Abstract
Pancreatic cancer is a severe disease with high morbidity and mortality. However, the primary molecular mechanisms of pancreatic tumor formation and progression remain unclear. The present study using sequencing technology revealed that the centromere protein M (CENPM) gene was overexpressed in pancreatic cancer tissues. CENPM is one of the components of a complex that plays a central role in kinetochore protein assembly, mitotic progression and chromosome segregation. However, the biological function of CENPM in pancreatic cancer has yet to be determined. Hence, two effective siRNAs were designed to knock down CENPM. Notably, downregulation of CENPM inhibited pancreatic cancer cell proliferation, altered the cell cycle and limited pancreatic cancer cell migration and invasion via the mTOR/p70S6K signaling pathway. This research provides new evidence that CENPM overexpression plays a significant role in the progression of pancreatic cancer. Overall, the present findings indicated that CENPM may be a significant biomarker for predicting the development and progression of pancreatic malignancy.
Keywords: pancreatic cancer, CENPM, metastasis, mTOR/p70S6K signaling pathway
Introduction
Pancreatic cancer is the one of the major lethal malignant neoplasms; it has a high incidence rate worldwide, and it can cause long hospital stays and high costs (1). The estimation of the 5-year survival rate for pancreatic cancer is approximately 5% (2). Certain risk factors leading to pancreatic cancer have been identified, such as cigarette smoking, positive family history, genetics, diabetes mellitus, obesity, dietary factors, alcohol use as well as physical inactivity (3).
Over the past several decades, emerging research has focused on identifying the underlying mechanisms of pancreatic cancer (4,5). However, to date, the investigations of the specific mechanisms, are not satisfactory (1,2,6,7). Therefore, there is an urgent need to improve the understanding of pancreatic cancer.
Centromere protein M (CENPM) has been reported as a novel biomarker of hepatocellular carcinoma (8), melanoma (9) and bladder cancer (10). However, the function of CENPM has not been studied in pancreatic cancer. The CENPM gene (11,12) encodes the centromere protein M. CENPM is recruited by CENPA nucleosomes and is assembled into CENPA nucleosome-associated complex (NAC) (13). CENPA NAC plays important roles in kinetochore assembly, mitotic progression regulation and chromatin complex formation (14). As is commonly known, the proliferation of cancer cells is closely related to mitosis and chromosome separation (12). Thus, the CENPM gene was selected as the target of our research.
In the present study, it was revealed that CENPM expression was upregulated in both pancreatic carcinoma and pancreatic ductal adenocarcinoma compared with normal tissues (data from the ‘Pei Pancreas’ and ‘Grutzmann Pancreas’ databases) (15,16). Whether the capacity of migration and invasion of pancreatic cancer would be influenced by CENPM is unknown. Hence, PANC-1 and CFPAC-1 were selected to determine the mechanism of CENPM in the proliferation and metastasis of pancreatic cancer.
Materials and methods
Bioinformatics analyses of CENPM
The expression data for the CENPM gene in pancreatic cancer patients were downloaded from the Oncomine online database (https://www.oncomine.org/). This database is a cancer microarray platform and can provide translational bioinformatics services (17). Oncomine gene expression array datasets include 715 independent datasets and >80,000 samples. The datasets extracted from Oncomine were the Pei Pancreas and Grutzmann Pancreas datasets (15,16). In the Pei Pancreas datasets, there were 16 normal and 36 pancreatic carcinoma samples. In the Grutzmann Pancreas datasets, there were 11 normal and 14 pancreatic ductal adenocarcinoma samples. Patient overall survival analysis and disease-free survival analyses were extracted from Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn/). Thanks to the work of Professor Zefang Tang from Peking University (18), RNA sequencing expression data of almost ten thousand cancers from TCGA and the GTEx datasets can be analyzed using this web tool. In this pancreatic adenocarcinoma dataset, the group cut-off level for CENPM expression was set as 50%. Samples with expression levels higher than this threshold were considered as the high-expression cohort. The hazards ratio (HR) was calculated based on the Cox PH Model. The dotted line shows the 95% confidence interval (CI).
Cell culture
hTERT-HPNE, BxPC-3, CFPAC-1, MIA PaCa-2, PANC-1 and PATU-8988 cell lines were used to perform the experiments in the present study. All cell lines were purchased from the Chinese Academy of Sciences Cell Bank (China). DMEM with 4.5 g/l glucose supplemented with 10% FBS, 100 U/ml penicillin and 100 µg/ml streptomycin (all from Gibco; Thermo Fisher Scientific, Inc.) was used to culture HTERT-HPNE, MIA PaCa-2 and PANC-1 cell lines. BxPC-3, CFPAC-1 and PATU-8988 cell lines were cultured in Roswell Park Memorial Institute (RPMI)-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS, 100 U/ml penicillin and 100 µg/ml streptomycin. Cells were incubated in a standard atmosphere with 5% CO2 and at a temperature of 37°C.
RNA isolation and real-time quantitative polymerase chain reaction (RT-qPCR)
Total RNA from cells was isolated according to the RNAprep cell kit instructions (Tiangen Biotech Co, Ltd.). The purity and concentration of RNA were examined by a nucleic acid protein detector (DeNovix, Inc.). RNA samples were reverse transcribed by a RevertAid First Strand kit (Thermo Fisher Scientific, Inc.). qRT-PCR was performed using a 7500 Fast Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.) with SYBR Green Master (ROX) reagent (Roche Diagnostics). The thermocycling conditions were as follows: Initial denaturation at 95°C for 10 min; 40 of cycles of denaturation for 15 sec at 95°C, annealing for 1 min at 60°C and elongation for 1 min at 72°C; and final extension for 10 min at 72°C. GAPDH was used for the normalization of CENPM expression levels. The sequences of the GAPDH primers used in the present study were forward 5′-GGACCTGACCTGCCGTCTAG-3′ and reverse 5′-GTAGCCCAGGATGCCCTTGA-3′. The sequences of the CENPM primers used in the present study were forward 5′-CTGGCGGACTCGATGCTCAAAG-3′ and reverse 5′-CGATTCACACTGGAGGGCAAAGG-3′. The results are presented with CT values and were analyzed by the ΔΔCq method (19). Each sample was performed in triplicate.
