Abstract
Background and Purpose
Stroke is a complex disease with multiple genetic and environmental risk factors. African Americans endure a nearly two-fold greater risk of stroke and are 2–3 times more likely to die from stroke than European Americans.
Methods
The Consortium of Minority Population genome-wide Association Studies of Stroke (COMPASS) has conducted a genome-wide association meta-analysis of stroke in more than 22,000 individuals of African ancestry (3,734 cases, 18,317 controls) from 13 cohorts.
Results
In meta-analyses, we identified one SNP (rs55931441) near the HNF1A gene that reached genome-wide significance (P = 4.62×10−8) and an additional 29 variants with suggestive evidence of association (P < 1×10−6), representing 24 unique loci. For validation, a look-up analysis for a 100Kb region flanking the COMPASS SNP was performed in SiGN Europeans, SiGN Hispanics, and METASTROKE (Europeans). Using a stringent Bonferroni correction P-value of 2.08 × 10−3 (0.05/24 unique loci), we were able to validate associations at the HNF1A locus in both SiGN (P = 8.18 × 10−4) and METASTROKE (P = 1.72 × 10−3) European populations. Overall, 16 of 24 loci showed evidence for validation across multiple populations. Previous studies have reported associations between variants in the HNF1A gene and lipids, C-reactive protein, and risk of coronary artery disease and stroke. Suggestive associations with variants in the SFXN4 and TMEM108 genes represent potential novel ischemic stroke loci.
Conclusion
These findings represent the most thorough investigation of genetic determinants of stroke in individuals of African descent, to date.
Keywords: stroke, meta-analysis, genome-wide association study, African American
SUMMARY
Despite its limitations, genetic studies such as COMPASS, that include minority populations have the huge potential to provide insight into the mechanisms underlying stroke disparities, such as the more than doubled incidence and mortality rates and younger age of onset for stroke observed in African Americans.5, 47 Our study identified novel associations for stroke that might not otherwise be detected in primarily European cohort studies. Collectively this highlights the critical nature and importance of genetic studies in a more diverse population with a high stroke burden, such as was the case in this study.
INTRODUCTION
Stroke is the second leading cause of death worldwide and a leading cause of long-term disability in the United States.1 Stroke is a heterogeneous disease encompassing multiple subtypes with unique etiologies and risk factors.2 Nearly 87% of the ~795,000 strokes that occur each year in the US are ischemic.1 Epidemiological studies suggest a substantial genetic component for stroke with overall heritability estimates of 38% for all ischemic strokes, and subtype-specific estimates of 20–25% for small-vessel disease3 and up to 40% for large-vessel disease.4 Compared to European Americans, African Americans have a nearly two-fold greater risk of incident stroke, more than two-fold increased risk of fatal stroke, strokes at younger ages, and higher frequency of post-stroke disability.5, 6 Despite this disproportionate burden, few attempts to map stroke susceptibility loci have focused on individuals of African ancestry.7 Recent genome-wide association studies (GWAS) have identified several stroke susceptibility loci8–14 primarily in individuals of European ancestry with little success replicating in non-European ancestry populations7, 13, 15–16 possibly due to differences in the genetic architecture of stroke among individuals of diverse ancestry.
This study represents a collective effort to investigate the genetic basis of stroke by mapping stroke susceptibility loci potentially unique to individuals of African ancestry. Using data obtained from the Consortium of Minority Population genome-wide Association Studies of Stroke (COMPASS), we expand upon our discovery GWAS meta-analysis of stroke in African-Americans7 using 1000 genomes (1000G) imputed data in 22,000 individuals.
METHODS
In order to minimize the possibility of unintentionally sharing information that can be used to re-identify private information, a subset of the data generated for this study are available at dbGaP and can be accessed at https://www.ncbi.nlm.nih.gov/gap/.
Study population
COMPASS included a total of 22,051 individuals of African descent with either a physician-adjudicated stroke (n= 3,734) or no history of stroke (n= 18,317) (Supplemental Table I) and genome-wide single nucleotide polymorphism (SNP) data. Participating studies include prospective cohorts[Atherosclerosis Risk in Communities (ARIC) study,17 Cardiovascular Health Study (CHS)18, Jackson Heart Study (JHS)19–20, the Women’s Health Initiative (WHI),21]; case-control studies [INTERSTROKE22, REasons for Geographic And Racial Differences in Stroke (REGARDS)23, Ischemic Stroke Genetics Study (ISGS),24 Vitamin Intervention for Stroke Prevention (VISP)25–26, South London Ethnicity and Stroke Study (SLESS)27, the Genetics of Early Onset Stroke (GEOS) Study28, the National Institute of Neurological Disorders and Stroke- Stroke Genetics Network (NINDS-SiGN)29, Healthy Aging in Neighborhoods of Diversity across the Life Span (HANDLS)30]; and an affected sibpair study--Siblings with Ischemic Stroke Study (SWISS).31 Race/ethnicity- and sex-matched controls were randomly selected from HANDLS and used as controls in the analyses of SWISS, ISGS and VISP, which lacked genotyped controls. All participants provided written, informed consent and institutional review boards approved each of the respective studies/institutions.
Outcomes
We defined stroke as a focal neurological deficit of presumed vascular cause with a sudden onset and lasting 24 hours or until death with clinical and/or radiological (CT/MRI) evidence with stroke diagnosis made when there is overwhelming clinical evidence in the absence of radiological evidence of a cerebral infarction. A lack of imaging data for all stroke cases does not increase the likelihood of false positives in our study. The cohort studies only considered first (incident) clinically validated ischemic strokes. Individuals with a baseline history of ischemic or hemorrhagic stroke were excluded.
Genotype data
All studies imputed SNPs using 1000G Phase I Version 3 Haplotypes (1KGp1v3), except SLESS and WHI, which used 1000G Phase III data (1KGp3) reference populations. We excluded SNPs if they had invalid or missing alleles, P-Values, or Beta values; had minor allele frequencies (MAF) < 1%; imputation quality (r2) <0.3; or were located on sex chromosomes. We analyzed SNPs available in two or more studies, for a total of ~16.9 million SNPs. The Supplement contains study-specific details about design, stroke definition, adjudication procedures, and genotyping.
Analysis
We used logistic regression (additive genetic model) analyses with a count of variant alleles (0, 1, or 2) for each genotyped SNP or allelic dose for imputed SNPs. To control for potential population stratification, we included estimated study specific principal components of global ancestry as covariates. As appropriate, we adjusted models for age, sex, and study site. We combined study-specific results in a fixed effects meta-analyses with inverse variance weighting (IVW) using METAL.32 We also performed sample size weighted (SSW) meta-analysis as an alternative approach to IVW (Supplemental Table II). We set a genome-wide significance (discovery) threshold of P<5×10−8 but investigated all SNPs with P<10−6.
