Abstract
Distal hereditary motor neuropathy (dHMN) is an inherited neuromuscular disease characterized by symmetric distal weakness and atrophy without sensory changes. There are about thirty known genes associated with dHMN, but together they explain only about a third of cases. Mutations in the sigma non-opioid intracellular receptor 1 gene (SIGMAR1) have been linked to autosomal recessive dHMN with pyramidal signs in several families. This phenotype can mimic amyotrophic lateral sclerosis (ALS). We report a 39-year-old man who was referred to our ALS clinic with distal motor weakness and hyperreflexia. Whole exome sequencing identified two novel variants in the SIGMAR1 gene in the proband. Targeted Sanger sequencing of asymptomatic family members confirmed that each carried one of these two variants. Our findings expand the number of known SIGMAR1 pathogenic variants associated with dHMN, which should be clinically distinguished from ALS.
1. Introduction:
Hereditary motor neuropathy (dHMN), often synonymous with distal spinal muscular atrophy (dSMA), is an inherited neuromuscular disease characterized by symmetric distal weakness and atrophy without sensory changes. Some forms of dHMN are accompanied by pyramidal signs. In dHMN, the degeneration of motor nerves leads to a length-dependent pattern of muscle weakness and atrophy. This phenotype is often misdiagnosed as ALS, however the two are distinguished by their weakness pattern as well as disease course. There are about thirty known genes associated with dHMN, but together they explain less than a third of cases [1]. We present a patient who displayed clinical signs of dHMN with pyramidal signs and describe two novel variants in the SIGMAR1 gene (SIGMAR1, OMIM 601978) found within his family.
2. Case report:
The subject is a 39-year-old man of European decent (German, French, Scottish and Dutch) without known familial consanguinity [Figure 1]. He experienced 5 years of progressive weakness beginning in the right hand and quickly spreading to the left hand. 4 years after onset, he noticed changes in his gait with slapping of his feet and weakness in the distal lower extremities. There was no weight loss, complaints of dyspnea, dysphagia or dysarthria. No other family members displayed similar symptoms.
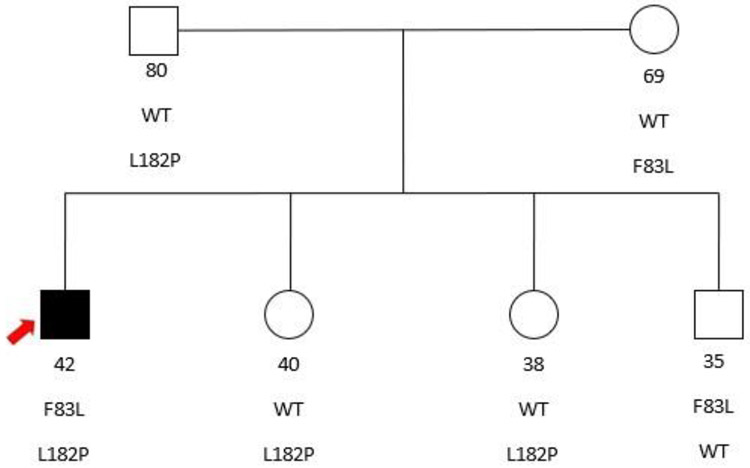
Figure 1.
Pedigree of the nuclear family with 2 unaffected parents and 3 unaffected siblings all heterozygous for either the F83L or the L182P mutations. The proband (arrow) is the only individual harboring both variants and manifesting disease.
Neurological examination demonstrated normal cognition and bulbar function. There was symmetric muscle wasting in the hands and feet, as well as high arches and thin calves [Figure 2]. Strength was 3 on the MRC scale in the intrinsic hand muscles, ankle dorsiflexors and great toe extensor, but normal in the proximal muscles. Sensation was intact to all modalities. Reflexes were normal in the arms but 3+ at the knees, absent at the ankles, and toes were upgoing.
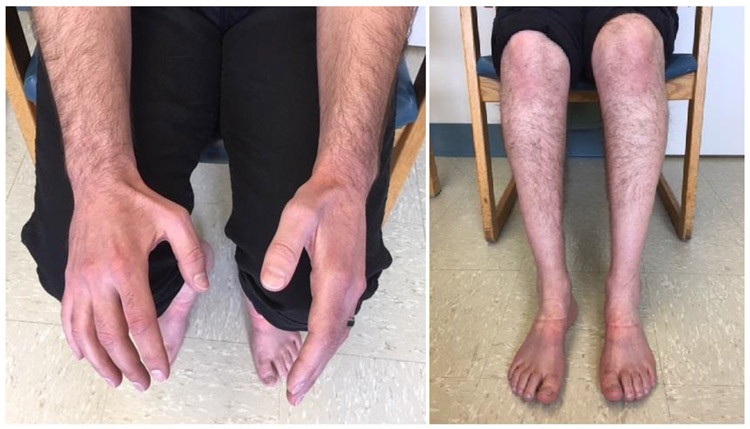
Figure 2.
