Abstract
COVID-19 pandemic outbreak is the most astounding scene ever experienced in the 21st century. It has been determined to be caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). With the global pandemic, the lack of efficient rapid and accurate molecular diagnostic testing tools has hindered the public opportunely response to the emerging viral threat. Herein, a DNA nanoscaffold hybrid chain reaction (DNHCR)-based nucleic acid assay strategy is reported for rapid detection of SARS-CoV-2 RNA. In this method, the DNA nanoscaffolds have been first constructed by the self-assembly of long DNA strands and self-quenching probes (H1). Then, the SARS-CoV-2 RNA will initiate the hybridization of H1 and free H2 DNA probes along the nanoscaffold, and an illuminated DNA nanostring is instantly obtained. By taking advantages of the localization design of the H1 probes and the temperature tolerance of the isothermal amplification, the proposed DNHCR method can detect target at short responding time (within 10 min) and mild condition (15 °C–35 °C). Moreover, the reliability of DNHCR method in serum and saliva samples have also been validated. Therefore, DNHCR-based method is expected to provide a simple and faster alternative to the traditional SARS-CoV-2 qRT-PCR assay.
Keywords: DNA self-Assembly, DNA nanoscaffold, Isothermal amplification, RNA detection, SARS-CoV-2
Highlights
-
•
A simple and specific method has proposed for the assay of SARS-CoV-2 RNAs.
-
•
Compared with the qRT-PCR, this new method is rapid (about 10 min) and has a wide temperature range (15°C–35°C).
-
•
This method can complete the detection in only one step, thereby avoiding possible secondary pollution.
-
•
This method proposed a trajectory hybrid chain reaction , which can improve the detection efficiency.
1. Introduction
In December 2019, a novel coronavirus (SARS-CoV-2) has caused an outbreak severe pneumonia disease COVID-19, and rapidly spread to produce a global pandemic (Wang et al., 2020). As of May, 150 000 people have died among the more than 2.2 million confirmed cases. A novel coronavirus, designated as SARS-CoV-2, has been implicated as the causative agent (Zhu et al., 2020). With the outbreak of COVID-19, the World Health Organization (WHO) has been urging the international community to perform massive diagnostic testing to fight against the transmission of the virus and decrease the number of undetected cases. Because detection tools can help researchers understand the epidemiology of this disease. And the screening of COVID-19 in the primary stage will block the larger-scale spread of pathogen and promote patients to receive treatment as soon as possible, improving the cure rate (Narváeza and Dincer, 2020; Cui and Zhou, 2020; Corman et al., 2020). Therefore, rapid detection technology has shown great application value in the clinical diagnosis of COVID-19 patients.
Assays using diagnostic quantitative real-time PCR (qRT-PCR) to detect SARS-CoV-2 virus have been played an important role in preventing and controlling COVID-19 outbreak (Read Online, 2020). QRT-PCR is also an emergency use authorization (EUA) approaches approved by the US Centers for Disease Control and Prevention (CDC) (Centers for Disease Control and Prevention, 2020; Chu et al., 2020; Shortage, 2020). However, qRT-PCR technology relies on expensive reagents and sophisticated instruments, and the steps are complicated and time-consuming. So it cannot meet the rapidly growing demand of suspected patients and asymptomatic infected patients (Bo-gyung, 2020; Bachman, 2013). For the COVID-19 testing, apart from the aforementioned viral RNA, the detection methods based on immunoglobulin (IgM/IgG) antibodies have expected to detect SARS-CoV-2 quickly and easily. But IgM antibodies are produced between 4 and 10 days after infection, while IgG response is produced around 2 weeks. Thus, in the early stage of infection, low-abundance antibodies in the sample will lead to false negative results (Grifoni et al., 2020; Zhang et al., 2020). However, with the occurrence of the asymptomatic infection and its transmission potential, the number of people who need to screen is greatly increase. And the SARS-CoV-2 RNA detection of COVID-19 patients can timely evaluate the patients’ treatment effect and prognosis (World Health Organization, 2020a; Bai et al., 2020). Therefore, there is an urgent need for diagnostic methods that can rapidly and conveniently detect SARS-CoV-2 infection.
To satisfy this need, currently, researchers have developed isothermal amplification methods for SARS-CoV-2 RNA analysis, such as recombinase polymerase amplification (RPA) and loop-mediated isothermal amplification (LAMP) (Craw and Balachandran, 2012; Notomi et al., 2000; Zaghloul and El-Shahat, 2014). Although these methods have been reported to rapidly and accurately detection of SARS-CoV-2, there are still some deficiencies in the cost and operability. For example, RPA amplification requires the participation of three enzymes, and the appropriate temperature of LAMP is about 63 °C.
