Abstract
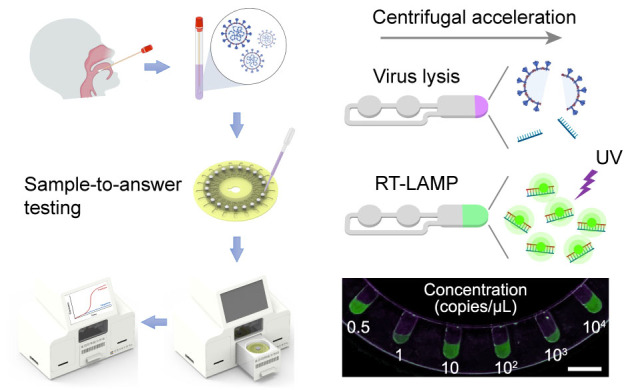
The outbreak of virus-induced infectious diseases poses a global public-health challenge. Nucleic acid amplification testing (NAAT) enables early detection of pandemic viruses and plays a vital role in preventing onward transmission. However, the requirement of skilled operators, expensive instrumentation, and biosafety laboratories has hindered the use of NAAT for screening and diagnosis of suspected patients. Here we report development of a fully automated centrifugal microfluidic system with sample-in-answer-out capability for sensitive, specific, and rapid viral nucleic acid testing. The release of nucleic acids and the subsequent reverse transcription loop-mediated isothermal amplification (RT-LAMP) were integrated into the reaction units of a microfluidic disc. The whole processing steps such as injection of reagents, fluid actuation by rotation, heating and temperature control, and detection of fluorescence signals were carried out automatically by a customized instrument. We validate the centrifugal microfluidic system using oropharyngeal swab samples spiked with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) armored RNA particles. The estimated limit of detection for armored RNA particles is 2 copies per reaction, the throughput is 21 reactions per disc, and the assay sample-to-answer time is approximately 70 min. This enclosed and automated microfluidic system efficiently avoids viral contamination of aerosol, and can be readily adapted for virus detection outside the diagnostic laboratory.
Electronic Supplementary Material
Supplementary material is available for this article at 10.1007/s11426-020-9800-6 and is accessible for authorized users.
Keywords: nucleic acid testing, virus, microfluidics, sample-to-answer, automation
Acknowledgements
This work was supported by the National Natural Science Foundation of China (91959101, 21904028), Chinese Academy of Sciences (YJKYYQ20180055, YJKYYQ20190068, ZDBS-LY-SLH025), and the Strategic Priority Research Program of Chinese Academy of Sciences (XDB36000000).
Supporting Information
A Fully Automated Centrifugal Microfluidic System for Sample-to-Answer Viral Nucleic Acid Testing
>Conflict of interest The authors declare no conflict of interest.
Footnotes
These authors contributed equally to this work.
Contributor Information
Lu Zhang, Email: zhangl@cnu.edu.cn.
Qinghua Chen, Email: chenqh@nanoctr.cn.
Jiashu Sun, Email: sunjs@nanoctr.cn.
