Abstract
Background
Rheumatoid arthritis (RA) is a chronic debilitating autoimmune disorder with a high prevalence, especially in industrialized countries. Dysbiosis of the intestinal microbiota has been observed in RA patients. For instance, new-onset untreated RA (NORA) is associated with the underrepresentation of the Clostridium cluster XIVa, including Lachnospiraceae, which are major butyrate producers, although the pathological relevance has remained obscure. Follicular regulatory T (TFR) cells play critical regulatory roles in the pathogenesis of autoimmune diseases, including RA. Reduced number of circulating TFR cells has been associated with the elevation of autoantibodies and disease severity in RA. However, the contribution of commensal microbe-derived butyrate in controlling TFR cell differentiation remains unknown.
Methods
We examined the contribution of microbe-derived butyrate in controlling autoimmune arthritis using collagen-induced arthritis (CIA) and SKG arthritis models. We phenotyped autoimmune responses in the gut-associated lymphoid tissues (GALT) in the colon and joint-draining lymph nodes in the CIA model. We developed an in vitro CXCR5+Bcl-6+Foxp3+ TFR (iTFR) cell culture system and examined whether butyrate promotes the differentiation of iTFR cells.
Findings
Microbe-derived butyrate suppressed the development of autoimmune arthritis. The immunization of type II collagen (CII) caused hypertrophy of the GALT in the colon by amplifying the GC reaction prior to the onset of the CIA. Butyrate mitigated these pathological events by promoting TFR cell differentiation. Butyrate directly induced the differentiation of functional TFR cells in vitro by enhancing histone acetylation in TFR cell marker genes. This effect was attributed to histone deacetylase (HDAC) inhibition by butyrate, leading to histone hyperacetylation in the promoter region of the TFR-cell marker genes. The adoptive transfer of the butyrate-treated iTFR cells reduced CII-specific autoantibody production and thus ameliorated the symptoms of arthritis.
Interpretation
Accordingly, microbiota-derived butyrate serves as an environmental cue to enhance TFR cells, which suppress autoantibody production in the systemic lymphoid tissue, eventually ameliorating RA. Our findings provide mechanistic insights into the link between the gut environment and RA risk.
Funding
This work was supported by AMED-Crest (16gm1010004h0101, 17gm1010004h0102, 18gm1010004h0103, and 19gm1010004s0104 to KH), the Japan Society for the Promotion of Science (JP17KT0055, JP16H01369, and JP18H04680 to KH; JP17K15734 to DT), Keio University Special Grant-in-Aid for Innovative Collaborative Research Projects (KH), Keio Gijuku Fukuzawa Memorial Fund for the Advancement of Education and Research (DT), the SECOM Science and Technology Foundation (KH), the Cell Science Research Foundation (KH), the Mochida Memorial Foundation for Medical and Pharmaceutical Research (DT), the Suzuken Memorial Foundation (KH and DT), the Takeda Science Foundation (KH and DT), The Science Research Promotion Fund, and The Promotion and Mutual Aid Corporation for Private Schools of Japan (KH).
Keywords: Rheumatoid arthritis, Intestinal microbiota, Butyrate, Follicular regulatory T cells, Autoimmunity
Research in context.
Evidence before this study
Accumulating evidence has demonstrated that intestinal dysbiosis is implicated in several autoimmune diseases, including RA. Overrepresentation of Prevotellaceae, particularly Prevotella copri and underrepresentation of the Clostridium cluster XIVa including Lachnospiraceae, which are major butyrate producers, are found in new-onset untreated RA (NORA) patients. Butyrate administration via drinking water, which is mostly absorbed in the upper small intestine, suppresses the development of autoimmune arthritis models in mice. Follicular regulatory T (TFR) cells play critical roles in the regulation of autoimmune diseases, including RA. The abundance of TFR cells is negatively correlated with disease activity in patients with RA.
Added value of this study
Here, we report that intestinal microbiota-derived butyrate serves as an environmental cue to induce the differentiation of functional TFR cells in the gut-associated lymphoid tissue (GALT). Intestinal microbiota plays an essential role in both the initiation and suppression of autoimmune arthritis by modifying the immune system in the GALT. We observed that immunization with collagen caused hypertrophy of the GALT in the colon by amplifying the GC reaction prior to the onset of collagen-induced arthritis, indicating that GALTs enhance the autoimmune response to circulating autoantigens. However, butyrate mitigated these pathological events by increasing TFR cells. We newly developed an in vitro CXCR5+Bcl-6+Foxp3+ TFR (iTFR) cell-inducing culture system, and confirmed that butyrate facilitates the differentiation of TFR cells directly. This effect was attributed to histone deacetylase (HDAC) inhibition by butyrate, leading to histone hyperacetylation in the promoter region of the TFR-cell marker genes. The adoptive transfer of the butyrate-treated T cells significantly reduced collagen-specific autoantibody production and thus ameliorated the symptoms of arthritis. Considering that butyrate production is affected in RA patients, this metabolite may play a key role in RA prevention.
Implications of all the available evidence
Our data and methods provide the basis for future studies allowing further mechanistic dissection of TFR cell differentiation. Administration of butyrate-producing bacteria or functional food to subjects genetically susceptible to RA could have therapeutic potential to prevent the disease onset or the development of following disease symptoms. Our findings provide a molecular basis for new prophylaxis and treatment approaches for systemic autoimmune disorders by targeting the intestinal environment and autoimmune responses in GALT.
Alt-text: Unlabelled box
1. Introduction
Rheumatoid arthritis (RA) is a systemic autoimmune disease characterized by synovial inflammation, cartilage lesions, and bone erosion. The generation of various autoantibodies through the germinal centre (GC) reaction is a characteristic of RA patients [1], [2], [3]. GCs form within secondary lymphoid tissues in response to T cell-dependent antigens to produce high-affinity class-switched antibodies. GCB cells interact with a specialized helper T cell subset, follicular helper T (TFH) cells, which activate GCB cells to promote differentiation into plasma or memory B cells. Lack of TFH cells affects antibody production [4], [5], [6], [7], and vice versa, the deregulation of TFH-cell activity could contribute to the production of autoantibodies and the development of autoimmune diseases [8], [9], [10], [11]. Furthermore, T-cell-specific deletion of CXC-chemokine receptor 5 (CXCR5), which hampers migration and retention of TFH cells in GC, reduces the generation of type II collagen (CII)-specific antibodies and alleviates the development of collagen-induced arthritis (CIA), a common autoimmune mouse model used to study RA [12]. A similar phenotype was observed in mice deficient in inducible T-cell costimulator (ICOS) or signalling lymphocytic activation molecule (SLAM)-associated protein (SAP), the functional molecules that facilitate the differentiation and function of TFH cells [13,14]. These observations suggest that dysregulation of GC reactions due to the activation of TFH cells may predispose to autoimmune diseases, including RA.
Recent studiesdefined follicular regulatory T (TFR) cells as regulators of the GC reaction [15], [16], [17]. TFR cells counteract the functions of TFH cells by downregulating the production of effector cytokines such as IL-4, IFN-γ, and IL-21 that are essential for B cell activation and class switch recombination [18], [19], [20]. Similar to TFH cells,TFR cells express the defining markers for follicular T cells, such as programmed death 1 (PD-1), ICOS, and CXCR5. [[15], [16], [17],20,21]. TFR cells can be subdivided into CD25+ and CD25− subpopulation without the difference in Foxp3 expression. In comparison to CD25+ TFR cells, CD25− TFR cells express PD-1, CXCR5, and Bcl-6 at a higher level, and are localized preferentially in GC [20,21]. Although transcription regulators Bcl-6 and T-cell factor 1 (TCF-1) are required for both TFH andTFR cells, TFR cells can be distinguished by the expression of specific transcriptional factor Foxp3 [[15], [16], [17],22] Importantly, TFR cells predominantly respond to self-, but not foreign, antigens, whereas TFH cells primarily react to foreign antigens [20,23].Indeed, selective depletion of TFR cells causes outgrowth of self-reactive B cells, leading to the production of anti-nuclear antibodies (ANA) following influenza infection [24]. These observations underscore the importance of the TFH/TFR balance in the maintenance of immune homeostasis by fine-tuning GC reactions in the context of host defence and autoimmunity [24,25]. Notably, a reduced number of circulating TFR cells has been associated with the elevation of autoantibodies and disease severity in RA, suggesting that the dysregulation of GC reactions due to impaired TFH/TFR balance may contribute to RA pathogenesis [26], [27], [28]. However, the molecular entities (e.g., genetic factors and environmental cues) regulating the TFH/TFR balance have yet to be fully elucidated.
Accumulating evidence has demonstrated that intestinal dysbiosis is implicated in several autoimmune diseases, including RA [29], [30], [31], [32], [33], [34], [35]. Overrepresentation of Prevotellaceae, particularly Prevotella copri and underrepresentation of the Clostridium cluster XIVa including Lachnospiraceae, which are major butyrate producers, are found in new-onset untreated RA (NORA) patients [29,30]. The RA-associated microbiota is considered as a causative factor for the development of RA because transplantation of intestinal microbiota from NORA patients to germ-free SKG mice accelerates autoimmune arthritis development [30]. The gut microbiota has emerged as a critical regulator for the establishment of the mucosal immune system by facilitating IgA response and differentiation of several helper T cell subsets like TH17, TREG, and TFH cells [36], [37], [38], [39], [40], [41], [42]. Germ-free (GF) K/BxN mice were reported to exhibit a decrease in spleen TFH cells and serum autoantibody levels compared to special-pathogen-free (SPF) K/BxN mice [43]. Gut commensal segmented filamentous bacteria (SFB) are responsible for the induction of TFH cells in Peyer's patches (PP), which migrate to the systemic lymphoid tissues and induce autoantibody production and arthritis development [42]. Meanwhile, TFR cells increase in the mesenteric lymph node of the mice fed a high fibre diet, which is fermented into short-chain fatty acids (SCFAs) in the large intestine [44]. However, the mechanism by which SCFAs increase TFR cells and the contribution of SCFAs-induced TFR cells in the regulation of autoimmune response remain obscure.
In this study, we report that a commensal bacteria-derived SCFAs, butyrate serves as an environmental cue to induce the differentiation of functional TFR cells in the gut-associated lymphoid tissue (GALT). We observed that systemic CII immunization caused hypertrophy of the GALT in the colon by amplifying the GC reaction prior to the onset of CIA, indicating that GALTs enhance the autoimmune response to circulating autoantigens. However, butyrate mitigated these pathological events by increasing TFR cells. This effect was attributed to histone deacetylase (HDAC) inhibition by butyrate, leading to histone hyperacetylation in the promoter region of the TFR-cell marker genes. The adoptive transfer of the butyrate-treated T cells significantly reduced CII-specific autoantibody production and thus ameliorated the symptoms of arthritis. Considering that butyrate production is affected in RA patients, this metabolite may play a key role in RA prevention.
2. Materials and methods
2.1. Mice
C57BL/6JJcl and SKG mice were purchased from CLEA Japan. DBA/1J Jms Slc mice were purchased from Sankyo Labo Service. Bcl6yfp mice [45] were described previously. Bcl6-tdTomato-creERT2 mice were provided by the Faculty of Pharmaceutical Sciences, Tokyo University of Science. Foxp3hCD2/hCD52 mice in which Foxp3+ cells express human CD2 (hCD2) and human CD52 fusion protein on the cell surface were described previously[46] and backcrossed onto the DBA/1J genetic background for collagen-induced arthritis experiments. SKG/Jcl mice were purchased from CLEA Japan and received a single intraperitoneal injection of 30 mg of laminarin (Wako Pure Chemical Industries, Osaka, Japan) to induce autoimmune arthritis. Tcrb−/−Tcrd−/− mice were purchased from Jackson Laboratories. KikGR mice were obtained from RIKEN BioResource Research Center (BRC) and described previously [47]. All mice were housed and bred at Keio University Faculty of Pharmacy under protocols approved by the Animal Studies Committees of Keio University. Mice were habituated to specific pathogen-free (SPF) or conventional vivarium conditions by feeding an AIN-93G diet (Oriental Yeast, Tokyo, Japan or Research Diets, New Brunswick, NJ, USA) for one week, followed by an AIN-93G diet containing 15% (w/w) high-amylose maize starch (HAMS), as a control, HAMS esterified with acetate (HAMSA), propionate (HAMSP) or butyrate (HAMSB). In some experiments, CE-2 chow (CLEA Japan, Tokyo, Japan) was used as a high fibre diet.
2.2. Human participants
Faecal samples were collected from 31 patients with NORA at the Graduate School of Medicine, Osaka University, National Hospital Organization (NHO) Osaka Minami Medical Center, and NTT West Osaka Hospital, and from 41 healthy controls (HC) at the Graduate School of Medicine. The inclusion criteria required that patients meet the American College of Rheumatology/European League Against Rheumatism 2010 criteria for RA and that they were between 20 and 80 years old (NORA, 55.6 ± 9.8 years old; HC, 62.4 ± 16.6 years old, respectively). NORA patients had been diagnosed for less than six months at the time of sample taking and had not previously been treated with disease-modifying antirheumatic drugs, biologic agents, or glucocorticoids (nonsteroidal anti-inflammatory drugs were allowed). The exclusion criteria for both groups were as follows: extreme diets (e.g., strict vegetarians), treatment with antibiotics for at least three months prior to sampling, and a known history of malignancy or severe disease of the heart, liver, or kidney. Studies were approved by the Keio University Faculty of Pharmacy, the Graduate School of Medicine, Osaka University, NHO Osaka Minami Medical Center, and NTT West Osaka Hospital following the Declaration of Helsinki; all patients signed informed consents. For Spearman's correlations between bacterial abundance and faecal butyrate concentration, we reanalyzed a previously published 16S rRNA sequencing data set [30] and newly measured butyrate concentration in the same faecal samples.
2.3. Collagen-induced arthritis (CIA) and collagen antibody-induced arthritis (CAIA)
CIA was induced by the immunization with an emulsion (0.1 ml) of 100 µg of immunization grade bovine type II collagen (CII; Chondrex, Redmond, WA, USA) and complete Freund's adjuvant (CFA) containing heat-killed Mycobacterium tuberculosisH37Ra at a final concentration of 1 mg ml−1 in incomplete Freund's adjuvant (IFA; Chondrex) into the tail of the DBA/1 J Jms Slc mice at 0.5 cm from the base. Three weeks after the primary immunization, mice were challenged with bovine CII and IFA emulsion (0.1 ml) as the booster immunization. CAIA was induced by the administration of a cocktail of monoclonal antibodies (Arthrogen-CIA 5-Clone Cocktail Kit, Chondrex) recognizing conserved epitopes within the LyC1 (CII 124–290) fragment of CB11 (CII 124–402), LyC2 (CII 291–374) fragment, followed by 25 μg Lipopolysaccharide (LPS from E.coli O111; FUJIFILM Wako Pure Chemical) injection on day 3. Arthritis development in the joints was assessed by a quantitative clinical score as follows: 0, no joint swelling; 1, mild but definite redness and swelling of the ankle or wrist, or apparent redness and swelling limited to individual digits, regardless of the number of affected digits; 2, moderate redness and swelling of ankle or wrist; 3, severe redness and swelling of the entire paw including digits; or 4, maximally inflamed limb with the involvement of multiple joints. The scores for all fingers of forepaws and hind paws, wrists, and ankles were added for each mouse (with a maximum possible score of 16 per mouse). The incidence of arthritis is defined by arthritic score per mouse > 3.
2.4. Immunization with human insulin
Foxp3hCD2 mice were immunized s.c. in the mouse flanks with an emulsion of 4.5 U of recombinant human insulin (FUJIFILM Wako Pure Chemical, Tokyo, Japan) and CFA containing heat-killed Mycobacterium tuberculosisH37Ra at a final concentration of 1 mg ml−1 in IFA. Mice were euthanized on day 10 for the analysis of follicular T cells in the inguinal lymph node (ILN).
