Abstract
Schizophrenia is a chronic, severe and disabling psychiatric disorder, whose treatment is based on psychosocial interventions and the use of antipsychotic drugs. While the effects of these drugs are well elucidated in neuronal cells, they are still not so clear in oligodendrocytes, which play a vital role in schizophrenia. Thus, we aimed to characterize biochemical profiles by proteomic analyses of human oligodendrocytes (MO3.13) which were matured using a protocol we developed and treated with either haloperidol (a typical antipsychotic), clozapine (an atypical antipsychotic) or a clozapine + d-serine co-treatment, which has emerged lately as an alternative type of treatment. This was accomplished by employing shotgun proteomics, using nanoESI-LC–MS/MS label-free quantitation. Proteomic analysis revealed biochemical pathways commonly affected by all tested antipsychotics were mainly associated to ubiquitination, proteasome degradation, lipid metabolism and DNA damage repair. Clozapine and haloperidol treatments also affected proteins involved with the actin cytoskeleton and with EIF2 signaling. In turn, metabolic processes, especially the metabolism of nitrogenous compounds, were a predominant target of modulation of clozapine + d-serine treatment. In this context, we seek to contribute to the understanding of the biochemical and molecular mechanisms involved in the action of antipsychotics on oligodendrocytes, along with their possible implications in schizophrenia.
Subject terms: Biochemistry, Molecular biology, Neuroscience, Systems biology
Introduction
Schizophrenia is a severe, chronic, and disabling psychiatric disorder characterized by behavioral disturbances, abnormal mental functions and a heterogeneous combination of symptoms1,2, being one of the most important public health problems worldwide1. The symptoms are categorized as positive, negative and cognitive1,2, that usually manifest around the age of 16–30 years3. Treatment is based on a combination of antipsychotic drugs and psychosocial interventions1,2. Antipsychotics are divided into typical (first-generation) and atypical (second-generation) drugs1. A common pharmacological property between them is that they both block the dopamine D2 receptor1.
First-generation antipsychotics, such as chlorpromazine and haloperidol, are effective in reducing positive symptoms; however, they are minimally efficient for negative and cognitive symptoms4,5. Second-generation antipsychotics, such as clozapine, olanzapine, quetiapine, and risperidone, in turn, are more effective in reducing negative symptoms and improving global cognition than typical drugs. Moreover, they are associated with a lower occurrence of extrapyramidal effects and lower rates of treatment discontinuation and long-term relapse1,4,5. However, they also have side effects, such as weight gain and sedation4–6, and are associated with a higher cardiometabolic risk when compared to typical antipsychotics1. For this study, haloperidol was selected as typical antipsychotic, since it has been widely used in the clinic to treat the positive symptoms7, 8. As other typical drugs, haloperidol treatment is associated with extrapyramidal side effects7,9. Clozapine, the selected atypical drug, has a limited clinic use due the agranulocytosis. However, its efficacy is superior to many antipsychotic drugs, both typical and atypical10. Clozapine can also reduce suicide risk and is indicated in refractory schizophrenia treatment11.
The administration of D-serine with antipsychotics has been shown to be more effective in relieving some symptoms of schizophrenia, especially negative symptoms, when compared to the administration of antipsychotics alone11. D-serine is an endogenous N-methyl-D-aspartate (NMDA) receptor co-agonist that plays an important modulatory role in binding to the receptor’s glycine sites (NR1/NR2 subunits)12. It is for this reason that this amino acid has been tested as a potential therapeutic agent for schizophrenia in combination with atypical antipsychotics13.
A widely accepted hypothesis for the origin of schizophrenia is that variations in several risk genes interact with environmental stimuli, affecting brain development and function, leading to abnormal information processing by the brain1,2,14. Studies have associated the development and establishment of schizophrenia with dysfunctions in neurotransmitter systems and oligodendrocytes15,16. Oligodendrocytes are cells that wrap around axons in specialized layers of cell membrane to form the myelin sheath. This sheath is important for electrical insulation, increasing axonal conduction velocity, and consequently, neural processing speed17.
The oligodendrocyte lineage is composed of a series of developing cells that progressively mature into myelinating postmitotic cells from progenitor cells (OPCs), in a process marked by morphological changes and sequential expression of stage-specific markers, such as 2′,3′-cyclic-nucleotide-3-phosphodiesterase (CNPase), myelin basic protein (MBP) and proteolipid protein (PLP)18. This process is characterized by distinct and serial phenotypic stages, such as OPCs, preoligodendrocytes, immature (or pre-myelinating) OLs and mature (or myelinating) OLs 18. Extrinsic molecules, such as growth factors, cytokines, hormones and neurotransmitters are fundamental in the regulation of OL maturation19.
Neural detachment, as well as other changes of the white matter in schizophrenia, appear to be related to abnormal oligodendrocytes. These alterations include changes in number, spatial distribution and variations in the morphology of these cells20. Furthermore, gene expression and proteome analyses suggest a dysregulation of oligodendrocyte and myelin related genes and proteins in patients with schizophrenia15. Neuropathological and neuroimaging studies have also found abnormalities and degeneration related to OLs, especially in the frontal and temporal lobes and in the corpus callosum of schizophrenia patients21. Moreover, studies have suggested that oligodendrocytes may be a target of antipsychotics16,21,22. Thus, studying the effects of these drugs on oligodendrocytes may contribute to the improvement of current treatments and to the development of new therapeutic approaches.
The elucidation of the pathophysiology and the development of more effective treatments are the main challenges in schizophrenia1. Therefore, proteomic techniques such as shotgun mass spectrometry have been used in research aiming to identify biomarkers and pathways related to schizophrenia23. One substantial goal in functional proteomics is to globally profile changes in protein expression of biological systems in response to different conditions, such as a disease or after drug treatment. Identifying and quantifying proteins in such highly complex mixtures requires accurate and sensitive assays24. One possibility to characterize and compare complex mixtures in a concentration-dependent manner is the use of chromatographic separation in combination with electrospray ionization mass spectrometry (ESI–MS)25. This approach, combined with label-free quantitation methods, is an extremely useful tool in large-scale proteomic analyses of complex biological samples, such as cell lines, bodily fluids and tissues26,27.
In this study, we analyzed the proteome of a matured human oligodendrocyte cell line (MO3.13 cells) treated with haloperidol (a first-generation antipsychotic), clozapine (a second-generation antipsychotic) and clozapine + d-serine, by a nanoESI-LC–MS/MS label-free analysis. MO3.13 cells are an immortalized hybrid cell line that expresses phenotypic characteristics of primary oligodendrocytes28. Techniques such as differentiation and maturation of cell lines and progenitor cells are crucial for the improvement and development of in vitro models that better represent the in vivo cells, and are also practical to the scientific community29. Therefore, we used a maturation model of MO3.13 cells, previously described in Seabra et al.,29, in order to study the effects of antipsychotics on the proteome of oligodendrocytes. The matured cells are similar to pre-myelinating OLs, with increased PLP expression in comparison to the initial MO3.13 cells. From this, we identified proteins and biochemical pathways that were affected by the presence of these drugs, investigating their influence on oligodendrocyte function, which may prove to be important for the development of more effective treatments for schizophrenia.
Results and discussion
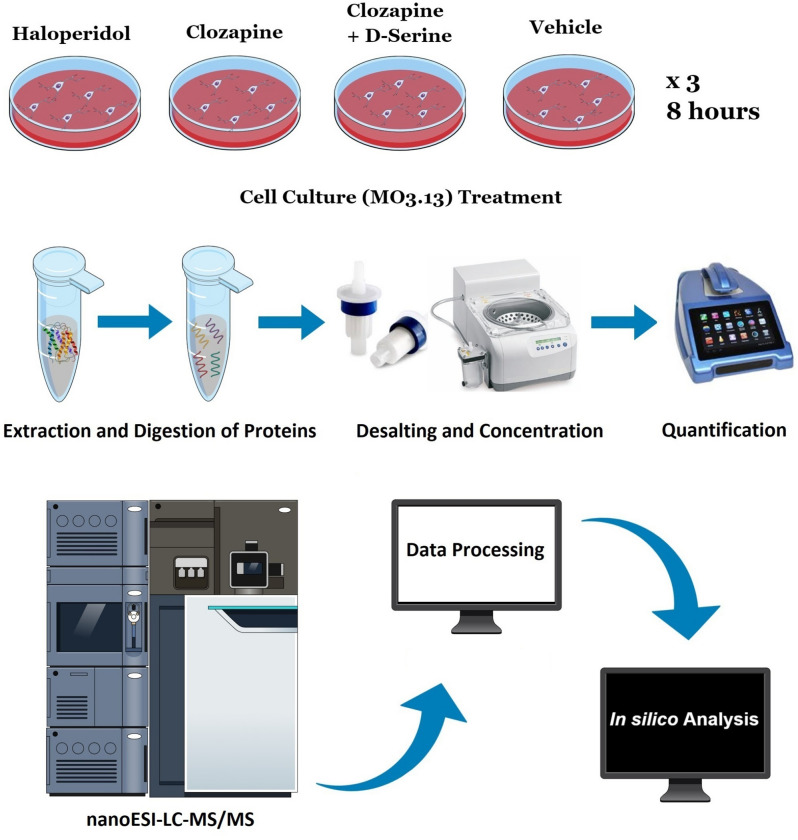
Using a three-fraction HDMSE, we identified a total of 1677 proteins. After applying stringent searching criteria (as described in 4.4. section) followed by a label-free method, we considered 1,427 proteins as the proteome of matured MO3.13 cells, with which we performed all systems biology analyses that follows (Fig. 1).
Figure 1.
Experimental steps from cell culture to in silico analysis. This figure was created using the Mind the Graph platform (https://www.mindthegraph.com).
