Abstract
Bacterial skin infections are a major societal health burden and are increasingly difficult to treat due to the emergence of antibiotic resistant strains such as community-acquired methicillin-resistant Staphylococcus aureus. Understanding the immunological mechanisms that provide durable protection against skin infections has the potential to guide the development of immunotherapies and vaccines to engage the host immune response to combat these antibiotic resistant strains. To this end, mouse skin infection models allow researchers to examine host immunity by investigating the timing, inoculum, route of infection and the causative bacterial species in different wildtype mouse backgrounds as well as in knockout, transgenic and other types of genetically engineered mouse strains. To recapitulate the various types of human skin infections, many different mouse models have been developed. For example, four models frequently used in dermatological research are based on route of infection, including: (i) subcutaneous infection models, (ii) intradermal infection models, (iii) wound infection models, and (iv) epicutaneous infection models. In this article, we will describe these skin infection models in detail along with their advantages and limitations. Additionally, we will discuss how humanized mouse models such as the human skin xenograft on immunocompromised mice might be used in bacterial skin infection research.
INTRODUCTION
The skin provides the first line of defense by providing a physical barrier with a low pH and temperature, an abundance of antimicrobial peptides and the normal healthy skin microbiome that protect against microbial invasion. However, when the protective skin barrier is damaged, breached, or develops a microbial dysbiosis, skin infections can arise. Staphylococcus aureus (S. aureus) is the leading cause of skin and soft tissue infections (SSTIs) in the U.S. (Dantes et al., 2013, Suaya et al., 2014). With the emergence of antibiotic-resistant bacterial clinical isolates such as community-acquired methicillin-resistant S. aureus (CA-MRSA), it is critical to understand the host immune responses that promote bacterial clearance in order to develop non-antibiotic immune-based therapies to prevent and/or treat skin infections. To investigate these immunological processes, mouse models that mimic various human skin infections have been developed and have been instrumental in identifying novel immunotherapeutic targets. This review will discuss different mouse skin infection models along with a human skin xenograft model, and the advantages and limitations of each model. Although we will focus on S. aureus and other bacterial pathogens to describe each mouse skin infection model, these models do not exclude or may not be representative of fungal, parasitic, or viral skin infections.
Mouse Models of Skin infection
Mouse skin infection models can be categorized into four groups based on the depth of infection: (i) subcutaneous infection in which the bacteria are inoculated below the dermis; (ii) intradermal infection in which the bacteria are inoculated into the dermis; (iii) wound infection in which the bacteria are inoculated into full-thickness incisional or excisional wounds; and (iv) epicutaneous infection in which the surface of the skin is exposed to the bacterial inoculum (Figure 1). These four models will be described in the context of S. aureus skin infections. Lastly, we will discuss the potential for translational studies with human skin xenografts. Understanding the strengths and weaknesses of each model will help provide key insights into which system is most appropriate to study specific immunologic responses as summarized in Table 1.
Figure 1. Graphical and photographic representations of bacterial skin infection models.
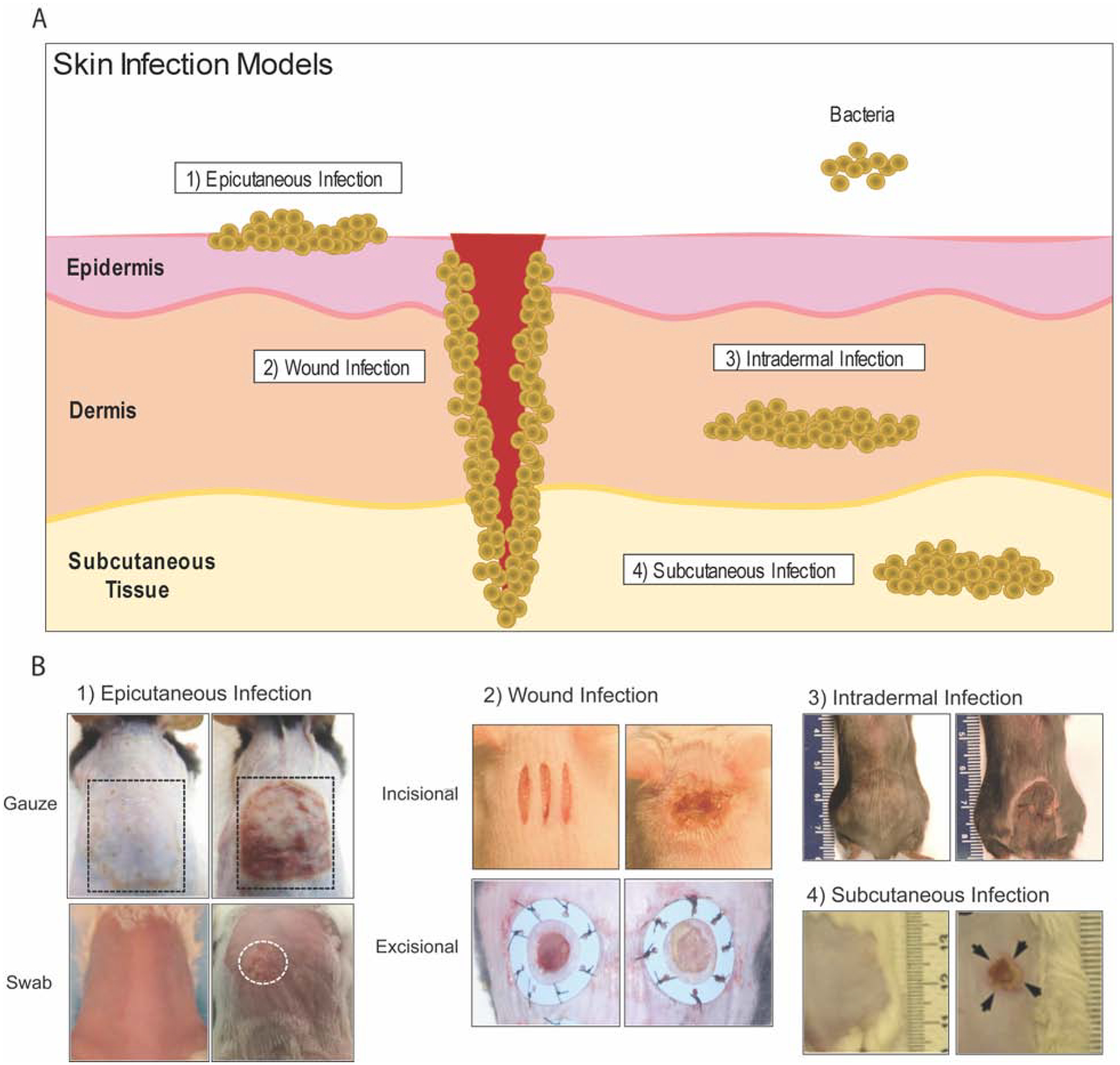
(A) Graphical representation of mouse skin infection models as defined by the depth of infection in the skin. (B) Representative clinical photographs of each of the following skin infection models (left panel: control; right panel: experimental): (1) epicutaneous infection where bacteria was inoculated on the surface of intact skin by applying a gauze soaked with bacteria or swabbing (Dai et al., 2011, Malhotra et al., 2016, Williams et al., 2019), (2) wound infection where S. aureus was inoculated on a full-thickness skin incisional or splint-sutured excisional wound (Archer et al., 2020, Morimoto et al., 2014), (3) intradermal infection model where S. aureus was inoculated into the dermis of the dorsal skin and developed dermonecrosis (Liu et al., 2017), (4) subcutaneous infection where S. aureus was inoculated into the subcutaneous tissue, which lead to dermonecrosis and muscle necrosis (Tseng et al., 2011).
