Abstract
Atrial fibrillation (AF) is a common heart rhythm disorder that leads to an increased risk for stroke and heart failure. AF is a complex disease with both environmental and genetic risk factors that contribute to the arrhythmia. Over the last decade, rapid progress has been made in identifying the genetic basis for this common condition. In this review we provide an overview of the primary types of genetic analyses performed for AF including linkage studies, genome-wide association studies (GWAS) and studies of rare coding variation. With these results in mind, we aim to highlighting the existing knowledge gaps and future directions for AF genetics research.
Keywords: Atrial fibrillation, genome wide association study, mutation, exome sequencing, genome sequencing, genetics
Introduction and Background
Atrial fibrillation (AF) is a common heart rhythm disorder with an estimated 33 million people affected worldwide[1]. Reported risk factors for AF include advancing age, obesity, hypertension, diabetes, and cardiovascular diseases[2]. Studies have also shown an increased risk for men to develop AF, compared to women[3]. As discussed in the accompanying article in this Compendium (formal reference provided by the journal), AF can lead to a number of serious medical consequences including stroke, heart failure, cognitive impairment, and increased mortality.
The treatment of AF remains challenging. While there are effective medications for anticoagulation to reduce the risk of stroke, antiarrhythmic medications are limited by lack of efficacy to reduce symptoms and have potential side effects. Alternatively, catheter ablation procedures can be effective in reducing the burden of AF, but these procedures are invasive, can be associated with complications and may require a repeat procedure for the long-term management of AF. Thus, there is a pressing need to develop new therapies for AF.
Similar to other common cardiovascular diseases such as hypertension and myocardial infarction, AF is a complex disease with shared environmental and genetic factors that contribute to disease pathogenesis. Over the last decade, multiple studies have observed familial aggregation of individuals with lone AF[4]. Similarly, the heritability of AF has been elegantly demonstrated in the Icelandic population[5]. Based on a study on monozygotic twins the heritability of AF has been estimated as high as 62%, indicating a strong genetic component[6]. In aggregate these studies have consistently observed an increased risk of AF particularly when a first-degree family member is affected, and among individuals with early-onset forms of the arrhythmia[7]. In more recent work, Weng and colleagues used genetic data to estimate that the heritability of AF based on common genetic variants in individuals of European ancestry is approximately 22%[8].
In the current review, we will provide an overview of multiple approaches used to examine the genetic basis of AF. We will present the most relevant results from these analyses and discuss emerging technological advances that could be leveraged to expand our understanding of the field. We discuss three broad genetic approaches applied to AF including: 1) linkage analysis using families with Mendelian forms of AF, 2) genome-wide association studies (GWAS) studies using genotyping array data, and 3) coding variation from genome sequence data (Figure 1). These approaches are not mutually exclusive but are helpful as a framework to consider when reviewing the genetic studies of AF published to date. We will subsequently describe the application of GWAS data to clinical risk prediction. Finally, we will discuss the knowledge gaps in the field of AF genetics and describe emerging technologies that may shape the future of the field.
Figure 1. Three primary types of genetic analyses for AF.
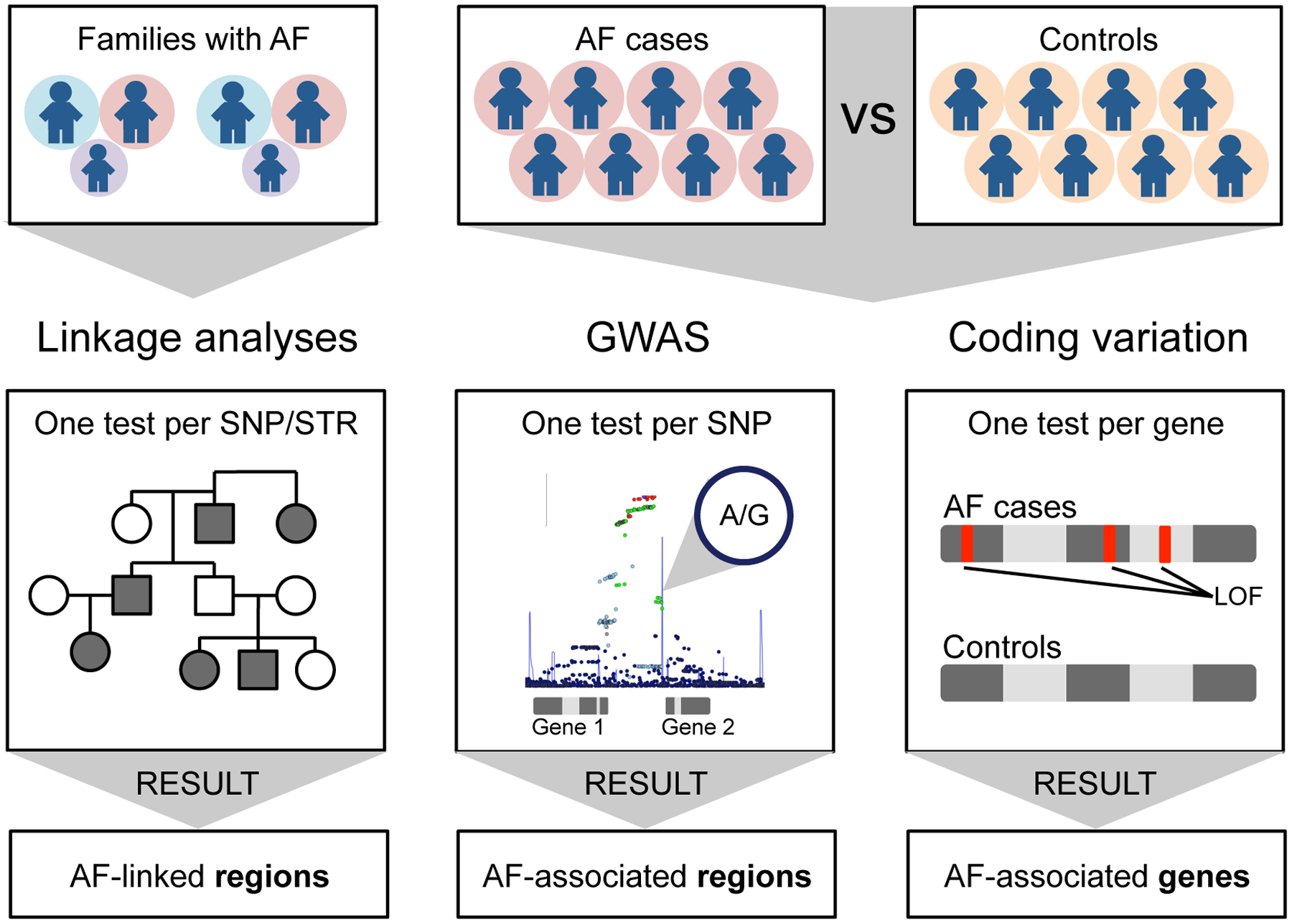
Linkage analyses primarily focus on large families with hereditary forms of AF. The disease associated linked regions can include multiple candidate genes one of which will contain a disease causing mutation. GWAS analyses are based on genotype array data that consists largely of non-coding variants that are presumed to regulate genes in the region or locus. Analyses of coding variation are derived from whole-exome or whole-genome sequencing data. Rare coding or loss-of-function variants are grouped and jointly tested in AF cases versus controls to identify specific disease-causing genes. Please note that these approaches are not mutually exclusive and are often combined depending upon the study design.
Abbreviations: AF, atrial fibrillation; GWAS, genome-wide association study; LOF, loss-of-function; SNP, single-nucleotide polymorphism; STR, short tandem repeat.
Three Broad Genetic Approaches Applied to AF
Familial AF and Linkage Analysis
Linkage analysis is typically performed in families with many affected individuals and a clear hereditary pattern. The analytic approach leverages genetic linkage or the tendency for a genetic marker near a disease-causing gene to be inherited together. The earliest application of this approach used a few hundred highly informative markers scattered throughout the genome. By matching transmission of the genetic marker with disease status in the family, a disease-causing region or locus can be linked to a given marker. In turn, the genes in this region can be sequenced to identify mutations that associate with disease. This approach has been elegantly used to identify many of the early genes implicated in hypertrophic cardiomyopathy and long QT syndrome. As summarized in Figure 2, a number of causative mutations for AF have been identified in large families or populations. Specific examples include the ion channel KCNQ1, the cardiac peptide NPPA, the transcription factor TBX5, and a motor protein MYL4.
Figure 2. Major AF-associated genes and lines of evidence.