2.5. Measurement of organic acids
Faecal and serum organic acids were measured by chromatography-mass spectrometry (GC–MS), as previously described [48], with some modifications. Faecal samples were homogenized and diluted 10-fold in water using a microtube homogenizer, and the precipitate was removed by centrifugation. Faecal homogenates and serum were spiked with one mM 2-ethyl butyric acid (2-EB; Sigma-Aldrich, St. Louis, MO, USA) as an internal standard and deproteinized with 10% (v/v) of 20% (w/v) 5-sulfosalicylic acid (Sigma-Aldrich) solution. The deproteinized extracts were acidified with 37% (w/v) hydrochloric acid (Sigma-Aldrich), and organic acids were extracted with diethyl ether by vortexing for 15 min. The samples were centrifuged for 5 min at 15,000 × g; the upper organic layer was added to a glass vial. After adding N‑tert-butyldimethylsilyl-N-methyltrifluoroacetamide (MTBSTFA; Sigma-Aldrich) as a derivatization reagent, the samples were incubated for 24 h at 25 °C in the dark. The derivatized samples were injected with a 5:1 split into a GCMS-QP2010 gas chromatography coupled with a mass spectrometer detector (SHIMADZU, Kyoto, Japan). Helium was used as a carrier gas. Analyses were performed using a ZB-5 capillary column (60 m× 0.25 mm, 0.25 µm film thickness; Shimadzu). The temperatures of the injector and source were 250 °C and 200 °C, respectively. The GC oven was programmed as follows: starting temperature 55 °C, increased to 70 °C at a rate of 10 °C/min, increased to 280 °C at a rate of 20 °C/min, and finally held for three minutes. Quantification was performed by selected ion monitoring at m/z 117 for acetic acid, m/z 131 for propionic acid, m/z 145 for butyric acid and isobutyric acid, m/z 159 for valeric acid and isovaleric acid, m/z 173 for 2-EB, m/z 261 for lactic acid, and m/z 289 for succinic acid. The extraction rate and derivatization rate were standardized using 2-EB acid, and quantification was performed using the corresponding external calibration curves.
2.6. Knee joint extract preparation
Knees of hind legs were collected and snap-frozen in liquid nitrogen. The knees were separately weighed then crushed under liquid nitrogen. Total protein was extracted by tissue homogenization by using a RIPA buffer containing protease inhibitor cocktail (nacalai tesque) at 5:1 v:v buffer to knee tissue. Samples were incubated on ice for 1 hour and centrifuged at 10,000 × g for 10 min at 4 °C. The supernatant was collected, and protein concentration was measured using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). Protein concentration was then adjusted to 3 mg ml−1, and samples were stored at−80 °C until use.
2.7. Preparation of synovial fibroblasts and knee joint cells
For the isolation of synovial fibroblasts, synovial tissues from the hind knee joints of DBA/1 J Jms Slc were minced and digested by 2 mg ml−1 type II collagenase (Worthington Biochemical, Lakewood, NJ, ISA) in DMEM (nacalai tesque) for 2.5 h, and then synvial cells were cultured in DMEM containing 20% v/v FCS for 2 days. Cells were subjected to TrypLE Express Enzyme (Thermo Fisher Scientific) digestion according to the manufacturer's instructions. CD45−Thy1+CD11b− cells were sorted as synovial fibroblasts. Sort purified synovial fibroblasts during the fourth to seventh passages were used for the experiments.
For knee joint cell preparation, synovial fluid and tissues from the hind knee joints of CIA mice were collected, minced and digested by 2 mg ml−1 type II collagenase (Worthington Biochemical, Lakewood, NJ, ISA) in DMEM (nacalai tesque) for 2.5 h.
2.8. Flow cytometry
Ileac and inguinal lymph nodes were isolated as DLNs from the site of CII immunization. Single leukocyte suspensions of the spleen, DLNs, and colonic patches (CoPs) were prepared by mechanically disrupting tissues through 100-µm nylon mesh cell strainers (Greiner Bio-One, Kremsmünster, Austria) in 2% FCS RPMI1640 media (nacalai tesque, Kyoto, Japan). Leukocytes were pre-incubated with a monoclonal antibody (mAb) against CD16/32 (93; BioLegend, San Diego, CA, USA) in 2% FCS and 0.1% NaN3 in PBS (nacalai tesque) before surface antigen staining. Follicular T cell staining was performed with mAbs including BUV737-conjugated anti-TCRβ-chain (H57–597; BD Biosciences, Franklin Lakes, NJ, USA), BUV395-conjugated anti-CD25 (PC61; BD Biosciences), Brilliant Violet 510 (BV510)-conjugated anti-CD45 (30-F11; BioLegend), PE-eFluor 610-conjugated anti-Nrp-1 (3DS304M; Thermo Fisher Scientific, Waltham, MA, USA), PE-conjugated anti-CXCR5 (L138D7; BioLegend), and redFluor710-conjugated anti-CD4 (RM4–5; Tonbo Biosciences, San Diego, CA, USA), followed by dead cell staining with Fixable Viability Stain 780 (FVS780; BD Biosciences). The cells were then fixed, permeabilized, and stained with mAbs, including eFluor 450-conjugated anti-Foxp3 (FJK-16 s; Thermo Fisher Scientific) and Alexa Fluor 647-conjugated anti-Bcl-6 (K112–91; BD Biosciences) using a transcription factor buffer set (BD Biosciences). B cell staining was performed with mAbs including BUV737-conjugated anti-CD19 (1D3; BD Biosciences), BUV395-conjugated anti-IgG1 (A85–1; BD Biosciences), BV510-conjugated anti-CD45, PerCP/Cy5.5-conjugated anti-I-A/I-E (M5/114.15.2; BioLegend), PE/Dazzle 594-conjugated anti-IgD (11–26c.2a; BioLegend), APC-R700-conjugated anti-CD95 (Jo2; BD Biosciences), eFluor 660-conjugated anti-GL7 (GL7; Thermo Fisher Scientific), and biotin-conjugated anti-IgG2a (5.7; BD Biosciences), followed by FVS780. For intracellular cytokine detection, DLNs cells (1 × 106) and hind knee joint cells (1 × 105) from CIA mice were resuspended in complete RPMI-1640 medium and stimulated with Cell Activation Cocktail (without Brefeldin A; BioLegend) for 2 h, followed by an additional 4 hour after addition of eBioscience Protein Transport Inhibitor Cocktail (Thermo Fisher Scientific). Stimulated cells were washed with staining buffer, and stained with mAbs includingBUV737-conjugated anti-TCRβ-chain, BV510-conjugated anti-CD45, and redFluor710-conjugated anti-CD4, followed by dead cell staining with FVS780. The cells were then fixed, permeabilized, and stained with mAbs, including Alexa Fluor 488-conjugated anti-IL-17A (TC11–18H10.1; BioLegend), PerCP/Cy5.5-conjugated anti-IFN-γ (XMG1.2; BioLegend) and PE-conjugated RORγt (Q31–378; BD Biosciences) using a transcription factor buffer set (BD Biosciences). Flow cytometry was performed using an LSR II or a FACSCelesta flow cytometer with DIVA v8.0 (BD Biosciences), and data were analyzed using FlowJo version 10.4 (FlowJo, Ashland, OR, USA).
2.9. Cell sorting
For the isolation of naïve T cells, CD4+ cells were enriched from the spleen by negative selection with the iMag cell separation system (BD Biosciences) according to the manufacturer's instructions. Briefly, single-cell suspension was incubate with a mixture of biotin-conjugated mAbs against CD8α (53–6.7; BioLegend), CD11b (M1/70; BioLegend), CD11c (N418; BioLegend), B220 (RA3–6B2; BioLegend), Gr-1 (RB6–8C5; BioLegend), and TER-119 (TER-119; BioLegend) in staining buffer (PBS containing 2% FCS with 2 mM EDTA) for 30 min at 4 °C. Cells were washed with staining buffer and incubated with Streptavidin Particle Plus-DM (BD Biosciences) for 30 min at 4 °C. The enriched CD4+ fraction was stained with 7-AAD (Tonbo Biosciences) and mAbs including BV605-conjugated anti-CD4 (RM4–5; BioLegend), BV510-conjugated anti-mouse CD44 (IM7; BD Biosciences), eFluor 450-conjugated anti-CD62L (MEL-14; Thermo Fisher Scientific), PE-Cy7-conjugated anti-CD25 (PC61; BioLegend), APC-eFluor 780-conjugated anti-NK1.1 (PK136; Thermo Fisher Scientific), and redFluor 710-conjugated anti-CD45 (30-F11; Tonbo Biosciences). Stained cells were subjected to cell sorting using a FACSAria III cell sorter to isolate live CD45+CD4+CD44loCD62hiCD25−NK1.1− naïve T cells.
For the isolation of resting B cells, DLN cells from CIA mice or C57BL/6JJcl mice were stained with 7-AAD and mAbs including BV510-conjugated anti-CD45, BV421-conjugated anti-CD3ε (145–2C11; BioLegend), PE-Cy7-conjugated anti-CD43 (1B11; BioLegend), PE-conjugated anti-IgG1 (A85–1; BD Biosciences), PE-conjugated anti-IgG2a (RMG2a-62; BioLegend), PE-conjugated anti-IgG2b (RMG2b-1; Biolegend), FITC-conjugated-anti-IgG3 (R40–82; BD Biosciences), FITC-conjugated-anti-IgE (RME-1; BioLegend), FITC-conjugated-anti-IgA (C10–3; BD Biosciences), and APC-eFluor 780-conjugated anti-CD19 (eBio1D3; Thermo Fisher Scientific). The stained cells were subjected to cell sorting using a FACSAria III cell sorter to isolate live CD45+CD3ε−CD19+CD43−Ig− resting B cells for in vitro type II collagen (CII)-specific assay and iTFR-cell suppression assay.
For the isolation of follicular T cells from CIA mice, DLN cells were stained with 7-AAD and mAbs, including BV605-conjugated anti-CD4, BV510-conjugated anti-CD45, PE-eFluor 610-conjugated anti-Nrp-1, PE-conjugated anti-CXCR5 (L138D7; BioLegend), APC-eFluor 780-conjugated anti-CD19, PE-Cy7-conjugated anti-ICOS (7E.17G9; Thermo Fisher Scientific), and APC-conjugated anti-GITR (DTA-1; Thermo Fisher Scientific). The stained cells were subjected to cell sorting using a FACSAria III cell sorter to isolate live CD45+CD19−CD4+ICOS+CXCR5+GITR− TFH cells and CD45+CD19−CD4+ICOS+CXCR5+GITR+ TFR cells for the type II collagen (CII)-specific in vitro assay.
For the isolation of follicular T cells from Foxp3hCD2 mice, the enriched CD4+ fraction was stained with 7-AAD and mAbs, including BV605-conjugated anti-CD4, BV510-conjugated anti-CD45, PE-eFluor 610-conjugated anti-Nrp-1, PE-conjugated anti-CXCR5, APC-eFluor 780-conjugated anti-CD19, PE-conjugated ICOS, and APC-conjugated anti-human CD2 (hCD2, RPA-2.10; BioLegend). The stained cells were subjected to cell sorting using a FACSAria III cell sorter to isolate CD45+CD19−CD4+ICOS+CXCR5+hCD2 (Foxp3)− TFH cells, and CD45+CD19−CD4+ICOS+CXCR5+hCD2+ TFR cells for in vitro iTFR-cell suppression assay.
2.10. In vitro T cell differentiation
For iTREG cell polarization, naïve CD4+T cells (5 × 105 cells ml−1) were stimulated with immobilized anti-TCRβ mAb (H57–597; BioXCell, West Lebanon, NH, USA, 5 µg ml−1) on a high-binding 96-well plate (Corning, Inc., Corning, NY, USA) and soluble anti-CD28 mAb (37.51; BioXCell, 2 µg ml−1) in complete advanced RPMI 1640 media (Thermo Fisher Scientific) containing 5% v/v fetal calf serum (FCS; MP Biomedicals, Santa Ana, CA, USA) supplemented with 0.1 ng ml−1 recombinant human (rh) TGF-β1 and 10 ng ml−1 recombinant mouse (rm) IL-2 (both from BioLegend) for two days. The stimulated T cells were expanded in complete media supplemented with 0.1 ng ml−1 rhTGF-β1 and 10 ng ml−1 rmIL-2 for another three days.
For induced follicular regulatory T (iTFR) cell polarization, naïve CD4+T cells (5 × 105 cells ml−1) were stimulated in iTREG cell culture conditions and expanded in complete media supplemented with 2 ng ml−1 rmIL-2, 25 ng ml−1 rmIL-6, 25 ng ml−1 rmIL-21 (both from BioLegend), 5 µg ml−1 anti-ICOS agonistic mAb (C398.4A; BioLegend), 10 µg ml−1 anti-IFN-γ (R4–6A2; BioXCell), and 10 µg ml−1 anti-IL-4 (11B11; BioXCell) with or without 0.1 ng ml−1 rhTGF-β1 for 3 days.
Follicular helper T (TFH) cell was induced using the conventional method with some modification [49]. Briefly, naïve CD4+T cells (5 × 105 cells ml−1) were stimulated with immobilized anti-TCRβ mAb on a high-binding 96-well plate and soluble anti-CD28 mAb in complete advanced RPMI 1640 media containing 5% v/v fetal calf serum supplemented with 25 ng ml−1 rmIL-6 and 10 ng ml−1 rmIL-12 (BioLegend) and 5 µg ml−1 anti-ICOS agonistic mAb, 10 µg ml−1 anti-IFN-γ, and 10 µg ml−1 anti-IL-4 for two days. The stimulated T cells were expanded in complete media supplemented with 25 ng ml−1 rmIL-6, 10 ng ml−1 rmIL-12, 5 µg ml−1 anti-ICOS agonistic mAb, 10 µg ml−1 anti-IFN-γ, and 10 µg ml−1 anti-IL-4 with or without 10 ng ml−1 rmIL-2 for three days.
2.11. In vitro co-culture assay
For in vitro type II collagen (CII)-specific assay, sort purified CD45+CD3ε−CD19+Ig−CD43− resting B cells and CD45+CD19−CD4+ICOS+CXCR5+GITR− TFH cells from DLNs of CIA mice were cultured in complete advanced RPMI 1640 media. B cells (5 × 104) and TFH cells (1.5 × 104) were cultured in 96-well round-bottom plates along with 100 µg ml−1 heat-denatured T-cell grade bovine CII (Chondrex) for six days. For the in vitro iTFR-cell suppression assay, CD45+CD3ε−CD19+Ig−CD43− resting B cells from C57BL/6JJcl mice, CD45+CD19−CD4+ICOS+CXCR5+hCD2 (Foxp3)− TFH cells, and CD45+CD19−CD4+ICOS+CXCR5+hCD2+ TFR cells were purified from Foxp3hCD2 mice by cell sorting. CD4+hCD2+CXCR5+Bcl-6-Eyfp+ iTFR cells were induced in vitro and purified by cell sorting. Resting B cells and TFH cells were cultured alone or with TFR cells or iTFR cells in complete advanced RPMI 1640 media. Resting B cells (5 × 104), TFH cells (1.5 × 104), and/or TFR or iTFR cells (1.5 × 104) were cultured in 96-well round-bottom plates with 2 µg ml−1 anti-CD3ε (145–2C11; BioLegend) and 5 µg ml−1 F(ab′)2 Goat anti-IgM μ chain (Thermo Fisher Scientific) for 6 days. B cells were stained with mAbs, including BUV737-conjugated anti-CD19, PerCP/Cy5.5-conjugated anti-I-A/I-E, and eFluor 660-conjugated anti-GL7, followed by dead cell staining with FVS780. The cells were fixed, permeabilized, and stained with mAbs, including BUV395-conjugated anti-IgG1 and PE-conjugated anti-IgG2a (RMG2a-62; BioLegend), using a transcription factor buffer set (BD Biosciences). Flow cytometry was performed using an LSR II or a FACSCelesta flow cytometer with DIVA v8.0 (BD Biosciences), and data were analyzed using FlowJo version 10.4 (FlowJo).