Table 1.
Summary of mouse skin infection models for immunity research
| Type | Human Relevance | Model Description | Limitations | References |
|---|---|---|---|---|
| Subcutaneous model |
|
|
|
(Berube et al., 2014, Jeong et al., 2019, Nippe et al., 2011, Tseng et al., 2011) |
| Intradermal model |
|
|
|
(Asai et al., 2010, Brown et al., 2009, Dillen et al., 2018) |
| Wound models | ||||
| • Incisional Wound |
|
|
|
(Guo et al., 2013, Ortines et al., 2018, Zolfaghari et al., 2009) |
| • Excisional Wound |
|
|
|
(Fila et al., 2016, Shi et al., 2007) |
| Epicutaneous models | ||||
| • Gauze Infection |
|
|
|
(Liu et al., 2017, Nakatsuji et al., 2016, Williams et al., 2019) |
| • Swab Infection |
|
|
|
(Kugelberg et al., 2005, Linehan et al., 2018, Malhotra et al., 2016, Pastagia et al., 2011) |
| • Human Skin Xenograft |
|
|
|
(Schulz et al., 2019) |
1. Subcutaneous Infection Models
The subcutaneous infection model mimics more invasive infections, such as subcutaneous abscesses and cellulitis (McCaig et al., 2006, Miller et al., 2005). Upon subcutaneous inoculation of S. aureus into the backs of mice, a deep abscess comprised of neutrophils forms around that bacteria. This abscess typically forms below the panniculus carnosus muscle in the deep dermis that primarily involves the subcutaneous fat above the deeper muscle layers (Liese et al., 2013, Tseng et al., 2011). Thus, this model has been widely used to elucidate immune mechanisms against deep soft tissue infections with various bacterial species such as S. aureus and S. pyogenes (Medina, 2010, Tseng et al., 2009). For instance, by subcutaneously inoculating different wild-type mouse strains with S. aureus, it was seen that resistance to the bacterial infection was associated with increasing number of infiltrating neutrophils at the site of infection (Nippe et al., 2011). The subcutaneous infection model has also been used to elucidate the role of antimicrobial peptides (AMPs) during S. aureus skin infections. The activities of hBD3 and LL-37 was shown to be essential for controlling subcutaneous skin infections by promoting the killing of S. aureus by either maintaining the anti-staphylococcal environment or permeabilizing the bacterial membrane, respectively (Cheung et al., 2018). Additionally, the subcutaneous model was used to discover an unexpected role for adipocyte-derived LL-37 in the control of S. aureus infection (Zhang et al., 2015). This model can also be used to investigate durable immune responses that protect the host from recurrent infections. For example, re-infected mice showed innate immune memory (e.g., trained memory) of macrophages against a recurrent S. aureus subcutaneous infection (Chan et al., 2018). However, obtaining the muscle lesion size, which is a common readout for the subcutaneous infection model, involves a more invasive procedure that requires sacrificing the mice (Tseng et al., 2011).
2. Intradermal Skin Infection Models
The intradermal skin infection model also recapitulates the hallmarks of human S. aureus skin infections, including dermonecrotic lesions and neutrophilic skin abscesses, which corresponds to the progression and severity of the infection (Asai et al., 2010, Mölne et al., 2000). In addition, bioluminescent bacterial strains and in vivo optical imaging systems can be used in conjunction to noninvasively and longitudinally monitor the dynamics of the bacterial infection (Miller et al., 2006). Genetically engineered mouse strains are also useful to study components of the host response required for protection against skin infections. For example, the critical role of the inflammasome and IL-1β/IL-1R signaling in promoting neutrophil recruitment and host defense against S. aureus skin infections was uncovered using mice deficient in ASC (apoptosis-associated speck-like protein containing a C-terminal caspase recruitment domain), IL-1β or IL-1R as well as IL-1β-DsRed reporter mice (Cho et al., 2012, Miller et al., 2006, Miller et al., 2007). In addition, mice lacking γδ T cells exhibited significant host defense defects due to impaired IL-17 production (Cho et al., 2010). Transgenic reporter mouse strains, such as the IL-17A-tdTomato/IL-17F-GFP dual-color reporter mice, can provide insights into the expression kinetics and relevant expressing cell types of host-derived cytokines that are important for protection against intradermal S. aureus infections (Marchitto et al., 2019). The intradermal model can also be modified to investigate the mechanisms of immunological memory by re-infecting mice at a different skin site from the original intradermal infection (Gaidamakova et al., 2012, Montgomery et al., 2014, Sampedro et al., 2014). Remarkably, using a S. aureus intradermal skin re-infection model, a clonal population of γδ T cells was found to expand in the draining lymph nodes and traffic to the site of infection to confer protection against a secondary S. aureus intradermal infection (Dillen et al., 2018). Despite the widespread use of S. aureus intradermal models of infection, inherent biological differences between mice and humans need to be considered such as the activity of specific S. aureus toxins that are highly active against human but not mouse cells, especially superantigens such as toxic shock syndrome toxin-1 (TSST1) (Salgado-Pabón and Schlievert, 2014). To overcome this limitation, humanized mice that express the human receptors (e.g., HLA-DR4 knock-in mice) targeted by these S. aureus toxins have been developed in which TSST1 has superantigen activity (Xu et al., 2014). Mouse immune cells are also less sensitive to the cytolytic activity of Panton-Valentine leukocidin (PVL) and α-hemolysin (Hongo et al., 2009, Spaan et al., 2013, Tseng et al., 2015). However, whereas α-hemolysin mainly has cytolytic activity against leukocytes and development of large purulent abscesses in humans, it induces keratinocyte cell death in mice that manifests as large dermonecrotic lesions (Kennedy et al., 2010). Additional interrogation of the pathophysiology and immunological responses can be done by histological, flow cytometric, or RNA/protein analyses to verify the relevance and validity phenotypic observations (Marchitto et al., 2019). Importantly, mice have an abundant population of γδ T cells called dendritic epidermal T cells (DETCs), which is not present in human epidermis (albeit 1–10% of resident T cells in the dermis of human skin are γδ T cells) (Nielsen et al., 2017). Mice also have more subsets of γδ T cells that reside at different layers of the skin with both conserved and distinct physiological functions as those in humans (Girardi, 2006, Suwanpradid et al., 2017). In addition to the differences in skin resident immune cells, there are other general immunological differences between the two species that could lead to discrepancies in infection outcomes between mouse and humans (McGovern et al., 2014). In humans, neutrophils make up the majority of circulating leukocytes whereas lymphocytes exist in higher percentages in mice (Mestas and Hughes, 2004). Furthermore, differences in hair follicles also contribute to differential protective mechanisms in mouse and human skin by influencing the accessibility, mobility, and communication of epithelial cells that initiate innate immune response against foreign pathogens (Al-Nuaimi et al., 2010, Bekeredjian-Ding et al., 2017, Oh et al., 2016). Therefore, it is important to consider a broad spectrum of differences between mouse and human when performing skin infection models.
