Abstract
Objective
To investigate the anti-proliferative and pro-apoptotic effects of curcumin on diffuse large B-cell lymphoma (DLBCL) cells and explore the mechanism.
Methods
OCI-LY7 cells were treated with curcumin (2.5, 5, 10, 20, and 40 μM) for 24, 48, or 72 hours. Cell viability and apoptosis were determined using the 3-(4, 5-dimethylthiazol-2-yl)-2, 5 diphenyl tetrazolium bromide assay and TdT-mediated dUTP nick-end labeling staining, respectively. MiR-28-5p expression was detected via qRT-PCR. The binding site of miR-28-5p was predicted using online databases and verified using the dual-luciferase reporter assay. MiR-28-5p overexpression and inhibition were achieved via transfection with an miR-28-5p mimic and inhibitor, respectively.
Results
Curcumin decreased the viability of OCI-LY7 cells in a concentration- and time-dependent manner, and these effects were attenuated by miR-28-5p inhibition. MiR-28-5p expression was upregulated by curcumin. Curcumin increased the numbers of apoptotic cells and upregulated cleaved caspase-3 expression, and these effects were attenuated by miR-28-5p inhibition. The dual-luciferase reporter assay confirmed that miR-28-5p directly targets the 3′-untranslated region of BECN1. Curcumin downregulated BECN1 and microtubule-associated protein 1 light chain 3 beta-II/I expression and upregulated p62 expression.
Conclusions
Our results described the curcumin exerted anti-proliferative and pro-apoptotic effects on OCI-LY7 cells through a mechanism potentially involving miR-28-5p.
Keywords: Diffuse large B-cell lymphoma, curcumin, anti-proliferation, apoptosis, miR-28-5p, BECN1, autophagy
Introduction
Currently, lymphoma is the seventh most common cancer, affecting more than a million people globally. There are more than 100 types of lymphoma,1 approximately 70% to 90% of which are B-cell derived.2 Diffuse large B-cell lymphoma (DLBCL) is one of the most common types of B-cell lymphoma, and it is associated with a high mortality rate.3 Although tremendous progress has been made in the treatment of this malignancy in recent decades, the average 5-year survival rate is less than 50%.4 Thus, the development of new therapeutic drugs is urgently needed.
Curcumin, a major bioactive component of turmeric, has been demonstrated to possess a wide range of effects against several malignancies, including gastric cancer,5 colorectal cancer,6 breast cancer,7 thyroid cancers,8 and leukemia9. Previous studies illustrated that curcumin inhibited the proliferation of non-Hodgkin’s lymphoma (NHL) and Hodgkin’s lymphoma (HL) cells.10–12 Moreover, Maribel et al.13 reported that curcumin can repress the growth of germinal center B-cell (GCB)-type DLBCL. DLBCL can be divided into two subtypes: activated B-cell–like and GCB-like.14 The mechanism by which curcumin inhibits the growth of GCB-type DLBCL remains unclear.
MicroRNAs (miRNAs) are small, single-stranded, non-coding RNAs of approximately 20 to 22 nucleotides in length. MiRNAs regulate physiological processes by binding recognition sites in the 3′-untranslated region (UTR) of target genes, thereby inhibiting translation. In previous research on miRNA expression profiles during human B-cell lymphomagenesis, MiR-28 was identified as an intragenic miRNA located on chromosome 3q28, which was specifically induced by the germinal center (GC) reaction.15 Evidence has revealed the potential involvement of miR-28 in the development of B-cell lymphoma.16 Schneider et al.17 found that miR-28 acts as a tumor suppressor and that it was downregulated in B-cell lymphomas. MiR-28 overexpression leads to the inhibition of lymphoma cell proliferation.18
In the present study, we hypothesized that curcumin exerts anti-proliferative effects on DLBCL cells by upregulating miR-28 expression. We investigated the effects of curcumin on miR-28 levels and clarified the underlying mechanism.
Materials and methods
Ethical approval
The present study was approved by the Experimental Ethics Committee of The Fourth Affiliated Hospital of Hebei Medical University (No. 2019021). This study was conducted according to the ethical guidelines of the Declaration of Helsinki. All studies were performed in cell levels, thus, informed consent was not required.
Cell culture and treatment
The human GCB-type DLBCL cell line OCI-LY7 and human embryonic kidney cell line HEK-293T were purchased from American Type Culture Collection (Manassas, VA, USA). OCI-LY7 cells were cultured in Iscove’s modified Dulbecco’s medium supplemented with 20% fetal bovine serum (FBS) (HyClone, Thermo Fisher Scientific, Waltham, MA, USA). HEK293T were cultured in Eagle’s minimum essential medium containing 10% FBS. All cell lines were supplemented with 100 IU/mL penicillin/streptomycin (HyClone) and incubated at 37°C and 5% CO2. Curcumin (98% purity) was purchased from Sigma-Aldrich (St. Louis, MO, USA). Curcumin was dissolved in 0.1% DMSO and diluted to final concentrations of 2.5, 5, 10, 20, and 40 μM.