Here, we present the development of a DNA nanoscaffold-based hybrid chain reaction (DNHCR) method for assay of SARS-CoV-2 RNA. Compared with previously reported techniques, this method has the following advantages: (1) high signal gain; (2) short reaction time with high specificity; (3) room temperature response and easily accessibility; (4) cost-effectiveness and readily available reagents. In addition, we have also verified the reliability of our method in complex samples, and thus we believe that this DNHCR-based technology may be of great potential in routine clinical diagnosis.
2. Experimental section
2.1. Chemicals and reagents
The DNA oligonucleotides used in this work (Table S1) were synthesized and purified (HPLC) by Sangon Biotech. Co. Ltd. (Shanghai, China). The RNA sequence was synthesized by Takara Bio. (Dalian China). T4 DNA ligase, exonuclease I (Exo I), exonuclease III (Exo III), phi29 DNA polymerase and RiboLock RNase Inhibitor were bought from Thermo Fisher Scientific (Waltham, USA). Deoxyribonucleoside 5′-triphosphate mixture (dNTPs) was bought from New England Biolabs (Beijing, China). Cell lysis solution were purchased from Sangon. Solutions used in all experiments were obtained from ultra-pure water purified with a Millipore system (> 18.0 MΩ). The human normal serum was obtained from First Affiliated Hospital of Nanjing Medical University. All other chemical agents were purchased from Sigma (St. Louis, MO, USA) unless otherwise indicated. The sequence that can trigger the reaction is the conserved regions of artificially synthesized SARS-CoV-2 RNA published by the Centers for Disease Control (CDC) (World Health Organization, 2020b; James et al., 2020).
2.2. Electrophoresis analysis
2% agarose gel electrophoresis was prepared using 1 × TBE buffer. The loading sample was prepared by mixing 1 μL nucleic acid sample, 1.5 μL 6 × loading buffer, 1 μL GelRed dye, and 6.5 μL H2O, then placed the mixture for 3 min before injected into agarose gel electrophoresis. The agarose gel electrophoresis was run at 110 V for 60 min in 1 × TBE buffer and visualized via a Molecular Imager Gel Doc XR.
2.3. Synthesis of long DNA strands by RCA
Firstly, 10 μL ultrapure water, 30 μL DNA ligation buffer (2 ×), 10 μL phosphorylated template (10 μM) and 50 μL primer (10 μM) were mixed in centrifuge tube to synthesize the circular DNA template. Then the mixture was heated at 95 °C for 5min, and the solution was cooled slowly to room temperature. After adding T4 DNA ligase (5 μL, 40 000 U/mL) to the above solution and mixing well, it was left at room temperature for 30 min. We used Exonuclease I and Exonuclease III to digest linear nucleic acid, and further purified the circular DNA template with PCR product purification. The 10 μL circular DNA template was added to 20 μL of the RCA reaction mixture containing: 2 μL Phi29 DNA polymerase (10 ×) reaction buffer, 4 μL dNTPs (10 mM), 2 μL BSA (10 ×), 1 μL DEPC water and 1 μL phi29 DNA polymerase (1 000 U/ml) incubating at 35 °C for 40 min and heating treated at 65 °C for 10 min to stop the reaction. Finally, the concentration of the RCA product treated with PCR purification kit was measured by Thermo NanoDrop 2000).
2.4. Design of DNA nanoscaffold for DNHCR
The DNA nanoscaffold consists of a long DNA single strand (RCA product) and some H1 probes. First, heating the long DNA strand and H1 probes were heated in Thermo Cell at 95 °C for 5 min respectively, and then slowly cooled down to room temperature (> 30 min). 25 μL H1 (10 μM), 25 μL long DNA strand (0.6 μM) and 125 μL Tris-MgCl2 buffer (500 mM, pH = 8.0) were mixed thoroughly in a 200 μL centrifuge tube to prepare the DNA nanoscaffolds. The mixture was incubated at room temperature for 30 min, and H1 probes were fully hybridized with long DNA strands to synthesis DNA nanoscaffolds.