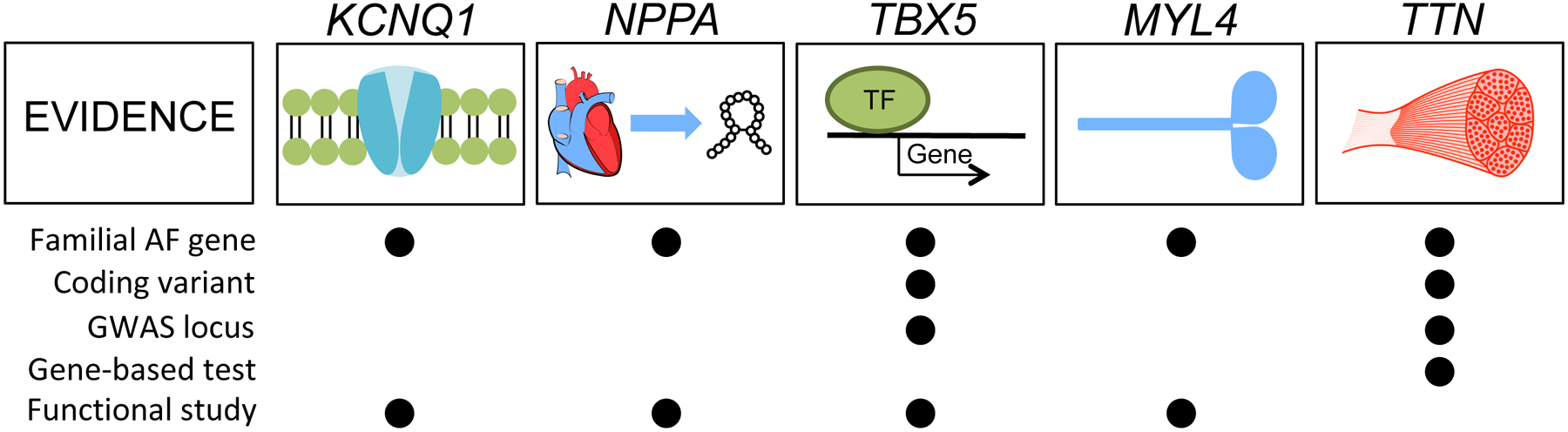
The figure illustrates AF-associated genes that were discovered through family based or gene-based studies. For each gene the lines of evidence are listed. The table includes the evidence from familial AF genetic analysis, whether coding variants in the gene are associated with AF, if the gene lies within an AF GWAS locus, whether loss-of-function variation is associated with AF, and functional evidence that has been reported for the gene in the context of AF.
Abbreviations: AF, atrial fibrillation; GWAS, genome-wide association study.
The first mutation linked to familial AF was found in the ion channel KCNQ1, a gene that encodes the alpha subunit of the IKs current. The AF related mutations result in a gain of channel function and likely shortening of the atrial refractory period, a finding that would make it easier for reentry to continue and increase the susceptibility to AF[9]. In a distinct family, a mutation was identified in NPPA, the gene encoding the atrial natriuretic peptide (ANP), a protein that is highly expressed in the heart[10]. A frame-shift mutation was found to remove a stop codon and lead to an extended mutant protein that had appeared to be protected from degradation resulting in greater circulating levels and increased activity[11]. In vivo experiments from the same study have shown that mutations in NPPA lead to a shortening of the monophasic action potential duration as well as the effective refractory period. These changes on atrial electrophysiology could increase susceptibility to AF.
A gain-of-function mutation in the transcription factor, TBX5, was associated with familial AF in the setting of Holt-Oram syndrome[12], a developmental disorder that leads to heart and limb malformations[13]. In vitro studies demonstrated that the mutated TBX5 had enhanced binding to DNA and could lead to up-regulation of downstream targets such as NPPA and CX40[12,14]. In a large-scale study of Icelanders, autosomal recessive mutations in MYL4 were identified among individuals with early-onset AF[15]. This atrial-specific myosin light chain was found to have a mutation in an F-actin binding region, and modeling the mutation in zebrafish resulted in disruption of the sarcomere and atrial enlargement.
Thus, individual families with hereditary forms of AF can be incredibly helpful in informing disease biology. However, the mutations identified to date are by nature rare and therefore have a small impact on the overall scope of this common arrhythmia.
Genome-Wide Association Studies for AF
In contrast to studies in families, GWAS permit the analysis of entire populations by comparing individuals with and without AF at a large scale. Each individual is genetically “fingerprinted” using a low cost, high throughput genotyping array. These arrays are used to determine the status of hundreds of thousands of genetic variants or single nucleotide polymorphisms (SNPs) throughout the genome. While individual SNPs contain relatively little information, in aggregate, the use of hundreds of thousands of markers can capture the majority of the genetic diversity between individuals. In order to compare the data from one genotyping platform to another, the data is imputed or harmonized to a common reference panel consisting of millions of genetic markers[16]. Comparisons of all the genetic markers are then made between cases and controls to identify regions associated with disease (Figure 1, middle). Importantly, unlike the analysis in families in which a single causative mutation is identified, in GWAS a region or locus is linked to disease. This region may or may not contain any genes. Many causative variants identified by GWAS are in non-coding regions of the genome and have an effect on the regulation of a nearby gene. In recent years, the cost of genotyping arrays has fallen to less than $50 per individual so it is now feasible undertake large-scale studies of common diseases and studies with more than 100,000 cases are increasingly common. Large sample sizes help to both ensure the validity of the results and to define the full extent of the genetic basis of the disease.
The first GWAS for AF was reported in 2007 and remarkably started with only a few hundred AF cases in the initial discovery[17]. The small number of individuals needed in this first report was a reflection of the strength of the genetic association which is considerably greater than that observed for most other genome wide studies. Carrying a single variant at this 4q25 locus near the gene PITX2 conferred over a 60% increased risk of disease in the general population and an even greater risk in younger individuals. As with many disease-associated regions identified by GWAS, the association with AF at the PITX2 locus is in a non-coding region of the genome. As reviewed in other manuscripts in this issue (formal reference to be provided by the journal), the AF risk variants modulate the expression of PITX2 and loss of this region or the PITX2 gene itself can results in AF[18,19].
In the ensuing years, many other GWAS for AF have been completed in predominantly European ancestry participants[20–24]. Initially novel loci were identified at 16q22 close to the gene ZFHX3 and 1q21 close to the KCNN3[25–27]. In two recent studies that both included over 60,000 cases[28,29] nearly 140 AF loci have been identified to date (Table 1). While rapid progress has been made in AF genetics, it is important to note that the vast majority of participants are of European descent (Figure 3). The two largest non-European GWAS were published in 2017 and report results from Korean[30] and Japanese[31] cohorts. At least three variants for AF show significant heterogeneity across different ancestries (rs2129977 at PITX2, rs11598047 at NEURL and rs2359171 at ZFHX3). Additionally, while a signal at the PITX2 locus can be found across ancestries, the top variant is not always the same nor strongly correlated[28]. Two AF loci from a GWAS in Japanese ancestry, at the genes NEBL and SH3PXD2A, do not replicate in European ancestry and may be ancestry specific[31].”
Table 1.
To date there are at least 138 AF loci identified in single variant testing with P-value < 5 × 10−8. The table below includes the most significant variant at each locus from the latest AF GWAS publications by Roselli et al.[28] or Nielsen et al.[29]. The listed genes are either the nearest gene(s) or the eGene. The eGene is defined as a gene with an eQTL to the variant at an AF GWAS locus and is highlighted in bold. The eGene is listed when it is the only eGene at that locus. Notably, only 18 out of 138 variants are associated with only one eGene.
| Rsid | Nearest Gene(s) or eGene | Rsid | Nearest Gene(s) or eGene | Rsid | Nearest Gene(s) or eGene |
|---|---|---|---|---|---|
| rs187585530 | UBE4B | rs716845 | KCNN2 | rs1822273 | NAV2 |
| rs880315 | CASZ1 | rs2012809 | FBN2, SLC27A6 | rs949078 | SORL1, MIR100HG |
| rs7529220 | HSPG2, CELA3B | rs34750263 | WNT8A, NME5 | rs76097649 | KCNJ5 |
| rs2885697 | SCMH1 | rs174048 | ARHGAP26, NR3C1 | rs6490029 | CUX2 |
| rs11590635 | AGBL4 | rs12188351 | SLIT3 | rs10842383 | LINC00477, BCAT1 |
| rs56202902 | FAF1 | rs6882776 | NKX2–5 | rs113819537 | SSPN |
| rs146518726 | C1orf185 | rs73366713 | ATXN1 | rs12809354 | PKP2 |
| rs12044963 | KCND3 | rs34969716 | KDM1B | rs7978685 | NACA |
| rs4484922 | CASQ2 | rs1307274 | C6orf1, NUDT3 | rs35349325 | BEST3 |
| rs79187193 | GJA5 | rs3176326 | CDKN1A | rs11180703 | KRR1, PHLDA1 |
| rs11264280 | KCNN3, PMVK | rs6907805 | CGA, ZNF292 | rs883079 | TBX5 |
| rs72700114 | METTL11B, LINC01142 | rs210632 | GOPC | rs12810346 | TBX5-AS1, TBX3 |
| rs608930 | GORAB, PRRX1 | rs17079881 | SLC35F1 | rs10773657 | HIP1R |
| rs10753933 | PPFIA4 | rs13191450 | GJA1, HSF2 | rs12298484 | DNAH10 |
| rs4951261 | NUCKS1 | rs12208899 | LINC00326, EYA4 | rs6560886 | FBRSL1 |
| rs6546620 | KIF3C | rs117984853 | UST | rs9580438 | LINC00540, BASP1P1 |
| rs6742276 | XPO1 | rs11768850 | SUN1 | rs35569628 | CUL4A |
| rs2540949 | CEP68 | rs55734480 | DGKB | rs28631169 | MYH7 |
| rs10165883 | SNRNP27 | rs6462078 | CREB5 | rs2145587 | AKAP6 |
| rs72926475 | REEP1, KDM3A | rs74910854 | PMS2P2 | rs73241997 | SNX6,CFL2 |
| rs28387148 | GYPC | rs11773884 | CDK6 | rs2738413 | SYNE2 |
| rs67969609 | TEX41 | rs62483627 | COG5 | rs74884082 | DPF3 |
| rs12992412 | MBD5 | rs11773845 | CAV1 | rs10873299 | LRRC74, IRF2BPL |
| rs56181519 | WIPF1 | rs55985730 | OPN1SW | rs147301839 | MYZAP |
| rs35504893 | FKBP7 | rs7789146 | KCNH2 | rs62011291 | USP3 |
| rs295114 | SPATS2L | rs35620480 | LINC00208, GATA4 | rs12591736 | TLE3, UACA |
| rs35544454 | ERBB4 | rs7508 | ASAH1 | rs74022964 | HCN4, REC114 |
| rs6810325 | MKRN2 | rs7846485 | XPO7 | rs12908004 | LINC00927, ARNT2 |
| rs73032363 | THRB | rs62521286 | FBXO32 | rs12908437 | IGF1R |
| rs6790396 | SCN10A | rs35006907 | MTSS1, LINC00964 | rs2286466 | RPL3L |
| rs2306272 | SLC25A26 | rs7460121 | MIR30B | rs2359171 | ZFHX3 |
| rs17005647 | FRMD4B | rs6993266 | PTK2 | rs7225165 | YWHAE, CRK |
| rs7632427 | EPHA3 | rs4977397 | SLC24A2, MLLT3 | rs8073937 | POLR2A, TNFSF12 |
| rs17490701 | PHLDB2 | rs4385527 | C9orf3 | rs72811294 | MYOCD |
| rs1278493 | PPP2R3A | rs4743034 | ZNF462 | rs11658278 | ZPBP2 |
| rs4855075 | GNB4 | rs10760361 | PSMB7 | rs242557 | MAPT |
| rs60902112 | XXYLT1 | rs2274115 | LHX3 | rs76774446 | GOSR2 |
| rs9872035 | PAK2 | rs2296610 | NEBL | rs7219869 | KCNJ2, CASC17 |
| rs3822259 | WDR1 | rs7919685 | NRBF2 | rs12604076 | CYTH1 |
| rs1458038 | PRDM8, FGF5 | rs7096385 | SIRT1 | rs9953366 | SMAD7 |
| rs3960788 | UBE2D3 | rs60212594 | SYNPO2L | rs8088085 | MEX3C |
| rs2129977 | PITX2, C4orf32 | rs11001667 | C10orf11 | rs2145274 | CASC20, BMP2 |
| rs55754224 | CAMK2D | rs1044258 | C10orf76 | rs7269123 | C20orf166 |
| rs10213171 | ARHGAP10 | rs11598047 | NEURL | rs2834618 | LOC100506385 |
| rs10520260 | HAND2-AS1 | rs2047036 | SH3PXD2A | rs465276 | TUBA8 |
| rs6596717 | LOC102467213, EFNA5 | rs10749053 | RBM20 | rs133902 | MYO18B |
Figure 3. Ancestry of the cases in genome-wide association studies for AF.