3. Wound Infection Models
S. aureus is the most common pathogen isolated from infected skin wounds, with diabetic patients being particularly susceptible to the development of chronic, non-healing wounds (Dunyach-Remy et al., 2016, Giurato et al., 2017, Tong et al., 2015). Pseudomonas aeruginosa is another invasive bacterial species commonly found in wounds that causes severe tissue damage (Mutluoglu and Uzun, 2011, Sivanmaliappan and Sevanan, 2011). Mouse wound infection models replicate multiple features of infected human wounds such as purulent drainage, necrotic debris and delayed wound healing. The mouse wound infection model is performed by inoculating bacteria into full-thickness incisional cuts or excisional wounds (Dai et al., 2011). For example, incisional wounds can be inoculated with a bioluminescent S. aureus strain in Lysozyme M-EGFP (LysM-EGFP) reporter mice to longitudinally monitor both the bacterial burden and neutrophil recruitment dynamics during the course of infection and wound healing (Figure 2A–D) (Anderson et al., 2019, Kim et al., 2008). Furthermore, histological analysis of the infected wound skin can be used to analyze neutrophil abscess area, bacterial band width, and the presence of specific cells (Figure 2E) (Cho et al., 2011). The benefits of these different wound infection models include the ability to replicate polymicrobial infections that typically occur in human wounds (Dalton et al., 2011, Pastar et al., 2013). By infecting the wounds of diabetic mice with polymicrobial isolates from human diabetic foot ulcers, Kalan et al. were able to correlate strain-specific S. aureus phenotypes in mice with patient outcomes (Kalan et al., 2019). With the availability of a new strain of bioluminescent S. aureus expressing click beetle red luciferase and a P. aeruginosa lux strain, it is now possible to longitudinally and noninvasively monitor the dynamics of each bacterial strain in the context of wound infection (Miller et al., 2019). Additionally, various different strains of genetically-engineered diabetic mice exist that exhibit impaired host defense against S. aureus wound infections, similar to human diabetics (Guo et al., 2013, Ortines et al., 2018). It is important to consider the route and depth of infection in the skin as these can affect the immunological processes involved. For instance, both IL-1α and IL-1β were found to be involved in neutrophil recruitment and immunity against a S. aureus wound infection, while IL-1β played a more predominant role against an intradermal S. aureus infection (Cho et al., 2011, Yan et al., 2016). Different cellular composition between mouse and human skin may lead to challenges in translating findings in mouse wound infection models. Unlike human skin, mouse skin is highly populated with DETCs, which are responsible for sensing skin injury and producing IL-17A to promote wound healing and to strengthen skin barrier function (MacLeod et al., 2013). Another limitation to this model is that wound contraction is much more pronounced in mouse skin than human skin. Some groups have tried to overcome this limitation by covering the wound bed with a transparent breathable film (that also keeps the wound open longer) or suturing a splint to prevent wound contracture (Griffin et al., 2015).
Figure 2. S. aureus skin infection in vivo imaging and histology.
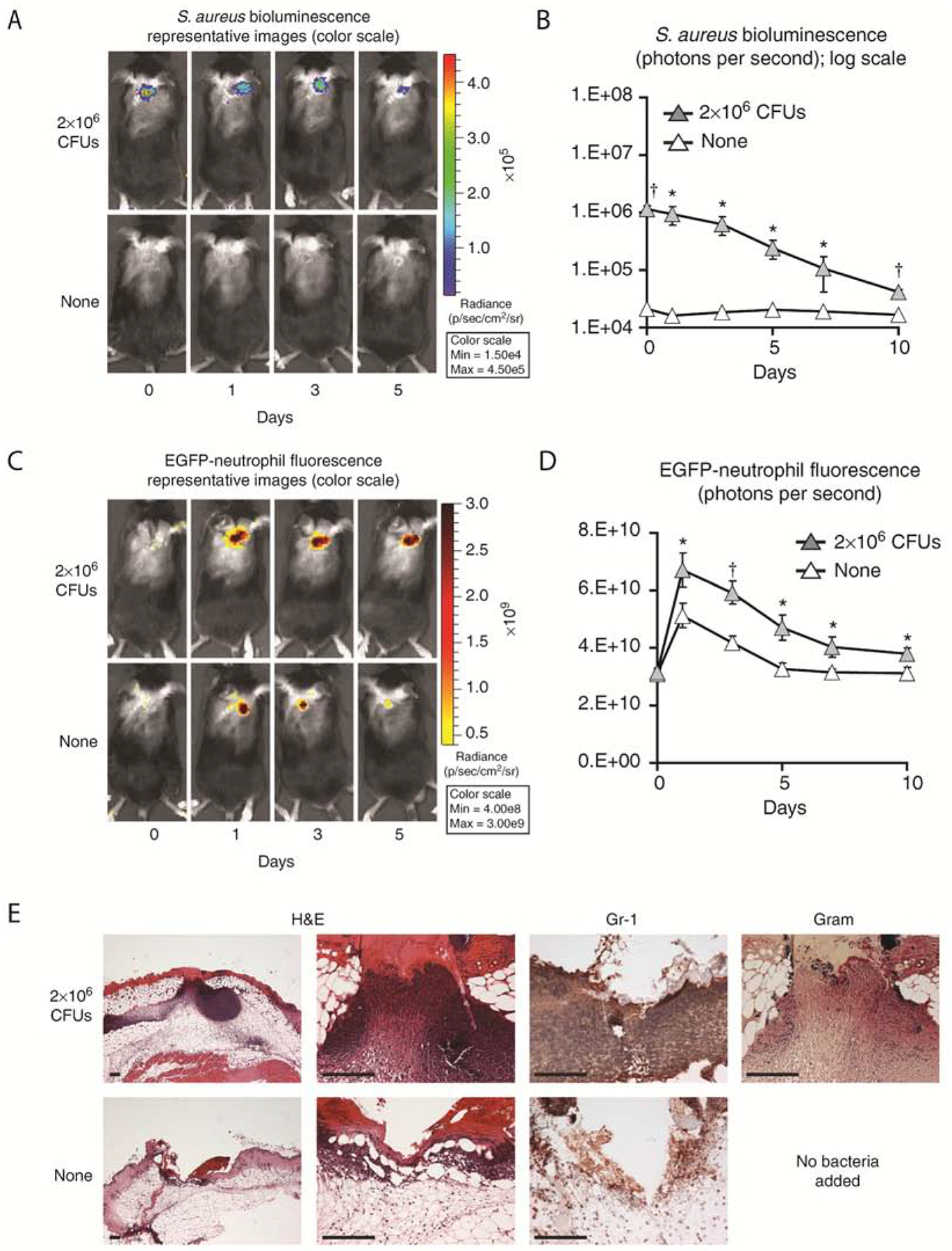
Three 8-mm in length, parallel scalpel wounds on the backs of (A-D) LysM-EGFP mice or (E) C57BL/6 mice inoculated with 2 × 106 colony-forming units (CFUs) per 10 μl of Staphylococcus aureus or no bacteria (none). (A) Representative photographs of in vivo S. aureus bioluminescence. (B) In vivo S. aureus burden as measured by in vivo bioluminescence imaging (mean total flux (photons per second) ± SEM) (logarithmic scale). (C) Representative photographs of in vivo EGFP-neutrophil fluorescence. (D) In vivo fluorescence imaging of EGFP-neutrophil infiltration (mean total flux (photons per second) ± SEM). (E) Representative photomicrographs of sections from skin punch biopsies at 1 day after wounding ± S. aureus infection labeled with hematoxylin and eosin (H&E) stain, anti-Gr-1 mAb (neutrophil marker), and Gram stain. Scale bars = 150 μm. This figure was derived from (Cho et al., 2011).
4. Epicutaneous Infection Models