2.5. Target RNA-initiated DNHCR reaction
First, a volume of 60 μL reaction mixture containing 6 μL DNA nanoscaffold (0.6 μM), 6 μL target RNA, 1 μL H2 probes (1 μM) and 37 μL Tris-MgCl2 buffer were mixed incubating in a thermal cycler. Here, we have verified the reaction efficiency under the different total volume and selected 50 μL as the appropriate volume (Fig. S1). The reaction mixture was then transferred to a black 384 well microplate (Fluotrac 200, Greiner, Germany) to fluorescence measurements. The measurement settings were excitation at 488 nm for FAM dye. The fluorescence emission spectra were measured with a fluorescence microplate reader (BioTek Instrument, Winooski, VT, USA) at different excitation wavelength.
3. Results and discussion
3.1. Principle of the method
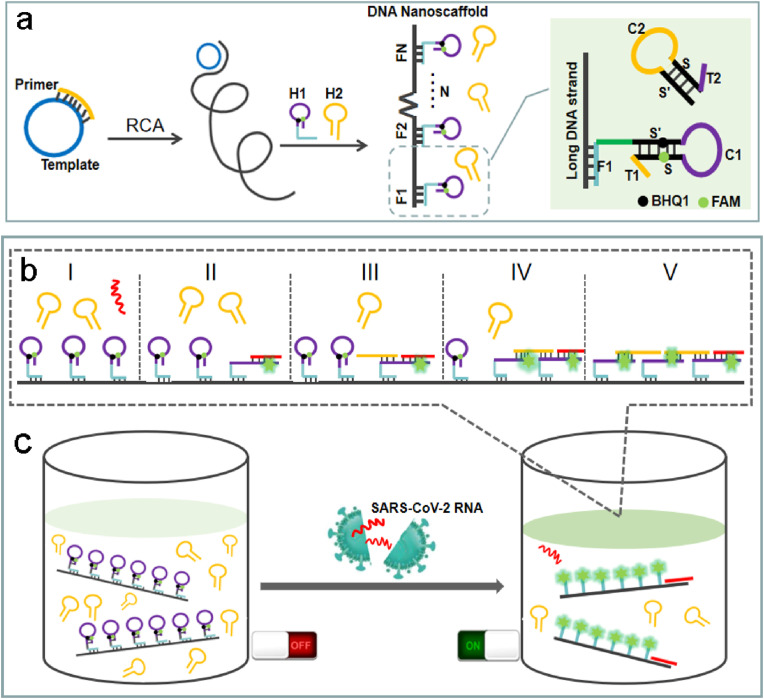
The designed principle for the rapid detection of SARS-CoV-2 RNA is displayed in Scheme 1 . Firstly, a long DNA strand containing repetitive fragments has been generated by RCA, which contains multiple H1 installation footholds, and the spacing distance of H1 can be programmed by changing the template sequence of RCA. RCA is an isothermal enzymatic process in which circular DNA templates are amplified using DNA or RNA primers to form long tandem repeats of single-stranded DNA or RNA. RCA technology is widely used in biomedical research and nanobiology research because of its simplicity and versatility (Ali et al., 2014; Tian et al., 2020; Michael et al., 2016). Then, the nanoscaffold is constructed by hybridizing the DNA hairpin probes (H1) with the long DNA strand (Scheme 1a). The DNA nanoscaffolds can be conveniently prepared in large quantities for subsequent detection. H1 is composed of foothold sequence F1 (anchored to DNA long strand), hairpin structure S’C1S and a tail T1, which is labeled with 5-carboxyfluorescein (FAM) dye and its black hole quencher (BHQ1) molecule to form a self-quenching probe and the fluorescence will recover upon hairpin opening. H2 is a hairpin structure (S’C2S) with a tail T2. Tail T1 in H1 is complementary to C2 in H2, and tail T2 in H2 is complementary to C1 in H1. When the target SARS-CoV-2 RNA is present, it triggers the cascade reaction along the DNA nanoscaffold (Scheme 1b). Firstly, the target RNA will hybridize with T1 and S, causing H1 to unfold to restore fluorescence (Scheme 1b II), and then H2 will hybridize with C1 and S′, causing H2 to unfold (Scheme 1b III) and continue to hybridize with adjacent H1 along the DNA nanoscaffold (Scheme 1b IV). Meanwhile, due to the compact arrangement of H1 on the DNA nanoscaffold, one target RNA can instantly light up the whole nanoscaffold with highly amplified signal gain (Scheme 1c).
Scheme 1.
A DNA nanoscaffold hybrid chain reaction (DNHCR)-based method for the detection of SARS-CoV-2 RNA. a, The synthesis of DNA nanoscaffolds based on the hybridization of H1 probes with long DNA strands produced by RCA. b, The procedure of DNHCR triggered by the SARS-CoV-2 RNA. c, The detection of the target SARS-CoV-2 RNA in the mixed solution of DNA nanoscaffolds and H2 probes.