References
- 1.Organization WH . World Health Statistics 2019: Monitoring Health for the SDGs, Sustainable Development Goals. Geneva: World Health Organization; 2019. [Google Scholar]
- 2.Fournier PE, Drancourt M, Colson P, Rolain JM, La Scola B, Raoult D. Nat Rev Microbiol. 2013;11:574–585. doi: 10.1038/nrmicro3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y, Ruan Q, Lei ZC, Lin SC, Zhu Z, Zhou L, Yang C. Anal Chem. 2018;90:5224–5231. doi: 10.1021/acs.analchem.8b00002. [DOI] [PubMed] [Google Scholar]
- 4.He P, Lv F, Liu L, Wang S. Sci China Chem. 2017;60:1567–1574. doi: 10.1007/s11426-017-9185-8. [DOI] [Google Scholar]
- 5.Land KJ, Boeras DI, Chen XS, Ramsay AR, Peeling RW. Nat Microbiol. 2019;4:46–54. doi: 10.1038/s41564-018-0295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang M, Li R, Wang J, Ling L. Sci China Chem. 2017;60:1468–1473. doi: 10.1007/s11426-017-9112-3. [DOI] [Google Scholar]
- 7.Dou M, Macias N, Shen F, Dien Bard J, Domínguez DC, Li XJ. EClinicalMedicine. 2019;8:72–77. doi: 10.1016/j.eclinm.2019.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dou M, Sanchez J, Tavakoli H, Gonzalez JE, Sun J, Dien Bard J, Li XJ. Anal Chim Acta. 2019;1065:71–78. doi: 10.1016/j.aca.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang B, Kong J, Fang X. Talanta. 2019;204:685–692. doi: 10.1016/j.talanta.2019.06.031. [DOI] [PubMed] [Google Scholar]
- 10.Yang B, Fan Y, Li Y, Yan J, Fang X, Kong J. Analyst. 2020;145:3814–3821. doi: 10.1039/C9AN02572C. [DOI] [PubMed] [Google Scholar]
- 11.Zhu Z, Zhang W, Leng X, Zhang M, Guan Z, Lu J, Yang CJ. Lab Chip. 2012;12:3907–3913. doi: 10.1039/c2lc40461c. [DOI] [PubMed] [Google Scholar]
- 12.Wu W, Zhang T, Han D, Fan H, Zhu G, Ding X, Wu C, You M, Qiu L, Li J, Zhang L, Lian X, Hu R, Mu Y, Zhou J, Tan W. Chem Sci. 2018;9:3050–3055. doi: 10.1039/C7SC05141G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu Y, Xu P, Luo J, He H, Du W. Anal Chem. 2017;89:745–750. doi: 10.1021/acs.analchem.6b03328. [DOI] [PubMed] [Google Scholar]
- 14.Broughton JP, Deng X, Yu G, Fasching CL, Servellita V, Singh J, Miao X, Streithorst JA, Granados A, Sotomayor-Gonzalez A, Zorn K, Gopez A, Hsu E, Gu W, Miller S, Pan CY, Guevara H, Wadford DA, Chen JS, Chiu CY. Nat Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu M, Chen X, Qu H, Ma L, Xu L, Lv W, Wang H, Ismagilov RF, Li M, Shen F. Anal Chem. 2019;91:8751–8755. doi: 10.1021/acs.analchem.9b01270. [DOI] [PubMed] [Google Scholar]
- 16.Dou M, Sanjay ST, Dominguez DC, Zhan S, Li XJ. Chem Commun. 2017;53:10886–10889. doi: 10.1039/C7CC03246C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang TZ, Zhang Y, Huang GL, Wang C, Xie L, Ma L, Li ZY, Luo XB, Tian H, Li Q, Li X, Lv ZY, Bao XF. Sci China Chem. 2012;55:508–514. doi: 10.1007/s11426-012-4543-8. [DOI] [Google Scholar]
- 18.Wu L, Ding H, Qu X, Shi X, Yang J, Huang M, Zhang J, Zhang H, Song J, Zhu L, Song Y, Ma Y, Yang C. J Am Chem Soc. 2020;142:4800–4806. doi: 10.1021/jacs.9b13782. [DOI] [PubMed] [Google Scholar]
- 19.Li X, Zhang D, Zhang H, Guan Z, Song Y, Liu R, Zhu Z, Yang C. Anal Chem. 2018;90:2570–2577. doi: 10.1021/acs.analchem.7b04040. [DOI] [PubMed] [Google Scholar]
