Abstract
Acute myelocytic leukemia (AML) is a frequent type of acute leukemia. The present study was performed to build a risk score system for the prognostic prediction of AML. AML RNA-sequencing data from samples from 111 children were downloaded from The Cancer Genome Atlas database. Using the DEseq and edgeR packages, the differentially expressed long non-coding RNAs (DE-lncRNAs) between bad and good prognosis groups were identified. A survival package was used to screen prognosis-associated lncRNAs and clinical factors. The optimal lncRNA combination was selected using the penalized package, and the risk-score system was built and evaluated. After the lncRNA-mRNA expression correlation network was constructed, the potential pathways involving the key lncRNAs were enriched using Gene Set Enrichment Analysis. Among the 61 DE-lncRNAs, 48 lncRNAs were significantly associated with prognosis. Relapse was an independent prognostic factor. The optimal 14-lncRNA risk score system was constructed. After 730 differentially expressed mRNAs were identified between the good and bad prognosis groups divided using a prognostic index, the lncRNA-mRNA expression correlation network was constructed. Enrichment analysis showed that semaphorin-3C [SEMA3C; regulated by probable leucine-tRNA ligase, mitochondrial (LARS2-AS1)] and secreted frizzled-related protein 5 [SFRP5; mediated by WASH complex subunit 5 (WASHC5)-antisense RNA 1 (AS1)] were involved in axon guidance and the Wnt signaling pathway, respectively. A 14-lncRNA (including paired box protein Pax8-AS1 and MYB AS1) risk-score system might be effective in predicting the prognosis of AML. Axon guidance (involving SEMA3C and LARS2-AS1) and the Wnt signaling pathway (involving SFRP5 and WASHC5-AS1) might be two important pathways affecting the prognosis of AML.
Keywords: acute myelocytic leukemia, long non-coding RNA, risk score system, survival analysis, expression correlation network, enrichment analysis
Introduction
Acute myelocytic leukemia (AML) is a tumor derived from the myeloid line of blood cells, in which abnormal cells in the bone marrow and blood are rapidly generated (1). AML is characterized by the proliferation and abnormal differentiation of cells of the hematopoietic system in bone marrow, blood and other tissues (2). The typical clinical symptoms of patients with AML are shortness of breath, fatigue, increased risk of infection, and easy bleeding and bruising (3). The risk factors of AML mainly include smoking, benzene exposure, myelodysplastic syndrome, and previous radiation therapy or chemotherapy (4,5). AML is more frequent in elderly men, and it progresses rapidly without immediate therapy (6). AML affects approximately one million individuals and resulted in 147,000 deaths globally in 2015(7). The pathogenesis of AML needs to be further investigated to decrease the prevalence and mortality of AML.
Long non-coding RNAs (lncRNAs) are transcripts with a length >200 nucleotides. lncRNAs have no protein coding potential but are implicated in various biological functions, including cell cycle, apoptosis, epigenetic regulation and imprinting (8). lncRNA expression is closely associated with recurrent mutations, and some lncRNAs play important roles in the AML therapeutic response and clinical survival (9). HOX transcript antisense RNA expression is upregulated in de novo patients with AML, which is strongly associated with the clinicopathological prognosis of patients with AML (10,11). Insulin-like growth factor type I receptor (IGF1R) antisense imprinted non-protein coding RNA is associated with long range IGF1R DNA interactions and upregulation of the IGF pathway in AML, and is a potential tumor suppressor and therapeutic target for the disease (12). lncRNA H19 (imprinted maternally expressed transcript) is an independent prognostic biomarker for patients with tumors, and its expression is significantly upregulated in AML and may play a role in AML cell proliferation (13). lncRNA RUNXOR [runt related transcription factor 1 (RUNX1) overlapping promoter-derived noncoding RNA) is located in the RUNX1 locus (14). Overexpressed RUNXOR may serve as a tumor suppressor through RUNX1 and scaffolding mediated translocation of DNA correlated in AML (15). However, the lncRNAs associated with the prognosis of AML have not been fully reported.
In the present study, prognosis-associated lncRNAs were screened to construct a risk score system for AML. Additionally, the potential functions of the key lncRNAs were predicted. The data could be valuable in clarifying the prognostic mechanisms of AML and could contribute to the prognostic prediction of the disease.
Materials and methods
Data source
All available RNA-sequencing data of AML samples were searched using The Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/) on March 17, 2018. In total, 187 AML samples with RNA-sequencing data were obtained from TCGA (16,17). According to the age information of the samples, 111 samples from patients under the age of 14 years were included. The 111 samples were randomly divided into two groups (56 samples in the training set and 55 samples in the validation set). The clinical information of the samples in each group is presented in Table I.
Table I.
Clinical information of the samples in the training set, the validation set and the entire set.
| Clinical characteristics | Training set, n=56 | Validation set, n=55 | Entire set, n=111 |
|---|---|---|---|
| Age, years, mean ± SD | 5.99±4.26 | 5.81±4.32 | 5.91±4.27 |
| Sex, male/female | 32/24 | 31/24 | 63/48 |
| WBC at diagnosis, 109/l, mean ± SD | 64.56±74.03 | 97.09±113.96 | 80.67±96.86 |
| Bone marrow leukemic blast percentage, %, mean ± SD | 69.51±20.44 | 72.99±20.39 | 71.19±20.39 |
| Peripheral blasts, %, mean ± SD | 49.61±28.76 | 57.7±30.09 | 53.62±29.58 |
| CNS disease, yes/no | 3/53 | 4/51 | 7/104 |
| Chloroma, yes/no/- | 3/53 | 6/48/1 | 9/101/1 |
| FAB Category, M0/M1/M2/M3/M4/M5/M6/M7/- | 0/3/9/0/18/12/1/6/7 | 3/11/14/0/10/11/1/1/4 | 3/14/23/0/28/23/2/7/11 |
| FLT3/ITD mutation, yes/no | 4/52 | 5/50 | 9/102 |
| NPM mutation, yes/no/- | 0/52/4 | 5/50/0 | 5/4/102 |
| CEBPA mutation, yes/no/- | 2/52/2 | 4/51 | 6/103/2 |
| WT1 mutation, yes/no | 2/50/4 | 4/51 | 6/101/4 |
| Relapse, yes/no | 37/19 | 40/15 | 77/34 |
| Relapse free survival time, months, mean ± SD | 33.01±34.84 | 31.52±32.42 | 32.27±33.52 |
| Death, dead/alive | 24/32 | 24/31 | 48/63 |
| Overall survival time, months, mean ± SD | 54.13±34.76 | 55.97±36.08 | 55.04±35.27 |
-, data unavailable; WBC, white blood cell; CNS, central nervous system; FAB, French-American-British; FLT3, receptor-type tyrosine-protein kinase FTL3; ITD, internal tandem duplications; NPM, nucleophosmin; CEBPA, CCAAT enhancer binding protein α; WTM, Wilms' tumor gene 1.
