of bias (n = 44) either due to the use of a multi‐group design with healthy or other disease controls (n = 26) or recruitment of only COVID‐19 cases (n = 19), inappropriate exclusions (n = 2) or inappropriate inclusions (n = 15). Numbers per group are not mutually exclusive. Eleven studies (20%) reported consecutive or random recruitment of participants.
We had high concerns about the applicability of the selection of participants in 44 studies (81%) meaning that the participants who were recruited were unlikely to be similar to those in whom the test would be used in clinical practice. This was largely because studies only recruited hospitalised, confirmed cases of COVID‐19, often with severe symptoms (18 studies) or recruited healthy or other disease non‐COVID‐19 groups (26 studies). We judged 10 (19%) studies likely to have selected an appropriate patient group, including the six studies that recruited participants suspected of COVID‐19 prior to definitive testing and four multi‐group studies that separately recruited COVID‐19 cases and suspected COVID‐19 control groups.
Index tests
Eight studies explicitly reported that they had undertaken the index test with knowledge of whether individuals did or did not have COVID‐19, and eight studies determined the threshold to define test positivity by analysing the data, rather than it being pre‐determined. In 37 studies, reporting of one or both of these issues was too unclear to be able to rule out the possibility of bias. These issues led to the index test performance in 14 studies being rated as at high risk of bias. We judged only three studies to have implemented the index test in a way that protected against the risk of bias.
In 34 studies (63%) we judged the test to be implemented as it would be in practice. Twenty‐two of these were evaluations of laboratory‐based, commercially available tests, and 12 were evaluations of lateral flow assays associated with commercial test manufacturers, primarily evaluated in an inpatient setting. Two of the 12 evaluated the assays as point‐of‐care tests in an emergency room setting. Sixteen studies raised concerns that the tests could not be purchased (high concerns for applicability). The remaining four studies provided inadequate information to make a judgement due to withholding of the names of the commercial tests (one additional study also withheld the names of the lateral flow assays evaluated but scored high concerns as it also reported results for an in‐house ELISA test).
Reference standards
We judged 13 studies (24%) to have used an appropriate reference standard and implemented it in ways that prevented bias. In six studies there was a risk of misclassification, as they had used a single, negative RT‐PCR result to define the absence of disease in people with suspected COVID‐19; eight studies did not report any RT‐PCR testing to confirm COVID‐19 status for contemporaneous healthy or other disease non‐COVID‐19 groups; and one study used serology results in part to determine the reference standard diagnosis, thus risking incorporation bias. We judged 24 studies as having unclear risk of bias due to lack of information about blinding of the reference standard to the index test (19/24) or unclear descriptions of the reference standards used (6/24).
We judged the reference standard to be equivalent to WHO or China CDC definitions of COVID‐19 in 15 studies (28%). We judged studies that used a definition based only on RT‐PCR‐positive results as high concern (32 (59%) of studies), and seven studies reported inadequate detail to assess the reference standard.
Flow and timing
Twenty‐nine (54%) studies were at high risk of bias due to using different reference standards to verify COVID‐19 and non‐COVID‐19 cases (n = 19), participants being excluded from the analysis (n = 15), or the inclusion of multiple samples per participant (n = 7). In 20 (37%) studies we could not make judgements on one or more of these issues, primarily due to lack of clarity around participant inclusion and exclusion from analyses. Five studies reported adequate detail to rule out these risks of bias. None of the included studies reported a Standards of Reporting Diagnostic Accuracy Studies (STARD)‐style participant flow diagram (Bossuyt 2015), and none mentioned that they aimed to report in line with STARD reporting recommendations for test accuracy studies.
In 39 studies all authors declared no conflicts of interest although four included co‐authors affiliated to test manufacturers. Ten studies did not provide a conflict of interest statement (two of these included co‐authors affiliated to test manufacturers or biotechnology companies); and in the five remaining studies at least one author declared conflicts of interest in relation to test manufacturers (four studies) or vaccine companies (one study).
Nine studies provided no funding statement, six reported no funding sources to declare, and 39 studies reported one or more funding sources. The reported funding sources were primarily public funding sources. Two studies reported receipt of equipment ‘in kind’ from test manufacturers and two studies reported private donors.
Findings
We included 54 different studies, which were reported in 57 publications. Fourteen of the 54 studies evaluated more than one test (Table 2), up to a maximum of 10 tests per study. To incorporate all results from all tests, in these analyses we have treated results from different tests of the same samples within a study as separate data points, such that data are available on 89 test‐study combinations. This leads to individual samples being included in some analyses multiple times where they have been evaluated using different tests. To identify where estimates are based on multiple assessments of the same sample sets, the tables include both the number of test‐study combinations and the number of studies. The numbers of true positives, false positives, COVID‐19 samples and non‐COVID samples are based on test result counts.
Overall analyses
We are unable to distinguish between studies that evaluated the accuracy of antibody tests to identify current infection from past infection. Whilst time since onset of symptoms is strongly related to whether an infection was current or past, few studies reported whether participants' symptoms had resolved (and thus they were in a convalescent state) when serology samples were taken. Whilst 21 days post‐symptom onset is assumed to be a point where COVID‐19 cases are likely to be convalescent, many participants in these studies were hospitalised for prolonged periods and likely to reflect those with more severe and long‐lasting symptoms.
A key aspect of interpreting the sensitivity of the tests is the relationship between accuracy and days since onset of symptoms. Sixteen (30%) studies only presented results aggregated over 0 to more than 35 days since onset, and did not present data (or provide datasets) that disaggregated data by week. The figures in Appendix 11 show forest plots of sensitivity and specificity estimates including these studies for IgG, IgM, and IgG/IgM (either positive), which clearly depict substantial heterogeneity in sensitivity, with estimates ranging from 0% to 100% for all three markers. Forest plots of results for IgA, total antibodies, IgA/IgG, IgA/IgM (Appendix 11), show similar heterogeneity with smaller numbers of studies. Given the heterogeneity and the known strong relationship of sensitivity with time, computation of an average estimate of sensitivity from these studies would be misleading and serves no purpose.
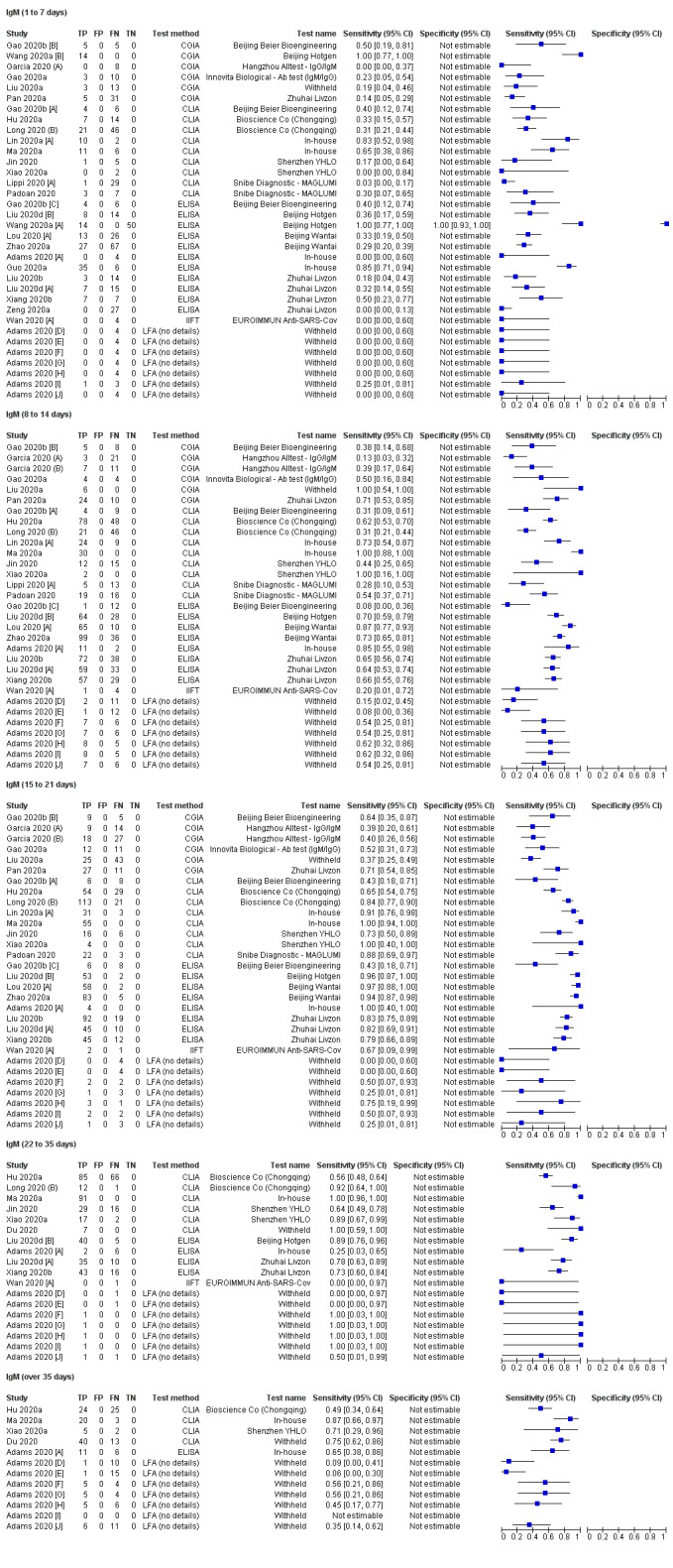
Sensitivity by time since onset of symptoms
Table 3 and Figure 3 present the results disaggregated by week of testing since onset of symptoms for IgG (from 23 studies), IgA (from 4 studies), IgM (from 24 studies), total antibodies (from 5 studies), combination of IgG/IgM (from 21 studies), and IgA/IgG (from 1 study; these results are based on a maximum of 12 participants per time period and we will not comment on them further). We did not find any data disaggregated by week for IgA/IgM. Forest plots of these data are given in Figure 4, Figure 5 and Figure 6. We have undertaken meta‐analyses of data stratified by week as heterogeneity, whilst still present, is substantially less. As indicated in Table 3, the strength of the relationship of time with sensitivity shows exceptionally high levels of statistical significance (P < 0.0005). All further analyses of sensitivity in this report are thus stratified by week since symptom onset.
2. Test sensitivity by time since onset of symptoms.
| Days 1‐7 | Days 8‐14 | Days 15‐21 | Days 22‐35 | Days > 35 | Comparison | |
|
Test groups [studies] (true positives/COVID cases) Sensitivity (95% CI) |
||||||
| IgG | 33 [23] (165/568) | 34 [22] (766/1200) | 34 [22] (974/1110) | 20 [12] (417/502) | 11 [4] (213/252) | |
| 29.7% (22.1 to 38.6) | 66.5% (57.9 to 74.2) | 88.2% (83.5 to 91.8) | 80.3% (72.4 to 86.4) | 86.7% (79.6 to 91.7) | P < 0.00005 | |
| IgM | 34 [24] (207/608) | 32 [21] (724/1171) | 32 [21] (800/1074) | 19 [11] (378/507) | 11 [4] 118/215 | |
| 23.2% (14.9 to 34.2) | 58.4% (45.5 to 70.3) | 75.4% (64.3 to 83.8) | 68.1% (55.0 to 78.9) | 53.9% (38.4 to 68.6) | P < 0.00005 | |
| IgA | 4 [4] (54/100) | 3 [3] (38/53) | 3 [3] (66/68) | 2 [2] (81/82) | 1 [1] (23/23) | |
| 28.4% (0.9 to 94.3) | 78.1% (9.5 to 99.2) | 98.7% (39.0 to 100) | 98.7% (91.9 to 99.8) | 100% (85.2 to 100) | * | |
| Total antibodies | 5 [4] (62/144) | 6 [5] (220/247) | 6 [5] (174/176) | 4 [3] (11/19) | 2 [1] (15/28) | |
| 24.5% (9.5 to 50.0) | 84.0% (64.1 to 93.9) | 98.1% (90.1 to 99.6) | 69.5% (34.8 to 90.7) | 79.0% (49.8 to 93.4) | P < 0.00005 | |
| IgG/IgM | 17 [9] (81/259) | 21 [9] (441/608) | 21 [9] (636/692) | 16 [5] (146/152) | 9 [2] (122/153) | |
| 30.1% (21.4 to 40.7) | 72.2% (63.5 to 79.5) | 91.4% (87.0 to 94.4) | 96.0% (90.6 to 98.3) | 77.7% (66.0 to 86.2) | P < 0.00005 | |
| IgA/IgG | 1 [1] (0/12) | 1 [1] (5/10) | 1 [1] (7/8) | 1 [1] (1/1) | 0 [0] | |
| 0% (0 to 26.5) | 50.0% (18.7 to 81.3) | 87.5% (47.3 to 99.6) | 100% (2.5 to 100) | * | ||
| IgA/IgM | 0 [0] | 0 [0] | 0 [0] | 0 [0] | 0 [0] | |
| CI: confidence interval; * inadequate data to make a formal statistical comparison | ||||||
3.

Meta‐analytical estimates of sensitivity (with 95% CI) by antibody class and time since onset of symptoms
4.

Forest plot of studies evaluating tests for detection of IgG according to week post‐symptom onset and type of test
5.
Forest plot of studies evaluating tests for detection of IgM according to week post‐symptom onset and type of test
6.

Forest plot of studies evaluating tests for detection of IgG/IgM according to week post‐symptom onset and type of test
The numbers of individuals contributing data within each study within each week are very small, thus by pooling these data across studies these meta‐analyses contribute clarity to the relationship between sensitivity and time, although the important limitations of these studies as described above should be considered when interpreting all findings.
Pooled results for IgG, IgM, IgA, total antibodies and IgG/IgM all show the same general pattern over the first three weeks, with sensitivity being low when tests were used in the first week since onset of symptoms, rising in the second week, and reaching their highest values in the third week. For IgG, sensitivity across the three weeks were 29.7% (95% confidence interval (CI) 22.1 to 38.6), 66.5% (95% CI 57.9 to 74.2) and 88.2% (95% CI 83.5 to 91.8); for IgM they were 23.2% (95% CI 14.9 to 34.2), 58.4% (95% CI 45.5 to 70.3) and 75.4% (95% CI 64.3 to 83.8); and for IgG/IgM they were 30.1% (95% CI 21.4 to 40.7), 72.2% (95% CI 63.5 to 79.5) and 91.4% (95% CI 87.0 to 94.4). Values for total antibodies and IgA are also given in Table 3.
It is important to note that these estimates are based on pooling multiple cross‐sectional studies, and are not based on tracking the same groups of participants over time or even using the same tests. The reasons why individuals are included at some particular time points and not at others is mostly not reported.
Estimates of sensitivity beyond three weeks are based on smaller sample sizes, with a maximum of 12 studies contributing data in weeks 4 and 5, and only four studies providing any follow‐up information beyond week 5. Estimates for IgA and total antibodies are based on fewer than 100 samples/participants and we will not comment upon them further. In weeks 4 and 5, pooled sensitivities of IgG were 80.3% (95% CI 72.4 to 86.4); IgM were 68.1% (95% CI 55.0 to 78.9); and for IgG/IgM were 96.0% (95% CI 90.6 to 98.3).
The data beyond week 5 gave sensitivity estimates of 86.7% (95% CI 79.6 to 91.7; IgG), 53.9% (95% CI 38.4 to 68.6; IgM) and 77.7% (95% CI 66.0 to 86.2; IgG/IgM). The expected decline in the sensitivity of IgM is evident.
Overall specificity
We estimated antibody test specificity from 35 studies. Specificity estimates for all studies are presented in Appendix 11 for IgG, IgM, IgG/IgM, IgA, total antibodies, and IgA/IgG. Results pooled across all studies are in Table 4 and show specificity exceeding 98% for all antibody types, with precise estimates (confidence intervals up to 2 percentage points wide), particularly for IgG, IgM, total antibodies and IgG/IgM, where estimates are based on several thousand non‐COVID samples. Inspection of the figures shows low heterogeneity in study estimates of specificity across studies. Nine studies provided some information on the cross‐reactivity of other infections, including other coronaviruses, with the SARS‐CoV‐2 antigens used in the assays (Table 5).
3. Specificity and impact of reference standard for non‐COVID cases.
| Overall specificitya | COVID suspects deemed negative | Current healthy or other disease | Pre‐pandemic | Comparison of control groups | |
|
Test groups [studies] (false positives/non‐COVID cases) Specificity (95% CI) |
|||||
| IgG | 62 [44] (159/6136) | 6 [6] (10/396) | 14 [10] (60/2614) | 19 [10] (88/2633) | |
| 99.1% (98.3% to 99.6%) | 98.0% (91.0% to 99.6%) | 99.2% (97.6% to 99.8%) | 99.2% (97.8% to 99.7%) | P = 0.56 | |
| IgM | 59 [41] (183/6103) | 5 [5] (12/384) | 14 [10] (89/3069) | 17 [9] (38/2075) | |
| 98.7% (97.4% to 99.3%) | 98.1% (89.9% to 99.7%) | 98.6% (96.0% to 99.5%) | 99.3% (98.0% to 99.8%) | P = 0.50 | |
| IgG/IgM | 34 [23] (78/5761) | 7 [7] (33/454) | 7 [5] (20/506) | 18 [6] (22/1104) | No formal comparison possible |
| 98.7% (97.2% to 99.4%) | 92.8% (89.7% to 95.0%) | 99.9% (65.2% to 100%) | 98.7% (96.6% to 99.5%) | ||
| Total antibodies | 16 [10] (41/3585) | ||||
| 99.2% (98.3% to 99.6%) | |||||
| IgA | 4 [4] (10/663) | ||||
| 98.5% (97.2% to 99.2%) | |||||
| IgA/IgGb | 2 [2] (1/528) | ||||
| 99.8% (98.9% to 100%) | |||||
| IgA/IgMb | 1 [1] (1/483) | ||||
| 99.8% (99.2% to 100%) | |||||
| CI: confidence interval | |||||
aIncludes studies that are categorised as mixed/other not included in the subgroups. bConfidence intervals computed using binomial exact on totals.
4. Reported cross‐reactivity with SARS‐CoV‐2 antigens.
| Study | Test(s) evaluated | What the study says about cross‐reactivity |
| Cai 2020 | In‐house CLIA | Reported no cross‐reactivity in 167 sera from patients with infection with other pathogens (influenza A virus (25), respiratory syncytial virus (7), parainfluenza virus (8), influenza B virus (5), adenovirus (6), Klebsiella pneumoniae (8), Streptococcus pneumoniae (3), mycoplasma (5), Acinetobacter baumannii (10), Candida albicans (2), Staphylococcus aureus (3), Mycobacterium tuberculosis (4), hepatitis B virus (33), hepatitis C virus (22), syphilis (23) and saccharomycopsis (3)). |
| Freeman 2020 | In‐house ELISA | Reported cross‐reactivity to SARS‐CoV‐2 spike protein in sera from patients with SARS‐1 and MERS‐CoV, and no cross‐reactivity with NL63, OC43, HKU1, 229E |
| Guo 2020a | In‐house ELISA | Reported Western Blot cross‐reactivity analysis in plasma samples positive for human CoV‐229E, ‐NL63, ‐OC43, ‐HKU1, and SARS‐CoV. Strong cross‐reactivity was observed only for SARS‐CoV. |
| Infantino 2020 | Shenzhen YHLO CLIA | Observed no cross‐reactivity in sample from blood donors from the COVID‐19 era (winter 2019) but positive results in two samples from people with CMV infections and 2 with rheumatic disease. |
| Lassauniere 2020 [A] | [A] Beijing Wantai ELISA [B] EUROIMMUN IgG ELISA [C] EUROIMMUN IgA ELISA [D] Dynamiker Biotechnology LFA [E] CTK Biotech ‐ OnSite LFA [F] Autobio Diagnostics LFA [G] Artron Laboratories LFA [H] Acro Biotech LFA [I] Hangzhou Alltest LFA |
Included sera from patients with acute viral respiratory tract infections caused by other coronaviruses (n = 5) or non‐coronaviruses (n = 45), and sera from patients positive for dengue virus (n = 9), CMV (n = 2) and Epstein Barr virus (n = 10). Cross reaction was observed for the EUROMIMMUN IgA ELISA (> 1 respiratory virus present, adenovirus, dengue virus) and for the EUROMIMMUN IgG ELISA (coronavirus HKU1 and adenovirus). Some cross‐reactivity also observed for CGIA tests. Study authors suggest related to antigen target and ELISA format. |
| Ma 2020a | In‐house CLIA | Limited detail but suggests limited cross‐reaction |
| Wang 2020a [A] | A. Beijing Hotgen IgM CGIA B. Beijing Hotgen IgM ELISA |
Demonstrated considerable cross‐reaction with rheumatoid factor IgM (22/36 false positive results). Other pathogens included influenza A virus (n = 5), influenza B virus (n = 5), Mycoplasma pneumoniae (n = 5), Legionella pneumophila (n = 5), HIV infection (n = 6), hypertension (n = 5) and diabetes mellitus (n = 5) |
| Zhang 2020b | Shenzhen YHLO CLIA | Observed false positive results in influenza A and B (2 each), adenovirus (n = 4) and Mycoplasma pneumoniae (n = 17). |
| Zhang 2020d | In‐house CGIA (co‐author Beijing Hotgen) | Appears to report a separate cross‐reactivity study for influenza A, influenza B, respiratory syncytial virus, Mycoplasma pneumoniae and Chlamydia pneumoniae. No cross reactions were observed. |
| CGIA: colloidal gold immunoassay; CLIA: Chemiluminescence immunoassay; CMV: cytomegalovirus; ELISA: enzyme‐linked immunosorbent assay; LFA: lateral flow assay; MERS: Middle East respiratory syndrome; SARS: severe acute respiratory syndrome | ||
Impact of reference standard for COVID‐19 cases on sensitivity
The majority of studies only included participants who were diagnosed with COVID‐19 based upon observing a positive RT‐PCR test. However, in clinical practice it is common to encounter patients from whom positive RT‐PCR results are never obtained, but who demonstrate clinical and imaging features of COVID‐19. Diagnostic criteria for COVID‐19 produced by WHO and the China CDC include definitions for suspected COVID‐19 in RT‐PCR‐negative patients. Twelve studies defined the presence of COVID‐19 using these criteria, thus including RT‐PCR‐negative patients in the COVID‐19 group as well as RT‐PCR‐positive patients. We compared estimates of sensitivity between studies using a RT‐PCR‐positive reference standard definition with a criteria‐based reference standard (including both RT‐PCR‐positives and RT‐PCR‐negatives; Table 6). We stratified the analysis for weeks since onset of symptoms. All the observed differences were within magnitudes expected by chance.
5. Investigation of impact of reference standard on sensitivity.
| RT‐PCR‐positive COVID‐19 cases | RT‐PCR‐negative COVID‐19 cases | Comparison | |
|
Test groups [studies] (true positives/COVID cases) Sensitivity (95% CI)a |
|||
| IgG | 26 [15] (1555/2280) | 8 [8] (925/1300) | |
| 87.9% (82.7 to 91.7) | 91.2% (83.9 to 95.4) | P = 0.36 | |
| IgM | 23 [13] (1368/2166) | 10 [9] (792/1292) | |
| 70.8% (56.3 to 82.0) | 87.5% (73.7 to 94.6) | P = 0.06 | |
| IgG/IgM | 17 [6] (966/1278) | 4 [4] (400/499) | |
| 90.6% (86.6 to 93.5) | 93.6% (88.9 to 96.4) | P = 0.22 | |
| CI: confidence interval; RT‐PCR: reverse transcription polymerase chain reaction | |||
aWe obtained sensitivity estimates from a model of all data stratified by week, estimating the average difference in sensitivity across follow‐up. The figures quoted correspond to the week 3 strata (15‐21 days) in the model.
In a further analysis, we separated COVID‐19 participants who were RT‐PCR‐positive from those who were RT‐PCR‐negative, where studies allowed, and subgrouped the results to investigate whether there is a difference in accuracy according to RT‐PCR status. Data from only three studies could be included in this analysis (Figure 7; Figure 8; Figure 9). Differences in estimates of sensitivity (pooled stratifying for weeks since onset of symptoms), varied in direction for IgG and IgM, and were very similar for IgG/IgM (Table 7). All differences were within magnitudes expected by chance. There was no consistent evidence that the accuracy of serology tests was lower in RT‐PCR‐positive patients, although there is high uncertainty in these findings.
7.
Sensitivity of IgG in PCR+ve and PCR‐ve COVID‐19 cases by week since onset of symptoms.
8.

