Abstract
Antibiotic resistance is a public health problem of growing concern. Animal manure application to soil is considered to be a main cause of the propagation and dissemination of antibiotic residues, antibiotic-resistant bacteria (ARB), and antibiotic resistance genes (ARGs) in the soil-water system. In recent decades, studies on the impact of antibiotic-contaminated manure on soil microbiomes have increased exponentially, in particular for taxonomical diversity and ARGs’ diffusion. Antibiotic resistance genes are often located on mobile genetic elements (MGEs). Horizontal transfer of MGEs toward a broad range of bacteria (pathogens and human commensals included) has been identified as the main cause for their persistence and dissemination. Chemical and bio-sanitizing treatments reduce the antibiotic load and ARB. Nevertheless, effects of these treatments on the persistence of resistance genes must be carefully considered. This review analyzed the most recent research on antibiotic and ARG environmental dissemination conveyed by livestock waste. Strategies to control ARG dissemination and antibiotic persistence were reviewed with the aim to identify methods for monitoring DNA transferability and environmental conditions promoting such diffusion.
Keywords: veterinary antibiotics, animal manure, antibiotic resistance genes, crop soils, antimicrobial resistance
Introduction
In recent decades, the overuse and misuse of antibiotics in human and veterinary medicine has become a serious public health issue (World Health Organization, 2014; Aidara-Kane et al., 2018). The increased number of resistant pathogens and commensal bacteria has been associated with the environmental spread of antibiotics and the propagation of antimicrobial resistant genes (ARGs; Levy, 1998; Witte, 1998; He et al., 2020). Furthermore, the environmental diffusion of antibiotics may lead to the change (Han et al., 2018) and loss (Chen et al., 2019) of microbial community diversity in soil (Kemper, 2008).
Antibiotics are used worldwide in livestock production, thus increasing the risk of antimicrobial resistance (AMR) spread. When administered for prophylactic treatments, antibiotics can directly increase selective pressure, thus favoring the generation of antibiotic-resistant bacteria (ARB; Pruden et al., 2013; Troiano et al., 2018; Blau et al., 2019). For these reasons, improved livestock and waste management strategies (i.e., diets, proximity between animals, waste treatment, use of additives, and operating conditions) should be adopted to limit the use of antibiotics in animal husbandry.
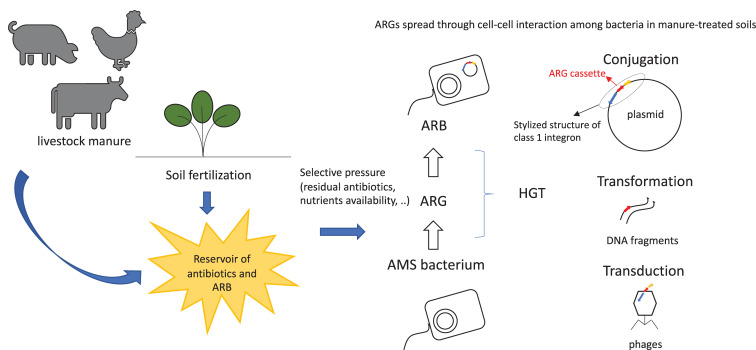
Antimicrobial resistant genes can enter and persist in ecosystem through multiple pathways. They spread across soil (Binh et al., 2007), crops (Su et al., 2015), and gut microbial communities of wild and livestock animals and of humans (Yadav and Kapley, 2019). Antimicrobial resistant genes’ spread occurs through horizontal gene transfer (HGT) of mobile genetic elements (MGEs), as phages, plasmids (Fondi and Fani, 2010), transposons, or integron gene cassettes (Figure 1). The acquisition of AMR by bacteria may be due to spontaneous mutations (Woodford and Ellington, 2007) or, more frequently, by gaining specific ARGs from other bacteria through HGT. High density of microbial cells in the presence of antimicrobial compounds and nutrients, as observable in manure (Blau et al., 2018), triggers HGT events among bacteria, thus conferring selective advantage to the hosts (Thomas and Nielsen, 2005). Mutations are essential for the continuous evolution of ARGs, producing hundreds of variants which are hardly identifiable and increasingly dangerous for the environment (Woodford and Ellington, 2007).
FIGURE 1.
Spread of ARGs and ARB in farm-related environments. ARB, antimicrobial resistant bacteria; ARG, antibiotic resistance gene; AMS, antimicrobial sensitive; HGT, horizontal gene transfer.
In this review, the effect of antibiotic occurrence in animal manure on the dissemination of AMR and ARGs in agricultural fields are discussed in a critical way. The main strategies to mitigate ARGs’ dissemination and to control antibiotic persistence are also reported. Methods monitoring changes in microbial communities and transferability and environmental diffusion of DNA were addressed as well.
The Dissemination Mechanisms of Environmental Resistome
The “resistome,” i.e., the total amount of resistance genes associated with an ecosystem (Finley et al., 2013), is generally mediated by conjugative plasmids. The resistome confers resistance of antibiotics and heavy metals to microorganisms, thus enhancing their survival in hostile environments (Bennett, 2008; Song et al., 2017). IncP-1, a common environmental plasmid group, is largely known for its efficient conjugative transferability potential and stable replication in a wide range of Gram-negative bacteria (Heuer et al., 2012). Conversely, plasmids IncF (Villa et al., 2010), IncI (Blau et al., 2018), and IncQ (Rawlings and Tietze, 2001) show a narrower host range. These plasmids are assumed to be important for the dissemination of ARG in Escherichia coli and other Enterobacteriaceae (Johnson and Nolan, 2009; Suzuki et al., 2010; Heuer et al., 2012; Van Houdt et al., 2013). As evidence, the study of the mechanisms of diffusion of these plasmids (Teuber, 2001) and compatibility evolution with broad or narrow host ranges should allow for ARG diffusion prediction.
Integrons play a key role in the fast spread of resistance determinants toward antibiotics. They are genetic elements composed of a gene encoding an integrase and an integration site for exogenous gene cassettes, which can be acquired and converted in functional and expressed genes (Mazel, 2006). Integrons can move horizontally in bacterial populations by frequent integration in plasmids or in transposons (Rowe-Magnus and Mazel, 2002). According to their aminoacidic sequence, integrases are divided into several classes. Classes 1, 2, and 3 (Inti1, Inti2, Inti3) were the first to be identified as associated with MGEs, while class 4 (Inti4) was associated with chromosomal integration (Deng et al., 2015). Among elements which facilitate DNA transfer, class 1 integron (int1) is the most frequently identified as responsible for spreading antibiotic resistance determinants amongst commensals and pathogens of humans and domesticated animals. Moreover, int1 cassette was found in different environments, such as fresh water, sediments, and sludge (Collis and Hall, 1995; Hall and Collis, 1998; Nardelli et al., 2012; Borruso et al., 2016), where it showed significantly positive correlations with the relative ARG abundance (Zhao et al., 2019).