Differential expression analysis
Among the 56 AML samples in the training set, the median survival time was 54.13 months. It was assumed that samples from patients with a short survival were high risk, and patients with longer survival being indicative of relatively low risk. Combining these assumptions with the median survival time established 54.13±18 months as the cut-off value to classify different prognostic samples. Samples with a survival time <36 months who had died were classified into the bad prognosis group (n=20). Samples with a survival time >72 months who were still alive were divided into the good prognosis group (n=21). The differentially expressed lncRNAs (DE-lncRNAs) between the two groups were selected using the DEseq package (version 1.30.0; https://bioconductor.org/packages/release/bioc/html/DESeq.html) (18) and edgeR package (version 3.20.9; http://bioconductor.org/packages/release/bioc/html/edgeR.html) (19) in R, which were also used in other previous studies (20-22). The false discovery rate (FDR) <0.05 and log fold change (FC) >1 were defined as the thresholds. By comparing the results of the two screening methods, the overlapped DE-lncRNAs were taken as the final DE-lncRNAs. Based on the expressions of the DE-lncRNAs in the training set, the bidirectional hierarchical clustering using the pheatmap package in R (version 1.0.8; https://cran.r-project.org/web/packages/pheatmap/index.html) (23) was performed.
Identification of prognosis-associated lncRNAs and clinical factors
Using the survival package in R (version 2.41.3; https://cran.r-project.org/web/packages/survival/index.html) (24), univariate and multivariate Cox regression analysis was conducted for the 56 AML samples in the training set to screen the prognosis-associated lncRNAs and clinical factors. The log-rank P<0.05 was set as the threshold for the significant results.
Construction and evaluation of risk score system
Based on the prognosis-associated lncRNAs, the optimal lncRNA combination was selected using the Cox-Proportional Hazards (Cox-PH) model in R package penalized (version 0.9.50; https://cran.r-project.org/web/packages/penalized/index.html) (25). The optimal parameter ‘λ’ in the Cox-PH model was obtained through the cross-validation likelihood (cvl) for 1,000 times. The risk score system was constructed for AML samples combined with the linear combination of the lncRNA expression with regression coefficient weighting. The formula for calculating the prognostic index (PI) of each sample was as follows:
PI=β lncRNA1 x exprlncRNA1 + β lncRNA2 x exprlncRNA2 + ··· + β lncRNAn x exprlncRNAn
The PI of each sample in the training set was calculated based on the aforementioned formula, and the median PI was utilized as the cut-off value of the good and bad prognosis samples. A Kaplan-Meier (KM) curve (26) was used to calculate the survival time in the two groups when stratified by different prognostic factors, based on which to evaluate the prognosis difference between the good and bad prognosis groups using a certain prognostic factor, and an area under the receiver operating characteristic (AUROC) curve (27) was applied to assess the performance of prognostic prediction effectiveness using the risk score system. Then, the risk score system was used to evaluate the prognosis of the samples in the validation set.
Construction of the lncRNA-mRNA expression correlation network
In the training set, the samples were divided into the good and bad prognosis groups according to their PIs in the predictive risk score system model. The differentially expressed mRNAs (DE-mRNAs) between the good and bad prognosis groups were analyzed using the edgeR package (19), with the thresholds of FDR <0.05 and log FC >1. Then, the Pearson correlation coefficients (PCCs) (28) of the DE-mRNAs and the lncRNAs involved in the risk score system were calculated. Finally, the DE-mRNAs with PPC >0.5 were obtained for constructing the lncRNA-mRNA expression correlation network.
Pathway enrichment analysis
Based on Gene Set Enrichment Analysis (GSEA; http://software.broadinstitute.org/gsea/index.jsp) (29), pathway enrichment analysis for the mRNAs in the network was performed to identify the significant pathways correlated with key lncRNAs. The pathways with nominal P<0.05 were considered as significant results.
Results
Differential expression analysis
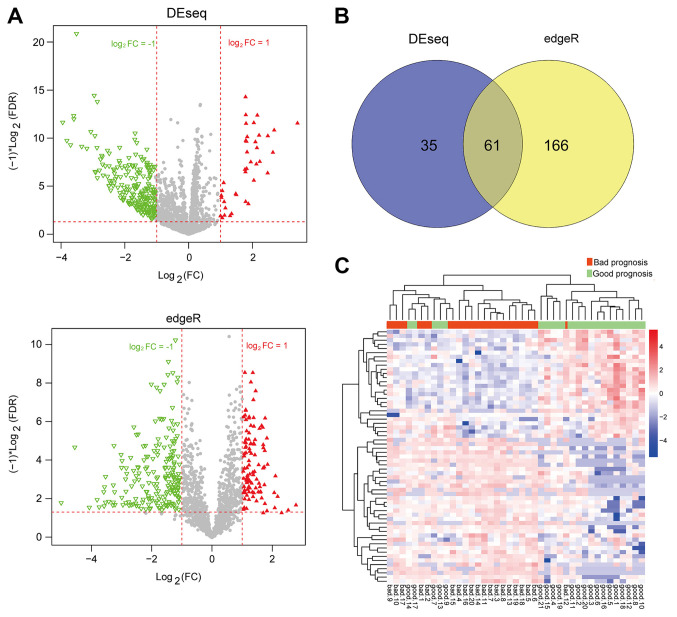
After the AML samples in the training set were classified into the bad and good prognosis groups, the DEseq package and edgeR package were used for the selection of DE-lncRNAs (Fig. 1A). A total of 61 overlapped DE-lncRNAs were obtained by comparing the results of the two screening methods (Fig. 1B). The clustering heatmap of the 61 DE-lncRNAs suggested the samples could be distinguished by these DE-lncRNAs (Fig. 1C).
