Abstract
Aims
Coronavirus disease 2019 (COVID-19), which is caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), is a major health concern worldwide. Due to the lack of specific medication and vaccination, drug-repurposing attempts has emerged as a promising approach and identified several human proteins interacting with the virus. This study aims to provide a comprehensive molecular profiling of the immune cell-enriched SARS-CoV-2 interacting protein USP13.
Materials and methods
The list of immune cell-enriched proteins interacting with SARS-CoV-2 was retrieved from The Human Protein Atlas. Genomic alterations were identified using cBioPortal. Survival analysis was performed via Kaplan-Meier Plotter. Analyses of protein expression and tumor infiltration levels were carried out by TIMER.
Key findings
14 human proteins that interact with SARS-CoV-2 were enriched in immune cells. Among these proteins, USP13 had the highest frequency of genomic alterations. Higher USP13 levels were correlated with improved survival in breast and lung cancers, while resulting in poor prognosis in ovarian and gastric cancers. Furthermore, copy number variations of USP13 significantly affected the infiltration levels of distinct subtypes of immune cells in head & neck, lung, ovarian and stomach cancers. Although our results suggested a tumor suppressor role for USP13 in lung cancer, in other cancers, its role seemed to be context-dependent.
Significance
It is critical to identify and characterize human proteins that interact with SARS-CoV-2 in order to have a better understanding of the disease and to develop better therapies/vaccines. Here, we provided a comprehensive molecular profiling the immune cell-enriched SARS-CoV-2 interacting protein USP13, which will be useful for future studies.
Keywords: COVID-19, Coronavirus, SARS-CoV-2 interacting proteins, Ubiquitin Specific Peptidase 13, USP13
1. Introduction
The recent pandemic of the coronavirus disease 2019 (COVID-19), which is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has become one of the major health concerns of the 21st century [1]. The disease has spread to more than 8 million people worldwide, killing over 450.000 individuals [2]. In addition to the shortcomings of public health systems in providing sufficient healthcare to such massive number of patients, another great challenge in fighting the disease has been the lack of specific medication and vaccination. Therefore, drug-repurposing attempts aimed at identification of efficient therapeutic agents for the treatment of COVID-19 has emerged as a promising approach to compensate for this urgent need. In line with this, a recent study has identified 332 human proteins as potential targets for drug repurposing that SARS-CoV-2 interact upon entry into the host cell [3].
As for any other viral infection, a well-coordinated immune response is key to battling the SARS-CoV-2 infection effectively. The innate and the adaptive immune systems of the host cooperate to fight the virus off by inhibiting viral replication and its further spread, through secretion of proinflammatory cytokines, induction of inflammation and activation of cytotoxic (CD8+) and helper (CD4+) T-cells [4,5]. Furthermore, host cells that are infected with the virus are targeted and cleared by the natural killer (NK) cells, which comprise a subset of innate immune cells [6]. Dendritic cells (DC) are also critical for the elimination of these infections via their antigen-presenting mechanisms [7]. The significance of immune system and immune cells in resolving SARS-CoV-2 infection is further emphasized by studies that suggest immune dysregulation and reduced levels of T-lymphocytes in patients infected with SARS-CoV-2 [8]. Likewise, a significant correlation between the host's immune system response and the severity of the disease has been proposed [9,10]. Thus, it is of utmost importance to identify and characterize proteins that might play a role in the proper functioning of the immune system during SARS-CoV-2 infection.
Pre-existing conditions such as diabetes, hypertension and cardiovascular diseases, as well as conditions that result in an immunocompromised state are considered as risk factors for increased susceptibility to SARS-CoV-2 infection [11,12]. Cancer patients are often in an immunocompromised state as their immune systems are damaged either by the cancer itself (i.e. lymphoma, leukemia) or by the treatment (chemotherapy or radiation therapy). In this study, we analyzed the immune cell-enriched SARS-CoV-2 interacting proteins that were recently identified in the context of cancer. USP13 (Ubiquitin Specific Peptidase 13), which is a SARS-CoV-2 interacting protein enriched in T-cells, is an important component of the ubiquitin-proteasome system that regulates the degradation of misfolded or damaged proteins in the cell. Ubiquitin-specific proteases (USPs) are involved in the regulation of several cellular mechanisms via deubiquitinating (removing the ubiquitin marks from) target proteins [13]. USP13 has previously been implicated in tumorigenesis and in interferon-induced signaling via targeting and deubiquitinating tumor suppressors p53/PTEN [14,15] and STAT1 [16], respectively. This study aims to provide a comprehensive molecular profiling of USP13, which might be useful in having a better understanding of the molecular mechanisms induced by SARS-CoV-2 infection in immune cells.