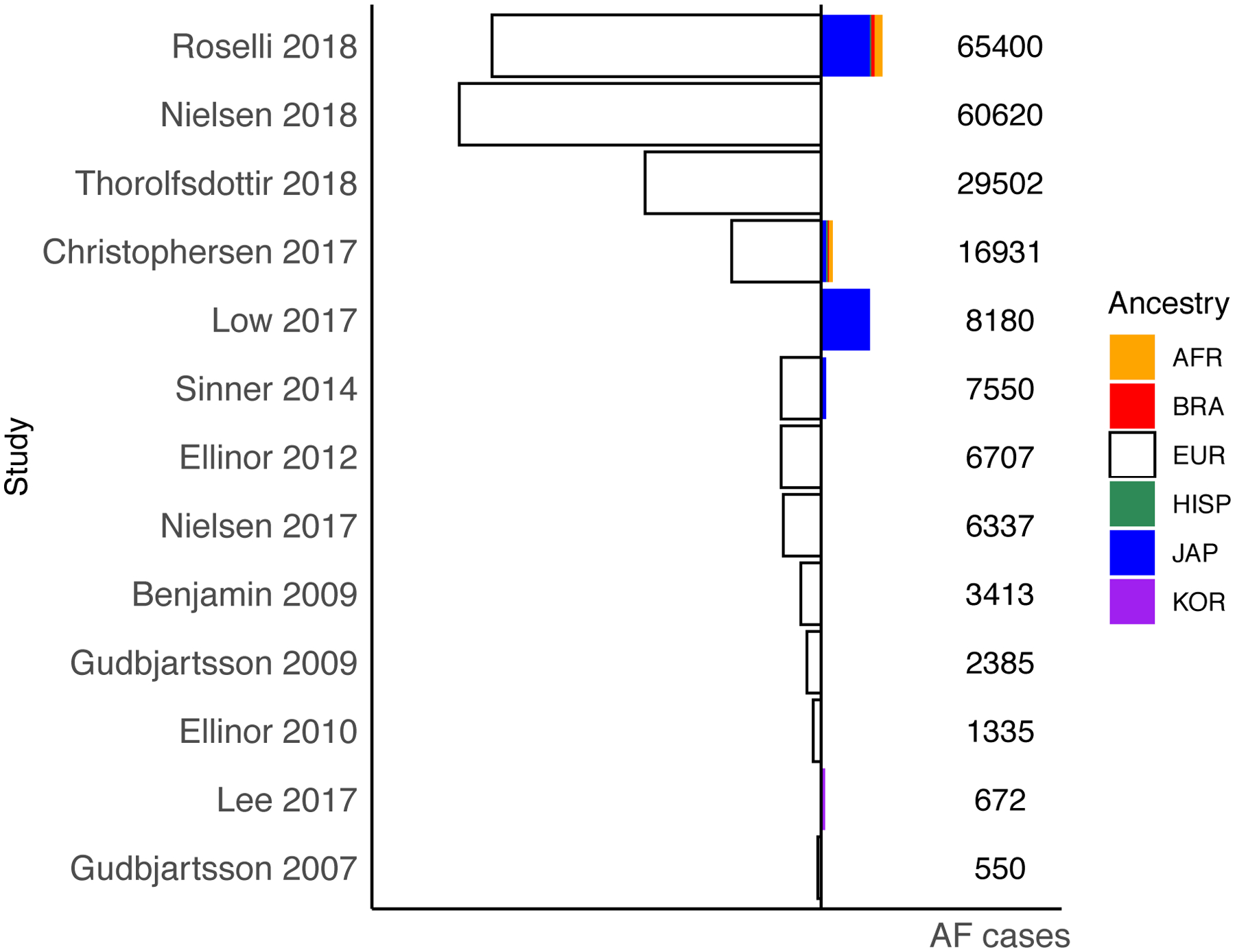
European ancestry sample is plotted towards the left in white and non-European ancestry is plotted towards the right highlighted in different colors. Plotted is the number of cases included in each published AF GWAS study or meta-analysis. 10 out of 12 studies include predominantly European ancestry samples, shown in white. Two studies are Japanese only and Korean only. Within the multi-ancestry meta-analyses Roselli et al.[28] included the largest proportion of non-European cases including Japanese, Brazilian, African American and Hispanic samples.
Abbreviations: AF, atrial fibrillation, AFR, African-American, BRA, Brazilian, EUR, European, HISP, Hispanic, JAP, Japanese, KOR, Korean.
Identifying genes at AF loci
Genetic variants identified in GWAS studies are largely located in non-coding regions of the genome. These non-coding variants are presumed to alter the activity of a transcriptional regulatory element such as an enhancer or repressor that in turn results in modifying the transcription of a nearby gene. Importantly, for most GWAS variants there is usually no straightforward path from an association by GWAS to a gene and a disease mechanism. A recent study used STARR-seq to identify regulatory elements and their target genes at multiple GWAS loci for AF. Further, they found that the loss of a regulatory element at the HCN4 locus led to reduced gene expression[32]. A myriad of analyses can follow a large scale GWAS ranging from computational analyses to the derivation of polygenic risk scores for AF risk prediction and the incorporation of GWAS data into other analyses (Table 2). In the subsequent sections we will touch on a few helpful approaches with respect to AF, but for other potential directions please also see these recent reviews[33–35]. One straightforward application of GWAS data is to perform a pathway analysis to evaluate the collective effect of the genetic association on different biological functions. Globally the AF associated genes represent distinct functional groups including those underlying cardiac development, cellular electrophysiology, cardiomyocyte contractility and structure[24,28,29]. A similar approach was taken in a GWAS of the Japanese population and implicated suppression of the mTOR signaling pathway in AF[36].
Table 2.
Overview of studies that integrated AF GWAS data with other clinical, epigenetic or genetic datasets.
| Study type | Description and reference |
|---|---|
| Mendelian Randomization | Obesity [72] |
| Thyroid Function [73] | |
| Serum albumin [74] | |
| Body mass index/body composition and cardiovascular conditions including AF [75] | |
| Body composition [76] | |
| Methylation | Genome-wide DNA methylation analysis [77] |
| Methylome-wide Association Study [78] | |
| Heritability | Heritability of AF [8] |
| Ancestry specific analyses | European ancestry as a risk factor for AF [79] |
| AF recurrence after ablation | Common variants predict AF recurrence [80] |
| Genetic Interaction | Gene-gene interactions [81] |
| Genetic interactions with risk factors [82] | |
| Associations with Mitochondrial DNA | Mitochondrial DNA and Incident AF [83] |
As noted above, a major challenge with GWAS is that the analyses usually identify a region of interest rather than a specific causative gene. Bridging the gap from variants to genes remains a major challenge in disease genetics, particularly for complex polygenic traits such as AF. One common and helpful approach is to use expression quantitative trait loci (eQTL) mapping. An eQTL analysis links the genotype of a SNP at an AF locus to the expression of genes in the region. If an AF associated SNP is strongly linked to the expression of a single gene, that gene is likely to be the causative gene at the locus. While this approach is simple in theory, practically there are two primary limitations. First, eQTL analyses are often tissue specific. Although there are terrific publicly available resources such as GTEx or the Genotype-Tissue Expression project (https://gtexportal.org) that includes gene expression profiles from many tissues, unfortunately the cardiac analyses were limited to the left ventricle and right atrial appendage[10]. Recent work in the Cleveland Clinic Atrial Tissue Bank[22,37] and the Myocardial Applied Genomics Network[28] have addressed this limitation by investigating the gene expression profiles of left atrial tissue. A second limitation of using eQTLs is that it requires large sample sizes and this is not always possible when tissues are hard to obtain such as the left atrium or the pulmonary veins in the case of AF. Finally, while eQTLs are powerful, they only explain a fraction of the disease loci. For example, in the latest two GWAS of AF only 13% of the variants at AF loci could be linked to the expression of a single gene, and at 22% of AF loci the variants linked to one or more genes. These results are summarized in Table 1.