Figure 1.
Differential expression analysis between good and bad prognosis groups. (A) Dysregulated lncRNAs identified separately by the DEseq package (top) and edgeR package (bottom). Red and green represent upregulated and downregulated lncRNAs in the good prognosis group, respectively. (B) Venn diagram for comparing the results of the two screening methods. (C) Clustering heatmap of the 61 intersected lncRNAs. Red and green indicate bad and good prognosis groups, respectively. FDR, false discovery rate; FC, fold change; lncRNA, long non-coding RNA.
Identification of prognosis-associated lncRNAs and clinical factors
In the training set, 61 lncRNAs were subjected to univariate Cox regression analysis. A total of 48 lncRNAs were significantly associated with prognosis (P<0.05; data not shown). Univariate and multivariate Cox regression analysis indicated that relapse was significantly associated with prognosis in the training set, the validation set and the entire set (Table II; P<0.05). Therefore, relapse was identified as an independent clinical factor with prognostic significance. The KM curves of relapse and the overall survival (OS) of the samples in the training set, the validation set and the entire set are presented in Fig. 2A. The results showed that the AML samples without relapse had better clinical prognosis, which was consistent with the actual situation.
Figure 2.
Kaplan-Meier curves and the selection of the optimal combination. (A) Kaplan-Meier curves for relapse in the training set (left), the validation set (middle), and the entire set (right). (B) Curve for selecting the optimal parameter ‘λ’ using cvl (the junction of the red dashed lines indicates the value of lambda is 24.34156 when the maximum value-108.177 is obtained for cvl). (C) Coefficient distribution diagram of the 14 optimal lncRNAs. cvl, cross-validation likelihood; lncRNA, long non-coding RNA.
Construction and evaluation of the risk score system
Based on the expression matrix of the 48 prognosis-associated lncRNAs in the training set, the optimal lncRNA combination was analyzed using the Cox-PH model using the penalized package. After performing the 1,000 times cvl algorithm, the maximum cvl value was-108.177 when the parameter ‘λ’ was set as 24.34156 (Fig. 2B). With the optimal parameter, the optimal lncRNA combination involving 14 lncRNAs [LINC00639, probable E3 ubiquitin-protein ligase makorin-3 (MKRN3)-antisense RNA 1 (AS1), LINC02060, LINC02133, LINC01726, LINC00240, paired box protein Pax8 (PAX8)-AS1, zinc finger protein 32 (ZNF32)-AS1, probable leucine-tRNA ligase, mitochondrial (LARS2)-AS1, Ras association domain family member 8 (RASSF8)-AS1, E74-like factor 3 (ELF3)-AS1, WASH complex subunit 5-antisense RNA 1 (WASHC5)-AS1, MYB-AS1 and FAR1 intronic transcript 1 (FAR1-IT1)] was screened (Fig. 2C; Table III) using the Cox-PH model.
Table III.
Information of the 14 optimal lncRNAs.
| lncRNA | Coef | HR (95% CI) | P-value |
|---|---|---|---|
| LINC00639 | 0.20989291 | 1.075 (1.023-1.130) | 0.0025 |
| MKRN3-AS1 | 0.06638359 | 1.054 (1.015-1.096) | 0.0038 |
| LINC02060 | 0.3139191 | 1.069 (1.024-1.117) | 0.0014 |
| LINC02133 | 0.42299789 | 1.152 (1.061-1.250) | <0.0001 |
| LINC01726 | -0.19338239 | 0.938 (0.898-0.979) | 0.0023 |
| LINC00240 | -0.2969476 | 0.945 (0.897-0.996) | 0.0302 |
| PAX8-AS1 | 0.73636164 | 1.093 (1.012-1.179) | 0.0224 |
| ZNF32-AS1 | 0.02132678 | 1.070 (1.023-1.119) | 0.0010 |
| LARS2-AS1 | 0.1805946 | 1.075 (1.019-1.133) | 0.0030 |
| RASSF8-AS1 | 0.53198533 | 1.128 (1.023-1.244) | 0.0142 |
| ELF3-AS1 | 0.41975344 | 1.223 (1.045-1.431) | 0.0205 |
| WASHC5-AS1 | 0.33392756 | 1.195 (1.030-1.387) | 0.0076 |
| MYB-AS1 | 0.62016856 | 1.175 (1.038-1.330) | 0.0247 |
| FAR1-IT1 | 0.02508178 | 1.135 (1.017-1.267) | 0.0325 |
Coef, coefficient; HR, hazard ratio; CI, confidence interval; lncRNA, long non-coding RNA.
The risk score system was constructed and the formula for calculating PIs was: PI=(0.20989291) x ExpLINC00639+ (0.06638359) x Exp MKRN3-AS1+(0.3139191) x ExpLINC02060 +(0.42299789) x ExpLINC02133+(-0.19338239) x ExpLINC01726 +(-0.2969476) x ExpLINC00240+(0.73636164) x ExpPAX8-AS1+ (0.02132678) x ExpZNF32-AS1+(0.1805946) x ExpLARS2-AS1+ (0.53198533) x ExpRASSF8-AS1+(0.41975344) x ExpELF3-AS1+ (0.33392756) x ExpWASHC5-AS1+(0.62016856) x ExpMYB-AS1+(0.02508178) x Exp FAR1-IT1.
Subsequently, the performance of the prognostic prediction of the risk score system for the samples in the training set, the validation set and the entire set were evaluated using KM curves and AUROC curves (Fig. 3). The expression levels of the 14 optimal lncRNAs in the aforementioned sets downloaded from TCGA are presented in Fig. 4, where all 14 lncRNAs exhibited distinct expression profiles between bad and good prognosis groups. The PIs, OS and expression heatmap of the 14 lncRNAs in the training set, the validation set and the entire set are presented in Fig. 5.