- 20.Feng Q, Zhang L, Liu C, Li X, Hu G, Sun J, Jiang X. Biomicrofluidics. 2015;9:052604. doi: 10.1063/1.4922957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng Q, Sun J, Jiang X. Nanoscale. 2016;8:12430–12443. doi: 10.1039/C5NR07964K. [DOI] [PubMed] [Google Scholar]
- 22.Zhang H, Liu Y, Wang J, Shao C, Zhao Y. Sci China Chem. 2019;62:87–94. doi: 10.1007/s11426-018-9340-y. [DOI] [Google Scholar]
- 23.Chen P, Chen C, Liu Y, Du W, Feng X, Liu BF. Sens Actuat B-Chem. 2019;283:472–477. doi: 10.1016/j.snb.2018.12.060. [DOI] [Google Scholar]
- 24.Li L, Geng Y, Xiang Y, Qiang H, Wang Y, Chang J, Zhao H, Zhang L. Anal Chim Acta. 2019;1062:102–109. doi: 10.1016/j.aca.2019.02.019. [DOI] [PubMed] [Google Scholar]
- 25.Yi Q, Cai D, Xiao M, Nie M, Cui Q, Cheng J, Li C, Feng J, Urban G, Xu YC, Lan Y, Du W. Biosens Bioelectron. 2019;135:200–207. doi: 10.1016/j.bios.2019.04.003. [DOI] [PubMed] [Google Scholar]
- 26.Feng Q, Liu J, Li X, Chen Q, Sun J, Shi X, Ding B, Yu H, Li Y, Jiang X. Small. 2017;13:1603109. doi: 10.1002/smll.201603109. [DOI] [PubMed] [Google Scholar]
- 27.Cai D, Xiao M, Xu P, Xu YC, Du W. Lab Chip. 2014;14:3917–3924. doi: 10.1039/C4LC00669K. [DOI] [PubMed] [Google Scholar]
- 28.Wei X, Zhou W, Sanjay ST, Zhang J, Jin Q, Xu F, Dominguez DC, Li XJ. Anal Chem. 2018;90:9888–9896. doi: 10.1021/acs.analchem.8b02055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dou M, Sanjay ST, Dominguez DC, Liu P, Xu F, Li XJ. Biosens Bioelectron. 2017;87:865–873. doi: 10.1016/j.bios.2016.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang X, Loeb JC, Manzanas C, Lednicky JA, Fan ZH. Angew Chem Int Ed. 2018;57:17211–17214. doi: 10.1002/anie.201809993. [DOI] [PubMed] [Google Scholar]
- 31.Song J, Mauk MG, Hackett BA, Cherry S, Bau HH, Liu C. Anal Chem. 2016;88:7289–7294. doi: 10.1021/acs.analchem.6b01632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahmed MG, Abate MF, Song Y, Zhu Z, Yan F, Xu Y, Wang X, Li Q, Yang C. Angew Chem Int Ed. 2017;56:10681–10685. doi: 10.1002/anie.201702675. [DOI] [PubMed] [Google Scholar]
- 33.Song J, Liu C, Mauk MG, Peng J, Schoenfeld T, Bau HH. Anal Chem. 2018;90:1209–1216. doi: 10.1021/acs.analchem.7b03834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Zhang L, Sun J, Liu Y, Ma X, Cui S, Ma L, Xi JJ, Jiang X. Anal Chem. 2014;86:7057–7062. doi: 10.1021/ac5014332. [DOI] [PubMed] [Google Scholar]
- 35.Zhang L, Zhang Y, Wang C, Feng Q, Fan F, Zhang G, Kang X, Qin X, Sun J, Li Y, Jiang X. Anal Chem. 2014;86:10461–10466. doi: 10.1021/ac503072a. [DOI] [PubMed] [Google Scholar]
- 36.Zhang L, Tian F, Liu C, Feng Q, Ma T, Zhao Z, Li T, Jiang X, Sun J. Lab Chip. 2018;18:610–619. doi: 10.1039/C7LC01234A. [DOI] [PubMed] [Google Scholar]
- 37.Zhang L, Ding B, Chen Q, Feng Q, Lin L, Sun J. TrAC Trends Anal Chem. 2017;94:106–116. doi: 10.1016/j.trac.2017.07.013. [DOI] [Google Scholar]
- 38.Ye X, Li Y, Fang X, Kong J. ACS Sens. 2020;5:1132–1139. doi: 10.1021/acssensors.0c00087. [DOI] [PubMed] [Google Scholar]
- 39.Kim D, Lee JY, Yang JS, Kim JW, Kim VN, Chang H. Cell. 2020;181:914–921.e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen JM, Huang PC, Lin MG. Microfluid Nanofluid. 2008;4:427–437. doi: 10.1007/s10404-007-0196-x. [DOI] [Google Scholar]