Cell viability assay
Cell viability was assessed using 3-(4, 5-dimethylthiazol-2-yl)-2, 5 diphenyl tetrazolium bromide (MTT) reagent. First, single-cell suspensions were collected after digestion with 0.5% trypsin. Cells at a density of 1 × 104/well were seeded in 96-well plates for 48 hours at 37°C. Curcumin was added at a final concentration of 2.5, 5, 10, 20, or 40 μM and incubated with the cells for 24, 48, or 72 hours. Then, 10 μL of MTT stock solution were added to each well, and the plate was incubated for 4 hours at 37°C to observe crystal formation. Finally, the crystals were dissolved by adding 100 μL of DMSO for 5 minutes. The absorbance at 570 nm was measured using a microplate reader (Bio-Tek, Winooski, VT, USA). Each well was duplicated three times.
TdT-mediated dUTP nick-end labeling (TUNEL) staining
OCI-LY7 cells were seeded into a six-well plate (1 × 105 cells/well) and cultured for 24 hours. Then, the cells were treated with curcumin (40 μM) or medium for 24 hours. After treatment, cells were washed with cold PBS and fixed with 4% paraformaldehyde at room temperature for 1 hour. DNA fragmentation was examined using an in situ cell death detection kit according to the manufacturer’s instructions (Roche, Basel, Switzerland). The stained cells were imaged via fluorescence microscopy (90i, Nikon, Tokyo, Japan). The number of TUNEL-positive cells was randomly counted.
Cell transfection
OCI-LY7 cells were cultured at 37°C with 5% CO2. In total, 2 × 106 cells were seeded per well in six-well plates for 12 hours. An miR-28-5p mimic or inhibitor was transfected into cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). A random sequence of oligonucleotides was used as a negative control (NC). The cells were collected 24 hours after transfection for the following experiments. The experiments were repeated three times. qRT-PCR was performed to verify the efficiency of transfection.
Dual-luciferase reporter assay
The luciferase reporter recombining the wild-type (wt) or mutant (mut) BECN1 3′-UTR containing the potential binding sites of miR-28-5p was purchased from Promega (Madison, WI, USA). The psiCHECK2 vector (Promega) containing the wt or mut BECN1 3′-UTR and internal control vector were co-transfected with the miR-28-5p mimic or NC into HEK293T cells using Lipofectamine 2000 reagent. After 48 hours, luciferase activity was measured using a dual-luciferase reporter assay kit according to the manufacturer’s protocol (Promega).
RNA isolation and qRT-PCR
Total RNA from the cultured cells was extracted using TRIzol reagent according to the manufacturer’s instructions (Invitrogen). cDNA was synthesized using an M-MLV reverse transcription kit (Invitrogen). MiRNA was extracted using an RNeasy Mini Kit (Tiangen, Beijing, China) and reversed-transcribed into cDNA using a poly (A) kit (Tiangen). qPCR analysis was performed using SYBR Green PCR mix (Tiangen) and the ABI Q6 system (Applied Biosystems, Thermo Fisher Scientific). All reactions were performed in triplicate. The gene-specific primers sequences are as follows: miR-28-5p sense, 5′-AACACGCAAGGAGCTCACAG-3′; U6 sense, 5′-AACAAGCCCTGC GCAAGGATGA-3′; BECN1 sense, 5′-GGAAGTTTTCCGG CGGCT-3′; BECN1 antisense, 5′-AGACCCTTCCATCCCTCAGC-3′; β-actin sense, 5′-GTCATTCCAAATATGA GATGCGT-3′; and β-actin antisense, 5′-GCTATCACCTCCCCTGTGTG-3′. The primers were synthesized by Invitrogen (Shanghai, China). β-actin and U6 were used as internal control genes for the relative quantities of mRNA and miRNA, respectively. The 2−△△Ct method was used to analyze the qPCR results.19
Western blotting
Cells were collected and lysed in RIPA lysis buffer containing 50 mM Tris-HCl (pH 7.4), 1 mM EDTA, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, and protease inhibitor cocktail (cOmplete™ ULTRA, Roche). The lysate was centrifuged at 14,000 × g for 20 minutes at 4°C, after which the supernatant was collected. The protein concentration was assayed using a BCA kit. Equal amounts of protein (10 μg) were separated via SDS-PAGE and transferred to PVDF membranes. After blocking with 5% non-fat milk for 2 hours, the membrane was incubated with primary monoclonal antibodies against BECN1 (1:1000, Cell Signaling Technology, Danvers, MA, USA), microtubule-associated protein 1 light chain 3 beta (LC3B, 1:4000, Sigma-Aldrich), p62 (1:1000, Cell Signaling Technology), caspase-3 (1:1000, Cell Signaling Technology), and cleaved caspase-3 (1:1000, Cell Signaling Technology) overnight at 4°C. After three washes with Tris-buffered saline with Tween 20 (TBST), the membranes were incubated with an HRP-labeled secondary antibody (1:30,000, Cell Signaling Technology). After three washes with TBST, enhanced chemiluminescence reagent (Merck, Darmstadt, Germany) was added, and the immunoreactive bands were captured using X-ray film (Kodak, Rochester, NY, USA). The relative protein expression was normalized using GAPDH as an internal reference.
