Abstract
The cellular prion protein (PrPC) is comprised of two domains – a globular C-terminal domain and an unstructured N-terminal domain. Recently, copper has been observed to drive tertiary contact in PrPC, inducing a neuroprotective cis interaction that structurally links the protein’s two domains. The location of this interaction on the C-terminus overlaps with the sites of human pathogenic mutations and toxic antibody docking. Combined with recent evidence that the N-terminus is a toxic effector regulated by the C-terminus, there is an emerging consensus that this cis interaction serves a protective role, and that the disruption of this interaction by misfolded PrP oligomers may be a cause of toxicity in prion disease. We demonstrate here that two highly conserved histidines in the C-terminal domain of PrPC are essential for the protein’s cis interaction, which helps to protect against neurotoxicity carried out by its N-terminus. We show that simultaneous mutation of these histidines drastically weakens the cis interaction and enhances spontaneous cationic currents in cultured cells - the first C-terminal mutant to do so. Whereas previous studies suggested that Cu2+ coordination was localized solely to the protein’s N-terminal domain, we find that both domains contribute equatorially coordinated histidine residue side chains, resulting in a novel bridging interaction. We also find that extra N-terminal histidines in pathological familial mutations involving octarepeat expansions inhibit this interaction by sequestering copper from the C-terminus. Our findings further establish a structural basis for PrPC’s C-terminal regulation of its otherwise toxic N-terminus.
Keywords: PrP, PrPC, Aβ42, NMR, EPR
Introduction
Prion diseases, or Transmissible Spongiform Encephalopathies (TSEs), are a class of contagious, fatal neurodegenerative illnesses brought on by the misfolding of the endogenous prion protein (PrP)[1,2]. These diseases, which are part of a larger group of protein aggregation disorders including Alzheimer’s and Parkinson’s diseases, are an ongoing threat to human health, as well as to agricultural and wildlife animals.
Prion diseases are initiated by the misfolding of the prion protein (PrP) from its cellular form (PrPC) into a beta sheet rich form (PrPSc)[1,2]. Misfolded PrPSc aggregates deposit as fibrillar structures in brain tissue. The self-propagating nature of these aggregates has been extensively studied[2–4], but the mechanism of their neuronal toxicity is still unknown. PrP null mice do not exhibit symptoms of prion disease, suggesting that the disease is due to the gain of a toxic function, rather than the loss of essential activity[5]. PrP null mice injected with PrPSc do not show symptoms of prion disease, demonstrating the requirement of PrPC as an essential precursor to PrPSc. In addition, PrPC has been implicated as a signal transducer that transmits the toxicity initiated by PrPSc[6–9].
Several kinds of experiments suggest that alterations in PrPC structure or conformation may mediate neurotoxic activities. For example, expression in transgenic mice of PrP molecules harboring deletions within the central domain of PrP, such as Δ105–125, causes a spontaneous neurodegenerative phenotype. In addition, monoclonal antibodies that bind the C-terminal domain of PrPC are profoundly neurotoxic in vivo and in cultured cells[10–12]. It is thought that these manipulations eliminate the necessity for PrP aggregation, and directly induce PrPC-mediated neurodegeneration. The effects of PrP deletions and anti-PrP antibodies can be measured in cell culture using patch-clamp recording, which detects the spontaneous, cationic currents that are induced[13].
In electrophysiological assays and in vivo, it has been observed that the flexible N-terminal domain, and in particular the first nine residues (23–31), are essential for neurotoxic activity[11,14]. Deletion of these residues, or treatment with antibodies and other ligands that bind this region, prevents the spontaneous currents and other neurotoxic activities[12,14]. In addition, cells expressing the N-terminal domain of PrP attached to another protein, EGFP, and entirely devoid of the C-terminal domain of PrP, exhibit spontaneous currents[11]. These results suggest that the N-terminal domain of PrP harbors a latent, toxic effector action, which is normally inhibited by interaction with the C-terminal domain.
The specific region on the C-terminus of PrPC where toxic antibodies bind is near the sites of pathogenic, familial mutations[15,16]. Recently, our lab and others have shown that there is a cis interaction between the N-terminal and C-terminal domains of PrPC, and that disruption of this interaction leads to neurotoxicity[15,17,18]. Taken together, these results suggest that an unregulated N-terminal domain of PrPC is responsible for neurotoxicity, and that toxic PrP oligomers (ranging in size from ~5–10 PrP units[19,20]) may interfere with a natural protective regulation of the N-terminal domain by the C-terminal domain[11,18]. Despite the importance of PrPC’s self-regulation, only limited information is known about the essential regulatory interface between the two domains, although important insights have recently come from a study using a combination of cross-linking mass spectrometry and NMR[21].
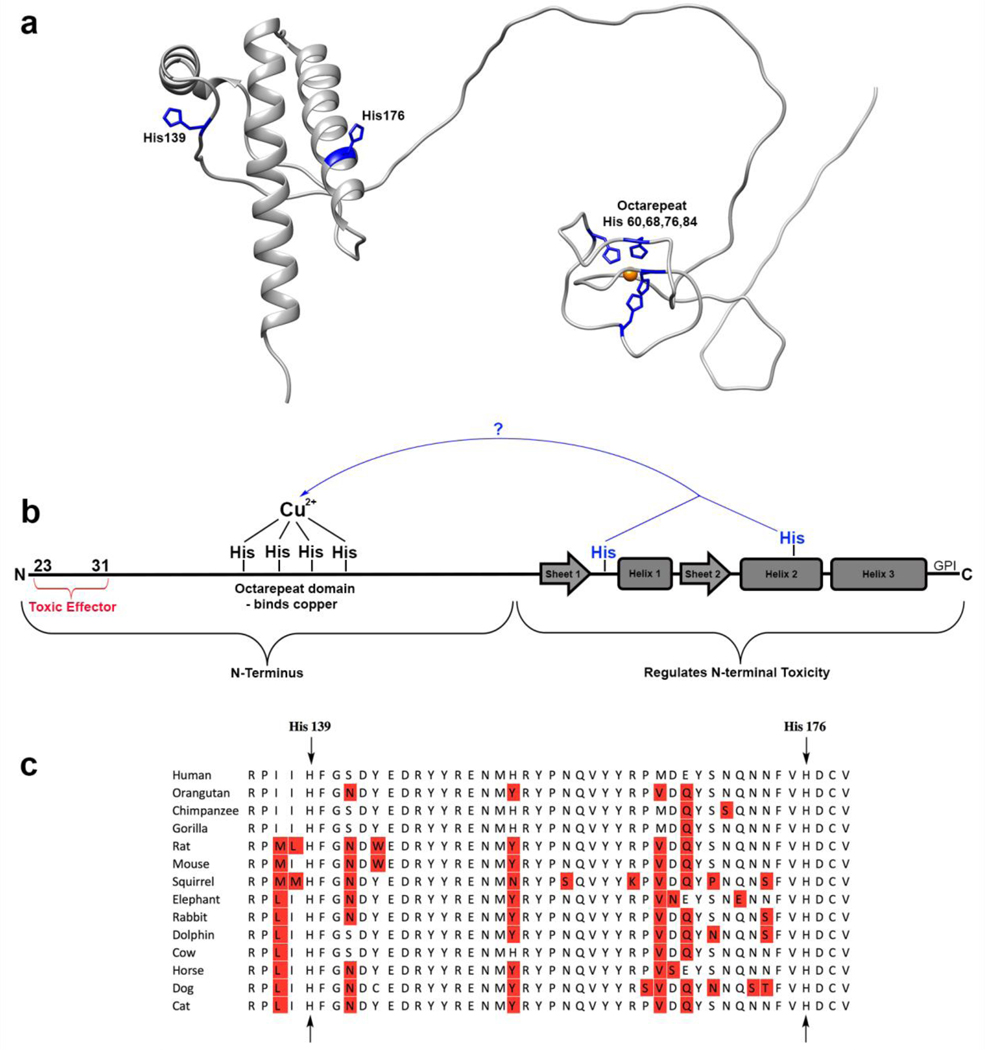
In order to understand the molecular details of PrPC’s self-regulation, we consider the role of the protein’s cognate metal ions. PrP physiology is highly dependent on the concentrations of divalent metal ions, particularly Cu2+. PrP has been shown to concentrate copper in brain tissue, and its expression is upregulated in response to increasing copper levels[22–24]. PrP is also known to regulate ion channels in a copper dependent manner[6,25]. Due to the strong link between copper and PrP function, the metal binding properties of the protein have been extensively studied[15,18,26–32]. In the N-terminus of the protein, there is an eight amino acid sequence repeated four times (octarepeat, OR; Figure 1). Each repeat contains one histidine, capable of coordinating the divalent metal ions Cu2+ and Zn2+. Cu2+ is found to bind the four histidine residues in the N-terminal domain’s octarepeat region with sub-nanomolar affinity[29].
Figure 1:
a) Our current understanding of the prion protein. b) The potential new mechanism of self-regulation investigated in this paper: C-terminal histidines coordinating an N-terminally bound copper ion. c) Conservation of the histidines hypothesized to be involved. Residues with sequence variation are noted in red.
Recently, N-terminal copper binding has been observed to drive tertiary structure in PrPC, inducing the aforementioned cis interaction between the N- and C-terminal domains of the protein[15,17,18,21]. The location of this interaction on the C-terminus overlaps with the sites of pathogenic mutations and toxic antibody docking. Copper delivered to cells has been shown to partially rescue toxicity resulting from deletion of a hydrophobic segment between the PrPC N- and C-terminal domains[11]. Together, these results suggest that PrPC’s neuroprotective self-regulation is in fact driven by copper, which directly promotes the cis interaction between its toxic N-terminal and regulatory C-terminal domains.
How does copper cause the two domains to co-localize? Upon examination of the region on the C-terminus of PrPC in which the copper-bound N-terminus docks, we were struck by the presence of two highly conserved histidine residues (Figure 1). The goal of this study is to determine if these C-terminal histidines facilitate the interdomain cis interaction, perhaps by acting as a tether to hold the two domains together. If so, a paradox is presented: if the copper ion is already coordinatively saturated by four N-terminal histidines, how could it bind additional C-terminal histidines?