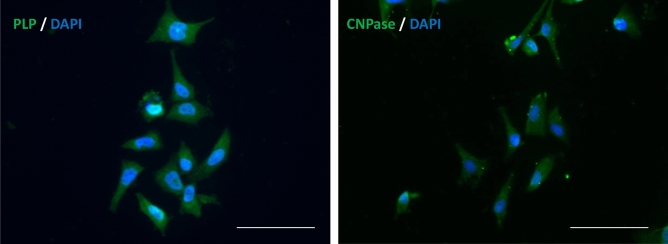
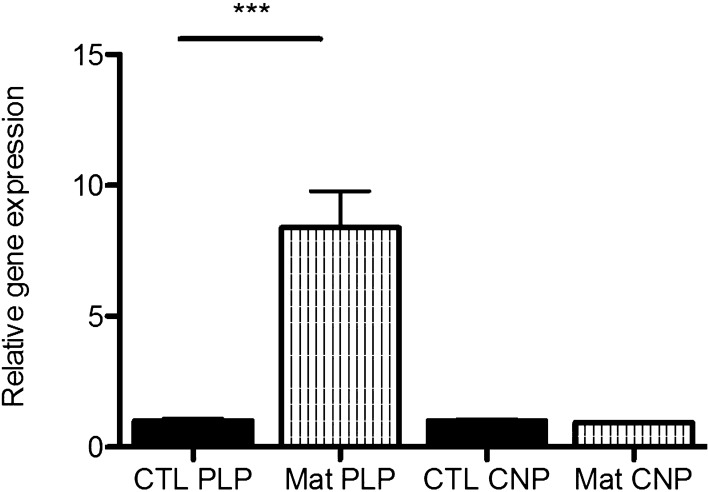
Figure 2 shows the MO3.13 cells labeled with anti-PLP (left) and anti-CNPase (right) antibodies after 15 days in maturation media. We also verified an eightfold increase in PLP expression in the cells by RT-qPCR (Fig. 3) after maturation protocol, while CNPase levels showed no significant changes. PLP is a marker for mature OLs, while CNPase is expressed both by immature and mature OLs18. CNP is highly expressed in maturing OLs to avoid myelin compaction in early stages30. In this context, there is a balance on CNP and MBP levels, as the latter is more expressed in myelinating OLs31. Thus, in advanced stages of maturation, CNP returns to basal levels allowing myelin ensheathment, in which MBP participates. Myelin sheath formation and compactation do not occur in our cell model, and this might be related to unchanged CNPase levels. Considering the conditions our cells were cultured in, we believe our cells have matured, since they might be similar to pre-myelinating OLs as proposed by Dawson et al.32.
Figure 2.
MO3.13 cells labeled with antibodies for PLP (left) and CNPase (right) (in green) and DAPI (in blue), after 15 days in maturation media. Scale bar: 200 µm.
Figure 3.
Relative gene expression of PLP and CNPase in MO3.13 cells after maturation protocol (Mat), compared to cells cultured in non-maturing media (CTL). These data were obtained by RT-qPCR and normalized to the expression of ACTB gene. Relative quantification value of each target gene was analyzed using a comparative CT method123. Statistical significance was determined by Student t-test, ***p < 0.001 compared to CTL group.
The number of differentially expressed proteins and their regulation are displayed in Table 1. Data were analyzed using in silico systems biology tools in order to identify biochemical pathways and biological processes in which differentially expressed proteins are involved in, as well as their interaction networks.
Table 1.
Number of differentially expressed proteins obtained from Hi3-based shotgun proteomic analyses per treatment (compared to vehicle).
| Treatment | Modulated proteinsa | Upregulated | Downregulated |
|---|---|---|---|
| Clozapine | 173 | 85 | 88 |
| Clozapine + d-serine | 84 | 45 | 39 |
| Haloperidol | 467 | 318 | 149 |
aInclude upregulated and downregulated proteins with ANOVA p-value < 0.1
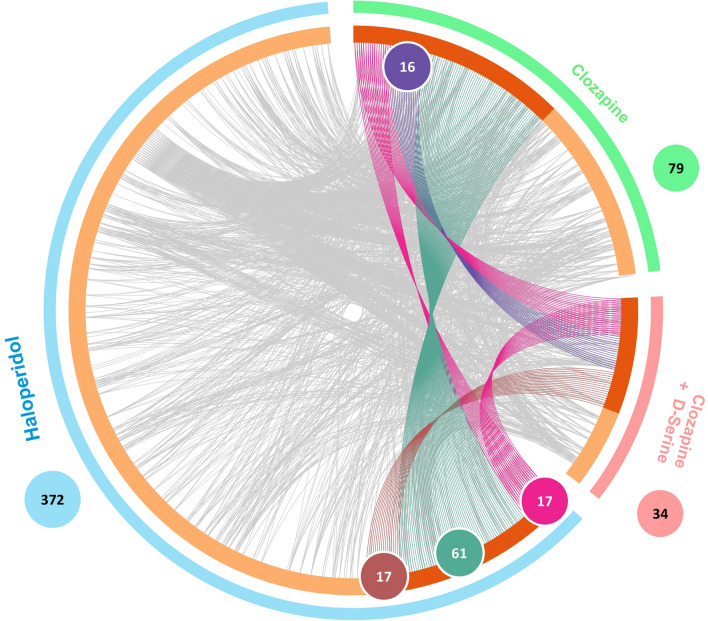
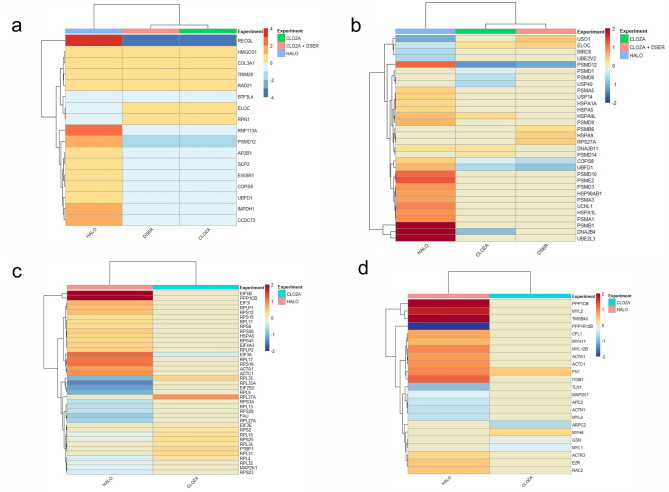
We found an overlap (Fig. 4) and treatment-specific modulation of the proteome by treatments. The 17 proteins modulated by all three treatments are displayed in Fig. 5a. Besides altering proteins involved in ubiquitination, proteasome degradation and lipid metabolism, all three treatments modulated DNA damage repair-related proteins, such as RECQL (ATP-dependent DNA helicase Q1), TRIM28 (Tripartite motif containing 28), RAD21 (Double-strand-break repair protein rad21 homolog) and RNF113A (Ring finger protein 113A). Therefore, these processes seem to be relevant in the oligodendrocyte response to antipsychotics.
Figure 4.
Chord diagram of the number of modulated proteins (ANOVA p-value ≤ 0.1) in matured MO3.13 cells treated with haloperidol, clozapine or clozapine + D-serine. This figure was made using Metascape122.
Figure 5.
Differentially expressed proteins in common for MO3.13 cells treated with haloperidol, clozapine and clozapine + d-serine (a). The UPS-related proteins modulated by those treatments are show in (b). Haloperidol and Clozapine also modulated proteins related to eIF2 signaling (c) and to the actin cytoskeleton (d). Proteins in shades of red are upregulated, while the ones in shades of blue are downregulated. This figure was made using Rstudio (version 1.2.5033)123, R (version 3.6.2)124, Pheatmap package (version 1.0.12)125.
Common Targets—Proteins related to ubiquitin–proteasome system (UPS)
Among the proteins modulated by all treatments are PSMD12 (26S proteasome non-ATPase regulatory subunit 12), UBFD1 (ubiquitin family domain containing 1) and COPS8 (COP9 signalosome complex subunit 8), all related to ubiquitination and degradation by proteasome. Other proteins related to the ubiquitin–proteasome system (UPS) were also modulated by these antipsychotics (Fig. 5b). The UPS plays a role in diverse cellular processes, such as protein labeling for degradation, vesicle and protein transport, and membrane receptor recycling33. Thus, if this mechanism has a functional or structural impairment, proteolytic activity is impaired, which may lead to an accumulation of damaged/dysfunctional proteins. This has already been associated with the pathogenesis and phenotypic characteristics of neurodegenerative diseases, including Alzheimer's34 and Parkinson's35.
Proteasomes also contribute to aspects of neural function that are abnormal in schizophrenia, including neurodevelopmental maintenance, dendritic morphology, energy homeostasis, neurotransmitter synthesis, receptor recycling and cytokine production and activation36–40. Several changes in the UPS have been related to schizophrenia through studies involving postmortem brains of patients. These studies have found changes in the levels of ubiquitinated proteins41,42, the expression of ubiquitinating, deubiquitinating and conjugating enzymes42–47 and a reduction in free ubiquitin levels47.
As discussed, changes in protein transport and turnover, and ubiquitination-related signaling pathways can lead to brain tissue abnormalities that may contribute to neurotoxicity and functional impairment41,48. Therefore, elements of the UPS may act as potential pharmacological targets41. Our data are in agreement with this hypothesis and suggest that the modulation of proteins involved in the UPS is a common effect of typical and atypical antipsychotics in oligodendrocytes.
Common targets—proteins related to lipid metabolism
Proteins related to lipid metabolism were also modulated by the three treatments, among them: SCP-2 (sterol carrier protein 2) (ANOVA p-value: 0.0695 for haloperidol; 0.0663 for clozapine; 0.0507 for clozapine + d-serine) and HMG-CoA synthase (hydroxymethylglutaryl-CoA synthase) (ANOVA p-value: 0.0629 for haloperidol; 0.0017 for clozapine; 0.0708 for clozapine + d-serine). SCP-2 binds and transports lipid ligands such as long chain fatty acids and their CoA thioesters49, endocannabinoids50 and phospholipids51. Thus, SCP-2 is an important component in maintaining lipid homeostasis. Meanwhile, HMG-CoA synthase catalyzes the condensation of acetyl-CoA with acetoacetyl-CoA to form 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA), an intermediate in cholesterol synthesis and ketogenesis52
Dysfunctions in lipid homeostasis can affect neural function, synaptic transmission, cell signaling, myelination and oligodendrocyte biology53, contributing to white matter dysfunction and disturbances in neural connectivity54. Therefore, changes in lipids have been associated with the pathophysiology of schizophrenia. Specifically, alterations related to fatty acids and ketone bodies55,56, phospholipids57,58 sphingolipids59, total lipids, triglycerides and cholesterol esters were observed in schizophrenia patients60–62.