3.2. Feasibility of target SARS-CoV-2 RNA triggered DNHCR
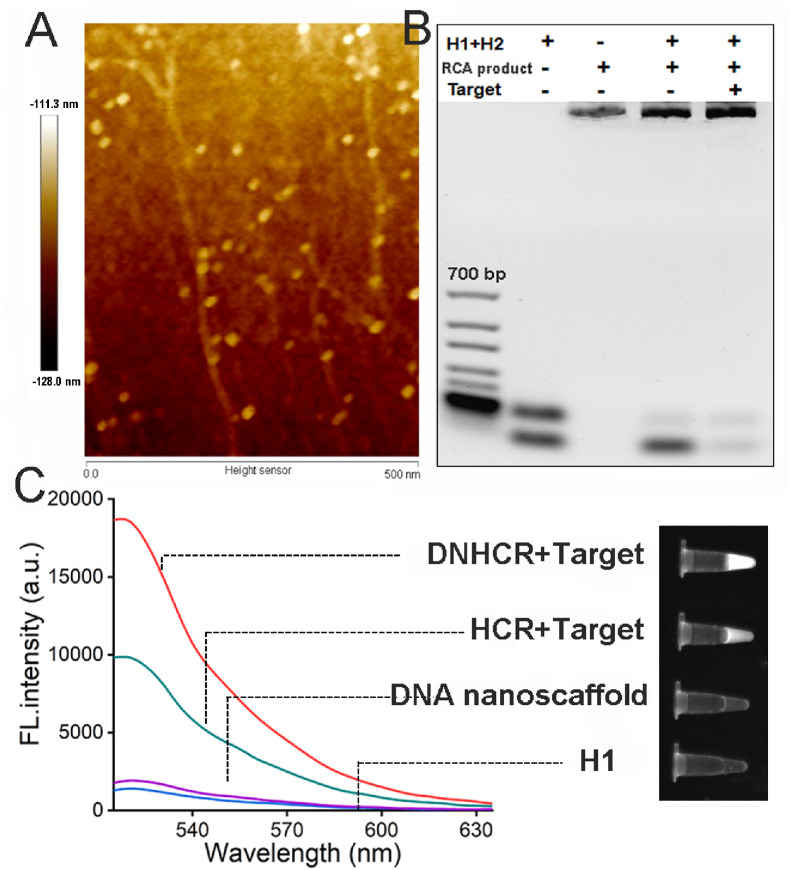
The elongation of template to form DNA long strand through RCA has been confirmed by 2% agarose gel electrophoresis experiment (Fig. S2). The successful formation of the nanoscaffold can be directly seen in the AFM phase image (Fig. 1 A). The result shows the long leaner structure with dispersed dots arranged along the side which is consistent with the leaner structure of the RCA product and the binding of large number of H1 probes. Then, the reaction process of proposed DNHCR-based system has been studied by gel electrophoresis. As can be seen in Fig. 1B, the mixture of H1 and H2 resulted in two bands in lane 2, which indicated the independent existence of the two probes in solution. After the addition of the nanowire formed via RCA to the mixture of H1 and H2, the band corresponding to H1 almost disappeared while the band of nanowire became brighter, which verified that the H1 has successfully assembled on the nanowire to form a nanoscaffold. Finally, the DNHCR reaction triggered by the target caused the consuming of H2 and the formation of an extra double helix leading to the darker of the H2 band and the brighter of the nanoscaffold band (lane 5). Meanwhile, the feasibility of the method has also confirmed by the result of florescent spectrophotometer (Fig. 1C). Without the addition of the target, both H1 and the nanoscalfold show low background fluorescence (purple and blue line). In the presence of the target, free H1 and H2 is capable of hybridizing to each other to trigger HCR, which led to the recovery of the fluorescence (green line). As we expected, with the help of the nanoscalffold, much stronger fluorescence intensity can be observed with the same concentration of the target. These results indicate that the DNHCR-based method could significantly improve the signal gain compared to the conventional HCR.
Fig. 1.
Feasibility of this method for the SARS-CoV-2 RNA detection. (A) AFM phase image of DNA nanoscaffold. (B) The agarose gel electrophoresis of the synthesized DNA nanoscaffolds and the DNHCR response to the target. (C) Left: fluorescence spectra of the DNHCR and HCR in the absence and presence of the target (Excitation, 494 nm). Right: UV imaging of the DNHCR and HCR in the absence and presence of the target.