A complimentary approach to eQTL analyses is to use the three-dimensional architecture of the genome to identify causative AF genes. Since many AF SNPs are in non-coding regions of the genome, they are presumed to alter regulatory elements such as enhancers or repressors that in turn bind to the promoter of a nearby gene to regulate its expression. The contact points between AF associated regulatory elements and gene promoters can be assessed genome-wide using chromosome conformation capture technologies such as HiC. Similar to eQTL analyses, enhancer-promoter contacts can be tissue specific and a recent HiC study from the human heart roughly doubled the number of AF associated genes derived from GWAS data[38]. Other analyses of AF GWAS data such as the incorporation of multi-omic datasets[39] or epigenetic analyses including STARR-seq[40] are described in detail in the accompanying review by XX and colleagues (formal reference to be provided by the journal). Ultimately, though any gene implicated by these methods will require further validation in vitro and in vivo.
Assessing polygenic risk from GWAS
Since we now have very dense GWAS datasets for AF, it is natural to wonder whether this data could be used to in a clinical setting to identify high risk individuals, stratify screening efforts or look for differential treatment outcomes. The polygenic nature of AF as captured in GWAS can be transformed into a genetic risk score for each individual. An AF polygenic risk score (PRS) summarizes the cumulative genetic risk and can be computed using anywhere from just a few variants at the top loci or the data from millions of SNPs across the entire genome (Figure 4).
Figure 4. Overview of polygenic risk scores (PRS) for AF.
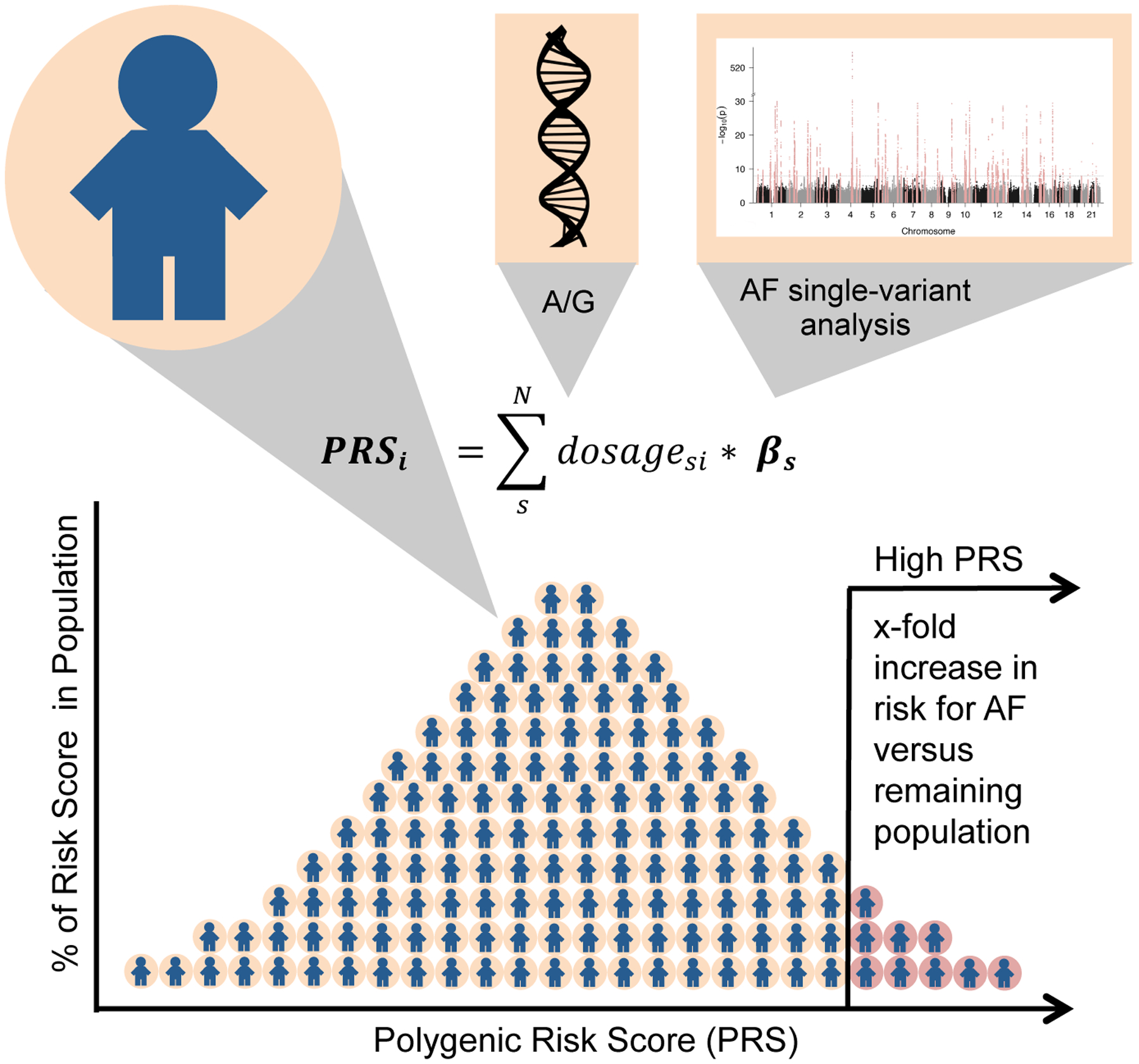
A polygenic risk score is calculated for each individual as a sum of the product of genetic dosage and a weight. The weights are derived from the effect estimates of a genome-wide association study. The PRS of individuals in a population follows a Gaussian distribution. Individuals in the highest percentile of the distributions show an increased risk for AF versus the remaining population. Potential applications of an AF PRS can include improving risk prediction, prioritizing high risk individuals for screening, and examining differential outcomes of AF.
Abbreviations: AF, atrial fibrillation, PRS, polygenic risk score.
An initial AF PRS scores used the top 12 genome-wide significant genetic variants[41], while more contemporary iterations that incorporate over 6.6 million variants[42]. In the later approach, the top 1.5% individuals with a high PRS had a more than a 4-fold increased risk for AF. The variants and weights included in the latest AF PRS are publicly available (http://broadcvdi.org). In one interesting application of an AF PRS, Weng and colleagues found that individuals in the highest tertile of polygenic risk had a higher lifetime risk for AF (47%) compared to the individuals in the lowest tertile (26%)[43]. The combination of clinical and genetic data permitted additional refinement of risk. For example, individuals with a low clinical risk but a high genetic risk had an overall lifetime risk for AF of 44%. A complementary review in this Compendium focuses on the potential clinical applications of AF genetics and polygenic risk scores (formal reference to be provided by the journal).
Exome and genome sequencing to identify AF genes
While GWAS has been deployed at great scale, a major limitation of the method is that it is only able to capture known variants in the genome. Yet many diseases arise from unique mutations in the coding region of genes that arise spontaneously in an individual of family. While these mutations can be identified using a targeted approach of a single or small number of genes, the availability of genome wide approaches has largely replaced these earlier studies. Currently, it is possible to sequence the entire protein-coding region of the genome for about $200 or the entire genome for less than $800, and prices are continuing to fall. These technological advancements and price reductions have started to enable the application of large scale sequencing studies for AF.
Once a population of cases and controls has been sequenced, common genetic variants are analyzed using the approach previously described for GWAS. In contrast, rare coding genetic markers, often defined as present in less than 1% of the population, are analyzed in gene-based tests. For a gene-based test, coding variants are analyzed jointly across a gene unit for an association with disease, as depicted in Figure 1. The most commonly used approach restricts the analysis to variants predicted to lead to a loss-of-function of the encoded protein. A considerable advantage of testing rare loss-of-function variants over GWAS is establishing a direct link from gene function to disease. In addition, this analysis provides a clear direction of effect between loss-of-function of the encoded protein and disease. While many sequencing analyses focus on loss-of-function variants, it is important to note that gain-of-function mutations such as those identified in families with TBX5 or KCNQ1 would not be identified with this approach.
In 2017 Thorolfsdottir and colleagues analyzed whole-genome sequencing data from the Icelandic population including 14,255 AF cases and 374,939 controls[44]. The Icelandic population provides a unique resource for genetic discovery, because it is a relatively homogeneous and genetically isolated population that is enriched with rare loss-of-function coding variants. The study identified a low-frequency missense mutation in the gene PLEC that encodes the cytoskeletal protein plectin. The missense mutation in PLEC is associated with increased risk for AF. Additionally, a missense mutation in the myosin gene MYH6 was significantly associated with AF, a gene that has previously been associated with sick sinus syndrome[45].
Within the last two years, at least four studies have identified loss-of-function mutations in TTN among individuals with AF. Ahlberg and colleagues found an enrichment of loss-of-function mutations in TTN among families and individuals in Denmark with early-onset AF[46]. Shortly thereafter, Choi and colleagues observed a similar finding among unrelated individuals with early-onset AF. Using nearly 2,800 AF cases and 5,000 controls, they found that TTN loss of function mutations were present in ~2% of individuals with an onset of AF before 65 years of age[47]. With younger ages of onset, the frequency of TTN mutations rose to a high of over 7% of individuals younger than 30 years of age. As with dilated cardiomyopathy, the association between TTN and AF had a stronger effect when the analysis was restricted to TTN exons that were highly expressed in cardiac tissue.