Sensitivity of IgM in PCR+ve and PCR‐ve COVID‐19 cases by week since onset of symptoms.
9.
Sensitivity of IgG/IgM in PCR+ve and PCR‐ve COVID‐19 cases by week since onset of symptoms.
6. Studies reporting sensitivity in both RT‐PCR‐positive and RT‐PCR‐negative subgroups.
| RT‐PCR‐positive COVID‐19 cases | RT‐PCR‐negative COVID‐19 cases | |||
|
Test groups [studies] (True positives/COVID‐19 cases) |
Sensitivity (95% CI) |
Test groups [studies] (True positives/COVID‐19 cases) |
Sensitivity (95% CI) | |
| IgG | ||||
| Days 1‐7b | 2 [2] (1/28) | 2 [2] (8/13) | ||
| Days 8‐14b | 2 [2] (21/33) | 3 [3] (25/30) | ||
| Days 15‐21b | 2 [2] (39/40) | 3 [3] (64/72) | ||
| Pooleda (stratified by time) | 72.6% (46.2% to 89.1%) | 84.0% (64.4% to 93.9%) | ||
| Test for difference in sensitivity between RT‐PCR‐positive and RT‐PCR‐negative groups: P = 0.18 | ||||
| IgM | ||||
| Days 1‐7b | 2 [2] (3/28) | 2 [2] (4/13) | ||
| Days 8‐14b | 2 [2] (25/33) | 3 [3] (11/30) | ||
| Days 15‐21b | 2 [2] (8/16) | 3 [3] (31/72) | ||
| Pooleda (stratified by time) | 64.6% (49.7% to 77.1%) | 49.0% (34.2% to 63.9%) | ||
| Test for difference in sensitivity between RT‐PCR‐positive and RT‐PCR‐negative group: P = 0.07 | ||||
| IgG/IgM | ||||
| Days 1‐7b | 2 [2] (8/36) | 2 [2] (4/17) | ||
| Days 8‐14b | 2 [2] (37/53) | 3 [3] (29/40) | ||
| Days 15‐21b | 2 [2] (141/150) | 3 [3] (104/113) | ||
| Pooleda (stratified by time) | 71.9% (58.7% to 82.2%) | 71.1% (57.0% to 82.0%) | ||
| Test for difference in sensitivity between RT‐PCR‐positive and RT‐PCR‐negative group: P = 0.90 | ||||
| CI: confidence interval; RT‐PCR: reverse transcription polymerase chain reaction | ||||
aThe sensitivity estimates are produced from a model that combines all data from both subgroups and time‐groups, stratifying by time‐group. The estimate corresponds to sensitivity in Days 15‐21. bRT‐PCR‐positive data have only been included here when the study includes a RT‐PCR‐negative subgroup as well.
Impact of reference standard for non‐COVID‐19 cases on specificity
We classified the reference standard used to verify non‐COVID cases into three main groups: pre‐pandemic controls (both healthy and with other diseases) who underwent no RT‐PCR testing, current controls from healthy or other disease groups (typically who also did not undergo RT‐PCR testing), and individuals who were investigated for COVID‐19 but deemed non‐COVID cases. Whilst results were similar for IgG and IgM, we noted more false positives for the IgG/IgM outcome in the studies using a COVID suspect group than in other studies (Table 4).
Sensitivity and specificity by assay type
We further investigated the heterogeneity in sensitivity estimates at any time point according to test technology type. We considered differences between CGIA, CLIAs, ELISAs and tests we can only describe as lateral flow assays due to lack of any names or detail (this group originate from the UK National COVID Testing Scientific Advisory Panel, which withheld names of the tests evaluated due to confidentiality clauses in the legal contracts with the manufacturers Adams 2020 [A]). There were inadequate numbers of studies evaluating FIAs and indirect immunofluorescence tests, luciferase immunoprecipitation assays and 'S‐flow' assays to analyse, and we were only able to assess IgG, IgM and IgG/IgM targets. In a sensitivity analysis we restricted the included studies to those that used commercial (rather than in‐house) tests.
We obtained estimates from a model that included all data stratified by weeks since onset of symptoms. The results presented in Table 8 and below correspond to estimates from the model of performance in week 3 post‐symptom onset.
7. Sensitivity and specificity by test technology.
| Test method | ||||||
| Test method | CGIA | CLIA | ELISA | LFA | Comparison | |
| IgG | ||||||
|
Test groups [studies] (True positives/COVID cases) |
6 [5] (268/397) | 10 [10] (1112/1432) | 12 [11] (1014/1552) | 7 [1] (133/238) | ||
| Sensitivity (95% CI)a | 87.3% (77.0 to 93.4) | 94.6% (90.7 to 97.0) | 85.8% (78.0 to 91.1) | 76.0% (61.0 to 86.5) | P = 0.004 | |
|
Test groups [studies] (True negatives/non‐COVID cases) |
11 [11] (409/415) | 12 [12] (318/322) | 18 [16] (2003/2102) | 6 [1] (354/360) | ||
| Specificity (95% CI)a | 99.5% (96.5 to 99.9) | 99.0% (91.6 to 99.9) | 98.8% (96.5 to 99.6) | 99.0% (95.3 to 99.8) | P = 0.85 | |
| IgM | ||||||
|
Test groups [studies] (True positives/COVID cases) |
7 [6] (109/411) | 10 [10] (884/1355) | 12 [11] (1083/1568) | 7 [1] (78/228) | ||
| Sensitivity (95% CI)a | 69.5% (44.3 to 86.7) | 80.9% (63.8 to 91.0) | 84.5% (70.7 to 92.5) | 51.4% (26.5 to 75.6) | P = 0.11 | |
|
Test groups [studies] (True negatives/non‐COVID cases) |
12 [11] (455/487) | 13 [13] (609/621) | 14 [12] (1674/1710) | 6 [1] (357/360) | ||
| Specificity (95% CI)a | 97.3 (90.0 to 99.3) | 98.5 (92.3 to 99.7) | 99.1 (97.2 to 99.7) | 99.6 (97.3 to 99.9) | P = 0.40 | |
| IgG/IgM | ||||||
|
Test groups [studies] (True positives/COVID cases) |
4 [3] (232/316) | 3 [3] (344/420) | 5 [4] (595/770) | 11 [2] (255/358) | ||
| Sensitivity (95% CI)a | 90.7% (82.7 to 95.2) | 97.5% (94.0 to 99.0) | 90.7% (83.3 to 95.0) | 88.6% (82.0 to 93.0) | P = 0.02 | |
|
Test groups [studies] (True negatives/non‐COVID cases) |
11 [11] (330/353) | 5 [4] (230/244) | 5 [4] (387/391) | 13 [3] (3797/3827) | ||
| Specificity (95% CI)a | 96.0 (90.1 to 98.5) | 94.1 (82.7 to 98.2) | 99.4 (97.4 to 99.9) | 98.2 (96.3 to 99.1) | P = 0.05 | |
| CGIA: colloidal gold immunoassay; CI: confidence interval; CLIA: chemiluminescence immunoassay; ELISA: enzyme‐linked immunosorbent assay; LFA: lateral flow assay (no further detail) | ||||||
aWe obtained sensitivity estimates from a model of all data stratified by week, estimating the average difference in sensitivity across follow‐up. The figures quoted correspond to the Week 3 (15‐21 days) strata in the model.
For IgG, there were clear differences in the sensitivity of assays, with CLIA (94.6%), CGIA (87.3%) and ELISA (85.8%) all outperforming the unknown lateral flow assay tests (76.0%). The differences between the groups was beyond that expected by chance (P = 0.004), but largely driven by the low value for lateral flow tests (all of the data coming from 40 COVID‐19 patients in the UK National COVID Testing Scientific Advisory Panel study tested multiple times).
For IgM, although laboratory‐based ELISA (84.5%) and CLIA (80.9%) outranked lateral flow CGIA (69.5%) and the unknown lateral flow assays (51.4%), the differences observed were in the realms of those expected by chance (P = 0.11).
In the smaller subset of studies that evaluated tests combining IgM/IgG, the performance of laboratory CLIA tests (97.3%) ranked above those of CGIA (91.4%), ELISA (90.5%) and unknown lateral flow tests (85.8%). These differences were beyond those expected by chance (P = 0.01)
Excluding the in‐house tests, and thus restricting the analysis to only commercial tests, made little difference to estimates of sensitivity.
Analyses of specificity presented by assay type are also given in Table 8. Differences in specificity of IgG and IgM between assay types were small, CLIA and CGIA tests showed lower specificity for IgG/IgM tests than ELISA and LFIA, but confidence intervals on all estimates are wide.
Sensitivity and specificity by brand
We have tabulated the results by brand for the 27 commercial tests: 15 tests for IgG Table 9; 14 tests for IgM Table 10; and nine tests for IgG/IgM Table 11. The study data for these estimates are provided in Figure 4, Figure 5 and Figure 6. Appendix 12 tabulates the information that we have been able to derive regarding the current availability of these commercially produced tests. Data for sensitivity are stratified by week of onset of symptoms and we present the numbers of studies and samples from which data are available for each time interval. Caution is required in the interpretation of these data as many are based only on single studies with small sample sizes. We present confidence intervals to quantify the uncertainty in the estimates. We would advise focusing on estimates based on at least 100 samples/participants per week further. Three tests have estimates of sensitivity based on more than 100 samples (Beijing Wantai ELISA, Bioscience Co. (Chongqing) CLIA, Zuhai Livzon ELISA). We evaluated the studies that we pooled to create these estimates as having multiple domains at risk of bias and having concerns about the applicability of the findings (all studies having at most 2 of the 7 ratings in the QUADAS‐2 assessment described as low risk or low concern).
8. Sensitivity and specificity by test brand (IgG).
| Test namea |
Test method |
IgG sensitivity by time since onset of symptoms Studies (true positives/COVID‐19 cases) Sensitivity (95% CI) |
IgG specificity Studies (false positives/COVID‐19 cases) Specificity (95% CI) |
||||
| 1‐7 days | 8‐14 days | 15‐21 days | 22‐35 days | > 35 days | |||
| Beijing Beier Bioengineering | CGIA | 1 (2/10) | 1 (6/13) | 1 (11/14) | |||
| 20.0% (2.5 to 55.6) | 46.2% (19.2 to 74.9) | 78.6% (49.2 to 95.3) | |||||
| Beijing Beier Bioengineering | CLIA | 1 (4/10) | 1 (6/13) | 1 (9/14) | |||
| 40.0% (12.2 to 73.8) | 46.2% (19.2 to 74.9) | 64.3% (35.1 to 87.2) | |||||
| Beijing Beier Bioengineering | ELISA | 1 (4/10) | 1 (8/13) | 1 (12/14) | |||
| 40.0% (12.2 to 73.8) | 61.5% (31.6 to 86.1) | 85.7% (57.2 to 98.2) | |||||
| Beijing Hotgen | ELISA | 1 (9/22) | 1 (60/92) | 1 (51/55) | 1 (39/45) | 2 (22/172) | |
| 40.9% (20.7 to 63.6) | 65.2% (54.6 to 74.9) | 92.7% (82.4 to 98.0) | 86.7% (73.2 to 94.9) | 87.2% (81.3 to 91.8) | |||
| Beijing Wantai | ELISA | 2 (31/133) | 2 (130/210) | 2 (127/149) | 2 (2/297) | ||
| 23.3% (16.4 to 31.4) | 61.9% (55.0 to 68.5) | 85.2% (78.5 to 90.5) | 99.3% (97.6 to 99.9) | ||||
| Beijing Wantai | CGIA | 1 (1/209) | |||||
| 99.5% (97.4 to 100) | |||||||
| Bioscience Co (Chongqing) | CLIA | 2 (43/92) | 2 (129/212) | 2 (208/244) | 2 (98/164) | 1 (75/76) | |
| 46.7% (36.3 to 57.4) | 60.8% (53.9 to 67.5) | 85.2% ( 80.2 to 89.4) | 59.8% (51.8 to 67.3) | 98.6% (92.9 to 100) | |||
| Darui Biotech | ELISA | 1 (0/64) | |||||
| 100% (94.4 to 100) | |||||||
| EUROIMMUN | ELISA | 1 (2/13) | 2 (13/25) | 2 (14/15) | 2 (98/164) | 2 (3/82) | |
| 15.4% (1.9 to 45.4) | 52.0% (31.3 to 72.2) | 93.3% (68.1 to 99.8) | 59.8% (51.8 to 67.3) | 96.3% (89.7 to 99.2) | |||
| EUROIMMUN Anti‐SARS‐Cov | IIFT | 1 (1/4) | 1 (3/5) | 1 (3/3) | 1 (1/1) | 1 (0/10) | |
| 25.0% (0.6 to 80.6) | 60.0% (14.7 to 94.7) | 100% (29.2 to 100) | 100% (2.5 to 100) | 100% (69.2 to 100) | |||
| EUROIMMUN Beta | ELISA | 1 (0/12) | 1 (3/10) | 1 (7/8) | 1 (1/1) | 1 (0/45) | |
| 0% (0 to 26.5) | 30%.0% (14.7 to 94.7) | 87.5% (47.3 to 99.7) | 100% (2.5 to 100) | 100% (92.1 to 100) | |||
| Hangzhou Alltest ‐ IgG/IgM | CGIA | 1 (1/8) | 2 (21/42) | 2 (57/68) | 2 (0/45) | ||
| 12.5% (0.3 to 52.7) | 50.0% (34.2 to 65.8) | 83.8% (72.9 to 91.6) | 100% (92.1 to 100) | ||||
| Innovita Biological ‐ Ab test (IgM/IgG) | CGIA | 1 (7/13) | 1 (7/8) | 1 (21/23) | |||
| 53.8% (25.1 to 80.8) | 87.5% (47.3 to 99.7) | 91.3% (72.0 to 98.9) | |||||
| Shenzhen YHLO | CLIA | 2 (2/8) | 2 (28/29) | 2 (25/26) | 2 (64/64) | 1 (7/7) | 7 (4/322) |
| 25.0% (3.2 to 65.1) | 96.6% (82.2 to 99.9) | 96.2% (80.4 to 99.9) | 100% (94.4 to 100) | 100% (59.0 to 100) | 98.8% (96.9 to 99.7) | ||
| Snibe Diagnostic ‐ MAGLUMI | CLIA | 2 (11/40) | 2 (35/48) | 25/25 | |||
| 27.5% (14.6 to 43.9) | 72.9% (58.2 to 84.7) | 100.0% (86.3 to 100) | |||||
| Vivachek ‐ VivaDiag IgM/IgG | CGIA | 2 (0/42) | |||||
| 100% (91.6 to 100) | |||||||
| Zhuhai Livzon | CGIA | 1 (5/36) | 1 (20/34) | 1 (35/38) | 2 (0/35) | ||
| 13.9% (4.7 to 29.5) | 58.8% (40.7 to 75.4) | 92.1% (78.6 to 98.3) | 100% ( 90.0 to 100) | ||||
| Zhuhai Livzon | ELISA | 4 (17/80) | 3 (163/288) | 3 (197/223) | 2 (91/104) | 5 (5/351) | |
| 21.3% (12.9 to 31.8) | 56.6% (50.7 to 62.4) | 88.3% (83.4 to 92.2) | 87.5% (79.6 to 93.2) | 98.6% (96.7 to 99.5) | |||
| CGIA: colloidal gold immunoassay; CI: confidence interval; CLIA: chemiluminescence immunoassay; ELISA: enzyme‐linked immunosorbent assay; FIA: fluorescence immunoassay; IIFT: indirect immunofluorescence assay; LFA: lateral flow assay | |||||||
aSee Appendix 12 for details of manufacturer product codes, where available.
9. Sensitivity and specificity by test brand (IgM).
| Test namea | Test method |
IgM sensitivity by time since onset of symptoms Studies (true positives/COVID‐19 cases) Sensitiivity (95% CI) |
IgM specificity Studies (false positives/COVID‐19 cases) Specificity (95% CI) |
||||
| 1‐7 days | 8‐14 days | 15‐21 days | 22‐35 days | > 35 days | |||
| Artron Laboratories IgM/IgG | CGIA | 1 (5/7) | 1 (12/15) | 1 (8/8) | |||
| 71.4% (29.0 to 96.3) | 80.0% (51.9 to 95.7) | 100% (63.1 to 100) | |||||
| Autobio Diagnostics IgM/IgG | CGIA | 1 (6/7) | 1 (14/15) | 1(8/8) | |||
| 85.7% (42.1 to 99.6) | 93.3% (68.1 to 99.8) | 100% (63.1 to 100) | |||||
| Beijing Hotgen | ELISA | 1 (10/22) | 1 (72/92) | 1 (72/92) | 1 (41/45) | 1 (0/100) | |
| 45.5% (24.4 to 67.8) | 78.3% (68.4 to 86.2) | 78.3% (68.4 to 86.2) | 91.1% (78.8 to 97.5) | 100% (96.4 to 100) | |||
| Beijing Hotgen | CGIA | 1 (22/72) | |||||
| 69.4% (57.5 to 79.8) | |||||||
| Beijing Wantai | ELISA | 1 (3/513) | |||||
| 99.4% (98.3 to 99.9) | |||||||
| Beijing Wantai | CGIA | 1 (4/209) | |||||
| 98.1% (95.2 to 99.5) | |||||||
| Bioscience Co (Chongqing) | CLIA | 1 (34/67) | 1 (34/67) | 1 (131/134) | 1 (13/13) | ||
| 50.7% (38.2 to 63.2) | 50.7% (38.2 to 63.2) | 97.8% (93.6 to 99.5) | 100% (75.3 to 100) | ||||
| CTK Biotech OnSite IgG/IgM | CGIA | 1 (5/7) | 1 (14/15) | 1 (8/8) | |||
| 71.4% (29.0 to 96.3) | 93.3% (68.1 to 99.8) | 100% (63.1 to 100) | |||||
| Darui Biotech | ELISA | 1 (14/64) | |||||
| 78.1% (66.0 to 87.5) | |||||||
| Dynamiker Biotechnology IgG/IgM | CGIA | 1 (5/7) | 1 (14/15) | 1 (8/8) | |||
| 71.4% (29.0 to 96.3) | 93.3% (68.1 to 99.8) | 100% (63.1 to 100) | |||||
| EUROIMMUN | ELISA | 1 (76/82) | |||||
| 92.7% (84.8 to 97.3) | |||||||
| EUROIMMUN Anti‐SARS‐Cov | IIFT | 1 (1/10) | |||||
| 90.0% (55.5 to 99.7) | |||||||
| Hangzhou Alltest ‐ IgG/IgM | CGIA | 1 (1/8) | 2 (23/42) | 2 (58/68) | 2 (0/45) | ||
| 12.5% (0.3 to 52.7) | 54.8% (38.7 to 70.2) | 85.3% (74.6 to 92.7) | 100% (92.1 to 100) | ||||
| Shenzhen YHLO | CLIA | 7 (10/321) | |||||
| 96.9% (94.3 to 98.5) | |||||||
| Vivachek ‐ VivaDiag IgM/IgG | CGIA | 2 (1/42) | |||||
| 97.6% (87.4 to 99.9) | |||||||
| Xiamen InnodDx Biotech | CLIA | 1 (2/300) | |||||
| 99.3% (97.6 to 99.9) | |||||||
| Zhuhai Livzon | CGIA | 1 (7/36) | 1 (31/34) | 1 (35/38) | 2 (0/35) | ||
| 19.4% (8.2 to 36.0) | 91.2% (76.3 to 98.1) | 92.1% (78.6 to 98.3) | 100% ( 90.0 to 100) | ||||
| Zhuhai Livzon | ELISA | 3 (14/66) | 2 (150/202) | 2 (159/166) | 1 (43/45) | 5 (3/351) | |
| 21.2% (12.1 to 33.0) | 74.3% (67.7 to 80.1) | 95.8% (91.5 to 98.3) | 95.6% (84.9 to 99.5) | 99.1% (97.5 to 99.8) | |||
| CGIA: colloidal gold immunoassay; CI: confidence interval; CLIA: chemiluminescence immunoassay; ELISA: enzyme‐linked immunosorbent assay; FIA: fluorescence immunoassay; IIFT: indirect immunofluorescence assay; LFA: lateral flow assay | |||||||
aSee Appendix 12 for details of manufacturer product codes, where available.
10. Sensitivity and specificity by test brand (IgG/IgM).
| Test namea | Test method |
IgG/IgM sensitivity by time since onset of symptoms Studies (true positives/COVID‐19 cases) Sensitiivity (95% CI) |
IgG/IgM specificity Studies (false positives/COVID‐19 cases) Specificity (95% CI) |
||||
| 1‐7 days | 8‐14 days | 15‐21 days | 22‐35 days | > 35 days | |||
| Acro Biotech ‐ IgG/IgM | CGIA | 1 (3/15) | |||||
| 80.0% (51.9 to 95.7) | |||||||
| Artron Laboratories IgM/IgG | CGIA | 1 (5/7) | 1 (12/15) | 1 (8/8) | 1 (0/17) | ||
| 71.4% (29.0 to 96.3) | 80.0% (51.9 to 95.7) | 100% (63.1 to 100) | 100% (80.5% to 100) | ||||
| Autobio Diagnostics IgM/IgG | CGIA | 1 (6/7) | 1 (14/15) | 1(8/8) | 1 (0/32) | ||
| 85.7% (42.1 to 99.6) | 93.3% (68.1 to 99.8) | 100% (63.1 to 100) | 100% (89.1 to 100) | ||||
| Beijing Hotgen | ELISA | 1 (10/22) | 1 (72/92) | 1 (72/92) | 1 (41/45) | 1 (0/100) | |
| 45.5% (24.4 to 67.8) | 78.3% (68.4 to 86.2) | 78.3% (68.4 to 86.2) | 91.1% (78.8 to 97.5) | 100% (96.4 to 100) | |||
| Bioscience Co (Chongqing) | CLIA | 1 (34/67) | 1 (34/67) | 1 (131/134) | 1 (13/13) | 2 (7/148) | |
| 50.7% (38.2 to 63.2) | 50.7% (38.2 to 63.2) | 97.8% (93.6 to 99.5) | 100% (75.3 to 100) | 95.3% (90.5 to 98.1) | |||
| CTK Biotech OnSite IgG/IgM | CGIA | 1 (5/7) | 1 (14/15) | 1 (8/8) | 1 (0/32) | ||
| 71.4% (29.0 to 96.3) | 93.3% (68.1 to 99.8) | 100% (63.1 to 100) | 100% (89.1 to 100) | ||||
| Dynamiker Biotechnology IgG/IgM | CGIA | 1 (5/7) | 1 (14/15) | 1 (8/8) | 1 (0/32) | ||
| 71.4% (29.0 to 96.3) | 93.3% (68.1 to 99.8) | 100% (63.1 to 100) | 100% (89.1 to 100) | ||||
| Hangzhou Alltest ‐ IgG/IgM | CGIA | 1 (1/8) | 2 (23/42) | 2 (58/68) | 3 (2/60) | ||
| 12.5% (0.3 to 52.7) | 54.8% (38.7 to 70.2) | 85.3% (74.6 to 92.7) | 96.7% (88.5 to 99.6) | ||||
| Shenzhen YHLO | CLIA | 2 (7/96) | |||||
| 92.7% (85.6 to 97.0) | |||||||
| Vivachek ‐ VivaDiag IgM/IgG | CGIA | 3 (14/162) | |||||
| 91.4% (85.9 to 95.2) | |||||||
| Zhuhai Livzon | CGIA | 1 (7/36) | 1 (31/34) | 1 (35/38) | 2 (0/35) | ||
| 19.4% (8.2 to 36.0) | 91.2% (76.3 to 98.1) | 92.1% (78.6 to 98.3) | 100% (90.0 to 100) | ||||
| Zhuhai Livzon | ELISA | 3 (14/66) | 2 (150/202) | 2 (159/166) | 1 (43/45) | 4 (4/291) | |
| 21.2% (12.1 to 33.0) | 74.3% (67.7 to 80.1) | 95.8% (91.5 to 98.3) | 95.6% (84.9 to 99.5) | 98.6% (96.5 to 99.6) | |||
| CGIA: colloidal gold immunoassay; CI: confidence interval; CLIA: chemiluminescence immunoassay; ELISA: enzyme‐linked immunosorbent assay; FIA: fluorescence immunoassay; IIFT: indirect immunofluorescence assay; LFA: lateral flow assay | |||||||
aSee Appendix 12 for details of manufacturer product codes, where available.
Eight tests have estimates of specificity based on more than 100 samples, with estimates over 98% for five tests (Bejing Hotgen ELISA, Beijing Wantai ELISA, Beijing Wantai CGIA, Xiamen InnodDx Biotech ELISA, Zhuhai Livzon ELISA). Again please note the concerns in the risk of bias and applicability of these findings.
Other sources of heterogeneity
Our protocol included additional planned analyses by:
current infection or past infection;
study design; and
setting.
We could not investigate these sources because of lack of variability across the studies in these features. Only two studies explicitly stated that they recruited only convalescent patients, and 48 (85%) studies recruited hospital inpatients. For study design only five out of 54 (11%) studies recruited a single group of suspected COVID‐19 patients, and did not use a 'COVID‐19 cases only' study, or a 'two‐group' study design.
Investigation of publication bias
We observed direct evidence of selective reporting through the withholding of names of the nine lateral flow assay testing brands from the UK National COVID Testing Scientific Advisory Panel study (Adams 2020 [A]). The paper states, "Individual manufacturers did not approve release of device‐level data, so device names are anonymised" (Adams 2020 [A]). The sensitivity estimates for the lateral flow assays in this study (which are most likely to be CGIA) were noted to be lower than estimates for CGIA tests from other studies. Four other studies also did not identify the test that they were evaluating.
Discussion
This is the first version of a Cochrane living review summarising the accuracy of antibody tests for detecting current or previous SARS‐CoV‐2infection. This version of the review is based on published studies or studies available as preprints up until the 27 April 2020. The speed of development and publication of studies for COVID‐19 antibody tests is unprecedented, and the content of this review will always be out of date. We are continuously identifying new published studies, and plan to update this review several times during the next few months.
The studies included in this version are largely from China, evaluating tests from Chinese universities and manufacturers. Many of the studies are the first that have been published for each test, and thus are early‐phase studies. Whilst there is no recognised stage classification of diagnostic studies, there are several common features of those undertaken during test development. These include multiple tests being described as 'in‐house', that thresholds for tests are determined from the data collected during the study, that all tests are undertaken by technical experts in laboratories, that the samples used are from collections easily available to the research team, and that multiple samples are used from the same participants. These limitations explain much of the rating for high risk of bias and concerns about applicability in this review. Many of these issues make it likely that the accuracy of tests when used in clinical care will be lower than that observed here. We did locate six evaluations recruiting patients identified in clinical pathways before it was established whether they had COVID‐19. This is more likely to produce results that reflect clinical practice, and we encourage future evaluations to consider this study design.
A concern with this review, and with its updates, is the high likelihood of selective reporting of results, particularly by manufacturers. We have already noted manufacturers being unwilling to be identified in the UK National COVID Testing Scientific Advisory Panel study (Adams 2020 [A]). Unlike randomised controlled trials of interventions, there are no requirements for test accuracy studies to be prospectively registered on study registers, nor to publish their findings. Many industry studies are only briefly described on 'Information for use' documents included with the tests, and study reports submitted to regulators are regarded as confidential. We are also aware that there are independent studies undertaken by National Public Health bodies, some of which have been submitted to FIND's data tracking tool for speedy data sharing. We plead for greater transparency and full publication in this field and continue to encourage laboratories to submit data and reports via FIND's portal. We request sharing of any unpublished reports for inclusion in future updates (please send to coviddta@contacts.bham.ac.uk). We have contacted test manufacturers to request full study reports which we will include in a future update of this review.
Summary of main results
We summarise 10 key findings from this review.
Evaluations of most antibody tests on the market are not available as publications or even as preprints. This review has evaluated data from 25 commercial tests and numerous in‐house assays. These represent a small fraction of the antibody assays currently available. We have identified 66 additional studies of antibody tests published or available as preprints up until 25 May 2020, which we will appraise for inclusion in the review update, but there still remain no published data for the majority of tests on the current FIND list.
The design and execution of the current studies limits the strength of conclusions that we are currently able to draw. Nearly all studies sampled COVID‐19 cases and non‐COVID cases separately, and methods for selecting participants were not described. Only four studies reported blinding reference standard and index tests, and some reference standards may misclassify individuals.
Many studies only applied tests in laboratory settings on plasma or serum, whilst they are also approved for use as point‐of‐care tests using whole blood. From these data it is not possible to ascertain the clinical accuracy of these tests in lower resource and more accessible settings.
Sensitivity varies with the time since of onset of symptoms. Figures from the studies showed the ability of antibody tests to detect SARS‐CoV‐2infection is very low in the first week (average sensitivity 30.1%, 95% CI 21.4 to 40.7) and only moderate (average sensitivity 72.2%, 95% CI 63.5 to 79.5) in the second week post‐symptom onset. These estimates are based on patients who have been hospitalised with COVID‐19, and remain in hospital at the time of sampling, and thus are likely to represent the more severe end of the disease spectrum and are potentially individuals with higher antibody responses.
Tests have higher sensitivity when done later in the course of the disease. The average sensitivity across all the included studies for IgG/IgM tests was estimated from the included studies as 91.4% (95% CI 87.0 to 94.4) for 15 to 21 days, and 96.0% (95% CI 90.6 to 98.3) for 22 to 35 days. Too few studies had evaluated tests beyond 35 days to estimate accuracy. These findings are expected given the delayed rise of IgG antibodies.
Studies estimate the specificity of tests precisely, and it appears to be high. The average from the studies for IgG/IgM is 98.7% (95% CI 97.2% to 99.4%). However, estimates of specificity are mainly based on testing pre‐pandemic, healthy people, or people known to have other disorders, and not those being investigated for possible COVID‐19.
From the limited evaluations studied, some differences were noted by test technology, CLIA methods appearing more sensitive (97.5%, 95% CI 94.0 to 99.0) than ELISA (90.7%, 95% CI 83.3 to 95.0) or CGIA‐based lateral flow assays (90.7%, 95% CI 82.7 to 95.2) for IgG/IgM, (there are also differences for IgG but no differences for IgM). There was little clear evidence of differences in specificity between technology types.
There is currently too little data on individual tests to be able to consider comparisons of their performance.
Study reports did not include many of the key items listed on the STARD reporting guideline for test accuracy studies (Bossuyt 2015), which has hindered assessment and data extraction. No study utilised a STARD participant flow diagram to enable identification of missing, indeterminate or unavailable test results.
We observed partial reporting (suppression of the identify of tests) in five studies, indicating the likelihood of publication bias.
Strengths and weaknesses of the review
Our review used a broad search screening all articles concerning COVID‐19. We undertook all screening and eligibility assessments, QUADAS‐2 assessments (Whiting 2011), and data extraction of study findings independently and in duplicate. Whilst we thus have reasonable confidence in the completeness and accuracy of the findings up until the search date, should errors be noted please inform us at coviddta@contacts.bham.ac.uk so that we can check and correct in our next update.
Weaknesses of the review primarily reflect the weaknesses in the primary studies and their reporting. Many studies omitted descriptions of sample recruitment, and key aspects of study design and execution. Some studies omit information that allows the tests to be identified. We have had to treat studies that describe their data as being based on 'samples' as if the samples were individual patients. We have been explicit about these issues where they arose.
More than half (28/54) of the studies we have included are currently only available as preprints, and as yet, have not undergone peer review. As published versions of these studies are identified in the future, we will double‐check study descriptions, methods and findings, and update the review as required.
We also did not make within‐study comparisons between tests. Two studies (Adams 2020 [A]; Lassauniere 2020 [A]), evaluated panels of nine or 10 tests, nine other studies evaluated two, three, or five tests. As we could not identify tests in Adams 2020 [A], and the sample of Lassauniere 2020 [A] was very small, it is not possible from the studies available at this time to make direct comparisons between alternative tests.
We identified only one study that included comparison of test results with a reference standard of a neutralisation assay in studies identified for inclusion in this first version of the review (Thompson 2020), but we did not include these data in this version of the review. We are aware of several more studies of these assays in more recent publications and will include this as a new target condition in the next update of the review.
In such a current and fast moving field searches will always be out of date. However we are committed to ongoing updates of this living review
Applicability of findings to the review question
In the background we outlined four main roles for antibody testing that would be addressed in this review.
In diagnosis of infection in patients presenting with symptoms of suspected COVID‐19, particularly where molecular testing had failed to detect the virus. Most studies included in the review collected data from patients in the acute phase of disease in hospital settings and thus provide evidence to address this question amongst hospitalised patients. The review showed that antibody tests had very low sensitivity in the first week following onset of symptoms, but sensitivity rose in the second week, and only exceeded 90% in the third week. In addition we saw no difference in sensitivity of tests according to RT‐PCR status. We had no data to inform the accuracy of the test in primary care and community settings for the purpose of diagnosis, where patients are likely to have milder symptoms.
In assessment of immune response in patients with severe disease. We stated in the Background that we would not cover this in this review. In any case, we found no studies that directly addressed this question. Assessment of the accuracy of a test used for assessment of immune response would involve comparison with a reference standard test of antibody response, rather than evidence of infection.
To assess whether individuals have had a SARS‐CoV‐2 infection. As above, we found no studies that directly addressed this question, and very few studies were undertaken in community settings in patients who had not undergone RT‐PCR testing during their symptomatic period. Conclusions about the likely value of tests for this purpose rely on the sensitivity of the tests being no different in mild disease than in severe disease that requires hospital admission.
In seroprevalence surveys for public health management purposes. We also found no studies that directly addressed this question (although Bendavid 2020 is a seroprevalence study, it did not evaluate the accuracy of the test in the seroprevalence sample). High specificity of tests is essential in seroprevalence testing, which appears likely for many of the tests included in this review. However, the suitability of pre‐pandemic samples to establish specificity requires further discussion. We found no difference in specificity between pre‐pandemic and current non‐COVID‐19 samples, but lower specificity in those where COVID‐19 was ruled out after initially being suspected. This either reflects misclassification, or a true lower specificity in those presenting with symptoms. As sensitivity of the tests was mainly evaluated in hospitalised patients it is also unclear whether the tests have the ability to detect lower antibody levels likely in non‐hospitalised COVID‐19 patients.
Authors' conclusions
Implications for practice.
Diagnosis of acute suspected COVID‐19 in symptomatic patients
Based on this analysis, in patients presenting with symptoms of acute suspected COVID‐19, antibody tests have no role on their own as the primary test to use in the diagnosis of COVID‐19 when patients present during the first week since onset of symptoms, as their sensitivity is too low.
A small number of studies showed that the sensitivity of antibody tests is no different in those who were reverse transcription polymerase chain reaction (RT‐PCR)‐negative rather than RT‐PCR‐positive. Thus in hospitalised patients where molecular tests have failed to detect virus, antibody tests have an increasing likelihood of detecting immune response to the infection as time since onset of symptoms progresses.
There may therefore be a role in using antibody tests in COVID‐19 RT‐PCR‐negative but strongly suspected patients where patients are more than two weeks since the onset of symptoms. This is in line with the most recent version of the China CDC (National Health Commission of the People's Republic of China) COVID‐19 case definition (Appendix 2).
Assessment of previous SARS‐CoV‐2 infection and immune response
The data analysed in the review suggest that antibody tests are likely to have a useful role for detecting previous SARS‐CoV‐2 infection if used at 15 days or more after the onset of symptoms. This conclusion needs to be cautioned by the poor study quality, the small sample sizes and restricted number of tests that have undergone evaluation. In addition, we have scant data to inform the accuracy of the test in non‐hospitalised patients with milder disease, and too little data to comment on accuracy beyond 35 days.
Using, for illustration the overall IgG/IgM data at days 15 to 21 (sensitivity 91.4%, 95% CI 87.0 to 94.4 and specificity 98.7%, 93% CI 97.2 to 99.4), we have computed predictive values, and the numbers of true positives, false positives, false negatives and true negatives in a sample of 1000, at a prevalence of 50% (a value seen in healthcare worker populations who have suffered respiratory symptoms in the past months). In this scenario, the positive predictive value is estimated as 99% (95% CI 97 to 99), the negative predictive value as 92% (95% CI 88 to 95), and of 1000 people undergoing testing we would anticipate 7 (95% CI 3 to 14) false positives and 43 (95% CI 28 to 65) false negatives.
Please note that it is not certain whether a detectable immune response indicates that a patient is immune nor no longer infectious.
Seroprevalence surveys for public health management purposes
The duration of antibody rises is not yet known, and this review contains very little data beyond 35 days post‐onset of symptoms. In the 'Summary of findings' table we present scenarios for the likely numbers of missed cases (false negatives) and false positive cases for prevalences of 2%, 5%, (likely values in national surveys), 10% and 20% (likely values in high‐risk settings such as healthcare workers), presuming that the performance of an IgG/IgM test would continue at the same level as for 14‐21 days. Again this conclusion needs to be cautioned by the poor study quality, the applicability of the study settings, the small sample sizes and restricted number of tests that have undergone evaluation. At a prevalence of 20%, a possible value in surveys in high‐risk settings, 17 (95% CI 11 to 26) would be missed per 1000 people tested and 10 (95% CI 5 to 22) would be falsely positive. At a lower prevalence of 5%, a likely value in national surveys, 4 (95% CI 3 to 7) would be missed per 1000 tested, and 12 (95% CI 6 to 27) would be falsely positive.
Implications for research.
Many more high‐quality evaluation studies of COVID‐19 antibody tests are needed in patients more than 21 days post‐symptom onset, and in people in the community, particularly those who experience milder symptoms, or who are asymptomatic (but known to be infected).
Future studies must report data on sensitivity disaggregated by time since onset of symptoms. In future updates of this review we will not include studies for analysis of sensitivity where this has not been done. We would suggest that studies standardise how they define time since symptom onset (not, for example, using time since positive RT‐PCR results since this has no biological basis) and present results using standard time groupings (we suggest initially by week up until 35 days and larger time intervals beyond). Studies that sample from the same patients at several time points over time are needed to fully understand how time since symptom onset directly affects performance – our current estimates are based on collation of multiple cross‐sectional studies, which has limitations.
Primary studies need to be undertaken for the many tests that are on the market but as yet have no independent evaluations. Future studies should evaluate test performance in consecutive individuals who are recruited in clinical care with suspected COVID‐19, to estimate both sensitivity and specificity, as this will estimate the likely performance of the tests in practice.
COVID‐19‐positive cases who are RT‐PCR‐negative should be included as well as those confirmed RT‐PCR, in accordance with the World Health Organization (WHO) and China CDC case definitions.
Studies should ensure that the test is used as it is intended to be used in clinical practice (i.e. being undertaken at point‐of‐care rather than in laboratories (where appropriate) on the right specimens, by the intended healthcare worker). However, when validating people with suspected COVID‐19 who do not have a positive identification of COVID‐19 by RT‐PCR, these studies need to take care to confirm or rule out COVID‐19 by obtaining standardised evidence from other sources (e.g. repeat RT‐PCR, CT scans, follow‐up). Future studies need to recruit larger sample sizes and consider recruiting from multiple centres. We did not find any multicentre studies for this review.
We would also encourage investigators to utilise blinding in their study designs, such that index tests are undertaken without knowledge of the reference standard diagnosis, and likewise, reference standards are determined without knowledge of the index test findings.
We need good data upon which to compare tests. The strongest comparisons are made by testing the same participants multiple times with different tests. Whilst it is possible for this to be undertaken in prospective studies, it is easier to undertake in laboratory‐based studies utilising serum banks, which will compromise on the applicability of the absolute estimates of test accuracy, but provide some information about comparability.
From these studies we can only draw limited conclusions about cross‐reactivity of COVID‐19 tests with other coronaviruses as these data are summarised in analytical accuracy studies. It would be of value for these results to be reviewed as well as clinical accuracy studies.
Study reporting requires substantial improvement. The STARD checklist outlines standard requirements for the reporting of a test accuracy study, which study investigators should take note of when planning their study to ensure the relevant information is collected and reported. No study was found that reported data using a STARD participant flow‐diagram (Bossuyt 2015).
Due to the speed of new publications in this field, frequent updates of this review are required. Future updates will not include data on tests that are not (or not likely to become) commercially available (thus we will exclude all in‐house assays).
History
Review first published: Issue 6, 2020
Acknowledgements
Members of the Cochrane COVID‐19 Diagnostic Test Accuracy Review Group include:
the project team (Deeks JJ, Dinnes J, Takwoingi Y, Davenport C, Leeflang MMG, Spijker R, Hooft L, Van den Bruel A, McInnes MDF, Emperador D, Dittrich S);
-
the systematic review teams for each review:
Molecular, antigen, and antibody tests (Adriano A, Beese S, Dretzke J, Ferrante di Ruffano L, Harris I, Price M, Taylor‐Phillips S)
Signs and symptoms (Stuyf T, Domen J, Horn S)
Routine laboratory markers (Yang B, Langendam M, Ochodo E, Guleid F, Holtman G, Verbakel J, Wang J, Stegeman I)
Imaging tests (Salameh JP, McGrath TA, van der Pol CB, Frank RA, Prager R, Hare SS, Dennie C, Jenniskens K, Korevaar DA, Cohen JF, van de Wijgert J, Damen JAAG, Wang J);
the wider team of systematic reviewers from University of Birmingham, UK who assisted with title and abstract screening across the entire suite of reviews for the diagnosis of COVID‐19 (Agarwal R, Baldwin S, Berhane S, Herd C, Kristunas C, Quinn L, Scholefield B).
We thank Dr Jane Cunningham (World Health Organization) for participation in technical discussions and comments on the manuscript.
The editorial process for this review was managed by Cochrane's EMD Editorial Service in collaboration with Cochrane Infectious Diseases. We thank Helen Wakeford, Anne‐Marie Stephani and Deirdre Walshe for their comments and editorial management. We thank Liz Bickerdike for comments on the Abstract. We thank Robin Featherstone for comments on the search and Mike Brown and Paul Garner for sign‐off comments. We thank Denise Mitchell for her efforts in copy‐editing this review.
Thank you also to peer referees Danny Altmann, Jessica Watson, Jeannette Guarner and Carlos Del Rio, consumer referee Dee Duckworth Shneiderman and methodological referees Danielle van der Windt and Matthew Grainge, for their insights.
The editorial base of Cochrane Infectious Diseases is funded by UK aid from the UK Government for the benefit of low‐ and middle‐income countries (project number 300342‐104). The views expressed do not necessarily reflect the UK Government’s official policies.
Jonathan Deeks is a UK National Institute for Health Research (NIHR) Senior Investigator Emeritus. Yemisi Takwoingi is supported by a NIHR Postdoctoral Fellowship. Jonathan Deeks, Jacqueline Dinnes, Yemisi Takwoingi, Clare Davenport and Malcolm Price are supported by the NIHR Birmingham Biomedical Research Centre. This paper presents independent research supported by the NIHR Birmingham Biomedical Research Centre at the University Hospitals Birmingham NHS Foundation Trust and the University of Birmingham. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health and Social Care.
Appendices
Appendix 1. Summary of World Health Organization and Chinese National Health Commission Guidelines for the diagnosis of SARS‐CoV‐2
Table A: World Health Organization guidelines for the diagnosis of SARS‐CoV‐2a
Includes laboratory testing guidelines and global surveillance guidelines
| Date range (2020) | Definition of confirmed case | Definition of confirmed non‐case | Definition of suspect case | Definition of probable case | Role of serology in testing |
| 10‐30 January | 10‐30 January: no documentation to define at this time (before first date of global guidelines)
31 January onwards: a confirmed case is a person with laboratory confirmation of COVID‐19 infection, irrespective of clinical signs and symptoms. No prescribed test in laboratory guidelines, suggested tests from 10 January include broad coronavirus RT‐PCR (with sequencing of precise virus in test positives), whole genome sequencing, broad coronavirus serology on paired samples, microscopy, culture. (Lab 10 January) Four suggested tests from 17 January: broad coronavirus RT‐PCR (with sequencing of precise virus in test positives), NAAT for SARS‐CoV‐2 when it becomes available, whole genome sequencing, and broad coronavirus serology on paired samples. States that once specific NAAT assays are developed and validated, confirmation will be based on specific detection of unique sequences of viral nucleic acid by RT‐PCR. |
None stated | No definition of 'suspect case' at this time, but case definitions for surveillance are defined as a combination of symptoms and exposure, with more severe symptoms requiring less evidence for exposure | No definition at this time | Serological testing may be useful to confirm immunologic response to a pathogen from a specific viral group, e.g. coronavirus. Best results from serologic testing requires the collection of paired serum samples (in the acute and convalescent phase) from cases under investigation. |
| 31 January‐26 February | None stated | Suspect case defined as combination of symptoms and exposure, with more severe symptoms requiring less evidence for exposure | A suspect case with inconclusive laboratory results or is test‐positive using a pan‐coronavirus assay without laboratory evidence of other respiratory pathogens. (global 31 January) | ||
| 27 February‐1 March | None stated | Suspect case defined as combination of symptoms and exposure, with more severe symptoms requiring less evidence for exposure, OR defined by symptoms requiring hospitalisation and an absence of alternative explanation. | A suspected case with inconclusive laboratory results (global 27 February) | ||
| 2 March‐19 March | A person with laboratory confirmation of COVID‐19 infection, irrespective of clinical signs and symptoms. (global 31 January, 27 February, 20 March)
Laboratory confirmation of cases by NAAT specific to SAR‐CoV‐2 such as real‐time reverse‐transcription polymerase chain reaction (rRT‐PCR) with confirmation by nucleic acid sequencing when necessary. The viral genes targeted so far include the N, E, S and RdRP genes. In areas with no known COVID‐19 virus circulation confirmation requires:
Discordant results should be resampled. In areas where COVID‐19 virus is widely spread a simpler algorithm might be adopted (e.g. RT‐PCR of a single discriminatory target) |
One or more negative results do not rule out the possibility of COVID‐19 virus infection. | In cases where NAAT assays are negative and there is a strong epidemiological link to COVID‐19 infection, paired serum samples (in the acute and convalescent phase) could support diagnosis once validated serology tests are available. Serological assays will play an important role in research and surveillance but are not currently recommended for case detection. |
||
| 19 March‐present | Probable case A suspect case for whom testing for the COVID‐19 virus is inconclusive. OR A suspect case for whom testing could not be performed for any reason. | ||||
| NAAT: nucleic acids amplification test; RT‐PCR: reverse transcription polymerase chain reaction; Source: WHO 2020. | |||||
aSource data from Laboratory testing of 2019 novel coronavirus (2019‐nCoV) in suspected human cases: interim guidance, World Health Organization. (2020), 10 January, 17January, 2nd March, 19 March, 21st March, and Global surveillance for COVID‐19 caused by human infection with COVID‐19 virus, interim guidance, 31st January, 27 February, and 20 March.
Table B: Summary of Chinese National Health Commission guidelines for diagnosis and treatment for novel coronavirus pneumonia (trial versions 1‐7)
| Dates in effect | Definition of confirmed case | Definition of confirmed non‐case | Definition of suspect case | Role of serology in testing |
| 16‐17 January 2020 (version 1) | Cases (not confirmed cases) defined as virus genome highly homologous to coronaviruses | Not defined | Observation cases: defined as combination of exposure in Wuhan and symptoms focused on pneumonia, leukopenia and lack of improvement. | No role |
| 18 January‐2 March (versions 2, 3, 4, 5, 5 revised, and 6) | Suspect cases with either
|
Suspect cases can be ruled out after 2 consecutive negative respiratory tract nucleic acid tests taken at least 24‐hours apart. | Suspect cases: combination of exposure (such as residence in/travel to Wuhan or exposure to a confirmed case within 14 days of onset) AND clinical features (such as symptoms: fever, respiratory symptoms, and tests: chest imaging, white blood cell and lymphocyte count). Exact definition varies slightly with version | No role |
| 3 March‐present (version 7) | Suspect cases with either
|
Suspect cases can be ruled out after 2 negative NAATs, taken at least 24‐hours apart, and the NCP virus‐specific IgM and IgG are negative after 7 days from onset. | Suspect cases: combination of exposure (such as residence in/travel to Wuhan or exposure to a confirmed case within 14 days of onset) AND clinical features (such as symptoms: fever, respiratory symptoms, and tests: chest imaging, white blood cell and lymphocyte count). | Part of definition of cases and confirmed non‐cases |
| NAAT: nucleic acids amplification test; NCP: novel coronavirus pneumonia; RT‐PCR: reverse transcription polymerase chain reaction; Source: CDC China 2020. | ||||
Appendix 2. Antibody test 'use case' scenarios
| Use Casea | Advantages | Limitations | Considerations | |
| Diagnosis | ||||
| Aid diagnosis of suspect cases, especially when RT‐PCR negative but X‐Ray/CT suggestive | May improve overall sensitivity of diagnosis Diagnosis of patients presenting late or for post‐infectious syndromes (low viral load) Diagnosis of patients when lower respiratory tract sampling not available |
Unlikely to catch early‐stage infection (< 7 days) May not detect asymptomatic cases Negative test cannot rule out infection IgM appears early, but is less specific |
Total antibody may have best sensitivity Should be confirmed by PCR, where possible Rising titres and seroconversion can improve sensitivity and specificity |
|
|
Aid diagnosis of suspect cases when PCR is not available (would require careful development of interpretive guidelines) |
As above and could enable decentralised/community testing in settings where the availability of PCR testing is limited. | |||
| Identification of individuals with protective immune status (conditional upon identifying correlates of protection for SARS‐CoV‐2) | ||||
| Identify convalescent plasma donors | Treatment for critically ill patients | Ideal timing of collection unknown to optimise efficaciousness | Preferentially patients recovered from moderate to severe disease (high titre). Theoretically may be derived from vaccinated donors | |
| CT: computed tomography; RT‐PCR: reverse transcription polymerase chain reaction; | ||||
| aTable from Cheng 2020b | ||||
Appendix 3. Cochrane COVID‐19 Study Register searches
| Source | Strategy |
| CT.gov | COVID‐19a |
| WHO ICTRP | Health topic: 2019‐nCov / COVID‐19 |
| PubMed | (("2019 nCoV"[tiab] OR 2019nCoV[tiab] OR "2019 novel coronavirus"[tiab] OR "COVID 19"[tiab] OR COVID19[tiab] OR "new coronavirus"[tiab] OR "novel coronavirus"[tiab] OR "novel corona virus"[tiab] OR "SARS CoV‐2"[tiab] OR (Wuhan[tiab] AND (coronavirus[tiab] OR "corona virus"[tiab])) OR "COVID‐19"[Supplementary Concept] OR "severe acute respiratory syndrome coronavirus 2"[Supplementary Concept]) NOT ("animals"[MeSH Terms] NOT "humans"[MeSH Terms])) NOT (editorial[pt] OR comment[pt] OR letter[pt] OR newspaper article[pt]) |
aAutomatic term mapping links results for 2019‐nCoV, 2019 novel coronavirus, SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
Appendix 4. Living search from the University of Bern
The following information is taken from the university of Bern website (see: ispmbern.github.io/covid-19/living-review/collectingdata.html).
The register is updated daily and CSV file downloads are made available.
1 April 2020
From 1 April 2020, we will retrieve the curated BioRxiv/MedRxiv dataset (connect.medrxiv.org/relate/content/181).
26 to 31 March 2020
MEDLINE: (\"Wuhan coronavirus\" [Supplementary Concept] OR \"COVID‐19\" OR \"2019 ncov\"[tiab] OR ((\"novel coronavirus\"[tiab] OR \"new coronavirus\"[tiab]) AND (wuhan[tiab] OR 2019[tiab])) OR 2019‐nCoV[All Fields] OR (wuhan[tiab] AND coronavirus[tiab])))))
Embase: (nCoV or 2019‐nCoV or ((new or novel or wuhan) adj3 coronavirus) or covid19 or covid‐19 or SARS‐CoV‐2).mp.
BioRxiv/MedRxiv: ncov or corona or wuhan or COVID or SARS‐CoV‐2
With the kind support of the Public Health & Primary Care Library PHC (www.unibe.ch/university/services/university_library/faculty_libraries/medicine/public_health_amp_primary_care_library_phc/index_eng.html), and following guidance of the Medical Library Association (www.mlanet.org/p/cm/ld/fid=1713).
1 January 2020 to 25 March 2020
MEDLINE: ("Wuhan coronavirus" [Supplementary Concept] OR "COVID‐19" OR "2019 ncov"[tiab] OR (("novel coronavirus"[tiab] OR "new coronavirus"[tiab]) AND (wuhan[tiab] OR 2019[tiab])) OR 2019‐nCoV[All Fields] OR (wuhan[tiab] AND coronavirus[tiab])))))
Embase: ncov OR (wuhan AND corona) OR COVID
BioRxiv/MedRxiv: ncov or corona or wuhan or COVID
Appendix 5. CDC Library, COVID‐19 Research Articles Downloadable Database
Embase records from the Stephen B. Thacker CDC Library, Covid‐19 Research articles Downloadable database
Records were obtained by the CDC library by searching Embase through Ovid using the following search strategy.
| Source | Strategy |
| Embase | coronavir* OR corona virus* OR betacoronavir* OR covid19 OR covid 19 OR nCoV OR novel CoV OR CoV 2 OR CoV2 OR sarscov2 OR 2019nCoV OR wuhan virus*).mp. OR ((wuhan OR hubei OR huanan) AND (severe acute respiratory OR pneumonia*) AND outbreak*).mp. OR Coronavirus infection/ OR coronavirinae/ OR exp betacoronavirus/ Limits: 2020‐ OR (novel coronavir* OR novel corona virus* OR covid19 OR covid 19 OR nCoV OR novel CoV OR CoV 2 OR CoV2 OR sarscov2 OR 2019nCoV OR wuhan virus*).mp. OR ((wuhan OR hubei OR huanan) AND (severe acute respiratory OR pneumonia*) AND outbreak*).mp. OR ((wuhan OR hubei OR huanan) AND (coronavir* OR betacoronavir*)).mp. Limits: 2019‐ |
Appendix 6. Data extraction items
| Patient sampling items | Patient characteristics and setting items | Index test items | Reference standard items | Flow and timing items | Notes items |
| A0 Test type (antibody/antigen etc) | COVID patients (or all patients if single group study) | ||||
| A1 Purpose | B1 Setting | D1.1 Test name | E1 Reference standard for cases including threshold | F1 What was the time interval between index and reference tests? | G1: Funding |
| A2 Design (and description of groups labelled [1] [2] …) | B2 Location (include name of institution if available) | D1.2 Manufacturer | E2 Samples used | F2 Did all patients receive the same reference standard? | G2: Publication status |
| A3 Recruitment | B3 Country | D1.3 Antibody targets | E3 Timing of reference standard (preferably since symptom onset only, if not from a different time points) | F3 Missing data | G3: Source (preprint or journal name) |
| A4 Were cases recruited prospectively or retrospectively? | B4 Dates | D1.4 Antigens used | E4 Was it blind to index test? | F4 Uninterpretable results | G4: Study author COI (including any manufacturer affiliations) |
| A5 Sample size (virus/COVID cases) | B5 Symptoms and severity | D1.5 Point‐of‐care or laboratory (is the test designed to be used at point‐of‐care or in laboratory, and was it used as point‐of‐care or in laboratory)? | E5 Did it incorporate index test? | F5 Indeterminate results | G5 Comment |
| A6 Inclusion and exclusion criteria | B6 Demographics | D1.6 Test method | F6 Samples or patients | ||
| A7 Comment | B7 Exposure history | D1.7 When were samples taken (preferably since symptom onset only, if not from a different time points)? | E6 Reference standard for non‐cases | F7 Comment | |
| B8 Comment | D1.8 Samples used | E7 Samples used | |||
| Non‐COVID patients (if additional groups) | D1.9 Who applied the test | E8 Timing of reference standard (preferably since symptom onset only, if not from a different time points) | |||
| C1.1 Group name | D1.10 How was positive defined? | E9 Was it blind to index test? | |||
| C1.2 Source and time | D1.11 Blinded to reference standard | E10 Did it incorporate index test? | |||
| C1.3 Characteristics | D1.12 Threshold predefined | E11 Comment | |||
| C2.1 Group name | D1.13 Comment | ||||
| C2.2 Source and time | |||||
| C2.3 Characteristics | |||||
| C4 Comment |
Appendix 7. Criteria for assessment of study quality (QUADAS‐2)
| DOMAIN: PARTICIPANT SELECTION | |
| Was a consecutive or random sample of patients enrolled? | This will be similar for all index tests, target conditions, and populations. YES: if a study explicitly stated that all participants within a certain time frame were included; that this was done consecutively; or that a random selection was done. NO: if it was clear that a different selection procedure was employed; for example, selection based on clinician's preference, or based on institutions. UNCLEAR: if the selection procedure was not clear or not reported. |
| Was a case‐control design avoided? | This will be similar for all index tests, target conditions, and populations. YES: if a study explicitly stated that all participants came from the same group of (suspected) patients. NO: if it was clear that a different selection procedure was employed for the participants depending on their COVID‐19 status or SARS‐CoV‐2 infection status; or if only participants with SARS‐CoV‐2 infection were included UNCLEAR: if the selection procedure was not clear or not reported. |
| Did the study avoid inappropriate exclusions? | Studies may have excluded patients, or selected patients in such a way that they avoided including those who were difficult to diagnose or likely to be borderline. Although the inclusion and exclusion criteria will be different for the different index tests, inappropriate exclusions and inclusions will be similar for all index tests: for example, only elderly patients excluded, or children (as sampling may be more difficult). This needs to be addressed on a case‐to‐case basis. YES: if a high proportion of eligible patients was included without clear selection. NO: if a high proportion of eligible patients was excluded without providing a reason; if, in a retrospective study, participants without index test or reference standard results were excluded. UNCLEAR: if the exclusion criteria were not reported. |
| Did the study avoid inappropriate inclusions? | Some laboratory studies may have intentionally included groups of patients in whom the accuracy was likely to differ, such as those with particularly low or high viral loads, or who had other diseases, such that the sample over‐represented these groups. This needs to be addressed on a case‐to‐case basis. Artificial spiked samples are a clear example. YES: if samples included were likely to be representative of the spectrum of disease. NO: if the study oversampled patients with particular characteristics likely to affect estimates of accuracy. UNCLEAR: if the exclusion criteria were not reported. |
| Could the selection of patients have introduced bias? | HIGH: if one or more signalling questions were answered with NO, as any deviation from the selection process may lead to bias. LOW: if all signalling questions were answered with YES. UNCLEAR: all other instances. |
| Is there concern that the included participants do not match the review question? | HIGH: for two‐group studies that included healthy or other disease controls, whether pre‐pandemic or contemporaneous; studies that only included people with COVID‐19 (whether reverse transcription polymerase chain reaction (RT‐PCR)‐confirmed only, participants meeting official guideline criteria); LOW: for single‐group studies recruiting participants with signs and symptoms of COVID‐19; or for two‐group studies where control groups suspected of COVID‐19 were separately recruited. UNCLEAR: if a description about the participants was lacking. |
| DOMAIN: INDEX TESTS | |
| Were the index test results interpreted without knowledge of the results of the reference standard? | YES: if blinding was explicitly stated or index test was recorded before the results from the reference standard were available. NO: if it was explicitly stated that the index test results were interpreted with knowledge of the results of the reference standard. UNCLEAR: if blinding was unclearly reported. |
| If a threshold was used, was it prespecified? | YES: if the test was dichotomous by nature, or if the threshold was stated in the methods section, or if study authors stated that the threshold as recommended by the manufacturer was used. NO: if a receiver operating characteristic curve was drawn or multiple threshold reported in the results section; and the final result was based on one of these thresholds. UNCLEAR: if threshold selection was not clearly reported. |
| Could the conduct or interpretation of the index test have introduced bias? | HIGH: if one or more signalling questions were answered with NO, as even in a laboratory situation knowledge of the reference standard may lead to bias. LOW: if all signalling questions were answered with YES. UNCLEAR: all other instances. |
| Is there concern that the index test, its conduct, or interpretation differ from the review question? | For evaluations of laboratory‐based tests, HIGH: if tests were built in‐house, or if commercially available tests using SARS‐Cov antigens instead of SARS‐CoV‐2‐specific antigens. LOW: most other laboratory‐evaluations UNCLEAR: name of the test was withheld For evaluations of lateral flow assays, HIGH: if tests were built in‐house; if only serum or plasma instead of fingerprick or whole blood samples were used; if test evaluated in laboratory settings rather than at the point of care LOW: commercially available tests, using whole blood or fingerprick samples, and that were conducted in the intended setting for the test (i.e. point‐of‐care). UNCLEAR: name of the test was withheld; mixed sample types; or did not report the evaluation setting |
| DOMAIN: REFERENCE STANDARD | |
| Is the reference standard likely to correctly classify the target condition? | We will define acceptable reference standards using a consensus process once the list of reference standards that have been used has been obtained from the eligible studies. For COVID‐19 cases YES: RT‐PCR; confirmed or suspected case using official criteria (WHO, CDC) or a clearly set out combination of signs/symptoms/exposure. NO: RT‐PCR not used, or if inadequate combination of clinical characteristics used in PCR negatives, e.g. computed tomography alone UNCLEAR: if definition of COVID‐19 was not reported For absence of COVID‐19 YES: if at least 2 negative RT‐PCR results reported if suspected COVID‐19 based on signs/symptoms; single negative RT‐PCR test for asymptomatic contacts or contemporaneous controls with no clinical suspicion of COVID‐19; only pre‐pandemic sources of control samples used. NO: single RT‐PCR or number of negative RT‐PCRs not reported for COVID‐19 suspects; no RT‐PCR reported (untested) for asymptomatic contacts or contemporaneous controls UNCLEAR: if timing of control samples (pre‐pandemic or contemporaneous) was not reported |
| Were the reference standard results interpreted without knowledge of the results of the index test? | YES: if it was explicitly stated that the reference standard results were interpreted without knowledge of the results of the index test, or if the result of the index test was obtained after the reference standard. NO: if it was explicitly stated that the reference standard results were interpreted with knowledge of the results of the index test or if the index test was used to make the final diagnosis. UNCLEAR: if blinding was unclearly reported. |
| Did the definition of the reference standard incorporate results from the index test(s)? | YES: if results from the index test were a component of the reference standard definition. NO: if the reference standard did not incorporate the index standard test. UNCLEAR: if it was unclear whether the results of the index test formed part of the reference standard. |
| Could the conduct or interpretation of the reference standard have introduced bias? | HIGH: if one or more signalling questions were answered with NO. LOW: if all signalling questions were answered with YES. UNCLEAR: all other instances. |
| Is there concern that the target condition as defined by the reference standard does not match the review question? | Applicability was judged primarily on the definition of disease‐positive. HIGH: if RT‐PCR alone used to define cases LOW: if clinical criteria, including RT‐PCR, were used to define cases, regardless of whether official criteria were used, as long as the criteria were explicitly described. UNCLEAR: if definition of COVID‐19 cases was not provided, including if some clinically diagnosed cases were included but the clinical criteria used were not described. |
| DOMAIN: FLOW AND TIMING | |
| Did all participants receive the same reference standard? | YES: if all participants received the same reference standard (clearly no differential verification). NO: if (part of) the index test‐positives or index test‐negatives received a different reference standard. UNCLEAR: if it was not reported. |
| Were all participants included in the analysis? | YES: if it is clear that all eligible participants were included in the analyses. NO: if after the inclusion/exclusion process, participants were removed from the analyses for different reasons: no reference standard done, no index test done, intermediate results of both index test or reference standard, indeterminate results of both index test or reference standard, samples unusable. UNCLEAR: if it is not possible to determine whether all participants were included (e.g. from a STARD style participant flow diagram) |
| Did all participants receive a reference standard? | YES: if all participants received a reference standard (clearly no partial verification). NO: if only (part of) the index test positives or index test negatives received the complete reference standard. UNCLEAR: if it was not reported. |
| Were results presented per participant? | YES: if either only one sample per participant (regardless of disaggregation of results over time), or if multiple samples per participant but results are disaggregated by time period (at least week by week) NO: if multiple samples per participant and results are not disaggregated by time period UNCLEAR: if it is not possible to tell whether results presented are per participant or per sample |
| Could the participantflow have introduced bias? | HIGH: if one or more signalling questions were answered with NO. LOW: if all signalling questions were answered with YES. UNCLEAR: all other instances. |
| CDC: Centers for Disease Control; ICU: intensive care unit; RT‐PCR: real‐time polymerase chain reaction; SARS‐CoV‐2: severe acute respiratory syndrome coronavirus 2; WHO: World Health Organization | |
Footnotes
Appendix 8. Summary details of study design and participants
This Appendix includes two tables. Please note that square brackets indicate different tests within one study.
Table A. Single‐group studies estimating sensitivity (and specificity)
Table B. Two‐group or more studies estimating sensitivity and specificity
Table A. Single‐group studies estimating sensitivity (and specificity)
| Study (source) |
Inclusion criteria
|
Instititution (recruitment dates) | Age (median) n, % male | Exposure history Symptoms/severity | Reference details (cases) | Missing or uninterpretable data |
| Single‐group studies estimating sensitivity and specificity | ||||||
|
Cassaniti 2020 (B) (published; letter) 50 participants (50 samples) |
COVID‐19‐suspected cases presenting at A&E with fever and respiratory syndrome (n = 50, including 38 RT‐PCR‐positive);
(Additional groups reported in Cassaniti 2020 (A)) |
A&E; Pavia, Italy (not stated) | 61.5 years 34, 68% |
Not stated Not stated |
RT‐PCR detecting RNA polymerase and E genes; nasal swab (On presentation at A&E) |
Weakly positive results counted as test positive |
|
Liu 2020a (preprint) 179 participants (179 samples) |
Inpatients and outpatients attending hospital during pandemic including COVID‐19‐suspected cases (all inpatient, n = 114) and outpatients (n = 64) with 'other disease'. (n = 179, including 90 PCR‐confirmed and 5 clinically confirmed cases)
|
Inpatient and outpatient; Wuhan, China (1 January‐12 March 2020) | [1] mean 76 years [2+3] mean 56 years [1] 60, 67% [2+3] 38, 43% |
Not reported Of 90 RT‐PCR+, 44, 49% severe/critical cases |
Clinical criteria (not clearly described) ≤ RT‐PCR; nasal and pharyngeal swabs (NR) |
Per sample data by time period is based on |
|
Long 2020 (A) (preprint) 164 participants (164 samples) |
Cohort of close contacts of 2 index cases (n = 164, 16 PCR‐positive cases)
|
Close contacts; Wanzhou, China (31 January‐9 February) | Not stated Not stated |
All exposed 151 (92%) asymptomatic |
RT‐PCR; NP (within 17 days of contact with confirmed cases) |
None stated |
|
Paradiso 2020a (preprint) 191 participants (191 samples) |
Symptomatic patients accessing A&E (n = 191, including 70 PCR‐positive)
|
A&E; Bari, Italy (23‐29 March) | 58.5 years 116, 61% |
Not stated 14, 9% asymptomatic |
RT‐PCR (Allplex 2019‐nCoV Assay; Seegene, Seoul, Republic of Korea); NP, OP (Simultaneous) |
1 D+ missing from results |
|
Zhang 2020b (preprint) 228 participants (228 samples) |
Suspected COVID‐19 cases admitted to fever clinic (n = 228, including 3 PCR‐positive)
(review team excluded additional reported groups) |
Inpatients, Shengjing, China (21 January‐16 February) | Mean 51 years (cases only) Not stated |
1, 33% Wuhan contact history (cases only) Not stated |
RT‐PCR (required presence of ORF1ab and N gene for positive result); NP, OP (Timing not stated) |
Not stated |
|
Zhang 2020d (preprint) 814 participants (814 samples) |
Participants suspected of harbouring COVID‐19 (n = 814, including 154 cases; 122 RT‐PCR‐positive and 32 clinically diagnosed by CT)
|
Samples from 5 hospitals (in/outpatient not stated); centres including Wuhan, Shenyang and Beijing, China (Not stated) | Not stated | Not stated | Real‐time PCR kit (no details on threshold); CT used in at least some PCR‐negative NP swabs. (Timing not stated) | None stated |
| Single group studies estimating sensitivity alone | ||||||
|
Du 2020 (published; letter) 60 participants (60 samples) |
Single group of convalescent inpatients 6‐7 weeks after symptom onset (n = 60)
|
Hospital inpatients; Wuhan, China (12 January‐5 February 2020) | Not stated | Not stated | Not described; not stated (During hospital stay) |
None described |
|
Gao 2020a (published; letter) 38 participants (38 samples) |
Inpatient cohort of COVID‐19 patients confirmed by Chinese Government‐issued guidelines (5th edition)
|
Inpatient; Fuyang, China (22 January‐28 February 2020) | 40.5 years 21, 55.3% |
Not stated 3, 8% severe or critical, 35 mild |
Chinese guideline (5th edition) | Not reported |
|
Gao 2020b [A] (accepted manuscript (peer reviewed; pre‐proof) 22 participants (37 samples) |
Confirmed COVID‐19 cases (n = 22)
|
Hospital inpatient; Shijiazhuang, Hebei, China (21 January‐24 February 2020) | 40 years 14, 64% |
11 (50%) recent travel to epidemic areas, 10 (45%) close contacts with confirmed COVID‐19 cases 22 (100%) typical CT findings; 'most' received oxygen therapy |
RT‐PCR assay (2019‐nCoV RNA Test Kit, Daan Gene Company, China); Nasal and pharyngeal swab specimens | None described |
|
Garcia 2020 (B) (preprint) 63 participants (63 samples) |
Patients admitted with a clinical and radiological diagnosis of pneumonia of unknown aetiology but RT‐PCR‐negative (n = 63)
Addional reported cohort extracted as Garcia 2020 (A) |
Inpatient hospital (9 February‐2 April) | 67 years 47, 74% |
Not stated | Clinical diagnosis of COVID‐19 (no further detail); all PCR‐negative | None reported |
|
Hu 2020a (preprint) 211 participants (993 samples) |
Confirmed COVID‐19 patients (221)
|
Inpatient; Chongqing, China (23 January‐3 March) | Mean 47.8 years 135, 64% |
Not stated 137, 62% with fever; 40, 19% severe |
Chinese Government guidelines (version 6); included RT‐PCR | None described; text states 993 samples but only 409 reported for IgM and 507 for IgG |
|
Jia 2020 (preprint) 57 participants (57 samples) |
1. RT‐PCR confirmed (24)
2. Clinical diagnosis for RT‐PCR negative (2 x negative results) according to Chinese Government guideline (6th ed) (33)
|
Inpatients; Shenzhen, China (Not stated) | Not stated | Not stated | 1. RT‐PCR 2. Chinese CDC guideline (6th ed). (1 to 34 days from exposure to first PCR test) | None described |
|
Li 2020a (accepted for publication and undergone full peer review) 525 participants (525 samples) |
COVID‐19 according to Chinese CDC guideline (5th ed) (525; 397 PCR‐positive))
|
Potentially inpatient and outpatient; 6 provinces, China (Not stated) |
Not stated | Not stated | Chinese CDC guideline (6th ed), including PCR; pharyngeal, sputum (Not stated) |
None stated |
|
Lippi 2020 [A] (published; letter) 48 participants (48 samples) |
Participants with suspected COVID‐19; subgroup of cases (48/131 patients) with available data on days post‐symptom onset data can be included
|
Inpatients; Verona, Italy (Not stated) | Totlal sample of 131: mean 56 years 60/131, 46% |
Not stated | RT‐PCR (Seegene Allplex 2019‐nCoV Assay. OP, NP swabs (During hospitalisation) | Excluded 83 patients with no time pso data |
|
Liu 2020c (preprint) 133 participants (133 samples) |
Patients diagnosed with SARS‐Cov‐2 according to Chinese CDC guideline (5th ed) (133)
|
Inpatients; Wuhan, China ( 17 February‐1 March) | Moderate 67.5 years; severe 68 years; critical 70 years 70, 53% |
Not stated moderate 44, 33%; severe 52, 39%; critical 37, 29% |
Clinical diagnosis (seems to be Chinese CDC guideline, 5th ed) Includes RT‐PCR (GeneoDx Biotech, Shanghai, China); 2 tests per participant; Table 2 refers to NP. | None described |
|
Long 2020 (B) (preprint) 262 participants (363 samples) |
RT‐PCR‐positive confirmed cases (n = 285). No further detail of inclusion or exclusion criteria.
Additional cohort extracted as Long 2020 (A); some additional cohorts excluded (see Characteristics of included studies)
|
Inpatients; Chongqing, China (38384) | 47 years 158, 55.4% |
103, 36% exposure to transmission sources 39, 14% severe or critical in ICU |
RT‐PCR; nasal and pharyngeal swabs (during hospital stay) | 23 patients with no information on time pso were excluded leaving 363 samples from 262 patients |
|
Padoan 2020 (peer reviewed; published) 37 participants (87 samples) |
Hospitalised patients with confirmed COVID‐19 (n = 37)
|
Inpatients; Padova, Italy (18 March‐26 March 2020) | Not stated | Not stated | RT‐PCR; NP (Not stated) |
None described |
|
Pan 2020a (peer reviewed; published) 105 participants (134 samples) |
COVID‐19 patients according to CDC guideline (5th ed); confirmed by PCR (67) or clinical diagnosis (37)
|
Inpatients; Wuhan, China (Not stated (symptom onset 7 January‐18 February)) | 58 years 48, 46% |
Not stated | RT‐PCR following WHO guidelines (BioGerm, Shanghai, China),
Clinical diagnosis according to CDC guideline (5th ed); throat swabs (Not stated) |
Data reported only for those with symptom onset information; 26 samples excluded |
|
To 2020a [A] (peer reviewed; published) 23 participants (108 serum samples) |
Confirmed COVID‐19 patients from 2 hospitals (n = 23, can only extract data for 16 with > 14‐day pso data)
|
Hospital inpatient, Hong Kong (22 January‐12 February) | Not stated 13/23 (57%) age: median 62 years (range 37–75) |
Not stated 10/23 (43%) severe; 5/23(22%) admitted to ICU, 3/23(13%) required intubation, 2/23(9%) died Fever in 22/23 (96%) patients, cough in 5/23 (22%), chills in 4/23 (17%), dyspnoea in 4/23 (17%) |
Laboratory‐confirmed ‐ not further described; NP or sputum (Unclear) |
7/23 (30%) were not tested between days 14 and 30 |
|
Xiao 2020a (accepted manuscript; pre‐proof) 34 participants (34 samples) |
Confirmed cases of COVID‐19 according to Chinese CDC (5th ed) (34)
|
Inpatients; Wuhan, China (1‐29 February) | 49 years (review team estimated) 22, 65% |
Not stated Not described |
COVID‐19 according to CDC diagnosis and treatment guideline (5th ed) | None reported |
|
Xie 2020a (accepted manuscript; pre‐proof) 56 participants (56 samples) |
Participants with suspected COVID‐19 based on Chinese CDC (5th ed) criteria (n = 56, including 16 PCR confirmed)
|
Inpatients; Wuhan, China (15‐25 February 2020) | 56.5 years 24, 43% |
Not stated 34, 61% severe |
[1] RT‐PCR QIAamp RNA virus kit (Qiagen, Heiden, Germany); NP and throat [2] clinical diagnosis (guideline, 5th edition | None reported |
|
Xu 2020a (preprint) 10 participants (10 samples) |
Confirmed (PCR) COVID‐19 cases (n = 10)
|
Hospital inpatients; Shanghai, China (Not stated) |
Not stated 6, 60% |
Not stated 10, 100% required oxygen |
RT‐PCR (cycle threshold value (Ct) < 37 defined as positive and Ct ≥ 40 defined as negative; pharyngeal swab (Not stated) |
None reported |
|
Yongchen 2020 (peer reviewed; published) 21 participants (≥ 42 samples) |
Participants with COVID‐19 (n = 16) and asymptomatic carriers (n = 5)
|
Mixed; Jiangsu, China (25 January‐18 March 2020) | 37 years 13, 62% |
Not stated 5, 24% severe; 5, 24% asymptomatic cases Illness severity defined according to the Chinese management guideline for COVID‐19 (version 6.0). |
RT‐PCR, confirmed after 2 sequential positive respiratory tract sample results; throat swabs | None described |
|
Zhang 2020a (preprint) 222 participants (222 samples) |
Confirmed COVID‐19 patients (RT‐PCR detection or antibody assay) (n = 222)
|
Inpatients; Wuhan, China (admitted 13 January 13‐1 March) | 62 years Not stated |
Not stated 87, 39% severe |
RT‐PCR or anti‐SARS‐CoV‐2 assay ; nasal or pharyngeal swabs (Not stated) |
None reported |
|
Zhang 2020c (peer reviewed; published) 16 participants (16 samples) |
RT‐PCR‐confirmed COVID‐19 patients (n = 139); included those with around 10 days of medical treatment after admission (n = 16)
|
Inpatients; Wuhan, China (Not stated) | Not stated | Not stated | RT‐PCR | < 10 days' medical treatment (n = 123) |
| A&E: Accident and Emergency Department; CDC: Center for Disease Control; CT: computed tomography; CGIA: colloidal gold immunoassay; D+: disease positive: D‐: disease negative; ed: edition; ELISA: enzyme‐linked immunosorbent assay; HCW: healthcare worker; ICU: intensive care unit; LFA: lateral flow assay; n: number; NP: nasopharyngeal; NR: not reported; OP: oropharyngeal; PCR: polymerase chain reaction; pso: post‐symptom onset; RNA: ribonucleic acid; RT‐PCR: reverse transcriptase polymerase chain reaction; SARS‐CoV‐2: severe acute respiratory syndrome coronavirus 2; suppl: supplementary; TB: tuberculosis | ||||||
Table B. Two‐group studies or more estimating sensitivity and specificity
| Study (source) | COVID‐19 cases (n) | Non‐COVID cases (n) (including method of verification) | Institution (Recruitment dates) | Age (median) n, % male | Exposure history Symptoms/severity | Reference details (cases) | Missing or uninterpretable data |
|
Adams 2020 [A] (preprint) 182 participants (40 samples) |
RT‐PCR confirmed COVID‐19 cases (n = 40) | Pre‐pandemic controls (n = 142); prior to December 2019 | Acute hospital (n = 16), recovering HCWs (n = 6), convalescent (n = 18); UK (Not stated) |
57y Not stated |
Not stated Asymptomatic (n = 1); mild (n = 26); severe (n = 4); critical (n = 9) |
RT‐PCR; nose/throat swabs (Not stated ) |
[B]–[J] tests evaluated in different numbers |
|
Bendavid 2020 (preprint) 3481 samples participants (3324 samples) |
COVID‐19 cases were obtained from 3 different sources (n = 157 specimens) Confirmed cases from manufacturer data (n = 85); local cases (PCR‐ and ELISA‐confirmed) (n = 37) or PCR‐confirmed (6‐10 days pso) (n = 35) |
Non‐COVID‐19 cases were obtained from 13 different sources (n = 3324 specimens), Pre‐pandemic (10 sources; n = 2811); pandemic era PCR‐negative (n = 202); not stated (n = 311) |
Multiple sources; USA, China, unclear (Not described) |
Not stated | Not stated | PCR‐positive only (n = 35); PCR and IgG or IgM confirmed by ELISA (n = 37); not stated (n = 85) | None described |
|
Burbelo 2020 [A] (preprint) 67 participants (140 samples) |
SARS‐CoV‐2 cases confirmed by PCR (n = 35 in results, n = 39 in methods) | Pre‐pandemic blood donors (n = 32); prior to 2018 [Review authors excluded 3rd group with no reference standard reported (n = 10)] | Hospital (unclear whether inpatient or outpatient); San Diego, Seattle, Washington, USA (Not stated) |
44 years 0.87 |
Not stated 13, 37% on a ventilator |
RT‐PCR; nasal and/or throat swabs (No information) |
none described |
|
Cai 2020 (preprint) 443 participants (443 samples) |
RT‐PCR confirmed (276) | Healthy, other controls; pre‐December 2019 (167) | Inpatient (cases only); Chongqing, China (Not stated) | 48 years 151, 55% |
99, 36% known exposure | RT‐PCR; no further details | None described |
|
Cassaniti 2020 (A) (published; letter) 60 participants (60 samples) |
COVID‐19‐positive patients in ICU (n = 30) [Additional cohort reported in Cassaniti 2020 (B)] | Healthy volunteers with negative RT‐PCR results (n = 30) | Infectious Diseases Unit or ICU, Tertiary hospital; Pavia, Italy (Not stated) |
73.5 years 25, 42% |
Not stated | RT‐PCR detecting RNA polymerase and E genes; Respiratory samples (During patient care) |
Weakly positive results counted as test positive |
|
Chen 2020a (Accepted manuscript; peer reviewed, pre‐proof) 19 participants (19 samples) |
RT–PCR positive samples (n = 7) | RT–PCR‐negative samples, but clinically suspicious for COVID‐19 (n = 12) [Additional group of 'normal' samples (n = 51) used to derive threshold] | Unclear, presumably inpatients; Guangzhou, China (Not described) | Not stated | Not stated | RT–PCR; not stated (Not stated; prior to LFA) |
None reported |
|
Dohla 2020 (peer reviewed; published) 49 participants (49 samples) |
[1] Attendees at community screening centre for COVID‐19 (n = 12 PCR‐positive), [2] Stored samples from 10 patients with confirmed diagnosis of COVID‐19 | [1] Attendees at community screening centre for COVID‐19 (n = 27 PCR‐negative) | Community screening centre [1] and unclear setting [2]; author institutions Bonn, Germany (Not stated) |
46 years 25, 51% |
Probable date of exposure identified in 22, 45% 5/49 (10%) asymptomatic. 71% dry cough; 65% fatigue; 46% runny nose (only %s reported) |
[1] RT–qPCR (Altona Diagnostics) [2] RT–qPCR (unknown if same kit); throat swabs group [1]; not stated for group [2]. ([1] same time as index test. [2] not stated) | Weak signals counted as positive; no missing data reported |
|
Freeman 2020 (preprint) 618 participants (618 samples) |
Confirmed COVID‐19 cases (n = 99) | Pre‐pandemic healthy (n = 377) + other infection (n = 142) | Convalescent; USA (reference to CDC National Center for Immunization and Respiratory Diseases) (Not stated) |
Not stated | Not stated | PCR; no further detail | None mentioned |
|
Garcia 2020 (A) (preprint) 100 participants (100 samples) |
Suspected COVID‐19 patients admitted to A&E; all RT‐PCR‐positive (n = 55) [Third cohort reported as Garcia 2020 (B)] | Pre‐pandemic healthy controls (n = 45); 1 October‐30 November, 2019 | Inpatient; Madrid, Spain (1 March‐6 April 2020) | 63 years 33, 60% |
Not stated | RT‐PCR | None described |
|
Grzelak 2020 [A] (preprint) 542 participants (652 samples) |
Hospitalized COVID‐19 patients (51) Review team excluded 2 additional cohorts with no reference standard (See COIS) | Pre‐pandemic sera from healthy individuals (491) | Inpatient; Paris, France (Not stated) |
Not described 47, 74% |
Not stated | Not stated | None reported |
|
Guo 2020a (accepted manuscript; corrected proof now available online) 275 participants (343 samples) |
Confirmed (82) or probable (58) COVID‐19 cases (provided 208 samples) | Pre‐pandemic acute lower respiratory tract infection (135) Healthy individuals (150) used to define threshold | Inpatients; Wuhan and Beijing, China (cases) (43,831) | Not stated | Not stated Confirmed cases ‐ 28, 34% severe Pobable cases ‐ 5, 9% severe |
Confirmed: deep sequencing or qPCR assay
Probable: cases ‐ clinical manifestation, chest X‐ray and epidemiology but no virus detected by deep sequencing or qPCR; throat (Not stated) |
None reported |
|
Infantino 2020 (accepted publication; peer reviewed; pre‐proof) 125 participants (125 samples) |
[1] confirmed COVID‐19 cases (n = 61) | [2] pre‐pandemic (2018‐19) control group with rheumatic and infectious diseases (n = 44) [3] blood donors (winter 2019) (n = 20) | Inpatient; Florence, Italy (Not stated) |
mean 59 years 26, 43% |
Not stated 30, 49% mild to moderate symptoms 31, 51% severe pneumonia requiring admission to ICU |
RT‐PCR (2 positive results required for confirmation); OP and NP swabs (Not stated) |
None reported |
|
Jin 2020 (peer reviewed; published) 76 participants (98 samples from 43 cases samples) |
Laboratory confirmed COVID‐19 patients (n = 43) [Review team excluded results for 34 participants after becoming PCR‐negative] |
COVID‐19 suspects, discharged with 2 x RT‐PCR‐negative results with an interval of 24 h and who quarantined at home (n = 33) | Hospital inpatients; Hangzhou, China (January to 4 Mar 2020) | 47 years 17, 40% |
Not stated [1] COVID‐19 patients: 27 (63%) fever; 26 (61%) cough [2] Non‐COVID‐19 patients: 24/43 (73%) fever; 15/33 (46%) cough |
RT‐PCR; oral swab or sputum specimens (During hospital stay) |
No data reported for 16 patients while PCR positive. |
|
Lassauniere 2020 [A] (preprint) 112 participants (112 samples) |
COVID‐19 PCR‐positive patients (n = 30) admitted to intensive care | Pre‐pandemic (n = 82) including blood donors (n = 10) and other infections (n = 72) | Intensive care; Hillerod, Denmark (Not stated) | Not stated | Not stated | Viral nucleic acid detection (no further detail) | Borderline results for tests [B] and [C] were considered test negative; for POC tests weak signals for IgM and IgG were considered positive. Some samples not tested with all assays. |
|
Lin 2020a [A] (preprint) 159 participants (159 samples) |
[1] Suspected COVID‐19 cases (epidemiological risk, clinical features and RT‐PCR respiratory specimen positive) from inpatient setting (specialised COVID‐19 hospital) (n = 79) | RT‐PCR negative controls (reportedly at least 3 x negative), including: [2] healthy volunteers; timing not reported, presumed contemporaneous (n = 29) [3] TB patients; timing not reported, presumed contemporaneous (n = 51) | Inpatients (specialised COVID hospital); Shenzhen, China (Not stated) | Not stated | Not stated | Epidemiological risk, clinical features and RT‐PCR respiratory specimen positive' 'GeneoDX kit (Taqman RT‐PCR method) | Only 65/79 D+ and 64/80 D‐ serum samples available for ELISA; reason not given. |
|
Liu 2020b (preprint) 358 participants (358 samples) |
Confirmed (153) or suspected (85) COVID‐19 | Ordinary patients (70) and randomly sampled healthy blood donors (50); timing not reported, presumed to be contemporaneous | Inpatients; Hubei, China (6 ‐14 February) | 55 years 138, 58% |
Not stated Fever (87%); dry cough (54%); fatigue (33%). 235/238 (99%) had CT ground glass opacity/patchy shadowing |
RT‐PCR (Daan Gene) targeting ORF1ab and N gene (≤ 40 Ct); Clinical diagnosis according to Chinese Government‐issued guideline (5th ed); pharyngeal swabs RT‐PCR sampling throughout inpatient stay | None reported |
|
Liu 2020d [A] (evaluation; accepted manuscript) 314 participants (314 samples) |
RT‐PCR‐confirmed COVID‐19 cases (n = 214) | Healthy blood donors, presumed to be contemporaneous (n = 100) | Inpatient, Hubei, China (18 January‐26 February) | Not stated | Not stated | RT‐PCR; pharyngeal swabs. Median 15 days pso (range 0–55 days) | None described |
|
Lou 2020 [A] (preprint) 380 participants (380 samples) |
Confirmed COVID‐19 cases according to Chinese Government‐issued guidelines (6th edition) (n = 80) | Healthy people enrolled from the community, presumed contemporaneous selection (n = 300) | inpatient; Hangzhou, China (19 January‐9 February 2020) | Mean 55 years 0.61 |
26, 33% critical | CDC guideline (6th ed); criteria described including PCR; deep sputum samples' (On admission) |
Not all control group participants were tested by all index tests (range 100‐300/300) |
|
Ma 2020a (preprint) 570 participants (216 samples from 87 cases samples) |
Confirmed (PCR‐positive) COVID‐19 patients (n = 87) | [2] Pre‐pandemic healthy donors (n = 330) [3] Contemporaneous 'other diseases' (no mention of PCR) (n = 138) [4] Suspected COVID pneumonia but negative PCR (n = 15) | Inpatient; Hefei, China (26 January‐5 March 2020) | Not stated | Not stated 56, 67% clinically moderate 17 severe 5 critical "few mild" [page 7] |
Chinese Government‐issued guidelines (7th edition) including RT‐qPCR; serum (During hospital admission as part of "routine clinical testing". Performed before index test) |
For comparison of sensitivity and specificity of 2 antigens only 20/total of 479 control sera were used (20/138 from 'other disease' group) |
| Okba 2020c (accepted manuscript; early release) 54 participants (76 samples) |
RT‐PCR‐confirmed SARS‐CoV‐2 cases (n = 9, 31 samples) | Contemporaneous healthy blood donors (n = 45) | Inpatient; Munich, Germany (occurred after 23 January, discovered (presume PCR‐positive) on 27 January 27) | Not stated | All identified through exposure to known cases Not stated |
RT‐PCR; OP, NP (day 1‐5 of symptoms) | Indeterminate or unclear index results on graphs considered negative by review team |
|
Qian 2020 (preprint) 2113 participants (2113 samples) |
[1] Confirmed COVID‐19 cases (RT‐PCR‐positive) (n = 503) and [2] suspected COVID‐19 cases based on epidemiological history, clinical symptoms and chest X‐ray but 3 x PCR‐negative (n = 52) | Apparently contemporaneous controls, including: [3] hospitalised with non‐COVID‐19 conditions (PCR testing not described) (n = 972) [4] healthy controls (n = 586) | Hospital inpatients; Hubei and other provinces, China (Unclear) | Not stated | Not stated | RT‐PCR; NP ("early onset of the symptoms of COVID‐19") | None described |
|
Wan 2020 [A] (preprint) 17 participants (36 samples) |
SARS‐Cov‐2 positive cases confirmed by RT‐PCR (n = 7, 26 samples) | Prepandemic sera (n = 5); plus controls SARS‐Cov‐2 negative on two occasions (n = 5) | Inpatients; Singapore (Not stated) | Not stated | Not stated | RT‐PCR | not stated |
|
Wang 2020a [A] (accepted manuscript) 86 participants (86 samples) |
COVID‐19 patients, meeting Chinese Government guideline criteria (14) | Contemporaneous patients with different pathogen infections and related chronic diseases with no clinical symptoms or imaging evidence of COVID‐19 (no PCR testing reported) (72) | Inpatient; Nanchong, China (25 January‐15 February) | Not stated | Not stated | Chinese CDC guideline (5th ed) | none described |
|
Xiang 2020a [A] (preprint) 98 participants (ELISA samples) , 126 participants (LFA samples) , 81 participants (PCR samples) |
COVID‐19 patients according to WHO interim guidance (suppl data reports PCR results for a subgroup); (n = 63 for ELISA, n = 91 for GICA, some overlap of cases) | Contemporaneous healthy individuals (n = 35) | Inpatient; Wuhan, China (admitted 1‐28 January; sampled 2‐4 February) | ELISA 65 years; LFA 61 years ELISA 35, 56% male LFA 49, 54% male |
Not stated ELISA 4, 6% severe LFA 4, 4% |
WHO interim guidance (subgroup of 82 also have PCR results); (PCR using throat swabs) (Not stated (PCR at 6‐37 days post‐admission)) |
Not stated |
|
Xiang 2020b (peer reviewed; published) 150 participants (216 samples from 85 cases samples) |
[1] RT‐PCR confirmed cases (n = 85) [2] Suspected cases with COVID‐19 pneumonia manifestations and ≥ 2 negative RT‐PCR (n = 24) classed as D+ for review purposes | [3] Contemporaneous control group of healthy blood donors (hospital staff) or patients with other diseases in the same hospital (all PCR‐negative) (n = 60) | Hospital patients (likely inpatients but not explicit); Wuhan, China (19 January‐2 March 2020) | 51 years 31, 26% |
Not stated 18/85 (21%) severe |
[1] RT‐PCR
[2] Clinical manifestations and PCR
; NP and/or OP (Unclear) |
Not stated |
|
Zeng 2020a (accepted manuscript; pre‐proof) 63 participants (63 samples) |
COVID‐19 cases (n = 27); no details of confirmation process | Healthy controls, presume contemporaneous but not stated (n = 36) | Hospital inpatient; Wuhan, China (Not stated) | 62 years 14, 52% |
Not stated 17, 63% severe |
No information; 'confirmed'; No information (No information) |
None reported |
|
Zhao 2020a (accepted manuscript; pre‐proof) 386 participants (386 samples) |
Confirmed RT‐PCR positive COVID‐19 cases (173) | Pre‐pandemic healthy individuals (213) | Inpatients; Shenzhen, China (11 January‐9 February) | 48 years 84, 49% |
126, 73% clear exposure 32, 18% critical |
RT‐PCR; respiratory (Not stated) |
inadequate plasma samples for 2 IgM tests and 1 IgG test |
|
Zhao 2020b (preprint) 481 participants (481 samples) |
Hospitalised and/or recovered COVID‐19 patients (n = 69) | Pre‐pandemic 'normal' samples ("strong negatives"); presumed healthy (n = 257) Contemporaneous 'normal' samples ("negatives"); presumed healthy (n = 155) |
Hospital (no detail); multiple author institutions, China (Not stated) | Not stated | Not stated | Not described; "hospitalized and/or recovered patients confirmed SARS‐CoV‐2 virus infection." | None reported |
|
Zhong 2020 [A] (published; letter) 347 participants (347 samples) |
PCR‐positive COVID‐19 patients (n = 47) | Pre‐pandemic healthy controls (n = 300) | Not stated; China (Not described (symptom onset 15 January‐13 February)) |
48 years 16, 34% |
Not stated 11, 24% severe (6) or critical (5) |
PCR | None reported |
| A&E: Accident and Emergency Department; CDC: Center for Disease Control; COIS: Characteristics of included studies table; CT: computed tomography; CGIA: colloidal gold immunoassay; D+: disease positive: D‐: disease negative; ed: edition; ELISA: enzyme‐linked immunosorbent assay; HCW: healthcare worker; ICU: intensive care unit; LFA: lateral flow assay; n: number; NP: nasopharyngeal; NR: not reported; OP: oropharyngeal; PCR: polymerase chain reaction; pso: post‐symptom onset; RNA: ribonucleic acid; RT‐PCR: reverse transcriptase polymerase chain reaction; RT‐qPCR: reverse transcriptase quantitative polymerase chain reaction; SARS‐CoV‐2: severe acute respiratory syndrome coronavirus 2; suppl: supplementary; TB: tuberculosis; | |||||||
Appendix 9. Summary details of index tests per study
| Study | Test type | Index test (manufacturer) | Antigen | Antibodies measured (threshold) | Sample used | Index sample timing (days pso) | Data by time pso |
| Adams 2020 [A] | [A]a Laboratory [B] to [J] LFAs | [A] ELISA (In‐house) [B] to [J] name withheld | [A] tri S‐based [B] to [J] details withheld | [A] IgG (> 0.4) and IgM (IgM > 0.07) (set in study) [B] and [C] not known [D] to [J] IgG and IgM (presumed from reported results) | Plasma | Day 4‐62 (median and range per group: acute 10 (4 to 27); recovering HCW 13 (8 to 19); convalescent 48 (31 to 62)) | Yes; per week |
| Bendavid 2020 | LFA | No details (Premier Biotech, Minneapolis, MN)) | Not specified | IgG, IgM (threshold not specified) | Serum, plasma, fingerstick blood, venous whole blood | Not described | No |
| Burbelo 2020 [A] | Laboratory | LIPS (in‐house) | [A] N‐based [B] S‐based | Appears to be total Ab (threshold set in healthy control sample) | plasma or serum | Day 2‐50 | Yes; ≤ 14 d, and > 14 d |
| Cai 2020 | Laboratory | CLIA (in‐house) | S‐based | IgM, IgG (both ≥ 0.7 CL) | Serum (day 2‐27 pso) | Day 2‐27 | No |
| Cassaniti 2020 (A) | LFA | CGIA: VivaDiag COVID‐19 IgM/IgG (VivaChek) | Not stated | IgM, IgG (visible line) | Serum or whole blood | On presentation at A&E | No |
| Cassaniti 2020 (B) | LFA | CGIA: VivaDiag COVID‐19 IgM/IgG (VivaChek) | Not stated | IgM, IgG (visible line) | Serum or blood | Median 7 days (IQR 4 to 11) after first test | No |
| Chen 2020a | LFA | FIA (in‐house; using lanthanide‐doped polystyrene nanoparticles) | N‐based | IgG (threshold: At (florescence peak of test line)/Ac (florescence peak of control line) ratio (R) > 0.0666 ) | Serum | Not stated | No |
| Dohla 2020 | LFA | CGIA (suspect this is a description of an anonymised test; no manufacturer stated) | SARS‐CoV‐2 antigen | IgG/IgM (weakly visible or clearly visible (strong positive) test line) | Fingerprick blood (n = 39 in cohort [1]); stored serum for cohort [2] | Median time from exposure–to–test 18.5 d (IQR 15 to 24) | No |
| Du 2020 | Laboratory | Not stated; coded as CLIA based on reported threshold in AU/mL (Manufacturer not reported) | Not stated | IgM, IgG (threshold > 10 AU/mL) | Not stated | day 22 to > 35 | Yes; by week from day 22 |
| Freeman 2020 | Laboratory | ELISA (in‐house) | S‐based | IgG and IgM (based on optical density signal) | Serum | All day ≥ 10 | No |
| Gao 2020a | LFA | CGIA (Innovita Biological Technology Co) | Not reported | IgG, IgM (coloured line) | Serum | day 0 to > 14 | Yes; by week from day 22 |
| Gao 2020b [A] | [A] Laboratory [B] CGIA [C] Laboratory | [A] CLIA [B] CGIA [C] ELISA (all Beier Bioengineering Company, Beijing) | [A], [B], and [C] all S‐ and N‐based | IgG and IgM ([A] ≥ 8 arbitrary unit (AU)/mL; [B] visible line; [C] method to calculate threshold reported ) | Serum | Day 1‐24 | Yes; by week |
| Garcia 2020 (A) | LFA | CGIA, AllTest COV‐19 IgG/IgM kit (AllTest Biotech, Hangzhou, China) | Not reported | IgG, IgM (visible line for either) | Serum | Day 0 to ≥ 14 | Yes; by week |
| Garcia 2020 (B) | LFA | CGIA, AllTest COV‐19 IgG / IgM kit (AllTest Biotech, Hangzhou, China)) | Not reported | IgG and IgM (visible line for either) | Serum | Day 8 to ≥ 14 | Yes; by week |
| Grzelak 2020 [A] | [A] to [E] all Laboratory | 5 tests evaluated: [A] and [B] LIPS (in‐house) [C] and [E] ELISA [D] S‐flow | [A] S1‐based [B] and [C] N‐based [D] S‐based [E] tri‐S‐based | a. IgG
b. total Ab
c. IgM or IgG
d. total Ab (Not clearly stated, plotted on Figure 1) |
Serum | Not stated; day 2‐18 for 5 patients | No |
| Guo 2020a | Laboratory | In‐house ELISA | N‐based | IgM, IgG, IgA (threshold set in healthy control sample) | Blood/plasma | Day 1‐39 | Yes; week 1 only |
| Hu 2020a | Laboratory | Magnetic MCLIA kit (Bioscience Co., Ltd (Chongqing, China)) | N‐ and S‐based | IgM, IgG (S/CO ≥ 1.0 considered positive (ratio of the chemiluminescence signal to the cut‐off value) | Serum | Day 1 to > 37 | Yes; by week |
| Infantino 2020 | Laboratory | SARS CoV‐2 IgM and IgG CLIA kits (Shenzhen YHLO Biotech Co) | N‐ and S‐based | IgM, IgG (multiple thresholds reported, including manufacturer recommended threshold ≥ 10 AU/mL ) | Blood (discussion mentions serum) | Day 8‐19 | No |
| Jia 2020 | LFA | FIA method (Beijing Diagreat Biotechnologies) | Not described | IgM (≥ 0.88 Flu) IgG (≥ 1.02 Flu) (threshold set in healthy control sample) | Not stated | Not stated | No |
| Jin 2020 | Laboratory | SARS CoV‐2 IgM and IgG CLIA kits (Shenzhen YHLO Biotech Co) | N‐ and S‐based | IgM, IgG (> 10 AU/mL) | Serum | Day 1‐55 | Yes; by week |
| Lassauniere 2020 [A] | [A] to [C] laboratory, [D] to [I] LFA | [A] ELISA (Beijing Wantai) [B] IgG ELISA (EUROIMMUN) [C] IgA ELISA ELISA (EUROIMMUN) [D] to [F] all presumed to be CGIA [D] Dynamiker Biotech 2019‐nCOV IgG/IgM Rapid Test [E] CTK Biotech ‐ OnSiteTM COVID‐19 IgG/IgM Rapid Test [F] Autobio Diagnostics Anti‐SARS‐CoV‐2 Rapid Test [G] Artron Labs Coronavirus Diseases 2019 (COVID‐19) IgM/IgG Antibody Test [H] Acro Biotech 2019‐nCoV IgG/IgM Rapid Test Cassette [I] Hangzhou All test 2019‐nCoV IgG/IgM Rapid Test Cassette | [A] S‐based [B] and [C] S1‐based [D] to [I] not stated | [A] Total Ab (calculated negative control value to 0.160) [B] IgG and [C] IgM (ratio < 0.8 is considered negative, ≥ 0.8 and < 1.1 borderline, and ≥ 1.1 positive) [D] to [I] IgG/IgM (Visual line change) | Serum | Day 7 to ≥ 21 | Yes |
| Li 2020a | LFA | CGIA (Jiangsu Medomics Medical Technologies) | S‐based | IgM, IgG (coloured line) | Serum, plasma | Not stated; for 1 site (n = 58), sampling day 8‐33 | No |
| Lin 2020a [A] | [A] and [B] laboratory | [A] In‐house CLIA [B] ELISA (Darui Biotech, China) | [A] and [B] N‐based | [A] IgM (RLU 162296); IgG (RLU 336697) (threshold set using ROC analysis) [B] IgM, IgG | Serum | Day 0 to ≥ 14 | Yes; by week |
| Lippi 2020 [A] | [A] and [B] laboratory | [A] MAGLUMI 2019‐nCoV CLIAs (Snibe Diagnostics ‐Shenzhen New Industries Biomedical Engineering Co., Ltd, ) [B] ELISAs (Euroimmun AG, Lübeck, Germany) | [A] N‐ and S‐based [B] Not stated | [A] IgM or IgG (≥ 1.10 AU/mL) [B] IgA or IgG (≥ 1.1 (absorbance of patient sample/absorbance of calibrator)) | [A] Serum or plasma [B] Not stated | Day < 5‐21 | Yes; 5 day intervals |
| Liu 2020a | LFA | CGIA (Not stated: 'Chinese biotechnology company') | Not stated | IgG, IgM (visible line) | Serum | Day 0 to ≥ 14 | Yes; by week |
| Liu 2020b | Laboratory | ELISA kit (Lizhu, Zhuhai, China) | N‐based | IgM, IgG (threshold set in healthy control sample) | Serum | Day 0 to ≥ 16 | Yes; 5‐day intervals |
| Liu 2020c | Laboratory | iFLash‐SARS‐CoV‐2 CLIA (Shenzhen YHLO Biotech) [based on company contact] | Not described | IgM, IgG (not stated) | Serum | Not stated | No |
| Liu 2020d [A] | Laboratory | [A] ELISA (Hotgen, Beijing, China) [B] ELISA (Lizhu, Zhuhai, China) | [A] S‐based [B] N‐based | IgM, IgG (threshold not stated but method of calculation reported) | Serum | Day 0‐30 | Yes; unequal intervals |
| Long 2020 (A) | Laboratory | Magnetic CLIA (Bioscience (Chongqing) Co., Ltd) | N‐ and S‐based | IgM, IgG (threshold not stated) | Serum | Not stated; 21‐31 days after PCR test | No |
| Long 2020 (B) | Laboratory | Magnetic CLIA (Bioscience (Chongqing) Co., Ltd) | N‐ and S‐based | IgM, IgG (threshold not described) | Serum | Day 2 to ≥ 23 | Yes; by week |
| Lou 2020 [A] | [A] and [C] laboratory [B] LFA | [A] ELISA (Beijing Wantai) [B] CGIA (Beijing Wantai) [C] CLIA (Xiamen InnoDx) | [A] N‐ and S‐based [B] and [C] not stated | IgG, IgM, Ab (thresholds as per manufacturer; NR) | Serum | Day 0‐29 | Yes; by week |
| Ma 2020a | Laboratory | CLIA (in‐house) | S‐based (RBD) | IgM, IgG, IgA (ROC analysis to determine optimal cut‐off in RLU, which is not stated | Serum | Day 4‐41 | Yes; 5‐day intervals |
| Okba 2020a | Laboratory | ELISA, beta version (EUROIMMUN) | Not stated | IgA, IgG IgM, IgG (threshold not stated but method of calculation reported) | Serum | Day 3 to > 23 | Yes; by week |
| Padoan 2020 | Laboratory | CLIA ‐ MAGLUMI 2000 Plus nCoV (Snibe Diagnostics) | Not stated | IgM (1.0 AU/mL); IgG (1.1 AU/mL) | Serum | Day 0 to ≥ 13 | Yes; by week |
| Pan 2020a | LFA | CGIA (Zhuhai Livzon Diagnositic Inc) | Not described | IgM, IgG (appearance of T line) | Serum or plasma | Day 1 to ≥ 15 | Yes; by week |
| Paradiso 2020a | LFA | VivaDiag (Jiangsu Medomics Medical Technologies) [VivaChek?] | S‐based | IgM, IgG (both indicated by presence of red/purple line) | Venous blood | Day 0 to > 15 | No |
| Qian 2020 | Laboratory | CLIA (states analysed using fully automated immune analyser from Shenzhen YHLO Biotech Co) | N‐ and S‐based | IgM and IgG (RLU ≥ 10 AU/mL) | Serum | Not stated | Patients |
| To 2020a [A] | Laboratory | EIAs (in‐house, considered with ELISA tests for analysis purposes) | [A] N‐based [B] S‐based | IgG, IgM (set as the mean value of 93 anonymous archived serum specimens from 2018, plus 3 SDs) | Used serum remnant from blood samples taken for routine biochemical testing | Day ≥ 14 (for subgroup with 2x2 data) | Samples |
| Wan 2020 [A] | [A] and [B] laboratory |
[A] IIFT (EUROIMMUN) [B] In‐house ELISA | [A] and [B] both SARS‐CoV | a. Total antibody (≥ 400) b. IgM, IgG (threshold not stated) | Serum | Day 3‐24 | Yes; per week |
| Wang 2020a [A] | [A] Laboratory [B] LFA | [A] ELISA (Beijing Hotgen Biotechnology Co) [B] CGIA (Beijing Hotgen Biotechnology Co) | Not stated | IgM ([A] not stated; [B] coloured line) | Serum | Day 3‐7 | Yes (week 1 only) |
| Xiang 2020a [A] | [A] Laboratory [B] LFA | [A] ELISA (Zhu Hai Livzon Diagnostics) [B] CGIA (Zhu Hai Livzon Diagnostics) | Not stated | IgM, IgG ([A] threshold not stated; [B] coloured line) | [A] Serum, [B] Plasma | Not stated (can be estimated as 5‐35 days post‐admission) | No |
| Xiang 2020b | Laboratory | ELISA (ELISA kits, Zuhai Livzon Inc) | N‐based | IgG, IgM (method to calculate threshold reported | Serum | day 0 to > 21 | Yes; per week |
| Xiao 2020a | Laboratory | CLIA (Shenzhen YHLO Biotechnology Co. Ltd) | Not described | IgM, IgG (≤ 10 AU/mL) | Blood | Day 1‐49 | Yes; per week |
| Xie 2020a | Laboratory | CLIA (Shenzhen YHLO Biological Technology) | N‐ and S‐based | IgG, IgM (≥ 10 AU/mL) | Serum | Day 0‐41 | No |
| Xu 2020a | LFA | CGIA (in‐house) | S‐based | IgG, IgM (coloured line) | Not stated | Day 15‐30 of observation | No |
| Yongchen 2020 | LFA | CGIA (Innovita Co. Ltd, China) | N‐ and S‐based | IgG, IgM (coloured line) | Serum | Day 8‐42 | Yes; by week |
| Zeng 2020a | Laboratory | ELISA (Zhuhai Livzon Diagnostics) | Not stated | IgG and IgM (OD = 0.105) | Serum | Day 3‐39; can extract for day 6 only | Yes; week 1 only |
| Zhang 2020a | Laboratory | CLIA ‐ iFlash‐SARS‐CoV‐2 IgG and iFlash‐SARS‐CoV‐2 (Shenzhen YHLO Biotech Co. Ltd.) | Not described | IgM, IgG (threshold not described) | Serum | Day 1‐35 | No |
| Zhang 2020b | Laboratory | CLIA ‐ iFlash‐SARS‐CoV‐2 (Shenzhen YHLO Biotechnology Co Ltd) [derived from company contact] | N‐ and S‐based | IgM, IgG (> 10.0 AU/mL); AU ‐ antibody concentration per mL | Serum (frozen until analysis) | Day 4‐18 | No |
| Zhang 2020c | Laboratory | ELISA (in‐house; anti‐SARSr‐CoV) | N‐based (SARS‐CoV) | IgM, IgG (threshold not described) | Serum | Day 0 and day 5 | Yes; week 1 only |
| Zhang 2020d | LFA | CGIA (in‐house) | S‐based | Total antibodies (IgM, IgG) (Visible test and control lines) | Serum | Not stated | No |
| Zhao 2020a | Laboratory | ELISA (Shenzhen YHLO Biotech Co) | N‐ and S‐based | Ab, IgM, IgG (Not stated) | Plasma | Day 1‐39 | Yes; week 1, 2 and 3+ |
| Zhao 2020b | Laboratory | ELISA (in‐house) | S1‐based | Total antibodies (IgG or IgM) (threshold calculation method reported) | Plasma | Not stated; n = 45 during week 1 | No |
| Zhong 2020 [A] | Laboratory | [A] and [B] in‐house ELISA [C] CLIA (author institution is Maccura Biotech) | [A] N‐based [B] S‐based [C] N‐ and S‐based (unclear) | IgM, IgG (optimal cut‐off based on ROC analysis) | Serum | Day 1‐29 | No |
| A&E: Accident and Emergency Department; Ab: antibody; AU: arbitrary units; CGIA: colloidal gold immunoassay; CL: chemilumonescence units; CLIA: chemiluminescence immunoassay; d: days; ELISA: enzyme‐linked immunosorbent assay; FIA: fluorescence immunoassay; Flu: fluorescence units; HCW: healthcare workers; IIFT: indirect immunofluorescence assay; IQR: interquartile range; LFA: lateral flow assay; LIPS: luciferase Immunoprecipitation System; mL: millilitre; N‐based: nucleocapsid protein; NR: not reported; OD: Optical density; pso: post‐symptom onset; RBD: receptor binding domain; RLU: relative light units; ROC: receiver operating characteristics; S‐based: spike protein; SD: standard deviation; S‐flow: flow‐cytometry based test; SARS‐CoV‐2: severe acute respiratory syndrome coronavirus 2; SD: standard deviation | |||||||
Footnotes
aPlease note that square brackets indicate different tests within one study.
Appendix 10. Study level assessments of study quality
10.