2.12. Adoptive transfer of Foxp3–hCD2+CD4+t cells
For the isolation of Foxp3+ cells, Foxp3–hCD2+CD4+T cells cultured under iTREG or iTFR conditions were sorted by the positive selection method using the iMag cell separation system (BD Biosciences) with PE-conjugated mAbs against hCD2 (RPA-2.10; BioLegend) and anti-PE magnetic particle (BD Biosciences). Purified Foxp3–hCD2+T cells (5 × 106; Foxp3hCD2 mice on the DBA/1 J background) cultured under iTFR conditions were injected intravenously into CIA mice one week before or after the primary immunization. Purified CD45.1+Foxp3–hCD2+T cells (1 × 106; Foxp3hCD2 mice on the C57BL/6 Jbackground) cultured under iTREG conditions and CD45.2+CD4+T cells isolated from the spleen of C57BL/6 J mice were injected intravenously into Tcrb−/−Tcrd−/− mice and TFR cells were analyzed eight weeks after transfer.
2.13. Cytokine and immunoglobulin measurements
Serum cytokines were measured using the LEGENDplex Mouse Th Cytokine Panel (13-plex) array. Data were collected using an LSR II flow cytometer and analyzed using the LEGENDplex V7.1 (BioLegend). In several experiments, serum cytokines were measured by enzyme-linked immunosorbent assay (ELISA) using Mouse IL-17A ELISA MAX Sets (BioLegend) or the IL-21 ELISA Kit (Thermo Scientific) according to the manufacturers’ protocols. Titres of CII-specific IgG in the serum or culture supernatant were measured by ELISA using the SBA Clonotyping system (SouthernBiotech, Birmingham, AL, USA). Briefly, 96-well Maxisorp Nunc-Immune Plates (Thermo Fisher Scientific) were coated with 5 µg ml−1 ELISA grade bovine CII (Chondrex) at 4 °C overnight. The coated plates were washed five times with PBS containing 0.05% Tween-20 (TPBS) and blocked with TPBS containing 2%w/v globulin-free bovine serum albumin (BSA; Nacalai Tesque) for 1 hour at room temperature. Diluted serum or culture supernatant samples were added and incubated for 2 h at room temperature. The plate was then washed five times with PBS and incubated with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG, IgG1, or IgG2a in TPBS containing 2% BSA for 1 hour at room temperature. The plate was further washed 5 times with TPBS and developed with ABTS 1-Component Microwell Peroxidase Substrate Kit (SeraCare Life Sciences, Milford MA, USA) for 10 min. Absorbance was measured at 405 nm using a ChroMate microplate reader (Awareness Technology, Palm City, FL, USA). OD405 values are presented after the subtraction of blanks.
2.14. Enzyme-Linked immunospot (ELISpot)
CII-specific antibody-forming cells were detected by ELISpot using a Mouse IgG ELISpotBASIC HRP Kit (Cat. #3825–2H; Mabtech, Nacka Strand, Sweden) according to the manufacturer's instructions. Briefly, a MultiScreenHTS IP filter plate (MSIPS4510; Merck, Kenilworth, NJ, USA) was activated with 35% v/v ethanol and then coated with 50 µg ml−1 ELISA grade bovine CII (Cat. # 2012; Chondrex) at 4 °C overnight. The coated plate was washed five times with PBS and blocked with PBS containing 10% v/v FCS for 30 min at room temperature. DLN cells (1.0 × 105) were then seeded and incubated in a 37 °C humidified incubator with 5% CO2 for 20 h. Plates were washed five times with PBS and incubated with 1 µg ml−1 biotin-conjugated anti-mouse IgG in PBS containing 0.5% FCS for 2 h at room temperature. Plates were washed 5 times with PBS and further incubated with streptavidin-HRP (1:500 dilution, in PBS containing 0.5% FCS) for 1 hour at room temperature, washed five times with PBS, and developed with TrueBlue Peroxidase Substrate (SeraCare) for 10 min. The plate was washed extensively with deionized water to stop colour development, air dried, and spots were counted using an ImmunoSpot S4 Analyzer (CTL) or a dissection microscope (Olympus, Tokyo, Japan).
2.15. Immunofluorescence
Ileac LN was frozen in OCT compound in liquid nitrogen and sectioned using a cryostat (Leica, Wetzlar, Germany). 6-μm thick tissue cross-sections were air-dried for 2 h at room temperature, fixed in ice-cold acetone for 10 min, rehydrated in TBS, and blocked with TBS containing 10% v/v normal goat serum and 5 µg ml−1 anti-CD16/32 for 1 hour at room temperature. The sections were then incubated with Alexa Fluor 488-conjugated Lectin PNA from Arachis hypogaea (Thermo Fisher Scientific), eFluor 570-conjugated anti-CD4 (RM4–5; Thermo Fisher Scientific), and Alexa Fluor 647-conjugated anti-IgD (11–26c.2a; BioLegend) in TBS containing 10% v/v normal goat serum at 4 °C overnight. The sections were mounted using ProLong Diamond Antifade Mountant (Thermo Fisher Scientific). Images were acquired using a BZ-9000 fluorescence microscope (KEYENCE, Osaka, Japan), and germinal centre areas were measured using a BZ-II Analyzer.
2.16. Chromatin immunoprecipitation (ChIP)
ChIP assays were performed using the SimpleChIP Enzymatic Chromatin IP Kit (Cell Signaling Technology, Danvers, MA, USA) according to the manufacturer's instructions, with a few modifications. Briefly, CD4+T cells were cultured under iTFR or iTREG conditions and harvested 24 h after a medium change. Cells (3.5 × 106) were fixed with 1% formaldehyde for 10 min at room temperature, and the reaction was stopped by adding glycine. Cells were washed twice and resuspended in ice-cold 1 × ChIP lysis buffer to obtain crude nuclei. Then, nuclei were treated with Micrococcal Nuclease for 20 min at 37 °C with frequent mixing to digest the DNA to 150 – 900 bp; digestion was stopped by adding 0.5 M EDTA. Nuclei were resuspended with 1 × ChIP buffer and sonicated by three sets of 20-s pulses to break nuclear membranes. Anti-acetyl histone H3 antibody or normal rabbit IgG (ChIP-grade; Cell Signaling Technology) was added to the digested, cross-linked chromatin lysate, and samples were incubated overnight at 4 °C with rotation. IP samples were immunoprecipitated with ChIP-Grade protein G magnetic beads (Cell Signaling Technology) for 2 h at 4 °C with rotation. After extensive washing, chromatin was eluted from the antibody/protein G magnetic beads with 1 × ChIP elution buffer for 30 min at 65 °C, and antibody/protein G magnetic beads were removed using a magnetic separation rack. Reverse crosslinking of chromatin samples was performed by treatment with proteinase K for 2 h at 65 °C. DNA was purified using ChIP DNA Clean & Concentrator Kits (Zymo Research, Irvine, CA, USA) according to the manufacturer's instructions. qPCR was performed using KOD SYBR qPCR Mix (TOYOBO Life Science) using promoter-specific primers. Reactions were run on a CFX Connect Real-time PCR System (Bio-Rad Laboratories, Hercules, CA, USA). Data were calculated using the percent input method. Primer sequences were as follows: Bcl6 promoter forward (5′-TGGGTTCGTTTCAAGGTCGT-3′), Bcl6 promoter reverse (5′-ACGCGCAGTATCTGTGATCC-3′); Cxcr5 promoter forward (5′-CAGTGCTTCGTCAGCTCCAGAC-3′), Cxcr5 promoter reverse (5′-CTCAGGTAGTCATGTTTGATGGC-3′); Tcf7 promoter forward (5′-GCCCAGGTGACTGACTAATCC-3′), Tcf7 promoter reverse (5′-ACTTGACAGGGAACAGCGAC-3′); Rpl30 promoter forward (5′-GCGAATGTCTAGCCTGTCCT-3′), Rpl30 promoter reverse (5′-GTTGCCTTGTAGAACACTGCG-3′); Vil1 promoter forward (5′-CCGATGTGCCATTCCTCAGATA-3′), Vil1 promoter reverse (5′-CCTGCATACTTGGGCACACTAA-3′).
2.17. Quantitative real-time PCR
RNA was obtained from synovial fibroblasts using iScript RT-qPCR Sample Preparation Reagent (Bio-Rad Laboratories) according to the manufacturer's instructions, and mRNA was isolated from Foxp3–hCD2+T cells using Dynabeads mRNA DIRECT Kit (ThermoFisher Scientific). cDNA was generated using the iScript Advanced cDNA Synthesis Kit for RT-qPCR (Bio-Rad Laboratories) according to the manufacturer's instructions. Real-time PCR was performed using SsoAdvanced Universal SYBR Green Supermax (Bio-Rad Laboratories) with specific primers. Reactions were run on a CFX Connect Real-time PCR System (Bio-Rad Laboratories). Ct values were determined using the CFX Manager Software (Bio-Rad Laboratories), and gene expression levels were determined according to the dCt method.Rpl32 was used as the housekeeping gene, and iTREG cell was used as the cell of reference. Primer sequences were as follows: Il6 forward (5′-TGATGCACTTGCAGAAAACA-3′), Il6 reverse (5′-ACCAGAGGAAATTTTCAATAGGC-3′); Pdcd1 forward (5′-CATTCACTTGGGCTGTGCT-3′), Pdcd1 reverse (5′-CAGGCTGGGTAGAAGGTGAG-3′); Cxcr5 forward (5′-TGGCCTTCTACAGTAACAGCA-3′), Cxcr5 reverse (5′-GCATGAATACCGCCTTAAAGGAC-3′); Bcl6 forward (5′-AGTTTCTAGGAAAGGCCGGA-3′), Bcl6 reverse (5′-GATACAGCTGTCAGCCGGG-3′); Tcf7 forward (5′-ACATTCTCAGGTCGCTCTGC-3′), Tcf7 reverse (5′-AGGTGTTCTGTGCCTAGCAA-3′); Rpl32 forward (5′-TTCCTGGTCCACAATGTCAA-3′), Rpl32 reverse (5′-GGCTTTTCGGTTCTTAGAGGA-3′).
2.18. Metagenomic 16S rRNA sequencing
A library of 16S rRNA was performed as described previously [50]. Approximately 50 mg of the sample was transferred into 2 mL tubes containing 0.1 mm zirconia/silica beads (BioSpec Products, Bartlesville, OK) and 3.0 mm zirconia beads (Biomedical Sciences, Tokyo, Japan). The stool samples were homogenized at 1,500 rpm for 10 min with Shake Master Neo (Biomedical Sciences, Tokyo, Japan) after adding the Inhibit EX buffer from the QIAamp Fast DNA Stool Mini Kit (Qiagen, Hilden, Germany). Genomic DNA was then extracted using the kit according to the manufacturer's instructions and was resuspended in 10 mM Tris–HCl buffer at 5 ng/μL. Each DNA sample was amplified by polymerase chain reaction (PCR) using KAPA HiFi HS ReadyMix (KAPA Biosystems, Wilmington, MA) and primers specific for variable regions 3 and 4 of the 16S rRNA gene. The PCR products were purified using Agencourt AMPure XP Beads (Beckman Coulter, Brea, CA) and appended by PCR using the Nextera XT index kit (Illumina, San Diego, CA). The libraries were further purified using Agencourt AMPure XP Beads, diluted to 4 nM with 10 mM Tris–HCl buffer, and pooled. The pooled samples were sequenced using the Miseq system (Illumina) with a 2 × 300-base pair protocol. All sequences analyzed in this study were deposited in the DNA Data Bank of Japan (DDBJ) database under the accession number DRA009247.
2.19. Bacterial composition analysis
Bacterial composition analysis was performed as described previously [50]. We trimmed the reads using Trimmomatic ver. 0.36 with the following parameter. We used the join_paired_ends.py QIIME script with the fastq-join method to join paired-end reads and converted the FASTQ files into FASTA files using the split_libraries_fastq.py QIIME script. We trimmed sequencing adaptor sequences using cutadapt and subsequently subsampled the reads to a depth of 9,000 reads per sample using seqtk (https://github.com/lh3/seqtk). Next, we removed chimaera reads using the identify_chimeric_seqs.py and filter_fasta.py QIIME scripts with usearch61. We concatenated the FASTA files of individual samples into one FASTA file and used the de_novo_otus.py QIIME script to pick OTUs. We assigned taxonomy by similarity searching against the publicly available 16S (RDP ver. 10.27 and CORE update 2 September 2012) and NCBI genome databases using BLASTN. The genus-level assignment was performed at 97% sequence identity thresholds. Diversity analyses of the microbiota (α- and β-diversity) were performed using the core_diversity_analyes.py QIIME script at a depth of 6,000 reads per sample.
2.20. Photoconversion of CoPs from KikGR mice
Photoconversion of the CoPs of KikGR mice was performed as described previously [47]. Briefly, KikGR mouse was anaesthetized with isoflurane, shaved with an electric razor, and antiseptically prepared with 10% povidone-iodine. The skin was incised anteriorly at the midline below the costal margin, and then the abdominal wall was incised. Each CoP was sequentially drawn out from the abdominal cavity, and the surgical site was covered by a piece of sterile aluminium foil with a 5 mm hole punched in it to leave only the CoP exposed. A Silver LED 430 with a high numerical aperture polymer optical fibre light guide, and fibre collimator (Prizmatix, Holon, Israel) was used as a 430-nm blue light source. Each CoP was exposed for 2 min and immediately replaced into the peritoneal cavity to avoid drying. The abdominal cavity and skin were closed with 4–0 nylon suture (Natsume Seisakusho, Tokyo, Japan). Photoconverted cells in the ileac lymph nodes were analyzed by a FACSAria III cell sorter(BD Biosciences) one day after exposure.
2.21. Statistical analysis
No statistical methods were used to predetermine the sample size. Prism ver. 8.4 (GraphPad Software) or R statistical environment was used for statistical analysis. Statistical comparisons were performed using the Mann-Whitney test, Welch's t-tests, unpaired two-tailed Student's t-tests, Log-rank tests, ANOVA, Wilcoxon rank-sum tests, orSpearman's correlations. P< 0.05 indicated statistical significance.
3. Results
3.1. Commensal bacteria-derived butyrate ameliorates autoimmune arthritis
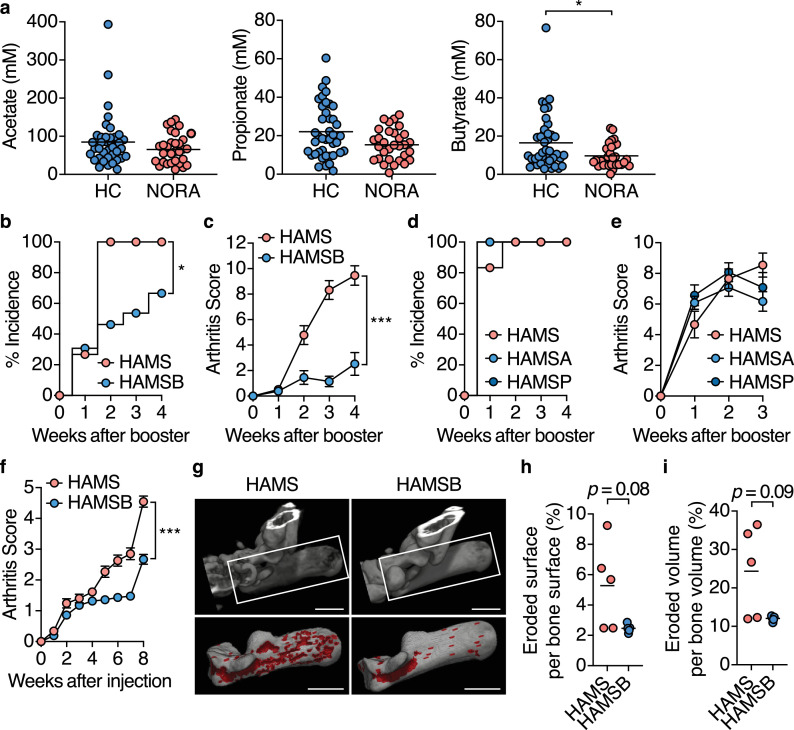
In NORA patients, the intestinal environment, such as the profile of intestinal metabolites, may be altered due to bacterial dysbiosis [29,30]. To better characterize the intestinal environment of NORA patients, we analyzed major organic acids in their stool samples by Gas chromatography-mass spectrometry (GC–MS). We observed that the stool concentrations of butyrate, but not other organic acids, were significantly lower in NORA patients than in healthy control (HC) subjects (Fig. 1a and S1a). To seek gut bacteria contributing to the butyrate production, we assessed correlations between faecal bacterial abundances and butyrate concentration reanalyzing a previously published data set of bacterial composition in HC and NORA patient subjects [30]. The abundance of unclassified Lachnospiraceae and Faecalibacterium showed a positive correlation withthe luminal concentration of butyrate (Figure S1b), both of which were underrepresented in NORA-associated microbiota (Figure S1c).