Validation of COMPASS Findings
Due to the absence of a comparable and adequately powered cohort of African Americans with GWAS and adjudicated stroke data, we performed a ‘look-up’ of COMPASS SNPs with P<10−6 in the SiGN European and Hispanic ischemic stroke populations and METASTROKE total ischemic stroke populations (Supplemental Table III). Additional METASTROKE subtype (cardio-embolic, large-vessel, and small vessel) specific look-up analyses were performed to further validate these findings. Given the known differences in linkage disequilibrium (LD) patterns between populations of European and African ancestry, we expanded the region of interest for each locus to include available SNPs ±100kb of the index COMPASS SNPs as previously described7 applying a Bonferroni correction to account for the number of loci tested.
RESULTS
Discovery of stroke-associated loci
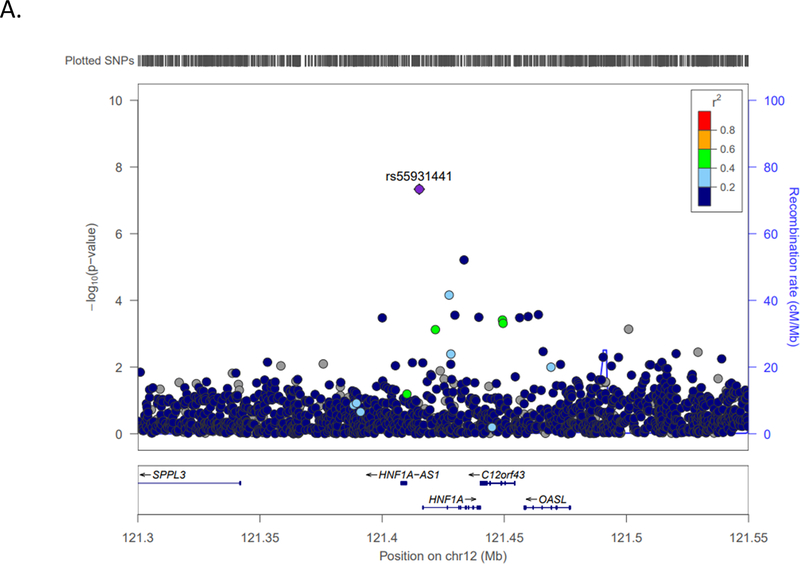
Using IVW meta-analyses (Table 1) we identified one genome-wide significant association (P<5×10−8) and an additional 29 variants with suggestive evidence of association (P<1×10−6), representing 24 unique loci in total. The genome-wide significant association was detected upstream of the HNF1 homeobox A (HNF1A) gene on chromosome 12 (rs55931441; P=4.62×10−8, odds ratio (OR)=1.68) (Figure 1A).
Table 1.
COMPASS Ischemic Stroke Suggestive and Genome-wide Significant Inverse Variance Weighted Associations
| Chr | Position* | Gene | SNP | Alleles (Coded/ Noncoded) | Beta | Standard Error | Odds Ratio (Confidence Interval) | Inverse Variance Weighted P-Value | Direction | Het P Value | Sample Size | Number of Studies |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 112853017 | CTTNBP2NL (nearest) | rs114947355 | T/C | 0.44 | 0.0902 | 1.56 (1.42–1.70) | 9.05×10−07 | ????-+?+??? | 0.1382 | 12610 | 3 |
| 1 | 112857084 | CTTNBP2NL (nearest) | rs147779128 | A/T | −0.46 | 0.0945 | 0.63 (0.57–0.69) | 9.61×10−07 | ?????-?-??? | 0.9293 | 9637 | 2 |
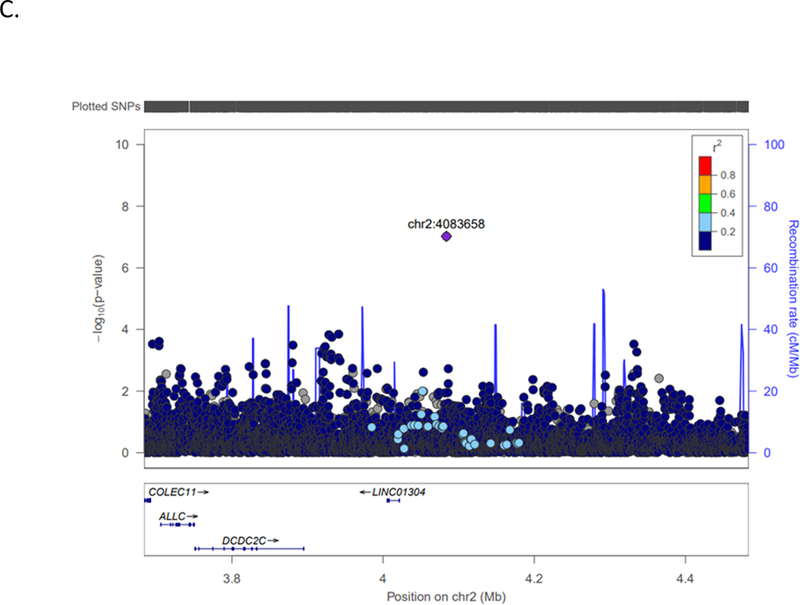
| 2 | 4083658 | NPM1P48 (nearest) | rs142655108 | A/C | 0.58 | 0.1089 | 1.79 (1.60–1.99) | 9.52×10−08 | ?????+?+??? | 0.2834 | 9637 | 2 |
| 2 | 198551159 | RFTN2 and MARS2 (nearest) | rs115670077 | T/G | 0.35 | 0.072 | 1.43 (1.33–1.53) | 8.48×10−07 | +?++++?+??? | 0.5735 | 16540 | 6 |
| 3 | 124048486 | KALRN | rs72976591 | A/C | 0.17 | 0.0342 | 1.18 (1.14–1.22) | 9.19×10−07 | +++++++++++ | 0.5356 | 22018 | 11 |
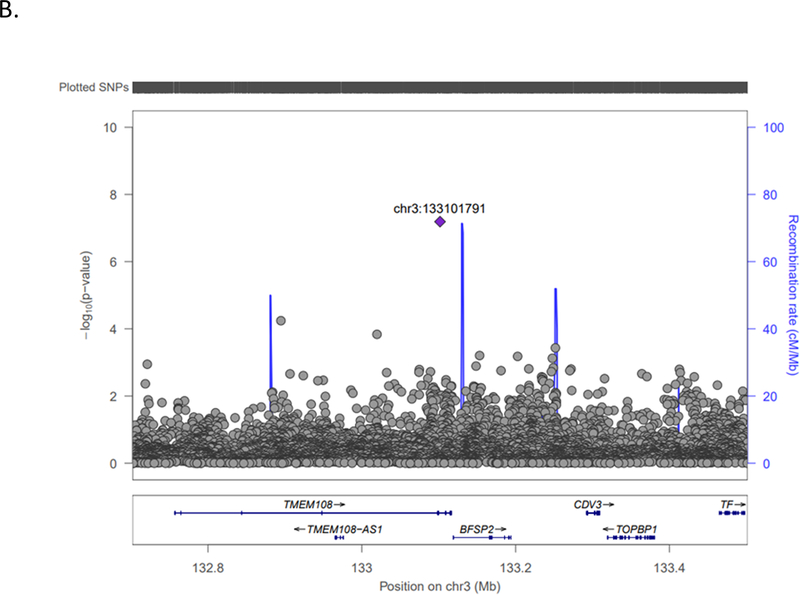
| 3 | 133101791 | TMEM108 | rs113509723 | -/AA | 0.45 | 0.0841 | 1.58 (1.45–1.71) | 6.46×10−08 | ?????+?+??? | 0.2014 | 9637 | 2 |
| 3 | 153125290 | AK092619 (nearest) | rs184221467 | A/G | 0.62 | 0.1246 | 1.85 (1.63–2.10) | 7.86×10−07 | ?????+?+??? | 0.468 | 9637 | 2 |
| 4 | 99435032 | TSPAN5 | rs138134155 | A/G | 0.36 | 0.0705 | 1.43 (1.33–1.53) | 3.94×10−07 | +?++++?++?? | 0.9442 | 18531 | 7 |
| 5 | 101123995 | OR7H2P (nearest) | rs77460585 | A/G | 0.59 | 0.1165 | 1.80 (1.60–2.02) | 4.36×10−07 | ????-??+??? | 0.004981 | 10940 | 2 |