Statistical analysis
All data are presented as the mean ± SEM. SPSS 19.0 software (IBM, Armonk, NY, USA) was used for statistical analysis. Student’s t-test was used to compare differences between the groups. P < 0.05 denoted statistical significance.
Results
Curcumin decreased the proliferative activity of OCI-LY7 cells
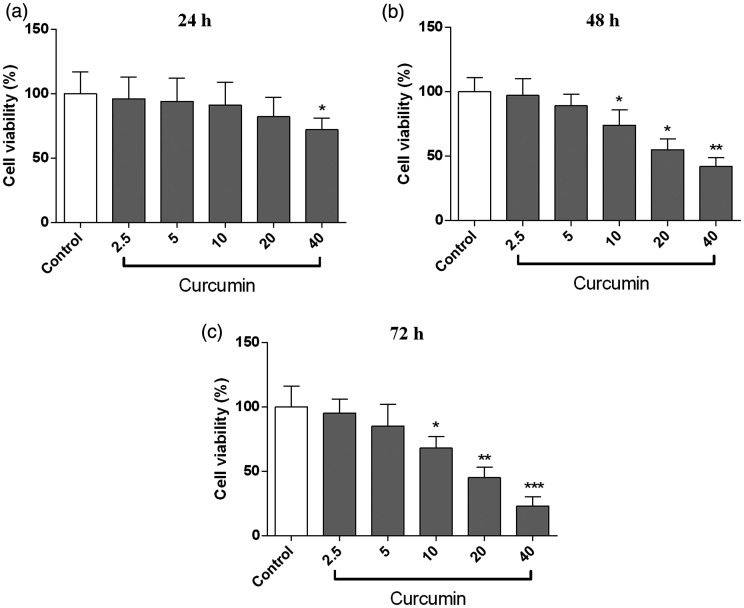
To investigate the effect of curcumin on proliferative activity, OCI-LY7 cells were treated with curcumin (2.5, 5, 10, 20, and 40 μM). As shown in Figure 1, cell viability was significantly decreased in the curcumin group (40 μM) at 24 hours compared with the findings in the control group (P < 0.05). In addition, curcumin (10–40 μM) significantly decreased cell viability at 48 and 72 hours (both P < 0.05).
Figure 1.
Curcumin reduced the proliferative activity of OCI-LY7 cells. The cells were incubated with curcumin (2.5, 5, 10, 20, and 40 μM) for 24 (a), 48 (b), and 72 hours (c). Cell viability was detected using the 3-(4, 5-dimethylthiazol-2-yl)-2, 5 diphenyl tetrazolium bromide assay. N = 6. Compared with the control group, *P < 0.05, **P < 0.01, ***P < 0.001.
Curcumin upregulated miR-28-5p expression in OCI-LY7 cells
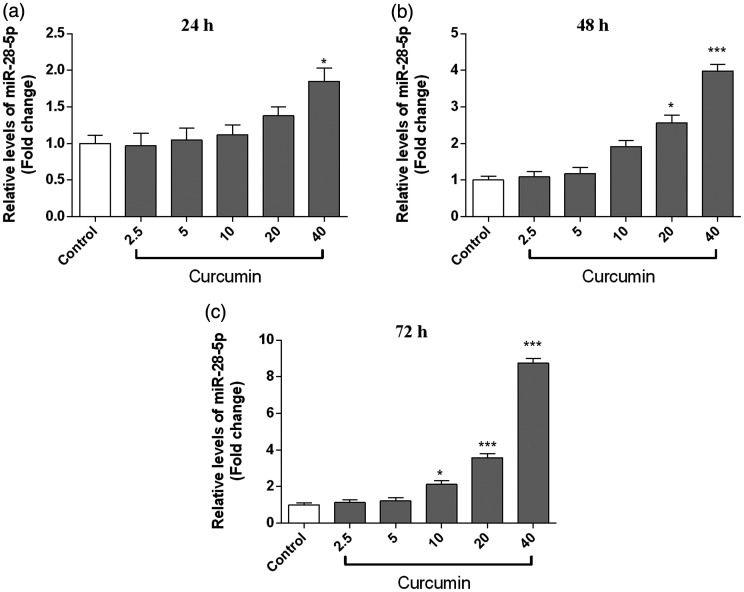
The effect of curcumin on miR-28-5p levels was detected by qRT-PCR. As shown in Figure 2, miR-28-5p expression was significantly upregulated in the curcumin group at 24 (40 μM, P < 0.05), 48 (20 μM: P < 0.05; 40 μM: P < 0.001), and 72 hours (10 μM: P < 0.05; 20 μM: P < 0.001; 40 μM: P < 0.001) compared with its levels in the control group.