Antibiotic residues, once entered into soil through manure application, can enhance persistence and HGT of ARGs (Binh et al., 2007; Zhao et al., 2019) through plasmids and integrons (Gotz and Smalla, 1997; Smalla et al., 2000; Sengeløv et al., 2003), promoting the spread of ARB in the environment and affecting the microbial community composition (Chen et al., 2019). Although manure-derived bacteria cannot always adapt to new environments, the antimicrobials can favor the enrichment of specific bacterial taxa in soils (through positive selection) and suppress others (Ding et al., 2014). In addition, the concentration of antibiotics in manure, usually at a sub-inhibitory level, can affect the interactions among strains and impact on gene expression and regulation (Gillings, 2013; Jechalke et al., 2014; Brüssow, 2015).
When manure is used as a fertilizer for crop production, both the increased ARB load and the antibiotic residues contained within may have negative effects on plant development and food product quality (Verraes et al., 2013; Mirza et al., 2020; Muhammad et al., 2020). In addition, antibiotic residues can persist and accumulate in the environment (Jechalke et al., 2014) by adsorption on soil solid phases (Du and Liu, 2012).
ARGs in the Environment
The majority of antibiotics are naturally produced by microbes as a self-protection mechanism against other microorganisms. ARGs have been always present in the environment. ARGs encoding resistance for a large set of antibiotics have been found in 30,000-year-old Beringian permafrost and in bacteria isolated from prehistoric caves (D’Costa et al., 2011; Berglund, 2015). When present in the environment at a sub-inhibitory concentration, antibiotics frequently play a role in transcription regulation and in the exchange of signals among cells (i.e., quorum sensing mechanism and conjugation) (Reygaert, 2018).
Antibiotic resistance consists of a large variety of mechanisms, such as inactivation by specific cleaving enzymes, exclusion from cells via efflux pumps, interference with protein synthesis, limitation of drug uptake, and modification of antibiotic target. Resistance acquired through MGEs and plasmids is responsible for the last two mechanisms in which the resistance extent depends on bacterial species and acquired ARGs (Reygaert, 2018; Kraemer et al., 2019). The antibiotic selective pressure driving the acquired resistance determines accurate ARGs’ specialization, thus making the environment a potential reservoir.
Anthropogenic activities affect antibiotic and ARGs’ spread with somewhat predictable effects (Vikesland et al., 2017). In livestock farming, the use of antibiotics varies depending on the farming type and location, having a considerable effect on ARGs’ concentration. Among the ARGs most frequently detected in livestock production, those related to sulfonamide resistance (sul) (Table 1) are particularly diffused in aquatic systems (Chen et al., 2015; Makowska et al., 2016). In surface and fresh waters, sul genes were found in IncQ plasmid group (Sköld, 2001; Berglund, 2015). Similarly, diaminopyrimidine genes (dfr), which confer resistance to antimicrobial trimethoprim, have been identified in both class 1 and class 2 integrons (Deng et al., 2015). Similarly, quinolone resistance qnr genes have been frequently associated with different plasmid groups. Both dfr and qnr genes easily disseminate in the environment, being found in surface waters (Berglund, 2015), wastewaters, and related irrigated soils (Dalkmann et al., 2012). Tetracycline resistance genes (tet) are widely diffused in different pathogenic and environmental bacteria (Roberts, 2005) and are often detected in sewage treatment plants, soil, and surface and ground water (Chee-Sanford et al., 2001; Berglund, 2015). In the same environments, erm genes, which are the most widespread macrolides resistance gene, were isolated.
TABLE 1.
The most commonly used antibiotics and the relative ARGs in livestock production (DHPS, dihydropteroate synthase; DHPR, dihydropyridine-resistant).
| Antibiotic family | Most used | Animal Farming | Use | Contrasted bacteria and recognized main targets | Resistance mechanism | Main ARGs |
| Macrolides | Tylosin | Cattle | Gastrointestinal and respiratory infections | Gram-positive bacteria. | Interference with protein synthesis (sequestration of mRNA ribosome-binding site) | erm, msr, mef genes |
| Erytromycin | Pig | Main target: Lawsonia intracellularis | ||||
| Clarithromycin | Poultry | Staphylococcus aureus | ||||
| Sulfonamides | Sulfamethazine | Cattle | Urinary tract infections | Gram-positive and Gram-negative bacteria. Main target: Enterobacteriaceae, Pasteurellaceae | Interference with folic acid synthesis competing for the enzyme DHPS | sulI, sulII genes |
| Pig | Respiratory infections | |||||
| Poultry | ||||||
| Tetracyclines | Chlortetracycline | Cattle | Systemic and local infections | Gram-positive and Gram-negative bacteria | Interference with efflux pump systems | tet genes |
| Oxytetracyclines | Pig | Gastrointestinal and respiratory infections | ||||
| Doxycycline | Poultry | |||||
| Quinolones | Fluoroquinolones (Enrofloxacin, Danofloxacin, Marbofloxacine) | Pig | Intestinal infections | Gram-positive and Gram-negative bacteria, including mycobacteria, and anaerobes | Mutations in the genes encoding quinolone target DNA gyrase and topoisomerase IV, interference with efflux pump systems | qnr genes |
| Cattle | ||||||
| β-lactams | Penicillins (Amoxycilline, Ampicillines) Cephalosporins, Carbapenems | Pig | Respiratory diseases | Gram-positive and Gram-negative bacteria | Interference with cell wall synthesis and permeability, inactivation through β-Lactamase enzyme | bla, amp, pen genes, |
| Cattle | Necrotic enteritis | |||||
| Poultry | ||||||
| Dog | ||||||
| Cat | ||||||
| Aminoglycosides | Streptomycin, Spectinomycin, Neomycin, Aspramycin, Gentamycin, Lincomycin | Pig | Intestinal infections | Gram-positive, and Gram-negative bacteria, if aerobic | Inhibition of protein synthesis (rhibosome interference) | aac, aad, aad aph genes |
| Poultry | ||||||
| Phenicols | Chloramphenicol | Pig | Respiratory disease, foot rot | Broad spectrum. Main target: Photobacterium, Salmonella, E. coli | Enzymatic modification of antibiotic molecules | cat, pp-flo, flo genes |
| Thiamphenicols (thiamphenicol, florfenicol) | ||||||
| Diaminopyrimidines | Trimethoprim | Horse | Post-weaning scours | Gram-positive and many Gram-negative bacteria. Main target: Enterobacteriaceae | Interference with folic acid synthesis by binding the enzyme DHFR | dfr genes |
| Pig | ||||||
| Polypeptides | Bacitracin, Colistin | Pig | Intestinal diseases | Gram-positive (Bacitracin) or Gram negative (Colistin) bacteria. Main Gram negative target: E. coli Salmonela spp. Pseudomonas aeruginosa, Klebsiella pneumoniae, or Acinetobacter. | LPS modification, efflux pump systems regulation | pmr,pho, mcr, kpn genes |
| Poultry | Main Gram positive target: Campylobacter | |||||
| Lincosamides | Lincomycin | Pig | Respiratory and Intestinal infections | Gram positive bacteria, most anaerobic and some mycolpasma species. Main target: Staphylococcus aureus | Alteration of the antibiotic target site | lnu, lin, erm genes |
| Poultry | ||||||
| Cattle | ||||||
| Pleuromutilins | Tiamulin | Pig | Respiratory and Intestinal infections | Pasteurellaceae, Brachyspira, Micoplasma | Alteration/protection of the antibiotic target site | vga, sal, lsa genes |
| Valnemulin | Poultry |
Essentially, ARGs’ diffusion is associated with a stress response activated by exposure to antibiotics as well as with the mobilization of several integrative and conjugative elements. ARGs’ maintenance depends on their considerably low fitness cost. In fact, once a specific ARG has been acquired by a bacterial cell, it must evolve to produce more benefits than costs in order for multiple copies of the same gene to be kept and to maintain the expression control of genes in MGEs (Bengtsson-Palme et al., 2017). Furthermore, as already mentioned, nutrient rich environments can positively influence the ARGs’ spread and facilitate cell–cell interactions (Manaia et al., 2018) (Figure 1).