Cells transfection
Small interfering RNA (siRNA) for CENPM was designed and generated by Shanghai GenePharma Co., Ltd. The sequences of the negative controls (si-NC) were as follows: Sense, 5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense, 5′-ACGUGACACGUUCGGAGAATT-3′. The sequences of the CENPM siRNAs were as follows: CENPM-homo-172 (si-RNA1): Sense, 5′-GGACUCGAUGCUCAAAGAGTT-3′ and antisense, 5′-CUCUUUGAGCAUCGAGUCCTT-3′; and CENPM-homo-206 (si-RNA2): Sense, 5′-CUGAAGGUCCACUUGGCAATT-3′ and antisense, 5′-UUGCCAAGUGGACCUUCAGTT-3′. Cell interference was performed according to the Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) protocol. Approximatelly 1×105 cells of the PANC-1 and CFPAC-1 cell lines were seeded into 6-well plates and cultured until they reached to 40–50% confluence. Lipofectamine 3000 reagent and siRNA were diluted in DMEM or RPMI-1640 medium and then mixed at a ratio of 1:1, and subsequently they were incubated at 37°C together for 15 min. The siRNA concentration of CENPM at 100 nmol/l was added to each well. Forty-eight hours later, cells were harvested for subsequent experiments. Each transfection was performed in triplicate.
Cell proliferation assay
Forty-eight hours after the siRNA transfection, approximately 5,000 cells/well of PANC-1 and CFPAC-1 cells were seeded into 96-well plates according to the Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.) protocol. Cell proliferation assays were detected at 0, 24, 48, 72 and 96 h by a Varioskan Flash multimode reader (Thermo Fisher Scientific, Inc.). Approximately 500 cells/well of siRNA-transfected PANC-1 and CFPAC-1 cells were seeded into 6-well plates and cultured with 10% FBS medium. Cells were fixed with 4% paraformaldehyde at 4°C (Solarbio Life Sciences) for 20 min when the colonies were evident enough to be observed macroscopically. Colonies (consisting of at least 50 cells) were observed by a light microscope at an ×100 magnification. The cells were then stained with 0.4% crystal violet (Beyotime Institute of Biotechnology) at 37°C for 20 min. Images of each well were captured using a camera (Canon). Each experiment was performed in triplicate.
Cell cycle assay
si-NC and si-CENPM cells obtained were washed twice using ice-cold PBS and were fixed in 70% ethanol at −20°C overnight. Then, the cells were washed again and resuspended in 0.1 ml of staining buffer (FBS) (BD Pharmingen; BD Biosciences). After washing the cells, 5 µl of 7-AAD (50 µg/ml) (BD Biosciences) was added into the cell suspension and incubated in a dark room for 10 min at 37°C. Finally, a cell cycle assay was performed using a flow cytometer (BD Accuri C6; BD Biosciences). The analysis software used was ModFit version 5 (Verity Software House, Inc.).
Cell migration and invasion assays
Approximately 5×104 cells/well of si-NC or si-CENPM cells were seeded into Transwell chambers (with pore size inserts of 8 µm) (Corning, Inc.). Due to the different migration capacities of cells, PANC-1 cells migrated for 1 day, while CFPAC-1 cells migrated for 3 days. In the invasion experiments, Matrigel (Corning, Inc.) was diluted with DMEM or RPMI-1640 medium at a ratio of 1:8 followed by incubation in the chambers overnight. The cells were cultured in the upper chambers and were then allowed to invade for 4 days for PANC-1 cells and 8 days for CFPAC-1 cells. All of the lower chambers were filled with 600 ml of DMEM or RPMI-1640 medium supplemented with 20% FBS. Upon completion of migration or invasion, the chambers were fixed with 4% paraformaldehyde at 4°C for 20 min and stained with 0.4% crystal violet at 37°C for 20 min carefully. Images were observed by light microscope at an ×200 magnification, and five fields were randomly selected for analysis. The analysis software uesd was VisionWorks version 8 (UVP, Inc.).
Protein extraction and western blot analysis
Seventy-two hours after siRNA transfection, cells were collected and lysed in RIPA buffer (Beyotime Institute of Biotechnology) supplemented with phenylmethanesulfonyl fluoride (Sigma-Aldrich; Merck KGaA), PhosSTOP (Roche Diagnostics) and DL-dithiothreitol (Beyotime Institute of Biotechnology). Cell protein concentrations were measured using a BCA protein assay kit (Beyotime Institute of Biotechnology) and equilibrated with PBS and loading buffer. A total of 50 µg boiled protein per lane was separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to a 0.45-µm polyvinylidene difluoride (PVDF) membrane (EMD Millipore). The membranes were incubated with 5% nonfat milk in Tris-buffered saline (TBS), which contained 0.05% Tween-20, for 1 h at room temperature. Each membrane was probed with various antibodies overnight at 4°C. The following primary antibodies were used: Anti-cyclin-dependent kinase (CDK)2 [product no. 18048S; Cell Signaling Technology (CST), Inc.], anti-CDK6 (product no. 3136S; CST), anti-cyclin D1 (product code ab134175; Abcam), anti-p21 (product no. 2947S; CST), anti-tubulin (product no. 2148S; CST), anti-CENPM (cat. no. DF2314; Affinity), anti-mammalian target of rapamycin (mTOR) (product code ab2732; Abcam), anti-phosphorylated (p)-mTOR (Ser2448) (product no. 5536S; CST), anti-p70S6K (cat. no. 14485; ProteinTech Group, Inc.), anti-p-p70S6K (Thr421/Ser424) (product no. 9204S; CST), and anti-GAPDH (product no. 2118S; CST). All the antibodies were used at a dilution of 1:1,000. Then, the membranes were incubated with HRP AffiniPure goat anti-rabbit IgG (product no. BL003A; Biosharp) (1:5,000) for 1 h at room temperature the following day. Protein bands were visualized using Amersham Imager 600 (General Electric Company) and enhanced with chemiluminescence (Thermo Fisher Scientific, Inc.). The analysis software used was VisionWorks version 8.