Figure 2.
Curcumin upregulated miR-28-5p expression in OCI-LY7 cells. The cells were incubated with curcumin (2.5, 5, 10, 20, and 40 μM) for 24 (a), 48 (b), and 72 hours (c). MiR-28-5p levels were detected via qRT-PCR. n = 6. Compared with the control group, *P < 0.05, **P < 0.01, ***P < 0.001.
MiR-28-5p inhibition attenuated the anti-proliferative activity of curcumin in OCI-LY7 cells
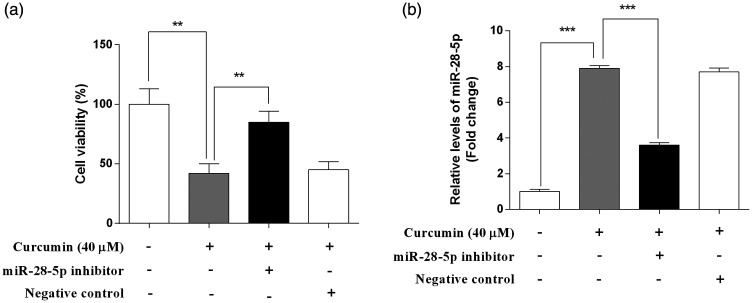
To evaluate whether the anti-proliferative ability of curcumin is related to the upregulation of miR-28-5p, the effect of curcumin on cell viability was detected in the presence or absence of an miR-28-5p inhibitor (Figure 3a). Compared with the findings for curcumin alone, cell viability was significantly increased in the presence of curcumin and the miR-28-5p inhibitor (P < 0.01). The NC failed to influence cell viability. In addition, the efficiency of transfection was verified by the decreased miR-28-5p levels following treatment with an miR-28-5p inhibitor (P < 0.001; Figure 3b).
Figure 3.
The anti-proliferative activity of curcumin was attenuated by an miR-28-5p inhibitor in OCI-LY7 cells. OCI-LY7 cells were transfected with an miR-28-5p inhibitor or negative control (NC). The cells were incubated with or without curcumin (40 μM) for 48 hours. (a) Cell viability was detected using the 3-(4, 5-dimethylthiazol-2-yl)-2, 5 diphenyl tetrazolium bromide assay. (b) The transfection efficiency was assessed by qRT-PCR. N = 6. *P < 0.05, **P < 0.01, ***P < 0.001.
MiR-28-5p inhibition attenuated the pro-apoptotic effects of curcumin in OCI-LY7 cells
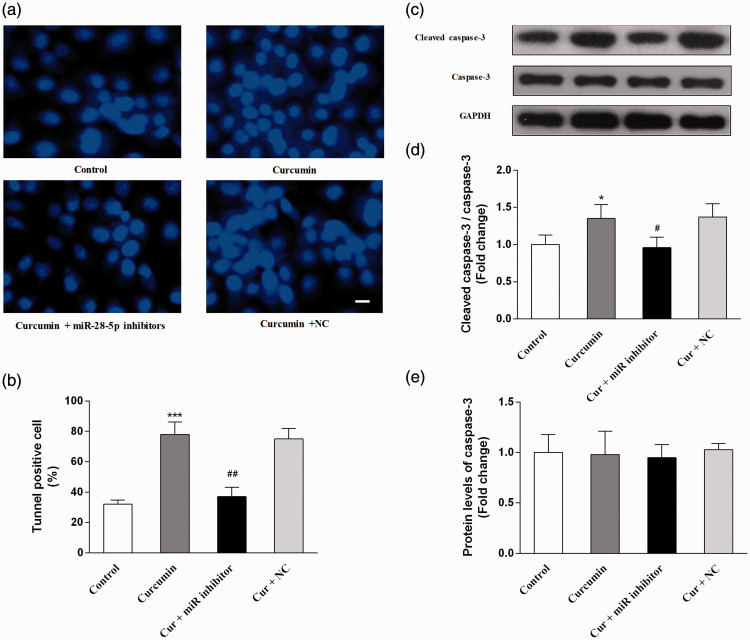
To investigate the effects of curcumin on cell apoptosis, TUNEL staining and the protein levels of caspase-3 were examined. Compared with the findings in the control group, curcumin significantly increased the percentage of apoptotic cells (P < 0.001) and the protein expression of cleaved caspase-3 (P < 0.05). Compared with the effects of curcumin alone, cell apoptosis was significantly attenuated by miR-28-5p inhibition (P < 0.05), whereas the NC did not induce significant changes (Figure 4).
Figure 4.