Risk of bias and applicability concerns summary: review authors' judgements about each domain for each included study
Appendix 11. Results of all studies across all time periods
11.
Forest plot of studies evaluating tests for detection of IgG at all time post‐symptom onset
12.
Forest plot of studies evaluating tests for detection of IgM at all time post‐symptom onset
13.
Forest plot of studies evaluating tests for detection of IgG/IgM at all time post‐symptom onset.
14.
Forest plot of tests: 19 IgA (all time points), 25 Total antibodies (Ab) (all time points), 38 IgA/IgM (all time points).
Appendix 12. Manufacturer product code details identified
| Test | Study | From paper | From company documentation/website |
| ELISA: Enzyme‐linked immunosorbent assay | |||
| Beijing Beier Bioengineering | Gao 2020b [C] | No product codes provided (Study author contacted 29 May 2020) |
No IFU and not on company website, no product code identified |
| Beijing Hotgen | Wang 2020a [A] | ELISA (20200101 and 20200201) | No IFU, no product code identified www.hotgen.com.cn/ky/upt.html Unclear if test on website is ELISA although the company do produce ELISAs |
| Liu 2020d [A] | No product code provided ; “The rS‐based ELISA kit (Hotgen, Beijing, China)” Study author contacted 1 June 2020 |
||
| Beijing Wantai | Lassauniere 2020 [A] | SARS‐CoV‐2 Ab ELISA (CE‐IVD) (WS‐1096) | www.sanbio.nl/ws-1096 |
| Lou 2020 [A] | IgM, IgG; no product code reported in paper Study author responded: ELISA‐Ab lot number NCOA20200201B ELISA‐IgM lot number NCOM20200202B ELISA‐IgG lot number NCOnG20200201B |
||
| Zhao 2020a | No product code reported; “(ELISA) kits supplied by Beijing Wantai Biological Pharmacy Enterprise Co” Study author contacted 1 June 2020 |
||
| Darui Biotechnology | Lin 2020a [A] | No product code reported; “commercial enzyme‐linked immunosorbent assay kit (Darui Biotech, CHINA)” Study author contacted 1 June 2020 |
No IFU, no product code on website (www.daruibiotech.com/eproduct/index_33.html) |
| EUROIMMUN | Lassauniere 2020 [B]; Lassauniere 2020 [C] | Anti‐SARS‐CoV‐2 IgG (EI 2668‐9601 G) Anti‐SARS‐CoV‐2 IgA (EI 2606‐9601 A) |
IFU: Anti‐SARS‐CoV‐2 ELISA (IgG) (EI 2606‐9601 G) IFU: Anti‐SARS‐CoV‐2 ELISA (IgA) (EI 2606‐9601 A) |
| Lippi 2020 [B] | No product code reported; “Anti‐SARS‐CoV‐2 IgA and IgG … ELISAs; Euroimmun AG, Lübeck, Germany” Study author replied but could not supply product code |
||
| Okba 2020a | No product code reported; “β‐versions of 2 commercial kits (EUROIMMUN)” (These are Beta versions) |
Not available | |
| Zhuhai Livzon | Xiang 2020a [A] | IgG/IgM antibody ELISA kits (lot number 2020010108) | No IFU identified; website only has the lateral flow assay en.livzon.com.cn/product/98.html No product code identified |
| Xiang 2020b | ELISA kits (lot numbers IgM 20200308, IgG 20200308) | ||
| Zeng 2020a | No product code reported: ELISA “assay kits (Zhuhai Livzon Diagnostics INC.)” Study author contacted 1 June 2020 |
||
| Zhuhai Lizhu | Liu 2020b | No product code reported: ELISA kit “(Lizhu, Zhuhai, China )” Study author contacted 1 June 2020 |
Presumed to be same as above, following contact with FIND |
| Liu 2020d [B] | No product code reported: ELISA kit (Lizhu, Zhuhai, China) Study author contacted 1 June 2020 |
||
| CLIA: chemiluminescence immunoassay | |||
| Beijing Beier Bioengineering | Gao 2020b [A] | No product codes provided (Study author contacted 29 May 2020) |
No IFU, not on website; no product code identified |
| Bioscience Co (Chongqing) | Hu 2020a | No product code reported; “(MCLIA) kit supplied by Bioscience Co., Ltd (Chongqing, China)“ Study author contacted 1 June 2020 |
Review authors unable to find this company; no product code identified |
| Long 2020 (A); Long 2020 (B) | No product code reported; “(MCLIA) kit supplied by Bioscience Co., Ltd China)“ Study author supplied NMA approval numbers only: MCLA IgG: China National Medical Products Administration approval number 20203400183; MCLA IgM: China National Medical Products Administration approval number20203400182 |
||
| Shenzhen YHLO | Infantino 2020 | No product code reported; “IgM and IgG CLIA kits were from Shenzhen YHLO Biotech Co., Ltd (China),” Study author provided details: IgG anti‐SARS Cov2 C86095G; IgM C86095M LOT NUMBER 207 |
Company flyer: iFlash‐SARS‐CoV‐2 IgG (C86095G) iFlash‐SARS‐CoV‐2 IgM (C86095G) |
| Jin 2020 | No product code reported; “(CLIA) kits used in this study were supplied by Shenzhen YHLO Biotech Co., Ltd (China)” Study author contacted 1 June 2020 |
||
| Liu 2020c | No product code reported; “SARS‐CoV‐2 antibody detection kit (YHLO Biotech, Shenzhen, China” Study author contacted 1 June 2020 |
||
| Xiao 2020a | No product code reported; “IgM and IgG were analyzed by … CLIA … (Shenzhen Yahuilong Biotechnology Co., Ltd). Study author contacted 1 June 2020 |
||
| Xie 2020a | No product code reported; “IgG and IgM assays were purchased from YHLO Biological Technology Co., Ltd., Shenzhen, China” Study author contacted 1 June 2020 |
||
| Zhang 2020a | No product code reported; “(CLIA) Assays panel (Shenzhen YHLO Biotech Co., Ltd., Shenzhen, China)” Study author contacted 1 June 2020 |
||
| Zhang 2020b | No product code reported; “CLIA detection kit from Shenzhen Yahuilong Biotechnology Co Ltd” Study author contacted 1 June 2020 |
||
| Snibe Diagnostic ‐ MAGLUMI | Lippi 2020 [A] | No product code reported; “MAGLUMI 2019‐nCoV IgG and IgM”. (IgM ‐ 130219016M; IgG – 130219015M) Study author contacted 1 June 2020 Study author replied 1 June 2020 with IFU (no lot numbers provided; code from IFU added above) |
From image on website: IgM ‐ Ref 130219016M; Lot 2712000501 IgG ‐ Ref 130219016M; Lot 2722000501 (www.snibe.com/zh_en/en_newsView.aspx?id=576) |
| Padoan 2020 | No product code reported; “MAGLUMI 2000 Plus (New Industries Biomedical Engineering Co., Ltd [Snibe], Shenzhen, China)” Study author supplied details: ‐ for SARS‐CoV‐2 IgG lot number used for all the 87 measurements: 2722000102, kit number 8131; ‐ for SARS‐CoV‐2 IgM lot number used for all the 87 measurements: 2712000201, kit number 5122 |
||
| Xiamen InnoDx Biotech | Lou 2020 [C] | No product code reported; “CMIA reagents were supplied by Xiamen InnoDx Biotech Co., Ltd., China Study author supplied following details: CMIA‐Ab product code CT0669 lot number 20200201 CMIA‐IgM product code CT0667 lot number 20200201 |
Review authors unable to find website for this company |
| Other laboratory‐based tests | |||
| EUROIMMUN | Wan 2020 [A] | No product code reported; “Anti‐SARS CoV Indirect 62 Immunofluorescence test (IIFT) (IgM & IgG) by Euroimmun (Germany)” (Test uses SARS‐Cov not SARS‐CoV‐2) |
|
| Lateral flow assays | |||
| Acro Biotech | Lassauniere 2020 [H] | 2019‐nCoV IgG/IgM Rapid Test Cassette (INCP‐402) | Code from IFU and Assay Genie website: INCP‐402 |
| Artron Laboratories | Lassauniere 2020 [G] | Coronavirus Diseases 2019 (COVID‐19) IgM/IgG Antibody Test (A03‐51‐322), | Brochure only; but no product code www.artronlab.com/products/CoVBrochure-ver3.pdf |
| Autobio Diagnostics | Lassauniere 2020 [F] | Anti‐SARS‐CoV‐2 Rapid Test (RTA0204) | Code from IFU: Anti‐SARS‐CoV‐2 Rapid Test (RTA0203) |
| Beijing Beier Bioengineering | Gao 2020b [B] | No product codes provided (Study author contacted 29 May 2020) |
No IFU and not on company website; Distributor: www.unifier.one/en/beier-new-2019-coronavirus-covid-19-rapid-test.html 2019‐ New Coronavirus IgM/IgG Rapid Test Casette (WB/S/P) No product code |
| Beijing Diagreat | Jia 2020 | COVID IgM/IgG antibodies kit, which have sent to Beijing Institute of Medical Device Testing (BIMT) for product verification (lot: 20200214) | Manual from 2019‐nCoV IgM Antibody Determination Kit (immunochromatographic Assay) Product No. P11802 2019‐nCoV IgG Antibody Determination Kit (Immunochromatographic Assay) Product No. P11801 |
| Beijing Hotgen | Wang 2020a [B] | Kit provided by Beijing Hotgen Biotechnology Co., Beijing, China: (lot number 20200208 and 105 20200229 for GICA) |
No IFU, on website but no product code www.hotgen.com.cn/ky/upt.html Coronavirus disease (COVID‐19) Antibody Test (Colloidal Gold) |
| Beijing Wantai | Lou 2020 [B] | No product code reported Study author provided: LFIA‐Ab lot number JNB20200202F LFIA‐IgM lot number JNM20200203F LFIA‐IgG lot number JNG20200201F |
Code from IFU: WJ‐2701, WJ‐2710, WJ‐2750 Website: Rapid test for coronavirus Ab (CE‐IVD) (WJ‐2750 www.sanbio.nl/wj-2750) |
| CTK Biotech ‐ OnSite | Lassauniere 2020 [E] | OnSite COVID‐19 IgG/IgM Rapid Test (R0180C) | IFU: OnSite COVID‐19 IgG/IgM Rapid Test R0180C |
| Dynamiker Biotechnology | Lassauniere 2020 [D] | 2019‐nCOV IgG/IgM Rapid Test (DNK‐1419‐1) | IFU: 2019‐nCOV IgG/IgM Rapid Test Catalogue No: DNK‐1419‐1 |
| Hangzhou Alltest | Lassauniere 2020 [I] | 2019‐nCoV IgG/IgM Rapid Test Cassette (INCP‐402) | IFU; 2019‐nCoV IgG/IgM Rapid Test Cassette (Whole blood/serum/plasma) Package Insert INCP‐402 |
|
Garcia 2020 (A) Garcia 2020 (B) |
AllTest COV‐19 IgG / IgM kit (no product code) Study author provided IFU (product code NCP‐402) |
||
| Innovita Biological | Gao 2020a | No product code reported Study author contacted 1 June 2020 |
IFU: 2019‐nCoV Ab Test (Colloidal gold); Catalogue No. YF 319C |
| Yongchen 2020 | No product code reported Study author provided lot number: 20200205 |
||
| Jiangsu Medomics | Li 2020a | No product code reported “SARS‐CoV‐2 rapid IgG‐IgM combined antibody test kit” Study author contacted 1 June 2020 |
Review authors were unable to find this company; study was also provided to review authors by Lomina (www.test-covid19.com/); no product code identified |
| Vivachek ‐ VivaDiag | Paradiso 2020a | No product code reported; “Viva‐DiagTM kit produced by Jiangsu Medomics Medical Technologies kit (https://www.vivachek.com/vivachek/English/prods/prod-covid19.html)”; link no longer active. test considered to be Vivachek test Study author contacted 1 June 2020 |
Package insert VivaDiag SARS‐CoV‐2 IgM/IgG Rapid Test (VID35‐08‐011 / VID35‐08‐012 / VID35‐08‐013 / VID35‐08‐014 / VID35‐08‐015) |
|
Cassaniti 2020 (A) Cassaniti 2020 (B) |
No product code reported; “VivaDiag COVID‐19 IgM/IgG from VivaChek”; study also provided by Vivachek following company contact Study author provided lot number E2002002, REF VID35‐08‐011 |
||
| Zhuhai Livzon | Pan 2020a | No product code reported Study author contacted 1 June 2020 |
Product flyer; Diagnostic Kit for IgM/IgG Antibody to Coronavirus (SARS‐CoV‐2); Catalogue number: 01040048 |
| Xiang 2020a [B] | IgG/IgM antibody GICA kits (lot number 2001010220) | ||
| FIND: Foundation for Innovative Diagnostics; IFU: instructions for use; NMA: China National Medical Products Administration | |||
Data
Presented below are all the data for all of the tests entered into the review.
Tests. Data tables by test.
1. Test.
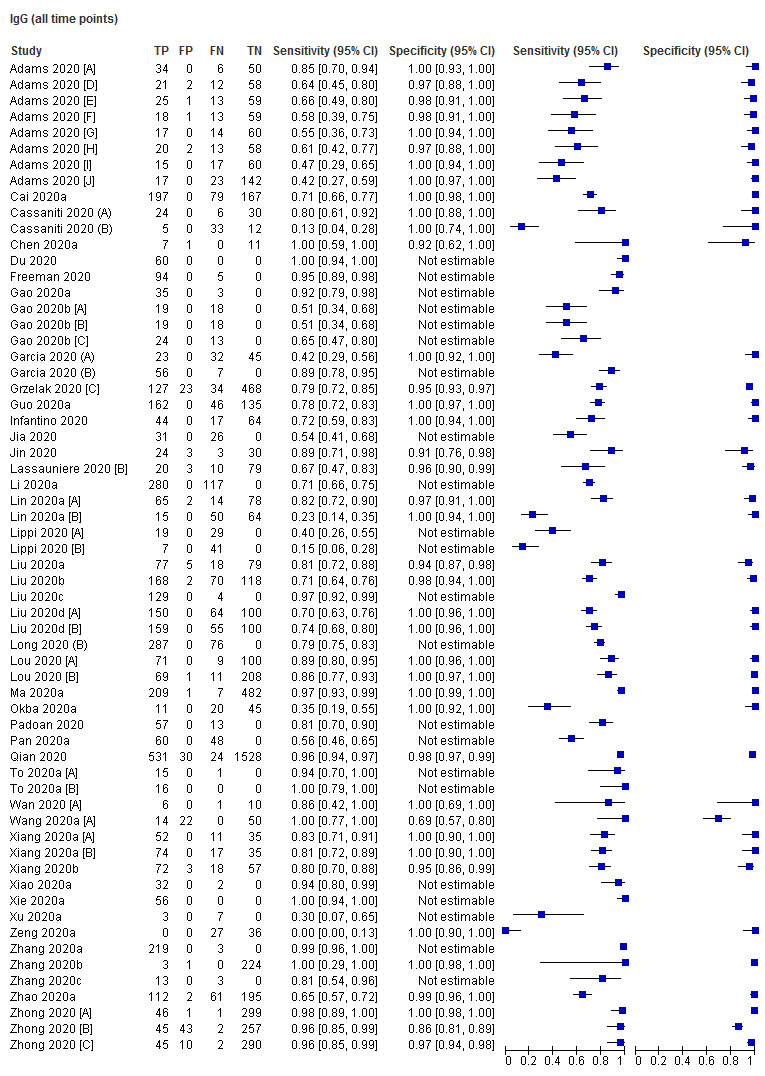
IgG (all time points)
2. Test.

IgG (1 to 7 days)
3. Test.

IgG (8 to 14 days)
4. Test.

IgG (15 to 21 days)
5. Test.

IgG (22 to 35 days)
6. Test.

IgG (over 35 days)
7. Test.
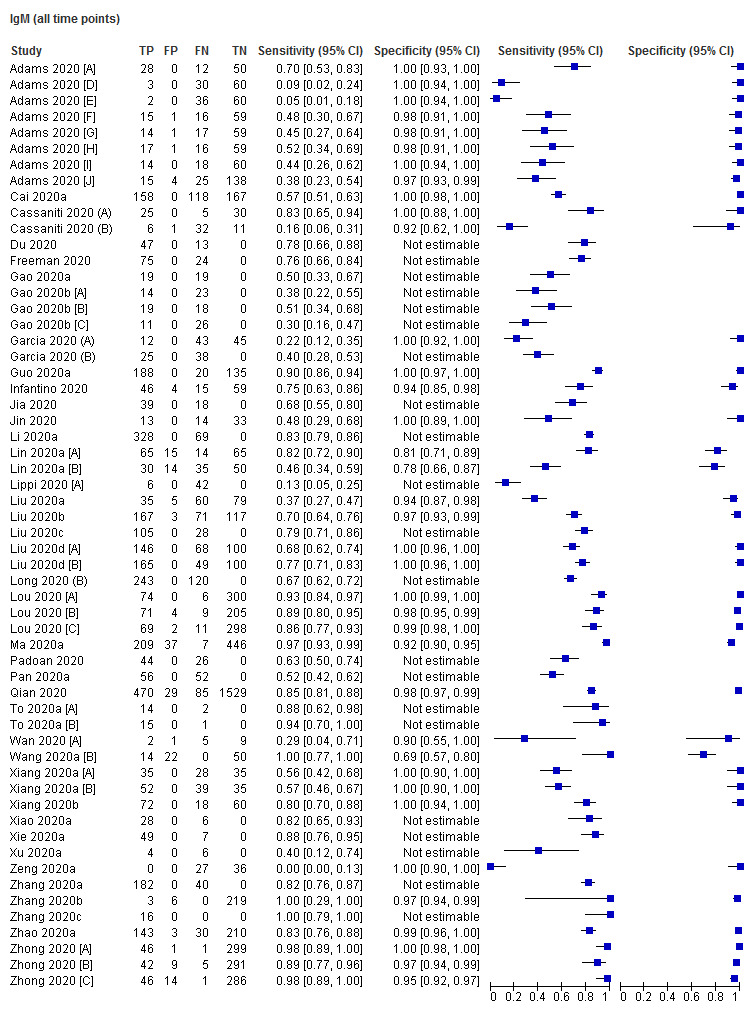
IgM (all time points)
8. Test.

IgM (8 to 14 days)
9. Test.

IgM (1 to 7 days)
10. Test.

IgM (15 to 21 days)
11. Test.

IgM (22 to 35 days)
12. Test.

IgM (over 35 days)
13. Test.

IgG/IgM (all time points)
14. Test.

IgG/IgM (1 to 7 days)
15. Test.

IgG/IgM (8 to 14 days)
16. Test.

IgG/IgM (15 to 21 days)
17. Test.

IgG/IgM (22 to 35 days)
18. Test.

IgG/IgM (over 35 days)
19. Test.

IgA (all time points)
20. Test.

IgA (1 to 7 days)
21. Test.

IgA (8 to 14 days)
22. Test.

IgA (15 to 21 days)
23. Test.

IgA (22 to 35 days)
24. Test.

IgA (over 35 days)
25. Test.

Total antibodies (Ab) (all time points)
27. Test.

Total antibodies (Ab) (1 to 7 days)
29. Test.
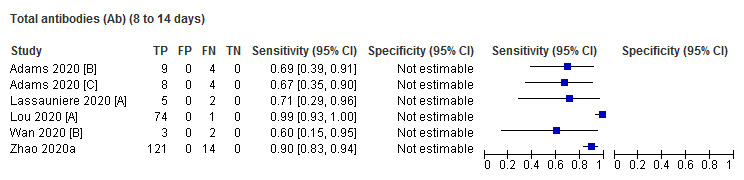
Total antibodies (Ab) (8 to 14 days)
30. Test.

Total antibodies (Ab) (15 to 21 days)
31. Test.

Total antibodies (Ab) (21 to 35 days)
32. Test.

Total antibodies (Ab) (over 35 days)
33. Test.

IgA/IgG (all time points)
34. Test.

IgA/IgG (1 to 7 days)
35. Test.

IgA/IgG (8 to 14 days)
36. Test.

IgA/IgG (15 to 21 days)
37. Test.

IgA/IgG (22 to 35 days)
38. Test.

IgA/IgM (all time points)
39. Test.

IgG in PCR+ve (all time points)
40. Test.

IgG in PCR +ve (1 to 7 days)
41. Test.

IgG in PCR+ve (8 to 14 days)
42. Test.

IgG in PCR+ve (15 to 21 days)
43. Test.

IgG in PCR‐ve (all time points)
44. Test.

IgG in PCR‐ve (1 to 7 days)
45. Test.

IgG in PCR‐ve (8 to 14 days)
46. Test.

IgG in PCR‐ve (15 to 21 days)
47. Test.

IgM in PCR+ve (all time points)
48. Test.

IgM in PCR+ve (1 to 7 days)
49. Test.

IgM in PCR+ve (8 to 14 days)
50. Test.

IgM in PCR+ve (15 to 21 days)
51. Test.

IgM in PCR‐ve (all time points)
52. Test.

IgM in PCR‐ve (1 to 7 days)
53. Test.

IgM in PCR‐ve (8 to 14 days)
54. Test.

IgM in PCR‐ve (15 to 21 days)
55. Test.

IgG/IgM in PCR+ve (all time points)
56. Test.

IgG/IgM in PCR+ve (1 to 7 days)
57. Test.

IgG/IgM in PCR+ve (8 to 14 days)
58. Test.

IgG/IgM in PCR+ve (15 to 21 days)
59. Test.
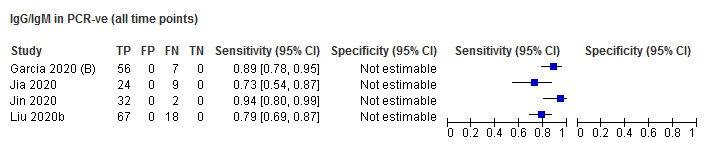
IgG/IgM in PCR‐ve (all time points)
60. Test.

IgG/IgM in PCR‐ve (1 to 7 days)
61. Test.

IgG/IgM in PCR‐ve (8 to 14 days)
62. Test.

IgG/IgM in PCR‐ve (15 to 21 days)
63. Test.

IgG (moderate)
64. Test.

IgG (severe)
65. Test.

IgG (critical)
66. Test.

IgM (moderate)
67. Test.

IgM (severe)
68. Test.

IgM (critical)
69. Test.

RT‐PCR (all time points ‐ throat)
70. Test.

RT‐PCR (1 to 7 days throat)
71. Test.

RT‐PCR (8 to 14 days ‐ throat)
72. Test.

RT‐PCR (15 to 21 days ‐ throat)
73. Test.

RT‐PCR (all time points ‐ sputum)
74. Test.

RT‐PCR (1 to 7 days ‐ sputum)
75. Test.

RT‐PCR (8 to 14 days ‐ sputum)
76. Test.