Statistical analysis
All data were obtained from at least three independent experiments. Data are presented as the mean values ± SEM and were performed using t-tests or one-way ANOVA. Multiple comparisons among the groups were performed using Sidak correction method. P-values <0.05 were considered to indicate a statistically significant difference. Statistical analyses were achieved using SPSS 23.0 software (IBM, Corp.) and GraphPad Prism version 7.01 (GraphPad Software, Inc.).
Results
CENPM is upregulated in pancreatic cancer and associated with the survival rate
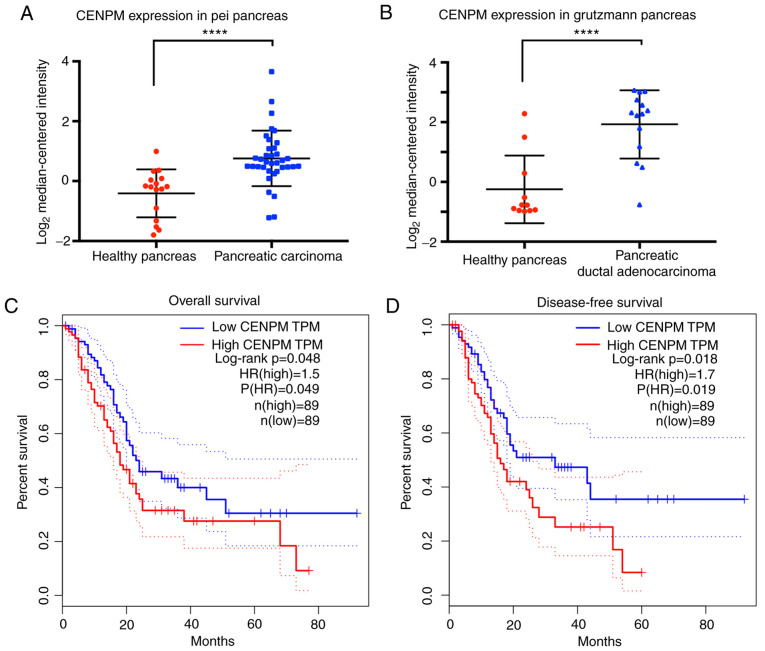
The data extracted from the Pei Pancreas database and the Grutzmann Pancreas database (15,16) revealed that the gene CENPM was significantly upregulated (P<0.0001) in 50 cases of cancer patients with pancreatic carcinoma or pancreatic ductal adenocarcinoma compared with 27 healthy cases (Fig. 1A and B).
Figure 1.
CENPM is upregulated in pancreatic cancer. (A and B) CENPM expression in pancreatic cancer is higher than in healthy tissue as determined using the ‘Pei Pancreas database’ and ‘Grutzmann Pancreas database’. (C) The overall survival analysis and (D) disease-free survival analysis revealed that patients with high expression of CENPM exhibited shorter survival time. ****P<0.0001 vs. healthy pancreas. CENPM, centromere protein M.
Overall survival and disease-free survival analyses (Fig. 1C and D) extracted from GEPIA revealed that patients with high CENPM expression presented decreased survival percentages (P<0.05). An HR value over 1.0 indicated that CENPM was positively associated to pancreatic cancer and suggested that the CENPM gene may act as a potential cancer therapeutic target in pancreatic cancer treatment.
CENPM expression levels in pancreatic cancer cell lines
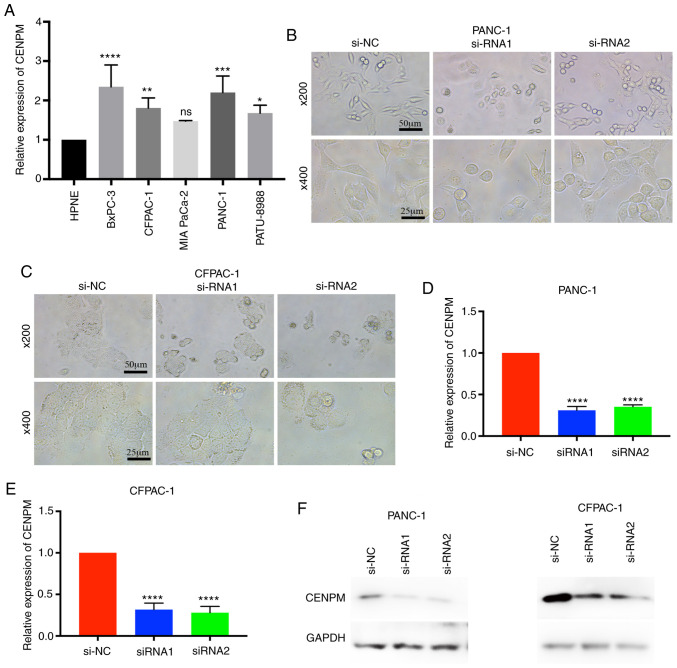
To determine whether CENPM was overexpressed in pancreatic cancer cell lines, the relative expression level of CENPM among various pancreatic cancer cell lines vs. the human pancreatic nestin-expressing (hTERT-HPNE) cell line were quantified. The data revealed that CENPM was overexpressed in BxPC-3 (P<0.0001), CFPAC-1 (P<0.001), PANC-1 (P<0.0005) and PATU-8988 (P<0.05) compared with hTERT-HPNE (Fig. 2A). In the present study, PANC-1 and CFPAC-1 were selected for further research due to their relatively higher expression levels and easy culture. Two effective siRNAs were transfected to interfere in the expression of CENPM in these two cell lines for subsequent experiments. With regard to the cell morphologies of PANC-1 and CFPAC-1 cells it was observed that the cells became round and underwent cell shrinkage when CENPM was knocked down by siRNA (Fig. 2B and C). The knockdown effect of the siRNA is presented in Fig. 2D-F. The inhibition levels of si-RNA1 and si-RNA2 in PANC-1 cells were 0.6908±0.01985 and 0.6486±0.02292 relative to si-NC (P<0.0001). The inhibition levels of si-RNA1 and si-RNA2 in CFPAC-1 were 0.6834±0.02452 and 0.7205±0.03211 relative to si-NC (P<0.0001).