While these studies pointed to an increased frequency of TTN mutations in individuals with early-onset AF, the question then arose whether TTN mutations were detectable in the general population with AF. To address this, Choi and colleagues used exome sequencing data in 1,400 AF cases and more than 40,000 controls from the UK Biobank. They observed a similar strong association between loss-of-function variation in TTN and AF[48]. Further, the penetrance of AF among TTN mutation carriers was markedly higher among individuals with an increased AF polygenic risk.
Future Directions in AF Genetics
While rapid progress has been made in our understanding of the genetic basis of AF over the last decade, it is important to realize that we are currently in the midst of an explosive growth in the scale of genetic data available worldwide. In the following sections we have sought to put these emerging resources in the context of future potential studies in AF genetic research (Figure 5). While there are many other potential avenues of exploration, hopefully this will serve as a broad framework for the reader.
Figure 5: Future directions in AF genetics.
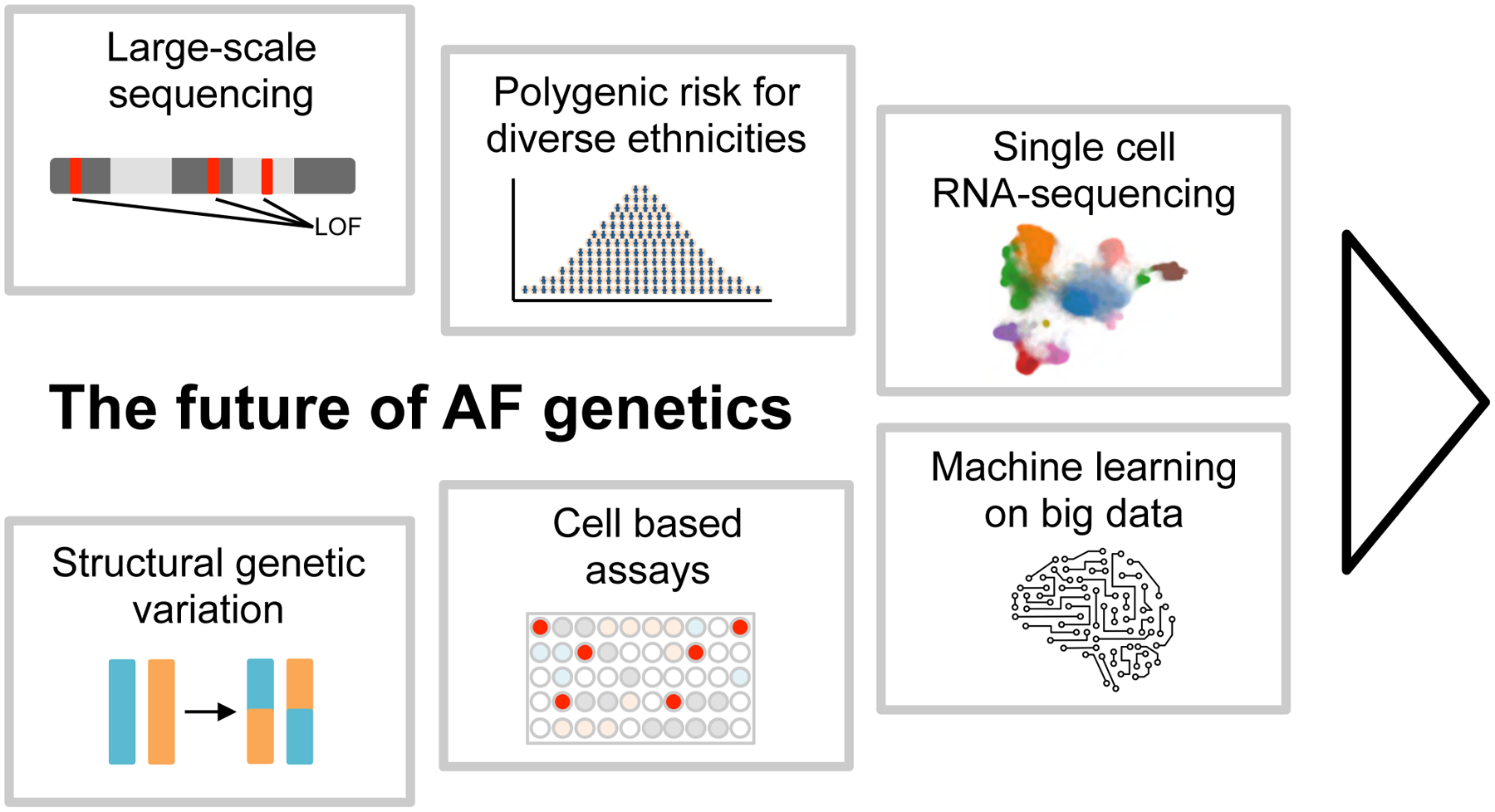
Overview of emerging technologies and analyses that could shape the field of AF genetics for the next decade. Large-scale rare coding sequence data: With dropping sequencing costs large-scale exome sequencing data sets will become available and accelerate the detection of rare and ultra rare coding variation that associate with AF. Structural genetic variation: whole-genome sequencing data allows the detection of structural variation such as inversions, translocations and large insertions and deletions. Methods to detect structural variation are improving and could lead to uncovering novel structural variant contributions to AF. Polygenic risk for diverse ethnicities: Increasing the contribution of non-European samples in AF GWAS will improve the polygenic risk prediction for diverse ethnicities. Functional cellular knockout assays: Gene knockout studies in relevant cell types, such as atrial cardiomyocytes, will enable the evaluation of AF candidate genes from GWAS loci in the context of functionally relevant readouts. Single cell RNA-sequencing: Next generation sequencing technologies such as the transcriptional profiling of individual cells from cardiac tissue will transform AF genetics and increase the resolution of gene expression profiles to a cell type specific level. Cell type specific expression quantitative trait loci could resolve the causal gene at AF GWAS loci. Machine learning on big data: Machine learning can facilitate the integration of big data sources such as gene expression profiles, proteomics data, protein-protein interaction networks, methylation data, regulatory regions and spatial organization of the DNA. Machine learning algorithms will support the goal to identify causal genes for AF, resolve regulatory mechanisms at AF GWAS loci and uncover patterns that imply disease mechanisms of AF.
Abbreviations: AF, atrial fibrillation, LOF, loss-of-function, RNA, ribonucleic acid.
Expanding genetic studies in non-Europeans
As poignantly illustrated in Figure 4, the vast majority of genetic analyses for AF have been performed in individuals of European descent. The historical tendency to focus on European populations has led to a non-representative distribution of ancestries in genetic studies compared to the real-world diversity. As we move forward it will be critical to expand our genetic resources throughout the world, not only for AF, but for all common diseases. Expanding beyond Europeans has two primary advantages. First, it is clear that there are unique lessons that can be learned about common diseases among different races and ethnicities. For example, in a large Japanese GWAS for AF, only 85% of the top hits overlapped with the results from individuals of European ancestry. Second, as we think about applying genetic risk scores to clinical care, it will be important that we don’t blindly apply a score developed in Europeans to other races and ethnicities. Such an approach may further exacerbate health care disparities[49].
Multiple programs in the United States are trying to address some of the disparity in genetic research including the NHLBI TOPMed Program (www.nhlbiwgs.org), Million Veteran Program (www.mvp.va.gov), and All of Us (www.allofus.nih.gov). The All of Us project is an ongoing longitudinal collection of over 1 million individuals and is being intentionally structured to ensure ethnic and racial diversity. The expansion of biobanks throughout the world including in China (www.ckbiobank.org), India[50] and Africa (https://h3africa.org) will also be essential to this effort.
Sequencing, sequencing, and more sequencing
An exciting development in AF genetics has been the continued expansion of the availability of exome and genome sequencing. As discussed above, the major advantage of sequencing over GWAS is that sequencing directly implicates the gene as causally related to AF. There are multiple international efforts that will coalesce to enhance our understanding of common disease genetics. These include ongoing work in the UK Biobank, deCODE, Estonia, Finland, Japanese Biobank, Million Veterans Program, the NHGRI CCDG and NHLBI TOPMed Programs among many others. As a result, within the next two years we can anticipate having datasets for AF consisting of more than 50,000 cases with either exome or genome sequencing. The power of such large datasets was nicely illustrated for autism where many additional disease-causing genes were identified with the enhanced power of these large-scale studies[51].
The utility in harnessing sequence data for AF can perhaps be exemplified best by the investigators at deCODE genetics[52]. The Icelandic population is relatively homogenous and can be traced back to a small set of common ancestors, making it one of the few bottleneck populations present in the current day world. The resulting enrichment for rare mutations can manifest in naturally occurring genetic knockouts in humans[53]. The uniqueness of the population structure combined with extensive whole genome sequencing has already led to the identification of multiple new AF genes. Similarly, in the coming years, sequencing of other bottleneck populations such as Finland[54] and Sardinia[55] will be a valuable resource for studying common diseases including AF.
Structural variation from whole-genome sequencing
Beyond gene-based tests, it is also clear that current whole-genome sequencing datasets have not been utilized to their fullest potential. Structural variants such as inversions, duplications, translocations as well as large deletions and insertions may be associated with AF. It has been estimated that structural variants can have a larger than expected impact on the genomic differences between individuals[56]. Identification of these structural modifications from the raw sequencing data is more challenging than identifying single variants, short insertions or short deletions. It requires data re-processing and the application of specialized algorithms to the sequencing data. To date, no single method can identify every type of structural variant with high confidence[57]. This suggests that a combination of methods may generate the highest yield. Structural variants can have a large impact on the function of genes and have been associated with diseases such as cancer[58], and complex diseases including Crohn’s disease, rheumatoid arthritis, diabetes[59]. We can anticipate that the systematic assessment of structural variation will identify novel genetic mechanisms for AF.