Regarding antipsychotics, studies have suggested that these drugs may compensate for or reverse lipid pathway dysregulation related to the early stage of the disorder55,59,63. Furthermore, fatty acid metabolism and cholesterol biosynthesis have been identified by gene expression studies as key elements in antipsychotic treatment response64,65. One hypothesis proposes that the modulation of cholesterol and the proportion of polyunsaturated and saturated fatty acids by antipsychotics may affect neural membrane fluidity, resulting in changes in neural connectivity65, which may be related to therapeutic effects.
In our data, HMG-CoA synthase, responsible for HMG-CoA synthesis, is upregulated. The up-regulation of the hydroxymethylglutaryl-CoA synthase 1 gene (HMGCS1) has been previously described by studies evaluating the effect of clozapine, olanzapine, haloperidol, chlorpromazine and risperidone on human glioma strains64 and epithelial cell retinal pigment (ARPE-19)65. The up-regulation of this enzyme may contribute to the synthesis of cholesterol, an important component of the myelin sheath and a key element of oligodendrocyte metabolism.
Haloperidol and clozapine—canonical pathways
Signaling by Rho GTPase proteins is evident in haloperidol treatment, while processes involved in actin cytoskeleton and eIF2 (eukaryotic initiation factor 2) signaling are modulated by both drugs. The proteins that were modulated by these treatments and are involved in eIF2 signaling and actin cytoskeleton are shown in Fig. 5c,d, respectively.
eIF2 is a transcription initiation factor critical for protein synthesis66. Furthermore, eIF2B plays a key role in maintaining the viability of oligodendrocytes, and its impaired activity can lead to a dysfunctional stress response66 in thes cells. eIF2B-related mutations are associated with the evanescent white matter disease, which causes severe loss of oligodendrocytes and astrocytes in early life67. We found that haloperidol treatment contributed to the downregulation of this pathway, while clozapine treatment upregulated the pathway. Thus, our findings suggest that typical and atypical antipsychotics may have distinct effects on protein synthesis, as well as on the maintenance of oligodendrocyte viability mediated by eIF2 signaling, in agreement with another study of our group68.
Actin cytoskeleton signaling was also modulated by haloperidol and clozapine treatments, being upregulated in both cases. Actin is critical in stabilizing and maintaining cell morphology and its dynamics underlies several central cellular processes, such as cell motility and intracellular protein traffic69. In the nervous system, this process is fundamental in maintaining the morphology and density of dendritic spines70. Furthermore, actin cytoskeleton regulation has been associated with schizophrenia71–74. Alterations in the expression of proteins involved in the regulation of actin polymerization and dynamics were observed in the dorsolateral prefrontal cortex (DLPFC)72 and in the anterior cingulate cortex (ACC) of schizophrenia patients71. Thus, it is possible that haloperidol and clozapine treatments help to modulate actin cytoskeleton signaling in oligodendrocytes, which might be related to their therapeutic effects.
Haloperidol
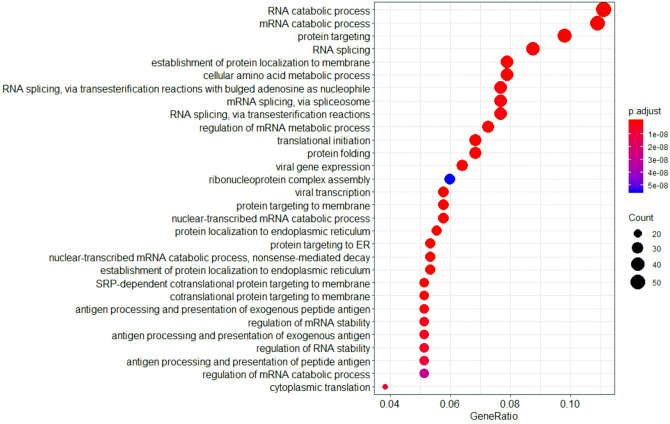
Haloperidol treatment affected, among others, pathways involved in amino acid metabolism, protein targeting and signal transduction (by Rho GTPases, for example). This treatment also modulated several proteins related to RNA splicing (via both transesterification reactions and spliceosome), and RNA metabolism (especially catabolism) and stability, as show in Fig. 6. A previous study of our group has shown that chlorpromazine, haloperidol, quetiapine and risperidone treatments affected proteins of spliceosome machinery in MO3.13 cells, especially belonging to nuclear heterogeneous ribonucleoprotein (hnRNP) family68.
Figure 6.
Dotplot of biological processes modulated by haloperidol treatment (TOP30), with protein count and adjusted p-value. This figure was made using R ClusterProfiler (version 3.14).
Several studies have identified global changes in alternative splicing in schizophrenia by comparing the transcriptomes of tissues from patients to healthy controls75. BICD2 and DLG3 exons, for example, were differentially expressed in brain and blood samples of schizophrenia patients76. Other genes that present abnormal splicing patterns in postmortem brain include DISC177, ERBB478, NRG179 e NRG380. Detailed data about alterations in alternative splicing in schizophrenia and other psychiatric disorders can be found in Morikawa and Manabe81 and Reble et al.75 reviews. Therefore, RNA splicing can be an important target for further studies in schizophrenia, considering the alterations previously described in literature and the modulation of related proteins by haloperidol and other antipsychotic treatments.
Furthermore, haloperidol treatment modulated proteins involved in the synthesis of selenocysteine, an important amino acid found in some proteins, conferring them specific properites; these proteins are often involved in cellular protection against oxidative stress, such as glutathione peroxidase82–84. On the other hand, there is evidence to suggest an increase in oxidative stress markers associated with antipsychotic use, especially typical drugs85–88. Deficits in defense mechanisms against free radicals were also related to the development of tardive dyskinesia, a notable extrapyramidal side effect of typical antipsychotics88. Tying these data together, we hypothesize that the impairment of selenocysteine synthesis may affect the functioning of enzymes involved in cellular protection against oxidative stress, contributing to an oxidative stress scenario, which might in turn related to the development of tardive dyskinesia.
In addition to the aforementioned proteins, several others modulated by haloperidol are involved in Rho-GTPase signaling, important to regulate the actin cytoskeleton. Postmortem studies reported a decreased expression of Rho-GTPase proteins in schizophrenia patients74,89. Rho GTPase Cdc42 mRNA levels, involved in actin polymerization regulation90,91, are decreased in the DLPFC74. Furthermore, a dysregulation of the RhoA pathway (crucial in controlling brain size and connectivity) is considered to be a risk factor for autism and for the development of schizophrenia92. In our study haloperidol upregulated RhoA and Cdc42 and downregulated RhoGDI, strengthening the previously found data. Therefore, this drug may have a role in regulating Rho-GTPase signaling in oligodendrocytes.
Clozapine
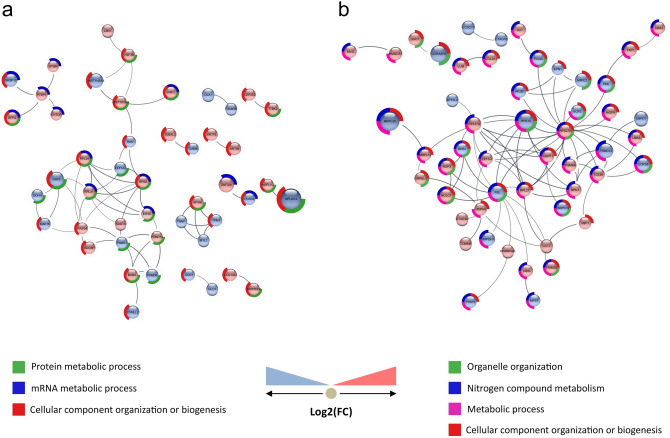
Clozapine treatment modulated proteins related to metabolism, gene expression regulation, RNA processing, axonal guidance, signal transduction and cellular response to stress, among others. The interactome of proteins modulated exclusively by clozapine treatment highlighted some processes, such as the organization or biogenesis of cellular components and mRNA metabolism (Fig. 7a). Proteins involved in protection against oxidative stress, such as glutaredoxin, superoxide dismutase (ANOVA p-value: 0.07893) and thioredoxin (TRX) (ANOVA p-value: 0.0891) were also affected.
Figure 7.
Interactome of proteins that were differentially expressed exclusively by clozapine treatment (a) and by clozapine + d-serine treatment (b). Known interactions between proteins are represented by lines connecting the nodes. The darker the line, the greater the degree of confidence about the interaction, calculated from evidence found in literature and databases. This figure was made using Cytoscape (version 3.7.2)126 and String app (version 1.5)127, with a minimum required interaction score of 0.700. (a) Blue bar indicates involvement in mRNA metabolism, red bar indicates involvement with in the organization or biogenesis of cellular components, and green bar indicates involvement in cellular protein metabolism. (b) Red bar indicates association with the organization or biogenesis of cellular components (especially organelles, such as ribosomes). Pink bar indicates participation in metabolic processes, both catabolic and anabolic. Blue bar indicates participation in the metabolism of nitrogenous compounds. Green bar indicates a relation to organelle organization.
TRX is a ubiquitous protein with oxide reductase activity93, associated with cognitive deficits in schizophrenia94,95. Increased TRX levels in serum have been observed in first-episode, psychotic-naïve patients, compared both to antipsychotic-treated and control subjects96. Thus, TRX serum levels are higher during acute psychotic episodes, tending to decrease after the remission of symptoms97. According to our data, TRX was downregulated by clozapine in oligodendrocytes, corroborating the previous findings that decreased levels of this protein are associated with antipsychotic treatment. However, further investigations are needed to understand the outcome for oligodendrocyte biology.
Furthermore, some proteins related to mitochondrial function and dynamics were affected by clozapine, such as cytochrome c oxidase, subunit 5B (upregulated; ANOVA p-value: 0.0752), acyl-CoA dehydrogenase of mid-chain fatty acids (downregulated; ANOVA p-value: 0.0031), TOM34 (outer mitochondrial membrane translocase 34; downregulated; ANOVA p-value: 0.0104) and TIM13 (inner mitochondrial membrane translocase 13; downregulated; ANOVA p-value: 0.0951). This treatment also modulated proteins related to the organization or biogenesis of cellular components, such as organelles, in MO3.13 cells.
Mitochondrial dysfunctions in schizophrenia include metabolic, enzymatic, anatomical and genetic abnormalities98. Studies have detected a reduced number of mitochondria in oligodendrocytes of the ACC99,100 and a reduction in mitochondrial size in caudate oligodendrocytes101. These alterations may affect the energy supply of these cells, and consequently the myelination process98. Antipsychotics, in turn, affect mitochondrial number, size and function, depending on the brain region, dose, length of use and routine of administration98. Thus, we hypothesize that the modulation of proteins involved in mitochondrial function and dynamics, as well as organelle organization and biogenesis, may modulate mitochondrial biology, both structurally and regarding the number of organelles. This can affect processes such as myelination and energy supply of neuronal axons, important functions of oligodendrocytes for maintaining neural connectivity.