The Use of Veterinary Antibiotics
In veterinary medicine, antimicrobials can be used as therapeutics and/or growth promoters. Antibiotic growth promoters (AGPs) are antimicrobial substances administered at a sub-therapeutic dose for a prolonged time with the main purpose being to improve the feed conversion rate, especially in young animals, raising the economical profit of farmers. Since 2006, both the European Union and Australia have forbidden the use of AGPs. Nevertheless, in most other countries the use of AGPs is still permitted (Guardabassi et al., 2009).
Among breeding farms, poultry and pig livestock have received the majority of antibiotics for therapeutic or prophylactic use (Ungemach, 2000; Kim et al., 2011), resulting in an abundance of ARGs greater than three orders of magnitude compared to other farming systems, such as fish and cattle farming. Several studies confirmed swine farms as a hot-spot for ARB and ARGs (Rosen, 1995; Cromwell, 2002; de Greeff et al., 2019; Petrin et al., 2019). Recently, the scientific community investigated prevalence, abundance, and possible mobilization of ARGs in pig farms and surrounding environments (Hölzel et al., 2010; Marti Serrano, 2014; Petrin et al., 2019; Van den Meersche et al., 2019; Wu et al., 2019).
Table 1 summarizes the main antibiotic families and the most used antimicrobics in livestock animals for therapeutic use. Nowadays, more than 150 antimicrobial compounds in livestock production are used. The residues inevitably end up in the environment because of manure application on agricultural lands (Baguer et al., 2000). In 2010, more than 63,000 tons of antimicrobials were consumed by livestock across the globe. The predicted growth of the world’s population allows for an estimated increase in antibiotic consumption of up to 105,000 tons by 2030 (Tasho and Cho, 2016). For this reason, specific action plans have been defined to reduce the use of antibiotics as therapeutics for livestock in several countries (i.e., the European One Health Action Plan against Antimicrobial Resistance, 2017; the National Strategy to Combat Antibiotic-Resistant Bacteria, proposed by the White House, 2014; the National Action Plan to Contain Antimicrobial Resistance issued by the Chinese National Health and Family Planning Commission, 2016–2020).
Manure Treatments
Besides direct collection into aerobic or anaerobic lagoons, animal manure can undergo drying and liquid-solid phase separation. Manure solid phase, as well as whole manure if shovellable, is traditionally composted to produce biofertilizer. Currently, anaerobic digestion and biological treatments of animal manure are often adopted on intensive animal farms (Van Epps and Blaney, 2016).
Composting can substantially reduce the antibiotic load, especially during the thermophilic phase (Zhang et al., 2019), but recalcitrant antibiotics accumulate in compost products and in amended soil (Bohrer et al., 2019; Zang et al., 2019). A general ARG abatement (0.7–2.0 log decrease) is obtained through thermophilic composting of swine, cattle, and poultry manure, depending on manure type and operational conditions (He et al., 2020).
Biological treatments of animal manure and wastewater, which are adopted to reduce the environmental input of nitrates, slightly decreases the levels of antibiotic residues and pathogenic bacteria (Van den Meersche et al., 2019). Antimicrobial resistant gene reduction of 0.1–3.3 log is observed in swine manure after treatment (He et al., 2020).
Anerobic digestion (AD) is adopted to stabilize manure with a final production of methane (Fubin et al., 2016, 2017). A 0.3–52 log decrease of ARGs was observed in digestate from swine wastewater (He et al., 2020). Interestingly, the higher the content of volatile solids in manure and the mixing rate, the higher the ARGs number in the digestate (Turker et al., 2018). The combined pasteurization and AD of swine manure reduced sole archaeal communities, whereas simple AD affected bacteria and archea (Fubin et al., 2020). Manure pretreatment with bacterial strains is effective in degrading antibiotics (Liu et al., 2019) and enhancing biogas production, but the overall effect on ARB and ARGs was not addressed.
Constructed wetlands are vegetated aquatic systems that can be adopted for the treatment of wastewater and agricultural drainage water (Lavrnic et al., 2018). Their ability to reduce ARGs in swine wastewater resulted in a 0.18–3 log decrease (He et al., 2020).
Oxidizing post-treatments, as ozonation or Fenton conditions, can be used on animal or treated wastewaters to degrade antibiotics and bacteria thanks to the activity of reactive oxygen species (Balcıoğlu and Ötker, 2003; Ikehata et al., 2006; Uslu and Balcıoğlu, 2009). Among advanced oxidation processes, highly costly ionizing radiations are known for their ability to destroy microbial DNA. Therefore, affordable combinations of ionizing radiation and oxidation allows for the degradation of antibiotics and ARGs in organic matrices, although with a high biological and environmental risk (Chu et al., 2019, 2020).
Different Approaches to Resistome Profiling Study
Even though AMRs introduced in the environment with animal manure have been largely explored (Dolliver et al., 2008; Selvam et al., 2012b), contradictory information exists regarding the fate of ARGs (Selvam et al., 2012a; Wang et al., 2015; Xie et al., 2016). The growing need for the control of ARGs’ spread prompted the scientific community to set up and to validate refined molecular methods for the study of ARGs’ dissemination dynamics among environmental microbial communities.
Both 16S rRNA amplicon and untargeted sequencing can be considered exhaustive methods for the exploration of microbial community structure in manure-fertilized soil and farm waste. Several studies on resistome diffusion in wastewater treatment plants (Yadav and Kapley, 2019), sewage sludge composting units (Su et al., 2015), and urban sewage support the metagenomic approach (Hendriksen et al., 2019) in monitoring ARGs’ level during treatments and seasonal changes. A recent work (Han et al., 2018) showed that the shift in soil bacterial communities caused by manure application leads to changes in the soil bacteria resistome.