2. Methods
2.1. Identification of immune cell-enriched SARS-CoV-2 interacting proteins
The list of immune cell-enriched proteins interacting with SARS-CoV-2 identified by Gordon et al. is retrieved from The Human Protein Atlas (https://www.proteinatlas.org) [3,17]. The Human Protein Atlas offers categorization of proteins according to their mRNA expression levels in specific cell types or compartments, based on RNA-seq data. The shortlist of immune cell-enriched SARS-CoV-2 interacting proteins was identified by narrowing down all SARS-CoV-2 interacting proteins by enrichment in immune cells.
2.2. Analysis of genomic alterations
Mutations and copy number alterations within the genes encoding immune cell-enriched SARS-CoV-2 interacting proteins across different cancer types were identified using The cBio Cancer Genomics Portal (http://cbioportal.org) [18,19]. The analyses were carried out on all TCGA PanCancer Atlas Studies available, which consisted of 32 studies and a total of 10,967 samples. TCGA PanCancer Studies were selected as they provide the latest curated data regarding the dataset.
2.3. Expression analysis
Protein expression levels across all TCGA tumors were analyzed using the DiffExp module of TIMER: Tumor IMmune Estimation Resource (http://timer.cistrome.org/) [20], which offers differential gene expression data (in log2 TPM-Transcript count Per Million) between tumor and adjacent normal tissues in the form of boxplots. Statistical significance between tumor/normal samples are automatically calculated by the tool using the Wilcoxon test.
2.4. Survival analysis
The effect of USP13 (Affymetrix ID: 226902_at) mRNA expression levels on overall survival was assessed via Kaplan-Meier Plotter (https://kmplot.com/analysis/) [21]. The analyses were run on 1764 breast cancer, 1144 lung cancer, 614 ovarian cancer and 631 gastric cancer patients using default settings. p-Values below 0.05 (%5) were considered significant.
2.5. Determination of tumor infiltration levels
The effect of somatic copy number alterations (SCNAs) in USP13 gene on tumor infiltration levels of B cells, CD4+ T cells, CD8+ T cells, macrophages, neutrophils and dendritic cells was analyzed using SCNA module of TIMER [20]. The algorithm defines SCNAs based on GISTIC 2.0 and the distributions of different immune cell types are depicted in relation to chromosomal deletions/amplifications. TIMER defines copy number variations on the basis of confidence levels between −2 to 2; as deep deletion (−2), arm-level deletion (−1), diploid/normal (0), arm-level gain (1), and high amplification (2). Statistical analysis uses a two-sided Wilcoxon rank-sum test. p-Values below 0.05 (%5) are considered significant.
3. Results
3.1. SARS-CoV-2 interacting proteins enriched in immune cells
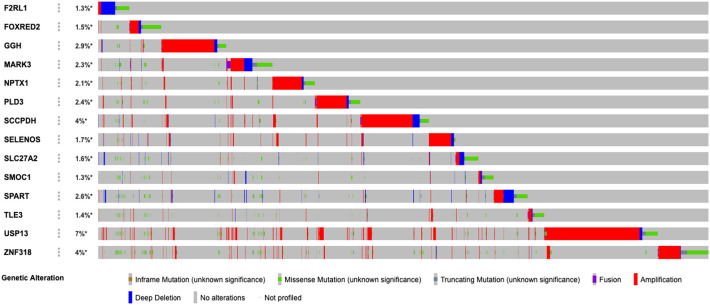
Among all SARS-CoV-2 interacting human proteins, 14 proteins namely F2RL1, FOXRED2, GGH, MARK3, NPTX1, PLD3, SCCPDH, SELENOS, SLC27A2, SMOC1, SPART, TLE3, USP13 and ZNF318 were identified as immune cell-enriched (Table 1 ). These proteins interacted with different viral proteins that were used as bait for detecting protein-protein interactions. The immune cell subtypes in which the SARS-CoV-2 interacting human proteins were enriched in included neutrophils, basophils, eosinophils, plasmacytoid DCs, NK-cells, T-cells and B-cells.