Clozapine + d-Serine
Clozapine + d-serine treatment modulated proteins related to metabolism, signal transduction, immune system and transport, among others. Some processes were shown in the interactome of affected proteins, such as the organization or biogenesis of cellular components (especially organelles), metabolic processes and the metabolism of nitrogenous compounds (Fig. 7b), a predominant target of modulation to this treatment.
Some studies have suggested a relationship between changes in nitrogen metabolism and symptomatic worsening and remission in schizophrenia102,103. A reduction in plasma nitrogenous amino acid levels has been observed in patients, especially during acute attacks, and an increase is seen with symptom remission103. Moreover, increased levels of N-acetylglutamine and reduced levels of N6-acetyl-l-lysine were observed in schizophrenia patients’ serum in comparison to control subjects104. Reduced concentrations of N-acetyl aspartate in the prefrontal cortex of chronic patients105 were also reported.
Other studies have found an increase in plasmatic nitric oxide (NO) levels in schizophrenia patients compared to control subjects106, 107. Furthermore, patients in the acute psychotic phase present extremely high levels of 3-nitrotyrosine (a marker of peroxynitrite production) on plasmatic proteins108. Therefore, peroxynitrite and other reactive nitrogen species, including NO and its metabolites, may be related to schizophrenia106–108. Thus, considering the expressive modulation of proteins involved in the metabolism of nitrogenous compounds, it is possible that co-treatment with clozapine + d-serine contributes to the normalization of the metabolism of these compounds. This hypothesis can be tested in future studies focusing on nitrogenous compounds levels and metabolism.
Decreased levels of d-serine have been reported in blood and cerebrospinal fluid (CSF) of schizophrenia patients12, 109–111, as well as disturbance of its metabolizing enzymes112. This amino acid administered in combination with usual antipsychotics was found to be more effective in relieving symptoms of schizophrenia, when compared to the administration of antipsychotics alone11, 13. d-Serine also seems to be related to neurophysiologic changes induced by cognitive training in schizophrenia, with increased levels being positively correlated with improvements in verbal learning and global cognition113. Further, good responders to clozapine presented an increase in d-serine plasma levels114. Therefore, the elucidation of proteins and pathways affected by d-Serine co-administration with antipsychotics is important to understand the mechanisms that might be related to symptom improvements associated to d-Serine use.
Conclusion
From this study, we identified proteins and biochemical pathways affected by antipsychotic treatment on oligodendrocytes through shotgun proteomics, using nanoESI-LC label-free shotgun mass spectrometry. Some affected proteins and processes were common to haloperidol, clozapine and clozapine + d-serine treatments. These were mainly related to ubiquitination, proteasome degradation, lipid metabolism and DNA damage repair. Clozapine and haloperidol also modulated proteins involved with the actin cytoskeleton and eIF2 signaling. Haloperidol treatment affected, among others, proteins involved in Rho GTPase signaling and in the synthesis of selenocysteine. Clozapine treatment modulated proteins related to metabolism, protection against oxidative stress (such as TRX) and organization or biogenesis of cellular components, among others. In turn, metabolic processes, especially the metabolism of nitrogenous compounds, were a predominant target of modulation of clozapine + d-serine treatment. With this data, we sought to contribute to the understanding of the biochemical and molecular mechanisms involved in the action of these drugs on oligodendrocytes and their possible implications in schizophrenia. Therefore, we aim to contribute to future studies focusing on the pathways and proteins highlighted by this work, and to studies dedicated to improving current treatments and developing new therapeutic approaches.
Methods
Cell culture and treatments
MO3.13 cells were initially cultured in Dulbecco’s modified eagle medium (DMEM) (Sigma-Aldrich), containing 0.5% penicillin/streptomycin (Gibco) and 10% fetal bovine serum (FBS) (Gibco), renewed every 2 days, as previously described in Brandão-Teles et al. (2017)115. For maturation, cells were cultured in 6-well plates (Corning, Inc.) containing DMEM (Sigma-Aldrich) supplemented with 0.5% N2 (Gibco), 0.5% B27 (Gibco), PDGFα (alpha-type platelet-derived growth factor) (10 ng/ml) (Thermo Fisher Scientific), 0.1% penicillin/streptomycin (Gibco) and triiodothyronine (T3—30 ng /ml) (Sigma-Aldrich) at 37 °C in 5% CO2. The medium was renewed every 3 days for a maturation period of 15 days. The procedures for thawing, culturing, counting, passaging, maturation and collection of cells are described in Seabra et al.29.
The cells were treated, in triplicate, with: (a) 50 μM clozapine (Cristália); (b) 50 μM haloperidol (Cristália); (c) vehicle solution (0.01 M HCl); or (d) 50 μM clozapine + D-serine (Sigma). After 8 h, the cells were collected in PBS (1 ×; Sigma-Aldrich) and scraped with a plastic scraper, according to Seabra et al.29. Afterwards, the samples were centrifuged at 200 × g for 5 min (FANEM 206BL Centrifuge), the supernatant was removed and the cellular pellets were stored at − 80 °C for later analyses.
Sample preparation
Cellular precipitates derived from the four conditions (clozapine, haloperidol, clozapine + d-serine or vehicle) in biological triplicate were homogenized in 100 μl of lysis buffer (containing 6 M urea [Sigma-Aldrich], 2 M thiourea [Synth], 10 mM dithiothreitol [DTT], 0.1 mM sodium pervanadate and 10 μL of 10 × protease inhibitor [Roche]). Then, they were vortexed and incubated for two hours at 37 °C. After incubation, samples were diluted ten times with 20 mM TEAB (triethylammonium bicarbonate buffer), pH 7.5 (Sigma-Aldrich) and sonicated on ice (Ultrasonic Homogenizer 4,710 Series, Cole-Parmer Instrument Co.) for 20 s (pulser on, duty cycle: 25%, micro-tip limit: 1.5). Next, samples were alkylated by incubation with 100 μl of 20 mM iodoacetamide (IAA) (Amersham Biosciences) for 20 min in the dark at room temperature (RT). Trypsin (sequencing grade; Sigma-Aldrich/Promega) was used for digestion, (1:50 enzyme: substrate), with overnight incubation (12–16 h) at 37 °C. After incubation, the reaction was stopped with formic acid (Sigma-Aldrich) (final concentration 5% v/v) and incubated for 5 min at RT. Subsequently, the samples were centrifuged for 45 min at 4 °C and 20,850×g (5430R Centrifuge—Eppendorf) and the supernatants were transferred to low-binding microtubes (Axygen).
Desalting and concentration of the peptides were done with HLB cartridges (Oasis-Waters) according to Brandão-Teles et al.115. Briefly, HLB (Oasis-Waters) cartridges were activated with 1 mL of 100% methanol (Merck), followed by 1 mL of 100% acetonitrile (ACN) (Merck), and equilibrated with 2 mL of 0.1% trifluoroacetic acid (TFA) (PerkinElmer). Then, samples were slowly added to the cartridges over two additions to load the full volume. The cartridge was washed twice with 2 mL of 0.1% TFA (PerkinElmer) and the peptides were eluted with 2 mL of 70% ACN/0.1% TFA. Then, the samples were concentrated (Eppendorf Concentrator Plus) and reconstituted in 20 mM ammonium formate, pH10. Next, the concentration of peptides was determined (DeNovix—DS11 Spectrophotometer) and samples were diluted to a final concentration of 1 μg/μL with 20 mM ammonium formate, pH10.
NanoESI-LC–MS/MS
The proteomic analyses were performed using a two-dimensional Acquity M-Class nanoUPLC system (Waters Corporation, Milford, MA), coupled to a Synapt G2-Si spectrometer (Waters Corporation, Milford, MA). The MS and MS/MS data were obtained using data-independent acquisition (DIA) and ion mobility separation. The peptide samples derived from the four conditions in biological triplicate (in 20 mM ammonium formate, pH10) were analyzed using an HDMSE (high-definition data-independent mass spectrometry) label-free method, with three fractions of 54 min in reverse-phase chromatography (steps of 13.7%, 18.4% and 50% of acetonitrile in the first dimension; and a linear gradient of 7–40% ACN in the second dimension). In the first-dimension, peptides were loaded onto an M-Class BEH C18 Column (130 Å, 5 μm, 300 μm × 50 mm, Waters Corporation, Milford, MA, United States), and a nanoACQUITY UPLC HSS T3 Column (100 Å, 1.8 μm, 75 μm × 150 mm, Waters Corporation, Milford, MA, United States) was used for the second dimension. Injections were performed using a nano-electrospray ionization source in positive ion mode (nanoESI (+), with a NanoLock-Spray (Waters, Manchester, United Kingdom) ionization source. A solution of [Glu1]-Fibrinopeptide B (Glu-Fib; Human) was used as the lock mass, and was sampled every 30 s. Biological triplicates were performed following the processing software specifications (Progenesis QI for Proteomics 3.0, Waters).
Data processing
MS and MS/MS spectra were processed with database search and a processing software. Progenesis QI for Proteomics 3.0 (Waters) was used to perform qualitative and quantitative, label-free analyses. In this process, the following parameters were considered: 5 as maximum peptide charge, maximum protein mass of 600 kDa, a maximum of 1 missed cleavage, at least 2 fragments per peptide, at least 5 fragments per protein, and at least one peptide per protein, with an FDR of < 1% and a mass error cutoff of 20 PPM. Hi-3 was used as the quantitation method, in which protein quantitation is based on the average integrated signal intensity of the top 3 most intense peptides116, 117. Cysteine carbamidomethylation and methionine oxidation were considered as fixed and variable modifications, respectively. The database used was reviewed human UNIPROT databank (version 2018/10). In the final list of proteins, identifications with at least one unique peptide were considered for in silico analysis. Quantitation differences that returned a p-value ≤ 0.1 from the ANOVA test were considered differentially expressed between the antipsychotics treated groups (haloperidol, clozapine and clozapine + d-serine) in comparison to the vehicle. The expression analysis was performed considering the three biological replicates available for each experimental condition, and each group as an independent variable.