Figure 3.
KM curves and AUROC curves. (A) KM curves showing the OS (left) and RFS (middle), and the AUROC curve (right) for the training set. (B) KM curves showing the OS (left) and RFS (middle) and the AUROC curve (right) for the validation set. (C) KM curves showing the OS (left) and RFS (middle) and the AUROC curve (right) for the entire set. KM, Kaplan-Meier; AUROC, area under the received operating characteristic; AUC, area under curve; OS, overall survival; RFS, relapse-free survival.
Figure 4.
Expression levels of the 14 optimal lncRNAs. (A) Expression levels of the 14 optimal lncRNAs in the training set. (B) Expression levels of the 14 optimal lncRNAs in the validation set. (C) Expression levels of the 14 optimal lncRNAs in the entire set. Green and red represent good and bad prognosis groups, respectively. *P<0.05; **P<0.01 and ***P<0.005 vs. respective good prognostic group. lncRNA, long non-coding RNA.
Figure 5.
PIs, OS and expression heatmap of the 14 optimal lncRNAs. (A) PIs (top), OS (middle) and expression heatmap (bottom) of the 14 optimal lncRNAs in the training set. (B) PIs (top), OS (middle) and expression heatmap (bottom) of the 14 optimal lncRNAs in the validation set. (C) PIs (top), OS (middle) and expression heatmap (bottom) of the 14 optimal lncRNAs in the entire set. PI, prognostic index; OS, overall survival; lncRNA, long non-coding RNA.
Construction of the lncRNA-mRNA expression correlation network
Based on the PIs of the samples in the training set, the samples were divided into the good and bad prognosis groups and 730 DE-mRNAs (718 upregulated and 12 downregulated) between the two groups were identified. The PCC of the expression level of each DE-mRNA and PI was calculated, and then the top 100 DE-mRNAs with highest absolute values of PCC were used for sample clustering (Fig. 6). Furthermore, the PCCs between the DE-mRNAs and the 14 optimal lncRNAs were calculated. The lncRNA-mRNA expression correlation network was constructed with the criterion of PCC <0.5 (Fig. 7).
Figure 6.
Clustering heatmap for the top 100 differentially expressed mRNAs with higher absolute value of Pearson correlation coefficient.
Figure 7.
lncRNA-mRNA expression correlation network. Circles and squares represent mRNAs and lncRNAs, respectively. The colors ranging from light to dark indicate the increased degree of upregulation in the good prognosis group. lncRNA, long non-coding RNA; FC, fold change.
Pathway enrichment analysis
Based on GSEA pathway enrichment analysis, axon guidance [involving semaphorin 3C (SEMA3C), semaphorin 6D and netrin 4] and the Wnt signaling pathway [involving Dickkopf-related protein 4, 1-phosphatidylinositol 4,5-biphosphate phosphodiesterase β 4 and secreted frizzled-related protein 5 (SFRP5)] were separately associated with the lncRNAs LARS2-AS1 and WASHC5-AS1, respectively (Table IV).
Table IV.
Pathways significantly associated with LARS2-AS1 and WASHC5-AS1.
| A, LARS2-AS1 | ||||
|---|---|---|---|---|
| Name | ES | NES | NOM P-val | Genes |
| Axon guidance | 0.842324 | 1.775851 | 0.0081 | SEMA3C, SEMA6D, NTN4 |
| B, WASHC5-AS1 | ||||
| Name | ES | NES | NOM P-val | Genes |
| Wnt signaling pathway | -0.6971 | -1.4588 | 0.0081 | DKK4, PLCB4, SFRP5 |
ES, enrichment score; NES, normalized enrichment score; NOM P-val, nominal P-value; AS1, antisense 1; LARS2, probable leucine-tRNA ligase, mitochondrial; WASHC5, WASH complex subunit 5; SEMA3C, semaphorin 3C; SEMA6D, semaphorin 6D; NTN4, netrin 4; DKK4, Dickkopf-related protein 4; PLCB4, 1-phosphatidylinositol 4,5-biphosphate phosphodiesterase β 4; secreted frizzled-related protein 5.
Discussion
In recent decades, bioinformatics methods have developed rapidly and are being applied for the analysis of gene expression profiles to determine the pathogenesis of human diseases (30). A total of six genes (triggering receptor expressed on myeloid cells like 2, solute carrier family 7 member 11, NLR family pyrin domain containing 2, DNA damage inducible transcript 4, lymphocyte specific protein 1 and C-type lectin domain containing 11A) have been identified as prognostic indicators of AML combining TCGA data with a Gene Expression Omnibus dataset (31). In total, two lncRNAs (NONHSAT027612.2 and NONHSAT134556.2) have been implicated in the pathogenesis of childhood acute lymphoblastic leukemia (ALL) (32). However, the prognosis has not been investigated (32). In another previous study, although both lncRNAs and their relationships with prognosis had been examined in AML, the authors focused on the competing endogenous RNAs regulation network (33). The predictive role of lncRNAs on the prognosis of AML remains obscure.
In the present study, 61 DE-lncRNAs between the good and bad prognosis groups were obtained. Of these, 48 lncRNAs were significantly associated with prognosis. Cox regression analysis demonstrated that relapse was significantly associated with the prognosis of patients with AML and was an independent prognostic factor. From the 48 prognosis-associated lncRNAs, the optimal lncRNA combination involving 14 lncRNAs was screened to construct the risk score system. Subsequently, 730 DE-mRNAs were identified for the good and bad prognosis groups divided by PIs, and the lncRNA-mRNA expression correlation network was constructed.
