Abstract
Endocrine disrupting chemicals (EDCs) can induce abnormalities in organisms via alteration of molecular pathways and subsequent disruption of the endocrine functions. Bisphenol A (BPA) and 17α-ethinylestradiol (EE2) are ubiquitous EDCs in the environment. Many aquatic organisms, including fish, are often exposed to varying concentrations of BPA and EE2 throughout their lifespan. Both BPA and EE2 can activate estrogenic signaling pathways and cause adverse effects on reproduction via alteration of pathways associated with steroidogenesis. However, transcriptional pathways that are affected by chronic exposure to these two ubiquitous environmental estrogens during embryonic, larval, and juvenile stages are not clearly understood. In the present study, we examined transcriptional alterations in the testis of medaka fish (Oryzias latipes) chronically exposed to a low concentration of BPA or EE2. Medaka were exposed to BPA (10 μg/L) or EE2 (0.01 μg/L) from 8 hours post-fertilization (as embryos) to adulthood 50 days post fertilization (dpf), and transcriptional alterations in the testis were examined by RNA sequencing (RNA-seq). Transcriptomic profiling revealed 651 differentially expressed genes (DEGs) between BPA-exposed and control testes, while 1475 DEGs were found between EE2-exposed and control testes. Gene ontology (GO) analysis showed a significant enrichment on “intracellular receptor signaling pathway”, “response to steroid hormone” and “hormone-mediated signaling pathway” in the BPA-induced DEGs, and on “cilium organization”, “microtubule-based process” and “organelle assembly” in the EE2-induced DEGs. Pathway analysis showed significant enrichment on “integrin signaling pathway” in both treatment groups, and on “cadherin signaling pathway”, “Alzheimer disease-presenilin pathway” in EE2-induced DEGs. Single nucleotide polymorphism (SNP) and insertion-deletion (Indel) analysis found no significant differences in mutation rates with either BPA or EE2 treatments. Global gene expression differences in testes of medaka during early stages of gametogenesis were responsive to chronic BPA and EE2 exposure.
Keywords: RNA sequencing, gametogenesis, Medaka fish, Male, BPA, EE2, chronic exposure
1. Introduction
Endocrine disrupting chemicals (EDCs) can perturb endocrine systems and consequently cause a variety of adverse health effects in organisms directly exposed, or in their progeny through transgenerational effects (Bhandari et al., 2015a). Many environmental EDCs are manmade chemicals (Diamanti-Kandarakis et al., 2009), including bisphenol A (BPA), one of the highest production chemicals (Galloway et al., 2010). BPA is primarily used as the monomer in polycarbonate plastics, which are used as food containers as well as epoxy resin in cans. BPA leaches out of these materials, while in use or after these materials have been disposed of in landfills, making BPA a ubiquitous groundwater contaminant near landfills (Masoner et al., 2014). Approximately, 30–40% of river and ground water samples tested were found to have detectable concentration of BPA ranging from 1–28 μg/L (Barnes et al., 2008; Heisterkamp et al., 2004; Jin et al., 2004; Kolpin et al., 2002; Loos et al., 2010; Rudel et al., 1998).
17α-ethinylestradiol (EE2) is a synthetic estrogen used in birth control pills, and approximately 16–68% of EE2 is excreted in the urine or feces of women taking these pills (Johnson and Williams, 2004). Thus, EE2 is commonly found in surface waters as current wastewater treatment methods cannot completely eliminate EE2 from effluents, making it one of the most frequently detected EDCs in aquatic environments (Kolpin et al., 2002). Both BPA and EE2 are present in surface water at concentrations that are sufficient to affect development, osmoregulation, and reproduction in aquatic organisms (Bhandari et al., 2015a; Bhandari et al., 2015b; Brown et al., 2009; Wang et al., 2019). Moreover, the pervasiveness of these chemicals predicts widespread and continued exposure of aquatic organisms, which may lead to reproductive impairment and potentially, population level effects (Kidd et al., 2007a; Schwindt et al., 2014).
An extensive literature exists with regards to EE2 effects on development and reproduction in fish and wildlife, suggesting that chronic exposure to low concentrations of EE2 can result in impairment of fertility in wild fish species, thereby risking future population of fish in the wild (Bhandari et al., 2015a; Kidd et al., 2007b; Schwindt et al., 2014). Pejerrey fish (Chilean silverside, Odonesthes regia) exposed to mixtures of 17 beta-estradiol (E2) and EE2 showed impaired sperm motility, reduced fertilization, and reduced embryo and larval survival (Gárriz et al., 2015). In zebrafish, early-life exposure to EE2 had both immediate and delayed impacts on adult phenotype (Santos et al., 2014). EE2 also caused cardiotoxicity in embryos and altered a set of genes related to oxidative stress and immune dysfunction (Colli-Dula et al., 2014). Additionally, EE2 caused transgenerational reproductive and neurobehavioral effects in subsequent, unexposed generations of fish (Bhandari et al., 2015b; Volkova et al., 2015).
BPA is known as a relatively weak estrogenic chemical compared to EE2, but BPA can still produce adverse effects in the male reproductive tract, since BPA circulates in human blood at levels sufficient to cause effects via estrogen receptors (Bhandari et al., 2015a; Flint et al., 2012; Vandenberg et al., 2009; vom Saal et al., 2012; Welshons et al., 2003). In mammals, BPA has been found to induce multiple reproductive abnormalities (Vandenberg et al., 2009; Whitacre et al., 2012), including alteration in prostate development (Timms et al., 2005), induction of obesity (vom Saal et al., 2012), and impairment of spermatogonial stem cell function (Vrooman et al., 2015). In fish and aquatic wildlife, BPA exposure causes a multitude of abnormalities (Bhandari et al., 2015a; Canesi and Fabbri, 2015; Flint et al., 2012), including precocious neurogenesis leading to hyperactivity (Kinch et al., 2015), and cardiovascular defects (Lombo et al., 2015). High concentration exposure (1–10 mg/L range) has been found to cause teratogenic effects in fish, while endocrine and pleiotropic effects have been observed at lower environmentally relevant concentrations within the low μg/L range (Flint et al., 2012).
Estrogenic effects of BPA are mediated by modulation of endogenous estradiol effects via direct interaction with estrogen receptors (Arase et al., 2011; Welshons et al., 2006; Zhao et al., 2017). BPA has been demonstrated not only to cause direct toxic effects in the generation of the exposed but also to future generations that were never directly exposed (Bhandari et al., 2015b; Drobna et al., 2017; Goldsby et al., 2017; Lombo et al., 2015; Wolstenholme et al., 2012; Wolstenholme et al., 2013). Mechanistically, epigenetic associations have been found with BPA-induced health effects, most of them in mammals (Dolinoy et al., 2007; Iqbal et al., 2015; Lombo et al., 2015; Manikkam et al., 2013).
In our previous work, exposure of medaka fish to a 100 μg/L concentration of BPA or 0.05 μg/L concentration of EE2 during first 7 days of early embryonic development induced transgenerational reproductive defects, such as a significant reduction in fertilization rates and an increased incidence of embryo mortality in three and four generations after the exposure was ceased (Bhandari et al., 2015b). It is not as yet clear what exposure-specific memory, and how it is established in developing embryos, that translates into altered reproductive and behavioral health later in life. While a wealth of information is available regarding direct effects of BPA and EE2 exposure on reproductively active tissues of adult fish (reproduction-centric), it is unclear whether chronic exposures to low concentrations of BPA or EE2 during embryonic, larval, and juvenile stages alter molecular pathways leading to later reproductive impairment. We, therefore, chronically exposed medaka to low concentrations of BPA and EE2 from day 0 through day 50 and examined alterations in transcriptional networks that are dysregulated by exposure to these two contaminants in sexually immature male medaka. Our objective was to examine whether reproductive and metabolic pathways in gene expression are altered before male medaka fish reach full adult reproductive age.
2. Materials and methods
2.1. Fish care
The study was conducted at USGS Columbia Environmental Research Center, Columbia, Missouri under approval of Institutional Animal Care and Use Committee (IACUC) and in accordance to the procedures described by the American Society of Ichthyologists and Herpetologists (ASIH), American Fisheries Society (AFS), and American Institute of Fishery Research Biologists (AIFRB), “Guidelines for Use of Fishes in Field Research” (ASIH, AFS, and AIFRB 1988); and CERC guidelines for the humane treatment of test organisms during culture and experimentation. Sexually mature broodfish were separately maintained on 14L:10D photoperiod and fed 3 times/day. During the first 10 days of post hatching development, juveniles were supplied with ground food and brine shrimp, and thereafter, with flake food once a day and brine shrimp twice a day. Tanks were siphon-cleaned periodically. Water temperature were maintained at 25±1°C.
2.2. Chemical exposure
All final dosing solutions were prepared in glass containers using CERC well water as previously described (Bhandari et al., 2015b). Briefly, water was brought to pH 11 for making stock solution for dissolving BPA, and subsequent dosing solutions of BPA was prepared with CERC well water at a concentration of 10 μg/L. The measured concentrations were determined for each replicate tanks as previously reported (Bhandari et al., 2015b). Briefly, EE2 was extracted from water samples using solid phase extraction (SPE) followed by detection with ELISA according to manufacturer protocols (Ecologiena EE2 ELISA kit, Tokiwa Chemical Industries, Tokyo, Japan). BPA was analyzed in water samples by liquid chromatography/mass spectrometry (LC-MS/MS) according to (Vandenberg et al., 2014). Briefly, BPA was extracted from 1 mL of water using Thermo Hypersen C18 cartridge (Thermoscientific) and eluates were dried under nitrogen and reconstituted in 50:50 methanol:20MM ammonium acetate for HPLC. BPA, in water was quantified by liquid chromatography with mass spectrometry (LCMS/MS) using a Thermo TSQ Quantum Access Max (Thermo Fisher Scientific, Waltham, MA) connected to an integrated Thermo-Accela LC system. Analytes were detected using electrospray ionization with negative polarity, and conditions (tube lens setting, collision energy) were optimized for each analyte using the instrument software. Separations were performed on a 100x4.6 mm 3-micron Hyperclone HPLC column (Phenomenex, Torrance, CA), at a flow rate of 350 μL/min. A gradient mobile phase was employed using 10:90 acetonitrile and acetonitrile, containing 0.01% ammonia. Thermo LCQuan software was used to autotune, acquire, and process the LC/MS data. BPA and C13-BPA were detected using selected reaction monitoring for m/z 227>212, m/z 239>224, respectively, and quantitation was made against standard curves of the analytes at concentrations ranging from 1–200 ng/ml. The limit of quantitation (LOQ) for BPA was 0.18 ug/L. Control water was prepared by mixing the required amount of pH 11 water with CERC well water. The final pH of exposure solution was within the acceptable range for medaka (7.0–7.4). EE2 was dissolved in methanol to make a concentrated stock solution and dosing solution was prepared at the concentration of 0.01 μg/L with CERC well water.