In silico analyses
The differentially expressed proteins were classified according to the biological processes in which they were involved using reference databases for human proteins. In order to visualize the network of altered proteins and affected biochemical pathways, we used Ingenuity Pathway Analysis (IPA) 2.4 (QIAGEN Inc.)118, which took into consideration the accession of differentially expressed proteins, ANOVA test p value (p ≤ 0.1) and Log2 fold change (considering the ratio between treatment and control). Online software was also used: Reactome (version 69)119, STRING (version 11.0)120, VENNY (version 2.1)121, Metascape122, Rstudio (version 1.2.5033)123, R (version 3.6.2)124, Pheatmap package (version 1.0.12)125, Cytoscape (version 3.7.2)126 and String app (version 1.5)127. The experimental steps described thus far are shown in Fig. 1.
Immunocytochemistry
MO3.13 cells were grown in 24-well plates (Corning Inc.) containing glass coverslips. For fixation, each well was incubated with 250 μL 4% paraformaldehyde (PFA) (Sigma-Aldrich)/PBS 1 × solution (Sigma-Aldrich) for 25 min at RT. The cells were washed 3 times with PBS 1 × (Sigma-Aldrich) and the coverslips were incubated with 0.1 M glycine (Sigma-Aldrich) for 20 min at RT (in a wet chamber), followed by a wash with 1 × PBS (Sigma-Aldrich). For the coverslips to be labeled by CNPase (Abcam—Ab28486), the membranes were permeabilized using 0.2% Triton X-100 (Sigma-Aldrich) in PBS 1 × (Sigma-Aldrich) for 2 min, with subsequent washing with 1 × PBS (Sigma-Aldrich). Blocking was performed with a 10% FBS solution (Gibco) in PBS 1 × (Sigma-Aldrich), incubating for 5 min at RT.
The coverslips were incubated with primary antibodies (1:400 dilution in 10% FBS/PBS 1 ×) against PLP (Abcam-Ab28486) and CNPase (Abcam-Ab6319), overnight at 4 °C, before being washed with PBS 1 × (Sigma-Aldrich). Then, samples were incubated with DAPI (Thermo Fisher Scientific) and secondary antibodies: anti-mouse (Alexa Fluor 488 goat anti-mouse—Thermo Fisher Scientific) and anti-rabbit (Alexa Fluor 488 goat anti-rabbit—Thermo Fisher Scientific) (both 1:400 in 10% FBS/PBS 1 ×), for 1 h in the dark at 37 °C. They were then rinsed three times with PBS 1 ×. For slide assembly, a drop of mounting medium (Dako) was added. The slides were dried at RT and sheltered from light, then read on a Cytation 5 (BioTek). The analyses were performed using ImageJ software (National Institutes of Health, NIH)128.
RNA isolation and cDNA synthesis
MO3.13 cells were lysed with trizol and chloroform was used for phase separation. For RNA precipitation, isopropanol 100% was added, and the RNA pellet was washed three times in ice-cold 75% ethanol by centrifugation. Then, RNA was resuspended in nuclease-free water, and quantified using a DeNovix spectrophotometer. RNA quality was tested with 1% agarose gel electrophoresis, checking 18S/28S band integrity. Finally, reverse transcription was performed using a kit (GoScript Reverse Transcriptase Kit, Promega, Madison, WI, USA) according to manufactures’ instructions in a 20-µL reaction wit 1,6 µg of total RNA.
Real-time quantitative polymerase chain reaction (RT-qPCR)
Quantitative PCR was performed on cDNA from MO3.13 cells diluted 1:10 using the qPCR Green Master Low Rox (Cellco) mastermix containing forward primer, reverse primer and diethyl pyrocarbonate (DEPC)-treated water. The RT-qPCR reaction was performed in a CFX384 Touch Real-Time PCR Detection System (Biorad). Cycling conditions were set as follows: after initial activation at 50 °C for 2 min and 40 cycles of 95 °C for 2 min, 95 °C for 15 s and 60 °C for 1 min, then melt curve analysis was performed by heating samples from 65 °C to 99 °C (1 °C increment changes at 5 s intervals), in order to evaluate primer specificity. All sample measurements were performed in duplicate.
Intron-spanning primers for RT-qPCR were designed with PrimerBlast, and used at 200 nM for PLP and CNP genes. Beta-actin (ACTB) was used as housekeeping gene (primer concentration: 100 nM). For primer sequences, see Supplementary Table S1. The efficiency of the primers used ranged from 95–100%. Data were normalized to the expression of ACTB gene and relative quantification value of each target gene was analyzed using a comparative CT method129.
Supplementary information
Acknowledgements
The authors would like to thank FAPESP (Sao Paulo Research Foundation; grants 2017/25588-1, 2014/10068-4, 2019/00098-7, 2017/18242-1, 2018/03673-0, 2019/03271-1, 2019/22398-2), CNPq (The Brazilian National Council for Scientific and Technological Development; Grant 302453/2017-2) and CAPES (Coordination for the Improvement of Higher Education Personnel; Grants 1656470 and 1691474) for financial support. The authors wish to thank Bradley J. Smith, MSc for English review support.
Abbreviations
- ACC
Anterior cingulate cortex
- ACN
Acetonitrile
- ACTB
Beta-actin
- CNPase
2′,3′-Cyclic-nucleotide-3-phosphodiesterase
- COPS8
COP9 signalosome complex subunit 8
- DIA
Data-independent acquisition
- DLPFC
Dorsolateral prefrontal cortex
- DMEM
Dulbecco’s modified eagle medium
- DTT
Dithiothreitol
- eIF2
Eukaryotic initiation factor -2
- ESI
Electrospray ionization
- FBS
Fetal bovine serum
- FC
Fold change
- HMG-CoA
3-Hydroxy-3-methylglutaryl-CoA
- hnRNP
Nuclear heterogeneous ribonucleoprotein
- IAA
Iodoacetamide
- IPA
Ingenuity pathway analysis
- MBP
Myelin basic protein
- MS
Mass spectrometry
- NMDAR
N-methyl-d-aspartate receptor
- NO
Nitric oxide
- OFC
Orbitofrontal cortex
- OPCs
Oligodendrocyte progenitor cells
- PBS
Phosphate buffered saline
- PDGFα
Alpha-type platelet-derived growth factor
- PFA
Paraformaldehyde
- PLP
Proteolipid protein
- PSMD12
26S proteasome non-ATPase regulatory subunit 12
- RAD21
Double-strand-break repair protein rad21 homolog
- RECQL
ATP-dependent DNA helicase Q1
- RNF113A
Ring finger protein 113A
- RT
Room temperature
- SCP-2
Sterol carrier protein 2
- T3
Triiodothyronine
- TEAB
Tetraethylammonium bromide
- TFA
Trifluoroacetic acid
- TIM13
Inner mitochondrial membrane translocase 13
- TOM34
Outer mitochondrial membrane translocase 34
- TRIM28
Tripartite motif-containing 28
- TRX
Thioredoxin
- UBFD1
Ubiquitin family domain containing 1
Author contributions
G.S. conceived the study, worked on data acquisition and interpretation, and wrote the manuscript. V.A. helped with data acquisition and manuscript revision. G.R.-d-O. assisted in data interpretation and designed Figs. 4, 5, 6 and 7. F.C. and A.S.L.M.A. performed RT-qPCR analysis. D.M.-d-S. conceived the study, supervised the process, and finalized the manuscript. All authors reviewed the manuscript
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-69543-5.
