Abstract
Aromatase catalyzes conversion of testosterone to estradiol and is expressed in a variety of tissues, including the brain. Suppression of aromatase adversely affects metabolism and physical activity behavior, but mechanisms remain uncertain. The hypothesis tested herein was that whole body aromatase deletion would cause gene expression changes in the nucleus accumbens (NAc), a brain regulating motivated behaviors such as physical activity, which is suppressed with loss of estradiol. Metabolic and behavioral assessments were performed in male and female wild-type (WT) and aromatase knockout (ArKO) mice. NAc-specific differentially expressed genes (DEGs) were identified with RNAseq, and associations between the measured phenotypic traits were determined. Female ArKO mice had greater percent body fat, reduced spontaneous physical activity (SPA), consumed less energy, and had lower relative resting energy expenditure (REE) than WT females. Such differences were not observed in ArKO males. However, in both sexes, a top DEG was Pts, a gene encoding an enzyme necessary for catecholamine (e.g., dopamine) biosynthesis. In comparing male and female WT mice, top DEGs were related to sexual development/fertility, immune regulation, obesity, dopamine signaling, and circadian regulation. SPA correlated strongly with Per3, a gene regulating circadian function, thermoregulation, and metabolism (r = −0.64, P = .002), which also correlated with adiposity (r = 0.54, P = .01). In conclusion, aromatase ablation leads to gene expression changes in NAc, which may in turn result in reduced SPA and related metabolic abnormalities. These findings may have significance to post-menopausal women and those treated with an aromatase inhibitor.
Keywords: Spontaneous physical activity, Brain, CYP19, Aromatase, Cognition, Metabolism, Sex differences
1. Introduction
Aromatase is required to convert testosterone to estradiol, and is the primary source of estrogen produced in both sexes across vertebrate species (Blakemore and Naftolin, 2016). Pharmacologic inhibition of aromatase is commonly used in both sexes to reduce estradiol levels in diseases involving estrogen dysregulation. Meanwhile, declining estradiol concentrations can result in metabolic dysfunction, and suppressed physical activity (Gibb et al., 2016; Muller et al., 2018; Park et al., 2016) although mechanisms are not fully understood. Moreover, although estradiol is produces in both sexes, sexual dimorphism may exist in its impact on behavior and physiology (Bowen et al., 2011; Muller et al., 2018; Park et al., 2016).
While highest concentrations of aromatase are in the gonads, aromatase is expressed in many organs, including brain and adipose tissue, where it acts in a paracrine manner (Nelson and Bulun, 2001). Ovary-intact female rodents are more active than males (Bartling et al., 2017; Klinker et al., 2017; Park et al., 2016), whereas genetic ablation of aromatase reduces locomotor activity even more dramatically than ovariectomy alone (Jones et al., 2000), suggesting that aromatase activity in the brain is essential to maintain a certain level of physical activity, even when ovarian estradiol production ceases.
Previous work from our laboratory has shown that the decrease in physical activity that occurs after ovariectomy correlates with suppression of dopamine-associated gene expression in the NAc (Park et al., 2016). This brain region is part of the mesolimbic pathway that plays a prominent role in the reward circuitry of the brain, relying on dopamine signaling to learn rewarding behaviors (e.g. food, sex, use of addictive drugs [e.g. cocaine and amphetamine]) and increases motivation to engage in those behaviors (Gorres-Martens et al., 2018; Tonn Eisinger et al., 2018). Thus, estradiol signaling in the NAc may modulate dopamine signaling; thereby, affecting motivation to engage in physical activity (Park et al., 2016).
Pharmacological aromatase inhibition is commonly used as adjuvant therapy for post-menopausal women with hormone-receptor positive breast cancer, with noteworthy increases in the incidence of cognitive impairments with such treatments (Collins et al., 2009; Jenkins et al., 2004). Indeed, the potential negative neuroendocrine consequences of aromatase inhibition are recently becoming more appreciated (Rosenfeld, 2017; Shay et al., 2018). Together with the metabolic dysfunction that already occurs post menopause, aromatase inhibition therapy may exacerbate these issues by decreasing motivation for engaging in physical activity. Indeed, aromatase inhibition therapy associates with reduced physical activity in humans (Brown et al. (2014); (de Paulo et al., 2018). In addition, males are also prescribed aromatase inhibitors, often to treat the decrease in endogenous testosterone that occurs with increasing age, but the cognitive effects of aromatase inhibition in men have not been thoroughly assessed (Dias et al., 2016). Thus, it is important to determine whether there are sex differences in risk for physical inactivity due to aromatase suppression and if so, the underlying molecular changes that might be driving such effects.
Although prior work has indicated that aromatase knock-out (ArKO) mice have suppressed physical activity (Jones et al., 2001), no studies to our knowledge have compared sexes in this regard. We hypothesized that physical activity would be significantly depressed in aromatase deficient male and female mice, but that female mice would show heightened sensitivity due to the fact that male motivation for physical activity may be primarily influenced by testosterone (Bowen et al., 2011). Additionally, we postulated that NAc gene expression patterns would be differentially regulated by sex and correlate with the degree of physical inactivity. To test these hypotheses, a systemic ArKO mouse model originally created by Ogawa et al. (Honda et al., 1998) and bred at our facilities was utilized to examine sex and genotype differences in spontaneous physical activity, as well as other behaviors (e.g., anxiety-like behaviors). RNA-seq was performed on the NAc brain region to examine for potential sex- and genotype-dependent associations between gene expression changes in the NAc and physical activity.
2. Materials and methods
2.1. Subjects
Male and female C57B16/J wild-type (WT) and ArKO mice were bred and housed at University of Missouri Animal Sciences Research Center (Columbia, MO, USA) under a normal 12–12 h light-dark cycle with room temperature maintained at 21.7 °C (71 °F) in standard polysulfone mouse cages (18.4 cm W × 29.2 cm D × 12.7 cm H; Alternative Design Manufacturing & Supply Co., Siloam Springs, AR, USA). Corn cob bedding (sterilized 1/4 in.; Andersons Lab Bedding Products, Maumee, OH, USA), food (standard chow, Purina 5008) and water were available ad libitum. All mice were weaned at 3 weeks of age and housed with siblings until genotyped. At 12 weeks of age, mice were divided based on sex and genotype (WT/ArKO) into the following groups: male (WT, n = 8; ArKO, n = 10) and female (WT, n = 11; ArKO, n = 9). Within these groups, mice were either housed singly [female (WT, n = 3; ArKO, n = 5); male (WT, n = 3; ArKO, n = 4)] or in pairs [female (WT, n = 8; ArKO, n = 4); male (WT, n = 5 {1 pair and 1 group of 3}; ArKO, n = 6)] to provide social enrichment, as recommended by our ACUC committee. Mice were singly housed, if they engaged in aggressive acts against other mice that manifested with age. All animal husbandry and experimental procedures were conducted in accordance with the NIH Guide for Care and Use of Laboratory Animals and approved by the University of Missouri Institutional Animal Care and Use Committee (ACUC, Protocol # 8573) prior to initiation of experiments.
2.2. Metabolic chambers
At 12 weeks of age, animals were placed in the indirect calorimetry (MET) chambers for a period of 72 h (Promethion; Sable Systems International, Las Vegas, Nevada) to assess metabolic activity parameters including total energy expenditure (TEE), resting energy expenditure (REE), and spontaneous physical activity (SPA) as quantified by the total distance (m) traveled within the home cage. This was the only period throughout the study mice were housed in the metabolic chamber cages (Supplementary Fig. 1). Data from the first 24 h was not used as this time was considered the habituation period. The following 48-h run captured at least two light and two dark cycles of REE. Mice were fed ad libitum for the duration of the study (standard chow, Purina 5008).
2.3. Body composition and food intake
Percent body fat and percent lean mass were measured within 24 h of removal from the metabolic chambers via a nuclear magnetic resonance imaging whole-body composition analyzer (EchoMRI 4in1/1100; Echo Medical Systems, Houston, TX). Total food consumed over a 5-day observation period (a time where mice were undisturbed in their home cage) was measured for each group following assessment of body composition and reported as average grams consumed per day (Supplementary Fig. 1).
2.4. Elevated plus maze
The elevated plus maze test was utilized to examine exploratory and anxiety-like behaviors in mice. The procedure was conducted as described previously when mice reached 15 weeks of age (Fountain et al., 2008; Jasarevic et al., 2011) (Supplementary Fig. 1). Briefly, each animal was placed in the center of the maze and was allowed to explore for a single 300 s trial. Each trial was recorded with a Sony HD Han-dycam HDR-CX440 (San Diego, CA) camcorder and videos were analyzed using Observer 11 software (Noldus Technologies, Leesburg, VA, USA). Behavioral indices measured that were considered to be exploratory, non-anxious behaviors included number of entries into the center of the maze, duration of time spent in the center (s), number of entries into the open arms of the maze, time spent in the open arms (s), number of rearing instances, duration of head dipping behavior below the level of the platform (s), and duration of time spent mobile. Behaviors examined that are considered more anxiety-like included number of entries and time spent (s) in the closed arms of the maze, number and duration (s) of grooming events, and time spent immobile. Male and female mice were assessed on the same day with all testing being performed between 8:00 am and 12:00 pm.
2.5. Brain collection & preparation
After animals were humanely euthanized in accordance with American Veterinary Medical Association guidelines for euthanasia of laboratory animals, and whole brain was collected and snap-frozen in liquid nitrogen and stored at −80 °C until transcriptome analyses. Brains were rapidly thawed at room temperature to obtain tissue punches of the NAc region. This brain region was located and dissected using the Allen mouse brain atlas (P56, coronal section) (Lein et al., 2007). Brain slices were obtained with a Zivic brain block (Adult mouse brain slicer matrix with 1.0 mm coronal section slice intervals; Zivic Instruments, Pittsburgh, PA) as a guide. Micro punch samples of the NAc were acquired using a 2 mm Harris micro punch (GE Whatman Harris micro punch, 2 mm; GE Healthcare Biosciences, Marlborough, MA). Bilateral NAc punches were combined and snap-frozen in liquid nitrogen and stored at −80 °C until further analyses.
2.6. RNA isolation of NAc region
NAc samples were homogenized in QIAzol solution using a tissue homogenizer (TissueLyser LT, Qiagen, Valencia, CA). Total RNA was isolated according to the Qiagen’s RNeasy lipid tissue protocol and assayed using a NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE) to assess purity and concentration. The five highest concentrations within each group in the young age-range were submitted to the University of Missouri DNA Core facility for RNA-seq analyses. Qubit fluorometer (Invitrogen, Carlsbad, CA) with the Qubit HS RNA assay kit was used to verify sample concentration. RNA integrity was assessed using the Fragment Analyzer (Advanced Analytical Technologies, Ankey, IA) automated electrophoresis system. Only those samples that had a RIN score above 7.0 were used for follow-up RNA-seq analysis.
2.7. RNA-seq
To investigate possible mechanisms for the robust decrease in physical activity in female ArKO mice, RNAseq data were collected from the NAc brain region for male and female WT and ArKO mice. The average total reads were 64,519,492.72 reads with an average of 56,945,785.76 reads after quality control (Supplementary Table 1). The average number of mapped reads was 56,501,086.96 translating to 99.214% of reads being mapped. We previously demonstrated this average number of reads to be sufficient for eukaryotic transcriptome data (Johnson et al., 2017). High-throughput sequencing was performed at the University of Missouri DNA Core Facility (Columbia, MO) as described previously (Ortega et al., 2019). Libraries were constructed following the manufacturer’s protocol with reagents supplied in Illimina’s TruSeq mRNA Stranded Library Preparation kit.
Briefly, the poly-A containing mRNA is purified from total RNA. RNA is fragmented and double-stranded cDNA is generated from fragmented RNA, and the index containing adapters are ligated to the ends. The final construct of each purified library was evaluated using the Fragment Analyzer automated electrophoresis system, quantified with the Qubit fluorometer using the Qubit HS dsDNA assay kit, and diluted according to Illumina’s standard sequencing protocol for sequencing on the NextSeq 500.
2.8. Analysis of RNA-seq data
RNA-seq data was processed and analyzed as described previously (Givan et al., 2012). Briefly, latent Illumina adapter sequence were identified and removed from input 100-mer RNA-Seq data using Cutadapt (Martin, 2011). Subsequently, input RNA-Seq reads were trimmed and filtered to remove low quality nucleotide calls and whole reads, respectively, using the Fastx-Toolkit (GitHub, 2010). To generate the final set of quality-controlled RNA-Seq reads, foreign or undesirable sequences were removed by similarity matching to the Phi-X genome (Genome Resource, 1993) (NC_001422.1), the relevant ribosomal RNA genes as downloaded from the National Center for Biotechnology Information (NCBI, 2015) or repeat elements in RepBase (Jurka et al., 2005), using Bowtie (Langmead et al., 2009). This final set of quality-controlled RNA-Seq reads was aligned to the Ensemble Mus musculus genome sequence, GRCm38.p5 (Ensembl, 2019), using STAR (Dobin et al., 2013) with the default settings, which also generates the initial expression estimates for each annotated gene. The R (The R Foundation, 2000) Bioconductor (Bioconductor, 2003) package DESeq2 (Love et al., 2014) is used to normalize the gene expression estimates across the samples and to analyze the differential expression of genes between sample types. A gene is identified as being differentially expressed between two conditions when the FDR-corrected p-value of its expression ratio is < 0.05. Subsequent data was reformatted, sorted and filtered using a variety of R commands (The R Foundation, 2000) and Bash command-line scripts (Gnu Operating System, 2019), which are available upon request.