RT‐PCR (15 to 21 days ‐ sputum)
Characteristics of studies
Characteristics of included studies [ordered by study ID]
Adams 2020 [A].
| Study characteristics | |||
| Patient Sampling | 2‐group study recruiting patients estimating sensitivity and specificity [1] Rt‐PCR confirmed COVID‐19 cases (n = 40) [2] Pre‐pandemic controls (n = 142) Recruitment: unclear Prospective or retrospective recruitment of cases: retrospective Sample size (virus/COVID cases): 182 (40) Inclusion and exclusion criteria: none stated | ||
| Patient characteristics and setting | Setting [1] Acute hospital samples (n = 16), recovering healthcare workers (n = 6), convalescent patients (n = 18) [2] Health blood donors (n = 60); organ donor samples (n = 50); pertussis vaccine study (BERT) (n = 32) Location [1] Acute hospital patients from Oxford University Hospitals NHS Foundation Trust (location of other groups NR) [2] National Health Service Blood and Transplant, UK National Quality in Organ Donation (QUOD) study, the ‘BERT’ study (A Study Exploring Whooping Cough Protection in Children and Adults), UK Country: UK Dates: [1] NR; [2] before December 2019 Symptoms and severity: [1] asymptomatic (n = 1); mild (n = 26); severe (n = 4); critical (n = 9) Sex: NR Age: [1] Median (range): 57 (22‐95) years; [2] NR Exposure history: NR |
||
| Index tests |
Adams 2020 [A] is test [A] from the following entry: Test name: [A] ELISA test [B]‐[J] LFIA names withheld Manufacturer: [A] in‐house [B]‐[J] manufacturer name withheld Ab targets: [A] IgG and IgM [B]‐[C] total antibodies [D]‐[J] IgG and IgM Antigens used: [A] SARS‐CoV‐2 trimeric S protein [B]‐[J] details withheld Test method: [A] ELISA [B]‐[J] LFIA further details withheld Timing of samples: 4‐62 days after onset of symptoms Samples used: plasma Test operators: laboratory staff Definition of test positivity: [A]‐[J] NR Blinded to reference standard: NR Threshold predefined: no for [A], unclear for [B] to [J] |
||
| Target condition and reference standard(s) | Reference standard for cases: RT‐PCR Samples used: nose or throat swabs Timing of reference standard: NR Blinded to index test: yes Incorporated index test: no Reference standard for non‐cases: pre‐pandemic | ||
| Flow and timing | Time interval between index and reference tests: NR Results presented by time period: computed from analysis of individual participant data All participants received the same reference standard: no Missing data: different tests were evaluted in different numbers of samples, no information on how sampling decisions were made Uninterpretable results: not mentioned Indeterminate results: not mentioned Unit of analysis: per patient | ||
| Comparative | |||
| Notes | Funding: NIHR, Oxford Biomedical Research Centre, the UK Government Department of Health and Social Care and grants from NIHR and the Medical Research Council Publication status: preprint (not peer reviewed) Source: medRxiv Study author COI: several authors declared relationships with companies for other work; funders were co‐authors | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | No | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | No | ||
| Could the conduct or interpretation of the index test have introduced bias? | High risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Adams 2020 [B].
| Study characteristics | |||
| Patient Sampling | See main entry for this study for characteristics and QUADAS‐2 assessment (Adams 2020 [A]) | ||
| Patient characteristics and setting | See main entry for this study for characteristics and QUADAS‐2 assessment (Adams 2020 [A]) | ||
| Index tests |
Adams 2020 [B] is test [B] from the following entry: Test name: [A] ELISA test [B]‐[J] LFIA names withheld Manufacturer: [A] in‐house [B]‐[J] manufacturer name withheld Ab targets: [A] IgG and IgM [B]‐[C] total antibodies [D]‐[J] IgG and IgM Antigens used: [A] SARS‐CoV‐2 trimeric S protein [B]‐[J] details withheld Test method: [A] ELISA [B]‐[J] LFIA further details withheld Timing of samples: 4‐62 days after onset of symptoms Samples used: plasma Test operators: laboratory staff Definition of test positivity: [A]‐[J] NR Blinded to reference standard: NR Threshold predefined: no for [A], unclear for [B] to [J] |
||
| Target condition and reference standard(s) | See main entry for this study for characteristics and QUADAS‐2 assessment (Adams 2020 [A]) | ||
| Flow and timing | See main entry for this study for characteristics and QUADAS‐2 assessment (Adams 2020 [A]) | ||
| Comparative | |||
| Notes | |||
Adams 2020 [C].
| Study characteristics | |||
| Patient Sampling | See main entry for this study for characteristics and QUADAS‐2 assessment (Adams 2020 [A]) | ||
| Patient characteristics and setting | See main entry for this study for characteristics and QUADAS‐2 assessment (Adams 2020 [A]) | ||
| Index tests |
Adams 2020 [C] is test [C] from the following entry: Test name: [A] ELISA test [B]‐[J] LFIA names withheld Manufacturer: [A] in‐house [B]‐[J] manufacturer name withheld Ab targets: [A] IgG and IgM [B]‐[C] total antibodies [D]‐[J] IgG and IgM Antigens used: [A] SARS‐CoV‐2 trimeric S protein [B]‐[J] details withheld Test method: [A] ELISA [B]‐[J] LFIA further details withheld Timing of samples: 4‐62 days after onset of symptoms Samples used: plasma Test operators: laboratory staff Definition of test positivity: [A]‐[J] NR Blinded to reference standard: NR Threshold predefined: no for [A], unclear for [B] to [J] |
||
| Target condition and reference standard(s) | See main entry for this study for characteristics and QUADAS‐2 assessment (Adams 2020 [A]) | ||
| Flow and timing | See main entry for this study for characteristics and QUADAS‐2 assessment (Adams 2020 [A]) | ||
| Comparative | |||
| Notes | |||
Adams 2020 [D].
| Study characteristics | |||
| Patient Sampling | See main entry for this study for characteristics and QUADAS‐2 assessment (Adams 2020 [A]) | ||
| Patient characteristics and setting | See main entry for this study for characteristics and QUADAS‐2 assessment (Adams 2020 [A]) | ||
| Index tests |
Adams 2020 [D] is test [D] from the following entry: Test name: [A] ELISA test [B]‐[J] LFIA names withheld Manufacturer: [A] in‐house [B]‐[J] manufacturer name withheld Ab targets: [A] IgG and IgM [B]‐[C] total antibodies [D]‐[J] IgG and IgM Antigens used: [A] SARS‐CoV‐2 trimeric S protein [B]‐[J] details withheld Test method: [A] ELISA [B]‐[J] LFIA further details withheld Timing of samples: 4‐62 days after onset of symptoms Samples used: plasma Test operators: laboratory staff Definition of test positivity: [A]‐[J] NR Blinded to reference standard: NR Threshold predefined: no for [A], unclear for [B] to [J] |
||
| Target condition and reference standard(s) | See main entry for this study for characteristics and QUADAS‐2 assessment (Adams 2020 [A]) | ||
| Flow and timing | See main entry for this study for characteristics and QUADAS‐2 assessment (Adams 2020 [A]) | ||
| Comparative | |||
| Notes | |||
Adams 2020 [E].
| Study characteristics | |||
| Patient Sampling | See main entry for this study for characteristics and QUADAS‐2 assessment (Adams 2020 [A]) | ||
| Patient characteristics and setting | See main entry for this study for characteristics and QUADAS‐2 assessment (Adams 2020 [A]) | ||
| Index tests |
Adams 2020 [E] is test [E] from the following entry: Test name: [A] ELISA test [B]‐[J] LFIA names withheld Manufacturer: [A] in‐house [B]‐[J] manufacturer name withheld Ab targets: [A] IgG and IgM [B]‐[C] total antibodies [D]‐[J] IgG and IgM Antigens used: [A] SARS‐CoV‐2 trimeric S protein [B]‐[J] details withheld Test method: [A] ELISA [B]‐[J] LFIA further details withheld Timing of samples: 4‐62 days after onset of symptoms Samples used: plasma Test operators: laboratory staff Definition of test positivity: [A]‐[J] NR Blinded to reference standard: NR Threshold predefined: no for [A], unclear for [B] to [J] |
||
| Target condition and reference standard(s) | See main entry for this study for characteristics and QUADAS‐2 assessment (Adams 2020 [A]) | ||
| Flow and timing | See main entry for this study for characteristics and QUADAS‐2 assessment (Adams 2020 [A]) | ||
| Comparative | |||
| Notes | |||
Adams 2020 [F].
| Study characteristics | |||
| Patient Sampling | See main entry for this study for characteristics and QUADAS‐2 assessment (Adams 2020 [A]) | ||
| Patient characteristics and setting | See main entry for this study for characteristics and QUADAS‐2 assessment (Adams 2020 [A]) | ||
| Index tests |
Adams 2020 [F] is test [F] from the following entry: Test name: [A] ELISA test [B]‐[J] LFIA names withheld Manufacturer: [A] in‐house [B]‐[J] manufacturer name withheld Ab targets: [A] IgG and IgM [B]‐[C] total antibodies [D]‐[J] IgG and IgM Antigens used: [A] SARS‐CoV‐2 trimeric S protein [B]‐[J] details withheld Test method: [A] ELISA [B]‐[J] LFIA further details withheld Timing of samples: 4‐62 days after onset of symptoms Samples used: plasma Test operators: laboratory staff Definition of test positivity: [A]‐[J] NR Blinded to reference standard: NR Threshold predefined: no for [A], unclear for [B] to [J] |
||
| Target condition and reference standard(s) | See main entry for this study for characteristics and QUADAS‐2 assessment (Adams 2020 [A]) | ||
| Flow and timing | See main entry for this study for characteristics and QUADAS‐2 assessment (Adams 2020 [A]) | ||
| Comparative | |||
| Notes | |||
Adams 2020 [G].
| Study characteristics | |||
| Patient Sampling | See main entry for this study for characteristics and QUADAS‐2 assessment (Adams 2020 [A]) | ||
| Patient characteristics and setting | See main entry for this study for characteristics and QUADAS‐2 assessment (Adams 2020 [A]) | ||
| Index tests | Adams 2020 [G] is test [G] from the following entry: Test name: [A] ELISA test [B]‐[J] LFIA names withheld Manufacturer: [A] in‐house [B]‐[J] manufacturer name withheld Ab targets: [A] IgG and IgM [B]‐[C] total antibodies [D]‐[J] IgG and IgM Antigens used: [A] SARS‐CoV‐2 trimeric S protein [B]‐[J] details withheld Test method: [A] ELISA [B]‐[J] LFIA further details withheld Timing of samples: 4‐62 days after onset of symptoms Samples used: plasma Test operators: laboratory staff Definition of test positivity: [A]‐[J] NR Blinded to reference standard: NR Threshold predefined: no for [A], unclear for [B] to [J] |
||
| Target condition and reference standard(s) | See main entry for this study for characteristics and QUADAS‐2 assessment (Adams 2020 [A]) | ||
| Flow and timing | See main entry for this study for characteristics and QUADAS‐2 assessment (Adams 2020 [A]) | ||
| Comparative | |||
| Notes | |||
Adams 2020 [H].
| Study characteristics | |||
| Patient Sampling | See main entry for this study for characteristics and QUADAS‐2 assessment (Adams 2020 [A]) | ||
| Patient characteristics and setting | See main entry for this study for characteristics and QUADAS‐2 assessment (Adams 2020 [A]) | ||
| Index tests |
Adams 2020 [H] is test [H] from the following entry: Test name: [A] ELISA test [B]‐[J] LFIA names withheld Manufacturer: [A] in‐house [B]‐[J] manufacturer name withheld Ab targets: [A] IgG and IgM [B]‐[C] total antibodies [D]‐[J] IgG and IgM Antigens used: [A] SARS‐CoV‐2 trimeric S protein [B]‐[J] details withheld Test method: [A] ELISA [B]‐[J] LFIA further details withheld Timing of samples: 4‐62 days after onset of symptoms Samples used: plasma Test operators: laboratory staff Definition of test positivity: [A]‐[J] NR Blinded to reference standard: NR Threshold predefined: no for [A], unclear for [B] to [J] |
||
| Target condition and reference standard(s) | See main entry for this study for characteristics and QUADAS‐2 assessment (Adams 2020 [A]) | ||
| Flow and timing | See main entry for this study for characteristics and QUADAS‐2 assessment (Adams 2020 [A]) | ||
| Comparative | |||
| Notes | |||
Adams 2020 [I].
| Study characteristics | |||
| Patient Sampling | See main entry for this study for characteristics and QUADAS‐2 assessment (Adams 2020 [A]) | ||
| Patient characteristics and setting | See main entry for this study for characteristics and QUADAS‐2 assessment (Adams 2020 [A]) | ||
| Index tests |
Adams 2020 [I] is test [I] from the following entry: Test name: [A] ELISA test [B]‐[J] LFIA names withheld Manufacturer: [A] in‐house [B]‐[J] manufacturer name withheld Ab targets: [A] IgG and IgM [B]‐[C] total antibodies [D]‐[J] IgG and IgM Antigens used: [A] SARS‐CoV‐2 trimeric S protein [B]‐[J] details withheld Test method: [A] ELISA [B]‐[J] LFIA further details withheld Timing of samples: 4‐62 days after onset of symptoms Samples used: plasma Test operators: laboratory staff Definition of test positivity: [A]‐[J] NR Blinded to reference standard: NR Threshold predefined: no for [A], unclear for [B] to [J] |
||
| Target condition and reference standard(s) | See main entry for this study for characteristics and QUADAS‐2 assessment (Adams 2020 [A]) | ||
| Flow and timing | See main entry for this study for characteristics and QUADAS‐2 assessment (Adams 2020 [A]) | ||
| Comparative | |||
| Notes | |||
Adams 2020 [J].
| Study characteristics | |||
| Patient Sampling | See main entry for this study for characteristics and QUADAS‐2 assessment (Adams 2020 [A]) | ||
| Patient characteristics and setting | See main entry for this study for characteristics and QUADAS‐2 assessment (Adams 2020 [A]) | ||
| Index tests |
Adams 2020 [J] is test [J] from the following entry: Test name: [A] ELISA test [B]‐[J] LFIA names withheld Manufacturer: [A] in‐house [B]‐[J] manufacturer name withheld Ab targets: [A] IgG and IgM [B]‐[C] total antibodies [D]‐[J] IgG and IgM Antigens used: [A] SARS‐CoV‐2 trimeric S protein [B]‐[J] details withheld Test method: [A] ELISA [B]‐[J] LFIA further details withheld Timing of samples: 4‐62 days after onset of symptoms Samples used: plasma Test operators: laboratory staff Definition of test positivity: [A]‐[J] NR Blinded to reference standard: NR Threshold predefined: no for [A], unclear for [B] to [J] |
||
| Target condition and reference standard(s) | See main entry for this study for characteristics and QUADAS‐2 assessment (Adams 2020 [A]) | ||
| Flow and timing | See main entry for this study for characteristics and QUADAS‐2 assessment (Adams 2020 [A]) | ||
| Comparative | |||
| Notes | |||
Bendavid 2020.
| Study characteristics | |||
| Patient Sampling | Multiple‐group study recruiting patients estimating sensitivity and specificity [1] Specimens from COVID‐19 cases recruited from 3 different sources (n = 157 specimens) [2] Specimens from non‐cases recruited from 13 different sources (n = 3308 specimens) Recruitment: unclear Prospective or retrospective recruitment of cases: unclear Sample size (virus/COVID cases): 3481 (3324) specimens Inclusion and exclusion criteria: none stated | ||
| Patient characteristics and setting | Setting: not described for most sample sets Location: not described for most sample sets Country: USA, China, but not described for most sample sets Dates: not described Symptoms and severity: not described Sex: not described Age: not described Exposure history: not described | ||
| Index tests | Test name: unnamed test Manufacturer: Premier Biotech, Minneapolis, USA (may be Hangzhou Alltest) Ab targets: IgG, IgM Antigens used: NR Test method: NR Timing of samples: NR Samples used: serum, plasma, fingerstick blood, venous whole blood (may be blood for majority of cases) Test operators: NR Definition of test positivity: NR Blinded to reference standard: no Threshold predefined: NR | ||
| Target condition and reference standard(s) | Reference standard for cases: various unclear, includes RT‐PCR‐pos Samples used: NR Timing of reference standard: NR Blinded to index test: yes Incorporated index test: serology tests were included in 1 cohort Reference standard for non‐cases: pre‐pandemic, RT‐PCR‐neg, healthy volunteers | ||
| Flow and timing | Time interval between index and reference tests: NR Results presented by time period: no All participants received the same reference standard: no Missing data: none mentioned Uninterpretable results: none mentioned Indeterminate results: none mentioned Unit of analysis: specimens | ||
| Comparative | |||
| Notes | Funding: individual donors Publication status: preprint (not peer reviewed) Source: medRxiv Study author COI: none declared | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | No | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | No | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | High risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Unclear | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Unclear | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| The reference standard does not incorporate the index test | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Unclear | ||
| Were results presented per patient? | No | ||
| Could the patient flow have introduced bias? | High risk | ||
Burbelo 2020 [A].
| Study characteristics | |||
| Patient Sampling | 2‐group study recruiting patients estimating sensitivity and specificity [1] 35 patients with COVID‐19 symptoms and RT‐PCR‐positive [2] 32 pre‐pandemic blood donors Recruitment: unclear Prospective or retrospective recruitment of cases: retrospective Sample size (virus/COVID cases): 67 (35) Inclusion and exclusion criteria: nothing additional The study included a 3rd group on COVID‐19 suspects (n = 10) who were not included as they had no reference standard diagnosis | ||
| Patient characteristics and setting | Setting [1] Hospital patients [2] Blood donors Location [1] University of California, San Diego; University of Washington, Seattle; EvergreenHealth, Kirkland, Washington; NIH Clinical Center, NIH [2] NIH Clinical Center, NIH Country: USA Dates: NR Symptoms and severity: [1] 37% (13/35) were on a ventilator Sex: [1] 87% (30/35) male [2] NR Age: [1] median age 44 years (range 32‐50 years) [2] NR Exposure history: NR |
||
| Index tests | This entry (Burbelo 2020 [A]) refers to the LIPS assay to detect antibodies to the nucleocapsid (N) protein Test name: LIPS Manufacturer: in‐house Ab targets: antibodies for the nucleocapsid and S proteins Antigens used: nucleocapsid and S proteins Test method: LIPS Timing of samples: 2‐50 days pso Samples used: plasma or serum Test operators: presumed laboratory researchers Definition of test positivity: 125,000 LU for nucleocapsid and 45,000 LU for S proteins Blinded to reference standard: unclear Threshold predefined: no, derived from analysis of group [2] to achieve 100% specificity |
||
| Target condition and reference standard(s) | Reference standard for cases: RT‐PCR Samples used: nasal or throat swabs Timing of reference standard: unclear Blinded to index test: yes Incorporated index test: no Reference standard for non‐cases: pre‐pandemic controls (no testing) | ||
| Flow and timing | Time interval between index and reference tests: unclear Results presented by time period: ≤ 14 days; > 14 days All participants received the same reference standard: no Missing data: yes ‐ unclear why there are different numbers for spike and nucleocapsid tests Uninterpretable results: not mentioned Indeterminate results: not mentioned | ||
| Comparative | |||
| Notes | Funding: intramural research programmes of the National Institute of Dental and Craniofacial Research, the National Institute of Allergy and Infectious Diseases, and the National Institute of Health Clinical Center Publication status: preprint (not peer reviewed) Source: medRxiv Study author COI: none declared | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | No | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | No | ||
| Could the conduct or interpretation of the index test have introduced bias? | High risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Unclear | ||
| Were results presented per patient? | No | ||
| Could the patient flow have introduced bias? | High risk | ||
Burbelo 2020 [B].
| Study characteristics | |||
| Patient Sampling | See main entry for this study for characteristics and QUADAS‐2 assessment (Burbelo 2020 [A]) | ||
| Patient characteristics and setting | See main entry for this study for characteristics and QUADAS‐2 assessment (Burbelo 2020 [A]) | ||
| Index tests | This entry (Burbelo 2020 [B]) refers to the LIPS assay to detect antibodies to the spike (S) protein; see (Burbelo 2020 [A] for further study characteristics and QUADAS‐2 assessments) Test name: LIPS Manufacturer: in‐house Ab targets: antibodies for the nucleocapsid and S proteins Antigens used: nucleocapsid and S proteins Test method: LIPS Timing of samples: 2‐50 days pso Samples used: plasma or serum Test operators: presumed laboratory researchers Definition of test positivity: 125,000 LU for nucleocapsid and 45,000 LU for S proteins Blinded to reference standard: unclear Threshold predefined: no ‐ derived from analysis of group [2] to achieve 100% specificity |
||
| Target condition and reference standard(s) | See main entry for this study for characteristics and QUADAS‐2 assessment (Burbelo 2020 [A]) | ||
| Flow and timing | See main entry for this study for characteristics and QUADAS‐2 assessment (Burbelo 2020 [A]) | ||
| Comparative | |||
| Notes | |||
Cai 2020a.
| Study characteristics | |||
| Patient Sampling | 2‐group study to estimate sensitivity and specificity for detection of active disease
[1] Laboratory‐confirmed COVID‐19 patients (n = 276)
[2] Controls with other infections (n = 167). A third group of healthy controls was used to set thresholds (n = 200) but not estimate accuracy. Recruitment method: NR if patients were consecutive Sample size (viral/COVID cases): 443 (276) Exclusion criteria: none stated |
||
| Patient characteristics and setting | [1] Hospital (inpatients); Chongqing Three Gorges Central Hospital, Yongchuan Hospital Affiliated to Chongqing Medical University (CQMU), and The Public Health Center, in Chongqing, China (recruitment dates NR). 168/276 (61%) had fever. Median age 48 (IQR 37‐56; range 0‐84) years, 151/276 (55%) male. 99/276 (36%) reported known exposure
[2] Controls with other infection (n = 167); Second Hospital Affiliated to CQMU and Children’s Hospital Affiliated to CQMU; time NR. Other infections included: influenza A virus (25), respiratory syncytial virus (7), parainfluenza 111 virus (8), influenza B virus (5), adenovirus (6), Klebsiella pneumoniae (8), Streptococcus pneumoniae (3), Mycoplasma (5), Acinetobacter baumannii (10), Candida albicans (2), Staphylococcus aureus (3), Mycobacterium tuberculosis (4), Hepatitis B virus (33), Hepatitis C virus (22), Syphilis (23) and Saccharomycopsis (3) [3] Healthy controls (n = 200), source NR; recruited > 1 year before the outbreak. No further details |
||
| Index tests | 1 Ab test, blinding NR Laboratory‐based in‐house luminescent immunoassay (CLIA) using serum samples Measured IgM +IgG. Antigen: peptide from SARS‐CoV‐2 S protein Test threshold: determined as the mean luminescence (CL) value of the 200 normal sera plus 5 folds of SD; cut‐off used ≥ 0.7 CL (for both IgG and IgM). (Determined in the healthy control group) Samples acquired day 2‐day 27 after symptoms. Person applying the test NR. | ||
| Target condition and reference standard(s) | 1. Real time RT‐PCR detection of virus RNA, samples not described. Reference threshold and timing NR. Blinded to index test 2. Healthy controls, pre‐December 2019 | ||
| Flow and timing | Time interval between index and reference: NR. Accuracy results were not disaggregated by time period since point of symptom onset. No missing data, uninterpretable or indeterminate results described Analysis participant‐based | ||
| Comparative | |||
| Notes | Funded by Emergency Project from the Science & Technology Commission of Chongqing; Major National S&T program grant from Science & Technology Commission of China; Grant from the National Natural Science Foundation of China, Grant from the Science & Technology Commission of Yuzhong district, Chongqing. COI (reported or derived): study author employed by BioScience Co. LTD, Tianjin, China Publication status (source): preprint (not peer reviewed) (medRxiv) NOTE: Study author institution reported as BioScience Co. LTD, Tianjin, China (www.bioscience‐tj.com/en/about.php) | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | No | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Unclear | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Cassaniti 2020 (A).
| Study characteristics | |||
| Patient Sampling | The report contains 3 different groups that fit with 2 different comparisons. 2‐group design with separate estimates of sensitivity and specificity [1] COVID‐19‐positive patients in ICU (n = 30) [2] Healthy volunteers with negative RT‐PCR results (n = 30) Recruitment: unclear Sample size (virus/COVID cases): 60 (30) Inclusion and exclusion criteria: not further described (Single group recruiting individuals presenting with symptoms extracted as Cassaniti 2020 (B)) | ||
| Patient characteristics and setting | Setting: hospital inpatients (Infectious Diseases Unit or ICU, Tertiary hospital) Location: Fondazione IRCCS Policlinico San Matteo, Pavia Country: Italy Dates: NR Symptoms and severity: [1] NR [2] NR Sex: [1] 83% male (25/30); [2] 55% Male (11/30) Age: [1] median age, 73.5; range 38‐86 years; [2] median age, 38.5; range 25‐69 years) Exposure history: [1] NR; [2] 10 (33.3%) previously infected with common OC43, 229E, HKU1, and NL63 coronavirus | ||
| Index tests | Test name: VivaDiag COVID‐19 IgM/IgG Manufacturer: VivaChek Ab targets: IgM, IgG Antigens used: NR Test method: LFIA Timing of samples: [1] median 7 days (IQR 4‐11) after first test; [2] NR Samples used: serum or blood Test operators: NR Definition of test positivity: visible line Blinded to reference standard: unclear Threshold predefined: yes | ||
| Target condition and reference standard(s) | Reference standard for cases: RT‐PCR targeting RNA‐dependent RNA polymerase and E genes were used to detect the presence of SARS‐CoV‐2 according to the WHO guidelines (2 negatives required for non‐cases) Samples used: respiratory samples Timing of reference standard: [1] during patient care [2] unclear Blinded to index test: yes Incorporated index test: no | ||
| Flow and timing | Time interval between index and reference tests: no information Results presented by time period: no information All participants received the same reference standard: yes Missing data: none mentioned Uninterpretable results: none mentioned Indeterminate results: none mentioned Unit of analysis: participants | ||
| Comparative | |||
| Notes | Funding: VivaDiag COVID‐19 IgM/IgG Rapid Test provided free of charge by the Italian Chinese community. Regional Health Authority of Lombardy, Milan, Italy and Italian Ministry of Health, Ricerca Finalizzata Publication status: published letter Source: academic journal Study author COI: none mentioned | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | No | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Cassaniti 2020 (B).
| Study characteristics | |||
| Patient Sampling | Single‐group study estimating sensitivity and specificity
Patients presenting to A&E with fever and respiratory symptoms indicative of COVID‐19 infection. 2 additional cohorts extracted as separate 2‐group study (Cassaniti 2020 (A)) Recruitment: unclear Sample size (virus/COVID cases): 50 (38) Inclusion and exclusion criteria: NR |
||
| Patient characteristics and setting | Setting: A&E Location: Fondazione IRCCS Policlinico San Matteo, Pavia Country: Italy Dates: NR Symptoms and severity: NR Sex: 68% male 34 male/16 female Age: median age, 61.50; range 33‐97 years Exposure history: NR | ||
| Index tests | Test name: VivaDiag COVID‐19 IgM/IgG
Manufacturer: VivaChek
Ab targets: IgM, IgG
Antigens used: NR
Test method: LFIA
Timing of samples: on presentation at A&E
Samples used: serum or blood
Test operators: NR
Definition of test positivity: visible line
Blinded to reference standard: yes on presentation Threshold predefined: yes |
||
| Target condition and reference standard(s) | Reference standard for cases: RT‐PCR targeting RNA‐dependent RNA polymerase and E genes were used to detect the presence of SARS‐CoV‐2 according to the WHO guidelines Samples used: nasal swab Timing of reference standard: on presentation Blinded to index test: yes Incorporated index test: no Reference standard for non‐cases: single negative RT‐PCR result | ||
| Flow and timing | Time interval between index and reference tests: done at the same time Results presented by time period: no (likely to be short as on admission) All participants received the same reference standard: yes Missing data: none mentioned Uninterpretable results: none mentioned Indeterminate results: not mentioned Unit of analysis: participants | ||
| Comparative | |||
| Notes | Funding: VivaDiag COVID‐19 IgM/IgG Rapid Test provided free of charge by the Italian Chinese community. Regional Health Authority of Lombardy, Milan, Italy and Italian Ministry of Health, Ricerca Finalizzata Publication status: published letter Source:Academic journal Study author COI: none mentioned | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Chen 2020a.
| Study characteristics | |||
| Patient Sampling | Unclear whether study recruited as 1 or 2 groups (we describe it as a 2‐group study), estimating sensitivity and specificity [1] RT–PCR‐positive samples, n = 7 samples [2] RT–PCR‐negative samples, but clinically suspicious for COVID‐19, n = 12 samples A 3rd group of 'normal' samples (n = 51), were used to derive test threshold and not included in the accuracy evaluation. Recruitment: unclear Prospective or retrospective recruitment of cases: unclear Sample size (virus/COVID cases): 19 (7) Inclusion and exclusion criteria: NR | ||
| Patient characteristics and setting | Setting: hospital samples Location: Guangzhou Eighth People’s Hospital and Nanfang Hospital, Guangzhou province Country: China Dates: NR Symptoms and severity: [1] NR; [2] fever: 12/12 (100%) Sex: NR Age: NR Exposure history: NR | ||
| Index tests | Test name: no name. LFIA that uses lanthanide‐doped polystyrene nanoparticles (LNPs) Manufacturer: in–house Ab targets: IgG Antigens used: recombinant nucleocapsid phosphoprotein of SARS‐CoV‐2 Test method: LFIA that uses lanthanide‐doped polystyrene nanoparticles (LNPs) Timing of samples: NR Samples used: serum Test operators: NR Definition of test positivity: At/Ac ratio (R) > 0.0666 Blinded to reference standard: NR Threshold predefined: defined from control samples | ||
| Target condition and reference standard(s) | Reference standard for cases: RT–PCR Samples used: NR Timing of reference standard: NR Blinded to index test: yes Incorporated index test: no Reference standard for non‐cases: RT–PCR single negative | ||
| Flow and timing | Time interval between index and reference tests: NR Results presented by time period: NR All participants received the same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results: none reported Unit of analysis: sample | ||
| Comparative | |||
| Notes | Funding: National Natural Science Foundation of China and China Postdoctoral Science Foundation Publication status: peer–reviewed early online Source: academic journal Study author COI: study authors state no competing financial interests | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | No | ||
| Could the patient flow have introduced bias? | High risk | ||
Dohla 2020.
| Study characteristics | |||
| Patient Sampling | 2‐group study recruiting patients estimating sensitivity and specificity [1] COVID‐19 suspects attending community screening (n = 39) [2] Confirmed COVID‐19 cases (n = 10) Recruitment: [1] random selection (no random sampling method stated); [2] unclear Prospective or retrospective recruitment of cases: [1] prospective; [2]retrospective Sample size (virus/COVID cases): 49 (22) Inclusion and exclusion criteria: NR | ||
| Patient characteristics and setting | Setting: [1] community screening centre; [2] NR Location: [1] German Red Cross COVID‐19 testing centre; [2] NR Country: Germany Dates: NR Symptoms and severity: (71%) with dry cough; (65%) with fatigue; (46%) with runny nose (only %s reported). 5/49 (10%) were asymptomatic Sex: 25/49 (51%) male Age: median 46 (IQR 28–72) years Exposure history: identified in 22/49 (45%): median time exposure–to–test of 18.5 days (IQR 15–24) | ||
| Index tests | Test name: NR Manufacturer: NR Ab targets: IgM and IgG Antigens used: SARS‐CoV‐2 antigen (not further described) Test method: CGIA Threshold: visible line ‐ weak and strong responses counted as positive Timing of median time exposure–to–test = 18.5 days (IQR 15–24) known (45%) for samples Samples used: [1] fingerprick blood [2] stored serum Test operators: NR Definition of test positivity: weakly clearly visible line Blinded to reference standard: unclear Threshold predefined: yes | ||
| Target condition and reference standard(s) | Reference standard for cases: [1] RT–qPCR (Altona Diagnostics), threshold NR; [2] RT–qPCR (unknown if same kit), threshold NR Samples used: [1] throat swab; [2] NR Timing of reference standard: [1] same time as index test. [2] NR. For 22 participants (unclear how many in group 1 or 2): median time exposure–to–test of 18.5 days (IQR 15–24) Blinded to index test: NR ‐ presumed Incorporated index test: no Reference standard for non‐cases: single negative RT‐qPCR | ||
| Flow and timing | Time interval between index and reference tests: simultaneous Results presented by time period: no All participants received the same reference standard: yes Missing data: none reported Uninterpretable results: reporting that there were none Indeterminate results: weak lines considered as test positive Unit of analysis: participant | ||
| Comparative | |||
| Notes | Funding: none declared Publication status: published paper (proof) Source: academic journal Study author COI: none declared | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Unclear | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Du 2020.
| Study characteristics | |||
| Patient Sampling | Single group estimating sensitivity in convalescent patients [1] Hospital COVID‐19 convalescent patients (n = 60) Recruitment: unclear Sample size (virus/COVID cases): 60 (60) Inclusion and exclusion criteria: NR | ||
| Patient characteristics and setting | Setting: hospital inpatients (convalescent) Location: Wuhan Tongji Hospital Country: China Dates: admitted 12 January 2020‐5 February 2020 Symptoms and severity: no information Sex: no information Age: no information Exposure history: no information | ||
| Index tests | Test name: NR
Manufacturer: NR
Ab targets: IgM, IgG
Antigens used: NR
Test method: NR but presumed to be CLIA based on reported threshold in AU/mL Timing of samples: during hospital stay (between 3 March 2020 and 14 March 2020) Samples used: NR Test operators: unclear Definition of test positivity: > 10 AU/mL Blinded to reference standard: unclear Threshold predefined: yes |
||
| Target condition and reference standard(s) | Reference standard for cases (including threshold): method NR Samples used: NR Timing of reference standard: diagnosed during initial hospital stay (6‐7 weeks previously) Blinded to index test: yes Incorporated index test: no | ||
| Flow and timing | Time interval between index and reference tests: 6‐7 weeks Results presented by time period: results presented by day since onset All participants received the same reference standard: presumed Missing data: none mentioned Uninterpretable results: none mentioned Indeterminate results: none mentioned Unit of analysis: participant | ||
| Comparative | |||
| Notes | Funding: Beijing Natural Science Foundation Publication status: published letter Source: academic journal Study author COI: none mentioned | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | No | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | High risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Unclear | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Unclear | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Freeman 2020.
| Study characteristics | |||
| Patient Sampling | 2‐group study recruiting patients estimating sensitivity and specificity
[1] confirmed COVID‐19 cases (n = 99)
[2] Healthy adults (n = 377) or with other infections (n = 142) Additionally reports a separate cross‐reactivity study using acute and convalescent paired sera from PCR confirmed commonly circulating coronavirus (229E, NL63, OC43, and HKU1)‐ infected patients Recruitment: unclear Prospective or retrospective recruitment of cases: unclear Sample size (virus/COVID cases): 618 (99) Inclusion and exclusion criteria: [1] convalescent PCR+ COVID‐19 cases sera collected at day 10 pso or later |
||
| Patient characteristics and setting | Setting: NR Location: NR Country: USA Dates: NR Symptoms and severity: [1] NR [2] healthy controls (n = 377); suspected hantavirus (n = 101); HIV (n = 21); hepatitis B virus (n = 10); hepatitis C virus‐positive (n = 10) Sex: NR Age: NR Exposure history: NR | ||
| Index tests | Test name: SARS‐CoV‐2 S protein ELISA Manufacturer: in‐house Ab targets: IgG, IGM and total antibodies Antigens used: pre‐fusion stabilised ectodomain of SARS‐CoV‐2 spike (S) Test method: ELISA Timing of samples: at day 10 pso or later Samples used: serum Test operators: laboratory staff Definition of test positivity: based on optical density signal Blinded to reference standard: NR Threshold predefined: NR | ||
| Target condition and reference standard(s) | Reference standard for cases: PCR Samples used: NR Timing of reference standard: NR Blinded to index test: yes, PCR was performed before index test (inferred) Incorporated index test: no Reference standard for non‐cases: pre‐pandemic (healthy controls or with other diseases) | ||
| Flow and timing | Time interval between index and reference tests: not clear Results presented by time period: no All participants received the same reference standard: no Missing data: not mentioned Uninterpretable results: not mentioned Indeterminate results: not mentioned Unit of analysis: per participant | ||
| Comparative | |||
| Notes | Funding: intramural funding from the National Institute of Allergy and Infectious Diseases Publication status: preprint (not peer‐reviewed) Source: bioRxiv Study author COI: NR | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | No | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Gao 2020a.
| Study characteristics | |||
| Patient Sampling | Single‐group study estimating sensitivity [1] Patients with confirmed COVID‐19 (n = 38) Recruitment: unclear Sample size (virus/COVID cases): 38 (38) Inclusion and exclusion criteria: COVID‐19 confirmed by New Coronavirus Pneumonia Prevention and Control Program (5th edition) published by the National Health Commission of China. | ||
| Patient characteristics and setting | Setting: hospital inpatient Location: Second People's Hospital of Fuyang Country: China Dates: 22 January 2020‐28 February 2020 Symptoms and severity: 3/38 described as in severe or critical conditions; 35/38 described as mild cases Sex: 55.3% (21/38) male Age: median age 40.5 years (IQR 31.0‐49.5years), range 15‐75 years Exposure history: NR | ||
| Index tests | Test name: Colloidal Gold Antibodies Test Manufacturer: Innovita Biological Technology Co., Ltd Ab targets: IgM, IgG Antigens used: NR Test method: CGIA Timing of samples: days 0‐15+ Samples used: serum Test operators: NR Definition of test positivity: visible line Blinded to reference standard: NR Threshold predefined: yes | ||
| Target condition and reference standard(s) | Reference standard for cases: participants met the criteria of the New Coronavirus Pneumonia Prevention and Control Program (5th edition) published by the National Health Commission of China Samples used: NR Timing of reference standard: NR Blinded to index test: yes Incorporated index test: no | ||
| Flow and timing | Time interval between index and reference tests: NR Results presented by time period: yes: 0‐7 days (n = 13), 8‐14 days (n = 8) and ≥ 15 days (n = 23) after onset of symptoms All participants received the same reference standard: yes Missing data: NR Uninterpretable results: NR Indeterminate results: NR Unit of analysis: results reported for participants. 38 participants included and 76 serum samples collected in total from these 38 participants. Median number of samples collected from each participant was 8 | ||
| Comparative | |||
| Notes | Funding: The Science and Technology Bureau of Fuyang Publication status: accepted manuscript (peer reviewed) Source: Journal of Medical Virology Study author COI: none reported | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| The reference standard does not incorporate the index test | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Gao 2020b [A].
| Study characteristics | |||
| Patient Sampling | Single‐group study recruiting patients estimating sensitivity [1] confirmed COVID‐19 cases Recruitment: consecutive (inferred). From all confirmed cases admitted to hospital Prospective or retrospective recruitment of cases: retrospectively (appears) Sample size (virus/COVID cases): 22 participants (corresponding to 37 samples) Inclusion and exclusion criteria: not clearly defined; describes all participants having typical ground‐glass opacity of the lung on CT but not clear if this was part of eligibility | ||
| Patient characteristics and setting | Setting: hospital inpatient Location: Fifth Hospital of Shijiazhuang Country: China Dates: from 21 January‐24 February 2020 Symptoms and severity: typical ground‐glass opacity in lung was observed in CT scan results of all participants. At the time the paper was written all participants had recovered and been discharged from hospital. Sex: 14/22 male (64%) Age: 40 (4‐72) years Exposure history: 11 participants had recent history of travel to epidemic areas, and the remaining 10 had close contacts with their family members, who were confirmed to be infected by 2019‐nCoV | ||
| Index tests |
Gao 2020b [A] is test [A] from the following entry: Test name: [A] CLIA; [B] GICA; [C] ELISA Manufacturer: Beier Bioengineering Company (Beijing, China) Ab targets: IgG and IgM Antigens used: spike (S) and nucleocapsid (N) proteins of 2019‐nCoV Test method: [A] CLIA; [B] GICA; [C] ELISA Timing of samples: [1] early stage (1‐7 days pso) 10/37 samples (27%), [2] middle stage (8‐14 days pso) 13/37 samples (35%); [3] late stage (14‐24 days pso) 14/37 samples (38%) Samples used: serum Test operators: laboratory staff Definition of test positivity: [A] samples with an concentration ≥ 8 arbitrary unit (AU)/mL were considered positive. [B] Visible line. [C] The absorbance at 450 nm (A450 nm) of each well was determined and the cut‐off value was 0.10+Anegative control. A value > cut‐off value was considered a positive result. Blinded to reference standard: NR Threshold predefined: [A] samples with an concentration ≥ 8 arbitrary unit (AU)/mL were considered positive. [B] Positive results showed the appearance of both control line and testing line. [C] The absorbance at 450 nm (A450 nm) of each well was determined and the cut‐off value was 0.10+Anegative control. A value > cut‐off value was considered a positive result. |
||
| Target condition and reference standard(s) | Reference standard for cases: RT‐PCR assay (2019‐nCoV RNA Test Kit, Daan Gene Company, China) Samples used: nasal and pharyngeal swab specimens Timing of reference standard: on admission (most likely) Blinded to index test: yes, index tests performed on already‐confirmed cases (inferred) Incorporated index test: no Reference standard for non‐cases: N/A | ||
| Flow and timing | Time interval between index and reference tests: NR Results presented by time period: yes All participants received the same reference standard: yes Missing data: timing of reference standard test Uninterpretable results: Indeterminate results: Unit of analysis: samples | ||
| Comparative | |||
| Notes | Funding: NR Publication status: published letter Source: Chinese Medical Journal Study author COI: none | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | No | ||
| Could the patient flow have introduced bias? | High risk | ||
Gao 2020b [B].
| Study characteristics | |||
| Patient Sampling | See main entry for this study for characteristics and QUADAS‐2 assessment (Gao 2020b [A]) | ||
| Patient characteristics and setting | See main entry for this study for characteristics and QUADAS‐2 assessment (Gao 2020b [A]) | ||
| Index tests |
Gao 2020b [B] is test [B] from the following entry: Test name: [A] CLIA; [B] GICA; [C] ELISA Manufacturer: Beier Bioengineering Company (Beijing, China) Ab targets: IgG and IgM Antigens used: spike (S) and nucleocapsid (N) proteins of 2019‐nCoV Test method: [A] CLIA; [B] GICA; [C] ELISA Timing of samples: [1] early stage (1‐7 days pso) 10/37 samples (27%), [2] middle stage (8‐14 days pso) 13/37 samples (35%); [3] late stage (14‐24 days pso) 14/37 samples (38%) Samples used: serum Test operators: laboratory staff Definition of test positivity: [A] samples with an concentration ≥ 8 arbitrary unit (AU)/mL were considered positive. [B] Visible line. [C] The absorbance at 450 nm (A450 nm) of each well was determined and the cut‐off value was 0.10+Anegative control. A value > cut‐off value was considered a positive result. Blinded to reference standard: NR Threshold predefined: [A] samples with an concentration ≥ 8 arbitrary unit (AU)/mL were considered positive. [B] Positive results showed the appearance of both control line and testing line. [C] The absorbance at 450 nm (A450 nm) of each well was determined and the cut‐off value was 0.10+Anegative control. A value > cut‐off value was considered a positive result. |
||
| Target condition and reference standard(s) | See main entry for this study for characteristics and QUADAS‐2 assessment (Gao 2020b [A]) | ||
| Flow and timing | See main entry for this study for characteristics and QUADAS‐2 assessment (Gao 2020b [A]) | ||
| Comparative | |||
| Notes | |||
Gao 2020b [C].
| Study characteristics | |||
| Patient Sampling | See main entry for this study for characteristics and QUADAS‐2 assessment (Gao 2020b [A]) | ||
| Patient characteristics and setting | See main entry for this study for characteristics and QUADAS‐2 assessment (Gao 2020b [A]) | ||
| Index tests |
Gao 2020b [C] is test [C] from the following entry: Test name: [A] CLIA; [B] GICA; [C] ELISA Manufacturer: Beier Bioengineering Company (Beijing, China) Ab targets: IgG and IgM Antigens used: spike (S) and nucleocapsid (N) proteins of 2019‐nCoV Test method: [A] CLIA; [B] GICA; [C] ELISA Timing of samples: [1] early stage (1‐7 days pso) 10/37 samples (27%), [2] middle stage (8‐14 days pso) 13/37 samples (35%); [3] late stage (14‐24 days pso) 14/37 samples (38%) Samples used: serum Test operators: laboratory staff Definition of test positivity: [A] samples with an concentration ≥ 8 arbitrary unit (AU)/mL were considered positive. [B] Visible line. [C] The absorbance at 450 nm (A450 nm) of each well was determined and the cut‐off value was 0.10+Anegative control. A value > cut‐off value was considered a positive result. Blinded to reference standard: NR Threshold predefined: [A] samples with an concentration ≥ 8 arbitrary unit (AU)/mL were considered positive. [B] Positive results showed the appearance of both control line and testing line. [C] The absorbance at 450 nm (A450 nm) of each well was determined and the cut‐off value was 0.10+Anegative control. A value > cut‐off value was considered a positive result. |
||
| Target condition and reference standard(s) | See main entry for this study for characteristics and QUADAS‐2 assessment (Gao 2020b [A]) | ||
| Flow and timing | See main entry for this study for characteristics and QUADAS‐2 assessment (Gao 2020b [A]) | ||
| Comparative | |||
| Notes | |||
Garcia 2020 (A).
| Study characteristics | |||
| Patient Sampling | 3‐group study estimating sensitivity and specificity
[1] COVID‐19 patients (n = 55)
[2] Pre‐pandemic healthy controls (n = 45) Third group of patients admitted with a clinical and radiological diagnosis of pneumonia of unknown etiology but RT‐PCR‐negative reported as Garcia 2020 (B) Recruitment: NR Sample size (virus/COVID cases): 100 (55) Inclusion and exclusion criteria: NR |
||
| Patient characteristics and setting | Setting: [1] hospital inpatient [2] pre‐pandemic controls Location: [1] Hospital Universitario Príncipe de Asturias, Madrid [2] Hospital Universitario Príncipe de Asturias Country: Spain Dates: [1] 1 March‐6 April 2020 [2] 1 October‐30 November 2019 Symptoms and severity: NR Sex: [1] male n = 33, 60% [2] male n = 27, 60% Age: [1] median age 63, IQR 50‐79 [2] median age 55, IQR 34‐66 Exposure history: NR | ||
| Index tests | Test name: AllTest COV‐19 IgG / IgM kit Manufacturer: AllTest Biotech, Hangzhou, China Ab targets: IgM, IgG Antigens used: NR Test method: immunochromatography Timing of samples: days 0‐14+ pso Samples used: serum Test operators: laboratory staff Definition of test positivity: visible line Blinded to reference standard: no Threshold predefined: yes | ||
| Target condition and reference standard(s) | Reference standard for cases: RT‐PCR Samples used: NR Timing of reference standard: NR Blinded to index test: yes Incorporated index test: no | ||
| Flow and timing | Time interval between index and reference tests: NR Results presented by time period: yes: < 7 days 15% (n = 8); 7‐13 days 44% (n = 24); ≥ 14 days 42% (n = 23) All participants received the same reference standard: no Missing data: NR Uninterpretable results: NR Indeterminate results: NR Unit of analysis: participants | ||
| Comparative | |||
| Notes | Funding: no funding received Publication status: preprint (not peer reviewed) Source: medRxiv Study author COI: none declared | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Unclear | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Garcia 2020 (B).
| Study characteristics | |||
| Patient Sampling | 3‐group study estimating sensitivity and specificity
[3] Patients admitted with a clinical and radiological diagnosis of pneumonia of unknown etiology but RT‐PCR‐negative (n = 63) 2 additional cohorts extracted as separate 2‐group study (Garcia 2020 (A)) Recruitment: NR Sample size (virus/COVID cases): 100 (55) Inclusion and exclusion criteria: NR |
||
| Patient characteristics and setting | Setting: hospital inpatient Location: Hospital Universitario Príncipe de Asturias, Madrid Country: Spain Dates: 9 February‐2 April 2020 Symptoms and severity: NR Sex: male n = 47, 74% Age: median age 67, IQR 57‐74 Exposure history: NR | ||
| Index tests | Test name: AllTest COV‐19 IgG / IgM kit Manufacturer: AllTest Biotech, Hangzhou, China Ab targets: IgM, IgG. Antigens used: NR Test method: immunochromatography Timing of samples: days 0‐14+ pso Samples used: serum Test operators: laboratory staff Definition of test positivity: visible line Blinded to reference standard: no Threshold predefined: yes | ||
| Target condition and reference standard(s) | Reference standard for cases: clinical diagnosis of COVID‐19 Criteria NR Timing of reference standard: NR Blinded to index test: yes Incorporated index test: no | ||
| Flow and timing | Time interval between index and reference tests: NR Results presented by time period: yes: 7‐13 days 29% (n = 18); ≥ 14 days 71% (n = 45) All participants received the same reference standard: yes Missing data: NR Uninterpretable results: NR Indeterminate results: NR Unit of analysis: participants | ||
| Comparative | |||
| Notes | Funding: no funding received Publication status: preprint (not peer reviewed) Source: medRxiv Study author COI: none declared | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Unclear | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| The reference standard does not incorporate the index test | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Grzelak 2020 [A].
| Study characteristics | |||
| Patient Sampling | 4‐group study to estimate sensitivity and specificity for diagnosing active disease. [1] Hospitalised COVID‐19 patients (51; 161 samples) [2] Pre‐pandemic sera (491) Recruitment: NR (appears retrospective); consecutive or otherwise NR Review team excluded: [3] Blood donors during pandemic (200) [4] Cohort of 209 pauci‐symptomatic suspected cases (mild signs compatible with COVID‐19 ‐fever, cough or dyspnea) who had been in contact with a confirmed case as no reference standard reported |
||
| Patient characteristics and setting | Setting: inpatient Location: Hôpital Bichat, Paris Country: France Dates: NR | ||
| Index tests | This entry (Grzelak 2020 [A]) refers to test [A] in the list below: 5 tests evaluated: A. LIPS (S1 protein) B. LIPS (N protein) C. ELISA (N protein) D. S‐Flow (unknown) E. ELISA tri‐S (S protein) Manufacturer: in‐house Ab targets: A. total Ab; B. total Ab; C. IgG; D IgM or IgG; E. total Ab Antigens used: A. S1; B. N‐based; C. full‐length SARS‐CoV‐2 N protein; D. S at the cell surface; E. trimeric S (recombinant S glycoprotein ectodomain) |
||
| Target condition and reference standard(s) | Reference standard for cases: NR. Described as confirmed COVID‐19 hospitalised cases only Samples used: not described Timing of reference standard: not described Was it blind to index test: not described | ||
| Flow and timing | Time interval between index and reference tests: NR
Results presented by time period: no
All participants received the same reference standard: no
Missing data: pre‐pandemic sera are missing from the evaluations of ELISA tri‐S (n = 391), S‐flow (n = 357) and LIPS S1 and N (n = 2) Sample‐based analysis |
||
| Comparative | |||
| Notes | Funding: OS lab is funded by Institut Pasteur, ANRS, Sidaction, the Vaccine Research Institute (ANR‐ 10‐LABX‐77), Labex IBEID (ANR‐10‐LABX‐62 IBEID), “TIMTAMDEN” ANR‐14‐CE14‐0029, “CHIKV‐Viro‐ Immuno” ANR‐14‐CE14‐0015‐01 and the Gilead HIV cure program. LG is supported by the French Ministry of Higher Education, Research and Innovation.ME lab is funded by Institut Pasteur, Labex IBEID (ANR‐10‐LABX‐62‐IBEID), Reacting, EU grant Recover, ANR Oh’ticks. HM received core grants from the G5 Institut Pasteur Program, the Milieu Intérieur Program (ANR‐10‐LABX‐69‐01) and INSERM. C.P. is supported by a fellowship from the Agence Nationale de Recherches sur le Sida et les Hépatites Virales (ANRS). SVDW lab is funded by Institut Pasteur, CNRS, Université de Paris, Santé publique France, Labex IBEID (ANR‐10‐LABX‐62‐IBEID), REACTing, EU grant Recover. Publication status: preprint Source: medRxiv Study author COI: PC is the founder and CSO of TheraVectys | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | No | ||
| Could the conduct or interpretation of the index test have introduced bias? | High risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Unclear | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| The reference standard does not incorporate the index test | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | No | ||
| Could the patient flow have introduced bias? | High risk | ||
Grzelak 2020 [B].
| Study characteristics | |||
| Patient Sampling | See main entry for this study for characteristics and QUADAS‐2 assessment (Grzelak 2020 [A]) | ||
| Patient characteristics and setting | See main entry for this study for characteristics and QUADAS‐2 assessment (Grzelak 2020 [A]) | ||
| Index tests | This entry (Grzelak 2020 [B]) refers to test [B] in the list below; see Grzelak 2020 [A] for further study characteristics and QUADAS‐2 assessments) 5 tests evaluated: A. LIPS (S1 protein) B. LIPS (N protein) C. ELISA (N protein) D. S‐Flow (unknown) E. ELISA tri‐S (S protein) Manufacturer: in‐house Ab targets: A. total Ab; B. total Ab; C. IgG; D IgM or IgG; E. total Ab Antigens used: A. S1; B. N‐based; C. full‐length SARS‐CoV‐2 N protein; D. S at the cell surface; E. trimeric S (recombinant S glycoprotein ectodomain) |
||
| Target condition and reference standard(s) | See main entry for this study for characteristics and QUADAS‐2 assessment (Grzelak 2020 [A]) | ||
| Flow and timing | See main entry for this study for characteristics and QUADAS‐2 assessment (Grzelak 2020 [A]) | ||
| Comparative | |||
| Notes | |||
Grzelak 2020 [C].
| Study characteristics | |||
| Patient Sampling | See main entry for this study for characteristics and QUADAS‐2 assessment (Grzelak 2020 [A]) | ||
| Patient characteristics and setting | See main entry for this study for characteristics and QUADAS‐2 assessment (Grzelak 2020 [A]) | ||
| Index tests | This entry (Grzelak 2020 [C]) refers to test [C] in the list below; see Grzelak 2020 [A] for further study characteristics and QUADAS‐2 assessments) 5 tests evaluated: A. LIPS (S1 protein) B. LIPS (N protein) C. ELISA (N protein) D. S‐Flow (unknown) E. ELISA tri‐S (S protein) Manufacturer: in‐house Ab targets: A. total Ab; B. total Ab; C. IgG; D IgM or IgG; E. total Ab Antigens used: A. S1; B. N‐based; C. full‐length SARS‐CoV‐2 N protein; D. S at the cell surface; E. trimeric S (recombinant S glycoprotein ectodomain) |
||
| Target condition and reference standard(s) | See main entry for this study for characteristics and QUADAS‐2 assessment (Grzelak 2020 [A]) | ||
| Flow and timing | See main entry for this study for characteristics and QUADAS‐2 assessment (Grzelak 2020 [A]) | ||
| Comparative | |||
| Notes | |||
Grzelak 2020 [D].
| Study characteristics | |||
| Patient Sampling | See main entry for this study for characteristics and QUADAS‐2 assessment (Grzelak 2020 [A]) | ||
| Patient characteristics and setting | See main entry for this study for characteristics and QUADAS‐2 assessment (Grzelak 2020 [A]) | ||
| Index tests | This entry (Grzelak 2020 [D]) refers to test [D] in the list below; see Grzelak 2020 [A] for further study characteristics and QUADAS‐2 assessments) 5 tests evaluated: A. LIPS (S1 protein) B. LIPS (N protein) C. ELISA (N protein) D. S‐Flow (unknown) E. ELISA tri‐S (S protein) Manufacturer: in‐house Ab targets: A. total Ab; B. total Ab; C. IgG; D IgM or IgG; E. total Ab Antigens used: A. S1; B. N‐based; C. full‐length SARS‐CoV‐2 N protein; D. S at the cell surface; E. trimeric S (recombinant S glycoprotein ectodomain) |