Table 1.
List of SARS-CoV-2 interacting human proteins enriched in immune cells.
| Gene name | Description | COVID-19 bait | Immune cell enrichment |
|---|---|---|---|
| F2RL1 | Proteinase-Activated Receptor 2 | SARS-CoV2 orf9c | Neutrophil |
| FOXRED2 | FAD Dependent Oxidoreductase Domain Containing 2 | SARS-CoV2 orf8 | Plasmacytoid DC |
| GGH | Gamma-Glutamyl Hydrolase | SARS-CoV2 orf8 | Plasmacytoid DC |
| MARK3 | Microtubule Affinity Regulating Kinase 3 | SARS-CoV2 orf9b | Eosinophil |
| NPTX1 | Neuronal Pentraxin 1 | SARS-CoV2 orf8 | NK-cell |
| PLD3 | Phospholipase D Family Member 3 | SARS-CoV2 orf8 | Basophil |
| SCCPDH | Saccharopine Dehydrogenase (Putative) | SARS-CoV2 nsp7 | Basophil |
| SELENOS | Selenoprotein S | SARS-CoV2 nsp7 | Plasmacytoid DC |
| SLC27A2 | Solute Carrier Family 27 Member 2 | SARS-CoV2 nsp2 | Basophil |
| SMOC1 | SPARC Related Modular Calcium Binding 1 | SARS-CoV2 orf8 | Plasmacytoid DC |
| SPART | Trans-Activated by Hepatitis C Virus Core Protein 1 | SARS-CoV2 nsp9 | Basophil |
| TLE3 | TLE Family Member 3, Transcriptional Corepressor | SARS-CoV2 nsp13 | Neutrophil |
| USP13 | Ubiquitin Specific Peptidase 13 | SARS-CoV2 nsp13 | T-cell |
| ZNF318 | Zinc Finger Protein 318 | SARS-CoV2 nsp12 | Naïve B-cell |
In order to have a glimpse of the potential roles of these shortlisted proteins, we analyzed genomic alterations (mutations, deletions, amplifications and fusions) of these genes in all TCGA PanCancer studies (Fig. 1 ). 26% of all cancer patients had at least one alteration in at least one of the immune cell-enriched SARS-CoV-2 interacting proteins. USP13 was identified as the most frequently altered gene (7%, 778 patients) among the others, which varied between 1.3 and 4%. Therefore, for future analyses, we focused on USP13.
Fig. 1.
Genomic alterations of immune cell-enriched SARS-CoV-2 interacting proteins in all TCGA PanCancer studies. Percentages of overall alteration for each gene are given on the left. Each bar represents a patient.