| 5 | 150981704 | FAT2 and SPARC (nearest) | rs114527838 | A/G | −0.28 | 0.055 | 0.76 (0.72–0.80) | 5.55×10−07 | -?-------?? | 0.7033 | 19032 | 8 |
| 6 | 97345991 | KLHL32 and NDUFA4 (nearest) | rs146522546 | -/CT | −0.45 | 0.0876 | 0.64 (0.58–0.69) | 2.22×10−07 | ???—?-??? | 0.3829 | 13353 | 4 |
| 7 | 83432409 | SEMA3A | rs6967981 | T/G | 0.15 | 0.0296 | 1.16 (1.12–1.19) | 7.57×10−07 | +-++++++++- | 0.1685 | 21970 | 11 |
| 8 | 1572874 | DLGAP2 | rs112455974 | A/C | 0.68 | 0.1336 | 1.97 (1.72–2.25) | 3.77×10−07 | ????+??+??? | 0.7366 | 10949 | 2 |
| 9 | 72475192 | C9orf135 | rs565295967 | T/C | 0.62 | 0.1199 | 1.86 (1.65–2.09) | 2.41×10−07 | ?????+?+??? | 0.1048 | 9637 | 2 |
| 10 | 53545098 | PRKG1 | rs140164788 | T/C | 0.52 | 0.1019 | 1.68 (1.52–1.86) | 3.37×10−07 | ????++?+??? | 0.7146 | 12618 | 3 |
| 10 | 53547264 | PRKG1 | rs74469072 | T/G | 0.52 | 0.1018 | 1.68 (1.52–1.86) | 3.50×10−07 | ????++?+??? | 0.7169 | 12618 | 3 |
| 10 | 120907173 | SFXN4 | rs150807690 | -/G | −0.20 | 0.0378 | 0.82 (0.79–0.85) | 9.67×10−08 | ?-?---?---- | 0.3014 | 18180 | 8 |
| 11 | 11360296 | GALNT18 | rs115825287 | T/C | 0.35 | 0.0696 | 1.43 (1.33–1.53) | 3.60×10−07 | ??++++?+??? | 0.6076 | 15673 | 5 |
| 11 | 75683895 | UVRAG | rs368167310 | T/C | −0.55 | 0.1085 | 0.58 (0.52–0.65) | 4.87×10−07 | ?????-?-??? | 0.8172 | 9637 | 2 |
| 12 | 29288407 | FAR2 (nearest) | rs113025543 | A/T | −0.27 | 0.0551 | 0.76 (0.72–0.81) | 9.23×10−07 | -+--------? | 0.7896 | 20224 | 10 |
| 12 | 29292793 | FAR2 (nearest) | rs142100833 | C/G | 0.24 | 0.0488 | 1.27 (1.21–1.34) | 8.65×10−07 | +-+++++++-? | 0.4482 | 20119 | 10 |
| 12 | 29341407 | FAR2 | - | -/?? | 0.65 | 0.1272 | 1.91 (1.68–2.17) | 3.79×10−07 | ???++???+?? | 0.9784 | 5542 | 3 |
| 12 | 119502791 | SRRM4 | rs531465435 | -/C | 0.59 | 0.1162 | 1.81 (1.61–2.03) | 3.39×10−07 | ?????+?+??? | 0.5809 | 9637 | 2 |
| 12 | 119542751 | SRRM4 | rs192977447 | A/T | 0.43 | 0.0816 | 1.53 (1.41–1.66) | 1.80×10−07 | ???+++?++?? | 0.1962 | 15333 | 5 |
| 12 | 121415209 | HNF1A (nearest) | rs55931441 | A/G | 0.52 | 0.0947 | 1.68 (1.53–1.84) | 4.62×10−08 | ?????+?+??? | 0.4599 | 9637 | 2 |
| 14 | 93788855 | BTBD7 | rs113949028 | -/G | 0.20 | 0.0396 | 1.22 (1.17–1.27) | 5.44×10−07 | ?+?+++?++++ | 0.948 | 18255 | 8 |
| 18 | 68475060 | GTSCR1 (nearest) | rs181095590 | A/G | 0.58 | 0.1138 | 1.78 (1.59–2.00) | 3.90×10−07 | ?????+?+??? | 0.4538 | 9637 | 2 |
| 19 | 29710081 | UQCRFS1 (nearest) | rs73923591 | A/G | 0.27 | 0.0548 | 1.31 (1.24–1.39) | 6.18×10−07 | ++++++++++? | 0.8774 | 20246 | 10 |
| 21 | 36442465 | RUNX1 | rs116262092 | A/T | −0.58 | 0.1174 | 0.56 (0.50–0.63) | 7.04×10−07 | ????–?-??? | 0.9789 | 12581 | 3 |
| 21 | 36443919 | RUNX1 | rs147867382 | C/G | −0.58 | 0.1174 | 0.56 (0.50–0.63) | 7.95×10−07 | ????–?-??? | 0.9792 | 12579 | 3 |
Chromosome (Chr) Position based on Human Genome (GRCh37/hg19)
Direction indicates the direction of the effect size: negative (−), neutral/unknown (./?), and positive (+) for each contributing cohort/population
Figure 1.