Fertilized eggs were collected and exposed to EE2 (0.01 μg/L) or BPA (10 μg/L) through the incubation water in petri dishes over a period of 12 dpf in 50 ml glass petri dishes. Embryo exposures were static with daily renewal of test chemicals from fertilization through day 12. Initial parameters (survival and success of hatching) were recorded. After hatching, on the 13th day the fish were transferred to a diluter system and the exposure was continued until 50 days post fertilization with food and aeration. Exposure solution in the diluter was replaced every day. Three fish tanks were assigned to each treatment and each tank contained 30 fish (including males and females). Individual fish in each tank represented a technical replicate, whereas each tank represented a biological replicate. In total, 3 exposure groups were used (Control, BPA-exposed, and EE2-exposed), and 3 biological replicates were used per exposure group. Treatment concentrations in the diluter were adjusted accordingly to meet the designated final concentration of the dosing chemical.
2.3. Sample collection
At 50 dpf, the sex of each fish was determined by the presence of male-specific orange coloration on the body and caudal fins, and subsequently by genomic PCR using the DNA from their caudal fin (Kinoshita et al., 2009). Total length, whole body weight, and gross morphological evaluations of the fish were conducted at this time to evaluate developmental anomalies of spinal curvature, fin, head and eye deformities. At 50 dpf, 3 males were randomly selected from each tank, and the whole testis was collected. In total, nine testes samples were collected from each exposure group (3 fish per biological replicate tank and 3 tanks per exposure group). Altogether, 27 testes samples (3 males x 3 tanks x 3 exposure groups) were used for RNA sequencing.
2.4. RNA extraction and RNA-seq library sequencing
To conduct a broad, detailed evaluation of the transcriptomic changes in testes undergoing early stage of spermatogenesis that coincides with epigenetic reprogramming of germ cells after chronic exposure to BPA or EE2, we performed RNA sequencing on testes samples to obtain gene transcription profiles. Total RNA from each testis sample was extracted using a commercial kit for RNA/DNA isolation (Qiagen, Carlsbad, CA) according to manufacturer’s instructions. The quality control and RNA-seq library preparation were conducted by the DNA sequencing Core facility of the University of Missouri Columbia. The RNA-seq libraries were sequenced on the Illumina Hiseq 2500 platform using the 100 bp single-end sequencing strategy.
2.5. Reads mapping and differentially expressed genes (DEGs) identification
Sequencing reads from each sample were mapped to medaka reference transcripts (Ensembl version 95) using HISAT2 software with default parameters (Kim et al., 2015; Kim et al., 2019). Only uniquely mapped reads were used for subsequent analysis. Cufflinks software (http://cole-trapnell-lab.github.io/cufflinks/, University of Washington) was used to conduct differentially expressed gene (DEG) analysis on the raw count data. Technical replicates were merged together using the publicly available SAMtools software (Latendresse et al., 2009). Differentially expressed genes were computed using Cuffdiff function of the Cufflinks package. Gene expression values were generated in the normalized form of fragments per kilobase of transcript per million mapped fragments (FPKM) values. The FDR-adjusted p-value < 0.05 (q-value in Suppl table 3 and Suppl table 3) was considered as significantly differentially expressed (Trapnell et al., 2012). A total of 27 samples was sequenced, which represent 3 technical replicates per tank (biological replicate), 3 biological replicates per exposure group, and 3 exposure groups. In the analysis, three technical replicates were merged together to reduce the sequence bias. We did this because the transcripts that are not sequenced in one library can be sequenced in the library of other replicates. Merging replicate sequence files reduces sequencing bias and increases sequencing depth. Finally, the DEGs were calculated from biological replicates and identified as chemical treated groups and control groups. Sequencing data reported in this paper have been deposited to the public database at NCBI Gene Expression Omnibus (GEO) under accession number GSE129727.
2.6. Gene enrichment analysis
Gene enrichment analysis was performed using the PANTHER Overrepresentation Test (Released 2019–07-11) with GO Ontology database (Released 2019–12-09) (Ashburner et al., 2000; Consortium, 2018; Mi et al., 2016). Gene Ontology terms in the biological processes (GO biological process complete), molecular functions (GO molecular function complete), cellular component (GO cellular component complete) and PANTHER Pathways annotation (Version 15, Released 2020–02-14) were selected. Fisher’s Exact Test followed by False Discovery Rate (FDR) correction were used to determine significance. FDR adjusted p-value < 0.05 indicated significantly enriched GO and pathway terms.
2.7. qRT-PCR validation
Quantitative RT-PCR was performed to quantify expression levels of 17 genes, including four DNA methyltransferases, three tet methylcytosine dioxygenases, six genes associated with male sex differentiation in medaka, and four genes randomly selected from RNA-Seq gene set.
Specific primer pairs used in the qRT-PCR were designed using Primer3web (Untergasser et al., 2012). The sequences of primers are shown in Supp Table 1. At least one junction of exon and intron was covered by primer or amplified region to avoid amplifying genomic DNA. β-actin was used as an endogenous reference gene to determine relative expression. Three technical replicates were used for each biological replicate. Since there were three biological replicates for each exposure group, the total of 9 testis samples were analyzed from each treatment group. qRT-PCR was performed by PowerUp SYBR™ Green Master Mix (Applied Biosystems) using a QuantStudio 3 Real-Time PCR System as the following process: 2 min at 50 °C and 10 min at 95 °C for pre-incubation, 40 cycles at 95 °C (15 s) and 60 °C (1 min). The PCR primer amplification efficiencies (E) for each PCR target sequence were calculated from the standard curve generated from five cDNA dilutions. The equation used for the efficiency calculation was E = [−1+10(−1/slope)]. Percent efficiency was calculated by dividing E by 2 and then multiplying by 100. A dissociation analysis was included for each primer pair to evaluate primer specificity for the target sequence. Target gene expression was analyzed by 2−ΔΔCt method (Livak and Schmittgen, 2001). PCR product produced by each primer pair produced a single band of expected size on 2% agarose gel, suggesting that the primers were specific to the target amplicon. Correlations between RNA-seq and qRT-PCR were conducted in Excel® software using the “PEARSON” function. Differences with p < 0.05 were considered as significant.
2.8. Single nucleotide polymorphism (SNP) and indel markers discovery
Samtools and VarScan (Koboldt et al., 2012) were used to identify potential single nucleotide polymorphisms (SNPs) and indels from transcriptomes, with the minimum read depth above 8 and p-value < 0.01.
3. Results
3.0. Water concentration of test solutions and fish morphology
The measured concentration of the exposure solution ranged from 95 to 98% of nominal concentration for EE2 and from 104% to 110% of nominal concentration for BPA. No differences in total length or whole-body wet weight were observed between treatment group and the control fish at 50 dpf, when the exposure was terminated (p < 0.98, One-way ANOVA followed by Bonferroni post-hoc test, see Supplemental Figure 1). Additionally, no gross abnormalities (skin lesions, spinal curvatures, fin erosions, cranial deformities) were observed at this time.
3.1. Next-generation sequencing and mapping to the reference genome
After quality control, next-generation sequencing generated 19,644,917 to 37,306,636 high quality clean sequence reads for each sample, and the reads uniquely mapping ratio ranged from 83.45% to 88.19% (Supplemental Table 2). Mapping results from the same biological replicates were merged together for the subsequent DEG analysis.
Correlation analysis of the gene expression of three groups showed that the BPA exposed samples is closer to EE2 exposed samples than control samples (Figure 1A). The Jensen-Shannon divergence analysis showed JS Distance between BPA exposed and control samples was 0.108, the JS Distance between EE2 exposed and control samples was 0.127, the JS Distance between BPA exposed and EE2 exposed samples was 0.103 (Figure 1B). Principal component analysis (PCA; Figure 1C) showed that treated groups were not clearly separated from the controls.