3.2. USP13 alterations in cancer
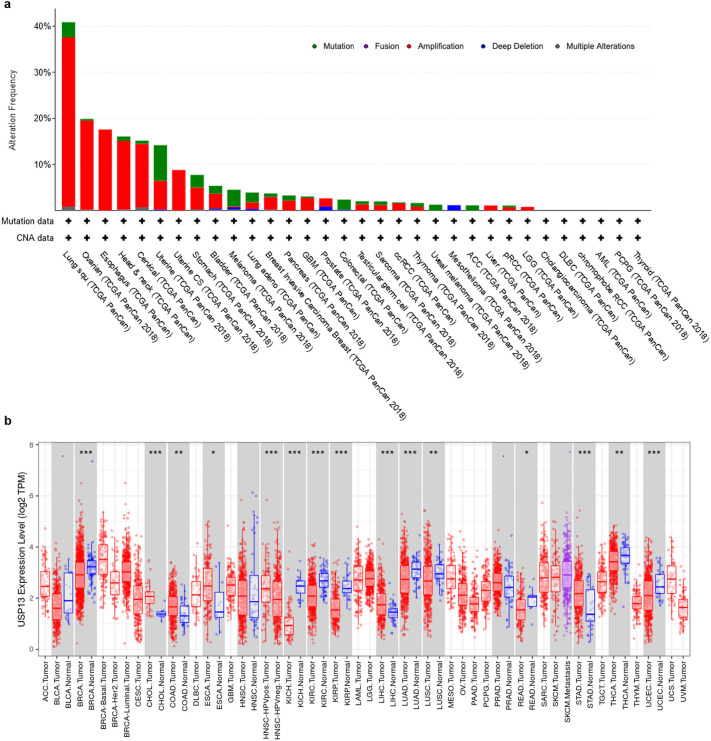
When we evaluated the genomic alterations in genes encoding for SARS-CoV-2 interacting immune-cell proteins on a single cancer type-basis, we observed that USP13 was altered in 40% of Lung Squamous Cell Carcinoma (LUSC) patients, which was followed by Ovarian Serous Cystadenoma (OV, 20%) and Esophageal Carcinoma (ESCA, 18%) (Fig. 2 .a). Most of these alterations were amplifications. While USP13 was highly altered, other SARS-CoV-2 interacting proteins had genomic alterations only up to 12% on a single cancer type-basis (data not shown).
Fig. 2.
Distribution of USP13 alterations across different cancer types and TCGA PanCancer studies (a). USP13 mRNA expression levels in tumor and adjacent normal tissues (***p < 0.001, **p < 0.01, *p < 0.05) (b).
The analysis on USP13 mRNA expression levels indicated that it was significantly overexpressed in some cancer types in comparison to the normal tissue, such as Cholangiocarcinoma (CHOL), Colon Adenocarcinoma (COAD), ESCA, Liver Hepatocellular Carcinoma (LIHC) and Stomach Adenocarcinoma (STAD) (Fig. 2.b). However, despite the high level of genomic amplifications of USP13 in lung cancer, it seems to be downregulated both in LUSC and Lung Adenocarcinoma (LUAD) tumors when compared to the adjacent normal tissue.
3.3. Effect of USP13 expression on survival
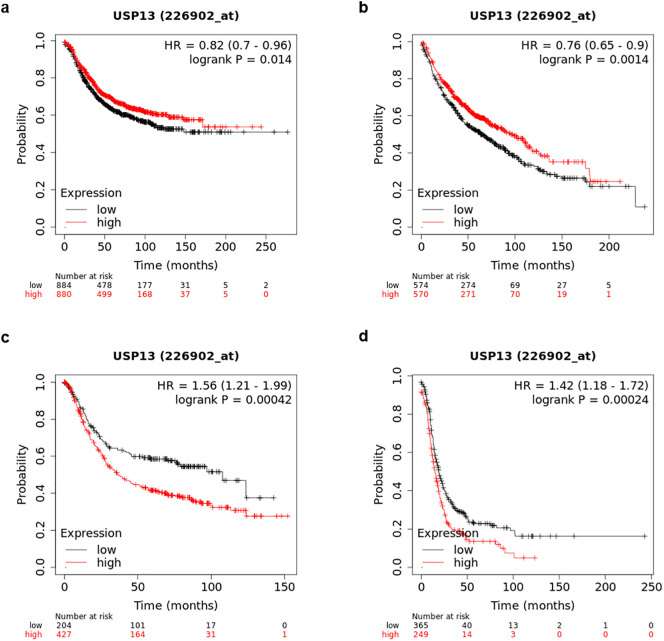
Kaplan-Meier analysis of overall survival indicated significant correlations between USP13 expression levels and prediction of the prognosis of the disease. Higher USP13 levels were correlated with longer survival periods in breast cancer (p < 0.05) and lung cancer (p < 0.01) (Fig. 3 ). Similar to the differential expression patterns of USP13 in different cancer types depicted in Fig. 2, higher levels of USP13 showed the opposite trend and correlated with poor prognosis in gastric cancer (p < 0.001) and ovarian cancers (p < 0.001).
Fig. 3.
Kaplan-Meier survival curves in relation to USP13 mRNA expression levels in breast (a), lung (b), gastric (c) and ovarian (d) cancers.