Recently, studies on the detection of genetic markers associated with AMR (transposases and class 1 integron-integrase genes) and ARGs have been markedly increasing. The quantification of ARGs in soils amended with livestock and swine manure (Brooks et al., 2014; Tao et al., 2014) was performed with high-throughput qPCR assay (Rocha et al., 2018; Blau et al., 2019). In a recent study, both intracellular and extracellular DNA containing ARGs were quantified in sludge at about 1010 and 1012 copies per gram, respectively (Dong et al., 2019). Here, the intracellular ARGs were assessed through conjugation with cell-cell contact, whereas the extracellular ARGs were assessed through natural transformation. Several works on different manure types focused on the quantification of targeted genes intI1 and intI2 for class 1 and 2 integron-integrase genes and korB gene, specific for IncP-1 plasmids, together with ARGs (Hu et al., 2016; Blau et al., 2018, 2019).
As already reported, plasmid-mediated ARGs’ diffusion is frequently used, especially for the role of plasmids in the rapid bacterial adaptation and fitness improvement (Smalla et al., 2000). Exogenous plasmid isolation techniques (Bale et al., 1988) clarified how plasmids diffuse in different environments. Recently, plasmids from municipal sewage sludge and recipient bacteria were analyzed for their transferability by exogenous isolation (Blau et al., 2018; Wolters et al., 2018). Referring to pig manure samples, four IncQ-like plasmids were isolated in recipient strains: Pseudomonas putida UWC1, Acinetobacter sp., Ralstonia eutropha, Agrobacterium tumefaciens, and E. coli. The plasmid transferability in E. coli strains was not efficient, underlying a broad but highly specific host range (Smalla et al., 2000).
Recently, simplified mathematical models have been applied to predict and quantify ARGs’ spread in livestock animal gut microbiomes (Andersen et al., 2020) and in agricultural waste (Baker et al., 2016). In such environments, the variables involved in the ARGs’ spread are countless and depend on a wide range of intrinsic and extrinsic factors, such as genetic mechanisms of ARB replication, HGT dynamics, environmental and stressor conditions, and microbiota composition. Therefore, future research should focus on the improvement of predictive models of ARGs’ dissemination mechanism, exploitable for targeted operations in livestock waste management.
Conclusion
Although a decrease in the use of antibiotics in livestock production is highly recommended, antibiotics’ overuse remains an important issue to solve. The uncontrolled spread of ARB and ARGs in the environment due to soil manuring is of serious concern. Many studies highlight ARGs’ presence in microbial communities of livestock manure and manured agricultural fields, despite the improved livestock and waste management strategies to contain in-farm ARGs’ spread. In the last thirty years, knowledge on pathways of ARGs’ diffusion from animal waste to the environment was enriched by multidisciplinary research approaches.
In light of the current knowledge, the study of the dynamics of AMR and ARGs’ spread in manure and environments surrounding livestock farms should combine molecular and functional genetics strategies with prediction models of the diffusion of MGEs (integrons and plasmids) and metagenomic data.
Author Contributions
AC: original draft preparation, figure and table conceptualization, review, and editing. PT, MM, and SB: review. DL: original draft preparation and table preparation. IB and PM: original draft preparation and review. All authors contributed to critically revising the manuscript and gave final approval for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This research was supported by Programma di Sviluppo Rurale 2014-2020 Regione Lombardia (Project REFLUA: Swine manure and environment).
References
- Abbas R. Z., Iqbal Z., Blake D., Khan M. N., Saleemi M. K. (2011). Anticoccidial drug resistance in fowl coccidia: the state of play revisited. Worlds. Poult. Sci. J. 67 337–350. 10.1017/s004393391100033x [DOI] [Google Scholar]
- Aghapour Z., Gholizadeh P., Ganbarov K., Bialvaei A. Z., Mahmood S. S., Tanomand A., et al. (2019). Molecular mechanisms related to colistin resistance in Enterobacteriaceae. Infect. Drug Resist. 12 965. 10.2147/idr.s199844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aidara-Kane A., Angulo F. J., Conly J. M., Minato Y., Silbergeld E. K., McEwen S. A., et al. (2018). World Health Organization (WHO) guidelines on use of medically important antimicrobials in food-producing animals. Antimicrob. Resist. Infect. Control 7:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen V. D., Aarestrup F. M., Munk P., Jensen M. S., de Knegt L. V., Bortolaia V., et al. (2020). Predicting effects of changed antimicrobial usage on the abundance of antimicrobial resistance genes in finisher’gut microbiomes. Prev. Vet. Med. 174:104853. 10.1016/j.prevetmed.2019.104853 [DOI] [PubMed] [Google Scholar]
- Baguer A. J., Jensen J., Krogh P. H. (2000). Effects of the antibiotics oxytetracycline and tylosin on soil fauna. Chemosphere 40 751–757. 10.1016/s0045-6535(99)00449-x [DOI] [PubMed] [Google Scholar]
- Baker M., Hobman J. L., Dodd C. E. R., Ramsden S. J., Stekel D. J. (2016). Mathematical modelling of antimicrobial resistance in agricultural waste highlights importance of gene transfer rate. FEMS Microbiol. Ecol. 92:fiw040. 10.1093/femsec/fiw040 [DOI] [PubMed] [Google Scholar]
- Balcıoğlu I. A., Ötker M. (2003). Treatment of pharmaceutical wastewater containing antibiotics by O3 and O3/H2O2 processes. Chemosphere 50 85–95. 10.1016/s0045-6535(02)00534-9 [DOI] [PubMed] [Google Scholar]