To investigate a potential role of C-terminal histidines in the prion protein’s cis interaction, we employed 1H-15N HSQC NMR analysis of the murine protein. We show that mutation of histidines 139 and 176 (equivalent to human residues 140 and 177) results in a drastic weakening of the cis interaction. Furthermore, when His176 is deleted and a new histidine is added one turn up or down on the same helix, the location of the cis interaction tracks the position of the histidine. We propose a model for this interaction in which copper acts as a tether – being simultaneously bound by both N and C-terminal histidine residues, analogous to the Cu(II) macro-chelate that occurs between His50 and Asp2 of α-Synuclein[33]. Using EPR spectroscopy, we find that the mechanism of this tether involves a histidine swap – one N-terminal histidine is removed from the copper ion and replaced with a C-terminal histidine. We show this model to be energetically plausible using molecular dynamics simulations and supporting density functional structure calculations. Together, these findings overturn the conventional view that Cu2+ is bound solely to the PrPC N-terminal domain. Instead, both domains participate in metal ion coordination.
We find that, in addition to its structural effects, disruption of the cis interaction by mutation of both C-terminal histidines H139 and H176 enhances spontaneous ionic currents in cultured cells – the first C-terminal mutant to do this. Lastly, we find that the insertional mutations of extra, histidine containing octarepeat segments, which are linked to familial prion diseases, inhibit the protein’s cis interaction by chelating copper away from the C-terminus. These findings advance our understanding of the toxicity caused by prion oligomers, toxic antibodies, and pathological mutations, and suggest that strengthening the cis interaction may be an effective therapeutic strategy for prion disease.
Results
Mutation of histidines 139 and 176 drastically weakens the prion protein’s cis interaction
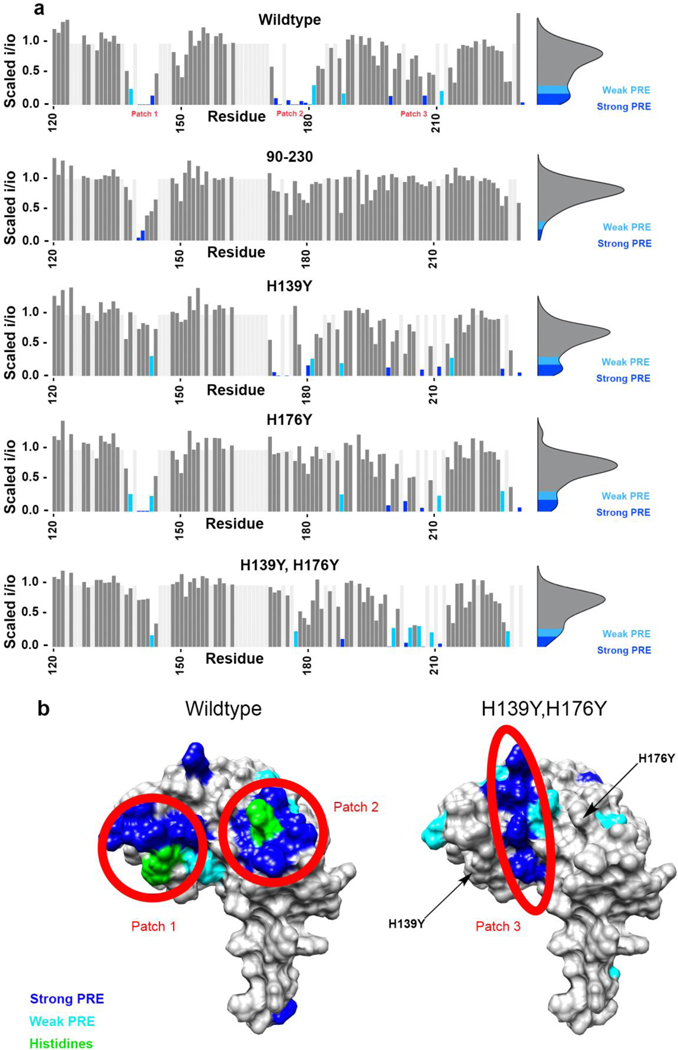
To examine whether C-terminal histidines contribute to the cis interaction, we mutated histidines 139, 176, and 186 of the murine prion protein to tyrosine, each in separate mutants. Because His186 is on the opposite face of the protein from the site of the cis interaction, we included it as a control. We chose tyrosine as a replacement amino acid because it is structurally similar to histidine, but cannot bind metals with similar affinity. At pH 7.4, the N-terminus of PrPC binds copper in three coordination modes[26,27,29]. At low copper : PrPC ratios, one copper ion is bound simultaneously by four histidines from the octarepeat region of PrPC. At higher copper levels, multiple copper ions bind to the protein. In order to simplify interpretation of the data, we performed our experiments at pH 6.0, which locks the coordination into the first mode. Using 1H-15N HSQC NMR at 37 °C and pH 6.0, we measured the peak intensities of C-terminal residues with no copper, and with one equivalent CuCl2. Dividing the intensity of each peak in the copper sample by its corresponding intensity in the apo sample, we calculated the intensity ratios (i/i0) associated with the addition of one equivalence of copper to the protein, and repeated this process for each mutant. We then scaled the i/i0 values to eliminate the effects of differential unspecific peak intensity reduction across mutants, and mapped the results onto the surface of the three-dimensional structure of the protein’s helical C-terminus.
The wild type protein showed peak intensity reduction due in part to the paramagnetic relaxation effect (PRE), and also to exchange, in three areas as reported previously[17,18]. We refer to these areas of the helical C-terminal domain as “patch 1”, which is centered around His139, “patch 2”, which is centered around His176, and “patch 3”, which is located on helix three, between patch 1 and patch 2 (Figure 2). The peak intensity reduction in patches 1 and 2 is stronger than in patch 3, suggesting that they are closer to the bound copper center of the OR. The H139Y mutation resulted in a much weaker peak intensity reduction at patch 1 than wild type, as did the H176Y mutation but at patch 2, while the H186Y mutation did not result in a difference to peak intensity reduction. The mutation of H186, which is the location of the pathogenic mutation H186R, appeared to affect C-terminal conformation of the protein and was therefore not further investigated. We varied protein and copper concentrations to test for the possibility of intermolecular interactions. We found that the peak linewidths were invariant as a function of protein concentration. In addition, the i/i0 profiles were not affected as would be expected if two PrPC molecules were forming a complex at higher concentrations. Together, these results suggest that there is only one PrPC molecule involved per copper, and that the interaction is strictly between its N and C termini.
Figure 2.
a) Bar plots showing the magnitude of the peak intensity reduction derived from the paramagnetic relaxation effect (PRE) on specific residues of the protein’s C-terminal domain, with 1 equivalent of CuCl2 at pH 6.0 in 10 mM MES buffer for all five protein samples. Patches 1 and 2, which are located around histidines 139 and 176 respectively, are much more strongly affected in the wild type protein than in (H139Y,H176Y) and in 90–230 PrPC. Unassigned residues are light grey and are drawn with no intensity reduction. b) Surface representations showing areas engaged in the cis interaction, as measured by the intensity reduction of NMR cross peaks, plotted on PDB: 1XYX.
With the mutation of only one C-terminal histidine at a time, the cis interaction persisted, centered around the remaining histidine. We then made the double mutant (H139Y,H176Y), to test the theory that either histidine was capable of driving the interaction. In this mutant we observed that both patches 1 and 2 exhibited significantly decreased peak intensity reduction compared to wild type, abolishing the primary contacts of the established cis interaction (Figure 2). Peak intensity in patch 3 was also slightly more reduced than in wild type, suggesting a backup, albeit weak, mode of cis interaction. The strength of this backup interaction is best visualized using the bar plot of peak intensity ratios in Figure 2. We note that residues N-terminal to residue 120 are largely unstructured, resulting in peak overlap that confounds extraction of peak intensities, and have not been included in this analysis.
In order to be sure that these effects were due to the removal of histidines, and not due to addition of tyrosines which could sterically hinder the cis interaction, we performed these same experiments with the mutants (H139N,H176N) and (H139A,H176A), and observed the same results (data not shown).
We also performed this experiment on 90–230 PrPC, which lacks the octarepeat histidines, to check that the observed C-terminal peak intensity reduction was due to a cis interaction rather than pure C-terminal copper binding. Although we observed slight intensity reduction around the C-terminal histidines in this mutant, the effect was much weaker than in the full-length protein. This observation provides convincing evidence that the reduction in peak intensity observed in the wild type protein upon the addition of copper results from the cis-interaction between its two domains, and not from transient copper binding solely to the C-terminal domain. Specifically, the loss of the octarepeat domain has a major effect on copper binding in the C-terminal domain, especially at His176.
Previous research by us and others has demonstrated the involvement of the non-octarepeat histidines H95 and H110 in copper binding[27,30,34]. At pH 7.4, copper coordination at these sites involves both the His side chain, as well as backbone amide nitrogens. To eliminate interference from coordination at these sites, we performed experiments at pH 6.0, which suppresses deprotonation of amides, thus reducing localized non-octarepeat copper binding. As a further control aimed at probing the involvement of histidines H95 and H110 in this interaction, we performed NMR on the construct (H95Y, H110Y). The strength and location of the cis interaction observed here is nearly identical to the wildtype protein (Figure S1). We also performed continuous wave EPR on the wildtype protein at both pH 6.0 and pH 7.4. In the pH 7.4 spectrum we observed a previously characterized signal that arises from copper coordination from His95 and His110 due to backbone protonation, but we do not see this coordination in the pH 6.0 spectrum (Figure S2). Based on this data, we can rule out the possibility that the C-terminus is interacting with copper bound solely to a central region histidine complex. However, we cannot rule out the possibility that the C-terminus is interacting with copper bound to a complex formed of both the octarepeat and central region histidines.
Mutation of C-terminal residues in the region of the cis interaction surrounding histidines 139 and 176 does not abolish the interdomain interaction
In order to show that the cis interaction was driven by histidine-copper interactions rather than by a specific interaction between the two domains of the protein, we mutated most of the other residues in patches 1 and 2 spatially adjacent to the histidines to alanine. Specifically, the mutants (137A,138A,140A) and (172A,173A,175A,179A,180A) were created, as these are the residues that are located most closely to His139 and H176, and examined by NMR. The mutant (137A,138A,140A) adopted a misfold, preventing the data from being analyzed. The mutant (172A,173A,175A,179A,180A), which mutates the majority of patch 2 but leaves its histidine intact, maintained its i/i0 profile consistent with the protein’s cis interaction (Figure S3). These results reaffirm the importance of this C-terminal histidine in the copper driven cis interaction.