The Wilms' tumor gene 1 (WT1) is overexpressed in AML and functions as an oncogene (34). Therefore, it is a therapeutic target of AML. PAX8 is a physiological regulator of WT1 that accounts for its upregulation (35). The lncRNA PAX8-AS1 located in the upstream region of the PAX8 gene regulates the expression of the gene (36). The rs4848320 and rs6726151 polymorphisms of PAX8-AS1 can significantly increase the risk of ALL (37). Therefore, PAX8-AS1 may act as a risk factor for childhood ALL (37). CREB-binding protein/P300 coactivation derived by MYB is necessary for ALL and AML, and MYB activity plays a critical role in maintaining AML (38). MYB expression is increased in T cell ALL, and the oncogene MYB could be a therapeutic target for the disease (39,40). These findings suggested that PAX8-AS1 and MYB-AS1 might be associated with the pathogenesis of patients with AML.
LINC00240-Kruppel-like factor 3 crosstalk is mediated by microRNA-26b-5p in esophageal squamous cell carcinoma (ESCC) and its loss may be involved in the oncogenesis of ESCC (41). ZNF32 is a transcription factor closely associated with oxidative stress and protects tumor cells against oxidative stress-induced cell death (42). Therefore, targeting ZNF32 may suppress multidrug resistance during chemotherapy and improve the clinical outcome of patients with lung adenocarcinoma (41,43). RASSF8 expression is negatively associated with lymph node metastasis and survival of patients with ESCC (44). Therefore, RASSF8 is a tumor suppressor and a candidate therapeutic target for the disease (44). The low expression of E74-like factor 3 (ELF3) is significantly correlated with decreased survival of patients with ovarian cancer, and ELF3 is a favorable prognostic biomarker for the tumor (45). Although LINC00240, ZNF32-AS1, RASSF8-AS1 and ELF3-AS1 have not been reported to be related to AML, their roles in other tumors were identified. Based on the findings of the present study, they could be prognostic factors for AML. Therefore, LINC00240, ZNF32-AS1, RASSF8-AS1 and ELF3-AS1 might also influence the prognosis of AML.
Dysregulated axon guidance molecules (AGMs) function as tumor suppressors or oncogenes in breast cancer, which may be promising targets for the prognosis and therapy of tumors (46). Some AGMs play roles in mediating cell migration and apoptosis in tumorigenic tissues, suggesting that promoting or inhibiting the activity of AGMs may be novel approaches for treating tumors (47). Semaphorins, which are one type of AGMs, serve as tumor suppressors (48). Of note, semaphorin 3B (SEMA3B) could act as a tumor inhibitor in the early stage of breast cancer, and could also promote metastasis during the late stage of the cancer (49). The role of SEMA3C has not been fully reported, but it belongs to the AGMs that promote tumor metastasis (46). In the present study, SEMA3C was significantly enriched in the axon guidance pathway, and it was associated with the lncRNA LARS2-AS1, suggesting that expression of SEMA3C may be regulated by LARS2-AS1, and the alteration of the gene might disturb the pathway, which might affect the pathogenesis of AML. However, these regulatory regulations remain to be experimentally validated.
The Wnt signaling pathway is overexpressed in most cases of AML and targeting the WNT/lymphoid enhancer-binding factor 1 signaling cascade may be used for developing novel therapies for AML (50,51). The Wnt signaling pathway is directly mediated by several secreted antagonist families, such as the SFRP family (52). In AML, this pathway is mediated by methylation of Wnt antagonists (53). In the present study, SFRP5 was significantly associated with the Wnt signaling pathway and was indirectly regulated by the lncRNA WASHC5-AS1 in the interaction network, suggesting the interactions between them might influence the signaling pathway, and regulate the development of AML. Nonetheless, these interactions need to be experimentally validated.
In the present study, relapse was identified as an independent prognostic factor in both the training and validation datasets, suggesting that relapse could affect the prognostic risk using this 14-lncRNA predictive system. However, more clinical evidence is required.
Although comprehensive bioinformatics analysis has been conducted, there were several limitations of the present study. The expressions of these lncRNAs and genes were not validated in vitro and the interactions were not confirmed. In addition, the predictions of prognostic roles of these lncRNAs also need the support of further clinical data. More experiments should be designed and conducted to support the present results.
In conclusion, a 14-lncRNA (including PAX8-AS1 and MYB-AS1) risk score system might be effective for the prediction of AML prognosis. There were two pathways identified from the GSEA pathway enrichment analysis; axon guidance, which might involve SEMA3C and LARS2-AS1, and the Wnt signaling pathway, which might include the interaction of SFRP5 and WASHC5-AS1. These two identified pathways might be important factors that affect the prognosis of childhood AML.
Table II.
Prognostic analysis of clinical factors for the samples in the training set, the validation set and the entire set.