In order to further understand the role of the identified DEGs, gene ontology (GO) terms were analyzed for molecular function and biological process, in addition to KEGG and Reactome biological pathways using gprofiler (Raudvere et al., 2019) in order to determine which biological pathways may be affected by differentially expressed genes based on sex or genotype. Benjamini-Hochberg false discovery rate (FDR) threshold was set at 0.02. Protein-protein interactions of the DEGs were analyzed using the Search Tool for the Retrieval of Interacting Genes (STRING database v.11.0) (Szklarczyk et al., 2019) to identify the most likely interactions between the observed DEGs and any potential downstream proteins of interest.
2.9. qPCR analysis of NAc to validate RNAseq
Following RNA extraction from the NAc (as described above), total RNA was assayed using a NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE) to assess purity and concentration. First-strand cDNA was synthesized from total RNA using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Carlsbad, CA). Quantitative real-time PCR was performed as previously described using the ABI StepOne Plus sequence detection system (Applied Biosystems) (Padilla et al., 2013; Roseguini et al., 2010). Primer sequences were designed using the NCBI Primer Design tool. All primers were purchased from Sigma-Aldrich (St. Louis, MO) or Integrated DNA Technologies (Carolville, IA). The internal house-keeping control gene used was 18 s and cycle threshold (CT) was not different among the groups of animals. mRNA expression was calculated by 2∆CT where ∆CT = 18 s CT - gene of interest CT and presented as fold-difference. mRNA levels were normalized to the female WT group which was set at 1. Primer sequences are provided in Table 1.
Table 1.
Primer sequences used for NAc qPCR. Gene name, forward and reverse primer sequence, supplier, and product size.
| Gene ID | Forward | Reverse | Company | Product size |
|---|---|---|---|---|
| 18s | TCAAGAACGAAAGTCGGAGG | GGACATCTAAGGGCATCAC | IDT | 488 |
| Cryab | GAACTCAAAGTCAAGGTTCTG | ATCAGATGACAGGGATGAAG | Sigma | 159 |
| Pts | CGATGAAGAGAACTTAAGAGTG | TGTAACAGGATCAATCTCTCC | Sigma | 106 |
| Dixdc | AAGAAATGGAGGAAGCAAAG | GCAGTTCTTTCTTAAGGTCC | Sigma | 195 |
| Gldn | CTATGATCACTTCCATTGGC | ACCATGATGCCTGAAAAATG | Sigma | 133 |
| Per3 | GAGAGTATGTCATTCTGGATTC | TCATTTAATGGACTCGTTCG | Sigma | 109 |
| Layn | AAACACAGAAAGAAGACACC | CTCGTTTTCTTCTACAGATCC | Sigma | 153 |
2.10. Statistics
Male WT (n = 8) and ArKO (n = 10) mice were compared to female WT (n = 11) and ArKO (n = 9) mice. Significant sex-by-genotype interactions were noted, as well as main effects of both sex and genotype using 2 × 2 ANOVA between groups on all metabolic and behavioral data. When an interaction occurred, Tukey’s post-hoc analyses were performed to determine group differences. All data are presented as mean ± SEM; n = 8–11/group and p values < .05 were considered statistically significant. Effect sizes were calculated using partial eta-squared for ANOVAs. Analyses were performed using SPSS version 25 (IBM, Armonk, New York).
To examine for potential associations between those genes that were identified by false discovery rate (FDR) to be differentially expressed and phenotypic changes, Pearson correlations were performed, as we have done previously (Manshack et al., 2017; Mao et al., 2020). This was done two ways: 1) values were calculated across all mice and 2) only based on female results, as these were the most pronounced. In each case, only the 7 DEGs as determined by FDR that were identified to be different in the female groups were considered in this analysis and p < .05 was considered significant. A strong correlation was defined as having an r-value > ± 0.7, a moderate correlation having an r-value of ± 0.5–0.7, and a weak correlation having an r-value < ± 0.5 (Mukaka, 2012). Since we only examined those genes identified by to be differentially expressed based on a FDR, this obviated the need to perform additional corrections for multiple comparisons.
3. Results
3.1. Body weight, fat mass, lean mass, and body composition
As expected, sex differences were observed among WT mice, such that males weighed more than females and had greater lean mass (S, p < .05 for both), although adiposity was not significantly different. A S × G interaction (F(3,34) = 47.79, p < .01, η2 = 0.58) was observed for body weight, such that ArKO females weighed more than WT females, resulting in the WT female weighing less than all other groups based on Tukey’s post hoc tests (p < .05) (Fig. 1A). Similarly, S × G interactions were observed for fat mass (F(3,34) = 9.47, p < .01, η2 = 0.21) and lean mass (F(3,34) = 50.43, p < .01, η2 = 0.60). ArKO female mice had significantly more fat than all other groups (p < .05) (Fig. 1B). Compared to respective WT, ArKO females had greater, whereas ArKO males had lower lean mass (Fig. 1C). These differences in fat and lean mass resulted in the ArKO mice of both sexes having significantly higher body fat percentage than respective WT mice (G, F(3,34) = 18.79, p < .01, η2 = 0.36) (Fig. 1D). Main effects of S (F(3,34) = 11.79, p < 0.01, η2 = 0.26) and G (F(3,34) = 19.32, p < .01, η2 = 0.36) were observed for lean mass percentage such that female and ArKO mice had lower percent lean mass (Fig. 1E).
Fig. 1.
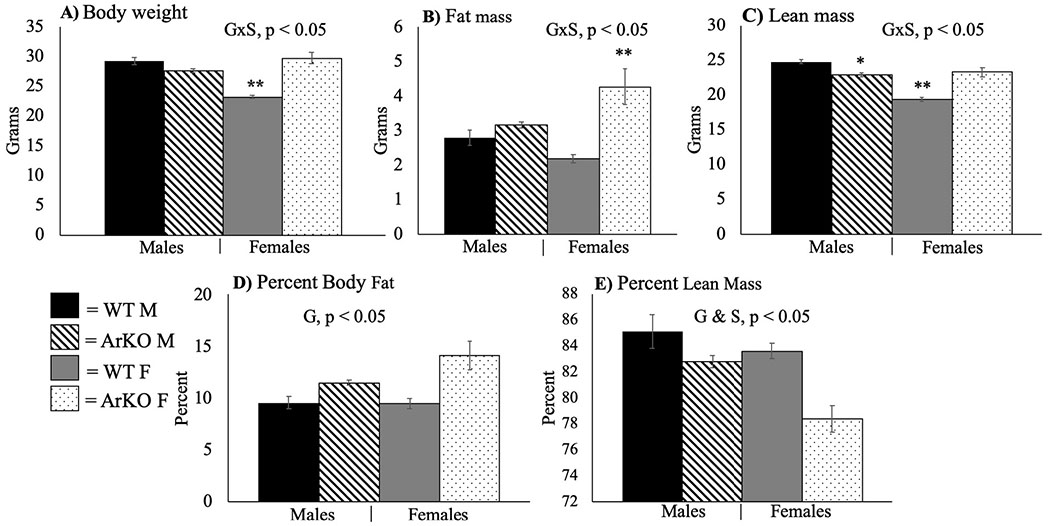
Aromatase ablation significantly increases adiposity in females. Body weight, fat and lean mass, and body composition. All values are expressed as mean ± SEM. S = main effect of sex; G = main effect of genotype. WT = wild-type; ArKO = knockout. M = male. F = female. Tukey’s post-hoc tests: * = p < .05 compared to WT within sex; ** = p < .05 compared to all other groups. Mice were 12–16 weeks old (n = 8–11/group).
3.2. Energy intake, expenditure, & fuel metabolism
Typical sex differences in energy intake and expenditure were also observed among WT mice. Despite being smaller, WT females consumed more relative energy than all other groups (p < .05) (Fig. 2A). However, a S × G interaction was observed for energy intake relative to total body weight (F(3,33) = 12.72, p < .01, η2 = 0.28), such that ArKO females had lower intake compared to WT, making them more similar to WT males (Fig. 2A). On the other hand, males were not affected by aromatase ablation in this regard.
Fig. 2.
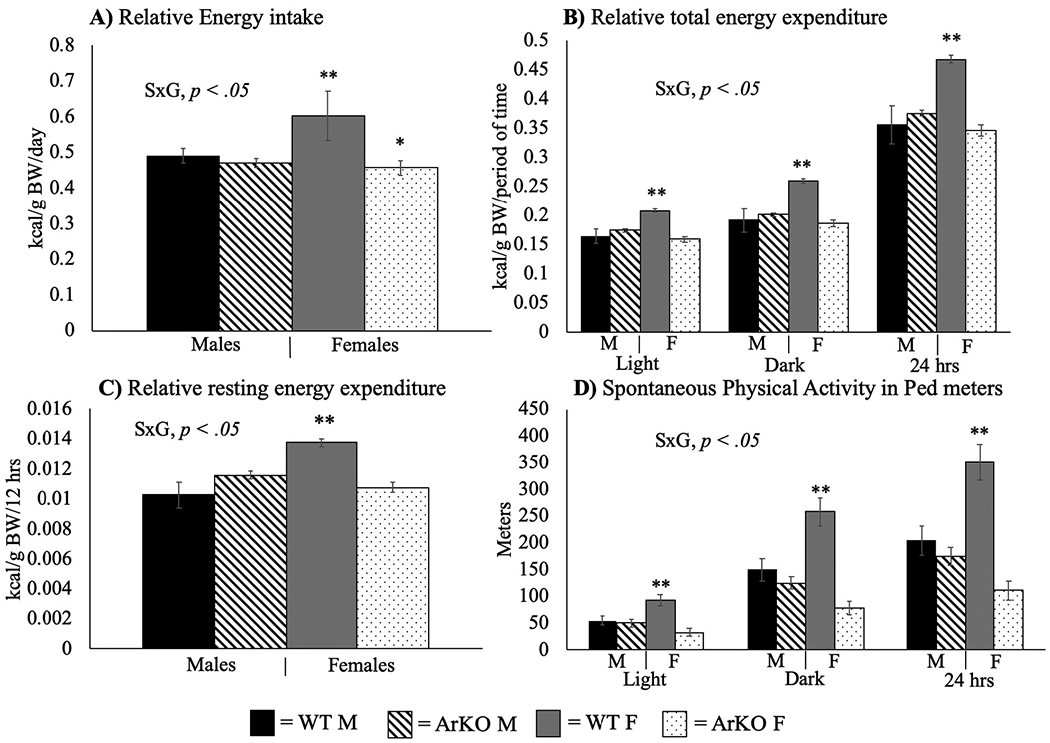
Aromatase ablation promotes a positive energy balance in females only via reductions in physical activity. Energy intake, energy expenditure (total and resting), and physical activity. All values are expressed as mean ± SEM. S × G = Sex by genotype interaction. WT = wild-type; ArKO = knockout. M = male. F = female. Tukey’s post-hoc tests: * = p < .05 compared to WT within sex; ** = p < .05 compared to all other groups. Mice were 12–16 weeks old (n = 8–11/group).
A S × G interaction was observed for total relative energy expenditure during the light (F(3,34) = 24.46, p < .01, η2 = 0.42), dark (F(3,34) = 18.12, p < .01, η2 = 0.35), and 24 h (F(3,34) = 21.09, p < .01, η2 = 0.38) cycles. Coinciding with energy intake, WT females had significantly higher total energy expenditure than all other groups (p < .01) (Fig. 2B). No significant differences were observed for fuel metabolism (i.e. respiratory quotient; data not shown); whereas, WT females had greater relative resting energy expenditure than all other groups (S × G, F (3,34) = 23.13, p < .01, η2 = 0.41) (Fig. 2C).
Spontaneous physical activity (SPA; quantified by total meters traveled in the home cage) revealed a S × G interaction (light cycle: F(3,34) = 10.13, p < .01, η2 = 0.23; dark cycle: F(3,34) = 15.96, p < .01, η2 = 0.32; 24-hour period: F(3,34) = 16.12, p < .01, η2 = 0.32). Post-hoc analyses revealed that WT females were significantly more active than all other groups (p < .05) (Fig. 2D), whereas ArKO females had a tendency (i.e., approaching statistical significance) to be even less active than WT males (dark cycle SPA: P = .08).
3.3. Sleep and anxiety-like behavior
Genotype and sex both affected sleep duration during the light (i.e., rodent inactive) cycle (S × G, p < .05). WT males and females did not differ in sleep quantity, while ArKO females slept more than WT (F(3,34) = 7.46, p < .05, η2 = 0.18) (Fig. 3A). During the dark (i.e., rodent active) cycle, WT females slept significantly less than males (F(3,34) = 18.96, p < .01, η2 = 0.33), while ArKO females slept more than all other groups (p < .05) (Fig. 3A). The same was true for the 24-hour period (F(3,34) = 15.15, p < .01, η2 = 0.33) (Fig. 3A).