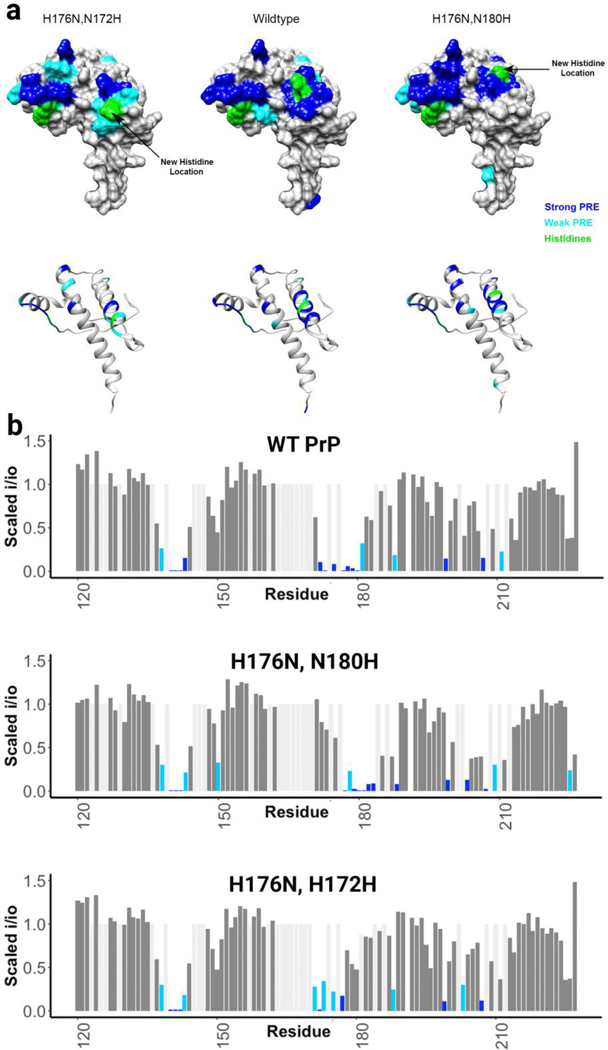
Deletion of histidine 176 and introduction of a new histidine one turn up or down its helix leads to a cis interaction localized to the new histidine
To show that the histidines were actively driving the cis interaction, rather than simply being positioned in the center of the involved areas, we deleted His176 and created two new mutants, one with a histidine one turn up and the other with a histidine one turn down its helix. As observed in Figure 3, the location of patch 2 follows the histidine. The ability of the histidine on helix 2 to control the location the cis interaction shows that it plays an active role in this docking event.
Figure 3:
a) Surface representations showing areas engaged in the cis interaction, as measured by the intensity reduction of NMR cross peaks, plotted on PDB: 1XYX. This figure shows that the cis interaction follows histidine b) Bar plots showing the magnitude of the peak intensity reduction derived from the paramagnetic relaxation effect (PRE) on specific residues of the protein’s C-terminal domain, with 1 equivalent of CuCl2 at pH 6.0 in 10 mM MES buffer for all protein samples.
Electron paramagnetic resonance (EPR) experiments show no change in copper coordination environment, suggesting an equatorial histidine swap
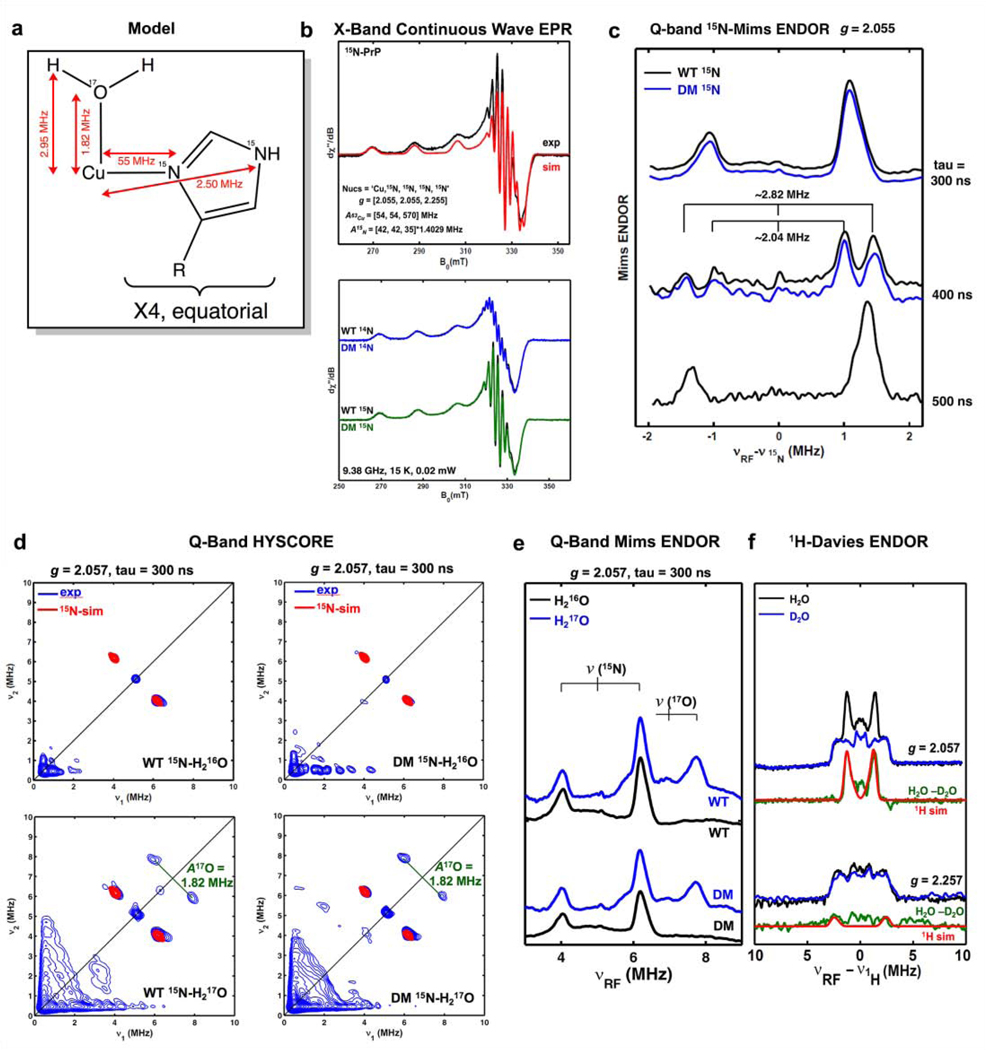
Copper(II) in aqueous solutions exists in octahedral or square planer coordination geometries. With the knowledge that the four histidines of the PrPC N-terminal octarepeat domain bind copper equatorially, this leaves only the axial binding sites available for a fifth, C-terminal histidine ligand. To investigate the coordination environment of the copper center, we employed X-band continuous wave (CW) EPR, Q-band electron nuclear double resonance (ENDOR), and hyperfine sub-level correlation (HYSCORE) spectroscopy at pH 6 (Figure 4).
Figure 4:
a) Representation of the copper site. b) X-band continuous wave EPR spectra of WT PrPC in agreement with simulation based on four equatorial nitrogen ligands and comparison of WT PrPC to (H139Y, H176Y) PrPC (labelled DM, for double mutant). c) Q-band 15N-Mims ENDOR showing weakly hyperfine interaction for both WT and DM. Varying tau values were used for confirming the Mims-hole effect. d) Q-band HYSCORE EPR spectra showing weakly coupled remote 15N from histidine and axial 17O from water. e) Q-band 17O Mims ENDOR, confirming axial water coordination. f) 1H-Davies ENDOR showing the disappearance of an exchangeable proton from water in when D2O is used as solvent.
As shown in Figure 4A, WT and the double mutant (H139Y, H176Y), which we refer to as “DM,” produces the same X-band CW EPR spectra, which are well simulated by employing four equatorial nitrogen nuclei (15N, I = 1/2) with a strong hyperfine coupling of aiso ca. 55 MHz. This strong 15N hyperfine coupling arises from directly coordinated nitrogens (15Nδ) of the equatorial His imidazole rings. The remote histidine nitrogens (15Nε), that are two bonds away from the 15Nδ, typically have a much smaller hyperfine coupling. This smaller coupling was probed by using Mims-ENDOR shown in Figure 4B. Both WT and the double mutant (H139Y, H176Y) show only one set of weakly coupled 15N, which is simulated using a hyperfine tensor A of [2.88, 2.88, 1.76] MHz, with aiso = 2.50 MHz. This hyperfine tensor is determined by the simulation of both X-band and Q-band 15N-HYSCORE spectra, as shown in Figure 4C and 4D (Note that both WT and DM show the same HYSCORE spectrum, see Figure 4D). The weak 15N hyperfine interactions are assigned to the remote nitrogens 15Nε of the equatorial imidazole rings,[35] as they are approximately 4.5% of that of the coordinated nitrogen, which is typical for imidazole-bound paramagnetic spin centers, such as imidazole-bound copper, vanadium and Rieske-type [2Fe-2S] clusters[35–37].
Besides the two sets of 15N hyperfine interactions arising from the 15Nδ and 15Nε of equatorially bound histidines, we did not observe other weak hyperfine interactions of 15N as might be expected from a possible axial histidine. This suggests that there is no axial histidine ligand in WT protein. Indeed, we were able to show that the axial ligand is an exchangeable water ligand for both WT and the double mutant, as is evident from 17O (I = 5/2) hyperfine interaction ca. 1.82 MHz observed from both 17O HYSCORE (Figure 4D) and ENDOR (Figure 4E) experiment on the samples enriched with H217O. This 17O hyperfine interaction is consistent with that of an axial water ligand binding a Cu(II) center in the literature[38,39]. The axial water ligand is further confirmed by observing the exchangeable proton by Q-band Davies ENDOR (Figure 4F), which is simulated by using the 1H hyperfine tensor A [−2.9, −2.8, 6.0] MHz, with the dipolar hyperfine T 2.95 MHz, corresponding to the distance of Cu-H ~2.99 Å.[40].