Fig. 1.
Commensal bacteria-derived butyrate ameliorates autoimmune arthritis
(a) The faecal concentration of acetate, propionate, and butyrate in healthy control and new-onset untreated rheumatoid arthritis (NORA) subjects (n= 41, 31).
(b-e) CIA incidence (b, d) and arthritis scores after booster immunization (c, e) in CIA mice fed the control HAMS, or HAMSB diet (n= 13–15; mean ± s.e.m. b, c), and CIA mice fed the control HAMS, HAMSA or HAMSP diet (n= 12; mean ± s.e.m. d, e). AUC was calculated in each mice for c, e.
(f) Arthritis scores for SKG mice fed the control HAMS or HAMSB diet after laminarin injection (n= 11; means ± s.e.m.). AUC was calculated in each mice.
(g–i) Micro-computed tomography (mCT) analysis of the calcaneus in ankle joint four weeks after booster immunization in CIA mice fed the control HAMS or HAMSB diet (n= 5). Representative mCT images (g). The eroded area is shown in Red. Scale bars: 1 mm. Eroded surface per bone surface (h,%) and eroded volume per bone volume (i,%).
Results show one representative experiment of at least two experiments for mouse study. *P< 0.05, **P< 0.01, ***P< 0.001 (A, Mann-Whitney test; c, f, h, i, Welch's t-test or unpaired two-tailed Student's t-test; b, d, Log-rank test; e, one-way ANOVA followed by Tukey's post-hoc test.
We and others previously reported that butyrate and, to a lesser extent, propionate facilitate the differentiation of peripherally generated Foxp3+ regulatory T (pTREG) cells in the colon to prevent inflammatory responses against the intestinal microbiota [38,51,52]. Furthermore, SCFAs, including acetate, propionate, and butyrate, have been reported to ameliorate autoimmune symptoms in a non-obese diabetic model and experimental autoimmune encephalomyelitis [33,53]. We, therefore, hypothesized that commensal microbe-derived SCFAs regulates local inflammatory responses as well as the systemic autoimmune response and that reduced SCFAs production is involved in RA pathogenesis. To test this hypothesis, we fed DBA/1J mice with an AIN-93G low-fibre diet containing high-amylose maize starch (HAMS), a resistant starch, esterified with acetate (HAMSA), propionate (HAMSP) or butyrate (HAMSB), which explicitly increases luminal concentrations of the corresponding SCFAs [38,54] and immunized CII to induce CIA development (Figure S2a-S2c). The HAMSB diet markedly suppressed the incidence and severity of arthritis (Fig. 1b and c).
In contrast, neither HAMSA nor HAMSP diet suppressed CIA development (Fig. 1d and e). The protective effect of the HAMSB diet on arthritis was also evident in SKG mice that develop T cell-mediated autoimmune arthritis (Fig. 1f).Micro-computed tomography of the calcaneus in the ankle joints also confirmed a reduction tendency in bone destruction in CIA mice fed the HAMSB diet (Fig. 1g-i). These data underscore the significance of commensal bacteria-derived butyrate, but not other SCFAs, in protecting against systemic autoimmunity leading to RA.
We further analyzed the microbial communities of HAMS and HAMSB-fed mice before the onset of the CIA to examine whether the protective effect of the HAMSB diet is attributable to alternation in the diversity and composition of the commensal microbiota. CII immunization gradually reduced operational taxonomic units (OTUs) and Chao1 richness estimator over three weeks before the booster immunization in both groups to a similar extent (Figure S3a and S3b). Moreover, principal coordinate analysis (PCoA) based on weighted UniFrac manifested that HAMS- and HAMSB-fed mice possess similar microbial communities (Figure S3c). Previous studies have demonstrated that bacterial genus Lactobacillus and Prevotella are implicated in arthritis development [30,55,56]. However, the prevalence of these genera was similar between the two groups (Figure S3d). Meanwhile, the intake of the HAMSB diet moderately increased the bacterial family Lachnospiraceae during the early phase of the CIA, while genus Bacteroides decreased 1.5-week after the first immunization. These data illustrated that the HAMSB-feeding exhibits a minor effect on the microbial community. It is therefore conceivable that the protective effect of the HAMSB-feeding on the CIA mainly results from the increment of luminal butyrate. However, we do not formally exclude the possibility that the minor alteration of the gut microbial community might also contribute to the alleviation of the CIA.
3.2. Butyrate inhibits CII-specific autoimmune responses in CIA mice
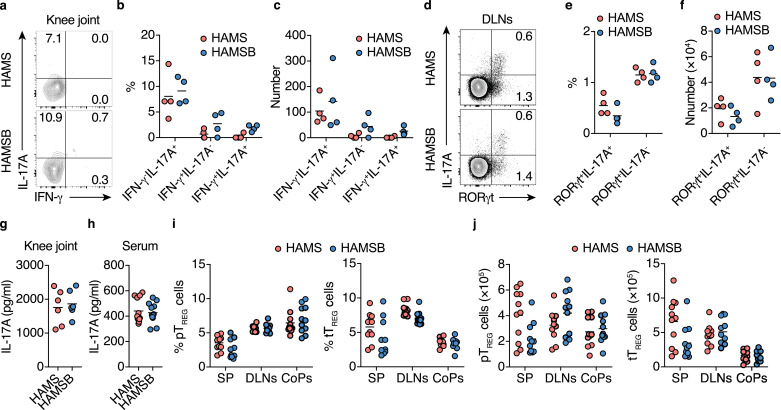
We subsequently explored the molecular mechanism by which butyrate suppresses systemic autoimmunity. Interleukin-17 (IL-17)-producing T helper (TH17) cells and serum IL-17A levels increase in patients with RA [57]. Since TH17 cells coordinate joint inflammation and induce bone destruction in the CIA model, we investigated the effect of butyrate on the local Th17 response. Unexpectedly, the HAMSB diet did not affect the frequency and number of TH17 cells in hind knee joints and their DLNs or the serum and joint tissue concentrations of IL-17A (Fig. 2a-h). We next analyzed pTREG cells and thymus-derived TREG (tTREG) cells according to the flow cytometry gating strategy shown in Figure S4.
Fig. 2.
HAMSB diet does not alter the TH17 response in CIA mice
(a–c) IFN-γ+ and IL-17A+ producing CD4+T cells in the hind knee joints of CIA mice fed the control HAMS or HAMSB diet two weeks after the initial immunization. Cells were restimulated with PMA and ionomycin for 6 h before staining. Representative flow cytometry contour plots of intracellular IFN-γ and IL-17A staining within CD4+TCRβ+ gate (a), and the frequency (b) and total number (c) of IL-17A+, IFN-γ+, and IFN-γ+IL-17A+ cells (n= 4).
(d–f) RORγt+ and IL-17A+ cells in the draining lymph nodes (DLNs) of CIA mice fed the control HAMS or HAMSB diet two weeks after the initial immunization. Cells were restimulated with PMA and ionomycin for 6 h. Representative flow cytometry contour plots of intracellular RORγt and IL-17A staining within CD4+TCRβ+ gate (d), and the frequency (e) and total number (f) of RORγt+IL-17A+ and RORγt−IL-17A+ cells (n= 4).
(g, h) IL-17A concentration in the hind knee joint extract (g, n= 6) and serum (h, n= 12) of CIA mice fed the control HAMS or HAMSB diet three weeks after the booster immunization.
(i, j) The frequency in CD4+T cells (i) and total number (j) of Nrp-1−Foxp3+ pTREG and Nrp-1+Foxp3+ tTREG cells in the spleen (SP), DLNs and colonic patches (CoPs) of CIA mice fed control HAMS or HAMSB diet two weeks after the initial immunization (n= 10). Gating strategy is depicted in Figure S4.
Results show one representative experiment of at least two experiments. (Welch's t-test or unpaired two-tailed Student's t-test).
Similarly, the HAMSB diet failed to increase the frequency and number of pTREG cells tTREG cells in the spleen, DLNs, and colonic patches (CoPs), which are the GALT in the colon, two weeks after the initial CII immunization (Fig. 2i and j). We previously found that approximately 100 µM butyrate is required to induce the differentiation of pTREG cells [38]. Although the butyrate concentration in colonic tissue exceeds this level in CIA mice fed the HAMSB diet, serum butyrate is maintained at much lower levels under physiological conditions; this was also true in the HAMSB group (Figure S2b, S2c and Furusawa et al. 2013 [38]). Of note, physiological concentrations of butyrate did not directly inhibit IL-6 production by synovial fibroblasts (Figure S5a and S5b), which induces local TH17 responses [58]. Collectively, the protective effect of butyrate on CIA development was not attributed to the regulation of the TH17/TREG cell balance.
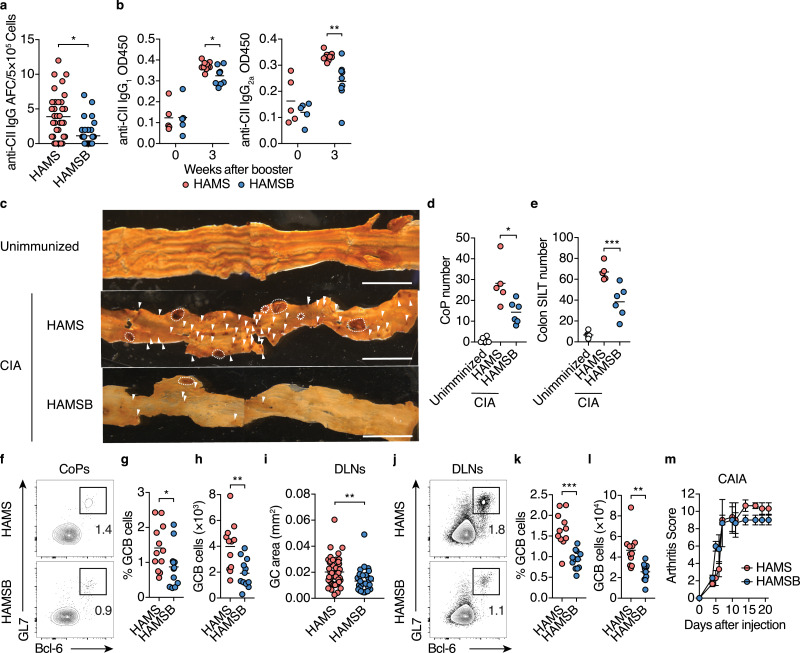
In the CIA mouse model, high titers of CII-specific IgG are essential for disease onset [59,60]. We detected a significant decrease in total IgG1+ and IgG2a+B cells, as well as CII-specific IgG-forming cells in DLNs of CIA mice fed the HAMSB diet than in mice fed with the control HAMS diet two weeks after initial immunization (Fig. 3a and S5c). Consistent with these observations, the levels of CII-specific total IgG, IgG1, and IgG2a in the serum and of CII-specific and IgG2a in knee joint extracts were significantly lower in the HAMSB diet group three weeks after booster immunization (Fig. 3b, S5d and S5e). CII-specific IgG formation depends on the GC reaction [12]. Strikingly, we found that systemic autoantigen immunization caused hypertrophy of CoPs and solitary intestinal lymphoid tissue (SILT) in the colon with the expansion of the GC reaction one week after the initial CII immunization (Fig. 3c-h). These data suggested that autoimmune GC reactions precede GALT formation, leading to autoantibody production at systemic lymphoid sites.
Fig. 3.
Butyrate suppresses the GC response and autoantibody production in CIA mice
(a, b) CII-specific IgG responses in CIA mice fed the control HAMS or HAMSB diet. The number of CII-specific IgG antibody-forming cells (AFC) cells per 5 × 105 DLN cells three weeks after booster immunization (a, each dot indicates data from 1 well, n= 36 from 6 mice). Statistical analysis was performed on the mean of each mouse. Serum levels of CII-specific IgG1 and IgG2a upon or three weeks after booster immunization (b, n= 5–10).
(c–e) Colonic patches (CoPs) and solitary intestinal lymphoid tissues (SILTs) of CIA mice fed the control HAMS or HAMSB diet. Representative whole-mount B220 immunostaining (brown) of colon tissues (c), and the number of CoPs (d) and SILTs (e) in unimmunized DBA1/J mice and CIA mice one week after the initial immunization (n= 5 or 6).Dashed line, CoP, arrowheads, SILT. Scale bars, 5 mm.
(f–h) GCB cells in CoPs from CIA mice fed the control HAMS or HAMSB diet. Representative flow cytometry contour plots of GL7 and intracellular Bcl-6 staining within CD19+ gate (f), and the frequency (g) and total number (h) of Bcl-6+GL7+ GCB cellstwo weeks after the initial immunization (n= 12).
(i) GC area (mm2) of DLNs from CIA mice fed the control HAMS, or HAMSB diet Data were obtained two weeks after the initial immunization. Each dot indicates a single GC area of DLNs on cross-sections (n= 43, 30). Statistical analysis was performed on the mean of each mouse.
(j–l) GCB cells in DLNs from CIA mice fed control HAMS or HAMSB diet. Representative flow cytometry contour plots of GL7 and intracellular Bcl-6 staining within CD19+ gate (j), and the frequency (k) and total number (l) of Bcl-6+GL7+ GCB cellstwo weeks after the initial immunization (n= 12).
(m) Arthritis scores after antibody cocktail injection of collagen antibody-induced arthritis (CAIA) mice fed control HAMS or HAMSB diet (n= 3; means ± s.e.m.). AUC was calculated for each mice.
Results show one representative experiment of at least two experiments. *P< 0.05, **P< 0.01, ***P< 0.001 (a, b, g–i, k, l, Welch's t-test or unpaired two-tailed Student's t-test; d, e, one-way ANOVA followed by Tukey's posthoc test; m, Log-rank test).
Nonetheless, the HAMSB diet conspicuously alleviated the hypertrophy of CoPs and SILTs (Fig. 3c-e). In addition to the reduction in CoP size and number, the frequency and number of GCB cells in CoPs of the HAMSB-fed mice were declined at two weeks after the initial immunization (Fig. 3f-h). Thus, butyrate prominently suppresses the autoantigen-driven GC development in the GALT. Furthermore, GC size and the frequency and number of GC B cells were also significantly reduced in the DLNs of CIA mice fed the HAMSB diet (Fig. 3i-l).
We further evaluated whether butyrate attenuated joint inflammation caused by the IgG-antigen immune complexes in a collagen antibody-induced arthritis (CAIA) model by injecting a cocktail of 5 monoclonal antibodies to CII [61]. In contrast with the observations in the CIA model, the HAMSB diet did not affect the development of CAIA (Fig. 3m), indicating that butyrate prevents the development of arthritis by diminishing autoantibody production rather than the immune complex-induced inflammation. This notion was supported by the fact that HAMSB failed to protect against the CIA after a booster immunization (Figure S5f and S5g). These observations show that the butyrate suppresses CII-specific IgG production by limiting GC development at the priming phase of CIA.
