Abstract
The quantification of cerebrospinal fluid (CSF) in the human brain has shown to play an important role in early postnatal brain development. Extra-axial fluid (EA-CSF), which is characterized by CSF in the subarachnoid space, is a promising marker for the early detection of children at risk for neurodevelopmental disorders, such as Autism Spectrum Disorder (ASD). Yet, non-ventricular CSF quantification, in particular extra-axial CSF quantification, is not supported in the major neuro-imaging software solutions, such as FreeSurfer. Most current structural image analysis packages mask out the extra-axial CSF space in one of the first pre-processing steps. A quantitative protocol was previously developed by our group to objectively measure the volume of total EA-CSF volume using a pipeline workflow implemented in a series of python scripts. While this solution worked for our specific lab, a graphical user interface-based tool is necessary to facilitate the computation of extra-axial CSF volume across a wide array of neuroimaging studies and research labs. This paper presents the development of a novel open-source, cross-platform, user-friendly software tool, called Auto-EACSF, for the automatic computation of such extra-axial CSF volume. Auto-EACSF allows neuroimaging labs to quantify extra-axial CSF in their neuroimaging studies in order to investigate its role in normal and atypical brain development.
1. INTRODUCTION
Produced by the brain, cerebrospinal fluid (CSF) is a clear, colorless fluid that circulates around the brain in the extra-axial space acting as a cushion or buffer for the brain, providing basic mechanical and immunological protection to the brain inside the skull. CSF also serves regulatory functions, including the distribution of growth factors critical to brain development. In addition, CSF circulation filters metabolic solutes from the brain interstitium, washing away waste particles that would otherwise build up. In fact, interruption of typical CSF circulation is shown to play a role in many diseases, including neurodegenerative conditions such as Alzheimer’s disease1.
Recent studies2-5 have also suggested a link between the amount of cerebrospinal fluid (CSF) in the subarachnoid space, called extra-axial CSF, and the development of autism spectrum disorder (ASD) in infants. The subjects of these studies who developed ASD had a significantly larger volume of CSF in this region of the brain (see Figure 1). The extent of extra-axial CSF enlargement also correlated with symptomology as those subjects with the most severe ASD symptoms showed larger extra-axial CSF. Thus, the quantification of extra-axial CSF could potentially be used in the future as part of a newer, earlier method of diagnosing ASD. It is noteworthy that prior to these recent studies, extra-axial CSF was commonly not quantified in neuroimaging studies and findings of enlarged extra-axial CSF space were considered incidental and not relevant in early development. That view has significantly shifted in recent years.
Figure 1:
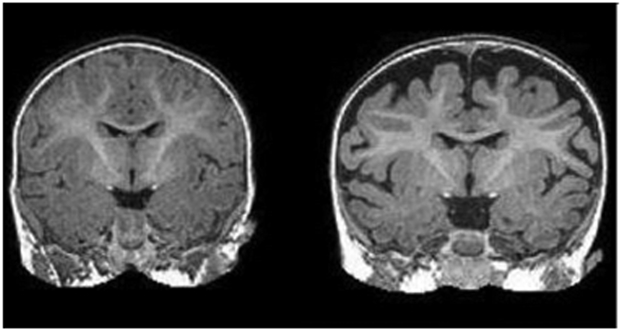
Right: MRI of a baby at 6 months who was diagnosed with autism at 2 years. The dark space between the brain folds and skull indicate increased amounts of cerebrospinal fluid. Left: MRI of a baby who was not diagnosed with autism at age 2.
Non-ventricular CSF quantification, in particular extra-axial CSF quantification, is not supported in any major neuro-imaging software solutions, such as FreeSurfer. Most current structural packages mask extra-axial CSF spaces away in one of the first pre-processing steps (skull stripping/brain masking). Also, accurate extra-axial CSF quantification necessitates the use of T2-weighted MR images, as the low intensity of both CSF and skeletal skull regions leads to little contrast to separate these two tissue compartments on T1-weighted MR images alone. In our lab, a quantitative protocol was developed to objectively measure the volume of the total EA-CSF in each participant using a pipeline workflow of a set of existing tools employing infant MR atlases and implemented in a series of python scripts 4. While this solution works in our lab and would be rather straightforward to disseminate to other technical neuroimaging labs, a user-focused tool is necessary in order to facilitate the computation of extra-axial CSF in neuroimaging studies performed in non-technical neuroimaging labs.
In this paper, we present a novel open-source, cross-platform, user-friendly software tool, called Auto-EACSF, for the automatic computation of extra-axial CSF. In the next sections, we will detail its pipeline, development and public dissemination at NITRC/GitHub.