S. aureus commonly colonizes the lesional skin of human atopic dermatitis (AD) patients and the level of colonization correlates with disease severity (Byrd et al., 2017, Kong et al., 2012). Patients with hyper-immunoglobulin E syndrome, which is often due to a dominant negative mutation in STAT3 gene, have been characterized by atopic manifestations and higher susceptibility to S. aureus and/or Candida cutaneous infections as a result of impaired Th17 development (Horváth et al., 2011, Milner et al., 2008). These clinical observations have caused intense interest in understanding the role of S. aureus in the immune pathogenesis of AD skin inflammation. To model this, a S. aureus-soaked gauze pad is applied to the shaved and depilated dorsal skin of mice. The erythematous skin inflammation that mimic human AD conditions in mice can be measured by disease scoring, epidermal thickening, and elevated serum IgE, while the increased skin barrier defect can be measured through transepidermal water loss (TEWL) (Alexander et al., 2018, Nakamura et al., 2013). The use of mouse genetic cre/lox systems provide another important tool for researchers to identify the cells involved in immune responses by targeting gene deletion in a specific cell type. For example, a cre/lox mouse with T cell specific deletion of MyD88 was used to uncover a novel role for IL-36-mediated IL-17 T cell responses in epicutaneous S. aureus-driven skin inflammation (Liu et al., 2017). Tape stripping of the skin can be performed prior to epicutaneous skin infection to recapitulate the barrier defect seen in AD skin. In this model, S. aureus -derived proteases and phenol-soluble modulin alpha (PSMα), which are under the regulation of the bacteria’s quorum sensing system, promoted skin inflammation by inducing epidermal proteolysis and skin barrier damage (Williams et al., 2019). Similarly, S. aureus was shown to exploit the barrier defect in filaggrin-deficient (ft/ft) mice to promote Th2 and Th22 cytokines that are associated with exacerbation of AD skin inflammation (Nakatsuji et al., 2016). Exploitation of skin barrier defects is not limited to S. aureus, but also vaccinia virus, which is the cause of a life-threatening condition called eczema vaccinatum in AD patients. Furthermore, cutaneous exposure to vaccinia virus in ft/ft mice through scarification, which recapitulates the route of exposure during smallpox vaccination in humans, showed IL-17A mediated dissemination of the virus in the skin (Oyoshi et al., 2015). Therefore, the epicutaneous infection model is useful in investigating the host and pathogen-derived factors that contribute to AD-like skin inflammation and AD-associated complications. Nonetheless, some of these models have used depilatory creams that result in baseline skin inflammation. Moreover, the models that wrap a pathogen-soaked gauze pad around the mouse to artificially expose the mouse skin to the pathogen of interest does not truly recapitulate the normal S. aureus colonization of uncovered skin in AD patients.
To investigate the cross-talk between the skin microbiome and host immune cells, an alternate epicutaneous infection model has been developed where a bacteria-soaked cotton swab is rubbed onto the shaved backs of mice (Belkaid and Segre, 2014, Kugelberg et al., 2005). This model was instrumental in understanding how skin discriminates between commensal and pathogenic skin microbes. In particular, the commensal S. epidermidis promoted T regulatory cell (Treg) expansion and skin immune tolerance in a crucial window in neonatal life (Scharschmidt et al., 2015). However, S. aureus manipulated IL-1β release to inhibit Treg expansion and induce skin inflammation (Leech et al., 2019). Furthermore, the model has been used to understand how the commensal bacterial strain S. epidermis promotes protection against pathogens as well as accelerate wound healing (Linehan et al., 2018). On the other hand, epicutaneous inoculation with Corynebacterium accolens promoted skin inflammation through activation of long-lasting skin T cells (Ridaura et al., 2018). Additionally, isolated S. aureus strains colonizing human AD induced more skin inflammation than laboratory strains isolated from other body sites (Byrd et al., 2017). Alternatively, Candida albicans was applied to the skin to interrogate a role for cutaneous sensory neurons in host defense (Kashem et al., 2015). Despite the usefulness of the swab epicutaneous model, it generally is done with multiple bacterial (or fungal) applications that might not fully replicate the normal colonization of commensal microbes on human skin.