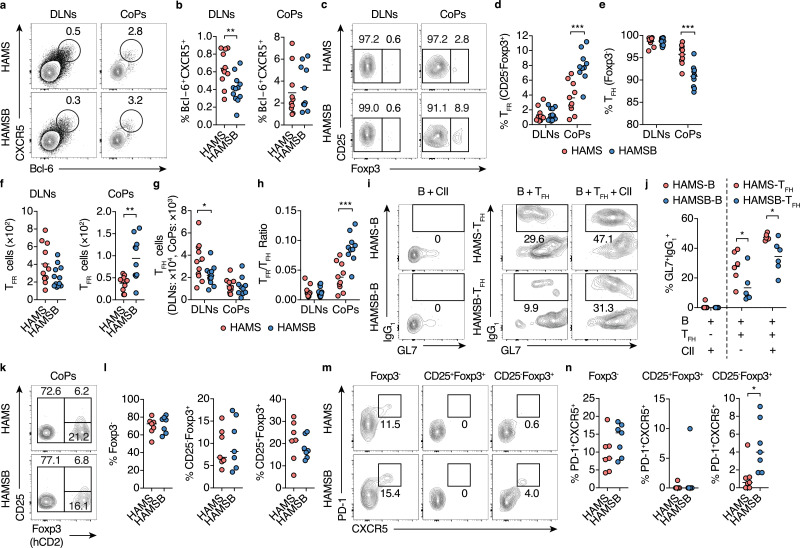
3.3. Butyrate increases Tfrcells in CoP in CIA mice
The magnitude and quality of the GC reaction are regulated by the balance between TFH cells and TFR cells [[15], [16], [17],[20], [21], [22],24], so we analyzed the abundance of these follicular T cell subsets. HAMSB prominently increased the frequency and number of CD25−Foxp3+CXCR5+PD-1+TFR cells, and thus elevated the TFR/TFH ratio by two-fold in CoPs at two weeks after the initial immunization (Fig. 4a-h). Meanwhile, the increase in TFR cells was not observed in DLNs (Fig. 4c-h). Nonetheless, TFH cells isolated from DLNs of CIA mice fed HAMSB diet exhibited attenuated induction of B cells to IgG1 class switching in the presence of CII, which reflects a decrease in CII-specific TFH cells in DLNs (Fig. 4i and j). Similar to the observation in the CIA model, the adoptive transfer of in vitro induced Foxp3+ TREG (iTREG, CD45.1+) cells with CD4+T cells (CD45.2+) into Tcrb−/−Tcrd−/− mice fed the HAMSB diet markedly increased CD45.1+CD25−Foxp3+CXCR5+PD-1+ TFR cells in CoP (Fig. 4k-n). Meanwhile, HAMSB did not affect TFH cells among CD45.1+ and CD45.2+ cells in CoPs (Fig. 4k-n). Based on these observations, we considered that butyrate directly induced the differentiation of TFR cells from TREG cells in CoPs, and butyrate-induced TFR cells limited the generation of autoreactive TFH and GCB cells in the gut and eventually reduced the provision of autoreactive cells to the DLNs. In support of this notion, a cell tracking study using photoconvertible KikGR transgenic mice demonstrated a substantial follicular Tand B cell migration from CoPs to a DLN (Figure S6a-c).
Fig. 4.
Butyrate increased TFR cells in CoPs
(a, b) Follicular T cells in the DLNs and CoPs from CIA mice fed the control HAMS or HAMSB diet two weeks after the initial immunization. Representative flow cytometry contour plots of CXCR5 and intracellular Bcl-6 staining (a), and the frequency (b) of Bcl-6+CXCR5+ follicular T cells within CD4+ TCRβ+ gate (n= 11, 10).
(c–g) TFR and TFH cells in the DLNs and CoPs from CIA mice fed the control HAMS or HAMSB diet two weeks after the initial immunization. Representative flow cytometry contour plots of CD25 and intracellular Foxp3 staining (c), and the frequency of CD25−Foxp3+ TFR (d) and Foxp3− TFH (e) cells within Bcl-6+CXCR5+ follicular T cell gate and the total number of CD25−Foxp3+ TFR (f) and Foxp3− TFH (g) cells (n= 11, 10). Gating strategy is depicted in Figure S4.
(h) TFR/TFH ratio calculated using the total number of CD25−Foxp3+ TFR and Foxp3− TFH cells in F and G.
(i, j) B cell class switching to IgG1 by TFH cells. IgG−CD19+B cells sort-purified from the DLNs of CIA mice fed the control HAMS or HAMSB diet (described as HAMS-B or HAMSB-B respectively) two weeks after the initial immunization were cultured alone with 100 µg/ml type II collagen (CII),and IgG−CD19+B cells sort-purified from the DLNs of CIA mice fed the control HAMS diet were co-cultured with CXCR5+ICOS+CD4+ TFH cells sort-purified from DLNs of CIA mice fed the control HAMS or HAMSB diet (described as HAMS-TFH or HAMSB-TFH respectively) in the presence or absence of CII. Representative flow cytometry contour plots of GL7 and intracellular IgG1 staining six days after cultivation (i, n= 6), and the frequency of GL7+IgG1+B cells within CD19+ gate (j, n= 6).
(k–n) Differentiation of TFR and TFH cells in CoPs of Tcrb−/−Tcrd−/− mice transferred with Foxp3-hCD2+iTREG cells. Sort-purified CD45.1+Foxp3-hCD2+T cells cultured for five days under iTREG conditions were injected intravenously with CD45.2+CD4+T cells into Tcrb−/−Tcrd−/− mice fed the control HAMS or HAMSB diet. Representative flow cytometry contour plots of hCD2 (Foxp3) and CD25 staining (k), and the frequency of Foxp3−, CD25+Foxp3+ and CD25−Foxp3+ cells (L) within CD45.1+CD4+ TCRβ+ gate (n= 7). Representative flow cytometry contour plots of CXCR5 and PD-1 staining among Foxp3−, CD25+Foxp3+ and CD25−Foxp3+ gate (m), and the frequency of CXCR5+PD-1+ cells (N) among Foxp3− (TFH cells), CD25+Foxp3+ (CD25+TFR cells) and CD25−Foxp3+(CD25−TFR cells) gates (n, n= 7).
Results show one representative experiment of at least two experiments. *P< 0.05, **P< 0.01, ***P< 0.001 (b, d–h, j, l, n, Welch's t-test or unpaired two-tailed Student's t-test).
3.4. Butyrate promotes Tfrcell differentiation thorough HDACi activity in vitro
Considering that butyrate decreased TFH cells in CIA mice, we wondered if butyrate directly suppressed the differentiation of TFH cells. To test this, we analyzed the expression of Bcl-6 and CXCR5 using in vitro TFH cell culture system [49]. The treatment of butyrate, however, did not affect Bcl-6+CXCR5+ TFH cells (Figure S7a and S7b). Thus, we hypothesized that butyrate induces the differentiation of TFR cells, thereby resulting in the reduction of TFH cells.
TFR cells can be subdivided into CD25+ and CD25− subpopulation without the difference in Foxp3 expression. Since the upregulation of PD-1, CXCR5, and Bcl-6 is associated with a loss of CD25 expression, CD25− TFR subpopulation is considered as a well-differentiated TFR cell [20,21]. Interestingly, in the present study, we found that butyrate increased CD25− TFR but not CD25+ TFR cells (Fig. 4f and n). To rigorously confirm whether butyrate directly induces the differentiation of CD25− TFR-cell differentiation, we newly established an in vitro TFR (iTFR)-cell-inducing culture system (Fig. 5a). We first analyzed the expression of TFR signature genes such as Pdcd1, Cxcr5, Bcl6, and Tcf7 in total Foxp3+ cells isolated from iTFR-cell and iTREG-cell cultures [[20], [21], [22],62]. Foxp3+ cells from the iTFR-cell culture express significantly higher levels of Pdcd1, Cxcr5, Bcl6, and Tcf7 compared to those from iTREG-cell culture (Fig. 5b). Flow cytometric analysis identified distinct CD25+Foxp3+ and CD25−Foxp3+ subpopulations in iTFR-cell culture, while the majority of the cells were CD25+Foxp3+ in iTREG-cell culture (Fig. 5c and d). We confirmed that both CD25+Foxp3+ and CD25−Foxp3+ subpopulations in iTFR-cell culture expressed PD-1, CXCR5, Bcl-6, and TCF-1 at a higher level than the corresponding iTREG subpopulation based on the binding of specific antibodies as well as Bcl-6-tdTomato reporter expression (Fig. 5e-h). Consistent with the previous reports, CD25−Foxp3+ subpopulation expressed higher levels of CXCR5 and Bcl-6 compared to CD25+Foxp3+ subpopulation in iTFR-cell culture, although PD-1 expression was similar between the two subpopulations (Fig. 5e-h). We also found that CD25−Foxp3+ subpopulation exhibited higher TCF-1 expression than CD25+Foxp3+ subpopulation (Fig. 5e and f). To examine the suppressive functions of iTFR cells, we isolated CD25−Foxp3+CXCR5+Bcl-6-Eyfp+ cells from iTFR-cell culture and mouse lymph nodes and cocultured with mouse lymph node-derived TFH and IgG−B cells. We observed that iTFR cells prominently suppressed B-cell survival and IgG class switching governed by TFH cells at a similar extent to in vivo generated TFR cells (Fig. 5i-l).
Fig. 5.
In vitro TFR (iTFR) cell differentiation
(a) Schematic of iTFR and iTREG cell differentiation culture.
(b) Relative mRNA expression of Pdcd1, Cxcr5, Bcl6, and Tcf7 in sort-purified TCRβ+CD4+Foxp3-hCD2+ cells fro, iTFR or iTREG cell culture conditions (n= 5). Sort-purified naïve CD4+T cells from Foxp3hCD2reporter mice were used for the culture.
(c, d) Expression of CD25 and Foxp3 by cells from iTFR or iTREG cultures. Representative flow cytometry contour plots of CD25 and Foxp3-hCD2 staining (c), and the frequency of CD25+Foxp3+ and CD25−Foxp3+ cells (d) within CD4+ TCRβ+ gate (n= 5).
(e, f) Expression of TFR cell phenotypic markers by cells from iTFR or iTREG cultures. Representative flow cytometry histograms of FSC, PD-1, CXCR5, Bcl-6, and TCF-1 expression (e), and the gMFI of PD-1, CXCR5, Bcl-6, and TCF-1 (f) within CD25+Foxp3+ and CD25−Foxp3+ gate (n= 5).
(g, h) Expression of Bcl-6-tdTomato reporter by cells from iTFR or iTREG cultures. Sort-purified naïve CD4+T cells from Bcl-6-tdTomato Foxp3hCD2 double reporter mice or Foxp3hCD2reporter mice (described as control) were used for the culture. Representative flow cytometry histograms of Bcl-6-tdTomato reporter and control expression (g), and Bcl-6-tdTomato gMFI (h) within CD25+Foxp3+ and CD25−Foxp3+gate (n= 5).
(i–l) B cell class switching to IgG1 and IgG2a in suppression assays. IgG−CD43−CD19+ resting B cells, Foxp3-hCD2−CXCR5+Bcl-6-Eyfp+ TFH cells, and CD25−Foxp3-hCD2+CXCR5+Bcl-6-Eyfp+ iTFR cells sort-purified from Bcl-6-Eyfp Foxp3hCD2 double reporter mice were used.The resting B cells from were co-cultured with TFH cells alone, TFH and TFR cells, or TFH and sort-purified CD25−Foxp3-hCD2+CXCR5+Bcl-6-Eyfp+ iTFR cells under the exsitance of 5 μg ml−1 anti-IgM and 2 μg ml−1 anti-CD3ε Abs. Representative flow cytometry histograms of Fixable Viability Stain 780 (FVS780) staining (i), and the frequency of FVS780− live cells (j) within GL7+CD19+B cells (n= 3–5). Representative flow cytometry histograms of intracellular IgG1 and IgG2a staining (k), and the frequency of IgG1+ and IgG2a+ cells (l) within GL7+ CD19+B gate (n= 3–5).
Results show one representative experiment of at least two experiments.***P< 0.001 (b, d, Welch's t-test or unpaired two-tailed Student's t-test; j, l, one-way ANOVA followed by Dunnett's post-hoc test; g, h, two-way ANOVA followed by Sidak's post-hoc test).
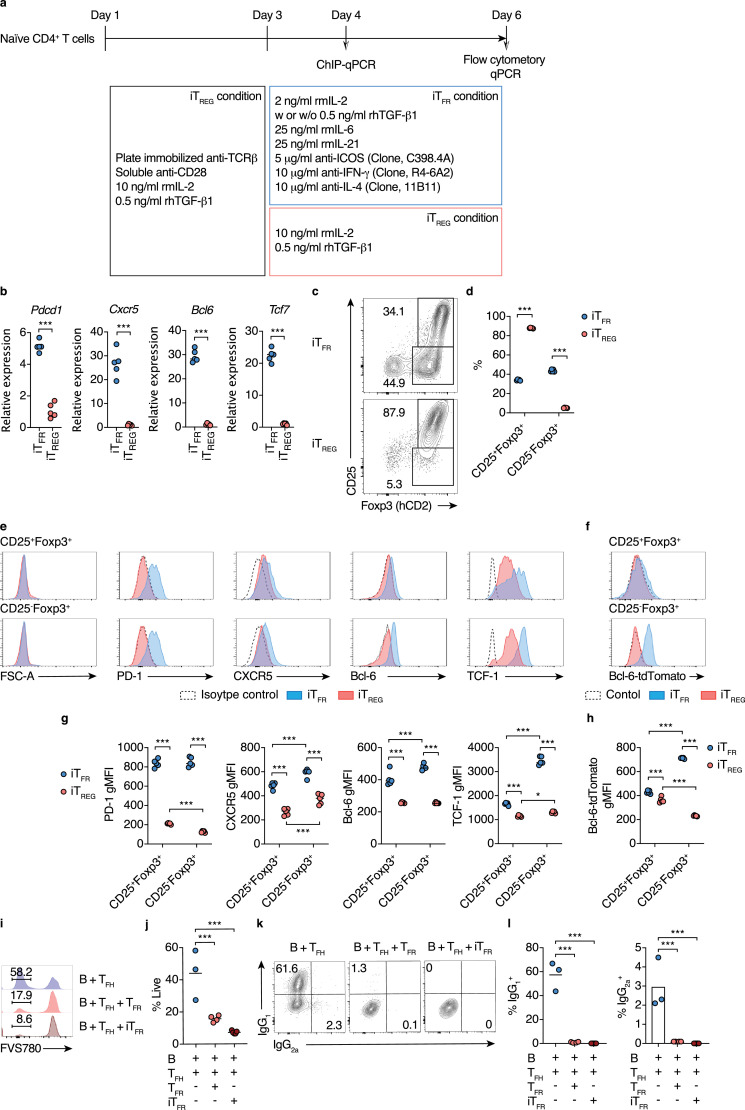
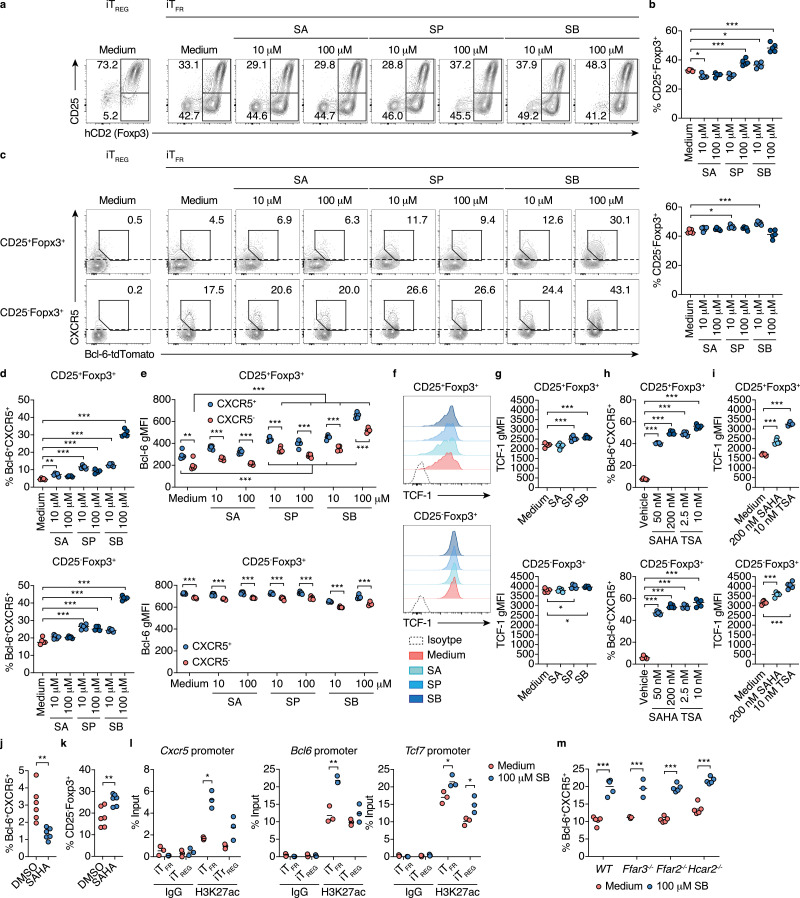
Using the iTFR-cell culture, we examined whether butyrate promotes the differentiation of CXCR5+Bcl-6+ iTFR cells. The treatment of sodium butyrate (SB) for five days increased CD25+Foxp3+ but not CD25−Foxp3+ population within CD4+T cells in a dose-dependent manner at physiological concentrations in the colonic tissue (Fig. 6a and b)[38]. Importantly, consistent with thein vivo observations, 100 μM SB treatment significantly promoted the differentiation into CXCR5+Bcl-6+ iTFR cells in the CD25−Foxp3+ population (Fig. 6c and d). Moreover, we also found that 100 μM SB treatment increased CXCR5+Bcl-6+ iTFR cells within the CD25+Foxp3+ population (Fig. 6c and d). Sodium propionate (SP) treatment also promoted the differentiation of CXCR5+Bcl-6+ iTFR cells among CD25+Foxp3+ and CD25−Foxp3+ populations to a lesser extent (Fig. 6c and d). In both CD25+Foxp3+ and CD25−Foxp3+ populations, CXCR5+ cells (over the dotted line in Fig. 6c) expressed a higher level of Bcl-6 compared to CXCR5− cells (less than the dotted line), indicating the parallel increase in these TFR-cell signature molecules (Fig. 6e). Additionally, SB and SP also slightly but significantly upregulated TCF-1 expression in both CD25+Foxp3+ and CD25−Foxp3+ populations (Fig. 6f and g). To test if butyrate directly promotes the differentiation of CXCR5+Bcl-6+ iTFR cells from Foxp3+ TREG cells, we added SB into or removed SB from the iTFR-cell culture on day three during 5-day culture (Figure S8a). The treatment of SB for the entire period of culture (day 1-day 6) exerted the maximal effect in the induction of CXCR5+Bcl-6+ iTFRcells in both CD25+Foxp3+ and CD25−Foxp3+ populations (Figure S8b and S8c). Importantly, SB treatment during the later stage of culture (day 3-day 6) was able to promote the differentiation of CXCR5+Bcl-6+ iTFRcells in the CD25−Foxp3+ populations, whereas SB treatment only at the early stage of culture (day 1-day 3) showed a moderate effect on promoting the differentiation CXCR5+Bcl-6+ iTFR cells in the CD25−Foxp3+ population(Figure S8b and S8c). These data suggest that butyrate directly promotes the differentiation of CXCR5+Bcl-6+ iTFR cells from TREG cells. However, the pretreatment of butyrate during iTREG-cell culture conditions is required for the maximum induction. Intriguingly, SB treatment conspicuously upregulated CXCR5 expression in both CD25+Foxp3+ and CD25−Foxp3+ populationseven under iTREG-cell culture conditions, although iTreg cells did not express Bcl-6 (Figure S9a, S9b, and 6c).