References
- 1.Owen J, Sawa A, Mortensen PB. Schizophrenia. Lancet. 2016;388(10039):86–97. doi: 10.1016/S0140-6736(15)01121-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kahn RS, Sommer IE, Murray RM, Meyer-Lindenberg A, Weinberger DR, et al. Schizophrenia. Nat. Rev. Dis Primers. 2015;1:15067. doi: 10.1038/nrdp.2015.67. [DOI] [PubMed] [Google Scholar]
- 3.Mueser KT, McGurk SR. Schizophrenia. Lancet. 2004;363(9426):2063–2072. doi: 10.1016/S0140-6736(04)16458-1. [DOI] [PubMed] [Google Scholar]
- 4.Zhang J-P, Gallego JA, Robinson DG, Malhotra AK, Kane JM, Correll CU. Efficacy and safety of individual second-generation vs first-generation antipsychotics in first-episode psychosis: a systematic review and meta-analysis. Int. J. Neuropsychopharmacol. 2013;16(6):1205–1218. doi: 10.1017/S1461145712001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leucht S, Corves C, Arbter D, Engel RR, Li C, Davis JM. Second-generation versus first-generation antipsychotic drugs for schizophrenia: a meta-analysis. Lancet. 2009;373(9657):31–41. doi: 10.1016/S0140-6736(08)61764-X. [DOI] [PubMed] [Google Scholar]
- 6.Tandon R. Antipsychotics in the treatment of Schizophrenia: an overview. J. Clin. Psychiatry. 2011;72:4–8. doi: 10.4088/JCP.10075su1.01. [DOI] [PubMed] [Google Scholar]
- 7.Essali A, Turkmani K, Aboudamaah S, AbouDamaah A, Diaa Aldeen MR, Marwa ME, et al. Haloperidol discontinuation for people with schizophrenia. Cochrane Database Syst. Rev. 2019;4(4):CD011408. doi: 10.1002/14651858.CD011408.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niu J, Mei F, Li N, Wang H, Li X, Kong J, et al. Haloperidol promotes proliferation but inhibits differentiation in rat oligodendrocyte progenitor cell cultures. Biochem Cell Biol. 2010;88(4):611–620. doi: 10.1139/O09-178. [DOI] [PubMed] [Google Scholar]
- 9.Tyler MW, Zaldivar-Diez J, Haggarty SJ. Classics in chemical neuroscience: haloperidol. ACS Chem. Neurosci. 2017;8(3):444–453. doi: 10.1021/acschemneuro.7b00018. [DOI] [PubMed] [Google Scholar]
- 10.Nucifora FC, Jr, Mihaljevic M, Lee BJ, Sawa A. Clozapine as a model for antipsychotic development. Neurotherapeutics. 2017;14(3):750–761. doi: 10.1007/s13311-017-0552-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horn MRV, Sild M, Ruthazer ES. D-serine as a gliotransmitter and its roles in brain development and disease. Front. Cell. Neurosci. 2013;7:39. doi: 10.3389/fncel.2013.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho SE, Na KS, Cho SJ, Kang SG. Low d-serine levels in schizophrenia: a systematic review and meta-analysis. Neurosci. Lett. 2016;634:42–51. doi: 10.1016/j.neulet.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 13.MacKay MB, Kravtsenyuk M, Thomas R, Mitchell ND, Dursun SM, Baker GB. D-Serine: potential therapeutic agent and/or biomarker in schizophrenia and depression? Front Psychiatry. 2019;10:25. doi: 10.3389/fpsyt.2019.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fatemi SH, Folsom TD. The neurodevelopmental hypothesis of schizophrenia, revisited. Schizophr Bull. 2009;35:528–548. doi: 10.1093/schbul/sbn187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cassoli JS, Guest PC, Malchow B, Schmitt A, Falkai P, Martins-de-Souza D. Disturbed macro-connectivity in schizophrenia linked to oligodendrocyte dysfunction: from structural findings to molecules. NPJ Schizophr. 2015;1:15034. doi: 10.1038/npjschz.2015.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raabe FJ, Slapakova L, Rossner MJ, Cantuti-Castelvetri L, Simons M, et al. Oligodendrocytes as a new therapeutic target in schizophrenia: from histopathological findings to neuron-oligodendrocyte interaction. Cells. 2019;8(12):E1496. doi: 10.3390/cells8121496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu H, Zhang H, Zhang J, Huang Q, Shen Z, Wu R. Evaluation of neuron-glia integrity by in vivo proton magnetic resonance spectroscopy: implications for psychiatric disorders. Neurosci. Biobehav. Rev. 2016;71:563–577. doi: 10.1016/j.neubiorev.2016.09.027. [DOI] [PubMed] [Google Scholar]
- 18.Barateiro A, Fernandes A. Temporal oligodendrocyte lineage progression: in vitro models of proliferation, differentiation and myelination. Biochim. Biophys. Acta Mol. Cell Res. 2014;1843(9):1917–1929. doi: 10.1016/j.bbamcr.2014.04.018. [DOI] [PubMed] [Google Scholar]
- 19.Santos AK, Vieira MS, Vasconcellos R, Goulart VAM, Kihara AH, Resende RR. Decoding cell signalling and regulation of oligodendrocyte differentiation. Semin. Cell Dev. Biol. 2019;95:54–73. doi: 10.1016/j.semcdb.2018.05.020. [DOI] [PubMed] [Google Scholar]
- 20.Bernstein HG, Steiner J, Guest PC, Dobrowolny H, Bogerts B. Glial cells as key players in schizophrenia pathology: recent insights and concepts of therapy. Schizophr. Res. 2015;161(1):4–18. doi: 10.1016/j.schres.2014.03.035. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Zhang H, Wang L, Jiang W, Xu H, et al. Quetiapine enhances oligodendrocyte regeneration and myelin repair after cuprizone-induced demyelination. Schizophr. Res. 2012;138(1):8–17. doi: 10.1016/j.schres.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 22.Cassoli JS, Iwata K, Steiner J, Guest PC, Turck CW, et al. Effect of MK-801 and clozapine on the proteome of cultured human oligodendrocytes. Front Cell Neurosci. 2016;10:52. doi: 10.3389/fncel.2016.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davalieva K, Maleva Kostovska I, Dwork AJ. Proteomics research in schizophrenia. Front Cell Neurosci. 2016;10:18. doi: 10.3389/fncel.2016.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, et al. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol Cell Proteomics. 2005;4(10):1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Moulder R, Goo YA, Goodlett DR. Label-free quantitation for clinical proteomics. Methods Mol. Biol. 2016;1410:65–76. doi: 10.1007/978-1-4939-3524-6_4. [DOI] [PubMed] [Google Scholar]
- 26.Wang M, You J, Bemis KG, Tegeler TJ, Brown DPG. Label-free mass spectrometry-based protein quantification technologies in proteomic analysis. Brief Funct. Genomic Proteomic. 2008;7(5):329–339. doi: 10.1093/bfgp/eln031. [DOI] [PubMed] [Google Scholar]
- 27.Megger DA, Bracht T, Meyer HE, Sitek B. Label-free quantification in clinical proteomics. Biochim. Biophys. Acta. 2013;1834(8):1581–1590. doi: 10.1016/j.bbapap.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 28.McLaurin J, Trudel GC, Shaw IT, Antel JP, Cashman NR. A human glial hybrid cell line differentially expressing genes subserving oligodendrocyte and astrocyte phenotype. J. Neurobiol. 1995;26(2):283–293. doi: 10.1002/neu.480260212. [DOI] [PubMed] [Google Scholar]
- 29.Seabra, G., de Almeida, V. & Martins-de-Souza, D. Maturation of a Human Oligodendrocyte Cell Line in Pre-Clinical Models. Methods in Molecular Biology (ed. Guest, P.) 1916, 113–121 (Humana Press, 2019) [DOI] [PubMed]
- 30.Snaidero N, Mobius W, Czopka T, Hekking LH, Mathisen C, et al. Myelin membrane wrapping of CNS axons by PI(3,4,5)P3-dependent polarized growth at the inner tongue. Cell. 2014;156:277–290. doi: 10.1016/j.cell.2013.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Domingues HS, Cruz A, Chan JR, Relvas JB, Rubinstein B, Pinto IM. Mechanical plasticity during oligodendrocyte differentiation and myelination. Glia. 2018;66(1):5–14. doi: 10.1002/glia.23206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dawson MRL, Levine JM, Reynolds R. NG2-expressing cells in the central nervous system: are they oligodendroglial progenitors? J. Neurosci. Res. 2000;61:471–479. doi: 10.1002/1097-4547(20000901)61:5<471::AID-JNR1>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 33.Haas KF, Broadie K. Roles of ubiquitination at the synapse. Biochim Biophys Acta. 2008;1779:495–506. doi: 10.1016/j.bbagrm.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Checler F, da Costa CA, Ancolio K, Chevallier N, Lopez-Perez E, Marambaud P. Role of the proteasome in Alzheimer's disease. Biochem. Biophys. Acta. 2000;1502(1):133–138. doi: 10.1016/s0925-4439(00)00039-9. [DOI] [PubMed] [Google Scholar]
- 35.Chung KK, Dawson VL, Dawson TM. The role of the ubiquitin-proteasomal pathway in Parkinson's disease and other neurodegenerative disorders. Trends Neurosci. 2001;24(11):7–14. doi: 10.1016/s0166-2236(00)01998-6. [DOI] [PubMed] [Google Scholar]
- 36.Santos AR, Mele M, Vaz SH, Kellermayer B, Grimaldi M, et al. Differential role of the proteasome in the early and late phases of BDNF-induced facilitation of LTP. J Neurosci. 2015;35:3319–3329. doi: 10.1523/JNEUROSCI.4521-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hamilton AM, Oh WC, Vega-Ramirez H, Stein IS, Hell JW, et al. Activity-dependent growth of new dendritic spines is regulated by the proteasome. Neuron. 2012;74:1023–1030. doi: 10.1016/j.neuron.2012.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Green MJ, Matheson SL, Shepherd A, Weickert CS, Carr VJ. Brain-derived neurotrophic factor levels in schizophrenia: a systematic review with meta-analysis. Mol Psychiatry. 2011;16:960–972. doi: 10.1038/mp.2010.88. [DOI] [PubMed] [Google Scholar]
- 39.Rajasekaran A, Venkatasubramanian G, Berk M, Debnath M. Mitochondrial dysfunction in schizophrenia: pathways, mechanisms and implications. Neurosci. Biobehav. Rev. 2015;48:10–21. doi: 10.1016/j.neubiorev.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 40.Moyer CE, Shelton MA, Sweet RA. Dendritic spine alterations in schizophrenia. Neurosci Lett. 2015;601:46–53. doi: 10.1016/j.neulet.2014.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bousman CA, Luza S, Mancuso SG, Kang D, Opazo CM, et al. Elevated ubiquitinated proteins in brain and blood of individuals with schizophrenia. Sci Rep. 2019;9(1):2307. doi: 10.1038/s41598-019-38490-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scott MR, Rubio MD, Haroutunian V, Meador-Woodruff JH. Protein expression of proteasome subunits in elderly patients with schizophrenia. Neuropsychopharmacology. 2016;41:896–905. doi: 10.1038/npp.2015.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Altar CA, Jurata LW, Charles V, Lemire A, Liu P, Bukhman Y, Young TA, Bullard J, Yokoe H, Webster MJ, Knable MB, Brockman JA. Deficient hippocampal neuron expression of proteasome, ubiquitin, and mitochondrial genes in multiple schizophrenia cohorts. Biol. Psychiatry. 2005;58:85–96. doi: 10.1016/j.biopsych.2005.03.031. [DOI] [PubMed] [Google Scholar]