3.3. Optimization of experimental conditions
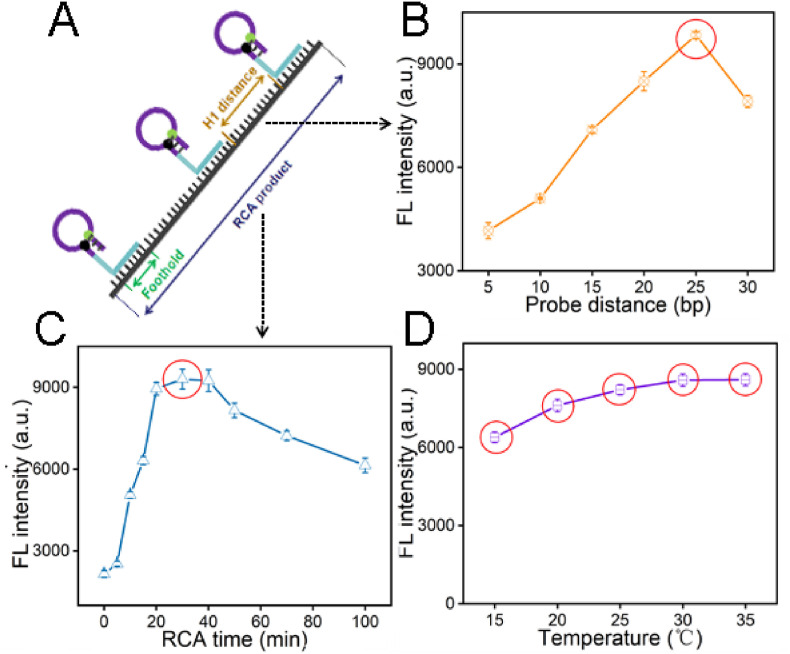
Some important variants of DNHCR method have been optimized for the better performance. Firstly, we have studied the effect of the stem length of the H1 probe and the length of H1 hybridization with DNA long strand (i.e. foothold) on background signal of DNHCR-based method. As shown in Fig. S3, the largest signal-to-noise ratio has generated in 14 bases stem length, and with the foothold length increases, the intensity of the background signal enhanced. Considering the background signal and the stability of the nanoscaffold, the stem with 14 bases and foothold with 12 base pairs are used in the subsequent research. We have optimized the distance between different H1 probes along the RCA products as well, 25 bases suggested to be the most proper distance due to the maximum fluorescent intensity (Fig. 2 B). RCA time may affect the efficiency of the DNHCR method by affecting the length of DNA scaffolds. As shown, the signal intensity increases as the RCA time increases from 0 to 30 min and then reduce when the time is prolonged from 40 to 100 min (Fig. 2C). Thus, 30 min has been selected as the best RCA time. Finally, the effect of detection temperature has been investigated by incubating target RNA with DNHCR at different temperatures. As shown in Fig. 2D, the florescent intensity is relatively stable at the temperature ranging from 15 °C to 35 °C. Therefore, compared with qRT-PCR, our method can be performed efficiently at room temperature without resorting to temperature change equipment.
Fig. 2.
Optimize of the experimental parameters of DNHCR. (A) The structure of DNA nanoscaffolds. (B) The effect of different separation distances of H1 probes assembled on long DNA strands. (C) The influence of the RCA time from 0 to 100 min. (D) The effect of the temperature from 15 °C to 35 °C. All the values selected by the red circle are the best condition. The data error bars indicate mean ± SD (n = 3). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.4. Kinetic analysis of accelerated response in DNHCR
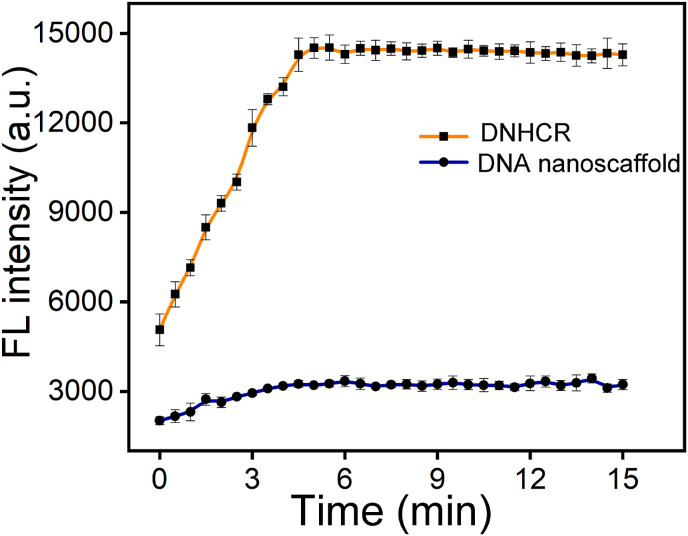
Under optimized conditions, the kinetic curve has been utilized to monitoring the whole process of the detection in homogeneous solution. As shown in Fig. 3 , in the absence of the target, almost no fluorescence signal is detected in both DNHCR and HCR systems. With the introduction of the target, in DNHCR-based method, the fluorescence intensity increased rapidly and reached a plateau at about 7 min. In comparison, it took more than 30 min for the conventional method to complete the detection. These results suggest that the nanoscalffold could distinctly reduce the response time, which is required for the rapid detection of the SARS-CoV-2. In addition, by quantitative analysis, the initial rate of DNHCR is 15.6 fold higher than HCR (Fig. S4). The reason may be that the hybridizing of large number of H1 to the nanowire in DNHCR increased the local concentration of H1 accelerating the random collision of H1 and H2.