Atrophy of intrinsic hand muscles especially prominent in the first dorsal interosseous muscles. Atrophy of bilateral calves more pronounced distally giving a “stork leg” appearance akin to Charcot-Marie-Tooth disease.
Blood chemistries and cell counts were normal. CSF studies demonstrated normal protein and glucose with one lymphocyte per microliter. MRI was performed on the brain and cervical spine, which showed age-related spondylosis without significant stenoses to explain his symptoms.
Electrodiagnostic testing was done with nerve conduction studies (NCS) performed on the upper and lower limbs, recording median, ulnar, fibular and tibial compound muscle action potentials (CMAPs) as well as median, ulnar, radial and sural sensory nerve action potential (SNAP). Motor nerve conduction studies demonstrated low amplitude to absent CMAPs in the distal hand and foot muscles without demyelinating features. SNAP amplitudes in the upper and lower limbs were normal. Needle electromyography (EMG) was performed on the limbs. This demonstrated both acute and chronic denervation changes in distal muscles of the upper and lower limbs to a similar severity and chronicity, normalizing in the proximal muscles.
Initial molecular genetic testing of the proband was performed through a commercial laboratory using a targeted next generation sequencing panel that interrogates the full coding regions of 19 known genes for dHMN. The panel did not include SIGMAR1. The result was negative.
Whole exome sequencing was then obtained through a commercial laboratory. Variants were identified using a proprietary joint-calling algorithm. Using a combination of public and proprietary databases, a search of phenotype-gene associations was made. This yielded two variants in the SIGMAR1 gene: c.247T>C (p.Phe83Leu) in exon 2 and c.545T>C (p.Leu182Pro) in exon 4 (RefSeq NM_005866.4). These amino acid substitutions are predicted to significantly alter protein structure, and algorithms assessed the potential impact to be “deleterious”. The variants affect evolutionarily conserved residues. Both variants are very rare in the population with allele frequencies of 0.0000129 and 0.000012 in gnomAD, respectively.
With informed consent, under a protocol approved by the University of Washington institutional review board, saliva samples were collected from five family members, including three siblings and both parents. Genomic DNA was extracted using a saliva DNA isolation kit. To confirm the reported exome variants in the proband and to investigate its co-segregation with the disease in the family, we performed Sanger sequencing using custom primers. We were able to confirm the above two variants in the proband and reveal that none of the asymptomatic family members carried both variants. Specifically, both sisters as well as the father carried the c.545T>C variant. The brother and the mother carried the c.247T>C variant. The proband was also tested for repeat expansion of the C9ORF72 gene as well as SOD1 gene mutations, which were negative.
3. Discussion:
dHMN is a disorder that presents with distal weakness and muscle wasting. Certain phenotypes – as seen here – also feature pyramidal signs, suggesting concurrent upper motor neuron dysfunction. This phenotype can mimic ALS and is often misdiagnosed as such. In fact, a 2011 paper published by Al-Saif, et al. described a consanguineous family from Saudi Arabia with 6 members harboring a homozygous SIGMAR1 mutation [2]. Clinical phenotype was juvenile onset of distal symmetric limb weakness and atrophy with spasticity. The authors diagnosed the affected individuals with juvenile ALS using the El Escorial criteria, and this genotype-phenotype combination was later coined ALS 16.
We question the designation of SIGMAR1-related motor disease as juvenile ALS and feel that this may create unnecessary ambiguity. The affected individuals described by Al-Saif, et al. clearly fit a dHMN phenotype. ALS, on the other hand, typically manifests in an asymmetric segmental fashion, often spreading to contiguous regions, and does not respect a length-dependent pattern. Furthermore, diagnostic criteria for ALS call for clinical progression. We reviewed 11 families/individuals in the literature with SIGMAR1-related motor disease (Table 1). Disease onset ranged from infancy to the early teenage years. After a period of progression, individuals tended to stabilize in terms of weakness. In contrast to the prognosis of ALS, no affected individual has been reported with a shortened life-span nor has anyone affected been reported to have progressed to respiratory failure. In our opinion, dHMN rather than juvenile ALS more accurately describes the SIGMAR1 phenotype.