2. METHODS
The extra-axial CSF quantification is performed using the workflow presented in Figure 2. Both T1 weighted and T2 weighted MRI data are recommended as input images to the workflow, although T2-weighted data is not strictly necessary. The MR data is first jointly co-registered and secondly registered into infant stereotaxic space. Conservative skull stripping is performed next with a focus on preserving all extra-axial CSF regions. In many existing neuroimaging software packages, these extra-axial CSF regions are removed in the brain masking process, whereas in our tool these regions are preserved. This is followed by tissue segmentation, selection of the CSF label, removal of ventricular CSF spaces and removal of CSF regions inferior to the AC-PC line, leading to the final extra-axial CSF segmentation. As the last step, the volume of the segmentation is computed and stored in a comma-separated value file. This workflow is implemented in the Auto-EACSF tool. The tool allows the user to specify input data, but also customize any of these steps, as detailed further below.
Figure 2:
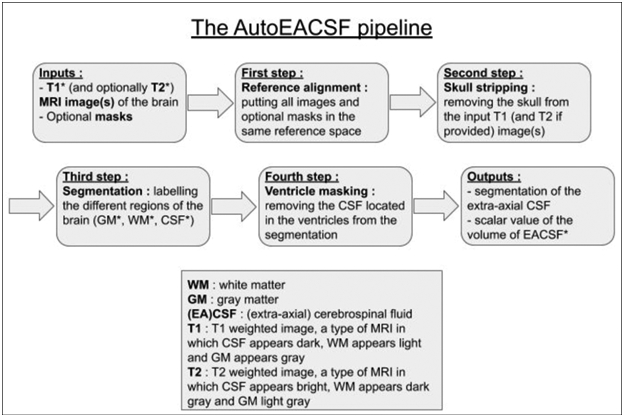
Diagram of the Auto-EACSF pipeline starting from DICOM converted, non-preprocessed MR data.
Input Data:
As mentioned above, this workflow is applicable to T1-weighted MR data only, yet we strongly suggest the use of T2-weighted MRI data as well. The high intensity of CSF spaces in T2-weighted images allow for an improved skull stripping and a much-improved segmentation of the extra-axial CSD spaces in particular. Input data is supported in any ITK-readable volumetric format, though there is currently no support for directly using DICOM data.
Reference alignment:
The first step of the processing is the reference alignment. This step places all MR data into the same reference space, commonly a pediatric stereotaxic space. Our tool provides the user with 12-24 month pediatric space on the tool’s website, and other templates can easily be specified in the user interface. This step can be skipped if the user provides pre-aligned data.
Skull stripping:
The brain mask necessary to perform skull stripping is performed using a multi-atlas approach that combines multiple candidate brain masks obtained via deformable registration of a prior set of atlases (each consisting of a T1-weighted, T2-weighted and brain mask label image). The deformable registration is computed via the ANTs registration toolkit6 using both T1 and T2 weighted data, if both images are available as input MR data. The fusion of the candidate brain masks is performed via straightforward majority vote. That set of prior atlases can be easily extended/adapted by the user to include additional MR datasets, for example, with different contrasts (e.g. adding FLAIR or contrast enhanced data), at different age groups (e.g.in studies of aging or Alzheimer disease) or different brain morphology (e.g. with severe ventricular enlargement). This step can be skipped if the user provides an existing brain mask.
Tissue segmentation:
Our workflow performs tissue segmentation via atlas moderated Expectation-Maximization based classification in ABC 7. The tissue segmentation process also estimates intensity inhomogeneity correction as part of the segmentation loop. A prior atlas with probabilistic maps for CSF, white matter (WM) and gray matter (GM) is employed and user configurable. The segmentation yields a label map with segmentations for CSF, WM and GM. This step can be skipped if the user provides an existing segmentation. In this case, the user also needs to provide the label number in the segmentation map that identifies the CSF segmentation.
Ventricle masking:
A major component of brain CSF is the ventricular CSF space consisting of the lateral ventricles, third and fourth ventricle. As these regions are not considered part of the extra-axial CSF, they need to be removed from the CSF segmentation map. For that purpose, we deformably co-register a single prior template with an existing ventricular area mask and use the registered mask to remove the ventricle. The user provides the information of the template location, which is disseminated on the tool’s website. Alternatively, the user can provide an existing ventricle mask.
Final extra-axial CSF segmentation:
As the final steps of the workflow, the ventricle-masked CSF segmentation is cropped at the axial level of the anterior-posterior commissure (AC-PC) line, preserving only superior CSF spaces. The location of the ACPC line is preset in the tool for the stereotaxic space. If the user employs pre-aligned input data or provides a separate reference alignment template, this information will need to be adjusted by the user.