||
| Target condition and reference standard(s) | See main entry for this study for characteristics and QUADAS‐2 assessment (Grzelak 2020 [A]) | ||
| Flow and timing | See main entry for this study for characteristics and QUADAS‐2 assessment (Grzelak 2020 [A]) | ||
| Comparative | |||
| Notes | |||
Grzelak 2020 [E].
| Study characteristics | |||
| Patient Sampling | See main entry for this study for characteristics and QUADAS‐2 assessment (Grzelak 2020 [A]) | ||
| Patient characteristics and setting | See main entry for this study for characteristics and QUADAS‐2 assessment (Grzelak 2020 [A]) | ||
| Index tests | This entry (Grzelak 2020 [E]) refers to test [E] in the list below; see Grzelak 2020 [A] for further study characteristics and QUADAS‐2 assessments) 5 tests evaluated: A. LIPS (S1 protein) B. LIPS (N protein) C. ELISA (N protein) D. S‐Flow (unknown) E. ELISA tri‐S (S protein) Manufacturer: in‐house Ab targets: A. total Ab; B. total Ab; C. IgG; D IgM or IgG; E. total Ab Antigens used: A. S1; B. N‐based; C. full‐length SARS‐CoV‐2 N protein; D. S at the cell surface; E. trimeric S (recombinant S glycoprotein ectodomain) |
||
| Target condition and reference standard(s) | See main entry for this study for characteristics and QUADAS‐2 assessment (Grzelak 2020 [A]) | ||
| Flow and timing | See main entry for this study for characteristics and QUADAS‐2 assessment (Grzelak 2020 [A]) | ||
| Comparative | |||
| Notes | |||
Guo 2020a.
| Study characteristics | |||
| Patient Sampling | 2‐group study estimating sensitivity and specificity for detection of active disease in people with suspected or confirmed SARS‐Cov‐2 infection and other infection controls. 1. Cases ‐ 101 inpatients from Wuhan (43 PCR confirmed and 58 probable) provided 169 paired throat and blood samples (69 from confirmed and 100 from probable) 2. Cases ‐ 39 inpatient confirmed cases from Beijing provided 39 samples (total of 208 samples) 3. Control samples provided by people with acute LRTI (135) Family cluster also recruited but does not contribute data. Healthy individuals (150) used to define threshold. Additional plasma samples positive for human CoV‐229E, ‐NL63, ‐OC43, ‐HKU1, and SARS‐CoV previously obtained were included for Western Blot cross‐reactivity analysis. Recruitment method NR | ||
| Patient characteristics and setting | 1. Inpatients at Wuhan hospitals. 43 confirmed cases (PCR or deep sequencing): 20 severe; 23 mild to moderate; 58 possible cases (test‐negative but with clinical signs, X‐ray evidence): 5 severe, 53 mild to moderate. Exposure history NR 2. Beijing hospitals, China (recruitment dates January 2020); 8 severe and 31 mild to moderate. No further details 3. Acute LRTI infection controls: 135 samples from adult patients. No further detail (Family cluster of 6; aged 2‐64, 3 male 3 female. Healthy control samples from Wuhan City adult health check‐ups, 2018‐19) | ||
| Index tests | 3 ELISA assays, blinding NR In‐house ELISA (indirect, laboratory‐based, using blood/plasma samples. Measured IgM, IgA, IgG. Antigen: rNPs (recombinant N protein) from SARS‐CoV‐2 virus Test threshold determined from mean values and SD of healthy individual plasma (calculated the mean absorbance at 450 nm (A450) of the negative sera plus 3 folds of the SD values which were 0.13, 0.1 and 0.30 for IgM, IgA, and IgG, respectively. Samples acquired 1‐39 days after disease onset (41/208 at 1‐7 days; 84/208 at 8‐14 days; 83 > 14 days pso). Person applying the test not described | ||
| Target condition and reference standard(s) | 1. and 2. Confirmed cases ‐ deep sequencing or a qPCR assay with a detection limit of 1 copy/μL, using throat swabs samples. Positivity threshold: NR. Probable cases ‐ clinical manifestation, chest radiography imaging and epidemiology but no virus detected by deep sequencing or qPCR. Timing NR. Not blinded to index test. 3. LRTI controls: pre‐pandemic samples (2018‐2019) | ||
| Flow and timing | Differential verification: all cases had RT‐PCR but some were negative, plus controls did not have RT‐PCR. Time interval between index and reference: presumed short. There are multiple samples for some participants (cases) but others contribute only sample with a range of days pso; only data for 1‐7 days pso can be disaggregated from the rest. Missing data, uninterpretable and indeterminate results not described Per participant and per sample data can be extracted |
||
| Comparative | |||
| Notes | Funded by Chinese Academy of Medical Sciences (CAMS) Innovation Fund for Medical Sciences, Non‐profit Central Research Institute Fund of CAMS, National Major Science & Technology Project for Control and Prevention of Major Infectious Diseases in China. No conflicts of interest reported Publication status: preprint | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | No | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Hu 2020a.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity for detecting active or prior infection Confirmed COVID‐19 patients (211) Recruitment: NR; likely retrospective. Consecutive or otherwise NR | ||
| Patient characteristics and setting | Setting: inpatient Location: Chongqing Three Gorges Central Hospital, Chongqing. Country: China Dates: 23 January‐3 March | ||
| Index tests | Test name: Magnetic Chemiluminescence Enzyme Immunoassay (MCLIA) kit Manufacturer: Bioscience Co., Ltd (Chongqing, China) Ab targets: IgM, IgG Antigens used: N and S (nucleoprotein and a peptide from the SARS SARS‐CoVCoV‐2 S protein) | ||
| Target condition and reference standard(s) | Reference standard for cases: Chinese CDC guidelines (Trial Version 6); included RT‐PCR Samples used: NR Timing of reference standard: unclear; appears that repeat PCR undertaken during hospitalisation; 74/211 met discharge criteria during study period (normal temperature, significantly improving respiratory symptoms and chest radiology plus 2 repeat negative PCRs with ≥ 1‐day interval) Was it blind to index test: unclear | ||
| Flow and timing | Time interval between index and reference tests: NR Results presented by time period: yes All participants received the same reference standard: yes Missing data: none described; however text states 993 samples but only 409 reported for IgM and 507 for IgG Uninterpretable results: none described | ||
| Comparative | |||
| Notes | Funding: funded by Chongqing Education Board “new coronavirus infection and prevention” emergency scientific research project (KYYJ202006YYJ202006). Chongqing Science and Technology Bureau “new crown pneumonia epidemic emergency science and technology special” the fourth batch of projects. Famous teacher project of Chongqing talent plan Publication status: preprint Source: medRxiv Study author COI: none declared | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| The reference standard does not incorporate the index test | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Infantino 2020.
| Study characteristics | |||
| Patient Sampling | 3‐group study recruiting patients estimating sensitivity and specificity [1] COVID‐19 confirmed [2] Rheumatic disease or infectious disease control group (2018‐19; pre COVID‐19 era) [3] Blood donor control group (November/December 2019) Recruitment: unclear Prospective or retrospective recruitment of cases: prospective Sample size (virus/COVID cases): 125 (51 COVID cases) Inclusion and exclusion criteria: COVID‐19 cases were confirmed by RT‐PCR | ||
| Patient characteristics and setting | Setting: hospital inpatient Location: San Giovanni di Dio Hospital, Florence Country: Italy Dates: NR Symptoms and severity: 30/61 (49%) mild to moderate symptoms 31/61 (51%) with severe pneumonia required admission to the ICU Sex: [1] 26/61 (43%) male [2] 26/61 (43%) male [3] 12/20 (60%) male Age: [1] mean 59 ± 23 years; [2] mean 49 ± 17 years; [3] 44 ± 11 years Exposure history: NR | ||
| Index tests | Test name: SARS CoV‐2 antibodies IgM and IgG CLIA kits (analysed with iFlash1800 fully automatic CLIA) Manufacturer: Shenzhen YHLO Biotech Co., Ltd (China) Ab targets: IgM or IgG Antigens used: N protein and S protein Test method: CLIA Timing of samples: NR Samples used: blood (discussion mentions serum) Test operators: NR Definition of test positivity: ≥ 10 AU/mL Blinded to reference standard: NR Threshold predefined: yes | ||
| Target condition and reference standard(s) | Reference standard for cases: RT‐PCR Samples used: OP and NP swabs Timing of reference standard: NR Blinded to index test: NR Incorporated index test: no Reference standard for non‐cases: [2] pre‐pandemic; [2] NR (contemporaneous blood donors) | ||
| Flow and timing | Time interval between index and reference tests: NR Results presented by time period: no All participants received the same reference standard: no Missing data: no Uninterpretable results: no Indeterminate results: no Unit of analysis: participant | ||
| Comparative | |||
| Notes | Funding: NR Publication status: accepted Source: Journal of Medical Virology Study author COI: NR | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Unclear | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Jia 2020.
| Study characteristics | |||
| Patient Sampling | Single‐group study estimating sensitivity for detection of active or recent infection in people with suspected COVID‐19.
Patients with highly suspected COVID‐19 (n = 57; 24 PCR‐positive) defined by exposure history, one of:
and by clinical manifestations, 2 of:
|
||
| Patient characteristics and setting | Inpatients at > 20 hospitals of ShenZhen, China (recruitment dates NR). Sample characteristics and exposure history not described | ||
| Index tests | 1 Ab test, blinding NR LFA. Time‐Resolved Immunofluorescence assay (needs fluorescence analyser); Beijing Diagreat Biotechnologies Co., Ltd,Lot: 20200214). Samples and timing of sampling not described. Measured IgM and IgG; antigen not described. Threshold (Flu) for IgM ≥ 0.88 Flu and IgG ≥ 1.02 Flu. Estimated from 242 healthy people without related diseases (95% of the values were negative) Person applying the test not described | ||
| Target condition and reference standard(s) | RT‐PCR using 2 kits from one of 6 companies (DAAN, Sansure Biotech, BGI, ShangHai ZJ Biotech, Geneodx, Biogerm) across 20 different hospitals. Each participant tested 3 times at different time points (24 positive on first test, all negative on 2nd and 3rd tests), using pharyngeal swabs (acquired 1‐34 days from exposure to first test). Negative on all PCR tests classed as D‐ for purposes of this review
For PCR‐negative, clinical diagnosis criteria required exposure history plus 2 (1) fever and (or) respiratory symptoms; (2) conforming to the imaging features; (3) white blood cells are normal or reduced in early stage of disease, and lymphocyte count is reduced. If there was no clear epidemiological history, 3 clinical manifestations required. No guideline cited but criteria clearly defined. Blinding to index test NR |
||
| Flow and timing | All received same reference standard but not all PCR‐positive; Time interval between index and reference not described. (Serology sample timing NR; PCR was 1‐34 days from exposure to confirmed case. Time pso NR) No missing data, uninterpretable or indeterminate results reported Participant‐based analysis | ||
| Comparative | |||
| Notes | No funding sources described COI: none described Publication status: preprint (not peer reviewed) | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Jin 2020.
| Study characteristics | |||
| Patient Sampling | 2‐group study recruiting patients estimating sensitivity and specificity [1] Laboratory‐confirmed COVID‐19 patients (n = 43); reported separately for 27 patients while still PCR‐positive and for 34 patients after becoming PCR‐negative (excluded from review) [2] Patients admitted with suspected SARS‐CoV‐2 infection, in whom the disease was eventually excluded in the hospital and who quarantined at home, were included as a control group (n = 33) Recruitment: unclear Sample size (virus/COVID cases): 76 (43) Inclusion and exclusion criteria: suspected SARS‐CoV‐2 infection (fever or any respiratory symptoms, especially in those with a history of travel to Wuhan or exposure to an infected case within 2 weeks) | ||
| Patient characteristics and setting | Setting: hospital inpatients Location: Xixi Hospital of Hangzhou, Zhejiang Province Country: China Dates: January 2020‐4 March 2020 Symptoms and severity: [1] COVID‐19 patients: 27/43 (63%) fever; 26/43 (61%) cough; [2] non‐COVID‐19 patients: 24/43 (73%) fever; 15/33 (46%) cough Sex: [1] COVID‐19 patients: 17/43 (40%) male. [2] Non‐COVID‐19 patients: 22/33 (67%) male Age: [1] COVID‐19 patients: median age 47 (IQR 34–59) years; [2] non‐COVID‐19 patients: median age 31 (IQR 26–38) years Exposure history: [1] NR; [2] NR | ||
| Index tests | Test name: The SARS‐CoV‐2 IgM and IgG CLIA kits
Manufacturer: Shenzhen YHLO Biotech Co., Ltd (China)
Ab targets: IgM, IgG
Antigens used: N protein, S protein
Test method: CLIA
Timing of samples: 1‐55 days pso whilst still in hospital
Samples used: serum
Test operators: laboratory
Definition of test positivity: > 10 AU/mL Blinded to reference standard: unclear Threshold predefined: yes |
||
| Target condition and reference standard(s) | Reference standard for cases: RT‐PCR testing at the Center for Disease Control of Hangzhou Samples used: oral swab or sputum Timing of reference standard: during patient care Blinded to index test: unclear Incorporated index test: no Reference standard for non‐cases: 2 consecutive negative RT‐PCR 24 h apart | ||
| Flow and timing | Time interval between index and reference tests: between 1 and 32 days Results presented by time period: days pso: 0‐5 6% (n = 6); 6‐10 12% (n = 12); 11‐15 15% (n = 15); 16‐20 22% (n = 22); 21‐25 22% (n = 22); 26‐30 15% (n = 15); 31‐55 8% (n = 8) All participants received the same reference standard: yes Missing data: review team excluded serology data for 34 participants after becoming PCR‐negative; no data reported for 16 participants while PCR‐positive Uninterpretable results: none mentioned Indeterminate results: none mentioned Unit of analysis: participants overall; samples by time period | ||
| Comparative | |||
| Notes | Funding: research Project on the Prevention and Treatment of COVID‐19 in Hangzhou (establishment of a clinical diagnosis and treatment system for COVID‐19 with treatment evaluation) Publication status: published paper Source: academic journal Study author COI: none mentioned | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Unclear | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Did the study avoid inappropriate inclusions? | No | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Lassauniere 2020 [A].
| Study characteristics | |||
| Patient Sampling | 2‐group design estimating sensitivity and specificity for 9 tests Groups: [1] COVID‐19‐positive group (n = 30) admitted to ICU; [2] non‐COVID‐19 group (n = 82) including pre‐pandemic (2017) blood donors (n = 10); acute viral respiratory tract infections with other coronaviruses (n = 5) or non‐coronaviruses (n = 45); dengue virus (n = 9), CMV; n = 2 and Epstein Barr virus (n = 10). 1 additional patient positive for both CMV and Epstein Barr virus Recruitment: [1] recruited consecutively (all cases in ICU on a single day); [2] unclear Sample size (virus/COVID cases): 112 (30) Inclusion and exclusion criteria: none stated | ||
| Patient characteristics and setting | Setting: [1] ICU; [2] biobank samples Location: [1] Hillerød Hospital Country: Denmark Dates: NR Symptoms and severity: NR Sex: 75% (24/32) male Age: median 67 years (IQR 52‐76) Exposure history: NR | ||
| Index tests | 9 tests evaluated, 3 ELISA and 6 LFIA; this entry (Lassauniere 2020 [A]), refers to test [A] in the list below: [A] test name: Wantai SARS‐CoV‐2 Ab ELISA Manufacturer: Beijing Wantai Biological Pharmacy Enterprise, Beijing, China; Cat # WS‐1096 Ab targets: total Ab Antigens used: SARS‐CoV‐2 S protein RBD Test method: ELISA Timing of samples: Samples used: serum Test operators: laboratory staff Definition of test positivity: calculated negative control value to 0.160 Blinded to reference standard: no Threshold predefined: yes [B] test name: Anti‐SARS‐CoV‐2 IgG ELISA Manufacturer: Euroimmun Medizinische Labordiagnostika, Lübeck, Germany; Cat # EI 2668‐9601 G Ab targets: IgG Antigens used: SARS‐CoV‐2 S protein subunit 1 (S1) Test method: ELISA Timing of samples: Samples used: serum Test operators: laboratory staff Definition of test positivity: ratio < 0.8 is considered negative, ≥ 0.8 and < 1.1 borderline, and ≥ 1.1 positive. For analysis 1.1 a more stringent cut‐off was used, and all values < 1.1 were considered negative. Blinded to reference standard: no Threshold predefined: yes [C] test name: Anti‐SARS‐CoV‐2 IgA ELISA Manufacturer: Euroimmun Medizinische Labordiagnostika, Lübeck, Germany; Cat # EI 2606‐9601 A Ab targets: IgA Antigens used: SARS‐CoV‐2 S protein subunit 1 (S1) Test method: ELISA Timing of samples: Samples used: serum Test operators: laboratory staff Definition of test positivity: ratio < 0.8 is considered negative, ≥ 0.8 and < 1.1 borderline, and ≥ 1.1 positive. For analysis 1.1 a more stringent cut‐off was used, and all values < 1.1 were considered negative. Blinded to reference standard: no Threshold predefined: yes [D] Test name: 2019‐nCOV IgG/IgM Rapid Test Manufacturer: Dynamiker Biotechnology, Tianjin, China Cat # DNK‐1419‐1 Ab targets: IgM, IgG Antigens used: NR Test method: CGIA Timing of samples: Samples used: serum Test operators: laboratory staff Definition of test positivity: visible line Blinded to reference standard: no Threshold predefined: yes [E] Test name: OnSiteTM COVID‐19 IgG/IgM Rapid Test Manufacturer: CTK Biotech, Poway, CA, USA; Cat # R0180C Ab targets: IgM, IgG Antigens used: NR Test method: CGIA Timing of samples: Samples used: serum Test operators: laboratory staff Definition of test positivity: visible line Blinded to reference standard: no Threshold predefined: yes [F] Test name: Anti‐SARS‐CoV‐2 Rapid Test Manufacturer: AutoBio Diagnostics, Zhengzhou, China; Cat # RTA0204 Ab targets: IgM, IgG Antigens used: NR Test method: CGIA Timing of samples: Samples used: serum Test operators: laboratory staff Definition of test positivity: visible line Blinded to reference standard: no Threshold predefined: yes [G] Test name: Coronavirus Diseases 2019 (COVID‐19) IgM/IgG Ab Test Manufacturer: Artron Laboratories, Burnaby, Canada; Cat # A03‐51‐322 Ab targets: IgM, IgG. Antigens used: NR Test method: CGIA Timing of samples: Samples used: serum Test operators: laboratory staff Definition of test positivity: visible line Blinded to reference standard: no Threshold predefined: yes [H] Test name: 2019‐nCoV IgG/IgM Rapid Test Cassette Manufacturer: Acro Biotech, Rancho Cucamonga, CA, USA; Cat # INCP‐402 Ab targets: IgM, IgG Antigens used: NR Test method: CGIA Timing of samples: Samples used: serum Test operators: laboratory staff Definition of test positivity: visible line Blinded to reference standard: no Threshold predefined: yes [I] Test name: 2019‐nCoV IgG/IgM Rapid Test Cassette Manufacturer: Hangzhou Alltest Biotech, Hangzhou, China; Cat # INCP‐402 Ab targets: IgM, IgG Antigens used: NR Test method: CGIA Timing of samples: Samples used: serum Test operators: laboratory staff Definition of test positivity: visible line Blinded to reference standard: no Threshold predefined: yes | ||
| Target condition and reference standard(s) | Reference standard for cases (including threshold): viral nucleic acid detection (no further detail) in hospital patients Samples used: respiratory Timing of reference standard: during hospital stay Blinded to index test: yes Incorporated index test: no Reference standard for non‐cases: pre‐pandemic (2017) | ||
| Flow and timing | Time interval between index and reference tests: unclear Results presented by time period: days since onset: 7‐13 (n = 7); 14‐20 (n = 15); ≥ 21 (n = 8) All participants received the same reference standard: no Missing data: some participant samples were not tested with all assays. Only 32 of the 80 control participants were tested with POC assays. Unclear how the 32 were selected Uninterpretable results: not mentioned Indeterminate results: borderline results for [2] and [3] were considered test‐negative. For POC tests, weak signals for IgM and IgG were considered positive. Unit of analysis: participants | ||
| Comparative | |||
| Notes | Funding: Danish National Biobank resource, supported by the Novo Nordisk Foundation Publication status: preprint (not peer reviewed) Source: medRxiv Study author COI: none declared | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Did the study avoid inappropriate inclusions? | No | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Lassauniere 2020 [B].
| Study characteristics | |||
| Patient Sampling | See main entry for this study for characteristics and QUADAS‐2 assessment (Lassauniere 2020 [A]) | ||
| Patient characteristics and setting | See main entry for this study for characteristics and QUADAS‐2 assessment (Lassauniere 2020 [A]) | ||
| Index tests | 9 tests evaluated, 3 ELISA and 6 LFIA; this entry (Lassauniere 2020 [B]) refers to test [B] [B] test name: Anti‐SARS‐CoV‐2 IgG ELISA Manufacturer: Euroimmun Medizinische Labordiagnostika, Lübeck, Germany; Cat # EI 2668‐9601 G Ab targets: IgG. Antigens used: SARS‐CoV‐2 S protein subunit 1 (S1) Test method: ELISA Timing of samples: Samples used: serum Test operators: laboratory staff Definition of test positivity: ratio < 0.8 is considered negative, ≥ 0.8 and < 1.1 borderline, and ≥ 1.1 positive. For analysis 1.1 a more stringent cut‐off was used, and all values < 1.1 were considered negative. Blinded to reference standard: no Threshold predefined: yes |
||
| Target condition and reference standard(s) | See main entry for this study for characteristics and QUADAS‐2 assessment (Lassauniere 2020 [A]) | ||
| Flow and timing | See main entry for this study for characteristics and QUADAS‐2 assessment (Lassauniere 2020 [A]) | ||
| Comparative | |||
| Notes | See main entry for this study for characteristics and QUADAS‐2 assessment (Lassauniere 2020 [A]) | ||
Lassauniere 2020 [C].
| Study characteristics | |||
| Patient Sampling | See main entry for this study for characteristics and QUADAS‐2 assessment (Lassauniere 2020 [A]) | ||
| Patient characteristics and setting | See main entry for this study for characteristics and QUADAS‐2 assessment (Lassauniere 2020 [A]) | ||
| Index tests | Nine tests evaluated, 3 ELISA and six LFIA; this entry (Lassauniere 2020 [C]) refers to test [C] [C] test name: Anti‐SARS‐CoV‐2 IgA ELISA Manufacturer: Euroimmun Medizinische Labordiagnostika, Lübeck, Germany; Cat # EI 2606‐9601 A Ab targets: IgA Antigens used: SARS‐CoV‐2 S protein subunit 1 (S1) Test method: ELISA Timing of samples: Samples used: serum Test operators: laboratory staff Definition of test positivity: ratio < 0.8 is considered negative, ≥ 0.8 and < 1.1 borderline, and ≥ 1.1 positive. For analysis 1.1 a more stringent cut‐off was used, and all values < 1.1 were considered negative. Blinded to reference standard: no Threshold predefined: yes |
||
| Target condition and reference standard(s) | See main entry for this study for characteristics and QUADAS‐2 assessment (Lassauniere 2020 [A]) | ||
| Flow and timing | See main entry for this study for characteristics and QUADAS‐2 assessment (Lassauniere 2020 [A]) | ||
| Comparative | |||
| Notes | |||
Lassauniere 2020 [D].
| Study characteristics | |||
| Patient Sampling | See main entry for this study for characteristics and QUADAS‐2 assessment (Lassauniere 2020 [A]) | ||
| Patient characteristics and setting | See main entry for this study for characteristics and QUADAS‐2 assessment (Lassauniere 2020 [A]) | ||
| Index tests | 9 tests evaluated, 3 ELISA and 6 LFIA; this entry (Lassauniere 2020 [D]) refers to test [D] [D] Test name: 2019‐nCOV IgG/IgM Rapid Test Manufacturer: Dynamiker Biotechnology, Tianjin, China Cat # DNK‐1419‐1 Ab targets: IgM, IgG Antigens used: NR Test method: CGIA Timing of samples: Samples used: serum Test operators: laboratory staff Definition of test positivity: visible line Blinded to reference standard: no Threshold predefined: yes |
||
| Target condition and reference standard(s) | See main entry for this study for characteristics and QUADAS‐2 assessment (Lassauniere 2020 [A]) | ||
| Flow and timing | See main entry for this study for characteristics and QUADAS‐2 assessment (Lassauniere 2020 [A]) | ||
| Comparative | |||
| Notes | |||
Lassauniere 2020 [E].
| Study characteristics | |||
| Patient Sampling | See main entry for this study for characteristics and QUADAS‐2 assessment (Lassauniere 2020 [A]) | ||
| Patient characteristics and setting | See main entry for this study for characteristics and QUADAS‐2 assessment (Lassauniere 2020 [A]) | ||
| Index tests | 9 tests evaluated, 3 ELISA and 6 LFIA; this entry (Lassauniere 2020 [E]) refers to test [E] [E] Test name: OnSiteTM COVID‐19 IgG/IgM Rapid Test Manufacturer: CTK Biotech, Poway, CA, USA; Cat # R0180C Ab targets: IgM, IgG Antigens used: NR Test method: CGIA Timing of samples: Samples used: serum Test operators: laboratory staff Definition of test positivity: visible line Blinded to reference standard: no Threshold predefined: yes |
||
| Target condition and reference standard(s) | See main entry for this study for characteristics and QUADAS‐2 assessment (Lassauniere 2020 [A]) | ||
| Flow and timing | See main entry for this study for characteristics and QUADAS‐2 assessment (Lassauniere 2020 [A]) | ||
| Comparative | |||
| Notes | |||
Lassauniere 2020 [F].
| Study characteristics | |||
| Patient Sampling | See main entry for this study for characteristics and QUADAS‐2 assessment (Lassauniere 2020 [A]) | ||
| Patient characteristics and setting | See main entry for this study for characteristics and QUADAS‐2 assessment (Lassauniere 2020 [A]) | ||
| Index tests | 9 tests evaluated, 3 ELISA and 6 LFIA; this entry (Lassauniere 2020 [F]) refers to test [F] [F] Test name: Anti‐SARS‐CoV‐2 Rapid Test Manufacturer: AutoBio Diagnostics, Zhengzhou, China; Cat # RTA0204 Ab targets: IgM, IgG Antigens used: NR Test method: CGIA Timing of samples: Samples used: serum Test operators: laboratory staff Definition of test positivity: visible line Blinded to reference standard: no Threshold predefined: yes | ||
| Target condition and reference standard(s) | See main entry for this study for characteristics and QUADAS‐2 assessment (Lassauniere 2020 [A]) | ||
| Flow and timing | See main entry for this study for characteristics and QUADAS‐2 assessment (Lassauniere 2020 [A]) | ||
| Comparative | |||
| Notes | |||
Lassauniere 2020 [G].
| Study characteristics | |||
| Patient Sampling | See main entry for this study for characteristics and QUADAS‐2 assessment (Lassauniere 2020 [A]) | ||
| Patient characteristics and setting | See main entry for this study for characteristics and QUADAS‐2 assessment (Lassauniere 2020 [A]) | ||
| Index tests | 9 tests evaluated, 3 ELISA and 6 LFIA; this entry (Lassauniere 2020 [G]) refers to test [G] [G] Test name: Coronavirus Diseases 2019 (COVID‐19) IgM/IgG Ab Test Manufacturer: Artron Laboratories, Burnaby, Canada; Cat # A03‐51‐322 Ab targets: IgM, IgG. Antigens used: NR Test method: CGIA Timing of samples: Samples used: serum Test operators: laboratory staff Definition of test positivity: visible line Blinded to reference standard: no Threshold predefined: yes |
||
| Target condition and reference standard(s) | See main entry for this study for characteristics and QUADAS‐2 assessment (Lassauniere 2020 [A]) | ||
| Flow and timing | See main entry for this study for characteristics and QUADAS‐2 assessment (Lassauniere 2020 [A]) | ||
| Comparative | |||
| Notes | |||
Lassauniere 2020 [H].
| Study characteristics | |||
| Patient Sampling | See main entry for this study for characteristics and QUADAS‐2 assessment (Lassauniere 2020 [A]) | ||
| Patient characteristics and setting | See main entry for this study for characteristics and QUADAS‐2 assessment (Lassauniere 2020 [A]) | ||
| Index tests | 9 tests evaluated, 3 ELISA and 6 LFIA; this entry (Lassauniere 2020 [H]) refers to test [H] [H] Test name: 2019‐nCoV IgG/IgM Rapid Test Cassette Manufacturer: Acro Biotech, Rancho Cucamonga, CA, USA; Cat # INCP‐402 Ab targets: IgM, IgG Antigens used: NR Test method: CGIA Timing of samples: Samples used: serum Test operators: laboratory staff Definition of test positivity: visible line Blinded to reference standard: no Threshold predefined: yes |
||
| Target condition and reference standard(s) | See main entry for this study for characteristics and QUADAS‐2 assessment (Lassauniere 2020 [A]) | ||
| Flow and timing | See main entry for this study for characteristics and QUADAS‐2 assessment (Lassauniere 2020 [A]) | ||
| Comparative | |||
| Notes | |||
Lassauniere 2020 [I].
| Study characteristics | |||
| Patient Sampling | See main entry for this study for characteristics and QUADAS‐2 assessment (Lassauniere 2020 [A]) | ||
| Patient characteristics and setting | See main entry for this study for characteristics and QUADAS‐2 assessment (Lassauniere 2020 [A]) | ||
| Index tests | 9 tests evaluated, 3 ELISA and 6 LFIA; this entry (Lassauniere 2020 [I]) refers to test [I] [I] Test name: 2019‐nCoV IgG/IgM Rapid Test Cassette Manufacturer: Hangzhou Alltest Biotech, Hangzhou, China; Cat # INCP‐402 Ab targets: IgM, IgG Antigens used: NR Test method: CGIA Timing of samples: Samples used: serum Test operators: laboratory staff Definition of test positivity: visible line Blinded to reference standard: no Threshold predefined: yes |
||
| Target condition and reference standard(s) | See main entry for this study for characteristics and QUADAS‐2 assessment (Lassauniere 2020 [A]) | ||
| Flow and timing | See main entry for this study for characteristics and QUADAS‐2 assessment (Lassauniere 2020 [A]) | ||
| Comparative | |||
| Notes | |||
Li 2020a.
| Study characteristics | |||
| Patient Sampling | Single‐group study estimating sensitivity for detection of active or recent infection Particpants with COVID‐19 according to guideline of diagnosis and treatment of COVID‐19 (9 Feb), 525 participants (397 PCR‐positive) Data comparing results using fingerstick blood, serum and plasma for COVID‐19 patients (7) and healthy volunteers (3) not extracted | ||
| Patient characteristics and setting | Samples from various hospitals and CDC testing laboratories (total 8) at 6 different provinces, China Recruitment dates NR Sample characteristics and exposure history not described | ||
| Index tests | 1 Ab test, blinding not described LFIA (colloidal gold). SARS‐CoV‐2 rapid IgG‐IgM combined Ab test kit, from Jiangsu Medomics Medical Technologies, Nanjing, China. Target: IgM and IgG, using recombinant antigen from SARS‐CoV‐2 S protein (MK201027) Threshold predefined, as per manufacturer Tests conducted using serum and plasma from venous blood. Samples acquired by clinical staff at each site. Timing not clearly described (during hospital stay for inpatients); detail provided for 1 site (n = 58), sampling between day 8 and 33 pso | ||
| Target condition and reference standard(s) | COVID‐19 clinically confirmed, according to guideline. (Prevention CCfDCa. The guideline of diagnosis and treatment of COVID‐19. 9 February 2020). PCR test using pharyngeal (throat) swab samples and sputum (threshold NR). Timing not described Presume blinded to index test | ||
| Flow and timing | Time interval between index and reference not described. No disaggregation of results by time pso No missing data, uninterpretable or indeterminate results reported Participant‐based analysis | ||
| Comparative | |||
| Notes | Funding not described Conflicts of interest: 4 co‐authors employed by Jiangsu Medomics Medical Technology Accepted for publication with full peer review | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Lin 2020a [A].
| Study characteristics | |||
| Patient Sampling | 3‐group study estimating sensitivity and specificity [1] COVID‐19 cases (n = 79) [2] Healthy volunteers (n = 29) [3] TB patients (n = 51) Recruitment: 'Random' for [1] (method not stated), no details given for [2] and [3] Sample size (virus/COVID cases): 159 (79) Inclusion and exclusion criteria: for [1]: "combinations of epidemiological risk, clinical features and RT‐PCR respiratory specimen positive" | ||
| Patient characteristics and setting | Setting: [1] specialist COVID hospital (inpatients); [2] university; [3] TB inpatient clinic Location: [1] Third People's Hospital, Shenzhen; [2] Shenzhen University; [3] Shenzhen Baoan Hospital Country: China Dates: NR Symptoms and severity: NR Sex: NR Age: [1] and [3] NR; [2] range 19‐72 Exposure history: NR | ||
| Index tests | 2 tests were evaluated; this entry (Lin 2020a [A]) refers to test [A] in the list below
Test name: [A] not named; [B] commercial ELISA kit
Manufacturer: [A] in‐house; [B] Darui Biotech, China
Ab targets: [A] and [B]: IgM, IgG
Antigens used: [A] recombinant nucleocapsid (YP_009724397.2); [B] SARS‐CoV‐2 N protein
Test method: [A] CLIA; [B] ELISA
Timing of samples: 0 to > 14 days (maximum NR) pso
Samples used: serum
Test operators: NR (assume laboratory staff)
Definition of test positivity: [A] IgM (RLU 162296); IgG (RLU 336697) [B] manufacturer's recommendation
Blinded to reference standard: not mentioned
Threshold predefined: [A] threshold derived from ROC curve; [B] yes (QUADAS ratings are for ELISA test) |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR: GeneoDX kit (Taqman RT‐PCR method, targeting the ORF1ab 101 and N genes) [2] and [3] were persistently negative in at least 3 tests. Samples used: respiratory Timing of reference standard: presume on presentation Blinded to index test: NR Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: unclear Results presented by time period:days 1‐7 (15%); 8‐13 (42%); 14+ (43%) All participants received the same reference standard: yes Missing data: 65/79 D+ serum samples available for ELISA; 64/80 D‐ serum samples available for ELISA; reason not given Uninterpretable results: NR Indeterminate results: NR Unit of analysis: participants | ||
| Comparative | |||
| Notes | Funding: Guangdong Provincial Science and Technology Program, National Natural Science Funds of China, Shenzhen University and the National Science and Technology Major Project Publication status: preprint (not peer reviewed) Source: medRxiv Study author COI: none declared | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | No | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | No | ||
| Could the conduct or interpretation of the index test have introduced bias? | High risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Lin 2020a [B].
| Study characteristics | |||
| Patient Sampling | See main entry for this study for characteristics and QUADAS‐2 assessment (Lin 2020a [A]) | ||
| Patient characteristics and setting | See main entry for this study for characteristics and QUADAS‐2 assessment (Lin 2020a [A]) | ||
| Index tests | 2 tests were evaluated; this entry (Lin 2020a [B]) refers to test [B] in the list below
Test name: [A] not named (CLIA); [B] commercial ELISA kit
Manufacturer: [A] in‐house; [B] Darui Biotech, China
Ab targets: [A] and [B]: IgM, IgG
Antigens used: [A] recombinant nucleocapsid (YP_009724397.2); [B] SARS‐CoV‐2 N protein
Test method: [A] CLIA; [B] ELISA
Timing of samples: 0 to > 14 days (maximum NR) pso
Samples used: serum
Test operators: NR (assume laboratory staff)
Definition of test positivity: [A] IgM (RLU 162296); IgG (RLU 336697) [B] manufacturer's recommendation
Blinded to reference standard: not mentioned
Threshold predefined: [A] threshold derived from ROC curve; [B] yes (QUADAS ratings are for ELISA test) |
||
| Target condition and reference standard(s) | See main entry for this study for characteristics and QUADAS‐2 assessment (Lin 2020a [A]) | ||
| Flow and timing | See main entry for this study for characteristics and QUADAS‐2 assessment (Lin 2020a [A]) | ||
| Comparative | |||
| Notes | See main entry for this study for characteristics and QUADAS‐2 assessment (Lin 2020a [A]) | ||
Lippi 2020 [A].
| Study characteristics | |||
| Patient Sampling | 1‐group study recruiting patients estimating sensitivity and specificity [1] Suspected COVID‐19; subgroup of confirmed cases included Recruitment: consecutive patients Prospective or retrospective recruitment of cases: prospective Sample size (virus/COVID cases): 131 (NR); subgroup of 48 confirmed cases included Inclusion and exclusion criteria: suspected COVID‐19 patients hospitalised, in whom NP and OP swabs were collected along with blood samples during hospital stay, for purposes of COVID‐19 diagnosis and/or monitoring | ||
| Patient characteristics and setting | Setting: hospital inpatients Location: University Hospital of Verona Country: Italy Dates: NR Symptoms and severity: NR Sex: 60/131 (46%) male Age: mean 56 ± 21 years Exposure history: NR | ||
| Index tests | 2 tests were evaluated; this entry (Lippi 2020 [A]) refers to test [A] in the list below Test name: [A] MAGLUMI 2019‐nCoV IgG and IgM (2 indirect tests) [B] Anti‐SARS‐CoV‐2 IgA and IgG ELISA Manufacturer: [A] SNIBE – Shenzhen New Industries Biomedical Engineering Co., Ltd, Shenzhen, China [B] Euroimmun AG, Lübeck, Germany Ab targets: [A] IgM or IgG ; [B] IgA or IgG Antigens used: [A] CoV‐S (spike) and e CoV‐N (nucleocapsid); [B] NR Test method: [A] CLIA; [B] ELISAs Timing of samples: NR Samples used: blood, serum or plasma Test operators: NR Definition of test positivity: [A] ≥ 1.10 AU/mL [B] ≥ 1.1 (absorbance of patient sample/absorbance of calibrator) Blinded to reference standard: NR Threshold predefined: yes by manufacturer |
||
| Target condition and reference standard(s) | Reference standard for cases: RT‐PCR (commercial RT‐PCR method, Seegene AllplexTM2019‐nCoV Assay) Samples used: venous blood Timing of reference standard: during hospital stay Blinded to index test: NR Incorporated index test: no Reference standard for non‐cases: same reference standard, single‐group | ||
| Flow and timing | Time interval between index and reference tests: both during hospital stay Results presented by time period: no All participants received the same reference standard: yes Missing data: NR Uninterpretable results: NR Indeterminate results: 36 Inconclusive results Unit of analysis: per patient | ||
| Comparative | |||
| Notes | Funding: none declared Publication status: published letter Source: Clinical Chemistry and Laboratory Medicine Study author COI: study authors state no conflict of interest | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Lippi 2020 [B].
| Study characteristics | |||
| Patient Sampling | See main entry for this study for characteristics and QUADAS‐2 assessment (Lippi 2020 [A]) | ||
| Patient characteristics and setting | See main entry for this study for characteristics and QUADAS‐2 assessment (Lippi 2020 [A]) | ||
| Index tests | 2 tests were evaluated; this entry (Lippi 2020 [B]) refers to test [B] in the list below Test name: [A] MAGLUMI 2019‐nCoV IgG and IgM (2 indirect tests) [B] Anti‐SARS‐CoV‐2 IgA and IgG ELISA Manufacturer: [A] SNIBE – Shenzhen New Industries Biomedical Engineering Co., Ltd, Shenzhen, China [B] Euroimmun AG, Lübeck, Germany Ab targets: [A] IgM or IgG ; [B] IgA or IgG Antigens used: [A] CoV‐S (spike) and e CoV‐N (nucleocapsid); [B] NR Test method: [A] CLIA (CLIAs); [B] ELISA Timing of samples: NR Samples used: blood, serum or plasma Test operators: NR Definition of test positivity: [A] ≥1.10 AU/mL [B] ≥1.1 (absorbance of patient sample/absorbance of calibrator Blinded to reference standard: NR Threshold predefined: yes by manufacturer |
||
| Target condition and reference standard(s) | See main entry for this study for characteristics and QUADAS‐2 assessment (Lippi 2020 [A]) | ||
| Flow and timing | See main entry for this study for characteristics and QUADAS‐2 assessment (Lippi 2020 [A]) | ||
| Comparative | |||
| Notes | |||
Liu 2020a.
| Study characteristics | |||
| Patient Sampling | Described as one group to estimate sensitivity and specificity (but unclear whether actually recruited as 2 groups) [1]. RT‐PCR confirmed COVID‐19 patients (n = 90) [2]. (a) COVID‐19 suspects with RT‐PCR‐negative results (n = 25) and [2]. (b) inpatients with 'other disease' with RT‐PCR‐negative results (n = 64) Recruitment: unclear Sample size (virus/COVID cases): 179 (90 confirmed; data for 5 clinically confirmed included as D+) Inclusion and exclusion criteria: |
||
| Patient characteristics and setting | All participants considered COVID‐19 suspects (criteria NR)
Setting: hospital (inpatients and outpatients)
Location: General Hospital of Central Theatre Command, Hubei Province
Country: China
Dates: 1 January to 12 March 2020
Group [1]
Symptoms and severity: 46 mild/common cases; 44 severe/critical cases
Sex: M/F: 60:30 (67%)
Age: Age: mean 76 (SD 15) years
Exposure history: NR.
Group [2]
[2a] Diagnoses: COVID‐19 diagnoses: 5 confirmed; 20 suspected. [2b] Non‐COVID‐19 diagnosis: n = 64 (10 cases of Sjogren's syndrome, 8 cases of diabetes, 6 cases of systemic lupus erythematosus, 5 cases of rheumatoid arthritis, 2 cases of dermatomyositis, 2 cases of connective tissue disease, 1 case of scleroderma, and 30 cases of common injuries with no underlying diseases) Sex: M/F: 38:51 (35%) Age: mean 56 (SD 21 ) years Exposure history: NR |
||
| Index tests | Test name: SARS‐CoV‐2 IgG/IgM Ab test kit Manufacturer: A 'Chinese biotechnology company" Ab targets: IgM, IgG Antigens used: NR Test method: LFA (CGIA) Timing of samples: time pso to sample collection mean (SD) (days): PCR‐postiive 30 (17), PCR‐negative 18 (14) Samples used: serum Test operators: NR, but suspect in laboratory (as serum was used) Definition of test positivity: visible line Blinded to reference standard: unclear Threshold predefined: yes | ||
| Target condition and reference standard(s) | Reference standard for cases (including threshold): RT‐PCR test positive or 'clinically confirmed' Samples used: nasal and pharyngeal swabs Timing of reference standard: NR Blinded to index test: NR Incorporated index test: NR Reference standard for non‐cases [2b]: RT‐PCR test negative and diagnosis of alternative condition | ||
| Flow and timing | Time interval between index and reference tests: NR Results presented by time period: known for 115 cases: 0‐7 days: n = 25 (22%); 8‐15 days n = 8 (7%); ≥ 16 days n = 82 (71%) All participants received the same reference standard: no Missing data: none mentioned Uninterpretable results: none mentioned Indeterminate results: none mentioned Unit of analysis: participants | ||
| Comparative | |||
| Notes | Funding: none reported Publication status: preprint (not peer reviewed) Source: medRxiv Study author COI: none reported | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | No | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Liu 2020b.
| Study characteristics | |||
| Patient Sampling | 2‐group study estimating sensitivity and specificity for diagnosing active disease. [1]. Consecutively recruited cohort of patients with confirmed or suspected COVID‐19 (n = 238; 153 PCR confirmed) [2]. Cohort of ordinary patients (n = 70); [3]. Cohort of randomly sampled healthy blood donors (n = 50) randomly sampled No further details | ||
| Patient characteristics and setting | [1]. Inpatients at General Hospital of Central Theater Command of People's Liberation Army (PLA), China (recruitment dates 6‐14 February 2020). Symptoms included fever (87%); dry cough (54%); fatigue (33%). 235/238 (99%) had CT ground glass opacity/patchy shadowing. Exposure history not described. Median age 55 [IQR 38.3‐65] years; 58% male [2]. Ordinary patients, characteristics not described. [3]. Healthy blood donors (n = 50), characteristics not described | ||
| Index tests | 2 Ab tests, blinding NR Both laboratory‐based a. ELISA kit (Lizhu, Zhuhai, China). Measured IgG and IgM detected using recombinant (rN) protein of SARS‐CoV‐2. Test threshold: NR, presumed as per manufacturer b. In‐house CLIA Serum samples acquired 17 (7%) day 0‐5; 41 (17%) day 6‐10; 21 (9%) day 11‐12; 48 (20%) day 13‐15; 111 (47%) day ≥ 16 | ||
| Target condition and reference standard(s) | 1. RT‐PCR (Daan Gene) targeting ORF1ab and N gene; Ct‐value ≤ 40 was defined as a positive test result. Pharyngeal swab specimens used Clinical diagnosis of highly‐suspected cases according to General Office of National Health Committee notice (General Office of National Health Committee. Office of State Administration of Traditional Chinese Medicine. Notice on the issuance of strategic guidelines for diagnosis and treatment of novel coronavirus (2019‐nCoV) infected pneumonia (Fifth edition draft) (2020‐02‐09) [EB/OL]) Timing: clinical diagnosis presumed on admission. RT‐PCR sampling ‐ 54 (23%) day 0‐5; 71 (30%) day 6‐10; 28 (12%) day 11‐12; 35 (15%) day 13‐15; 50 (21%) day ≥ 16 2. No reference standard described for 'ordinary' patients or healthy controls |
||
| Flow and timing | Time interval between index and reference NR, but within hospital stay. Data are disaggregated by time pso but different participants contributed samples at each time. No missing data, uninterpretable or indeterminate results described. Basis for analysis: participants | ||
| Comparative | |||
| Notes | Funded by National Natural Science Foundation of China; National Key Research and Development Program of China; and the China Postdoctoral Science Foundation. Wuhan Institute of Virology of Chinese Academy of Sciences and Zhuhai Lizhu Diagnostics Inc. for providing assistance in ELISA detection. Conflicts of interest: Zhuhai Lizhu Diagnostics Inc. acknowledged in Funding statement. Preprint (not peer reviewed): medRxiv | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Liu 2020c.
| Study characteristics | |||
| Patient Sampling | Single‐group study to determine sensitivity in acute phase sera. Cohort of 133 patients diagnosed with SARS‐Cov‐2 according to the "pneumonia diagnosis protocol for novel coronavirus infection (trial version 5)". Inclusion and exclusion criteria not further described | ||
| Patient characteristics and setting | Inpatients at Renmin Hospital (Wuhan University), China (recruitment dates 17 February‐1 March 2020). Severity of condition classified as moderate 44, 33%; severe 52, 39%; critical 37, 29%. Median age (range) per group: moderate 67.5 years (64 to 71.5 years); severe 68 years (61.25 to 74); critical 70 years (60 to 76.5). Male 70, 53% Exposure history not described | ||
| Index tests | One Ab test, blinding NR Laboratory‐based evaluation of CLIA (details as per company contact) to measure IgG and IgM ‐ SARS‐CoV‐2 Ab detection kit (iFLash‐SARS‐CoV‐2 IgG/IgM CLIA) (YHLO Biotech, Shenzhen), using serum samples. Antigen used NR Sample timing not described | ||
| Target condition and reference standard(s) | 1. Clinical diagnosis according to established protocol (not cited but appears to be Chinese Government‐issued ‐ National Health Commission of the People’s Republic of China, pneumonia diagnosis protocol for novel coronavirus detection (trial version 5)) 2. RT‐PCR (ORF1ab/N qPCR detection kit from GeneoDx Biotech, Shanghai, China). 2 tests per participant but number of positive tests required NR. Samples not described, but Table 2 refers to 'NP', which could be NP samples. Positivity threshold not described | ||
| Flow and timing | Time interval between index and reference standard not described; time pso not described No missing data, uninterpretable or indeterminate results described Basis for analysis: participants | ||
| Comparative | |||
| Notes | Funded by National Natural Science Foundation of China (81672079 to CZ and 31800147 to ZL), the Open Research Fund Program of the State Key Laboratory of Virology of China (2019KF001 to ZL), the Outstanding Leaders Training Program of Pudong Health Bureau of Shanghai (PWR12018‐05 to XL), and the Key Disciplines Group Construction Project of Pudong Health Bureau of Shanghai (PWZxq2017‐15 to XL) No conflicts of interest declared Preprint (not peer reviewed): medRxiv | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Liu 2020d [A].
| Study characteristics | |||
| Patient Sampling | 2‐group study to estimate sensitivity and specificity in acute and convalescent phase sera 1. RT‐PCR confirmed COVID‐19 cases (n = 214) 2. Healthy blood donors (n = 100) Retrospective design; recruitment method NR. No further detail | ||
| Patient characteristics and setting | [1] Inpatients at General Hospital of the Central Theater Command of the People’s Liberation Army (PLA), China (recruitment dates 18 January‐26 February). Exposure history and participant characteristics not described [2] Healthy blood donors; not further described | ||
| Index tests | 2 Ab tests, blinding NR; this entry (Liu 2020d [A]) refers to test [A] in the list below Laboratory‐based evaluations of ELISA assays measuring IgM and IgG using serum samples: A. rN‐based ELISA (Lizhu, Zhuhai, China), using recombinant N protein B. rS‐based ELISA (Hotgen, Beijing, China), using receptor‐binding domain of the recombinant S polypeptide (rS) Test thresholds: A. cut‐off calculated by summing 0.100 (IgM) or 0.130 (IgG) and the average A450 of negative control replicates. When A450 < cut‐off value, the test was considered negative, and when A450 was ≥ cut‐off value, the test was considered positive. B. cut‐off values (IgM and IgG) calculated by summing 0.250 and the average A450 of negative control replicates. When A450 < cut‐off value, the test was considered negative, and when A450 was ≥ cut‐off value, the test was considered positive. Samples acquired 0‐5 d 22, 10%; 6‐10 d 38, 18%; 11‐15 d 54, 25%; 16‐20 d 55, 26%; ≥ 21 d 45, 21% (32/45 are d 21‐30). Person applying the test not described | ||
| Target condition and reference standard(s) | [1] RT‐PCR (no further detail), using pharyngeal swabs samples. Positivity threshold NR. Samples acquired at a median of 15 d pso (range 0–55 days) 2. Healthy blood donors; no description of timing of serum sample collection | ||
| Flow and timing | Sampling for index and reference for cases was conducted within same time frame. No missing data, uninterpretable or indeterminate results described Basis for analysis: participants. Includes a single sample per participant with results disaggregated by time pso, but different participants contributed data to each time period. |
||
| Comparative | |||
| Notes | Supported by the National Natural Science Foundation, the China Postdoctoral Science Foundation (2019M664008), and the Wuhan Young and Middle‐aged Medical Backbone Talents Training Project (Wuweitong [2019] 87th266) Accepted manuscript (Journal of Clinical Microbiology) No conflicts of interest declared |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Liu 2020d [B].
| Study characteristics | |||
| Patient Sampling | See main entry for this study for characteristics and QUADAS‐2 assessment (Liu 2020d [A]) | ||
| Patient characteristics and setting | See main entry for this study for characteristics and QUADAS‐2 assessment (Liu 2020d [A]) | ||
| Index tests | 2 Ab tests, blinding NR; this entry (Liu 2020d [B]) refers to test [B] in the list below Laboratory‐based evaluations of ELISA assays measuring IgM and IgG using serum samples A. rN‐based ELISA ( Lizhu, Zhuhai, China), using recombinant N protein B. rS‐based ELISA (Hotgen, Beijing, China), using receptor‐binding domain of the recombinant S polypeptide (rS) Test thresholds: A. cut‐off calculated by summing 0.100 (IgM) or 0.130 (IgG) and the average A450 of negative control replicates. When A450 < cut‐off value, the test was considered negative, and when A450 was ≥ cut‐off value, the test was considered positive. B. cut‐off values (IgM and IgG) calculated by summing 0.250 and the average A450 of negative control replicates. When A450 < cut‐off value, the test was considered negative, and when A450 was ≥ cut‐off value, the test was considered positive. Samples acquired 0‐5 d 22, 10%; 6‐10 d 38, 18%; 11‐15 d 54, 25%; 16‐20 d 55, 26%; ≥ 21 d 45, 21% (32/45 are d 21‐30). Person applying the test not described | ||
| Target condition and reference standard(s) | See main entry for this study for characteristics and QUADAS‐2 assessment (Liu 2020d [A]) | ||
| Flow and timing | See main entry for this study for characteristics and QUADAS‐2 assessment (Liu 2020d [A]) | ||
| Comparative | |||
| Notes | |||
Long 2020 (A).
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity for detection of active or prior infection
Cohort of close contacts (n = 164, 23 cases) of 2 index cases (diagnosis confirmed 4 February 2020; contacts between 20 January‐6 February 2020 identified and PCR tested)
Additional cohorts reported but not extracted included:
a. follow‐up cohort in RT‐PCR‐positive confirmed cases sampling every 3 days (n = 63 subset of cross‐sectional study); does not provide accuracy data
b. cohort of RT‐PCR‐negative suspects (n = 52); did not provide full accuracy data (specificity only could be extracted) c. extracted as Long 2020 (B) |
||
| Patient characteristics and setting | Close contacts identified by Chongqing CDC in Wanzhou (n = 164), China PCR testing conducted 31 January‐9 February; serum samples collected 2 March 2020 13 (8%) symptomatic, 151 asymptomatic; no further details | ||
| Index tests | One Ab test, blinding NR Laboratory‐based evaluated of magnetic CLIA kit (Bioscience (Chongqing) Co., Ltd), measuring IgM and IgG in serum samples, using recombinant antigen containing nucleoprotein and a peptide from S protein. Test threshold not described; presume interpretation according to manufacturer's instructions. Sample timing: 21‐31 days after PCR test | ||
| Target condition and reference standard(s) | RT‐PCR using nasal and pharyngeal swab specimens during hospital stay. No further detail. Theshold for positivity NR Timing of reference standard sampling: within 17‐day period after contact with confirmed cases | ||
| Flow and timing | Time interval between index and reference: index 21‐30 days after PCR test, potential for repeat exposure during this time. No missing data, uninterpretable or indeterminate results reported Participant‐based analysis | ||
| Comparative | |||
| Notes | Funded by Emergency Project from the Science & Technology Commission of Chongqing; The Major National S&T programme grant from Science & Technology Commission of China. No conflicts of interest reported; 1 author from BioScience Co. Ltd, Chongqing, China Preprint (not peer reviewed) | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | No | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Long 2020 (B).
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity for diagnosing acute phase infection
RT‐PCR‐positive confirmed cases (n = 285). No further detail of inclusion or exclusion criteria.
Additional cohorts reported but not extracted included:
a. follow‐up cohort in RT‐PCR‐positive confirmed cases sampling every 3 days (n = 63 subset of cross‐sectional study); does not provide accuracy data
b. cohort of RT‐PCR‐negative suspects (n = 52); did not provide full accuracy data (specificity only could be extracted) c. cohort of asymptomatic contacts of 2 confirmed cases extracted as Long 2020 (A) |
||
| Patient characteristics and setting | Inpatients at 3 hospitals, Chongqing Three Gorges Central Hospital (TGH) (n = 158), Yongchuan Hospital Affiliated to Chongqing Medical University (YCH) (n = 75), and The Public Health Center of Chongqing (PHCC), China (n = 52), recruited 5 February 2020 Median age 47 years (IQR 34‐56 years); 55.4% male. 39/285 (14%) severe or critical in ICU. 103/285 (36%) patients had an history of exposure to transmission sources | ||
| Index tests | One Ab test, blinding NR Laboratory‐based evaluated of magnetic CLIA kit (Bioscience (Chongqing) Co., Ltd), measuring IgM and IgG in serum samples, using recombinant antigen containing nucleoprotein and a peptide from S protein. Test threshold not described; presume interpretation according to manufacturer's instructions Sample timing: 67/363 (18%) day 2‐7 from symptom onset; 149 (41%) day 8‐13; and 147 (40%) day 14+ | ||
| Target condition and reference standard(s) | RT‐PCR using nasal and pharyngeal swab specimens during hospital stay. No further detail. Theshold for positivity NR Timing of reference standard sampling NR | ||
| Flow and timing | Time interval between index and reference NR. Data are disaggregated by time period but different participants contributed samples at each time pso Missing data: 23 participants with no information on time pso were excluded leaving 363 samples from 262 participants No uninterpretable or indeterminate results reported Basis for analysis: samples | ||
| Comparative | |||
| Notes | Funded by Emergency Project from the Science & Technology Commission of Chongqing; The Major National S&T programme grant from Science & Technology Commission of China No conflicts of interest declared; 1 study author from BioScience Co. Ltd, Chongqing, China Preprint paper (not peer reviewed) | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Lou 2020 [A].
| Study characteristics | |||
| Patient Sampling | 2‐group study recruiting patients estimating sensitivity and specificity [1] n = 80 confirmed COVID cases [2] n = 300 healthy people enrolled from the community Recruitment: Prospective or retrospective recruitment of cases: Sample size (virus/COVID cases): 380 (80) Inclusion and exclusion criteria: willing to donate blood | ||
| Patient characteristics and setting | Setting: inpatient Location: First affiliated hospital of Zhejiang University Country: China Dates: 19 January‐9 February 2020 Symptoms and severity: n = 26. Critical case = any one of a) ARDS or oxygen saturation < 93% and needing mechanical ventilation invasively or non‐invasively; b) shock; c) complication of organ failure requiring ICU support N= 54 non‐critical case (not meeting criteria a) or b) or c) above Sex: 38.7% female Age: 55 years (IQR 45‐64) Exposure history: for 45/80: incubation period (defined as interval between earliest date of SARS‐Cov‐2 exposure (unambiguous close contact with confirmed COVID‐19 case) and earliest date of symptom onset) range 0‐23 days, median 5 (IQR 2–10) | ||