The pro-apoptotic effects of curcumin were attenuated using an miR-28-5p inhibitor in OCI-LY7 cells. OCI-LY7 cells were transfected with an miR-28-5p inhibitor or negative control (NC). The cells were incubated with or without curcumin (40 μM) for 48 hours. (a) TdT-mediated dUTP nick-end labeling (TUNEL) staining. (b) The percentage of TUNEL-positive cells. (c) The immunoblot bands of caspase-3 and cleaved caspase-3. (d) The relative protein levels of cleaved caspase-3. (e) The relative protein levels of caspase-3. N = 6. Compared with the control group, *P < 0.05, **P < 0.01, ***P < 0.001. Compared with the curcumin group, #P < 0.05, ##P < 0.01. Scale bar equal to 25 μm.
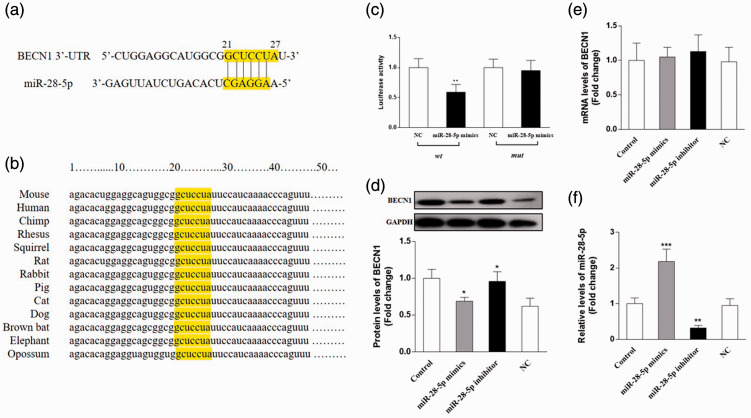
Identification of BECN1 as a direct target of miR-28-5p
To explore the potential target gene of miR-28, we predicted the binding sites of miR-28-5p using TargetScan, miRanda, and miRDB. We identified a putative miR-28-5p binding site in the 3′-UTR of the BECN1 gene at a site between nucleotides 21 and 26, and this region is highly conserved in various species (Figure 5a and 5b). The dual-luciferase reporter assay revealed that the miR-28-5p mimic significantly reduced luciferase activity in the wt BECN1 group compared with the findings in the NC group (P < 0.01); however, the miR-28-5p mimic did not affect luciferase activity in the mut BECN1 group (Figure 5c).
Figure 5.
BECN1 was a direct target of miR-28-5p. (a) Complementarity between miR-28-5p and the 3′-untranslated region (UTR) of BECN1. (b) The conserved binding sites in 3′-UTR of the BECN1 gene in various species. (c) Dual-luciferase reporter assay of the interaction between miR-28-5p and BECN1. (d) Protein expression of BECN1 in OCI-LY7 cells following transfection with an miR-28-5p mimic, miR-28-5p inhibitor, or negative control (NC). (e) The transfection efficiency was assessed by qRT-PCR. (f) The mRNA levels of BECN1 in OCI-LY7 cells. N = 6. Compared with the control group, *P < 0.05, **P < 0.01, ***P < 0.001. Cir, curcumin.
To further validate that BECN1 is the direct target of miR-28-5p, an miR-28-5p mimic, miR-28-5p inhibitor, or NC were transfected into OCI-LY7 cells. The mRNA and protein levels of BECN1 were detected by qRT-PCR and western blotting, respectively. As shown in Figure 5d and 5f, the miR-28-5p mimic significantly decreased the protein levels of BECN1 (P < 0.05), whereas the miR-28-5p inhibitor upregulated its expression (P < 0.05). Neither the miR-28-5p mimic nor inhibitor affected the mRNA levels of BECN1. The efficiency of transfection is displayed in Figure 5e.
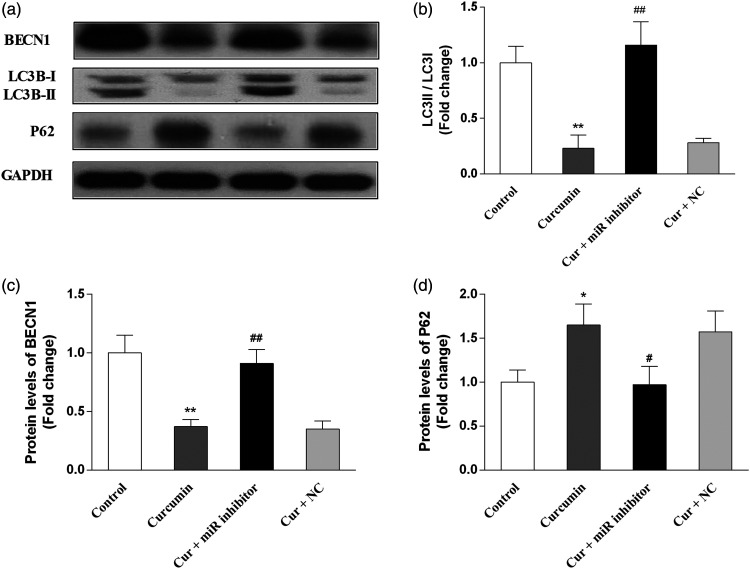
Curcumin downregulated BECN1 expression and inhibited autophagy in OCI-LY7 cells
Concerning the effects of curcumin on autophagy-related protein levels, the results demonstrated that curcumin significantly decreased the protein levels of BECN1 (P < 0.01) and the LC3B-II/LC3B-I ratio (P < 0.01) as well as increased P62 expression (P < 0.05) in OCI-LY7 cells. Moreover, the miR-28-5p inhibitor significantly prevented these curcumin-induced changes (P < 0.05), whereas the NC had no effects. The results are presented in Figure 6.