Figure 2.
Expression level of CENPM in pancreatic cancer cell lines. (A) The relative expression levels of CENPM in pancreatic cancer cell lines were increased compared to the HPNE cell line. *P<0.05, **P<0.001, ***P<0.0005, ****P<0.0001 compared to HPNE. (B and C) The cell morphologies of PANC-1 and CFPAC-1 were altered when CENPM was knocked down by si-RNAs. (D-F) The effect of CENPM-knockdown by si-RNA in PANC-1 and CFPAC-1 cells. The data were obtained from at least three independent experiments. ****P<0.0001 compared to si-NC. CENPM, centromere protein M; si-RNA, small interfering RNA.
CENPM gene knockdown inhibits pancreatic cancer cell proliferation
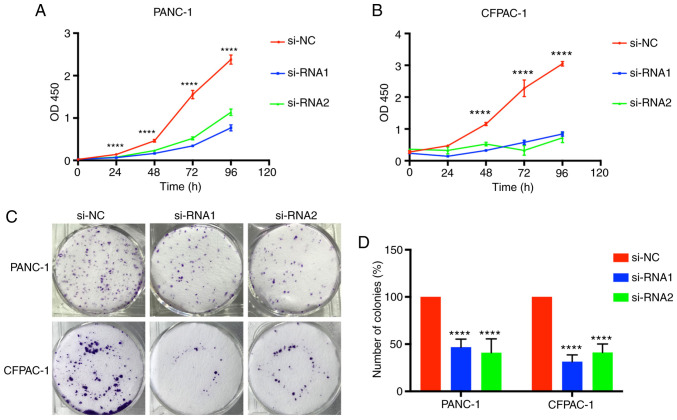
Tumor growth was associated with limitless proliferation of tumor cells; therefore, considering the function of CENPM in tumor cell proliferation was valuable. Time-dependent proliferation of cells was calculated by CCK-8 assay, and the values were analyzed at OD450 nm (Fig. 3A and B). Colony formation assays were also performed to detect cell proliferation (Fig. 3C and D). These results indicated that cell lines transfected with si-RNA1 and si-RNA2 had significantly lower values (P<0.0001), demonstrating that downregulation of CENPM could effectively inhibit PANC-1 and CFPAC-1 proliferation compared with the si-NC group.
Figure 3.
CENPM gene-knockdown inhibits pancreatic cancer cell proliferation. (A and B) PANC-1 and CFPAC-1 cells were cultured for 4 days and the OD450 nm values were detected using CCK-8 assays. (C) PANC-1 and CFPAC-1 colony formation assays. (D) Statistical analysis of the number of colonies. The data were obtained from at least three independent experiments. ****P<0.0001 compared to si-NC. CENPM, centromere protein M; si-RNA, small interfering RNA; CCK-8, Cell Counting Kit-8.
CENPM gene knockdown affects the cell cycle in pancreatic cancer cells
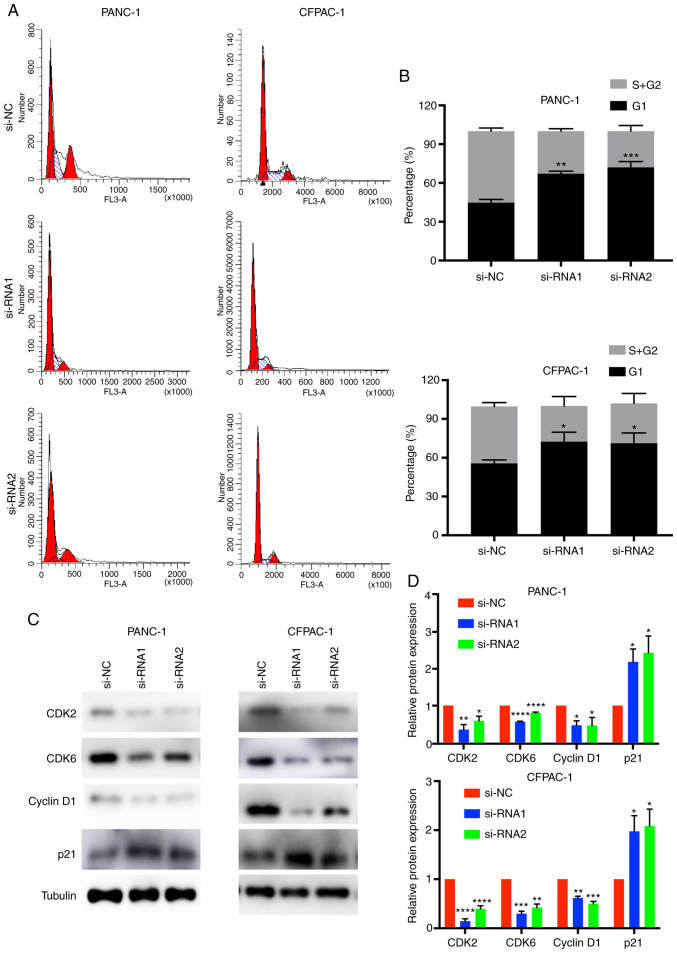
In order to better understand the effect of CENPM in pancreatic cancer cells, cell cycle analysis was performed (Fig. 4A and B). The results of flow cytometric analysis revealed that the percentages of cells in the G0/G1 phase of the CENPM-knockdown PANC-1 and CFPAC-1 cells were higher than in the si-NC groups (P<0.05). In addition, the percentages of the S+G2/M phase were decreased in the siRNA groups (P<0.05).