Fig. 3.
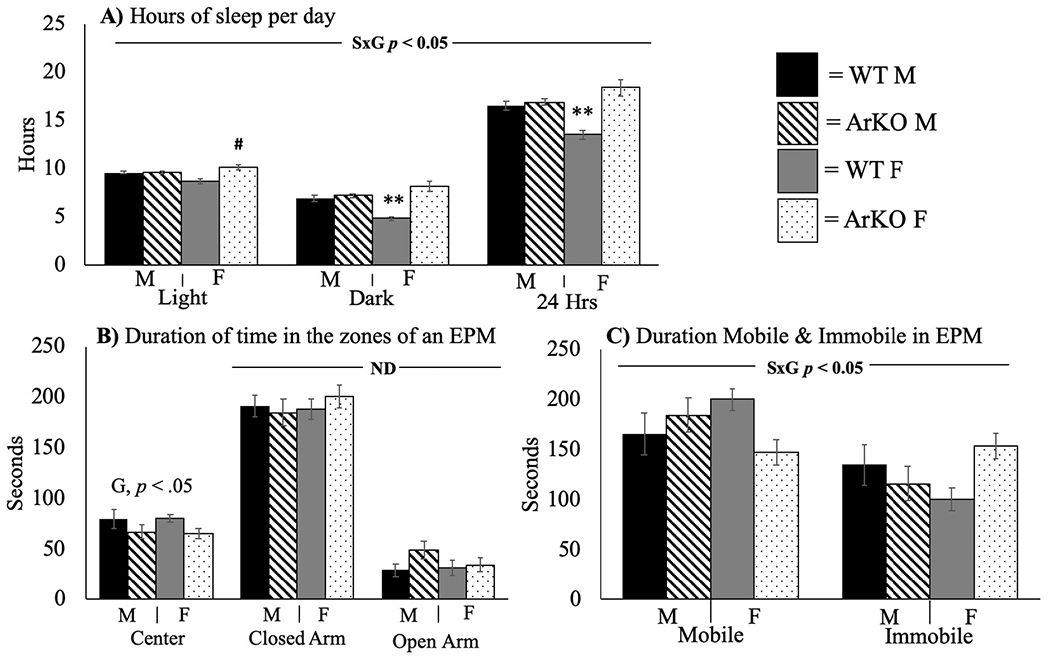
Aromatase ablation alters sleep duration in females but has no effect on anxiety-like behavior in either sex. Sleep, anxiety-like behavior in the Elevated Plus Maze (EPM). All values are expressed as mean ± SEM. S × G = Sex by genotype interaction (p < .05). G = Main effect of genotype. ND = No differences were observed between groups. WT = wild-type; ArKO = knockout. M = male. F = female. Tukey’s post-hoc tests: # = p < .05 when compared to WT female; ** = p < .05 compared to all other groups. Mice were 12–16 weeks old (n = 8–11/group).
The elevated plus maze was used to assess anxiety-like behavior. A main effect of genotype was observed for the duration of time spent in the center of the maze (F(3,33) = 2.43, p < .05, η2 = 0.17) such that ArKO mice of both sexes spent less time in the center compared to WT, but no differences were observed between groups for the duration in the closed and open arms of the maze (Fig. 3B). In addition, a S × G interaction was observed for duration mobile and immobile, such that ArKO male mice spent more time mobile, while ArKO females spent less time mobile compared to respective WT (F(3,33) = 2.42, p < .05, η2 = 0.13) (Fig. 3C). However, there were no significant post hoc differences between groups (p = .08) (Fig. 3C). No differences were observed regarding the number of entries into each zone of the elevated plus maze, frequency or duration of grooming events, rearing, or head dips while in the maze (data not shown). Together, these findings suggest that aromatase deletion does not affect anxiety-like behavior. The locomotor differences in exploratory behavior during this test align with the sex-specific effects of aromatase deletion on SPA behavior.
3.4. Sex differences in NAc gene expression profiles
Among WT mice, a total of 48 DEGs were identified by RNAseq based on sex (i.e., FDR-adjusted p < .05), 35 of which are indicated and described in Fig. 4a. Among those, six known genes showed a fold-change > 2 (i.e., were at least 2-fold different in females compared to males in either the positive or negative direction), including: Xist (FCWT = −11.37, FCArKO = −11.99), Ebf2 (FCWT = −2.85), Eif2s3y (FCWT = +12.24, FCArKO = +13.76), KdmSd (FCWT = +13.77, FCArKO = +13.50), and Ddx3y (FCWT = +13.91, FCArKO = +13.73), and Uty (FCWT = +12.07, FCArKO = +11.90). These genes, with the exception of Ebf2, have all previously been associated with sex chromosome activity. Other notable DEGs between sexes included those associated with estrogen signaling and locomotor activity (Esrl (FCWT = −1.27)), cellular metabolism (Ngfr (FCWT = −1.75)), circadian rhythm (Per3 (FCWT = +0.33), dopamine synthesis (Prkg2 (FCWT = −0.50)), and obesity and associated cancers (Ccnd1 (FCWT = −0.52)) (Fig. 4a). Full gene descriptions can be found in Table 2.
Fig. 4.
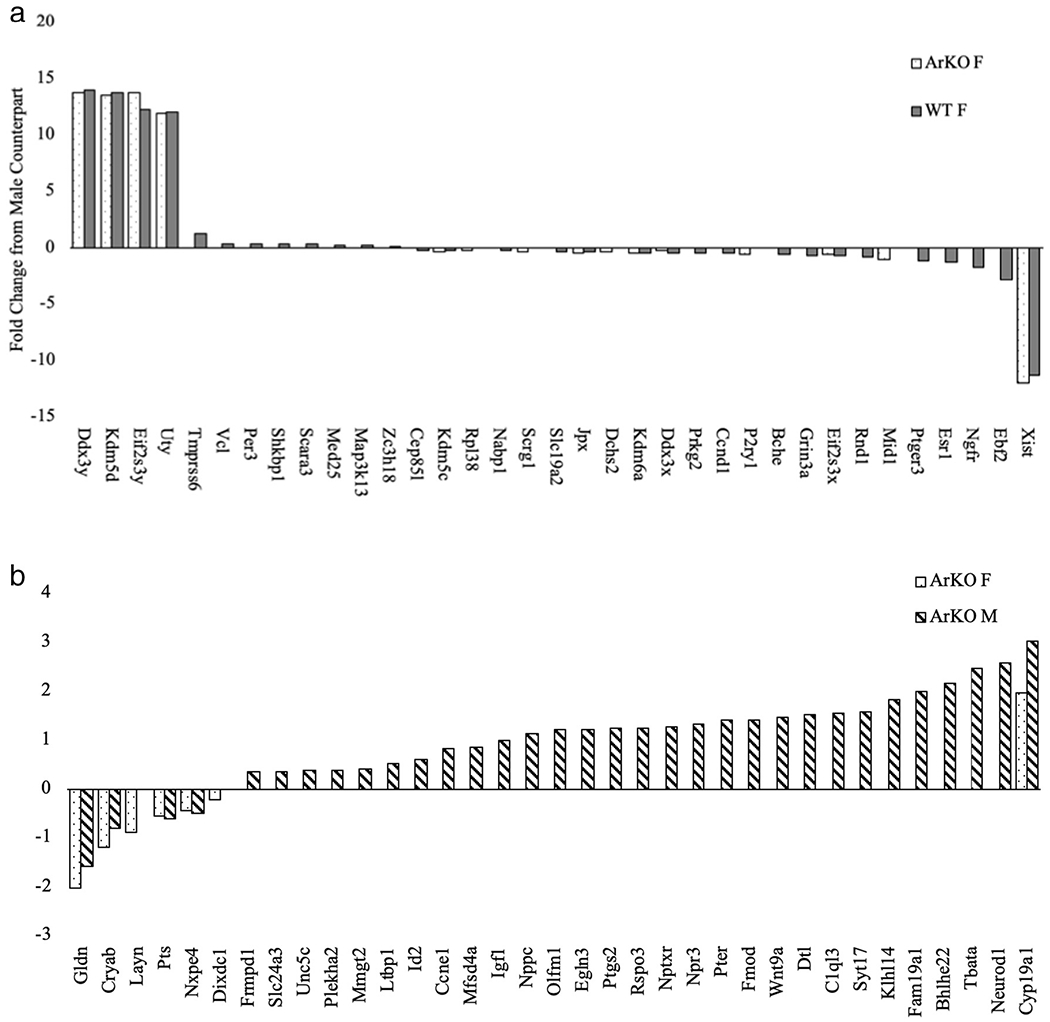
a: Sex differences in NAc gene expression. The top 35 DEGs expressed between sexes. Each bar represents the fold change for that gene in the female compared to her male counterpart. All DEGs are significant (p < .05). Solid gray bars are WT mice. Patterned bars are ArKO mice.
b: Genotype differences in Nac gene expression. The top 35 DEGs expressed between genotypes. Each bar represents the fold-change of the identified gene in the ArKO mouse compared to WT counterparts. All DEGs are significant (p < .05). Striped bars are male mice. Dotted bars are female mice.
Table 2.
Gene descriptions of top 35 DEGs between WT & ArKO mice of both sexes. Gene name, protein name, description, and fold change in female from male counterpart for each genotype for genes expressed in Fig. 4a.
| Gene | Protein | Description | Fold change in WT | Fold change in ArKO |
|---|---|---|---|---|
| Ddx3y | ATP-dependent RNA helicase Ddx3y | Involved in ATP binding, hydrolysis, RNA binding, and the formation of intramolecular interactions. Mutations may result in male infertility. | 13.91 | 13.73 |
| Kdm5d | Lysine Demthylase 5D | Linked with spermatogenic failure. | 13.77 | 13.50 |
| Eif2s3y | Eukaryotic translation initiation factor 2 subunit 3, Y-linked | Initiator of protein synthesis. | 12.24 | 13.76 |
| Uty | Histone Demethylase UTY | Male-specific histone demethylase. Associated with Kabuki syndrome, a rare autosomal dominant disorder characterized by skeletal abnormalities, short stature, heart defects, and intellectual disability, and has been affiliated with obesity. | 12.07 | 11.90 |
| Tmprss6 | Transmembrane Serine Protease 6 | May be involved in matrix remodeling in the liver. Hydrolyzes a range of proteins including: type I collagen, fibronectin, and fibrinogen. | 1.31 | |
| Vcl | Vinculin | Actin filamnet binding protein invovled in cell-matrix and cell-cell adhesion. | 0.34 | |
| Per3 | Period Circadian Regulator 3 | CLOCK gene; regulates circadian rhythm impacting metabolism, sleep, body temperature, blood pressure, endocrine, immune, cardiovascular, renal function, and locomotor activity. | 0.33 | |
| Shkbpl | SH3KBP1 Binding Protein 1 | Positive regulation of epidermal growth factor receptor signaling pathway. | 0.31 | |
| Scara3 | Scavenger Receptor Class A Member 3 | Macrophage scavenger receptor-like protein. Has been shown to deplete reactive oxygen species and protect the cell from oxidative stress. | 0.29 | |
| Med25 | Mediator Complex Subunit 25 | CC; Plays a role in chromatin modification and in preinitiation complex assembly. | 0.21 | |
| Map3k13 | Mitogen-Activated Protein Kinase Kinase Kinase 13 | Can phosphorylate and activate MAPK8/JNK, MAP2K7/MKK7, which suggests a role in the JNK signaling pathway. | 0.20 | |
| Zc3h18 | Zinc Finger CCCH-Type Containing 18 | Necessary for RNA binding. | 0.15 | |
| Cep851 | Centrosomal Protein 85 Like | A breast cancer antigen. | −0.21 | |
| Kdm5c | Lysine-specific demethylase 5C | Demethylates Lysine-4 of histone H3. Associated with two rare forms of congenital mental retardation. Related pathways include chromatin organization and chromatin regulation/acetylation. | −0.21 | −0.35 |
| Rpl38 | Ribosomal protein L38 | Componenet of the 60S subunit of the ribosome. | −0.24 | |
| Nabpl | Nucleic Acid Binding Protein 1 | Essential for a variety of DNA metabolic processes, including replication, recombination, and detection and repair of damage. | −0.26 | |
| Scrg1 | Scrapie responsive gene 1 | Associated with neurodegenerative changes observed in transmissible spongiform encephalopathies. | −0.34 | |
| Slc19a2 | Thiamine Transporter 1 | Mutations cause thiamin-responsive megaloblastic anemia syndrome (TRMA), an autosomal recessive disorder characterized by diabetes mellitus, megaloblastic anemia, and deafness. | −0.35 | |
| Jpx | JPX Transcript, XIST Activator | Involved with X-chromosome inactivation. | −0.38 | −0.42 |
| Dchs2 | Dachsous cadherin-related 2 | Likely functions in cell adhesion and may be involved in Alzheimer’s disease, compressive strength index, and appendicular lean mass. | −0.40 | |
| Kdm6a | Lysine-specific demethylase 6A | Demethylates Lysine-27 of histone H3. Associated with Kabuki Syndrome. Related pathways are activation of PKN1 to stimulate androgen receptor regulated genes KLK2 and KLK3. | −0.41 | −0.45 |
| Ddx3x | ATP-dependent RNA helicase Ddx3x | ,In mice expressed in oocytes. Ubiquitously found in 9 days post-conception embryo, at later stages it is restricted to brain and kidney. Associated with neutrophil degranulation in pathway analyses. | −0.43 | −0.27 |
| Prkg2 | CGMP-Dependent Protein Kinase II | Regulates intestinal secretion, bone growth and renin secretion. Necessary for BH4 synthesis and thus, catecholamine synthesis. | −0.50 | |
| Ccnd1 | Cyclin D1 | Function as regulator of CDK kinases; interacts with tumor suppressor protein Rb; misregulated in cancer. | −0.52 | |
| P2ry1 | Purinergic Receptor P2Y1 | Functions as a receptor for extracellular ATP and ADP. | −0.52 | |
| Bche | Butyrylcholinesterase | Aids in the detoxification of poisons including organophosphate nerve agents, pesticides, and the metabolism of cocaine, heroin and aspirin. | −0.56 | |
| Grin3a | Glutamate Ionotropic Receptor NMDA Type Subunit 3A | Subunit of the N-methyl-D-aspartate (NMDA) receptors; functions in physiological and pathological processes in the central nervous system | −0.67 | |
| Eif2s3x | Eukaryotic translation initiation factor 2 subunit 3, X-linked | Involved in the early steps of protein synthesis. Associated with Mehmo Syndrome, an x-linked intellectual disability characterized by epilepsy, mental retardation, microcephaly, hypogenitalism, and obesity. | −0.74 | −0.60 |
| Rnd1 | Rho-related GTP-binding protein Rho6 | Regulates rearrangements of the actin cytoskeleton in response to extracellular growth factors | −0.75 | |
| Mid1 | E3 ubiquitin-protein ligase Midline-1 | Involved in formation of microtubules in the cytoplasm. Associated with Opitz Gbbb Syndrome, a congenital midline malformation syndrome. | −1.01 | |
| Ptger3 | Prostaglandin E Receptor 3 | Involved in many biological processes including: digestion, kidney reabsorption, and uterine contractions. Also active in the nervous system, blood coagulation, and fever generation. | −1.17 | |
| Esr1 | Estrogen Receptor 1 | Major isoform of the etrogen receptor (i.e., alpha), a nuclear hormone receptor. Essential for sexual development and reproductive function; present in both sexes across species. | −1.27 | |
| Ngfr | Tumor Necrosis Factor Receptor Superfamily Member 16 | Low-affinity receptor. Can bind NGF, BDNF, NT-3, and NT-4. Signaling pathways include: Apoptosis modulation and signaling and beta-adrenergic signaling. | −1.75 | |
| Ebf2 | EBF Transcription Factor 2 | May be required for the development of dopaminergic neurons. | −2.85 | |
| Xist | X Inactive Specific Transcript | Involved in X-chromosome inactivation. | −11.37 | −11.99 |
In order to gain insight into the role played by aromatase on those sex differences in NAc gene expression, we determined which of those genes were no longer differentially expressed when comparing female and male ArKO mice. Interestingly, with the exception of Ebf2 (gene encoding a transcription factor associated with dopamine neurogenesis (Yang et al., 2015) all of the genes with a FC > 2 described above were also differentially expressed between sexes within ArKO mice. However, for all significant DEGs with fold change < 2 noted above, sex differences disappeared with deletion of the aromatase gene. (Fig. 4a). A Venn diagram depicting the number of DEGs based on sex for each genotype, including those that were consistent across genotypes, is provided in Fig. 5a.