- Bale M. J., Day M. J., Fry J. C. (1988). Novel method for studying plasmid transfer in undisturbed river epilithon. Appl. Environ. Microbiol. 54 2756–2758. 10.1128/aem.54.11.2756-2758.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson-Palme J., Kristiansson E., Larsson D. G. J. (2017). Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiol. Rev. 42:fux053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett P. M. (2008). Plasmid encoded antibiotic resistance: acquisition and transfer of antibiotic resistance genes in bacteria. Br. J. Pharmacol. 153 S347–S357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund B. (2015). Environmental dissemination of antibiotic resistance genes and correlation to anthropogenic contamination with antibiotics. Infect. Ecol. Epidemiol. 5:28564. 10.3402/iee.v5.28564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binh C. T. T., Heuer H., Gomes N. C. M., Kotzerke A., Fulle M., Wilke B. M., et al. (2007). Short-term effects of amoxicillin on bacterial communities in manured soil. FEMS Microbiol. Ecol. 62 290–302. 10.1111/j.1574-6941.2007.00393.x [DOI] [PubMed] [Google Scholar]
- Blau K., Bettermann A., Jechalke S., Fornefeld E., Vanrobaeys Y., Stalder T., et al. (2018). The transferable resistome of produce. mBio 9:e01300-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau K., Samuel J., Su J.-Q., Zhu Y.-G., Smalla K., Jechalke S. (2019). Manure and doxycycline affect the bacterial community and its resistome in lettuce rhizosphere and bulk soil. Front. Microbiol. 10:725. 10.3389/fmicb.2019.00725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohrer R. E. G., Carissimi E., Lopez D. A. R., Wolff D. B., da Silva D. M., Prestes O. D. (2019). Composting of swine waste in the treatment of veterinary drug residues. Semin. Ciências Agrárias 40 2813–2830. [Google Scholar]
- Borruso L., Harms K., Johnsen P. J., Nielsen K. M., Brusetti L. (2016). Distribution of class 1 integrons in a highly impacted catchment. Sci. Total Environ. 566 1588–1594. 10.1016/j.scitotenv.2016.06.054 [DOI] [PubMed] [Google Scholar]
- Brooks J. P., Adeli A., McLaughlin M. R. (2014). Microbial ecology, bacterial pathogens, and antibiotic resistant genes in swine manure wastewater as influenced by three swine management systems. Water Res. 57 96–103. 10.1016/j.watres.2014.03.017 [DOI] [PubMed] [Google Scholar]
- Brüssow H. (2015). Microbiota and the human nature: know thyself. Environ. Microbiol. 17 10–15. 10.1111/1462-2920.12693 [DOI] [PubMed] [Google Scholar]
- Chee-Sanford J. C., Aminov R. I., Krapac I. J., Garrigues-Jeanjean N., Mackie R. I. (2001). Occurrence and diversity of tetracycline resistance genes in lagoons and groundwater underlying two swine production facilities. Appl. Environ. Microbiol. 67 1494–1502. 10.1128/aem.67.4.1494-1502.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Hao L., Guo X., Wang N., Ye B. (2015). Prevalence of antibiotic resistance genes of wastewater and surface water in livestock farms of Jiangsu Province. China. Environ. Sci. Pollut. Res. 22 13950–13959. 10.1007/s11356-015-4636-y [DOI] [PubMed] [Google Scholar]
- Chen Q.-L., An X.-L., Zheng B.-X., Gillings M., Peñuelas J., Cui L., et al. (2019). Loss of soil microbial diversity exacerbates spread of antibiotic resistance. Soil Ecol. Lett. 1 3–13. 10.1007/s42832-019-0011-0 [DOI] [Google Scholar]
- Chu L., Chen D., Wang J., Yang Z., Shen Y. (2019). Degradation of antibiotics and antibiotic resistance genes in erythromycin fermentation residues using radiation coupled with peroxymonosulfate oxidation. Waste Manag. 96 190–197. 10.1016/j.wasman.2019.07.031 [DOI] [PubMed] [Google Scholar]
- Chu L., Chen D., Wang J., Yang Z., Yang Q., Shen Y. (2020). Degradation of antibiotics and inactivation of antibiotic resistance genes (ARGs) in Cephalosporin C fermentation residues using ionizing radiation, ozonation and thermal treatment. J. Hazard. Mater. 382:121058. 10.1016/j.jhazmat.2019.121058 [DOI] [PubMed] [Google Scholar]
- Collis C. M., Hall R. M. (1995). Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob. Agents Chemother. 39 155–162. 10.1128/aac.39.1.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromwell G. L. (2002). Why and how antibiotics are used in swine production. Anim. Biotechnol. 13 7–27. 10.1081/abio-120005767 [DOI] [PubMed] [Google Scholar]
- Dalkmann P., Broszat M., Siebe C., Willaschek E., Sakinc T., Huebner J., et al. (2012). Accumulation of pharmaceuticals, Enterococcus, and resistance genes in soils irrigated with wastewater for zero to 100 years in central Mexico. PLoS One 7:e45397. 10.1371/journal.pone.0045397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Costa V. M., King C. E., Kalan L., Morar M., Sung W. W. L., Schwarz C., et al. (2011). Antibiotic resistance is ancient. Nature 477:457. [DOI] [PubMed] [Google Scholar]
- de Greeff S. C., Mouton J. W., Schoffelen A. F., Verduin C. M. (2019). NethMap 2019: Consumption of antimicrobial agents and antimicrobial resistance among medically important bacteria in the Netherlands/MARAN 2019: Monitoring of Antimicrobial Resistance and Antibiotic Usage in Animals in the Netherlands in 2018. Bilthoven: National Institute for Public Health and the Environment, 10.21945/RIVM-2019-0038 [DOI] [Google Scholar]
- Deng F., Wang H., Liao Y., Li J., Feßler A. T., Michael G. B., et al. (2017). Detection and genetic environment of pleuromutilin-lincosamide-streptogramin A resistance genes in staphylococci isolated from pets. Front. Microbiol. 8:234. 10.3389/fmicb.2017.00234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Bao X., Ji L., Chen L., Liu J., Miao J., et al. (2015). Resistance integrons: class 1, 2 and 3 integrons. Ann. Clin. Microbiol. Antimicrob. 14:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding G.-C., Radl V., Schloter-Hai B., Jechalke S., Heuer H., Smalla K., et al. (2014). Dynamics of soil bacterial communities in response to repeated application of manure containing sulfadiazine. PLoS One 9:e92958. 10.1371/journal.pone.0092958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolliver H., Gupta S., Noll S. (2008). Antibiotic degradation during manure composting. J. Environ. Qual. 37 1245–1253. 10.2134/jeq2007.0399 [DOI] [PubMed] [Google Scholar]
- Dong P., Wang H., Fang T., Wang Y., Ye Q. (2019). Assessment of extracellular antibiotic resistance genes (eARGs) in typical environmental samples and the transforming ability of eARG. Environ. Int. 125 90–96. 10.1016/j.envint.2019.01.050 [DOI] [PubMed] [Google Scholar]
- Du L., Liu W. (2012). Occurrence, fate, and ecotoxicity of antibiotics in agro-ecosystems. A review. Agron. Sustain. Dev. 32 309–327. 10.1007/s13593-011-0062-9 [DOI] [Google Scholar]