Taken together, these data show that there is no axial histidine binding in either the WT or the double mutant (H139Y, H176Y), and that copper coordination was unchanged by this mutation. This suggests that C-terminal histidines are in fact not acting as a fifth, axial ligand, but are instead displacing an equatorially bound N-terminal octarepeat histidine, giving equivalent coordination environments in both WT and DM proteins. We then produced the mutant (H84Y), which only has three octarepeat histidines. This mutant produced equivalent CW spectra to wild type, with four equatorial histidine ligands, and by NMR produced a slightly stronger cis interaction than in wild type, presumably because N-terminal histidines could no longer fully coordinate copper on their own (Figure S4). Combined with NMR evidence that a C-terminal histidine is driving the cis interaction, these data suggest that an N-terminal histidine is swapped out for a C-terminal histidine during the protein’s cis interaction.
Pathogenic insertion and deletion mutation of histidine-containing octarepeat segments are explained by this model and supported by NMR experiments
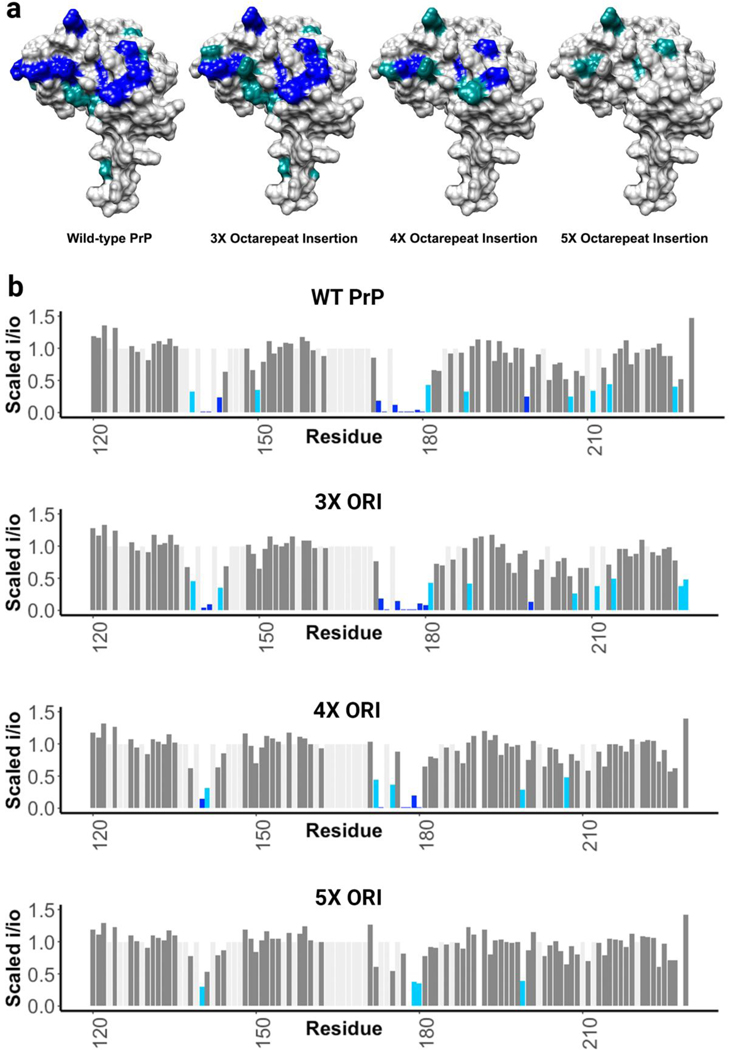
An important class of pathogenic mutations in the prion protein involves the insertion of one or more extra copies of the histidine containing octarepeat segment. Mutations leading to extra octarepeats are associated with familial prion disease, and there is a drastic and sudden shift towards early onset disease when there are five or more extra octarepeat insertions[41]. To study these mutations, we employed the same NMR experiments listed above with one equivalent of copper. As shown in Fig. 5, insertion of three extra histidine containing octarepeats did not inhibit the observed cis interaction, four extra histidines slightly weakened it, and five extra histidines almost completely inhibited the cis interaction. This suggests that insertion mutations sufficient to cause early onset prion disease may act by chelating copper away from C-terminal histidines and preventing the otherwise neuroprotective cis interaction. Alternatively, it is possible that the interaction is prevented by an increased entropic penalty due to the longer distance between the termini. In addition to insertional mutations, there are also deletion mutations in this region: deletion of one octarepeat segment does not cause prion disease, but deletion of two segments does[42–44]. This is consistent with the notion that three histidines from the octarepeat and one histidine from the C-terminus are involved in the cis interaction.
Figure 5.
a) Surface representations showing areas engaged in the cis interaction, as measured by the intensity reduction of NMR cross peaks, plotted on PDB: 1XYX. Five additional octarepeat regions results in a strong decrease in the cis interaction. b) Bar plots showing the magnitude of the peak intensity reduction derived from the paramagnetic relaxation effect (PRE) on specific residues of the protein’s C-terminal domain, with 1 equivalent of CuCl2 at pH 6.0 in 10 mM MES buffer for all protein samples.
Mutation of C-terminal histidines enhances spontaneous currents
Certain mutations in the central region of PrP, such as Δ105–125 (referred to as ΔCR) have been observed to cause spontaneous cationic currents in a variety of cell lines and in primary neurons[13]. The currents caused by CR mutations are suppressed by coexpression of wild type PrP, which mirrors the ability of coexpressed WT PrP to suppress the neurotoxicity of these mutations in transgenic mice[10]. C-terminal antibodies that are neurotoxic when injected into the mouse brain also cause spontaneous currents. We attribute the spontaneous currents and neurotoxicity observed in these scenarios to a weakening of the N-C inter-domain interactions within the PrP molecule[11,21]. We therefore tested whether the H139Y,H176Y mutations, by weakening the cis interaction, caused spontaneous currents in cultured cells.
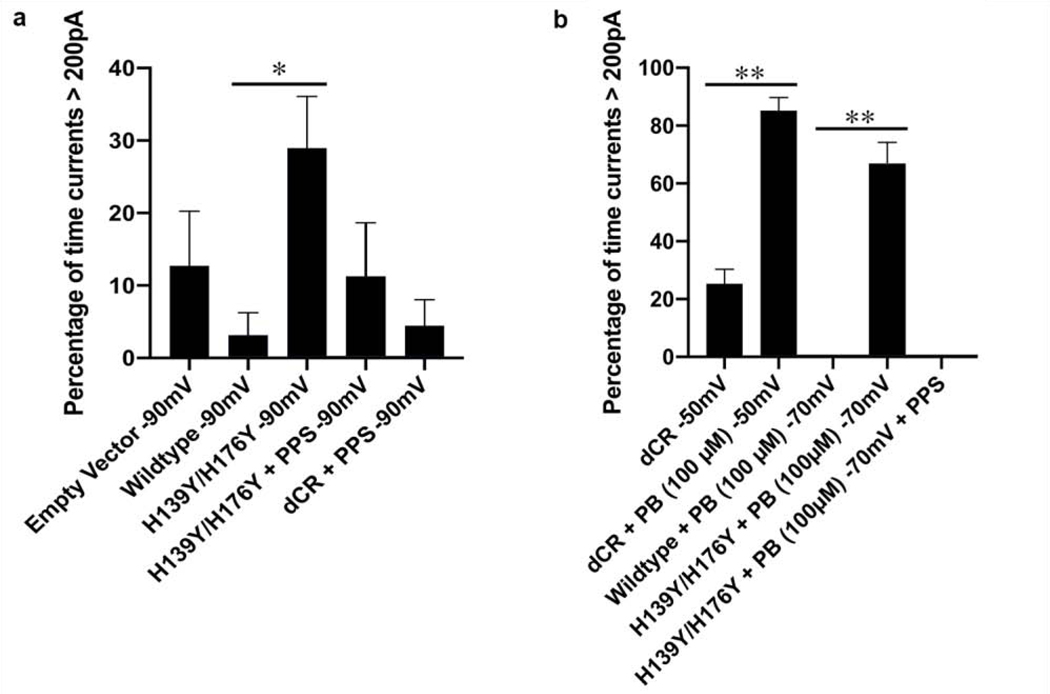
Using whole-cell patch-clamping of N2a cells held at −90 mV, we observed spontaneous currents for (H139Y,H176Y), and not for a wild-type control (Figure 6). We used a −90 mV holding potential, which is more hyper-polarized than the normal resting potential of N2a cells (−70 mV), to enhance our ability to detect spontaneous currents; we have shown that these currents are voltage-dependent, being greater at more hyper-polarized membrane potential[11]. Consistent with studies of CR PrP mutants, the currents caused by H139Y,H176Y were suppressed by the addition of pentosan polysulfate (PPS), a polymer that sequesters the N-terminal domain of PrP[11].
Figure 6.
a) Comparison of cells transfected with empty vector (N=10), wild-type PrP (N=5), (H139Y,H176Y) PrP (N=20), and ΔCR PrP (N=10) showing that the H139Y,H176Y mutations significantly enhance spontaneous currents at a hyperpolarized holding potential (−90 mv) (*p=0.045, α=0.05, one tailed two sample t-test), and these currents are suppressed by the addition of pentosan polysulfate (PPS). b) Comparison of cells expressing wild-type PrP (N=10), (H139Y,H176Y) PrP (N=10), and ΔCR PrP (N=9) showing that the H139Y,H176Y mutations significantly enhance spontaneous currents at −70 mV in the presence of pyrenebutyrate (**p<0.00001) , and these currents are suppressed by pentosan polysulfate. The ΔCR mutation enhances currents in the presence of pyrenebutyrate, even at −50 mV (**p=<0.00001). Error bars show the standard error of the mean.
Pyrenebutyrate (PB) is a polyaromatic, negatively charged small molecule that binds to positively charged groups such as the side chains of arginine and lysine. By shielding the positive charges, PB facilitates the ability of polybasic peptide regions, like those found at the N-terminus of PrP, to penetrate cell membranes[45–47]. We found (Figure 6b) that the presence of PB greatly enhanced the spontaneous currents caused the 105–125 mutation, even when recorded at −50 mV, a potential at which currents cannot normally be observed[11]. By using PB to enhance the detectability of spontaneous currents, we found that the (H139Y, H176Y) mutation caused currents at −70 mV, the normal resting of N2a cells, whereas no currents were detected at this potential for WT PrP (Figure 6b).