Figure 6.
Curcumin downregulated the protein expression of BECN1 and inhibited autophagy in OCI-LY7 cells. OCI-LY7 cells were transfected with an miR-28-5p inhibitor or negative control (NC). Then, the protein expression of BECN1, microtubule-associated protein 1 light chain 3 beta (LC3B), and p62 were examined via western blotting. (a) Immunoblot bands. (b) The relative protein levels of BECN1. (c) The relative protein levels of LC3B-II/LC3B-I. (d) The relative protein levels of p62. N = 6. Compared with the control group, *P < 0.05, **P < 0.01, ***P < 0.001. Compared with the curcumin group, #P < 0.05, ##P < 0.01, ###P < 0.001. Cir, curcumin.
Discussion
Previous studies found that curcumin exerts anti-proliferative effects on B-lymphoma cells including the Raji B-NHL cell line and HL cells. In the present study, we investigated the anti-proliferative effects of curcumin on OCI-LY7 cells, a GCB-type DLBCL cell line. Our data indicated that curcumin could inhibit the proliferation of OCI-LY7 cells in a concentration- and time-dependent manner. The present results suggested that curcumin may have beneficial effects in the treatment of GCB-type DLBCL.
Numerous studies have focused on the regulatory function of miRNA in the pathogenesis and progression of B-cell lymphomas.20,21 The abnormality of miRNA expression was highly correlated with the development of B-cell lymphoma. Many miRNAs have been identified as biomarkers of B-cell lymphoma.22,23 Schneider et al.17 revealed that miR-28 expression is downregulated in B-cell lymphomas, suggesting that silencing of miR-28 will contribute to lymphomagenesis. Another study confirmed that miR-28 is a key regulator of the GC reaction and that it could inhibit B cell proliferation, suggesting that miR-28 has anti-tumor activity in Burkitt’s lymphoma and DLBCL.16 To investigate whether the anti-proliferative effects of curcumin were related to the upregulation of miR-28, we detected miR-28-5p levels via qRT-PCR. Our results illustrated that curcumin significantly upregulated miR-28-5p expression in OCI-LY7 cells. Additionally, we found that miR-28-5p inhibitors attenuated the anti-proliferative activity of curcumin, further demonstrating that this anti-proliferative activity was related to the upregulation of miR-28-5p. To investigate whether curcumin affects apoptosis in OCI-LY7 cells, TUNEL staining was performed, and the results revealed that curcumin treatment significantly increased the number of apoptotic cells. Moreover, curcumin also upregulated the protein expression of cleaved caspase-3, which more clearly confirmed the pro-apoptotic effects of curcumin on OCI-LY7 cells. However, these effects were blocked by an miR-28-5p inhibitor, but not by the NC, further suggesting that the anti-proliferative and pro-apoptotic effects of curcumin were partly mediated by miR-28-5p upregulation.