- Finley R. L., Collignon P., Larsson D. G. J., McEwen S. A., Li X.-Z., Gaze W. H., et al. (2013). The scourge of antibiotic resistance: the important role of the environment. Clin. Infect. Dis. 57 704–710. 10.1093/cid/cit355 [DOI] [PubMed] [Google Scholar]
- Fondi M., Fani R. (2010). The horizontal flow of the plasmid resistome: clues from inter-generic similarity networks. Environ. Microbiol. 12 3228–3242. 10.1111/j.1462-2920.2010.02295.x [DOI] [PubMed] [Google Scholar]
- Fubin Y., Dong H., Zhang W., Zhu Z., Shang B. (2020). Additional function of pasteurisation pretreatment in combination with anaerobic digestion on antibiotic removal. Bioresour. Technol. 297:122414. 10.1016/j.biortech.2019.122414 [DOI] [PubMed] [Google Scholar]
- Fubin Y., Ji C., Dong H., Tao X., Chen Y. (2016). Research progress on effect of antibiotic on anaerobic digestion treatment in animal manure. J. Agric. Sci. Technol. 18 171–177. [Google Scholar]
- Fubin Y., Zifu L., Saino M., Hongmin D. (2017). Performance of alkaline pretreatment on pathogens inactivation and sludge solubilization. Int. J. Agric. Biol. Eng. 10 216–223. [Google Scholar]
- Gillings M. R. (2013). Evolutionary consequences of antibiotic use for the resistome, mobilome and microbial pangenome. Front. Microbiol. 4:4. 10.3389/fmicb.2013.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz A., Smalla K. (1997). Manure enhances plasmid mobilization and survival of Pseudomonas putida introduced into field soil. Appl. Environ. Microbiol. 63 1980–1986. 10.1128/aem.63.5.1980-1986.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guardabassi L., Jensen L. B., Kruse H. (2009). Guide to Antimicrobial Use in Animals. New York, NY: John Wiley & Sons. [Google Scholar]
- Hall R. M., Collis C. M. (1998). Antibiotic resistance in gram-negative bacteria: the role of gene cassettes and integrons. Drug Resist. Updat. 1 109–119. 10.1016/s1368-7646(98)80026-5 [DOI] [PubMed] [Google Scholar]
- Han X.-M., Hu H.-W., Chen Q.-L., Yang L.-Y., Li H.-L., Zhu Y.-G., et al. (2018). Antibiotic resistance genes and associated bacterial communities in agricultural soils amended with different sources of animal manures. Soil Biol. Biochem. 126 91–102. 10.1016/j.soilbio.2018.08.018 [DOI] [Google Scholar]
- He Y., Yuan Q., Mathieu J., Stadler L., Senehi N., Sun R., et al. (2020). Antibiotic resistance genes from livestock waste: occurrence, dissemination, and treatment. NPJ Clean Water 3 1–11. [Google Scholar]
- Hendriksen R. S., Munk P., Njage P., Van Bunnik B., McNally L., Lukjancenko O., et al. (2019). Global monitoring of antimicrobial resistance based on metagenomics analyses of urban sewage. Nat. Commun. 10:1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuer H., Binh C. T. T., Jechalke S., Kopmann C., Zimmerling U., Krögerrecklenfort E., et al. (2012). IncP-1ε plasmids are important vectors of antibiotic resistance genes in agricultural systems: diversification driven by class 1 integron gene cassettes. Front. Microbiol. 3:2. 10.3389/fmicb.2012.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hölzel C. S., Schwaiger K., Harms K., Küchenhoff H., Kunz A., Meyer K., et al. (2010). Sewage sludge and liquid pig manure as possible sources of antibiotic resistant bacteria. Environ. Res. 110 318–326. 10.1016/j.envres.2010.02.009 [DOI] [PubMed] [Google Scholar]
- Van Houdt R., Toussaint A., Ryan M. P., Pembroke J. T., Mergeay M., Adley C. C. (2013). “The Tn4371 ICE family of bacterial mobile genetic elements,” in Bacterial Integrative Mobile Genetic Elements, eds Roberts A. P., Mullany P. (Milton Park: Taylor & Francis; ). [Google Scholar]
- Hu H.-W., Han X.-M., Shi X.-Z., Wang J.-T., Han L.-L., Chen D., et al. (2016). Temporal changes of antibiotic-resistance genes and bacterial communities in two contrasting soils treated with cattle manure. FEMS Microbiol. Ecol. 92:fiv169. 10.1093/femsec/fiv169 [DOI] [PubMed] [Google Scholar]
- Ikehata K., Jodeiri Naghashkar N., Gamal El-Din M. (2006). Degradation of aqueous pharmaceuticals by ozonation and advanced oxidation processes: a review. Ozone Sci. Eng. 28 353–414. 10.1080/01919510600985937 [DOI] [Google Scholar]
- Jechalke S., Heuer H., Siemens J., Amelung W., Smalla K. (2014). Fate and effects of veterinary antibiotics in soil. Trends Microbiol. 22 536–545. 10.1016/j.tim.2014.05.005 [DOI] [PubMed] [Google Scholar]
- Johnson T. J., Nolan L. K. (2009). Pathogenomics of the virulence plasmids of Escherichia coli. Microbiol. Mol. Biol. Rev. 73 750–774. 10.1128/mmbr.00015-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper N. (2008). Veterinary antibiotics in the aquatic and terrestrial environment. Ecol. Indic. 8 1–13. 10.1016/j.ecolind.2007.06.002 [DOI] [Google Scholar]
- Kim K.-R., Owens G., Kwon S.-I., So K.-H., Lee D.-B., Ok Y. S. (2011). Occurrence and environmental fate of veterinary antibiotics in the terrestrial environment. Water Air Soil Pollut. 214 163–174. 10.1007/s11270-010-0412-2 [DOI] [Google Scholar]
- Kraemer S. A., Ramachandran A., Perron G. G. (2019). Antibiotic pollution in the environment: from microbial ecology to public policy. Microorganisms 7:180. 10.3390/microorganisms7060180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavrnic S., Braschi I., Anconelli S., Blasioli S., Solimando D., Mannini P., et al. (2018). Long-term monitoring of a surface flow constructed wetland treating agricultural drainage water in northern Italy. Water 10:644 10.3390/w10050644 [DOI] [Google Scholar]
- Levy S. B. (1998). Multidrug resistance—a sign of the times in Multidrug resistance—a sign of the times. N. Engl. J. Med. 338 1376–1378. 10.1056/nejm199805073381909 [DOI] [PubMed] [Google Scholar]
- Li L., Sun J., Liu B., Zhao D., Ma J., Deng H., et al. (2013). Quantification of lincomycin resistance genes associated with lincomycin residues in waters and soils adjacent to representative swine farms in China. Front. Microbiol. 4:364. 10.3389/fmicb.2013.00364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Ni H., Yang L., Chen G., Yan X., Leng X., et al. (2019). Pretreatment of swine manure containing β-lactam antibiotics with whole-cell biocatalyst to improve biogas production. J. Clean. Prod. 240:118070 10.1016/j.jclepro.2019.118070 [DOI] [Google Scholar]