Figure 4.
Downregulation of CENPM affects the cell cycle in pancreatic cancer cells. (A and B) Flow cytometric analysis of the cell cycle in PANC-1 and CFPAC-1 cells. (C and D) The expression levels of CDK2, CDK6, cyclin D1 and p21 in PANC-1 and CFPAC-1 cells. The data were obtained from at least three independent experiments. *P<0.05, **P<0.001, ***P<0.0005, ****P<0.0001 compared to si-NC. CENPM, centromere protein M; si-RNA, small interfering RNA; CDK, cyclin-dependent kinase.
CDKs are proteins involved in the cell cycle of tumor cells (20). Cyclin D1 is a cell cycle-related protein. p21 is a type of cyclin-dependent kinase inhibitor (21). CDK2, CDK6, cyclin D1 and p21 were detected by western blotting (Fig. 4C and D). The results revealed that knockdown of CENPM decreased the expression levels of CDK2 (P<0.05), CDK6 (P<0.001), cyclin D1 (P<0.05) and increase the expression levels of p21 (P<0.05). Collectively, low expression of CENPM induced cell cycle arrest at the G1 phase in pancreatic tumor cells.
Downregulation of CENPM inhibits pancreatic cancer cell migration and invasion
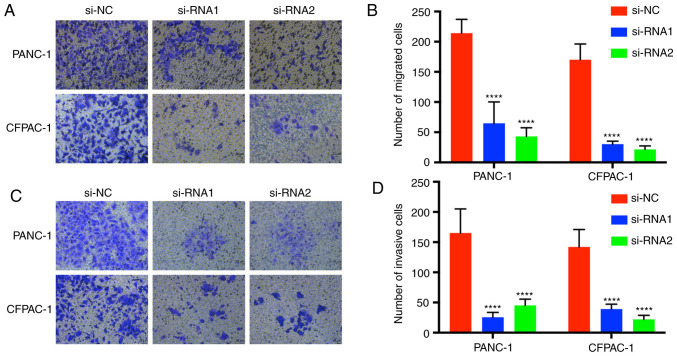
Transwell assays were performed to demonstrate the migration and invasion function of CENPM in pancreatic tumor cells (Fig. 5). Cells with migration capacity can move from the upper chambers without FBS to the lower chambers with a high concentration of FBS. The present results revealed that knockdown of CENPM could efficiently hinder the migration capacity of PANC-1 and CFPAC-1 cells compared with the si-NC group (P<0.0001). The invasion assays also revealed a similar trend.
Figure 5.
Downregulation of CENPM inhibits pancreatic cancer cell migration and invasion. (A and C) Migration and invasion abilities of PANC-1 and CFPAC-1 cells with knockdown of CENPM in pancreatic tumor cells. Statistical analysis of the number of (B) migrated cells and (D) invasive cells. The data were obtained from at least three independent experiments. ****P<0.0001 compared to si-NC. CENPM, centromere protein M; si-RNA, small interfering RNA.
CENPM function may be regulated via the mTOR/p70S6K signaling pathway
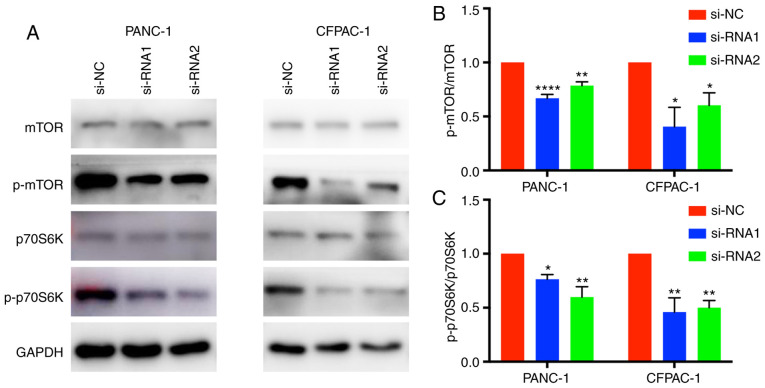
Western blotting was applied to elucidate the mechanism by which CENPM regulates the migration and invasion capacities of tumor cells. The phosphorylation levels of p70S6K and mTOR were significantly reduced when CENPM was knocked down by si-RNA-1 and si-RNA-2 (Fig. 6). The mTOR/p70S6K pathway had an important role in cancer cell metastasis, which may influence cell migration and invasion. These results indicated that downregulation of CENPM inhibited phosphorylation of the mTOR/p70S6K pathway thus reducing the malignant degree of pancreatic tumors.
Figure 6.
CENPM function may be regulated via the mTOR/p70S6K signaling pathway. (A) The phosphorylation levels of p70S6K and mTOR were significantly reduced when CENPM was knocked down by si-RNA1 and si-RNA2. (B and C) Densitometric analysis of the protein bands in A. The data were obtained from at least three independent experiments. *P<0.05, **P<0.001, ****P<0.0001 compared to si-NC. CENPM, centromere protein M; si-RNA, small interfering RNA; mTOR, mammalian target of rapamycin.
Discussion
Pancreatic carcinoma, a fatal disease with high morbidity, has long been a tribulation for patients and doctors (1). Pancreatic cancer is the seventh cause of tumor-induced deaths and has a high mortality/incidence ratio of up to 98% worldwide (3). In addition, its 5% five-year survival rate makes this cancer even more serious (2,3).