- 44.Middleton FA, Mirnics K, Pierri JN, Lewis DA, Levitt P. Gene expression profiling reveals alterations of specific metabolic pathways in schizophrenia. J Neurosci. 2002;22:2718–2729. doi: 10.1523/JNEUROSCI.22-07-02718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vawter MP, Barrett T, Cheadle C, Sokolov BP, Wood WH, 3rd, et al. Application of cDNA microarrays to examine gene expression differences in schizophrenia. Brain Res Bull. 2001;55:641–650. doi: 10.1016/s0361-9230(01)00522-6. [DOI] [PubMed] [Google Scholar]
- 46.Andrews JL, Goodfellow FJ, Matosin N, Snelling MK, Newell KA, et al. Alterations of ubiquitin related proteins in the pathology and development of schizophrenia: evidence from human and animal studies. J. Psychiatr. Res. 2017;90:31–39. doi: 10.1016/j.jpsychires.2017.01.009. [DOI] [PubMed] [Google Scholar]
- 47.Rubio M, Wood K, Haroutunian V, et al. Dysfunction of the ubiquitin proteasome and ubiquitin-like systems in schizophrenia. Neuropsychopharmacol. 2013;38:1910–1920. doi: 10.1038/npp.2013.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scott MR, Meador-Woodruff JH. Intracellular compartment-specific proteasome dysfunction in postmortem cortex in schizophrenia subjects. Mol. Psychiatry. 2020;25:776–790. doi: 10.1038/s41380-019-0359-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dansen TB, Westerman J, Wouters F, Wanders RJ, van Hoek A, et al. High affinity binding of very long chain fatty acyl CoA esters to the peroxisomal non-specific lipid transfer protein (sterol carrier protein-2) Biochem J. 1999;339:193–199. [PMC free article] [PubMed] [Google Scholar]
- 50.Martin GG, Chung S, Landrock D, Landrock KK, Huang H, et al. FABP1 gene ablation impacts brain endocannabinoid system in male mice. J. Neurochem. 2016;138:407–422. doi: 10.1111/jnc.13664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schroeder F, Zhou M, Swaggerty CL, Atshaves BP, Petrescu AD, et al. Sterol carrier protein-2 functions in phosphatidylinositol transfer and signaling. Biochemistry. 2003;42:3189–3202. doi: 10.1021/bi026904+. [DOI] [PubMed] [Google Scholar]
- 52.Shimazu T, Hirschey MD, Hua L, Dittenhafer-Reed KE, Schwer B, et al. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell. Metab. 2010;12(6):654–661. doi: 10.1016/j.cmet.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Narayan S, Head SR, Gilmartin TJ, Dean B, Thomas EA. Evidence for disruption of sphingolipid metabolism in schizophrenia. J. Neurosci. Res. 2009;87:278–288. doi: 10.1002/jnr.21822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ghosh S, Dyer RA, Beasley CL. Evidence for altered cell membrane lipid composition in postmortem prefrontal white matter in bipolar disorder and schizophrenia. J. Psychiatr. Res. 2017;95:135–142. doi: 10.1016/j.jpsychires.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 55.Kriisa K, Leppik L, Balõtšev R, Ottas A, Soomets U, et al. Profiling of acylcarnitines in first episode psychosis before and after antipsychotic treatment. J. Proteome Res. 2017;16:3558–3566. doi: 10.1021/acs.jproteome.7b00279. [DOI] [PubMed] [Google Scholar]
- 56.Yang J, Chen T, Sun L, Zhao Z, Qi X, et al. Potential metabolite markers of schizophrenia. Mol. Psychiatry. 2013;18(1):67–78. doi: 10.1038/mp.2011.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang J. Metabolic profiling identifies phospholipids as potential serum biomarkers for schizophrenia. Psychiatry Res. 2019;272:18–29. doi: 10.1016/j.psychres.2018.12.008. [DOI] [PubMed] [Google Scholar]
- 58.Matsumoto J, Sugiura Y, Yuki D, Hayasaka T, Goto-Inoue N, et al. Abnormal phospholipids distribution in the prefrontal cortex from a patient with schizophrenia revealed by matrix-assisted laser desorption/ionization imaging mass spectrometry. Anal. Bioanal. Chem. 2011;400:1933. doi: 10.1007/s00216-011-4909-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leppik L, Parksepp M, Janno S, Koido K, Haring L, et al. Profiling of lipidomics before and after antipsychotic treatment in first-episode psychosis. Eur. Arch. Psychiatry Clin. Neurosci. 2020;270(1):59–70. doi: 10.1007/s00406-018-0971-6. [DOI] [PubMed] [Google Scholar]
- 60.Taha AY, Cheon Y, Ma K, Rapoport SI, Rao JS. Altered fatty acid concentrations in prefrontal cortex of schizophrenic patients. J. Psychiatr. Res. 2013;47(5):636–643. doi: 10.1016/j.jpsychires.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Misiak B, Stanczykiewicz B, Łaczmanski Ł, Frydecka D. Lipid profile disturbances in antipsychotic-naive patients with first-episode non-affective psychosis: a systematic review and meta-analysis. Schizophr Res. 2017;190:18–27. doi: 10.1016/j.schres.2017.03.031. [DOI] [PubMed] [Google Scholar]
- 62.Aquino A, Alexandrino GL, Guest PC, Augusto F, Gomes AF, et al. Blood-based lipidomics approach to evaluate biomarkers associated with response to olanzapine, risperidone, and quetiapine treatment in schizophrenia patients. Front Psychiatry. 2018;9:209. doi: 10.3389/fpsyt.2018.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McEvoy J, Baillie RA, Zhu H, Buckley P, Keshavan MS, et al. Lipidomics reveals early metabolic changes in subjects with schizophrenia: effects of atypical antipsychotics. PLoS ONE. 2013;8:e68717. doi: 10.1371/journal.pone.0068717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ferno J, Skrede S, Vik-Mo AO, Havik B, Steen VM. Drug-induced activation of SREBP-controlled lipogenic gene expression in CNS-related cell lines: marked differences between various antipsychotic drugs. BMC Neurosci. 2006;7:69. doi: 10.1186/1471-2202-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Polymeropoulos MH, Licamele L, Volpi S, Mack K, Mitkus SN, et al. Common effect of antipsychotics on the biosynthesis and regulation of fatty acids and cholesterol supports a key role of lipid homeostasis in schizophrenia. Schizophr. Res. 2009;108(1–3):134–142. doi: 10.1016/j.schres.2008.11.025. [DOI] [PubMed] [Google Scholar]
- 66.Carter CJ. EIF2B and oligodendrocyte survival: where nature and nurture meet in bipolar disorder and schizophrenia? Schizophr Bull. 2007;33:1343–1353. doi: 10.1093/schbul/sbm007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pronk JC, van Kollenburg B, Scheper GC, van der Knaap MS. Vanishing white matter disease: a review with focus on its genetics. Ment. Retard. Dev. Disabil. Res. Rev. 2006;12:123–128. doi: 10.1002/mrdd.20104. [DOI] [PubMed] [Google Scholar]
- 68.Brandão-Teles, C., de Almeida, V., Cassoli, J. S. & Martins-de-Souza, D. Biochemical pathways triggered by antipsychotics in human [corrected] oligodendrocytes: potential of discovering new treatment targets [published correction appears in Front Pharmacol. 2019; 10:344]. Front Pharmacol. 10, 186 (2019). [DOI] [PMC free article] [PubMed]
- 69.Dominguez R, Holmes KC. Actin structure and function. Ann. Rev. Biophys. 2011;40:169–186. doi: 10.1146/annurev-biophys-042910-155359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Glausier JR, Lewis DA. Dendritic spine pathology in schizophrenia. Neuroscience. 2013;22:90–107. doi: 10.1016/j.neuroscience.2012.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bhambhvani HP, Mueller TM, Simmons MS, Meador-Woodruff JH. Actin polymerization is reduced in the anterior cingulate cortex of elderly patients with schizophrenia. Transl Psychiatry. 2017;7(12):1278. doi: 10.1038/s41398-017-0045-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yan Z, Kim E, Datta D, Lewis DA, Soderling SH. Synaptic actin dysregulation, a convergent mechanism of mental disorders? J Neurosci. 2016;36(45):11411–11417. doi: 10.1523/JNEUROSCI.2360-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao Z, Xu J, Chen J, Kim S, Reimers M, et al. Transcriptome sequencing and genome-wide association analyses reveal lysosomal function and actin cytoskeleton remodeling in schizophrenia and bipolar disorder. Mol. Psychiatry. 2015;20(5):563–572. doi: 10.1038/mp.2014.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hill JJ, Hashimoto T, Lewis DA. Molecular mechanisms contributing to dendritic spine alterations in the prefrontal cortex of subjects with schizophrenia. Mol. Psychiatry. 2006;11:557–566. doi: 10.1038/sj.mp.4001792. [DOI] [PubMed] [Google Scholar]
- 75.Reble E, Dineen A, Barr CL. The contribution of alternative splicing to genetic risk for psychiatric disorders. Genes Brain Behav. 2018;17:e12430. doi: 10.1111/gbb.12430. [DOI] [PubMed] [Google Scholar]
- 76.Oldmeadow C, Mossman D, Evans TJ, Holliday EG, Tooney PA, et al. Combined analysis of exon splicing and genome wide polymorphism data predict schizophrenia risk loci. J. Psychiatr. Res. 2014;52:44–49. doi: 10.1016/j.jpsychires.2014.01.011. [DOI] [PubMed] [Google Scholar]
- 77.Nakata K, Lipska BK, Hyde TM, Ye T, Newburn EN, et al. DISC1 splice variants are upregulated in schizophrenia and associated with risk polymorphisms. Proc. Natl. Acad. Sci. USA. 2009;106:15873–15878. doi: 10.1073/pnas.0903413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Law AJ, Kleinman JE, Weinberger DR, Weickert CS. Disease-associated intronic variants in the ErbB4 gene are related to altered ErbB4 splice-variant expression in the brain in schizophrenia. Hum. Mol. Genet. 2007;16:129–141. doi: 10.1093/hmg/ddl449. [DOI] [PubMed] [Google Scholar]
- 79.Uezato A, Yamamoto N, Iwayama Y, Hiraoka S, Hiraaki E, et al. Reduced cortical expression of a newly identified splicing variant of the DLG1 gene in patients with early-onset schizophrenia. Transl. Psychiatry. 2015;5:e654. doi: 10.1038/tp.2015.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kao WT, Wang Y, Kleinman JE, Lipska BK, Hyde TM, et al. Common genetic variation in Neuregulin 3 (NRG3) influences risk for schizophrenia and impacts NRG3 expression in human brain. Proc. Natl. Acad. Sci. USA. 2010;107:15619–15624. doi: 10.1073/pnas.1005410107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Morikawa T, Manabe T. Aberrant regulation of alternative pre-mRNA splicing in schizophrenia. Neurochem. Int. 2010;57(7):691–704. doi: 10.1016/j.neuint.2010.08.012. [DOI] [PubMed] [Google Scholar]
- 82.Flohe L, Günzler WA, Schock HH. Glutathione peroxidase: A selenoenzyme. FEBS Lett. 1973;32(1):132–134. doi: 10.1016/0014-5793(73)80755-0. [DOI] [PubMed] [Google Scholar]