- Makowska N., Koczura R., Mokracka J. (2016). Class 1 integrase, sulfonamide and tetracycline resistance genes in wastewater treatment plant and surface water. Chemosphere 144 1665–1673. 10.1016/j.chemosphere.2015.10.044 [DOI] [PubMed] [Google Scholar]
- Manaia C. M., Rocha J., Scaccia N., Marano R., Radu E., Biancullo F., et al. (2018). Antibiotic resistance in wastewater treatment plants: tackling the black box. Environ. Int. 115 312–324. 10.1016/j.envint.2018.03.044 [DOI] [PubMed] [Google Scholar]
- Marti Serrano E. (2014). Bacteriophages as a reservoir of extended-spectrum ß-lactamase and fluoroquinolone resistance genes in the environment. Clin. Microbiol. Infect. 20 O456–O459. [DOI] [PubMed] [Google Scholar]
- Mazel D. (2006). Integrons: agents of bacterial evolution. Nat. Rev. Microbiol. 4 608–620. 10.1038/nrmicro1462 [DOI] [PubMed] [Google Scholar]
- Mirza S. A., Afzaal M., Begum S., Arooj T., Almas M., Ahmed S., et al. (2020). “Uptake mechanism of antibiotics in plants,” in Antibiotics and Antimicrobial Resistance Genes in the Environment, Ed. Hashmi M. Z. (Amsterdam: Elsevier; ), 183–188. 10.1016/b978-0-12-818882-8.00011-5 [DOI] [Google Scholar]
- Muhammad J., Khan S., Su J. Q., Hesham A. E. L., Ditta A., Nawab J., et al. (2020). Antibiotics in poultry manure and their associated health issues: a systematic review. J. Soils Sediments 20 486–497. 10.1007/s11368-019-02360-0 [DOI] [Google Scholar]
- Nardelli M., Scalzo P. M., Ramírez M. S., Quiroga M. P., Cassini M. H., Centrón D. (2012). Class 1 integrons in environments with different degrees of urbanization. PLoS One 7:e39223. 10.1371/journal.pone.0039223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petinaki E., Guérin-Faublée V., Pichereau V., Villers C., Achard A., Malbruny B., et al. (2008). Lincomycin resistance gene lnu (D) in Streptococcus uberis. Antimicrob. Agents Chemother. 52 626–630. 10.1128/aac.01126-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrin S., Patuzzi I., Di Cesare A., Tiengo A., Sette G., Biancotto G., et al. (2019). Evaluation and quantification of antimicrobial residues and antimicrobial resistance genes in two Italian swine farms. Environ. Pollut. 255:113183. 10.1016/j.envpol.2019.113183 [DOI] [PubMed] [Google Scholar]
- Pruden A., Larsson D. J., Amézquita A., Collignon P., Brandt K. K., Graham D. W., et al. (2013). Management options for reducing the release of antibiotics and antibiotic resistance genes to the environment. Environ. Health Perspect. 121 878–885. 10.1289/ehp.1206446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings D. E., Tietze E. (2001). Comparative biology of IncQ and IncQ-like plasmids. Microbiol. Mol. Biol. Rev. 65 481–496. 10.1128/mmbr.65.4.481-496.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reygaert W. C. (2018). An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. 4:482. 10.3934/microbiol.2018.3.482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M. C. (2005). Update on acquired tetracycline resistance genes. FEMS Microbiol. Lett. 245 195–203. 10.1016/j.femsle.2005.02.034 [DOI] [PubMed] [Google Scholar]
- Rocha J., Cacace D., Kampouris I., Guilloteau H., Jäger T., Marano R. B. M., et al. (2018). Inter-laboratory calibration of quantitative analyses of antibiotic resistance genes. J. Environ. Chem. Eng. 8:102214 10.1016/j.jece.2018.02.022 [DOI] [Google Scholar]
- Rosen G. D. (1995). “Antibacterials in poultry and pig nutrition,” in Biotechnology in Animal Feeds and Animal Feeding, eds Wallace R. J., Chesson A. (Hoboken, NJ: Wiley; ), 172. [Google Scholar]
- Rowe-Magnus D. A., Mazel D. (2002). The role of integrons in antibiotic resistance gene capture. Int. J. Med. Microbiol. 292 115–125. 10.1078/1438-4221-00197 [DOI] [PubMed] [Google Scholar]
- Schwarz S., Kehrenberg C., Walsh T. R. (2001). Use of antimicrobial agents in veterinary medicine and food animal production. Int. J. Antimicrob. Agents 17 431–437. 10.1016/s0924-8579(01)00297-7 [DOI] [PubMed] [Google Scholar]
- Selvam A., Xu D., Zhao Z., Wong J. W. C. (2012a). Fate of tetracycline, sulfonamide and fluoroquinolone resistance genes and the changes in bacterial diversity during composting of swine manure. Bioresour. Technol. 126 383–390. 10.1016/j.biortech.2012.03.045 [DOI] [PubMed] [Google Scholar]
- Selvam A., Zhao Z., Wong J. W. C. (2012b). Composting of swine manure spiked with sulfadiazine, chlortetracycline and ciprofloxacin. Bioresour. Technol. 126 412–417. 10.1016/j.biortech.2011.12.073 [DOI] [PubMed] [Google Scholar]
- Sengeløv G., Agersø Y., Halling-Sørensen B., Baloda S. B., Andersen J. S., Jensen L. B. (2003). Bacterial antibiotic resistance levels in Danish farmland as a result of treatment with pig manure slurry. Environ. Int. 28 587–595. 10.1016/s0160-4120(02)00084-3 [DOI] [PubMed] [Google Scholar]
- Shang R., Wang J., Guo W., Liang J. (2013). Efficient antibacterial agents: a review of the synthesis, biological evaluation and mechanism of pleuromutilin derivatives. Curr. Top. Med. Chem. 13 3013–3025. 10.2174/15680266113136660217 [DOI] [PubMed] [Google Scholar]
- Sköld O. (2001). Resistance to trimethoprim and sulfonamides. Vet. Res. 32 261–273. 10.1051/vetres:2001123 [DOI] [PubMed] [Google Scholar]
- Smalla K., Heuer H., Götz A., Niemeyer D., Krögerrecklenfort E., Tietze E. (2000). Exogenous isolation of antibiotic resistance plasmids from piggery manure slurries reveals a high prevalence and diversity of IncQ-like plasmids. Appl. Environ. Microbiol. 66 4854–4862. 10.1128/aem.66.11.4854-4862.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Rensing C., Holm P. E., Virta M., Brandt K. K. (2017). Comparison of metals and tetracycline as selective agents for development of tetracycline resistant bacterial communities in agricultural soil. Environ. Sci. Technol. 51 3040–3047. 10.1021/acs.est.6b05342 [DOI] [PubMed] [Google Scholar]
- Su J.-Q., Wei B., Ou-Yang W.-Y., Huang F.-Y., Zhao Y., Xu H.-J., et al. (2015). Antibiotic resistome and its association with bacterial communities during sewage sludge composting. Environ. Sci. Technol. 49 7356–7363. 10.1021/acs.est.5b01012 [DOI] [PubMed] [Google Scholar]