Expanding gene expression and epigenetic analyses to tissues relevant to AF
To move from associations to mechanisms for the AF GWAS loci, it will be essential to expand our repertoire of disease relevant eQTL and epigenetic datasets. Currently there are modestly sized transcriptional datasets from the human left atrium and very limited data from pulmonary venous tissues. Expanding these data from the 100–200 available samples by an order of magnitude will dramatically increase the availability of eQTLs that link AF disease variants to causative genes.
It will also be essential to move beyond the analyses of bulk tissues and to focus on the analyses of single cells for transcriptional and epigenetic profiling[60], techniques which have only rarely been applied to AF to date. Recent work by Tucker and colleagues in which they performed single nucleus RNA sequencing of the healthy human heart provides an example of the benefit from this approach. In a study of nearly 280,000 single nuclei from the four chambers of the human heart, a number of findings emerged that are relevant to AF. They were able to identify at least 10 major cell types in the heart, chamber specific transcription in non-myocyte populations, and an cardiomyocyte enrichment of the genes at AF GWAS loci[61].
A logical extension of this work will be to compare the single cell transcriptional and epigenetic profiles of left atrial and pulmonary vein samples from AF cases versus controls. These results would also enable the discovery of changes of the cellular compositions, transcription and cell type specific eQTLs in AF versus healthy individuals.
Developing large scale functional screens for AF genes
Even with additional expression and epigenetic datasets, it will be critical to expand our throughput for the functional assessment of the genes at the AF GWAS loci. The current state of our AF GWAS results can illustrate the scope of the problem. At present there are close to 140 genetic loci for AF and within these loci there are more than 1000 genes or transcripts. Of these many loci, only a minority have a single gene that can be convincingly linked by eQTL or HiC analyses. This disconnect between our expansive knowledge of disease associated variants and limited understanding of the mechanisms, is not unique to AF, but is present for essentially all common diseases.
To help address this challenge, the International Common Disease Alliance was founded in 2019. This partnership between academia, governments, pharmaceutical and technology companies is a collaborative initiative with the goal to improve diagnosis, prevention and treatment of disease through accelerating research that focuses on translating genetic findings into disease biology. While a lofty goal, the implementation of high throughput functional studies to elucidate the missing link between non-coding genetic variants, causal gene and gene function was identified as one of the key priorities by this effort (www.icda.bio).
How could such functional screens be implemented for AF? It will clearly be impossible to characterize more than a small number of mouse or even zebrafish models and as a result, we will have to turn to cell-based models as an intermediate step. While cell-based models have many potential limitations, the scale of screening is the primary strength of this approach. Combining stem cell derived cardiomyocytes and CRISPR-Cas9 technology will facilitate high throughput gene knockout studies for cellular assays. Potential cell readouts could include electrophysiological measurements of calcium signaling or action potential duration[62,63], structural assessments of sarcomere assembly[64], contractility[65] and transcription[66,67]. Given that current AF GWAS loci represent a cross section of transcription factors, ion channels and sarcomeric proteins, cell-based screening will likely require multiple readouts.
It will also be important that more consideration is given to the development of cell models that more fully recapitulate the diverse etiologies of AF. For example, there are at least 10 major cell types in the human atria and the study of myocytes will not be helpful if an AF gene is predominantly expressed in another cell type such as fibroblasts[68] and in a cell subtype arising from the conduction system or pulmonary veins.
Implementing machine learning to identify endophenotypes of AF
The rapid evolution in machine learning methods has already started to transform the medical field. The availability of multi-dimensional datasets ranging from electronic health records, imaging data, clinical measurements and genetics is providing the basis for algorithm-based clinical research that has the power to improve risk prediction, response to treatment and clinical diagnostics for AF. Particularly exciting recent work has applied machine learning to the electrocardiogram to identify subclinical markers or endophenotypes that predict the future development of AF[69,70]. In the future it will be interesting to study the genetics of these AF endophenotypes and to use machine learning to enhance the prediction of loss-of function impact of coding variants[71]. Further, it is clear that novel computational approaches will be required to integrate the existing and emerging data sources in AF genetics. Predicting the most likely causal genes at an AF GWAS locus or identify gene networks relevant for AF will require the integration of large data sources. The combination of GWAS results with gene expression profiles, proteomics data, protein-protein interaction networks, methylation data, regulatory regions and spatial organization of the DNA requires efficient computational solutions that can deal with multi-dimensional data. For a broader discussion on this topic please see the accompanying review in this Compendium (formal reference to be provided by the journal).
Conclusions
AF is a complex disease with a combination of both environmental and genetic factors that contribute to the pathogenesis of the arrhythmia. Rapid progress has been made in identifying many common variant loci in GWAS for AF, yet major challenges remain in moving from disease associations to specific mechanisms. Recent genome and exome-based sequencing studies have identified TTN as the gene most commonly associated with mutations in individuals with AF. Future studies will seek to explore the application of polygenic risk scores to clinical care, build out genetic studies in non-European populations, and further expand single cell sequencing and omic technologies in cells and tissues relevant to AF. Further refinement of the genetic basis of AF will ultimately facilitate the identification of new therapeutic targets and enable more precise risk stratification for this common arrhythmia.
Sources of funding
Dr. Ellinor is supported by the National Institutes of Health (1RO1HL092577, R01HL128914, K24HL105780), the American Heart Association (18SFRN34110082), and by the Foundation Leducq (14CVD01).
Disclosures
Dr. Ellinor is supported by a grant from Bayer AG to the Broad Institute focused on the genetics and therapeutics of cardiovascular disease. Dr. Ellinor has consulted for Bayer AG, Novartis, MyoKardia and Quest Diagnostics. Roselli is supported by a grant from Bayer AG to the Broad Institute focused on the development of therapeutics for cardiovascular disease. Dr. Rienstra has no disclosures.
Non-standard Abbreviations and Acronyms
- AF
Atrial fibrillation
- BRA
Brazilian
- eQTL
Expression quantitative trait locus
- EUR
European
- GWAS
Genome-wide association study
- HISP
Hispanic
- JAP
Japanese
- KOR
Korean
- LOF
Loss-of-function
- PRS
Polygenic risk score
- RNA
Ribonucleic acid
- SNP
Single nucleotide polymorphism
- STR
Short tandem repeat
References
- [1].Chugh SS, Havmoeller R, Narayanan K, Singh D, Rienstra M, Benjamin EJ, et al. Worldwide epidemiology of atrial fibrillation: a Global Burden of Disease 2010 Study. Circulation 2014;129:837–847. doi: 10.1161/CIRCULATIONAHA.113.005119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Huxley RR, Lopez FL, Folsom AR, Agarwal SK, Loehr LR, Soliman EZ, et al. Absolute and Attributable Risks of Atrial Fibrillation in Relation to Optimal and Borderline Risk Factors. Circulation 2011;123:1501–1508. doi: 10.1161/CIRCULATIONAHA.110.009035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Magnussen C, Niiranen TJ, Ojeda FM, Gianfagna F, Blankenberg S, Njølstad I, et al. Sex Differences and Similarities in Atrial Fibrillation Epidemiology, Risk Factors, and Mortality in Community Cohorts. Circulation 2017:1. doi: 10.1161/CIRCULATIONAHA.117.028981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ellinor PT, Yoerger DM, Ruskin JN, MacRae CA. Familial aggregation in lone atrial fibrillation. Hum Genet 2005;118:179–184. doi: 10.1007/s00439-005-0034-8. [DOI] [PubMed] [Google Scholar]
- [5].Arnar DO, Thorvaldsson S, Manolio TA, Thorgeirsson G, Kristjansson K, Hakonarson H, et al. Familial aggregation of atrial fibrillation in Iceland. Eur Heart J 2006;27:708–712. doi: 10.1093/eurheartj/ehi727. [DOI] [PubMed] [Google Scholar]
- [6].Christophersen IE, Ravn LS, Budtz-Joergensen E, Skytthe A, Haunsoe S, Svendsen JH, et al. Familial aggregation of atrial fibrillation: a study in Danish twins. Circ Arrhythmia Electrophysiol 2009;2:378–383. doi: 10.1161/CIRCEP.108.786665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Alzahrani Z, Ornelas-Loredo A, Darbar SD, Farooqui A, Mol D, Chalazan B, et al. Association Between Family History and Early-Onset Atrial Fibrillation Across Racial and Ethnic Groups. JAMA Netw Open 2018;1:e182497. doi: 10.1001/jamanetworkopen.2018.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Weng L-C, Choi SH, Klarin D, Smith JG, Loh P-R, Chaffin M, et al. Heritability of Atrial Fibrillation. Circ Cardiovasc Genet 2017;10:e001838. doi: 10.1161/CIRCGENETICS.117.001838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chen Y-H, Xu S-J, Bendahhou S, Wang X-L, Wang Y, Xu W-Y, et al. KCNQ1 Gain-of-Function Mutation in Familial Atrial Fibrillation. Science (80-) 2003;299:251–254. doi: 10.1126/science.1077771. [DOI] [PubMed] [Google Scholar]