LocusZoom plots, with linkage disequilibrium based on hg19/1000 Genomes Nov 2014 AFR, depicting the top (P= 10−8) three associations with ischemic stroke in COMPASS individuals of African descent. A.) HNF1A (rs55931441) chromosome 12 locus; B.) TMEM108 (rs113509723) chromosome 3 locus; C.) Chromosome 2 (rs142655108) locus nearest NPM1P48.
Validation of COMPASS SNPs in SiGN and METASTROKE
Expanding to the flanking regions and using a stringent Bonferroni correction of ⍺=2.08×10−3 for replication (0.05/24 unique loci), our most significant locus, HNF1A, was validated in both SiGN and METASTROKE European ancestry cohorts and approached significance in SiGN Hispanics (Supplemental Figure I). Overall, 16 of 24 loci showed evidence for validation across multiple populations (Table 2).
Table 2.
Genome-wide and suggestive COMPASS associations with lookups in European and Hispanic populations from SiGN and METASTROKE
| Chr | Unique Locus | Top SiGN European SNP | Alleles | Z Score | P-Value | Direction | Top SiGN Hispanic SNP | Alleles | Z Score | P-Value | Direction | Metastroke Top SNP | Alleles | Effect | P-Value | Direction |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | CTTNBP2NL (nearest) | rs186896391 | C/A | −3.28 | 0.0010 | ------+++-. | rs3121986 | A/G | −2.79 | 0.0052 | - | rs10158830 | C/G | 0.073 | 0.0019 | ++++++-+++++ |
| 2 | NPM1P48 (nearest) | 2–4077298 (rs527602504) | TC/T | 2.56 | 0.0104 | ......+.... | rs60037207 | T/C | −2.21 | 0.0268 | - | rs114152357 | A/T | −0.186 | 0.0048 | --+-++------ |
| 2 | RFTN2 and MARS2 (nearest) | 2–198592085 (rs543821034) | C/T | 2.98 | 0.0029 | ....+...... | rs150235598 | G/A | −2.74 | 0.0061 | - | rs191948652 | A/T | 0.513 | 0.005 | +-+???+++?+? |
| 3 | KALRN | rs2034173 | T/C | 2.99 | 0.0027 | +....+..... | rs185731506 | C/G | −3.11 | 0.0019 | - | rs73188175 | T/C | 0.300 | 0.0019 | --++++++++++ |
| 3 | TMEM108 | rs13087036 | C/A | −2.52 | 0.0116 | --+----+--- | rs139695007 | G/C | 3.09 | 0.0020 | + | rs2699882 | A/G | 0.053 | 0.0096 | +-+--++-++++ |
| 3 | AK092619 (nearest) | rs183598421 | T/C | −2.36 | 0.0185 | ....-...... | rs200248409 | GT/G | −2.86 | 0.0043 | - | rs7427054 | T/C | 0.093 | 0.0015 | ++---++++++- |
| 4 | TSPAN5 | rs28392914 | T/G | −3.16 | 0.0016 | +---------+ | rs1045655 | G/C | −2.87 | 0.0041 | - | rs12509107 | A/G | −0.445 | 0.0168 | --???-?????? |
| 5 | OR7H2P (nearest) | rs139061870 | GT/G | 2.80 | 0.0052 | ........+.. | rs73776672 | T/C | −3.43 | 0.0006 | - | rs62386289 | T/C | −0.117 | 0.0039 | -+------+-++ |
| 5 | FAT2 and SPARC (nearest) | rs141575897 | G/A | −3.03 | 0.0024 | ----+..---- | rs80009114 | A/G | 2.53 | 0.0113 | + | rs6579892 | A/T | 0.075 | 0.00095 | +++++++++++- |
| 6 | KLHL32 and NDUFA4 (nearest) | rs200056339 | C/CA | −2.68 | 0.0074 | ..-..-.--.. | rs78235656 | G/A | −2.77 | 0.0057 | - | rs117804808 | T/C | 0.250 | 0.0099 | +++-+++++-++ |
| 7 | SEMA3A | rs151172774 | T/C | 2.76 | 0.0058 | +++++++-+++ | rs6955094 | A/G | 3.18 | 0.0015 | + | rs150770834 | A/G | 0.494 | 0.0108 | -?????++++++ |
| 8 | DLGAP2 | rs117175403 | G/A | 2.79 | 0.0053 | -+++++--++. | rs184526444 | A/T | −2.90 | 0.0037 | - | rs11998452 | A/G | −0.218 | 0.0021 | +-+?-?------ |
| 9 | C9orf135 | rs56179412 | C/T | −2.13 | 0.0330 | ----++-+-+- | rs77797545 | A/G | 2.29 | 0.0220 | + | rs143862820 | T/C | 0.289 | 0.0055 | ?++-?+++-+?? |
| 10 | PRKG1 | rs10999787 | C/A | −2.70 | 0.0069 | ----++----. | rs10998992 | C/T | −2.81 | 0.0049 | - | rs192204676 | A/G | 0.332 | 0.016 | +---??++++++ |
| 10 | SFXN4 | rs143931152 | T/G | −3.64 | 0.0003 | ----------. | rs56095167 | G/A | −3.21 | 0.0013 | - | rs188855777 | T/C | −0.653 | 0.0032 | ??????-?-??- |
| 11 | GALNT18 | rs117835740 | C/T | −2.45 | 0.0142 | ----.--.--- | rs11021735 | C/T | 2.90 | 0.0037 | + | rs4909989 | A/G | −0.080 | 0.0033 | ---------++- |
| 11 | UVRAG | 11–75761242 (rs565239444) | T/G | −2.76 | 0.0058 | ......-.... | rs138825035 | A/G | −3.39 | 0.0007 | - | rs139079454 | T/C | 0.233 | 0.0043 | +++?++++-+++ |
| 12 | FAR2 (nearest) | rs151183596 | T/A | −2.70 | 0.0070 | -+----.+--- | rs141911197 | T/G | −3.50 | 0.0005 | - | rs12311115 | A/G | −0.119 | 0.00031 | +-------+--+ |
| 12 | SRRM4 | rs61937966 | C/T | 3.37 | 0.0007 | +++++-+++++ | rs4767761 | A/G | −3.40 | 0.0007 | - | rs78381318 | A/G | 0.194 | 0.000013 | ++-+-+++++-+ |
| 12 | HNF1A (nearest) | rs182546302 | T/A | −3.35 | 0.0008 | -+---+..-+. | rs80019595 | C/T | −2.62 | 0.0087 | - | rs117548270 | A/G | −0.312 | 0.0017 | ---+?------- |
| 14 | BTBD7 | rs112848587 | C/T | −2.19 | 0.0284 | ....---.-.- | rs76789831 | C/G | −2.77 | 0.0057 | - | rs111650311 | T/C | 0.072 | 0.0228 | ++++-++-++?+ |
| 18 | GTSCR1 (nearest) | rs11151610 | T/C | −3.27 | 0.0011 | ----------- | rs75968601 | C/T | 2.98 | 0.0029 | + | rs146227033 | C/G | −0.245 | 0.00068 | ----?---+--- |
| 19 | UQCRFS1 (nearest) | rs148613358 | T/C | 3.22 | 0.0013 | .....++..+. | rs12608817 | C/A | −3.12 | 0.0018 | - | rs2160742 | A/G | 0.074 | 0.0047 | ++-++++-+++- |
| 21 | RUNX1 | rs7280028 | T/C | −3.42 | 0.0006 | -------+--- | rs9981811 | G/A | 2.92 | 0.0035 | + | rs2247822 | T/C | 0.071 | 0.00055 | +---+++++++- |
Likely due to the inclusion of ischemic stroke cases only, we were not able to replicate the novel association for rs4471613, which was associated with total (ischemic and hemorrhagic) stroke in our prior COMPASS HapMap imputation report (IVW P=0.85)7. Additionally, we found no evidence of replication for loci previously associated with stroke in European-Ancestry populations (P ranging from 0.02 to 0.95; Supplemental Tables IV–V).