We next predicted the target genes of miR-28-5p using online databases and identified BECN1 as a potential target gene of miR-28-5p. We confirmed the direct binding of miR-28-5p to the 3′-UTR of BECN1 using the dual-luciferase reporter assay. To further verify the interaction between miR-28-5p and BECN1, an miR-28-5p mimic or inhibitor was transfected into OCI-LY7 cells, and subsequently, mRNA and protein levels were examined. The protein levels of BECN1 were significantly decreased in the miR-28-5p mimic group and increased in the miR-28-5p inhibitor group. However, mRNA expression did not differ between the groups. The present results demonstrated that the protein levels of BECN1 were negatively correlated with miR-28-5p, which is consistent with the common mechanism of animal miRNA function.24
BECN1 is an important promoter of autophagy. Autophagy, also known as macroautophagy, is an intracellular degradation pathway activated in response to stress stimuli, such as hunger and hypoxia.25,26 Cells degrade damaged or unnecessary organelles suing lysosomes to enhance their survival.27 Although the role of autophagy in tumor suppression has been widely studied,28,29 many researchers proposed that autophagy was beneficial for the survival of cancer cells.30,31 The development of malignant cells is challenged by the boosted demand for nutrients. In such circumstances, several key mechanisms are activated, such as hypoxia-induced factor 1α and autophagy. The activation of autophagy in tumor cells promotes survival, especially by increasing resistance to nutrient changes, low oxygen availability, and chemotherapy-induced toxicity.32,33 Chloroquine, an autophagy inhibitor, impaired spontaneous lymphomagenesis in a Burkitt’s lymphoma model and provoked the clonogenic death of myc-induced lymphoma cells.34 Therefore, autophagy is more likely a double-edged sword, especially in lymphomas.35 Our present results uncovered that BECN1 was a direct target of miR-28-5p, which suggested the participation of miR-28-5p in the regulation of autophagy. To investigate whether the anti-proliferative effects of curcumin were related to the inhibition of autophagy, we detected the protein levels of BECN1, LC3B, and p62. LC3B is the most important biomarker of the autophagic process. When autophagy is initiated, cytoplasmic LC3B-I is converted into its phosphatidylethanolamine-conjugated form LC3B-II.36 LC3B-II participates in the process of autophagosome membrane expansion and fusion.37 An elevated LC3B-II/LC3B-I ratio reflects the increased activity of autophagy. However, these effects may have also resulted from the decreased degradation of LC3B-II in autolysosomes. Therefore, to exclude this possibility, p62 expression should be measured. p62 is an intracellular longevity protein that is mainly degraded in autolysosomes via an interaction with LC3B-II.38 The increased levels of p62 reflect the inhibition of degradation in autolysosomes. The present results demonstrated that BECN1 and LC3B-II/I protein expression was downregulated by curcumin, whereas p62 expression was upregulated. These results confirmed that curcumin inhibited autophagy and reduced the degradation of p62 in autolysosomes. Additionally, these effects of curcumin were attenuated by an miR-28-5p inhibitor, which further suggested that the inhibition of autophagy by curcumin is related to the upregulation of miR-28-5p.
Conclusion
Taken together, the present study demonstrated that curcumin reduced the proliferation of OCI-LY7 cells by upregulating the expression of miR-28-5p. MiR-28-5p overexpression resulted in the inhibition of autophagy through direct effects on BECN1. Our findings suggested that miR-28-5p is a potential target through which curcumin inhibits the proliferation of GCB-type DLBCL cells. However, further experiments are necessary to verify these findings in vivo.
Funding
The present study was funded by the Medical Science Research Program of Hebei Province (20190504) And the Key Research and Development Project of the Hebei Provincial Science and Technology Department (203777104D).
Declaration of conflicting interest
The authors declare that there is no conflict of interest.
ORCID iD
Tian Kang https://orcid.org/0000-0003-4766-8545
References
- 1.Abd El-Haleim EA, Bahgat AK, Saleh S. Resveratrol and fenofibrate ameliorate fructose-induced nonalcoholic steatohepatitis by modulation of genes expression. World J Gastroenterol 2016; 22: 2931–2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander DD, Mink PJ, Adami HO, et al. The non-Hodgkin lymphomas: a review of the epidemiologic literature. Int J Cancer 2007; 120: 1–39. [DOI] [PubMed] [Google Scholar]
- 3.Yatomi Y. From FAB classification to WHO classification of tumors of hematopoietic and lymphoid tissue. Rinsho Byori 2012; 60: 550–552. [PubMed] [Google Scholar]
- 4.Gerrard M, Waxman IM, Sposto R, et al. Outcome and pathologic classification of children and adolescents with mediastinal large B-cell lymphoma treated with FAB/LMB96 mature B-NHL therapy. Blood 2013; 121: 278–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jakubek M, Kejik Z, Kaplanek R, et al. Strategy for improved therapeutic efficiency of curcumin in the treatment of gastric cancer. Biomed Pharmacother 2019; 118: 109278. [DOI] [PubMed] [Google Scholar]
- 6.Ismail NI, Othman I, Abas F, et al. Mechanism of Apoptosis Induced by Curcumin in Colorectal Cancer. Int J Mol Sci 2019; 20: 2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Yu J, Cui R, et al. Curcumin in Treating Breast Cancer: a Review. J Lab Autom 2016; 21: 723–731. [DOI] [PubMed] [Google Scholar]