Table 1.
Reported cases of Sigmar1-related motor disease.
| Genotypea | Proteina | Ethnic Origin | Age at Onset | Distal Weaknessb |
Knee Jerksc |
Babinski Response |
Citation |
|---|---|---|---|---|---|---|---|
| C.151+1G>T | p.Gly31_Ala50del | Chinese | 9-12 yrs | + | +++ | + | 8 |
| C.151+1G>T | p.Gly31_Ala50del | Afghani | 10 yrs | + | +++ | + | 9 |
| c,194T>A | p.Leu65Gln | French/British | 3 yrs | + | +++ | + | 12 |
| c.238C>T | p.Gln80* | Omani | 13 mo – 11 yrs | + | +++ | + | 3 |
| c.247T>C/c.545T>C | p.Phe83Leu/p.Leu182Pro | German/French | 34 yrs | + | +++ | + | This report |
| c.283dupC | p.Leu95Profs*29 | Hispanic | 5 yrs | + | +++ | + | 14d |
| c.304G>C | p.Glu102Gln | Saudi | 1-2 yrs | + | ++++ | NA | 13d |
| c.412G>C | p.Glu138Gln | Italian | School age | + | +++ | + | 10 |
| c.448G>A | p.Glu150Lys | Italian | Infancy | + | +++ | + | 10 |
| c.500A>T | p.Asn167Ile | Jordani | 6-10 yrs | + | +++ | + | 1 |
| c.561_576del | p.Asp188Profs*69 | Portuguese | 4 yrs | + | − | − | 15 |
| c.505T>A/c.622C>T | p.Thr169Arg/p.Arg208Trp | Japanese | 80 yrs | + | +++ | + | 7d |
The table with 11 previous reported cases of SIGMAR1-related motor disease as well as our subject (bold) was adapted and extended from Nandhagopal 2017 [3]. Note that the two compound heterozygous individuals present significantly later in life compared with homozygous individuals.
Homozygous unless otherwise denoted.
+, present; −, absent
−, absent/lost; +, sluggish; ++, normal; +++, brisk; ++++, exaggerated (with clonus). NA, not available.
Initially reported as ALS.
Some authors have also suggested that SIGMAR1 mutation can cause a more classic adult onset ALS-FTD phenotype. In 2010, Luty et al. reported that a mutation in the 3'-untranslated region (UTR) of the SIGMAR1 gene was found in three families with ALS-FTD [16]. However, it was later determined that one of these families possessed a repeat expansion in C9ORF72 [17]. This and other examples have prompted authors to argue that any evidence linking SIGMAR1 mutation and familial ALS is merely circumstantial [18].
Sigma non-opioid intracellular receptor 1 (Sig-1R) is found ubiquitously on the endoplasmic reticulum and is prominently expressed in motor neurons. Sig-1R is located on the mitochondria-associated ER membrane (MAM), which is the interface between the ER and mitochondria. In response to ER stress, Sig-1R can translocate from the MAM to the plasma membrane [5]. The exact role of Sig-1R in dHMN is uncertain but in its normal environment, it functions to regulate calcium homeostasis and ER-mitochondria interactions. SIGMAR1(−/−) mice display locomotor deficits associated with muscle weakness, axonal degeneration and motor neuron loss [6].
In cultured primary motor neurons, inactivation of SIGMAR1 led to axonal degeneration and cell death. An impairment of mitochondria dynamics and transport seems to underly this phenomenon [6]. It may not be coincidental that in most cases, SIGMAR1-related dHMN tends to manifest as childrens’ limbs are growing, and axons are required to lengthen. The pattern of length-dependent weakness suggests axonopathy rather than neuronopathy as the pathophysiology, again in contrast with ALS.
Ten of the eleven published affected individuals with SIGMAR1-related dHMN possessed homozygous mutations within consanguineous families. Our subject was from a nonconsanguineous family and carried two different novel SIGMAR1 variants each inherited from an unaffected heterozygous parent. The substitution of proline for leucine at residue 182 in the C-terminal hydrophobic region is predicted to reduce the hydrophobicity (hydropathy index of leucine and proline = 3.8 and −1.6, respectively) [19]. This region presents a hydrophobic membrane-proximal surface and is suggested to have an intimate association with the cytosolic surface of the endoplasmic reticulum membrane [20]. Decreased hydrophobicity may destabilize such an association. The second variant, p.Phe83Leu, is not in a known functional domain. We can speculate that it has a milder effect on the protein function than some other reported pathogenic variants, perhaps accounting for the later age of onset. There is a recent report of another individual with SIGMAR1 compound heterozygous variants who presented to care at the age of 89 with similar symmetric distal weakness [7]. Interestingly, one of these variants is also located in the C-terminal hydrophobic region and the other is in a region without known function. We Acknowledge that it is difficult to determine with any certainty that the Phe83Leu substitution is pathogenic, and the variant remains one of unclear significance. With more cases reported, the possibility of genotype-phenotype correlation may become apparent and the functional mechanism can be further explored.