DISCUSSION
This new COMPASS meta-analysis of ischemic stroke only identified 24 unique loci with suggestive (n=23) or genome-wide (n=1) evidence for association with ischemic stroke. The most significantly associated HNF1A variant, rs55931441 (G/A), is monomorphic in European populations (G allele present only), with a 2% minor allele frequency (allele A) reported in sub-Saharan and 1000G African populations, and 3.8% frequency in COMPASS. This SNP was present in the only two studies imputed to 1KGp3 (WHI and SLESS). Collectively, WHI and SLESS account for 9,637 subjects (1,147 stroke cases and 8,490 controls). We were unable to assess the association for rs55931441 directly in our cross-ethnic look-up, however SNPs in a 100kb flanking region were significant (Supplemental Figure I) in SiGN Europeans (top SNP rs182546302; P=8.18×10−4), METASTROKE ischemic stroke phenotype (top SNP rs117548270; P=1.72×10−3), and METASTROKE cardioembolic stroke phenotype (top SNP rs184865012; P=9.98×10−4); while SNP rs80019595 approached significance (P=8.74×10−3) in the SiGN Hispanic cohort. Previous studies have reported associations between variants in HNF1A and lipids,33 C-reactive protein,34–35 and risk of coronary artery disease and stroke.33, 35 Taken together, these findings may provide greater insight regarding subtype specific influences and potential mechanism of HNF1A variants in stroke risk.
Three additional variants reached suggestive associations at the P≤10−8 level (rs113509723 in TMEM108 (Figure 1B); rs142655108 near NPM1P48 (Figure 1C); rs150807690 in SFXN4). The NPM1P48 locus showed no evidence for replication in the cross-ethnic look-up while TMEM108 was replicated in SiGN Hispanics only (top SiGN Hispanic SNP rs139695007; P= 0.002). The SFXN4 SNP, rs150807690, is a G insertion (-/G) with a 22% minor allele frequency (G insertion) in the 1000G African population and 24% frequency in COMPASS. Variant rs150807690 did not replicate in SiGN Hispanic (P=0.796) or SiGN Europeans (P=0.696) analyses and was not present in the METASTROKE look-up, however nearby SNPs with evidence of replication in a 100kb flanking region were detected in SiGN Europeans (top SNP rs143931152; P=2.68×10−4) and SiGN Hispanics (top SNP rs56095167; P=1.31×10−3), located 35,540 bp and 97,388 bp from the indexed COMPASS variant, respectively. The SFXN4 gene has not been previously implicated in stroke. The protein encoded by SFXN4 is critical for mitochondrial respiration and erythropoiesis.36–37 Recent clinical trials suggest that erythropoiesis-stimulating agents effectively treat anemia associated with chronic kidney disease but increase the risk of stroke possibly due to hyper-viscosity.38
Of the 23 loci with suggestive association in COMPASS, 15 showed evidence for replication in one or more look-up analysis. One locus was replicated in SiGN Europeans only, four loci were replicated in SiGN Hispanics only, two loci were replicated in METASTROKE ischemic stroke only, while eight loci had evidence for replication in two or more look-ups. Two loci, SFXN4 and UQCRFS1, were replicated in both the SiGN Europeans and Hispanics, two loci were replicated in SiGN Hispanics and METASTROKE ischemic stroke (KALRN and FAR2), and three loci were replicated in SiGN Europeans and METASTROKE ischemic stroke (CTTNBP2L, GTSCR1, and RUNX1). Most notably, one locus (SRRM4) was replicated in all three look-ups. Evidence for association across multiple ethnicities might indicate stroke susceptibility loci with a global impact. For example, the KALRN locus which was replicated in SiGN Hispanics and METASTROKE has been implicated in coronary artery disease risk across multiple populations39–41 and was recently associated with ischemic stroke and lacunar stroke in a Han Chinese population.42 Although the SRRM4 locus, which was replicated in all three look-ups, has not previously been implicated in stroke, the gene is important for neurogenesis43 and has shown associations with neurological conditions including Alzheimer’s disease44 and epilepsy.45
Although this effort represents the largest stroke GWAS meta-analysis in individuals of African descent, the modest sample size of 3,734 stroke cases limits our power to detect associations for variants with a MAF of ≤3%. Only two cohorts used the most recent imputation panel limiting our ability, and thus power, to detect novel variants only present in 1KGp3 and not 1KGp1v3. Furthermore, individuals of African descent suffer ischemic strokes of small vessel etiology more frequently. Therefore, due to the increased genetic diversity of this COMPASS population combined with the greater prevalence of small vessel stroke, we are not surprised at a lack of validation of previous European-ancestry associations. Failure to replicate associations across ethnicities is a common occurrence in genetic studies of various diseases and therefore does not threaten the validity of our current study. Moreover, the lack of availability of an adequate replication cohort consisting of individuals of African descent suffering a stroke that have genome-wide SNP genotype data remains a substantial global challenge. Likewise, due to smaller LD blocks and increased genetic diversity in populations of African descent, larger sample sizes would help alleviate limitations of statistical power, challenges associated with imputing genotypes, and allow for more detailed stroke subtype analyses. A recent analysis showed that although the number of GWAS conducted as of 2016 has increased more than 6-fold since 2009, African descent participants increased by only 2.5%.46 Therefore, our study will help advance precision medicine applications by identifying genetic loci (and subsequent polygenic risk scores) for stroke prediction and risk stratification in diverse populations.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the staff and participants of the ARIC, CHS, VISP, HANDLS, INTERSTROKE, ISGS, JHS, SIGNET-REGARDS, GEOS, SiGN, SLESS, SWISS, and WHI studies for their dedication, and willingness to participate in the respective research studies, which made this work possible.