- [10].GTEx Consortium {fname}. The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hodgson-Zingman DM, Karst ML, Zingman LV., Heublein DM, Darbar D, Herron KJ, et al. Atrial Natriuretic Peptide Frameshift Mutation in Familial Atrial Fibrillation. N Engl J Med 2008;359:158–165. doi: 10.1056/NEJMoa0706300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Postma AV, van de Meerakker JBA, Mathijssen IB, Barnett P, Christoffels VM, Ilgun A, et al. A gain-of-function TBX5 mutation is associated with atypical Holt-Oram syndrome and paroxysmal atrial fibrillation. Circ Res 2008;102:1433–1442. doi: 10.1161/CIRCRESAHA.107.168294. [DOI] [PubMed] [Google Scholar]
- [13].HOLT M, ORAM S. Familial heart disease with skeletal malformations. Br Heart J 1960;22:236–242. doi: 10.1136/hrt.22.2.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ma J-F, Yang F, Mahida SN, Zhao L, Chen X, Zhang ML, et al. TBX5 mutations contribute to early-onset atrial fibrillation in Chinese and Caucasians. Cardiovasc Res 2016;109:442–450. doi: 10.1093/cvr/cvw003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Orr N, Arnaout R, Gula LJ, Spears DA, Leong-Sit P, Li Q, et al. A mutation in the atrial-specific myosin light chain gene (MYL4) causes familial atrial fibrillation. Nat Commun 2016;7:11303. doi: 10.1038/ncomms11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kowalski MH, Qian H, Hou Z, Rosen JD, Tapia AL, Shan Y, et al. Use of >100,000 NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium whole genome sequences improves imputation quality and detection of rare variant associations in admixed African and Hispanic/Latino populations. BioRxiv 2019:683201. doi: 10.1101/683201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gudbjartsson DF, Arnar DO, Helgadottir A, Gretarsdottir S, Holm H, Sigurdsson A, et al. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature 2007;448:353–357. doi: 10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- [18].Kirchhof P, Kahr PC, Kaese S, Piccini I, Vokshi I, Scheld H-H, et al. PITX2c Is Expressed in the Adult Left Atrium, and Reducing Pitx2c Expression Promotes Atrial Fibrillation Inducibility and Complex Changes in Gene Expression. Circ Cardiovasc Genet 2011;4:123–133. doi: 10.1161/CIRCGENETICS.110.958058. [DOI] [PubMed] [Google Scholar]
- [19].Syeda F, Kirchhof P, Fabritz L. PITX2 -dependent gene regulation in atrial fibrillation and rhythm control. J Physiol 2017;595:4019–4026. doi: 10.1113/JP273123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sinner MF, Tucker NR, Lunetta KL, Ozaki K, Smith JGD, Trompet S, et al. Integrating genetic, transcriptional, and functional analyses to identify 5 novel genes for atrial fibrillation. Circulation 2014;130:1225–1235. doi: 10.1161/CIRCULATIONAHA.114.009892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ellinor PT, Lunetta KL, Albert CM, Glazer NL, Ritchie MD, Smith AV, et al. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat Genet 2012;44:670–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Christophersen IE, Rienstra M, Roselli C, Yin X, Geelhoed B, Barnard J, et al. Large-scale analyses of common and rare variants identify 12 new loci associated with atrial fibrillation. Nat Genet 2017;49:946–952. doi: 10.1038/ng.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Thorolfsdottir RB, Sveinbjornsson G, Sulem P, Nielsen JB, Jonsson S, Halldorsson GH, et al. Coding variants in RPL3L and MYZAP increase risk of atrial fibrillation. Commun Biol 2018;1:68. doi: 10.1038/s42003-018-0068-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Nielsen JB, Fritsche LG, Zhou W, Teslovich TM, Holmen OL, Gustafsson S, et al. Genome-wide Study of Atrial Fibrillation Identifies Seven Risk Loci and Highlights Biological Pathways and Regulatory Elements Involved in Cardiac Development. Am J Hum Genet 2018;102:103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gudbjartsson DF, Holm H, Gretarsdottir S, Thorleifsson G, Walters GB, Thorgeirsson G, et al. A sequence variant in ZFHX3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat Genet 2009;41:876–878. doi: 10.1038/ng.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Benjamin EJ, Rice KM, Arking DE, Pfeufer A, van Noord C, Smith AV, et al. Variants in ZFHX3 are associated with atrial fibrillation in individuals of European ancestry. Nat Genet 2009;41:879–881. doi: 10.1038/ng.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ellinor PT, Lunetta KL, Glazer NL, Pfeufer A, Alonso A, Chung MK, et al. Common variants in KCNN3 are associated with lone atrial fibrillation. Nat Genet 2010;42:240–244. doi: 10.1038/ng.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Roselli C, Chaffin MD, Weng L-C, Aeschbacher S, Ahlberg G, Albert CM, et al. Multi-ethnic genome-wide association study for atrial fibrillation. Nat Genet 2018;50:1225–1233. doi: 10.1038/s41588-018-0133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Nielsen JB, Thorolfsdottir RB, Fritsche LG, Zhou W, Skov MW, Graham SE, et al. Biobank-driven genomic discovery yields new insight into atrial fibrillation biology. Nat Genet 2018;50:1234–1239. doi: 10.1038/s41588-018-0171-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lee J-Y, Kim T-H, Yang P-S, Lim HE, Choi E-K, Shim J, et al. Korean atrial fibrillation network genome-wide association study for early-onset atrial fibrillation identifies novel susceptibility loci. Eur Heart J 2017;38:2586–2594. doi: 10.1093/eurheartj/ehx213. [DOI] [PubMed] [Google Scholar]
- [31].Low S-K, Takahashi A, Ebana Y, Ozaki K, Christophersen IE, Ellinor PT, et al. Identification of six new genetic loci associated with atrial fibrillation in the Japanese population. Nat Genet 2017;49:953–958. doi: 10.1038/ng.3842. [DOI] [PubMed] [Google Scholar]
- [32].van Ouwerkerk AF, Bosada F, Liu J, Zhang J, van Duijvenboden K, Chaffin M, et al. Identification of Functional Variant Enhancers Associated with Atrial Fibrillation. Circ Res 2020:CIRCRESAHA.119.316006. doi: 10.1161/CIRCRESAHA.119.316006. [DOI] [PubMed] [Google Scholar]
- [33].Gallagher MD, Chen-Plotkin AS. The Post-GWAS Era: From Association to Function. Am J Hum Genet 2018;102:717–730. doi: 10.1016/j.ajhg.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Broekema RV., Bakker OB, Jonkers IH. A practical view of fine-mapping and gene prioritization in the post-genome-wide association era. Open Biol 2020;10:190221. doi: 10.1098/rsob.190221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Tucker NR, Clauss S, Ellinor PT. Common variation in atrial fibrillation: navigating the path from genetic association to mechanism. Cardiovasc Res 2016;109:493–501. doi: 10.1093/cvr/cvv283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ebana Y, Sun Y, Yang X, Watanabe T, Makita S, Ozaki K, et al. Pathway analysis with genome-wide association study (GWAS) data detected the association of atrial fibrillation with the mTOR signaling pathway. Int J Cardiol Hear Vasc 2019;24:100383. doi: 10.1016/j.ijcha.2019.100383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Deshmukh A, Barnard J, Sun H, Newton D, Castel L, Pettersson G, et al. Left atrial transcriptional changes associated with atrial fibrillation susceptibility and persistence. Circ Arrhythm Electrophysiol 2015;8:32–41. doi: 10.1161/CIRCEP.114.001632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Bianchi V, Geeven G, Tucker N, Hilvering CRE, Hall AW, Roselli C, et al. Detailed Regulatory Interaction Map of the Human Heart Facilitates Gene Discovery for Cardiovascular Disease. BioRxiv 2019:705715. doi: 10.1101/705715. [DOI] [Google Scholar]
- [39].Wang B, Lunetta KL, Dupuis J, Lubitz SA, Trinquart L, Yao L, et al. Integrative Omics Approach to Identifying Genes Associated With Atrial Fibrillation. Circ Res 2020;126:350–360. doi: 10.1161/CIRCRESAHA.119.315179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].van Ouwerkerk AF, Bosada FM, van Duijvenboden K, Hill MC, Montefiori LE, Scholman KT, et al. Identification of atrial fibrillation associated genes and functional non-coding variants. Nat Commun 2019;10:4755. doi: 10.1038/s41467-019-12721-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lubitz SA, Lunetta KL, Lin H, Arking DE, Trompet S, Li G, et al. Novel genetic markers associate with atrial fibrillation risk in Europeans and Japanese. J Am Coll Cardiol 2014;63:1200–1210. doi: 10.1016/j.jacc.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Khera AV., Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet 2018;50:1219–1224. doi: 10.1038/s41588-018-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Weng L-C, Preis SR, Hulme OL, Larson MG, Choi SH, Wang B, et al. Genetic Predisposition, Clinical Risk Factor Burden, and Lifetime Risk of Atrial Fibrillation. Circulation 137. doi: 10.1161/CIRCULATIONAHA.117.031431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Thorolfsdottir RB, Sveinbjornsson G, Sulem P, Helgadottir A, Gretarsdottir S, Benonisdottir S, et al. A Missense Variant in PLEC Increases Risk of Atrial Fibrillation. J Am Coll Cardiol 2017;70:2157–2168. doi: 10.1016/j.jacc.2017.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Holm H, Gudbjartsson DF, Sulem P, Masson G, Helgadottir HT, Zanon C, et al. A rare variant in MYH6 is associated with high risk of sick sinus syndrome. Nat Genet 2011;43:316–320. doi: 10.1038/ng.