Tool customization:
Apart from the parameter setting of the existing tool set, different tools can be specified by the user to replace the existing one. For example, the user can replace the current segmentation via ABC with the segmentation performed with FSL-FIRST by modifying the corresponding executable location information in the tool settings. This would necessitate the writing of a “wrapper” script as Auto-EACSF would still use ABC’s command line settings. More sophisticated tool customization is possible as the tool’s code is publicly available.
4. RESULTS
We implemented the above workflow and customization in a C++ tool using Qt for the graphical user interface (see Figure 3 for a screenshot of the tool). All settings are saved in corresponding parameter files and the correspondingly generated workflow is written into a set of python scripts. The Auto-EACSF tool runs those scripts and provides the user with a visual log of the processing progress. Alternatively, an advanced user can also run the tool without the graphical user interface, but rather using its command line interface. That command line interface usage allows the efficient computation of larger datasets.
Figure 3:
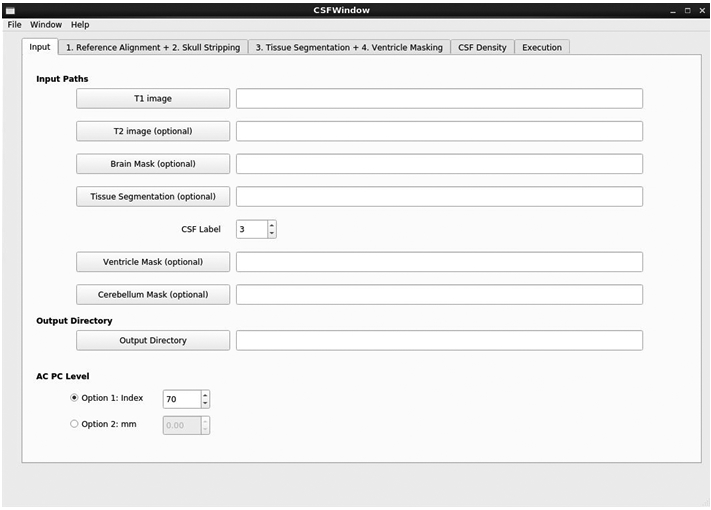
Main window of Auto-EA-CSF
Auto-EACSF is being publicly disseminated on NITRC: https://www.nitrc.org/projects/auto_eacsf/ with binary disseminations, user forums, and support. The code and issue tracker are hosted on GitHub as opensource: https://github.com/NIRALUser/auto_EACSF and are directly accessible through the NITRC project page.
Since its development and first release in January 2019, Auto-EACSF has been applied to over 2000 MR datasets of pediatric subjects ranging from a few weeks of age to 6 years of age, as well as aging subjects from 40 to 90 years of age. It is currently being employed in several developmental neuroimaging studies at the University of North Carolina, and at collaborating laboratories at University of California-Davis and Purdue University.
The most limiting step is the automatic skull stripping that can fail in cases of highly enlarged CSF spaces, particularly visible in late stage degenerative disorders. Manual corrections of the computed brain masks are often necessary in such cases. Alternatively, the user can provide manual brain masks as input in the multi-atlas based brain mask computation in corresponding tab (see Figure 5)
Figure 5:
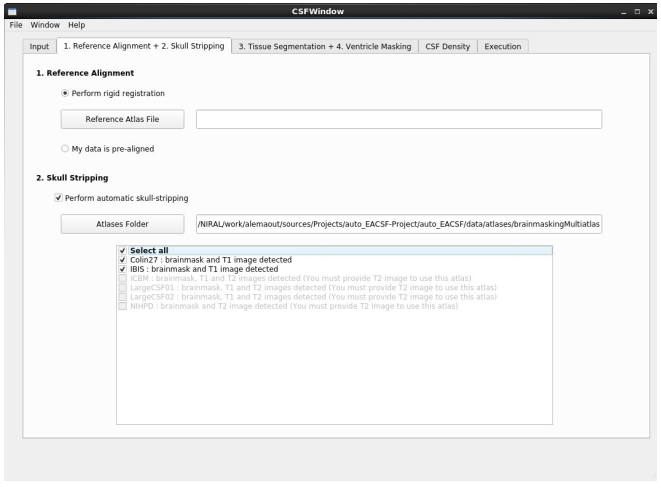
Template registration and brain masking parameter tab. The user easily can extend the brain mask database.