| Index tests | 3 tests evaluated, this entry (Lou 2020 [A]) refers to test [A] Test name: [A] ELISA; [B] CGIA; [C] CLIA Manufacturer: NR Ab targets: Ab; IgM; IgG Antigens used: IgM and Ab: RBD of the SARS‐CoV‐2 S protein IgG: indirect immunoassays using recombinant nucleoprotein of SARS‐CoV‐2 Test method: ELISA, CLIA; LFIA Timing of samples: between 0 and 29 days pso Samples used: serum Test operators: NR Definition of test positivity: NR Blinded to reference standard: unclear Threshold predefined: yes |
||
| Target condition and reference standard(s) | Reference standard for cases: confirmed case should meet 3 criteria: 1) fever and/or respiratory symptoms; 2) abnormal lung imaging findings; and 3) positive result of the nucleic acid of SARS‐CoV‐2
Samples used: deep sputum
Timing of reference standard: on admission
Blinded to index test: unclear
Incorporated index test: unclear Reference standard for non‐cases: NR |
||
| Flow and timing | Time interval between index and reference tests: NR Results presented by time period: yes All participants received the same reference standard: unclear Missing data: [1] 36, 71 and 58/80 contributed to 0‐7, 8‐14 and 15‐29 days pso estimates of sensitivity for tests [A], [B] and [C] only [2] Not all control group participants were tested by all index tests (range 100‐300/300) Uninterpretable results: NR Indeterminate results: NR Unit of analysis: participant | ||
| Comparative | |||
| Notes | Funding: China National Mega‐Projects for Infectious Diseases and the Science and Technology Major Project of Xiamen Publication status: preprint Source:Pre print server (medRxiv) Study author COI: none declared | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | No | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| The reference standard does not incorporate the index test | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Unclear | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Unclear | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Lou 2020 [B].
| Study characteristics | |||
| Patient Sampling | See main entry for this study for characteristics and QUADAS‐2 assessment (Lou 2020 [A]) | ||
| Patient characteristics and setting | See main entry for this study for characteristics and QUADAS‐2 assessment (Lou 2020 [A]) | ||
| Index tests | 3 tests evaluated, this entry (Lou 2020 [B]) refers to test [B] Test name: [A] ELISA; [B] CGIA; [C] CLIA Manufacturer: NR Ab targets: Ab; IgM; IgG Antigens used: IgM and Ab: RBD of the SARS‐CoV‐2 S protein IgG: indirect immunoassays using recombinant nucleoprotein of SARS‐CoV‐2 Test method: ELISA, CLIA; LFIA Timing of samples: between 0 and 29 days pso Samples used: serum Test operators: NR Definition of test positivity: NR Blinded to reference standard: unclear Threshold predefined: yes |
||
| Target condition and reference standard(s) | See main entry for this study for characteristics and QUADAS‐2 assessment (Lou 2020 [A]) | ||
| Flow and timing | See main entry for this study for characteristics and QUADAS‐2 assessment (Lou 2020 [A]) | ||
| Comparative | |||
| Notes | |||
Lou 2020 [C].
| Study characteristics | |||
| Patient Sampling | See main entry for this study for characteristics and QUADAS‐2 assessment (Lou 2020 [A]) | ||
| Patient characteristics and setting | See main entry for this study for characteristics and QUADAS‐2 assessment (Lou 2020 [A]) | ||
| Index tests | 3 tests evaluated, this entry (Lou 2020 [C]) refers to test [C] Test name: [A] ELISA; [B] CGIA; [C] CLIA Manufacturer: NR Ab targets: Ab; IgM; IgG Antigens used: IgM and Ab: RBD of the SARS‐CoV‐2 S protein IgG: indirect immunoassays using recombinant nucleoprotein of SARS‐CoV‐2 Test method: ELISA, CLIA; LFIA Timing of samples: between 0 and 29 days pso Samples used: serum Test operators: NR Definition of test positivity: NR Blinded to reference standard: unclear Threshold predefined: yes |
||
| Target condition and reference standard(s) | See main entry for this study for characteristics and QUADAS‐2 assessment (Lou 2020 [A]) | ||
| Flow and timing | See main entry for this study for characteristics and QUADAS‐2 assessment (Lou 2020 [A]) | ||
| Comparative | |||
| Notes | |||
Ma 2020a.
| Study characteristics | |||
| Patient Sampling | 4‐group study recruiting patients estimating sensitivity and specificity [1] n = 87 confirmed COVID‐19 (216 samples) [2] n = 330 healthy donors pre‐October 2019 [3] n = 138 'other diseases' (no mention of PCR) [4] n = 15 suspected COVID pneumonia but negative PCR Recruitment: cases admitted between 26 January‐5 March 2020 Prospective or retrospective recruitment of cases: retrospective Sample size (virus/COVID cases): 570 (87) Inclusion and exclusion criteria: NR | ||
| Patient characteristics and setting | Setting: inpatient Location: First Affiliated Hospital of USTC Hospital and the First Affiliated Hospital of Anhui Medical University Country: China Dates: 26 January‐5 March 2020 Symptoms and severity: 56/87 clinically moderate, 17 severe, 5 critical, "few mild" Sex: NR Age: NR Exposure history: NR | ||
| Index tests | Test name: CLIA RBD Manufacturer: in‐house Ab targets: IgM;IgG;IgA Antigens used: SARS CoV‐2 RBD protein (S‐based) Test method: CLIA Timing of samples: during 'routine inpatient testing' Samples used: serum Test operators: NR Definition of test positivity: NR Blinded to reference standard: unclear Threshold predefined: no | ||
| Target condition and reference standard(s) | Reference standard for cases: New Coronavirus Pneumonia Prevention and Control Program (7th edition) published by the National Health Commission of China, RT‐qPCR was used to confirm COVID‐19 (all cases were RT‐PCR‐positive) Samples used: serum Timing of reference standard: during 'routine inpatient testing' Blinded to index test: unclear Incorporated index test: no Reference standard for non‐cases: [2] Pre‐pandemic [3] NR [4] NR | ||
| Flow and timing | Time interval between index and reference tests: NR Results presented by time period: yes All participants received the same reference standard: no Missing data: for comparison of sensitivity and specificity of 2 antigens only 20/total of 479 control sera were used (20/138 from 'other disease' group Uninterpretable results: NR Indeterminate results: NR Unit of analysis: samples | ||
| Comparative | |||
| Notes | Funding: T.J. is supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB29030104), National Natural Science Fund (Grant No.: 31870731 and U1732109), the Fundamental Research Funds for the Central Universities (WK2070000108). TJ and XLM is supported by a COVID‐19 special task grant supported by Chinese Academy of Science Clinical Research Hospital (Hefei) with Grant No. YD2070002017 and YD2070002001, respectively. M.H. is supported by the new medical science fund of USTC (WK2070000130). Publication status: preprint Source: preprint server: medRxiv Study author COI: 3 study authors are employees of Kangrun Biotech LTD (Guangzhou, 308 China). 4 study authors have jointly applied for a patent related to the Ab detecting kits. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | No | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | No | ||
| Could the conduct or interpretation of the index test have introduced bias? | High risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | Yes | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | No | ||
| Could the patient flow have introduced bias? | High risk | ||
Okba 2020a.
| Study characteristics | |||
| Patient Sampling | 2‐group design estimating sensitivity and specificity in acute disease [1] SARS‐CoV‐2 cases confirmed by RT‐PCR (n = 9, 31 samples) [2] Healthy blood donors (n = 45) date NR Recruitment method and exclusion criteria NR Third group of RT‐PCR confirmed cases from France (n = 3, 10 samples excluded by review author team) | ||
| Patient characteristics and setting | [1] Inpatient (plus initial testing prior to admission) at hospital in Munich, Germany. Cases are epidemiologically linked, identified through exposure to known cases, and occurred after 23 January 2020, discovered on 27 January (Woelfel 2020). Symptoms and severity, and demographics NR [2] Sanquin Blood Bank, Netherlands, date not specified | ||
| Index tests | Beta version of commercial EuroImmun IgA and IgG ELISA Ab test, from EUROIMMUN Medizinische Labordiagnostika AG. Targets IgA and IgG. Threshold not pre‐defined: in‐house threshold of mean background reactivity of all SARS‐CoV‐2–negative serum samples in the study multiplied by 3. Blinding NR | ||
| Target condition and reference standard(s) | [1] All positive on RT‐PCR between days 1‐5 of symptom onset, using OP or NP swab. Blind to index test [2] Blood bank samples, reported as negative but date of sampling NR | ||
| Flow and timing | Different reference standard for cases and controls, and cases were from 2 separate cohorts. Limited details available for each cohort. Results available by case, but only in graph format Indeterminate or unclear index results on graphs considered negative by review team | ||
| Comparative | |||
| Notes | No information provided on study author conflicts. Published as early release (not final). Report the following funding "Zoonoses Anticipation and Preparedness Initiative (project Innovative Medicines Initiative grant no. 115760), the Innovative Medicines Initiative; the European Commission, and partners of the European Federation of Pharmaceutical Industries and Associations" | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | No | ||
| If a threshold was used, was it pre‐specified? | No | ||
| Could the conduct or interpretation of the index test have introduced bias? | High risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Padoan 2020.
| Study characteristics | |||
| Patient Sampling | 1‐group study recruiting patients estimating sensitivity [1] Hospitalised patients with confirmed COVID‐19 Recruitment: cases with residual serum samples collected between 18 March‐26 March 2020 Prospective or retrospective recruitment of cases: retrospective Sample size (virus/COVID cases): 37 (37) Inclusion and exclusion criteria: | ||
| Patient characteristics and setting | Setting: inpatient Location: University Hospital of Padova Country: Italy Dates: 18 March‐26 March 2020 Symptoms and severity: NR Sex: NR Age: NR Exposure history: NR | ||
| Index tests | Test name: MAGLUMI 2000 Plus nCoV IgM and IgG Manufacturer: New Industries Biomedical Engineering Co., Ltd [Snibe], Shenzhen, China Ab targets: IgM; IgG Antigens used: NR Test method: CLIA Timing of samples: days since symptom onset: ≤ 5 days 4/37 (11%) 6‐7 days 6/37 (16%) 0‐7 days: 10/37 (27%) 8‐9 days 12/37 (32%) 10‐11 days 14/37 (38%) 12‐13 days 9/37 (24%) 8‐13 days: 35/37 (95%) > 13 days 25/37 (68%) Samples used: serum Test operators: NR Definition of test positivity: [A] IgM 1.0 AU/mL [B] IgG 1.1 AU/mL Blinded to reference standard: no Threshold predefined: yes | ||
| Target condition and reference standard(s) | Reference standard for cases: PCR Samples used: NP Timing of reference standard: NR Blinded to index test: yes Incorporated index test: no Reference standard for non‐cases: N/A | ||
| Flow and timing | Time interval between index and reference tests: NR Results presented by time period: yes All participants received the same reference standard: yes Missing data: text describes 87 samples from 37 participants but only 70 samples reported per time period and no per participant data are reported Uninterpretable results: NR Indeterminate results: NR Unit of analysis: sample | ||
| Comparative | |||
| Notes | Funding: none declared Publication status: published Source: academic journal Study author COI: none declared | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | No | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | High risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | No | ||
| Could the patient flow have introduced bias? | High risk | ||
Pan 2020a.
| Study characteristics | |||
| Patient Sampling | Single group of cases to estimate sensitivity in acute disease SARS‐CoV‐2‐positive cases (n = 105, 134 samples) of which 67 cases (86 samples) confirmed by RT‐PCR, and 37 patients (39 samples) clinically diagnosed (RT‐PCR‐negative, radiography‐positive) Recruitment method NR Exclusion criteria NR | ||
| Patient characteristics and setting | Inpatients in Zhongnan hospital (Wuhan University, China). Testing 6 February‐23 February 2020, symptom onset 7 January‐18 February 2020 (for subgroup of 108) 48 male, 57 female, median age 58 years (range 20‐96). Symptoms and severity and exposure status NR | ||
| Index tests | Commercial Ab test LFA (conducted in laboratory setting). Colloidal gold‐based immunochromatographic strip assay (Zhuhai Livzon Diagnositic Inc) to detect IgM, IgG. Antigen used NR (as per manufacturer) Presence of T line indicating positive Serum or plasma samples used (includes comparison with whole blood for subgroup; not extracted). No information on timing or who read the test results. | ||
| Target condition and reference standard(s) | 1. RT‐PCR following WHO guidelines for qRT‐PCR, using throat swabs (Chinese CDC recommended kit used, BioGerm, Shanghai, China) 2. clinically diagnosed as SARS‐CoV‐2 infection according to the 5th edition of guideline on diagnosis and treatment of the novel coronavirus pneumonia. Specifically, the clinical diagnosis means the suspected cases were negative to the real‐time RT‐PCR test but presented viral pneumonia by radiography Samples taken during inpatient stay but no details about timing or personnel for test interpretation | ||
| Flow and timing | All participants received a reference standard, but there was differential verification with some patients confirmed by RT‐PCR and others RT‐PCR‐negative but confirmed by radiography. Subset who were RT‐PCR‐positive are reported separately. Timing of index tests and reference standard unclear. Data reported only for those with symptom onset information; 26 samples excluded. No reporting of test failures or indeterminate results. Per‐sample analysis; multiple samples (2 or 3) per participant disaggregated over time | ||
| Comparative | |||
| Notes | Funding from the National Key Research and Development Program of China (2018YFE0204500) Declared no conflict of interest Published in the Journal of Infection | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Paradiso 2020a.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity for diagnosing SARS‐Cov‐2 [1] Cohort of patients attending A&E with COVID‐19‐like symptoms Recruitment: consecutive Sample size (virus/COVID cases): 191 (70) Inclusion and exclusion criteria: no further details | ||
| Patient characteristics and setting | Setting: A&E Location: Ospedale Policlinico Consorziale of Bari Country: Italy Dates: 23‐29 March 2020 Symptoms and severity: 14/160 (9%) asymptomatic; symptoms not available for 31/191 Sex: 116, 60.6% male Age: median 58.5 years Exposure history: NR | ||
| Index tests | Test name: VivaDiag Manufacturer: Jiangsu Medomics Medical Technologies Ab targets: IgM, IgG Antigens used: surface antigen from SARSCoV‐2 POC or laboratory: POC Test method: LFA (CGIA) Timing of samples: on presentation; time from symptom onset varied from asymptomatic 14, 9%; d 0‐5 97, 61%; d 6‐8 17, 11%; d 9‐10 21, 13%; d 11‐15 5, 3%, > 15 d 6, 5%; NR 31, 19%) Samples used: venous blood Test operators: 2 operators in the laboratory (operators obtained images of the device and disagreements evaluated by a third party) Definition of test positivity: presence of red/purple line in the specific region indicated on the device Blinded to reference standard: yes Threshold predefined: as per manufacturer | ||
| Target condition and reference standard(s) | Reference standard for cases including threshold: RT‐PCR (Allplex2019‐nCoV Assay; Seegene, Seoul, Republic of Korea); target genes E gene, RdRP gene and N gene; threshold NR Single PCR‐negative for D‐ presumed (NR) Samples used: NP/OP swabs Timing of reference standard: obtained simultaneously with blood samples (on presentation) Blinded to index test: yes Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous testing All participants received the same reference standard: yes Missing data: 1 participant missing from 2x2 table with no explanation although all participants had the reference standard and index test. No data on time pso for 31/191 Uninterpretable results: none stated Indeterminate results: none stated Unit of analysis: participant. A considerable range in time pso was reported however, and results were not disaggregated by time pso. |
||
| Comparative | |||
| Notes | Funding: none stated Publication status: preprint (not peer reviewed) Source: medRxiv Study author COI: none stated | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Qian 2020.
| Study characteristics | |||
| Patient Sampling | 4‐group study recruiting patients estimating sensitivity and specificity [1] Confirmed COVID‐19 cases (RT‐PCR‐positive) (n = 503) and [2] suspected COVID‐19 cases based on epidemiological history, clinical symptoms and chest X‐ray but 3 x PCR‐negative (n = 52) Apparently contemporaneous controls, including: [3] hospitalised with non‐COVID‐19 conditions (PCR testing not described) (n = 972) [4] healthy controls (n = 586) Recruitment: unclear Prospective or retrospective recruitment of cases: prospective Sample size (virus/COVID cases): 2113 (555) Inclusion and exclusion criteria: NR |
||
| Patient characteristics and setting | Setting: hospital inpatients (cases) Location: 10 hospitals Country: China Dates: unclear Symptoms and severity: NR Sex: NR Age: NR Exposure history: NR | ||
| Index tests | Test name: NR Manufacturer: in‐house Ab targets: IgG, IgM Antigens used: recombinant antigen from viral N protein and S protein Test method: CLIA Timing of samples: NR Samples used: serum Test operators: unclear Definition of test positivity: ≥ 10 AU/mL Blinded to reference standard: unclear Threshold predefined: yes | ||
| Target condition and reference standard(s) | Reference standard for cases: RT‐PCR for confirmed cases; suspected cases according to National Health Commission guideline (version 5)
Samples used: unclear
Timing of reference standard: during hospitalisation
Blinded to index test: yes
Incorporated index test: no Reference standard for non‐cases: unclear |
||
| Flow and timing | Time interval between index and reference tests: unclear Results presented by time period: no All participants received the same reference standard: unclear Missing data: NR Uninterpretable results: NR Indeterminate results: NR Unit of analysis: participant | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | No | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | Yes | ||
| Did all participants receive a reference standard? | Unclear | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
To 2020a [A].
| Study characteristics | |||
| Patient Sampling | Single‐group study recruiting patients estimating sensitivity and specificity [1] Confirmed COVID‐19 patients from 2 hospitals (n = 23) Recruitment: consecutive cases between 22 January‐12 February, but excluding people with insufficient stored material Prospective or retrospective recruitment of cases: retrospective Sample size (virus/COVID cases): 108 serum samples from 23 participants (23 cases). Only extractable > 14‐day subset of 16 cases Inclusion and exclusion criteria: confirmed cases | ||
| Patient characteristics and setting | Setting: hospital inpatients Location: Princess Margaret Hospital and Queen Mary Hospital, Hong Kong Country: Hong Kong, China Dates: 22 January‐12 February Symptoms and severity: 10/23 (43%) severe Sex: 13/23 (57%) male Age: median 62 years (range 37‐75) Exposure history: NR | ||
| Index tests | 2 tests evaluated, this entry (To 2020a [A]) refers to test [A] Test name: EIAs for [A] SARS‐CoV‐2 nucleoprotein and [B] S protein RBD Manufacturer: in‐house Ab targets: IgG IgM Antigens used: [A] nucleoprotein and [B] S protein RBD Test method: EIA (considered with ELISA tests for analysis purposes) Timing of samples: 3‐30 days pso Samples used: serum remnant from blood samples Test operators: NR Definition of test positivity: mean of 93 archived serum samples plus 3 x SD Blinded to reference standard: NR Threshold predefined: yes |
||
| Target condition and reference standard(s) | Reference standard for cases: laboratory confirmed ‐ exact test unclear Samples used: NP or sputum specimens Timing of reference standard: NR Blinded to index test: yes Incorporated index test: no Reference standard for non‐cases: n/a | ||
| Flow and timing | Time interval between index and reference tests: NR Results presented by time period: no, not extractable All participants received the same reference standard: yes Missing data: 7/23 (30%) were not tested between days 14 and 30 Uninterpretable results: NR Indeterminate results: NR Unit of analysis: unclear | ||
| Comparative | |||
| Notes | Funding: this study was partly supported by the Consultancy Service for Enhancing Laboratory Surveillance of Emerging Infectious Diseases and Research Capability on Antimicrobial Resistance for the Department of Health of Hong Kong; the Theme‐Based Research Scheme (T11/707/15) of the Research Grants Council, Hong Kong Special Administrative Region; Sanming Project of Medicine in Shenzhen, China (SZSM201911014); the High Level‐Hospital Program, Health Commission of Guangdong Province, China; and donations from the Shaw Foundation Hong Kong, Richard Yu and Carol Yu, May Tam Mak Mei Yin, Michael Seak‐Kan Tong, Respiratory Viral Research Foundation, Hui Ming, Hui Hoy and Chow Sin Lan Charity Fund Limited, Chan Yin Chuen Memorial Charitable Foundation, Marina Man‐Wai Lee, and the Hong Kong Hainan Commercial Association South China Microbiology Research Fund Publication status: published paper Source: Lancet Infectious Diseases Study author COI: declare they have none | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | No | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | No | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | High risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Unclear | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Unclear | ||
| Could the patient flow have introduced bias? | High risk | ||
To 2020a [B].
| Study characteristics | |||
| Patient Sampling | See main entry for this study for characteristics and QUADAS‐2 assessment (To 2020a [A]) | ||
| Patient characteristics and setting | See main entry for this study for characteristics and QUADAS‐2 assessment (To 2020a [A]) | ||
| Index tests | 2 tests evaluated, this entry (To 2020a [B]) refers to test [B] Test name: EIAs for [A] SARS‐CoV‐2 nucleoprotein and [B] S protein RBD Manufacturer: in‐house Ab targets: IgG IgM Antigens used: [A] nucleoprotein and [B] S protein RBD Test method: EIA (considered with ELISA tests for analysis purposes) Timing of samples: 3‐30 days pso Samples used: serum remnant from blood samples Test operators: NR Definition of test positivity: mean of 93 archived serum samples plus 3 x SD Blinded to reference standard: NR Threshold predefined: yes |
||
| Target condition and reference standard(s) | See main entry for this study for characteristics and QUADAS‐2 assessment (To 2020a [A]) | ||
| Flow and timing | See main entry for this study for characteristics and QUADAS‐2 assessment (To 2020a [A]) | ||
| Comparative | |||
| Notes | |||
Wan 2020 [A].
| Study characteristics | |||
| Patient Sampling | 2‐group design estimating sensitivity and specificity in acute disease [1] SARS‐Cov‐2‐positive cases confirmed by RT‐PCR (n = 7, 26 samples) [2] prepandemic sera (n = 5) and controls SARS‐Cov‐2 negative on 2 occasions (n = 5) Recruitment method NR Exclusion criteria NR | ||
| Patient characteristics and setting | [1] Inpatients at Singapore General Hospital, Singapore. Recruitment dates NR. Symptoms and severity, demographics and exposure history NR [2] Archived controls (n = 5) from Singapore General Hospital from 2015; recent patients with pneumonia investigated for COVID‐19 but RT‐PCR‐negative twice and not meeting the criteria for suspected SARS‐Cov‐2 (n = 5) | ||
| Index tests | 2 Ab tests used on serology samples, this entry (Wan 2020 [A]) refers to test [A]
[A] in‐house SARS‐CoV total Ab ELISA laboratory assay (not a SARS‐CoV‐2‐specific test). Measured total Ab; antigens NR. Positive defined as ≥ 400 [B] anti‐SARS CoV IIFT laboratory assay from Euroimmun (Germany) (not a SARS‐CoV‐2‐specific test). Measured IgM and IgG; antigens NR. Threshold NR Samples anonymised and blinded |
||
| Target condition and reference standard(s) | [1] Confirmed COVID‐19 determined by RT‐PCR; samples and methods NR. Tests undertaken during inpatient stay; blind to the index test [2] Confirmed not COVID‐19 by chronology in n = 5, and by 2 repeated RT‐PCR‐negative results and not fulfilling criteria for suspected COVID‐19 in n = 5; samples and methods NR. Tests undertaken during inpatient stay; blind to the index test | ||
| Flow and timing | All participants received a reference standard, but different reference standards were used in [1] and [2]. Multiple samples were included per participant; however these were disaggregated by time pso. Timing of reference standard and index tests NR Uninterpretable results not mentioned; 1/10 samples in group [2] indeterminate due to non‐specific fluorescence. 5/26 samples in group [1] excluded due to narrow interval between tests or close proximity to the date of onset of illness. In group [1] between 1 and 9 samples per participant (mean = 3.7). Results for all samples per participant are presented allowing participant‐ and sample‐based analyses. | ||
| Comparative | |||
| Notes | No funding declared No conflicts of interest noted Report from a medRxiv preprint (not peer reviewed) |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Wan 2020 [B].
| Study characteristics | |||
| Patient Sampling | See main entry for this study for characteristics and QUADAS‐2 assessment (Wan 2020 [A]) | ||
| Patient characteristics and setting | See main entry for this study for characteristics and QUADAS‐2 assessment (Wan 2020 [A]) | ||
| Index tests | 2 Ab tests used on serology samples, this entry (Wan 2020 [B]) refers to test [B]
[A) in‐house SARS‐CoV total Ab ELISA laboratory assay (not a SARS‐CoV‐2‐specific test). Measured total Ab; antigens NR. Positive defined as ≥ 400 [B] anti‐SARS CoV IIFT laboratory assay from Euroimmun (Germany) (not a SARS‐CoV‐2 specific test). Measured IgM and IgG; antigens NR. Threshold NR Samples anonymised and blinded |
||
| Target condition and reference standard(s) | See main entry for this study for characteristics and QUADAS‐2 assessment (Wan 2020 [A]) | ||
| Flow and timing | See main entry for this study for characteristics and QUADAS‐2 assessment (Wan 2020 [A]) | ||
| Comparative | |||
| Notes | |||
Wang 2020a [A].
| Study characteristics | |||
| Patient Sampling | Purpose: diagnosis of active infection Design: 2‐group study to estimate sensitivity and specificity for diagnosis of acute infection [1] COVID‐19 patients, meeting diagnostic criteria a to Chinese Government guidelines (fifth edition) (n = 14) [2] Sera from patients with different pathogen infections and related chronic diseases with no clinical symptoms or imaging evidence of COVID‐19 (n = 72), (with deliberate selection of rheumatoid factor IgM‐positive sera) Recruitment: NR Sample size (virus/COVID cases): 86 (14) Inclusion and exclusion criteria: not described | ||
| Patient characteristics and setting | Setting: inpatient Location: affiliated Hospital of North Sichu, Chinaan Medical College and Nanchong Central Hospital Country: China Dates: 25 January ‐15 February 2020 Symptoms and severity: NR Demographics: NR Exposure history: NR Non‐COVID patients group: other infection/chronic disease controls Source and time: 25 January‐15 February 2020 Characteristics: IgM‐positive sera from patients with different pathogen infections and related chronic diseases with no clinical symptoms or imaging evidence of COVID‐19 (n = 72); flu A (n = 5), flu B (n = 5), Mycoplasma pneumoniae (n = 5), Legionella pneumophila (n = 5), positive rheumatoid factor (n = 36), HIV infection (n = 6), hypertension (n = 5) and diabetes mellitus (n = 5) | ||
| Index tests | 2 tests evaluated, this entry (Wang 2020a [A]) refers to test [A] A. SARS‐CoV‐2 IgM detection kit CGIA (Beijing Hotgen Biotechnology Co., Beijing, China) (POC test, evaluation appears to be laboratory‐based) B. ELISA (Beijing Hotgen Biotechnology Co., Beijing, China) (laboratory test) Ab targets: IgM Antigens used: NR Timing of samples: within 3‐7 days after the beginning of the clinical symptoms for COVID‐19 cases Samples used: serum Test operators: NR Definition of test positivity: A. as per manufacturer, colloidal gold colour reaction occurs at both T‐line and C‐line positions B. not described Blinded to reference standard: NR Threshold predefined: yes, as per manufacturer |
||
| Target condition and reference standard(s) | Reference standard for cases including threshold: diagnostic criteria from "Notice on the Issuance of Strategic Guidelines for Diagnosis and Treatment of Novel Coronavirus (SARS‐CoV‐2) Infected Pneumonia (Fifth Edition Version)
Samples used: NR/ N/A
Timing of reference standard: during hospital stay
Blinded to index test: NR
Incorporated index test: no
Reference standard for controls: no clinical symptoms or imaging evidence of COVID‐19 Samples used: NR Timing of reference standard: NR Blinded to index test: NR Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: NR, but all serology samples acquired within first week pso All participants received the same reference standard: yes Missing data: none stated Uninterpretable results: none stated Indeterminate results: none stated Unit of analysis: participant | ||
| Comparative | |||
| Notes | Funding: none declared Publication status: accepted manuscript Source: Journal of Clinical Microbiology Study author COI: none declared | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Wang 2020a [B].
| Study characteristics | |||
| Patient Sampling | See main entry for this study for characteristics and QUADAS‐2 assessment (Wang 2020a [A]) | ||
| Patient characteristics and setting | See main entry for this study for characteristics and QUADAS‐2 assessment (Wang 2020a [A]) | ||
| Index tests | 2 tests evaluated, this entry(Wang 2020a [B]) refers to test [B] A. SARS‐CoV‐2 IgM detection kit CGIA (Beijing Hotgen Biotechnology Co., Beijing, China) (POC test, evaluation appears to be laboratory‐based) B. ELISA (Beijing Hotgen Biotechnology Co., Beijing, China) (laboratory test) Ab targets: IgM Antigens used: NR Timing of samples: within 3‐7 days after the beginning of the clinical symptoms for COVID‐19 cases Samples used: serum Test operators: NR Definition of test positivity: A. as per manufacturer, colloidal gold colour reaction occurs at both T‐line and C‐line positions B. not described Blinded to reference standard: NR Threshold predefined: yes, as per manufacturer |
||
| Target condition and reference standard(s) | See main entry for this study for characteristics and QUADAS‐2 assessment (Wang 2020a [A]) | ||
| Flow and timing | See main entry for this study for characteristics and QUADAS‐2 assessment (Wang 2020a [A]) | ||
| Comparative | |||
| Notes | |||
Xiang 2020a [A].
| Study characteristics | |||
| Patient Sampling | 2‐group design estimating sensitivity and specificity in acute disease [1] SARS‐Cov‐2 diagnosed patients (n = 63 for ELISA, n = 91 for GICA, some overlap of cases) [2] Healthy individuals (n = 35) Group [1] were recruited as a consecutive series and were inpatients with confirmed COVID‐19 diagnosed according to WHO interim guidance | ||
| Patient characteristics and setting | [1] Inpatients at Wuhan Jinyintan Hospital, China, admitted 1‐28 January 2020. Samples taken 2‐4 February 2020. For ELISA 4/63 (6%) and GICA 4/91 (4%) classified as severe; 35/63 (56%) and 49/91 (54%) male. Median (IQR) age 65 (55‐74) (n = 63) and 61 (48.5‐67) years. Exposure NR [2] Healthy controls (n = 35). 17/35 (49%) male. Median (IQR) age 44 (39‐49.5) years. No other detail given | ||
| Index tests | 2 tests evaluated, this entry (Xiang 2020a [A]) refers to test [A] [A] novel coronavirus IgG/IgM Ab ELISA kits (laboratory kit manufactured by Zhu Hai Livzon Diagnostics). Measured IgM and IgG; antigen reported as "Enzyme‐labelled antibody‐linked antigen" for IgM and "recombinant antigen of new coronavirus" for IgG. Threshold NR [B] novel coronavirus IgG/IgM Ab GICA kits (POC test strips manufactured by Zhu Hai Livzon Diagnostics). Measured IgM and IgG; antigens NR. Threshold based on observing a coloured band turning red. A subset of participants who provided throat swab samples were also re‐tested with a qRT‐PCR test. Discussion states "that the new type of coronavirus antibody of the kit (doesn’t specify which kit though) is against the severe acute respiratory syndrome (SARS)‐like coronavirus, not only against SARS‐CoV‐2" | ||
| Target condition and reference standard(s) | [1] Confirmed COVID‐19 determined according to WHO interim guidance; tests, samples and methods NR. Diagnosis made during inpatient stay; prior to the index test [2] No description given | ||
| Flow and timing | Unclear which participants received a reference standard, and the form of the reference standard Timing of reference standard and index tests NR Uninterpretable, indeterminate and missing results not mentioned One sample tested by each test per participant, unstated overlap of participants | ||
| Comparative | |||
| Notes | Supported by the Zhongnan Hospital of Wuhan University Science, Technology and Innovation Seed Fund. No conflicts of interest noted. Report from a medRxiv preprint (not peer reviewed) | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | No | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | High risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Xiang 2020a [B].
| Study characteristics | |||
| Patient Sampling | See main entry for this study for characteristics and QUADAS‐2 assessment (Xiang 2020a [A]) | ||
| Patient characteristics and setting | See main entry for this study for characteristics and QUADAS‐2 assessment (Xiang 2020a [A]) | ||
| Index tests | 2 tests evaluated, this entry (Xiang 2020a [B] refers to test [B] [A] novel coronavirus IgG/IgM Ab ELISA kits (laboratory kit manufactured by Zhu Hai Livzon Diagnostics) Measured IgM and IgG; antigen reported as "Enzyme‐labelled antibody‐linked antigen" for IgM and "recombinant antigen of new coronavirus" for IgG. Threshold NR [B] novel coronavirus IgG/IgM Ab GICA kits (POC test strips manufactured by Zhu Hai Livzon Diagnostics) Measured IgM and IgG; antigens NR. Threshold based on observing a coloured band turning red. A subset of participants who provided throat swab samples were also re‐tested with a qRT‐PCR test. Discussion states "that the new type of coronavirus antibody of the kit (doesn’t specify which kit though) is against the severe acute respiratory syndrome (SARS)‐like coronavirus, not only against SARS‐CoV‐2" | ||
| Target condition and reference standard(s) | See main entry for this study for characteristics and QUADAS‐2 assessment (Xiang 2020a [A]) | ||
| Flow and timing | See main entry for this study for characteristics and QUADAS‐2 assessment (Xiang 2020a [A]) | ||
| Comparative | |||
| Notes | |||
Xiang 2020b.
| Study characteristics | |||
| Patient Sampling | 2‐group study recruiting patients estimating sensitivity and specificity PCR conducted for patients presenting with a history of travel to or residence in Wuhan or local endemic areas; contact history with confirmed or suspected COVID‐19 patients or part of a clustering outbreak, combined with clinical manifestation of 1) fever and/or respiratory symptoms, or 2) positive findings similar to COVID‐19 pneumonia on chest CT scan, or 3) laboratory tests showing reduced lymphocytes and white blood cell counts in the early stage. Resulted in inclusion of [1] 85 RT‐PCR‐confirmed cases [2] 24 suspected cases with ≥ 2 negative RT‐PCR and none positive (and protocol is to retest RT‐PCR negatives every 1‐2 days) [3] 60 control group of healthy blood donors (hospital staff) or from patients with other lung diseases in the same hospital (all PCR‐negative) Recruitment: NR Prospective or retrospective recruitment of cases: unclear Sample size (virus/COVID cases): 169 (109; data for 66 lab‐confirmed and 24 suspected cases extracted as D+ group) Inclusion and exclusion criteria: unclear |
||
| Patient characteristics and setting | Setting: hospital inpatients Location: Wuhan Country: China Dates: 19 January‐2 March 2020 Symptoms and severity: [1] severe 18/85 (21%) [2] 2/24 (8%) severe Sex: [1] female 54/85 (64%) [2] female 12/24 (50%) [3] 35/60 (58%) female Age: [1] median 51 (IQR 32‐65) [2] median 44 (IQR 36‐61) [3] median 34 (IQR 29‐51) Exposure history: NR | ||
| Index tests | Test name: ELISA Livzon Manufacturer: ELISA kits, Livzon Inc, Zhuhai, P.R.China, lot number of IgM: 20200308, IgG: 20200308 Ab targets: IgG IgM Antigens used: N protein? Test method: ELISA Timing of samples: NR Samples used: serum Test operators: NR Definition of test positivity: unclear "The optical density of each well was determined by a microplate reader set to 450 nm within 30 min. The ratio of optical density to the cut off value (optical density of the blank well + 0.1) was reported as the Ab concentration. For detection of IgG, the dilution factor was changed (1:20) and the cut off value was modified (optical density of the blank well + 0.13)." Blinded to reference standard: no Threshold predefined: unclear | ||
| Target condition and reference standard(s) | Reference standard for cases: [1] RT‐PCR [2] Symptoms and PCR‐negative (no guideline cited but criteria clearly elaborated) Samples used: NP and/or OP swabs Timing of reference standard: NR Blinded to index test: yes Incorporated index test: no Reference standard for non‐cases: (no exposure or symptoms) and RT‐PCR‐negative | ||
| Flow and timing | Time interval between index and reference tests: NR Results presented by time period: no All participants received the same reference standard: Missing data: data per sample are provided for the 85 confirmed cases, however per participant data are available only for 66/85 confirmed cases plus 24/24 suspected cases (total number of cases reported = 90) Uninterpretable results: NR Indeterminate results: NR Unit of analysis: reports both samples and participants | ||
| Comparative | |||
| Notes | Funding: this work is funded by National Natural Science Foundation of China (No. 81973990, 91643101), and Science Foundation of Huazhong University of Science and Technology (No. 2020kfyXGYJ100) Publication status: published in journal Source:Infectious Disease Society of America Study author COI: declare that they have none | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | No | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | High risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Xiao 2020a.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity for diagnosing active or prior infection Confirmed cases of COVID‐19 (n = 34) according to the diagnosis and treatment guideline for SARS‐CoV‐2 from Chinese National Health Committee (Version 5) and the interim guidance from Centers for Disease Control and Prevention Recruitment method not clearly reported, likely convenience sample | ||
| Patient characteristics and setting | Inpatients, presumably at study authors' institution (Tongji Hospital) Wuhan, China. Admission date: 1‐29 February 2020; final follow‐up date 3 March 2020
NR Exposure history Sex: 12 female, 22 male Median age (review team estimated) 49 years (range 26‐87), 22 (65%) male. Exposure history not described |
||
| Index tests | 1 Ab test, blinding NR Laboratory‐based CLIA (Shenzhen Yahuilong Biotechnology Co. Ltd.) measuring IgM and IgG. Antigen used not described. Threshold ≤ 10 AU/mL (describes following manufacturer protocol, but unclear if this includes threshold setting) Blood samples acquired ≥ 2 weeks after symptoms onset for 32/34 participants; and on day 2 and day 3 for remaining 2 participants | ||
| Target condition and reference standard(s) | COVID‐19 according to diagnosis and treatment guideline for SARS‐CoV‐2 from Chinese National Health Committee (Version 5) and the interim guidance from Centers for Disease Control and Prevention; no further detail Timing and blinding to index test not described | ||
| Flow and timing | Time interval between index and reference not described. Study provides a breakdown in results by time pso but is different participants in each time period rather than multiple samplings for same participants No missing data, uninterpretable or indeterminate results described | ||
| Comparative | |||
| Notes | No funding sources declared No conflicts of interest declared Pre‐proof paper accepted for publication (Journal of Infection) | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Unclear | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Xie 2020a.
| Study characteristics | |||
| Patient Sampling | Single‐group study recruiting patients estimating sensitivity to detect active disease [1] 16 confirmed COVID‐19 using RT‐PCR [2] 40 suspected cases using Chinese criteria but PCR‐negative Recruitment: upon admission between 15‐25 February 2020, unclear if consecutive Prospective or retrospective recruitment of cases: prospective Sample size (virus/COVID cases): 56 (56) of which 16 confirmed by RT‐PCR Inclusion and exclusion criteria: unclear | ||
| Patient characteristics and setting | Setting: hospital inpatient Location: Unit Z6 at the Cancer Center of Wuhan Union Hospital Country: China Dates: enrolled 15‐25 February 2020 Symptoms and severity: 34 severe, 22 not severe (more details on data extraction) Sex: 32/56 (57% female) Age: median age was 56.5 years (IQR 49.25‐64.75) Exposure history: NR | ||
| Index tests | Test name: CLIA Manufacturer: YHLO Biological Technology Co., Ltd., Shenzhen, China Ab targets: IgG IgM Antigens used: envelope (E) protein and N protein Test method: CLIA Timing of samples: upon admission to hospital (with questionnaire to determine how many days prior to this symptom onset) Samples used: serum Test operators: NR Definition of test positivity: ≥ 10 AU/mL Blinded to reference standard: yes (upon admission) Threshold predefined: yes | ||
| Target condition and reference standard(s) | Reference standard for cases: [1] RT‐PCR QIAamp RNA virus kit (Qiagen, Heiden, Germany), 1ab (ORF1ab) and N protein [2] diagnosed according to the 5th edition of the Guideline on diagnosis and treatment of COVID‐19 established by China’s National Health Commission, including patient’s epidemic history, clinical characteristics, chest CT scan and laboratory findings ‐ RT‐PCR‐negative Samples used: NP and throat swabs Timing of reference standard: NR Blinded to index test: NR Incorporated index test: no Reference standard for non‐cases: N/A |
||
| Flow and timing | Time interval between index and reference tests: NR Results presented by time period: no All participants received the same reference standard: yes Missing data: NR Uninterpretable results: NR Indeterminate results: NR Unit of analysis: participant | ||
| Comparative | |||
| Notes | Funding: this work was funded by the Special Project for Emergency Scientific and Technological Research on New Coronavirus Infection | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Xu 2020a.
| Study characteristics | |||
| Patient Sampling | Single‐group study recruiting patients estimating sensitivity [1] confirmed COVID‐19 cases Recruitment: unclear Prospective or retrospective recruitment of cases: retrospective Sample size (virus/COVID cases): 10 (10) patients Inclusion and exclusion criteria: unclear | ||
| Patient characteristics and setting | Setting: hospital inpatients Location: Affiliated hospitals of Shanghai University of Medicine & Health Sciences Country: China Dates: NR Symptoms and severity: 2/10 died, 10/10 required oxygen Sex: 6/10 (60%) male Age: NR Exposure history: NR | ||
| Index tests | Test name: COVID‐19 IgG and IgM LFA Manufacturer: in‐house Ab targets: IgG and IgM Antigens used: recombinant antigen (R18850) Test method: NR, lateral flow type Timing of samples: day 15‐30 of observation Samples used: NR Test operators: NR Definition of test positivity: NR Blinded to reference standard: NR Threshold predefined: NR | ||
| Target condition and reference standard(s) | Reference standard for cases: RT‐PCR Samples used: NR Timing of reference standard: NR Blinded to index test: NR Incorporated index test: no Reference standard for non‐cases: N/A | ||
| Flow and timing | Time interval between index and reference tests: unclear Results presented by time period: no All participants received the same reference standard: yes Missing data: NR Uninterpretable results: NR Indeterminate results: NR Unit of analysis: participant | ||
| Comparative | |||
| Notes | Funding: NR Publication status: preprint Source: medRxiv Study author COI: NR | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Yongchen 2020.
| Study characteristics | |||
| Patient Sampling | 1‐group study recruiting patients estimating sensitivity
[1] 11 non‐severe COVID‐19 patients
[2] 5 severe COVID‐19 patients
[3] 5 asymptomatic carriers
Recruitment:
Prospective or retrospective recruitment of cases: retrospective Sample size (virus/COVID cases): 21 (21) Inclusion and exclusion criteria: no more details available |
||
| Patient characteristics and setting | Setting: hospital Location: 2 medical centres ‐ Second Hospital of Nanjing and the Affiliated Hospital of Xuzhou Medical University in Jiangsu Province Country: China Dates: 25 January‐18 March 2020 Symptoms and severity: 5 severe, 11 non‐severe and 5 asymptomatic cases. Illness severity defined according to the Chinese management guideline for COVID‐19 (version 6.0). Severe cases defined as having any of the following: (a) respiratory distress; (b) hypoxia (SpO2 ≤ 93%); (c) abnormal blood gas analysis (PaO2/FiO2 ≤ 300 mm Hg); or (d) severe disease complications including respiratory failure, which requires mechanical ventilation, septic shock, or non‐respiratory organ failure. Asymptomatic carriers were defined as individuals who were positive for COVID‐19 nucleic acid but without any symptoms during screening of close contacts. Sex: 13/21 (62%) male Age: median (range) = 37 (10‐73) Exposure history: NR | ||
| Index tests | Test name: no commercial name stated Manufacturer: Innovita Co., Ltd, China Ab targets: IgG and IgM Antigens used: SARS‐CoV‐2 S protein and N protein Test method: GICA Timing of samples: NR Samples used: serum Test operators: NR Definition of test positivity: NR Blinded to reference standard: NR and no assumptions made based on timing of the test Threshold predefined: NR | ||
| Target condition and reference standard(s) | Reference standard for cases: RT‐PCR ‐ confirmed after 2 sequential positive respiratory tract sample results Samples used: throat swabs Timing of reference standard: throat swab samples collected every 1‐2 days Blinded to index test: yes (serum samples for serological evaluation were stored for later evaluation) Incorporated index test: no Reference standard for non‐cases: N/A | ||
| Flow and timing | Time interval between index and reference tests: NR
Results presented by time period: yes All participants received the same reference standard: yes Missing data: NR Uninterpretable results: NR Indeterminate results: NR Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: supported by the National Natural Science Foundation of China, Jiangsu Provincial Medical Talent, Six talent peaks project of Jiangsu Province, Advanced health talent of six‐one project of Jiangsu Province, Nanjing Medical Science and Technique Development Foundation Publication status: published paper Source: Emerging Microbes & Infections Study author COI: none was declared | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Zeng 2020a.
| Study characteristics | |||
| Patient Sampling | 2‐group study recruiting patients estimating sensitivity and specificity [1] COVID‐19 cases [2] Healthy controls Recruitment: NR Prospective or retrospective recruitment of cases: prospective Sample size (virus/COVID cases): 63 (27) Inclusion and exclusion criteria: no details available | ||
| Patient characteristics and setting | Setting: hospital inpatient Location: Zhongnan Hospital, Wuhan Country: China Dates: NR Symptoms and severity: 17 severe cases. No further details Sex: 14/27 (52%) male Age: cases only ‐ median (range) 62 (29‐87) years; IQR 46‐67 years Exposure history: NR | ||
| Index tests | Test name: none Manufacturer: Zhuhai Livzon Diagnostics INC Ab targets: IgG and IgM Antigens used: NR Test method: ELISA Timing of samples: 3‐39 days for cases Samples used: serum Test operators: NR Definition of test positivity: OD = 0.105 Blinded to reference standard: NR Threshold predefined: unclear | ||
| Target condition and reference standard(s) | Reference standard for cases: NR Samples used: NR Timing of reference standard: NR Blinded to index test: NR Incorporated index test: NR Reference standard for non‐cases: healthy controls; no indication of timing, PCR testing | ||
| Flow and timing | Time interval between index and reference tests: NR Results presented by time period: yes but only average Ab levels All participants received the same reference standard: NR Missing data: NR Uninterpretable results: NR Indeterminate results: NR Unit of analysis: participant | ||
| Comparative | |||
| Notes | Funding: supported by National Key Research and Development Program of China and Emergency Science and Technology Project of Hubei Province Publication status: Journal pre‐proof Source: Journal of Infection Study author COI: none | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| The reference standard does not incorporate the index test | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Unclear | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Unclear | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Zhang 2020a.
| Study characteristics | |||
| Patient Sampling | Single‐group design estimating sensitivity in acute and convalescent phase sera SARS‐Cov‐2 laboratory (RT‐PCR detection or Ab assay) confirmed patients (n = 222) Participants were identified retrospectively, likely as a consecutive series | ||
| Patient characteristics and setting | Inpatients at Renmin Hospital of Wuhan University, China, admitted 13 January‐1 March 2020. Samples dates not known. 87/222 (39%) classified as severe; 35/63 (56%) male. Median (IQR) age 62 (52‐69) years. Exposure NR | ||
| Index tests | 2 Ab tests used on serology samples iFlash‐SARS‐CoV‐2 IgG and iFlash‐SARS‐CoV‐2 IgM (laboratory tests manufactured by Shenzhen YHLO Biotech Co., Ltd.,). Measured IgM and IgG; antigens NR. Thresholds NR. Serum taken between day 1 and 35, 148/222 (67%) from day 21 onwards. |
||
| Target condition and reference standard(s) | COVID‐19 determined with laboratory RT‐PCR or anti‐SARS‐CoV‐2 assay from nasal or pharyngeal swabs. No further detail given (coded as Chinese government guideline, 7th Ed) guideline. Diagnosis made during inpatient stay; prior to the index test | ||
| Flow and timing | Unclear which participants received which test (RT‐PCR or Ab test as the reference standard). Samples acquired over considerable period pso; only disaggregation is for day 21 and over Timing of reference standard and index tests NR Uninterpretable, indeterminate and missing results not mentioned One sample tested by each test per participant | ||
| Comparative | |||
| Notes | No funding declared. No conflicts of interest noted Report from a medRxiv preprint (not peer reviewed) | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | No | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| The reference standard does not incorporate the index test | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Zhang 2020b.
| Study characteristics | |||
| Patient Sampling | Multi‐group design estimating sensitivity and specificity in acute phase sera First group included as single cohort to estimate sensitivity and specificity [1] Suspected COVID‐19 cases (n = 228) admitted to fever clinic with RT‐PCR testing for COVID‐19 Other groups recruited but not analysed as part of this review: [2] Controls ‐ outpatients with other diseases (n = 222) [3] Controls ‐ medical staff working for the fever clinic (n = 63) [4] Controls ‐ pre‐pandemic healthy physical examinees (n = 223) No information about recruitment |
||
| Patient characteristics and setting | [1] Inpatients at Fever Clinic, Shengjing Hospital of China Medical University, China, admitted between 21 January‐16 February 2020, samples dates not known. Median (range) age 35 (1‐86). 124/225 (55%) male. Exposure NR [2] Outpatients at Shengjing Hospital of China Medical University, China, admitted between 21 January‐16 February 2020, samples dates not known. Median (range) age 50 (27‐85). 62/222 (28%) male [3] Medical staff at Fever Clinic, Shengjing Hospital of China Medical University, China, samples dates not known. Median (range) age 40 (25‐61). 7/63 (11%) male [4] Healthy controls. Physical examinees in 2018. No setting stated. Median (range) age 59 (29‐95). 77/223 (35%) male | ||
| Index tests | 1 Ab test, no mention of blinding Unnamed IgG and IgM CLIA assay (laboratory tests manufactured by Shenzhen YHLO Biotech Co., Ltd). Measured IgM and IgG in sera; 2019‐nCoV S protein S and N protein N antigen. Thresholds > 10.0 AU/mL (Ab concentration per mL. Sample timing only described for 3 cases (tests repeated every 1‐3 days until between day 11 and day 17 (from Figure 1) | ||
| Target condition and reference standard(s) | [1] Virus detected with RT‐PCR from NP/OP swabs. Ct value according to manufacturers instructions (NR); one of ORF1ab and N gene were required to be positive in same sample. Tests repeated once in negatives. Timing of swabs unclear Excluded cohorts: [2] No reference standard stated [3] No reference standard stated [4] Reference standard based on being pre‐pandemic samples |
||
| Flow and timing | Timing of reference standard NR. Time pso reported only for the 3 confirmed cases Uninterpretable, indeterminate and missing results not mentioned 1 sample tested per participant | ||
| Comparative | |||
| Notes | Funded by National Science and Technology Major Project of China, Liaoning Province Natural Science Foundation Project, Liaoning Province Central Government's special project to guide local scientific and technological development, Guangdong Province Major key projects of indusTentative technology, Major Special Project of Construction Program of China Medical University in 2018 and 345 talent project of Shengjing Hospital of China Medical University No conflicts of interest noted Report from a preprint (not peer reviewed) | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Zhang 2020c.
| Study characteristics | |||
| Patient Sampling | Single‐group design estimating sensitivity in acute phase sera. RT‐PCR positive confirmed patients (n = 139) who received around 10 days of medical treatment after admission (n = 16) were identified. No information about recruitment The study includes a separate group of patients reporting multiple RT‐PCR results | ||
| Patient characteristics and setting | Inpatients at Wuhan pulmonary hospital, China, admission dates NR, samples dates not known. No demographic or clinical information. Exposure NR | ||
| Index tests | One Ab test, blinding not described Anti‐SARSr‐CoV IgG and IgM ELISA kits (in‐house laboratory method). Measured IgM and IgG in serum from samples on day 0 and day 5; antigen: SARSr‐CoV Rp3 nucleoprotein. Threshold NR | ||
| Target condition and reference standard(s) | COVID‐19 confirmed with laboratory RT‐PCR. No further detail given. Diagnosis made during inpatient stay; prior to the index test | ||
| Flow and timing | Timing of reference standard NR Excluded if < 10 days medical treatment (n = 123) Uninterpretable, indeterminate results not mentioned One sample tested per participant at each time point; samples obtained on same days pso and all participants had ≥ 10 days medical Rx post admission | ||
| Comparative | |||
| Notes | Supported by the Mega‐Project for Infectious Disease from Minister of Science and Technology of the People’s Republic of China, China Natural Science Foundation for excellent scholars, Strategic Priority Research Program of the CAS, Youth innovation promotion association of CAS No conflicts of interest noted Report from a published peer reviewed paper |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Zhang 2020d.
| Study characteristics | |||
| Patient Sampling | Single‐group study recruiting patients estimating sensitivity and specificity [1] Patients with suspected COVID‐19 (n = 824, 154 cases) Recruitment: unclear | ||
| Patient characteristics and setting | Setting: hospital Location: 5 hospitals ‐ Huoshenshan Hospital (Wuhan), General Hospital of Central 19 Threater Command of the PLA (Wuhan), the Sixth People’s Hospital of Shenyang, Peking Union Medical College Hospital, and Shijiazhuang Fifth Hospital. Country: China Dates: no details Symptoms and severity: no details Sex: no details Age: no details Exposure history: no details | ||
| Index tests | Test name: colloidal GICA Manufacturer: in‐house Ab targets: total Abs (IgG and IgM) Antigens used: rS1 and rS‐RBD‐mFc S proteins Test method: CGIA Timing of samples: NR Samples used: serum Test operators: NR Definition of test positivity: visible line Blinded to reference standard: unclear Threshold predefined: yes | ||