Fig. 5.
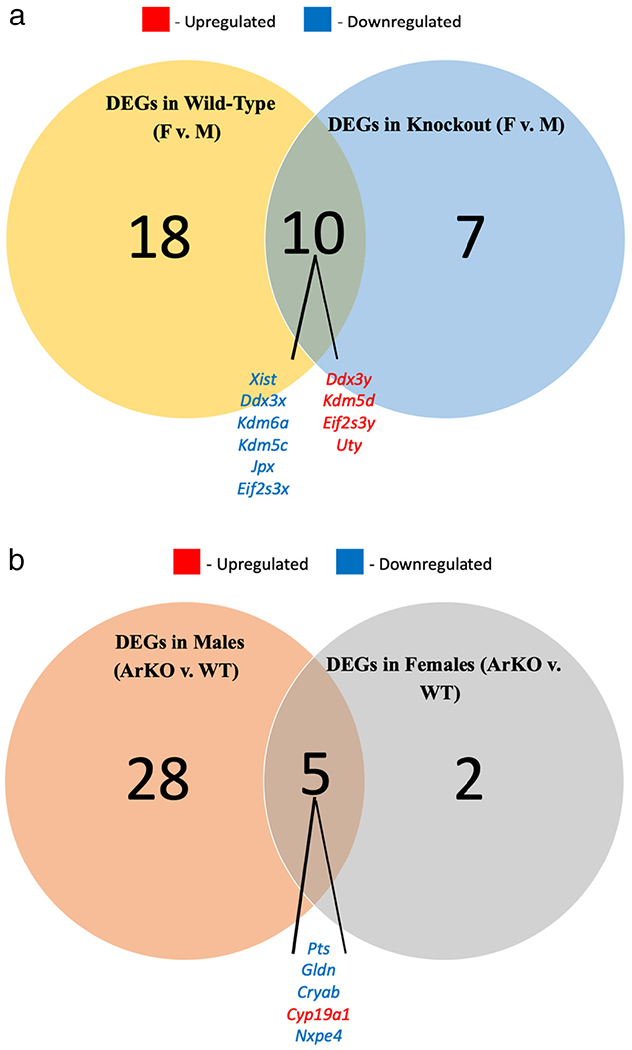
a: Venn diagram of the number differentially expressed genes between sexes. F = Female. M = Male. DEG = differentially expressed genes. Ddx3y, Kdm5d, Eif2s3y, and Uty are upregulated in females of both genotypes compared to males, while Eif2s3x, Ddx3x, Kdm6a, Kdm5c, Jpx, and Xist are downregulated.
b: Venn diagram of the number of differentially expressed genes between genotypes. ArKO = Aromatase knockout. WT = Wild-type. DEG = differentially expressed gene. Pts, Gldn, Cryab, and Nxpe4 are downregulated in ArKO of both sexes, compared to WT, while Cyp19a1 is upregulated.
3.5. Genotype differences in NAc gene expression profiles
Once we characterized major sex differences in WT mice, and determined which of those were dependent upon aromatase, we then determined how aromatase ablation affected gene expression patterns in the NAc within each sex. The top 35 DEGs based on genotype were identified for males and females and are depicted in Fig. 4b. Descriptions of those DEGs are provided in Table 3. A Venn diagram depicting the numbers of DEGs for each sex, highlighting those that were consistent across sexes is provided in Fig. 5b.
Table 3.
Gene descriptions of top 35 DEGs between male and female mice of both genotypes. Gene name, protein name, description, fold change in ArKO from WT counterpart for each sex for genes expressed in Fig. 4b.
| Gene | Protein | Description | Fold change F | Fold change M |
|---|---|---|---|---|
| Gldn | Gliomedin | Required for the proper clustering of sodium channels on the Nodes of Ranvier in the peripheral nervous system. | −2.04 | −1.58 |
| Cryab | Crystallin Alpha B | Member of the small heat shock protein family; elevated expression occurs in many neuropogical diseases; has chaperone-like activity; prevents the aggregation of various proteins under stress conditions. | −1.21 | −0.81 |
| Layn | Layilin | Involved in binding of carbohydrate and hyaluronic acid. | −0.91 | |
| Pts | 6-Pyruvoyltetrahydropterin Synthase | Catalyzes the second and irreversible step in the formation of tetrahydropterin (BH4), an essential cofactor in catecholamine biosynthesis. Mutations result in hyperphenylalanemia. | −0.57 | −0.62 |
| Nxpe4 | Neurexophilin And PC-Esterase Domain Family Member 4 | Associated with glycosalation of amino acids. | −0.45 | −0.51 |
| Dixdc1 | Dixin | Positive regulator of the Wnt pathway. | −0.22 | |
| Frmpd1 | FERM And PDZ Domain Containing 1 | Regulation of G protein-coupled receptor signaling pathway. | 0.34 | |
| Slc24a3 | Sodium/Potassium/Calcium Exchanger 3 | Believed to transport intracellular calcium and potassium ion in exchange. | 0.36 | |
| Unc5c | Unc-5 Netrin Receptor C | Belongs to a family of receptors that bind proteins that direct axon extension and cell migration during neural development. | 0.38 | |
| Plekha2 | Pleckstrin homology domain-containing family A member | Binds phosphatidylinositol 3,4-diphosphate (PtdIns3,4P2); gene ontology related to lipid binding and metabolic pathways. | 0.39 | |
| Mmgt2 | Membrane Magnesium Transporter 1 | Involved in the transport of glucose and other sugars, bile salts, and organic acids, metal ions, and amine compounds. | 0.41 | |
| Ltbp1 | Latent Transforming Growth Factor Beta Binding Protein 1 | Involved in cAMP-dependent activation of PKA. | 0.52 | |
| Id2 | DNA-binding protein inhibitor ID-2 | Negatively regulates the basic helix-loop-helix (bHLH) transcription factors by forming heterodimers and inhibiting their DNA binding and transcriptional activity. Implicated in regulating a variety of cellular processes including growth, differentiation, and apoptosis. Inhibits skeletal muscle and cardiac myocyte differentiation Regulates the circadian clock by modulating the magnitude of photic entrainment and contributes to the regulation of a variety of liver clock-controlled genes involved in lipid metabolism. | 0.61 | |
| Ccne1 | Cyclin E1 | Implicated in various carcinomas, including breast, gastric, stomach and colorectal. | 0.82 | |
| Mfsd4a | Major Facilitator Superfamily Domain Containing 4A | Implicated in glucose transmembrane transporter activity. | 0.85 | |
| Igf1 | Insulin Like Growth Factor 1 | Similar to insulin in structure and function and involved in mediating growth and development. | 0.98 | |
| NPPC | Natriuretic Peptide C | Cardiac natriuretic peptides which exhibit vasorelaxation in laboratory animals and are elevated in humans with chronic heart failure. | 1.14 | |
| Olfm1 | Olfactomedin 1 | Abundant in rodent brains. Exact function is not known but is associated with ectopic pregnancy in humans. | 1.20 | |
| Egln3 | Egl-9 Family Hypoxia Inducible Factor 3 | Cellular oxygen sensor that catalyzes, under normoxic conditions, the post-translational formation of 4-hydroxyproline in hypoxia-inducible factor (HIF) alpha proteins. In neurons, has an NGF-induced proapoptotic effect, probably through regulating CASP3 activity. | 1.21 | |
| Ptgs2 | COX-2 | Key enzyme in prostaglandin biosynthesis. PTGS2 is responsible for production of inflammatory prostaglandins. | 1.23 | |
| RspoS | R-Spondin 3 | The encoded protein plays a role in the regulation of Wnt signaling pathways, which are involved in development, cell growth and disease pathogenesis. | 1.23 | |
| Nptxr | Neuronal Pentraxin Receptor | May be involved in mediating uptake of synaptic material during synapse remodeling or in mediating the synaptic clustering of AMPA glutamate receptors at a subset of excitatory synapses. | 1.26 | |
| Npr3 | Natriuretic Peptide Receptor C/Guanylate Cyclase C | Involved in regulation of blood volume and pressure, pulmonary hypertension, and cardiac function as well as some metabolic and growth processes. | 1.32 | |
| Pter | Phosphotriesterase-Related Protein | Cofactor binding 2 divalent metal cations per subunit; acting on ester bonds. | 1.40 | |
| Fmod | Fibromodulin | Encoded protein may play a role in the assembly of extracellular matrix | 1.40 | |
| Wnt9a | Protein Wnt-9a | Implicated in oncogenesis and in several developmental processes, including regulation of cell fate and patterning during embryogenesis; is expressed in gastric cancer cell lines | 1.48 | |
| Dtl | Denticleless E3 Ubiquitin Protein Ligase Homolog | Required for cell cycle control, DNA damage response and translesion DNA synthesis. | 1.53 | |
| C1ql3 | C1q And Tumor Necrosis Factor-Related Protein 13 | May regulate the number of excitatory synapses but has no effect on inhibitory synapses. Plays a role in glucose homeostasis via AMPK signaling pathway. Stimulates glucose uptake in adipocytes, myotubes and hepatocytes and enhances insulin-stimulated glucose uptake. In a hepatoma cell line, reduces the expression of gluconeogenic enzymes G6PC and PCK1 and decreases de novo glucose production. | 1.54 | |
| Syt17 | Synaptotagmin 17 | Plays a role in dendrite formation by melanocytes. | 1.57 | |
| Klhl14 | Kelch-Like Protein 14 | Associated with Central Nervous System Hematologic Cancer. | 1.83 | |
| Fam19a1 | TAFA Chemokine Like Family Member 1 | Postulated to function as brain-specific chemokines or neurokines that act as regulators of immune and nerve cells. | 1.98 | |
| Bhlhe22 | Class E basic helix-loop-helix protein 22 | Regulates cell fate determination, proliferation, and differentiation; plays a crucial role in retinogenesis. | 2.16 | |
| Tbata | Thymus, Brain And Testes-Associated Protein | Regulates thymic epithelial cell proliferation and thymus size. May also play a role in spermatid differentiation, as well as in neuronal morphogenesis and synaptic plasticity. | 2.47 | |
| Neurod1 | Neuronal Differentiation 1 | Regulates expression of the insulin gene, and mutations in this gene result in type II diabetes mellitus. | 2.58 | |
| Cyp19a1 | Aromatase | Localizes to the endoplasmic reticulum and catalyzes the final steps in estrogen biosynthesis; mutations can increase or decrease gene expression; associated phenotypes suggest that estrogen functions as both a sex steroid hormone and in growth/differentiation. | 1.96 | 3.01 |
The following 5 DEGs were consistently different between WT and ArKO mice within each sex: Pts (FCF = −0.57, FCM = −0.62), Gldn (FCF = −2.04, FCM = −1.58), Cryab (FCF = −1.21, FCM = −0.81), Nxpe4 (FCF = −0.45, FCM = −0.51), and Cypl9al (FCF = +1.96, FCM = +3.01). Notably, Pts is a gene necessary for tetrahydrobiopterin (BH4) synthesis, and thus production of catecholamines, such as dopamine (Stelzer et al., 2016).