Fig. 3.
Time-dependent fluorescence spectra of DNHCR in response to 50 nM target RNA, respectively. The data error bars indicate mean ± SD (n = 3).
3.5. Analytical performance of DNHCR method
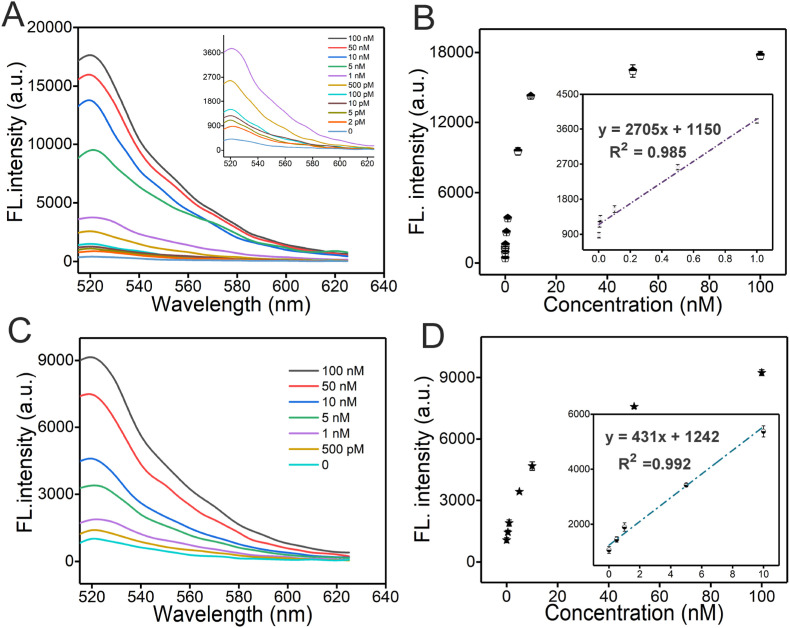
The amplification efficiency of the proposed method in quantitative analysis of target RNA has been studied by recording the fluorescence intensity. Fig. 4 A illustrates the changes in fluorescence intensity in responding to the different target RNA concentrations, it increased gradually upon enhancement of the concentration of the target. To obtain a leaner relationship between the FL intensity and the concentration, the peak value is selected. As shown in Fig. 4B, for DNHCR method, the fluorescence intensity is linearly dependent on the RNA concentration with a correlation equation of y = 2705 X + 1150 (R2 = 0.985). The limit of detection (LOD) is approximately 0.96 pM. In addition, the LOD of HCR method is about at 232 pM with a correlation equation of y = 431 X + 1242 (R2 = 0.992) (Fig. 4D). Comparing with the conventional HCR method the fluorescence intensity of DNHCR increased about 2-fold at the same concentration of the target (Fig. 4C).
Fig. 4.
The sensitivity analysis of DNHCR method and traditional HCR method. (A), (C) Fluorescence spectra upon addition of target SARS-CoV-2 RNA with various concentrations in DNHCR and HCR. Inset: Fluorescence responses to target at low concentrations. (B), (D) Scatter plot of fluorescence intensity as a function of the SARS-CoV-2 RNA concentrations. Inset: Linear relationship between the fluorescence intensity and the concentrations of SARS-CoV-2 RNA.