| Target condition and reference standard(s) | Reference standard for cases: real‐time PCR kit, included patients PCR‐negative but clinically diagnosed by CT as D+; D‐ are RT‐PCR negative but unclear if all had CT to confirm absence
Samples used: nasal/pharyngeal swab
Timing of reference standard: unclear
Blinded to index test: unclear
Incorporated index test: no Reference standard for non‐cases: single PCR‐negative |
||
| Flow and timing | Time interval between index and reference tests: no information Results presented by time period: no All participants received the same reference standard: yes Missing data: no information Uninterpretable results: no information Indeterminate results: no information Unit of analysis: participant | ||
| Comparative | |||
| Notes | Funding: The National Key Research and Development Program of China, and The National Science and Technology Major Project Publication status: preprint Source:preprint server (medRxiv) Study author COI: report no COI but 1 author from a company (Beijing Hotgen Biotechnology Inc., Beijing) | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Zhao 2020a.
| Study characteristics | |||
| Patient Sampling | 2‐group design estimating sensitivity and specificity in acute phase sera [1] Confirmed COVID‐19 cases (n = 173) with positive RT‐PCR testing for COVID‐19 [2] Controls ‐ pre‐pandemic healthy individuals (n = 213) No information about recruitment | ||
| Patient characteristics and setting | [1] Inpatients at Shenzhen Third People’s Hospital, Shenzhen, China, admitted between 11 January‐9 February 2020, samples between day 1 and day 39. Median (IQR) age 48 (35‐61). 84/173 (49%) male. 126/173 (73%) clear exposure identified. 32/173 (18%) considered critical (presence of ARDS or oxygen saturation < 93%, requiring mechanical ventilation) [2] No information given | ||
| Index tests | One Ab test, no mention of blinding ELISA double antigen sandwich immunoassay (laboratory tests manufactured by Shenzhen YHLO Biotech Co.,Ltd). Measured total Ab, IgM and IgG in plasma; Ab and IgM ‐ RBD of the S protein of SARS‐CoV‐2; IgG ‐ recombinant nucleoprotein antigen. Thresholds NR. Sample timing described for all participants Results from repeat RT‐PCR test mentioned, but no details given |
||
| Target condition and reference standard(s) | [1] Virus detected with RT‐PCR from respiratory swabs. Timing of swabs unclear but precedes serology tests [2] Reference standard based on being pre‐pandemic samples | ||
| Flow and timing | Timing of reference standard NR, all within hospital stay Inadequate plasma samples for 2 IgM tests and 1 IgG test Uninterpretable and indeterminate results not mentioned 535 samples tested from 173 participants; data disaggregated over time. Overall sensitivity and specificity defined as positive at any time point. Accuracy in different time periods based on fewer repeat samples (numbers not known) | ||
| Comparative | |||
| Notes | Supported by Bill & Melinda Gates Foundation No conflicts of interest noted Report from a preprint (not peer reviewed) | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | No | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | High risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Zhao 2020b.
| Study characteristics | |||
| Patient Sampling | 3‐group study recruiting patients estimating sensitivity and specificity [1] Pre‐pandemic controls (n = 257) [2] Controls selected during pandemic (n = 155) [3] Cases from hospitalised or recovered patients (n = 69) Recruitment: various sources and locations Prospective or retrospective recruitment of cases: prospective Sample size (virus/COVID cases): 481 (69) Inclusion and exclusion criteria: no details | ||
| Patient characteristics and setting | Setting: hospital inpatient (cases), community/hospital/clinical lab (controls) Location: for cases various hospitals (including 2 in Beijing and one in Wuhan); group 2 controls from Beijing (N = 15) and Zheiiang province (N = 140) Country: China Dates: NR Symptoms and severity: cases‐at different clinic stages. No more detail Sex: no overall details Age: no details Exposure history: no details | ||
| Index tests | Test name: SARS‐CoV‐2 virus serology ELISA kit
Manufacturer: in‐house
Ab targets: total Ab (IgG + IgM)
Antigens used: SARS‐CoV‐2‐S1 protein
Test method: ELISA
Timing of samples: during hospitalisation
Samples used: plasma
Test operators: NR
Definition of test positivity: standard ELISA method
Blinded to reference standard: unclear Threshold predefined: as per controls supplied with ELISA |
||
| Target condition and reference standard(s) | Reference standard for cases: unclear Samples used: unclear Timing of reference standard: unclear Blinded to index test: yes Incorporated index test: unclear Reference standard for non‐cases: group 1‐pre‐pandemic, group 2‐unclear | ||
| Flow and timing | Time interval between index and reference tests: unclear Results presented by time period: no but possible for a subset of the cases All participants received the same reference standard: no Missing data: no details Uninterpretable results: no details Indeterminate results: no details Unit of analysis: unclear | ||
| Comparative | |||
| Notes | Funding: research Grants from Beijing Science and Technology Commission, Bill & Melinda Gates Foundation, National Natural Science Foundation of China (NSFC) and the National Science and Technology Major Project Publication status: preprint Source: preprint server medRxiv Study author COI: no details but 3 authors are from 3 different companies (AnyGo Technology Co., Ltd, Beijing; AbMax Biotechnology Co., LTD, Beijing; Zhenge Biotechnology Co., LTD, Shanghai) | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Unclear | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| The reference standard does not incorporate the index test | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Zhong 2020 [A].
| Study characteristics | |||
| Patient Sampling | 2‐group study to estimate sensitivity and specificity for diagnosis of active infection [1] PCR‐positive COVID‐19 patients (n = 47) [2] Healthy controls (n = 300) No further details of inclusion or exclusion criteria | ||
| Patient characteristics and setting | [1] Source of cases not described. Study conducted in China. Recruitment period not described (symptom onset for cases dates from 15 January‐13 February 2020; sampling dates from 28 January‐21 February 2020)
Severity (cases only): mild = 22 (47%); moderate = 14 (30%); severe = 6 (13%); critical 5 (11%) Sex: 34% male Age: median 48 (range 18‐82) years. Exposure history not described [2] Healthy controls not described in regard to timing of sampling or characteristics |
||
| Index tests | 3 tests evaluated, this entry (Zhong 2020 [A]) refers to test [A]
Both laboratory‐based evaluations to detect IgM and IgG
A. ELISA using N gene of the SARS‐CoV‐2 cloned into a pET28a vector (rN‐based assay) B ELISA using S gene cloned into a pMFcIg vector‐based (rS‐based assay) C. CLIA (not clearly described; potentially uses both of above described antigens) Thesholds defined retrospectively in regard to optimal cut‐off on ROC curve |
||
| Target condition and reference standard(s) | [1] PCR (no further details); positivity threshold not described. Symptom onset 15 January‐13 February, with serology sampling up to 21 February 2020. RT‐PCR probably SARS‐Cov‐2 specific, but not certain [2] No description of healthy controls provided | ||
| Flow and timing | Time interval between index and reference not described. Results not disaggregated by time period pso No missing data, uninterpretable or indeterminate results described Patient‐based analysis | ||
| Comparative | |||
| Notes | Work was supported by the grants from Sichuan Science and Technology Program (2020YFS0014 and 2020YFS0558), the Chinese Academy of Medical Sciences (2019‐I2M‐5‐032) and Technology & Science & Technology Bureau of Chengdu (2020‐YF05‐00060‐SN and 2020‐YF05‐00075‐SN)
Authors declare no COI present; 3 co‐authors employed by Maccura Biotech Published letter to Editor (Sci China Life Sci) |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| DOMAIN 2: Index Test (Antibody tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | No | ||
| Could the conduct or interpretation of the index test have introduced bias? | High risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| The reference standard does not incorporate the index test | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Were results presented per patient? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Zhong 2020 [B].
| Study characteristics | |||
| Patient Sampling | See main entry for this study for characteristics and QUADAS‐2 assessment (Zhong 2020 [A]) | ||
| Patient characteristics and setting | See main entry for this study for characteristics and QUADAS‐2 assessment (Zhong 2020 [A]) | ||
| Index tests | 3 tests evaluated, this entry (Zhong 2020 [B]) refers to test [B]
Both laboratory‐based evaluations to detect IgM and IgG
A. ELISA using N gene of the SARS‐CoV‐2 cloned into a pET28a vector (rN‐based assay) B ELISA using S gene cloned into a pMFcIg vector‐based (rS‐based assay) C. CLIA (not clearly described; potentially uses both of above described antigens) Thesholds defined retrospectively in regard to optimal cut‐off on ROC curve |
||
| Target condition and reference standard(s) | See main entry for this study for characteristics and QUADAS‐2 assessment (Zhong 2020 [A]) | ||
| Flow and timing | See main entry for this study for characteristics and QUADAS‐2 assessment (Zhong 2020 [A]) | ||
| Comparative | |||
| Notes | |||
Zhong 2020 [C].
| Study characteristics | |||
| Patient Sampling | See main entry for this study for characteristics and QUADAS‐2 assessment (Zhong 2020 [A]) | ||
| Patient characteristics and setting | See main entry for this study for characteristics and QUADAS‐2 assessment (Zhong 2020 [A]) | ||
| Index tests | 3 tests evaluated, this entry (Zhong 2020 [C]) refers to test [C]
Both laboratory‐based evaluations to detect IgM and IgG
A. ELISA using N gene of the SARS‐CoV‐2 cloned into a pET28a vector (rN‐based assay) B ELISA using S gene cloned into a pMFcIg vector‐based (rS‐based assay) C. CLIA (not clearly described; potentially uses both of above described antigens) Thesholds defined retrospectively in regard to optimal cut‐off on ROC curve |
||
| Target condition and reference standard(s) | See main entry for this study for characteristics and QUADAS‐2 assessment (Zhong 2020 [A]) | ||
| Flow and timing | See main entry for this study for characteristics and QUADAS‐2 assessment (Zhong 2020 [A]) | ||
| Comparative | |||
| Notes | |||
A&E: Accident and Emergency Department; Ab: antibody; ARDS: acute respiratory distress syndrome; AU: arbitrary unit; CDC: Center for Disease Control; CMV: cytomegalovirus; CT: computed tomography; CGIA: colloidal gold immunoassay; CLIA: chemiluminescence immunoassay; COI: conflict of interest; D‐: disease negative; D+: disease positive; EIA: enzyme immunoassay; ELISA: enzyme‐linked immunosorbent assay; Flu: fluorescence intensity; GICA: gold immunochromatography assay; HCW: healthcare worker; ICU: intensive care unit; IIFT: indirect Immunofluorescence test; LFA: lateral flow assay; LFIA: lateral flow immunoassay; LIPS: luciferase immunoprecipitation system; LRTI: lower respiratory tract infection; N protein: nucleocapsid protein; N/A: not applicable; NAAT: nucleic acid amplification test; NIH: National Institues of Health; NIHR: National Institute for Health Research; NP: nasopharyngeal; NR: not reported; OP: oropharyngeal; PCR: polymerase chain reaction; POC: point‐of‐care; pso: post‐symptom onset; RBD: receptor binding domain; RNA: ribonucleic acid; ROC: receiver operating characteristic; RT‐PCR: reverse transcriptase polymerase chain reaction; RT‐qPCR: reverse transcriptase quantitative polymerase chain reaction; SARS‐CoV‐2: severe acute respiratory syndrome coronavirus 2; S‐flow: flow‐cytometry based test; S protein: spike protein; SD: standard deviation; TB: tuberculosis; WHO: World Health Organization
Characteristics of excluded studies [ordered by study ID]
| Study | Reason for exclusion |
|---|---|
| Ai 2020 | Ineligible reference standard |
| Aitken 2020 | Ineligible study design |
| Amanat 2020 | Accuracy data cannot be extracted |
| Annamalai 2020 | Ineligible study design |
| Argenziano 2020 | Ineligible index test |
| Arons 2020 | Ineligible index test |
| Arumugam 2020 | Ineligible study design |
| Baggett 2020 | Ineligible study design |
| Bai 2020 | Inadequate sample size |
| Bajema 2020 | Ineligible study design |
| Barra 2020 | Ineligible study design |
| Batista 2020 | Ineligible index test |
| Beltran Corbellini 2020 | Ineligible index test |
| Beltran Pavez 2020 | Inadequate sample size |
| Ben Ami 2020 | Ineligible index test |
| Bhadra 2020 | Ineligible study design |
| Bordi 2020 | Ineligible study design |
| Burhan 2020 | Ineligible study design |
| Cai 2020 | Ineligible study design |
| Callahan 2020 | Accuracy data cannot be extracted |
| Chan 2020 | Ineligible study design |
| Chandler Brown 2020 | Ineligible study design |
| Chen 2020b | Ineligible target condition |
| Chen 2020c | Ineligible study design |
| Cheng 2020a | Ineligible reference standard |
| Chu 2020 | Ineligible study design |
| Colson 2020 | Inadequate sample size |
| Comar 2020 | Ineligible reference standard |
| Corman 2020 | Ineligible study design |
| Cui 2020 | Ineligible study design |
| Curti 2020 | Ineligible study design |
| Dahlke 2020 | Inadequate sample size |
| Ding 2020 | Ineligible study design |
| Fang 2020z | Ineligible index test |
| Farfan 2020 | Ineligible study design |
| Feng 2020 | Ineligible index test |
| Fontanet 2020 | Ineligible study design |
| Fu 2020a | Ineligible population |
| Fu 2020b | Accuracy data cannot be extracted |
| Fumeaux 2020 | Ineligible study design |
| Gao 2020 | Ineligible index test |
| Giamarellos Bourboulis 2020 | Ineligible study design |
| Gietema 2020 | Ineligible index test |
| Gonzalez Gonzalez 2020a | Ineligible study design |
| Gonzalez Gonzalez 2020b | Ineligible study design |
| Guan 2020 | Ineligible study design |
| Guo 2020b | Ineligible study design |
| Guo 2020c | Ineligible population |
| Han 2020 | Ineligible index test |
| Hao 2020 | Ineligible study design |
| Hass 2020 | Ineligible target condition |
| Hirotsu 2020 | Ineligible study design |
| Hogan 2020 | Ineligible index test |
| Holland 2020 | Ineligible study design |
| Hu 2020b | Ineligible study design |
| Hu 2020c | Author contact needed |
| Hu 2020d | Ineligible index test |
| Huang 2020a | Ineligible study design |
| Jenkins 2020 | Ineligible study design |
| Jiang 2020a | Ineligible study design |
| Jiang 2020b | Accuracy data cannot be extracted |
| Jung 2020 | Ineligible study design |
| Khan 2020 | Ineligible study design |
| Kim 2019 | Ineligible study design |
| Kong 2020 | Ineligible study design |
| Konrad 2020 | Ineligible study design |
| Kurstjens 2020 | Ineligible index test |
| Lamb 2020 | Ineligible study design |
| Lan 2020 | Ineligible population |
| Lechien 2020 | Ineligible index test |
| Lei 2020 | Ineligible study design |
| Li 2020c | Inadequate sample size |
| Li 2020d | Ineligible study design |
| Li 2020e | Ineligible index test |
| Li 2020f | Ineligible population |
| Li 2020g | Ineligible index test |
| Liang 2020a | Ineligible index test |
| Liang 2020b | Ineligible study design |
| Ling 2020 | Ineligible target condition |
| Liu 2020e | Ineligible index test |
| Liu 2020f | Ineligible index test |
| Liu 2020g | Ineligible index test |
| Liu 2020h | Ineligible study design |
| Lo 2020 | Ineligible index test |
| Lopez‐Rincon 2020 | Ineligible study design |
| Lu 2020 | Ineligible study design |
| Ma 2020b | Ineligible study design |
| Mahari 2020 | Ineligible study design |
| Mardani 2020 | Ineligible index test |
| Marzinotto 2020 | Accuracy data cannot be extracted |
| McKay 2020 | Ineligible study design |
| McRae 2020 | Ineligible index test |
| Mei 2020 | Ineligible index test |
| Meng 2020 | Ineligible index test |
| Mercurio 2020 | Ineligible study design |
| Metsky 2020 | Ineligible study design |
| Nelson 2020 | Ineligible study design |
| Nemati 2020 | Ineligible study design |
| Nie 2020 | Ineligible study design |
| Nunez Bajo 2020 | Ineligible study design |
| Okba 2020b | Ineligible study design |
| Paden 2020 | Ineligible study design |
| Pan 2020b | Ineligible index test |
| Pan 2020c | Ineligible study design |
| Pan 2020d | Ineligible index test |
| Pan 2020e | Ineligible study design |
| Paradiso 2020b | Accuracy data cannot be extracted |
| Park 2020 | Ineligible study design |
| Peng 2020 | Ineligible index test |
| Pfefferle 2020 | Ineligible study design |
| Rauch 2020 | Ineligible study design |
| Scallan 2020 | Accuracy data cannot be extracted |
| Seo 2020 | Accuracy data cannot be extracted |
| Shental 2020 | Ineligible study design |
| Shi 2020 | Ineligible index test |
| Shirato 2020 | Ineligible study design |
| Song 2020 | Ineligible index test |
| Su 2020 | Ineligible index test |
| Sun 2020a | Ineligible index test |
| Sun 2020b | Ineligible index test |
| Sun 2020c | Ineligible study design |
| Tagarro 2020 | Ineligible index test |
| Tan 2020a | Ineligible study design |
| Tan 2020b | Ineligible index test |
| Tan 2020c | Ineligible study design |
| Toptan 2020 | Ineligible study design |
| Tsang 2003 | Ineligible target condition |
| Vermeiren 2020 | Accuracy data cannot be extracted |
| Viehweger 2020 | Ineligible study design |
| Vogels 2020 | Ineligible study design |
| Waghmare 2020 | Ineligible population |
| Wang 2020b | Ineligible index test |
| Wang 2020c | Accuracy data cannot be extracted |
| Wang 2020d | Accuracy data cannot be extracted |
| Wang 2020e | Accuracy data cannot be extracted |
| Wang 2020f | Ineligible study design |
| Wang 2020g | Retracted study |
| Wang 2020h | Accuracy data cannot be extracted |
| Wang 2020i | Ineligible index test |
| Wee 2020 | Ineligible study design |
| Weiss 2020 | Accuracy data cannot be extracted |
| Woelfel 2020 | Ineligible reference standard |
| Won 2020 | Ineligible study design |
| Woo 2020 | Ineligible study design |
| Wu 2020a | Ineligible index test |
| Wu 2020b | Ineligible study design |
| Wu 2020c | Accuracy data cannot be extracted |
| Xia 2020a | Ineligible index test |
| Xia 2020b | Ineligible study design |
| Xie 2020b | Ineligible population |
| Xie 2020c | Accuracy data cannot be extracted |
| Xing 2020a | Inadequate sample size |
| Xing 2020b | Ineligible reference standard |
| Xu 2020b | Ineligible study design |
| Xu 2020c | Inadequate sample size |
| Xu 2020d | Ineligible index test |
| Yan 2020 | Ineligible study design |
| Yang 2020a | Ineligible reference standard |
| Yang 2020b | Ineligible study design |
| Yelin 2020 | Ineligible study design |
| Yuan 2020 | Accuracy data cannot be extracted |
| Yun 2020 | Ineligible study design |
| Zeng 2020b | Accuracy data cannot be extracted |
| Zhang 2020e | Ineligible study design |
| Zhang 2020f | Ineligible study design |
| Zhang 2020g | Accuracy data cannot be extracted |
| Zhang 2020h | Accuracy data cannot be extracted |
| Zhao 2020c | Ineligible study design |
| Zhao 2020d | Ineligible study design |
| Zhifeng 2020 | Ineligible reference standard |
| Zhou 2020 | Accuracy data cannot be extracted |
| Zhuang 2020 | Retracted study |
Characteristics of studies awaiting classification [ordered by study ID]
Li 2020b.
| Patient Sampling | Foreign language study awaiting translation | ||
| Patient characteristics and setting | |||
| Index tests | |||
| Target condition and reference standard(s) | |||
| Flow and timing | |||
| Comparative | |||
| Notes | |||
Thompson 2020.
| Patient Sampling | Study of neutralising antibodies; to be assessed for inclusion in review update | ||
| Patient characteristics and setting | |||
| Index tests | |||
| Target condition and reference standard(s) | |||
| Flow and timing | |||
| Comparative | |||
| Notes | |||
Xiong 2020.
| Patient Sampling | Foreign language study awaiting translation | ||
| Patient characteristics and setting | |||
| Index tests | |||
| Target condition and reference standard(s) | |||
| Flow and timing | |||
| Comparative | |||
| Notes | |||
Characteristics of ongoing studies [ordered by study ID]
ChiCTR2000029625.
| Study name | Construction of early warning and prediction system for patients with severe / critical novel coronavirus pneumonia (COVID‐19) |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
ChiCTR2000029695.
| Study name | Early detection of novel coronavirus pneumonia (COVID‐19) based on a novel high‐throughput mass spectrometry analysis with exhaled breath |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
ChiCTR2000029810.
| Study name | Clinical study of a novel high sensitivity nucleic acid assay for novel coronavirus pneumonia (COVID‐19) based on CRISPR‐cas protein |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
ChiCTR2000029870.
| Study name | Evaluation of rapid diagnostic kit (IgM/IgG) for novel coronavirus pneumonia (COVID‐19) |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
ChiCTR2000029883.
| Study name | A comparative study on the sensitivity of nasopharyngeal and oropharyngeal swabbing for the detection of SARS‐CoV‐2 by real‐time PCR |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
ChiCTR2000029982.
| Study name | Study for using multiomics in the diagnosis and treatment of novel coronavirus pneumonia (COVID‐19 |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
ChiCTR2000030005.
| Study name | Nucleic acid analysis of novel coronavirus pneumonia (COVID‐19) in morning sputum samples and pharyngeal swabs‐a prospectively diagnostic test |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
ChiCTR2000030085.
| Study name | Cancelled by the investigator Study for the false positive rate of IgM / IgG antibody test kit for novel coronavirus pneumonia (COVID‐19) in different inpatients |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
ChiCTR2000030185.
| Study name | The value of critical care ultrasound in rapid screening, diagnosis, evaluation of effectiveness and intensive prevention of novel coronavirus pneumonia (COVID‐19) |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
ChiCTR2000030253.
| Study name | Exploration and research for a new method for detection of novel coronavirus (COVID‐19) nucleic acid |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
ChiCTR2000030334.
| Study name | MicroRNA as a marker for early diagnosis of novel coronavirus infection (COVID‐19) |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
ChiCTR2000030542.
| Study name | A clinical study about the diagnosis and prognosis evaluation of novel coronavirus pneumonia (COVID‐19) based on viral genome, host genomic sequencing, relative cytokines and other laboratory indexes |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
ChiCTR2000030543.
| Study name | Detection of coronavirus in simultaneously collecting tears and throat swab samples collected from the patients with novel coronavirus pneumonia (COVID‐19) |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
ChiCTR2000030558.
| Study name | Cancelled by the investigator Epidemiological research of novel coronavirus pneumonia (COVID‐19) suspected cases based on virus nucleic acid test combined with low‐dose chest CT screening in primary hospital |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
ChiCTR2000030706.
| Study name | Cancelled by the investigator Application of cas13a‐mediated RNA detection in the assay of novel coronavirus nucleic acid (COVID‐19) |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
ChiCTR2000030721.
| Study name | A comparative study for the sensitivity of induced sputum and throat swabs for the detection of SARS‐CoV‐2 by real‐time PCR in patients with novel coronavirus pneumonia (COVID‐19) |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
ChiCTR2000030754.
| Study name | Medical records based study for the accuracy of SARS‐CoV‐2 IgM antibody screening for diagnosis of novel coronavirus pneumonia (COVID‐19) |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
ChiCTR2000030833.
| Study name | Clinical validation and application of high‐throughput novel coronavirus (2019‐nCoV) screening detection kit |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
ChiCTR2000030834.
| Study name | Epidemiological characteristics and antibody levels of novel coronavirus pneumonia (COVID‐19) of pediatric medical staff working in quarantine area |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
ChiCTR2000030838.
| Study name | Development of warning system with clinical differential diagnosis and prediction for severe type of novel coronavirus pneumonia (COVID‐19) patients based on artificial intelligence and CT images |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
ChiCTR2000030856.
| Study name | An artificial intelligence assistant system for suspected novel coronavirus pneumonia (COVID‐19) based on chest CT |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
ChiCTR2000030859.
| Study name | A medical based analysis for influencing factors of death of novel coronavirus pneumonia (COVID‐19) patients in Wuhan Third Hospital |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
ChiCTR2000030860.
| Study name | A medical records based study for investigation of dynamic profile of RT‐PCR test for SARS‐CoV‐2 nucleic acid of novel coronavirus pneumonia (COVID‐19) patients |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
ChiCTR2000030862.
| Study name | Correlation analysis of blood eosinophil cell levels and clinical type category of novel coronavirus pneumonia (COVID‐19): a medical records based retrospective study |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
NCT04245631.
| Study name | Development of a simple, fast and portable recombinase aided amplification assay for 2019‐nCoV |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
NCT04259892.
| Study name | Viral excretion in contact subjects at high/moderate risk of coronavirus 2019‐nCoV infection |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
NCT04279795.
| Study name | Detection of 2019 novel coronavirus in multiple organ system and its relationship with clinical manifestations |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
NCT04304690.
| Study name | SARS‐CoV2 seroconversion among front line medical and paramedical staff in emergency, intensive care units and infectious disease departments during the 2020 epidemic |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
NCT04311398.
| Study name | Development and verification of a new coronavirus multiplex nucleic acid detection system |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
NCT04316728.
| Study name | Clinical performance of the VivaDiag ™ COVID‐19 lgM IgG rapid test in early detecting the infection of COVID‐19 |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
NCT04321369.
| Study name | Impact of swab site and sample collector on testing sensitivity for SARS‐CoV‐2 virus in symptomatic individuals |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
NCT04322279.
| Study name | Factors associated with a positive SARS‐CoV‐2 serology in contact subjects at high/moderate risk of coronavirus SARS‐CoV‐2 infection (CoV‐CONTACT‐SERO) |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
NCT04322513.
| Study name | Biomarkers for identification of SARS‐COV‐2 infection |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
Sullivan 2020.
| Study name | Detection of SARS‐CoV‐2 RNA and antibodies in diverse samples: protocol to validate the sufficiency of provider‐observed home‐collected blood, saliva and oropharyngeal samples |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
Differences between protocol and review
As we explained in the review, due to poor reporting, we were unable to identify studies that evaluated the test in patients who were symptomatic (active disease) separately from those who had recovered from their symptoms (convalescent). Our stratification of results according to days since onset of symptoms will in part be related to these categorisations.
We planned to check the following websites for eligible index tests, however these did not prove to be very accessible or easy to use and, after initial review, were not further considered:
National Institute for Health Research (NIHR) Innovation Observatory (www.io.nihr.ac.uk/)
We planned to check the following evidence repository for additional eligible studies however, the EPPI‐Centre and Norwegian Institute of Public Health resources proved to be more accessible therefore we decided to prioritise our other sources of evidence.
Meta‐evidence (meta-evidence.co.uk/the-role-of-evidence-synthesis-in-covid19/)
QUADAS‐2 (Whiting 2011), item "Was there an appropriate interval between index test(s) and reference standard?" was dropped from assessment because for antibody tests, the body's immune response to SARS‐CoV‐2 infection tends to increase over time such that the time between confirmation of the presence of SARS‐CoV‐2 an the index test is less relevant than the time from symptom onset to the application of the index test.
Contributions of authors
JJD was the contact person with the editorial base. JJD co‐ordinated contributions from the co‐authors and wrote the final draft of the review. JJD, JDi, YT, CD, STP, IH, AA, LFR, MP, JDr, SB screened papers against eligibility criteria. RS conducted the literature searches JJD, JDi, YT, CD, STP, IH, AA, LFR, MP, JDr, SB appraised the quality of papers. JJD, JDi, YT, CD, STP, IH, AA, LFR, MP, JDr, SB extracted data for the review and sought additional information about papers. JJD and JDi entered data into Review Manager 2014. JJD and JDi analysed and interpreted data. JJD, JDi, YT, CD, STP, RS, ML, LH, AVB, DE, SD worked on the methods sections. JJD and JDi responded to the comments of the referees. JJD.is the guarantor of the update.
Sources of support
Internal sources
Liverpool School of Tropical Medicine, UK
University of Birmingham, UK
External sources
-
Department for International Development, UK
Project number: 300342‐104
National Institute for Health Research (NIHR), UK
NIHR Birmingham Biomedical Research Centre at the University Hospitals Birmingham NHS Foundation Trust and the University of Birmingham, UK
Declarations of interest
Jonathan J Deeks: none known
Jacqueline Dinnes: none known
Yemisi Takwoingi: none known
Clare Davenport: none known
René Spijker: the Dutch Cochrane Centre (DCC) has received grants for performing commissioned systematic reviews. In no situation, the commissioner had any influence on the results of the work.
Sian Taylor‐Phillips: none known
Ada Adriano: none known
Sophie Beese: none known
Janine Dretzke: none known
Lavinia Ferrante di Ruffano: none known
Isobel Harris: none known
Malcolm Price: none known
Sabine Dittrich: is employed by FIND with funding from DFID and Australian Aid. FIND is a global non‐for profit product development partnership and WHO Diagnostic Collaboration Centre. It is FIND’s role to accelerate access to high quality diagnostic tools for low resource settings and this is achieved by supporting both R&D and access activities for a wide range of diseases, including COVID‐19. .FIND has several clinical research projects to evaluate multiple new diagnostic tests against published Target Product Profiles that have been defined through consensus processes. These studies are for diagnostic products developed by private sector companies who provide access to know‐how, equipment/reagents, and contribute through unrestricted donations as per FIND policy and external SAC review.
Devy Emperador: is employed by FIND with funding from DFID and KFW. FIND is a global non‐for profit product development partnership and WHO Diagnostic Collaboration Centre. It is FIND’s role to accelerate access to high quality diagnostic tools for low resource settings and this is achieved by supporting both R&D and access activities for a wide range of diseases, including COVID‐19. .FIND has several clinical research projects to evaluate multiple new diagnostic tests against published Target Product Profiles that have been defined through consensus processes. These studies are for diagnostic products developed by private sector companies who provide access to know‐how, equipment/reagents, and contribute through unrestricted donations as per FIND policy and external SAC review.
Lotty Hooft: none known
Mariska MG Leeflang: none known
Ann Van den Bruel: none known
New
References
References to studies included in this review
Adams 2020 [A] {published data only}
- Adams ER, Anand R, Andersson MI, Auckland K, Baillie JK, Barnes E, et al. Evaluation of antibody testing for SARS-Cov-2 using ELISA and lateral flow immunoassays. medRxiv [Preprint] 2020. [www.medrxiv.org/content/10.1101/2020.04.15.20066407v1.full.pdf] [Google Scholar]
Adams 2020 [B] {published data only}
Adams 2020 [C] {published data only}
Adams 2020 [D] {published data only}
Adams 2020 [E] {published data only}
Adams 2020 [F] {published data only}
Adams 2020 [G] {published data only}
Adams 2020 [H] {published data only}
Adams 2020 [I] {published data only}
Adams 2020 [J] {published data only}
Bendavid 2020 {published data only}
- Bendavid E, Mulaney B, Sood N, Shah S, Ling E, Bromley-Dulfano R, et al. COVID-19 antibody seroprevalence in Santa Clara County, California. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Burbelo 2020 [A] {published data only}
- Burbelo PD, Riedo FX, Morishima C, Rawlings S, Smith D, Das S, et al. Detection of nucleocapsid antibody to SARS-CoV-2 is more sensitive than antibody to spike protein in COVID-19 patients. MedRxiv [Preprint] 2020. [Google Scholar]
Burbelo 2020 [B] {published data only}
Cai 2020a {published data only}
- Cai X, Chen J, Hu J, Long Q, Deng H, Fan K, et al. A peptide-based magnetic chemiluminescence enzyme immunoassay for serological diagnosis of corona virus disease 2019 (COVID-19). MedRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Cassaniti 2020 (A) {published data only}
- Cassaniti I, Novazzi F, Giardina F, Salivaro F, Sachs M, Perlini S, et al. Performance of VivaDiagTM COVID-19 IgM/IgG Rapid Test is inadequate for diagnosis of COVID-19 in acute patients referring to emergency room department. Journal of Medical Virology 2020 Mar 30 [Epub ahead of print]. [DOI: 10.1002/jmv.25800] [DOI] [PMC free article] [PubMed] [Google Scholar]
Cassaniti 2020 (B) {published data only}
Chen 2020a {published data only}
- Chen Z, Zhang Z, Zhai X, Li Y, Lin L, Zhao H, et al. Rapid and sensitive detection of anti-SARS-CoV-2 IgG using lanthanide-doped nanoparticles-based lateral flow immunoassay. Analytical Chemistry 2020;92(10):7226-31. [DOI] [PubMed] [Google Scholar]
Dohla 2020 {published data only}
- Dohla M, Boesecke C, Schulte B, Diegmann C, Sib E, Richter E, et al. Rapid point-of-care testing for SARS-CoV-2 in a community screening setting shows low sensitivity. Public Health 2020;182:170-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Du 2020 {published data only}
- Du Z, Zhu F, Guo F, Yang B, Wang T. Detection of antibodies against SARS-CoV-2 in patients with COVID-19. Journal of Medical Virology 2020 Apr 3 [Epub ahead of print]. [DOI: 10.1002/jmv.25820] [DOI] [PMC free article] [PubMed] [Google Scholar]
Freeman 2020 {published data only}
- Freeman B, Lester S, Mills L, Rasheed MA, Moye S, Abiona O, et al. Validation of a SARS-CoV-2 spike ELISA for use in contact investigations and serosurveillance. bioRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Gao 2020a {published data only}
- Gao Y, Yuan Y, Li T T, Wang W X, Li Y X, Li A, et al. Evaluation the auxiliary diagnosis value of antibodies assays for detection of novel coronavirus (SARS-Cov-2) causing an outbreak of pneumonia (COVID-19). medRxiv [Preprint] 2020. [Google Scholar]
- Gao Y, Yuan Y, Li TT, Wang WX, Li YX, Li A, et al. Evaluation of the auxiliary diagnostic value of antibody assays for the detection of novel coronavirus (SARS-CoV-2). Journal of Medical Virology 2020. [DOI: 10.1002/jmv.25919] [DOI] [PMC free article] [PubMed] [Google Scholar]
Gao 2020b [A] {published data only}
- Gao HX, Li YN, Xu ZG, Wang YL, Wang HB, Cao JF, et al. Detection of serum immunoglobulin M and immunoglobulin G antibodies in 2019-novel coronavirus infected cases from different stages. Chinese Medical Journal (Engl) 2020 Mar 26 [Epub ahead of print]. [DOI: 10.1097/CM9.0000000000000820] [DOI] [PMC free article] [PubMed] [Google Scholar]
Gao 2020b [B] {published data only}
Gao 2020b [C] {published data only}
Garcia 2020 (A) {published data only}
- Garcia FP, Perez Tanoira R, Romanyk Cabrera JP, Arroyo Serrano T, Gomez Herruz P, Cuadros Gonzalez J. Rapid diagnosis of SARS-CoV-2 infection by detecting IgG and IgM antibodies with an immunochromatographic device: a prospective single-center study. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Garcia 2020 (B) {published data only}
Grzelak 2020 [A] {published data only}
- Grzelak L, Temmam S, Planchais C, Demeret C, Huon C, Guivel F, et al. SARS-CoV-2 serological analysis of COVID-19 hospitalized patients, pauci-symptomatic individuals and blood donors. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Grzelak 2020 [B] {published data only}
Grzelak 2020 [C] {published data only}
Grzelak 2020 [D] {published data only}
Grzelak 2020 [E] {published data only}
Guo 2020a {published data only}
- Guo L, Ren L, Yang S, Xiao M, Chang, Yang F, et al. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19). Clinical Infectious Diseases 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Hu 2020a {published data only}
- Hu Q, Cui X, Liu X, Peng B, Jiang J, Wang X, et al. The production of antibodies for SARS-CoV-2 and its clinical implication. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Infantino 2020 {published data only}
- Infantino M, Grossi V, Lari B, Bambi R, Perri A, Manneschi M, et al. Diagnostic accuracy of an automated chemiluminescent immunoassay for anti-SARS-CoV-2 IgM and IgG antibodies: an Italian experience. Journal of Medical Virology 2020 Apr 24 [Epub ahead of print]. [DOI: 10.1002/jmv.25932] [DOI] [PMC free article] [PubMed] [Google Scholar]
Jia 2020 {published data only}
- Jia X, Zhang P, Tian Y, Wang J, Zeng H, Wang J, et al. Clinical significance of IgM and IgG test for diagnosis of highly suspected COVID-19 infection. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Jin 2020 {published data only}
- Jin Y, Wang M, Zuo Z, Fan C, Ye F, Cai Z, et al. Diagnostic value and dynamic variance of serum antibody in coronavirus disease 2019. International Journal of Infectious Diseases 2020;94:49-52. [DOI: 10.1016/j.ijid.2020.03.065] [DOI] [PMC free article] [PubMed] [Google Scholar]
Lassauniere 2020 [A] {published data only}
- Lassauniere R, Frische A, Harboe ZB, Nielsen AC, Fomsgaard A, Krogfelt KA, et al. Evaluation of nine commercial SARS-CoV-2 immunoassays. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Lassauniere 2020 [B] {published data only}
Lassauniere 2020 [C] {published data only}
Lassauniere 2020 [D] {published data only}
Lassauniere 2020 [E] {published data only}
Lassauniere 2020 [F] {published data only}
Lassauniere 2020 [G] {published data only}
Lassauniere 2020 [H] {published data only}
Lassauniere 2020 [I] {published data only}
Li 2020a {published data only}
- Li Z, Yi Y, Luo X, Xiong N, Liu Y, Li S, et al. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. Journal of Medical Virology 2020 Feb 27 [Epub ahead of print]. [DOI: 10.1002/jmv.25727] [DOI] [PMC free article] [PubMed] [Google Scholar]
Lin 2020a [A] {published data only}
- Lin D, Liu L, Zhang M, Hu Y, Yang Q, Guo J, et al. Evaluations of serological test in the diagnosis of 2019 novel coronavirus (SARS-CoV-2) infections during the COVID-19 outbreak. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Lin 2020a [B] {published data only}
Lippi 2020 [A] {published data only}
- Lippi G, Salvagno GL, Pegoraro M, Militello V, Caloi C, Peretti A, et al. Assessment of immune response to SARS-CoV-2 with fully automated MAGLUMI 2019-nCoV IgG and IgM chemiluminescence immunoassays. Clinical Chemistry and Laboratory Medicine 2020 Apr 16 [Epub ahead of print]. [DOI: 10.1515/cclm-2020-0473] [DOI] [PubMed] [Google Scholar]
Lippi 2020 [B] {published data only}
Liu 2020a {published data only}
- Liu Y, Liu Y, Diao B, Ren F, Wang Y, Ding J, et al. Diagnostic indexes of a rapid IgG/IgM combined antibody test for SARS-CoV-2. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Liu 2020b {published data only}
- Liu L, Liu W, Wang S, Zheng S. A preliminary study on serological assay for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in 238 admitted hospital patients. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Liu 2020c {published data only}
- Liu R, Liu X, Han H, Shereen MA, Niu Z, Li D, et al. The comparative superiority of IgM-IgG antibody test to real-time reverse transcriptase PCR detection for SARS-CoV-2 infection diagnosis. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Liu 2020d [A] {published data only}
- Liu W, Liu L, Kou G, Zheng Y, Ding Y, Ni W, et al. Evaluation of nucleocapsid and spike protein-based ELISAs for detecting antibodies against SARS-CoV-2. Journal of Clinical Microbiology 2020. [DOI: 10.1128/JCM.00461-20] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Liu L, Kou G, Zheng Y, Ding Y, Ni W, et al. Evaluation of nucleocapsid and spike protein-based ELISAs for detecting antibodies against SARS-CoV-2. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Liu 2020d [B] {published data only}
Long 2020 (A) {published data only}
- Long Q, Deng H, Chen J, Hu J, Liu B, Liao P, et al. Antibody responses to SARS-CoV-2 in COVID-19 patients: the perspective application of serological tests in clinical practice. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Long 2020 (B) {published data only}
Lou 2020 [A] {published data only}
- Lou B, Li T, Zheng S, Su Y, Li Z, Liu W, et al. Serology characteristics of SARS-CoV-2 infection since the exposure and post symptoms onset. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Lou 2020 [B] {published data only}
Lou 2020 [C] {published data only}
Ma 2020a {published data only}
- Ma H, Zeng W, He H, Zhao D, Yang Y, Jiang D, et al. COVID-19 diagnosis and study of serum SARS-CoV-2 specific IgA, IgM and IgG by a quantitative and sensitive immunoassay. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Okba 2020a {published data only}
- Okba NM, Muller MA, Li W, Wang C, GeurtsvanKessel CH, Corman VM, et al. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease 2019 patients. Emerging Infectious Diseases 2020 Apr 8 [Epub ahead of print];26(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woelfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Muller MA, et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020;581:465-9. [DOI] [PubMed] [Google Scholar]
Padoan 2020 {published data only}
- Padoan A, Cosma C, Sciacovelli L, Faggian D, Plebani M. Analytical performances of a chemiluminescence immunoassay for SARS-CoV-2 IgM/IgG and antibody kinetics. Clinical Chemistry and Laboratory Medicine 2020. [DOI: doi.org/10.1515/cclm-2020-0443 ] [DOI] [PubMed] [Google Scholar]
Pan 2020a {published data only}
- Pan Y, Li X, Yang G, Fan J, Tang Y, Zhao J, et al. Serological immunochromatographic approach in diagnosis with SARS-CoV-2 infected COVID-19 patients. Journal of Infection (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Li X, Yang G, Fan J, Tang Y, Zhao J, et al. Serological immunochromatographic approach in diagnosis with SARS-CoV-2 infected COVID-19 patients. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Paradiso 2020a {published data only}
- Paradiso AV, De Summa S, Loconsole D, Procacci V, Sallustio A, Centrone F, et al. Clinical meanings of rapid serological assay in patients tested for SARS-Co2 RT-PCR. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Qian 2020 {published data only}
- Qian C, Zhou M, Cheng F, Lin X, Gong Y, Xie X, et al. Development and multicenter performance evaluation of the first fully automated SARS-CoV-2 IgM and IgG immunoassays. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PubMed] [Google Scholar]
To 2020a [A] {published data only}
- To KK, Tsang OT, Leung WS, Tam AR, Wu TC, Lung DC, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infectious Diseases 2020;20(5):565-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
To 2020a [B] {published data only}
Wan 2020 [A] {published data only}
- Wan W Y, Lim S H, Seng E H. Cross-reaction of sera from COVID-19 patients with SARS-CoV assays. medRxiv [Preprint] 2020. [DOI: ] [PubMed] [Google Scholar]
Wan 2020 [B] {published data only}
Wang 2020a [A] {published data only}
- Wang Q, Du Q, Guo B, Guo Y, Fang L, X Guo. Performance of urea-mediated dissociation in reducing false-positive of 2019-nCoV IgM test. Chinese Journal of Laboratory Medicine 2020. [DOI: 10.3760/cma.j.cn114452-20200219-00091] [DOI] [Google Scholar]
- Wang Q, Du Q, Guo B, Mu D, Lu X, Ma Q, et al. A method to prevent SARS-CoV-2 IgM false positives in gold immunochromatography and enzyme-linked immunosorbent assays. Journal of Clinjcal Microbiology 2020. [DOI: 10.1128/JCM.00375-20] [DOI] [PMC free article] [PubMed] [Google Scholar]
Wang 2020a [B] {published data only}
Xiang 2020a [A] {published data only}
- Xiang J, Yan M, Li H, Liu T, Lin C, Huang S, et al. Evaluation of enzyme-linked immunoassay and colloidal gold- immunochromatographic assay kit for detection of novel coronavirus (SARS-Cov-2) causing an outbreak of pneumonia (COVID-19). medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Xiang 2020a [B] {published data only}
Xiang 2020b {published data only}
- Xiang F, Wang X, He X, Peng Z, Yang B, Zhang J, et al. Antibody detection and dynamic characteristics in patients with COVID-19. Clinical Infectious Diseases 2020;ciaa461. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Xiao 2020a {published data only}
- Xiao DA, Gao DC, Zhang DS. Profile of specific antibodies to SARS-CoV-2: the first report. Journal of Infection 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Xie 2020a {published data only}
- Xie J, Ding C, Li J, Wang Y, Guo H, Lu Z, et al. Characteristics of patients with coronavirus disease (COVID-19) confirmed using an IgM-IgG antibody test. Journal of Medical Virology 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Xu 2020a {published data only}
- Xu Y. Dynamic profile of severe or critical COVID-19 cases. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Yongchen 2020 {published data only}
- Yongchen Z, Shen H, Wang X, Shi X, Li Y, Yan J, et al. Different longitudinal patterns of nucleic acid and serology testing results based on disease severity of COVID-19 patients. Emerging Microbes and Infections 2020;9(1):833-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Zeng 2020a {published data only}
- Zeng Z, Chen L, Pan Y, Deng Q, Ye G, Li Y, et al. Re: Profile of specific antibodies to SARS-CoV-2: the first report. Journal of Infection 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Zhang 2020a {published data only}
- Zhang B, Zhou X, Zhu C, Feng F, Qiu Y, Feng J, et al. Immune phenotyping based on neutrophil-to-lymphocyte ratio and IgG predicts disease severity and outcome for patients with COVID-19. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Zhang 2020b {published data only}
- Zhang J, Liu J, Li N, Liu Y, Ye R, Qin X, et al. Serological detection of 2019-nCoV respond to the epidemic: a useful complement to nucleic acid testing. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Zhang 2020c {published data only}
- Zhang W, Du RH, Li B, Zheng XS, Yang X L, Hu B, et al. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerging Microbes and Infections 2020;9(1):386-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Zhang 2020d {published data only}
- Zhang P, Gao Q, Wang T, Ke Y, Mo F, Jia R, et al. Evaluation of recombinant nucleocapsid and spike proteins for serological diagnosis of novel coronavirus disease 2019 (COVID-19). medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Zhao 2020a {published data only}
- Zhao J, Yuan Q, Wang H, Liu W, Liao X, Su Y, et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clinical Infectious Diseases 2020;ciaa344. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Yuan Q, Wang H, Liu W, Liao X, Su Y, et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Zhao 2020b {published data only}
- Zhao R, Li M, Song H, Chen J, Ren W, Feng Y, et al. Serological diagnostic kit of SARS-CoV-2 antibodies using CHO-expressed full-length SARS-CoV-2 S1 proteins. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Zhong 2020 [A] {published data only}
- Zhong L, Chuan J, Gong BO, Shuai P, Zhou Y, Zhang Y, et al. Detection of serum IgM and IgG for COVID-19 diagnosis. Science China. Life Sciences 2020;63(5):777-80. [DOI: 10.1007/s11427-020-1688-9] [DOI] [PMC free article] [PubMed] [Google Scholar]
Zhong 2020 [B] {published data only}
Zhong 2020 [C] {published data only}
References to studies excluded from this review
Ai 2020 {published data only}
- Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W, et al. Correlation of Chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology 2020. [DOI: 10.1148/radiol.2020200642 10.1148/radiol.2020200642] [DOI] [PMC free article] [PubMed] [Google Scholar]
Aitken 2020 {published data only}
- Aitken J, Ambrose K, Barrell S, Beale R, Bineva-Todd G, Biswas D, et al. Scalable and resilient SARS-CoV2 testing in an academic centre. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.19.20071373] [DOI] [Google Scholar]
Amanat 2020 {published data only}
- Amanat F, Nguyen T, Chromikova V, Strohmeier S, Stadlbauer D, Javier A, et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.17.20037713] [DOI] [PMC free article] [PubMed] [Google Scholar]
Annamalai 2020 {published data only}
- Annamalai P, Kanta M, Ramu P, Ravi B, Veerapandian K, Srinivasan R. A simple colorimetric molecular detection of novel coronavirus (COVID-19), an essential diagnostic tool for pandemic screening. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.10.20060293] [DOI] [Google Scholar]
Argenziano 2020 {published data only}
- Argenziano MG, Bruce SL, Slater CL, Tiao JR, Baldwin MR, Barr RG. Characterization and clinical course of 1000 patients with COVID-19 in New York: retrospective case series. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.20.20072116] [DOI] [PMC free article] [PubMed] [Google Scholar]
Arons 2020 {published data only}
- Arons MM, Hatfield KM, Reddy SC, Kimball A, James A, Jacobs JR. Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. New England Journal of Medicine 2020. [DOI: 10.1056/NEJMoa2008457] [DOI] [PMC free article] [PubMed] [Google Scholar]
Arumugam 2020 {published data only}
- Arumugam A, Wong SS. The potential use of unprocessed sample for RT-qPCR detection of COVID-19 without an RNA extraction step. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.06.028811] [DOI] [Google Scholar]
Baggett 2020 {published data only}
- Baggett TP, Keyes H, Sporn N, Gaeta JM. COVID-19 outbreak at a large homeless shelter in Boston: implications for universal testing. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.12.20059618] [DOI] [Google Scholar]
Bai 2020 {published data only}
- Bai SL, Wang JY, Zhou YQ, Yu DS, Gao XM, Li LL, et al. Analysis of the first cluster of cases in a family of novel coronavirus pneumonia in Gansu Province. Zhonghua Yu Fang Yi Xue Za Zhi 2020;54(0):E005. [DOI: 10.3760/cma.j.issn.0253-9624.2020.0005 10.3760/cma.j.issn.0253-9624.2020.0005.] [DOI] [PubMed] [Google Scholar]
Bajema 2020 {published data only}
- Bajema KL, Oster AM, McGovern OL, Lindstrom S, Stenger MR, Anderson TC, et al. Persons evaluated for 2019 novel coronavirus - United States, January 2020. MMWR. Morbidity and Mortality Weekly Report 2020;69(6):166-70. [DOI: 10.15585/mmwr.mm6906e1 10.15585/mmwr.mm6906e1] [DOI] [PMC free article] [PubMed] [Google Scholar]
Barra 2020 {published data only}
- Barra GB, Santa Rita TH, Mesquita PG, Jacomo RH, Nery LF. Analytical sensibility and specificity of two RT-qPCR protocols for SARS-CoV-2 detection performed in an automated workflow. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.07.20032326] [DOI] [PMC free article] [PubMed] [Google Scholar]
Batista 2020 {published data only}
- Batista, AF, Miraglia JL, Donato TH, Chiavegatto Filho AD. COVID-19 diagnosis prediction in emergency care patients: a machine learning approach. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.04.20052092] [DOI] [Google Scholar]
Beltran Corbellini 2020 {published data only}
- Beltran-Corbellini NA, Chico-Garcia JL, Martinez-Poles J, Rodriguez-Jorge F, Natera-Villalba E, Gomez-Corral J, et al. Acute-onset smell and taste disorders in the context of COVID-19: a pilot multicenter PCR-based case-control study. European Journal of Neurology 2020 Apr 22 [Epub ahead of print]. [DOI: 10.1111/ene.14273] [DOI] [PMC free article] [PubMed] [Google Scholar]
Beltran Pavez 2020 {published data only}
- Beltran-Pavez C, Marquez CL, Munoz G, Valiente-Echeverria F, Gaggero A, Soto-Rifo R, et al. SARS-CoV-2 detection from nasopharyngeal swab samples without RNA extraction. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.28.013508] [DOI] [Google Scholar]
Ben Ami 2020 {published data only}
- Ben-Ami R, Klochendler A, Seidel M, Sido T, Gurel-Gurevich O, Yassour M, et al. Pooled RNA extraction and PCR assay for efficient SARS-CoV-2 detection. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.17.20069062] [DOI] [PMC free article] [PubMed] [Google Scholar]
Bhadra 2020 {published data only}
- Bhadra S, Maranhao AC, Ellington AD. A one-enzyme RT-qPCR assay for SARS-CoV-2, and procedures for reagent production. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.29.013342] [DOI] [Google Scholar]
Bordi 2020 {published data only}
- Bordi L, Nicastri E, Scorzolini L, Di Caro A, Capobianchi MR, Castilletti C, et al, on behalf of Inmi COVID-Study Group Collaborating Centers. Differential diagnosis of illness in patients under investigation for the novel coronavirus (SARS-CoV-2), Italy, February 2020. Eurosurveillance 2020;25(8):pii=2000170. [DOI: 10.2807/1560-7917.Es.2020.25.8.2000170 10.2807/1560-7917.ES.2020.25.8.2000170] [DOI] [PMC free article] [PubMed] [Google Scholar]
Burhan 2020 {published data only}
- Burhan E, Prasenohadi P, Rogayah R, Isbaniyah F, Reisa T, Dharmawan I. Clinical progression of COVID-19 patient with extended incubation period, delayed RT-PCR time-to-positivity, and potential role of chest CT-scan. Acta Medica Indonesiana 2020;52(1):80-3. [PubMed] [Google Scholar]
Cai 2020 {published data only}
- Cai J, Xu J, Lin D, Yang Z, Xu L, Qu Z, et al. A case series of children with 2019 novel coronavirus infection: clinical and epidemiological features. Clinical Infectious Diseases 2020. [DOI: 10.1093/cid/ciaa198 10.1093/cid/ciaa198] [DOI] [PMC free article] [PubMed] [Google Scholar]
Callahan 2020 {published data only}
- Callahan CJ, Lee R, Zulauf K, Tamburello L, Smith KP, Previtera J, et al. Rapid open development and clinical validation of multiple new 3D-printed nasopharyngeal swabs in response to the COVID-19 pandemic. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.14.20065094] [DOI] [PMC free article] [PubMed] [Google Scholar]
Chan 2020 {published data only}
- Chan JF, Yuan S, Kok KH, To KK, Chu H, Yang J, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 2020;395(10223):514-23. [DOI: 10.1016/s0140-6736(20)30154-9 10.1016/S0140-6736(20)30154-9. Epub 2020 Jan 24.] [DOI] [PMC free article] [PubMed] [Google Scholar]
Chandler Brown 2020 {published data only}
- Chandler-Brown D, Bueno AM, Atay O, Tsao DS. A highly scalable and rapidly deployable RNA extraction-free COVID-19 assay by quantitative Sanger sequencing. BioRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.07.029199] [DOI] [Google Scholar]
Chen 2020b {published data only}
- Chen X, Ling J, Mo P, Zhang Y, Jiang Q, Ma Z, et al. Restoration of leukomonocyte counts is associated with viral clearance in COVID-19 hospitalized patients. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.03.20030437] [DOI] [Google Scholar]
Chen 2020c {published data only}
- Chen C, Li J, Di L, Jing Q, Du P, Song C, et al. MINERVA: A facile strategy for SARS-CoV-2 whole genome deep sequencing of clinical samples. BioRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.25.060947] [DOI] [PMC free article] [PubMed] [Google Scholar]
Cheng 2020a {published data only}
- Cheng Z, Lu Y, Cao Q, Qin L, Pan Z, Yan F, et al. Clinical features and chest CT manifestations of coronavirus disease 2019 (COVID-19) in a single-center study in Shanghai, China. American Journal of Roentgenology 2020:1-6. [DOI: 10.2214/ajr.20.22959 10.2214/AJR.20.22959] [DOI] [PubMed] [Google Scholar]
Chu 2020 {published data only}
- Chu DK, Pan Y, Cheng SM, Hui KP, Krishnan P, Liu Y, et al. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clinical Chemistry 2020. [DOI: 10.1093/clinchem/hvaa029 10.1093/clinchem/hvaa029.] [DOI] [PMC free article] [PubMed] [Google Scholar]
Colson 2020 {published data only}
- Colson P, Lagier JC, Baudoin JP, Bou Khalil J, La Scola B, Raoult D. Ultrarapid diagnosis, microscope imaging, genome sequencing, and culture isolation of SARS-CoV-2. European Journal of Clinical Microbiology & Infectious Diseases 2020. [DOI: 10.1007/s10096-020-03869-w] [DOI] [PMC free article] [PubMed] [Google Scholar]
Comar 2020 {published data only}
- Comar M, Brumat M, Concas MP, Argentini G, Bianco A, Bicego L, et al. COVID-19 experience: first Italian survey on healthcare staff members from a mother-child research hospital using combined molecular and rapid immunoassays test. MedRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.19.20071563] [DOI] [Google Scholar]
Corman 2020 {published data only}
- Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020;25(3). [DOI: 10.2807/1560-7917.Es.2020.25.3.2000045 10.2807/1560-7917.ES.2020.25.3.2000045.] [DOI] [PMC free article] [PubMed] [Google Scholar]