Protein molecular dynamics (MD) simulations and density functional structure calculations of copper binding in PrPC
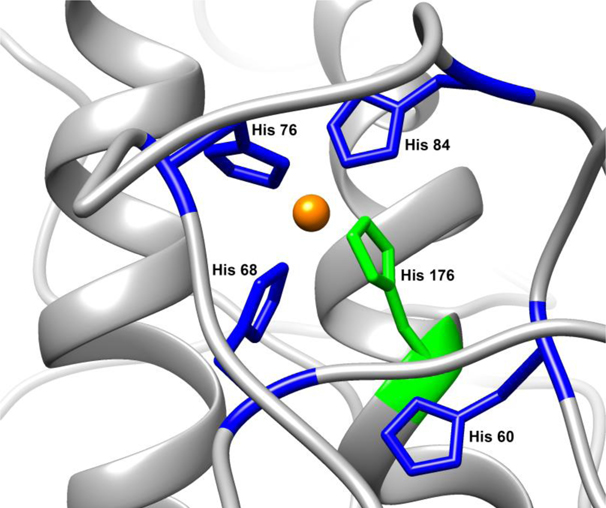
Histidines from both domains appear to be capable of cooperatively coordinating a copper ion, however, the specific His residues involved in this interaction and their location in the Cu2+ coordination sphere is not yet known. Starting from an initial coordination state where Cu2+ is bound by the four N-terminal His residues in a square planar coordination environment we identified eight additional possible configurations if one of the OR His residues is swapped for either His139 or His176. Using all-atom molecular dynamics simulations, we acquired a total of 675 ns of simulation time over all of the Cu-PrPC configurations considered. We performed an additional seven simulations where two of the OR His residues are exchanged for His139 and His176 simultaneously (for a total of 285 ns of simulation time). While the specific atoms that comprise the Cu2+ coordination environment are different between each simulation, the number of atoms and the type of coordinating atoms immediately surrounding the Cu2+ center remain unchanged. We used the Cu2+ center and the imidazole heavy atoms (non-H-atoms) of the His residues that are involved in Cu coordination in each simulation along with the protein backbone heavy atoms of helix 2 and helix 3 as a common reference for accessing conformational stability. A clustering analysis was performed (details in methods section). Although there were no explicitly-bound water molecules at the Cu2+ axial positions, water molecules tended to occupy these sites for the majority of the simulation time, typically with no exchange or 1–2 waters exchanging during the course of the simulation. A persistent H-bond interaction was observed in several simulations between the interacting C-terminal Glu residues and one of these axially-retained waters. These results suggest that a His exchange between the N- and C-terminal domains holds the N-terminal OR region close to the C-terminal domain and that additional stabilization is afforded through electrostatic interactions between the cationic copper site and anionic residues in the C-terminus. Importantly, however, we also observe stable close-approach and these same stabilizing electrostatic interactions in the absence of His exchange, with the Cu2+ center bound by only the OR regions. This observation confirms that initial close approach of the Cu2+-bound OR region is already a favorable event and that His exchange then occurs as a further stabilizing consequence of the close association. An equilibrated, energy-minimized example of this interaction involving three OR His residues and C-terminal His176 simultaneously coordinating Cu2+ is shown in Figure 7.
Figure 7:
A structural explanation for the results of this paper. An N-terminal histidine (blue) swaps out for C-terminal His176 histidine (green) during the protein’s cis interaction, creating a copper tether between the two domains.
To further characterize the His exchange event within the Cu2+ coordination environment we employed density functional structure calculations to assess the free energy differences between several potential exchange mechanisms. The His residues were modeled as 4-methylimidazole, representing the side chain, with the Nε-position protonated and the Nδ-position available for metal coordination. Axial waters were excluded from the [Cu2+ (4-methylimidazole)4]2+ structure as these are only weakly bound and tend to form H-bonds instead of coordinate covalent interactions. We assessed two potential exchange mechanisms: i) an associative route, whereby the Cu2+ center coordinates a 5th imidazole moiety in an axial position, which we hypothesize would then displace an equatorial imidazole in a subsequent step, or ii) a dissociative route where one of the four imidazole moieties is first exchanged for a water molecule and then this water is subsequently exchanged for another imidazole moiety (representing coordination of a C-terminal His residue). Comparing the relative free energies ( G(aq)) for the associative vs dissociative route, we find that both are relatively low, at +30.5 and +24.2 kJ/mol, respectively, suggesting that both mechanisms could contribute to ligand exchange. These estimates are from model coordination environments, however, and we anticipate that within the protein, the protein is likely to contribute significantly. While electrostatic interaction tends to draw the Cu2+-bound N-terminal OR region toward the C-terminal domain, the steric bulk of the protein is likely to preclude formation of a crowded 5-His intermediate.
Discussion
Recent evidence shows that the flexible N-terminus of the prion protein acts as a toxic effector, and that this toxicity is down-regulated by interaction with the globular C-terminus[11,14]. Consistent with this model, deletions within the central region linking the two domains, as well as antibodies recognizing the region in the C-terminal domain that docks with the N-terminus, produce toxic effects in cultured neurons and in brain tissue. We hypothesize that these effects are caused by a disruption of the protein’s self-regulation, which allows its N-terminus to extend away from the C-terminal domain, allowing aberrant interactions with the plasma membrane or membrane-bound receptors. NMR, EPR, and chemical cross-linking/MS studies strongly support a physical interaction between the N- and C-terminal domains of PrP[11,15,18,21].
Copper ions have been suggested to play a key role in mediating the interaction between the two domains of PrP, although the molecular details are not fully characterized. It is known that Cu2+ binds histidines in the octarepeat region of the protein’s N-terminus, and that Cu2+ drives the cis interaction, but how does Cu2+ interact with individual amino acids to carry out this function? In this work, we show that two highly conserved histidines in the C-terminal domain of PrP are essential for the protein’s cis interaction, and that their substitution with residues that do not bind copper diminishes the interaction, and also induces spontaneous ionic currents associated with an unregulated N-terminal domain. N- and C-terminal histidines work in synchrony to stabilize the cis interaction, suggesting that together they co-bind a copper ion, which serves as an inter-domain tether. Because the coordination environment of the copper is unchanged by this event, our data suggest that an N-terminal histidine is swapped with a C-terminal histidine, thereby stabilizing the cis interaction. An energy minimized example of this interaction is shown in Figure 7. Whereas previous PrPC structural models suggested that Cu2+ coordination was limited to the protein’s N-terminal domain, we find instead that both domains participate in binding the metal ion.
Similar, but rare, intra- and inter-domain copper-sharing mechanisms are observed in other protein systems. In blue copper proteins, copper links two domains, using three ligands from one domain and one ligand from another[48]. The copper chaperone Atx1 transfers copper to the Ccc2 ATPase through a transient intermediate in which the copper is co-bound by cysteines from both proteins[49]. A similar copper handoff occurs in the CusCBAF export pump in E.coli, and is mediated by methionine ligands[50]. Despite the existence of these interdomain and interprotein copper bridges in other protein systems, PrPC has not previously been known to display these structures. It is established that the N-terminus of PrPC binds copper[26,27,29,30,51,52], and there is evidence in the literature that the C-terminus can as well[53–56]. However, this work is the first to demonstrate a non-local interaction, with a copper ion bridging N- and C-terminal histidine ligands and thereby stabilizing an inter-domain interaction. In this model, the affinity of Cu2+ for histidine residues in the N-terminal octarepeat region (Kd=0.1 nM), although high, is nevertheless weak enough to allow for exchange with histidine residues in the C-terminus.
The details of the cis interaction observed here provide a new explanation for a specific set of genetic prion diseases involving octarepeat variations. Individuals with at least five extra inserts of the octarepeat segment (nine total repeats) develop early onset prion disease. Based on the exceptionally high binding affinity between copper and this expanded OR segment (Kd < 0.01 nM), early onset disease was previously hypothesized to be due to either a loss of a copper dependent function of PrPC, or a loss of copper mediated protection against misfolding[41]. In light of the new findings presented here, we suggest an alternative interpretation: the toxicity of these mutations may be due to the extra octarepeat histidines chelating copper away from C-terminal histidines, thereby disrupting the regulatory cis interaction. In addition, deletion of one octarepeat segment is nontoxic in humans, but deletion of two or more results in prion disease[42–44]. This is consistent with the notion that three histidines from the octarepeat and one histidine from the C-terminus are necessary for the cis interaction. Lastly, specific C-terminal point mutations that reduce negative charge and increase positive charge, such as D177N and E199K in mice (D178N and E200K in humans), are located close to H176, and therefore likely to reduce or prevent C-terminal copper binding, which would also impair the cis interaction.
The experiments described in this study have utilized a 1:1 Cu2+ : PrPC stoichiometry. This stoichiometry is probably physiologically relevant, given the subnanomolar affinity of PrPC for Cu2+ and the Cu2+ concentrations thought to be present at the synapse. However, we have shown previously that coordination of Cu2+ by PrPC changes with concentration[29]. Studies into the nature of this interaction as a function of Cu2+ concentration may provide further insight into the cis interaction, PrPC function and familial disease. Additionally, future investigations will consider zinc, another metal known to bind to PrPC.
In summary, we have shown that two histidine residues in the C-terminal domain of PrPC drive a protective cis interaction between the N- and C-terminal domains. We have designed a novel mutation, involving simultaneous deletion of these two histidines, that impairs N-C interaction and enhances spontaneous currents associated with neurotoxicity. Thus, our work furthers the body of evidence for a neuroprotective cis interaction in the protein. These findings suggest a novel PrPC-mediated neurodegenerative mechanism whereby toxic prion oligomers, antibodies, and mutations elicit their effects through disruption of this interaction. Therefore, small molecule drugs or antibodies that stabilize and reinforce the cis interaction are potential therapeutic avenues to protect against toxicity in prion disease.