The pathogenesis of pancreatic cancer has been studied for several years, identifying contributing factors such as smoking, heavy alcohol intake, high BMI, type II diabetes and chronic pancreatitis (22). Unfortunately, once pancreatic cancer is diagnosed, the only treatment to cure the cancer is surgery. Further compounded, a locally resectable tumor can result in post-surgery recurrence (7).
Due to the limited options for therapy with pancreatic cancer, it is imperative to explore new targeted therapies and to provide different directions for clinical treatment. Currently, gene editing technology is considered as a curable treatment for pancreatic cancer (23,24).
The kinetochore is a surface structure on the centromeric region of DNA and can be observed during every cell cycle mitosis. The function of the kinetochore is to ensure the correct segregation of chromosomes. CENPA is a leading marker in the assembly of CENPA NAC, which is the beginning of kinetochore construction. CENPM, along with CENPH, CENPC, CENPN, CENPU/50 and CENPT, is physically closer to the CENPA nucleosome compared to CENPA NAC (11). CENPM is highly expressed in proliferating cells, such as activated lymphoid cells and tumor cells (25). It is also known as proliferation-associated nuclear element 1 (PANE1) (25,26). Moreover, mitotic aberrations and aneuploidy have been reported in CENPM-deficient cells (13,14,27).
Recently, Yu et al defined CENPM as a complement to AFP in the diagnosis of hepatocellular carcinoma (8); Xiao et al further confirmed that CENPM was highly associated with hepatocellular carcinoma progression and could be a candidate novel biomarker for hepatocellular carcinoma (28). Investigations of the underlying mechanisms of melanoma highlighted that CENPM may play an important role in the metastases of melanoma (9). Chen et al suggested that CENPM may contribute to bladder cancer development and cancer recurrence (12). In fact, bladder cancer patients with high CENPM expression had markedly shorter progression-free survival than those with low expression (10).
However, the mechanism between CENPM and pancreatic carcinoma has not been explored. The present study aimed to determine the function and mechanism of CENPM in the proliferation and metastasis of pancreatic carcinoma.
CENPM, identified as ‘pseudo G-protein’, is structurally related to Rab-family GTPases (29). The Rab GTPase family-associated factors have been recognized as major regulators of the activity of signaling pathways regulating cell growth and survival (30). A study identified Rab as promoting oncogenesis by direct interaction with mTOR, resulting in activation of the pathway (31).
mTOR (32) is a protein that can influence cell growth via the stimulation of amino acids, insulin, IGF-1, and ATP among other factors (33). Once mTOR is activated, mTORC1 can regulate proteins, including ribosomal protein p70S6 kinases (S6Ks) (34). p70S6K1 and p70S6K2 share a large proportion of their kinase domains. p70S6K1, with 502 amino acids, has been investigated more than p70S6K2. p70S6K has been revealed to be highly expressed and activated in several breast cancer cells (35). It has also been revealed to play crucial roles in tumor angiogenesis, cellular proliferation, migration and motility metastasis (36–38). Moreover, the mTOR/p70S6K signaling pathway was revealed to suppress autophagy through phosphorylation of transcription factor EB and restrain the expression of autophagy genes (39,40). Holz et al revealed that p70S6K can directly regulate the phosphorylation of mTOR at threonine 2446/serine 2448, which is a portion of the regulatory repressor domain (41). In addition, the mTOR/S6K signaling pathway has been demonstrated to be essential in several diseases, such as cancer, diabetes and obesity (32).
In the present study, the CENPM gene was revealed to be regulated in both pancreatic carcinoma and pancreatic ductal adenocarcinoma compared with healthy tissues. The results revealed that cells with low expression of CENPM could significantly inhibit pancreatic cancer cell proliferation, cause cell cycle arrest at the G1 phase and hinder pancreatic cancer cell migration and invasion. Among total cellular protein, the expression of p-p70S6K and p-mTOR was reduced due to low CENPM levels. This indicated that the mTOR/p70S6K signaling pathway may be the underlying mechanism of CENPM. CENPM may regulate the activity of the kinase by interacting with the kinase domain and play an important role in the phosphorylation of mTOR and p70S6K. However, the specific molecular mechanisms remain unclear. Various cancer-related signaling pathways, such as JAK/STAT (42), PI3K/Akt (43), JNK (44), Wnt (45), and MAPK/ERK (46), have been revealed to play pivotal roles in tumor cell invasion and migration. The effect of CENPM in the aforementioned pathways will be explored in future studies.