Cui 2020 {published data only}
- Cui P, Chen Z, Wang T, Dai J, Zhang J, Ding T, et al. Clinical features and sexual transmission potential of SARS-CoV-2 infected female patients: a descriptive study in Wuhan, China. medRxiv [preprint] 2020. [DOI: 10.1101/2020.02.26.20028225] [DOI] [Google Scholar]
Curti 2020 {published data only}
- Curti L, Pereyra-Bonnet F, Gimenez C. An ultrasensitive, rapid, and portable coronavirus SARS-CoV-2 sequence detection method based on CRISPR-Cas12. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.02.29.971127] [DOI] [Google Scholar]
Dahlke 2020 {published data only}
- Dahlke C, Heidepriem J, Kobbe R, Santer R, Koch T, Fathi A. Distinct early IgA profile may determine severity of COVID-19 symptoms: an immunological case series. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.14.20059733] [DOI] [Google Scholar]
Ding 2020 {published data only}
- Ding X, Yin K, Li Z, Liu C. All-in- one dual CRISPR-Cas12a (AIOD-CRISPR) assay: a case for rapid, ultrasensitive and visual detection of novel coronavirus SARS-CoV-2 and HIV virus. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.19.998724] [DOI] [PMC free article] [PubMed] [Google Scholar]
Fang 2020z {published data only}
- Fang D, Ma J, Guan J, Wang M, Song Y, Tian D, et al. Manifestations of digestive system in hospitalized patients with novel coronavirus pneumonia in Wuhan, China: a single-center, descriptive study. Chinese Journal of Digestion 2020. [DOI: 10.3760/cma.j.issn. 0254-1432.2020.03.000] [Google Scholar]
Farfan 2020 {published data only}
- Farfan MJ, Torres JP, Oryan M, Olivares M, Gallardo P, Salas C. Optimizing RT-PCR detection of SARS-CoV-2 for developing countries using pool testing. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.15.20067199] [DOI] [PubMed] [Google Scholar]
Feng 2020 {published data only}
- Feng K, Yun YX, Wang XF, Yang GD, Zheng YJ, Lin CM, et al. Analysis of CT features of 15 children with 2019 novel coronavirus infection. Zhonghua Er Ke Za Zhi 2020;58(0):E007. [DOI: 10.3760/cma.j.issn.0578-1310.2020.0007 10.3760/cma.j.issn.0578-1310.2020.0007.] [DOI] [PubMed] [Google Scholar]
Fontanet 2020 {published data only}
- Fontanet A, Tondeur L, Madec Y, Grant R, Besombes C, Jolly N, et al. Cluster of COVID-19 in northern France: a retrospective closed cohort study. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.18.20071134] [DOI] [Google Scholar]
Fu 2020a {published data only}
- Fu H, Xu H, Zhang N, Xu H, Li Z, Chen H, et al. Association between clinical, laboratory and CT characteristics and RT-PCR results in the follow-up of COVID-19 patients. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.19.20038315] [DOI] [Google Scholar]
Fu 2020b {published data only}
- Fu YQ, Sun YL, Lu SW, Yang Y, Wang Y, Xu F. Impact of blood analysis and immune function on the prognosis of patients with COVID-19. medRxiv [preprint] 2020. [DOI: 10.1101/2020.04.16.20067587] [DOI] [PMC free article] [PubMed] [Google Scholar]
Fumeaux 2020 {published data only}
- Fumeaux T. COVID-19: Que sait-on des patients admis en soins intensifs? Revue Medicale Suisse 2020;16(690):756. [PubMed] [Google Scholar]
Gao 2020 {published data only}
- Gao Y, Li T, Han M, Li X, Wu D, Xu Y, et al. Diagnostic utility of clinical laboratory data determinations for patients with the severe COVID-19. Journal of Medical Virology 2020. [DOI: 10.1002/jmv.25770 10.1002/jmv.25770] [DOI] [PMC free article] [PubMed] [Google Scholar]
Giamarellos Bourboulis 2020 {published data only}
- Giamarellos-Bourboulis EJ, Netea MG, Rovina N, Akinosoglou K, Antoniadou A, Antonakos N, et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host & Microbe 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Gietema 2020 {published data only}
- Gietema HA, Zelis N, Nobel JM, Lambriks LJ, Van Alphen LB, Oude Lashof AM, et al. CT in relation to RT-PCR in diagnosing COVID-19 in the Netherlands: a prospective study. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.22.20070441] [DOI] [PMC free article] [PubMed] [Google Scholar]
Gonzalez Gonzalez 2020a {published data only}
- Gonzalez-Gonzalez E, Lara-Mayorga IM, Garcia-Rubio A, Garciamendez-Mijares CE, Guerra-Alvarez GE, Garcia-Martinez G, et al. Scaling diagnostics in times of COVID-19: rapid prototyping of 3D-printed water circulators for loop-mediated isothermal amplification (LAMP) and detection of SARS-CoV-2 virus. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.09.20058651] [DOI] [Google Scholar]
Gonzalez Gonzalez 2020b {published data only}
- Gonzalez-Gonzalez E, Trujillo-de Santiago G, Lara-Mayorga IM, Martinez-Chapa SO, Alvarez MM. Portable and accurate diagnostics for COVID-19: combined use of the miniPCR thermocycler and a well-plate reader for SARS-Co2 virus detection. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.03.20052860] [DOI] [PMC free article] [PubMed] [Google Scholar]
Guan 2020 {published data only}
- Guan WD, Chen LP, Ye F, Ye D, Wu SG, Zhou HX, et al. High-throughput sequencing for confirmation of suspected 2019-nCoV infection identified by fluorescence quantitative polymerase chain reaction. Chinese Medical Journal 2020. [DOI: 10.1097/cm9.0000000000000792 10.1097/CM9.0000000000000792.] [DOI] [PMC free article] [PubMed] [Google Scholar]
Guo 2020b {published data only}
- Guo L, Sun X, Wang X, Liang C, Jiang H, Gao Q, et al. SARS-CoV-2 detection with CRISPR diagnostics. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.10.023358] [DOI] [PMC free article] [PubMed] [Google Scholar]
Guo 2020c {published data only}
- Guo X, Guo Z, Duan C, Chen Z, Wang G, Lu Y, et al. Long- term persistence of IgG antibodies in SARS-CoV infected healthcare workers. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.02.12.20021386] [DOI] [Google Scholar]
Han 2020 {published data only}
- Han R, Huang L, Jiang H, Dong J, Peng H, Zhang D. Early clinical and CT manifestations of coronavirus disease 2019 (COVID-19) pneumonia. American Journal of Roentgenology 2020:1-6. [DOI: 10.2214/ajr.20.22961 10.2214/AJR.20.22961.] [DOI] [PubMed] [Google Scholar]
Hao 2020 {published data only}
- Hao W, Li M. Clinical diagnostic value of CT imaging in COVID-19 with multiple negative RT-PCR testing. Travel Medicine and Infectious Disease 2020:101627. [DOI: 10.1016/j.tmaid.2020.101627] [DOI] [PMC free article] [PubMed] [Google Scholar]
Hass 2020 {published data only}
- Hass K, Bao M, He Q, Park M, Qin P, Du K. Integrated micropillar polydimethylsiloxane accurate CRISPR detection (IMPACT) system for rapid viral DNA sensing. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.17.994137] [DOI] [PMC free article] [PubMed] [Google Scholar]
Hirotsu 2020 {published data only}
- Hirotsu Y, Mochizuki H, Omata M. Double- quencher probes improved the detection sensitivity of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by One-Step RT-PCR. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.17.20037903] [DOI] [PMC free article] [PubMed] [Google Scholar]
Hogan 2020 {published data only}
- Hogan CA, Sahoo MK, Pinsky BA. Sample pooling as a strategy to detect community transmission of SARS-CoV-2. JAMA 2020. [DOI: 10.1001/jama.2020.5445] [DOI] [PMC free article] [PubMed] [Google Scholar]
Holland 2020 {published data only}
- Holland M, Negron D, Mitchell S, Dellinger N, Ivancich M, Barrus T, et al. BioLaboro: a bioinformatics system for detecting molecular assay signature erosion and designing new assays in response to emerging and reemerging pathogens. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.08.031963] [DOI] [Google Scholar]
Hu 2020b {published data only}
- Hu F, Chen F, Wang Y, Xu T, Tang X, Li F. Failed detection of the full-length genome of SARS-CoV-2 by ultra-deep sequencing from the recovered and discharged patients retested viral PCR positive. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.27.20043299] [DOI] [Google Scholar]
Hu 2020c {published data only}
- Hu, X, An, T, Situ, B, Hu, Y, Ou, Z, Li, Q, et al. Heat inactivation of serum interferes with the immunoanalysis of antibodies to SARS-CoV-2. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.12.20034231] [DOI] [PMC free article] [PubMed] [Google Scholar]
Hu 2020d {published data only}
- Hu Z, Song C, Xu C, Jin G, Chen Y, Xu X, et al. Clinical characteristics of 24 asymptomatic infections with COVID-19 screened among close contacts in Nanjing, China. Science China. Life Sciences 2020 Mar 4 [Epub ahead of print]. [DOI: 10.1007/s11427-020-1661-4] [DOI] [PMC free article] [PubMed] [Google Scholar]
Huang 2020a {published data only}
- Huang WH, Teng LC, Yeh TK, Chen YJ, Lo WJ, Wu MJ, et al. 2019 novel coronavirus disease (COVID-19) in Taiwan: reports of two cases from Wuhan, China. Journal of Microbiology, Immunology and Infection 2020;53(3):481-4. [DOI: 10.1016/j.jmii.2020.02.009 10.1016/j.jmii.2020.02.009.] [DOI] [PMC free article] [PubMed] [Google Scholar]
Jenkins 2020 {published data only}
- Jenkins C, Orsburn B. In silico approach to accelerate the development of mass spectrometry-based proteomics methods for detection of viral proteins: application to COVID-19. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.08.980383] [DOI] [Google Scholar]
Jiang 2020a {published data only}
- Jiang DM. Recurrent PCR positivity after hospital discharge of people with coronavirus disease 2019 (COVID-19). Journal of Infection 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Jiang 2020b {published data only}
- Jiang HW, Li Y, Zhang HN, Wang W, Men D, Yang X, et al. Global profiling of SARS-CoV-2 specific IgG/ IgM responses of convalescents using a proteome microarray. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.20.20039495] [DOI] [PMC free article] [PubMed] [Google Scholar]
Jung 2020 {published data only}
- Jung YJ, Park GS, Moon JH, Ku K, Beak SH, Kim S, et al. Comparative analysis of primer-probe sets for the laboratory confirmation of SARS-CoV-2. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.02.25.964775] [DOI] [Google Scholar]
Khan 2020 {published data only}
- Khan S, Nakajima R, Jain A, Assis RR, Jasinskas A, Obiero JM, et al. Analysis of serologic cross-reactivity between common human coronaviruses and SARS-CoV-2 using coronavirus antigen microarray. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.24.006544] [DOI] [Google Scholar]
Kim 2019 {published data only}
- Kim JH, Kang M, Park E, Chung DR, Kim J, Hwang ES. A simple and multiplex loop-mediated isothermal amplification (LAMP) assay for rapid detection of SARS-CoV. BioChip Journal 2019;13(4):341-51. [DOI: 10.1007/s13206-019-3404-3] [DOI] [PMC free article] [PubMed] [Google Scholar]
Kong 2020 {published data only}
- Kong WH, Li Y, Peng MW, Kong DG, Yang XB, Wang L, Liu MQ. SARS-CoV-2 detection in patients with influenza-like illness. Nature Microbiology 2020;5:675-8. [DOI: 10.1038/s41564-020-0713-1] [DOI] [PubMed] [Google Scholar]
Konrad 2020 {published data only}
- Konrad R, Eberle U, Dangel A, Treis B, Berger A, Bengs K, et al. Rapid establishment of laboratory diagnostics for the novel coronavirus SARS-CoV-2 in Bavaria, Germany, February 2020. Eurosurveillance 2020;25(9). [DOI: 10.2807/1560-7917.Es.2020.25.9.2000173 10.2807/1560-7917.ES.2020.25.9.2000173.] [DOI] [PMC free article] [PubMed] [Google Scholar]
Kurstjens 2020 {published data only}
- Kurstjens S, Van der Horst A, Herpers R, Geerits MW, Kluiters-de Hingh YC, Göttgens EL, et al. Rapid identification of SARS-CoV-2-infected patients at the emergency department using routine testing. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.20.20067512] [DOI] [PubMed] [Google Scholar]
Lamb 2020 {published data only}
- Lamb LE, Bartolone SN, Ward E, Chancellor MB. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.02.19.20025155] [DOI] [Google Scholar]
Lan 2020 {published data only}
- Lan L, Xu D, Ye G, Xia C, Wang S, Li Y, et al. Positive RT-PCR test results in patients recovered from COVID-19. JAMA 2020. [DOI: 10.1001/jama.2020.2783 10.1001/jama.2020.2783.] [DOI] [PMC free article] [PubMed] [Google Scholar]
Lechien 2020 {published data only}
- Lechien J, Cabaraux P, Chiesa-Estomba C, Khalife M, Plzak J, Hans S, et al. Objective olfactory testing in patients presenting with sudden onset olfactory dysfunction as the first manifestation of confirmed COVID-19 infection. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.15.20066472] [DOI] [Google Scholar]
Lei 2020 {published data only}
- Lei Z, Cao H, Jie Y, Huang Z, Guo X, Chen J, et al. A cross-sectional comparison of epidemiological and clinical features of patients with coronavirus disease (COVID-19) in Wuhan and outside Wuhan, China. Travel Medicine and Infectious Disease 2020:101664. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Li 2020c {published data only}
- Li Y, Hu Y, Zhang X, Yu Y, Li B, Wu J, et al. Follow-up testing of viral nucleic acid in discharged patients with moderate type of 2019 coronavirus disease (COVID-19). Zhejiang Da Xue Xue Bao Yi Xue Ban 2020;49(1):0. [DOI: 10.3785/j.issn.1008-9292.2020.03.11] [DOI] [PMC free article] [PubMed] [Google Scholar]
Li 2020d {published data only}
- Li C, Debruyne D, Spencer J, Kapoor V, Liu LY, Zhang B, et al. High sensitivity detection of SARS-CoV-2 using multiplex PCR and a multiplex-PCR-based metagenomic method. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.12.988246] [DOI] [Google Scholar]
Li 2020e {published data only}
- Li Y, Xia L. Coronavirus disease 2019 (COVID-19): role of chest CT in diagnosis and management. American Journal of Roentgenology 2020:1-7. [DOI: 10.2214/ajr.20.22954 10.2214/AJR.20.22954.] [DOI] [PubMed] [Google Scholar]
Li 2020f {published data only}
- Li D, Wang D, Dong J, Wang N, Huang H, Xu H, et al. False- negative results of real-time reverse-transcriptase polymerase chain reaction for severe acute respiratory syndrome coronavirus 2: role of deep-learning-based CT diagnosis and insights from two cases. Korean Journal of Radiology 2020. [DOI: 10.3348/kjr.2020.0146 10.3348/kjr.2020.0146.] [DOI] [PMC free article] [PubMed] [Google Scholar]
Li 2020g {published data only}
- Li YY, Wang WN, Lei Y, Zhang B, Yang J, Hu JW, et al. Comparison of the clinical characteristics between RNA positive and negative patients clinically diagnosed with 2019 novel coronavirus pneumonia. Zhonghua Jie He He Hu Xi Za Zhi 2020;43(0):E023. [DOI: 10.3760/cma.j.cn112147-20200214-00095 10.3760/cma.j.cn112147-20200214-00095.] [DOI] [PubMed] [Google Scholar]
Liang 2020a {published data only}
- Liang Y, Liang J, Zhou Q, Li X, Lin F, Deng Z, et al. Prevalence and clinical features of 2019 novel coronavirus disease (COVID-19) in the fever clinic of a teaching hospital in Beijing: a single-center, retrospective study. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.02.25.20027763] [DOI] [Google Scholar]
Liang 2020b {published data only}
- Liang C, Cao J, Liu Z, Ge F, Cang J, Miao C, et al. Positive RT-PCR test results after consecutively negative results in patients with COVID-19. Infectious Diseases 2020;52(7):517-9. [DOI: 10.1080/23744235.2020.1755447] [DOI] [PubMed] [Google Scholar]
Ling 2020 {published data only}
- Ling Y, Xu SB, Lin YX, Tian D, Zhu ZQ, Dai FH, et al. Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chinese Medical Journal 2020. [DOI: 10.1097/cm9.0000000000000774 10.1097/CM9.0000000000000774.] [DOI] [PMC free article] [PubMed] [Google Scholar]
Liu 2020e {published data only}
- Liu J, Li S, Liu J, Liang B, Wang X, Wang H, et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EbioMedicine 2020;55:102763. [DOI: 10.1016/j.ebiom.2020.102763] [DOI] [PMC free article] [PubMed] [Google Scholar]
Liu 2020f {published data only}
- Liu W, Tao ZW, Lei W, Ming-Li Y, Kui L, Ling Z, et al. Analysis of factors associated with disease outcomes in hospitalized patients with 2019 novel coronavirus disease. Chinese Medical Journal 2020. [DOI: 10.1097/cm9.0000000000000775 10.1097/CM9.0000000000000775.] [DOI] [PMC free article] [PubMed] [Google Scholar]
Liu 2020g {published data only}
- Liu R, Han H, Liu F, Lv Z, Wu K, Liu Y, et al. Positive rate of RT-PCR detection of SARS-CoV-2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020. Clinica Chimica Acta; International Journal of Clinical Chemistry 2020;505:172-5. [DOI: 10.1016/j.cca.2020.03.009 10.1016/j.cca.2020.03.009.] [DOI] [PMC free article] [PubMed] [Google Scholar]
Liu 2020h {published data only}
- Liu J, Xiao Y, Shen Y, Shi C, Chen Y, Shi P, et al. Detection of SARS-CoV-2 by RT-PCR in anal from patients who have recovered from coronavirus disease 2019. Journal of Medical Virology 2020. [DOI: 10.1002/jmv.25875] [DOI] [PMC free article] [PubMed] [Google Scholar]
Lo 2020 {published data only}
- Lo IL, Lio CF, Cheong HH, Lei CI, Cheong TH, Zhong X, et al. Evaluation of SARS-CoV-2 RNA shedding in clinical specimens and clinical characteristics of 10 patients with COVID-19 in Macau. International Journal of Biological Sciences 2020;16(10):1698-707. [DOI: 10.7150/ijbs.45357] [DOI] [PMC free article] [PubMed] [Google Scholar]
Lopez‐Rincon 2020 {published data only}
- Lopez-Rincon A, Tonda A, Mendoza-Maldonado L, Claassen E, Garssen J, Kraneveld AD. Accurate identification of SARS-CoV-2 from viral genome sequences using deep learning. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.13.990242] [DOI] [Google Scholar]
Lu 2020 {published data only}
- Lu R, Wu X, Wan Z, Li Y, Zuo L, Qin J, et al. Development of a novel reverse transcription loop-mediated isothermal amplification method for rapid detection of SARS-CoV-2. Virologica Sinica 2020. [DOI: 10.1007/s12250-020-00218-1] [DOI] [PMC free article] [PubMed] [Google Scholar]
Ma 2020b {published data only}
- Ma SY, Luo YM, Hu TY, You ZC, Sun JG, Yu SY, et al. Clinical application effect of modified nasopharyngeal swab sampling for 2019 novel coronavirus nucleic acid detection. Zhonghua Shao Shang Za Zhi 2020;36(0):E009. [DOI: 10.3760/cma.j.cn501120-20200312-00153] [DOI] [PubMed] [Google Scholar]
Mahari 2020 {published data only}
- Mahari S, Roberts A, Shahdeo D, Gandhi S. eCovSens-ultrasensitive novel in-house built printed circuit board based electrochemical device for rapid detection of nCOVID-19. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.24.059204] [DOI] [Google Scholar]
Mardani 2020 {published data only}
- Mardani R, Ahmadi Vasmehjani A, Zali F, Gholami A, Mousavi Nasab SD, Kaghazian H, et al. Laboratory parameters in detection of COVID-19 patients with positive RT-PCR; a diagnostic accuracy study. Archives of Academic Emergency Medicine 2020;8(1):e43. [PMC free article] [PubMed] [Google Scholar]
Marzinotto 2020 {published data only}
- Marzinotto S, Mio C, Cifu A, Verardo R, Pipan C, Schneider C, et al. A streamlined approach to rapidly detect SARS-CoV-2 infection, avoiding RNA extraction. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.06.20054114] [DOI] [PMC free article] [PubMed] [Google Scholar]
McKay 2020 {published data only}
- McKay PF, Hu K, Blakney AK, Samnuan K, Bouton CR, Rogers P, et al. Self-amplifying RNA SARS-CoV-2 lipid nanoparticle vaccine induces equivalent preclinical antibody titers and viral neutralization to recovered COVID-19 patients. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.22.055608] [DOI] [Google Scholar]
McRae 2020 {published data only}
- McRae MP, Simmons GW, Christodoulides NJ, Lu Z, Kang SK, Fenyo D, et al. Clinical decision support tool and rapid point-of-care platform for determining disease severity in patients with COVID-19. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.16.20068411] [DOI] [PMC free article] [PubMed] [Google Scholar]
Mei 2020 {published data only}
- Mei X, Lee HC, Diao K, Huang M, Lin B, Liu C, et al. Artificial intelligence for rapid identification of the coronavirus disease 2019 (COVID-19). medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.12.20062661] [DOI] [Google Scholar]
Meng 2020 {published data only}
- Meng Z, Wang M, Song H, Guo S, Zhou Y, Li W, et al. Development and utilization of an intelligent application for aiding COVID-19 diagnosis. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.18.20035816] [DOI] [Google Scholar]
Mercurio 2020 {published data only}
- Mercurio I, Tragni V, Busto F, De Grassi A, Pierri CL. Protein structure analysis of the interactions between SARS-CoV-2 spike protein and the human ACE2 receptor: from conformational changes to novel neutralizing antibodies. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.17.046185] [DOI] [PMC free article] [PubMed] [Google Scholar]
Metsky 2020 {published data only}
- Metsky HC, Freije CA, Kosoko-Thoroddsen TS, Sabeti PC, Myhrvold C. CRISPR-based COVID-19 surveillance using a genomically-comprehensive machine learning approach. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.02.26.967026] [DOI] [Google Scholar]
Nelson 2020 {published data only}
- Nelson AC, Auch B, Schomaker M, Gohl DM, Grady P, Johnson D, et al. Analytical validation of a COVID-19 qRT-PCR detection assay using a 384-well format and three extraction methods. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.02.022186] [DOI] [Google Scholar]
Nemati 2020 {published data only}
- Nemati S, Najari HR, Eftekharzadeh A, Kazemifar AM, Qandian A, Fattahi P, et al. Association between rRT-PCR test results upon admission and outcome in hospitalized chest CT-Positive COVID-19 patients; a provincial retrospective cohort with active follow-up. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.21.20074641] [DOI] [Google Scholar]
Nie 2020 {published data only}
- Nie J, Li Q, Wu J, Zhao C, Hao H, Liu H, et al. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerging Microbes and Infections 2020;9(1):680-6. [DOI: 10.1080/22221751.2020.1743767 10.1080/22221751.2020.1743767.] [DOI] [PMC free article] [PubMed] [Google Scholar]
Nunez Bajo 2020 {published data only}
- Nunez-Bajo E, Kasimatis M, Cotur Y, Asfour T, Collins A, Tanriverdi U, et al. Ultra-low-cost integrated silicon-based transducer for on-site, genetic detection of pathogens. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.23.002931] [DOI] [Google Scholar]
Okba 2020b {published data only}
- Okba N, Muller MA, Li W, Wang C, GeurtsvanKessel CH, Corman VM, et al. SARS-CoV-2 specific antibody responses in COVID-19 patients. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.18.20038059] [DOI] [Google Scholar]
Paden 2020 {published data only}
- Paden CR, Tao Y, Queen K, Zhang J, Li Y, Uehara A, et al. Rapid, sensitive, full genome sequencing of severe acute respiratory syndrome virus coronavirus 2 (SARS-CoV-2). bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.22.055897] [DOI] [PMC free article] [PubMed] [Google Scholar]
Pan 2020b {published data only}
- Pan Y, Yu X, Du X, Li Q, Li X, Qin T, et al. Epidemiological and clinical characteristics of 26 asymptomatic SARS-CoV-2 carriers. Journal of Infectious Diseases 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Pan 2020c {published data only}
- Pan Y, Zhang D, Yang P, Poon LL, Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infectious Diseases 2020. [DOI: 10.1016/S1473-3099(20)30113-4] [DOI] [PMC free article] [PubMed] [Google Scholar]
Pan 2020d {published data only}
- Pan F, Ye T, Sun P, Gui S, Liang B, Li L, et al. Time course of lung changes on chest CT during recovery from 2019 novel coronavirus (COVID-19) pneumonia. Radiology 2020:200370. [DOI: 10.1148/radiol.2020200370 10.1148/radiol.2020200370.] [DOI] [PMC free article] [PubMed] [Google Scholar]
Pan 2020e {published data only}
- Pan Y, Long L, Zhang D, Yan T, Cui S, Yang P, et al. Potential false-negative nucleic acid testing results for severe acute respiratory syndrome coronavirus 2 from thermal inactivation of samples with low viral loads. Clinical Chemistry 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Paradiso 2020b {published data only}
- Paradiso AV, De Summa S, Silvestris N, Tommasi S, Tufaro A, De Palma G, et al. Rapid serological tests have a role in asymptomatic health workers COVID-19 screening. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.15.20057786] [DOI] [Google Scholar]
Park 2020 {published data only}
- Park GS, Ku K, Baek SH, Kim SJ, Kim SI, Kim BT, et al. Development of reverse transcription loop-mediated isothermal amplification (RT-LAMP) assays targeting SARS-CoV-2. Journal of Molecular Diagnostics 2020;22(6):729-35. [DOI: 10.1016/j.jmoldx.2020.03.006] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park GS, Ku K, Beak SH, Kim SJ, Kim SI, Kim BT, et al. Development of reverse transcription loop-mediated isothermal amplification (RT-LAMP) assays targeting SARS-CoV-2. bioRxiv [Preprint] 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Peng 2020 {published data only}
- Peng L, Liu J, Xu W, Luo Q, Deng K, Lin B, et al. 2019 novel coronavirus can be detected in urine, blood, anal swabs and oropharyngeal swabs samples. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.02.21.20026179] [DOI] [PMC free article] [PubMed] [Google Scholar]
Pfefferle 2020 {published data only}
- Pfefferle S, Reucher S, Nörz D, Lütgehetmann M. Evaluation of a quantitative RT-PCR assay for the detection of the emerging coronavirus SARS-CoV-2 using a high throughput system. Eurosurveillance 2020;25(9). [DOI: 10.2807/1560-7917.ES.2020.25.9.2000152] [DOI] [PMC free article] [PubMed] [Google Scholar]
Rauch 2020 {published data only}
- Rauch JN, Valois E, Solley SC, Braig F, Lach RS, Baxter NJ, et al. A scalable, easy-to-deploy, protocol for Cas13-based detection of SARS-CoV-2 genetic material. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.20.052159] [DOI] [PMC free article] [PubMed] [Google Scholar]
Scallan 2020 {published data only}
- Scallan MF, Dempsey C, MacSharry J, O'Callaghan I, O'Connor PM, Hogan CP, et al. Validation of a lysis buffer containing 4 M guanidinium thiocyanate (GITC)/ Triton X-100 for extraction of SARS-CoV-2 RNA for COVID-19 testing: comparison of formulated lysis buffers containing 4 to 6 M GITC, Roche external lysis buffer and Qiagen RTL lysis buffer. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.05.026435] [DOI] [Google Scholar]
Seo 2020 {published data only}
- Seo G, Lee G, Kim MJ, Baek SH, Choi M, Ku KB, et al. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano 2020. [DOI: 10.1021/acsnano.0c02823] [DOI] [PubMed] [Google Scholar]
Shental 2020 {published data only}
- Shental N, Levy S, Skorniakov S, Wuvshet V, Shemer-Avni Y, Porgador A, et al. Efficient high throughput SARS-CoV-2 testing to detect asymptomatic carriers. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.14.20064618] [DOI] [PMC free article] [PubMed] [Google Scholar]
Shi 2020 {published data only}
- Shi Y, Wang X, Liu G, Zhu Q, Wang J, Yu H, et al. A quickly, effectively screening process of novel corona virus disease 2019 (COVID-19) in children in Shanghai, China. Annals of Translational Medicine 2020;8(5). [DOI: 10.21037/atm.2020.03.22] [DOI] [PMC free article] [PubMed] [Google Scholar]
Shirato 2020 {published data only}
- Shirato K, Nao N, Katano H, Takayama I, Saito S, Kato F, et al. Development of genetic diagnostic methods for novel coronavirus 2019 (nCoV-2019) in Japan. Japanese Journal of Infectious Diseases 2020. [DOI: 10.7883/yoken.JJID.2020.061 10.7883/yoken.JJID.2020.061.] [DOI] [PubMed] [Google Scholar]
Song 2020 {published data only}
- Song C, Wang Y, Li W, Hu B, Chen G, Xia P, et al. Detection of 2019 novel coronavirus in semen and testicular biopsy specimen of COVID-19 patients. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.31.20042333] [DOI] [Google Scholar]
Su 2020 {published data only}
- Su L, Ma X, Yu H, Zhang Z, Bian P, Han Y, et al. The different clinical characteristics of corona virus disease cases between children and their families in China - the character of children with COVID-19. Emerging Microbes and Infections 2020;9(1):707-13. [DOI: 10.1080/22221751.2020.1744483 10.1080/22221751.2020.1744483.] [DOI] [PMC free article] [PubMed] [Google Scholar]
Sun 2020a {published data only}
- Sun X, Zhang X, Chen X, Chen L, Deng C, Zou X, et al. The infection evidence of SARS-COV-2 in ocular surface: a single-center cross-sectional study. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.02.26.20027938] [DOI] [PMC free article] [PubMed] [Google Scholar]
Sun 2020b {published data only}
- Sun Y, Koh V, Marimuthu K, Ng OT, Young B, Vasoo S, et al. Epidemiological and clinical predictors of COVID-19. Clinical Infectious Diseases 2020. [DOI: 10.1093/cid/ciaa322 10.1093/cid/ciaa322.] [DOI] [PMC free article] [PubMed] [Google Scholar]
Sun 2020c {published data only}
- Sun F, Ganguli A, Nguyen J, Brisbin R, Shanmugam K, Hirschberg DL, et al. Smartphone-based multiplex 30-minute nucleic acid test of live virus from nasal swab extract. Lab on a Chip 2020. [DOI: 10.1039/d0lc00304b] [DOI] [PubMed] [Google Scholar]
Tagarro 2020 {published data only}
- Tagarro A, Epalza C, Santos M, Sanz-Santaeufemia FJ, Otheo E, Moraleda C, et al. Screening and severity of coronavirus disease 2019 (COVID-19) in children in Madrid, Spain. JAMA Pediatrics 2020. [DOI: 10.1001/jamapediatrics.2020.1346] [DOI] [PMC free article] [PubMed] [Google Scholar]
Tan 2020a {published data only}
- Tan W, Lu Y, Zhang J, Wang J, Dan Y, Tan Z, et al. Viral kinetics and antibody responses in patients with COVID-19. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.24.20042382] [DOI] [Google Scholar]
Tan 2020b {published data only}
- Tan X, Huang J, Zhao F, Zhou Y, Li JQ, Wang XY. Clinical features of children with SARS-CoV-2 infection: an analysis of 13 cases from Changsha, China. Zhongguo Dang Dai Er Ke Za Zhi 2020;22(4):294-8. [DOI: 10.7499/j.issn.1008-8830.2003199] [DOI] [PMC free article] [PubMed] [Google Scholar]
Tan 2020c {published data only}
- Tan X, Lin C, Zhang J, Khaing Oo MK, Fan X. Rapid and quantitative detection of COVID-19 markers in micro-liter sized samples. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.20.052233] [DOI] [Google Scholar]
Toptan 2020 {published data only}
- Toptan T, Hoehl S, Westhaus S, Bojkova D, Berger A, Rotter B, et al. Optimized qRT-PCR approach for the detection of intra- and extracellular SARS-CoV-2 RNAs. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.20.052258] [DOI] [PMC free article] [PubMed] [Google Scholar]
Tsang 2003 {published data only}
- Tsang OT, Chau TN, Choi KW, Tso EY, Lim W, Chiu MC, et al. Coronavirus-positive nasopharyngeal aspirate as predictor for severe acute respiratory syndrome mortality. Emerging Infectious Diseases 2003;9(11):1381-7. [DOI: 10.3201/eid0911.030400 10.3201/eid0911.030400.] [DOI] [PMC free article] [PubMed] [Google Scholar]
Vermeiren 2020 {published data only}
- Vermeiren C, Marchand-Senécal X, Sheldrake E, Bulir D, Smieja M, Chong S, et al. Comparison of Copan Eswab and FLOQswab for COVID-19 PCR diagnosis: working around a supply shortage. Journal of Clinical Microbiology 2020. [DOI: 10.1128/JCM.00669-20] [DOI] [PMC free article] [PubMed] [Google Scholar]
Viehweger 2020 {published data only}
- Viehweger A, Kühnl F, Brandt C, König B. Increased PCR screening capacity using a multi-replicate pooling scheme. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.16.20067603] [DOI] [Google Scholar]
Vogels 2020 {published data only}
- Vogels CB, Brito AF, Wyllie AL, Fauver JR, Ott IM, Kalinich CC, et al. Analytical sensitivity and efficiency comparisons of SARS-COV-2 qRT-PCR assays. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.30.20048108] [DOI] [PMC free article] [PubMed] [Google Scholar]
Waghmare 2020 {published data only}
- Waghmare A, Krantz EM, Baral S, Vasquez E, Loeffelholz T, Chung EL, et al. Reliability of self-sampling for accurate assessment of respiratory virus viral and immunologic kinetics. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.03.20051706] [DOI] [PMC free article] [PubMed] [Google Scholar]
Wang 2020b {published data only}
- Wang F, Hou H, Luo Y, Tang G, Wu S, Huang M, et al. The laboratory tests and host immunity of COVID-19 patients with different severity of illness. JCI Insight 2020. [DOI: 10.1172/jci.insight.137799] [DOI] [PMC free article] [PubMed] [Google Scholar]
Wang 2020c {published data only}
- Wang Z, Li H, Li J, Yang C, Guo X, Hu Z, et al. Elevated serum IgM levels indicate poor outcome in patients with coronavirus disease 2019 pneumonia: a retrospective case-control study. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.22.20041285] [DOI] [Google Scholar]
Wang 2020d {published data only}
- Wang X, Guo X, Xin Q, Chu Y, Li J, Pan Y, et al. Neutralizing antibodies responses to SARS-CoV-2 in COVID-19 inpatients and convalescent patients. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.15.20065623] [DOI] [Google Scholar]
Wang 2020e {published data only}
- Wang B, Wang L, Kong X, Geng J, Xiao D, Ma C. Long-term co-existence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) with antibody response in non-severe coronavirus disease 2019 (COVID-19) patients. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.13.20040980] [DOI] [Google Scholar]
Wang 2020f {published data only}
- Wang X, Yao H, Xu X, Zhang P, Zhang M, Shao J, et al. Limits of detection of six approved RT-PCR kits for the novel SARS-coronavirus-2 (SARS-CoV-2). Clinical Chemistry 2020. [DOI: 10.1093/clinchem/hvaa099] [DOI] [PMC free article] [PubMed] [Google Scholar]
Wang 2020g {published data only}
- Wang XF, Yuan J, Zheng YJ, Chen J, Bao YM, Wang YR, et al. Retracted: Clinical and epidemiological characteristics of 34 children with 2019 novel coronavirus infection in Shenzhen. Zhonghua Er Ke Za Zhi 2020;58(0):E008. [DOI: 10.3760/cma.j.issn.0578-1310.2020.0008 10.3760/cma.j.issn.0578-1310.2020.0008.] [DOI] [PubMed] [Google Scholar]
Wang 2020h {published data only}
- Wang QX, Zeng XH, Zheng SL. The nucleic acid test of induced sputum should be used for estimation of patients cure with 2019-nCov. European Review for Medical and Pharmacological Sciences 2020;24(7):3437. [DOI: ] [DOI] [PubMed] [Google Scholar]
Wang 2020i {published data only}
- Wang Z, Weng J, Li Z, Hou R, Zhou L, Ye H, et al. Development and validation of a diagnostic nomogram to predict COVID-19 pneumonia. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.03.20052068] [DOI] [Google Scholar]
Wee 2020 {published data only}
- Wee SK, Sivalingam SP, Yap EP. Rapid direct nucleic acid amplification test without RNA extraction for SARS-CoV-2 using a portable PCR thermocycler. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.17.042366] [DOI] [PMC free article] [PubMed] [Google Scholar]
Weiss 2020 {published data only}
- Weiss S, Klingler J, Hioe C, Amanat F, Baine I, Kojic EM, et al. A high through-put assay for circulating antibodies directed against the S protein of severe acute respiratory syndrome corona virus 2. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.14.20059501] [DOI] [PMC free article] [PubMed] [Google Scholar]
Woelfel 2020 {published data only}
- Woelfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Mueller MA, et al. Clinical presentation and virological assessment of hospitalized cases of coronavirus disease 2019 in a travel-associated transmission cluster. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.05.20030502] [DOI] [Google Scholar]
Won 2020 {published data only}
- Won J, Lee S, Park M, Kim TY, Park MG, Choi BY, et al. Development of a laboratory-safe and low-cost detection protocol for SARS-CoV-2 of the coronavirus disease 2019 (COVID-19). Experimental Neurobiology 2020. [DOI: 10.5607/en20009 10.5607/en20009.] [DOI] [PMC free article] [PubMed] [Google Scholar]
Woo 2020 {published data only}
- Woo CH, Jang S, Shin G, Jung GY, Lee JW. Sensitive one-step isothermal detection of pathogen-derived RNAs. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.05.20031971] [DOI] [Google Scholar]
Wu 2020a {published data only}
- Wu F, Wang A, Liu M, Wang Q, Chen J, Xia S, et al. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.30.20047365] [DOI] [Google Scholar]
Wu 2020b {published data only}
- Wu Q, Xing Y, Shi L, Li W, Gao Y, Pan S, et al. Epidemiological and clinical characteristics of children with coronavirus disease 2019. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.19.20027078] [DOI] [Google Scholar]
Wu 2020c {published data only}
- Wu X, Fu B, Chen L, Feng Y. Serological tests facilitate identification of asymptomatic SARS-CoV-2 infection in Wuhan, China. Journal of Medical Virology 2020. [DOI: 10.1002/jmv.25904] [DOI] [PMC free article] [PubMed] [Google Scholar]
Xia 2020a {published data only}
- Xia J, Tong J, Liu M, Shen Y, Guo D. Evaluation of coronavirus in tears and conjunctival secretions of patients with SARS-CoV-2 infection. Journal of Medical Virology 2020. [DOI: 10.1002/jmv.25725 10.1002/jmv.25725.] [DOI] [PMC free article] [PubMed] [Google Scholar]
Xia 2020b {published data only}
- Xia XY, Wu J, Liu HL, Xia H, Jia B, Huang WX. Epidemiological and initial clinical characteristics of patients with family aggregation of COVID-19. Journal of Clinical Virology 2020;127:104360. [DOI: 10.1016/j.jcv.2020.104360] [DOI] [PMC free article] [PubMed] [Google Scholar]
Xie 2020b {published data only}
- Xie X, Zhong Z, Zhao W, Zheng C, Wang F, Liu J. Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology 2020:200343. [DOI: 10.1148/radiol.2020200343 10.1148/radiol.2020200343] [DOI] [PMC free article] [PubMed] [Google Scholar]
Xie 2020c {published data only}
- Xie C, Lu J, Wu D, Zhang L, Zhao H, Rao B, et al. False negative rate of COVID-19 is eliminated by using nasal swab test. Travel Medicine and Infectious Disease 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Xing 2020a {published data only}
- Xing Y, Ni W, Wu Q, Li W, Li G, Wang W, et al. Dynamics of faecal SARS-CoV-2 in infected children during the convalescent phase. Journal of Infection 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Xing 2020b {published data only}
- Xing Y, Ni W, Wu Q, Li W, Li G, Tong J, et al. Prolonged presence of SARS-CoV-2 in feces of pediatric patients during the convalescent phase. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.11.20033159] [DOI] [Google Scholar]
Xu 2020b {published data only}
- Xu SP, Kuang D, Hu Y, Liu C, Duan YQ, Wang GP. Detection of 2019-nCoV in the pathological paraffin embedded tissue. Zhonghua Bing Li Xue Za Zhi 2020;49(0):E004. [DOI: 10.3760/cma.j.cn112151.20200219.00001 10.3760/cma.j.cn112151.20200219.00001.] [DOI] [PubMed] [Google Scholar]
Xu 2020c {published data only}
- Xu Y, Xiao M, Liu X, Xu S, Du T, Xu J, et al. Significance of serology testing to assist timely diagnosis of SARS-CoV-2 infections: implication from a family cluster. Emerging Microbes and Infection 2020:01/Dec. [DOI: 10.1080/22221751.2020.1752610] [DOI] [PMC free article] [PubMed] [Google Scholar]
Xu 2020d {published data only}
- Xu K, Chen Y, Yuan J, Yi P, Ding C, Wu W, et al. Factors associated with prolonged viral RNA shedding in patients with COVID-19. Clinical Infectious Diseases 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Yan 2020 {published data only}
- Yan S, Song X, Lin F, Zhu H, Wang X, Li M, et al. Clinical characteristics of coronavirus disease 2019 in Hainan, China. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.19.20038539] [DOI] [Google Scholar]
Yang 2020a {published data only}
- Yang Y, Yang M, Shen C, Wang F, Yuan J, Li J, et al. Evaluating the accuracy of different respiratory specimens in the laboratory diagnosis and monitoring the viral shedding of 2019-nCoV infections. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.02.11.20021493] [DOI] [Google Scholar]
Yang 2020b {published data only}
- Yang P, Shao FL, Wang GJ. Clinical observation on increasing the positive rate of novel coronavirus nucleic acid tests by sputum excretion induced by nebulizer therapy. Zhonghua Jie He He Hu Xi Za Zhi 2020;43(4):335-6. [DOI: 10.3760/cma.j.cn112147-20200212-00076] [DOI] [PubMed] [Google Scholar]
Yelin 2020 {published data only}
- Yelin I, Aharony N, Shaer-Tamar E, Argoetti A, Messer E, Berenbaum D, et al. Evaluation of COVID-19 RT-qPCR test in multi-sample pools. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.26.20039438] [DOI] [PMC free article] [PubMed] [Google Scholar]
Yuan 2020 {published data only}
- Yuan J, Zou R, Zeng L, Kou S, Lan J, Li X, et al. The correlation between viral clearance and biochemical outcomes of 94 COVID-19 infected discharged patients. Inflammation Research 2020. [DOI: 10.1007/s00011-020-01342-0] [DOI] [PMC free article] [PubMed] [Google Scholar]
Yun 2020 {published data only}
- Yun H, Sun Z, Wu J, Tang A, Hu M, Xiang Z. Laboratory data analysis of novel coronavirus (COVID-19) screening in 2510 patients. Clinica Chimica Acta 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Zeng 2020b {published data only}
- Zeng F, Dai C, Cai P, Wang J, Xu L, Li J, et al. A comparison study of SARS-CoV-2 IgG antibody between male and female COVID-19 patients: a possible reason underlying different outcome between gender. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.26.20040709] [DOI] [PMC free article] [PubMed] [Google Scholar]
Zhang 2020e {published data only}
- Zhang X, Wu X, Wang D, Lu M, Hou X, Wang H, et al. Proteome-wide analysis of differentially-expressed SARS-CoV-2 antibodies in early COVID-19 infection. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.14.20064535] [DOI] [Google Scholar]
Zhang 2020f {published data only}
- Zhang G, Zhang J, Wang B, Zhu X, Wang Q, Qiu S. Analysis of clinical characteristics and laboratory findings of 95 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a retrospective analysis. Respiratory Research 2020;21(1):74. [DOI: 10.1186/s12931-020-01338-8] [DOI] [PMC free article] [PubMed] [Google Scholar]
Zhang 2020g {published data only}
- Zhang L, Pang R, Xue X, Bao J, Ye S, Dai Y, et al. Anti-SARS-CoV-2 virus antibody levels in convalescent plasma of six donors who have recovered from COVID-19. Aging (Albany NY) 2020;12. [DOI: 10.18632/aging.103102] [DOI] [PMC free article] [PubMed] [Google Scholar]
Zhang 2020h {published data only}
- Zhang N, Gong Y, Meng F, Bi Y, Yang P, Wang F. Virus shedding patterns in nasopharyngeal and fecal specimens of COVID-19 patients. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.28.20043059] [DOI] [PMC free article] [PubMed] [Google Scholar]
Zhao 2020c {published data only}
- Zhao D, Yao F, Wang L, Zheng L, Gao Y, Ye J, et al. A comparative study on the clinical features of COVID-19 pneumonia to other pneumonias. Clinical Infectious Diseases 2020. [DOI: 10.1093/cid/ciaa247 10.1093/cid/ciaa247] [DOI] [PMC free article] [PubMed] [Google Scholar]
Zhao 2020d {published data only}
- Zhao Z, Cui H, Song W, Ru X, Zhou W, Yu X. A simple magnetic nanoparticles-based viral RNA extraction method for efficient detection of SARS-CoV-2. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.02.22.961268] [DOI] [Google Scholar]
Zhifeng 2020 {published data only}
- Zhifeng J, Feng A, Li T. Consistency analysis of COVID-19 nucleic acid tests and the changes of lung CT. Journal of Clinical Virology 2020;127:104359. [DOI: 10.1016/j.jcv.2020.104359] [DOI] [PMC free article] [PubMed] [Google Scholar]
Zhou 2020 {published data only}
- Zhou J, Bai Z, Liu X, Guo Y, Jiang N, Li X, et al. Flocked swab might be one main reason causing the high false-negative rate in COVID-19 screening----the advantages of a novel silicone swab. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.29.014415] [DOI] [Google Scholar]
Zhuang 2020 {published data only}
- Zhuang GH, Shen MW, Zeng LX, Mi BB, Chen FY, Liu WJ, et al. Potential false-positive rate among the 'asymptomatic infected individuals' in close contacts of COVID-19 patients. Zhonghua Liu Xing Bing Xue Za Zhi 2020;41(4):485-8. [DOI: 10.3760/cma.j.cn112338-20200221-00144 10.3760/cma.j.cn112338-20200221-00144.] [DOI] [PubMed] [Google Scholar]
References to studies awaiting assessment
Li 2020b {published data only}
- Li H, Li Y, Zhang Z, Lu Z, Wang Y, Lin G, et al. Establishment and clinical performance evaluation of 2019 novel coronavirus antibody colloidal gold detection method. Chinese Journal of Infectious Diseases 2020. [Google Scholar]
Thompson 2020 {published data only}
- Thompson C, Grayson N, Paton R, Lourenco J, Penman B, Lee LN, et al. Neutralising antibodies to SARS coronavirus 2 in Scottish blood donors - a pilot study of the value of serology to determine population exposure. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.13.20060467] [DOI] [PMC free article] [PubMed] [Google Scholar]
Xiong 2020 {published data only}
- Xiong Z, Fu L, Zhou H, Liu JK, Wang AM, Huang, Y, et al. Construction and evaluation of a novel diagnosis process for 2019-corona virus disease. Zhonghua Yi Xue Za Zhi 2020;100(0):E019. [DOI: 10.3760/cma.j.cn112137-20200228-00499 10.3760/cma.j.cn112137-20200228-00499.] [DOI] [PubMed] [Google Scholar]
References to ongoing studies
ChiCTR2000029625 {published data only}
- ChiCTR2000029625. Construction of early warning and prediction system for patients with severe / critical novel coronavirus pneumonia (COVID-19). apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000029625 (first received 27 April 2020).
ChiCTR2000029695 {published data only}
- ChiCTR2000029695. Early detection of novel coronavirus pneumonia (COVID-19) based on a novel high-throughput mass spectrometry analysis with exhaled breath. apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000029695 (first received 27 April 2020).
ChiCTR2000029810 {published data only}
- ChiCTR2000029810. Clinical study of a novel high sensitivity nucleic acid assay for novel coronavirus pneumonia (COVID-19) based on CRISPR-cas protein. apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000029810 (first received 27 April 2020).
ChiCTR2000029870 {published data only}
- ChiCTR2000029870. Evaluation of rapid diagnostic kit (IgM/IgG) for novel coronavirus pneumonia (COVID-19). apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000029870 (first received 27 April 2020).
ChiCTR2000029883 {published data only}
- ChiCTR2000029883. A comparative study on the sensitivity of nasopharyngeal and oropharyngeal swabbing for the detection of SARS-CoV-2 by real-time PCR. apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000029883 (first received 27 April 2020).
ChiCTR2000029982 {published data only}
- ChiCTR2000029982. Study for using multiomics in the diagnosis and treatment of novel coronavirus pneumonia (COVID-19). apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000029982 (first received 27 April 2020).
ChiCTR2000030005 {published data only}
- ChiCTR2000030005. Nucleic acid analysis of novel coronavirus pneumonia (COVID-19) in morning sputum samples and pharyngeal swabs-a prospectively diagnostic test. apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000030005 (first received 27 April 2020).
ChiCTR2000030085 {published data only}
- ChiCTR2000030085. Cancelled by the investigator Study for the false positive rate of IgM / IgG antibody test kit for novel coronavirus pneumonia (COVID-19) in different inpatients. apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000030085 (first received 27 April 2020).
ChiCTR2000030185 {published data only}
- ChiCTR2000030185. The value of critical care ultrasound in rapid screening, diagnosis, evaluation of effectiveness and intensive prevention of novel coronavirus pneumonia (COVID-19). apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000030185 (first received 27 April 2020).
ChiCTR2000030253 {published data only}
- ChiCTR2000030253. Exploration and research for a new method for detection of novel coronavirus (COVID-19) nucleic acid. apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000030253 (first received 27 April 2020).
ChiCTR2000030334 {published data only}
- ChiCTR2000030334. MicroRNA as a marker for early diagnosis of novel coronavirus infection (COVID-19). apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000030334 (first received 27 April 2020).
ChiCTR2000030542 {published data only}
- ChiCTR2000030542. A clinical study about the diagnosis and prognosis evaluation of novel coronavirus pneumonia (COVID-19) based on viral genome, host genomic sequencing, relative cytokines and other laboratory indexes. apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000030542 (first received 27 April 2020).
ChiCTR2000030543 {published data only}
- ChiCTR2000030543. Detection of coronavirus in simultaneously collecting tears and throat swab samples collected from the patients with novel coronavirus pneumonia (COVID-19). apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000030543 (first received 27 April 2020).
ChiCTR2000030558 {published data only}
- ChiCTR2000030558. Cancelled by the investigator Epidemiological research of novel coronavirus pneumonia (COVID-19) suspected cases based on virus nucleic acid test combined with low-dose chest CT screening in primary hospital. apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000030558 (first received 27 April 2020).
ChiCTR2000030706 {published data only}
- ChiCTR2000030706. Cancelled by the investigator Application of cas13a-mediated RNA detection in the assay of novel coronavirus nucleic acid (COVID-19). apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000030706 (first received 27 April 2020).
ChiCTR2000030721 {published and unpublished data}
- ChiCTR2000030721. A comparative study for the sensitivity of induced sputum and throat swabs for the detection of SARS-CoV-2 by real-time PCR in patients with novel coronavirus pneumonia (COVID-19). apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000030721 (first received 27 April 2020).
ChiCTR2000030754 {published data only}
- ChiCTR2000030754. Medical records based study for the accuracy of SARS-CoV-2 IgM antibody screening for diagnosis of novel coronavirus pneumonia (COVID-19). apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000030754 (first received 27 April 2020).
ChiCTR2000030833 {published data only}
- ChiCTR2000030833. Clinical validation and application of high-throughput novel coronavirus (2019-nCoV) screening detection kit. apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000030833 (first received 27 April 2020).
ChiCTR2000030834 {published data only}
- ChiCTR2000030834. Epidemiological characteristics and antibody levels of novel coronavirus pneumonia (COVID-19) of pediatric medical staff working in quarantine area. apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000030834 (first received 27 April 2020).
ChiCTR2000030838 {published data only}
- ChiCTR2000030838. Development of warning system with clinical differential diagnosis and prediction for severe type of novel coronavirus pneumonia (COVID-19) patients based on artificial intelligence and CT images. apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000030838 (first received 27 April 2020).
ChiCTR2000030856 {published data only}
- ChiCTR2000030856. An artificial intelligence assistant system for suspected novel coronavirus pneumonia (COVID-19) based on chest CT. apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000030856 (first received 27 April 2020).
ChiCTR2000030859 {published data only}
- ChiCTR2000030859. A medical based analysis for influencing factors of death of novel coronavirus pneumonia (COVID-19) patients in Wuhan Third Hospital. apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000030859 (first received 27 April 2020).
ChiCTR2000030860 {published data only}
- ChiCTR2000030860. A medical records based study for investigation of dynamic profile of RT-PCR test for SARS-CoV-2 nucleic acid of novel coronavirus pneumonia (COVID-19) patients. apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000030860 (first received 27 April 2020).
ChiCTR2000030862 {published data only}
- ChiCTR2000030862. Correlation analysis of blood eosinophil cell levels and clinical type category of novel coronavirus pneumonia (COVID-19): a medical records based retrospective study. apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000030862 (first received 27 April 2020).
NCT04245631 {published data only}
- NCT04245631. Development of a simple, fast and portable recombinase aided amplification assay for 2019-nCoV. clinicaltrials.gov/ct2/show/NCT04245631 (first received 27 April 2020).
NCT04259892 {published data only}
- NCT04259892. Viral excretion in contact subjects at high/moderate risk of coronavirus 2019-nCoV infection. ClinicalTrials.gov/show/NCT04259892 (first received 27 April 2020).
NCT04279795 {published data only}
- NCT04279795. Detection of 2019 novel coronavirus in multiple organ system and its relationship with clinical manifestations. ClinicalTrials.gov/show/NCT04279795 (first received 27 April 2020).
NCT04304690 {published data only}
- NCT04304690. SARS-CoV2 seroconversion among front line medical and paramedical staff in emergency, intensive care units and infectious disease departments during the 2020 epidemic. clinicaltrials.gov/ct2/show/NCT04304690 (first received 27 April 2020).
NCT04311398 {published data only}
- NCT04311398. Development and verification of a new coronavirus multiplex nucleic acid detection system. ClinicalTrials.gov/show/NCT04311398 (first received 27 April 2020).
NCT04316728 {published data only}
- NCT04316728. Clinical performance of the VivaDiag ™ COVID-19 lgM IgG rapid test in early detecting the infection of COVID-19. ClinicalTrials.gov/show/NCT04316728 (first received 27 April 2020).
NCT04321369 {published data only}
- NCT04321369. Impact of swab site and sample collector on testing sensitivity for SARS-CoV-2 virus in symptomatic individuals. clinicaltrials.gov/ct2/show/NCT04321369 (first received 27 April 2020).
NCT04322279 {published data only}
- NCT04322279. Factors associated with a positive SARS-CoV-2 serology in contact subjects at high/moderate risk of coronavirus SARS-CoV-2 infection (CoV-CONTACT-SERO). clinicaltrials.gov/ct2/show/NCT04322279 (first received 27 April 2020).
NCT04322513 {published data only}
- NCT04322513. Biomarkers for identification of SARS-COV-2 infection. clinicaltrials.gov/ct2/show/NCT04322513 (first received 27 April 2020).
Sullivan 2020 {published data only}
- Sullivan PS, Sailey C, Guest JL, Guarner J, Siegler AJ, Valentine-Graves, et al. Detection of SARS-CoV-2 RNA and antibodies in diverse samples: protocol to validate the sufficiency of provider-observed home-collected blood, saliva and oropharyngeal samples. JMIR Public Health and Surveillance 2020. [DOI: 10.2196/19054] [DOI] [PMC free article] [PubMed] [Google Scholar]
Additional references
Bossuyt 2015
- Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig L, et al. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. BMJ 2015;351:h5527. [DOI: 10.1136/bmj.h5527] [PMID: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
CDC China 2020
- National Health Commission of the People's Republic of China. Release of 7th edition of case definitions. www.nhc.gov.cn/yzygj/s7653p/202003/46c9294a7dfe4cef80dc7f5912eb1989.shtml (accessed prior to 23 June 2020).
Cheng 2020b
- Cheng MP, Yansouni CP, Basta NE, Desjardins M, Kanjilal S, Paquette K, et al. Serodiagnostics for severe acute respiratory syndrome-related coronavirus-2: a narrative review. Annals of Internal Medicine (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
Covidence [Computer program]
- Veritas Health Innovation Covidence. Version accessed 27 April 2020. Melbourne, Australia: Veritas Health Innovation.Available at covidence.org.
Macaskill 2010
- Macaskill P, Gatsonis C, Deeks JJ, Harbord RM, Takwoingi Y. Chapter 10: analysing and presenting results. In: Deeks JJ, Bossuyt PM, Gatsonis C editor(s). Cochrane Handbook for Systematic Reviews of Diagnostic Test Accuracy Version 1.0. The Cochrane Collaboration, 2010. Available from methods.cochrane.org/sdt/handbook-dta-reviews.
McInnes 2018
- McInnes MDF, Moher D, Thombs BD, McGrath TA, Bossuyt PM and the PRISMA-DTA Group. Preferred Reporting Items for a Systematic Review and Meta-analysis of Diagnostic Test Accuracy Studies: The PRISMA-DTA Statement. JAMA 2018;319(4):388-396. [DOI: 10.1001/jama.2017.19163] [PMID: ] [DOI] [PubMed] [Google Scholar]
McInnes 2020
- McInnes M, Leeflang MM, Salameh J-P, McGrath T, Van der Pol CB, Frank RA, et al. Imaging tests for the diagnosis of COVID-19. Cochrane Database of Systematic Reviews 2020, Issue 4. [DOI: 10.1002/14651858.CD013639] [DOI] [PubMed] [Google Scholar]
Moher 2009
- Moher D, Liberati A, Tetzlaff J, Altman DG, The PRISMA Group (2009). Preferred reporting items for systematic reviews and meta-analyses: the PRISMA Statement. PLoS Medicine 6;7:1000097. [DOI: 10.1371/journal.pmed1000097] [DOI] [PMC free article] [PubMed] [Google Scholar]
Review Manager 2014 [Computer program]
- Nordic Cochrane Centre, The Cochrane Collaboration Review Manager 5 (RevMan 5). Version 5.3. Copenhagen: Nordic Cochrane Centre, The Cochrane Collaboration, 2014.
Stata [Computer program]
- Stata. Version 15. College Station, TX, USA: StataCorp, 2017.Available at www.stata.com.
Takwoingi 2017
- Takwoingi Y, Guo B, Riley RD, Deeks JJ. Performance of methods for meta-analysis of diagnostic test accuracy with few studies or sparse data. Statistical Methods in Medical Research 2017;26(4):1896-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
Whiting 2011
- Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS‐2: a revised tool for the quality assessment of diagnostic accuracy studies. Annals of Internal Medicine 2011;155(8):529-36. [DOI] [PubMed] [Google Scholar]
WHO 2020
- World Health Organization (WHO). Global surveillance for COVID-19 caused by human infection with COVID-19 virus Interim guidance. www.who.int/publications/i/item/global-surveillance-for-human-infection-with-novel-coronavirus-(2019-ncov) (accessed prior to 23 June 2020). [https://www.who.int/publications/i/item/global-surveillance-for-human-infection-with-novel-coronavirus-(2019-ncov)]
References to other published versions of this review
Deeks 2020
- Deeks JJ, Dinnes J, Takwoingi Y, Davenport C, Leeflang MM, Spijker R, et al. Diagnosis of SARS-CoV-2 infection and COVID-19: accuracy of signs and symptoms; molecular, antigen, and antibody tests; and routine laboratory markers. Cochrane Database of Systematic Reviews 2020, Issue 4. [DOI: 10.1002/14651858.CD013596] [DOI] [Google Scholar]