Figure 1:
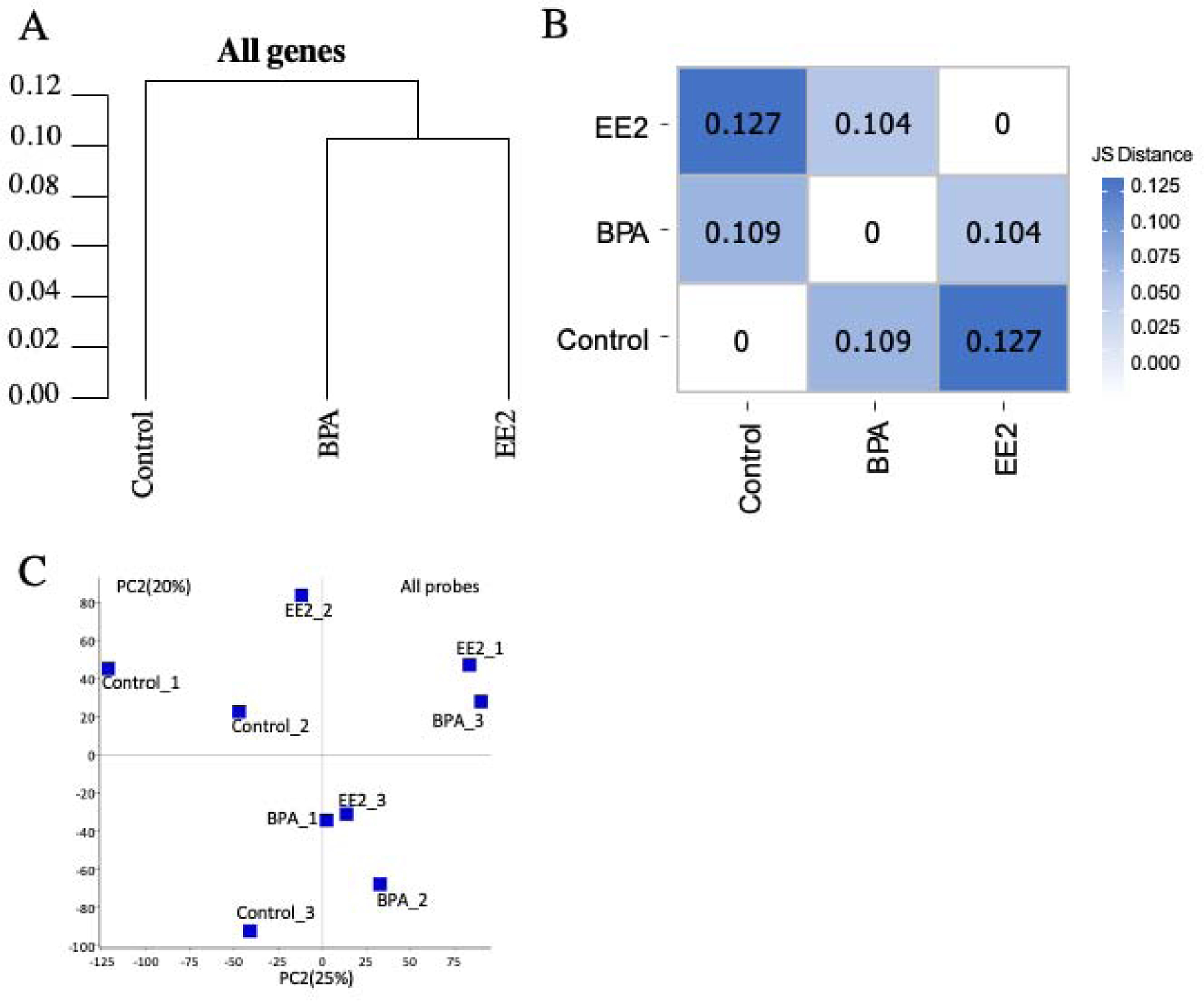
Distance and distance heatmap between each sample. (A): distance between each sample; (B): JS distance heatmap between each sample; (C): Principal component analysis (PCA): plot showing the first 2 principal components (PC1, PC2).
3.2. Identification of differentially expressed genes in medaka testes
Significant DEGs were identified in each treatment group. Genes with a fold change (FC) > 2 and FDR < 0.05 in comparisons between BPA-exposed or EE2-exposed and control treatment were identified as DEGs (Table 1). Briefly, there were 651 genes annotated by the medaka reference genome (Hd-rR strain, Ensembl release 95) found to be significantly differentially expressed between BPA exposed and control groups (Supplemental Table 3). Within these DEGs, 282 genes were upregulated, and 364 genes downregulated in the BPA-exposed group (Figure 2A). Three genes were specifically expressed in controls and 2 genes were specifically expressed in testes of the BPA treatment group.
Table 1:
Summaries of DEGs between different sample comparation
| Exposure vs. | Exposure | Up-regulated in sample_1 | Down-regulated in sample_1 | Specific in sample_1 | Specific in sample_2 | Total |
|---|---|---|---|---|---|---|
| BPA | control | 300 | 376 | 3 | 3 | 682 |
| EE2 | control | 682 | 866 | 7 | 7 | 1562 |
| BPA | EE2 | 183 | 420 | 4 | 2 | 609 |
Figure 2:
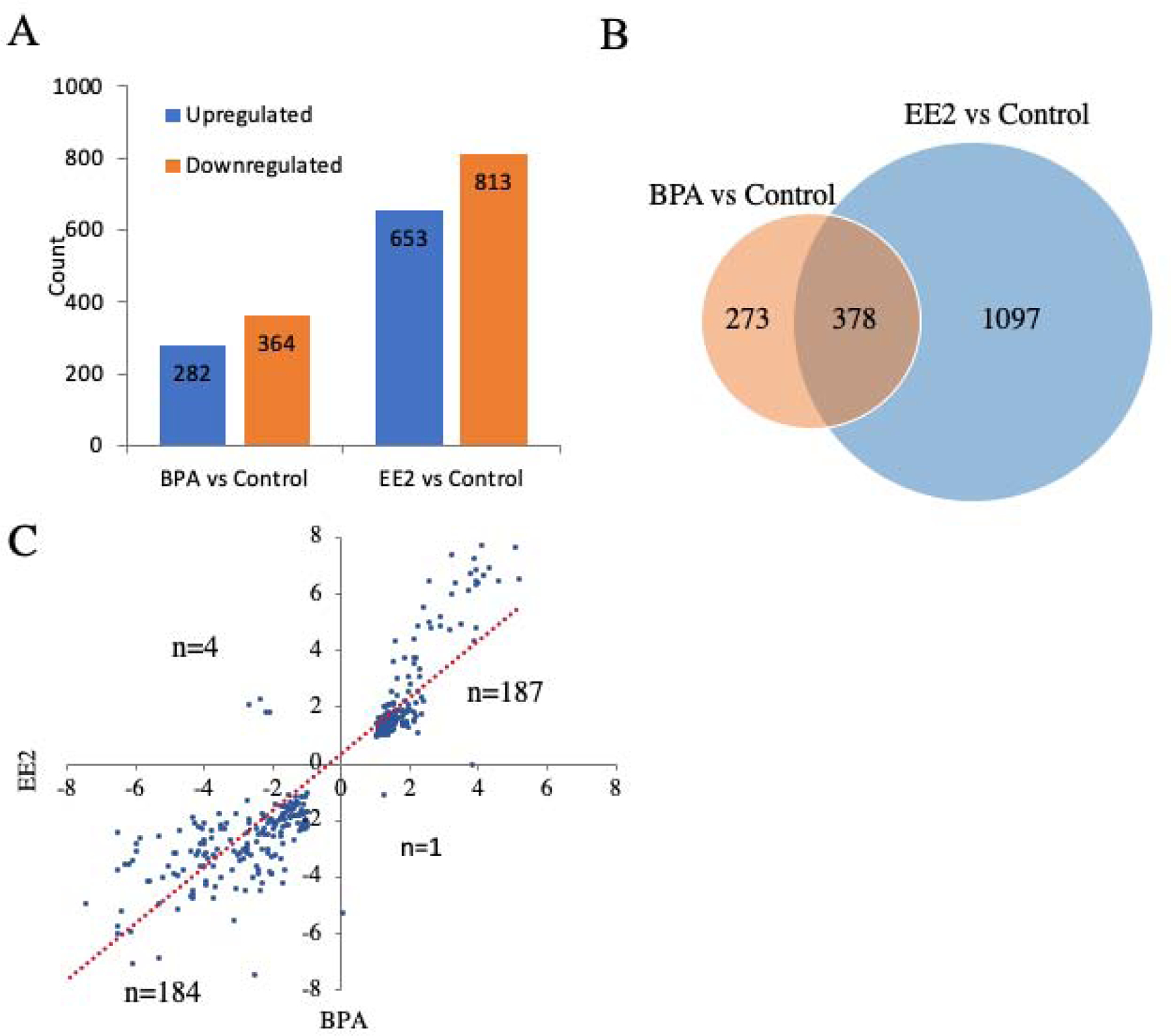
Differentially expressed genes (DEGs) identified between BPA or EE2 exposed and control samples. (A): Upregulated or downregulated genes between BPA or EE2 exposed and control samples; (B): Venn diagrams for DEG relationships between two groups showing the shared DEGs and specific DEGs in each treatment; (C): DEGs shared by two treatment groups were consistent.
A total of 1475 genes was found to be significantly differentially expressed between EE2 and control groups (Supplemental Table 4). Within them, 653 genes were upregulated, and 813 genes were downregulated (Figure 2A). Seven genes were specifically expressed in controls and 2 genes were specifically expressed in EE2 treatment group testes.
A total of 378 genes was shared by the BPA and EE2 treatment groups (Figure 2B). Interestingly, 187 DEGs were upregulated in both BPA and EE2 treatment groups and 184 DEGs were downregulated in both BPA and EE2 treatment (Figure 2C).
3.3. qRT-PCR validation
qRT-PCR results were consistent with RNA-seq results for both BPA (Figure 3A and Supplemental Figure 2), and EE2 treatment groups (Figure 3B and Supplemental Figure 3). For the BPA-exposed group, the correlation coefficient of RNA-seq results and the qRT-PCR confirmation was 0.72 (Figure 3A), for the EE2-exposed group the correlation was 0.97 (Figure 3B). Overall, the results of qRT-PCR validated the results of RNA-seq and offered support for the reliability of the observed DEGs.