4. Conclusions:
Our findings expand the number of known SIGMAR1 gene variants that cause autosomal recessive distal hereditary motor neuropathy. Clinically, SIGMAR1-related dHMN can mimic ALS with both upper and lower motor neuron signs. However, factors that distinguish dHMN include its younger age of onset, distal symmetric weakness pattern, and a more indolent course. The nosology of hereditary motor neuropathy continues to evolve in the age of neurogenetics. It is important for neurologists to distinguish SIGMAR1-related dHMN from ALS in the appropriate clinical setting given their important phenotypic and prognostic differences.
Highlights:
A 39-year-old man presents to ALS clinic with progressive symmetric distal weakness
Genetic studies find 2 novel variants in the SIGMAR1 gene
The SIGMAR1 gene has been associated with a distal hereditary motor (dHMN)
Unlike ALS, dHMN has a symmetric distal pattern and more indolent disease course
dHMN is a more accurate name for SIGMAR1-related disease than juvenile ALS
Acknowledgement:
The authors would like to thank the patient and his family for allowing us to participate in their care and conduct these research activities. We also acknowledge Dr. Anusha Mannava MD for supplying the images.
Funding sources: This work was supported by the National Institutes of Health [grant number 1R01NS069719] and Department of Veterans Affairs Research Funds.
ABBREVIATIONS:
- ALS
amyotrophic lateral sclerosis
- C9ORF72
chromosome 9 open reading frame 72
- CMAP
compound muscle action potential
- CSF
cerebrospinal fluid
- dHMN
distal hereditary motor neuropathy
- dSMA
distal spinal muscular atrophy
- EMG
electromyography
- ER
endoplasmic reticulum
- FTD
frontotemporal dementia
- gnomAD
genome aggregation database
- MAM
mitochondria-associated endoplasmic reticulum membrane
- MRC
medical research council
- MRI
magnetic resonance imagining
- NCS
nerve conduction study
- OMIM
online mendelian inheritance in man
- Sig-1R
Sigma non-opioid intracellular receptor
- SNAP
sensory nerve action potential
- SOD1
Superoxide dismutase 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Ethical publication statement: We confirm that we have read the Journal’s position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
Statement of Disclosure: None of the authors has any conflict of interest to disclose.
CONFLICT OF INTEREST STATEMENT:
Maxwell Ma, M.D. – No conflicts of interest
Dong-Hui Chen, M.D. Ph.D. – No conflicts of interest
Wendy H. Raskind, M.D. Ph.D. – Licensing fees from Athena Diagnostics
Thomas D. Bird, M.D. – Licensing fees from Athena Diagnostics; Consultant for Genentech
References:
- [1].Ververis A, Dajani R, Koutsou P, Aloqaily A, Nelson-Williams C, Loring E et al. Distal hereditary motor neuronopathy of the Jerash type is caused by a novel SIGMAR1 c.500A>T missense mutation. J Med Genet 2020; 57:178–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Al-Saif A, Al-Mohanna F, Bohlega S. A mutation in sigma-1 receptor causes juvenile amyotrophic lateral sclerosis. Ann Neurol. 2011. December;70(6):913–9. doi: 10.1002/ana.22534. Epub 2011 Aug 12. [DOI] [PubMed] [Google Scholar]
- [3].Nandhagopal R, Meftah D, Al-Kalbani S, Scott P. Recessive distal motor neuropathy with pyramidal signs in an Omani kindred: underlying novel mutation in the SIGMAR1 gene . Eur J Neurol. 2018. February;25(2):395–403. doi: 10.1111/ene.13519. Epub 2017 Dec 12. [DOI] [PubMed] [Google Scholar]
- [4].Almendra L, Laranjeira F, Fernández-Marmiesse A, Negrão L. SIGMAR1 gene mutation causing Distal Hereditary Motor Neuropathy in a Portuguese family. Acta Myol. 2018. May 1;37(1):2–4. eCollection 2018 May. [PMC free article] [PubMed] [Google Scholar]