FUNDING
Atherosclerosis Risk in Communities (ARIC) Study. Supported in whole or in part with Federal funds from the National Heart, Lung, and Blood Institute, National Institutes of Health, Department of Health and Human Services (contract numbers HHSN268201700001I, HHSN268201700002I, HHSN268201700003I, HHSN268201700004I and HHSN268201700005I), R01HL087641, R01HL059367 and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. The authors thank the staff and participants of the ARIC study for their important contributions. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research.
Cardiovascular Health Study (CHS). Supported by National Heart, Lung, and Blood Institute contracts HHSN268201200036C, HHSN268200800007C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086; and grants U01HL080295, R01HL087652, R01HL105756, R01HL103612, and R01HL120393 with contributions from the National Institute of Neurological Disorders and Stroke. Additional support was provided through R01AG023629 from the National Institute on Aging. A full list of principal CHS investigators and institutions can be found at CHS-NHLBI.org. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, grant UL1TR000124, and the National Institute of Diabetes and Digestive and Kidney Disease Research Center grant DK063491 to the Southern California Diabetes Endocrinology Research Center.
Vitamin Intervention for Stroke Prevention (VISP). Funded by the National Institute of Neurological Disorders and Stroke (R01-NS34447). GWAS data for a subset of VISP participants was supported by the National Human Genome Research Institute (U01-HG005160), as part of the Genomics and Randomized Trials Network (GARNET) (PI: MM Sale and BB Worrall).
INTERSTROKE. INTERSTROKE has received unrestricted grants from the Canadian Institutes of Health Research, Heart and Stroke Foundation of Canada, Canadian Stroke Network, Pfizer Cardiovascular Award, Merck, AstraZeneca, and Boehringer Ingelheim.
Jackson Heart Study (JHS). The Jackson Heart Study (JHS) is supported and conducted in collaboration with Jackson State University (HHSN268201800013I), Tougaloo College (HHSN268201800014I), the Mississippi State Department of Health (HHSN268201800015I) and the University of Mississippi Medical Center (HHSN268201800010I, HHSN268201800011I and HHSN268201800012I) contracts from the National Heart, Lung, and Blood Institute (NHLBI) and the National Institute for Minority Health and Health Disparities (NIMHD).
The Sea Islands Genetics Network (SIGNET)- REasons for Geographic And Racial Differences in Stroke (REGARDS) (SIGNET-REGARDS). The Sea Islands Genetics Network (SIGNET) was supported by R01 DK084350 (MM Sale) and consists of data from the REasons for Geographic And Racial Differences in Stroke (REGARDS) cohort, (U01 NS041588; G Howard).
The Genetics of Early Onset Stroke (GEOS) Study. The Genetics of Early Onset Stroke (GEOS) Study was supported by the NIH Genes, Environment, and Health Initiative grant U01 HG004436, as part of the Gene Environment Association Studies consortium, with additional support provided by the Mid-Atlantic Nutrition and Obesity Research Center (P30 DK072488) and the Office of Research and Development, Medical Research Service, and the Baltimore Geriatrics Research, Education, and Clinical Center of the Department of Veterans Affairs. Genotyping services were provided by the Johns Hopkins University Center for Inherited Disease Research (CIDR), which is fully funded through a federal contract from the NIH to the Johns Hopkins University (contract No. HHSN268200782096C). Assistance with data cleaning was provided by the Gene Environment Association Studies Consortium Coordinating Center (U01 HG 004446; PI Bruce S Weir). Study recruitment and assembly of data sets were supported by a Cooperative Agreement with the Division of Adult and Community Health, Centers for Disease Control and by grants from the NINDS and the NIH Office of Research on Women’s Health (R01 NS45012, U01 NS069208-01).
The National Institute of Neurological Disorders and Stroke (NINDS) Stroke Genetics Network (NINDS-SiGN) Groups 4. The Stroke Genetics Network (SiGN) study was funded by a cooperative agreement grant from the National Institute of Neurological Disorders and Stroke (NINDS) U01 NS069208. Genotyping services were provided by the Johns Hopkins University CIDR, which is fully funded through a federal contract from the National Institutes of Health (NIH) to the Johns Hopkins University (contract No.HHSN268200782096C). The Biostatistics Department Genetics Coordinating Center at the University of Washington (Seattle) provided more extensive quality control of the genotype data through a subcontract with CIDR. Additional support to the Administrative Core of SiGN was provided by the Dean’s Office, University of Maryland School of Medicine.
South London Ethnicity and Stroke Study (SLESS). Recruitment to SLESS was supported by a program grant from the Stroke Association. This study represents independent research part-funded by the National Institute for Health Research Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London, and the National Institute for Health Research Biomedical Research Centre at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London. Hugh Markus is supported by a NIHR Senior Investigator award, and his work is supported by NIHR Comprehensive Biomedical Research Unit funding awarded to Cambridge University Hospitals Trust.
Ischemic Stroke Genetics Study (ISGS) and Siblings with Ischemic Stroke Study (SWISS). ISGS and SWISS were supported by the National Institute of Neurological Disorders and Stroke grants (R01 NS42733; PI Meschia) and (R01NS39987; PI Meschia), respectively. Both studies received additional support, in part, from the Intramural Research Program of the National Institute of Aging (Z01 AG000954-06; PI Singleton), and used samples and clinical data from the NIH-NINDS Human Genetics Resource Center DNA and Cell Line Repository (http://ccr.cori-ell.org/ninds), human subjects protocol numbers 2003-081 and 2004-147.
The Women’s Health Initiative (WHI). Supported by the National Heart, Lung, and Blood Institute, NIH, and the U.S. Department of Health and Human Services through contracts N01WH22110, 24152, 32100-2, 32105-6, 32108-9, 32111-13, 32115, 32118-32119, 32122, 42107-26, 42129-32, and 44221. The funders had no role in study design, data collection and analysis, decision to publish, or manuscript preparation. H.I.H is supported by NIH/NHLBI grants U01HL117721, R01HL138423 and R56HL136210.
Healthy Aging in Neighborhoods of Diversity across the Life Span (HANDLS). Supported by the Intramural Research Program of the NIH, National Institute of Aging (project# Z01-AG000513 and human subjects protocol# 09AGN248).