Human Skin Xenograft Model
Given the inherent differences between human and mouse skin, human skin xenografts can be used to validate and translate the findings in mouse models to human skin (Parker, 2017). To prevent graft rejection, human skin biopsies are sutured onto immunodeficient mice that include NSG (NOD.Cg-Prkdcscid IL2rgtm1Wjl), NOG (NOD.cg-Prkdcscid IL2rgtm1Sug), and NRG (NOD.Cg-Rag1tm1Mom IL2rgtm1Wj) mice, all of which lack T, B and NK cells (Kenney et al., 2016). Moreover, it is possible to perform human skin xenografts in combination with engraftment of CD34+ stem cells (allowing the development of human immune cells in the same mice) to provide the new in vivo capability to study the human immune system in the context of a human skin infection (Brehm et al., 2012). There are numerous advantages for the use of human skin, including healthy samples that are readily available and engrafted skin tissue with epidermal and dermal layers and vascularized skin that closely resembles normal human skin. For example, Soong et al. demonstrated toxin-deficient, agr mutants of S. aureus are able to persist on the human skin by stimulating autophagy (Soong et al., 2015). In addition, epicutaneously swabbed S. aureus on human skin xenografts led to local production of IL-8, which induced neutrophil migration to the skin to promote bacterial clearance (Schulz et al., 2019). Studies involving human skin xenograft infections are not widely used, and thus represent an exciting opportunity in the dermatology field to translate the immunological findings from mouse skin infection models to human skin.
CONCLUSION
Mouse models of skin infection remain the most commonly used model of skin infections due to their relatively inexpensive experimental costs as well as the opportunity to take advantage of genetically engineered mice and in vivo optical imaging techniques. Currently, a great variety of skin infection models and genetically engineered mice are readily available, which serve as extremely valuable tools for noninvasive and longitudinal monitoring of the underlying immune responses and host-pathogen interactions that occur during skin infections. Mouse skin infection models will continue to be essential for better understanding skin immunological responses in different contexts, including skin colonization, impetiginization, abscesses and wounds as well as in the setting of diseases such as atopic dermatitis and diabetes. Unfortunately, mouse models cannot completely replicate the pathogenesis of human disease. Therefore, these limitations need to be considered when translating the results to cutaneous immune responses in human skin (summarized in Table 1). Further advancements in humanized skin xenografts in immunocompromised mice are continually being developed to help validate and improve the discrepancies between the species.
MULTIPLE CHOICE QUESTIONS
- Which of the following would be the most immunologically relevant purpose to re-infect mice in a skin infection model?
- To study primary T cell responses to skin infection.
- To examine memory T cell responses to skin infection.
- To study innate immune responses during initial skin infection.
- To study polymicrobial infections.
Answer: B. To examine memory T cell responses to skin infection
- Which of the genetically engineered mouse strains can be used to monitor cytokine expression kinetics during skin infections?
- IL-17A/F KO mouse
- Mouse with specific IL-17A/F deletion in T cells
- IL-17A-tdTomato/IL-17F-GFP dual-color reporter mice
- All of the above
Answer: C. IL-17A-tdTomato/IL-17F-GFP dual-color reporter mice
- Which of the following skin infection models has the potential to be used with human skin xenografts?
- Epicutaneous model
- Intradermal model
- Wound model
- All of the above
Answer: D. All of the above
- The epicutaneous skin infection model replicates which type of skin inflammation?
- Atopic dermatitis
- Psoriasis
- Vitiligo
- Alopecia areata
Answer: A. Atopic dermatitis
- Which bacteria is the leading cause of skin infections in humans?
- Staphylococcus epidermidis
- Pseudomonas aeruginosa
- Staphylococcus aureus
- Corynebacterium accolens
Answer: C. Staphylococcus aureus
Detailed Answer: Memory T cells are involved in the secondary response to skin infection.
Detailed Answer: The IL-17A-tdTomato/IL-17F-GFP dual-color reporter mouse allows for in vivo visualization of IL-17A and IL-17F with an In Vivo Imaging System.
Detailed Answer: Human skin xenografts can be adapted to work with any of the skin infection models.
Detailed Answer: The epicutaneous model replicates S. aureus colonization and skin inflammation on atopic dermatitis skin.
Detailed Answer: S. aureus is the leading cause of skin and soft tissue infections in humans.
SUMMARY.
Advantages
Mouse skin infection models are powerful tools to elucidate immune mechanisms of protection and identify therapeutic targets against skin infections.
Human skin xenografts on immunocompromised mice provide the potential to validate findings from mouse infection models in human skin.
Limitations
Immune responses can differ against the same infectious agent depending on the skin infection model used and should be verified in each model separately.
There are inherent immunological and physiological differences between mouse and human skin.
ACKNOWLEDGMENTS
We apologize to the many researchers in the field whose work could not be included due to space and citation limitations by the journal for this research techniques made simple manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST
L.S.M. is a full-time employee at Janssen Research and Development and may own Johnson & Johnson stock and stock options. L.S.M. has received grant support from AstraZeneca, MedImmune (a subsidiary of AstraZeneca), Pfizer, Boerhinger Ingelheim, Regeneron Pharmaceuticals, and Moderna Therapeutics, is a shareholder of Noveome Biotherapeutics, was a paid consultant for Armirall and Janssen Research and Development and was on the scientific advisory board of Integrated Biotherapeutics, which are all developing therapeutics against infections (including S. aureus and other pathogens) and/or inflammatory conditions. N.K.A. has received grant support from Pfizer.