Figure 3:
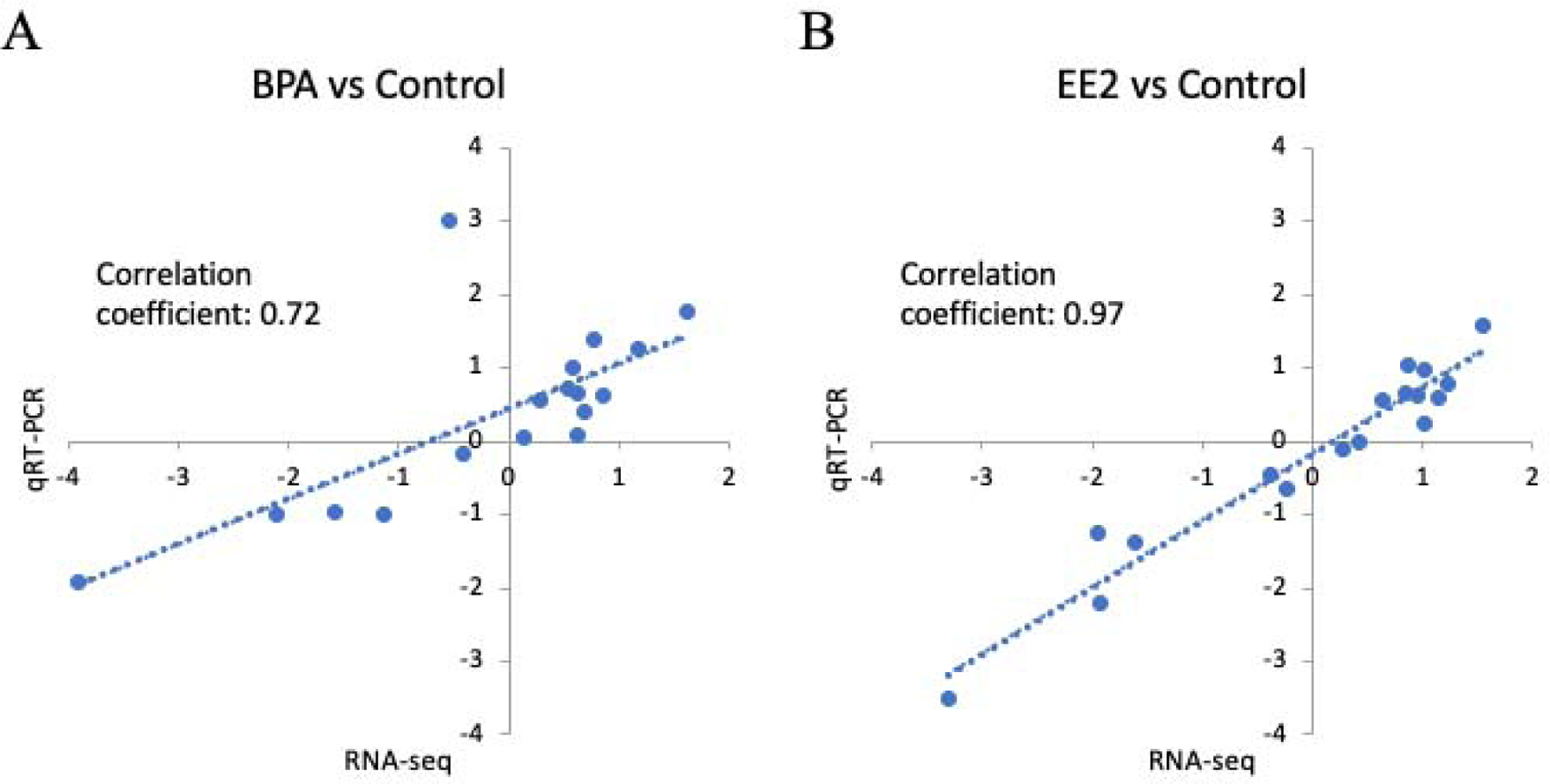
Validation of the RNA-Seq data using qRT-PCR. Consistency of log2(FC) between RNA-Seq data (X-axis) and qRT-PCR assay (Y-axis) is high based on the selected genes. (A): BPA exposed group; (B): EE2 exposed group.
3.4. Gene ontology analysis
The functional enrichment of DEGs identified in the BPA-exposed group included 647 out of the 651 DEGs assigned to GO categories (Supplemental Table 5). In biological processes, the most enriched GO terms included “chitin metabolic process”, “intracellular receptor signaling pathway”, “cellular response to steroid hormone stimulus”, “steroid hormone mediated signaling pathway”, “response to steroid hormone” and “hormone-mediated signaling pathway” (Figure 4A and Supplemental Table 5). In cellular components, the most enriched GO terms included “extracellular region”, “intrinsic component of membrane”, “integral component of membrane”, “membrane” and “cellular anatomical entity” (Figure 4B and Supplemental Table 5). In molecular function, the most enriched GO terms included “bile acid binding”, “serine-type carboxypeptidase activity”, “serine-type exopeptidase activity”, “oxidoreductase activity”, “ephrin receptor activity” and “nuclear receptor activity” (Figure 4C and Supplemental Table 5).
Figure 4:
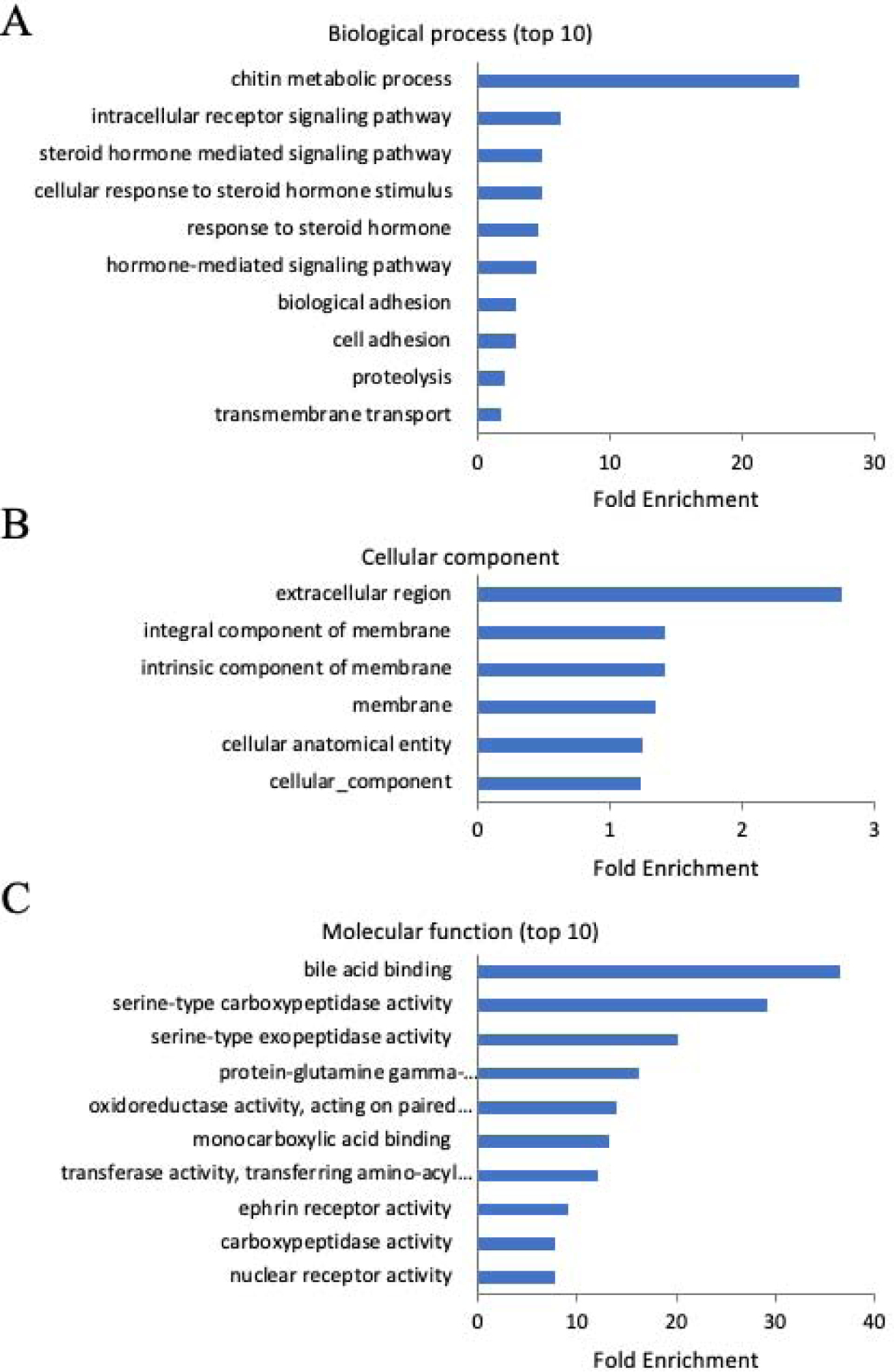
Gene Ontology (GO) enrichment of DEGs in BPA exposed sample. (A): Biological process; (B): Cellular component; (C): Molecular function.