Materials and Methods
15N Labeled Protein Expression
Starting with the full length wild type mouse prion protein in the pJ414 vector backbone (DNA 2.0), we used QuickChange site-directed mutagenesis PCR to construct mutants, with primers generated manually and by www.primerdesigner.com. DNA was transformed into E. coli BL21Star (DE3) cells (Invitrogen) and expressed in M9 minimal media supplemented with 1 g/L 15N ammonium chloride (Cambridge Isotope Labs). Cells were grown at 37 °C until they reached an OD600 of 1.0, at which point the temperature was dropped to 30 C and protein expression induced with 1 mM isopropyl-1-thio-D-galactopyranoside (IPTG). Cells were allowed to express for 20 hours. Inclusion bodies were washed, then treated with 8 M guanidine HCl in pH 8 tris-acetate buffer to extract proteins. The extracted proteins were purified by immobilized metal affinity chromatography, allowed to fold overnight at pH 8, then transferred into pH 4.5 sodium acetate buffer by size exclusion chromatography. The proteins were then dialyzed into Milli-Q ultrapure water and purified further by reverse phase HPLC using C4 silica resin. Identity and purity were checked using a Sciex X500b mass spectrometer. The proteins were lyophilized, and quantified by UV-Vis spectrophotometry before use.
Nuclear Magnetic Resonance Spectroscopy
All samples were made to pH 6.0 in 10mM 2-(N-morpholino)ethanesulfonic acid (MES) buffer (Sigma), using potassium as a counterion and containing 10% D2O. For all samples, the protein was added to a concentration of 300 μM. For samples with copper, CuCl2 was used at 300 μM. 1H-15N HSQC spectra were recorded at 37 °C on a Varian INOVA 600 MHz spectrometer (Varian, Santa Clara, CA) at UCSC NMR facility (Santa Cruz, CA), and on a Bruker 800 MHz spectrometer also at UCSC. NMR spectra were analyzed with NMR Pipe and Sparky using assignments transferred from previous experiments by visual inspection, and figures were made using Chimera and R. To determine a cutoff i/i0 value to separate the residues involved in the cis interaction from the rest of the protein, we performed a kernel density estimation on the data using a Gaussian smoothing kernel. To eliminate the effects of differential unspecific peak intensity reduction across mutants, the data were scaled so that the center values of each mutant’s group of unaffected peaks were aligned. We divided the residues into three categories based on their i/i0 values: strongly affected (dark blue), weakly affected (light blue) and unaffected (grey). These divisions were created by using the local minimum separating the affected from unaffected residues in the wild type protein (i/i0 = 0.35), and dividing the affected peaks into two groups (i/i0 = 0 to 0.175, and i/i0 = 0.175 to 0.35).
Electron Paramagnetic Resonance Spectroscopy
The X-band (9.38 GHz) continuous-wave (CW) EPR spectra were recorded on a Bruker (Billerica, MA) EleXsys E500 spectrometer equipped with a super-high Q resonator (ER4122SHQE). Cryogenic temperatures were achieved and controlled using an ESR900 liquid helium cryostat in conjunction with a temperature controller (Oxford Instruments ITC503) and gas flow controller. CW EPR data were collected under slow-passage, non-saturating conditions. The spectrometer settings were as follows: temperature = 15 K, conversion time = 40 ms, modulation amplitude = 0.8 mT, and modulation frequency = 100 kHz; other settings are given in corresponding figure captions. Simulations of the CW spectra and the following pulsed EPR spectra were performed using EasySpin 5.1.10 toolbox[57,58] within the Matlab 2014a software suite (The Mathworks Inc., Natick, MA).
Q-band (~34.0 GHz) pulse ENDOR experiments were performed on a Bruker Biospin EleXsys 580 spectrometer equipped with a 10 W amplifier and a R.A. Isaacson cylindrical TE011 resonator in an Oxford CF935 cryostat. ENDOR measurements were performed at 20 K by employing the Mims pulse sequence (π/2-τ-π/2-RF-π/2-τ-echo) for small hyperfine couplings[59] or Davies pulse sequence (π-RF-π/2-τ-π-τ-echo) for larger hyperfine couplings[60]. ENDOR spectra were collected stochastically by randomly hopping the RF excitation frequency[61]. Pulse sequences were programmed with the PulseSPEL programmer via the Xepr interface.
For Mims-ENDOR experiments,[59] the ENDOR intensities are modulated by the response factor (R) which is a function of the hyperfine coupling A and the time interval (τ) between the first and the second π/2 microwave pulse in the three-pulse sequence: R ~ [1-cos(2πAτ)]. When Aτ = n (n = 0, 1, 2, 3 …), this factor will be zero, corresponding to a minima in the ENDOR response, i.e., the hyperfine “suppression holes” in Mims-ENDOR spectra. This Mims-hole effect can be avoided by adjusting the τ value, as shown in Figure 4B.
X-band and Q-band HYSCORE spectra were recorded on the Bruker Biospin EleXsys 580 spectrometer with a split-ring (MS5) resonator at 20 K using the pulse sequence π/2-τ-π/2-t1-π-t2-π/2-τ-echo. The pulse length for inversion pulse (tπ) and the π/2 pulse (tπ/2) was 32 ns and 16 ns, respectively. Eight-step phase cycling was used. Time-domain spectra were baseline-corrected (third-order polynomial), apodized with a hamming window, zero-filled to eight-fold points, and fast Fourier-transformed to yield the frequency-domain spectra.
Whole-Cell Patch Clamp Experiments
Recordings were made from N2a cells 24–48 hr. after transfection. Transfected cells were recognized by green fluorescence resulting from co-transfection with pEGFP-N1. Whole-cell patch clamp recordings were collected using standard techniques. Pipettes were pulled from borosilicate glass and polished to an open resistance of 2–5 megaohms. Experiments were conducted at room temperature with the following solutions: internal, 140 mM Cs-glucuronate, 5 mM CsCl, 4 mM MgATP, 1 mM Na2GTP, 10 mM EGTA, and 10 mM HEPES (pH 7.4 with CsOH); external, 150 mM NaCl, 4 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 10 mM glucose, and 10 mM HEPES (pH 7.4 with NaOH). Current signals were collected from a Multiclamp 700B amplifier (Molecular Devices, Sunnyvale, CA), digitized with a Digidata 1440 interface (Molecular Devices), and saved to disc for analysis with PClamp 10 software.
Molecular dynamics simulations
The model of the Cu2+-bound mouse PrPC protein was constructed from a representative structure from the NMR-derived dataset for 1XYX[62] (mouse PrPC fragment 121–231). Residues 23–120 were added, with the ϕ and ψ angles for residues 57–91, the OR region, were taken from Pushie and Vogel[63]. Both Cys residues were maintained in a disulfide cross linkage, and all His side chains were singly protonated at the Nδ ring position, leaving Nε available to potentially serve as a donor atom for copper. An initial simulation maintained Cu2+ coordination to each of the 4 OR His residues (Nε bound), in a square planar geometry, parameterized based on extended X-ray absorption fine structure (EXAFS) spectroscopy and DFT calculations[63,64]. MD simulations were performed with GROMACS 2016.3[65] using the OPLS-AA force field[66] modified to include parameterization of the square planar Cu2+ center[63]. The initial structure was subjected to steepest-decent energy minimization in the presence of explicit solvent, using 32360 TIP4P waters, without constraints. The starting Cu-MoPrPC (23–231) structure was simulated for 100 ns to allow the N-terminal region to equilibrate. While the N-terminal domain remained disordered is was observed to collapse and remain relatively compact. During this initial equilibration the N-terminally-bound Cu2+ site was observed to approach the face of the C-terminal domain identified in experiments (vide infra), similar to the interactions described previously in Evans et al.[18]. At the end of the equilibration phase the Cu2+ site was modified to allow each of the OR His residues to exchange with either H139 or H176. After the coordinating His parameters were swapped the steepest descent minimization was re-run to obtain a new starting structure for each of the His-exchanged models. Trajectories in the simulations were recalculated every 2 fs and structures were saved every 10 ps. Each simulation, including a simulation were no C-terminal exchange was performed (only coordination by the OR His residues), was run for 80 ns with an NPT ensemble, at 300 K and 1 atm, with each preceded by a 5 ns equilibration phase which was not included in analysis.
Conformational stability of the interacting Cu2+-binding environment and the C-terminal domain of the protein was analyzed by grouping the Cu2+ center and the heavy atoms from each coordinating His imidazole ring with the backbone heavy atoms from Helix 2 and Helix 3. This allowed each of the exchange models to be compared using the same number and type of atoms: Helix 2, Helix 3, and Cu(imidazole)4. The RMSD of these atoms were used in cluster analysis (with a 0.045 nm cutoff using gmx cluster) to quantify the variability in conformational stability. We hypothesize that if a given Cu/His configuration has a low number of clusters and if the dominant cluster represents a stable close association between the OR region and the C-terminal domain then these were likely to be more favorable configurations of the system as opposed to configurations that gave rise to broad ranges of low-population conformational states. We found that all of the configurations with clusters that represented more than 50% of the sampled structures during the simulation also had stabilizing electrostatic interactions between the Cu2+ coordination environment and anionic residues in the C-terminal domain. Importantly, of all the configurations that included two His exchanges only the simulation where His61 and His69 (the first two OR His residues) were swapped for His176 and His139 respectively resulted in a stable conformation, all other two-His exchange configurations resulted in disordered interactions between the Cu2+ coordination environment and the C-terminal domain. Of the single His exchange simulations those that involved exchange of the first or last OR His residue (His61 or His85) tended to be dominated by a single stable conformation that represented 80% or more of the sampled conformations, while the simulation with the 4th OR His exchanged for His176 had two dominant and closely-related conformations that represented 72% of all structures sampled. Close inspection of the dominant clusters revealed that most simulations formed stabilizing electrostatic interactions between the Cu2+ coordination site and Glu207 and Glu211 in the C-terminal domain.
Density functional structure calculations
Density functional calculations employed Gaussian 16, revision A.03[67], with the B3LYP functional method[68] employing the 6–311+G(d,p) basis set on all atoms, and included the Grimme-D3 dispersion correction[69] for geometry optimizations and subsequent harmonic frequency calculations. The long-range stabilizing influence of solvation was calculated from the gas-phase optimized geometries using a self-consistent reaction field dielectric continuum (IEFPCM)[70] with ε = 78.36 (for water) and employing the united atom topological model radii (UAHF). Relative free energies between calculated structures followed previously-published methods[64].