REFERENCES
- Al-Nuaimi Y, Baier G, Watson RE, Chuong CM, Paus R. The cycling hair follicle as an ideal systems biology research model. Exp Dermatol 2010;19(8):707–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander H, Brown S, Danby S, Flohr C. Research Techniques Made Simple: Transepidermal Water Loss Measurement as a Research Tool. J Invest Dermatol 2018;138(11):2295–300.e1. [DOI] [PubMed] [Google Scholar]
- Anderson LS, Reynolds MB, Rivara KR, Miller LS, Simon SI. A Mouse Model to Assess Innate Immune Response to Staphylococcus aureus Infection. J Vis Exp 2019(144). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer NK, Wang Y, Ortines RV, Liu H, Nolan SJ, Liu Q, et al. Preclinical Models and Methodologies for Monitoring Staphylococcus aureus Infections Using Noninvasive Optical Imaging. Methods Mol Biol 2020;2069:197–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai A, Tsuda Y, Kobayashi M, Hanafusa T, Herndon DN, Suzuki F. Pathogenic role of macrophages in intradermal infection of methicillin-resistant Staphylococcus aureus in thermally injured mice. Infect Immun 2010;78(10):4311–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekeredjian-Ding I, Stein C, Uebele J. The Innate Immune Response Against Staphylococcus aureus. Curr Top Microbiol Immunol 2017;409:385–418. [DOI] [PubMed] [Google Scholar]
- Belkaid Y, Segre JA. Dialogue between skin microbiota and immunity. Science 2014;346(6212):954–9. [DOI] [PubMed] [Google Scholar]
- Berube BJ, Sampedro GR, Otto M, Bubeck Wardenburg J. The psmα locus regulates production of Staphylococcus aureus alpha-toxin during infection. Infect Immun 2014;82(8):3350–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm MA, Racki WJ, Leif J, Burzenski L, Hosur V, Wetmore A, et al. Engraftment of human HSCs in nonirradiated newborn NOD-scid IL2rγ null mice is enhanced by transgenic expression of membrane-bound human SCF. Blood 2012;119(12):2778–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EL, Dumitrescu O, Thomas D, Badiou C, Koers EM, Choudhury P, et al. The Panton-Valentine leukocidin vaccine protects mice against lung and skin infections caused by Staphylococcus aureus USA300. Clin Microbiol Infect 2009;15(2):156–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd AL, Deming C, Cassidy SKB, Harrison OJ, Ng WI, Conlan S, et al. Staphylococcus aureus and Staphylococcus epidermidis strain diversity underlying pediatric atopic dermatitis. Sci Transl Med 2017;9(397). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan LC, Rossetti M, Miller LS, Filler SG, Johnson CW, Lee HK, et al. Protective immunity in recurrent Staphylococcus aureus infection reflects localized immune signatures and macrophage-conferred memory. Proc Natl Acad Sci U S A 2018;115(47):E11111–E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JS, Guo Y, Ramos RI, Hebroni F, Plaisier SB, Xuan C, et al. Neutrophil-derived IL-1β is sufficient for abscess formation in immunity against Staphylococcus aureus in mice. PLoS Pathog 2012;8(11):e1003047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JS, Pietras EM, Garcia NC, Ramos RI, Farzam DM, Monroe HR, et al. IL-17 is essential for host defense against cutaneous Staphylococcus aureus infection in mice. J Clin Invest 2010;120(5):1762–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JS, Zussman J, Donegan NP, Ramos RI, Garcia NC, Uslan DZ, et al. Noninvasive in vivo imaging to evaluate immune responses and antimicrobial therapy against Staphylococcus aureus and USA300 MRSA skin infections. J Invest Dermatol 2011;131(4):907–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai T, Kharkwal GB, Tanaka M, Huang YY, Bil de Arce VJ, Hamblin MR. Animal models of external traumatic wound infections. Virulence 2011;2(4):296–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton T, Dowd SE, Wolcott RD, Sun Y, Watters C, Griswold JA, et al. An in vivo polymicrobial biofilm wound infection model to study interspecies interactions. PLoS One 2011;6(11):e27317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantes R, Mu Y, Belflower R, Aragon D, Dumyati G, Harrison LH, et al. National burden of invasive methicillin-resistant Staphylococcus aureus infections, United States, 2011. JAMA Intern Med 2013;173(21):1970–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillen CA, Pinsker BL, Marusina AI, Merleev AA, Farber ON, Liu H, et al. Clonally expanded γδ T cells protect against Staphylococcus aureus skin reinfection. J Clin Invest 2018;128(3):1026–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunyach-Remy C, Ngba Essebe C, Sotto A, Lavigne JP. Staphylococcus aureus Toxins and Diabetic Foot Ulcers: Role in Pathogenesis and Interest in Diagnosis. Toxins (Basel) 2016;8(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fila G, Kasimova K, Arenas Y, Nakonieczna J, Grinholc M, Bielawski KP, et al. Murine Model Imitating Chronic Wound Infections for Evaluation of Antimicrobial Photodynamic Therapy Efficacy. Front Microbiol 2016;7:1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidamakova EK, Myles IA, McDaniel DP, Fowler CJ, Valdez PA, Naik S, et al. Preserving immunogenicity of lethally irradiated viral and bacterial vaccine epitopes using a radio- protective Mn2+-Peptide complex from Deinococcus. Cell Host Microbe 2012;12(1):117–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardi M. Immunosurveillance and immunoregulation by gammadelta T cells. J Invest Dermatol 2006;126(1):25–31. [DOI] [PubMed] [Google Scholar]
- Giurato L, Meloni M, Izzo V, Uccioli L. Osteomyelitis in diabetic foot: A comprehensive overview. World J Diabetes 2017;8(4):135–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin DR, Weaver WM, Scumpia PO, Di Carlo D, Segura T. Accelerated wound healing by injectable microporous gel scaffolds assembled from annealed building blocks. Nat Mater 2015;14(7):737–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Ramos RI, Cho JS, Donegan NP, Cheung AL, Miller LS. In vivo bioluminescence imaging to evaluate systemic and topical antibiotics against community-acquired methicillin-resistant Staphylococcus aureus-infected skin wounds in mice. Antimicrob Agents Chemother 2013;57(2):855–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongo I, Baba T, Oishi K, Morimoto Y, Ito T, Hiramatsu K. Phenol-soluble modulin alpha 3 enhances the human neutrophil lysis mediated by Panton-Valentine leukocidin. J Infect Dis 2009;200(5):715–23. [DOI] [PubMed] [Google Scholar]