3.6. Specificity and stability of DNHCR method
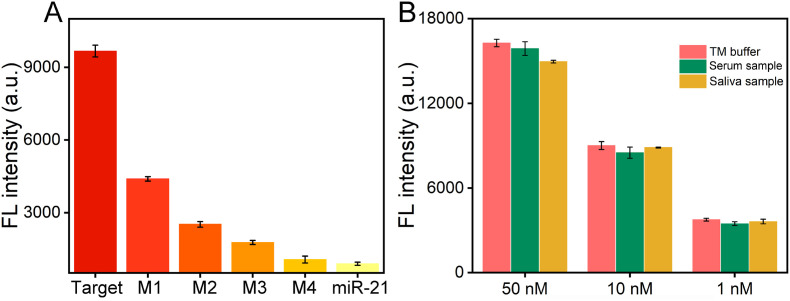
A significant challenge for SARS-CoV-2 RNA analysis is the specificity, which is of great importance in accurately detecting target and avoiding false positives. Here, the specificity has been investigated by the comparison with the control RNA, miR-21 and single to four bases mismatched target RNA. As shown in Fig. 5 A, the sample fluorescence intensity dropped dramatically (2.4 fold) upon the addition of the sequence with 1 mismatched base. With the increasing of the number of mismatched bases, the fluorescence reduced rapidly. It decreases to be similar to that of the sample at the present of random sequence when 4 mismatched bases occurred in the sequence. This result indicates the ability of this method to discriminate the mismatched sequences. As can be seen in Fig. 5B, tests in serum and saliva samples have verified the practical ability of our method. In addition, the precision and accuracy of DNHCR method have been studied. As shown in Table S2, for SARS-CoV-2 RNA, the coefficient of variation (CV) at intra-assay level and inter-assay level range from 0.09% to 0.8% and 1%–4%, respectively. Furthermore, the recovery values range from 89% to 108.7%. The results demonstrated good repeatability and stability of the DNHCR method at different environments and target concentrations, which indicates that our method has the good propensity for clinical application.
Fig. 5.
(A) Selectivity response of this method against mutation probes (M1~M4 refer to 1, 2, 3 and 4 base mutation) and the random sequence miR-21. The concentration of the SARS-CoV-2 RNA is 10 nM, the concentrations of mutation probes and random sequence miR-21 are 100 nM. The data error bars indicate mean ± SD (n = 3). (B) Detection results in TM buffer, 10% serum and 10% saliva sample by our method, respectively. The data error bars indicate mean ± SD (n = 3).
4. Conclusion
In summary, a DNHCR method has been successfully developed for rapid detection of COVID-19 based on SARS-CoV-2 RNA triggered isothermal amplification. Due to the localized acceleration of the DNA probes, the reaction time reduced to within about 10 min. And high signal gain is obtained in wide temperature range of 15 °C–35 °C. Compared to previously reported biosensors for SARS-CoV-2 RNA detection, the proposed approach achieves a significantly simplified operation and saves the cost. However, in our method, the output of the fluorescent signal also requires the assistance of the equipment, so further efforts need to be made in point of care testing to meet the more convenient self-test requirements. We are currently investigating the potential to constructing a lateral flow assay device to achieve a portable and intuitive detection of SARS-CoV-2.