Within males, 5 DEGs showed a fold change > 2, including: Neurodl (FCM = +2.58)), Tbata (FCM = +2.47)), and Bhlhe22 (FCM = +2.16)). Notably, Neurod1 is associated with glucose homeostasis by regulating the expression of the insulin gene within the brain (Lee et al., 2016).
In addition, various genes associated with lipid metabolism (Plekha2 (FCM = +0.34)), glucosemetabolism (Mfsd4a (FCM = +0.85); Clql3 (FCM = +1.54)), circadian rhythm (Id2 (FCM = +0.61)), and cancer (Ccnd1 (FCM = +0.82); Klhl14 (FCM = +1.83)) were also differentially expressed in males only, but with a smaller fold change (Fig. 4b).
Two additional DEGs were unique to the female and were associated with binding of carbohydrate and hyaluronic acid (Layn, FCF = −0.91) and regulation of the Wnt signaling pathway (Dixdc1, FCF = −0.22). Key DEGs were validated via qPCR. These data are provided in Supplementary Fig. 2.
3.6. Correlations between gene expression and phenotypical traits and behaviors
Because of the particularly robust genotype differences detected among female cohorts, we chose the top 6 DEGs and assessed correlations between those DEGs and key physiological and behavioral phenotypic characteristics. Those DEGs (Pts, Layn, Dixdc1, Per3, Cryab, Nxpe4, Gldn) were chosen for correlation analyses across all mice (Fig. 6a) and in female mice only (Fig. 6b). Key correlations with Pts (a necessary gene for catecholamine synthesis (Korner et al., 2016)), Per3 (a gene known to regulate circadian rhythm (Dewandre, 2018)), and Cryab (a member of the small heat shock protein family that may associate with cellular stress resistance (Stelzer et al., 2016)) are highlighted below.
Fig. 6.
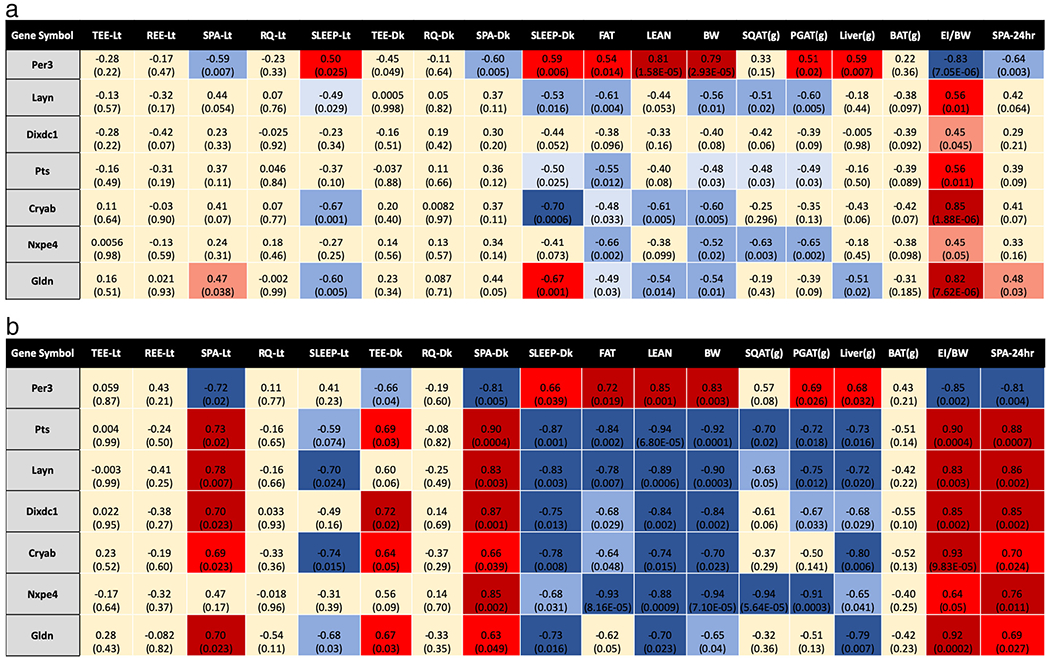
a: Correlations between differentially expressed genes and selected phenotypic traits across all mice. Significance was set at p < .05. The top number in each box is the r-value for that correlation. The bottom number in parentheses is the associated p-value. Positive correlations are in red with the weakest correlations being the lightest shade and the strongest correlations being the darkest shade. The same trend applies to the negative correlations in blue. Correlation strength: strong correlation r > ± 0.7; moderate correlation r = ± 0.5–0.7; weak correlation r < ± 0.5. TEE/REE = Total/Resting energy expenditure. SPA = spontaneous physical activity. RQ = Respiratory quotient. SLEEP = Sleep duration. FAT = Fat mass (g). LEAN = Lean mass (g). BW = Bodyweight (g). SQAT(g) = Subcutaneous fat mass (g). PGAT(g) = Perigonadal adipose tissue mass (g). Liver(g) = Liver weight (g). BAT(g) = Brown adipose tissue mass (g). EI/BW = Relative energy intake (g food/g bodyweight). Lt = Light cycle. Dk = Dark cycle. 24 h = 24-hour total.
b: Correlations between differentially expressed genes and selected phenotypic traits in female mice only. Significance was set at p < .05. Positive correlations are in red with the weakest correlations being the lightest shade and the strongest correlations being the darkest shade. The same trend applies to the negative correlations in blue. Correlation strength: strong correlation, r > ± 0.7; moderate correlation, r = ± 0.5–0.7; weak correlation, r < ± 0.5. TEE/REE = Total/Resting energy expenditure. SPA = spontaneous physical activity. RQ = Respiratory quotient. SLEEP = Sleep duration. FAT = Fat mass (g). LEAN = Lean mass (g). BW = Bodyweight (g). SQAT(g) = Subcutaneous fat mass (g). PGAT(g) = Perigonadal adipose tissue mass (g). Liver(g) = Liver weight (g). BAT(g) = Brown adipose tissue mass (g). EI/BW = Relative energy intake (g food/g bodyweight). Lt = Light cycle. Dk = Dark cycle. 24 h = 24-h total. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
When analyzing the entire cohort, energy intake, which was significantly reduced in ArKO mice, significantly correlated with Cryab (r = 0.85, p < .01) and Per3 (r = −0.83, p < .01). Interestingly, Pts exhibited a strong positive correlation with spontaneous physical activity (r = 0.9, p < .01), a negative correlation with body fat (r = −0.55, p < .05), and a positive correlation with energy intake (r = 0.55, p < .05) (Fig. 6a).
Within females, Per3 (i.e, a clock gene) correlated negatively with physical activity (r24 = −0.81, p24 < 0.01), and energy intake (r = −0.85, p < .01) and positively with sleep during the rodent active cycle (r = 0.66, p < .05) (Fig. 6b). Interestingly, Pts (i.e., gene associated with dopamine synthesis) correlated with all indices of physical activity (rLt = 0.73, p < .05; rDk = 0.90, p < .01; r24 = 0.88, p < .01), but correlated negatively with sleep during the active cycle (r = −0.87,p < .05) (Fig. 5b). Finally, Cryab (i.e., a stress response gene) correlated positively with energy intake (r = 0.85, p < .01) and negatively with sleep during the active cycle (r = −0.70, p < .01).
3.7. Pathway analysis and protein-protein interaction networks
Gene ontology analysis of DEGs in M v. F WT and ArKO mice were performed and the top biological pathways were identified and provided in Tables 4–7. The major molecular function pathways between male and female mice of both genotypes involved histone demethylation activity, steroid and nuclear hormone receptor binding, estrogen response element binding, and estrogen receptor activity. Protein-protein interaction networks of pair-wise comparisons between groups are presented in Supplementary Fig. 3A–D.
Table 4.
Significantly enriched interactions of DEGs indicating the gene ontology and biological pathways (KEGG & Reactome) in WT males v. WT females.
| Description | FDR | |
|---|---|---|
| GO: molecular function | Histone demethylase activity | 2.10845E–05 |
| Steroid hormone receptor binding | 0.003656028 | |
| Estrogen response element binding | 0.006161726 | |
| Nuclear hormone receptor binding | 0.006985698 | |
| Estrogen receptor activity | 0.00720369 | |
| hormone receptor binding | 0.00854285 | |
| GO: biological processes | Histone demethylation | 4.97867E–05 |
| Regulation of macromolecule metabolic process | 0.000254213 | |
| Regulation of phospholipase C activity | 0.0008599 | |
| Regulation of lipase activity | 0.003559273 | |
| Response to steroid hormone | 0.004524849 | |
| Response to estrogen | 0.004724675 | |
| Regulation of circadian rhythm | 0.006080162 | |
| Intracellular steroid hormone receptor signaling pathway | 0.007111892 | |
| Steroid hormone mediated signaling pathway | 0.010956526 | |
| chromatin organization | 0.012145584 | |
| Hormone-mediated signaling pathway | 0.013819596 | |
| Negative regulation of triglyceride metabolic process | 0.015261211 | |
| Cellular response to steroid hormone stimulus | 0.017041363 | |
| Biological pathways (Reactome) | HDMs demethylate histones | 6.88327126245209E–07 |
| Chromatin organization | 1.42112E–06 | |
| RUNX1 regulates estrogen receptor mediated transcription | 0.017513604 | |
| Biological pathways (KEGG) | Transcriptional misregulation in cancer | 0.002297158 |
| Melanoma | 0.004863766 | |
| Prostate cancer | 0.006409001 | |
| Endocrine resistance | 0.006409001 | |
| Thyroid hormone signaling pathway | 0.008216614 | |
| Breast cancer | 0.01121615 | |
| MicroRNAs in cancer | 0.01121615 |
Table 7.
Significantly enriched interactions of DEGs indicating the gene ontology and biological pathways (KEGG & Reactome) in Female WT v. ArKO mice.
| Description | FDR | |
|---|---|---|
| GO: molecular | 6-Pyruvoyltetrahydropterin synthase | 0.006792819 |
| function | activity | |
| aromatase activity | 0.015984047 | |
| Biological pathways | Tetrahydrobiopterin (BH4) synthesis, | 0.010725654 |
| (Reactome) | recycling, salvage and regulation | |
| Metabolism of cofactors | 0.010725654 | |
| Estrogen biosynthesis | 0.010725654 | |
| HSF1-dependent transactivation | 0.011254654 | |
| Endogenous sterols | 0.011254654 | |
| Metabolism of steroid hormones | 0.014391637 | |
| Biological pathways | Folate biosynthesis | 0.013072049 |
| (KEGG) | Steroid hormone biosynthesis | 0.013072049 |
| Ovarian steroidogenesis | 0.013072049 | |
| Metabolic pathways | 0.013072049 | |
| Longevity regulating pathway-multiple species | 0.013072049 |
Among WT mice, the top GO biological processes that were different between males and females involved steroid hormones, lipid metabolism, regulation of sleep/circadian rhythm, and DNA methylation (Table 4). Among ArKO mice, the top GO biological processes that were different between sexes involved steroid hormones, metabolism, sexual differentiation, growth and development, regulation of circadian rhythm, and ovulation (Table 5).
Table 5.