In the EE2-exposed group, 1470 out of 1475 DEGs were assigned to GO categories (Supplemental Table 6). In biological process, the most enriched GO terms included “cilium organization”, “microtubule-based movement”, “microtubule-based process”, “organelle assembly”, “plasma membrane bounded cell projection organization” and “cell projection organization” (Figure 5A and Supplemental Table 6). In cellular components, the most enriched GO terms included “condensed chromosome”, “cilium”, “plasma membrane bounded cell projection cytoplasm”, “plasma membrane bounded cell projection”, “cell projection”, and “polymeric cytoskeletal fiber” (Figure 5B and Supplemental Table 6). In molecular function, the enriched GO terms included “nucleic acid binding” and “calcium ion binding” (Figure 5C and Supplemental Table 6);
Figure 5:
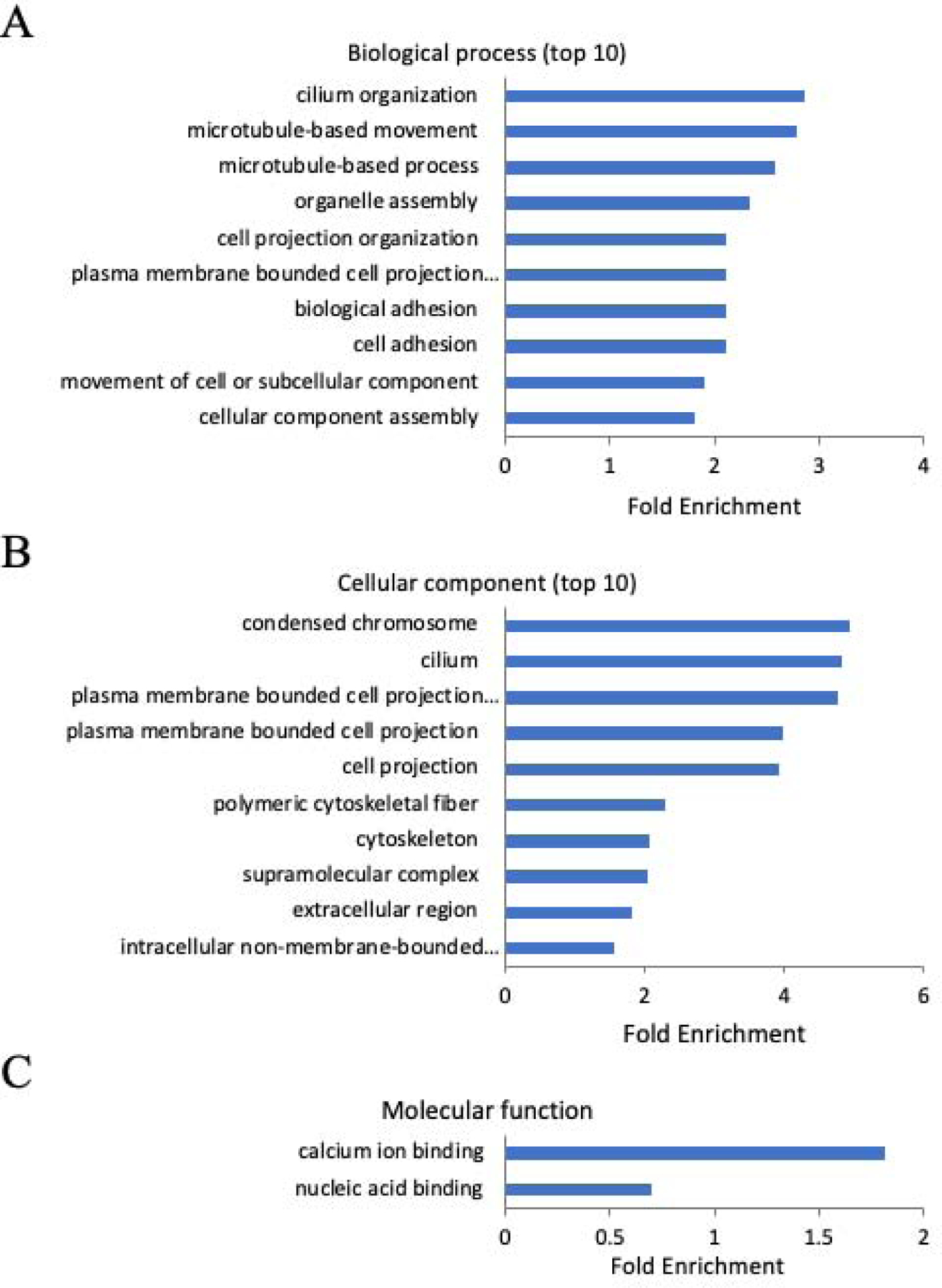
Gene Ontology (GO) enrichment of DEGs in EE2 exposed sample. (A): Biological process; (B): Cellular component; (C): Molecular function.
Out of the 378 DEGs shared by the BPA and EE2 treatment groups (Figure 2B), 376 genes were assigned to GO categories. In biological process, the most enriched GO terms included “chitin metabolic process”, “glucosamine-containing compound metabolic process”, “peptide cross-linking”, “amino sugar metabolic process” and “cell migration” (Supplemental Figure 4A), In cellular components, those genes enriched in terms “extracellular region” only (Supplemental Figure 4B). In molecular function, the enriched GO terms included “serine-type carboxypeptidase activity”, “protein-glutamine gamma-glutamyltransferase activity”, “serine-type exopeptidase activity”, “chitin binding”, “transferase activity, transferring amino-acyl groups” and “ephrin receptor activity” (Supplemental Figure 4C).
3.5. Pathway enrichment analysis
In the BPA-exposed group, upregulated DEGs were enriched with “Integrin signaling pathway” (Supplemental Table 7), while no significant enrichment was observed in downregulated DEGs. In the EE2-exposed group, upregulated DEGs were enriched at “Cadherin signaling pathway”, “Alzheimer disease-presenilin pathway”, “Integrin signaling pathway” and “Inflammation mediated by chemokine and cytokine signaling pathway” (Supplemental Table 8); no significant enrichment was observed in downregulated DEGs.
3.6. SNP markers identification
To investigate if BPA or EE2 exposure induced any genetic variations, we searched for SNPs and Indels in each sample. A total of 98,239 SNPs was identified in the control group, 97,826 SNPs were identified in the BPA-exposed group, and 100,774 SNPs in the EE2-exposed group (Figure 6 and Supplemental Figure 5). No significant differences were found in SNP numbers among different groups. For indels, a total of 6,739 were identified in control group, 6,822 in BPA-exposed group, and 6,919 in EE2-exposed group (Figure 6 and Supplemental Figure 6). No significant differences were found in indel numbers among different groups either. These results suggest that BPA and EE2 treatment did not induce genetic variation in coding regions of the genome analyzed.
Figure 6:
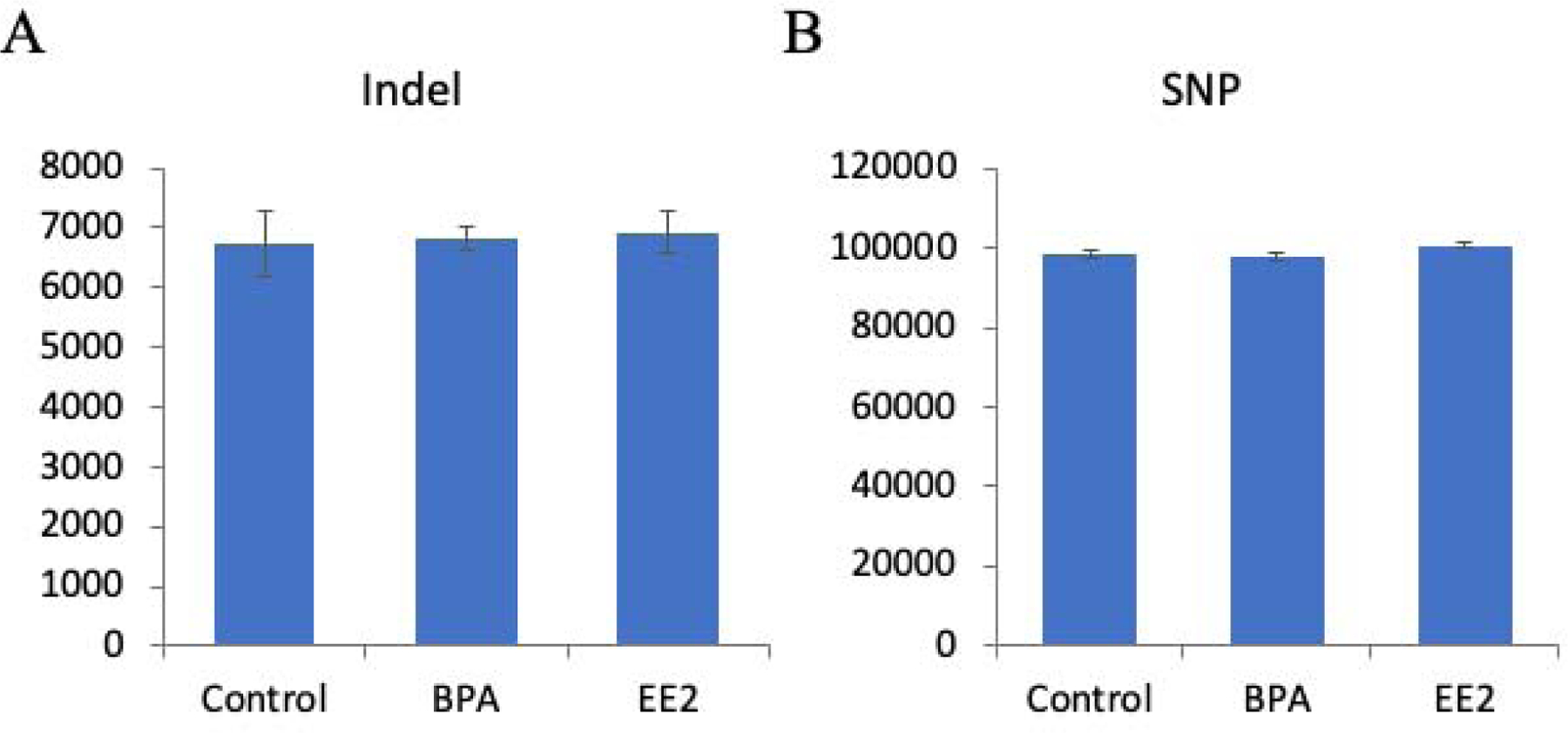
SNPs and indels identified in each sample
4. Discussion
Environmental estrogens can induce organizational effects in developing embryos, including sex reversal, altered reproductive functions, neurobehavioral alterations, and cardiovascular/metabolic defects. However, the molecular pathways that are initially programmed in the developing testis and are integral to adult onset reproductive abnormalities from exposure to estrogenic chemicals are largely unknown. The present study identified a suite of molecular pathways in the testes affected by chronic BPA or EE2 exposures from the day of fertilization to early adult stage initiation of gametogenesis (50 dpf) in medaka.