- 83.Gonzalez-Flores JN, Shetty SP, Dubey A, Copeland PR. The molecular biology of selenocysteine. Biomol. Concepts. 2013;4(4):349–365. doi: 10.1515/bmc-2013-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Steinbrenner H, Sies H. Protection against reactive oxygen species by selenoproteins. Biochim. Biophys. Acta. 2009;1790(11):1478–1485. doi: 10.1016/j.bbagen.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 85.Dakhale G, Khanzode S, Khanzode S, Saoji A, Khobragade L, Turankar A. Oxidative damage and schizophrenia: the potential benefit by atypical antipsychotics. Neuropsychobiology. 2004;49:205–209. doi: 10.1159/000077368. [DOI] [PubMed] [Google Scholar]
- 86.Pillai A, Parikh V, Terry AV, Jr, Mahadik SP. Long-term antipsychotic treatments and crossover studies in rats: differential effects of typical and atypical agents on the expression of antioxidant enzymes and membrane lipid peroxidation in rat brain. J. Psychiatr. Res. 2007;41:372–386. doi: 10.1016/j.jpsychires.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 87.Martins MR, Petronilho FC, Gomes KM, Dal-Pizzol F, Streck EL, Quevedo J. Antipsychotic-induced oxidative stress in rat brain. Neurotox. Res. 2008;13:63–69. doi: 10.1007/BF03033368. [DOI] [PubMed] [Google Scholar]
- 88.Cho CH, Lee HJ. Oxidative stress and tardive dyskinesia: Pharmacogenetic evidence. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2013;46:207–213. doi: 10.1016/j.pnpbp.2012.10.018. [DOI] [PubMed] [Google Scholar]
- 89.Hayashi-Takagi A, Takaki M, Graziane N, Seshadri S, Murdoch H, et al. Disrupted-in-Schizophrenia 1 (DISC1) regulates spines of the glutamate synapse via Rac1. Nat. Neurosci. 2010;13:327–332. doi: 10.1038/nn.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Scott EK, Reuter JE, Luo L. Small GTPase Cdc42 is required for multiple aspects of dendritic morphogenesis. J. Neurosci. 2003;23:3118–3123. doi: 10.1523/JNEUROSCI.23-08-03118.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Saneyoshi T, Fortin DA, Soderling TR. Regulation of spine and synapse formation by activity-dependent intracellular signaling pathways. Curr. Opin. Neurobiol. 2010;20:108–115. doi: 10.1016/j.conb.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lin GN, Corominas R, Lemmens I, Yang X, Tavernier J, et al. Spatiotemporal 16p11.2 protein network implicates cortical late mid-fetal brain development and KCTD13-Cul3-RhoA pathway in psychiatric diseases. Neuron. 2015;85:742–754. doi: 10.1016/j.neuron.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Holmgren A. Thioredoxin. Annu Rev Biochem. 1985;54(1):237–271. doi: 10.1146/annurev.bi.54.070185.001321. [DOI] [PubMed] [Google Scholar]
- 94.Bitanihirwe BK, Woo TUW. Oxidative stress in schizophrenia: an integrated approach. Neurosci. Biobehav. Rev. 2011;35(3):878–893. doi: 10.1016/j.neubiorev.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yao JK, Keshavan MS. Antioxidants, redox signaling, and pathophysiology in schizophrenia: an integrative view. Antioxid. Redox Signal. 2011;15(7):2011–2035. doi: 10.1089/ars.2010.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang XY, Xiu MH, Wang F, Qi LY, Sun HQ, et al. The novel oxidative stress marker thioredoxin is increased in first-episode schizophrenic patients. Schizophr Res. 2009;113(2):151–157. doi: 10.1016/j.schres.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 97.Bas A, Gultekin G, Incir S, Bas TO, Emul M, Duran A. Level of serum thioredoxin and correlation with neurocognitive functions in patients with schizophrenia using clozapine and other atypical antipsychotics. Psychiatry Res. 2017;247:84–89. doi: 10.1016/j.psychres.2016.11.021. [DOI] [PubMed] [Google Scholar]
- 98.Roberts RC. Postmortem studies on mitochondria in schizophrenia. Schizophr. Res. 2017;187:17–25. doi: 10.1016/j.schres.2017.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Uranova NA, Vostrikov VM, Vikhreva OV, Zimina IS, Kolomeets NS, Orlovskaya DD. The role of oligodendrocyte pathology in schizophrenia. Int. J. Neuropsychopharmacol. 2007;10(4):537–545. doi: 10.1017/S1461145707007626. [DOI] [PubMed] [Google Scholar]
- 100.Vikhreva OV, Rakhmanova VI, Orlovskaya DD, Uranova NA. Ultrastructural alterations of oligodendrocytes in prefrontal white matter in schizophrenia: a postmortem morphometric study. Schizophr Res. 2016;177(1–3):28–36. doi: 10.1016/j.schres.2016.04.023. [DOI] [PubMed] [Google Scholar]
- 101.Uranova NA, Orlovskaya D, Vikhreva O, Zimina I, Kolomeets N, et al. Electron microscopy of oligodendroglia in severe mental illness. Brain Res Bull. 2001;55:597–610. doi: 10.1016/s0361-9230(01)00528-7. [DOI] [PubMed] [Google Scholar]
- 102.Gjessing R. Disturbances of somatic functions in catatonia with a periodic course, and their compensation. J. Ment. Sci. 1938;84:608. [Google Scholar]
- 103.Man EB, Bettcher PG, Brown WT. Variations in plasma-amino acid nitrogen of schizophrenics. Yale J Biol Med. 1947;20(2):167–174. [PMC free article] [PubMed] [Google Scholar]
- 104.Huang N, Cao B, Brietzke E, Park C, Cha D, et al. A pilot case-control study on the association between N-acetyl derivatives in serum and first-episode schizophrenia. Psychiatry Res. 2019;272:36–41. doi: 10.1016/j.psychres.2018.11.064. [DOI] [PubMed] [Google Scholar]
- 105.Molina V, Sanchez J, Sanz J, Reig S, Benito C, et al. Dorsolateral prefrontal N-acetyl-aspartate concentration in male patients with chronic schizophrenia and with chronic bipolar disorder. Eur. Psychiatry. 2007;22(8):505–512. doi: 10.1016/j.eurpsy.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 106.Yilmaz N, Herken H, Cicek HK, Celik A, Yureklin M, Akyol O. Increased levels of nitric oxide, cortisol and adrenomedullin in patients with chronic schizophrenia. Med. Princ. Pract. 2007;6:137–141. doi: 10.1159/000098367. [DOI] [PubMed] [Google Scholar]
- 107.Taneli F, Pirildar S, Akdeniz F, Uyanik BS, Ari Z. Serum nitric oxide metabolite levels and the effect of antipsychotic therapy in schizophrenia. Arch. Med. Res. 2004;35:401–405. doi: 10.1016/j.arcmed.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 108.Dietrich-Muszalska A, Olas B, Głowacki R, Bald E. Oxidative/nitrative modifications of plasma proteins and thiols from patients with schizophrenia. Neuropsychobiology. 2009;59(1):1–7. doi: 10.1159/000202822. [DOI] [PubMed] [Google Scholar]
- 109.Hashimoto K, Engberg G, Shimizu E, Nordin C, Lindstrom LH, Iyo M. Reduced d-serine to total serine ratio in the cerebrospoinal fluid of drug naïve schizophrenic patients. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2005;29:767–769. doi: 10.1016/j.pnpbp.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 110.Bendikov I, Nadri C, Amar S, Panizzutti R, De Miranda J, et al. A CSF and postmortem brain study of D-serine metabolic parameters in schizophrenia. Schizophr. Res. 2007;90:41–51. doi: 10.1016/j.schres.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 111.Calcia MA, Madeira C, Alheira FV, Silva TC, Tannos FM, et al. Plasma levels of D-serine in Brazilian individuals with schizophrenia. Schizophr. Res. 2012;142:83–87. doi: 10.1016/j.schres.2012.09.014. [DOI] [PubMed] [Google Scholar]
- 112.El-Tallawy HN, Saleem TH, El-Ebidi AM, Hassan MH, Gabra RH, et al. Clinical and biochemical study of d-serine metabolism among schizophrenia patients. Neuropsychiatr. Dis. Treatm. 2017;13:1057–1063. doi: 10.2147/NDT.S126979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Panizzutti R, Fisher M, Garrett C, Man WH, Sena W, et al. Association between increased serum d-serine and cognitive gains induced by intensive cognitive training in schizophrenia. Schizophr. Res. 2019;207:63–69. doi: 10.1016/j.schres.2018.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yamamori H, Hashimoto R, Fujita Y, Numata S, Yasuda Y, et al. Changes in plasma d-serine, l-serine, and glycine levels in treatment-resistant schizophrenia before and after clozapine treatment. Neurosci. Lett. 2014;582:93–98. doi: 10.1016/j.neulet.2014.08.052. [DOI] [PubMed] [Google Scholar]
- 115.Brandão-Teles, C., Martins-de-Souza, D., Guest, P.C. & Cassoli, J.S. MK-801-Treated Oligodendrocytes as a Cellular Model to Study Schizophrenia in Proteomic Methods in Neuropsychiatric Research. Advances in Experimental Medicine and Biology (ed. Guest, P.) 974, 269–277 (Springer Cham, 2017). [DOI] [PubMed]
- 116.Silva JC, Gorenstein MV, Li GZ, Vissers JPC, Geromanos SJ. Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol. Cell. Proteomics. 2006;5(1):144–156. doi: 10.1074/mcp.M500230-MCP200. [DOI] [PubMed] [Google Scholar]
- 117.Chevreux G, Tilly N, Bihoreau N. Quantification of proteins by data independent acquisition: performance assessment of the Hi3 methodology. Anal. Biochem. 2018;549:184–187. doi: 10.1016/j.ab.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 118.Krämer A, Green J, Pollard J, Jr, Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30(4):523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2018;46(D1):D649–D655. doi: 10.1093/nar/gkx1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Szklarczyk, D., Gable, A.L., Lyon, D., Junge, A., Wyder, S., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res.47, 607–613 (2019). [DOI] [PMC free article] [PubMed]
- 121.Oliveros, J.C. VENNY. An interactive tool for comparing lists with Venn Diagrams. https://bioinfogp.cnb.csic.es/tools/venny/index.html (2007).
- 122.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.RStudio Team. RStudio: Integrated Development for R. RStudio. https://www.rstudio.com (2015).
- 124.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing. Vienna, Austria. https://www.R-project.org (2019).
- 125.Kolde, R. Pheatmap: Pretty Heatmaps. R package version 1.0.12. https://CRAN.R-project.org/package=pheatmap (2019)
- 126.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Doncheva NT, Morris JH, Gorodkin J, Jensen LJ. Cytoscape StringApp: network analysis and visualization of proteomics data. J. Proteome Res. 2019;18(2):623–632. doi: 10.1021/acs.jproteome.8b00702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, et al. Image J2: ImageJ for the next generation of scientific image data. BMC Bioinf. 2017;18:529. doi: 10.1186/s12859-017-1934-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.