Significantly enriched interactions of DEGs indicating the gene ontology and biological pathways (KEGG & Reactome) in KO males v. ArKO females.
| Description | FDR | |
|---|---|---|
| GO: molecular function | Histone demethylase activity | 2.10845E–05 |
| Steroid hormone receptor binding | 0.003656028 | |
| Type 1 metabotropic glutamate receptor binding | 0.003724315 | |
| Estrogen response element binding | 0.006161726 | |
| Nuclear hormone receptor binding | 0.006985698 | |
| Estrogen receptor activity | 0.00720369 | |
| Hormone receptor binding | 0.00854285 | |
| GO: biological processes | Histone demethylation | 4.97867E–05 |
| Cellular macromolecule biosynthetic process | 8.52132E–05 | |
| Macromolecule metabolic process | 0.000153116 | |
| Aromatic compound biosynthetic process | 0.000348629 | |
| Development of primary male sexual characteristics | 0.000563921 | |
| Regulation of primary metabolic process | 0.000733981 | |
| Male sex differentiation | 0.000815365 | |
| Regulation of phospholipase C activity | 0.0008599 | |
| Developmental growth | 0.002098432 | |
| Regulation of lipase activity | 0.003559273 | |
| Response to steroid hormone | 0.004524849 | |
| Ovulation cycle | 0.004537605 | |
| Cellular hormone metabolic process | 0.005381536 | |
| Histone modification | 0.005591953 | |
| Regulation of circadian rhythm | 0.006080162 | |
| Development of primary female sexual characteristics | 0.007830935 | |
| Female sex differentiation | 0.009139693 | |
| Response to estradiol | 0.009148897 | |
| Biological pathways (Reactome) | HDMs demethylate histones | 6.88327126245209E–07 |
| Chromatin modifying enzymes | 1.42112E–06 | |
| Chromatin organization | 1.42112E–06 | |
| Negative regulation of the PI3K/AKT network | 0.01699868 | |
| NFG and proNGF binds to p75NTR | 0.01699868 | |
| PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling | 0.01699868 | |
| Signal transduction | 0.01699868 | |
| RUNX1 regulates estrogen receptor mediated transcription | 0.017513604 | |
| Neutrophil degranulation | 0.019035096 | |
| Axonal growth stimulation | 0.019035096 | |
| Biological pathways (KEGG) | Focal adhesion | 0.002297158 |
| Pathways in cancer | 0.002297158 | |
| Transcriptional misregulation in cancer | 0.002297158 | |
| Human cytomegalovirus infection | 0.003347549 | |
| Glioma | 0.004863766 | |
| Melanoma | 0.004863766 | |
| PI3K-Akt signaling pathway | 0.004863766 | |
| Prolactin signaling pathway | 0.004863766 | |
| Endocrine resistance | 0.006409001 | |
| Thyroid hormone signaling pathway | 0.008216614 | |
| MicroRNAs in cancer | 0.01121615 | |
| JAK-STAT signaling pathway | 0.012700471 | |
| RNA transport | 0.012700471 | |
| Calcium signaling pathway | 0.015053718 | |
| Proteoglycans in cancer | 0.015382804 | |
| Rap1 signaling pathway | 0.01612003 | |
| Regulation of actin cytoskeleton | 0.01612003 | |
| Viral carcinogenesis | 0.01612003 | |
| Ras signaling pathway | 0.017199112 |
Reactome analysis identified 3 biological pathways that were differentially regulated by sex independent of genotype: HDMs demethylase histones, chromatin organization, and RUNX1 regulation of estrogen receptor mediated transcription (Tables 4 & 5). The KEGG biological pathways in WT mice were associated with cancer, endocrine resistance, or thyroid hormone signaling pathways, while these same pathways were identified between sexes in ArKO mice, suggesting that those sex differences may be independent of aromatase (i.e., estrogen availability) (Tables 4 & 5).
As described in Table 6, gene ontology analysis of DEGs in WT v. ArKO males indicated involvement in molecular function of lipid binding, prostaglandin binding, 6-pyruvoyltetrahydropterin synthase (Pts) activity, neuropeptide Y receptor, and peptide YY receptor activity. The top GO biological processes associated with these DEGs involved neurogenesis, steroid hormone signaling, metabolism, growth and development of the CNS, locomotor behavior, BH4 synthesis and metabolism, and aging. Of those, Reactome identified four processes: Class A/1 rhodopsin-like receptors, metabolism, Got signaling events, and GPCR ligand binding. One KEGG biological pathway was identified as a longevity regulating pathway (Table 6).
Table 6.
Significantly enriched interactions of DEGs indicating the gene ontology and biological pathways (KEGG & Reactome) in Male WT v. ArKO mice.
| Description | FDR | |
|---|---|---|
| GO: Molecular Function | ||
| lipid binding | 0.003086564 | |
| prostaglandin binding | 0.003452799 | |
| 6-pyruvoyltetrahydropterin synthase activity | 0.003452799 | |
| neuropeptide Y receptor activity | 0.018157161 | |
| peptide YY receptor activity | 0.010641921 | |
| GO: Biological Process | ||
| glial cell migration | 0.000348694 | |
| neurogenesis | 0.000686558 | |
| locomotion | 0.000792012 | |
| metabolic process | 0.0014716 | |
| nervous system development | 0.002011761 | |
| regulation of hormone levels | 0.002758 | |
| regulation of astrocyte differentiation | 0.003388127 | |
| cell-cell adhesion involved in neuronal-glial interactions involved in cerebral cortex radial glia guided migration | 0.006503613 | |
| neuron differentiation | 0.006503613 | |
| aging | 0.00761918 | |
| hormone secretion | 0.008085701 | |
| hormone transport | 0.008424296 | |
| regulation of glial cell differentiation | 0.008963381 | |
| neuronal-glial interaction involved in cerebral cortex radial glia guided migration | 0.009730318 | |
| cerebellum structural organization | 0.009730318 | |
| hindbrain structural organization | 0.009730318 | |
| regulation of primary metabolic process | 0.009730318 | |
| cellular hormone metabolic process | 0.009959859 | |
| estradiol secretion | 0.011722735 | |
| response to drug | 0.011818741 | |
| feeding behavior | 0.012703111 | |
| aromatic compound biosynthetic process | 0.013689053 | |
| androgen catabolic process | 0.013769906 | |
| motor neuron migration | 0.017303974 | |
| Post-synapse organization | 0.017605926 | |
| tetrahydrobiopterin biosynthetic process | 0.018216943 | |
| tetrahydrobiopterin metabolic process | 0.019688381 | |
| Biological Pathways (Reactome) | ||
| Class A/1 (Rhodopsin-like receptors) | 0.016216258 | |
| Metabolism | 0.016216258 | |
| G alpha (i) signalling events | 0.018546984 | |
| GPCR ligand binding | 0.018546984 | |
| Biological Pathways (KEGG) | ||
| Longevity Regulating Pathway-multiple species | 0.014854072 |
In Table 7, GO analysis of DEGs in female WT v. ArKO mice indicated involvement in molecular function of Pts activity and aromatase activity. Six biological pathways were identified via Reactome: BH4 synthesis, recycling, salvage, and regulation; metabolism of cofactors; estrogen biosynthesis; HSF1-dependent transactivation; endogenous sterols; and metabolism of steroid hormones. Five KEGG biological pathways were identified as associated with: folate biosynthesis, steroid hormone biosynthesis, ovarian steroidogenesis, metabolic pathways, and longevity regulating pathways (Table 7).
4. Discussion
Male and female WT and ArKO mice were compared for metabolic and behavioral traits and their relation to differences in gene expression profiles in the NAc brain region obtained via RNAseq. Gene ontology pathway enrichment analyses were then performed to identify the potential biological pathways most likely to be different between sex and genotype.
Previous research established that ArKO mice are obese even provided a standard chow diet (Fisher et al., 1998; Honda et al., 1998; Jones et al., 2000, 2001) and have impaired lipid metabolism (Amano et al., 2017; Hewitt et al., 2003; Hewitt et al., 2004; Jones et al., 2000; Nemoto et al., 2000), disrupted sexual behavior (Dalla et al., 2004; Honda et al., 1998), and reduced locomotor activity (Jones et al., 2000). Gonadotropin assays consistently show elevated testosterone levels in male mice, but mixed results in females (Amano et al., 2017; Fisher et al., 1998; Harada et al., 2009; Liew et al., 2010; Takeda et al., 2003; Toda et al., 2001).
In the current study, observed sex differences and genotypic effects support previous findings, while reduced physical activity and obesity was only observed in mutant females. For example, ArKO females had greater fat mass compared to all other groups. Our findings indicate that this is due to reduced physical activity, rather than increases in energy intake or decreased resting energy expenditure, similar to what has been previously shown in another ArKO model (Jones et al., 2001). Unlike females, ArKO males did not differ in body weight or fat mass from WT males, neither did they differ in spontaneous physical activity. A 2011 study by Mahoney et al. examined wheel-running behavior in the same mouse model and discovered similar genotype effects as our findings in female mice. Interestingly, Mahoney’s group also noted a decrease in wheel-running in male ArKO mice, contrasting our findings (Brockman et al., 2011). Paralleling previous literature (Jones et al., 2001; Jones et al., 2000), we determined that ArKO males had lower lean mass compared to WT counterparts. This finding is intriguing since testosterone is known to increase lean mass (Mouser et al., 2016) and endogenous testosterone is increased in ArKO males (Amano et al., 2017); we confirmed this in our mice (data not shown).
In addition to energy intake and physical activity, sleep patterns are now recognized as profoundly influencing metabolic health and are known to be regulated by sex steroids (Brockman et al., 2011). Importantly, aromatase inhibition therapy in humans is known to cause insomnia (Bhave et al., 2018; Karatas et al., 2015), but the mechanisms are not fully understood. We assessed sleep patterns in the ArKO and WT mice and found that ArKO females slept more during the dark cycle (i.e., when mice are most typically active) and light (i.e., typical sleep/ inactive) cycle relative to WT. These differences associated with their physical inactivity, while male sleep patterns did not differ by genotype. Similar findings have been reported where ArKO females slept more during the dark cycle, although the total sleep duration in 24 h did not differ between ArKO and WT females in that study (Vyazovskiy et al., 2006). This may be due to differences in the genetic ArKO model, as they used a model with a neomycin disruption in exon IX of the Cypl9 gene, whereas our model had an exon I and II disruption, as well as the brain promoter region (Fisher et al., 1998; Honda et al., 1998).
Interestingly, our findings in relation to anxiety-like and learning behaviors did not significantly differ with loss of aromatase in males or females. This is in contrast to previous findings. Impaired short-term spatial reference memory in male and female ArKO mice has been previously demonstrated via Y-maze testing (Martin et al., 2003). While we utilized a Barnes maze in this study to analyze short-term learning, mice did not effectively perform in the maze, providing null results (data not shown). The different types of mazes to measure spatial learning and memory may account for these conflicting results. Another group reported analogous findings with regards to anxiety-like behavior when ArKO males and females were tested in the elevated plus maze, in that no significant effects of genotype were observed in either sex (Dalla et al., 2004, 2005).
In addition to phenotypic differences, our current work builds on original findings by identifying gene expression differences in the NAc region, which we hypothesize may partially account for differences in SPA and ensuing obesity. Within WT mice, notable sex differences in gene expression included those with a FC > 2 (i.e. Xist, Ebf2, Uty, Eif2s3y, Kdm5d, and Ddx3y). Of these genes, Ebf2 is a transcription factor for dopamine neurogenesis (Yang et al., 2015) and is the only DEG between WT sexes that disappears when comparing ArKO mice. All other genes with fold change > 2 were similarly differentially expressed between sexes within WT and ArKO mice and are all associated with sex-chromosomes (UniProt, 2019).
Several DEGs between WT males and females were not present between sexes in the ArKO mouse, possibly indicating that these changes are dependent on the presence of estradiol. For instance, Esr1 (the gene for estrogen receptor alpha (UniProt, 2019)) disappears when comparing male and female ArKO mice. Interestingly, estrogen receptor beta (i.e., Esr2) was not different between sexes or genotypes. Other notable gene differences that disappeared with aromatase ablation include the CLOCK gene, Per3, Prkg2 (associated with circadian regulation (Oster et al., 2003), as well as BH4, and thus, dopamine synthesis (Scherer-Oppliger et al., 1999)), Ccnd1 (a potential mechanistic link between obesity and breast cancer (Adams et al., 2018)), and Ngfr (associated with circadian rhythms of glucose and lipid homeostasis genes (Baeza-Raja et al., 2013)).
When assessing the effect of aromatase ablation on gene expression of each sex, Pts (essential for BH4, and thus catecholamine synthesis (Elzaouk et al., 2003)), Gldn (involved in sodium channel function on the Nodes of Ranvier in the peripheral nervous system (Eshed et al., 2005)), Cryab (gene coding for a heat shock protein (Zhang et al., 2019)), and Nxpe4 (associated with glycosylation of amino acids (UniProt, 2019)) were differentially expressed within males and females. It is noteworthy that a previous study genetically downregulated systemic Pts expression in C57B1/6J mice, resulting in significantly impaired BH4 synthesis in liver and brain (Korner et al., 2016). While systemic ablation of Pts is not a viable mutation, mice with significantly reduced Pts expression were obese with impaired glucose and lipid metabolism. In particular, these mice had elevated intrabdominal fat, particularly in males (Korner et al., 2016). Their findings determined that downregulation of Pts contributed to increased body weight and elevated intrabdominal fat, particularly in males, with alterations in glucose and lipid metabolism in both sexes potentially via reduced eNOS function (Korner et al., 2016). Unfortunately, analyses of behaviors that may have contributed to obesity, namely reduced physical activity, were not analyzed in this previous work. Since Pts is necessary for dopamine production, and prior work has shown that dopamine signaling in the NAc brain region is a critical determinant of voluntary physical activity (Ruegsegger et al., 2016; Ruegsegger et al., 2017), it is tempting to speculate that the low physical activity among ArKO mice is due to aromatase’s inhibitory actions on Pts expression in the NAc. Furthermore, our prior work has shown that ovariectomy in rats both leads to reduced voluntary wheel running, and reduces genes associated with dopamine signaling in the NAc brain region. Furthermore, a strong correlation was noted between positive dopamine signaling in this brain region and voluntary wheel running (Park et al., 2016).