During the experimental exposure period, no morphological deformities, including histological alterations and body somatic indices, were found in fish exposed to BPA and EE2. In our previous study, embryonic BPA or EE2 exposure at a concentration slightly higher than used in the present study also did not induce morphological deformities and reproductive impairment in adulthood in the exposed generation (Bhandari et al., 2015b). However, embryonic exposure alone lead to a transgenerational phenotype of reduced fertilization and increased embryo mortality in the F2 and F3 generation of medaka, respectively (Bhandari et al., 2015b). No effects of BPA exposure on morphological end points have been reported for directly exposed fish of several species (Bhandari et al. 2015a). For example, zebrafish fed with BPA at the concentration of 500 mg/kg feed did not have effects on development of embryos (Drastichova et al., 2005); whereas 60 days exposure to 7 μg/L BPA reduced tail length in swordtail fish (Kwak et al., 2001). With exposure to concentrations such as 837 μg /L or higher, BPA induced feminization in medaka (Kang et al., 2002). Together, observations suggested that low concentration of BPA do not impose directly observable acute toxic effects in medaka and other fish species, but it is not clearly understood whether the changes in gene expression reported in the present study could affect physiological endpoints later in life.
Given the estrogenic nature of both BPA and EE2, a degree of similarity was expected in the transcriptomic response to exposure, based on our attempt to use doses of BPA and EE2 predicted to result in similar effects on phenotype. Over 58% of DEGs identified in the BPA-exposed group overlapped with DEGs in the EE2-exposed group, suggesting that BPA and EE2 may affect testis development through at least some common mechanisms. Most biological processes altered in the EE2-exposed testis included cell adhesion, microtubules, and assembly suggesting mitotic cell activities (Vagnarelli, 2012); whereas BPA exposure resulted in regulation of biological processes involving steroid hormone-mediated signaling and response pathways. Specifically, there was a significant increase in the expression pattern of steroidogenic acute regulatory protein (star) gene in the BPA-exposed group. StAR is an enzyme that catalyzes the production of pregnenolone from cholesterol, the first step of sex steroid biosynthesis. StAR is responsible for the transport of cholesterol across the mitochondrial membrane for it to serve as substrate for sex steroid production. Alteration in expression of star suggests BPA exposure effects on the steroidogenic pathway in the testes of fish that are not reproductively active yet.
We also examined if downstream pathways are affected by BPA or EE2 exposure in the same samples. No changes in expression of reproduction related genes were observed. Testis development and male reproduction in fish is a precisely coordinated event involving timely expression and repression of genes (Devlin and Nagahama, 2002). At the juvenile stage (50 dpf), no reproductive genes, particularly those in downstream steroidogenic pathway, are expected to be expressed. That being said, exposure did not induce precocious expression of these genes. A number of studies in medaka have demonstrated alteration in expression pattern of sex determining genes in response to the varying concentrations of BPA or EE2; generally at concentrations greater than those used in the present study (Kang et al., 2002; Sun et al., 2014). However, we did not find changes in any major male sex determining genes, such as gsdf, sox9a, dmrt1 and dmy. Interestingly, we observed upregulation of r-spondins (an ovarian differentiation related gene (Zhou et al., 2012) in the BPA exposed group, indicating that r-spondins may be an upstream target of BPA, which may lead to male-female sex reversal or impairment of male reproduction in medaka (Horie et al., 2020). On other hand, estrogen receptor 2 (esr2) was significantly upregulated in both treatment groups, suggesting the role of esr2 in mediation of BPA/EE2-induced reproductive impairment in males, which requires further studies. Esr2 has been demonstrated to be essential for female reproduction in medaka and zebrafish (Kayo et al., 2019; Lu et al., 2017). Taken together, the present results suggest that the effects of exposure at these concentrations of BPA or EE2 on steroid pathways and reproductive pathways in the testes are not sex-specific in the exposed generation. It was not investigated if this exposure could lead to transgenerational effects in the future generations.
Single nucleotide polymorphisms (SNPs) and indels are the most common type of genetic variations among individuals, which are caused by point mutations or deficiencies of DNA repair pathways. To determine if developmental exposure to BPA or EE2 at early stages could induce genetic variation in testes, we determined the SNPs and indels from each of these treatments. There was no change in total number of SNPs and indels after exposure to either BPA or EE2, suggesting that exposure to BPA or EE2 did not induce genetic mutations in the genome sequence. In a previous study, exposure of medaka fish to a 100 μg/L concentration of BPA or 0.05 μg/L concentration of EE2 during first 7 days of early embryonic development induced transgenerational reproductive defects (Bhandari et al., 2015b). Present results support our hypothesis that the transgenerational phenotypes caused by BPA or EE2 exposure, which we previously reported (Bhandari et al., 2015a), are mediated by epigenetic rather than genetic modifications.
Among the cellular components altered by BPA exposure involved cell membrane and cell adhesion molecules; whereas EE2 exposure resulted in alterations in pathways to condensed chromosome suggesting epigenetic processes being perturbed (Vagnarelli, 2012). BPA can alter molecular pathways known to be important in epigenetic programming and potentially involved in transmission of transgenerational effects in fish. BPA exposure decreased global DNA methylation in zebrafish testes (Laing et al., 2016; Liu et al., 2016) and suppressed anti-mullerian hormone in ovaries through recruitment of histone H3K4me3 and H3K27me3 marks together with increased CpG methylation of promoter regulatory regions (Santangeli et al., 2019). We investigated DNA methyltransferases in the RNA-seq results and verified them with qRT-PCR to determine if there were changes in gene expression of epigenetic modification-related genes. DNA methylation is one of the major forms of genome apparent modification in transcriptional regulation and inhibition of retroviral factors, gene imprinting and X chromosome inactivation (Jaenisch and Bird, 2003). We did not observe any significant changes in DNA methyltransferase or TET methylcytosine dioxygenase gene expressions in the RNA-seq data, and only dnmt3bb.1 was significantly upregulated according to the qRT-PCR results. However, expression of mbd3b was decreased in both treated groups. mbd3b gene encodes a Methyl-CpG binding domain (MBD) containing proteins, which specifically binds methylated-CpG nucleotides and regulates transcription of target genes by changing accessibility of transcription factors to their binding sites and altering chromatin structure. The current observations are derived from whole organ, which is heterogenous in cell population. It is thus not possible to conclude that the expression patterns of the genes that regulate epigenetic processes truly reflect BPA-induced epigenetic alterations in the target cells. Also, the measurement of global DNA methylation profile of the whole testis will not yield any plausible information. Future studies, therefore, require isolation of a pure population of germ or somatic cells from the testis for epigenetic assessments.
Primordial germ cells (PGCs) are one of the cell types that give rise to future generations and undergo epigenetic programming during the period of sex determination and testis differentiation (Bhandari, 2016; Nilsson et al., 2019; Seisenberger et al., 2012; Wang and Bhandari, 2020b). Exposure window in the present study overlapped with the sensitive windows of epigenetic reprogramming of germ cells in medaka (Wang and Bhandari, 2020a); however, it is not clear if any of the observed DEGs are involved in the promotion of long-term transgenerational effects in male germ cells. It is not yet clearly understood if environmentally induced effects on developing germ cells persist in sperm and eggs in adulthood. Studies in various animal models suggest that environmental chemicals may alter epigenetic reprogramming of developing PGCs, leading to no apparent reproductive effects in the exposed generation, yet multiple health problems in future generations. BPA or EE2 can induce transgenerational health effects in fish, including impairment of male reproduction, female reproductive defects, cardiac deformities, as well as anxiety behavior. Additionally, several endocrine disrupting chemicals have been found to induce transgenerational health abnormalities in fish if exposed during early embryogenesis or gametogenesis (Baker et al., 2014; Carvan et al., 2017; Cheng et al., 2017; Knecht et al., 2017; Seemann et al., 2015; Sun et al., 2015; Wang et al., 2016; Wang and Bhandari, 2019; Wang and Bhandari, 2020b). All transgenerational studies agree in that chemicals-induced molecular changes are programmed in such a way that they affect gene expression and physiological processes long past when the exposure ceases. These initially programmed networks of genes may not necessarily be the same as those involved in subsequent adverse endpoints observed. Hence, a better understanding is required between the initial molecular signatures altered by environmental chemical stressors, and whether they contribute to the emergence of phenotypes.
In conclusion, the present study provides gene expression profiles in response to early life stage exposures to BPA or EE2 in medaka. There was significant overlap (58%) of DEGs in the testes between the BPA and EE2 treatments, as might be expected from two estrogens, suggesting disruption of some common pathways. No morphological measurements were altered by either treatment, and no SNPs were observed in the testes of either treatment group. Exposure-induced molecular signatures in the reproductively active adult testis may be different from the signatures that are established developmentally. Future studies, therefore, should focus on connection of molecular pathways specially from pre-reproductive to reproductively active states.
Supplementary Material
Table 2:
Statistics of single nucleotide polymorphism types
| Transition | Transversion | ||||||
|---|---|---|---|---|---|---|---|
| C/T | A/G | A/T | A/C | T/G | C/G | Total | |
| Control | 1958 | 2141 | 573 | 644 | 589 | 563 | 6468 |
| BPA | 1901 | 2052 | 529 | 604 | 546 | 513 | 6145 |
| EE2 | 2065 | 2186 | 551 | 675 | 623 | 567 | 6667 |
Highlights.
Gene networks affected by BPA prior to gonadal maturation were investigated
BPA and EE2 altered gene networks involved in receptor-mediated endocytosis
Expression of genes involved in male reproduction are not altered by BPA exposure prior to sexual maturation
Acknowledgments
The funding for this study was provided by University of Missouri, Mizzou Advantage Fund to DET, FVS, and RKB and National Institute of Environmental Health Sciences (R21ES027123) to RKB. Authors would like to acknowledge the technical assistance of Diane Nicks, Rachel Claunch, James Candrl, and Vanessa Velez from U.S. Geological Survey, Columbia Environmental Research Center, and Pooja Bhandari from Division of Biological Sciences, University of Missouri Columbia (Currently address: Biology Department, University of North Carolina Greensboro).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Publisher's Disclaimer: Disclaimer
Publisher's Disclaimer: Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Disclosures
All authors have no competing financial interests.