NON-STANDARD ABBREVIATIONS and ACRONYMS
- 1000G
1000 genomes
- 1KGp1v3
1000G Phase I Version 3
- 1KGp3
1000G Phase III
- ARIC
Atherosclerosis Risk in Communities
- Chr
Chromosome
- CHS
Cardiovascular Health Study
- CIDR
Center for Inherited Disease Research
- COMPASS
Consortium of Minority Population genome-wide Association Studies of Stroke
- GEOS
Genetics of Early Onset Stroke
- GWAS
Genome-wide association study
- HANDLS
Healthy Aging in Neighborhoods of Diversity across the Life Span
- HNF1A
HNF1 homeobox A
- ISGS
Ischemic Stroke Genetics Study
- IVW
inverse variance weighted
- JHS
Jackson Heart Study
- LD
Linkage disequilibrium
- MAF
Minor allele frequency
- NINDS
National Institute of Neurological Disorders and Stroke
- OR
Odds ratio
- REGARDS
REasons for Geographic And Racial Differences in Stroke
- SiGN
Stroke Genetics Network
- SIGNET
Sea Islands Genetics Network
- SLESS
South London Ethnicity and Stroke Study
- SNP
Single nucleotide polymorphism
- SSW
Sample size weighted
- SWISS
Siblings with Ischemic Stroke Study
- VISP
Vitamin Intervention for Stroke Prevention
- WHI
Women’s Health Initiative
Footnotes
DISCLOSURES
MAN’s participation is supported by a consulting contract between Data Tecnica International and the National Institute on Aging, NIH. MAN consults for the Michael J. Fox Foundation, Vivid Genomics, Lysosomal Therapies Inc., Illumina Inc. and SK Therapeutics. KLF is a Deputy Editor for Stroke journal. BMP serves on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. CC receives research support from Biogen, EISAI, Alector and Parabon. CC is a member of the advisory board of Vivid genetics, Halia Therapeutics and ADx Healthcare. BBW serves as Deputy Editor for the journal Neurology. HSM has received personal fees from BIBA. RG is a former Associate Editor for the journal Neurology. JML consults for Regenera.
REFERENCES
- 1.Benjamin EJ, Muntner P, Alonso A, Bittencourt MS, Callaway CW, Carson AP, et al. Heart Disease and Stroke Statistics-2019 Update: A Report From the American Heart Association. Circulation. 2019;139:e56–e528. [DOI] [PubMed] [Google Scholar]
- 2.Hajat C, Dundas R, Stewart JA, Lawrence E, Rudd AG, Howard R, et al. Cerebrovascular risk factors and stroke subtypes: Differences between ethnic groups. Stroke. 2001;32:37–42. [DOI] [PubMed] [Google Scholar]
- 3.Traylor M, Bevan S, Baron J-C, Hassan A, Lewis CM, Markus HS. Genetic architecture of lacunar stroke. Stroke. 2015;46:2407–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bevan S, Traylor M, Adib-Samii P, Malik R, Paul NL, Jackson C, et al. Genetic heritability of ischemic stroke and the contribution of previously reported candidate gene and genomewide associations. Stroke. 2012;43:3161–3167. [DOI] [PubMed] [Google Scholar]
- 5.Howard G, Moy CS, Howard VJ, McClure LA, Kleindorfer DO, Kissela BM, et al. Where to focus efforts to reduce the black-white disparity in stroke mortality: Incidence versus case fatality? Stroke. 2016;47:1893–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.White H, Boden-Albala B, Wang C, Elkind MS, Rundek T, Wright CB, et al. Ischemic stroke subtype incidence among whites, blacks, and hispanics: The northern manhattan study. Circulation. 2005;111:1327–1331. [DOI] [PubMed] [Google Scholar]
- 7.Carty CL, Keene KL, Cheng YC, Meschia JF, Chen WM, Nalls M, et al. Meta-analysis of genome-wide association studies identifies genetic risk factors for stroke in african americans. Stroke. 2015;46:2063–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gretarsdottir S, Thorleifsson G, Manolescu A, Styrkarsdottir U, Helgadottir A, Gschwendtner A, et al. Risk variants for atrial fibrillation on chromosome 4q25 associate with ischemic stroke. Ann Neurol. 2008;64:402–409. [DOI] [PubMed] [Google Scholar]
- 9.Dichgans M, Malik R, Konig IR, Rosand J, Clarke R, Gretarsdottir S, et al. Shared genetic susceptibility to ischemic stroke and coronary artery disease: A genome-wide analysis of common variants. Stroke. 2014;45:24–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bellenguez C, Bevan S, Gschwendtner A, Spencer CC, Burgess AI, Pirinen M, et al. Genome-wide association study identifies a variant in hdac9 associated with large vessel ischemic stroke. Nature genetics. 2012;44:328–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holliday EG, Maguire JM, Evans TJ, Koblar SA, Jannes J, Sturm JW, et al. Common variants at 6p21.1 are associated with large artery atherosclerotic stroke. Nature genetics. 2012;44:1147–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ikram MA, Seshadri S, Bis JC, Fornage M, DeStefano AL, Aulchenko YS, et al. Genomewide association studies of stroke. The New England journal of medicine. 2009;360:1718–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kilarski LL, Achterberg S, Devan WJ, Traylor M, Malik R, Lindgren A, et al. Meta-analysis in more than 17,900 cases of ischemic stroke reveals a novel association at 12q24.12. Neurology. 2014;83:678–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Traylor M, Farrall M, Holliday EG, Sudlow C, Hopewell JC, Cheng YC, et al. Genetic risk factors for ischaemic stroke and its subtypes (the metastroke collaboration): A meta-analysis of genome-wide association studies. The Lancet Neurology. 2012;11:951–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malik R, Traylor M, Pulit SL, Bevan S, Hopewell JC, Holliday EG, et al. Low-frequency and common genetic variation in ischemic stroke: The metastroke collaboration. Neurology. 2016;86:1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malik R, Chauhan G, Traylor M, Sargurupremraj M, Okada Y, Mishra A, et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nature genetics. 2018;50:524–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.The ARIC Investigators. The atherosclerosis risk in communities (aric) study: Design and objectives. Am J Epidemiol. 1989;129:687–702. [PubMed] [Google Scholar]
- 18.Fried LP, Borhani NO, Enright P, Furberg CD, Gardin JM, Kronmal RA, et al. The cardiovascular health study: Design and rationale. Annals of epidemiology 1991;1:263–276. [DOI] [PubMed] [Google Scholar]
- 19.Sempos CT, Bild DE, Manolio TA. Overview of the jackson heart study: A study of cardiovascular diseases in african american men and women. The American journal of the medical sciences. 1999;317:142–146. [DOI] [PubMed] [Google Scholar]
- 20.Wilson JG, Rotimi CN, Ekunwe L, Royal CD, Crump ME, Wyatt SB, et al. Study design for genetic analysis in the jackson heart study. Ethnicity & disease. 2005;15:S6–30–37. [PubMed] [Google Scholar]