3.4. Tumor infiltration analysis
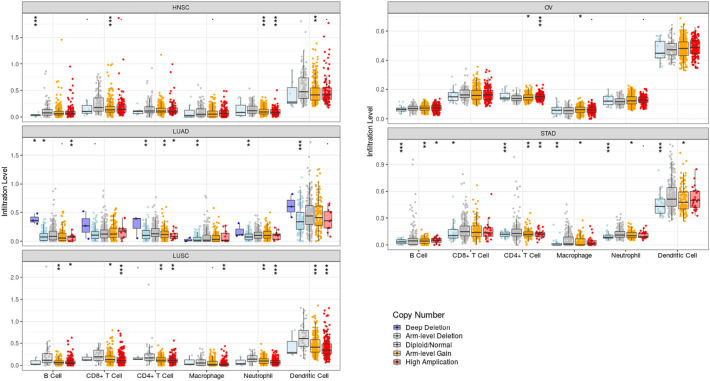
Tumor-infiltrating lymphocytes (TILs) are important components of the immune system in killing tumor cells and are potentially considered as prognostic biomarkers [22]. In order to analyze the effect of USP13 somatic copy number alterations on immune cell infiltration, TIMER analysis was performed on all TCGA datasets, which showed significant correlation in Head-Neck Squamous Cell Carcinoma (HNSC), LUAD, LUSC, STAD and OV tumors (Fig. 4 ).
Fig. 4.
Tumor infiltration levels in HNSC, LUAD, LUSC, STAD and OV. USP13 amplifications are significantly correlated with increased infiltration levels in immune cell subtypes. Only the cancer types with most statistically significant effects on tumor infiltration due to USP13 copy number alterations are shown (***p < 0.001, **p < 0.01, *p < 0.05).
In particular, USP13 arm level gains were correlated with CD8+ T-cell, neutrophil and DC infiltration in HNSC, while USP13 high amplifications indicated significant correlation with tumor infiltration in the neutrophils. In LUAD, deep deletions and arm level deletions of USP13 appeared as the predominant effectors of tumor infiltration in all immune cell subtypes analyzed except for CD8+ T-cells. In addition, in B-cells and CD4+ T-cells, high amplifications and arm level gains of USP13 resulted in increased tumor infiltration. In LUSC, high amplifications and to a lesser extent, arm level deletions of USP13 significantly correlated with immune cell infiltration in all cell types. CD4+ T-cells and macrophages were the only immune cell subtypes in which USP13 high amplifications and arm level gains correlated with enhanced infiltration. Lastly, both arm level deletions and arm level gains affected tumor infiltration in B-cells, CD4+ T-cells, macrophages, neutrophils and DCs. High amplifications were also correlated with B-cells and CD4+ T-cells infiltration.
4. Discussion
COVID-19 is currently one of the most urgent health issues that need immediate attention worldwide. Attempts directed at developing specific treatments for Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) have identified some promising target proteins. In this study, we surveyed the immune cell-enriched SARS-CoV-2 interacting human proteins using an in silico PanCancer analysis approach. Due to their immunocompromised state, it was interesting to study these immune cell-enriched proteins in cancer patients. We identified USP13 (Ubiquitin Specific Peptidase 13), which is an important component of the ubiquitin-proteasome system, as the most frequently altered gene among all SARS-CoV-2 interacting immune cell-enriched proteins. Modification of proteins via the covalent addition of ubiquitin moieties is an important regulatory mechanism in the cell. Ubiquitination is mediated by a cascade of ATP-dependent enzymes comprising E1, E2 and E3 ligases [23]; while deubiquitinases (DUBs) are responsible for the removal of these marks [24]. Recently, it was reported that several viral proteins of SARS-CoV-2 interact with human proteins involved in the ubiquitin proteasome system, such as E3 ligases TRIM59 and MIB1, Cullin 2 (CUL2) RING E3 ligase complex, and USP13 [3]. E3 ubiquitin ligases are crucial mediators of activating the host's antiviral immune response (through NF-KB and IFN pathways) [25,26] and are often taken over by the virus upon infection to facilitate its own replication and pathogenesis [27]. Similarly, USP13 has previously been implicated in downregulation of innate immune response against both DNA and RNA viruses through deubiquitination of a protein called STING [28,29].