Conflicts of interest
All authors have no conflicts of interests.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Arase S, Ishii K, Igarashi K, Aisaki K, Yoshio Y, Matsushima A, Shimohigashi Y, Arima K, Kanno J, Sugimura Y, 2011. Endocrine disrupter bisphenol a increases in situ estrogen production in the mouse urogenital sinus. Biol Reprod 84, 734–742. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, 2000. Gene ontology: tool for the unification of biology. Nature genetics 25, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TR, Peterson RE, Heideman W, 2014. Using zebrafish as a model system for studying the transgenerational effects of dioxin. Toxicol Sci 138, 403–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes KK, Kolpin DW, Furlong ET, Zaugg SD, Meyer MT, Barber LB, 2008. A national reconnaissance of pharmaceuticals and other organic wastewater contaminants in the United States—I) Groundwater. Science of the Total Environment 402, 192–200. [DOI] [PubMed] [Google Scholar]
- Bhandari RK, 2016. Medaka as a model for studying environmentally induced epigenetic transgenerational inheritance of phenotypes. Environmental Epigenetics 2, dvv010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari RK, Deem SL, Holliday DK, Jandegian CM, Kassotis CD, Nagel SC, Tillitt DE, vom Saal FS, Rosenfeld CS, 2015a. Effects of the environmental estrogenic contaminants bisphenol A and 17α-ethinyl estradiol on sexual development and adult behaviors in aquatic wildlife species. General and comparative endocrinology 214, 195–219. [DOI] [PubMed] [Google Scholar]
- Bhandari RK, Vom Saal FS, Tillitt DE, 2015b. Transgenerational effects from early developmental exposures to bisphenol A or 17α-ethinylestradiol in medaka, Oryzias latipes. Scientific reports 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KH, Schultz IR, Nagler JJ, 2009. Lack of a heritable reproductive defect in the offspring of male rainbow trout exposed to the environmental estrogen 17α-ethynylestradiol. Aquatic toxicology 91, 71–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canesi L, Fabbri E, 2015. Environmental effects of BPA: focus on aquatic species. Dose-Response 13, 1559325815598304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvan MJ 3rd, Kalluvila TA, Klingler RH, Larson JK, Pickens M, Mora-Zamorano FX, Connaughton VP, Sadler-Riggleman I, Beck D, Skinner MK, 2017. Mercury-induced epigenetic transgenerational inheritance of abnormal neurobehavior is correlated with sperm epimutations in zebrafish. PLoS One 12, e0176155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Yan W, Wu Q, Liu C, Gong X, Hung TC, Li G, 2017. Parental exposure to microcystin-LR induced thyroid endocrine disruption in zebrafish offspring, a transgenerational toxicity. Environ Pollut 230, 981–988. [DOI] [PubMed] [Google Scholar]
- Colli-Dula R-C, Martyniuk CJ, Kroll KJ, Prucha MS, Kozuch M, Barber DS, Denslow ND, 2014. Dietary exposure of 17-alpha ethinylestradiol modulates physiological endpoints and gene signaling pathways in female largemouth bass (Micropterus salmoides). Aquatic toxicology 156, 148–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium GO, 2018. The Gene Ontology resource: 20 years and still GOing strong. Nucleic acids research 47, D330–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin RH, Nagahama Y, 2002. Sex determination and sex differentiation in fish: an overview of genetic, physiological, and environmental influences. Aquaculture 208, 191–364. [Google Scholar]
- Diamanti-Kandarakis E, Bourguignon J-P, Giudice LC, Hauser R, Prins GS, Soto AM, Zoeller RT, Gore AC, 2009. Endocrine-disrupting chemicals: an Endocrine Society scientific statement. Endocrine reviews 30, 293–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinoy DC, Huang D, Jirtle RL, 2007. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci U S A 104, 13056–13061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drastichova J, Svobodova Z, Groenland M, Dobšíková R, Žlábek V, Weissova D, Szotkowska M, 2005. Effect of exposure to bisphenol A on the sex differentiation in zebrafish (danio rerio). Acta Veterinaria Brno 74, 287–291. [Google Scholar]
- Drobna Z, Henriksen AD, Wolstenholme JT, Montiel C, Lambeth PS, Shang S, Harris EP, Zhou C, Flaws JA, Adli M, Rissman EF, 2017. Transgenerational effects of Bisphenol A on gene expression and DNA methylation of imprinted genes in brain. Endocrinology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint S, Markle T, Thompson S, Wallace E, 2012. Bisphenol A exposure, effects, and policy: a wildlife perspective. Journal of environmental management 104, 19–34. [DOI] [PubMed] [Google Scholar]
- Galloway T, Cipelli R, Guralnik J, Ferrucci L, Bandinelli S, Corsi AM, Money C, McCormack P, Melzer D, 2010. Daily bisphenol A excretion and associations with sex hormone concentrations: results from the InCHIANTI adult population study. Environmental health perspectives 118, 1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gárriz Á, Menéndez-Helman RJ, Miranda LA, 2015. Effects of estradiol and ethinylestradiol on sperm quality, fertilization, and embryo–larval survival of pejerrey fish (Odontesthes bonariensis). Aquatic toxicology 167, 191–199. [DOI] [PubMed] [Google Scholar]
- Goldsby JA, Wolstenholme JT, Rissman EF, 2017. Multi- and Transgenerational Consequences of Bisphenol A on Sexually Dimorphic Cell Populations in Mouse Brain. Endocrinology 158, 21–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisterkamp I, Gandrass J, Ruck W, 2004. Bioassay-directed chemical analysis utilizing LC–MS: a tool for identifying estrogenic compounds in water samples? Analytical and bioanalytical chemistry 378, 709–715. [DOI] [PubMed] [Google Scholar]
- Horie Y, Kanazawa N, Takahashi C, Tatarazako N, Iguchi T, 2020. Bisphenol A induces a shift in sex differentiation gene expression with testis‐ova or sex reversal in Japanese medaka (Oryzias latipes). Journal of Applied Toxicology. [DOI] [PubMed] [Google Scholar]
- Iqbal K, Tran DA, Li AX, Warden C, Bai AY, Singh P, Wu X, Pfeifer GP, Szabo PE, 2015. Deleterious effects of endocrine disruptors are corrected in the mammalian germline by epigenome reprogramming. Genome Biol 16, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Bird A, 2003. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature genetics 33, 245–254. [DOI] [PubMed] [Google Scholar]
- Jin X, Jiang G, Huang G, Liu J, Zhou Q, 2004. Determination of 4-tert-octylphenol, 4-nonylphenol and bisphenol A in surface waters from the Haihe River in Tianjin by gas chromatography–mass spectrometry with selected ion monitoring. Chemosphere 56, 1113–1119. [DOI] [PubMed] [Google Scholar]
- Johnson AC, Williams RJ, 2004. A model to estimate influent and effluent concentrations of estradiol, estrone, and ethinylestradiol at sewage treatment works. Environmental science & technology 38, 3649–3658. [DOI] [PubMed] [Google Scholar]
- Kang I, Yokota H, Oshima Y, Tsuruda Y, Oe T, Imada N, Tadokoro H, Honjo T, 2002. Effects of bisphenol A on the reproduction of Japanese medaka (Oryzias latipes). Environmental Toxicology and Chemistry 21, 2394–2400. [PubMed] [Google Scholar]
- Kayo D, Zempo B, Tomihara S, Oka Y, Kanda S, 2019. Gene knockout analysis reveals essentiality of estrogen receptor β1 (Esr2a) for female reproduction in medaka. Scientific reports 9, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd KA, Blanchfield PJ, Mills KH, Palace VP, Evans RE, Lazorchak JM, Flick RW, 2007a. Collapse of a fish population after exposure to a synthetic estrogen. Proceedings of the National Academy of Sciences of the United States of America 104, 8897–8901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd KA, Blanchfield PJ, Mills KH, Palace VP, Evans RE, Lazorchak JM, Flick RW, 2007b. Collapse of a fish population after exposure to a synthetic estrogen. Proceedings of the National Academy of Sciences 104, 8897–8901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL, 2015. HISAT: a fast spliced aligner with low memory requirements. Nature methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL, 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nature biotechnology 37, 907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinch CD, Ibhazehiebo K, Jeong J-H, Habibi HR, Kurrasch DM, 2015. Low-dose exposure to bisphenol A and replacement bisphenol S induces precocious hypothalamic neurogenesis in embryonic zebrafish. Proceedings of the National Academy of Sciences 112, 1475–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita M, Murata K, Naruse K, Tanaka M, 2009. Medaka: biology, management, and experimental protocols. John Wiley & Sons. [Google Scholar]
- Knecht AL, Truong L, Marvel SW, Reif DM, Garcia A, Lu C, Simonich MT, Teeguarden JG, Tanguay RL, 2017. Transgenerational inheritance of neurobehavioral and physiological deficits from developmental exposure to benzo[a]pyrene in zebrafish. Toxicol Appl Pharmacol 329, 148–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK, 2012. VarScan 2: somatic mutation and copy number alteration discovery in c ncer by exome sequencing. Genome research 22, 568–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolpin DW, Furlong ET, Meyer MT, Thurman EM, Zaugg SD, Barber LB, Buxton HT, 2002. Pharmaceuticals, hormones, and other organic wastewater contaminants in US streams, 1999–2000: A national reconnaissance. Environmental science & technology 36, 1202–1211. [DOI] [PubMed] [Google Scholar]