- 8.Shin HJ, Hwang KA, Choi KC. Antitumor Effect of Various Phytochemicals on Diverse Types of Thyroid Cancers. Nutrients 2019; 11: 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kouhpeikar H, Butler AE, Bamian F, et al. Curcumin as a therapeutic agent in leukemia. J Cell Physiol 2019; 234: 12404–12414. [DOI] [PubMed] [Google Scholar]
- 10.Liu HL, Chen Y, Cui GH, et al. Curcumin, a potent anti-tumor reagent, is a novel histone deacetylase inhibitor regulating B-NHL cell line Raji proliferation. Acta Pharmacol Sin 2005; 26: 603–609. [DOI] [PubMed] [Google Scholar]
- 11.Li ZX, Ouyang KQ, Jiang X, et al. Curcumin induces apoptosis and inhibits growth of human Burkitt's lymphoma in xenograft mouse model. Mol Cells 2009; 27: 283–289. [DOI] [PubMed] [Google Scholar]
- 12.Guorgui J, Wang R, Mattheolabakis G, et al. Curcumin formulated in solid lipid nanoparticles has enhanced efficacy in Hodgkin's lymphoma in mice. Arch Biochem Biophys 2018; 648: 12–19. [DOI] [PubMed] [Google Scholar]
- 13.Cotto M, Cabanillas F, Tirado M, et al. Epigenetic therapy of lymphoma using histone deacetylase inhibitors. Clin Transl Oncol 2010; 12: 401–409. [DOI] [PubMed] [Google Scholar]
- 14.Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000; 403: 503–511. [DOI] [PubMed] [Google Scholar]
- 15.Basso K, Sumazin P, Morozov P, et al. Identification of the human mature B cell miRNome. Immunity 2009; 30: 744–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartolome-Izquierdo N, De Yebenes VG, Alvarez-Prado AF, et al. miR-28 regulates the germinal center reaction and blocks tumor growth in preclinical models of non-Hodgkin lymphoma. Blood 2017; 129: 2408–2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneider C, Setty M, Holmes AB, et al. MicroRNA 28 controls cell proliferation and is down-regulated in B-cell lymphomas. Proc Natl Acad Sci U S A 2014; 111: 8185–8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Almeida MI, Nicoloso MS, Zeng L, et al. Strand-specific miR-28-5p and miR-28-3p have distinct effects in colorectal cancer cells. Gastroenterology 2012; 142: 886–896.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 20.Zheng B, Xi Z, Liu R, et al. The Function of MicroRNAs in B-Cell Development, Lymphoma, and Their Potential in Clinical Practice. Front Immunol 2018; 9: 936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larrabeiti-Etxebarria A, Lopez-Santillan M, Santos-Zorrozua B, et al. Systematic Review of the Potential of MicroRNAs in Diffuse Large B Cell Lymphoma. Cancers (Basel) 2019; 11: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopez-Santillan M, Larrabeiti-Etxebarria A, Arzuaga-Mendez J, et al. Circulating miRNAs as biomarkers in diffuse large B-cell lymphoma: a systematic review. Oncotarget 2018; 9: 22850–22861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sole C, Arnaiz E, Lawrie CH. MicroRNAs as Biomarkers of B-cell Lymphoma. Biomark Insights 2018; 13: 1177271918806840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tetreault N, De Guire V. miRNAs: their discovery, biogenesis and mechanism of action. Clin Biochem 2013; 46: 842–845. [DOI] [PubMed] [Google Scholar]
- 25.Eskelinen EL. Autophagy: supporting cellular and organismal homeostasis by self-eating. Int J Biochem Cell Biol 2019; 111: 1–10. [DOI] [PubMed] [Google Scholar]
- 26.Tang D, Kang R, Berghe TV, et al. The molecular machinery of regulated cell death. Cell Res 2019; 29: 347–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuma A, Hatano M, Matsui M, et al. The role of autophagy during the early neonatal starvation period. Nature 2004; 432: 1032–1036. [DOI] [PubMed] [Google Scholar]
- 28.Ding ZB, Shi YH, Zhou J, et al. Association of autophagy defect with a malignant phenotype and poor prognosis of hepatocellular carcinoma. Cancer Res 2008; 68: 9167–9175. [DOI] [PubMed] [Google Scholar]
- 29.Takamura A, Komatsu M, Hara T, et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev 2011; 25: 795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell 2011; 147: 728–741. [DOI] [PubMed] [Google Scholar]
- 31.Janku F, McConkey DJ, Hong DS, et al. Autophagy as a target for anticancer therapy. Nat Rev Clin Oncol 2011; 8: 528–539. [DOI] [PubMed] [Google Scholar]
- 32.Zhang H, McCarty N. Tampering with cancer chemoresistance by targeting the TGM2-IL6-autophagy regulatory network. Autophagy 2017; 13: 627–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang S, Wang X, Contino G, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev 2011; 25: 717–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maclean KH, Dorsey FC, Cleveland JL, et al. Targeting lysosomal degradation induces p53-dependent cell death and prevents cancer in mouse models of lymphomagenesis. J Clin Invest 2008; 118: 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang H. Targeting autophagy in lymphomas: a double‑edged sword? Int J Hematol 2018; 107: 502–512. [DOI] [PubMed] [Google Scholar]
- 36.Sou YS, Tanida I, Komatsu M, et al. Phosphatidylserine in addition to phosphatidylethanolamine is an in vitro target of the mammalian Atg8 modifiers, LC3, GABARAP, and GATE-16. J Biol Chem 2006; 281: 3017–3024. [DOI] [PubMed] [Google Scholar]
- 37.Klionsky DJ, Abdelmohsen K, Abe A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016; 12: 1–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy 2011; 7: 279–296. [DOI] [PMC free article] [PubMed] [Google Scholar]