- 21.Women’s Health Initiative Study Group. Design of the women’s health initiative clinical trial and observational study. Control Clin Trials. 1998;19:61–109. [DOI] [PubMed] [Google Scholar]
- 22.O’Donnell M, Xavier D, Diener C, Sacco R, Lisheng L, Zhang H, et al. Rationale and design of interstroke: A global case-control study of risk factors for stroke. Neuroepidemiology. 2010;35:36–44. [DOI] [PubMed] [Google Scholar]
- 23.Howard VJ, Cushman M, Pulley L, Gomez CR, Go RC, Prineas RJ, et al. The reasons for geographic and racial differences in stroke study: Objectives and design. Neuroepidemiology. 2005;25:135–143. [DOI] [PubMed] [Google Scholar]
- 24.Meschia JF, Brott TG, Brown RD Jr., Crook RJ, Frankel M, Hardy J, et al. The ischemic stroke genetics study (isgs) protocol. BMC neurology. 2003;3:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spence JD, Howard VJ, Chambless LE, Malinow MR, Pettigrew LC, Stampfer M, et al. Vitamin intervention for stroke prevention (visp) trial: Rationale and design. Neuroepidemiology. 2001;20:16–25. [DOI] [PubMed] [Google Scholar]
- 26.Williams SR, Hsu FC, Keene KL, Chen WM, Nelson S, Southerland AM, et al. Shared genetic susceptibility of vascular-related biomarkers with ischemic and recurrent stroke. Neurology. 2016;86:351–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Traylor M, Rutten-Jacobs L, Curtis C, Patel H, Breen G, Newhouse S, et al. Genetics of stroke in a uk african ancestry case-control study: South london ethnicity and stroke study. Neurology Genetics. 2017;3:e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng YC, O’Connell JR, Cole JW, Stine OC, Dueker N, McArdle PF, et al. Genome-wide association analysis of ischemic stroke in young adults. G3 (Bethesda). 2011;1:505–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meschia JF, Arnett DK, Ay H, Brown RD Jr., Benavente OR, Cole JW, et al. Stroke genetics network (sign) study: Design and rationale for a genome-wide association study of ischemic stroke subtypes. Stroke. 2013;44:2694–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans MK, Lepkowski JM, Powe NR, LaVeist T, Kuczmarski MF, Zonderman AB. Healthy aging in neighborhoods of diversity across the life span (handls): Overcoming barriers to implementing a longitudinal, epidemiologic, urban study of health, race, and socioeconomic status. Ethn dis. 2010;20:267–275. [PMC free article] [PubMed] [Google Scholar]
- 31.Meschia JF, Brown RD Jr., Brott TG, Chukwudelunzu FE, Hardy J, Rich SS. The siblings with ischemic stroke study (swiss) protocol. BMC medical genetics. 2002;3:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Willer CJ, Li Y, Abecasis GR. Metal: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou YJ, Yin RX, Hong SC, Yang Q, Cao XL, Chen WX. Association of the hnf1a polymorphisms and serum lipid traits, the risk of coronary artery disease and ischemic stroke. The journal of gene medicine. 2017;19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lopez-Mejias R, Genre F, Remuzgo-Martinez S, Gonzalez-Juanatey C, Robustillo-Villarino M, Llorca J, et al. Influence of elevated-crp level-related polymorphisms in non-rheumatic caucasians on the risk of subclinical atherosclerosis and cardiovascular disease in rheumatoid arthritis. Scientific reports. 2016;6:31979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi H, Leng S, Liang H, Zheng Y, Chen L. Association study of c-reactive protein associated gene hnf1a with ischemic stroke in chinese population. BMC medical genetics. 2016;17:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hildick-Smith GJ, Cooney JD, Garone C, Kremer LS, Haack TB, Thon JN, et al. Macrocytic anemia and mitochondriopathy resulting from a defect in sideroflexin 4. Am J Hum Genet. 2013;93:906–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng H, Ji C, Zou X, Wu M, Jin Z, Yin G, et al. Molecular cloning and characterization of a novel human putative transmembrane protein homologous to mouse sideroflexin associated with sideroblastic anemia. DNA sequence. 2003;14:369–373. [DOI] [PubMed] [Google Scholar]
- 38.Seliger SL, Zhang AD, Weir MR, Walker L, Hsu VD, Parsa A, et al. Erythropoiesis-stimulating agents increase the risk of acute stroke in patients with chronic kidney disease. Kidney international. 2011;80:288–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horne BD, Hauser ER, Wang L, Muhlestein JB, Anderson JL, Carlquist JF, et al. Validation study of genetic associations with coronary artery disease on chromosome 3q13–21 and potential effect modification by smoking. Annals of Human Genetics. 2009;73:551–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang L, Hauser ER, Shah SH, Pericak-Vance MA, Haynes C, Crosslin D, et al. Peakwide mapping on chromosome 3q13 identifies the kalirin gene as a novel candidate gene for coronary artery disease. The American Journal of Human Genetics. 2007;80:650–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boroumand M, Ziaee S, Zarghami N, Anvari MS, Cheraghi S, Abbasi SH, et al. The kalirin gene rs9289231 polymorphism as a novel predisposing marker for coronary artery disease. Laboratory Medicine. 2014;45:302–308. [DOI] [PubMed] [Google Scholar]
- 42.Dang M, Wang Z, Zhang R, Li X, Peng Y, Han X, et al. Kalrn rare and common variants and susceptibility to ischemic stroke in chinese han population. NeuroMolecular Medicine. 2015;17:241–250. [DOI] [PubMed] [Google Scholar]
- 43.Raj B, Irimia M, Braunschweig U, Sterne-Weiler T, O’Hanlon D, Lin Z-Y, et al. A global regulatory mechanism for activating an exon network required for neurogenesis. Molecular Cell. 2014;56:90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chung J, Wang X, Maruyama T, Ma Y, Zhang X, Mez J, et al. Genome-wide association study of alzheimer’s disease endophenotypes at prediagnosis stages. Alzheimer’s & Dementia. 2018;14:623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rusconi F, Paganini L, Braida D, Ponzoni L, Toffolo E, Maroli A, et al. Lsd1 neurospecific alternative splicing controls neuronal excitability in mouse models of epilepsy. Cerebral Cortex. 2015;25:2729–2740. [DOI] [PubMed] [Google Scholar]
- 46.Popejoy AB, Fullerton SM. Genomics is failing on diversity. Nature. 2016;538:161–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Howard G, Kissela BM, Kleindorfer DO, McClure LA, Soliman EZ, Judd SE, et al. Differences in the role of black race and stroke risk factors for first vs recurrent stroke. Neurology. 2016;86:637–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.