The data in the literature regarding the role of USP13 in carcinogenesis is controversial. Some studies propose that USP13 favors tumor progression and plays an oncogenic role in glioma [30], melanoma [31], ovarian cancer [32] and lung cancer [33] while others claim that it functions as a tumor suppressor in oral squamous cell carcinoma [34], breast cancer [15] and bladder cancer [35]. Collectively, these data suggest a context-dependent role for USP13 in cancer. In line with this, our expression analysis for USP13 indicated that in some cancer types it was overexpressed in comparison to the adjacent normal tissue, whereas, in others it was downregulated. Likewise, USP13 expression levels seemed to affect the prognosis and survival differentially, depending on the cancer type.
Lastly, as USP13 is an immune-cell enriched protein, we analyzed its potential impact in tumor infiltration. Tumor infiltrating lymphocytes (TILs) are crucial for the immune system to effectively fight against cancer. Lymphocyte infiltration serves as a good indication of whether a patient would respond to immunotherapy and is often associated with improved clinical outcome [22]. Th1 type CD4+ and CD8+ T-cell infiltration is correlated with tumor cell death and better prognosis [36], while Th-2 type T-cells and B-cells are implicated in promotion of tumor growth, suggesting poor survival [37]. In lung cancer, infiltration of the tumor by CD8+ T-cells, NK-cells, DCs and macrophages were correlated with improved survival [38]. CD8+ T-cell infiltration was associated with better outcome in ovarian cancer, although it was limited only to high grade serous ovarian carcinomas [39, 40]. Head and neck cancer has different subtypes, which show different levels of lymphocyte infiltration; however, T-lymphocytes rather than B-lymphocytes, in particular CD8+ T-cells are linked with improved outcome [41].
Here we showed that copy number variations of USP13 significantly affected the infiltration levels of distinct subtypes of immune cells in HNSC, LUAD, LUSC, OV and STAD. These are also the cancer types, which had the highest levels of USP13 amplifications. Specifically, in LUSC, USP13 amplifications were correlated with higher infiltration of CD4+ T cells, CD8+ T cells, macrophages, neutrophils and dendritic cells, which is associated in the literature with better prognosis [38]. It is also noteworthy to point out that USP13 amplifications did not seem to increase B-cell infiltration in LUSC, which has the opposite effect on disease outcome [37]. Furthermore, the Kaplan-Meier survival analysis showed that higher levels of USP13 expression resulted in improved survival. Therefore, we can claim that immune cell-enriched SARS-CoV-2 interacting protein USP13 might have a tumor suppressor role in lung cancer, especially in LUSC subtype. On the other hand, although USP13 arm-level gains and high amplifications significantly increased lymphocyte infiltration in HNSC, OV and STAD, higher USP13 levels were correlated with shorter survival in ovarian and gastric cancers. One explanation for this unexpected result could be that different analyses might have been run on different datasets for the same cancer type. This is also why we were not able to perform survival analysis on HNSC. The other explanation is that the role of USP13 in cancer is context-dependent, as suggested by others in the literature.
5. Conclusion
In conclusion, it is critical to identify and characterize human proteins that interact with SARS-CoV-2 to the best of our current knowledge in order to have a better understanding of the disease itself and to develop better therapies/vaccines to fight it. In this paper, we provided a comprehensive molecular profiling of the immune cell-enriched SARS-CoV-2 interacting protein USP13, which we believe will be useful for future studies.
Author contributions
BBS performed the conceptualization of the study, data mining analyses, interpretation of the results, and writing of the manuscript.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of competing interest
The author declares no financial or non-financial conflict of interest.