Supplementary Material
Table 1:
Summary of couplings from EPR experiments and their meanings.
| Hyperfine Coupling | Origin | Conclusion |
|---|---|---|
| aiso: 55 MHz in CW | Coordinated nitrogen on equatorial histidine | |
| aiso: 2.50 MHz in Mims ENDOR and HYSCORE | Remote nitrogen on equatorial histidine | - |
| T: 2.95 MHZ in Davies ENDOR | Exchangeable proton of axial water | - |
| A: 1.82 MHz in HYSCORE and Mims ENDOR | Oxygen from H217O | Presence of axial water |
| Absence of weak coupling of 15N in Mims ENDOR | Axial histidine ligand | No axial histidine |
Table 2:
Constructs tested by NMR in this paper
| Construct | Cis Interaction |
|---|---|
| Wild type Mouse PrPC | Cis Interaction, four his ligands |
| H139Y | Interaction shifted towards H176 |
| H176Y | Interaction shifted towards H139 |
| H186Y | Interaction maintained |
| H186R | Interaction maintained |
| H139Y,H176Y | Strongly reduced interaction |
| H139N,H176N | Strongly reduced interaction |
| H139A,H176A | Strongly reduced interaction |
| M139A,I138A,F140A | Misfold / no data on interaction |
| N172A,N173A,V175A,V179A,N180A | Interaction maintained |
| Segment 90–230 | No interaction |
| H176N,N172H | Interaction moved down helix |
| H176N,N180H | Interaction moved up helix |
| 3XORI | Very weakly reduced interaction |
| 4XORI | Weakly reduced interaction |
| 5XORI | Strongly reduced interaction |
| H84Y | Slightly stronger cis interaction than WT |
| H95Y, H110Y | No difference in cis interaction from WT |
Highlights.
The two domains of the cellular prion protein are tethered together by a copper ion
This tethering is the basis of the protein’s neuroprotective cis interaction
Disruption of this interaction explains the toxicity of prion diseases
Disruption may explain prion protein mediated toxicity in Alzheimer’s disease
Acknowledgements
We thank NIH grants R01GM065790, R35GM131781 (GLM), R35GM126961 (RDB) and R01NS065244 (DAH) for financial support, and NIH instrumentation grant S10OD018455 for acquisition of the 800 MHz NMR spectrometer and S10OD024980 for acquisition of the pulsed EPR spectrometer. This research was enabled in part by support provided by WestGrid (www.westgrid.ca) and Compute Canada Calcul Canada (www.computecanada.ca).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Prusiner SB (1982). Novel proteinaceous infectious particles cause scrapie. Science 216, 136–144 [DOI] [PubMed] [Google Scholar]
- 2.Prusiner SB (1998). Prions. Proc. Natl. Acad. of Sci. USA 95, 13363–13383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Riesner D. (2003). Biochemistry and structure of PrPC and PrPSc. Br. Med. Bull 66, 21–33 [DOI] [PubMed] [Google Scholar]
- 4.Wille H, & Requena J. (2018). The Structure of PrPSc Prions. Pathogens 7, 20–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mallucci GR, Ratté S, Asante EA, Linehan J, Gowland I, Jefferys JGR, & Collinge J. (2002). Post-natal knockout of prion protein alters hippocampal CA1 properties, but does not result in neurodegeneration. EMBO J. 21, 202–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biasini E, Turnbaugh JA, Unterberger U, & Harris DA (2012). Prion protein at the crossroads of physiology and disease. Trends Neurosci. 35, 92–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brandner S, Isenmann S, Raeber A, Fischer M, Sailer A, Kobayashi Y, Marino S, Weissmann C, & Aguzzi A. (1996). Normal host prion protein necessary for scrapie-induced neurotoxicity. Nature 379, 339–343 [DOI] [PubMed] [Google Scholar]
- 8.Chesebro B, Trifilo M, Race R, Meade-White K, Teng C, LaCasse R, Raymond L, Favara C, Baron G, Priola S, Caughey B, Masliah E, & Oldstone M. (2005). Anchorless prion protein results in infectious amyloid disease without clinical scrapie. Science 308, 1435–1439 [DOI] [PubMed] [Google Scholar]
- 9.Mallucci G, Dickinson A, Linehan J, Klöhn P-C, Brandner S, & Collinge J. (2003). Depleting neuronal PrP in prion infection prevents disease and reverses spongiosis. Science 302, 871–874 [DOI] [PubMed] [Google Scholar]
- 10.Li A, Christensen HM, Stewart LR, Roth KA, Chiesa R, & Harris DA (2007). Neonatal lethality in transgenic mice expressing prion protein with a deletion of residues 105–125. EMBO J. 26, 548–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu B, McDonald AJ, Markham K, Rich CB, McHugh KP, Tatzelt J, Colby DW, Millhauser GL, & Harris DA (2017). The N-terminus of the prion protein is a toxic effector regulated by the C-terminus. eLife 6:e23473, 1–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reimann RR, Sonati T, Hornemann S, Herrmann US, Arand M, Hawke S, & Aguzzi A. (2016). Differential Toxicity of Antibodies to the Prion Protein. PLoS Pathog. 12, e1005401, 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Solomon IH, Huettner JE, & Harris DA (2010). Neurotoxic mutants of the prion protein induce spontaneous ionic currents in cultured cells. J. Biol. Chem 285, 26719–26726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sonati T, Reimann RR, Falsig J, Baral PK, O’Connor T, Hornemann S, Yaganoglu S, Li B, Herrmann US, Wieland B, Swayampakula M, Rahman MH, Das D, Kav N, Riek R, Liberski PP, James MNG, & Aguzzi A. (2013). The toxicity of antiprion antibodies is mediated by the flexible tail of the prion protein. Nature 501, 102–106 [DOI] [PubMed] [Google Scholar]
- 15.Spevacek AR, Evans EGB, Miller JL, Meyer HC, Pelton JG, & Millhauser GL (2013). Zinc Drives a Tertiary Fold in the Prion Protein with Familial Disease Mutation Sites at the Interface. Structure 21, 236–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen L, & Ji H-F (2011). Mutation directional selection sheds light on prion pathogenesis. Biochem. Biophys. Res. Commun 410, 159–163 [DOI] [PubMed] [Google Scholar]
- 17.Thakur AK, Srivastava AK, & Srinivas V. Chary KVR, Rao CM (2011). Copper alters aggregation behavior of prion protein and induces novel interactions between its N-and C-terminal regions. J. Biol. Chem 286, 38533–38545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evans EGB, Pushie MJ, Markham KA, Lee H-W, & Millhauser GL (2016). Interaction between Prion Protein’s Copper-Bound Octarepeat Domain and a Charged C-Terminal Pocket Suggests a Mechanism for N-Terminal Regulation. Structure. 24, 1057–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silveira JR, Raymond GJ, Hughson AG, Race RE, Sim VL, Hayes SF, & Caughey B. (2005). The most infectious prion protein particles. Nature 437, 257–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masel J, Genoud N, & Aguzzi A. (2005). Efficient Inhibition of Prion Replication by PrP-Fc2 Suggests that the Prion is a PrPSc Oligomer. J. Mol. Bio 345, 1243–1251 [DOI] [PubMed] [Google Scholar]
- 21.McDonald AJ, Leon DR, Markham KA, Wu B, Heckendorf CF, Schilling K, Showalter HD, Andrews PC, McComb ME, Pushie MJ, Costello CE, Millhauser GL, & Harris DA (2019). Altered Domain Structure of the Prion Protein Caused by Cu2+ Binding and Functionally Relevant Mutations: Analysis by Cross-Linking, MS/MS, and NMR. Structure. 27, 907–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pushie MJ, Pickering IJ, Martin GR, Tsutsui S, Jirik FR, & George GN (2011). Prion protein expression level alters regional copper, iron and zinc content in the mouse brain. Metallomics 3, 206–214 [DOI] [PubMed] [Google Scholar]
- 23.Armendariz AD, Gonzalez M, Loguinov AV, & Vulpe CD (2004). Gene expression profiling in chronic copper overload reveals upregulation of Prnp and App. Physiol. Genomics 20, 45–54 [DOI] [PubMed] [Google Scholar]
- 24.Varela-Nallar L, Toledo EM, Larrondo LF, Cabral ALB, Martins VR, & Inestrosa NC (2006). Induction of cellular prion protein gene expression by copper in neurons. Am. J. Physiol. Cell Physiol 290, C271–C281 [DOI] [PubMed] [Google Scholar]
- 25.Linden R, Martins VR, Prado MAM, Cammarota M, Izquierdo I, & Brentani RR (2008). Physiology of the Prion Protein. Physiol. Rev 88, 673–728 [DOI] [PubMed] [Google Scholar]
- 26.Aronoff-Spencer E, Burns CS, Avdievich NI, Gerfen GJ, Peisach J, Antholine WE, Ball HL, Cohen FE, Prusiner SB, & Millhauser GL (2000). Identification of the Cu2+ Binding Sites in the N-Terminal Domain of the Prion Protein by EPR and CD Spectroscopy. Biochemistry 39, 13760–13771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burns CS, Aronoff-Spencer E, Legname G, Prusiner SB, Antholine WE, Gerfen GJ, Peisach J, & Millhauser GL (2003). Copper Coordination in the Full-Length, Recombinant Prion Protein. Biochemistry 42, 6794–6803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burns CS, Aronoff-Spencer E, Dunham CM, Lario P, Avdievich NI, Antholine WE, Olmstead MM, Vrielink A, Gerfen GJ, Peisach J, Scott WG, & Millhauser GL (2002). Molecular features of the copper binding sites in the octarepeat domain of the prion protein. Biochemistry 41, 3991–4001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chattopadhyay M, Walter ED, Newell DJ, Jackson PJ, Aronoff-Spencer E, Peisach J, Gerfen GJ, Bennett B, Antholine WE, & Millhauser GL (2005). The Octarepeat Domain of the Prion Protein Binds Cu(II) with Three Distinct Coordination Modes at pH 7.4. J. Am. Chem. Soc 127, 12647–12656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jackson GS, Murray I, Hosszu LLP, Gibbs N, Waltho JP, Clarke AR, & Collinge J. (2001). Location and properties of metal-binding sites on the human prion protein. Proc. Natl. Acad. of Sci. USA 98, 8531–8535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salzano G, Giachin G, & Legname G. (2019). Structural Consequences of Copper Binding to the Prion Protein. Cells 8, 770–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Millhauser GL (2004). Copper Binding in the Prion Protein. Acc. Chem. Res 37, 79–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dudzik CG, Walter ED, Abrams BS, Jurica MS, & Millhauser GL (2012). Coordination of Copper to the Membrane-Bound Form of α-Synuclein. Biochemistry 52, 53–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walter ED, Stevens DJ, Spevacek AR, Visconte MP, Dei Rossi A, & Millhauser GL (2009). Copper binding extrinsic to the octarepeat region in the prion protein. Curr. Protein Pept. Sci 10, 529–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dikanov SA, Spoyalov AP, & Hüttermann J. (1994). Exploiting the properties of line‐shape singularities in orientationally selected electron spin echo envelope modulation spectra of Cu2+(15N‐imidazole)4 for the determination of hyperfine coupling with the remote imidazole nitrogen. J. Chem. Phys 100, 7973–7983 [Google Scholar]