| A, Training set, n=56 | ||||
|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | |||
| Variables | HR | P-value | HR | P-value |
| Age, years, mean ± SD | 1.103 | 0.045 | 1.010 | 0.881 |
| Sex, male/female | 0.966 | 0.934 | 1.062 | 0.905 |
| WBC at diagnosis, 109/l, mean ± SD | 0.996 | 0.296 | 0.997 | 0.530 |
| Bone marrow leukemic blast percentage, %, mean ± SD | 1.011 | 0.321 | 1.020 | 0.245 |
| Peripheral blasts, %, mean ± SD | 0.987 | 0.077 | 0.992 | 0.527 |
| Chloroma, yes/no/- | 0.702 | 0.728 | 0.594 | 0.732 |
| FAB Category, M0/M1/M2/M3/M4/M5/M6/M7/- | 1.103 | 0.471 | 1.095 | 0.692 |
| FLT3/ITD mutation, yes/no | 2.480 | 0.130 | 1.297 | 0.840 |
| WT1 mutation, yes/no | 6.876 | 0.005 | 4.549 | 0.186 |
| Relapse, yes/no | 8.259 | 0.001 | 12.462 | 0.016 |
| B, Validation set, n=55 | ||||
| Univariate analysis | Multivariate analysis | |||
| Variables | HR | P-value | HR | P-value |
| Age, years, mean ± SD | 0.864 | 0.008 | 0.766 | 0.003 |
| Sex, male/female | 1.018 | 0.966 | 1.249 | 0.691 |
| WBC at diagnosis, 109/l, mean ± SD | 1.001 | 0.409 | 0.998 | 0.608 |
| Bone marrow leukemic blast percentage, % mean ± SD | 0.992 | 0.417 | 0.961 | 0.217 |
| Peripheral blasts, %, mean ± SD | 1.001 | 0.879 | 1.029 | 0.057 |
| Chloroma, yes/no/- | 0.574 | 0.447 | 0.633 | 0.688 |
| FAB Category, M0/M1/M2/M3/M4/M5/M6/M7/- | 0.852 | 0.213 | 0.812 | 0.255 |
| FLT3/ITD mutation, yes/no | 2.412 | 0.145 | 9.222 | 0.073 |
| WT1 mutation, yes/no | 2.584 | 0.113 | 0.656 | 0.591 |
| Relapse, yes/no | 9.150 | 0.003 | 5.828 | 0.011 |
| C, Entire set, n=111 | ||||
| Univariate analysis | Multivariate analysis | |||
| Variables | HR | P-value | HR | P-value |
| Age, years, mean ± SD | 0.981 | 0.566 | 0.943 | 0.173 |
| Sex, male/female | 0.991 | 0.976 | 1.031 | 0.929 |
| WBC at diagnosis, 109/l, mean ± SD | 1.001 | 0.959 | 0.998 | 0.537 |
| Bone marrow leukemic blast percentage, %, mean ± SD | 1.001 | 0.925 | 0.999 | 0.911 |
| Peripheral blasts, %, mean ± SD | 0.995 | 0.281 | 1.005 | 0.537 |
| Chloroma, yes/no/- | 0.619 | 0.416 | 0.462 | 0.301 |
| FAB Category, M0/M1/M2/M3/M4/M5/M6/M7/- | 0.967 | 0.705 | 1.072 | 0.528 |
| FLT3/ITD mutation, yes/no | 2.496 | 0.031 | 2.268 | 0.215 |
| WT1 mutation, yes/no | 3.388 | 0.007 | 2.019 | 0.185 |
| Relapse, yes/no | 9.227 | <0.0001 | 11.048 | 0.001 |
HR, hazard ratio; WBC, white blood cell; FAB, French-American-British; FLT3, receptor-type tyrosine-protein kinase FTL3; ITD, internal tandem duplications; WTM, Wilms' tumor gene 1.
Acknowledgements
Not applicable.
Funding
The present study was supported by Medical Scientific Research Project of Henan Province (grant no. 2018020905), Medical Scientific Research Project of Henan Province (grant no. SB201903035), Medical and Health Program of Luoyang Science & Technology Project (grant no. 1910014A).
Availability of data and materials
All data used in this study are available in TCGA database (https://portal.gdc.cancer.gov/).
Authors' contributions
HRW designed this study; SLG, BL, XYX, WLW, SYW and TL performed the data analysis. The final version of the manuscript was read and approved by all authors.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Clarkson BD. Acute myelocytic leukemia in adults. Cancer. 2015;30:1572–1582. doi: 10.1002/1097-0142(197212)30:6<1572::aid-cncr2820300624>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 2.Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373:1136–1152. doi: 10.1056/NEJMra1406184. [DOI] [PubMed] [Google Scholar]
- 3.Kootte AM, Thompson J, Bruijn JA. Acute myelocytic leukemia with acute aortic occlusion as presenting symptom. Acta Haematol. 2009;75:120–121. doi: 10.1159/000206100. [DOI] [PubMed] [Google Scholar]
- 4.Pyatt DW, Aylward LL, Hays SM. Is age an independent risk factor for chemically induced acute myelogenous leukemia in children? J Toxicol Environ Health B Crit Rev. 2007;10:379–400. doi: 10.1080/15287390600975061. [DOI] [PubMed] [Google Scholar]
- 5.Belson M, Kingsley B, Holmes A. Risk factors for acute leukemia in children: A review. Environ Health Perspect. 2007;115:138–145. doi: 10.1289/ehp.9023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deschler B, Lübbert M. Acute myeloid leukemia: Epidemiology and etiology. Cancer. 2010;107:2099–2107. doi: 10.1002/cncr.22233. [DOI] [PubMed] [Google Scholar]
- 7. doi: 10.1016/S0140-6736(16)31012-1. GBD 2015 Mortality and Causes of Death Collaborators: Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980-2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet 388: 1459-1544, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Angrand PO, Vennin C, Le Bourhis X, Adriaenssens E. The role of long non-coding RNAs in genome formatting and expression. Front Genet. 2015;6(165) doi: 10.3389/fgene.2015.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garzon R, Volinia S, Papaioannou D, Nicolet D, Kohlschmidt J, Yan PS, Mrózek K, Bucci D, Carroll AJ, Baer MR, et al. Expression and prognostic impact of lncRNAs in acute myeloid leukemia. Proc Natl Acad Sci USA. 2014;111:18679–18684. doi: 10.1073/pnas.1422050112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hao S, Shao Z. HOTAIR is upregulated in acute myeloid leukemia and that indicates a poor prognosis. Int J Clin Exp Pathol. 2015;8:7223–7228. [PMC free article] [PubMed] [Google Scholar]