- Kwak HI, Bae MO, Lee MH, Lee YS, Lee BJ, Kang KS, Chae CH, Sung HJ, Shin JS, Kim JH, Mar WC, Sheen YY, Cho MH, 2001. Effects of nonylphenol, bisphenol A, and their mixture on the viviparous swordtail fish (Xiphophorus helleri). Environ Toxicol Chem 20, 787–795. [PubMed] [Google Scholar]
- Laing L, Viana J, Dempster E, Trznadel M, Trunkfield L, Uren Webster T, van Aerle R, Paull G, Wilson R, Mill J, 2016. Bisphenol A causes reproductive toxicity, decreases dnmt1 transcription, and reduces global DNA methylation in breeding zebrafish (Danio rerio). Epigenetics 11, 526–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latendresse JR, Bucci TJ, Olson G, Mellick P, Weis CC, Thorn B, Newbold RR, Delclos KB, 2009. Genistein and ethinyl estradiol dietary exposure in multigenerational and chronic studies induce similar proliferative lesions in mammary gland of male Sprague–Dawley rats. Reproductive toxicology 28, 342–353. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang Y, Tao S, Guan Y, Zhang T, Wang Z, 2016. Global DNA methylation in gonads of adult zebrafish Danio rerio under bisphenol A exposure. Ecotoxicology and environmental safety 130, 124–132. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD, 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lombo M, Fernandez-Diez C, Gonzalez-Rojo S, Navarro C, Robles V, Herraez MP, 2015. Transgenerational inheritance of heart disorders caused by paternal bisphenol A exposure. Environ Pollut 206, 667–678. [DOI] [PubMed] [Google Scholar]
- Loos R, Locoro G, Comero S, Contini S, Schwesig D, Werres F, Balsaa P, Gans O, Weiss S, Blaha L, 2010. Pan-European survey on the occurrence of selected polar organic persistent pollutants in ground water. Water research 44, 4115–4126. [DOI] [PubMed] [Google Scholar]
- Lu H, Cui Y, Jiang L, Ge W, 2017. Functional analysis of nuclear estrogen receptors in zebrafish reprod ction by genome editing approach. Endocrinology 158, 2292–2308. [DOI] [PubMed] [Google Scholar]
- Manikkam M, Tracey R, Guerrero-Bosagna C, Skinner MK, 2013. Plastics derived endocrine disruptors (BPA, DEHP and DBP) induce epigenetic transgenerational inheritance of obesity, reproductive disease and sperm epimutations. PLoS One 8, e55387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoner JR, Kolpin DW, Furlong ET, Cozzarelli IM, Gray JL, Schwab EA, 2014. Contaminants of emerging concern in fresh leachate from landfills in the conterminous United States. Environmental science. Processes & impacts 16, 2335–2354. [DOI] [PubMed] [Google Scholar]
- Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD, 2016. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic acids research 45, D183–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson E, Maamar MB, Skinner MK, 2019. Definition of epigenetic transgenerational inheritance and biological impacts, Transgenerational Epigenetics. Elsevier, pp. 13–24. [Google Scholar]
- Rudel RA, Melly SJ, Geno PW, Sun G, Brody JG, 1998. Identification of alkylphenols and other estrogenic phenolic compounds in wastewater, septage, and groundwater on Cape Cod, Massachusetts. Environmental Science & Technology 32, 861–869. [Google Scholar]
- Santangeli S, Consales C, Pacchierotti F, Habibi HR, Carnevali O, 2019. Transgenerational effects of BPA on female reproduction. Sci Total Environ [DOI] [PubMed] [Google Scholar]
- Santos D, Matos M, Coimbra AM, 2014. Developmental toxicity of endocrine disruptors in early life stages of zebrafish, a genetic and embryogenesis study. Neurotoxicology and teratology 46, 18–25. [DOI] [PubMed] [Google Scholar]
- Schwindt AR, Winkelman DL, Keteles K, Murphy M, Vajda AM, 2014. An environmental oestrogen disrupts fish population dynamics through direct and transgenerational effects on survival and fecundity. Journal of applied ecology 51, 582–591. [Google Scholar]
- Seemann F, Peterson DR, Witten PE, Guo BS, Shanthanagouda AH, Ye RR, Zhang G, Au DW, 2015. Insight into the transgenerational effect of benzo[a]pyrene on bone formation in a teleost fish (Oryzias latipes). Comp Biochem Physiol C Toxicol Pharmacol 178, 60–67. [DOI] [PubMed] [Google Scholar]
- Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, Popp C, Thienpont B, Dean W, Reik W, 2012. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Molecular cell 48, 849–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Lin X, Jin R, Peng T, Peng Z, Fu Z, 2014. Toxic Effects of Bisphenol A on Early Life Stages of Japanese Medaka (Oryzias latipes). Bulletin of Environmental Contamination and Toxicology 93, 222–227. [DOI] [PubMed] [Google Scholar]
- Sun L, Zuo Z, Chen M, Chen Y, Wang C, 2015. Reproductive and transgenerational toxicities of phenanthrene on female marine medaka (Oryzias melastigma). Aquat Toxicol 162, 109–116. [DOI] [PubMed] [Google Scholar]
- Timms BG, Howdeshell KL, Barton L, Bradley S, Richter CA, vom Saal FS, 2005. Estrogenic chemicals in plastic and oral contraceptives disrupt development of the fetal mouse prostate and urethra. Proceedings of the National Academy of Sciences of the United States of America 102, 7014–7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L, 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols 7, 562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG, 2012. Primer3—new capabilities and interfaces. Nucleic acids research 40, e115–e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagnarelli P, 2012. Mitotic chromosome condensation in vertebrates. Experimental Cell Research 318, 1435–1441. [DOI] [PubMed] [Google Scholar]
- Vandenberg LN, Gerona RR, Kannan K, Taylor JA, van Breemen RB, Dickenson CA, Liao C, Yuan Y, Newbold RR, Padmanabhan V, Vom Saal FS, Woodruff TJ, 2014. A round robin approach to the analysis of bisphenol A (BPA) in human blood samples. Environ Health 13, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberg LN, Maffini MV, Sonnenschein C, Rubin BS, Soto AM, 2009. Bisphenol-A and the great divide: a review of controversies in the field of endocrine disruption. Endocrine reviews 30, 75–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkova K, Reyhanian Caspillo N, Porseryd T, Hallgren S, Dinnetz P, Olsen H, Porsch Hallstrom I, 2015. Transgenerational effects of 17alpha-ethinyl estradiol on anxiety behavior in the guppy, Poecilia reticulata. Gen Comp Endocrinol 223, 66–72. [DOI] [PubMed] [Google Scholar]
- vom Saal FS, Nagel SC, Coe BL, Angle BM, Taylor JA, 2012. The estrogenic endocrine disrupting chemical bisphenol A (BPA) and obesity. Molecular and cellular endocrinology 354, 74–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrooman LA, Oatley JM, Griswold JE, Hassold TJ, Hunt PA, 2015. Estrogenic exposure alters the spermatogonial stem cells in the developing testis, permanently reducing crossover levels in the adult. PLoS genetics 11, e1004949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SY, Lau K, Lai KP, Zhang JW, Tse AC, Li JW, Tong Y, Chan TF, Wong CK, Chiu JM, Au DW, Wong AS, Kong RY, Wu RS, 2016. Hypoxia causes transgenerational impairments in reproduction of fish. Nat Commun 7, 12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Bhandari RK, 2019. The dynamics of DNA methylation during epigenetic reprogramming of primordial germ cells in medaka (Oryzias latipes). Epigenetics, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Bhandari RK, 2020a. DNA methylation reprogramming in medaka fish, a promising animal model for environmental epigenetics research. Environmental Epigenetics, In press. [DOI] [PMC free article] [PubMed]
- Wang X, Bhandari RK, 2020b. The dymanics of DNA methylation during epigenetic reprogramming of primordial germ cells in medaka (Oryzias latipes). Epigenetics 15, 483–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hill D, Tillitt DE, Bhandari RK, 2019. Bisphenol A and 17alpha-ethinylestradiol induced transgenerational differences in expression of osmoregulatory genes in the gill of medaka fish (Oryzias latipes). Aquatic Toxicology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welshons WV, Nagel SC, vom Saal FS, 2006. Large effects from small exposures. III. Endocrine mechanisms mediating effects of bisphenol A at levels of human exposure. Endocrinol. 147, S56–S69. [DOI] [PubMed] [Google Scholar]
- Welshons WV, Thayer KA, Judy BM, Taylor JA, Curran EM, vom Saal FS, 2003. Large effects from small exposures. I. Mechanisms for endocrine-disrupting chemicals with estrogenic activity. Environ. Health Perspect. 111, 994–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitacre DM, Fernanda M, Gunther FA, 2012. Reviews of environmental contamination and toxicology. Springer. [Google Scholar]
- Wolstenholme JT, Edwards M, Shetty SR, Gatewood JD, Taylor JA, Rissman EF, Connelly JJ, 2012. Gestational exposure to bisphenol a produces transgenerational changes in behaviors and gene expression. Endocrinology 153, 3828–3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolstenholme JT, Goldsby JA, Rissman EF, 2013. Transgenerational effects of prenatal bisphenol A on social recognition. Horm Behav 64, 833–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F, Wei P, Wang J, Yu M, Zhang X, Tian H, Wang W, Ru S, 2017. Estrogenic effects associated with bisphenol a exposure in male zebrafish (Danio rerio) is associated with changes of endogenous 17beta-estradiol and gene specific DNA methylation levels. Gen Comp Endocrinol 252, 27–35. [DOI] [PubMed] [Google Scholar]
- Zhou L, Charkraborty T, Yu X, Wu L, Liu G, Mohapatra S, Wang D, Nagahama Y, 2012. R-spondins are involved in the ovarian differentiation in a teleost, medaka (Oryzias latipes). BMC developmental biology 12, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