- Horváth R, Rožková D, Lašťovička J, Poloučková A, Sedláček P, Sedivá A, et al. Expansion of T helper type 17 lymphocytes in patients with chronic granulomatous disease. Clin Exp Immunol 2011;166(1):26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S, Kim HY, Kim AR, Yun CH, Han SH. Propionate Ameliorates Staphylococcus aureus Skin Infection by Attenuating Bacterial Growth. Front Microbiol 2019;10:1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalan LR, Meisel JS, Loesche MA, Horwinski J, Soaita I, Chen X, et al. Strain- and Species-Level Variation in the Microbiome of Diabetic Wounds Is Associated with Clinical Outcomes and Therapeutic Efficacy. Cell Host Microbe 2019;25(5):641–55.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashem SW, Riedl MS, Yao C, Honda CN, Vulchanova L, Kaplan DH. Nociceptive Sensory Fibers Drive Interleukin-23 Production from CD301b+ Dermal Dendritic Cells and Drive Protective Cutaneous Immunity. Immunity 2015;43(3):515–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney LL, Shultz LD, Greiner DL, Brehm MA. Humanized Mouse Models for Transplant Immunology. Am J Transplant 2016;16(2):389–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MH, Liu W, Borjesson DL, Curry FR, Miller LS, Cheung AL, et al. Dynamics of neutrophil infiltration during cutaneous wound healing and infection using fluorescence imaging. J Invest Dermatol 2008;128(7):1812–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res 2012;22(5):850–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugelberg E, Norström T, Petersen TK, Duvold T, Andersson DI, Hughes D. Establishment of a superficial skin infection model in mice by using Staphylococcus aureus and Streptococcus pyogenes. Antimicrob Agents Chemother 2005;49(8):3435–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leech JM, Dhariwala MO, Lowe MM, Chu K, Merana GR, Cornuot C, et al. Toxin-Triggered Interleukin-1 Receptor Signaling Enables Early-Life Discrimination of Pathogenic versus Commensal Skin Bacteria. Cell Host Microbe 2019;26(6):795–809.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liese J, Rooijakkers SH, van Strijp JA, Novick RP, Dustin ML. Intravital two-photon microscopy of host-pathogen interactions in a mouse model of Staphylococcus aureus skin abscess formation. Cell Microbiol 2013;15(6):891–909. [DOI] [PubMed] [Google Scholar]
- Linehan JL, Harrison OJ, Han SJ, Byrd AL, Vujkovic-Cvijin I, Villarino AV, et al. Non-classical Immunity Controls Microbiota Impact on Skin Immunity and Tissue Repair. Cell 2018;172(4):784–96.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Archer NK, Dillen CA, Wang Y, Ashbaugh AG, Ortines RV, et al. Staphylococcus aureus Epicutaneous Exposure Drives Skin Inflammation via IL-36-Mediated T Cell Responses. Cell Host Microbe 2017;22(5):653–66.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod AS, Hemmers S, Garijo O, Chabod M, Mowen K, Witherden DA, et al. Dendritic epidermal T cells regulate skin antimicrobial barrier function. J Clin Invest 2013;123(10):4364–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra N, Yoon J, Leyva-Castillo JM, Galand C, Archer N, Miller LS, et al. IL-22 derived from γδ T cells restricts Staphylococcus aureus infection of mechanically injured skin. J Allergy Clin Immunol 2016;138(4):1098–107.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchitto MC, Dillen CA, Liu H, Miller RJ, Archer NK, Ortines RV, et al. Clonal Vγ6 + Vδ4 + T cells promote IL-17–mediated immunity against Staphylococcus aureus skin infection. Proc Natl Acad Sci U S A 2019;116(22):10917–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaig LF, McDonald LC, Mandal S, Jernigan DB. Staphylococcus aureus-associated skin and soft tissue infections in ambulatory care. Emerg Infect Dis 2006;12(11):1715–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern N, Schlitzer A, Gunawan M, Jardine L, Shin A, Poyner E, et al. Human dermal CD14+ cells are a transient population of monocyte-derived macrophages. Immunity 2014;41(3):465–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina E. Murine model of cutaneous infection with Streptococcus pyogenes. Methods Mol Biol 2010;602:395–403. [DOI] [PubMed] [Google Scholar]
- Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. J Immunol 2004;172(5):2731–8. [DOI] [PubMed] [Google Scholar]
- Miller LG, Perdreau-Remington F, Rieg G, Mehdi S, Perlroth J, Bayer AS, et al. Necrotizing fasciitis caused by community-associated methicillin-resistant Staphylococcus aureus in Los Angeles. N Engl J Med 2005;352(14):1445–53. [DOI] [PubMed] [Google Scholar]
- Miller LS, O’Connell RM, Gutierrez MA, Pietras EM, Shahangian A, Gross CE, et al. MyD88 mediates neutrophil recruitment initiated by IL-1R but not TLR2 activation in immunity against Staphylococcus aureus. Immunity 2006;24(1):79–91. [DOI] [PubMed] [Google Scholar]
- Miller LS, Pietras EM, Uricchio LH, Hirano K, Rao S, Lin H, et al. Inflammasome-mediated production of IL-1beta is required for neutrophil recruitment against Staphylococcus aureus in vivo. J Immunol 2007;179(10):6933–42. [DOI] [PubMed] [Google Scholar]
- Miller RJ, Crosby HA, Schilcher K, Wang Y, Ortines RV, Mazhar M, et al. Development of a Staphylococcus aureus reporter strain with click beetle red luciferase for enhanced in vivo imaging of experimental bacteremia and mixed infections. Sci Rep 2019;9(1):16663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner JD, Brenchley JM, Laurence A, Freeman AF, Hill BJ, Elias KM, et al. Impaired T(H)17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature 2008;452(7188):773–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery CP, Daniels M, Zhao F, Alegre ML, Chong AS, Daum RS. Protective immunity against recurrent Staphylococcus aureus skin infection requires antibody and interleukin-17A. Infect Immun 2014;82(5):2125–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto K, Ozawa T, Awazu K, Ito N, Honda N, Matsumoto S, et al. Photodynamic therapy using systemic administration of 5-aminolevulinic acid and a 410-nm wavelength light-emitting diode for methicillin-resistant Staphylococcus aureus-infected ulcers in mice. PLoS One 2014;9(8):e105173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutluoglu M, Uzun G. Pseudomonas infection in a postoperative foot wound. CMAJ 2011;183(8):E499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mölne L, Verdrengh M, Tarkowski A. Role of neutrophil leukocytes in cutaneous infection caused by Staphylococcus aureus. Infect Immun 2000;68(11):6162–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Oscherwitz J, Cease KB, Chan SM, Muñoz-Planillo R, Hasegawa M, et al. Staphylococcus δ-toxin induces allergic skin disease by activating mast cells. Nature 2013;503(7476):397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuji T, Chen TH, Two AM, Chun KA, Narala S, Geha RS, et al. Staphylococcus aureus Exploits Epidermal Barrier Defects in Atopic Dermatitis to Trigger Cytokine Expression. J Invest Dermatol 2016;136(11):2192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen MM, Witherden DA, Havran WL. γδ T cells in homeostasis and host defence of epithelial barrier tissues. Nat Rev Immunol 2017;17(12):733–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nippe N, Varga G, Holzinger D, Löffler B, Medina E, Becker K, et al. Subcutaneous infection with S. aureus in mice reveals association of resistance with influx of neutrophils and Th2 response. J Invest Dermatol 2011;131(1):125–32. [DOI] [PubMed] [Google Scholar]