- 11.Wu S, Zheng C, Chen S, Cai X, Shi Y, Lin B, Chen Y. Overexpression of long non-coding RNA HOTAIR predicts a poor prognosis in patients with acute myeloid leukemia. Oncol Lett. 2015;10:2410–2414. doi: 10.3892/ol.2015.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun J, Hu J, Li W, Sun Y. A novel suppressive long noncoding RNA within the IGF1R Gene locus is imprinted in acute myelocytic leukemia. Blood. 2014;124(3592) doi: 10.1093/nar/gku549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao TF, Jia HZ, Zhang ZZ, Zhao XS, Zou YF, Zhang W, Wan J, Chen XF. LncRNA H19 regulates ID2 expression through competitive binding to hsa-miR-19a/b in acute myelocytic leukemia. Mol Med Rep. 2017;16:3687–3693. doi: 10.3892/mmr.2017.7029. [DOI] [PubMed] [Google Scholar]
- 14.Nie Y, Zhou L, Wang H, Chen N, Jia L, Wang C, Wang Y, Chen J, Wen X, Niu C, et al. Profiling the epigenetic interplay of lncRNA RUNXOR and oncogenic RUNX1 in breast cancer cells by gene in situ cis-activation. Am J Cancer Res. 2019;9:1635–1649. [PMC free article] [PubMed] [Google Scholar]
- 15.Wang H, Guo R, Hoffman AR, Hu J, Li W. Effect of long noncoding RNA RUNXOR on the epigenetic regulation of RUNX1 in acute myelocytic leukemia. J Brazilian Soc Mechanical Sci Engineering. 2015;25:341–346. [Google Scholar]
- 16.Wang F, Tian X, Zhou J, Wang G, Yu W, Li Z, Fan Z, Zhang W, Liang A. A three-lncRNA signature for prognosis prediction of acute myeloid leukemia in patients. Mol Med Rep. 2018;18:1473–1484. doi: 10.3892/mmr.2018.9139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee SH, Chiu YC, Li YH, Lin CC, Hou HA, Chou WC, Tien HF. High expression of dedicator of cytokinesis 1 (DOCK1) confers poor prognosis in acute myeloid leukemia. Oncotarget. 2017;8:72250–72259. doi: 10.18632/oncotarget.19706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(R106) doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson M, Mccarthy D, Smyth GK. edgeR: Differential expression analysis of digital gene expression data. J Hospice Palliative Nurs. 2010;4:206–207. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen LP, Wang H, Zhang Y, Chen QX, Lin TS, Liu ZQ, Zhou YY. Robust analysis of novel mRNA-lncRNA cross talk based on ceRNA hypothesis uncovers carcinogenic mechanism and promotes diagnostic accuracy in esophageal cancer. Cancer Manag Res. 2018;11:347–358. doi: 10.2147/CMAR.S183310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perron AM, Girolimetti G, Procaccini M, Marchio L, Livi A, Borghese G, Porcelli AM, De Iaco P, Gasparre G. Potential for mitochondrial DNA sequencing in the differential diagnosis of gynaecological malignancies. Int J Mol Sci. 2018;19(pii: E2048) doi: 10.3390/ijms19072048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He JH, Han ZP, Zou MX, Wang L, Lv YB, Zhou JB, Cao MR, Li YG. Analyzing the LncRNA, miRNA, and mRNA regulatory Network in prostate cancer with Bioinformatics Software. J Comput Biol. 2018;25:146–157. doi: 10.1089/cmb.2016.0093. [DOI] [PubMed] [Google Scholar]
- 23.Wang L, Cao C, Ma Q, Zeng Q, Wang H, Cheng Z, Zhu G, Qi J, Ma H, Nian H, Wang Y. RNA-seq analyses of multiple meristems of soybean: Novel and alternative transcripts, evolutionary and functional implications. BMC Plant Biol. 2014;14(169) doi: 10.1186/1471-2229-14-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang P, Wang Y, Hang B, Zou X, Mao JH. A novel gene expression-based prognostic scoring system to predict survival in gastric cancer. Oncotarget. 2016;7:55343–55351. doi: 10.18632/oncotarget.10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goeman JJ. L1 penalized estimation in the Cox proportional hazards model. Biom J. 2010;52:70–84. doi: 10.1002/bimj.200900028. [DOI] [PubMed] [Google Scholar]
- 26.Ranstam J, Cook JA. Kaplan-meier curve. Br J Surg. 2017;104(442) doi: 10.1002/bjs.10238. [DOI] [PubMed] [Google Scholar]
- 27.Kaushal B, Jain K, Sharma SK. Estimation of area under receiver operating characteristic curve for Bi-Pareto and Bi-Two parameter exponential models. Open J Statistics. 2014;4:1–10. [Google Scholar]
- 28.Hauke J, Kossowski T. Comparison of values of Pearson's and Spearman's correlation coefficients on the same sets of data. Quaestiones Geographicae. 2011;30:87–93. [Google Scholar]
- 29.Tilford CA, Siemers NO. Gene set enrichment analysis. Methods Mol Biol. 2009;563:99–121. doi: 10.1007/978-1-60761-175-2_6. [DOI] [PubMed] [Google Scholar]
- 30.Servant N, Roméjon J, Gestraud P, La Rosa P, Lucotte G, Lair S, Bernard V, Zeitouni B, Coffin F, Jules-Clément G, et al. Bioinformatics for precision medicine in oncology: Principles and application to the SHIVA clinical trial. Front Genet. 2014;5(152) doi: 10.3389/fgene.2014.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao X, Li Y, Wu H. A novel scoring system for acute myeloid leukemia risk assessment based on the expression levels of six genes. Int J Mol Med. 2018;42:1495–1507. doi: 10.3892/ijmm.2018.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li S, Bian H, Cao Y, Juan C, Cao Q, Zhou G, Fang Y. Identification of novel lncRNAs involved in the pathogenesis of childhood acute lymphoblastic leukemia. Oncol Lett. 2019;17:2081–2090. doi: 10.3892/ol.2018.9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yin X, Huang S, Zhu R, Fan F, Sun C, Hu Y. Identification of long non-coding RNA competing interactions and biological pathways associated with prognosis in pediatric and adolescent cytogenetically normal acute myeloid leukemia. Cancer Cell Int. 2018;18(122) doi: 10.1186/s12935-018-0621-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rossi G, Minervini MM, Carella AM, Melillo L and Cascavilla N: Wilms' tumor gene (WT1) expression and minimal residual disease in acute myeloid leukemia. In: van den Heuvel-Eibrink MM (ed), Wilms Tumor, Brisbane (AU), Codon Publications, 2016. [PubMed] [Google Scholar]