- [5].Benarroch EE. Sigma-1 receptor and amyotrophic lateral sclerosis. Neurology. 2018. October 16;91(16):743–47. doi: 10.1212/WNL.0000000000006347. Epub 2018 Sep 21. [DOI] [PubMed] [Google Scholar]
- [6].Bernard-Marissal N, Médard JJ, Azzedine H, Chrast R. Dysfunction in endoplasmic reticulum-mitochondria crosstalk underlies SIGMAR1 loss of function mediated motor neuron degeneration. Brain. 2015. April;138(Pt 4):875–90. doi: 10.1093/brain/awv008. Epub 2015 Feb 11. [DOI] [PubMed] [Google Scholar]
- [7].Izumi Y, Morino H, Miyamoto R, Matsuda Y, Ohsawa R, Kurashige T et al. Compound heterozygote mutations in the SIGMAR1 gene in an oldest-old patient with amyotrophic lateral sclerosis. Geriatr Gerontol Int. 2018. October;18(10):1519–20. doi: 10.1111/ggi.13506. [DOI] [PubMed] [Google Scholar]
- [8].Li X, Hu Z, Liu L, et al. SIGMAR1 splice-site mutation causes distal hereditary motor neuropathy. Neurology 2015; 84: 2430–37. [DOI] [PubMed] [Google Scholar]
- [9].Lee JJY, van Karnebeek CDM, Drögemoller B, et al. Further validation of the SIGMAR1 c.151+1G>T mutation as cause of distal hereditary motor neuropathy. Child Neurol Open 2016; 3: 2329048X16669912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gregianin E, Pallafacchina G, Zanin S, et al. Loss-of-function mutations in the SIGMAR1 gene cause distal hereditary motor neuropathy by impairing ER-mitochondria tethering and Ca2+ signalling. Hum Mol Genet 2016; 25: 3741–53. [DOI] [PubMed] [Google Scholar]
- [11].Christodoulou K, Zamba E, Tsingis M, et al. A novel form of distal hereditary motor neuronopathy maps to chromosome 9p21.1–pl2. Ann Neurol 2000; 48: 877–84 [PubMed] [Google Scholar]
- [12].Horga A, Tomaselli PJ, Gonzalez MA, et al. SIGMAR1 mutation associated with autosomal recessive Silver-like syndrome. Neurology 2016; 87: 1607–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Al-Saif A, Al-Mohanna F, Bohlega S. A mutation in sigma-1 receptor causes juvenile amyotrophic lateral sclerosis. Ann Neurol 2011; 70: 913–9. [DOI] [PubMed] [Google Scholar]
- [14].Watanabe S, Ilieva H, Tamada H, et al. Mitochondria-associated membrane collapse is a common pathomechanism in SIGMAR1- and SOD1-linked ALS. EMBO Mol Med 2016; 8: 1421–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Almendra L 1, Laranjeira F 2, Fernández-Marmiesse A 3, Negrão L 1 SIGMAR1 gene mutation causing Distal Flereditary Motor Neuropathy in a Portuguese family. Acta Myol. 2018. May 1;37(1):2–4. eCollection 2018 May. [PMC free article] [PubMed] [Google Scholar]
- [16].Luty AA1, Kwok JB, Dobson-Stone C, Loy CT, Coupland KG, Karlström H, Sobow T, et al. Sigma nonopioid intracellular receptor 1 mutations cause frontotemporal lobar degeneration-motor neuron disease. Ann Neurol. 2010. November;68(5):639–49. doi: 10.1002/ana.22274. [DOI] [PubMed] [Google Scholar]
- [17].C1 Dobson-Stone, Hallupp M, Loy CT, Thompson EM, Haan E, Sue CM, et al. C9ORF72 repeat expansion in Australian and Spanish frontotemporal dementia patients. PLoS One. 2013;8(2):e56899. doi: 10.1371/journal.pone.0056899. Epub 2013 Feb 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Pickering-Brown S, Hardy J. Is SIGMAR1 a confirmed FTD/MND gene? Brain. 2015. November;138(Pt 11):e393. doi: 10.1093/brain/awv173. Epub 2015 Jun 18 [DOI] [PubMed] [Google Scholar]
- [19].A simple method for displaying the hydropathic character of a protein. Kyte J, Doolittle RF. J Mol Biol. 1982; 157(1): 105–32. [DOI] [PubMed] [Google Scholar]
- [20].Crystal structure of the human σ1 receptor. Schmidt HR, Zheng S, Gurpinar E, Koehl A, Manglik A, Kruse AC. Nature. 2016;532(7600):527–30. [DOI] [PMC free article] [PubMed] [Google Scholar]