It is also notable that Per3, a CLOCK gene and known regulator of circadian rhythm, may impact metabolism, sleep, body temperature, and locomotor activity, and was expressed higher in F vs. M WT, yet this sex difference went away with aromatase deletion. This may suggest that estrogen-mediated regulation of CLOCK genes in the NAc is implicated in sex differences in sleep and physical activity patterns, and that physical activity and sleep disruptions that occur following menopause are related to changes in such genes in the NAc brain region.
Pathway enrichment and functional analysis supported our hypothesis and identified locomotion as a GO:BP pathway between WT and ArKO mice. Although this pathway was identified as different between WT and KO males, no significant differences were observed in our study regarding male physical activity. The lack of physical activity differences in males may indicate that this behavior is age-dependent in a sexually dimorphic manner. Previous work has identified dopamine in the NAc as differentially regulated with changes in physical activity (Bonansco et al., 2018; Jardi et al., 2018; Park et al., 2016). This brain region may be sexually dimorphic in that females reduce physical activity after loss of estradiol, while males rely on testosterone for the same behavioral changes, an idea which is supported by studies of addiction and locomotor activity, behaviors that are also associated with the NAc and have been shown to be sexually dimorphic (Jardi et al., 2018; Wissman et al., 2011; Zhu et al., 2016).
Limitations of this study include the use of a systemic aromatase knock-out mouse model and the diet used in our facility as well as the use of single/pair housing. The rodent chow available to these mice included phytoestrogens, which may have confounded the effects; nevertheless, all groups received the same diet. Regarding the model, these mice lack the aromatase enzyme throughout fetal development, so it is not possible to determine if the observed effects of aromatase are organizational or activational in nature. In addition, mice were not singly-housed or pair-housed consistently, contributing to possible variations in behavioral outcomes as social interactions are known to impact mouse behavior (Ma et al., 2011). Mice used in these studies systemically lack Cyp19 throughout their lifespan, and thus, it is not clear if similar gene expression changes may be evident in the NAc region of adult females treated with AI. To address this issue, future studies in WT mice may examine aromatase action in the NAc by pharmacologic manipulation via localized cranial injection of pharmacological aromatase inhibitor or use of Designer Receptors Exclusively Activated by Designer Drugs (DREADD) technology targeting dopaminergic neurons in order to assess the organizational versus activational role of aromatase in this brain region (Zhu et al., 2016). In addition, metabolic and behavioral analyses will need be performed in order to evaluate the physiologic effects of the changes in aromatase and dopamine signaling on these outcomes. These findings likely have important implications for clinical treatments of various diseases associated with estrogen signaling, in particular, the metabolic dysfunction and insulin resistance associated with obesity. Future studies in humans could also utilize fluorescent techniques such as positron emission tomography (PET) imaging to analyze aromatase expression and activity in vivo (Hurria et al., 2014), allowing scientists to investigate which brain regions are most affected by changes in aromatase.
In conclusion, our findings of ArKO phenotype sex differences support previous work in this model. Novel findings of correlations between NAc gene expression (e.g. Pts, Cryab, & Per3) and physical activity, sleep, and body composition provide insight to possible mechanisms of the role of estrogen in the brain via aromatase. Future studies should examine the underpinning mechanisms leading to sex differences in physical activity that may be regulated by the aromatase activity in the NAc.
Supplementary Material
Acknowledgements
We appreciate helpful comments and assistance from Julia C. Hatzigeorgiou and Kevin L. Fritsche.
Funding
CSR is supported by NIEHS 1R01ES025547. DAS (mentor VVP) was supported by a University of Missouri DNA Core Research Facility internal grant.
Footnotes
Declaration of competing interest
None.
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://doi.org/10.1016/j.yhbeh.2020.104719.
References
- Adams BD, Arem H, Hubal MJ, Cartmel B, Li F, Harrigan M, Sanft T, Cheng CJ, Pusztai L, Irwin ML, 2018. Exercise and weight loss interventions and miRNA expression in women with breast cancer. Breast Cancer Res. Treat. 170, 55–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano A, Kondo Y, Noda Y, Ohta M, Kawanishi N, Machida S, Mitsuhashi K, Senmaru T, Fukui M, Takaoka O, Mori T, Kitawaki J, Ono M, Saibara T, Obayashi H, Ishigami A, 2017. Abnormal lipid/lipoprotein metabolism and high plasma testosterone levels in male but not female aromatase-knockout mice. Arch. Biochem. Biophys 622, 47–58. [DOI] [PubMed] [Google Scholar]
- Baeza-Raja B, Eckel-Mahan K, Zhang L, Vagena E, Tsigelny IF, Sassone-Corsi P, Ptacek LJ, Akassoglou K, 2013. p75 neurotrophin receptor is a clock gene that regulates oscillatory components of circadian and metabolic networks. J. Neurosci 33, 10221–10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartling B, Al-Robaiy S, Lehnich H, Binder L, Hiebl B, Simm A, 2017. Sex-related differences in the wheel-running activity of mice decline with increasing age. Exp. Gerontol 87, 139–147. [DOI] [PubMed] [Google Scholar]
- Bhave MA, Speth KA, Kidwell KM, Lyden A, Alsamarraie C, Murphy SL, Henry NL, 2018. Effect of aromatase inhibitor therapy on sleep and activity patterns in early-stage breast Cancer. Clin. Breast Cancer 18 (168–174), e162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bioconductor, 2003. Bioconductor.
- Blakemore J, Naftolin F, 2016. Aromatase: contributions to physiology and disease in women and men. Physiology (Bethesda) 31, 258–269. [DOI] [PubMed] [Google Scholar]
- Bonansco C, Martinez-Pinto J, Silva RA, Velasquez VB, Martorell A, Selva MV, Espinosa P, Moya PR, Cruz G, Andres ME, Sotomayor-Zarate R, 2018. Neonatal exposure to estradiol increases dopaminergic transmission in nucleus Accumbens and morphine-induced conditioned place preference in adult female rats. J. Neuroendocrinol 30, e12574. [DOI] [PubMed] [Google Scholar]
- Bowen RS, Ferguson DP, Lightfoot JT, 2011. Effects of aromatase inhibition on the physical activity levels of male mice. J Steroids Horm Sci 1, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockman R, Bunick D, Mahoney MM, 2011. Estradiol deficiency during development modulates the expression of circadian and daily rhythms in male and female aromatase knockout mice. Horm. Behav 60, 439–447. [DOI] [PubMed] [Google Scholar]
- Brown JC, Mao JJ, Strieker C, Hwang WT, Tan KS, Schmitz KH, 2014. Aromatase inhibitor associated musculoskeletal symptoms are associated with reduced physical activity among breast cancer survivors. Breast J. 20, 22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins B, Mackenzie J, Stewart A, Bielajew C, Verma S, 2009. Cognitive effects of hormonal therapy in early stage breast cancer patients: a prospective study. Psychooncology 18, 811–821. [DOI] [PubMed] [Google Scholar]
- Dalla C, Antoniou K, Papadopoulou-Daifoti Z, Balthazart J, Bakker J, 2004. Oestrogen-deficient female aromatase knockout (ArKO) mice exhibit depressive-like symptomatology. Eur. J. Neurosci 20, 217–228. [DOI] [PubMed] [Google Scholar]
- Dalla C, Antoniou K, Papadopoulou-Daifoti Z, Balthazart J, Bakker J, 2005. Male aromatase-knockout mice exhibit normal levels of activity, anxiety and “depressive-like” symptomatology. Behav. Brain Res 163, 186–193. [DOI] [PubMed] [Google Scholar]
- Dewandre D, M., A., Sanchez-Espinosa MP, Cantero JL, 2018. Effects of PER3 clock gene polymorphisms on aging-related changes of the cerebral cortex. Brain Struct Funct 223, 597–607. [DOI] [PubMed] [Google Scholar]
- Dias JP, Melvin D, Simonsick EM, Carlson O, Shardell MD, Ferrucci L, Chia CW, Basaria S, Egan JM, 2016. Effects of aromatase inhibition vs. testosterone in older men with low testosterone: randomized-controlled trial. Andrology 4, 33–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elzaouk L, Leimbacher W, Turri M, Ledermann B, Burki K, Blau N, Thony B, 2003. Dwarfism and low insulin-like growth factor-1 due to dopamine depletion in Pts−/− mice rescued by feeding neurotransmitter precursors and H4-biopterin. J. Biol. Chem 278, 28303–28311. [DOI] [PubMed] [Google Scholar]
- Ensembl, 2019. Mus musculm - Ensembl Release 90.
- Eshed Y, Feinberg K, Poliak S, Sabanay H, Sarig-Nadir O, Spiegel I, Bermingham JR Jr., Peles E, 2005. Gliomedin mediates Schwann cell-axon interaction and the molecular assembly of the nodes of Ranvier. Neuron 47, 215–229. [DOI] [PubMed] [Google Scholar]
- Fisher CR, Graves KH, Parlow AF, Simpson ER, 1998. Characterization of mice deficient in aromatase (ArKO) because of targeted disruption of the cypl9 gene. Proc. Natl. Acad. Sci. U. S. A 95, 6965–6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fountain ED, Mao J, Whyte JJ, Mueller KE, Ellersieck MR, Will MJ, Roberts RM, Macdonald R, Rosenfeld CS, 2008. Effects of diets enriched in omega-3 and omega-6 polyunsaturated fatty acids on offspring sex-ratio and maternal behavior in mice. Biol. Reprod 78, 211–217. [DOI] [PubMed] [Google Scholar]
- Genome Resource, N., 1993. Enterobacteria Phage phiX174 Sensu Lato (ID 4241).
- Gibb FW, Homer NZ, Faqehi AM, Upreti R, Livingstone DE, Mclnnes KJ, Andrew R, Walker BR, 2016. Aromatase inhibition reduces insulin sensitivity in healthy men. J. Clin. Endocrinol. Metab 101, 2040–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GitHub, 2010. FASTX toolkit.