- Suzuki H., Yano H., Brown C. J., Top E. M. (2010). Predicting plasmid promiscuity based on genomic signature. J. Bacteriol. 192 6045–6055. 10.1128/jb.00277-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao C.-W., Hsu B.-M., Ji W.-T., Hsu T.-K., Kao P.-M., Hsu C.-P., et al. (2014). Evaluation of five antibiotic resistance genes in wastewater treatment systems of swine farms by real-time PCR. Sci. Total Environ. 496 116–121. 10.1016/j.scitotenv.2014.07.024 [DOI] [PubMed] [Google Scholar]
- Tasho R. P., Cho J. Y. (2016). Veterinary antibiotics in animal waste, its distribution in soil and uptake by plants: a review. Sci. Total Environ. 563 366–376. 10.1016/j.scitotenv.2016.04.140 [DOI] [PubMed] [Google Scholar]
- Teuber M. (2001). Veterinary use and antibiotic resistance. Curr. Opin. Microbiol. 4 493–499. 10.1016/s1369-5274(00)00241-1 [DOI] [PubMed] [Google Scholar]
- Thomas C. M., Nielsen K. M. (2005). Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3:711. 10.1038/nrmicro1234 [DOI] [PubMed] [Google Scholar]
- Troiano E., Beneduce L., Gross A., Ronen Z. (2018). Antibiotic-resistant bacteria in greywater and greywater-irrigated soils. Front. Microbiol. 9:2666. 10.3389/fmicb.2018.02666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turker G., Akyol Ç, Ince O., Aydin S., Ince B. (2018). Operating conditions influence microbial community structures, elimination of the antibiotic resistance genes and metabolites during anaerobic digestion of cow manure in the presence of oxytetracycline. Ecotoxicol. Environ. Saf. 147 349–356. 10.1016/j.ecoenv.2017.08.044 [DOI] [PubMed] [Google Scholar]
- Ungemach F. R. (2000). Figures on quantities of antibacterials used for different purposes in the EU countries and interpretation. Acta Vet. Scand. 93 89–97. [PubMed] [Google Scholar]
- Uslu M. Ö, Balcıoğlu I. A. (2009). Comparison of the ozonation and Fenton process performances for the treatment of antibiotic containing manure. Sci. Total Environ. 407 3450–3458. 10.1016/j.scitotenv.2009.01.045 [DOI] [PubMed] [Google Scholar]
- Van den Meersche T., Rasschaert G., Haesebrouck F., Van Coillie E., Herman L., Van Weyenberg S., et al. (2019). Presence and fate of antibiotic residues, antibiotic resistance genes and zoonotic bacteria during biological swine manure treatment. Ecotoxicol. Environ. Saf. 175 29–38. 10.1016/j.ecoenv.2019.01.127 [DOI] [PubMed] [Google Scholar]
- Van Epps A., Blaney L. (2016). Antibiotic residues in animal waste: occurrence and degradation in conventional agricultural waste management practices. Curr. Pollut. Rep. 2 135–155. 10.1007/s40726-016-0037-1 [DOI] [Google Scholar]
- Van Hoek A. H. A. M., Mevius D., Guerra B., Mullany P., Roberts A. P., Aarts H. J. M. (2011). Acquired antibiotic resistance genes: an overview. Front. Microbiol. 2:203. 10.3389/fmicb.2011.00203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verraes C., Van Boxstael S., Van Meervenne E., Van Coillie E., Butaye P., Catry B., et al. (2013). Antimicrobial resistance in the food chain: a review. Int. J. Environ. Res. Public Health 10 2643–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vikesland P. J., Pruden A., Alvarez P. J. J., Aga D., Bürgmann H., Li X., et al. (2017). Toward a comprehensive strategy to mitigate dissemination of environmental sources of antibiotic resistance. Environ. Sci. Technol. 51 13061–13069. 10.1021/acs.est.7b03623 [DOI] [PubMed] [Google Scholar]
- Villa L., García-Fernández A., Fortini D., Carattoli A. (2010). Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J. Antimicrob. Chemother. 65 2518–2529. 10.1093/jac/dkq347 [DOI] [PubMed] [Google Scholar]
- Wang J., Ben W., Zhang Y., Yang M., Qiang Z. (2015). Effects of thermophilic composting on oxytetracycline, sulfamethazine, and their corresponding resistance genes in swine manure. Environ. Sci. Process. Impacts 17 1654–1660. 10.1039/c5em00132c [DOI] [PubMed] [Google Scholar]
- Witte W. (1998). Medical consequences of antibiotic use in agriculture. Science 279 996–997. 10.1126/science.279.5353.996 [DOI] [PubMed] [Google Scholar]
- Wolters B., Fornefeld E., Jechalke S., Su J.-Q., Zhu Y.-G., Sørensen S. J., et al. (2018). Soil amendment with sewage sludge affects soil prokaryotic community composition, mobilome and resistome. FEMS Microbiol. Ecol. 95:fiy193. [DOI] [PubMed] [Google Scholar]
- Woodford N., Ellington M. J. (2007). The emergence of antibiotic resistance by mutation. Clin. Microbiol. Infect. 13 5–18. 10.1111/j.1469-0691.2006.01492.x [DOI] [PubMed] [Google Scholar]
- World Health Organization (2014). Antimicrobial Resistance: Global Report on Surveillance. Geneva: WHO. [Google Scholar]
- Wu B., Qi Q., Zhang X., Cai Y., Yu G., Lv J., et al. (2019). Dissemination of Escherichia coli carrying plasmid-mediated quinolone resistance (PMQR) genes from swine farms to surroundings. Sci. Total Environ. 665 33–40. 10.1016/j.scitotenv.2019.01.272 [DOI] [PubMed] [Google Scholar]
- Xie W.-Y., Yang X.-P., Li Q., Wu L.-H., Shen Q.-R., Zhao F.-J. (2016). Changes in antibiotic concentrations and antibiotic resistome during commercial composting of animal manures. Environ. Pollut. 219 182–190. 10.1016/j.envpol.2016.10.044 [DOI] [PubMed] [Google Scholar]
- Yadav S., Kapley A. (2019). Exploration of activated sludge resistome using metagenomics. Sci. Total Environ. 692 1155–1164. 10.1016/j.scitotenv.2019.07.267 [DOI] [PubMed] [Google Scholar]
- Zang J., Shih J. C. H., Cheng J. J., Liu Z., Liu Y., Lu W. (2019). Thermophilic solid state anaerobic digestion of switchgrass for liquid digestate reuse and organic fertilizer production. Renew. Agric. Food Syst. 1–10. 10.1017/s1742170519000139 [DOI] [Google Scholar]
- Zhang M., He L.-Y., Liu Y.-S., Zhao J.-L., Liu W.-R., Zhang J.-N., et al. (2019). Fate of veterinary antibiotics during animal manure composting. Sci. Total Environ. 650 1363–1370. 10.1016/j.scitotenv.2018.09.147 [DOI] [PubMed] [Google Scholar]
- Zhao X., Wang J., Zhu L., Wang J. (2019). Field-based evidence for enrichment of antibiotic resistance genes and mobile genetic elements in manure-amended vegetable soils. Sci. Total Environ. 654 906–913. 10.1016/j.scitotenv.2018.10.446 [DOI] [PubMed] [Google Scholar]