References
- 1.World Health Organization Naming the coronavirus disease (COVID-19) and the virus that causes it. 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it
- 2.World Health Organization Coronavirus disease (COVID-2019) situation report -151. 19 June 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200619-covid-19-sitrep-151.pdf?sfvrsn=8b23b56e_2
- 3.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020 doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ivashkiv L.B., Donlin L.T. Regulation of type I interferon responses. Nat. Rev. Immunol. 2014;14(1):36–49. doi: 10.1038/nri358132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li G., Fan Y., Lai Y., Han T., Li Z. Coronavirus infections and immune responses. J. Med. Virol. 2020;92(4):424–432. doi: 10.1002/jmv.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paul S., Lal G. The molecular mechanism of natural killer cells function and its importance in cancer immunotherapy. Front. Immunol. 2017;8:1124. doi: 10.3389/fimmu.2017.01124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pollara G., Kwan A., Newton P.J., Handley M.E., Chain B.M., Katz D.R. Dendritic cells in viral pathogenesis: protective or defective? Int. J. Exp. Pathol. 2005;86(4):187–204. doi: 10.1111/j.0959-9673.2005.00440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin C., Zhou L., Hu Z., Zhang S., Yang S. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wan S., Yi Q., Fan S., Lv J., Zhang X. 2020. Characteristics of Lymphocyte Subsets and Cytokines in Peripheral Blood of 123 Hospitalized Patients With 2019 Novel Coronavirus Pneumonia (NCP) MedRxiv. [DOI] [Google Scholar]
- 10.Yang Y., Shen C., Li J., Yuan J., Yang M. 2020. Exuberant Elevation of IP-10, MCP-3 and IL-1ra during SARS-CoV-2 Infection is Associated With Disease Severity and Fatal Outcome. MedRxiv. [DOI] [Google Scholar]
- 11.Zhou F., Yu T., Du R. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo Y.R., Cao Q.D., Hong Z.S. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Mil. Med. Res. 2020;7:11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D'Arcy P., Linder S. Proteasome deubiquitinases as novel targets for cancer therapy. Int. J. Biochem. Cell Biol. 2012;44(11):1729–1738. doi: 10.1016/j.biocel.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 14.Liu J., Xia H., Kim M., Xu L., Li Y., Zhang L. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell. 2011;147(1):223–234. doi: 10.1016/j.cell.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J., Zhang P., Wei Y., Piao H.L., Wang W., Maddika S. Deubiquitylation and stabilization of PTEN by USP13. Nat. Cell Biol. 2013;15(12):1486–1494. doi: 10.1038/ncb2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeh H.M., Yu C.Y., Yang H.C., Ko S.H., Liao C.L., Lin Y.L. Ubiquitin-specific protease 13 regulates IFN signaling by stabilizing STAT1. J. Immunol. 2013;191(6):3328–3336. doi: 10.4049/jimmunol.1300225. [DOI] [PubMed] [Google Scholar]
- 17.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 18.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6(269):l1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li T., Fan J., Wang B. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagy Á., Lánczky A., Menyhárt O., Győrffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci. Rep. 2018;8(1):9227. doi: 10.1038/s41598-018-27521-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hendry S., Salgado R., Gevaert T. Assessing tumor-infiltrating lymphocytes in solid tumors: a practical review for pathologists and proposal for a standardized method from the International Immuno-Oncology Biomarkers Working Group: part 2: TILs in melanoma, gastrointestinal tract carcinomas, non-small cell lung carcinoma and mesothelioma, endometrial and ovarian carcinomas, squamous cell carcinoma of the head and neck, genitourinary carcinomas, and primary brain tumors. Adv. Anat. Pathol. 2017;24(6):311–335. doi: 10.1097/PAP.0000000000000161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng N., Shabek N. Ubiquitin ligases: structure, function, and regulation. Annu. Rev. Biochem. 2017;86:129–157. doi: 10.1146/annurev-biochem-060815-014922. [DOI] [PubMed] [Google Scholar]