- Givan SA, Bottoms CA, Spollen WG, 2012. Computational analysis of RNA-seq. Methods Mol. Biol 883, 201–219. [DOI] [PubMed] [Google Scholar]
- Gnu Operating System, 2019. GNU Bash. Free Software Foundation. [Google Scholar]
- Gorres-Martens BK, Field TJ, Schmidt ER, Munger KA, 2018. Exercise prevents HFD- and OVX-induced type 2 diabetes risk factors by decreasing fat storage and improving fuel utilization. Physiol Rep 6, e13783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada N, Wakatsuki T, Aste N, Yoshimura N, Honda SI, 2009. Functional analysis of neurosteroidal oestrogen using gene-disrupted and transgenic mice. J. Neuroendocrinol 21, 365–369. [DOI] [PubMed] [Google Scholar]
- Hewitt KN, Boon WC, Murata Y, Jones ME, Simpson ER, 2003. The aromatase knockout mouse presents with a sexually dimorphic disruption to cholesterol homeostasis. Endocrinology 144, 3895–3903. [DOI] [PubMed] [Google Scholar]
- Hewitt KN, Pratis K, Jones ME, Simpson ER, 2004. Estrogen replacement reverses the hepatic steatosis phenotype in the male aromatase knockout mouse. Endocrinology 145, 1842–1848. [DOI] [PubMed] [Google Scholar]
- Honda S, Harada N, Ito S, Takagi Y, Maeda S, 1998. Disruption of sexual behavior in male aromatase-deficient mice lacking exons 1 and 2 of the cypl9 gene. Biochem. Biophys. Res. Commun 252, 445–449. [DOI] [PubMed] [Google Scholar]
- Hurria A, Patel SK, Mortimer J, Luu T, Somlo G, Katheria V, Ramani R, Hansen K , Feng T, Chuang C, Geist CL, Silverman DH, 2014. The effect of aromatase inhibition on the cognitive function of older patients with breast cancer. Clin. Breast Cancer 14, 132–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardi F, Laurent MR, Kim N, Khalil R, De Bundel D, Van Eeckhaut A, Van Helleputte L, Deboel L, Dubois V, Schollaert D, Decallonne B, Carmeliet G, Van den Bosch L, D’Hooge R, Claessens F, Vanderschueren D, 2018. Testosterone boosts physical activity in male mice via dopaminergic pathways. Sci. Rep 8, 957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasarevic E, Sieli PT, Twellman EE, Welsh TH Jr., Schachtman TR, Roberts RM, Geary DC, Rosenfeld CS, 2011. Disruption of adult expression of sexually selected traits by developmental exposure to bisphenol a. Proc. Natl. Acad. Sci. U. S. A 108, 11715–11720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins V, Shilling V, Fallowfield L, Howell A, Hutton S, 2004. Does hormone therapy for the treatment of breast cancer have a detrimental effect on memory and cognition? A pilot study. Psychooncology 13, 61–66. [DOI] [PubMed] [Google Scholar]
- Johnson SA, Spollen WG, Manshack LK, Bivens NJ, Givan SA, Rosenfeld CS, 2017. Hypothalamic transcriptomic alterations in male and female California mice (Peromyscus californicus) developmentally exposed to bisphenol a or ethinyl estradiol. Physiol Rep 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones ME, Thorburn AW, Britt KL, Hewitt KN, Wreford NG, Proietto J, Oz OK, Leury BJ, Robertson KM, Yao S, Simpson ER, 2000. Aromatase-deficient (ArKO) mice have a phenotype of increased adiposity. Proc. Natl. Acad. Sci. U. S. A 97, 12735–12740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones ME, Thorburn AW, Britt KL, Hewitt KN, Misso ML, Wreford NG, Proietto J, Oz OK, Leury BJ, Robertson KM, Yao S, Simpson ER, 2001. Aromatase-deficient (ArKO) mice accumulate excess adipose tissue. J. Steroid Biochem. Mol. Biol 79, 3–9. [DOI] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J, 2005. Repbase update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 110, 462–467. [DOI] [PubMed] [Google Scholar]
- Karatas F, Sahin S, Babacan T, Akin S, Sever AR, Altundag K,, 2015. Insomnia caused by aromatase inhibitors; a therapeutic challenge in patients with breast cancer. J. BUON 20, 1640. [PubMed] [Google Scholar]
- Klinker F, Kohnemann K, Paulus W, Liebetanz D, 2017. Dopamine D3 receptor status modulates sexual dimorphism in voluntary wheel running behavior in mice. Behav. Brain Res 333, 235–241. [DOI] [PubMed] [Google Scholar]
- Korner G, Scherer T, Adamsen D, Rebuffat A, Crabtree M, Rassi A, Scavelli R, Homma D, Ledermann B, Konrad D, Ichinose H, Wolfram C, Horsch M, Rathkolb B, Klingenspor M, Beckers J, Wolf E, Gailus-Durner V, Fuchs H, Hrabe de Angelis M, Blau N, Rozman J, Thony B, 2016. Mildly compromised tetrahydrobiopterin cofactor biosynthesis due to Pts variants leads to unusual body fat distribution and abdominal obesity in mice. J. Inherit. Metab. Dis 39, 309–319. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL, 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kim K, Yu SW, Kim EK, 2016. Wnt3a upregulates brain-derived insulin by increasing NeuroD1 via Wnt/beta-catenin signaling in the hypothalamus. Molecular brain 9, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, Desaki AL, Desta T, Diep E, Dolbeare TA, Donelan MJ, Dong HW, Dougherty JG, Duncan BJ, Ebbert AJ, Eichele G, Estin LK, Faber C, Facer BA, Fields R, Fischer SR, Fliss TP, Frensley C, Gates SN, Glattfelder KJ, Halverson KR, Hart MR, Hohmann JG, Howell MP, Jeung DP, Johnson RA, Karr PT, Kawal R, Kidney JM, Knapik RH, Kuan CL, Lake JH, Laramee AR, Larsen KD, Lau C, Lemon TA, Liang AJ, Liu Y, Luong LT, Michaels J, Morgan JJ, Morgan RJ, Mortrud MT, Mosqueda NF, Ng LL, Ng R, Orta GJ, Overly CC, Pak TH, Parry SE, Pathak SD, Pearson OC, Puchalski RB, Riley ZL, Rockett HR, Rowland SA, Royall JJ, Ruiz MJ, Sarno NR, Schaffnit K, Shapovalova NV, Sivisay T, Slaughterbeck CR, Smith SC, Smith KA, Smith BI, Sodt AJ, Stewart NN, Stumpf KR, Sunkin SM, Sutram M, Tam A, Teemer CD, Thaller C, Thompson CL, Varnam LR, Visel A, Whitlock RM, Wohnoutka PE, Wolkey CK, Wong VY, Wood M, Yaylaoglu MB, Young RC, Youngstrom BL, Yuan XF, Zhang B, Zwingman TA, Jones AR, 2007. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176. [DOI] [PubMed] [Google Scholar]
- Liew SH, Drummond AE, Jones ME, Findlay JK, 2010. The lack of estrogen and excess luteinizing hormone are responsible for the female ArKO mouse phenotype. Mol. Cell. Endocrinol 327, 56–64. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S, 2014. Moderated estimation of fold change and dis-persion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XC, Jiang D, Jiang WH, Wang F, Jia M, Wu J, Hashimoto K, Dang YH, Gao CG, 2011. Social isolation-induced aggression potentiates anxiety and depressive-like behavior in male mice subjected to unpredictable chronic mild stress. PLoS One 6, e20955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manshack LK, Conard CM, Bryan SJ, Deem SL, Holliday DK, Bivens NJ, Givan SA, Rosenfeld CS, 2017. Transcriptomic alterations in the brain of painted turtles (Chrysemys picta) developmentally exposed to bisphenol a or ethinyl estradiol. Physiol. Genomics 49, 201–215. [DOI] [PubMed] [Google Scholar]
- Mao J, Jain A, Denslow ND, Nouri M-Z, Chen S, Wang T, Zhu N, Koh J, Sarma SJ, Sumner BW, Lei Z, Sumner LW, Bivens NJ, Roberts RM, Tuteja G, Rosenfeld CS, 2020. Bisphenol A and bisphenol S disruptions of the mouse placenta and potential effects on the placenta-brain axis. Proc. Natl. Acad. Sci. U. S. A 10.1073/pnas.1919563117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, 2011. Cutadapt removes adaptor sequences from high-throughput sequencing reads. EMBnet.journal 17, 10. [Google Scholar]
- National Center for Biotechnology Information, Accessed 2015, https://www.ncbi.nlm.nih.gov/
- Martin S, Jones M, Simpson E, van den Buuse M, 2003. Impaired spatial reference memory in aromatase-deficient (ArKO) mice. Neuroreport 14, 1979–1982. [DOI] [PubMed] [Google Scholar]
- Mouser JG, Loprinzi PD, Loenneke JP, 2016. The association between physiologic testosterone levels, lean mass, and fat mass in a nationally representative sample of men in the United States. Steroids 115, 62–66. [DOI] [PubMed] [Google Scholar]
- Mukaka MM, 2012. Statistics corner: a guide to appropriate use of correlation coefficient in medical research. Malawi Med. J 24, 69–71. [PMC free article] [PubMed] [Google Scholar]
- Muller ST, Keiler AM, Kraker K, Zierau O, Bernhardt R, 2018. Influence of estrogen on individual exercise motivation and bone protection in ovariectomized rats. Lab. Anim 23677218756455. [DOI] [PubMed] [Google Scholar]
- Nelson LR, Bulun SE, 2001. Estrogen production and action. J. Am. Acad. Dermatol 45, S116–S124. [DOI] [PubMed] [Google Scholar]
- Nemoto Y, Toda K, Ono M, Fujikawa-Adachi K, Saibara T, Onishi S, Enzan H, Okada T, Shizuta Y, 2000. Altered expression of fatty acid-metabolizing enzymes in aromatase-deficient mice. J. Clin. Invest 105, 1819–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega MT, Bivens NJ, Jogahara T, Kuroiwa A, Givan SA, Rosenfeld CS, 2019. Sexual dimorphism in brain transcriptomes of Amami spiny rats (Tokudaia osimensis): a rodent species where males lack the Y chromosome. BMC Genomics 20, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oster H, Werner C, Magnone MC, Mayser H, Feil R, Seeliger MW, Hofmann F, Albrecht U, 2003. cGMP-dependent protein kinase II modulates mPerl and mPer2 gene induction and influences phase shifts of the circadian clock. Curr. Biol 13, 725–733. [DOI] [PubMed] [Google Scholar]
- Padilla J, Jenkins NT, Roberts MD, Arce-Esquivel AA, Martin JS, Laughlin MH, Booth FW, 2013. Differential changes in vascular mRNA levels between rat iliac and renal arteries produced by cessation of voluntary running. Exp. Physiol 98, 337–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YM, Kanaley JA, Padilla J, Zidon T, Welly RJ, Will MJ, Britton SL, Koch LG, Ruegsegger GN, Booth FW, Thyfault JP, Vieira-Potter VJ, 2016. Effects of intrinsic aerobic capacity and ovariectomy on voluntary wheel running and nucleus accumbens dopamine receptor gene expression. Physiol. Behav 164, 383–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Paulo TRS, Winters-Stone KM, Viezel J, Rossi FE, Simoes RR, Tosello G, Freitas IFJ, 2018. Effects of resistance plus aerobic training on body composition and metabolic markers in older breast cancer survivors undergoing aromatase inhibitor therapy. Exp. Gerontol 111, 210–217. [DOI] [PubMed] [Google Scholar]
- Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, Vilo J, 2019. G:profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47, W191–W198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseguini BT, Mehmet Soylu S., Whyte JJ, Yang HT, Newcomer S, Laughlin MH, 2010. Intermittent pneumatic leg compressions acutely upregulate VEGF and MCP-1 expression in skeletal muscle. Am. J. Physiol. Heart Circ. Physiol 298, H1991–H2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld CS, 2017. Sex-dependent differences in voluntary physical activity. J. Neurosci. Res 95, 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegsegger GN, Brown JD, Kovarik MC, Miller DK, Booth FW, 2016. Mu-opioid receptor inhibition decreases voluntary wheel running in a dopamine-dependent manner in rats bred for high voluntary running. Neuroscience 339, 525–537. [DOI] [PubMed] [Google Scholar]
- Ruegsegger GN, Grigsby KB, Kelty TJ, Zidon TM, Childs TE, Vieira-Potter VJ, Klinkebiel DL, Matheny M, Scarpace PJ, Booth FW, 2017. Maternal Western diet age-specifically alters female offspring voluntary physical activity and dopamine-and leptin-related gene expression. FASEB J. 31, 5371–5383. [DOI] [PubMed] [Google Scholar]
- Scherer-Oppliger T, Leimbacher W, Blau N, Thony B, 1999. Serine 19 of human 6-pyruvoyltetrahydropterin synthase is phosphorylated by cGMP protein kinase II. J. Biol. Chem 274, 31341–31348. [DOI] [PubMed] [Google Scholar]
- Shay DA, Vieira-Potter VJ, Rosenfeld CS, 2018. Sexually dimorphic effects of aromatase on neurobehavioral responses. Front. Mol. Neurosci 11, 374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, Stein TI, Nudel R, Lieder I, Mazor Y, Kaplan S, Dahary D, Warshawsky D, Guan-Golan Y, Kohn A, Rappaport N, Safran M, Lancet D, 2016. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr Protoc Bioinformatics 54, 1 30 31–31 30 33. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M , Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV, 2019. STRING vll: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K, Toda K, Saibara T, Nakagawa M, Saika K, Onishi T, Sugiura T, Shizuta Y, 2003. Progressive development of insulin resistance phenotype in male mice with complete aromatase (CYP19) deficiency. J. Endocrinol. 176, 237–246. [DOI] [PubMed] [Google Scholar]
- The R Foundation, 2000. The R Project for Statistical Computing. The R Project for Statistical Computing. [Google Scholar]
- Toda K, Takeda K, Okada T, Akira S, Saibara T, Kaname T, Yamamura K, Onishi S, Shizuta Y, 2001. Targeted disruption of the aromatase P450 gene (Cyp19) in mice and their ovarian and uterine responses to 17betaõestradiol. J. Endocrinol 170, 99–111. [DOI] [PubMed] [Google Scholar]
- Tonn Eisinger KR, Larson EB, Boulware MI, Thomas MJ, Mermelstein PG, 2018. Membrane estrogen receptor signaling impacts the reward circuitry of the female brain to influence motivated behaviors. Steroids 133, 53–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt C, 2019. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyazovskiy VV, Kopp C, Wigger E, Jones ME, Simpson ER, Tobler I, 2006. Sleep and rest regulation in young and old oestrogen-deficient female mice. J. Neuroendocrinol 18, 567–576. [DOI] [PubMed] [Google Scholar]
- Wissman AM, McCollum AF, Huang GZ, Nikrodhanond AA, Woolley CS, 2011. Sex differences and effects of cocaine on excitatory synapses in the nucleus accumbens. Neuropharmacology 61, 217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Liu S, Yin M, Yin Y, Zhou G, Zhou J, 2015. Ebf2 is required for development of dopamine neurons in the midbrain periaqueductal gray matter of mouse. Dev. Neurobiol 75, 1282–1294. [DOI] [PubMed] [Google Scholar]
- Zhang J, Liu J, Wu J, Li W, Chen Z, Yang L, 2019. Progression of the role of CRYAB in signaling pathways and cancers. Onco Targets Ther 12, 4129–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Ottenheimer D, DiLeone RJ, 2016. Activity of Dl/2 receptor expressing neurons in the nucleus Accumbens regulates running, locomotion, and food intake. Front. Behav. Neurosci 10, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