Although the biological function of CENPM in pancreatic cancer was elucidated, the present study has some limitations. First, in vivo experiments should be performed to confirm the present conclusions. Next, clinical data, such as serum or tissue from pancreatic cancer resections, need to be collected for further research. In conclusion, it suggested that CENPM may positively affect the tumorigenesis of pancreatic tumors and could become a potential marker and a target for gene therapy in pancreatic cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
CZ performed most of the experiments of this study. TZ and DL designed the work as well as acquired, analyzed and interpreted the data. CH and HT performed some of the experiments. XN conceived the study and performed some of the experiments. BC contributed to the writing of the manuscript and revised it critically for intellectual content. All authors read and approved the final version of the manuscript and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ducreux M, Seufferlein T, Van Laethem JL, Laurent-Puig P, Smolenschi C, Malka D, Boige V, Hollebecque A, Conroy T. Systemic treatment of pancreatic cancer revisited. Semin Oncol. 2019;46:28–38. doi: 10.1053/j.seminoncol.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 2.Ilic M, Ilic I. Epidemiology of pancreatic cancer. World J Gastroenterol. 2016;22:9694–9705. doi: 10.3748/wjg.v22.i44.9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGuigan A, Kelly P, Turkington RC, Jones C, Coleman HG, McCain RS. Pancreatic cancer: A review of clinical diagnosis, epidemiology, treatment and outcomes. World J Gastroenterol. 2018;24:4846–4861. doi: 10.3748/wjg.v24.i43.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J, Wang B, Ren H, Chen W. miR-9-5p inhibits pancreatic cancer cell proliferation, invasion and glutamine metabolism by targeting GOT1. Biochem Biophys Res Commun. 2019;509:241–248. doi: 10.1016/j.bbrc.2018.12.114. [DOI] [PubMed] [Google Scholar]
- 5.Liang J, Liu Y, Zhang L, Tan J, Li E, Li F. Overexpression of microRNA-519d-3p suppressed the growth of pancreatic cancer cells by inhibiting ribosomal protein S15A-mediated Wnt/beta-catenin signaling. Chem Biol Interact. 2019;304:1–9. doi: 10.1016/j.cbi.2019.02.026. [DOI] [PubMed] [Google Scholar]
- 6.Lin QJ, Yang F, Jin C, Fu DL. Current status and progress of pancreatic cancer in China. World J Gastroenterol. 2015;21:7988–8003. doi: 10.3748/wjg.v21.i26.7988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goess R, Friess H. A look at the progress of treating pancreatic cancer over the past 20 years. Expert Rev Anticancer Ther. 2018;18:295–304. doi: 10.1080/14737140.2018.1428093. [DOI] [PubMed] [Google Scholar]
- 8.Yu Z, Wang R, Chen F, Wang J, Huang X. Five novel oncogenic signatures could be utilized as AFP-related diagnostic biomarkers for hepatocellular carcinoma based on next-generation sequencing. Dig Dis Sci. 2018;63:945–957. doi: 10.1007/s10620-018-4961-3. [DOI] [PubMed] [Google Scholar]
- 9.Chen J, Wu F, Shi Y, Yang D, Xu M, Lai Y, Liu Y. Identification of key candidate genes involved in melanoma metastasis. Mol Med Rep. 2019;20:903–914. doi: 10.3892/mmr.2019.10314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim WT, Seo SP, Byun YJ, Kang HW, Kim YJ, Lee SC, Jeong P, Song HJ, Choe SY, Kim DJ, et al. The anticancer effects of garlic extracts on bladder cancer compared to cisplatin: A common mechanism of action via centromere protein M. Am J Chin Med. 2018;46:689–705. doi: 10.1142/S0192415X18500362. [DOI] [PubMed] [Google Scholar]
- 11.Perpelescu M, Fukagawa T. The ABCs of CENPs. Chromosoma. 2011;120:425–446. doi: 10.1007/s00412-011-0330-0. [DOI] [PubMed] [Google Scholar]
- 12.Chen Q, Hu J, Deng J, Fu B, Guo J. Bioinformatics analysis identified key molecular changes in bladder cancer development and recurrence. Biomed Res Int. 2019;2019:3917982. doi: 10.1155/2019/3917982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR, III, Cleveland DW. The human CENP-A centromeric nucleosome-sassociated complex. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 14.Izuta H, Ikeno M, Suzuki N, Tomonaga T, Nozaki N, Obuse C, Kisu Y, Goshima N, Nomura F, Nomura N, Yoda K. Comprehensive analysis of the ICEN (Interphase Centromere Complex) components enriched in the CENP-A chromatin of human cells. Genes Cells. 2006;11:673–684. doi: 10.1111/j.1365-2443.2006.00969.x. [DOI] [PubMed] [Google Scholar]
- 15.Grützmann R, Pilarsky C, Ammerpohl O, Lüttges J, Böhme A, Sipos B, Foerder M, Alldinger I, Jahnke B, Schackert HK, et al. Gene expression profiling of microdissected pancreatic ductal carcinomas using high-density DNA microarrays. Neoplasia. 2004;6:611–622. doi: 10.1593/neo.04295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pei H, Li L, Fridley BL, Jenkins GD, Kalari KR, Lingle W, Petersen G, Lou Z, Wang L. FKBP51 affects cancer cell response to chemotherapy by negatively regulating Akt. Cancer Cell. 2009;16:259–266. doi: 10.1016/j.ccr.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daniel R, Rhodes JY, Arul M. Chinnaiyan, ONCOMINE-A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Malumbres M. Cyclin-dependent kinases. Genome Biol. 2014;15(122) doi: 10.1186/gb4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park EY, Woo Y, Kim SJ, Kim DH, Lee EK, De U, Kim KS, Lee J, Jung JH, Ha KT, et al. Anticancer effects of a new SIRT inhibitor, MHY2256, against Human Breast Cancer MCF-7 cells via regulation of MDM2-p53 binding. Int J Biolo Sci. 2016;12:1555–1567. doi: 10.7150/ijbs.13833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bosetti C, Bertuccio P, Negri E, La Vecchia C, Zeegers MP, Boffetta P. Pancreatic cancer: Overview of descriptive epidemiology. Mol Carcinog. 2012;51:3–13. doi: 10.1002/mc.20785. [DOI] [PubMed] [Google Scholar]