Fig. 6.
Butyrate induces iTFR cell differentiation
(a-d) iTFR cell differentiation with SCFAs treatment. Sort-purified naïve CD4+T cells from Bcl-6-tdTomato Foxp3hCD2 double reporter mice were cultivated under iTFR-cell culture conditions in the presence of sodium acetate (SA), sodium propionate (SP), or sodium butyrate (SB) at 10 μM or 100 μM. Representative flow cytometry contour plots of hCD2 (Foxp3) and CD25 staining (a), and the frequency of CD25+Foxp3+ (upper panel) and CD25−Foxp3+ (lower panel) cells within CD45+CD4+ TCRβ+ gate (b, n= 5). Representative flow cytometry contour plots of Bcl-6-tdTomato reporter signal and CXCR5 staining among CD25+Foxp3+ and CD25−Foxp3+ gate (c), and the frequency of Bcl-6-tdTomato+CXCR5+ cells among CD25+Foxp3+ (upper panel) and CD25−Foxp3+(lower panel) gates in (b) (d, n= 5).
(e) gMFI of Bcl-6-tdTomato reporter in CXCR5+ and CXCR5− populations among CD25+Foxp3+(upper panel) and CD25−Foxp3+(lower panel) gates of iTFRculture in (c). CXCR5+and CXCR5− populations are shown as areas equal to or higher than, or less than the dotted lines in (c), respectively (n= 5).
(f, g) Expression of TCF-1 among CD25+Foxp3+(upper panel) and CD25−Foxp3+(lower panel) gates of iTFRculture in (c). iTFRculture was treated with 100 μM SA, SP or SB. Representative flow cytometry histograms of TCF-1 expression (f), and the gMFI of TCF-1 (g, n= 5).
(h, i) iTFR cell differentiation with pan-HDAC inhibitor treatment. Sort-purified naïve CD4+T cells from Bcl-6-tdTomato Foxp3hCD2 double reporter mice were cultivated under iTFR-cell culture conditions with the treatment of suberoylanilide hydroxamic acid (SAHA, 50 or 200 nM), trichostatin A (TSA, 2.5 or 10 nM), or vehicle control 0.01% dimethyl sulfoxide (DMSO). The frequency of Bcl-6-tdTomato+CXCR5+ cells (h) and the gMFI of TCF-1 (i) among CD25+Foxp3+CD4+ TCRβ+ (upper panel) and CD25−Foxp3+CD4+TCRβ+(lower panel) gates (n= 5).
(j, k) Differentiation of TFR cells in the inguinal lymph node (InLN) of mice treated with SAHA. C57BL/6 J mice were subcutaneously immunized with human insulin and intravenously injected with SAHA (20 mg/kg body weight) or vehicle control DMSO every day after immunization. The frequency of Bcl-6+CXCR5+ follicular T cells within CD4+ TCRβ+ gate (j, n= 6), and CD25+Foxp3+ TFR cells within Bcl-6+CXCR5+ gate (k, n= 6) from InLN 10 days after immunization.
(l) Accumulation of H3K27 acetylation (ac) at the promoter region of TFRcell-related genes in Foxp3+ cells from iTFR or iTREG cultures. ChIP quantitative RT-PCR (qPCR) analysis of H3K27ac levels in the promoters of Bcl6, Cxcr5, and Tcf7 in sort purified Foxp3+T cells cultured for the total three days under iTFR or iTREG-cell polarizing conditions (see Fig. 5a) with or without the treatment of 100 µM SB.
(m) iTFR cell differentiation using G-protein coupled receptor-deficient mice. Sort-purified naïve CD4+T cells from Ffar3−/−, Ffar2−/−, or Hcar2−/−mice were cultivated under iTFR-cell culture conditions with or without the treatment of 100 µM S. The frequency of Bcl-6+CXCR5+ cells within CD25−Foxp3+CD4+ TCRβ+gate (n= 5).
Results show one representative experiment of at least two experiments.*P< 0.05, **P< 0.01, ***P< 0.001 (b, c, d, f-h, one-way ANOVA followed by Dunnett's post-hoc test; l-m Welch's t-test or unpaired two-tailed Student's t-test).
Butyrate functions as an HDAC inhibitor (HDACi) and a ligand for G-protein coupled receptors (Gpr) 41, 43, and 109a, and both pathways contribute to the differentiation and functional maturation of pTREG cells [38,52,63]. We found that pan-HDACi, i.e., suberoylanilide hydroxamic acid (SAHA, Vorinostat) and trichostatin A (TSA) also promoted the differentiation of CXCR5+Bcl-6+ iTFR cells in a dose-dependent manner (Fig. 6h). SAHA and TSA also increased TCF-1 expression (Fig. 6i).Moreover, in vivo SAHA treatment increased CD25− TFR cells within CXCR5+Bcl-6+ cells with the reduction of TFH cells in the DLN of mice immunized with an autoantigen (Fig. 6j and k). On the other hand, the treatment of anacardic acid, a histone acetyltransferase inhibitor (HATi), which promotes histone deacetylation, nullified butyrate-driven CXCR5+Bcl-6+ iTFR cell differentiation (Figure S10a). Histone H3K27 was highly acetylated at the promoter regions of Cxcr5,Bcl6, and Tcf7 upon butyrate exposure (Fig. 6l and S10b). In contrast, Gpr41, 43, and 109a are dispensable for iTFR cell induction by butyrate (Fig. 6m). Thus, our results unequivocally revealed that butyrate facilitates the differentiation of CXCR5+Bcl-6+ iTFR cells by upregulating TFR cell signature molecules via histone acetylation.
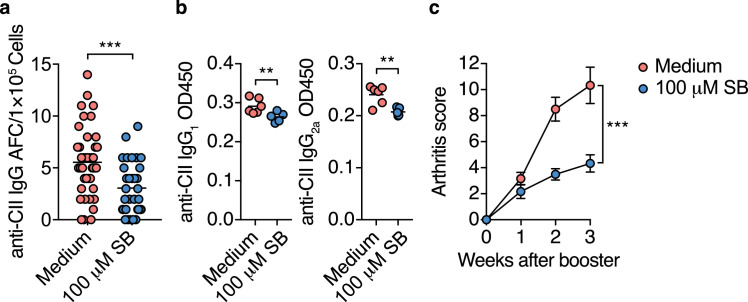
3.5. iTFR cells prevent autoimmune responses and CIA development
Finally, we examined whether butyrate-induced CXCR5+Bcl-6+ iTFR cells could prevent the development of CIA. Sort purified Foxp3–hCD2+CD4+T cells cultured with or without 100 μM SB under iTFR-cell culture conditions were adoptively transferred to DBA/1 J mice one week before and after the initial immunization of CII. SB-treated Foxp3–hCD2+CD4+T cell population contains much higher numbers of CXCR5+Bcl-6+ iTFR cells compared to butyrate-untreated CXCR5+Bcl-6+ iTFR cells, as shown in Fig. 6a-d. The adoptive transfer of the SB-treated cells significantly reduced CII-specific IgG-forming cells and serum levels of CII-specific IgG1 and IgG2 three weeks after the booster immunization, and thus ameliorated the symptoms of arthritis (Fig. 7a-c). These data suggest that butyrate-induced CXCR5+Bcl-6+ iTFR cells inhibit the autoimmune responses against CII and that TFR cells play a critical role in preventing autoimmune arthritis in CIA mice.
Fig. 7.
iTFR cells prevent autoimmune responses and CIA development
(a, b) CII-specific IgG responses in CIA mice inoculated with Foxp3-hCD2+ cells cultured under iTFR-cell conditions in the absence or presence of 100 µM SB. Sort-purified 5 × 106 of Foxp3-hCD2+CD4+T cells were injected intravenously into DBA/1 J mice one week before the initial CII immunization.All mice were fed the control HAMS diet (n= 10, 11). The number of CII-specific IgG AFC cells per 1 × 105 DLN cells (a, each dot indicates data from 1 well, n= 44, 32, statistical analysis was performed on the mean of each mouse.), and serum levels of CII-specific IgG1 and IgG2a three weeks after booster immunization (b, n= 5, 6).
(c) Arthritis scores after booster immunization of CIA mice inoculated with Foxp3-hCD2+ cells from iTFR cell culture (n= 10, 11). AUC was calculated in each mice.
Results show one representative experiment of at least two experiments.**P< 0.01, ***P< 0.001 (a-c, Welch's t-test or unpaired two-tailed Student's t-test).
4. Discussion
Here we found that butyrate production is considerably affected in NORA patients due to the underrepresentation of Lachnospiraceae and Faecalibacterium belonging to the Clostridium clusters XIVa and IV, respectively. The intake of the HAMSB diet before or one week after the initial immunization ameliorated CIA symptoms after the booster immunization by suppressing the generation of autoantibodies against CII, supporting the notion that the decrease in luminal butyrate may be implicated in RA development. Since autoantibodies are detected several years before the onset of arthritis in some RA patients [64], [65], [66], we consider that an intervention to raise luminal butyrate during these period could help reduce RA incidence if the presence of autoantiboy could be monitored easily in NORA patients or people who have a genetic risk of developing RA in the future. Additionally, a recent study revealed that feacal level of butyrate was significantly reduced during the remission phase of RA [67]. Thus, the raising luminal butyrate during this phase may reduce the risk of relapses in patients with RA. We also detected a reduction tendency of luminal acetate and propionate in NORA patients, and another larger cohort study may find the diminished production of these SCFAs in NORA patients. However, the intake of either HAMSA or HAMSP diet, which increased acetate or propionateconcentration in the colon, failed to alleviate the incidence and development of CIA. Therefore, the decrease in acetate and propionate is unlikely to influence RA development.
The intake of the HAMSB diet increased TFR cells in CoP of CIA mice at the early phase after the initial immunization with CII. Moreover, the transfer of Foxp3+ TREG cells with CD4+T cells into TCR-deficient mice showed clear evidence of the promotion of TFR cell differentiation from TREG cells in CoP by commensal bacteria-derived butyrate. In vitro experiments with primary CD4+T cell culture illustrated that butyrate facilitates the differentiation of not only iTREG cells but also iTFR cells in the presence of IL-6 and IL-21. Given that RA-associated inflammatory conditions lead to massive production of inflammatory cytokines, including IL-6 and IL-21, the induction of TFR cells over TREG cells by butyrate deems to be context-dependent and could be enhanced under inflammatory conditions like the CIA model. Our data also illustrated that the effect of butyrate on TFR cell differentiation mainly depends on its HDACi activity, but not activation of GPCRs. Several lines of evidence support this view; other HDAC inhibitors such as SAHA and TSA also promote TFR differentiation, and vice versa, HATi cancelled the TFR cell-inducing activity of butyrate.
Furthermore, a ChIP assay confirmed that treatment with butyrate induced hyperacetylation status on the promoter regions of TFR cell-signature genes, namely, Bcl6, Cxcr5, and Tcf7 in T cells cultured under iTFR-skewed conditions. Importantly, such epigenetic modifications preceded iTFR cell differentiation.The IC50 value of butyrate for HDACs is approximately 20–60 µM [68,69], which is sufficient to promote iTFR cell differentiation. The concentrations of butyrate are much higher (approximately 1 mM) than the IC50 value in the colonic tissue but decline to 8.3 µM in the serum [68,69]. It is, therefore, reasonable to think that commensal-derived butyrate facilitates TFR differentiation, mainly in CoP. Besides having HDACi activity, butyrate also serves as a ligand for Gpr41, Gpr43, and Gpr109a [70], [71], [72]. However, naive T cells lacking Gpr41, Gpr43, or GPR109a normally differentiate into iTFR cells in the presence of butyrate, indicating that these receptors are dispensable for butyrate-mediated iTFR cell differentiation.
In this study, we newly established the culture conditions to induce iTFR cell differentiation. According to current knowledge, TFR cells primarily generate from CD25+ TREG cells, but not TFH cells [[15], [16], [17],21]. Therefore, we initially treated naïve T cells with TGF-β1 and IL-2 to induce CD25+ iTREG cells and then stimulated the cells with IL-21, IL-6, and anti-ICOS Abs with a lower concentration of IL-2 to dictate terminal differentiation into iTFR cells [20,21,38,73]. IL-2 is essential for the development and function of TREG cells, but conversely inhibits TFR cell development [20,21]. Therefore, TFR cells downregulate IL-2Rα/CD25 during differentiation. Likewise, a part of iTFR cells lacked CD25 expression, and the CD25− iTFR cells express higher levels of TFR-cell maker molecules such as CXCR5 and Bcl-6 compared to CD25+ iTFR cells. Additionally, CD25− iTFR cells potently suppress proliferation and class switching of B cells cocultured with TFH cells. Thus, CD25− iTFR cells phenocopied the functionality of bona fide TFR cells. Indeed, the adoptive transfer of the butyrate-treated iTFR cells reduced CII-specific autoimmune responses and attenuated CIA pathogensis. Nontheless, future effort should be made to examine if iTFR cell transfer can treat CIA even after the onset of arthritis. Moreover, it is important to investigate whether CD25+ or CD25− iTFR cells are differentially involved in preventing CIA development.