5. CONCLUSSIONS
Here we presented the development of a novel, publicly available tool for the computation of extra-axial CSF to be used in neuroimaging studies of infant brain development with application to early developmental disorders. Our next steps focus on extending this tool to include our local EA-CSF quantification approach 8.
Figure 4:
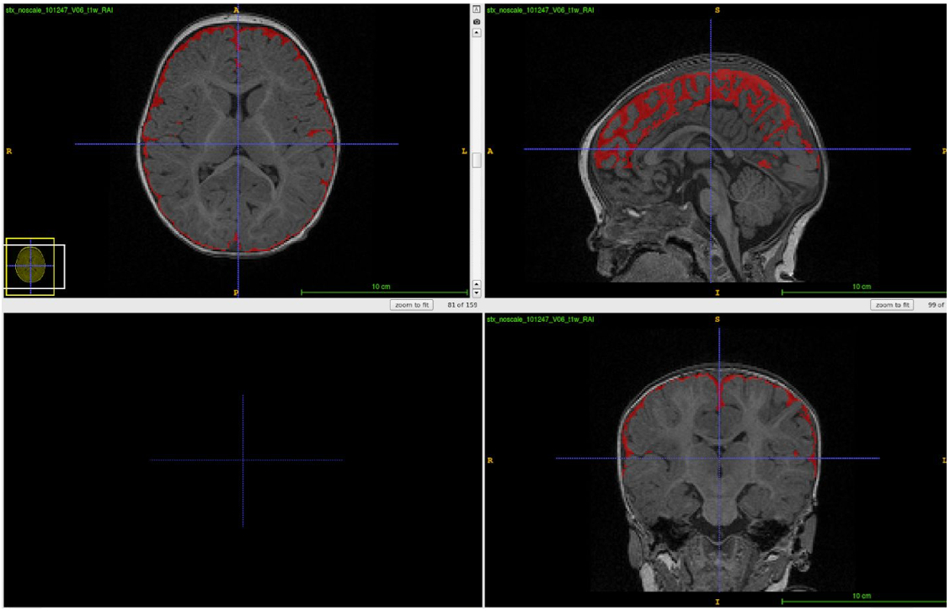
Typical segmentation result on a pediatric 6 month old dataset
Acknowledgement:
Funding was provided by the IBIS (Infant Brain Imaging Study) Network, an NIH funded Autism Center of Excellence (HDO55741) that consists of a consortium of 7 Universities in the U.S. and Canada, and the NIH grants U54HDO79124, R01EB021391, and K12-HD001441.
References
- [1].Rasmussen MK, Mestre H, Nedergaard M, “The glymphatic pathway in neurological disorders.,” Lancet Neurol 17(11), 1016–1024 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Shen MD, Nordahl CW, Young GS, Wootton-Gorges SL, Lee A, Liston SE, Harrington KR, Ozonoff S, Amaral DG, “Early Brain Enlargement and Elevated Extra-Axial Fluid in Infants Who Develop Autism Spectrum Disorder”. Brain, 136(9), 2825–2835. (2013). PMID: 23838695; PMCID: PMC3754460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Shen MD, Nordahl CW, Li DD, Lee A, Angkustsiri K, Emerson RW, Rogers SJ, Ozonoff S, Amaral DG, “Extra-axial cerebrospinal fluid in high-risk and normal-risk children with autism aged 2–4 years: a case-control study.,” The Lancet Psychiatry 5(11), 895–904 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Shen MD, Kim SH, McKinstry RC, Gu H, Hazlett HC, Nordahl CW, Emerson RW, Shaw D, Elison JT, et al. , “Increased Extra-axial Cerebrospinal Fluid in High-Risk Infants Who Later Develop Autism.,” Biol. Psychiatry 82(3), 186–193 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hussain ZB, Hussain AB, Mitchell P, “Extra-axial cerebrospinal fluid spaces in children with benign external hydrocephalus: A case-control study.,” Neuroradiol J 30(5), 410–417 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC, “A reproducible evaluation of ANTs similarity metric performance in brain image registration.,” NeuroImage 54(3), 2033–2044 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Prastawa M, Gilmore JH, Lin W, Gerig G, “Automatic segmentation of MR images of the developing newborn brain.,” Med Image Anal 9(5), 457–466 (2005). [DOI] [PubMed] [Google Scholar]
- [8].Mostapha M, Shen MD, Kim S, Swanson M, Collins DL, Fonov V, Gerig G, Piven J, Styner M, et al. , “A Novel Framework for the Local Extraction of Extra-Axial Cerebrospinal Fluid from MR Brain Images.,” Proc SPIE Int Soc Opt Eng 10574 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]