- 23.Chu QD, Sun G, Pope M, Luraguiz N, Curiel DT, Kim R, Li BD, Mathis JM. Virotherapy using a novel chimeric oncolytic adenovirus prolongs survival in a human pancreatic cancer xenograft model. Surgery. 2012;152:441–448. doi: 10.1016/j.surg.2012.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamamoto Y, Hiraoka N, Goto N, Rin Y, Miura K, Narumi K, Uchida H, Tagawa M, Aoki K. A targeting ligand enhances infectivity and cytotoxicity of an oncolytic adenovirus in human pancreatic cancer tissues. J Control Release. 2014;192:284–293. doi: 10.1016/j.jconrel.2014.07.053. [DOI] [PubMed] [Google Scholar]
- 25.Bierie B, Edwin M, Melenhorst JJ, Hennighausen L. The proliferation associated nuclear element (PANE1) is conserved between mammals and fish and preferentially expressed in activated lymphoid cells. Gene Expr Patterns. 2004;4:389–395. doi: 10.1016/j.modgep.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 26.Renou JP, Bierie B, Miyoshi K, Cui Y, Djiane J, Reichenstein M, Shani M, Hennighausen L. Identification of genes differentially expressed in mouse mammary epithelium transformed by an activated beta-catenin. Oncogene. 2003;22:4594–4610. doi: 10.1038/sj.onc.1206596. [DOI] [PubMed] [Google Scholar]
- 27.Okada M, Cheeseman IM, Hori T, Okawa K, McLeod IX, Yates JR, III, Desai A, Fukagawa T. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat Cell Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- 28.Xiao Y, Najeeb RM, Ma D, Yang K, Zhong Q, Liu Q. Upregulation of CENPM promotes hepatocarcinogenesis through mutiple mechanisms. J Exp Clin Cancer Res. 2019;38:458. doi: 10.1186/s13046-019-1444-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Basilico F, Maffini S, Weir JR, Prumbaum D, Rojas AM, Zimniak T, De Antoni A, Jeganathan S, Voss B, van Gerwen S, et al. The pseudo GTPase CENP-M drives human kinetochore assembly. Elife. 2014;3:e02978. doi: 10.7554/eLife.02978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gopal Krishnan PD, Golden E, Woodward EA, Pavlos NJ, Blancafort P. Rab GTPases: Emerging oncogenes and tumor suppressive regulators for the editing of survival pathways in cancer. Cancers (Basel) 2020;12:259. doi: 10.3390/cancers12020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomas JD, Zhang YJ, Wei YH, Cho JH, Morris LE, Wang HY, Zheng XF. Rab1A is an MTORC1 activator and a colorectal oncogene. Cancer Cell. 2014;26:754–769. doi: 10.1016/j.ccell.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Magnuson B, Ekim B, Fingar DC. Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem J. 2012;441:1–21. doi: 10.1042/BJ20110892. [DOI] [PubMed] [Google Scholar]
- 33.Tavares MR, Pavan IC, Amaral CL, Meneguello L, Luchessi AD, Simabuco FM. The S6K protein family in health and disease. Life Sci. 2015;131:1–10. doi: 10.1016/j.lfs.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 34.Hall MN. mTOR-what does it do? Transplant Proc. 2008;40(Suppl 10):S5–S8. doi: 10.1016/j.transproceed.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 35.Holz MK, Blenis J. Identification of S6 kinase 1 as a novel mammalian target of rapamycin (mTOR)-phosphorylating kinase. J Biol Chem. 2005;280:26089–26093. doi: 10.1074/jbc.M504045200. [DOI] [PubMed] [Google Scholar]
- 36.Pende M, Um SH, Mieulet V, Sticker M, Goss VL, Mestan J, Mueller M, Fumagalli S, Kozma SC, Thomas G. S6K1(−/-)/S6K2(−/-) mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol Cell Biol. 2004;24:3112–3124. doi: 10.1128/MCB.24.8.3112-3124.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skinner HD, Zhong XS, Gao N, Shi X, Jiang BH. Arsenite induces p70S6K1 activation and HIF-1alpha expression in prostate cancer cells. Mol Cell Biochem. 2004;255:19–23. doi: 10.1023/B:MCBI.0000007257.67733.3b. [DOI] [PubMed] [Google Scholar]
- 38.Athamneh K, Alneyadi A, Alsamri H, Alrashedi A, Palakott A, El-Tarabily KA, Eid AH, Al Dhaheri Y, Iratni R. Origanum majorana essential Oil triggers p38 MAPK-mediated protective autophagy, apoptosis, and caspase-dependent cleavage of P70S6K in colorectal cancer cells. Biomolecules. 2020;10:412. doi: 10.3390/biom10030412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J, Ren Y, Hou Y, Zhang C, Wang B, Li X, Sun R, Liu J. Dihydroartemisinin induces endothelial cell autophagy through suppression of the Akt/mTOR pathway. J Cancer. 2019;10:6057–6064. doi: 10.7150/jca.33704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8:913–914. doi: 10.4161/auto.19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holz MK, Ballif BA, Gygi SP, Blenis J. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell. 2005;123:569–580. doi: 10.1016/j.cell.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 42.Ashrafizadeh M, Rafiei H, Mohammadinejad R, Afshar EG, Farkhondeh T, Samarghandian S. Potential therapeutic effects of curcumin mediated by JAK/STAT signaling pathway: A review. Phytother Res. 2020 Mar 10; doi: 10.1002/ptr.6642. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 43.Xing J, Bhuria V, Bui KC, Nguyen MLT, Hu Z, Hsieh CJ, Wittstein K, Stadler M, Wilkens L, Li J, et al. Haprolid inhibits tumor growth of hepatocellular carcinoma through Rb/E2F and Akt/mTOR inhibition. Cancers (Basel) 2020;12:615. doi: 10.3390/cancers12030615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ichimaru Y, Sano M, Kajiwara I, Tobe T, Yoshioka H, Hayashi K, Ijichi H, Miyairi S. Indirubin 3′-oxime inhibits migration, invasion, and metastasis InVivo in mice bearing spontaneously occurring pancreatic cancer via blocking the RAF/ERK, AKT, and SAPK/JNK Pathways. Transl Oncol. 2019;12:1574–1582. doi: 10.1016/j.tranon.2019.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han F, Xu Q, Zhao J, Xiong P, Liu J. ERO1L promotes pancreatic cancer cell progression through activating the Wnt/catenin pathway. J Cell Biochem. 2018;119:8996–9005. doi: 10.1002/jcb.27155. [DOI] [PubMed] [Google Scholar]
- 46.Yang K, Li Y, Lian G, Lin H, Shang C, Zeng L, Chen S, Li J, Huang C, Huang K, Chen Y. KRAS promotes tumor metastasis and chemoresistance by repressing RKIP via the MAPK-ERK pathway in pancreatic cancer. Int J Cancer. 2018;142:2323–2334. doi: 10.1002/ijc.31248. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.