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ahlberg G, Refsgaard L, Lundegaard PR, Andreasen L, Ranthe MF, Linscheid N, et al. Rare truncating variants in the sarcomeric protein titin associate with familial and early-onset atrial fibrillation. Nat Commun 2018;9:4316. doi: 10.1038/s41467-018-06618-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Choi SH, Weng L-C, Roselli C, Lin H, Haggerty CM, Shoemaker MB, et al. Association Between Titin Loss-of-Function Variants and Early-Onset Atrial Fibrillation. JAMA 2018;320:2354. doi: 10.1001/jama.2018.18179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Choi SH, Jurgens SJ, Weng L-C, Pirruccello JP, Roselli C, Chaffin M, et al. Monogenic and Polygenic Contributions to Atrial Fibrillation Risk: Results From a National Biobank. Circ Res 2020;126:200–209. doi: 10.1161/CIRCRESAHA.119.315686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Duncan L, Shen H, Gelaye B, Meijsen J, Ressler K, Feldman M, et al. Analysis of polygenic risk score usage and performance in diverse human populations. Nat Commun 2019;10:3328. doi: 10.1038/s41467-019-11112-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Chatterjee S, Majumder PP. Kalyani cohort - the first platform in Eastern India for longitudinal studies on health and disease parameters in peri-urban setting. Glob Heal Epidemiol Genomics 2017;2:e2. doi: 10.1017/gheg.2016.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Satterstrom FK, Kosmicki JA, Wang J, Breen MS, De Rubeis S, An J-Y, et al. Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell 2020;180:568–584.e23. doi: 10.1016/j.cell.2019.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Gudbjartsson DF, Helgason H, Gudjonsson SA, Zink F, Oddson A, Gylfason A, et al. Large-scale whole-genome sequencing of the Icelandic population. Nat Genet 2015;47:435–444. doi: 10.1038/ng.3247. [DOI] [PubMed] [Google Scholar]
- [53].Chheda H, Palta P, Pirinen M, McCarthy S, Walter K, Koskinen S, et al. Whole-genome view of the consequences of a population bottleneck using 2926 genome sequences from Finland and United Kingdom. Eur J Hum Genet 2017;25:477–484. doi: 10.1038/ejhg.2016.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Lim ET, Würtz P, Havulinna AS, Palta P, Tukiainen T, Rehnström K, et al. Distribution and Medical Impact of Loss-of-Function Variants in the Finnish Founder Population. PLoS Genet 2014;10:e1004494. doi: 10.1371/journal.pgen.1004494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Sidore C, Busonero F, Maschio A, Porcu E, Naitza S, Zoledziewska M, et al. Genome sequencing elucidates Sardinian genetic architecture and augments association analyses for lipid and blood inflammatory markers. 2015;47. doi: 10.1038/ng.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, et al. Detection of large-scale variation in the human genome. Nat Genet 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- [57].Kosugi S, Momozawa Y, Liu X, Terao C, Kubo M, Kamatani Y. Comprehensive evaluation of structural variation detection algorithms for whole genome sequencing. Genome Biol 2019;20:117. doi: 10.1186/s13059-019-1720-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Wellcome Trust Case Control Consortium, Craddock N, Hurles ME, Cardin N, Pearson RD, Plagnol V, et al. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature 2010;464:713–720. doi: 10.1038/nature08979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Method of the Year 2019: Single-cell multimodal omics. Nat Methods 2020;17:1–1. doi: 10.1038/s41592-019-0703-5. [DOI] [PubMed] [Google Scholar]
- [61].Tucker NR, Chaffin M, Fleming SJ, Hall AW, Parsons VA, Bedi K, et al. Transcriptional and Cellular Diversity of the Human Heart n.d doi: 10.1101/2020.01.06.896076. [DOI] [PMC free article] [PubMed]
- [62].Leyton-Mange JS, Mills RW, Macri VS, Jang MY, Butte FN, Ellinor PT, et al. Rapid cellular phenotyping of human pluripotent stem cell-derived cardiomyocytes using a genetically encoded fluorescent voltage sensor. Stem Cell Reports 2014;2:163–170. doi: 10.1016/j.stemcr.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Bedut S, Seminatore-Nole C, Lamamy V, Caignard S, Boutin JA, Nosjean O, et al. High-throughput drug profiling with voltage- and calcium-sensitive fluorescent probes in human iPSC-derived cardiomyocytes. Am J Physiol Circ Physiol 2016;311:H44–H53. doi: 10.1152/ajpheart.00793.2015. [DOI] [PubMed] [Google Scholar]
- [64].Bray M-A, Singh S, Han H, Davis CT, Borgeson B, Hartland C, et al. Cell Painting, a high-content image-based assay for morphological profiling using multiplexed fluorescent dyes. Nat Protoc 2016;11:1757–1774. doi: 10.1038/nprot.2016.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Toepfer CN, Sharma A, Cicconet M, Garfinkel AC, Mücke M, Neyazi M, et al. SarcTrack. Circ Res 2019;124:1172–1183. doi: 10.1161/CIRCRESAHA.118.314505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X, et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 2017;171:1437–1452.e17. doi: 10.1016/j.cell.2017.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Dixit A, Parnas O, Li B, Chen J, Fulco CP, Jerby-Arnon L, et al. Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell 2016;167:1853–1866.e17. doi: 10.1016/j.cell.2016.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Palano G, Jansson M, Backmark A, Martinsson S, Sabirsh A, Hultenby K, et al. A high-content, in vitro cardiac fibrosis assay for high-throughput, phenotypic identification of compounds with anti-fibrotic activity. J Mol Cell Cardiol 2020;142:105–117. doi: 10.1016/j.yjmcc.2020.04.002. [DOI] [PubMed] [Google Scholar]
- [69].Attia ZI, Kapa S, Lopez-Jimenez F, McKie PM, Ladewig DJ, Satam G, et al. Screening for cardiac contractile dysfunction using an artificial intelligence–enabled electrocardiogram. Nat Med 2019;25:70–74. doi: 10.1038/s41591-018-0240-2. [DOI] [PubMed] [Google Scholar]
- [70].Raghunath S, Pfeifer JM, Cerna AEU, Nemani A, Carbonati T, Jing L, et al. Deep Neural Networks can Predict Incident Atrial Fibrillation from the 12-lead Electrocardiogram and may help Prevent Associated Strokes. MedRxiv 2020:2020.04.23.20067967. doi: 10.1101/2020.04.23.20067967. [DOI] [Google Scholar]
- [71].Pagel KA, Pejaver V, Lin GN, Nam H-J, Mort M, Cooper DN, et al. When loss-of-function is loss of function: assessing mutational signatures and impact of loss-of-function genetic variants. Bioinformatics 2017;33:i389–i398. doi: 10.1093/bioinformatics/btx272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Chatterjee NA, Giulianini F, Geelhoed B, Lunetta KL, Misialek JR, Niemeijer MN, et al. Genetic Obesity and the Risk of Atrial Fibrillation. Circulation 2017;135:741–754. doi: 10.1161/CIRCULATIONAHA.116.024921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Ellervik C, Roselli C, Christophersen IE, Alonso A, Pietzner M, Sitlani CM, et al. Assessment of the Relationship Between Genetic Determinants of Thyroid Function and Atrial Fibrillation. JAMA Cardiol 2019;4:144. doi: 10.1001/jamacardio.2018.4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Liao L, Zhang S, Li W, Liu Y, Li J, Zhuang X, et al. Serum albumin and atrial fibrillation: insights from epidemiological and mendelian randomization studies. Eur J Epidemiol 2019. doi: 10.1007/s10654-019-00583-6. [DOI] [PubMed] [Google Scholar]
- [75].Larsson SC, Bäck M, Rees JMB, Mason AM, Burgess S. Body mass index and body composition in relation to 14 cardiovascular conditions in UK Biobank: a Mendelian randomization study. Eur Heart J 2020;41:221–226. doi: 10.1093/eurheartj/ehz388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Tikkanen E, Gustafsson S, Knowles JW, Perez M, Burgess S, Ingelsson E. Body composition and atrial fibrillation: a Mendelian randomization study. Eur Heart J 2019;40:1277–1282. doi: 10.1093/eurheartj/ehz003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Zhao G, Zhou J, Gao J, Liu Y, Gu S, Zhang X, et al. Genome-wide DNA methylation analysis in permanent atrial fibrillation. Mol Med Rep 2017;16:5505–5514. doi: 10.3892/mmr.2017.7221. [DOI] [PubMed] [Google Scholar]
- [78].Lin H, Yin X, Xie Z, Lunetta KL, Lubitz SA, Larson MG, et al. Methylome-wide Association Study of Atrial Fibrillation in Framingham Heart Study. Sci Rep 2017;7:40377. doi: 10.1038/srep40377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Marcus GM, Alonso A, Peralta CA, Lettre G, Vittinghoff E, Lubitz SA, et al. European Ancestry as a Risk Factor for Atrial Fibrillation in African Americans. Circulation 2010;122:2009. doi: 10.1161/CIRCULATIONAHA.110.958306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Zhao L, Zhang G, Wen Z, Huang C, Wu H, Xu J, et al. Common variants predict recurrence after nonfamilial atrial fibrillation ablation in Chinese Han population. Int J Cardiol 2017;227:360–366. doi: 10.1016/j.ijcard.2016.11.057. [DOI] [PubMed] [Google Scholar]
- [81].Lin H, Mueller-Nurasyid M, Smith AV., Arking DE, Barnard, Bartz TM, et al. Gene-gene Interaction Analyses for Atrial Fibrillation. Sci Rep 2016;6:35371. doi: 10.1038/srep35371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Weng L-C, Lunetta KL, Müller-Nurasyid M, Smith AV, Thériault S, Weeke PE, et al. Genetic Interactions with Age, Sex, Body Mass Index, and Hypertension in Relation to Atrial Fibrillation: The AFGen Consortium. Sci Rep 2017;7:11303. doi: 10.1038/s41598-017-09396-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Zhao D, Bartz TM, Sotoodehnia N, Post WS, Heckbert SR, Alonso A, et al. Mitochondrial DNA Copy Number and Incident Atrial Fibrillation. BioRxiv 2019:848085. doi: 10.1101/848085. [DOI] [PMC free article] [PubMed] [Google Scholar]