- 24.Kaushal K., Antao A.M., Kim K.S., Ramakrishna S. Deubiquitinating enzymes in cancer stem cells: functions and targeted inhibition for cancer therapy. Drug Discov. Today. 2018;23(12):1974–1982. doi: 10.1016/j.drudis.2018.05.035. [DOI] [PubMed] [Google Scholar]
- 25.Kondo T., Watanabe M., Hatakeyama S. TRIM59 interacts with ECSIT and negatively regulates NF-κB and IRF-3/7-mediated signal pathways. Biochem. Biophys. Res. Commun. 2012;422(3):501–507. doi: 10.1016/j.bbrc.2012.05.028. [DOI] [PubMed] [Google Scholar]
- 26.Li S., Wang L., Berman M., Kong Y.Y., Dorf M.E. Mapping a dynamic innate immunity protein interaction network regulating type I interferon production. Immunity. 2011;35(3):426–440. doi: 10.1016/j.immuni.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahon C., Krogan N.J., Craik C.S., Pick E. Cullin E3 ligases and their rewiring by viral factors. Biomolecules. 2014;4(4):897–930. doi: 10.3390/biom4040897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun H., Zhang Q., Jing Y.Y. USP13 negatively regulates antiviral responses by deubiquitinating STING. Nat. Commun. 2017;8 doi: 10.1038/ncomms15534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holm C.K., Rahbek S.H., Gad H.H. Influenza A virus targets a cGAS-independent STING pathway that controls enveloped RNA viruses. Nat. Commun. 2016;7 doi: 10.1038/ncomms10680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fang X., Zhou W., Wu Q. Deubiquitinase USP13 maintains glioblastoma stem cells by antagonizing FBXL14-mediated Myc ubiquitination. J. Exp. Med. 2017;214(1):245–267. doi: 10.1084/jem.20151673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao X., Fiske B., Kawakami A., Li J., Fisher D.E. Regulation of MITF stability by the USP13 deubiquitinase. Nat. Commun. 2011;2:414. doi: 10.1038/ncomms1421. [DOI] [PubMed] [Google Scholar]
- 32.Zhang S., Zhang M., Jing Y. Deubiquitinase USP13 dictates MCL1 stability and sensitivity to BH3 mimetic inhibitors. Nat. Commun. 2018;9(1):215. doi: 10.1038/s41467-017-02693-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu Y., Zhang Y., Liu C. Amplification of USP13 drives non-small cell lung cancer progression mediated by AKT/MAPK signaling. Biomed. Pharmacother. 2019;114:108831. doi: 10.1016/j.biopha.2019.108831. [DOI] [PubMed] [Google Scholar]
- 34.Qu Z., Zhang R., Su M., Liu W. USP13 serves as a tumor suppressor via the PTEN/AKT pathway in oral squamous cell carcinoma. Cancer Manag. Res. 2019;11:9175–9183. doi: 10.2147/CMAR.S186829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Man X., Piao C., Lin X., Kong C., Cui X., Jiang Y. USP13 functions as a tumor suppressor by blocking the NF-kB-mediated PTEN downregulation in human bladder cancer. J. Exp. Clin. Cancer Res. 2019;38(1):259. doi: 10.1186/s13046-019-1262-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zitvogel L., Galluzzi L., Kepp O., Smyth M.J., Kroemer G. Type I interferons in anticancer immunity. Nat. Rev. Immunol. 2015;15(7):405–414. doi: 10.1038/nri3845. [DOI] [PubMed] [Google Scholar]
- 37.Tan A.H., Goh S.Y., Wong S.C., Lam K.P. T helper cell-specific regulation of inducible costimulator expression via distinct mechanisms mediated by T-bet and GATA-3. J. Biol. Chem. 2008;283(1):128–136. doi: 10.1074/jbc.M707693200. [DOI] [PubMed] [Google Scholar]
- 38.Remark R., Becker C., Gomez J.E. The non-small cell lung cancer immune contexture. A major determinant of tumor characteristics and patient outcome. Am. J. Respir. Crit. Care Med. 2015;191:377–390. doi: 10.1164/rccm.201409-1671PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hwang W.T., Adams S.F., Tahirovic E. Prognostic significance of tumor-infiltrating T cells in ovarian cancer: a meta-analysis. Gynecol. Oncol. 2012;124:192–198. doi: 10.1016/j.ygyno.2011.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Milne K., Kobel M., Kalloger S.E. Systematic analysis of immune infiltrates in high-grade serous ovarian cancer reveals CD20, FoxP3 and TIA-1 as positive prognostic factors. PLoS One. 2009;4 doi: 10.1371/journal.pone.0006412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Russell S.M., Angell T.E., Lechner M.G. Immune cell infiltration patterns and survival in head and neck squamous cell carcinoma. Head Neck Oncol. 2013;5:24. [PMC free article] [PubMed] [Google Scholar]