- 36.Iwasaki T, Kounosu A, Uzawa T, Samoilova RI, & Dikanov SA (2004). Orientation-Selected 15N-HYSCORE Detection of Weakly Coupled Nitrogens around the Archaeal Rieske [2Fe−2S] Center. J. Am. Chem. Soc 126, 13902–13903 [DOI] [PubMed] [Google Scholar]
- 37.Hoff AJ (1989). Advanced EPR Applications in Biochemistry and Biology. Elsevier; Amsterdam [Google Scholar]
- 38.Getz D, & Silver BL (1974). ESR of Cu2+(H2O)6. I. The oxygen‐17 superhyperfine tensors in 63Cu2+ doped zinc Tutton’s salt at 20 °K. J. Chem. Phys 61, 630–637 [Google Scholar]
- 39.Kim D, Kim NH, & Kim SH (2012). 34 GHz Pulsed ENDOR Characterization of the Copper Coordination of an Amyloid β Peptide Relevant to Alzheimer’s Disease. Angew. Chem. Int. Ed 52, 1139–1142 [DOI] [PubMed] [Google Scholar]
- 40.Atherton NM, & Horsewill AJ (2006). Proton ENDOR of Cu(H2O)62+ in Mg(NH4) 2(SO2)4 . 6H2O. Mol. Phys 37, 1349–1361 [Google Scholar]
- 41.Stevens DJ, Walter ED, Rodríguez A, Draper D, Davies P, Brown DR, & Millhauser GL (2009). Early Onset Prion Disease from Octarepeat Expansion Correlates with Copper Binding Properties. PLoS Pathog. 5, e1000390 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beck JA, Mead S, Campbell TA, Dickinson A, Wientjens DP, Croes EA, Van Duijn CM, & Collinge J. (2001). Two-octapeptide repeat deletion of prion protein associated with rapidly progressive dementia. Neurology 57, 354–356 [DOI] [PubMed] [Google Scholar]
- 43.Capellari S, Parchi P, Wolff BD, Campbell J, Atkinson R, Posey DM, Petersen RB, & Gambetti P. (2002). Creutzfeldt–Jakob disease associated with a deletion of two repeats in the prion protein gene. Neurology 59, 1628–1630 [DOI] [PubMed] [Google Scholar]
- 44.Piazza M, Prior TW, Khalsa PS, & Appleby B. (2020). A case report of genetic prion disease with two different PRNP variants. Mol. Genet. Genomic. Med 8, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katayama S, Nakase I, Yano Y, Murayama T, Nakata Y, Matsuzaki K, & Futaki S. (2013). Effects of pyrenebutyrate on the translocation of arginine-rich cell-penetrating peptides through artificial membranes: Recruiting peptides to the membranes, dissipating liquid-ordered phases, and inducing curvature. BBA. - Biomembranes 1828, 2134–2142 [DOI] [PubMed] [Google Scholar]
- 46.Guterstam P, Madani F, Hirose H, Takeuchi T, Futaki S, Andaloussi SE, Gräslund A, & Langel Ü (2009). Elucidating cell-penetrating peptide mechanisms of action for membrane interaction, cellular uptake, and translocation utilizing the hydrophobic counter-anion pyrenebutyrate. Biochim. Biophys. Acta, Biomembr 1788, 2509–2517 [DOI] [PubMed] [Google Scholar]
- 47.Madani F, Lindberg S, Langel Ü, Futaki S, & Gräslund A. (2011). Mechanisms of Cellular Uptake of Cell-Penetrating Peptides. J. Biophys 2011, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Canters GW, Kolczak U, Armstrong F, Jeuken LJC, Camba R, & Sola M. (2000). The effect of pH and ligand exchange on the redox properties of blue copper proteins. Faraday Discuss. 116, 205–220 [DOI] [PubMed] [Google Scholar]
- 49.Banci L, Bertini I, Cantini F, Felli IC, Gonnelli L, Hadjiliadis N, Pierattelli R, Rosato A, & Voulgaris P. (2006). The Atx1-Ccc2 complex is a metal-mediated protein-protein interaction. Nat. Chem. Biol 2, 367–368 [DOI] [PubMed] [Google Scholar]
- 50.Chacón KN, Perkins J, Mathe Z, Alwan K, Ho EN, Ucisik MN, Merz KM, & Blackburn NJ (2018). Trapping intermediates in metal transfer reactions of the CusCBAF export pump of Escherichia coli. Commun. Biol 1, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garnett AP, & Viles JH (2003). Copper binding to the octarepeats of the prion protein. Affinity, specificity, folding, and cooperativity: insights from circular dichroism. J. Biol. Chem 278, 6795–6802 [DOI] [PubMed] [Google Scholar]
- 52.Qin K, Yang Y, Mastrangelo P, & Westaway D. (2002). Mapping Cu(II) Binding Sites in Prion Proteins by Diethyl Pyrocarbonate Modification and Matrix-assisted Laser Desorption Ionization-Time of Flight (MALDI-TOF) Mass Spectrometric Footprinting. J. Biol. Chem 277, 1981–1990 [DOI] [PubMed] [Google Scholar]
- 53.Cereghetti GM, Schweiger A, Glockshuber R, & Van Doorslaer S. (2001). Electron paramagnetic resonance evidence for binding of Cu2+ to the C-terminal domain of the murine prion protein. Biophys. J 81, 516–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Colombo MC, VandeVondele J, Van Doorslaer S, Laio A, Guidoni L, & Rothlisberger U. (2007). Copper binding sites in the C-terminal domain of mouse prion protein: A hybrid (QM/MM) molecular dynamics study. Proteins 70, 1084–1098 [DOI] [PubMed] [Google Scholar]
- 55.Van Doorslaer S, Cereghetti GM, Glockshuber R, & Schweiger A. (2001). Unraveling the Cu2+ Binding Sites in the C-Terminal Domain of the Murine Prion Protein: A Pulse EPR and ENDOR Study. J. Phys. Chem. B 105, 1631–1639 [Google Scholar]
- 56.Van Doorslaer S, & Vinck E. (2007). The strength of EPR and ENDOR techniques in revealing structure–function relationships in metalloproteins. Phys. Chem. Chem. Phys 9, 4620–4638 [DOI] [PubMed] [Google Scholar]
- 57.Stoll S, & Schweiger A. (2006). EasySpin, a comprehensive software package for spectral simulation and analysis in EPR. J. Magn. Reson 178, 42–55 [DOI] [PubMed] [Google Scholar]
- 58.Stoll S, & Britt RD (2009). General and efficient simulation of pulse EPR spectra. Phys. Chem. Chem. Phys 11, 6614–6625 [DOI] [PubMed] [Google Scholar]
- 59.Mims WB (1965). Pulsed Endor Experiments. Proc. R. Soc. London, Ser. A 283, 452–457 [Google Scholar]
- 60.Davies ER (1974). A new pulse endor technique. Phys. Let. A 47, 1–2 [Google Scholar]
- 61.Brüggemann W, & Niklas JR (1994). Stochastic ENDOR. J. Magn. Reson 108, 25–29 [Google Scholar]
- 62.Gossert AD, Bonjour S, Lysek DA, Fiorito F, Wüthrich K. (2005) Prion protein NMR structures of elk and of mouse/elk hybrids. Proc. Natl. Acad. of Sci. USA 102, 646–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pushie MJ, & Vogel HJ (2008). Modeling by Assembly and Molecular Dynamics Simulations of the Low Cu2+ Occupancy Form of the Mammalian Prion Protein Octarepeat Region: Gaining Insight into Cu2+-Mediated β-Cleavage. Biophys. J 95, 5084–5091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pushie MJ, Nienaber KH, McDonald A, Millhauser GL, & George GN (2014). Combined EXAFS and DFT Structure Calculations Provide Structural Insights into the 1:1 Multi-Histidine Complexes of CuII, CuI, and ZnII with the Tandem Octarepeats of the Mammalian Prion Protein. Chem. Eur. J 20, 9770–9783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, & Lindahl E. (2015). GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1-2, 19–25 [Google Scholar]
- 66.Jorgensen WL, Maxwell DS, & Tirado-Rives J. (1996). Development and Testing of the OPLS All-Atom Force Field on Conformational Energetics and Properties of Organic Liquids. J. Am. Chem. Soc 118, 11225–11236 [Google Scholar]
- 67.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Petersson GA, Nakatsuji H, Li X, Caricato M, Marenich AV, Bloino J, Janesko BG, Gomperts R, Mennucci B, Hratchian HP, Ortiz JV, Izmaylov AF, Sonnenberg JL, Williams, Ding F, Lipparini F, Egidi F, Goings J, Peng B, Petrone A, Henderson T, Ranasinghe D, Zakrzewski VG, Gao J, Rega N, Zheng G, Liang W, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Throssell K, Montgomery JA Jr, Peralta JE, Ogliaro F, Bearpark MJ, Heyd JJ, Brothers EN, Kudin KN, Staroverov VN, Keith TA, Kobayashi R, Normand J, Raghavachari K, Rendell AP, Burant JC, Iyengar SS, Tomasi J, Cossi M, Millam JM, Klene M, Adamo C, Cammi R, Ochterski JW, Martin RL, Morokuma K, Farkas O, Foresman JB, & Fox DJ (2016). Gaussian 16 Rev. A.03. [Google Scholar]
- 68.Becke AD, (1993). Density‐functional thermochemistry. III. The role of exact exchange. J. Chem. Phys 98, 5648–5652 [Google Scholar]
- 69.Grimme S, Antony J, Ehrlich S, & Krieg H. (2010). A consistent and accurate ab initioparametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J. Chem. Phys 132, 154104–20 [DOI] [PubMed] [Google Scholar]
- 70.Scalmani G, & Frisch MJ (2010). Continuous surface charge polarizable continuum models of solvation. I. General formalism. J. Chem. Phys 132, 114110–16 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.