- 35.Siehl JM, Thiel E, Heufelder K, Snarski E, Schwartz S, Mailänder V, Keilholz U. Possible regulation of Wilms' tumour gene 1 (WT1) expression by the paired box genes PAX2 and PAX8 and by the haematopoietic transcription factor GATA-1 in human acute myeloid leukaemias. Br J Haematol. 2003;123:235–242. doi: 10.1046/j.1365-2141.2003.04622.x. [DOI] [PubMed] [Google Scholar]
- 36.Han J, Zhou W, Jia M, Wen J, Jiang J, Shi J, Zhang K, Ma H, Liu J, Ren J, et al. Expression quantitative trait loci in long non-coding RNA PAX8-AS1 are associated with decreased risk of cervical cancer. Mol Genet Genomics. 2016;291:1743–1748. doi: 10.1007/s00438-016-1217-9. [DOI] [PubMed] [Google Scholar]
- 37.Bahari G, Hashemi M, Naderi M, Sadeghi-Bojd S, Taheri M. Long non-coding RNA PAX8-AS1 polymorphisms increase the risk of childhood acute lymphoblastic leukemia. Biomed Rep. 2018;8:184–190. doi: 10.3892/br.2017.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramaswamy K, Forbes L, Minuesa G, Gindin T, Brown F, Kharas MG, Krivtsov AV, Armstrong SA, Still E, de Stanchina E, et al. Peptidomimetic blockade of MYB in acute myeloid leukemia. Nat Commun. 2018;9(110) doi: 10.1038/s41467-017-02618-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lahortiga I, De Keersmaecker K, Van Vlierberghe P, Graux C, Cauwelier B, Lambert F, Mentens N, Beverloo HB, Pieters R, Speleman F, et al. Duplication of the MYB oncogene in T cell acute lymphoblastic leukemia. Nat Genet. 2007;39:593–595. doi: 10.1038/ng2025. [DOI] [PubMed] [Google Scholar]
- 40.Mets E, Van der Meulen J, Van Peer G, Boice M, Mestdagh P, Van de Walle I, Lammens T, Goossens S, De Moerloose B, Benoit Y, et al. MicroRNA-193b-3p acts as a tumor suppressor by targeting the MYB oncogene in T-cell acute lymphoblastic leukemia. Leukemia. 2015;29:798–806. doi: 10.1038/leu.2014.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shuang Y, Ning Q, Zhang G, Hong S, Zhen W, Li Y. Construction of differential mRNA-lncRNA crosstalk networks based on ceRNA hypothesis uncover key roles of lncRNAs implicated in esophageal squamous cell carcinoma. Oncotarget. 2016;7:85728–85740. doi: 10.18632/oncotarget.13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li K, Gao B, Li J, Chen H, Li Y, Wei Y, Gong D, Gao J, Zhang J, Tan W, et al. ZNF32 protects against oxidative stress-induced apoptosis by modulating C1QBP transcription. Oncotarget. 2015;6:38107–38126. doi: 10.18632/oncotarget.5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kai L, Bo G, Jun L, Chen H, Li Y, Wei Y, Gong D, Gao J, Zhang J, Tan W, et al. ZNF32 protects against oxidative stress-induced apoptosis by modulating C1QBP transcription. Oncotarget. 2015;6:38107–38126. doi: 10.18632/oncotarget.5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang L, Wang JH, Liang RX, Huang ST, Xu J, Yuan LJ, Huang L, Zhou Y, Yu XJ, Wu SY, et al. RASSF8 downregulation promotes lymphangiogenesis and metastasis in esophageal squamous cell carcinoma. Oncotarget. 2015;6:34510–34524. doi: 10.18632/oncotarget.5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yeung TL, Leung CS, Wong KK, Gutierrez-Hartmann A, Kwong J, Gershenson DM, Mok SC. ELF3 is a negative regulator of epithelial-mesenchymal transition in ovarian cancer cells. Oncotarget. 2017;8:16951–16963. doi: 10.18632/oncotarget.15208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harburg GC, Hinck L. Navigating breast cancer: Axon guidance molecules as breast cancer tumor suppressors and oncogenes. J Mammary Gland Biol Neoplasia. 2011;16:257–270. doi: 10.1007/s10911-011-9225-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kerjan G, Moreaufauvarque C. The brain within the tumor: New roles for axon guidance molecules in cancers. Cell Death Differ. 2005;12:1044–1056. doi: 10.1038/sj.cdd.4401707. [DOI] [PubMed] [Google Scholar]
- 48.Neufeld G, Mumblat Y, Smolkin T, Toledano S, Nir-Zvi I, Ziv K, Kessler O. The role of the semaphorins in cancer. Cell Adh Migr. 2016;10:652–674. doi: 10.1080/19336918.2016.1197478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rolny C, Capparuccia L, Casazza A, Mazzone M, Vallario A, Cignetti A, Medico E, Carmeliet P, Comoglio PM, Tamagnone L. The tumor suppressor semaphorin 3B triggers a prometastatic program mediated by interleukin 8 and the tumor microenvironment. J Exp Med. 2008;205:1155–1171. doi: 10.1084/jem.20072509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Minke KS, Staib P, Puetter A, Gehrke I, Gandhirajan RK, Schlösser A, Schmitt EK, Hallek M, Kreuzer KA. Small molecule inhibitors of WNT signaling effectively induce apoptosis in acute myeloid leukemia cells. Eur J Haematol. 2010;82:165–175. doi: 10.1111/j.1600-0609.2008.01188.x. [DOI] [PubMed] [Google Scholar]
- 51.Mikesch JH, Steffen B, Berdel WE, Serve H, Müller-Tidow C. The emerging role of Wnt signaling in the pathogenesis of acute myeloid leukemia. Leukemia. 2007;21:1638–1647. doi: 10.1038/sj.leu.2404732. [DOI] [PubMed] [Google Scholar]
- 52.Pehlivan M, Çalışkan C, Yüce Z, Sercan HO. Secreted Wnt antagonists in leukemia: A road yet to be paved. Leuk Res. 2018;69:24–30. doi: 10.1016/j.leukres.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 53.Valencia A, Romángómez J, Cervera J, Such E, Barragán E, Bolufer P, Moscardó F, Sanz GF, Sanz MA. Wnt signaling pathway is epigenetically regulated by methylation of Wnt antagonists in acute myeloid leukemia. Leukemia. 2009;23:1658–1666. doi: 10.1038/leu.2009.86. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data used in this study are available in TCGA database (https://portal.gdc.cancer.gov/).