- Oh JW, Kloepper J, Langan EA, Kim Y, Yeo J, Kim MJ, et al. A Guide to Studying Human Hair Follicle Cycling In Vivo. J Invest Dermatol 2016;136(1):34–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortines RV, Liu H, Cheng LI, Cohen TS, Lawlor H, Gami A, et al. Neutralizing Alpha-Toxin Accelerates Healing of Staphylococcus aureus-Infected Wounds in Nondiabetic and Diabetic Mice. Antimicrob Agents Chemother 2018;62(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyoshi MK, Beaupré J, Venturelli N, Lewis CN, Iwakura Y, Geha RS. Filaggrin deficiency promotes the dissemination of cutaneously inoculated vaccinia virus. J Allergy Clin Immunol 2015;135(6):1511–8.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker D Humanized Mouse Models of Staphylococcus aureus Infection. Front Immunol 2017;8:512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastagia M, Euler C, Chahales P, Fuentes-Duculan J, Krueger JG, Fischetti VA. A novel chimeric lysin shows superiority to mupirocin for skin decolonization of methicillin-resistant and -sensitive Staphylococcus aureus strains. Antimicrob Agents Chemother 2011;55(2):738–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastar I, Nusbaum AG, Gil J, Patel SB, Chen J, Valdes J, et al. Interactions of methicillin resistant Staphylococcus aureus USA300 and Pseudomonas aeruginosa in polymicrobial wound infection. PLoS One 2013;8(2):e56846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridaura VK, Bouladoux N, Claesen J, Chen YE, Byrd AL, Constantinides MG, et al. Contextual control of skin immunity and inflammation by Corynebacterium. J Exp Med 2018;215(3):785–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado-Pabón W, Schlievert PM. Models matter: the search for an effective Staphylococcus aureus vaccine. Nat Rev Microbiol 2014;12(8):585–91. [DOI] [PubMed] [Google Scholar]
- Sampedro GR, DeDent AC, Becker RE, Berube BJ, Gebhardt MJ, Cao H, et al. Targeting Staphylococcus aureus α-toxin as a novel approach to reduce severity of recurrent skin and soft-tissue infections. J Infect Dis 2014;210(7):1012–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharschmidt TC, Vasquez KS, Truong HA, Gearty SV, Pauli ML, Nosbaum A, et al. A Wave of Regulatory T Cells into Neonatal Skin Mediates Tolerance to Commensal Microbes. Immunity 2015;43(5):1011–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz A, Jiang L, de Vor L, Ehrström M, Wermeling F, Eidsmo L, et al. Neutrophil Recruitment to Noninvasive MRSA at the Stratum Corneum of Human Skin Mediates Transient Colonization. Cell Rep 2019;29(5):1074–81.e5. [DOI] [PubMed] [Google Scholar]
- Shi CM, Nakao H, Yamazaki M, Tsuboi R, Ogawa H. Mixture of sugar and povidone-iodine stimulates healing of MRSA-infected skin ulcers on db/db mice. Arch Dermatol Res 2007;299(9):449–56. [DOI] [PubMed] [Google Scholar]
- Sivanmaliappan TS, Sevanan M. Antimicrobial Susceptibility Patterns of Pseudomonas aeruginosa from Diabetes Patients with Foot Ulcers. Int J Microbiol 2011;2011:605195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soong G, Paulino F, Wachtel S, Parker D, Wickersham M, Zhang D, et al. Methicillin-resistant Staphylococcus aureus adaptation to human keratinocytes. mBio 2015;6(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaan AN, Henry T, van Rooijen WJM, Perret M, Badiou C, Aerts PC, et al. The staphylococcal toxin Panton-Valentine Leukocidin targets human C5a receptors. Cell Host Microbe 2013;13(5):584–94. [DOI] [PubMed] [Google Scholar]
- Suaya JA, Mera RM, Cassidy A, O’Hara P, Amrine-Madsen H, Burstin S, et al. Incidence and cost of hospitalizations associated with Staphylococcus aureus skin and soft tissue infections in the United States from 2001 through 2009. BMC Infect Dis 2014;14:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwanpradid J, Holcomb ZE, MacLeod AS. Emerging Skin T-Cell Functions in Response to Environmental Insults. J Invest Dermatol 2017;137(2):288–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev 2015;28(3):603–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng CW, Biancotti JC, Berg BL, Gate D, Kolar SL, Müller S, et al. Increased Susceptibility of Humanized NSG Mice to Panton-Valentine Leukocidin and Staphylococcus aureus Skin Infection. PLoS Pathog 2015;11(11):e1005292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng CW, Kyme P, Low J, Rocha MA, Alsabeh R, Miller LG, et al. Staphylococcus aureus Panton-Valentine leukocidin contributes to inflammation and muscle tissue injury. PLoS One 2009;4(7):e6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng CW, Sanchez-Martinez M, Arruda A, Liu GY. Subcutaneous infection of methicillin resistant Staphylococcus aureus (MRSA). J Vis Exp 2011(48). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams MR, Costa SK, Zaramela LS, Khalil S, Todd DA, Winter HL, et al. Quorum sensing between bacterial species on the skin protects against epidermal injury in atopic dermatitis. Sci Transl Med 2019;11(490). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu SX, Gilmore KJ, Szabo PA, Zeppa JJ, Baroja ML, Haeryfar SM, et al. Superantigens subvert the neutrophil response to promote abscess formation and enhance Staphylococcus aureus survival in vivo. Infect Immun 2014;82(9):3588–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Gao N, Sun H, Yin J, Lee P, Zhou L, et al. Targeting Imbalance between IL-1β and IL-1 Receptor Antagonist Ameliorates Delayed Epithelium Wound Healing in Diabetic Mouse Corneas. Am J Pathol 2016;186(6):1466–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolfaghari PS, Packer S, Singer M, Nair SP, Bennett J, Street C, et al. In vivo killing of Staphylococcus aureus using a light-activated antimicrobial agent. BMC Microbiol 2009;9:27. [DOI] [PMC free article] [PubMed] [Google Scholar]