We propose that the iTFR culture system is useful to understand the molecular machinery of TFR cell differentiation fully. Treatment with butyrate increased the frequencies of Bcl-6+CXCR5+ populations within CD25+ iTFR cells. Thus, butyrate most likely facilitates the expression of Bcl-6 and CXCR5 at the early to middle stage of Tfr cell differentiation, leading to the development of fully matured CD25− iTFR cells. It is noteworthy that even under conventional iTREG conditions with a higher concentration of IL-2, the exposure to butyrate generated iTFR-like cells characterized by CXCR5 expression. In line with this observation, butyrate substantially raised the histone acetylation status on the TFR cell-signature gene locus under iTREG conditions. These facts imply that butyrate may play a critical role in the early-stages of cell-fate decision from TREG cells to differentiated TFR cells. As mentioned earlier, butyrate upregulates the expression of genes responsible for TFR cell differentiation such as Cxcr5, Tcf7, and Bcl6; however, these molecules are also essential for differentiation of TFH cells. Nevertheless, HAMSB intake preferentially induced the development of TFR, but not TFH cells in GALT. Consistent with the in vivo observations, treatment with butyrate did not promote in vitro differentiation of TFH cells. Previous ChIP-seq analyses demonstrated the accumulation of DNase I hypersensitivity sites (DHS) and active histone modification, H3K4me3, around the Bcl6 locus even in naive T cells, suggesting that the Bcl6 gene locus is accessible [49,74]. Hence, treatment with butyrate is unlikely to affect the already accessible chromatin status on the Bcl6 promoter region in naïve T cells and consequently failed to induce TFH cell differentiation. In sharp contrast, the Bcl6 promoter region was repressed in TREG cells, indicating that butyrate-mediated histone acetylation could be essential to establish accessible chromatin and subsequently upregulate Bcl6 in the TFR precursor upon STAT3 activation. Although such differences in epigenetic status among CD4+T cell subsets at least partly account for TFR cell-prone differentiation upon exposure to butyrate, further investigations are required to clarify the underlying mechanism.
HAMSB intake did not influence the number of TFR cells in the DLNs and SP of CII-immunized mice. Importantly, systemic immunization with CII prominently activated GC reactions in distal GALTs like CoPs and colonic SILT and induced GALT hypertrophy. The intake of HAMSB efficiently suppressed these pathological events and eventually mitigated arthritis development after booster immunization. These observations raise the possibility that autoimmune GC reactions are initially induced in the distal GALT. This possibility is supported by the fact that the elimination of gut microbiota attenuates CIA development [75]. Our cell trafficking analysis using KikGR mice corroborated that both TFH and B cells migrate from CoP to the DLNs after immunization. Thus, the distal GALT is considered as a primary site to initiate the autoimmune response in the CIA model, and gut microbiota may provide an assortment of GALT adjuvants. It remains unclear how gut microbiota-derived butyrate suppresses the systemic autoimmune response. Our study exhibited that butyrate from the intestinal lumen prevented the development of autoimmune arthritis, including the CIA and SKG model; however, it failed to suppress CAIA after injection with a cocktail of autoantibodies and CIA with supplementation of butyrate after booster immunization. These data suggest that the butyrate suppresses the initial phase of TFH cell-mediated autoimmune responses rather than the effector phase of arthritis development. We observed a substantial decrease in IgG2a subtype of anti-CII autoantibodies both in the serum and knee joint extract, but a slight reduction of IgG1 subtype in CIA mice fed HAMSB, raising the possibility that butyrate-mediated suppression of autoantibody production may be more specific for IgG2a. Given that IgG2a, rather than IgG1 subtype of anti-CII autoantibody, plays a critical role in the pathology of the CIA model [76], the reduction of IgG2a subtype of anti-CII autoantibodies in HAMSB-fed mice most likely explain the attenuation of CIA development. Moreover, a recent report demonstrated that butyrate indirectly promotes the differentiation of regulatory B cells that inhibits the differentiation of GC B cells and plasmablasts in antigen-induced arthritis models [67]. Thus, butyrate might suppress the induction of CII-specific antibody-producing B cells by inducing regulatory B cells in addition to inhibiting TFH-cell-mediated anti-CII responses in the CIA model. However, further investigations will be necessary to clarify this point.
The intraperitoneal administration of sodium butyrate at a high dose (100 mg/kg) suppresses the CIA by decreasing the TH17/TREG ratio in DLNs [77]. In contrast, the TH17/TREG ratio was not affected by the HAMSB diet in our study. The difference in the administration route explains this apparent discrepancy. The intraperitoneal injection of high-dose sodium butyrate may exert an artificial effect by increasing serum butyrate to non-physiological levels. The serum level of butyrate is usually maintained at a very low level (ca. 3–4 µM) in healthy subjects [78,79]. To mimic physiological conditions, we took advantage of a low-fibre diet supplemented with HAMSB [38], which resists small intestinal amylolysis and reaches the colon, where commensal microbes release esterified butyrate, raising the luminal concentration to approximately 20 mM, equivalent to the level in human stool [80].
Meanwhile, the serum concentration was maintained at a much lower level (approximately 8 µM) in HAMSB-fed mice, because butyrate generated in the gut lumen is primarily utilized as an energy source by colonocytes and possibly metabolized by hepatocytes after transport via the portal vein. Considering the fact that commensal bacteria-derived butyrate did not suppress CAIA development, these data suggest that the physiological level of butyrate does not directly impede the effector phase of arthritis development driven by osteoclast activation and TH17 cell response. However, supplementation of high-dose (150 mM) SCFAs, including butyrate in the drinking water are reported to inhibit osteoclast differentiation and bone destruction in the CIA model [81]. Considering that orally administrated SCFAs are readily absorbed in the upper intestine and aberrantly elevate their serum concentrations, the protective effect on osteoclast differentiation may be an artificial rather than physiological event. Nonetheless, these data provide evidence about the therapeutic potential of oral administration with high-dose SCFAs to ameliorate bone destruction in RA patients.
5. Conclusion
This study suggests that commensal bacteria-derived butyrate is a crucial environmental factor promoting the differentiation of TFR cells in the GALT. Commensal bacteria play an essential role in both the initiation and suppression of autoimmune arthritis by modifying the immune system in the GALT. Given that a commensal bacteria-driven autoimmune response in the GALT precedes systemic autoimmune responses, the induction of GALT TFR cells might be a new target for preventing RA. Indeed, the abundance of TFR cells is negatively correlated with disease activity in patients with RA [45]. However, for the better understanding of TFR cell-mediated attenuation of RA, the study using TFR cell-deficient mice is required in the future. Administration of butyrate-producing bacteria or HAMSB to subjects genetically susceptible to RA could have therapeutic potential to prevent the disease onset or the development of following disease symptoms. Furthermore, a combination of butyrate-producing agents (e.g., HAMSB) and pre-existing antirheumatic drugs such as anti-TNF-α antibody may lead to more effective treatments even after the onset of RA. However, further investigations will be necessary to ascertain these notion. It is also a future task to investigate whether TFR cells generated in GALT also suppress the development of other autoimmune diseases like systemic lupus erythematosus. Our findings provide a molecular basis for new prophylaxis and treatment approaches for systemic autoimmune disorders by targeting the intestinal environment and autoimmune responses in GALT.
Funding sources
This work was supported by AMED-Crest (16gm1010004h0101 and 17gm1010004h0102 to KH), the Japan Society for the Promotion of Science (JP17KT0055, JP16H01369, and JP18H04680 to KH; JP17K15734 to DT), Keio University Special Grant-in-Aid for Innovative Collaborative Research Projects (KH), Keio Gijuku Fukuzawa Memorial Fund for the Advancement of Education and Research (DT), the SECOM Science and Technology Foundation (KH), the Cell Science Research Foundation (KH), the Mochida Memorial Foundation for Medical and Pharmaceutical Research (DT), the Suzuken Memorial Foundation (KH and DT), the Takeda Science Foundation (KH and DT), The Science Research Promotion Fund, and The Promotion and Mutual Aid Corporation for Private Schools of Japan (KH). The funders had no role in study design, data collection, data analysis, interpretation, writing of the report nor in the decision to publish.
Author contributions
D.T., N.H., and K.H. conceived the project. D.T. performed most experiments, data, and statistical analyses. D.T. and K.H. wrote the manuscript. K.H. supervised the study. N.H. conducted CIA experiments and data analyses. A.S., Y.K., H.T., K.M., Y.Y., Y.F, A.S., R.M., and H.A. supported animal experiments. Y.K., J.I., and A.N. performed the GC–MS analysis. M.U. and M.T. performed cell trafficking analysis. Y.M. Y.K. and T.Y. performed the 16S rRNA amplicon sequencing and data analysis. J.M.C. provided the modified starches. Y.H., N.K., T.O., and H.T. provided the mice. Y.M., J.I, E.U., and K.T. collected and provided the human faecal samples.
Declaration of Competing Interest
The authors declare no competing financial interests. Correspondence should be addressed to K.H. (hase-kj@pha.keio.ac.jp).
Acknowledgements
We would like to thank Kouya Hattori, Tatsuki Kimizuka, Ryo Aoki, Yun-Gi Kim, Shunsuke Kimura, and Tatsuya Morita for the valuable discussion; Mitsuharu Matsumoto and Miho Nakamura for the technical help; Trevor Lockett for providing HAMSB (Starplus B®). We would also like to acknowledge the Animal Facility at the Keio University Faculty of Pharmacy for the breeding and maintenance of our mouse strains and the Instrument Management Division. We are grateful to editage (https://www.editage.jp)for editing a draft of this manuscript.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102913.
Appendix. Supplementary materials
References
- 1.McInnes I.B., Schett G. The pathogenesis of rheumatoid arthritis. N Engl J Med. 2011;365:2205–2219. doi: 10.1056/NEJMra1004965. [DOI] [PubMed] [Google Scholar]
- 2.McInnes I.B., Schett G. Pathogenetic insights from the treatment of rheumatoid arthritis. Lancet. 2017;389:2328–2337. doi: 10.1016/S0140-6736(17)31472-1. [DOI] [PubMed] [Google Scholar]
- 3.Firestein G.S., McInnes I.B. Immunopathogenesis of Rheumatoid Arthritis. Immunity. 2017;46:183–196. doi: 10.1016/j.immuni.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hollister K., Kusam S., Wu H., Clegg N., Mondal A., Sawant D V. et al. Insights into the Role of Bcl6 in Follicular Th Cells Using a New Conditional Mutant Mouse Model. J Immunol. 2013;191:3705–3711. doi: 10.4049/jimmunol.1300378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu X., Chen X., Zhong B., Wang A., Wang X., Chu F. Transcription factor achaete-scute homologue 2 initiates follicular T-helper-cell development. Nature. 2014;507:513–518. doi: 10.1038/nature12910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu L., Cao Y., Xie Z., Huang Q., Bai Q., Yang X. The transcription factor TCF-1 initiates the differentiation of T<inf>FH</inf>cells during acute viral infection. Nat Immunol. 2015;16:991–999. doi: 10.1038/ni.3229. [DOI] [PubMed] [Google Scholar]
- 7.Yang J., Lin X., Pan Y., Wang J., Chen P., Huang H. Critical roles of mTOR complex 1 and 2 for t follicular helper cell differentiation and germinal center responses. Elife. 2016;5:e17936. doi: 10.7554/eLife.17936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Silva D.G., Daley S.R., Hogan J., Lee S.K., Teh C.E., Hu D.Y. Anti-Islet Autoantibodies Trigger Autoimmune Diabetes in the Presence of an Increased Frequency of Islet-Reactive CD4 T Cells. Diabetes. 2011;60:2102–2111. doi: 10.2337/db10-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCarron M.J., Marie J.C. TGF-β prevents T follicular helper cell accumulation and B cell autoreactivity. J Clin Invest. 2014;124:4375–4386. doi: 10.1172/JCI76179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim Y.U., Lim H., Jung H.E., Wetsel R.A., Chung Y. Regulation of autoimmune germinal center reactions in lupus-prone BXD2 mice by follicular helper T cells. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0120294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ueno H., Banchereau J., Vinuesa C.G. Pathophysiology of T follicular helper cells in humans and mice. Nat Immunol. 2015;16:142–152. doi: 10.1038/ni.3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moschovakis G.L., Bubke A., Friedrichsen M., Falk C.S., Feederle R., Förster R. T cell specific Cxcr5 deficiency prevents rheumatoid arthritis. Sci Rep. 2017;7:8933. doi: 10.1038/s41598-017-08935-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhong M.C., Veillette A. The adaptor molecule signaling lymphocytic activation molecule (SLAM)-associated protein (SAP) is essential in mechanisms involving the Fyn tyrosine kinase for induction and progression of collagen-induced arthritis. J Biol Chem. 2013;288:31423–31436. doi: 10.1074/jbc.M113.473736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Panneton V., Bagherzadeh Yazdchi S., Witalis M., Chang J., Suh W.-.K. ICOS Signaling Controls Induction and Maintenance of Collagen-Induced Arthritis. J Immunol. 2018;200:3067–3076. doi: 10.4049/jimmunol.1701305. [DOI] [PubMed] [Google Scholar]
- 15.Chung Y., Tanaka S., Chu F., Nurieva R.I., Martinez G.J., Rawal S. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nat Med. 2011;17:983–988. doi: 10.1038/nm.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Linterman M.A., Pierson W., Lee S.K., Kallies A., Kawamoto S., Rayner T.F. Foxp3+ follicular regulatory T cells control the germinal center response. Nat Med. 2011;17:975–982. doi: 10.1038/nm.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wollenberg I., Agua-Doce A., Hernández A., Almeida C., Oliveira V.G., Faro J. Regulation of the Germinal Center Reaction by Foxp3 + Follicular Regulatory T Cells. J Immunol. 2011;187:4553–4560. doi: 10.4049/jimmunol.1101328. [DOI] [PubMed] [Google Scholar]
- 18.Wu H., Chen Y., Liu H., Xu L.L., Teuscher P., Wang S. Follicular regulatory T cells repress cytokine production by follicular helper T cells and optimize IgG responses in mice. Eur J Immunol. 2016;46:1152–1161. doi: 10.1002/eji.201546094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sage P.T., Alvarez D., Godec J., Von Andrian U.H., Sharpe A.H. Circulating T follicular regulatory and helper cells have memory-like properties. J Clin Invest. 2014;124:5191–5204. doi: 10.1172/JCI76861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ritvo P.-.G.G., Churlaud G., Quiniou V., Florez L., Brimaud F., Fourcade G. T fr cells lack IL-2Rα but express decoy IL-1R2 and IL-1Ra and suppress the IL-1–dependent activation of T fh cells. Sci Immunol. 2017;2 doi: 10.1126/sciimmunol.aan0368. eaan0368. [DOI] [PubMed] [Google Scholar]