CRediT authorship contribution statement
Jin Jiao: Conceptualization, Methodology, Software, Investigation, Validation, Data curation, Writing - original draft. Chengjie Duan: Methodology, Investigation, Data curation. Lan Xue: Data curation, Validation. Yunfei Liu: Software, Validation. Weihao Sun: Resources, Validation, Supervision. Yang Xiang: Funding acquisition, Supervision, Writing - review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work is supported by the National Natural Science Foundation of China (Grant No. 81672570 and 81370926 to Yang Xiang).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2020.112479.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Ali M.M., Li F., Zhang Z., Zhang K., Kang D., Ankrum J.A., Le X.C., Zhao W. Rolling circle amplification: a versatile tool for chemical biology, materials science and medicine. Chem. Soc. Rev. 2014;43:3324–3341. doi: 10.1039/c3cs60439j. [DOI] [PubMed] [Google Scholar]
- Bachman J. Reverse-transcription PCR (RT-PCR) Methods Enzymol. 2013;530:67–74. doi: 10.1016/B978-0-12-420037-1.00002-6. [DOI] [PubMed] [Google Scholar]
- Bai Y., Yao L.S., Wei T., Tian F., Jin D.Y., Chen L.J., Wang M.Y. Presumed asymptomatic carrier transmission of COVID-19. JAMA. 2020;323:1406–1407. doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bo-gyung K. The Korea Herald; 2020. Tests in Recovered Patients Found False Positives, Not Reinfections, Experts Say.http://www.koreaherald.com/view.php?ud=20200429000724 [Google Scholar]
- Centers for Disease Control and Prevention . 2020. Real-time RT-PCR Panel for Detection 2019-nCoV (US Centers for Disease Control and Prevention)https://www.cdc.gov/coronavirus/2019-ncov/lab/ [Google Scholar]
- Chu D.K.W., Pan Y., Cheng S.M.S., Hui K.P.Y., Krishnan P., Liu Y., Ng D.Y.M., Wan C.K.C., Yang P., Wang Q., Peiris M., Poon L.L.M. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin. Chem. 2020;66:549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V., Bleicker T., Brünink S., Charité C.D. 2020. Diagnostic Detection of Wuhan Coronavirus 2019 by Real-Time RT PCR, Virology.https://virologie-ccm.charite.de/en/ christian.drosten@charite.de. [Google Scholar]
- Craw P., Balachandran W. Isothermal nucleic acid amplification technologies for point-of-care diagnostics: a critical review. Lab Chip. 2012;12:2469–2486. doi: 10.1039/c2lc40100b. [DOI] [PubMed] [Google Scholar]
- Cui F.Y., Zhou H.S. Diagnostic methods and potential portable biosensors for coronavirus disease. Biosens. Bioelectron. 2020;165:112349–112356. doi: 10.1016/j.bios.2020.112349. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to. SARS-CoV-2 Cell Host Microbe. 2020;27:671–680. doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P.B., Deng X.D., Yu G.X., Clare L.F., Venice S., Jasmeet S., Miao X., Jessica A.S., Andrea G., Alicia S.G., Kelsey Z., Allan G., Elaine H., Gu W., Steve M., Pan C.Y., Hugo G., Debra A.W., Janice S.C., Charles Y.C. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael G., Kool Mohsen, Eric T. The discovery of rolling circle amplification and rolling circle transcription. Acc. Chem. Res. 2016;49:2540–2550. doi: 10.1021/acs.accounts.6b00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narváeza E.M., Dincer C. The impact of biosensing in a pandemic outbreak: COVID-19, Biosens. Bioelectron. 2020;163:112274–112280. doi: 10.1016/j.bios.2020.112274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:63–73. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read Online-Handbook of COVID-19 Prevention and Treatment. Pdf. 2020. https://www.alibabacloud.com/universal-service/pdf_ [WWW Document] [Google Scholar]
- Shortage of RNA Extraction Kits Hampers Efforts to Ramp up COVID-19 Coronavirus Testing. Chemical & Engineering News; 2020. https://cen.acs.org/analytical-chemistry/diagnostics/Shortage-RNA-extraction-kits-hampers/98/web/2020/03 [WWW Document] [Google Scholar]
- Tian Bo, Gao Fei, Fock Jeppe, Dufva Martin, Hansen Mikkel Fougt. Homogeneous circle-to-circle amplification for real-time optomagnetic detection of SARS-CoV-2 RdRp coding sequence. Biosens. Bioelectron. 2020;165:112356–112363. doi: 10.1016/j.bios.2020.112356. [DOI] [PubMed] [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . 2020. Coronavirus Disease 2019 (COVID-19) Situation Report 44. [Google Scholar]
- World Health Organization . 2020. Diagnostic Detection of Wuhan Coronavirus 2019 by Real-Time RT-PCR.https://www.who.int/docs/default-source/coronaviruse/wuhan-virus-assay [Google Scholar]
- Zaghloul H., El-Shahat M. Recombinase polymerase amplification as a promising tool in hepatitis C virus diagnosis. World J. Hepatol. 2014;6:916–922. doi: 10.4254/wjh.v6.i12.916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Du R.H., Li B., Zheng X.S., Yang X.L., Hu B., Wang Y.Y., Xiao G.F., Yan B., Shi Z.L., Zhou P. Molecular and serological investigation of 2019-nCoV infected patients: implications of multiple shedding routes. Emerg. Microb. Infect. 2020;9:386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D.Y., Wang W.L., Li X.W., Yang B., Song J.D., Zhao X., Huang B.Y., Shi W.F., Lu R.J., Niu P.H., Zhan F.X., Ma X.J., Wang D.Y., Xu W.B., Wu G.Z., Gao G.F., Tan W.J. A novel coronavirus from patients with pneumonia in China. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.