- 21.Wing J.B., Kitagawa Y., Locci M., Hume H., Tay C., Morita T. A distinct subpopulation of CD25 − T-follicular regulatory cells localizes in the germinal centers. Proc Natl Acad Sci. 2017;114:E6400–E6409. doi: 10.1073/pnas.1705551114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu L., Huang Q., Wang H., Hao Y., Bai Q., Hu J. The Kinase mTORC1 Promotes the Generation and Suppressive Function of Follicular Regulatory T Cells. Immunity. 2017;47 doi: 10.1016/j.immuni.2017.08.011. 538-551.e5. [DOI] [PubMed] [Google Scholar]
- 23.Ritvo P.-.G., Saadawi A., Barennes P., Quiniou V., Chaara W., El Soufi K. High-resolution repertoire analysis reveals a major bystander activation of Tfh and Tfr cells. Proc Natl Acad Sci. 2018;115:9604–9609. doi: 10.1073/pnas.1808594115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Botta D., Fuller M.J., Marquez-Lago T.T., Bachus H., Bradley J.E., Weinmann A.S. Dynamic regulation of T follicular regulatory cell responses by interleukin 2 during influenza infection. Nat Immunol. 2017;18:1249–1260. doi: 10.1038/ni.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu W., Liu X., Lin X., Feng H., Sun L., Li S. Deficiency in T follicular regulatory cells promotes autoimmunity. J Exp Med. 2018;215:815–825. doi: 10.1084/jem.20170901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma J., Zhu C., Ma B., Tian J., Baidoo S.E., Mao C. Increased frequency of circulating follicular helper T cells in patients with rheumatoid arthritis. Clin Dev Immunol. 2012;2012:827480–827487. doi: 10.1155/2012/827480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu C., Wang D., Lu S., Xu Q., Zhao L., Zhao J. Increased Circulating Follicular Treg Cells Are Associated With Lower Levels of Autoantibodies in Patients With Rheumatoid Arthritis in Stable Remission. Arthritis Rheumatol. 2018;70:711–721. doi: 10.1002/art.40430. [DOI] [PubMed] [Google Scholar]
- 28.Niu Q., Huang Z chun, Wu X juan, Jin Y xiong, An Y fei, Li Y mei. Enhanced IL-6/phosphorylated STAT3 signaling is related to the imbalance of circulating T follicular helper/T follicular regulatory cells in patients with rheumatoid arthritis. Arthritis Res Ther. 2018;20:200. doi: 10.1186/s13075-018-1690-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scher J.U., Sczesnak A., Longman R.S., Segata N., Ubeda C., Bielski C. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife. 2013;2013:e01202. doi: 10.7554/eLife.01202.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maeda Y., Kurakawa T., Umemoto E., Motooka D., Ito Y., Gotoh K. Dysbiosis contributes to arthritis development via activation of autoreactive T cells in the intestine. Arthritis Rheumatol (Hoboken, NJ) 2016;68:2646–2661. doi: 10.1002/art.39783. [DOI] [PubMed] [Google Scholar]
- 31.De Paiva C.S., Jones D.B., Stern M.E., Bian F., Moore Q.L., Corbiere S. Altered Mucosal Microbiome Diversity and Disease Severity in Sjögren Syndrome. Sci Rep. 2016;6 doi: 10.1038/srep23561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levy M., Kolodziejczyk A.A., Thaiss C.A., Elinav E. Dysbiosis and the immune system. Nat Rev Immunol. 2017;17:219–232. doi: 10.1038/nri.2017.7. [DOI] [PubMed] [Google Scholar]
- 33.Mariño E., Richards J.L., McLeod K.H., Stanley D., Yap Y.A., Knight J. Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nat Immunol. 2017;18:552–562. doi: 10.1038/ni.3713. [DOI] [PubMed] [Google Scholar]
- 34.Tsigalou C., Stavropoulou E., Bezirtzoglou E. Current insights in microbiome shifts in Sjogren'S syndrome and possible therapeutic interventions. Front Immunol. 2018;9:162. doi: 10.3389/fimmu.2018.01106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Opazo M.C., Ortega-Rocha E.M., Coronado-Arrázola I., Bonifaz L.C., Boudin H., Neunlist M. Intestinal microbiota influences non-intestinal related autoimmune diseases. Front Microbiol. 2018;9:432. doi: 10.3389/fmicb.2018.00432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Macpherson A.J., Yilmaz B., Limenitakis J.P., Ganal-Vonarburg S.C. IgA Function in Relation to the Intestinal Microbiota. Annu Rev Immunol. 2018;36:359–381. doi: 10.1146/annurev-immunol-042617-053238. [DOI] [PubMed] [Google Scholar]
- 37.Tanoue T., Atarashi K., Honda K. Development and maintenance of intestinal regulatory T cells. Nat Rev Immunol. 2016;16:295–309. doi: 10.1038/nri.2016.36. [DOI] [PubMed] [Google Scholar]
- 38.Furusawa Y., Obata Y., Fukuda S., Endo T.A., Nakato G., Takahashi D. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 39.Ohnmacht C., Park J.H., Cording S., Wing J.B., Atarashi K., Obata Y. The microbiota regulates type 2 immunity through RORγt+ T cells. Science (80-) 2015;349:989–993. doi: 10.1126/science.aac4263. [DOI] [PubMed] [Google Scholar]
- 40.Atarashi K., Tanoue T., Shima T., Imaoka A., Kuwahara T., Momose Y. Induction of colonic regulatory T cells by indigenous Clostridium species. Science (80-) 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ivanov I.I., Atarashi K., Manel N., Brodie E.L., Shima T., Karaoz U. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teng F., Klinger C.N., Felix K.M., Bradley C.P., Wu E., Tran N.L. Gut Microbiota Drive Autoimmune Arthritis by Promoting Differentiation and Migration of Peyer’s Patch T Follicular Helper Cells. Immunity. 2016;44:875–888. doi: 10.1016/j.immuni.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu H.J., Darce J., Hattori K., Shima T., Umesaki Y. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan J., McKenzie C., Vuillermin P.J., Goverse G., Vinuesa C.G., Mebius R.E. Dietary Fiber and Bacterial SCFA Enhance Oral Tolerance and Protect against Food Allergy through Diverse Cellular Pathways. Cell Rep. 2016;15:2809–2824. doi: 10.1016/j.celrep.2016.05.047. [DOI] [PubMed] [Google Scholar]
- 45.Kitano M., Moriyama S., Ando Y., Hikida M., Mori Y., Kurosaki T. Bcl6 Protein Expression Shapes Pre-Germinal Center B Cell Dynamics and Follicular Helper T Cell Heterogeneity. Immunity. 2011;34:961–972. doi: 10.1016/j.immuni.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 46.Miyao T., Floess S., Setoguchi R., Luche H., Fehling H.J., Waldmann H. Plasticity of Foxp3 + T Cells Reflects Promiscuous Foxp3 Expression in Conventional T Cells but Not Reprogramming of Regulatory T Cells. Immunity. 2012;36:262–275. doi: 10.1016/j.immuni.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 47.Tomura M., Hata A., Matsuoka S., Shand F.H.W., Nakanishi Y., Ikebuchi R. Tracking and quantification of dendritic cell migration and antigen trafficking between the skin and lymph nodes. Sci Rep. 2014;4:6030. doi: 10.1038/srep06030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moreau N.M., Goupry S.M., Antignac J.P., Monteau F.J., Le Bizec B.J., Champ M.M. Simultaneous measurement of plasma concentrations and13C-enrichment of short-chain fatty acids, lactic acid and ketone bodies by gas chromatography coupled to mass spectrometry. J Chromatogr B Anal Technol Biomed Life Sci. 2003;784:395–403. doi: 10.1016/S1570-0232(02)00827-9. [DOI] [PubMed] [Google Scholar]
- 49.Nakayamada S., Kanno Y., Takahashi H., Jankovic D., Lu K.T., Johnson T.A. Early Th1 Cell Differentiation Is Marked by a Tfh Cell-like Transition. Immunity. 2011;35:919–931. doi: 10.1016/j.immuni.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamada T., Hino S., Iijima H., Genda T., Aoki R., Nagata R. Mucin O-glycans facilitate symbiosynthesis to maintain gut immune homeostasis. EBioMedicine. 2019;48:513–525. doi: 10.1016/j.ebiom.2019.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arpaia N., Campbell C., Fan X., Dikiy S., Van Der Veeken J., Deroos P. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith P.M., Howitt M.R., Panikov N., Michaud M., Gallini C.A., Bohlooly-Y M. The microbial metabolites, short-chain fatty acids, regulate colonic Tregcell homeostasis. Science (80-) 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haghikia A., Jörg S., Duscha A., Berg J., Manzel A., Waschbisch A. Dietary Fatty Acids Directly Impact Central Nervous System Autoimmunity via the Small Intestine. Immunity. 2015;43:817–829. doi: 10.1016/j.immuni.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 54.Clarke J.M., Young G.P., Topping D.L., Bird A.R., Cobiac L., Scherer B.L. Butyrate delivered by butyrylated starch increases distal colonic epithelial apoptosis in carcinogen-treated rats. Carcinogenesis. 2012;33:197–202. doi: 10.1093/carcin/bgr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu X., Zeng B., Zhang J., Li W., Mou F., Wang H. Role of the Gut Microbiome in Modulating Arthritis Progression in Mice. Sci Rep. 2016;6:1–11. doi: 10.1038/srep30594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abdollahi-Roodsaz S., Joosten L.A.B., Koenders M.I., Devesa I., Roelofs M.F., Radstake T.R.D.J. Stimulation of TLR2 and TLR4 differentially skews the balance of T cells in a mouse model of arthritis. J Clin Invest. 2008;118:205–216. doi: 10.1172/JCI32639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Al-Saadany H.M., Hussein M.S., Gaber R.A., Zaytoun H.A. Th-17 cells and serum IL-17 in rheumatoid arthritis patients: correlation with disease activity and severity. Egypt Rheumatol. 2016;38:1–7. doi: 10.1016/j.ejr.2015.01.001. [DOI] [Google Scholar]
- 58.Komatsu N., Okamoto K., Sawa S., Nakashima T., Oh-Hora M., Kodama T. Pathogenic conversion of Foxp3 + T cells into TH17 cells in autoimmune arthritis. Nat Med. 2014;20:62–68. doi: 10.1038/nm.3432. [DOI] [PubMed] [Google Scholar]
- 59.Svensson L., Jirholt J., Holmdahl R., Jansson L. B cell-deficient mice do not develop type II collagen-induced arthritis (CIA) Clin Exp Immunol. 1998;111:521–526. doi: 10.1046/j.1365-2249.1998.00529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Díaz De Ståhl T., Andrén M., Martinsson P., Verbeek J.S., Kleinau S. Expression of FcγRIII is required for development of collagen-induced arthritis. Eur J Immunol. 2002;32:2915–2922. doi: 10.1002/1521-4141(2002010)32:10<2915::AID-IMMU2915>3.0.CO;2-4. https://doi.org/10.1002/1521-4141(2002010)32:10<2915::AID−IMMU2915>3.0.CO. [DOI] [PubMed] [Google Scholar]
- 61.Hutamekalin P., Saito T., Yamaki K., Mizutani N., Brand D.D., Waritani T. Collagen antibody-induced arthritis in mice: development of a new arthritogenic 5-clone cocktail of monoclonal anti-type II collagen antibodies. J Immunol Methods. 2009;343:49–55. doi: 10.1016/j.jim.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 62.Yang B.H., Wang K., Wan S., Liang Y., Yuan X., Dong Y. TCF1 and LEF1 Control Treg Competitive Survival and Tfr Development to Prevent Autoimmune Diseases. Cell Rep. 2019;27 doi: 10.1016/j.celrep.2019.05.061. 3629-3645.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Singh N., Gurav A., Sivaprakasam S., Brady E., Padia R., Shi H. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014;40:128–139. doi: 10.1016/j.immuni.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rantapaa-Dahlqvist S., de Jong B.A.W., Berglin E., Hallmans G., Wadell G., Stenlund H. Antibodies against cyclic citrullinated peptide and IgA rheumatoid factor predict the development of rheumatoid arthritis. Arthritis Rheum. 2003;48:2741–2749. doi: 10.1002/art.11223. [DOI] [PubMed] [Google Scholar]
- 65.Nielen M.M.J., Van Schaardenburg D., Reesink H.W., Van De Stadt R.J., Van Der Horst-Bruinsma I.E., De Koning M.H.M.T. Specific Autoantibodies Precede the Symptoms of Rheumatoid Arthritis: a Study of Serial Measurements in Blood Donors. Arthritis Rheum. 2004;50:380–386. doi: 10.1002/art.20018. [DOI] [PubMed] [Google Scholar]
- 66.Van De Stadt L.A., De Koning M.H.M.T., Van De Stadt R.J., Wolbink G., Dijkmans B.A.C., Hamann D. Development of the anti-citrullinated protein antibody repertoire prior to the onset of rheumatoid arthritis. Arthritis Rheum. 2011;63:3226–3233. doi: 10.1002/art.30537. [DOI] [PubMed] [Google Scholar]
- 67.Rosser E.C., Piper C.J.M., Matei D.E., Blair P.A., Rendeiro A.F., Orford M. Microbiota-Derived Metabolites Suppress Arthritis by Amplifying Aryl-Hydrocarbon Receptor Activation in Regulatory B Cells. Cell Metab. 2020;31 doi: 10.1016/j.cmet.2020.03.003. 837-851.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thangaraju M., Gopal E., Martin P.M., Ananth S., Smith S.B., Prasad P.D. SLC5A8 triggers tumor cell apoptosis through pyruvate-dependent inhibition of histone deacetylases. Cancer Res. 2006;66:11560–11564. doi: 10.1158/0008-5472.CAN-06-1950. [DOI] [PubMed] [Google Scholar]
- 69.Thangaraju M., Carswell K.N., Prasad P.D., Ganapathy V. Colon cancer cells maintain low levels of pyruvate to avoid cell death caused by inhibition of HDAC1/HDAC3. Biochem J. 2009;417:379–389. doi: 10.1042/BJ20081132. [DOI] [PubMed] [Google Scholar]
- 70.Brown A.J., Goldsworthy S.M., Barnes A.A., Eilert M.M., Tcheang L., Daniels D. The orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem. 2003;278:11312–11319. doi: 10.1074/jbc.M211609200. [DOI] [PubMed] [Google Scholar]
- 71.Le Poul E., Loison C., Struyf S., Springael J.Y., Lannoy V., Decobecq M.E. Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. J Biol Chem. 2003;278:25481–25489. doi: 10.1074/jbc.M301403200. [DOI] [PubMed] [Google Scholar]
- 72.Thangaraju M., Cresci G.A., Liu K., Ananth S., Gnanaprakasam J.P., Browning D.D. GPFM 09A is a G-protein-coupled receptor for the bacterial fermentation product butyrate and functions as a tumor suppressor in colon. Cancer Res. 2009;69:2826–2832. doi: 10.1158/0008-5472.CAN-08-4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nurieva R.I., Chung Y., Hwang D., Yang X.O., Kang H.S., Ma L. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. 2008;29:138–149. doi: 10.1016/j.immuni.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lu K.T., Kanno Y., Cannons J.L., Handon R., Bible P., Elkahloun A.G. Functional and Epigenetic Studies Reveal Multistep Differentiation and Plasticity of In Vitro-Generated and In Vivo-Derived Follicular T Helper Cells. Immunity. 2011;35:622–632. doi: 10.1016/j.immuni.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rogier R., Evans-Marin H., Manasson J., Van Der Kraan P.M., Walgreen B., Helsen M.M. Alteration of the intestinal microbiome characterizes preclinical inflammatory arthritis in mice and its modulation attenuates established arthritis. Sci Rep. 2017;7:15613. doi: 10.1038/s41598-017-15802-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Watson W.C. Genetic susceptibility to murine collagen II autoimmune arthritis. Proposed relationship to the IgG2 autoantibody subclass response, complement C5, major histocompatibility complex (MHC) and non-MHC loci. J Exp Med. 1985;162:1878–1891. doi: 10.1084/jem.162.6.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim D.S., Kwon J.E., Lee S.H., Kim E.K., Ryu J.G., Jung K.A. Attenuation of rheumatoid inflammation by sodium butyrate through reciprocal targeting of HDAC2 in osteoclasts and HDAC8 in T cells. Front Immunol. 2018;9:1525. doi: 10.3389/fimmu.2018.01525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Khor B., Gagnon J.D., Goel G., Roche M.I., Conway K.L., Tran K. The kinase DYRK1A reciprocally regulates the differentiation of Th17 and regulatory T cells. Elife. 2015;4:1–27. doi: 10.7554/eLife.05920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gotfryd L., Becker K., Pietek K.G., Szolomicki Z., Piwowonska J. Direct measurements of reaction time for extractive processes. Physicochem Probl Miner Process. 2016;52:909–919. doi: 10.5277/ppmp160230. [DOI] [Google Scholar]
- 80.Bajka B.H., Clarke J.M., Cobiac L., Topping D.L. Butyrylated starch protects colonocyte DNA against dietary protein-induced damage in rats. Carcinogenesis. 2008;29:2169–2174. doi: 10.1093/carcin/bgn173. [DOI] [PubMed] [Google Scholar]
- 81.Lucas S., Omata Y., Hofmann J., Böttcher M., Iljazovic A., Sarter K. Short-chain fatty acids regulate systemic bone mass and protect from pathological bone loss. Nat Commun. 2018;9:e73. doi: 10.1038/s41467-017-02490-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.